Abstract
A 249-nucleotide coding region instability determinant (CRD) destabilizes c-myc mRNA. Previous experiments identified a CRD-binding protein (CRD-BP) that appears to protect the CRD from endonuclease cleavage. However, it was unclear why a CRD-BP is required to protect a well-translated mRNA whose coding region is covered with ribosomes. We hypothesized that translational pausing in the CRD generates a ribosome-deficient region downstream of the pause site, and this region is exposed to endonuclease attack unless it is shielded by the CRD-BP. Transfection and cell-free translation experiments reported here support this hypothesis. Ribosome pausing occurs within the c-myc CRD in tRNA-depleted reticulocyte translation reactions. The pause sites map to a rare arginine (CGA) codon and to an adjacent threonine (ACA) codon. Changing these codons to more common codons increases translational efficiency in vitro and increases mRNA abundance in transfected cells. These data suggest that c-myc mRNA is rapidly degraded unless it is (i) translated without pausing or (ii) protected by the CRD-BP when pausing occurs. Additional mapping experiments suggest that the CRD is bipartite, with several upstream translation pause sites and a downstream endonuclease cleavage site.
mRNA abundance is determined by the rates of transcription and mRNA decay. For example, changes in mRNA half-life affect the abundance of proto-oncogene and cytokine mRNAs during cell growth, differentiation, and neoplastic transformation. mRNA decay rates are regulated by cis-acting sequence determinants, mRNA-binding proteins, degradative endo- and exoribonucleases, and translation (4, 8, 11, 34, 42, 53, 64).
The link between translation and mRNA stability is the focus of this work. Six observations demonstrate this link (10, 22, 39, 45, 54, 58). (i) Most mRNAs are stabilized in cells exposed to translation inhibitors. (ii) Mutations in the coding region can affect both translation and mRNA half-life. (iii) c-myc, c-fos, interleukin-2, and other mRNAs contain instability determinants in their coding regions; these determinants modify mRNA half-life in a translation-dependent manner. (iv) Factors that influence mRNA decay (ribonucleases and mRNA-binding proteins) can associate with ribosomes. (v) Poly(A) tract length, mRNA stability, and translation are interdependent. (vi) Premature translational termination enhances mRNA decay.
Translational elongation can be interrupted when ribosomes reach a translation pause site (55, 60). Pausing can be induced by several factors, including mRNA structure (48), the translation product itself (24), mRNA-binding proteins (18), signal recognition particle binding (30), and tRNA abundance and rare codons (55). Rare codons are underrepresented in the total mRNA population of an organism, and the tRNAs for rare codons are also relatively scarce (20). Pausing occurs as a ribosome awaits entry of the scarce aminoacyl-tRNA into its “A site” (55). Pausing correlates with mRNA instability in the yeast Saccharomyces cerevisiae (7, 17).
Three observations suggested that translational pausing might also influence the stability of c-myc mRNA. (i) The mRNA contains an instability sequence within its coding region. This sequence, the coding region instability determinant (CRD), is located in the last 249 nucleotides of the coding region (59). It functions independently of the AU-rich element to make the mRNA unstable (26, 27, 59, 62). For example, when the c-myc CRD is inserted in frame within the coding region of β-globin mRNA, the resulting chimeric mRNA is destabilized (19). (ii) The CRD contains a higher than usual percentage of rare codons. (iii) The CRD must be translated to destabilize the mRNA (19, 59). Placing a translational stop codon upstream of the CRD stabilizes the chimeric mRNA (19).
Most cis-acting mRNA stability/instability determinants are also binding sites for proteins, and the c-myc CRD is no exception. The CRD interacts with a 68-kDa CRD-binding protein (CRD-BP), which contains two RNA recognition motifs and four hnRNP K homology domains. When the CRD-BP is bound to c-myc mRNA, the CRD of the mRNA is shielded from endonucleolytic attack (2, 12, 38). The mRNA is then degraded only by an AU-rich element-dependent deadenylation pathway (5). If the CRD-BP dissociates from the mRNA, the CRD becomes exposed to the endonuclease. The mRNA is then rapidly degraded by endonucleolytic cleavage within the CRD. This scenario would account, at least in part, for the rapid changes in c-myc mRNA abundance observed in starved or differentiating cells (13, 46, 50). Since the CRD-BP is abundant during fetal life but is scarce or absent in adults, the CRD-BP might have a special role in regulating c-myc mRNA in utero (29).
This CRD/CRD-BP protection model is consistent with in vitro and in vivo data but is difficult to reconcile with an efficiently translated mRNA. Why would such an mRNA require a protein to protect its coding region if ribosomes were actively translating that region? In fact, how could a CRD-BP even bind to a CRD that was covered with ribosomes? One answer might be related to the known links between c-myc mRNA translation and stability (see above). Perhaps c-myc mRNA is not efficiently translated because ribosomes pause within the CRD. As a result of pausing, a ribosome-free mRNA segment would be created between the pause site and the translation termination site. This unprotected segment could be cleaved by the endonuclease unless it was protected by the CRD-BP during the pause interval. After elongation resumed, the CRD-BP would dissociate from the mRNA, and translation would be completed.
Data presented here support this model. First, we show that CRD-containing mRNA substrates are translated inefficiently in tRNA-depleted reticulocyte extracts. Translation is increased by adding excess tRNA or by shifting the translational reading frame. Second, toeprint assays confirm that translational inefficiency results from ribosome pausing. Third, we show that mRNA abundance and, by inference, mRNA stability correlate with translational efficiency in cells.
MATERIALS AND METHODS
Plasmids.
The plasmids pSP6β/c′cGMG and pSP6β/c′cGGG, containing globin-Myc-globin (GMG) and globin-glyceraldehyde phosphate dehydrogenase (GAPDH)-globin (GGG) cDNAs, respectively, have been described (2, 19). For in vivo expression assays, HindIII-XbaI fragments of the globin-Myc-globin and globin-GAPDH-globin genes (2) were inserted into plasmid pcDNA3 (Invitrogen), a eukaryotic expression vector. The resulting clones were named pcDNA3-GMG and pcDNA3-GGG, respectively.
All mutant CRD fragments were created by PCR and were cloned into pSP6β/c′cGMG and pcDNA3-GMG to construct the cDNA and the gene, respectively, using the EcoRI sites that flank the CRD. PCR mutagenesis was performed as follows. Two fragments of the CRD segment with overlapping 3′ and 5′ ends were produced by PCR using primers containing altered nucleotides. The fragments were then hybridized with each other and extended. A second PCR was then used to amplify the extended fragments. We refer to this procedure as PCR ligation. The resulting DNA products were used for cloning.
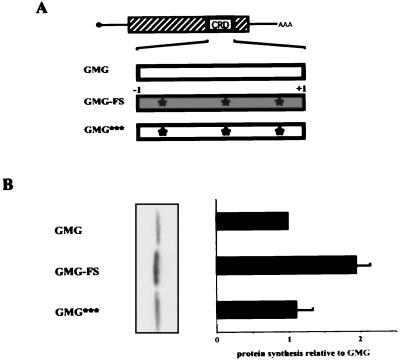
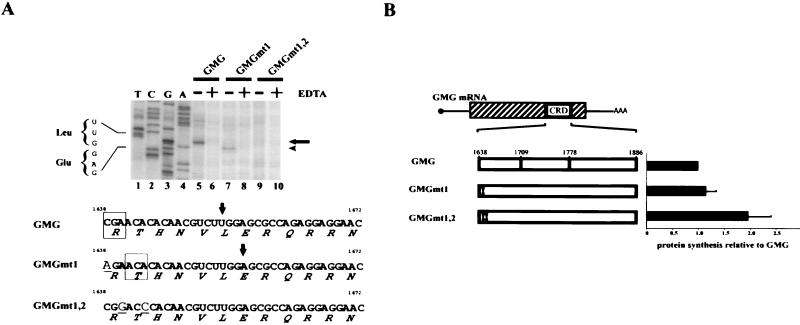
The following mRNAs were synthesized from these DNAs. GMG-FS, carrying a frameshift of only the CRD segment, was made by deleting the first C and adding an extra G at the 3′ end of the CRD segment (diagrammed in Fig. 2A). Three additional point mutations were introduced to avoid premature translation termination at nonsense codons within the frameshifted segment. GMG*** is a GMG mRNA containing the same point mutations as GMG-FS. GMG*** mRNA was not translationally frameshifted. GMGmut1 has a change in c-myc codon 357 (CGA→AGA; R). GMGmt1,2 carries changes in c-myc codons 357 (CGA→CGG; R) and 358 (ACA→ACC; T).
