Abstract
The general transcription factor TFIID is a multisubunit complex of TATA-binding protein (TBP) and 14 distinct TBP-associated factors (TAFs). Although TFIID constituents are required for transcription initiation of most mRNA encoding genes, the mechanism of TFIID action remains unclear. To gain insight into TFIID function, we sought to generate a proteomic catalogue of proteins specifically interacting with TFIID subunits. Toward this end, TFIID was systematically immunopurified by using polyclonal antibodies directed against each subunit, and the constellation of TBP- and TAF-associated proteins was directly identified by coupled multidimensional liquid chromatography and tandem mass spectrometry. A number of novel protein-protein associations were observed, and several were characterized in detail. These interactions include association between TBP and the RSC chromatin remodeling complex, the TAF17p-dependent association of the Swi6p transactivator protein with TFIID, and the identification of three novel subunits of the SAGA acetyltransferase complex, including a putative ubiquitin-specific protease component. Our results provide important new insights into the mechanisms of mRNA gene transcription and demonstrate the feasibility of constructing a complete proteomic interaction map of the eukaryotic transcription apparatus.
One of the most important challenges facing modern biology is to define the native context in which a given protein functions. It has become increasingly clear that most proteins work not alone but within large multisubunit complexes. Typically, multiprotein factors are defined by first subjecting a chromatographically purified fraction to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) separation and individual gel-separated protein bands are then excised and subjected to mass spectrometric protein identification (48a). Although certainly proven and powerful, in the postgenomic era novel approaches to protein identification are needed which do not require tedious protein band excision and are also amenable to systematic large-scale analyses of macromolecular complexes isolated under a range of physiological conditions. A recently developed multidimensional mass spectrometry approach, direct analysis of large protein complexes (DALPC), offers such capabilities by coupling multidimensional liquid chromatography with tandem mass spectrometry (MS/MS) (35).
DALPC differs from conventional mass spectrometric protein identification in one important aspect. Instead of identification of gel-separated proteins, complex protein mixtures are proteolyzed directly, and the resulting peptides are fractionated twice by chromatography (Fig. 1B). In the first chromatographic step peptides are fractionated over a strong-cation-exchange (SCX) microcapillary column. The resulting individual SCX column fractions are then applied to a reversed-phase high-pressure liquid chromatography (RP-HPLC) column that is eluted with a gradient of acetonitrile. The peptide content of the eluate of this RP-HPLC column is analyzed directly by electrospray ionization (ESI)-MS/MS. The protein composition of the original sample is then deduced from the thousands of individual MS/MS spectra collected (per initial protein sample; 8,000 to 10,000 MS/MS spectra/sample in this study) by genome-assisted computer analyses (15). The two-dimensional chromatographic prefractionation prior to ESI-MS/MS analysis dramatically reduces the peptide (protein) complexity of any protein mixture analyzed by MS/MS, consequently allowing for the analysis of more complex samples, protein samples comprised of hundreds to thousands of distinct proteins in a single DALPC run (35, 55, 56).
FIG. 1.
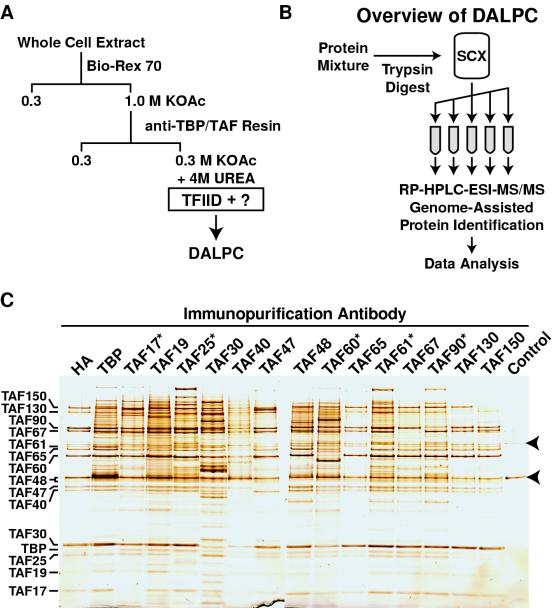
Strategy for the identification of proteins associated with TBP and TAF subunits of TFIID. (A) Outline of Bio-Rex 70 chromatographic fractionation and immunopurification procedure. (B) DALPC schematic. See the text for details. The number of fractions collected from the SCX microcapillary column can be increased depending on the complexity of the initial tryptic peptide (i.e., protein) mixture. (C) SDS-PAGE determination of the complexity of protein fractions immunopurified by using antigen-affinity-purified antibodies raised against all 15 TFIID subunits. A 5- to 10 μl-aliquot of each immunopurified fraction was subjected to SDS-PAGE on a 10% NuPAGE gel run with morpholinepropanesulfonic acid buffer (Invitrogen), and the resolved proteins were visualized by silver staining. Antibodies used for immunopurification are listed across the top, and TFIID subunits are marked on the left. A sample of TFIID immunopurified with anti-HA (HA) MAb IgG from a Bio-Rex 70 fraction derived from an HA-TAF130-tagged strain as described in Materials and Methods serves as a positive control and indicates the mobility of the 15 known TFIID subunits. At this level of analysis this anti-HA immunopurified fraction contains only TFIID subunits, except for the presence of two contaminating proteins (see negative control fraction, labeled “Control,” and polypeptides indicated by arrowheads). The asterisk indicates the five TAFs that are shared between TFIID and SAGA. Note that the anti-TAF40p IgG immunopurified fraction required more sample (20 μl) for SDS-PAGE due to low immunopurification efficiency.
Because protein fractions are analyzed directly without band excision, DALPC offers the ability to characterize both stoichiometric and substoichiometric components of a complex (53). Indeed, even nonabundant proteins that fail to stain and hence would be impossible to reproducibly excise from an SDS-PAGE gel are detected by DALPC. Thus, by combining DALPC with systematic purification, the possibility exists to define a proteomic map of potential protein-protein associations between all of the components that comprise complex multisubunit, multicomponent cellular machines such as the eukaryotic transcription apparatus. Despite this high potential, however, it has yet to be demonstrated that DALPC (or any other protein identification technology) can be reliably utilized on a large scale to systematically define multiprotein complexes and characterize proteins that associate with such assemblies.
The general transcription factor (GTF) TFIID plays a central role in the initiation of mRNA gene transcription. TFIID is the only GTF with specific TATA box-binding activity and the expression of many RNA polymerase II (RNAP II)-transcribed genes is dependent upon TFIID function (1, 21). Saccharomyces cerevisiae TFIID is composed of 15 subunits: TATA-binding protein (TBP) and 14 distinct TBP-associated factors (TAFs) (45), all of which have been highly evolutionarily conserved (5, 16). TBP is responsible for the TATA box-binding activity of TFIID, but the precise roles played by TAFs within TFIID are still an area of intense investigation. Initial models argued that TAFs could function as either core promoter selectivity factors or as general coactivators, serving as receptors integrating signals between DNA-bound gene-specific trans-acting factors and the general transcription machinery (1). The coactivator model of TAF function, which was based primarily upon in vitro studies with either individual recombinant subunits or subcomplexes of TFIID subunits, was called into question by early studies in yeast (1). Although recent in vivo studies are consistent with the coactivator function of TAFs (34), a functionally relevant, mutationally sensitive in vivo interaction between a transactivator and a native TFIID complex has yet to be demonstrated by combined genetic and biochemical means.
In addition to the core promoter recognition and coactivator functions of TFIID, evidence has accumulated that suggests TFIID may interact with a variety of gene regulatory factors. First, studies have established a link between TFIID and RNA processing as recruitment of cleavage-polyadenylation specificity factor (CPSF) to promoters is mediated by interaction with TFIID in mammalian cells (12). Second, our own yeast two-hybrid analyses (29), as well as global two-hybrid screening (52), suggests that the TAF subunits of TFIID may interact with a variety of factors. Finally, mass spectrometric analyses of TFIID subunits indicates several TFIID components are subject to a number of posttranslational modifications such as phosphorylation, methylation, and acetylation (45; A. Link, unpublished observations). These data suggest that TFIID may be the target of, and interact with, components of cellular signaling pathways. Although our studies have revealed only a single major yeast TFIID complex, multiple TFIID and non-TFIID TAF-containing complexes have been characterized in metazoan cells (4, 5, 47). TBP is a component of multiple transcription factors (33), and five yeast TAFs are shared between TFIID and the SAGA histone acetyltransferase complex (20). Together, these data suggest the possibility of additional proteins capable of associating with TBP and/or TFIID-TAFs in the yeast S. cerevisiae.
Here we report our efforts to gain insight into the function of TFIID by identifying proteins that can physically associate with the components of this important transcription factor. TBP- and TAF-containing complexes were systematically immunopurified, and proteins that specifically associated with these TFIID subunits were directly identified by DALPC. Our results reveal a number of novel connections between TBP, TAFs, and other factors, including interactions between TBP and the RSC chromatin remodeling complex. We also identified several previously uncharacterized subunits of the RSC and SAGA chromatin remodeling complexes. Finally, we showed that the yeast transactivator Swi6p was a TFIID-interacting factor and that a C-terminal truncation mutation in TAF17 known to abrogate Swi6p-dependent gene transcription in vivo (37) disrupts TFIID-Swi6p interaction. Together, these results illustrate the general applicability of systematic immunopurification-DALPC while simultaneously providing important new insights into the function of TBP and TAFs, and thus TFIID, for the initiation of mRNA gene transcription. Further, our work provides an important first step toward the generation of a comprehensive catalogue of protein-protein associations within the RNAP II transcription machinery.
MATERIALS AND METHODS
Yeast methods.
Relevant yeast strains are listed in Table 1, and standard laboratory procedures for yeast cell growth and manipulation were utilized (22). Epitope tagging was performed as described previously (36) with strain BY4741 or BY4742.
TABLE 1.
Yeast strains
| Strain | Genotype | Source or reference |
|---|---|---|
| BY4741 | MATaura3 leu2 his3 LYS2 met15 | 7 |
| BY4742 | MATα ura3 leu2 his3 lys2 MET15 | 7 |
| YEK20 | MATaura3 lys2 ade2 trpl his3 leu2 suc2 taf25::TRP1 pRS413-HA3-TAF25 | 30 |
| YSLS18 | MATaura3 lys2 ade2 trp1 his3 leu2 tafl30::TRP1 pRS313-HA1-TAF130 | 45 |
| YSLS104 | MATaura3 leu2 his3 LYS2 met15 RSC6::HA3-KANr | This study |
| YSLS137 | MATaura3 leu2 his3 LYS2 met15 SGF73::HA3-KANr | This study |
| YSLS140 | MATaura3 leu2 his3 LYS2 met15 NPL6::HA3-KANr | This study |
| YSLS145 | MATaura3 leu2 his3 LYS2 met15 RSC58::HA3-KANr | This study |
| YSLS149 | MATα ura3 leu2 his3 Lys2 MET15 SGF29::HA1-KANr | This study |
| YSLS155 | MATaura3 leu2 his3 LYS2 met15 UBP8::HA3-KANr | This study |
| YSLS163 | MATα TAF17 ura3 lys2 ade his3 leu2 SW16::HA3-KANr | This study |
| YSLS164 | MATα taf17W133amber (slm7-1) ura3 lys2 ade his3 leu2 SW16::HA3-KANr | This study |
| YTW802 | MATalys2 ade2 leu2 trp1 ura3 his3 rsc1::HIS3 rsc2::LEU2 pRS316-HA2-RSC2 pRS424-MYC-RSC1 | 8 |
| INO80-Flag | MATaura3 leu2 his3 met15 trp1 INO80-FLAG2 | 48 |
| INO80-Flag/RVB2-HA | MATaura3 leu2 his3 met15 trp1 INO80-FLAG2 pRVB2-HA | 48 |
Biochemical methods.
