Abstract
Pancreas duodenum homeobox 1 (PDX-1) is absolutely required for pancreas development and the maintenance of islet β-cell function. Temporal and cell-type-specific transcription of the pdx-1 gene is controlled by factors acting upon sequences found within its 5′-flanking region. Critical cis-acting transcriptional control elements are located within a nuclease hypersensitive site that contains three conserved subdomains, termed areas I, II, and III. We show that area II acts as a tissue-specific regulatory region of the pdx-1 gene, directing transgene expression to a subpopulation of islet cells. Mutation of the area II hepatocyte nuclear factor 3 (HNF3) binding element in the larger area I- and area II- containing PstBst fragment also decreases PBhsplacZ transgene penetrance. These two results indicate possible ontogenetic and/or functional heterogeneity of the β-cell population. Several other potential positive- and negative-acting control elements were identified in area II after mutation of the highly conserved sequence blocks within this subdomain. Pax6, a factor essential for islet α-cell development and islet hormone gene expression, was shown to bind in area II in vitro. Pax6 and HNF3β were also found to bind to this region in vivo by using the chromatin immunoprecipitation assay. Collectively, these data suggest an important role for both HNF3β and Pax6 in regulating pdx-1 expression in β cells.
The pancreas duodenum homeobox 1 (PDX-1) homeodomain containing transcription factor is required for pancreas development (25, 32) and the maintenance of functional islet β cells (1). Strikingly, the pancreas is not formed in humans (49) or mice that are homozygous for an inactivating mutation in PDX-1 (25, 32), apparently due to a block in the proliferation and differentiation of precursors in the endocrine and exocrine compartments of the pancreas. In the adult pancreas, PDX-1 is primarily expressed in endocrine islet β cells, where it is a key transcriptional regulator of many genes functionally associated with this cell type, including insulin (33, 34), glucose transporter type 2 (Glut2) (54), islet amyloid polypeptide (5, 6, 55), and glucokinase (56).
The diabetic phenotype that is induced by selectively removing PDX-1 from β cells in mice by using the Cre/loxP system appears to result from the limited expression of target genes associated with glucose sensing (i.e., Glut2) and hormone production (i.e., insulin) (1). The reduced expression of PDX-1 responsive genes also results in the compromised glucose sensing found in mice (1, 11) and humans (46) carrying only one functional pdx-1 allele. In addition, a form of maturity onset diabetes of the young is found in humans who are heterozygous for an inactivating mutation in PDX-1 (46, 48).
Because PDX-1 is essential for islet cell development and function, studies have focused on identifying transcription factors that mediate pdx-1 expression. Sequences that control appropriate developmental and adult specific expression are located in the 5′-flanking region (40, 47, 60). Three nuclease hypersensitive sites, termed HSS1 (bp −2560 to −1880), -2 (bp −1330 to −880), and -3 (bp −260 to +180), were identified within the endogenous mouse pdx-1 gene. However, only HSS1 sequences were capable of directing pancreatic β-cell-selective expression in transfection studies (60). Sequence alignment of the mouse, chicken, and human pdx-1 genes revealed that the HSS1 region also represented the only extensive area of significant identity within 4.5 kb of the transcription start site (15). Nucleotide sequence conservation was used to divide the HSS1 region into area I (bp −2761 to −2457), area II (bp −2153 to −1923), and area III (bp −1879 to −1600) (15). In contrast to areas I and III, which were conserved in mice, rats, chickens, and humans, area II is only found within mammalian pdx-1 genes (15), indicating that this subdomain may serve a species-specific function. However, areas I and II, but not area III, were independently capable of effectively directing β-cell-selective reporter gene activity in transfection assays. Moreover, a transgene spanning areas I and II (−2917bp/PstI to −1918bp/BstEII, termed PBhsplacZ) was expressed in the majority of islet β cells in vivo (14, 60).