FIG. 2.
Effect of a single-base frameshift on translation of CRD-containing mRNA. (A) Diagrams of translated mRNAs. GMG, unmodified GMG mRNA. GMG-FS, 1 nucleotide was deleted at the 5′ end of the CRD insert, and 1 nucleotide was added at the 3′ end of the insert to maintain the reading frame to the end of the mRNA coding region. This −1/+1 frameshift created three stop codons within the frameshifted segment. To avoid premature translational termination, each nonsense codon was modified to a sense codon by changing a single nucleotide. These changes are indicated by asterisks. GMG***, mRNA containing the three internal, single nucleotide changes of GMG-FS. GMG*** mRNA is read in the same frame as GMG mRNA. (B) SDS-PAGE and data quantification. Translation reactions were performed with reticulocyte lysate treated with micrococcal nuclease as for Fig. 1. The translation efficiencies of GMG-FS and GMG*** mRNAs were normalized to that of GMG mRNA. The graph represents data from five experiments. Error bars show the standard deviation.
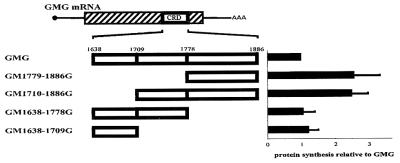
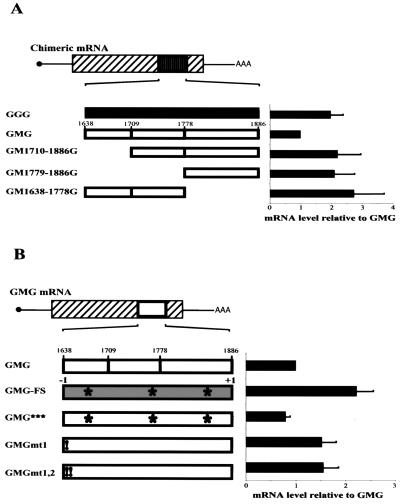
The following mRNAs contained the indicated c-myc mRNA segments inserted into β-globin mRNA (diagrammed in Fig. 3): GM1779-1886G, nucleotides 1779 to 1886; GM1638-1709G, nucleotides 1638 to 1709; GM1710-1886G, nucleotides 1710 to 1886; and GM1779-1886G, nucleotides 1779 to 1886. The orientation and sequence of all clones were verified using a Big-Dye sequencing kit (Amersham). Translation of all mRNAs was initiated at the β-globin start codon and was terminated at the β-globin stop codon.
FIG. 3.
Mapping translational inefficiency sequences to the first 72 nucleotides of the c-myc CRD segment. Chimeric mRNAs containing the indicated CRD segments were analyzed by in vitro translation in reticulocyte extracts treated with micrococcal nuclease as for Fig. 1. The results of 10 independent experiments with 10 different mRNA preparations are summarized in the graph on the right. The translational efficiency of each transcript was normalized to that of GMG mRNA. Error bars show the standard deviation.
Plasmids used to produce antisense RNA probes for the primer extension assay were created as follows. Exon 1 and intron 1 of the β-globin gene were amplified from cgenglobin (2) by PCR and cloned into pcDNA3 (Invitrogen) to create globinE1-I1. To create the neomycin antisense construct Neo-pcDNA3, the first 300 nucleotides of the neomycin phosphotransferase coding sequence were amplified by PCR and inserted into pcDNA3 (Invitrogen). The actin probe was synthesized from the pTRI-β-actin-mouse antisense template (Ambion).
Sequences of all oligonucleotides used for PCR amplification, mutagenesis, and sequencing are available upon request.
Reticulocyte lysate.
Reticulocytes were obtained from New Zealand White rabbits made anemic with phenylhydrazine (21). Cells were washed three times with ice-cold buffered saline and lysed with 1.5 volumes of water. The lysate was centrifuged at 2°C, 15,000 × g, and the membrane-free supernatant was supplemented with hemin to a final concentration of 0.02 mM, creatine kinase to a final concentration of 0.05 mg/ml, and CaCl2 to a final concentration of 1 mM. Micrococcal nuclease was then added to a final concentration of 150, 300, 600, or 900 U/ml. The lysate was incubated for 20 min at 20°C, and EGTA was added to a final concentration of 4 mM to block further nuclease digestion. The lysate was aliquoted and stored at −70°C.
RNA synthesis.
Capped mRNAs were synthesized with the mMessage Machine kit from Ambion using HindIII-linearized pSP6β/c-based plasmids as templates. The quality and concentration of each mRNA were analyzed by electrophoresis in 6% denaturing polyacrylamide gels and by measuring the optical density (OD) at 260 and 280 nm. To generate antisense RNA probes for RNase protection assays, Neo-pcDNA3, glo-E1-I1, and pTRI-β-actin-mouse were transcribed using the SP6-Maxiscript kit (Ambion) in reactions with 50 μCi of [α-32P]UTP (800 Ci/mmol; Amersham). Each probe was gel purified prior to use.
In vitro translation.
mRNA (200 ng) was incubated for 30 min at 30°C in a 20-μl reaction mixture containing 1 μl of KM solution (2 M KCl, 10 mM MgCl2), 1 μl of creatine phosphate (0.2 M), 2 μl of amino acid mix without methionine (1 mM; Promega), 1 μl of l-[35S]methionine (1,000 Ci/mmol; Amersham), and 14 μl of reticulocyte lysate. The reaction was stopped by adding 3 μl of 0.05 M EDTA and 2.5 μl of RNase A (1 mg/ml). Radiolabeled proteins were separated by electrophoresis in a 4 to 12% NuPAGE Bis-Tris gel (Novex).
For toeprint assays, the translation reaction was performed with complete, nonradioactive amino acid mixture for 0 or 15 min at 25°C. After the incubation, cycloheximide was added to a final concentration of 320 μM. To study the effect of inhibitors using the toeprint assay, cycloheximide (final concentration, 320 μM) or EDTA (final concentration, 5 mM) was added to the translation mixture. When reaction mixtures were incubated with EDTA, additional MgCl2 (final concentration, 5 mM) was added back prior to performing the primer extension reaction.
Toeprint analysis.
The toeprint extension inhibition assay was modified from previously described procedures (6, 16, 56). 32P-end-labeled primer (2 × 106 cpm; complementary to c-myc nucleotides 1764 to 1739 or to GAPDH nucleotides 949 to 923) was added to 2 μl of the translation reaction mixture. The reaction mixture was incubated for 5 min at 37°C, after which reverse transcription buffer (Gibco-BRL), deoxynucleoside triphosphate mix (final concentration, 0.25 mM), and dithiothreitol (final concentration, 10 mM) were added. The reaction was heated to 50°C for 2 min and immediately placed on ice. Superscript II reverse transcriptase (50 U; Gibco-BRL) was added, and the reaction mixture was incubated for 30 min at 30°C. cDNA was extracted using phenol-chloroform, followed by ethanol precipitation, and analyzed in a 6% denaturing polyacrylamide sequencing gel. Primer extension products were compared with a dideoxynucleotide sequence ladder obtained from the same 32P-end-labeled primer and the same mRNA used for the toeprint assay. The sequencing reaction contained 0.16 mM dideoxynucleotide.
Cell culture and transfection.
H4IIE cells, a rat hepatoma cell line containing small amounts of CRD-BP (unpublished observation), were cultured in α-minimal essential medium (MEM) (Gibco-BRL) supplemented with 10% fetal bovine serum (Gibco-BRL), 2 mM glutamine (Gibco-BRL), and 10 U of penicillin-streptomycin (Gibco-BRL) per ml at 37°C and 5% CO2. Then 5 × 105 cells were seeded to a 60-mm dish 24 h before transfection. Just prior to transfection, cells were washed twice with 37°C phosphate-buffered saline (PBS) and overlaid with 1 ml of Optimem. Plasmid DNA (1 μg) was mixed with 4.0 μl of Plus reagent (Gibco-BRL) and 25 μl of Optimem, and the mixture was incubated for 15 min at room temperature. Lipofectamine (4 μl; Gibco-BRL) and Optimem (25 μl) were added, and the mixture was incubated for another 15 min at room temperature. The DNA-Optimem solution was then added dropwise to the cells, which were incubated for 3 h at 37°C and 5% CO2. The DNA-Optimem solution was removed and replaced with growth medium, and the cells were harvested 36 to 48 h later.