Affinity-purified polyclonal antibodies have been described previously (45, 46), and immunoprecipitations (IPs) were performed as detailed earlier (45). Both preparation of yeast whole-cell extracts (WCE) (from strains YSLS18 and BY4741) and Bio-Rex 70 chromatography of WCE were performed as described previously (46). All manipulations were performed at 4°C unless otherwise stated. For immunopurification, typically 200 μg of affinity purified polyclonal antibody or control antibody were first cross-linked to 100 μl of protein A-Sepharose (Sigma). As controls, two nonspecific rabbit and two mouse monoclonal antibodies (MAbs), anti-HA (12CA5; Roche) and anti-Flag (M2; VWR Scientific), were utilized. To a portion of the 1 M Bio-Rex 70 fraction, Buffer A/0 (20 mM HEPES-KOH [pH 7.6], 10% [vol/vol] glycerol, 1 mM dithiothreitol, 0.1 mM phenylmethylsulfonyl fluoride, 1 mM benzamidine) and Nonidet P-40 (NP-40 [Surfact-Amps grade]; Pierce Chemical) were added such that after dilution with Buffer A/0 (BA/0) the conductivity of the protein solution was equivalent to BA containing 300 mM potassium acetate (BA/300) (45) and the final concentration of NP-40 was 0.1% (vol/vol). To prevent protein precipitation, NP-40 was added prior to dilution with BA/0. Ethidium bromide (Bio-Rad) was then added to the diluted 1 M Bio-Rex 70 fraction to a final concentration of 10 μg/ml (45), and an amount (typically 2 to 2.5 ml) of this diluted 1 M Bio-Rex 70 fraction equivalent to 1 ml of the initial undiluted 1 M Bio-Rex 70 fraction was then slurried with the bead bound antibody and mixed 12 to 14 h on a tiltboard at 4°C. The resin was collected in a microspin column (Bio-Rad) and washed extensively with BA/300 plus 0.1% NP-40. To elute proteins from the bead-bound antibody-antigen complex, the resin was slurried with 200 μl of BA/300 plus 6 M urea and 0.1% NP-40 (to achieve a final concentration of 4 M urea), followed by incubation at room temperature for 30 min on a tiltboard. Eluted material was collected with a brief spin in a microcentrifuge, frozen on dry ice, and stored at −80°C. Typically, 5 to 15 independent immunopurifications were simultaneously processed.
DALPC.
Proteins in the immunopurified protein complexes (50 to 100 μl) were precipitated with freshly prepared trichloroacetic acid (TCA; Sigma). First, 0.11× volume of 100% (wt/vol) TCA was added with mixing; samples were then incubated on ice for 10 min, 500 μl of cold 10% (wt/vol) TCA added, and incubation was continued for 10 min on ice. Precipitated proteins were collected by centrifugation (15,000 × g, 10-min spin), pellets were rinsed with 500 μl of acetone and centrifuged as described above, and the acetone was aspirated away. A Speed-Vac was used to remove the residual acetone, and the protein pellets were resuspended in a 100 mM ammonium bicarbonate-5% acetonitrile. Proteins were then reduced by the addition of a 0.1× volume of 50 mM dithiothreitol, followed by incubation at 65°C for 30 min. Cysteines were then alkylated by the addition of a 0.1× volume of 100 mM iodoacetamide, followed by incubation at 30°C for 30 min in the dark. Reduced and alkylated proteins were digested with 0.5 μg of modified sequencing-grade trypsin (Promega) by overnight digestion at 37°C. Tryptic peptides were desalted by using an RP cartridge (Michrom Bioresources, Auborn, Calif.), lyophilized, and resolubilized in 0.5% acetic acid. The entire peptide mixture was loaded onto a 75-μm-inner-diameter SCX column (Partisphere SCX; Whatman) equilibrated in 0.5% acetic acid-2% acetonitrile, and iterative peptide-containing fractions were displaced by using an increasing salt step gradient of 0, 10, 20, 30, 40, 60, 80, and 100% Solution B (250 mM ammonium acetate, 0.5% acetic acid, 2% acetonitrile). This elution regimen was followed by two additional step elutions: first with 500 mM KCl-0.5% acetic acid-2% acetonitrile and then with 1 M NaCl-0.5% acetic acid-2% acetonitrile. The column elution flow rate was maintained throughout the run at 1 μl/min. By using an autosampler (FAMOS; LC Packings), each fraction (5 of 6 μl) was separately loaded onto a 75-μm-inner-diameter RP-HPLC column (Poros R2; Perceptive Biosystems) equilibrated in a 0.5% acetic acid, and peptides were eluted by using a linear gradient of 0 to 40% acetonitrile for 60 min, followed by elution with 40 to 60% acetonitrile for 10 min at a flow rate of 0.5 μl/min. Peptides in the RP-HPLC column eluate were directly analyzed by ESI-MS/MS by using an ion trap mass spectrometer (LCQ Deca; ThermoFinnigan) equipped with a microelectrospray source (James A. Hill Instrument Services, Arlington, Mass.). All tandem spectra were searched against the S. cerevisiae Open Reading Frame database (SGD, Stanford University) by using the SEQUEST algorithm (15).
Data analysis.
Data processing of the SEQUEST output files into a list of proteins has been previously described (35). The complete data set is available as a tab-delimited text file (http://linkdata.mc.vanderbilt.edu). Proteins were scored and ranked as indicated in the text and figure legends. Proteins identified only in control antibody-immunopurified samples were automatically filtered out. Note that the cognate fractions for each TFIID subunit were not utilized for calculating the appropriate score due to the deficiency of the antigen (Fig. 1 and 2). Statistical analyses were performed at the Quantitative Service Core, Kennedy Center, Vanderbilt University. For each protein a t-value comparing the standardized mean difference number of peptide hits from each of the appropriate fraction sets was calculated. A step-down multivariate permutation test was then used to determine significant t-values (19). For each protein presented, the difference in peptide hit distribution between the appropriate fraction sets was further judged significant by another nonparametric statistical test, the Wilcoxon rank sum test (57).
FIG. 2.
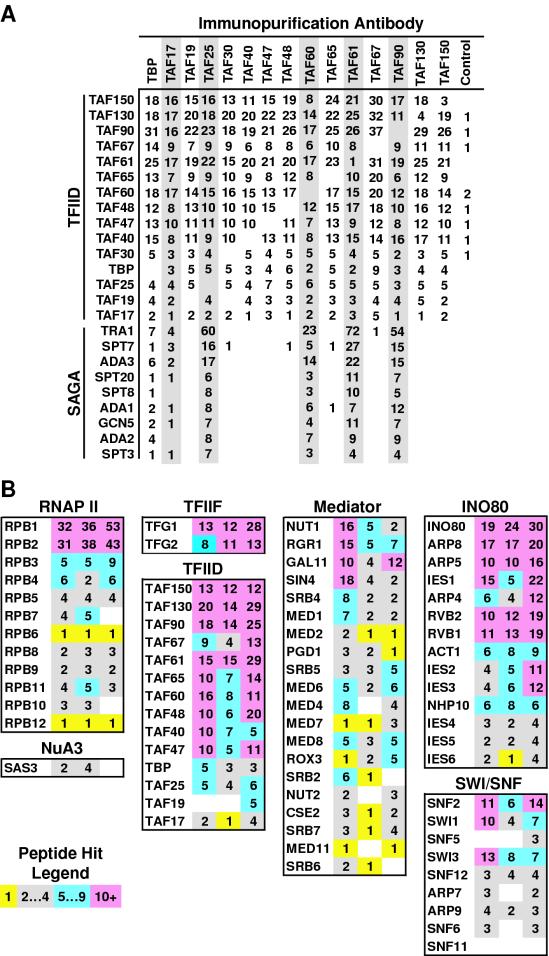
Accuracy, specificity, and precision of the DALPC method. (A) Identification of TFIID and SAGA subunits. Representative peptide hit data from a single set of immunopurifications (antibodies listed top) are shown listing the TFIID and SAGA subunits (left) identified by DALPC. All peptides were uniquely assigned to the corresponding protein. Shared TAF fractions are shaded; all subunits are listed in descending order of molecular mass within their respective complexes. (B) Identification of proteins immunopurified with antibodies directed against TAF30p. Three independently derived anti-TAF30p IgG immunopurified preparations were subjected to DALPC. Peptide hit data (see the legend for the abundance color code) for the identification of protein subunits (listed on the left of each box) of known TAF30p-containing complexes (listed across the top of each box) from each analysis is listed across the columns. Mediator subunits Srb8p, Srb9p, Srb10p, and Srb11p were not identified in any fraction.
RESULTS
Immunopurification of TBP- and TAF-containing complexes.
We previously described the generation and use of affinity-purified polyclonal antibodies raised against each full-length recombinant TFIID subunit (29, 45, 46). Utilizing this collection of reagents, complexes containing these 15 TFIID subunits were immunopurified from a fractionated yeast cell extract enriched in TBP and TAFs (Fig. 1A). Consistent with the existence of a single yeast TFIID complex, immunoblotting (not shown) and direct SDS-PAGE analyses indicated that immunopurification through any one TFIID subunit specifically copurified all of the other known TFIID subunits (Fig. 1C). Compare the overall polypeptide patterns of the nonimmune, control immunopurification (labeled “Control” in Fig. 1) with the polypeptide profiles of the lanes labeled TBP and TAF17* to TAF150 (Fig. 1C). At this level of sensitivity it is clear that the signal-to-noise ratio (i.e., nonspecifically precipitated polypeptide to specifically precipitated polypeptides) is quite high, a finding consistent with the results of our previous studies (29, 30, 45, 46) (see also Fig. 3, 4 and 7). Note that the elution conditions we utilized (4 M urea) generally did not disrupt the antibody-antigen interaction; hence, the protein targeted for purification was either deficient or not present in the cognate fraction. Immunoblotting (not shown) also indicated SAGA subunits were present in the appropriate shared TAF immunopurified fractions (see the five gel lanes marked with an asterisk in Fig. 1C). The overall complexity of the immunopurified fractions varied from sample to sample. Some fractions, such as the anti-TAF65p immunoglobulin G (IgG) purified fraction, contained only TFIID subunits as the major polypeptides (Fig. 1C). Other fractions, such as the anti-TAF48p IgG immunopurified fraction, the anti-TBP and anti-TAF30p IgG purified fractions, or the TFIID/SAGA shared TAF IgG immunopurified fractions all displayed a fairly large number of polypeptides in addition to TFIID subunits (Fig. 1C). Each of these immunopurified preparations was subjected to DALPC protein identification.
FIG. 3.
