Abstract
The ubiquitin-proteasome pathway regulates the turnover of many transcription factors, including steroid hormone receptors such as the estrogen receptor and progesterone receptor. For these receptors, proteasome inhibition interferes with steroid-mediated transcription. We show here that proteasome inhibition with MG132 results in increased accumulation of the glucocorticoid receptor (GR), confirming that it is likewise a substrate for the ubiquitin-proteasome degradative pathway. Using the mouse mammary tumor virus (MMTV) promoter integrated into tissue culture cells, we found that proteasome inhibition synergistically increases GR-mediated transactivation. This increased activation was observed in a number of cell lines and on various MMTV templates, either as transiently transfected reporters or stably integrated into chromatin. These observations suggest that the increase in GR-mediated transcription due to proteasome inhibition may occur downstream of the initial chromatin remodeling step. In support of this concept, the increase in transcription did not correlate with an increase in chromatin remodeling, as measured by restriction enzyme hypersensitivity, or transcription factor loading, as exemplified by nuclear factor 1. To investigate the relationship between GR turnover, transcription, and subnuclear trafficking, we examined the effect of proteasome inhibition on the mobility of the GR within the nucleus and association of the GR with the nuclear matrix. Blocking GR turnover reduced the mobility of the GR within the nucleus, and this correlated with increased association of the receptor with the nuclear matrix. As a result of proteasome inhibition, GR mobility within the nucleus was reduced while its association with the nuclear matrix was increased. Thus, while altered nuclear mobility of steroid receptors may be a common feature of proteasome inhibition, GR is unique in its enhanced transactivation activity that results when proteasome function is compromised. Proteasomes may therefore impact steroid receptor action at multiple levels and exert distinct effects on individual receptor types.
Glucocorticoids mediate diverse cellular processes such as differentiation, development, and homeostasis through the ligand-activated glucocorticoid receptor (GR) (24). The GR is a versatile transcriptional regulator, and it can activate or repress a large variety of natural promoters in multiple tissues. This versatility is due in part to the numerous mechanisms utilized by the GR to regulate transcription, depending on the cellular and promoter context. These mechanisms include GR modification of chromatin structure through interactions with chromatin remodeling complexes, differential binding of the receptor to both canonical and noncanonical response elements, binding to other regulatory factors such as AP-1 and NF-κB, and interaction with coactivators (2, 11, 46).
The activity of the GR is also subject to regulation that occurs at least in part by changes in the half-life of the receptor protein (6). Turnover of the GR protein, and of several other SHRs such as estrogen receptor alpha (ERα), progesterone receptor (PR), and androgen receptor, occurs via degradation by the 26S proteasome (21, 23, 39, 45). Other transcriptional activators and coactivators are also regulated by proteasomal degradation, including GRIP1, Jun, p53, E2F-1, p300, SRC-1, and BRCA1 (5, 7, 9, 12, 33, 36, 43, 48). For many transcription factors, the sequence responsible for activating transcription overlaps with the sequence which targets the protein for ubiquitin-mediated proteolysis (36). These observations support a model in which transcription factors such as SHRs, whose activity is under precise temporal control, may be targeted for degradation upon their engagement in transcriptional regulation (42).
The mechanism(s) by which ubiquitin-mediated proteasomal regulation of receptors affects transcription is currently under investigation. Because proteasome inhibition leads to decreased transcription from a transiently introduced template, it has been suggested that receptor turnover is required for transcriptional ability of some, but not all, steroid hormone receptors (23). However, targeting of the GR to the proteasome degradation pathway by overexpression of the cochaperone CHIP reduces steroid-binding activity and the transactivation ability of the GR (13). Treatment of tissue culture cells with proteasome inhibitors has also been shown to modify the intracellular localization of nuclear receptors, in addition to altering protein levels, and this change in localization may also alter receptor-mediated transcription. Proteasome inhibition in the absence of ligand causes translocation of the aryl hydrocarbon receptor to the nucleus in embryonic primary fibroblasts (37). This translocation results in elevated transcription from endogenous promoters. For the ER, proteasome inhibition leads to immobilization of the ER to the nuclear matrix, resulting in a loss of receptor mobility as visualized by FRAP (fluorescence recovery after photobleaching) (40). Thus, proteasomal degradation may also be required to release ER from the nuclear matrix and increase its availability to target sites within chromatin.
The effect of proteasomal degradation on GR-mediated transcription and nuclear matrix association is not well understood. In addition, what impact GR turnover has on gene transcription in the context of chromatin has not been explored. Therefore, we determined whether the proteasomal degradation pathway regulates GR turnover and, if so, what effect stabilization of GR levels has on chromatin remodeling and transcription of a stably integrated glucocorticoid-responsive promoter. In addition, we investigated whether proteasome inhibitors affect GR mobility within the nucleus and its targeting to the nuclear matrix.
We have found that inhibition of GR turnover by the proteasome inhibitor MG132 in human breast cancer cells robustly increases GR protein levels, and this increase correlates with elevation of RNA and reporter activity levels of a stably integrated mouse mammary tumor virus (MMTV) promoter. However, unlike the case for other steroid receptors, increasing GR levels upon proteasome inhibition, altering GR mobility within the nucleus as assessed by FRAP analysis, and nuclear matrix targeting do not correlate with reduced transcriptional activity. Thus, GR trafficking within the nucleus may not be strictly coupled to its degradation and engagement with the transcription machinery.
MATERIALS AND METHODS
Cells.
A1-2 cells were derived previously from T47D breast cancer cells by stable transfection with pGRneo and MMTV-LTR-luc plasmids as described previously (30). GR2 cells were also previously generated by stably transfecting a GR expression vector into M10 cells. M10 cells are a T47D-based cell line lacking both PR and GR but containing a stably integrated MMTV-CAT reporter (20). Both A1-2 and GR2 cells were grown at 37°C with 5% CO2 in modified Eagle's medium supplemented with 2 mM glutamine, 100 μg of penicillin-streptomycin/ml, 10 mM HEPES, and 10% fetal bovine serum and maintained with 1 μg of puromycin/ml (GR2) or 160 μg of G418/ml (A1-2). 2963.1 cells are derived from a T47D parental cell line and contain an MMTV-CAT reporter (27). 2963.1 cells were grown at 37°C with 5% CO2 in Dulbecco's modified Eagle's medium supplemented with 2 mM glutamine, 100 μg of penicillin-streptomycin/ml, 10 mM HEPES, and 10% fetal bovine serum.
Western blot analysis.
The protocol for preparing nuclear and cytoplasmic extracts was described previously (38). Cells were plated on 100-mm dishes and grown until approximately 80% confluence before preparation of extracts. Ten to 40 μg of nuclear or cytoplasmic extracts was separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) on NOVEX Tris-glycine gels, transferred to a polyvinylidene difluoride membrane, and immunoblotted with the following antibodies: GR (Clontech); mSin3A (sc-994), β-tubulin (sc-5274), nuclear factor 1 (NF1; sc-870), and Oct-1 (sc-232; Santa Cruz Biotechnologies); TATA-binding protein (TBP; catalog no. 160-0070; Geneka Biotechnologies); SRC-1 and pCIP/RAC3 (kind gift of J. Torchia); BRG1 and BAF170 (kind gift of W. Wang).
CAT assays. (i) GR2 and 2963.1 cells.