Hepatocyte nuclear factor 3β (HNF3β), a forkhead transcription factor important in endodermal cell development (reviewed in reference 63), has been proposed to be a common activator of area I (15, 29), area II (15, 29, 60), and the rat distal islet-specific control region located between bp −6200 and −5670 (39). Mutagenesis of the conserved HNF3 binding elements in areas I or II resulted in a profound decrease in reporter gene (PstBst:pTKCAT) expression in β-cell lines, suggesting that both sites have a role in pdx-1 expression. However, they both were not absolutely essential, since the double mutant was as active as single mutants (15). Three HNF3 proteins (α, β, and γ) are actually found in islet β cells (53). However, since antisera that specifically recognize HNF3β quantitatively shifted the HNF3 site gel shift complex, HNF3β is believed to be the activator (15, 29, 60). Unfortunately, a direct role for HNF3β in pdx-1 transcription in vivo cannot be assessed in hnf3β−/− mice because of early embryonic lethality (3, 57). However, a decrease in pdx-1 mRNA levels observed in embryoid bodies from hnf3β−/− embryonic stem cells supports a role for HNF3β in pdx-1 transcription in vivo (15).
In the present study, we examined by transgenic analysis the activity of the individual area I and area II subdomains and the effect of the mutation in the area II HNF3 site on PBhsplacZ expression. In addition, cultured cell lines were used to identify regulators of area II. Area IIhsplacZ expression was observed in islet β cells but only in a subfraction of those expressing PDX-1. The area II HNF3 binding site mutant of PBhsplacZ was also only detected in a subfraction of the β-cell population. In contrast, the area I-driven transgene had no islet activity. Analysis of a series of individual area II block mutants in transfected β-cell lines defined a number of other potential positive- and negative-acting control elements. Pax6 was shown to be capable of specifically binding to an area II control element that is involved in transcriptional activation. Chromatin immunoprecipitation (ChIP) assays performed on the area II region demonstrated that Pax6 and HNF3β also bind in vivo. Thus, our data imply that area II plays a principal role in regulating pdx-1 expression in β cells, in part through the actions of Pax6 and HNF3β.
MATERIALS AND METHODS
pdx-1 transgene constructs and generation of transgenic mice.
Human area I (bp −2839 to −2520) and area II (bp −2141 to −1890) sequences were PCR amplified, and transgenes were constructed as previously described (60). The area II HNF3β binding site mutant transgene was created by subcloning PstBst m5 sequences from PstBst:pTK m5 into the transgene vector. The constructs were confirmed by restriction mapping and by partial sequence analysis. Transgenic lines were subsequently generated for each construct as previously described (60).
Genotyping.
Adults were genotyped by PCR with nested primers for lacZ. DNA from neonatal brain tissue or adult tails was prepared and analyzed as described previously (21). Southern blotting was performed with PvuII-digested genomic DNA and a 726-bp PvuII lacZ fragment as a probe to estimate the transgene copy number in each line.
Detection of β-Gal.
Abdominal organs and pancreata from 1- to 2-day-old pups were dissected and placed in phosphate-buffered saline on ice. Tissues were stained for β-galactosidase (β-Gal) activity and then paraffin embedded as previously described (60). Serial 7-μm sections were mounted on glass slides for immunohistochemical analysis.
Immunohistochemistry.
The Vectastain ABC (Vector Laboratories) and Histomouse SP (Zymed Lab-SA System; Zymed Laboratories, Inc.) kits were used for glucagon and insulin immunohistochemical staining, respectively. The guinea pig antibovine insulin serum (Linco) and rabbit antiglucagon serum (Linco) were used at a 1:1,000 dilution. Immunoperoxidase activity was detected by incubation with diaminobenzidine-H2O2 substrate (Vector Laboratories).
Transfection constructs.
The area II and PstBst reporter constructs were made by using human sequence from bp −2141 to −1961) and mouse (Pst/−2917bp:Bst/−1890bp [60]) pdx-1 sequences, which were cloned directly upstream of the herpes simplex virus thymidine kinase (TK) promoter in the chloramphenicol acetyltransferase (CAT) vector, pTK(An) (23). Block transversion and single base mutations were created in area II:pTK and PstBst:pTK by using the QuickChange mutagenesis kit (Stratagene). Mutagenesis primers were designed according to the kit protocol. The human −2141/−1961 sequence was generated by the PCR and cloned into pTK(An). All construct sequences were confirmed by DNA sequencing.
Cell transfections.
Monolayer cultures of the pancreatic islet β-cell (βTC-3, HIT-T15, and INS-1) and non-β-cell (BHK, HeLa, H4IIE, and NIH 3T3) lines were maintained as described previously (59). Rous sarcoma virus (RSV) enhancer-driven luciferase (LUC) activity from pRSVLUC was used to normalize CAT activity in each transfection. HIT-T15 and BHK cells were transfected by the calcium phosphate coprecipitation procedure (10 μg, total DNA) with 1 μg of area II:pTK and pRSVLUC and treated 4 h after addition of the DNA precipitates with 20% glycerol for 2 min. The Lipofectamine reagent (Gibco-BRL) was used to transfect 1 μg each of area II or PstBst:pTK and pRSVLUC into βTC-3 and INS-1 cells. Extracts were prepared 40 to 48 h after transfection, and LUC (9) and CAT (31) enzymatic assays were performed. Each experiment was carried out at least three independent times.