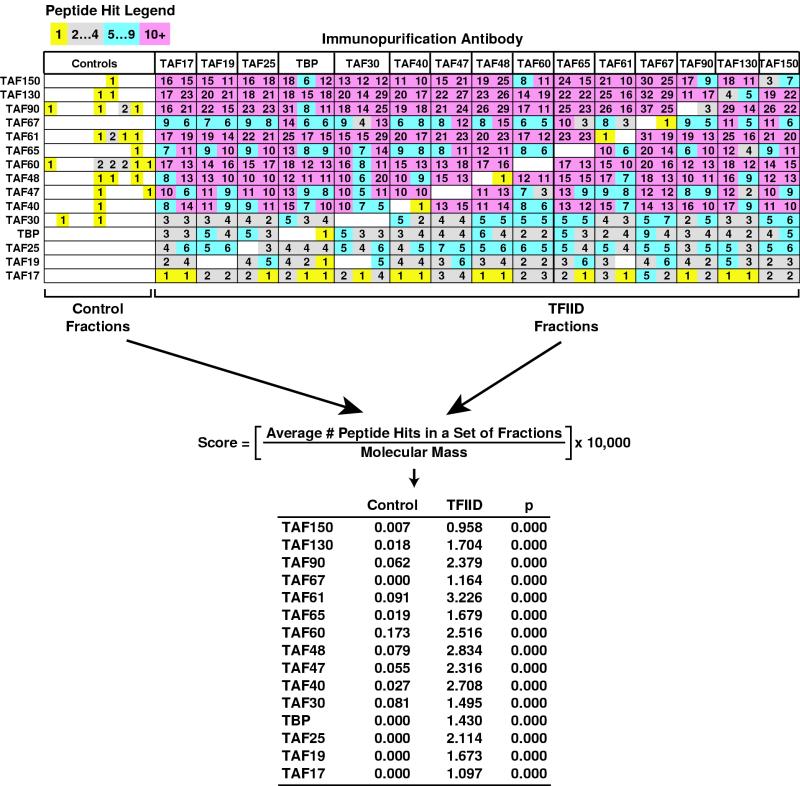
Protein scoring by DALPC. The peptide hit data (top; see legend for color coding) and scoring (bottom) for the identification of TFIID subunits by DALPC are shown. Antibodies used for immunopurification are listed across the top, and boxed columns directly under each label correspond to peptide hit data from DALPC analyses of immunopurified fractions independently generated with the corresponding antibody. TFIID subunit proteins identified by DALPC are indicated on the left, listed in order of descending molecular mass, and the number of peptide hits corresponding to each protein is listed across the diagram. All peptide hits were uniquely assigned to the corresponding protein. The scoring equation and the fractions used to calculate scores are indicated in the middle. A t-value comparing TFIID scores versus control scores was calculated and the significance (P) of the t-values were determined by step-down multivariate permutation test (17), (n1 = 9, n2 = 32, α = 0.05). Scores for the identification of TFIID subunits by DALPC in either the control or TFIID fractions are shown in tabular form at the bottom. Note that with this type of statistical analysis (i.e., the step-down multivariate permutation test) P values can be zero.
Validation of DALPC approach.
DALPC analysis of 41 independent immunopurification reactions generated 354,700 MS/MS spectra. Upon analysis, these MS/MS spectra identified 1,272 distinct open reading frames (ORFs). These data were compiled for analysis as detailed in Materials and Methods. To ensure a reliable data set, multiple preparations of immunopurified proteins were generated for each antibody (4 control antibodies and 15 distinct anti-TFIID subunit IgGs; 19 different antibodies in all), and immunopurification reactions were derived from independently generated 1 M Bio-Rex 70 fractions. Figure 2A presents a summary of a single complete DALPC analysis performed on the proteins immunopurified with the entire collection of 15 anti-TFIID subunit-specific antibodies. This tabular data listing is the mass spectrometry correlate of the SDS-PAGE analysis of Fig. 1C.
To demonstrate the validity of the approach, we first analyzed our DALPC protein data for specific and accurate identification of proteins known (45) and therefore predicted to be present in the immunopurified fractions (Fig. 2). The initial DALPC output is reported as the number of independent peptides that correlated significantly to each ORF and is here referred to as peptide hits (35). Peptide hits corresponding to TFIID subunits were reproducibly identified in all of the anti-TBP, anti-TAF IgG immunopurified fractions in a manner that strongly correlated with the molecular mass of each protein (Fig. 2A and Fig. 3). In contrast, TFIID subunits were only sporadically identified in control fractions, a finding consistent with the high specificity of immunopurification, with at most one or two peptide hits being observed (Fig. 3 and below). Importantly, the difference in peptide hits between the anti-TBP IgG, anti-TAF IgG and control IgG immunopurified fractions was statistically significant for all TFIID subunits (see below and Fig. 3). SAGA subunits were specifically identified in the shared TAF (TAF90p, TAF61p TAF60p, TAF25p, and TAF17p), and to a lesser extent, in the anti-TBP fractions (Fig. 2A and Fig. 4B). Together, our DALPC and immunoblot data (not shown) represent the first direct demonstration that endogenous TBP can stably associate with all of the known components of the native SAGA holocomplex. The slight reduction in the amounts of SAGA in the anti-TAF17p and anti-TBP IgG purified fractions (Fig. 2A and 4B) is consistent with immunoblotting (data not shown).
FIG. 4.
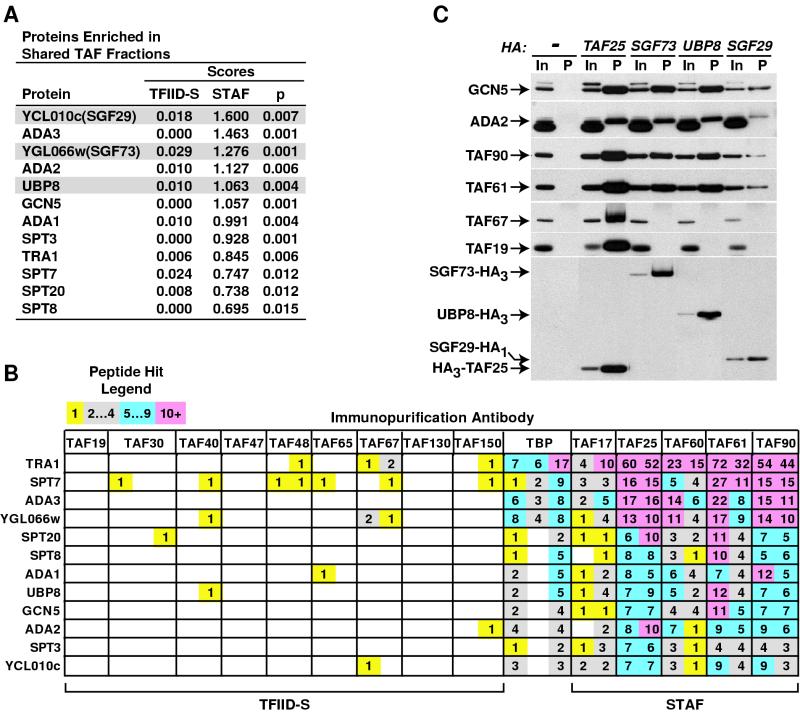
Identification of novel SAGA subunits. (A) Proteins identified by DALPC in STAF fractions were ranked in order by their score. The significance (P) of the calculated t-value comparing STAF versus TFIID-S scores for each protein was determined by step-down multivariate permutation test (n1 = 10, n2 = 19, α = 0.05). Only proteins with significant t-values are shown; candidate SAGA subunits are shaded. Identical results were obtained with α values of 0.05 (>95% confidence; shown) and 0.1 (>90% confidence), suggesting that there is a low likelihood that there are additional novel SAGA subunits besides those listed. (B) DALPC peptide hit output for the identification of proteins enriched in the shared TAF fractions illustrating the specificity (i.e., TFIID-S hits; left) and reproducibility (STAF) of these data. Peptide hit abundance legend is as for Fig. 3. (C) Candidate subunits encoded by YCL010C, YGL066W, and UBP8 are SAGA associated. Anti-HA IPs were performed with WCE prepared from either an untagged wild-type strain (−) or strains harboring the indicated HA-tagged allele (top). A fraction of each input (In, 2%) and precipitate (P, 25%) was subjected to immunoblotting to detect the indicated proteins (left). Individual HA-tagged proteins were detected from four independent exposures of the same blot (bottom); detection of the input and precipitate for each protein (top) is from the same exposure.
To directly test the precision of the DALPC method, three independent anti-TAF30p IgG immunopurified preparations were analyzed for the presence of known TAF30p-containing complexes including TFIID, RNAP II holoenzyme, and the Swi/Snf, NuA3, and INO80 chromatin remodeling complexes (8, 24, 27, 48; X. Shen and C. Wu, unpublished data; see also below). The data presented in Fig. 2B demonstrate that of the listed TAF30p-associated proteins 85% were identified in three of three independent analyses, and 97% were identified in two of three independent analyses. Again, there is a strong correlation between the molecular mass of a protein and the number of peptide hits for that protein within a particular sample. We concluded from these data that our approach could be utilized to accurately, precisely, and specifically define TBP- and TAF-associated factors.
Scoring of DALPC data.
Having established the validity of our approach, we next sought to develop a system that would allow for an unbiased analysis of the DALPC data by identifying proteins whose enrichment in one set of immunopurified fractions versus a second set of fractions was statistically significant (Fig. 3). Such an unbiased scoring system would enable us to more efficiently focus our efforts to independently authenticate any novel findings by identifying proteins with the highest probability of being associated. To score proteins, we first utilized our observation of the strong relationship between molecular mass and peptide hits described above. Proteins identified by DALPC in a specific set of immunopurified fractions were first given and ranked by a score that incorporated the average number of peptide hits normalized to the molecular mass of the cognate protein. Typically, two scores are calculated. In this example, an aggregate control IgG score is generated, followed by calculation of an aggregate TFIID-IgG score. Thus, as shown in Fig. 3, we calculated the control score for the identification of each of the fifteen TFIID subunits within the nine control immunopurification reactions. Similarly, a TFIID score was calculated (Fig. 3, formula and table listing all scores) for each of the same 15 proteins. A statistical analysis of the two scores (or alternatively when n, the sample size, is too small, the ratio of the two scores; see below) was then performed to identify proteins with significantly different scores and thus the proteins that were statistically significantly enriched in the one set of fractions (i.e., TFIID-specific score) relative to a second set (i.e., control-score) (see Fig. 3, table with P values).
Several important observations can be made about this protein scoring system. First, all of the known subunits of the TFIID complex (and other complexes analyzed; see Fig. 4A and 7A and Table 2) scored similarly. Second, consistent with being the major polypeptides detected by SDS-PAGE (Fig. 1C), TFIID subunits are generally the highest-scoring proteins within each individual anti-TBP and anti-TAF IgG immunopurified fraction. Third, the scores for individual TFIID subunits across the anti-TBP and anti-TAF fractions are reflective of our estimates of TFIID subunit stoichiometry (S. L. Sanders, K. A. Garbett, and P. A. Weil, submitted for publication). Thus, although the score for a particular protein from a specific set of fractions is not an absolute measure of its abundance, the score is in general reflective of its relative abundance within that specific (set of) fraction(s).