GR2 or 2963.1 cells were plated in duplicate on 100-mm dishes with 700,000 cells/dish and either left untreated or treated with dexamethasone (Dex), MG132, or RU486 for 20 h as described in the figure legends. Chloramphenicol acetyltransferase (CAT) activity was determined by a kinetic assay and normalized for total protein (4). All assays were carried out twice in duplicate.
(ii) A1-2 cells transiently transfected with MMTV-CAT.
A1-2 cells were plated and transfected the next day with an MMTV-CAT construct by using the Lipofectamine Plus protocol (Invitrogen). The cells were allowed to recover for 24 h and then treated as indicated in the figure legends. CAT activity was assayed as described for GR2 cells (above).
Restriction enzyme hypersensitivity analysis.
Cells were either untreated or treated as described in the figure legends. Nuclei were digested in vivo with 30 U of SstI or 50 U of DpnII as described previously (1). After purification of genomic DNA, samples were recut with BamHI. DNA fragments were analyzed using linear Taq polymerase amplification with a 32P-labeled single-stranded primer corresponding to the +60 to +84 region of the MMTV coding region (MMTV-22). Purified extended products were analyzed on 8% polyacrylamide denaturing gels and quantified using a Molecular Dynamics PhosphorImager.
Exonuclease III footprinting.
Cells were either untreated or treated as described in the figure legends. Nuclei were isolated and subjected to exonuclease III footprinting analysis as described previously (16). BamHI was used as an in vivo entry site for exonuclease. After purification of genomic DNA, the samples were redigested to completion with BamHI. Fifteen micrograms of genomic DNA was analyzed using linear Taq polymerase amplification with the 32P-labeled MMTV-22 primer. Purified extended products were analyzed on an 8% polyacrylamide denaturing gel and quantified using the Molecular Dynamics PhosphorImager.
Reverse transcription-PCR (RT-PCR).
Cells were left untreated or treated as described in the figure legends. Total cellular RNA was isolated using TRIzol (Invitrogen) according to the manufacturer's instructions. cDNA was synthesized as described previously (16). For PCR of the cDNA, two separate reactions were run for each experimental condition. The cDNA was combined with 5 U of Taq DNA polymerase in a final volume of 50 μl. The PCR mixture contained 20 mM Tris-HCl (pH 8.4), 50 mM KCl, 1.5 mM MgCl2, a 100 μM concentration of each deoxynucleoside triphosphate, 10 pmol of MMTV-619, and 0.25 pmol of MMTV-22. MMTV-22 was end labeled with T4 polynucleotide kinase to generate a 32P-labeled single-stranded primer. Human β2-microglobulin was similarly amplified using sequences previously described (16). PCR products were analyzed on 8% polyacrylamide gels and exposed to Molecular Dynamics PhosphorImager screens or autoradiography film for analysis.
Nuclear matrix preparation.
Nuclear matrices were prepared from cells grown on tissue culture plates as described elsewhere (18). Briefly, cells were washed three times with ice-cold phosphate-buffered saline and then treated for 5 min with ice-cold CSK buffer [10 mM piperazine-N,N′-bis(2-ethanesulfonic acid), pH 6.8; 100 mM NaCl; 300 mM sucrose; 3 mM MgCl2; 1 mM EGTA; 4 mM vanadyl riboside complex; 0.5% Triton X-100; and protease inhibitors]. A complete nuclear matrix preparation was obtained by subjecting CKS buffer-extracted cells to a DNase I digestion and ammonium sulfate extraction. Proteins were extracted from the resultant nuclear matrix pellet by using SDS sample buffer and subjected to SDS-PAGE and Western blotting, using previously described conditions to visualize GR (47). Blots were also stained with an antibody to the nuclear matrix protein, lamin B, to provide an internal control for recovery of nuclear matrix protein. ATP depletion of A1-2 cells was performed under established conditions (41). Briefly, cells were cultured in glucose-free medium containing 6 mM 2-deoxyglucose and 10 mM sodium azide for 60 min prior to harvesting.
FRAP analysis.
A1-2 cells were plated and transfected the next day with a GR-green fluorescent protein (GFP) construct using the Lipofectamine Plus protocol (Invitrogen) (10). The cells were allowed to recover for 24 h and then treated with corticosterone (1 μM) and/or MG132 as indicated in the figure legends. Live cells expressing nuclear GR-GFP were subjected to a 16 s photobleaching in a designated nuclear zone using a Molecular Dynamics confocal microscope. Fluorescence recovery was followed at various times following the photobleaching as indicated in the figure legends. Twenty to 30 individual cells were examined for each condition.
RESULTS
Proteasome inhibition elevates steady-state GR protein levels.
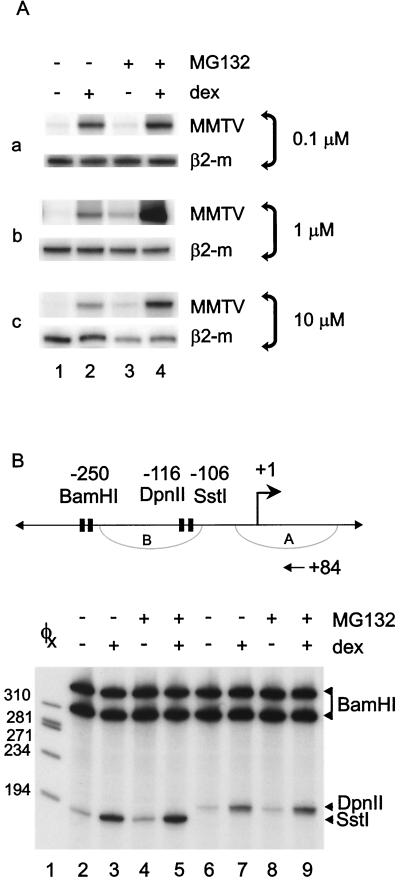
The metabolic turnover of several SHRs, including the GR, has been attributed at least in part to ubiquitylation and degradation by the 26S proteasome (6, 45). We initially determined if GR turnover was regulated by proteasomal degradation in A1-2 breast cancer cells that contain the rat GR and the promoter for the steroid-responsive MMTV promoter (4). To determine what concentration of the peptide aldehyde MG132 would produce optimal GR levels for subsequent analysis, we analyzed steady-state GR levels in A1-2 cells treated with various doses of MG132 (22). As shown in Fig. 1A, steady-state levels of GR increased upon treatment with doses of MG132 that ranged from 0.1 to 10 μM. As a control, we measured the level of β-tubulin under the same conditions and, as predicted, there was no significant change in β-tubulin levels. Since optimal effects on GR transactivation occurred at 1 μM MG132 (Fig. 2A), we chose this dose for many subsequent studies. A time course analysis of the effects of 1 μM MG132 treatment on GR levels was also performed. Treatment of A1-2 cells with MG132 for 1 to 24 h demonstrated that GR levels increased twofold at 2 h (Fig. 1B). Because the most substantial increase in GR protein (approximately 20-fold) occurred at 16 to 24 h, cells were subjected to 1 μM MG132 for 16 or 24 h in our subsequent experiments. In contrast to the changes in the GR, the steady state levels of other transcription factors, exemplified by TBP and NF1 were unchanged under these conditions. (Fig. 1B). To confirm that proteasome inhibition led to an increase in GR levels in the presence of ligand, we examined GR protein levels and subcellular localization by Western blotting (Fig. 1C). In the presence of the synthetic glucocorticoid Dex, we again observed a substantial increase in nuclear GR protein levels due to proteasome inhibition above that with Dex treatment alone (Fig. 1C, compare lanes 3 and 7).
FIG. 1.