Electrophoretic mobility shift assays.
Nuclear extract from cell lines (38) and human islets (6) were prepared as described previously. The area II block 8 sequence (bp −2072 to −2054) was excised from −2072/−2054:pBluescript and Klenow labeled with [α-32P]dATP. Binding reactions (20 μl, total volume) were conducted with 5 to 10 μg of nuclear extract protein and probe to either bp −2072 to −2054 or P6Con, a consensus Pax6 binding element (8 × 104 cpm) (12). The conditions for the competition analyses were the same, except that a 50- to 100-fold excess of the specific competitor DNA was included in the mixture prior to addition of extract. The TNT-Coupled Reticulocyte Lysate system (Promega) was used to in vitro transcribe and translate Pax6 (pKW10-Pax6 [37]), Pax4 [pBluescript II KS(+)-Pax4; Beatriz Sosa-Pineda, St. Jude's Childrens Research Hospital, Memphis, Tenn.], Nkx6.1 [pBluescript II SK(+)-Nkx6.1 (26)], and Hblx9 [pBluescript II KS(+)-Hblx9; John Kehrl, National Institutes of Health (NIH), Bethesda, Md.]. Translation reactions conducted with [35S]methionine were used to determine the relative amount of protein in each reaction. Anti-Pax6 monoclonal antibody or a nonrelated Nkx2.2 monoclonal antibody was preincubated with extract protein for 10 min prior to the addition of the DNA probe.
DNase I footprinting.
The Core Footprinting System kit (Promega) was used to analyze the human region from bp −2141 to −1952 with some minor modifications. The labeled probe was prepared by linearizing area II:Bluescript with NotI (5′) or XhoI (3′), dephosphorylated with calf intestinal alkaline phosphatase, and labeled with T4 polynucleotide kinase and [γ-32P]ATP. The isolated bp −2141 to −1952 probe (7.5 × 104 cpm) was incubated with 50 μg of nuclear extract in a buffer containing 10 mM Tris-HCl (pH 7.4), 50 mM NaCl, 1 mM EDTA, 25 μg of poly(dA-dT), 10% glycerol, and 1 mM dithiothreitol (50 μl, total). The DNA pellets were resuspended in loading buffer, heat denatured, and applied to a 6% DNA sequencing gel.
SDS-PAGE fractionation.
βTC-3 nuclear extract (30 μg) was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on a 10% polyacrylamide gel and then electrotransferred onto an Immobilon polyvinylidene difluoride membrane (Millipore). The βTC-3 extract lane was cut horizontally into 4-mm slices representing different molecular weight fractions as determined by comparison with colored Rainbow protein markers (Amersham). The proteins from each fraction were eluted and renatured as previously described (64). Each fraction was analyzed for binding to the bp −2072 to −2054 probe.
ChIP assays.
βTC-3 cells (∼0.5 × 108 to 1.0 × 108) were formaldehyde cross-linked, and the sonicated protein-DNA complexes were isolated under conditions described previously (16). Pax6 (20 μl; Covance) or HNF3β (15 μl; Santa Cruz Biotechnology) antibody, normal rabbit immunoglobulin G (IgG; 10 μg; Santa Cruz Biotechnology), or no antibody was added to the sonicated chromatin, followed by incubation for 1 h at 4°C. Antibody-protein-DNA complexes were isolated by incubation with A/G-agarose (Santa Cruz Biotechnology). PCR was performed on one-tenth of the purified, immunoprecipitated DNA by using Ready-to-Go PCR beads (Amersham Pharmacia Biotech) and 15 pmol of each primer. The primers used for amplification of mouse pdx-1 area II were −2208 (5′-GGTGGGAAATCCTTCCCTCAAG-3′) and −1927 (5′-CCTTAGGGATAGACCCCCTGC-3′). The primers used for amplification of mouse PEPCK (phosphoenolpyruvate carboxykinase) were −434 (5′-GAGTGACACCTCACAGCTGTGG-3′) and −96 (5′-GGCAGGCCTTTGGATCATAGCC-3′). The PCR products were confirmed by sequencing. Amplified products were electrophoresed through a 1.4% agarose gel in TAE buffer and visualized by ethidium bromide staining.