RNA isolation and RNase protection analysis.
Total RNA was isolated using RNAwiz solution (Ambion) and analyzed by RNase protection using the RPAIII kit from Ambion. The Glo-E1-I1 probe was used to detect chimeric globin mRNAs. Neomycin phosphotransferase RNA and actin RNA were analyzed in parallel as transfection and gel loading controls. RNase-protected RNA fragments were separated in a 6% denaturing polyacrylamide gel. RNA abundance was calculated based on the GMG or GGG signal and normalized using the actin and neomycin signals.
RESULTS
Reduced in vitro translation of mRNAs containing the coding region instability determinant of c-myc mRNA.
c-myc mRNA is unstable in vivo, with a half-life of 30 to 60 min. Two segments of the mRNA account for its instability, an AU-rich element in the 3′ untranslated region and a CRD comprising the last 249 nucleotides of the coding region (nucleotides 1638 to 1886). A protein (CRD-BP) binds to the CRD and might protect the mRNA from rapid degradation during translational pausing. To test this model, we first compared the in vitro translation of two chimeric mRNAs, GMG and GGG. Each mRNA has a β-globin backbone into which a 249-nucleotide coding segment from c-myc or GAPDH mRNA was inserted (Fig. 1A). The c-myc insert corresponds to the CRD. We chose to analyze chimeric mRNAs rather than full-length c-myc mRNA in order to focus solely on the coding sequences that destabilize c-myc mRNA.
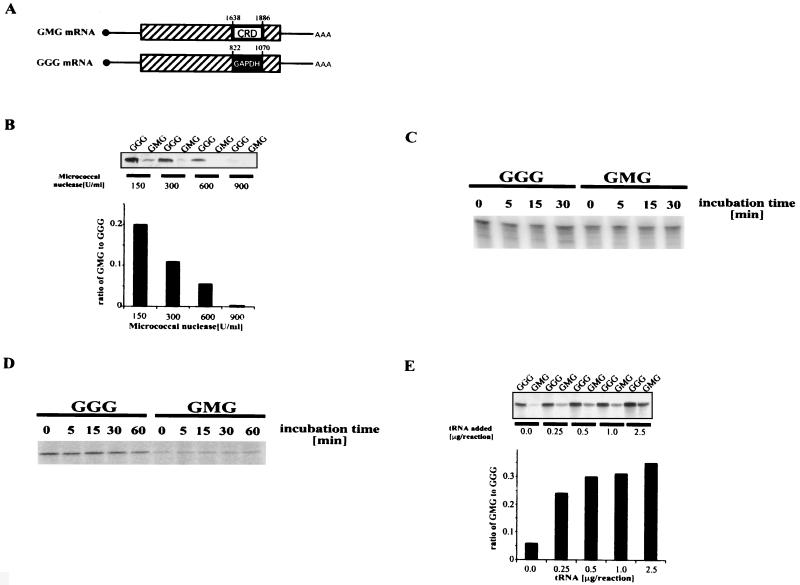
FIG. 1.
c-myc CRD and translational inefficiency. (A) Diagram of chimeric mRNAs GMG and GGG. Solid circle, cap site. Lines, 5′ and 3′ untranslated regions. Rectangle, coding region. Sequences (249 nucleotides) from the coding regions of c-myc and GAPDH mRNAs were inserted into the EcoRI site of human β-globin mRNA to generate GMG and GGG, respectively. Numbers above the inserted segments refer to the nucleotide number from each mRNA. (B) In vitro translation of GMG and GGG mRNAs under conditions of limiting tRNA. Reticulocyte lysate was treated with the indicated amount of micrococcal nuclease to reduce the levels of endogenous mRNA and tRNA. The nuclease was inactivated, and GGG and GMG mRNAs were added and translated in reaction mixtures containing [35S]methionine. Proteins were electrophoresed and visualized with a PhosphorImager. Top: GMG and GGG protein synthesis in nuclease-treated extracts. Bottom: data quantification. Translation efficiencies were adjusted for the number of methionines in each protein. (C) Stability of GGG and GMG mRNA under translation conditions. Radiolabeled, capped GGG and GMG mRNA were incubated in reticulocyte lysate treated with micrococcal nuclease (600 U/ml). At the indicated times, total RNA was extracted and analyzed by electrophoresis in a polyacrylamide gel. (D) Stability of GGG and GMG proteins under translation conditions. GGG and GMG mRNAs were translated in reticulocyte lysate treated with micrococcal nuclease (600 U/ml). Translation was halted after 30 min by adding 2.5 μl of RNase A (1 mg/ml) and incubating for 15 min at 37°C. Incubation was then continued for another 60 min at 30°C, during which time aliquots were harvested and analyzed by SDS-PAGE. Time zero indicates the start of the 30°C incubation. Radiolabeled proteins were visualized with a PhosphorImager. (E) Effect of additional tRNA on the GMG-to-GGG synthesis ratio. The indicated amounts of calf liver tRNA were added to reticulocyte lysate pretreated with 600 U of micrococcal nuclease/ml, and translation was performed as for panel B. Top: GMG and GGG protein synthesis in extracts supplemented with the indicated amounts of tRNA. Bottom: data quantification.
To assess the translation capacity of GGG and GMG mRNAs under limiting tRNA conditions, each mRNA was translated in a “homemade” reticulocyte lysate that had been treated with micrococcal nuclease and was thus deficient in tRNA. GMG mRNA was inefficiently translated relative to GGG mRNA, and the ratio of GMG to GGG protein synthesis was inversely proportional to the nuclease concentration (Fig. 1B). Three mechanisms could account for this result: GMG mRNA instability, GMG protein instability, or translational pausing.
To test the first possibility, radiolabeled, capped GGG and GMG mRNAs were incubated under translation conditions in reticulocyte lysate treated with 600 U of micrococcal nuclease/ml. RNA was isolated and analyzed in a polyacrylamide gel. Both mRNAs were stable during the 30-min incubation (Fig. 1C). To determine whether GMG protein was less stable than GGG protein, each mRNA was translated for 30 min under the same conditions as in Fig. 1A. The translation reaction was stopped by adding RNase A (see figure legend), and the reactions were then incubated for an additional 60 min at 30°C. GMG and GGG proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and both remained stable during the 60-min incubation (Fig. 1D). Therefore, the inefficient production of GMG protein observed in Fig. 1B did not result from the instability of GMG mRNA or protein.
To determine if tRNA depletion lowered GMG mRNA translation, tRNA-depleted extracts were supplemented with additional tRNA. The GMG-to-GGG synthesis ratio increased at least sixfold, from 0.06 to 0.36, when 0.5 μg of tRNA was added (Fig. 1E). Therefore, the reduced translation of GMG mRNA observed in Fig. 1B resulted from a deficiency of tRNA in the reactions. This result implied that GMG mRNA was undergoing translational pausing in the tRNA-depleted extracts. GMG mRNA is still translated less efficiently than GGG mRNA in tRNA-supplemented reactions, for reasons we do not understand.
Rare codons and ribosome pausing in the c-myc CRD.
The data in Fig. 1 suggested that ribosomes pause when they encounter rare codons in the c-myc CRD. It is unlikely that mRNA secondary structure reduced GMG translation, because tRNA supplementation would not be expected to alter mRNA structure (Fig. 1E). To test the ribosome-pausing hypothesis further, we shuffled the codons in the CRD by generating a −1 frameshift mutant, GMG-FS. The frameshift was made by removing a single nucleotide at the 5′ end of the CRD and adding a single nucleotide at the 3′ end (Fig. 2A). The globin segments were unchanged. Therefore, the reading frame of the GMG-FS CRD was drastically modified with minimal change in overall mRNA structure. The −1 frameshift also generated three in-frame stop codons. To avoid premature termination in the frameshifted mRNA, each stop codon was changed at one base each to create a sense codon (Fig. 2A).