Proteasome inhibition enhances steady-state GR protein levels. (A) A1-2 cells were untreated (lane 1) or treated for 22 h with 0.1, 1, or 10 μM MG132 (lanes 2 to 4). Cytoplasmic extracts were examined by Western blotting with antibodies specific to the GR or β-tubulin. (B) A1-2 cells were untreated (lane 1) or treated with MG132 (1 μM) for 1, 2, 8, 16, or 24 h (lanes 2 to 6). Nuclear and cytoplasmic extracts were examined by Western blotting with antibodies specific for GR, TBP, or NF1. (C) A1-2 cells were untreated (lanes 1 and 2) or treated with Dex (10−7 M) for 6 h (lanes 3 and 4), MG132 (1 μM) for 22 h (lanes 5 and 6), or MG132 (1 μM) for 16 h after which Dex (10−7 M) was added to the MG132-containing medium for 6 h (lanes 7 and 8). Nuclear and cytoplasmic extracts were examined by Western blotting by using the same GR antibody as used for panels A and B.
FIG. 2.
The proteasome inhibitor MG132 increases glucocorticoid-activated transcription but not chromatin remodeling of the MMTV promoter. (A) A1-2 cells were untreated (lane 1) or treated with Dex (10−7 M) for 6 h (lane 2), MG132 for 22 h (lane 3), or MG132 for 16 h followed by Dex (10−7 M) for 6 h (lane 4). RNA was prepared and analyzed by RT-PCR with primers specific for MMTV or β2-microglobulin. (B) A1-2 cells were untreated (lanes 2 and 6) or treated with Dex (10−7 M) for 1 h (lanes 3 and 7), MG132 (1 μM) for 17 h (lanes 4 and 8), or MG132 (1 μM) for 16 h followed by Dex (10−7 M) for 1 h (lanes 5 and 9). Isolated nuclei were digested in vivo with SstI or DpnII and recut after DNA purification with BamHI. 32P-labeled DNA fragments generated by primer extension were analyzed by 8% denaturing PAGE followed by autoradiography.
Proteasome inhibition enhances GR-mediated transactivation.
For ERα and PR, blocking receptor turnover by treatment with proteasome inhibitors compromises transcription from transiently introduced promoters in HeLa cells (23). To investigate the effect of proteasome inhibition on GR-mediated transcription in the context of chromatin, we used the stably integrated MMTV promoter as our glucocorticoid-responsive target promoter. Cells were pretreated with 0.1, 1, and 10 μM MG132 and stimulated with Dex, and RNA levels were analyzed by RT-PCR (Fig. 2A). Blocking GR turnover synergistically increased the elevation in MMTV RNA levels due to Dex at all concentrations of MG132 tested (Fig. 2A, compare lanes 2 and 4). β2-Microglobulin RNA levels were unaffected by 0.1 and 1.0 μM MG132. However, 10 μM MG132 reduced β2-microglobulin RNA levels significantly, which further supported the choice of 1 μM MG132 as the treatment for our other experiments (Fig. 2A, panel c).
Glucocorticoid activation of the MMTV promoter requires remodeling of the promoter chromatin to allow binding of other transcription factors and for transcription to proceed (14). We have shown previously that restriction endonuclease hypersensitivity is an excellent readout for hormone-dependent chromatin remodeling (1). This glucocorticoid-mediated chromatin remodeling is evident in increased promoter sensitivity to restriction endonucleases after steroid treatment. In A1-2 cells, the MMTV promoter chromatin is “closed” and requires glucocorticoid treatment to be “opened” (4). We hypothesized that the dramatic amplification of Dex-induced MMTV RNA levels as a result of proteasome inhibition might also result in a substantial increase in chromatin remodeling (Fig. 2B). We used the restriction enzyme hypersensitivity assay as a measure of chromatin remodeling of the MMTV promoter. Inhibiting proteasome function had no significant effect on the increase in hypersensitivity due to glucocorticoid treatment (Fig. 2B, lanes 3 versus 5 and lanes 7 versus 9). These results suggest that the elevated levels of GR due to proteasome inhibition may enhance MMTV transcription at a step downstream of chromatin remodeling.
The increase in MMTV transcription due to proteasome inhibition occurs on open and closed chromatin templates.
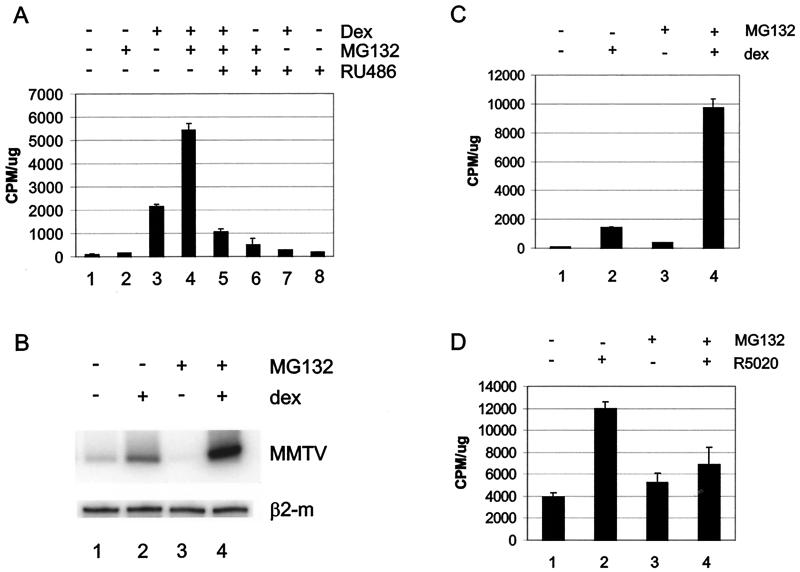
If the MG132-mediated increase in MMTV RNA levels occurs independently of chromatin remodeling, then activation of an open MMTV template or a transiently introduced template should respond similarly to the closed promoter in A1-2 cells. To test this hypothesis, we took advantage of another cell line, GR2, which contains both the GR and an MMTV-CAT reporter and which was derived from the same T47D parental cell line as A1-2 cells (20). In this cell line, MMTV-CAT is open or constitutively remodeled but requires ligand for transactivation. Consistent with this hypothesis, we observed a synergistic increase in both CAT activity (Fig. 3A, compare lanes 3 and 4) and RNA levels (Fig. 3B, compare lanes 2 and 4) in GR2 cells treated with Dex and MG132 in combination. This increase required the hormone-activated GR, since cotreatment with the antiglucocorticoid RU486 blocked the synergistic effect produced by proteasome inhibitor treatment in the presence of Dex (Fig. 3A, compare lanes 4 and 5). We tested this hypothesis further by observing the impact of proteasome inhibitor treatment on an MMTV-CAT promoter transiently introduced into A1-2 cells (Fig. 3C). The transiently introduced MMTV promoter does not acquire the regularly phased nucleosomes that are characteristic of the MMTV promoter when integrated into tissue culture cells (3). Again, inhibiting proteasome function significantly enhanced Dex activation of the transient MMTV-CAT template in A1-2 cells (Fig. 3C).
FIG. 3.