RESULTS
A sequence-conserved domain of the PstBst region imparts β-cell-selective activity in vivo.
The PstBst transgene (PBhsplacZ) spans conserved areas I and II and is expressed at high levels in the islet. To test whether these subdomains also possess intrinsic tissue-specific regulatory activity, area IhsplacZ and area IIhsplacZ transgenic mice were generated. Twelve lines of area I transgenic mice and nine lines of area II transgenic mice were analyzed. None of the area IhsplacZ mice expressed β-Gal in the pancreas (data not shown). In contrast, pancreas expression was observed in seven of the nine area IIhsplacZ lines, with β-Gal activity principally limited to islet β cells (Fig. 1). Each of the area IIhsplacZ lines gave a similar expression pattern in the pancreas that was independent of transgene copy number (one to five copies/line) or chromosomal integration site. However, extrapancreatic staining varied between lines, presumably reflecting regulation imposed by nonrelated sequences at the integration site.
FIG. 1.
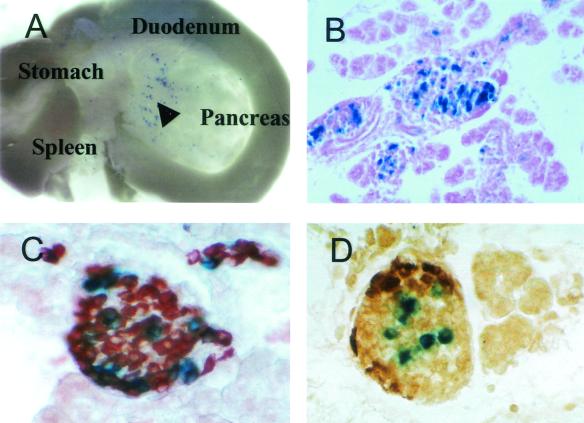
Area II directs islet β-cell-specific transgene expression in mice. (A) Abdominal organs of a 1-day-old area IIhsplacZ pup showed expression (blue) in the pancreas (arrowhead) and not in the stomach, spleen, or duodenum. Magnification, ×3.5. (B to D) β-Gal activity (blue) was only detected in islets (B) and overlapped with the brown immunostaining observed for insulin (C) and not for glucagon (D) in sections of 1-day-old pancreas. Magnification, ×100.
Area II-driven transgene expression was detected in a subfraction of the islet cells and primarily in the insulin-producing cell population (Fig. 1). In comparison to PBhsplacZ (see Fig. 2A), the limited expression of the area II transgene within islet β cells presumably reflects the loss of critical regulatory contributions by non-area II sequences within the PstBst region. Nonetheless, these results directly demonstrate that area II contains control elements sufficient for islet β-cell-selective transcription of pdx-1.
FIG. 2.
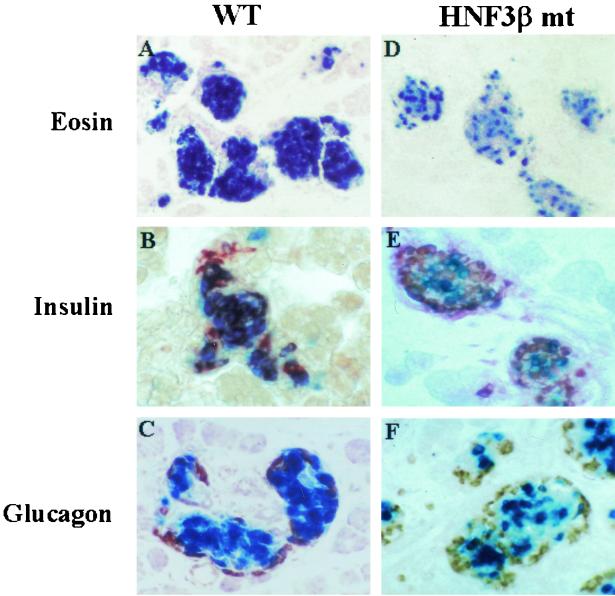
The HNF3β binding site mutant in area II limits PBhsplacZ transgene expression to a subpopulation of β cells. Sections of neonatal pancreas were stained for β-Gal activity (blue) and eosin counterstained (A and D) and with insulin (B and E) or glucagon antibodies (C and F) in the wild-type (WT) and HNF3β site mutant (HNF3β mt) transgenic PBhsplacZ mice. The β-Gal activity in wild-type mice seen in panel A is found in the majority of islet β cells but is limited to a subfraction in the HNF3β mt seen in panel D. β-Gal expression colocalizes with insulin immunolabeling in either wild type (in panel B) or HNF3β mt (in panel E) but rarely with glucagon (in panels C and F). Magnification, ×100.