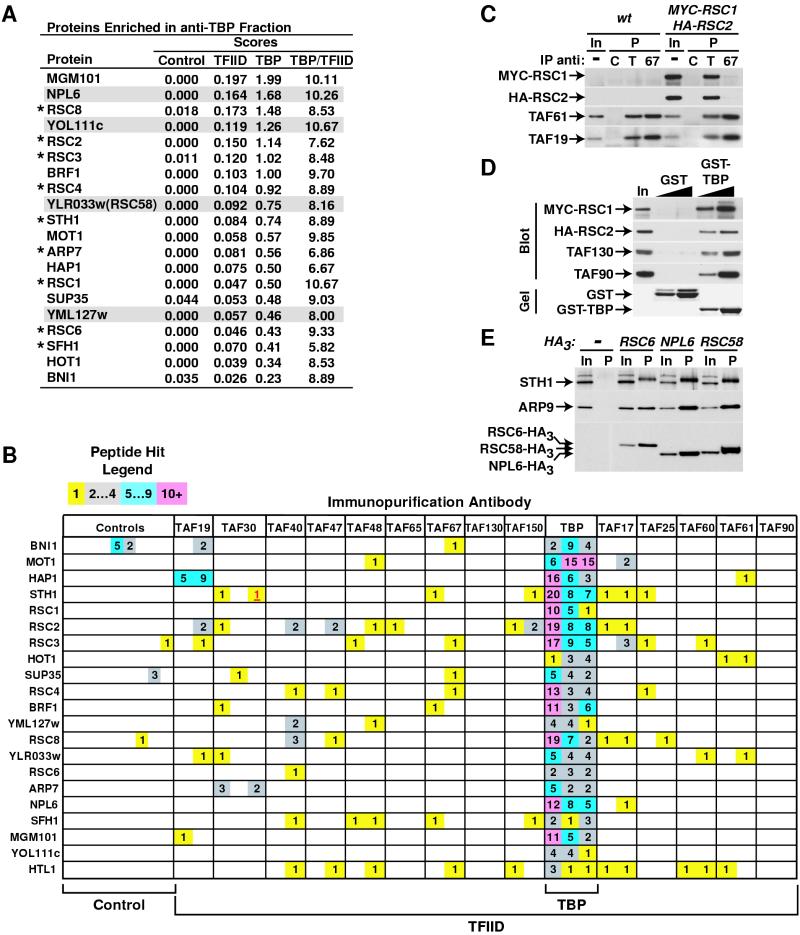
FIG. 7.
Association of TBP with the RSC chromatin remodeling complex. (A) Proteins identified by DALPC in three of three independently generated anti-TBP IgG immunopurified preparations were ranked in descending order by their TBP score; those with a TBP/TFIID score ratio of >4 are shown. The asterisk indicates known RSC subunits; novel RSC candidate subunits are shaded. (B) DALPC peptide hit output for the identification of proteins enriched in the anti-TBP IgG purified fractions. Proteins are listed in order of decreasing molecular weight. The legend is as described for Fig. 3. (C) Association of TBP with RSC. IPs were performed with WCE prepared from the strain with relevant genotype indicated across the top and either control (lanes C), anti-TBP (lanes T), or anti-TAF67p (lanes 67) antibodies. A percentage of the input (In, 2%) and precipitate (P, 20%) was subjected to immunoblotting to detect the indicated proteins (left). (D) Interaction of TBP with RSC. A WCE prepared from a yeast strain carrying MYC-RSC1 and HA-RSC2 genes was utilized for pulldowns with 2 or 5 μg of Escherichia coli-derivedrecombinant GST or GST-TBP bound to glutathione agarose. 25% of each precipitate was visualized by Coomassie blue staining (Gel) or immunoblotting (Blot) to detect the proteins indicated. Input equals 2%. (E) Several of the candidate subunits are RSC associated. IPs with anti-HA MAb 12CA5 IgG were performed as described in the legend of Fig. 4B with WCE prepared from yeast strains expressing the indicated HA-tagged genes (top). Input equals 4%, and precipitate equals 20%. Note that Sth1p migrated more slowly in the precipitate versus the input; the reason(s) for this behavior is unknown.
TABLE 2.
Putative TAF-associated factorsa
| Immunopurification antibody | Protein identified | Regulatory complex | Scores
|
|||
|---|---|---|---|---|---|---|
| Control | TFIID | TAFx | TAFx/TFIID | |||
| TAF150 | FIP1 | 0.000 | 0.087 | 1.398 | 16.00 | |
| PAP1 | 0.000 | 0.073 | 1.084 | 14.93 | ||
| MPE1 | 0.000 | 0.038 | 0.604 | 16.00 | ||
| PTA1 | Polyadenylation factor I complex | 0.000 | 0.035 | 0.565 | 16.00 | |
| CFT1 | 0.000 | 0.037 | 0.554 | 15.11 | ||
| CFT2 | 0.000 | 0.029 | 0.468 | 16.00 | ||
| YSH1 | 0.000 | 0.014 | 0.228 | 16.00 | ||
| REF2 | 0.000 | 0.042 | 0.585 | 14.00 | ||
| SSU72 | 0.000 | 0.027 | 0.426 | 16.00 | ||
| MSN2 | 0.000 | 0.040 | 0.257 | 6.40 | ||
| GCR1 | 0.000 | 0.020 | 0.106 | 5.33 | ||
| LEU3 | 0.000 | 0.012 | 0.100 | 8.00 | ||
| TAF130 | SHE3 | 0.000 | 0.066 | 0.527 | 8.00 | |
| IOC3 | ISWI complex | 0.000 | 0.024 | 0.385 | 16.00 | |
| ISW1 | 0.000 | 0.024 | 0.381 | 16.00 | ||
| HIR3 | 0.000 | 0.060 | 0.365 | 6.05 | ||
| SUA7 | 0.000 | 0.049 | 0.262 | 5.33 | ||
| TAF90 | RPC17 | RNAP III | 0.000 | 0.252 | 1.075 | 4.27 |
| RPC82 | 0.015 | 0.063 | 0.270 | 4.27 | ||
| TAF67 | RTG3 | 0.000 | 0.092 | 0.462 | 5.00 | |
| MCM1 | 0.034 | 0.086 | 0.457 | 5.33 | ||
| RRP3 | 0.018 | 0.046 | 0.328 | 7.11 | ||
| CRZ1 | 0.000 | 0.016 | 0.131 | 8.00 | ||
| TAF61 | CBF1 | 0.000 | 0.048 | 0.762 | 16.00 | |
| MRS1 | 0.054 | 0.045 | 0.605 | 13.33 | ||
| SMB1 | 0.000 | 0.042 | 0.447 | 10.67 | ||
| NOT5 | CCR4-NOT complex | 0.000 | 0.033 | 0.152 | 4.57 | |
| CDC39 | 0.000 | 0.023 | 0.208 | 8.89 | ||
| ARG81 | 0.000 | 0.028 | 0.150 | 5.33 | ||
| REB1 | 0.000 | 0.010 | 0.109 | 10.67 | ||
| RGT1 | 0.000 | 0.007 | 0.117 | 16.00 | ||
| TAF65 | PHO4 | 0.000 | 0.119 | 0.880 | 7.38 | |
| TAF60 | NOP1 | 0.032 | 0.172 | 0.870 | 5.05 | |
| TFC5 | 0.000 | 0.060 | 0.665 | 11.08 | ||
| MCM1 | 0.034 | 0.086 | 0.457 | 5.33 | ||
| PAB1 | 0.000 | 0.053 | 0.311 | 5.82 | ||
| NOT5 | 0.000 | 0.033 | 0.152 | 4.57 | ||
| TAF48 | HOS3 | 0.014 | 0.150 | 2.146 | 14.32 | |
| SPP2 | 0.000 | 0.106 | 1.695 | 16.00 | ||
| PRP2 | 0.000 | 0.088 | 1.403 | 16.00 | ||
| HPC2 | 0.000 | 0.204 | 1.111 | 5.45 | ||
| ASK10 | 0.000 | 0.108 | 1.104 | 10.18 | ||
| HIR2 | Associated | 0.000 | 0.206 | 0.965 | 4.68 | |
| HIR1 | 0.000 | 0.103 | 0.746 | 7.23 | ||
| MSL1 | 0.000 | 0.049 | 0.779 | 16.00 | ||
| STB4 | 0.000 | 0.122 | 0.728 | 5.95 | ||
| RPD3 | RPD3-SIN3 HDAC | 0.000 | 0.070 | 0.716 | 10.18 | |
| SIN3 | 0.006 | 0.046 | 0.458 | 9.85 | ||
| SAP30 | 0.000 | 0.027 | 0.434 | 16.00 | ||
| ELP4 | 0.000 | 0.086 | 0.391 | 4.57 | ||
| HIR3 | 0.000 | 0.060 | 0.339 | 5.62 | ||
| CCR4 | 0.000 | 0.056 | 0.264 | 4.71 | ||
| SFL1 | 0.000 | 0.053 | 0.240 | 4.57 | ||
| TAF25 | SRB7 | 0.000 | 0.331 | 1.867 | 5.65 | |
| NUT2 | 0.000 | 0.262 | 1.675 | 6.40 | ||
| CSE2 | 0.000 | 0.234 | 1.151 | 4.92 | ||
| MED4 | 0.000 | 0.213 | 0.932 | 4.36 | ||
| SRB6 | Mediator | 0.000 | 0.135 | 0.721 | 5.33 | |
| MED7 | 0.000 | 0.134 | 0.586 | 4.36 | ||
| MED1 | 0.000 | 0.092 | 0.545 | 5.89 | ||
| PGD1 | 0.000 | 0.093 | 0.534 | 5.71 | ||
| PAF1 | PAF complex | 0.000 | 0.066 | 0.772 | 11.64 | |
| CDC73 | 0.000 | 0.049 | 0.225 | 4.57 | ||
| CTR9 | 0.000 | 0.025 | 0.241 | 9.60 | ||
| DAL81 | 0.000 | 0.043 | 0.504 | 11.73 | ||
| SUA7 | 0.000 | 0.049 | 0.262 | 5.33 | ||
| TAF17 | RRN6 | 0.000 | 0.009 | 0.147 | 16.00 | |
| ELP1 | 0.007 | 0.086 | 1.111 | 12.95 | ||
| ELP2 | 0.000 | 0.056 | 0.895 | 16.00 | ||
| ELP3 | 0.000 | 0.093 | 1.492 | 16.00 | ||
| ELP4 | Elongator | 0.000 | 0.086 | 0.782 | 9.14 | |
| ELP5 | 0.000 | 0.027 | 0.284 | 10.67 | ||
| ELP6 | 0.000 | 0.031 | 0.327 | 10.67 | ||
| RPP1 | RNase P and RNase MRP | 0.000 | 0.107 | 1.241 | 11.64 | |
| POP1 | 0.000 | 0.019 | 0.249 | 13.33 | ||
| SKO1 | 0.000 | 0.062 | 0.997 | 16.00 | ||
Proteins identified by DALPC in two of two independent anti-TAF immunopurified fractions were ranked by their TAF-specific score (TAFx). The ratio of the TAF-specific score divided by the total TFIID score (TAFx/TFIID) was used to identify proteins that appear to be preferentially associated with the 10 indicated TAFs. Only known transcription-related and RNA processing-related factors with a TAFx score of ≥0.1, a control score of ≤0.1, and a ratio of >4 are shown. Note that SAGA components that were enriched in the shared TAF fractions are not shown. Results for proteins preferentially associated with TBP and TAF30p are also not shown again here but are presented in Fig. 2 and Fig. 6, respectively. Results for the anti-TAF19p and anti-TAF40p immunopurified fractions are not shown (see the text for discussion). No transcription or RNA processing or modifying factors were preferentially associated with TAF47p. Regulatory complexes (as defined by the Yeast Proteome Database [11]) for which two or more of the known components were associated with a specific TAF are indicated by the brackets. This list is derived from the complete list available at http://linkdata.mc.vanderbilt.edu. For the apparent TAF-specific lists above, the proteins shown (n1) were derived from a total listing of proteins (n2) that scored within the criteria listed above. For each TAF, n1 and n2 are, respectively, as follows: TAF150p, n1 = 12 of n2 = 25; TAF130p, 5 of 18; TAF90p, 5 of 24, of which SAGA subunits (not listed here) represent 12 of the 24 total; TAF67p, 4 of 61; TAF65p, 1 of 9; TAF61p, 8 of 38, of which SAGA subunits (not listed here) represent 12 of the 38 total; TAF60p, 5 of 52, TAF48p, 16 of 77; TAF25p, 14 of 33, of which SAGA subunits (not listed here) represent 12 of the 33 total; and TAF17p, 9 of 21.