The increase in ligand-mediated transcription of MMTV is independent of promoter architecture. (A) GR2 cells were untreated (lane 1) or treated with Dex (10−8 M) (lanes 3, 4, 5, and 7), MG132 (1 μM) (lanes 2, 4, 5, and 6), or RU486 (10−7 M) (lanes 5 to 8) for 24 h. Cellular extracts from duplicate samples were analyzed for CAT activity and normalized against total protein. (B) GR2 cells were untreated (lane 1) or treated with Dex (10−7 M) for 6 h (lane 2), MG132 (1 μM) for 22 h (lane 3), or MG132 (1 μM) for 16 h followed by Dex (10−7 M) for 6 h (lane 4). RNA was prepared and analyzed by RT-PCR with primers specific for MMTV (top panel) or β2-microglobulin (bottom panel). (C) A1-2 cells transfected with an MMTV-CAT promoter were untreated or treated with Dex (10−7 M), MG132 (1 μM), or MG132 plus Dex for 24 h, analyzed for CAT activity, and normalized against total protein. (D) 2963.1 T47D cells were treated with R5020 (10−8 M), MG132 (1 μM), or MG132 plus R5020 for 24 h, analyzed for CAT activity, and normalized against total protein. Data were obtained from triplicate samples and are representative of two to three independent experiments.
We next tested the possibility that MMTV RNA levels may be enhanced by MG132 if proteasome inhibition increases the stability of the MMTV RNA message. For these experiments we turned to a third T47D-based MMTV-CAT reporter-containing cell line, 2963.1, which express the PR but not the GR (27). Treatment with proteasome inhibitors has been shown to increase PR levels in T47D cells (21). In contrast to glucocorticoid treatment, blocking PR turnover inhibited MMTV activation by the synthetic progestin R5020 (Fig. 3D, compare lanes 2 and 4). This result supports the hypothesis that the MG-Dex synergism observed in A1-2 and GR2 cells is not likely due to stabilization of MMTV RNA levels by MG132. Therefore, because MG-Dex synergism occurs on transient, closed (A1-2) and open (GR2) integrated templates, the pool of GR that results from proteasome inhibition may enhance transcription downstream of the chromatin remodeling step of transcriptional activation.
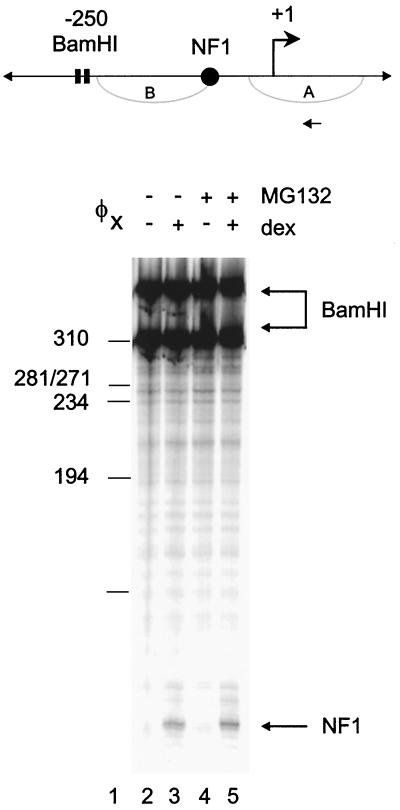
Glucocorticoid-induced chromatin reorganization of the MMTV promoter is concomitant with the loading of transcription factors onto the promoter to form a stable preinitiation complex (PIC) (3). To ascertain if the increased availability of the GR due to proteasome inhibition would influence Dex-induced transcription factor loading, we used an in vivo exonuclease III footprinting assay to investigate binding of the NF1 transcription factor to the promoter (Fig. 4). Addition of Dex promoted NF1 binding to the chromatin template (Fig. 4, compare lanes 2 and 3). MG132 had little effect on the amount of NF1 bound in the presence of Dex (Fig. 4, compare lanes 3 and 5). Therefore, while proteasome inhibition increases MMTV RNA levels induced by Dex, neither chromatin remodeling nor NF1 loading is affected by this treatment.
FIG. 4.
Effect of proteasome inhibition on glucocorticoid-mediated transcription factor loading. A1-2 cells were untreated (lane 2) or treated with Dex (10−7 M) for 6 h (lane 3), MG132 (1 μM) for 22 h (lane 4), or MG132 (1 μM) for 16 h followed by Dex (10−7 M) for 6 h (lane 5). Isolated nuclei were digested with BamHI and ExoIII to detect specific stops corresponding to the 5′ boundaries of bound factors (1). The lower arrow indicates the boundary for NF1.
Effect of proteasome inhibition on steady-state levels of transcriptional cofactors.
We have observed that the ubiquitin-proteasome pathway regulates GR levels. Thus, we next wanted to determine if other transcriptional regulators are similarly modulated by proteasomal inhibition and might contribute to the enhanced glucocorticoid-mediated transcription we observed. In particular, we investigated the effect of MG132 treatment on the expression of the well-known coregulatory proteins mSin3A, BRG1, SRC-1, BAF170, and pCIP/RAC3 by Western blotting, as well as the transcription factor Oct-1 (Fig. 5). Treatment of A1-2 cells under conditions previously used for RNA analysis had no significant impact on nuclear levels or nuclear localization of these factors (Fig. 5, compare lanes 1, 3, 5, and 7). However, BAF170 levels were slightly elevated and there was also a slight increase in cytoplasmic mSin3A levels observed after MG132 treatment (Fig. 5, compare lanes 1 and 7 for BAF170 and lanes 2 and 6 for mSin3A). Thus, in these cells, regulation of protein levels by the ubiquitin-proteasome pathway occurs for the GR, but this is not ubiquitous for all transcriptional cofactors examined here.
FIG. 5.
A subset of transcriptional cofactors is unaffected by inhibiting proteasome function. A1-2 cells were treated as described for Fig. 1C and nuclear and cytoplasmic levels were isolated and analyzed by Western blotting for mSin3A, BRG1, pCIP/RAC3, SRC-1, BAF170, and Oct-1 proteins.
Proteasome inhibition limits GR mobility within the nucleus.
Inhibiting proteasome-dependent ERα turnover also reduces the mobility of this receptor by immobilizing it to the nuclear matrix (40). Because we observed that proteasome inhibition enhanced rather than inhibited glucocorticoid-mediated transcription, we next determined whether disrupting GR turnover would affect GR mobility within the nucleus by using both FRAP and biochemical analysis of GR targeting to the nuclear matrix. Because increased GR accumulation and potentiation of transactivation occurred at doses ranging from 0.1 to 10 μM MG132, we assessed mobility and matrix binding at these three concentrations.