Mutation of the HNF3 element in area II affects PBhsplacZ activity in vivo.
Mutational and functional analysis of the conserved HNF3 binding sites located within area I and area II demonstrated that a large fraction of the PstBst region activity in transfected β cells was dependent upon it (15, 29, 60). To determine the importance of HNF3 function, a mutation that selectively eliminated binding in area II was incorporated into PBhsplacZ (15, 60).
Five independent transgenic lines of the area II HNF3 binding site mutant PBhsplacZ were generated, of which three yielded an islet cell-specific expression pattern. The remaining two showed no activity possibly due to integration into transcriptionally inactive chromatin. The β-Gal expression in the HNF3 binding site mutant was compared to islets from PBhsplacZ. Insulin and glucagon immunohistochemical analysis demonstrated that β-Gal activity from the PBhsplacZ HNF3 mutant was primarily found in islet cells (Fig. 2). Because HNF3 is expressed in all islet cells (60), we expected to see a general decrease in β-Gal activity in the HNF3 site mutant relative to PBhsplacZ-expressing cells. Instead, we observed that the PBhsplacZ HNF3 mutant activity was confined to a subfraction of the β cells.
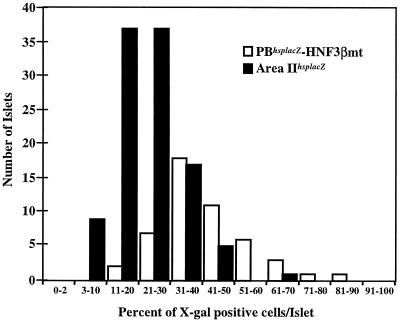
The level of islet cell penetrance of the area II HNF3 site mutant PBhsplacZ and area IIhsplacZ transgenes was compared. The fraction of expressing cells per islet ranged between 10 to 90% and 3 to 70% for the PBhsplacZ HNF3 site mutant and the area IIhsplacZ mice, respectively (Fig. 3). The lower level of expression in the area IIhsplacZ mice suggests that area I factors are necessary for expression throughout the β-cell population. In addition, these and the above data suggest that the comprehensive islet expression of PBhsplacZ requires key cis-acting element(s) within area II.
FIG. 3.
Fraction of β-Gal-expressing cells in area IIhsplacZ and HNF3β site mutant PBhsplacZ islets. The total islet cell number was determined by counting nuclei on eosin-counterstained sections from HNF3βhsplacZ site mutant (49 islets) and area IIhsplacZ (106 islets) neonatal mice.
Conserved area II sequences are important for β-cell activity.
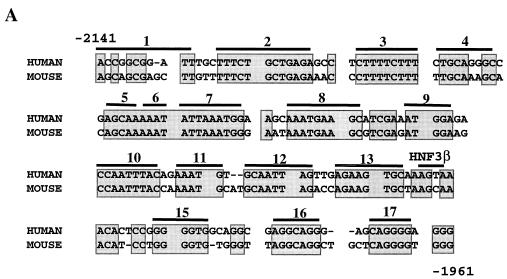
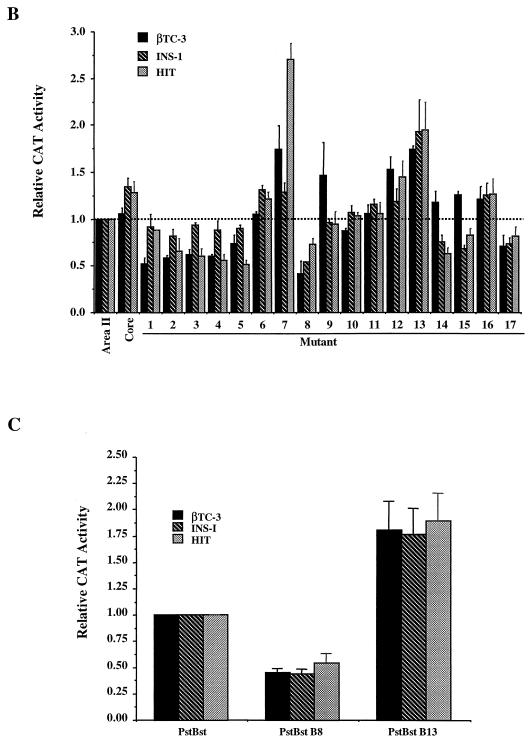
In transfection assays, area II reporter constructs containing either human or mouse sequence display β-cell-selective activity (15). Conserved area II sequences appear to be the most important for activity, since the region of maximal identity between human and mouse sequence (bp −2141 to −1961, 80% identity; termed Core; Fig. 4 ) can stimulate reporter gene expression as effectively as the larger area II reporter construct (bp −2141 to −1890, 59% identity) in HIT-T15, βTC-3, and INS-1 β-cell lines (compare Core to area II; Fig. 4B). To characterize the contribution of the conserved sequences to activity, each block (B1 to B17) of identity in human area II was independently mutated, and activity was measured in transfected β-cell lines (Fig. 4).