As a control for these changes, the same three internal codon changes were introduced into the wild-type CRD reading frame to generate GMG*** mRNA. GMG*** has essentially the same CRD sequence as GMG-FS mRNA, but GMG*** mRNA is not translated out of frame. GMG and GMG*** mRNAs were both translated relatively inefficiently (Fig. 2B). Therefore, the three internal codon changes in GMG*** mRNA did not upregulate translation. In contrast, the translation efficiency of GMG-FS mRNA increased almost twofold (Fig. 2B). This result confirms that rare codons, not extensive secondary structure, account for the translational inefficiency of GMG mRNA. GMG-FS translation in tRNA-depleted extracts (Fig. 2B) was not as efficient as GMG translation in tRNA-supplemented extracts (Fig. 1E). A likely explanation for this result is that GMG-FS mRNA still contains a relatively high percentage of rare codons. (Rare codons comprise 29% and 25% of the CRD segments of GMG and GMG-FS mRNAs, respectively. In contrast, the GAPDH segment contains only 7% rare codons.) The fact that GMG-FS is more efficiently translated than GMG mRNA implies that at least some critical pause-inducing codons were altered in GMG-FS. This conclusion is supported by the experiments described below.
Role of the first 72 nucleotides in inefficient translation of CRD-containing mRNAs.
To identify which portion(s) of the CRD contributed to translational pausing, chimeric globin-myc mRNAs containing different CRD segments were translated in vitro. GM1779-1886G and GM1710-1886G mRNAs lack the first 141 nucleotides and the first 72 nucleotides of the CRD, respectively, and were translated 2.5-fold more efficiently than wild-type GMG (Fig. 3). In contrast, GM1638-1778G and GM1638-1709G mRNAs, which lack the last 108 nucleotides and the last 177 nucleotides of the CRD, respectively, were translated as inefficiently as GMG mRNA (Fig. 3). Thus, the codons primarily responsible for GMG translational inefficiency are located within the first 72 nucleotides of the CRD.
Translational pausing at the first arginine codon of the CRD.
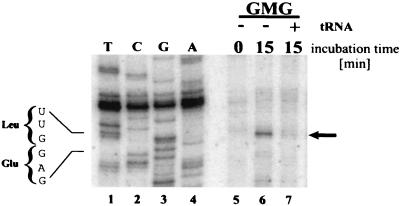
To map the location of at least some GMG mRNA pause sites, we exploited ribosome toeprinting assays (translation-coupled primer extension) (16). GMG and GGG mRNAs were translated for 0 or 15 min in tRNA-depleted reticulocyte extracts. Oligodeoxynucleotide primers for each mRNA were added, and the primers were extended by using reverse transcriptase. If ribosomes were paused upstream of the primer binding site, the primer would be extended until the polymerase encountered the stalled ribosome. The size of the extension product would then indicate the pausing site. Primers were used that annealed at the same relative positions within the GAPDH and CRD segments of GGG and GMG mRNAs, respectively.
Primer extension with deproteinized GMG and GGG mRNAs generated predominantly full-length transcripts, showing that the mRNAs themselves do not contain blocks to reverse transcription (data not shown). In contrast, a strong toeprint signal was observed with translating GMG mRNA (Fig. 4, top). A signal of comparable intensity was not observed with GGG mRNA. Three observations confirmed that the GMG toeprint signal was specific and was translation dependent. (i) The signal appeared only after translation had commenced (Fig. 4, compare lanes 8 and 9). (ii) The signal was dependent on the association of GMG mRNA with polysomes. Reaction mixtures incubated for 15 min were exposed to EDTA to disrupt polysomes. Magnesium was then added, and primer extension was performed. Under these conditions, the toeprint signal was eliminated (Fig. 4, lane 10). Therefore, the toeprint was dependent on polysome integrity and was not generated by mRNA cleavage products or by mRNA secondary structure. (iii) A toeprint signal was not observed when the translation elongation inhibitor cycloheximide was added to the reactions at the start of the 15-min incubation (data not shown).
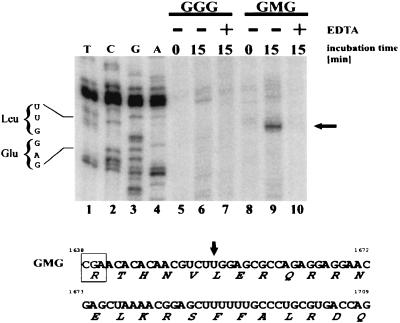
FIG. 4.
Mapping a ribosomal pause site in GMG mRNA via primer extension inhibition analysis (toeprint). GGG and GMG mRNAs were translated for 0 min (ice) or for 15 min at 25°C in reticulocyte extracts treated with micrococcal nuclease. For each mRNA, two reaction mixtures were incubated for 15 min. One was analyzed immediately by toeprint analysis, as per Materials and Methods (lanes 6 and 9). To the other reaction, EDTA was added to a final concentration of 5 mM (lanes 7 and 10) to dissociate polysomes. Magnesium was then added to allow the reverse transcriptase to function, after which primer extension was performed. Primer extension products were electrophoresed and visualized by phosphorimager. Lanes 1 to 4, a sequencing reaction with intact, deproteinized GMG mRNA as the substrate. The sequencing and toeprinting primers were identical. The sequence in the toeprint region is noted on the left. Arrow on right, prominent toeprint signal observed with GMG mRNA. The sequence on the bottom corresponds to the first 72 nucleotides of the c-myc CRD (1638 to 1709). Amino acids are indicated below each codon. Arrow, position of the toeprint signal (lane 9). Boxed codon, proposed ribosome pause site.
The GMG toeprint signal mapped to a UUG leucine codon located 15 to 18 nucleotides downstream from the start of the CRD (Fig. 4, top, lanes 1 to 4, and bottom). A single ribosome protects 30 to 35 nucleotides of mRNA, and the decoding site is located approximately in the middle of the ribosome (25, 51). Therefore, the toeprint for a paused ribosome should map 15 to 18 nucleotides downstream from the pause site (6), which places the pause at the rare arginine codon CGA (Fig. 4, bottom) (the CGA frequency is 6.3 per 103 human codons [35; http://www.kazusa.or.jp/codon/]). By exposing the autoradiograms for long periods, we observed additional pause sites downstream of the CGA (data not shown). These signals were neither as strong nor as reproducible as that resulting from the arginine codon. We conclude that ribosomes can pause at different sites within the CRD, but the first arginine codon is a particularly strong pause site. Further confirmation of this conclusion is that the CGA codon is eliminated in the frameshift mutant GMG-FS, which, in turn, is translated more efficiently than GMG mRNA (Fig. 2).
Elimination of the toeprint signal by excess tRNA and codon dependence of the toeprint signal.
Since excess tRNA enhanced the translation of CRD-containing mRNAs (Fig. 1E), we asked whether tRNA supplementation would also eliminate the GMG toeprint signal. The prominent UUG toeprint was eliminated when 2.5 μg of tRNA was added to the translation reaction mixtures (Fig. 5, lane 7). We then asked whether changing the rare arginine codon (CGA) to a more common codon (AGA) would also eliminate the UUG toeprint. GMGmt1 mRNA, which contained the CGA-to-AGA change, did not generate the UUG toeprint (Fig. 6A, lane 7). Therefore, the rare CGA codon is a true ribosome pause site. However, GMGmt1 translation did generate a new toeprint signal that mapped one codon downstream of the UUG (Fig. 6A, lane 7). This signal corresponds to a ribosome pause at threonine ACA, which is the second CRD codon (Fig. 6A, middle sequence).
FIG. 5.
Elimination of ribosome pausing by added tRNA. GMG mRNA was translated for the indicated times at 25°C in reticulocyte lysate treated with micrococcal nuclease. Calf liver tRNA (2.5 μg) was added to the indicated reaction prior to the 15-min incubation (lane 7). Primer extension inhibition analysis (toeprinting) was performed as in Fig. 4. Symbols are as in Fig. 4.
FIG. 6.