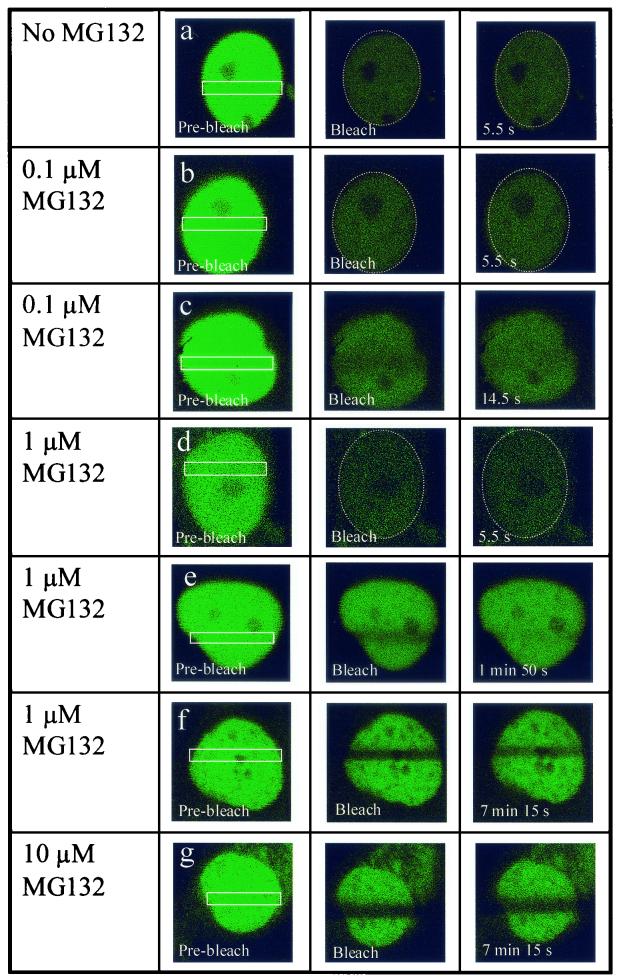
We investigated trafficking of a functional human GR-GFP chimera at various doses of MG132 in live cells using FRAP (10). Consistent with a previously published report, we observed a very rapid movement of GR-GFP within the nucleus of Dex-treated A1-2 cells (Fig. 6, row a) (25). In fact, GR-GFP mobility is so rapid in these cells that a 15-s photobleaching of only a narrow zone within the nucleus reduced the fluorescence intensity of the entire nucleus. This rapid nuclear bleaching occurs, as described previously (40), due to the very rapid passage of bulk nuclear receptors through the narrow bleach zone. Given the rapid mobility of nuclear GR-GFP, we were also unable to detect a bleached zone when fluorescence recovery was examined 5.5 s after the initial photobleaching (Fig. 6, row a). This rapid mobility of GR-GFP within the nucleus is relatively unaffected by treatment of A1-2 cells with 0.1 μM MG132 (Fig. 6, rows b and c), although we could sometimes observe a faint bleached zone that recovers fluorescence within 14.5 s (Fig. 6, row c). In contrast, treatment of A1-2 cells with 10 μM MG132 led to a dramatic reduction in GR mobility within the nucleus, as recovery of the photobleached zone was incomplete, even when examined over 7 min after photobleaching (Fig. 6, row g). This is analogous to the effects of 10 μM MG132 on ERα-GFP mobility within the nucleus (40). Interestingly, we observed a mixed phenotype in A1-2 cells that were treated with 1 μM MG132 under the identical conditions used to show potentiating effects of MG132 on GR transactivation. Specifically, in 57% of the cells examined (n = 30), nuclear GR-GFP showed a rapid to modest recovery from photobleaching with fluorescence signal completely recovered within the photobleached zone within approximately 2 min (Fig. 6, rows d and e). Within this group, for some of the cells (20% of total), GR mobility is so rapid that the GFP signal within the entire nucleus is bleached (Fig. 6, row d; see explanation above), while in other cells (37% of total) a faint bleach zone could be detected that was rapidly recovered (Fig. 6, row e). The remaining cells in the population, 43% (n = 30), showed a dramatic reduction in GR-GFP migration with incomplete recovery of the photobleached zone even after over 7 min of recovery (Fig. 6, row f). This mixed phenotype was observed in three separate experiments that utilized cells plated on three separate days and was not correlated with relative GR-GFP expression levels. Since we have not assessed the transactivation activity of GR within individual cells, definitive conclusions cannot be made regarding the relationship between proteasome involvement in GR mobility versus transcription. Nonetheless, these experiments establish that GR mobility within the nucleus is indeed linked to appropriate functioning of the proteasome machinery.
FIG. 6.
Differential effects of various MG132 doses on GR nuclear mobility. A1-2 cells transiently transfected with GR-GFP were either treated with corticosterone (10−6 M) for 6 h (no MG132), or with various doses of MG132 (0.1, 1, or 10 μM) for 16 h followed by corticosterone (10−6M) for 6 h. The zone of the nucleus that was subjected to a 15-s photobleaching is demarked in the Pre-bleach panels. Additional panels show individual cells either immediately after the photobleaching (Bleach) or at the various times indicated following the photobleaching.
Proteasome inhibition targets the GR to the nuclear matrix.
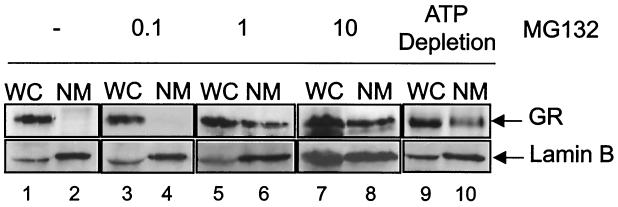
In addition, biochemical extractions were used to measure the binding of endogenous GRs in A1-2 cells to the nuclear matrix under the same conditions used to assess GR mobility by FRAP. Unlike other steroid receptors, hormone-dependent association of GR with the nuclear matrix is inefficient and requires chemical cross-linking (44) or ATP depletion (41) for detection. The molecular basis for the apparent differential affinities of individual steroid receptors for the nuclear matrix is unclear. In our experiments, GR association with the nuclear matrix was examined by Western blot analysis using the matrix-associated protein lamin B as a positive control (Fig. 7). The exposures of enhanced chemiluminescence-developed Western blots were varied in Fig. 7 to better illustrate the ratio of whole-cell to nuclear matrix-bound GR. Fig. 1A shows how GR levels increased in response to MG132 treatment. As shown in Fig. 7, no increase in GR binding to the nuclear matrix, relative to control hormone-treated cells, was observed in A1-2 cells treated with 0.1 μM MG132 (Fig. 7, lanes 3 and 4). In contrast, GR binding to the nuclear matrix was enhanced approximately fivefold (to 15% of total GR) when A1-2 cells were treated with 1 or 10 μM MG132 (Fig. 7, lanes 5 to 8). As a control, GR binding to the nuclear matrix was also increased by ATP depletion of A1-2 cells (Fig. 7, lanes 9 and 10), which agrees with previous results obtained using other cell types (41). Thus, increased nuclear matrix binding of GR in A1-2 cells treated with a proteasome inhibitor correlated with reduced GR mobility as assessed by FRAP, particularly with cells exposed to 10 μM MG132. Although effects on GR mobility are variable with 1 μM MG132, GR transactivation activity is significantly increased at this dose of MG132. Enhanced GR nuclear matrix binding of bulk GR is therefore not indicative of a reduced-capacity receptor transactivation, as suggested from recent results with ERα-GFP.
FIG. 7.
Differential effects of various MG132 doses on GR association with the nuclear matrix. A1-2 cells were either treated with corticosterone (10−6 M) for 6 h (lanes 1 and 2) or various doses of MG132 (0.1, 1, or 10 μM) for 16 h followed by corticosterone (10−6 M) for 6 h (lanes 3 to 8). As a control, cells were untreated but subjected to an ATP-depletion paradigm (lanes 9 and 10) (41). Whole-cell extracts were Western analyzed (WC) and nuclear matrix-associated proteins were subjected to blot analysis using an anti-GR (top) or anti-lamin B (bottom) antibody. The amount of whole-cell extract protein loaded was only 20% of the nuclear matrix fraction in all lanes. Exposures of enhanced chemiluminescence-developed Western blots for GR were varied to better illustrate the ratio of whole-cell extract to nuclear matrix-bound GR.
DISCUSSION
The protein levels of many steroid hormone receptors are regulated by ubiquitin-mediated degradation via the 26S proteasome (6, 21, 28, 29, 45). How this degradation influences transcription is poorly understood, but it appears to vary between individual steroid receptors (23). Since the relationship between GR-mediated transcription and turnover of the GR by the proteasome has not been extensively characterized, we investigated the effect of proteasomal inhibition on GR-mediated transcription in the context of chromatin by using the MMTV promoter.