Identification of novel SAGA subunits.
To test the validity and utility of our scoring system, we attempted to identify novel subunits of the SAGA complex, since several unknown polypeptides were present in a highly purified SAGA preparation (20). Candidate SAGA subunits were identified by first ranking each protein scored by DALPC in the five shared TAF immunopurified fractions (763 ORFs). Within this listing only 12 proteins were statistically significantly enriched (>95% confidence) in the shared TAF (STAF) versus the TFIID-specific (TFIID-S) TAF fractions (compare STAF and TFIID-S scores and P values Fig. 4A). As expected, all nine non-TAF SAGA subunits scored highly in the analysis. However, three additional proteins, those encoded by two uncharacterized ORFs, YCL010C and YGL066W, encoding putative 29- and 73-kDa proteins (referred to here as Sgf29p and Sgf73p for SAGA-associated factors of 29 and 73 kDa, respectively), and the putative ubiquitin-specific protease (or deubiquitylating enzyme) Ubp8p (2) scored as well as, or higher than, known SAGA subunits. These data suggested that these three proteins might be novel components of the SAGA complex (Fig. 4A and B). The identification of a putative ubiquitin-protease subunit of SAGA is of particular interest given recent studies linking protein ubiquitylation and transcriptional control (26, 51). Characteristic of non-TAF SAGA components (except TRA1) none of the genes encoding the three candidate proteins are essential for yeast cell viability (11).
To independently test the association of these candidate subunits with SAGA, we generated strains expressing HA (influenza hemagglutinin) epitope-tagged genomic alleles of each of the genes encoding putative SAGA subunits, prepared WCE from these strains and conducted coimmunoprecipitation (Co-IP) assays. IP of each tagged candidate protein with the anti-HA MAb 12CA5 specifically coimmunoprecipitated SAGA subunits Tra1p, Gcn5p, Ada2p, TAF61p, TAF17p, and TAF90p but not TFIID-specific subunits TAF130p, TAF67p, or TAF19p (Fig. 4C and data not shown). Further when an antibody specific for Gcn5p was utilized for immunopurification-DALPC, all three candidate proteins again scored similarly to known SAGA subunits (not shown). Together, these data strongly argue that Sgf73p, Sgf29p, and Ubp8p are indeed novel components of the yeast SAGA complex. Additional work will be required to elucidate the exact role that these novel subunits might play in SAGA function and gene regulation.
Association of DNA-binding transcription factors with TFIID.
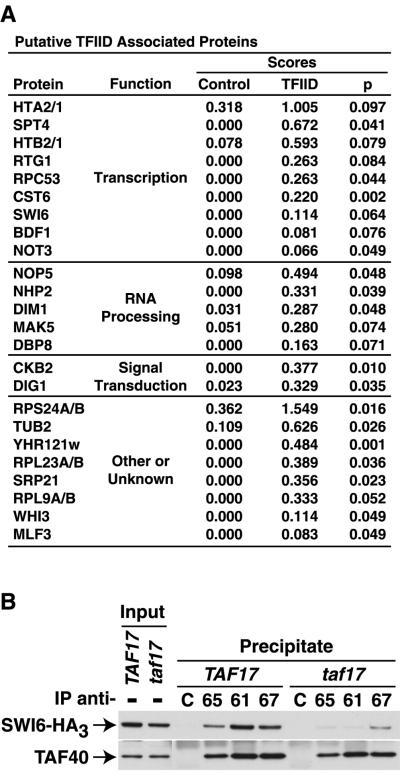
Analysis of the DALPC data presented here failed to indicate the existence of any previously unknown integral, stoichiometric TFIID subunits (data not shown). However, a number of lower-scoring, presumably nonintegral and/or more loosely associated proteins were identified as potential TFIID-associated proteins by immunopurification-DALPC (Fig. 5A). Among these candidate TFIID-associated factors were proteins involved in signal transduction, RNA processing, and transcription, including the known TFIID-associated, bromodomain kinase Bdf1p (38) (Fig. 5A). Further characterization of a number of these novel associations is ongoing. Of particular interest, given the coactivator model of TFIID function (see the introduction above), was the identification of several DNA-binding transactivator proteins (Rtg1p, Cst6p, and Swi6p). We focused on Swi6p for more detailed analyses because it has been shown that a temperature-conditional allele of TAF17 (taf17W133amber or taf17) was lethal in yeast cells when combined with a deletion of the normally nonessential SWI6 (37) gene. Together with our DALPC results, these data suggested that Swi6p functions in part by interacting with TAF17p-containing complexes. Importantly, even at the permissive temperature, taf17 cells display defects in Swi6p-dependent gene transcription (37). We reasoned, based upon this behavior, that the interaction between Swi6p and the C-terminally truncated TAF17p-containing TFIID complexes in the taf17 mutant cells might be compromised.
FIG. 5.
Putative TFIID-associated proteins. (A) Proteins identified by DALPC in immunopurified preparations generated with anti-TBP and anti-TAF IgGs were ranked by TFIID score. The significance (P) of the calculated t-value comparing TFIID versus control scores was determined by step-down multivariate permutation test (n1 = 9, n2 = 32, α = 0.1). Only non-TFIID subunit proteins with significant t-values (90% confidence level) are shown; background proteins identified in eight or nine (of nine) control fractions are not presented. The functional classification as listed in the Yeast Proteome Database (11) is indicated. (B) Swi6p-TFIID association is TAF17p dependent. Immunoprecipitates were formed with control (lanes C), anti-TAF65p (lanes 65), anti-TAF61p (lanes 61), or anti-TAF67p (lanes 67) antibodies with WCE prepared from TAF17 SWI6-HA3 (TAF17) cells or taf17 SWI6-HA3 (taf17) cells grown at a permissive temperature (30°C). A fraction of the input (2%) and precipitate (67%) was subjected to immunoblotting with appropriate antibodies to detect the proteins indicated (left). Detection of TAF40p in the input and precipitate are from independent exposures of the same blot.
To directly test the hypothesis that Swi6p interacts with TFIID, we quantitated the interaction of Swi6p with TFIID in TAF17 wild-type cells and taf17 mutant cells by Co-IP assay. For this purpose we generated two yeast strains, genotype TAF17 or taf17, that both carried an HA3-tagged, genomic copy of SWI6. WCE were prepared from these two yeast strains and utilized for Co-IP studies (Fig. 5B). Both Swi6p and the TAF40p subunit of TFIID were specifically coimmunoprecipitated from TAF17 WCE with anti-TFIID subunit-specific antibodies. In contrast, reproducibly 50 to 80% less Swi6p, relative to the amount of TAF40p coimmunoprecipitated, was found to be associated with TFIID in taf17 cells (Fig. 5B). The loss of association between Swi6p and TFIID is not due to a general, equivalent decrease in TFIID content or integrity in the taf17 strain since holo-TFIID levels, including normal amounts of TAF60p (cf. reference 17), were similar in TAF17 and taf17 cells, as judged by additional Co-IP studies (data not shown). It is likely that the interaction between TFIID and Swi6p is direct, although this hypothesis bears further investigation. These data provide the first demonstration of a mutationally sensitive interaction between a transactivator and a native TFIID complex in vivo and further argue that Swi6p transactivation is, at least in part, mediated by interaction with the TFIID complex on some Swi6p-dependent genes.
Novel TBP- and TAF-associated factors.
We next analyzed our DALPC data in order to attempt to discover proteins specifically associated with only one TFIID subunit. Because of the relatively small number of individual anti-TBP and anti-TAF immunopurified fractions (n = 2 or 3), a meaningful statistical analysis of the data was not possible. Therefore, we utilized the ratio of a protein's specific anti-TBP fraction score or specific anti-TAF IgG immunopurified fraction score versus that protein's total TFIID score to identify proteins potentially preferentially associated with 1 of these 15 proteins (either TBP or one of the multiple TAFs). A subset of the results of this analysis, highlighting only transcription and RNA-processing factors preferentially associated with a specific TAF, are shown in Table 2 The comparable data for TBP is presented below (see Fig. 7). Depending upon the particular antibody used, a number of other, non-transcription-related proteins also appeared to be TFIID subunit-enriched but are not shown (see http://linkdata.mc.vanderbilt.edu). The number of other proteins that scored within the ranges of the transcription-related proteins listed in the figure are detailed in the footnote for Table 2. For example, in the case of the two anti-TAF150p IgG immunopurified samples, the 12 transcription-related proteins noted exhibited TAF150p/TFIID ratio scores ranging from a low of 5.33 (Gcr1p) to a high of 16.00 (Fip1p, Mpe1p, Pta1p, Cft2p, Ysh1p, and Ssu72p). Only 13 other, distinct proteins scored within this TAF150/TFIID ratio score range (not shown).
These data suggest a number of potentially important and novel interactions between TAF proteins and other proteins or protein complexes. Of particular note is the apparent association of TAF48p with two histone deacetylases, Rpd3p and Hos3p, given the recent characterization of TAFs, including the fly ortholog of TAF48p (dTAFII110), as components of the multisubunit Drosophila polycomb group repression complex (47). Our data also suggests that TAF150p may provide either a direct or indirect link between TFIID and transcription initiation and polyadenylation in yeast since the mammalian equivalent of CPSF, Ysh1p and other components of polyadenylation factor I complex, were found to be specifically associated with TAF150p by immunopurification-DALPC (Table 2). Additionally, we observed that a number of mediator subunits were preferentially associated with TAF25p. Further analysis revealed that 16 of 20 mediator subunits (as well as RNAP II subunits; Rpb1p to Rpb11p) were present in the anti-TAF25p fraction. The association of TAF25p with mediator most likely reflects an interaction between the SAGA and mediator complexes since mediator was also found to be Gcn5p-associated by DALPC analysis (not shown). These biochemical data suggest that previously characterized genetic interactions between SAGA and mediator (13, 42) may be due to direct physical interaction.