Analysis of GMG mRNAs with rare to common codon changes. (A) Toeprinting of GMG, GMGmt1, and GMGmt1,2 mRNAs. mRNAs were translated for the indicated times at 25°C in reticulocyte lysate treated with micrococcal nuclease. Toeprinting analysis was performed as for Fig. 4. Arrow on right, GMG mRNA toeprint signal. Arrowhead on right, GMGmt1 toeprint signal. Bottom, sequences of the first 36 nucleotides of the c-myc CRD segment of GMG, GMGmt1, and GMGmt1,2 mRNAs. In GMGmt1, the first arginine codon CGA (rare) is changed to AGA (common). In GMGmt1,2, the first arginine codon CGA (rare) and the second threonine codon ACA were changed to more common codons (CGG and ACC, respectively). The altered nucleotides are in larger type and are underlined. Arrows, positions of toeprint signals. Boxed codons, proposed ribosome pause sites. (B) Translation efficiencies of GMG, GMGmt1, and GMGmt1,2 mRNAs. Each mRNA was translated in vitro as for Fig. 1C. Codon changes are indicated by exclamation points. The translation efficiencies of the GMGmt1 and GMGmt1,2 mRNAs were normalized to that of GMG mRNA. The experiment was performed 10 times. Error bars show the standard deviation.
To confirm that the ACA codon is also a pause site, we translated mutant GMGmt1,2 mRNA, in which the arginine and threonine codons were both changed. No toeprint signal was observed with this mRNA (Fig. 6A, lane 9). Therefore, both codons can function as pause sites, although ACA is not a particularly rare codon in humans (14.9 per thousand human codons [35; http://www.kazusa.or.jp/codon/]). To confirm that the first two codons of the CRD depress translational efficiency, the in vitro translation rates of each mutant mRNA were compared. GMGmt1 and GMG mRNAs were translated with comparable efficiencies (Fig. 6B). This result was not surprising, because GMGmt1 mRNA contains the second (threonine) pause site, which should reduce translation efficiency even though the first rare codon was changed. GMGmt1,2 mRNA was translated almost twice as efficiently as GMG mRNA, confirming that both the arginine and threonine codons induce translational pausing and reduce translational efficiency.
Codon usage and mRNA expression in vivo.
At least two codons within the first 72 nucleotides of the c-myc CRD contribute to translational inefficiency and ribosome pausing in vitro. The CRD is also an mRNA instability element in vivo (19). Therefore, it was important to determine whether mRNA half-life in cells correlated with CRD translation in extracts. The prediction was that more efficiently translated mRNAs would be more stable and would accumulate to higher levels in cells. CRD-containing mRNAs were expressed in transiently transfected H4IIE cells (a rat hepatoma cell line). These cells were chosen because they express little or no CRD-BP (data not shown) and thus allow us to assess translational pausing and mRNA stability in an environment deficient in the CRD-shielding protein. Each transgene contained the same backbone, the human β-globin gene. These transgenes are actively transcribed in cells and have been used successfully to compare mRNA half-lives in cells (19). mRNA levels were assayed by RNase protection. The probe spanned the first globin intron, so that any signal would derive from mRNA, not pre-mRNA.
GGG mRNA was more abundant (more stable) than GMG mRNA (Fig. 7A), consistent with previous data (19). GM1710-1886G and GM1779-1886G mRNAs were translated more efficiently in vitro and were also more abundant than GMG mRNA in cells (Fig. 7A). This result is also consistent with the hypothesis that ribosome pausing exposes the CRD to endonuclease attack, leading to mRNA destruction. GM1638-1778G mRNA contains the translation pause site but was slightly more abundant than GGG mRNA (Fig. 7A). This observation was not peculiar to H4IIE cells, because GM1638-1778G mRNA was also relatively abundant in HO-15 cells, which express neither c-myc mRNA (32) nor the CRD-BP (data not shown). Therefore, the GM1638-1778G result seems inconsistent with a ribosome-pausing model, because GM1638-1778G mRNA contains the ribosome pause region and was translated inefficiently in vitro (Fig. 3). This apparent inconsistency likely results from the fact that mRNA instability requires two processes, ribosome pausing and recognition of the CRD by an mRNA-degrading RNase. c-myc mRNA is cleaved in vivo at several sites within the CRD, two of which map close to or just downstream of nucleotide 1778 (15). Therefore, GM1638-1778G mRNA lacks several nuclease cleavage sites, which could explain why it is more stable than GMG mRNA (see Discussion).
FIG. 7.
Abundance (stability) of GMG and modified mRNAs in transfected H4IIE cells. Transiently transfected H4IIE cells expressing the indicated chimeric mRNAs were harvested 36 to 48 h after transfection. Total cytoplasmic RNA was prepared and analyzed via RNase protection assay with probes for GMG, GGG, neomycin phosphotransferase, and actin mRNAs (Materials and Methods). The signal intensity of each protected fragment was quantified with a phosphorimager. The abundance of each mRNA was then adjusted to the internal controls (neomycin phosphotransferase and actin mRNAs). The histogram shows the relative abundance of each mRNA, with the GMG level set at 1. The histograms reflect the results of at least five independent transfection experiments. Error bars show the standard deviation. (A) Abundance of chimeric globin mRNAs containing CRD segments. (B) Abundance of mutant GMG mRNAs.
GMG-FS and GMGmt1,2 mRNAs were translated relatively efficiently in vitro and were more stable than GMG mRNA in cells (Fig. 7B). GMG*** and GMG mRNAs were translated inefficiently and were relatively unstable in cells. These data are consistent with a link between translational pausing and c-myc mRNA instability. GMGmt1 mRNA seems to be an exception. It was translated as inefficiently as GMG mRNA in vitro (Fig. 6B) but was more abundant than GMG mRNA in cells (Fig. 7B). We suggest that this result is readily explained by differences in tRNA abundance between cells and tRNA-depleted translation extracts. The first CRD codon, CGA (Arg), is rare in human and rat cells (6.3 and 6.8 per 103 codons, respectively). Therefore, tRNAArg(CGA) is probably rare in H4IIE cells, which explains why GMG and GMG*** mRNAs, both of which contain the Arg (CGA) codon, were unstable. In contrast, ACA is not a very rare codon in humans (14.9 per 103) or in rats (14.6 per 103). Therefore, H4IIE cells likely contain abundant tRNAThr(ACA), which would reduce or preclude ACA site pausing and account for the relative stability of GMGmt1 mRNA in cells.
DISCUSSION
Our data help to explain the function of a coding region instability determinant in regulating c-myc mRNA half-life. Previous work indicated that the CRD is an endonuclease target site unless it is shielded by the CRD-BP (28, 38). This model is particularly attractive for an mRNA such as c-myc, which must be degraded very rapidly during periods of starvation or differentiation. However, the model failed to explain how a protein could bind to the coding region if the mRNA was efficiently translated and the coding region was covered with ribosomes. The CRD-BP should be superfluous for an efficiently translated mRNA. Work reported here provides a rationale for the CRD-BP related to the observation that ribosomes pause within the 5′ segment of the CRD. Pausing would generate a ribosome-free downstream region, which would be attacked by an endonuclease unless it was protected by the CRD-BP. After elongation resumed, ribosomes would displace the CRD-BP from the mRNA, and c-Myc protein synthesis would be completed.
Ribosome pausing accounts for three observations. (i) CRD-containing mRNAs are translated relatively inefficiently (Fig. 1). (ii) Placing a stop codon upstream of the CRD stabilizes the mRNA in cells (19, 59). This observation is probably related to the association of both the CRD-BP and the endonuclease with ribosomes (12, 28). Thus, ribosomes might need to reach the 5′ segment of the CRD in order for either protein to gain access to the downstream CRD (see also below). (iii) With one exception, CRD mutations that increase translational efficiency and reduce ribosome pausing in vitro also lead to increased mRNA stability in cells (Fig. 7B). As noted above, this exception is readily explained by differences in tRNA abundance between cells and tRNA-depleted extracts.
We recognize that pausing seems to have only modest effects (twofold or so) on GMG mRNA translation. However, even a brief pause could have a major effect on the mRNA. We imagine pausing to be the starting gun for a race between the endonuclease and the CRD-BP for access to the (unprotected) CRD. The winner determines whether the mRNA is degraded or preserved, respectively. In either case, even the briefest pause might be sufficient for one of the proteins to gain access to the CRD. Therefore, our data showing a twofold change in translational efficiency are compatible with a measurable effect on c-myc mRNA metabolism.