In the absence of hormone, treatment with a proteasome inhibitor significantly increased steady-state GR protein levels (Fig. 1A to C), suggesting that even in resting cells the GR is continuously degraded. In the absence of ligand, this treatment increased MMTV RNA levels and had a synergistic effect on transcription when cotreated with glucocorticoid (Fig. 2A). The potentiating effect of MG132 on glucocorticoid-mediated transcription corresponds to what has previously been observed on a transiently introduced glucocorticoid-responsive promoter (45). Thus, the high GR levels resulting from proteasome inhibition correlated with increased transcription from a glucocorticoid-responsive gene. This correlation between GR number and cellular response to hormone has been previously demonstrated in lymphoma cells (8, 17). Since we observed that blocking GR turnover synergistically enhanced the glucocorticoid-dependent increase in MMTV RNA levels, we wanted to determine if a synergistic increase in chromatin remodeling would also be observed. In A1-2 cells, the MMTV promoter acquires a closed chromatin conformation that requires glucocorticoid to open the chromatin and render it more accessible to restriction enzymes. The resulting two-step model involves opening of chromatin structure by the GR, which then allows access of other transcription factors to the DNA and formation of the PIC (14). Using an enzyme hypersensitivity assay as a measure of chromatin remodeling, we found that MG132 treatment had no significant effect on either basal or glucocorticoid-mediated hypersensitivity of the MMTV promoter (Fig. 2B). These results suggest that the pool of GR protein that accumulates upon MG132 treatment affects MMTV transactivation at a step downstream of chromatin remodeling, as indicated by hypersensitivity to restriction endonucleases. The increase in MMTV RNA levels that we observe in the presence of proteasome inhibitors is not simply due to increased stability of the mRNA, since cotreatment with the synthetic progestin R5020 inhibited rather than enhanced MMTV promoter activity (Fig. 3D) (27). The results are consistent with inhibition of the progestin-mediated transcription by proteasome inhibitors observed in other cell lines (28).
If the enhanced MMTV activation due to compromise of proteasome function occurs independently of chromatin remodeling, then activation of a nonchromatin template, or an open MMTV template should yield the same MG132-glucocorticoid synergism observed on a closed template. As the open template, we took advantage of a second T47D-based cell line, GR2, which contains the GR and an integrated MMTV-CAT reporter (Fig. 3A and B). In this cell line, the MMTV promoter is constitutively open but requires ligand and GR-coactivator binding for transcription (4). As our nonchromatin template, we transiently introduced an MMTV-CAT reporter into A1-2 cells (Fig. 3C). Transiently introduced MMTV reporters do not acquire the nucleosomal organization characteristic of integrated constructs, and they are constitutively hypersensitive, as is the promoter in GR2 cells (3). We found that both the open MMTV-CAT in GR2 cells and a transiently introduced MMTV-CAT exhibited the same synergistic increase in activity and RNA due to MG132 treatment as observed with the closed MMTV promoter in A1-2 cells. In addition, cotreatment with the antiglucocorticoid RU486 blocked the observed MG132-Dex synergism, indicating that this effect required the GR (Fig. 3A, compare lanes 4 and 5). These results further support the idea that the effect of MG132 on GR-mediated transcription of the MMTV promoter occurs downstream of chromatin remodeling.
How might the large pool of GR resulting from MG132 treatment significantly increase MMTV RNA levels without similarly affecting chromatin remodeling? One possibility is that the higher levels of GR increase the rate of transcription from the MMTV promoter. Thus, although chromatin remodeling may not increase, the higher levels of the GR may increase the rate of PIC formation and therefore increase transcriptional initiation. This hypothesis is consistent with the observation that antagonism of glucocorticoid activation of MMTV transcription by 8-bromo-cyclic AMP is not accompanied by changes in promoter hypersensitivity or NF1 loading (32). Instead, it was shown that 8-bromo-cyclic AMP reduces the rate of transcription from the promoter in the absence of any change in chromatin remodeling. Thus, preventing GR degradation may increase the rate of MMTV transcription in the absence of increased chromatin remodeling. The observation that hormone-dependent structural changes in the promoter chromatin can be dissociated from activated transcription is also supported by studies in two human breast cancer cell lines in which the integrated MMTV promoter acquires an open chromatin structure that is constitutively remodeled. In both cases, steroid treatment does not increase promoter hypersensitivity but is required for transcription (20, 27).
In addition to SHRs, the ubiquitin-proteasome pathway can also regulate transcriptional coactivator and corepressor levels. Coactivators that are targets of ubiquitin-proteasome degradation include the coactivators GRIP1, p300/CBP, SRC-1, TIF-2, and pCIP/RAC3 (5, 23, 34). Levels of the corepressor NCoR are also regulated in this way (48). We were therefore interested in what impact proteasome inhibition might have on various corepressors and coactivators in the A1-2 cell line. While we were unable to look at all possible coregulators, we found that the levels of the cofactors pCIP/RAC3, mSin3A, BRG1, BAF170, or SRC-1 were not affected by MG132 treatment, as was seen with the GR (Fig. 5). Although pCIP/RAC3 and SRC-1 have been shown to be degraded via the proteasome in other cell lines, we did not find similar regulation in the T47D-based cell line used in this study, suggesting that coactivator regulation by ubiquitylation may be cell-type specific (23).
What characteristics of a particular receptor determine whether its degradation is required for, or inhibits, transcription? One mechanism that may contribute to this difference involves a novel function of the ubiquitin pathway—regulation of SHR mobility by association with the nuclear matrix. Treatment of tissue culture cells with proteasome inhibitors was recently shown to inhibit ERα mobility through targeting of the receptor to the nuclear matrix (40). We thus wanted to determine if blocking the degradation of the GR would also reduce the GR's mobility and/or enhance its targeting to the nuclear matrix. At relatively high doses of MG132 (i.e., 10 μM), GR and ERα behave similarly and are protected from proteasome-mediated degradation and limited in their internuclear mobility as assessed in live cells. At intermediate doses of MG132 (e.g., 1 μM) GR association with the nucleus is increased, but receptor transactivation is enhanced rather than inhibited as observed with ERα (40). GR mobility is variable in A1-2 cells treated with 1 μM MG132, with the receptor displaying rapid to modest mobility within the nuclei of some cells but severely retarded movement in the others. Given this variability, we cannot establish a strict correlation between GR nuclear mobility and transactivation in A1-2 cells. In fact, the variability within individual cells treated with 1 μM MG132 might be indicative of multiple roles for proteasomes in GR function. Thus, reducing GR turnover through proteasome inhibition could enhance GR transactivation by increasing the effective concentration of receptors at the PIC. Prolonged or more-effective proteasome inhibition might reveal a requirement for some proteasome function in maintaining appropriate receptor trafficking within the nucleus.
A number of model studies have suggested that the potency of specific transactivation domains is correlated with their rate of degradation by the ubiquitin-proteasome system (26). Proteasome-mediated degradation of transcriptional activators correlates with activation domain potency in vivo (36). Importantly, not all transactivation domains tested in this study are affected by alterations in proteasome activity (36). Furthermore, even though transactivation activity of the retinoid X receptor is linked to its proteasome-dependent degradation, some mutants in the AF-2 transactivation domain of this receptor uncouple the link between transactivation activity and degradation (31). Thus, the differential effects of proteasome inhibitors on GR versus those on ERα may reveal unique features of subnuclear processing or trafficking of these highly related receptors (23).