TAF30p was found to be a component of the INO80 chromatin remodeling complex as all known INO80 subunits (48; Shen and Wu, unpublished) were specifically identified in the anti-TAF30p immunopurified fractions (Fig. 2B). The association of TAF30p with INO80 was verified by Co-IP experiments (Fig. 6). Wu and colleagues independently characterized TAF30p as a component of the INO80 complex (Shen and Wu, unpublished). Further investigation of the multiple putative protein-protein interactions described in Fig. 6 and Table 2 is clearly needed in order to authenticate these intriguing, novel connections.
FIG. 6.
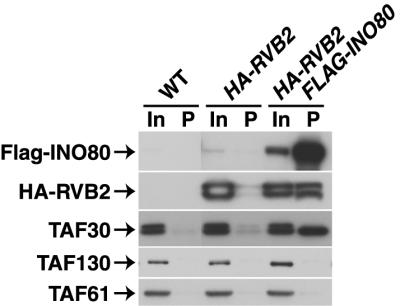
TAF30p is an integral subunit of the INO80 chromatin remodeling complex. IPs were performed with anti-Flag MAb and with WCE prepared from the strains with the indicated relevant genotype listed across the top. A fraction of the input (In, 0.6%) and of the precipitate (P, 25%) was subjected to immunoblotting with appropriate antibodies to detect the proteins indicated at the left.
We next turned our attention to those proteins specifically enriched in the anti-TBP IgG immunopurified fractions (Fig. 7A and B). Consistent with our previous studies, the known TBP-associated factors Mot1p and Brf1p (39, 40), were highly enriched in the anti-TBP IgG fraction, as were two transcription-related proteins (i.e., Hap1p and Hot1p). Interestingly, 9 of 11 known subunits of the RSC chromatin remodeling complex (3) were specifically immunopurified with anti-TBP IgG and identified by DALPC (Fig. 7A and B). A less-stringent ranking of these data revealed that the two other known RSC subunits, Arp9p and Rsc30p, were also enriched in the anti-TBP IgG purified fractions (data not shown). By utilizing yeast strains expressing epitope-tagged alleles as the sole source of Rsc1p and Rsc2p, we independently verified the association of TBP with both forms of the RSC complex marked by Rsc1p and Rsc2p (8) by performing Co-IP and glutathione S-transferase (GST) pulldown assays (Fig. 7C and D). The experimental data presented here obviously do not address whether or not the interaction between TBP and RSC is direct but clearly did demonstrate that the association is independent of TFIID [Fig. 7B and C, compare anti-TBP(T) precipitate (P) versus anti-TAF67p(67) precipitate (P)]. Preliminary DALPC analyses performed with anti-Mot1p and anti-Brf1p IgGs suggest that this TBP-RSC interaction does not involve Mot1p-TBP or Brf1p-TBP complexes (data not shown) and therefore appears not to involve several of the known, abundant TBP-TAF complexes (TFIID, TFIIIB, or Mot1p [33]).
Importantly, the association between TBP and the RSC chromatin remodeling complex that we have observed (as well as all other associations characterized) was not DNA dependent since all immunopurification, IP, and pulldown experiments were performed in the presence of ethidium bromide. As with the SAGA complex, we utilized our DALPC data to identify four potentially novel RSC components (Fig. 7A) (3). Co-IP studies with appropriate epitope-tagged strains indicated that two of the candidate RSC subunits, Npl6p and Rsc58p (encoded by YLR033W); indeed, appeared to be RSC associated (Fig. 7E). We were unable to generate an epitope-tagged allele of YML127W, and we did not further pursue characterization of YOL111C. As with the genes encoding most other RSC subunits, NPL6 and RSC58(YLR033W) are essential for yeast cell viability; moreover, Npl6p has been reported to interact with Rsc8p in yeast two-hybrid assays (11). Collectively, these data strongly argue that both Npl6p and Rsc58p are bona fide RSC subunits.
DISCUSSION
Utility of systematic, controlled immunopurification-DALPC for characterization of multisubunit assemblies.
In contrast to two other studies that used DALPC for multisubunit complex characterization (35, 53), we combined DALPC with systematic, controlled immunopurification coupled with unbiased data analysis to identify novel protein-protein interactions. Our controlled, systematic scoring, ranking, and statistical approach has allowed us to quickly and efficiently focus our efforts on just a few putative TBP- and TAF-interacting proteins culled from a very large pool of candidates. The utility and robustness of this approach has been illustrated by our definition of several previously unknown and potentially important components of the yeast SAGA, INO80, and RSC chromatin-modifying complexes. Additionally, we characterized a novel association between TBP and the RSC complex and provided the first demonstration of a mutationally sensitive, functionally relevant transactivator-TFIID interaction in yeast. Collectively, these data illustrate that DALPC, appropriately applied, exhibits both the requisite accuracy and precision necessary for proteomics-based protein discovery and provides the first proteomic catalogue of protein-protein associations for a eukaryotic transcription factor. The methods developed here can be readily applied to the entire collection of components comprising the RNAP II transcription machinery to define and catalogue the multitude of protein-protein associations required for transcriptional control of mRNA gene expression.
Appropriate interpretation of immunopurification-DALPC data.
We generated here a catalogue of proteins that can associate with components of the GTF TFIID. It has been quite gratifying that each of the interactions identified by immunopurification-DALPC that we extensively investigated by independent means has been validated. However, despite this success, it is important to recognize that it is absolutely essential both to perform such additional biochemical and, where possible, genetic tests of interaction (i.e., Fig. 4C, 5B, 6, and 7C to E) and to include current knowledge of protein function in evaluating and interpreting immunopurification-DALPC data. For example our analyses suggested Spt4p might be a putative TFIID-interacting factor (Fig. 5A). Spt4p and Spt5p together compose the DSIF elongation factor (23, 54); thus, we might have expected Spt5p to score as a TFIID-interacting factor. However, we found that Spt5p nonspecifically immunopurified with essentially every antibody we tested; Spt5p typically generated control scores and TFIID scores of 0.855 and 1.208 (P > 0.3). Although it remains possible that a fraction of Spt4p interacts with TFIID independent of Spt5p, validation of such an interaction would require further detailed investigation and at present is viewed as unlikely. Clearly then, as with other high-throughput, broad-based screening approaches such as global two-hybrid screening (52) or global chromatin IP assays (49), additional biochemical and genetic filters must be applied to immunopurification-DALPC data in order to fully elucidate the functional importance of any novel connections identified. However, it should be noted that our approach has a significant advantage over screening methods such as protein chips (58) or global two-hybrid analyses in that protein-protein interactions are defined within the native context of multisubunit assemblies rather than by pairwise interactions between individual components.
General applicability of immunopurification-DALPC methodology.
The methods used here to identify novel TBP- and TAF-associated proteins in S. cerevisiae should be generally applicable to a wide variety of biological problems and systems regardless of the availability of genetic tools. Although here we utilized a fractionated extract for immunopurification, initial experiments with immunopurification-DALPC directly from WCE have yielded results quite similar to those presented (not shown). The relative ease with which polyclonal antibodies can be generated and the large number of commercially available antibodies against known factors makes the application of immunopurification routine. In our laboratory, we readily generate milligram quantities of highly specific purified IgG of sufficient quality for immunopurification with a single affinity purification step directly from immunized serum utilizing immobilized antigen columns. An obvious alternative to polyclonal antibody-based immunopurification is to utilize epitope or other affinity tags. However, we have found that each MAb has its own unique pattern of (sometimes high) nonspecific protein binding. Additionally, we have observed that tag accessibility can vary widely from protein to protein and tag to tag, thereby complicating both purification logistics and data interpretation. In contrast, of the more than 25 different polyclonal antibodies generated and tested in our laboratory, only two (one of which is anti-TAF40p IgG) do not efficiently (>70%) immunoprecipitate the appropriate antigen. Moreover, in metazoan systems (unlike yeast) tagged proteins are often over expressed and/or not the only source of the protein in the cell. These are conditions that potentially complicate characterization of truly native complexes. Finally, since most eukaryotic systems do not have facile methods of complementation testing, it is difficult if not impossible to establish that tagging per se does not inactivate protein function. For these reasons, we feel our polyclonal immunopurification approach can be most universally applied.
We have found that the most critical component to successful immunopurification-DALPC analysis is the quality of the antibody utilized for immunopurification. As discussed above, we feel that polyclonal antibodies generated against full-length recombinant proteins offer the most generally applicable immunopurification approach. However, when polyclonal antibodies are used it is absolutely essential to ensure the specificity of the antibody and to take this into consideration when the immunopurification-DALPC data are interpreted. For example, SDS-PAGE (Fig. 1) and DALPC analysis of the anti-TAF19p IgG immunopurified fractions suggested that a large number of non-TFIID proteins were associated with TAF19p. However, further biochemical characterization of TAF19p and TAF19p-containing complexes did not support this hypothesis (data not shown). To address this discrepancy, we simultaneously performed immunopurification with either polyclonal anti-TAF19p IgG or MAb anti-HA IgG utilizing a Bio-Rex 70 1 M fraction prepared from a yeast strain in which the genomic copy of TAF19 was HA3 tagged. SDS-PAGE and DALPC analyses of the resulting two immunopurified fractions indicated that immunopurification with anti-HA, in contrast to immunopurification with anti-TAF19p polyclonal IgG, purified primarily just TFIID (data not shown). The difference in the complement of apparent TAF19p-associated proteins was not due to epitope accessibility since ∼70% of the total TAF19p was immunopurified from the Bio-Rex 70 fraction with each antibody. Thus, it appears that, despite affinity purification, this particular polyclonal anti-TAF19p IgG does not display sufficient specificity toward TAF19p. Perhaps this antibody recognizes the histone fold domain, a key structural element of TAF19p, several other TAFs, and myriad other proteins (50) in a pan-specific fashion; alternatively, this particular antibody may simply display a lower specificity than our other antibodies. We conclude that highly specific antibodies are essential to reliable immunopurification-DALPC analyses.
It is important to note, however, that our polyclonal anti-TAF19p IgG preparation is almost certainly a special case. First, all of our other antibodies ‘behaved’ much more specifically and reproducibly (cf. Fig. 2 to 7 and Table 2) (29, 46; data not shown). Second, we independently tested the specificity of two additional polyclonal antibodies by the method outlined above for polyclonal TAF19p IgG. Thus, we compared the constellations of proteins TFIID associated when TFIID was immunopurified via polyclonal antibodies directed against the two TFIID-specific subunits TAF130p and TAF48p versus the associated proteins detected when TFIID was immunopurified with a MAb, the anti-HA MAb 12CA5. For these experiments, we used two congenic yeast strains: one expressing HA-tagged TAF130p as the sole source of TAF130p and the other expressing an HA3-tagged version of TAF48p. Importantly, the proteins detected by the DALPC analyses of the four derived immunopurified protein fractions were essentially the same (not shown). These data clearly demonstrate both that our polyclonal antibodies display appropriate high specificity and selectivity and also underscore the power and validity of the immunopurification-DALPC approach.
Comparison of immunopurification-DALPC method to other proteomics approaches.
While this study was in review, two other reports appeared that described global proteomics approaches to systematically identify and catalog yeast multiprotein complexes (18, 25). These investigators used either tandem affinity purification (TAP [41]) or Flagx3-tagging strategies for the identification and purification of proteins or protein complexes from yeast. Several points regarding the results of these studies relative to our DALPC approach are notable.