Translational pausing or inefficiency in vitro maps to two codons, arginine CGA and threonine ACA (Fig. 4 and 6). Deleting or modifying these codons increases translation efficiency (Fig. 3 and 6B). The influence of rare codons on gene expression is well documented in lower organisms such as Escherichia coli, in which rare arginine codons downregulate translation significantly (9, 23, 49, 63). Less is understood about the function(s) of rare codons and tRNA pools in higher eukaryotes, although it is known that amino acid deprivation can slow protein synthesis and either increase or decrease mRNA half-lives. For example, albumin mRNA is destabilized in a tryptophan-depleted rat hepatoma cell line (31), while amino acid depletion stabilizes cationic amino acid transporter-1 mRNA (1).
We do not fully understand why the 5′ segment of the c-myc CRD influences translation and mRNA abundance to such an extent (Fig. 2, 3, and 7), because other segments of the CRD also contain rare codons. Perhaps pausing results from some special property of the arginine and threonine codons, such as their contiguity. Codon contiguity can accentuate translational pausing in lower organisms (40).
c-myc CRD as a bipartite mRNA instability element: bipartite CRDs in other mRNAs.
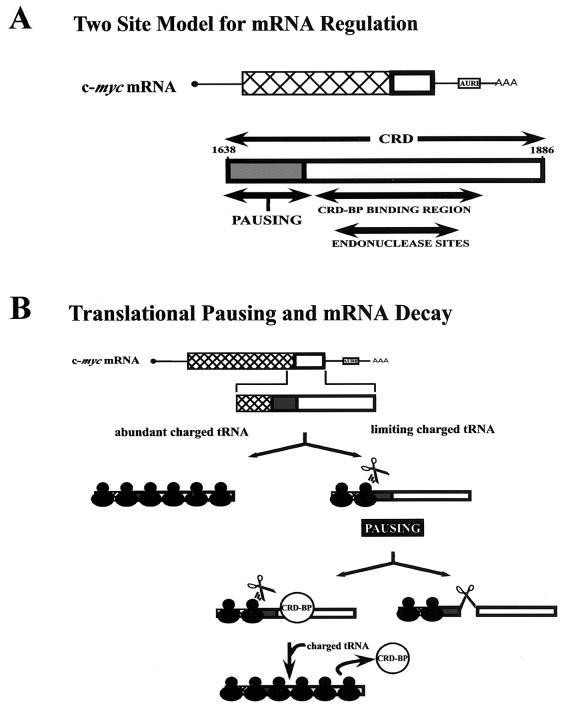
Our data also suggest that the CRD is bipartite (Fig. 8A). The first 72 nucleotides contain the codons that induce pausing. The downstream 177 nucleotides contain sites for CRD-BP binding and endonuclease cleavage. The bipartite nature of the CRD could explain why GM1638-1778G mRNA is relatively abundant in cells (Fig. 7A). This mRNA includes the translation pause sites but lacks almost half of the CRD. It might undergo translational pausing but not rapid degradation, because it is a relatively poor endonuclease substrate. Two observations support this idea. (i) The endonuclease cleavage sites appear to be compromised in GM1638-1778G mRNA. A major in vitro cleavage site maps upstream of nucleotide 1778 (28). The same cleavage site plus four additional sites were observed in murine erythroleukemia cells (15). Two of the additional sites (at nucleotides 1754 and 1771) are close to the 3′ end of the 1638 to 1778 segment, while the third site (nucleotide 1819) is deleted from GM1638-1778G mRNA. (ii) The secondary structure of the CRD, which might play a role in endonuclease recognition, is disrupted in GM1638-1778G mRNA. A stem-loop element comprising nucleotides 1705 to 1790 was identified by chemical and enzymatic mapping of the CRD (unpublished observations). This stem-loop is essential for CRD-BP binding and, as determined by the Mfold RNA-folding program (33, 65), is disrupted in GM1638-1778G mRNA. Disruption of this secondary structure might also reduce endonuclease cleavage and thereby stabilize the mRNA.
FIG. 8.
Model for the link between c-myc mRNA translation and instability. (A) Bipartite nature of the CRD. The diagrams are based on data in Fig. 1 to 7 plus published observations. The CRD of c-myc mRNA is proposed to contain two functional segments. The upstream segment can undergo translational pausing, depending on tRNA and amino acid abundance. The downstream segment contains both a binding site for a shielding protein, the CRD-BP, and an endonuclease cleavage site. Nucleotide 1886 is the last coding nucleotide of the mRNA. (B) Kinetic model for endonucleolytic degradation of c-myc mRNA. The model suggests that c-myc mRNA is efficiently translated when charged tRNAs are abundant. Under conditions such as starvation (see Discussion), the abundance of charged tRNAs becomes limiting. Ribosomes pause within the first 72 nucleotides of the CRD, and the mRNA is then subject to either of two fates. (i) It is degraded by a ribosome-associated endonuclease (scissors). The endonuclease attacks the ribosome-free segment of the CRD located downstream of the paused ribosome. (ii) The CRD segment is shielded from endonuclease attack by the CRD-BP, helping to stabilize the mRNA. When the rare tRNA at last reaches the ribosome, elongation resumes. Translating ribosomes displace the CRD-BP from the mRNA and complete c-Myc protein synthesis.
Other mRNAs contain bipartite coding region stability and translation determinants. MATα1 mRNA of S. cerevisiae contains a 65-nucleotide instability element with rare codons in its 5′ half and an AU-rich element in its 3′ half (7, 17, 36). The rare codons, the AU-rich element, and translation of the determinant are necessary for maximal mRNA destabilization. Therefore, the MATα1 and c-myc CRDs share several important properties, but they function in different ways. The MATα1 CRD mediates mRNA deadenylation and decapping (7, 17). The c-myc CRD does not accelerate deadenylation and decapping (59) but is a proposed endonuclease target (28). c-fos mRNA contains a ≈320-nucleotide CRD (47) that functions as an instability element only if it is translated (44). However, the c-fos CRD differs in several ways from the c-myc CRD: (i) it is recognized by virtue of its sequence or structure (57), and (ii) it promotes accelerated deadenylation, not endonucleolytic decay (14).
AU-rich element, CRD, and alternative pathways for c-myc mRNA decay.
We and others have described two alternative pathways for c-myc mRNA destruction, either 3′ to 5′ (deadenylation dependent) or endonucleolytic (3, 5, 19, 27, 52, 59, 62). Each pathway presumably depends on an instability determinant, the AU-rich element or CRD, respectively. We do not understand the conditions required for activating either pathway or for regulating the association of the CRD-BP with c-myc mRNA. Presumably, the CRD-BP functions primarily or exclusively in fetal and neoplastic cells, because the CRD-BP is abundant in fetal tissues, many cell lines, and some tumors but is scarce or absent in normal adult tissues (29, 43).
Results reported here and elsewhere suggest three situations during which the CRD-BP might play an important role: starvation, embryonic cell differentiation, and mitosis.
(i) Starvation.
Arginine/threonine scarcity might induce ribosome pausing in starved fetal or neoplastic cells. If the CRD-BP blocked rapid c-myc mRNA destruction, cells would maintain a pool of c-myc mRNA to allow growth and differentiation to resume without a significant lag when amino acids were replenished.
(ii) Differentiation.
c-myc mRNA is degraded via a deadenylation-dependent pathway in undifferentiated HL60 cells but is degraded quite rapidly by an endonucleolytic pathway when cells are induced to differentiate (52). When myoblasts differentiate into myotubes, c-myc mRNA is downregulated in a translation-dependent manner that requires determinants in coding regions from exons 2 and 3 (61), which include the CRD. These data suggest that undifferentiated cells require abundant c-myc mRNA to replicate but must degrade c-myc mRNA rapidly to differentiate. Perhaps the CRD-BP plays a role in rapid c-myc mRNA degradation during fetal cell differentiation.
(iii) Mitosis.
Transcription and translation are inhibited by approximately 75% between prometaphase and telophase (37). In late telophase or shortly thereafter, transcription and translation rates return to normal (G1) levels. Perhaps the CRD-BP is essential to stabilize c-myc mRNA during mitosis and thereby to preserve it as cells enter the next G1 phase. Since the c-myc mRNA half-life is 30 to 60 min, its levels might decrease by 10-fold or more by the end of mitosis were the mRNA not stabilized through the transcriptional-translational block of M phase (41). The deficiency in c-myc mRNA and, by inference, c-Myc protein might have deleterious effects as cells entered the next G1 phase.