FIG. 4.
Conserved sequences within area II contribute to transcriptional activation in β cells. (A) Conserved sequences within the core region of area II are shaded. The nucleotides are numbered relative to the mouse pdx-1 gene. Each bar spans a block of mutated sequence; the HNF3β binding site (B14) is labeled. (B) Wild-type and mutant area II:pTK CAT constructs were transfected into βTC-3, INS-1, and HIT-T15 cells. The mutations were made within the area II construct spanning bp −2141 to −1890; the Core construct spans bp −2141 to −1961. The mutant area II:pTK activity ± the standard error is presented relative to area II:pTK. (C) The activity of wild-type and B8 and B13 mutations in PstBst is shown as a ratio of the mutant to wild-type PstBst:pTK activity ± the standard error.
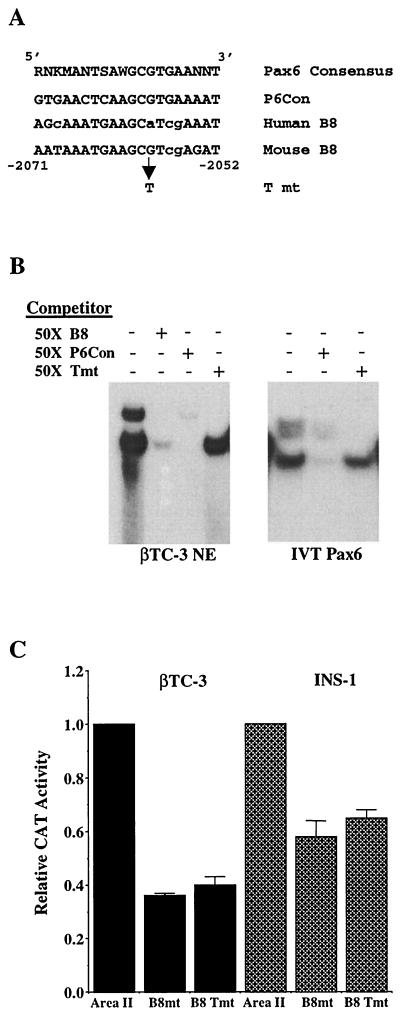
Mutations in B7, B8, and B13 affected (with B8 decreasing and B7 and B13 increasing) area II-driven activity in all β-cell lines used, whereas mutations in B2 to B5, B12, B14, B15, and B17 affected activity in only two of the three β-cell lines (Fig. 4B). In contrast, the mutations in B1, B6, B9 to B11, and B16 had little effect on area II activation. The B8 site and a representative negative control site (i.e., B13) were mutated, and their activity was analyzed within the context of the area I- and area II-containing PstBst construct. The B8 and B13 mutations also affected PstBst stimulation (Fig. 4C), since the B8 mutant activity was reduced by 50%, a level comparable to that of the HNF3β binding site mutants (15), and the B13 mutation activity increased by 75%. We conclude that B8 (bp −2068 to −2060) defines a site for a β-cell activator, B13 (bp −2016 to −2009), and B7 (bp −2081 to −2073), a β-cell repressor.
Identification of potential area II transcription factor binding sites.