First, in the more comprehensive study (18), which attempted to tag 1,739 distinct genes, only 589 genes or proteins were successfully tagged and purified. This result indicates that <34% of the genes targeted for analysis could be successfully tagged and subsequently purified. Presumably, these authors obtained such a low success rate for the reasons outlined above, namely, tag lethality and (TAP−) tag inaccessibility; with the use of polyclonal antibodies, as in our DALPC method, neither of these concerns is relevant. Second, both groups used SDS-PAGE fractionation and band excision to generate samples for protein identification by mass spectrometry. This approach dramatically increased the number of mass spectrometry runs required to analyze either 589 TAP-tagged genes or proteins (20,946 individual samples for mass spectrometry [18]) or 600 Flag-tagged genes or proteins (15,683 samples for mass spectrometry [25]). With immunopurification-DALPC, only 589 or 600 samples, respectively, would have had to be analyzed (although most likely in duplicate = ∼1,200 samples). Thus, DALPC is also more efficient. Third, the constellation of background, contaminating polypeptides, comprised primarily of very abundant cellular proteins (glycolytic and general metabolic enzymes, single-strand nucleic-acid-binding proteins, and ribosomal and related translation proteins) was very similar in both studies and were essentially the same set of proteins that we detected in our immunopurification-DALPC analyses. Fourth, although we have not made a complete comparative analysis of the two data sets, it appears that in most cases where both groups tagged and analyzed the same genes or proteins, there is, on average, <50% agreement between them in the identification of the same sets of putative bait-interacting proteins. This statistic suggests, in genetic parlance, that neither protein interaction screen is yet saturated. Finally, when these workers happened to have tagged and analyzed genes encoding TFIID or SAGA subunits (in Ho et al. [25], GCN5; in Gavin et al. [18], TAF150/TSM1, TAF130, TAF90, TAF60, TAF25, SPT15 [TBP], SPT7, and SPT8), neither group identified the totality of TFIID (15 subunits) or SAGA (12 subunits) components that we identified and characterized by immunopurification-DALPC. (Gavin et al. [18] did, however, come much closer in this regard than Ho et al. [25].) Together, these observations argue that immunopurification-DALPC, as detailed and utilized in our report, has superior sensitivity and accuracy compared to tagging approaches.
Identification of the interaction of TBP with chromatin remodeling complexes.
Recent studies have shown that recruitment of TBP to the promoters of certain mRNA encoding genes can apparently occur independent of TFIID (31, 34). Evidence suggested that TBP could interact with components of the SAGA complex (14, 43) and that SAGA may provide an alternative mechanism for either escorting or recruiting or retaining TBP on the promoters of appropriate SAGA-target genes (6, 32). The data presented here support this hypothesis by providing the first direct demonstration that endogenous TBP can stably associate with the native holo-SAGA complex (Fig. 2). We further suggest that the interaction between RSC and TBP observed here (Fig. 7) could provide a similar function. Although it can clearly associate with both RSC and SAGA, TBP apparently does not copurify as an integral stoichiometric subunit of either complex (3, 20). These results are consistent with our findings that, compared to TFIID, a relatively small fraction of the total TBP is associated with either SAGA or RSC (S. A. Sanders and P. A. Weil, unpublished observation). These data further emphasize the power of our methodology, since a nonstoichiometric association previously unseen by conventional purification methods was readily demonstrated with our immunopurification-DALPC approach.
Yeast TFIID appears to serve as a transcriptional coactivator for Swi6p.
The coactivator model of TFIID function argues that interaction between TFIID and transactivating factors is critical for the recruitment to, or stabilization of, TFIID on promoters and subsequent initiation of mRNA gene transcription (1). Despite an extensive body of in vitro studies to support this hypothesis, direct biochemical-genetic evidence of a functionally relevant interaction between TFIID and a transactivator in vivo has been lacking. Here we have provided such evidence by directly demonstrating a mutationally sensitive association of the Swi6p transactivator with the TFIID complex in vivo (Fig. 5). Deletion of the C-terminal 26 amino acids of TAF17p is synthetically lethal when combined with a deletion of SWI6, and such mutant taf17 cells display defects in Swi6p-dependent gene transcription (37). Importantly, we have demonstrated that the interaction between TFIID and Swi6p is disrupted in taf17 cells, and our data argue that the interaction is direct. Together, these studies suggest that the TFIID-Swi6p interaction is critical for mediating Swi6p function and may further be required for the recruitment of TFIID to at least some Swi6p-dependent promoters. Swi6p functions as part of the MBF (Swi6p+Mbp1p) and SBF (Swi6p+Swi4p) transcription factor complexes to control the expression of genes required for the G1-to-S-phase transition (49). It is unclear from our results whether Swi6p is interacting with TFIID in the context of only one, both, or neither of these heteromeric trans-factor complexes. Further studies will be required to address this and other aspects of the TFIID-Swi6p interaction.
Deubiquitylation, SAGA, and transcriptional regulation.
It is interesting to speculate on the possible target(s) of a deubiquitylating enzyme (Ubp8p) within the SAGA complex. Obviously, nucleosomal histones could serve as a target for this enzymatic activity given that histone tails are known to be ubiquitylated (26). A more intriguing possibility is that Ubp8p directly targets transcription factors and that SAGA, interacting with DNA-bound trans-factors, actively directs deubiquitylation by Ubp8p. Deubiquitylation of promoter bound yeast Met4p is required for transactivation by Met4p (28) and phosphorylation by the Srb10p component of mediator targets the yeast Gcn4p and Msn2p transactivators for ubiquitination and subsequent degradation (10). It has recently been shown that transactivation domains themselves serve as direct ubiquitylation targets (44) and that this ubiquitylation is an obligatory step leading both to gene activation and trans-factor degradation. Modulation of transactivation domain ubiquitylation levels by chromatin and/or transactivation domain-bound SAGA could provide an important mechanism for the regulation of gene transcription. Further biochemical and genetic studies will be required to address this and other possible roles for Ubp8p, as well as the roles of Sgf73p and Sgf29p, in the function of the SAGA complex.
] ]
Acknowledgments
We are grateful to B. Cairns and B. Andrews for gifts of valuable reagents and to X. Shen and C. Wu both for communication of results regarding the composition of the INO80 complex prior to publication and for providing INO80-specific reagents. We are especially grateful to J. Blackford for help and advice with the statistical analyses of DALPC data. Finally, we thank all of the members of the Weil and Link laboratories for helpful discussions.
This work was supported by National Institutes of Health Grant GM52461 (P.A.W.) and by Vanderbilt-Ingram Cancer Support grant 5P30CA68485, HHMI, and Ingram Family gift (A.J.L.).
REFERENCES
- 1.Albright, S. R., and R. Tjian. 2000. TAFs revisited: more data reveal new twists and confirm old ideas. Gene 242:1-13. [DOI] [PubMed] [Google Scholar]
- 2.Amerik, A. Y., S. J. Li, and M. Hochstrasser. 2000. Analysis of the deubiquitinating enzymes of the yeast Saccharomyces cerevisiae. Biol. Chem. 381:981-992. [DOI] [PubMed] [Google Scholar]
- 3.Angus-Hill, M. L., A. Schlichter, D. Roberts, H. Erdjument-Bromage, P. Tempst, and B. R. Cairns. 2001. A Rsc3/Rsc30 zinc cluster dimer reveals novel roles for the chromatin remodeler RSC in gene expression and cell cycle control. Mol. Cell 7:741-751. [DOI] [PubMed] [Google Scholar]
- 4.Bell, B., and L. Tora. 1999. Regulation of gene expression by multiple forms of TFIID and other novel TAFII-containing complexes. Exp. Cell Res. 246:11-19. [DOI] [PubMed] [Google Scholar]
- 5.Bell, B., E. Scheer, and L. Tora. 2001. Identification of hTAF(II)80δ links apoptotic signaling pathways to transcription factor TFIID function. Mol. Cell 8:591-600. [DOI] [PubMed] [Google Scholar]
- 6.Bhaumik, S. R., and M. R. Green. 2001. SAGA is an essential in vivo target of the yeast acidic activator Gal4p. Genes Dev. 15:1935-1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brachmann, C. B., A. Davies, G. J. Cost, E. Caputo, J. Li, P. Hieter, and J. D. Boeke. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14:115-132. [DOI] [PubMed] [Google Scholar]
- 8.Cairns, B. R., A. Schlichter, H. Erdjument-Bromage, P. Tempst, R. D. Kornberg, and F. Winston. 1999. Two functionally distinct forms of the RSC nucleosome-remodeling complex, containing essential AT hook, BAH, and bromodomains. Mol. Cell 4:715-723. [DOI] [PubMed] [Google Scholar]
- 9.Cairns, B. R., N. L. Henry, and R. D. Kornberg. 1996. TFG/TAF30/ANC1, a component of the yeast SWI/SNF complex that is similar to the leukemogenic proteins ENL and AF-9. Mol. Cell. Biol. 16:3308-3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chi, Y., M. J. Huddleston, X. Zhang, R. A. Young, R. S. Annan, S. A. Carr, and R. J. Deshaies 2001. Negative regulation of Gcn4 and Msn2 transcription factors by Srb10 cyclin-dependent kinase. Genes Dev. 15:1078-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Costanzo, M. C., M. E. Crawford, J. E. Hirschman, J. E. Kranz, P. Olsen, L. S. Robertson, M. S. Skrzypek, B. R. Braun, K. L. Hopkins, P. Kondu, C. Lengieza, J. E. Lew-Smith, M. Tillberg, and J. I. Garrels. 2001. YPD, PombePD and WormPD: model organism volumes of the BioKnowledge library, an integrated resource for protein information. Nucleic Acids Res. 29:75-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dantonel, J. C., K. G. Murthy, J. L. Manley, and L. Tora. 1997. Transcription factor TFIID recruits factor CPSF for formation of 3′ end of mRNA. Nature 389:399-402. [DOI] [PubMed] [Google Scholar]
- 13.Drysdale, C. M., B. M. Jackson, R. McVeigh, E. R. Klebanow, Y. Bai, T. Kokubo, M. Swanson, Y. Nakatani, P. A. Weil, and A. G. Hinnebusch. 1998. The Gcn4p activation domain interacts specifically in vitro with RNA polymerase II holoenzyme, TFIID, and the Adap-Gcn5p coactivator complex. Mol. Cell. Biol. 18:1711-1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eisenmann, D. M., K. M. Arndt, S. L. Ricupero, J. W. Rooney, and F. Winston. 1992. SPT3 interacts with TFIID to allow normal transcription in Saccharomyces cerevisiae. Genes Dev. 6:1319-1331. [DOI] [PubMed] [Google Scholar]
- 15.Eng, J. K., A. L. McCormack, and J. R. Yates III. 1994. An approach to correlate tandem mass-spectral data of peptides with amino-acid-sequences in a protein database. J. Am. Soc. Mass Spectrom. 5:976-989. [DOI] [PubMed] [Google Scholar]
- 16.Gangloff, Y. G., J. C. Pointud, S. Thuault, L. Carre, C. Romier, S. Muratoglu, M. Brand, L. Tora, J. L. Couderc, and I. Davidson. 2001. The tfiid components human taf(ii)140 and Drosophila bip2 (taf(ii)155) are novel metazoan homologues of yeast taf(ii)47 containing a histone fold and a phd finger. Mol. Cell. Biol. 21:5109-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gangloff, Y., C. Romier, S. Thuault, S. Werten, and I. Davidson. 2001. The histone fold is a key structural motif of transcription factor TFIID. Trends Biochem. Sci. 26:250-257. [DOI] [PubMed] [Google Scholar]
- 18.Gavin, A. C., M. Bosche, R. Krause, P. Grandi, M. Marzioch, A. Bauer, J. Schultz, J. M. Rick, A. M. Michon, C. M. Cruciat, M. Remor, C. Hofert, M. Schelder, M. Brajenovic, H. Ruffner, A. Merino, K. Klein, M. Hudak, D. Dickson, T. Rudi, V. Gnau, A. Bauch, S. Bastuck, B. Huhse, C. Leutwein, M. A. Heurtier, R. R. Copley, A. Edelmann, E. Querfurth, V. Rybin, G. Drewes, M. Raida, T. Bouwmeester, P. Bork, B. Seraphin, B. Kuster, G. Neubauer, and G. Superti-Furga. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415:141-147. [DOI] [PubMed] [Google Scholar]
- 19.Good, P. 2000. Permutation tests: a pratical guide to resampling methods for testing hypotheses, 2nd ed. Springer-Verlag, New York, N.Y.