Finally, it may be necessary to consider the possibility that ubiquitylation of distinct transactivators, even closely related members of the steroid receptor family, impacts multiple receptor functions independent of proteasome targeting. The linkage of a nonremovable ubiquitin moiety to the VP16 transactivation domain restores its transactivation activity in yeast cells defective for a specific ubiquitin ligase (35). This restoration of transactivation occurred in the absence of any effects on VP16 transactivation domain degradation (35). Thus, ubiquitylation, particularly monoubiquitylation, may provide a unique signal to specific transactivation domains that enhances their effectiveness in the absence of direct proteasome involvement. Monoubitquitylation is becoming recognized as a posttranslational modification that regulates protein trafficking and activity in a variety of contexts (19). In fact, it has been postulated that such effects of ubiquitylation may be manifested at the level of transcription elongation, as a ubiquitin-binding component of the proteasome has been found to interact with an RNA polymerase II elongation factor and affect its function (15). Such a mechanism would be consistent with our findings that proteasome inhibition enhances GR transactivation without noticeable changes in chromatin remodeling and PIC formation at the proximal promoter. Inhibition of proteasomes by MG132 treatment of A1-2 cells could impact the ratio of monoubiquitylated versus polyubiquitylated GRs and thereby affect the recruitment of limiting RNA polymerase II elongation factors. Clearly, the link between GR subnuclear trafficking, processing, and transactivation is complex and is likely to include pathways that intersect and affect each other's activity in ways that might be unique for each member of the nuclear receptor superfamily.
Acknowledgments
We thank W. Wang for the anti-BRG1 and anti-BAF170 antibodies and J. Torchia for the SRC-1 and anti-pCIP/RAC3 antibodies. We also thank the members of the Archer Lab for their helpful advice and comments. N. Burke and S. Watkins of the Center for Biologic Imaging at the University of Pittsburgh School of Medicine are thanked for their help with FRAP analysis.
A Medical Research Council of Canada Doctoral Scholarship supported B.J.D.
REFERENCES
- 1.Archer, T. K., M. G. Cordingley, R. G. Wolford, and G. L. Hager. 1991. Transcription factor access is mediated by accurately positioned nucleosomes on the mouse mammary tumor virus promoter. Mol. Cell. Biol. 11:688-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Archer, T. K., B. J. Deroo, and C. J. Fryer. 1997. Chromatin modulation of glucocorticoid and progesterone receptor activity. Trends Endocrinol. Metab. 8:384-390. [DOI] [PubMed] [Google Scholar]
- 3.Archer, T. K., P. Lefebvre, R. G. Wolford, and G. L. Hager. 1992. Transcription factor loading on the MMTV promoter: a bimodal mechanism for promoter activation. Science 255:1573-1576. (Erratum, 256:161.) [DOI] [PubMed]
- 4.Archer, T. K., E. Zaniewski, M. L. Moyer, and S. K. Nordeen. 1994. The differential capacity of glucocorticoids and progestins to alter chromatin structure and induce gene expression in human breast cancer cells. Mol. Endocrinol. 8:1154-1162. [DOI] [PubMed] [Google Scholar]
- 5.Baumann, C. T., H. Ma, R. Wolford, J. C. Reyes, P. Maruvada, C. Lim, P. M. Yen, M. R. Stallcup, and G. L. Hager. 2001. The glucocorticoid receptor interacting protein 1(GRIP1) localizes in discrete nuclear foci that associate with ND10 bodies and are enriched in components of the 26S proteasome. Mol. Endocrinol. 15:485-500. [DOI] [PubMed] [Google Scholar]
- 6.Bellingham, D. L., M. Sar, and J. A. Cidlowski. 1992. Ligand-dependent down-regulation of stably transfected human glucocorticoid receptors is associated with the loss of functional glucocorticoid responsiveness. Mol. Endocrinol. 6:2090-2102. [DOI] [PubMed] [Google Scholar]
- 7.Blagosklonny, M. V., W. G. An, G. Melillo, P. Nguyen, J. B. Trepel, and L. M. Neckers. 1999. Regulation of BRCA1 by protein degradation. Oncogene 18:6460-6468. [DOI] [PubMed] [Google Scholar]
- 8.Bourgeois, S., and R. F. Newby. 1979. Correlation between glucocorticoid receptor and cytolytic response of murine lymphoid cell lines. Cancer Res. 39:4749-4751. [PubMed] [Google Scholar]
- 9.Campanero, M. R., and E. K. Flemington. 1997. Regulation of E2F through ubiquitin-proteasome-dependent degradation: stabilization by the pRB tumor suppressor protein. Proc. Natl. Acad. Sci. USA 94:2221-2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carey, K. L., S. A. Richards, K. M. Lounsbury, and I. G. Macara. 1996. Evidence using a green fluorescent protein-glucocorticoid receptor chimera that the Ran/TC4 GTPase mediates an essential function independent of nuclear protein import. J. Cell Biol. 133:985-996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cato, A. C., and E. Wade. 1996. Molecular mechanisms of anti-inflammatory action of glucocorticoids. Bioessays 18:371-378. [DOI] [PubMed] [Google Scholar]
- 12.Chowdary, D. R., J. J. Dermody, K. K. Jha, and H. L. Ozer. 1994. Accumulation of p53 in a mutant cell line defective in the ubiquitin pathway. Mol. Cell. Biol. 14:1997-2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Connell, P., C. A. Ballinger, J. Jiang, Y. Wu, L. J. Thompson, J. Hohfeld, and C. Patterson. 2001. The co-chaperone CHIP regulates protein triage decisions mediated by heat-shock proteins. Nat. Cell Biol. 3:93-96. [DOI] [PubMed] [Google Scholar]
- 14.Deroo, B. J., and T. K. Archer. 2001. Glucocorticoid receptor-mediated chromatin remodeling in vivo. Oncogene Rev. 20:3039-3046. [DOI] [PubMed] [Google Scholar]
- 15.Ferdous, A., F. Gonzalez, L. Sun, T. Kodadek, and S. A. Johnston. 2001. The 19S regulatory particle of the proteasome is required for efficient transcription elongation by RNA polymerase II. Mol. Cell 7:981-991. [DOI] [PubMed] [Google Scholar]
- 16.Fryer, C. J., S. K. Nordeen, and T. K. Archer. 1998. Antiprogestins mediate differential effects on glucocorticoid receptor remodeling of chromatin structure J. Biol. Chem. 273:1175-1183. [DOI] [PubMed] [Google Scholar]
- 17.Gehring, U., K. Mugele, and J. Ulrich. 1984. Cellular receptor levels and glucocorticoid responsiveness of lymphoma cells. Mol. Cell. Endocrinol. 36:107-113. [DOI] [PubMed] [Google Scholar]
- 18.He, D. C., J. A. Nickerson, and S. Penman. 1990. Core filaments of the nuclear matrix. J. Cell Biol. 110:569-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hicke, L. 2001. Protein regulation by monoubiquitin. Nat. Rev. Mol. Cell. Biol. 2:195-201. [DOI] [PubMed] [Google Scholar]
- 20.Kinyamu, H. K., C. J. Fryer, K. B. Horwitz, and T. K. Archer. 2000. The MMTV promoter adopts distinct chromatin structures in human breast cancer cells with and without glucocorticoid receptor. J. Biol. Chem. 275:20061-20068. [DOI] [PubMed] [Google Scholar]