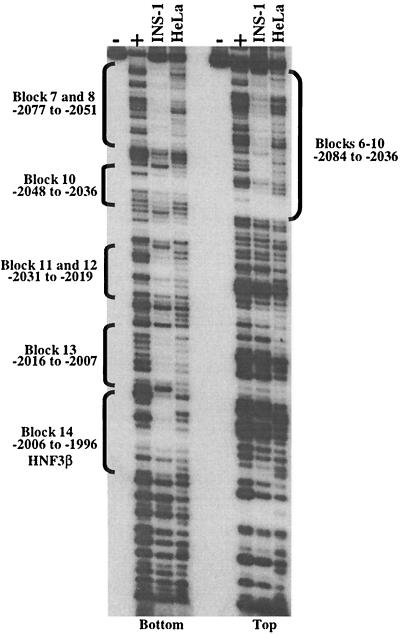
To identify possible area II regulatory protein binding sites, DNase I footprinting analysis was performed over the region from bp −2141 to −1961 with β-cell (INS-1) and non-β-cell (HeLa) nuclear extracts. The protected regions that were specific to the β-cell extract were presumed to represent cell-type-enriched DNA-binding activities. Using a labeled bottom-strand probe, five distinct sequence segments displayed β-cell-specific footprints. These included B7 and B8 (bp −2077 to −2051), B10 (bp −2048 to −2036), B11 and B12 (bp −2031 to −2019), B13 (bp −2016 to −2007), and B14 (bp −2006 to −1996) (Fig. 5). The B14 footprint results from HNF3β binding (15, 29, 60). The footprint observed with the area II top-strand probe was continuous over a large region extending from bp −2084 to −2036, spanning B6 to B10 (Fig. 5). Highly similar digestion patterns were observed with βTC-3, HIT-T15, and Min6 nuclear extracts (data not shown). Because the B8 activator bound in a β-cell-selective fashion, the resulting studies focused on its identification.
FIG. 5.
DNase I footprinting reveals multiple β-cell-specific binding sites in area II. The human area II fragment extending from bp −2141 to −1961 was end labeled on the top or bottom strand and subjected to DNase I digestion after incubation with INS-1 or HeLa nuclear extracts (50 μg). The DNase I-produced fragments were run along side Maxam-Gilbert sequencing reactions (G and G+A) of the same fragment. Note the INS-1 selective protection pattern observed over B7 and B8 (bp −2077 to −2051), B10 (bp −2048 to −2036), B11 and B12 (bp −2031 to −2019), B13 (bp −2016 to −2007), and the HNF3β binding site (B14; bp −2006 to −1996) with the bottom-strand probe and over B6 through B10 with the top-strand probe.
Pax6 binds to the B8 activator element.
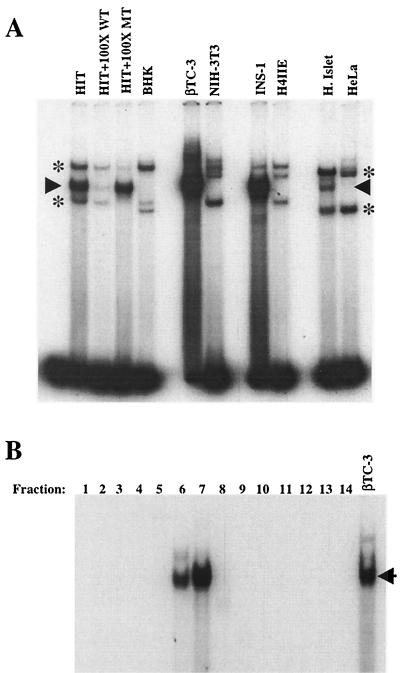
The specificity of protein-DNA binding to a bp −2072 to −2054 B8 probe was determined by wild-type and mutant B8 competition in nuclear extracts prepared from β-cell lines (HIT-T15, βTC-3, and INS-1), human islets, and non-β-cell lines (BHK [kidney], NIH 3T3, H4IIE [liver], and HeLa). A single, common protein-DNA complex was detected within β-cell and human islet extracts but not within the non-β-cell extracts (Fig. 6A). Furthermore, only the wild-type B8 competitor efficiently reduced the binding levels of this complex. However, the B8 block mutant competitor prevented formation of the ubiquitous complexes (indicated by asterisks in the figure), a finding consistent with the conclusion that they are unrelated to B8 activation. Together with the DNase I footprinting analysis, this experiment suggests that the B8 activator is a nuclear protein enriched in islet β cells.
FIG. 6.