In summary, stabilizing the mRNA during periods of translation arrest induced by starvation or mitosis might be a significant benefit to cells. Additional experiments are required to test these models.
Acknowledgments
We thank Andreas Kuhn and Brad Berberet for critical reading of the manuscript, Jia Qian for assistance with the initial phases of this study, and Brad Berberet, Anisa Kaenjak-Angeletti, and Charles Tessier for helpful suggestions. We also thank John Sedivy for providing the HO-15 cells.
This work was supported by National Cancer Institute grants R01-CA78710 (to J.R.) and P30-CA07175 (Cancer Center Support Grant to the McArdle Laboratory) and by a grant from the University of Wisconsin Robert Draper Technology Innovation Fund (to J.R.).
REFERENCES
- 1.Aulak, K. S., R. Mishra, L. Zhou, S. L. Hyatt, W. de Jonge, W. Lamers, M. Snider, and M. Hatzoglou. 1999. Post-transcriptional regulation of the arginine transporter Cat-1 by amino acid availability. J. Biol. Chem. 274:30424-30432. [DOI] [PubMed] [Google Scholar]
- 2.Bernstein, P. L., D. J. Herrick, R. D. Prokipcak, and J. Ross. 1992. Control of c-myc mRNA half-life in vitro by a protein capable of binding to a coding region stability determinant. Genes Dev. 6:642-654. [DOI] [PubMed] [Google Scholar]
- 3.Bonnieu, A., P. Roux, L. Marty, P. Jeanteur, and M. Piechaczyk. 1990. AUUUA motifs are dispensable for rapid degradation of the mouse c-myc RNA. Oncogene 5:1585-1588. [PubMed] [Google Scholar]
- 4.Brewer, G. 1991. An A+U-rich element RNA-binding factor regulates c-myc mRNA stability in vitro. Mol. Cell. Biol. 11:2460-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brewer, G., and J. Ross. 1988. Poly(A) shortening and degradation of the 3′ A+U-rich sequences of human c-myc mRNA in a cell-free system. Mol. Cell. Biol. 8:1697-1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cao, J., and A. P. Geballe. 1996. Coding sequence-dependent ribosomal arrest at termination of translation. Mol. Cell. Biol. 16:603-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caponigro, G., D. Muhlrad, and R. Parker. 1993. A small segment of the MATα 1 transcript promotes mRNA decay in Saccharomyces cerevisiae: a stimulatory role for rare codons. Mol. Cell. Biol. 13:5141-5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, C. Y., and A. B. Shyu. 1995. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem. Sci. 20:465-470. [DOI] [PubMed] [Google Scholar]
- 9.Chen, G. T., and M. Inouye. 1994. Role of the AGA/AGG codons, the rarest codons in global gene expression in Escherichia coli. Genes Dev. 8:2641-2652. [DOI] [PubMed] [Google Scholar]
- 10.Day, D. A., and M. F. Tuite. 1998. Post-transcriptional gene regulatory mechanisms in eukaryotes: an overview. J. Endocrinol. 157:361-371. [DOI] [PubMed] [Google Scholar]
- 11.Decker, C. J., and R. Parker. 1994. Mechanisms of mRNA degradation in eukaryotes. Trends Biochem. Sci. 19:336-340. [DOI] [PubMed] [Google Scholar]
- 12.Doyle, G. A., N. A. Betz, P. F. Leeds, A. J. Fleisig, R. D. Prokipcak, and J. Ross. 1998. The c-myc coding region determinant-binding protein: a member of a family of KH domain RNA-binding proteins. Nucleic Acids Res. 26:5036-5044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Evan, G. I., A. H. Wyllie, C. S. Gilbert, T. D. Littlewood, H. Land, M. Brooks, C. M. Waters, L. Z. Penn, and D. C. Hancock. 1992. Induction of apoptosis in fibroblasts by c-myc protein. Cell 69:119-128. [DOI] [PubMed] [Google Scholar]
- 14.Grosset, C., C. Y. Chen, N. Xu, N. Sonenberg, H. Jacquemin-Sablon, and A. B. Shyu. 2000. A mechanism for translationally coupled mRNA turnover: interaction between the poly(A) tail and a c-fos RNA coding determinant via a protein complex. Cell 103:29-40. [DOI] [PubMed] [Google Scholar]
- 15.Hanson, M. N., and D. R. Schoenberg. 2001. Identification of in vivo mRNA decay intermediates corresponding to sites of in vitro cleavage by polysomal ribonuclease 1. J. Biol. Chem. 276:12331-12337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hartz, D., D. S. McPheeters, R. Traut, and L. Gold. 1988. Extension inhibition analysis of translation initiation complexes. Methods Enzymol. 164:419-425. [DOI] [PubMed] [Google Scholar]
- 17.Hennigan, A. N., and A. Jacobson. 1996. Functional mapping of the translation-dependent instability element of yeast MATα1 mRNA. Mol. Cell. Biol. 16:3833-3843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hentze, M. W., and L. C. Kuhn. 1996. Molecular control of vertebrate iron metabolism: mRNA-based regulatory circuits operated by iron, nitric oxide, and oxidative stress. Proc. Natl. Acad. Sci. USA 93:8175-8182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herrick, D. J., and J. Ross. 1994. The half-life of c-myc mRNA in growing and serum-stimulated cells: influence of the coding and 3′ untranslated regions and role of ribosome translocation. Mol. Cell. Biol. 14:2119-2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ikemura, T. 1982. Correlation between the abundance of yeast transfer RNAs and the occurrence of the respective codons in protein genes. Differences in synonymous codon choice patterns of yeast and Escherichia coli with reference to the abundance of isoaccepting transfer RNAs. J. Mol. Biol. 158:573-597. [DOI] [PubMed] [Google Scholar]
- 21.Jackson, R. J., and T. Hunt. 1983. Preparation and use of nuclease-treated rabbit reticulocyte lysates for the translation of eukaryotic messenger RNA. Methods Enzymol. 96:50-74. [DOI] [PubMed] [Google Scholar]
- 22.Jacobson, A., and S. W. Peltz. 1996. Interrelationships of the pathways of mRNA decay and translation in eukaryotic cells. Annu. Rev. Biochem. 65:693-739. [DOI] [PubMed] [Google Scholar]
- 23.Kawakami, K., S. Pande, B. Faiola, D. P. Moore, J. D. Boeke, P. J. Farabaugh, J. N. Strathern, Y. Nakamura, and D. J. Garfinkel. 1993. A rare tRNA-Arg(CCU) that regulates Ty1 element ribosomal frameshifting is essential for Ty1 retrotransposition in Saccharomyces cerevisiae. Genetics 135:309-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim, J., P. G. Klein, and J. E. Mullet. 1991. Ribosomes pause at specific sites during synthesis of membrane-bound chloroplast reaction center protein D1. J. Biol. Chem. 266:14931-14938. [PubMed] [Google Scholar]
- 25.Kozak, M. 1983. Comparison of initiation of protein synthesis in prokaryotes, eukaryotes, and organelles. Microbiol. Rev. 47:1-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lachman, H. M., G. H. Cheng, and A. I. Skoultchi. 1986. Transfection of mouse erythroleukemia cells with myc sequences changes the rate of induced commitment to differentiate. Proc. Natl. Acad. Sci. USA 83:6480-6484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lavenu, A., S. Pistoi, S. Pournin, C. Babinet, and D. Morello. 1995. Both coding exons of the c-myc gene contribute to its posttranscriptional regulation in the quiescent liver and regenerating liver and after protein synthesis inhibition. Mol. Cell. Biol. 15:4410-4419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee, C. H., P. Leeds, and J. Ross. 1998. Purification and characterization of a polysome-associated endoribonuclease that degrades c-myc mRNA in vitro. J. Biol. Chem. 273:25261-25271. [DOI] [PubMed] [Google Scholar]
- 29.Leeds, P., B. T. Kren, J. M. Boylan, N. A. Betz, C. J. Steer, P. A. Gruppuso, and J. Ross. 1997. Developmental regulation of CRD-BP, an RNA-binding protein that stabilizes c-myc mRNA in vitro. Oncogene 14:1279-1286. [DOI] [PubMed] [Google Scholar]
- 30.Lipp, J., B. Dobberstein, and M. T. Haeuptle. 1987. Signal recognition particle arrests elongation of nascent secretory and membrane proteins at multiple sites in a transient manner. J. Biol. Chem. 262:1680-1684. [PubMed] [Google Scholar]
- 31.Marten, N. W., E. J. Burke, J. M. Hayden, and D. S. Straus. 1994. Effect of amino acid limitation on the expression of 19 genes in rat hepatoma cells. FASEB J. 8:538-544. [DOI] [PubMed] [Google Scholar]
- 32.Mateyak, M. K., A. J. Obaya, S. Adachi, and J. M. Sedivy. 1997. Phenotypes of c-Myc-deficient rat fibroblasts isolated by targeted homologous recombination. Cell. Growth Differ. 8:1039-1048. [PubMed] [Google Scholar]
- 33.Mathews, D. H., J. Sabina, M. Zuker, and D. H. Turner. 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 288:911-940. [DOI] [PubMed] [Google Scholar]
- 34.Muhlrad, D., and R. Parker. 1994. Premature translational termination triggers mRNA decapping. Nature 370:578-581. [DOI] [PubMed] [Google Scholar]
- 35.Nakamura, Y., T. Gojobori, and T. Ikemura. 1999. Codon usage tabulated from the international DNA sequence databases: its status 1999. Nucleic Acids Res. 27:292.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parker, R., and A. Jacobson. 1990. Translation and a 42-nucleotide segment within the coding region of the mRNA encoded by the MATα1 gene are involved in promoting rapid mRNA decay in yeast. Proc. Natl. Acad. Sci. USA 87:2780-2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prescott, D. M. (ed.). 1976. Reproduction of eukaryotic cells. Academic Press, New York, N.Y.