- 21.Lange, C. A., T. Shen, and K. B. Horwitz. 2000. Phosphorylation of human progesterone receptors at serine-294 by mitogen-activated protein kinase signals their degradation by the 26S proteasome. Proc. Natl. Acad. Sci. USA 97:1032-1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee, D., and A. Goldberg. 1998. Proteasome inhibitors: valuable new tools for cell biologists. Trends Cell Biol. 8:397.. [DOI] [PubMed] [Google Scholar]
- 23.Lonard, D. M., Z. Nawaz, C. L. Smith, and B. W. O'Malley. 2000. The 26S proteasome is required for estrogen receptor-alpha and coactivator turnover and for efficient estrogen receptor-alpha transactivation. Mol. Cell 5:939-948. [DOI] [PubMed] [Google Scholar]
- 24.McKay, L. I., and J. A. Cidlowski. 1999. Molecular control of immune/inflammatory responses: interactions between nuclear factor-kappa B and steroid receptor-signaling pathways. Endocr. Rev. 20:435-459. [DOI] [PubMed] [Google Scholar]
- 25.McNally, J. G., W. G. Muller, D. Walker, R. Wolford, and G. L. Hager. 2000. The glucocorticoid receptor: rapid exchange with regulatory sites in living cells. Science 287:1262-1265. [DOI] [PubMed] [Google Scholar]
- 26.Molinari, E., M. Gilman, and S. Natesan. 1999. Proteasome-mediated degradation of transcriptional activators correlates with activation domain potency in vivo. EMBO J. 18:6439-6447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mymryk, J. S., D. Berard, G. L. Hager, and T. K. Archer. 1995. Mouse mammary tumor virus chromatin in human breast cancer cells is constitutively hypersensitive and exhibits steroid hormone-independent loading of transcription factors in vivo. Mol. Cell. Biol. 15:26-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nawaz, Z., D. M. Lonard, A. P. Dennis, C. L. Smith, and B. W. O'Malley. 1999. Proteasome-dependent degradation of the human estrogen receptor. Proc. Natl. Acad. Sci. USA 96:1858-1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nomura, Y., T. Nagaya, Y. Hayashi, F. Kambe, and H. Seo. 1999. 9-cis-Retinoic acid decreases the level of its cognate receptor, retinoid X receptor, through acceleration of the turnover. Biochem. Biophys. Res. Commun. 260:729-733. [DOI] [PubMed] [Google Scholar]
- 30.Nordeen, S. K., B. Kuhnel, J. Lawler-Heavner, D. A. Barber, and D. P. Edwards. 1989. A quantitative comparison of dual control of a hormone response element by progestins and glucocorticoids in the same cell line. Mol. Endocrinol. 3:1270-1278. [DOI] [PubMed] [Google Scholar]
- 31.Osburn, D. L., G. Shao, H. M. Seidel, and I. G. Schulman. 2001. Ligand-dependent degradation of retinoid X receptors does not require transcriptional activity or coactivator interactions. Mol. Cell. Biol. 21:4909-4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pennie, W. D., G. L. Hager, and C. L. Smith. 1995. Nucleoprotein structure influences the response of the mouse mammary tumor virus promoter to activation of the cyclic AMP signaling pathway. Mol. Cell. Biol. 15:2125-2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Poizat, C., V. Sartorelli, G. Chung, R. A. Kloner, and L. Kedes. 2000. Proteasome-mediated degradation of the coactivator p300 impairs cardiac transcription. Mol. Cell. Biol. 20:8643-8654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reyes, J. C., C. Muchardt, and M. Yaniv. 1997. Components of the human SWI/SNF complex are enriched in active chromatin and are associated with the nuclear matrix. J. Cell Biol. 137:263-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Salghetti, S. E., A. A. Caudy, J. G. Chenoweth, and W. P. Tansey. 2001. Regulation of transcriptional activation domain function by ubiquitin. Science 293:1651-1653. [DOI] [PubMed] [Google Scholar]
- 36.Salghetti, S. E., M. Muratani, H. Wijnen, B. Futcher, and W. P. Tansey. 2000. Functional overlap of sequences that activate transcription and signal ubiquitin-mediated proteolysis. Proc. Natl. Acad. Sci. USA 97:3118-3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Santiago-Josefat, B., E. Pozo-Guisado, S. Mulero-Navarro, and P. M. Fernandez-Salguero. 2001. Proteasome inhibition induces nuclear translocation and transcriptional activation of the dioxin receptor in mouse embryo primary fibroblasts in the absence of xenobiotics. Mol. Cell. Biol. 21:1700-1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scheinman, R. I., A. A. Beg, and A. S. Baldwin, Jr. 1993. NF-κB p100 (Lyt-10) is a component of H2TF1 and can function as an IκB-like molecule. Mol. Cell. Biol. 13:6089-6101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sheflin, L., B. Keegan, W. Zhang, and S. W. Spaulding. 2000. Inhibiting proteasomes in human HepG2 and LNCaP cells increases endogenous androgen receptor levels. Biochem. Biophys. Res. Commun. 276:144-150. [DOI] [PubMed] [Google Scholar]
- 40.Stenoien, D. L., K. Patel, M. G. Mancini, M. Dutertre, C. L. Smith, B. W. O'Malley, and M. A. Mancini. 2001. FRAP reveals that mobility of oestrogen receptor-alpha is ligand- and proteasome-dependent. Nat. Cell Biol. 3:15-23. [DOI] [PubMed] [Google Scholar]
- 41.Tang, Y., and D. B. DeFranco. 1996. ATP-dependent release of glucocorticoid receptors from the nuclear matrix. Mol. Cell. Biol. 16:1989-2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thomas, D., and M. Tyers. 2000. Transcriptional regulation: kamikaze activators. Curr. Biol. 10:R341-R343. [DOI] [PubMed] [Google Scholar]
- 43.Treier, M., L. M. Staszewski, and D. Bohmann. 1994. Ubiquitin-dependent c-Jun degradation in vivo is mediated by the delta domain. Cell 78:787-798. [DOI] [PubMed] [Google Scholar]
- 44.van Steensel, B., G. Jenster, K. Damm, A. O. Brinkmann, and R. van Driel. 1995. Domains of the human androgen receptor and glucocorticoid receptor involved in binding to the nuclear matrix. J. Cell. Biochem. 57:465-478. [DOI] [PubMed] [Google Scholar]
- 45.Wallace, A. D., and J. A. Cidlowski. 2001. Proteasome mediated glucocorticoid receptor degradation restricts transcriptional signaling by glucocorticoids. J. Biol. Chem. 276:42714-42721. [DOI] [PubMed] [Google Scholar]
- 46.Wallberg, A. E., A. Wright, and J. A. Gustafsson. 2001. Chromatin-remodeling complexes involved in gene activation by the glucocorticoid receptor. Vitam. Horm. 60:75-122. [DOI] [PubMed] [Google Scholar]
- 47.Yang, J., and D. B. DeFranco. 1994. Differential roles of heat shock protein 70 in the in vitro nuclear import of glucocorticoid receptor and simian virus 40 large tumor antigen. Mol. Cell. Biol. 14:5088-5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang, J., M. G. Guenther, R. W. Carthew, and M. A. Lazar. 1998. Proteasomal regulation of nuclear receptor corepressor-mediated repression. Genes Dev. 12:1775-1780. [DOI] [PMC free article] [PubMed] [Google Scholar]