- 20.Grant, P. A., D. Schieltz, M. G. Pray-Grant, D. J. Steger, J. C. Reese, J. R. Yates III, and J. L. Workman. 1998. A subset of TAF(II)s are integral components of the SAGA complex required for nucleosome acetylation and transcriptional stimulation. Cell 94:45-53. [DOI] [PubMed] [Google Scholar]
- 21.Green, M. R. 2000. TBP-associated factors (TAFIIs): multiple, selective transcriptional mediators in common complexes. Trends Biochem. Sci. 25:59-63. [DOI] [PubMed] [Google Scholar]
- 22.Guthrie, C., and G. R. Fink. 1991. Guide to yeast genetics and molecular biology. Methods Enzymol. 194:1-933. [PubMed] [Google Scholar]
- 23.Hartzog, G. A., T. Wada, H. Handa, and F. Winston. 1998. Evidence that Spt4, Spt5, and Spt6 control transcription elongation by RNA polymerase II in Saccharomyces cerevisiae. Genes Dev. 12:357-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Henry, N. L., A. M. Campbell, W. J. Feaver, D. Poon, P. A. Weil, and R. D. Kornberg. 1994. TFIIF-TAF-RNA polymerase II connection. Genes Dev. 8:2868-2878. [DOI] [PubMed] [Google Scholar]
- 25.Ho, Y., A. Gruhler, A. Heilbut, G. D. Bader, L. Moore, S. L. Adams, A. Millar, P. Taylor, et al. 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415:180-183. [DOI] [PubMed] [Google Scholar]
- 26.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 293:1074-1080. [DOI] [PubMed] [Google Scholar]
- 27.John, S., L. Howe, S. T. Tafrov, P. A. Grant, R. Sternglanz, and J. L. Workman. 2000. The something about silencing protein, Sas3, is the catalytic subunit of NuA3, a yTAF(II)30-containing HAT complex that interacts with the Spt16 subunit of the yeast CP (Cdc68/Pob3)-FACT complex. Genes Dev. 14:1196-1208. [PMC free article] [PubMed] [Google Scholar]
- 28.Kaiser, P., K. Flick, C. Wittenberg, and S. I. Reed. 2000. Regulation of transcription by ubiquitination without proteolysis: Cdc34/SCF(Met30)-mediated inactivation of the transcription factor Met4. Cell 102:303-314. [DOI] [PubMed] [Google Scholar]
- 29.Kirchner, J., S. L. Sanders, E. Klebanow, and P. A. Weil. 2001. Molecular genetic dissection of TAF25, an essential yeast gene encoding a subunit shared by TFIID and SAGA multiprotein transcription factors. Mol. Cell. Biol. 21:6668-6680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klebanow, E. R., D. Poon, S. Zhou, and P. A. Weil. 1996. Isolation and characterization of TAF25, an essential yeast gene that encodes an RNA polymerase II-specific TATA-binding protein-associated factor. J. Biol. Chem. 271:13706-13715. [DOI] [PubMed] [Google Scholar]
- 31.Kuras, L., P. Kosa, M. Mencia, and K. Struhl. 2000. TAF-containing and TAF-independent forms of transcriptionally active TBP in vivo. Science 288:1244-1248. [DOI] [PubMed] [Google Scholar]
- 32.Larschan, E., and F. Winston. 2001. The Saccharomyces cerevisiae SAGA complex functions in vivo as a coactivator for transcriptional activation by Gal4. Genes Dev. 15:1946-1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee, T. I., and R. A. Young. 1998. Regulation of gene expression by TBP-associated proteins. Genes Dev. 12:1398-1408. [DOI] [PubMed] [Google Scholar]
- 34.Li, X. Y., S. R. Bhaumik, and M. R. Green. 2000. Distinct classes of yeast promoters revealed by differential TAF recruitment. Science 288:1242-1244. [DOI] [PubMed] [Google Scholar]
- 35.Link, A. J., J. Eng, D. M. Schieltz, E. Carmack, G. J. Mize, D. R. Morris, B. M. Garvik, and J. R. Yates III. 1999. Direct analysis of protein complexes using mass spectrometry. Nat. Biotechnol. 17:676-682. [DOI] [PubMed] [Google Scholar]
- 36.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 37.Macpherson, N., V. Measday, L. Moore, and B. Andrews. 2000. A yeast taf17 mutant requires the Swi6 transcriptional activator for viability and shows defects in cell cycle-regulated transcription. Genetics 154:1561-1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matangkasombut, O., R. M. Buratowski, N. W. Swilling, and S. Buratowski 2000. Bromodomain factor 1 corresponds to a missing piece of yeast. TFIID Genes Dev. 14:951-962. [PMC free article] [PubMed] [Google Scholar]
- 39.Poon, D., and P. A. Weil. 1993. Immunopurification of yeast TATA-binding protein and associated factors: presence of transcription factor IIIB transcriptional activity. J. Biol. Chem. 268:15325-15328. [PubMed] [Google Scholar]
- 40.Poon, D., A. M. Campbell, Y. Bai, and P. A. Weil. 1994. Yeast Taf170 is encoded by MOT1 and exists in a TATA box-binding protein (TBP)-TBP-associated factor complex distinct from transcription factor IID. J. Biol. Chem. 269:23135-23140. [PubMed] [Google Scholar]
- 41.Rigaut, G., A. Shevchenko, B. Rutz, M. Wilm, M. Mann, and B. Seraphin. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17:1030-1032. [DOI] [PubMed] [Google Scholar]
- 42.Roberts, S. M., and F. Winston. 1997. Essential functional interactions of SAGA, a Saccharomyces cerevisiae complex of Spt, Ada, and Gcn5 proteins, with the Snf/Swi and Srb/mediator complexes. Genetics 147:451-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saleh, A., V. Lang, R. Cook, and C. J. Brandl. 1997. Identification of native complexes containing the yeast coactivator/repressor proteins NGG1/ADA3 and ADA2. J. Biol. Chem. 272:5571-5578. [DOI] [PubMed] [Google Scholar]
- 44.Salghetti, S. E., A. A. Caudy, J. G. Chenoweth, and W. P. Tansey. 2001. Regulation of transcriptional activation domain function by ubiquitin. Science 293:1651-1653. [DOI] [PubMed] [Google Scholar]
- 45.Sanders, S. L., and P. A. Weil. 2000. Identification of two novel TAF subunits of the yeast Saccharomyces cerevisiae TFIID complex. J. Biol. Chem. 275:13895-13900. [DOI] [PubMed] [Google Scholar]
- 46.Sanders, S. L., E. R. Klebanow, and P. A. Weil. 1999. TAF25p, a non-histone-like subunit of TFIID and SAGA complexes, is essential for total mRNA gene transcription in vivo. J. Biol. Chem. 274:18847-18850. [DOI] [PubMed] [Google Scholar]
- 47.Saurin, A. J., Z. Shao, H. Erdjument-Bromage, P. Tempst, and R. E. Kingston. 2001. A Drosophila polycomb group complex includes Zeste and dTAFII proteins. Nature 412:655-660. [DOI] [PubMed] [Google Scholar]
- 48.Shen, X., G. Mizuguchi, A. Hamiche, and C. Wu. 2000. A chromatin remodeling complex involved in transcription and DNA processing. Nature 406:541-544. [DOI] [PubMed] [Google Scholar]
- 48a.Shevchenko, A., M. Wilm, O. Vorm, and M. Mann. 1996. Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels. Anal. Chem. 68:850-858. [DOI] [PubMed] [Google Scholar]
- 49.Simon, I., J. Barnett, N. Hannett, C. T. Harbison, N. J. Rinaldi, T. L. Volkert, J. J. Wyrick, J. Zeitlinger, D. K. Gifford, T. S. Jaakkola, and R. A. Young. 2001. Serial regulation of transcriptional regulators in the yeast cell cycle. Cell 106:697-708. [DOI] [PubMed] [Google Scholar]
- 50.Sullivan, S., D. W. Sink, K. L. Trout, I. Makalowska, P. M. Taylor, A. D. Baxevanis, and D. Landsman. 2002. The histone database. Nucleic Acids Res. 30:341-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tansey, W. P. 2001. Transcriptional activation: risky business. Genes Dev. 15:1045-1050. [DOI] [PubMed] [Google Scholar]
- 52.Uetz, P., L. Giot, G. Cagney, T. A. Mansfield, R. S. Judson, J. R. Knight, D. Lockshon, V. Narayan, M. Srinivasan, P. Pochart, A. Qureshi-Emili, Y. Li, B. Godwin, D. Conover, T. Kalbfleisch, G. Vijayadamodar, M. Yang, M. Johnston, S. Fields, and J. M. Rothberg. 2000. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403:623-627. [DOI] [PubMed] [Google Scholar]
- 53.Verma, R., S. Chen, R. Feldman, D. Schieltz, J. Yates, J. Dohmen, and R. J. Deshaies. 2000. Proteasomal proteomics: identification of nucleotide-sensitive proteasome-interacting proteins by mass spectrometric analysis of affinity-purified proteasomes. Mol. Biol. Cell 11:3425-3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wada, T., T. Takagi, Y. Yamaguchi, A. Ferdous, T. Imai, S. Hirose, S. Sugimoto, K. Yano, G. A. Hartzog, F. Winston, S. Buratowski, and H. Handa. 1998. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev. 12:343-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Washburn, M. P., and J. R. Yates III. 2000. Analysis of the microbial proteome. Curr. Opin. Microbiol. 3:292-297. [DOI] [PubMed] [Google Scholar]
- 56.Washburn, M. P., D. Wolters, and J. R. Yates III. 2001. Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 19:242-247. [DOI] [PubMed] [Google Scholar]
- 57.Wilcoxon, F. 1945. Individual comparisons by ranking methods. Biometrics 1:50-83. [PubMed] [Google Scholar]
- 58.Zhu, H., M. Bilgin, R. Bangham, D. Hall, A. Casamayor, P. Bertone, N. Lan, R. Jansen, S. Bidlingmaier, T. Houfek, T. Mitchell, P. Miller, R. A. Dean, M. Gerstein, and M. Snyder. 2001. Global analysis of protein activities using proteome chips. Science 293:2101-2105. [DOI] [PubMed] [Google Scholar]