- 38.Prokipcak, R. D., D. J. Herrick, and J. Ross. 1994. Purification and properties of a protein that binds to the C-terminal coding region of human c-myc mRNA. J. Biol. Chem. 269:9261-9269. [PubMed] [Google Scholar]
- 39.Ragheb, J. A., M. Deen, and R. H. Schwartz. 1999. CD28-mediated regulation of mRNA stability requires sequences within the coding region of the IL-2 mRNA. J. Immunol. 163:120-129. [PubMed] [Google Scholar]
- 40.Rosenberg, A. H., E. Goldman, J. J. Dunn, F. W. Studier, and G. Zubay. 1993. Effects of consecutive AGG codons on translation in Escherichia coli, demonstrated with a versatile codon test system. J. Bacteriol. 175:716-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ross, J. 1997. A hypothesis to explain why translation inhibitors stabilize mRNAs in mammalian cells: mRNA stability and mitosis. Bioessays 19:527-529. [DOI] [PubMed] [Google Scholar]
- 42.Ross, J. 1995. mRNA stability in mammalian cells. Microbiol. Rev. 59:423-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ross, J., I. Lemm, and B. Berberet. 2001. Overexpression of an mRNA-binding protein in human colorectal cancer. Oncogene 20:6544-6550. [DOI] [PubMed] [Google Scholar]
- 44.Schiavi, S. C., C. L. Wellington, A. B. Shyu, C. Y. Chen, M. E. Greenberg, and J. G. Belasco. 1994. Multiple elements in the c-fos protein-coding region facilitate mRNA deadenylation and decay by a mechanism coupled to translation. J. Biol. Chem. 269:3441-3448. [PubMed] [Google Scholar]
- 45.Schwartz, D. C., and R. Parker. 1999. Mutations in translation initiation factors lead to increased rates of deadenylation and decapping of mRNAs in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:5247-5256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shichiri, M., K. D. Hanson, and J. M. Sedivy. 1993. Effects of c-myc expression on proliferation, quiescence, and the G0 to G1 transition in nontransformed cells. Cell. Growth Differ. 4:93-104. [PubMed] [Google Scholar]
- 47.Shyu, A. B., J. G. Belasco, and M. E. Greenberg. 1991. Two distinct destabilizing elements in the c-fos message trigger deadenylation as a first step in rapid mRNA decay. Genes Dev. 5:221-231. [DOI] [PubMed] [Google Scholar]
- 48.Somogyi, P., A. J. Jenner, I. Brierley, and S. C. Inglis. 1993. Ribosomal pausing during translation of an RNA pseudoknot. Mol. Cell. Biol. 13:6931-6940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Spanjaard, R. A., K. Chen, J. R. Walker, and J. van Duin. 1990. Frameshift suppression at tandem AGA and AGG codons by cloned tRNA genes: assigning a codon to argU tRNA and T4 tRNA(Arg). Nucleic Acids Res. 18:5031-5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Spencer, C. A., and M. Groudine. 1991. Control of c-myc regulation in normal and neoplastic cells. Adv. Cancer Res. 56:1-48. [DOI] [PubMed] [Google Scholar]
- 51.Steitz, J. A. 1980. RNA-RNA interactions during polypeptide chain initiation, p. 479-495. In G. Chambliss, J. Davies, K. Davis, L. Kahan, and M. Nomura (ed.), Ribosomes: structure, function, and genetics. University Park Press, Baltimore, Md.
- 52.Swartwout, S. G., H. Preisler, W. D. Guan, and A. J. Kinniburgh. 1987. Relatively stable population of c-myc RNA that lacks long poly(A). Mol. Cell. Biol. 7:2052-2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tharun, S., and R. Parker. 1997. Mechanisms of mRNA turnover in eukaryotic cells, p. 181-199. In J. B. Harford and D. R. Morris (ed.), mRNA metabolism and post-transcriptional gene regulation. Wiley-Liss, Inc., New York, N.Y.
- 54.Theodorakis, N. G., and D. W. Cleveland. 1996. Translationally coupled degradation of mRNA in eukaryotes, p. 631-652. In M. B. Mathews, J. W. B. Hershey, and N. Sonenberg (ed.), Translational control. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 55.Varenne, S., J. Buc, R. Lloubes, and C. Lazdunski. 1984. Translation is a non-uniform process. Effect of tRNA availability on the rate of elongation of nascent polypeptide chains. J. Mol. Biol. 180:549-576. [DOI] [PubMed] [Google Scholar]
- 56.Wang, Z., and M. S. Sachs. 1997. Ribosome stalling is responsible for arginine-specific translational attenuation in Neurospora crassa. Mol. Cell. Biol. 17:4904-4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wellington, C. L., M. E. Greenberg, and J. G. Belasco. 1993. The destabilizing elements in the coding region of c-fos mRNA are recognized as RNA. Mol. Cell. Biol. 13:5034-5042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wells, S. E., P. E. Hillner, R. D. Vale, and A. B. Sachs. 1998. Circularization of mRNA by eukaryotic translation initiation factors. Mol. Cell 2:135-140. [DOI] [PubMed] [Google Scholar]
- 59.Wisdom, R., and W. Lee. 1991. The protein-coding region of c-myc mRNA contains a sequence that specifies rapid mRNA turnover and induction by protein synthesis inhibitors. Genes Dev. 5:232-243. [DOI] [PubMed] [Google Scholar]
- 60.Wolin, S. L., and P. Walter. 1988. Ribosome pausing and stacking during translation of a eukaryotic mRNA. EMBO J. 7:3559-3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yeilding, N. M., W. N. Procopio, M. T. Rehman, and W. M. Lee. 1998. c-myc mRNA is down-regulated during myogenic differentiation by accelerated decay that depends on translation of regulatory coding elements. J. Biol. Chem. 273:15749-15757. [DOI] [PubMed] [Google Scholar]
- 62.Yeilding, N. M., M. T. Rehman, and W. M. Lee. 1996. Identification of sequences in c-myc mRNA that regulate its steady-state levels. Mol. Cell. Biol. 16:3511-3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zahn, K. 1996. Overexpression of an mRNA dependent on rare codons inhibits protein synthesis and cell growth. J. Bacteriol. 178:2926-2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zubiaga, A. M., J. G. Belasco, and M. E. Greenberg. 1995. The nonamer UUAUUUAUU is the key AU-rich sequence motif that mediates mRNA degradation. Mol. Cell. Biol. 15:2219-2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zuker, M., D. H. Mathews, and D. H. Turner. 1999. Algorithms and thermodynamics for RNA secondary structure prediction: a practical guide, p. 11-43. In J. Barciszewski and B. F. C. Clark (ed.), RNA biochemistry and biotechnology. Kluwer Academic Publishers, Dordrecht, The Netherlands.