A β-cell-specific protein binds to B8. (A) Nuclear extracts from β-cell (HIT-T15, βTC-3, and INS-1) and non-β-cell (BHK, NIH 3T3, H4IIE, and HeLa) lines and human islets were analyzed for B8 binding in gel shift assays. A 100-fold molar excess of unlabeled wild type or block mutant B8 competitor to probe was used to distinguish the B8 activator complex from the nonspecific binding complexes. The arrow denotes the specific B8 binding activity, and the asterisks mark the nonspecific complexes. (B) βTC-3 nuclear extract was fractionated (see Materials and Methods) and assayed for specific B8 binding. Each fraction (i.e., fractions 1 to 14) represents a different molecular size range. A binding complex was detected in fractions 6 (44-kDa lower limit) and 7 (58-kDa upper limit) that comigrated with the β-cell-enriched B8 binding complex in unfractionated βTC-3 nuclear extract. The arrow points to the β-cell-enriched binding complex.
To characterize the B8 activator further, the molecular weight of the protein(s) binding to the bp −2072 to −2054 probe was estimated by separation and renaturation of βTC-3 nuclear proteins (Fig. 6B). The major binding activity in fractions 6 and 7 comigrated with the principal B8 complex found in unfractionated βTC-3 extracts. The binding specificity of this complex corresponded to the B8 activator as binding was selectively eliminated by the wild-type competitor (data not shown). The molecular size range for fractions 6 and 7 (44 to 58 kDa) indicated that the B8 activator complex is composed of a protein(s) of roughly 50 kDa.
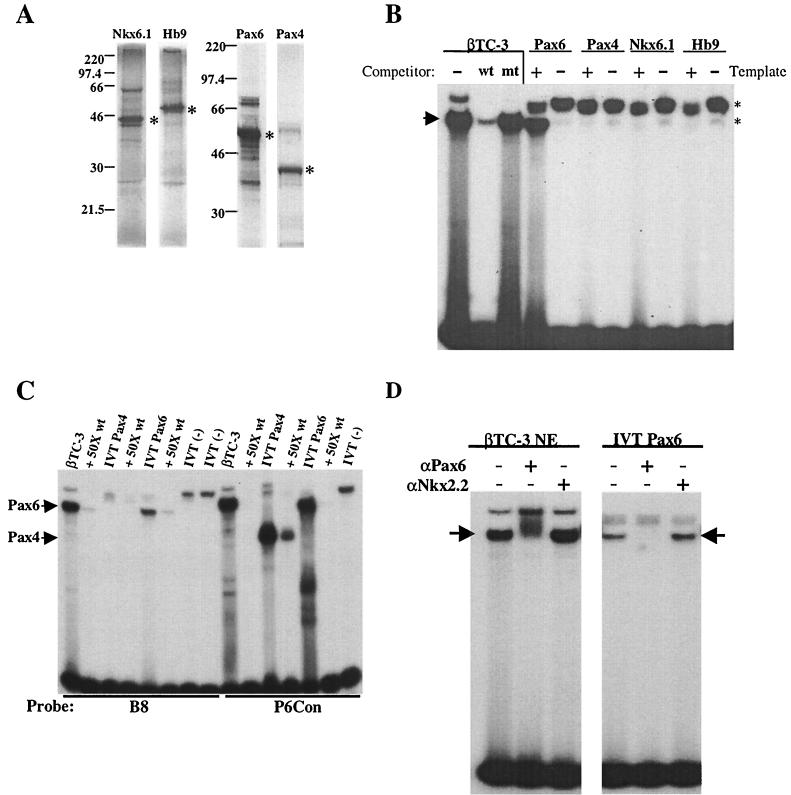
The molecular size of several β-cell-enriched transcription factors lies within this range, although no candidate was suggested by Transfac analysis (20). We therefore determined directly whether in vitro-translated (IVT) forms of Hb9 (41 kDa [19]), Nkx6.1 (44 to 46 kDa [26]), Pax4 (38 kDa [22, 44]), Pax6 (46.6 kDa [51]), and PDX-1 (47 kDa [34; data not shown]) could bind to the B8 element. Each of these proteins was translated at similar levels in our reactions (Fig. 7A). However, binding to the B8 probe was only observed with Pax6, and this complex comigrated with the complex detected in βTC-3 extracts (Fig. 7B). Because Pax4 and Pax6 have similar binding properties (12, 27), gel shifts were also conducted with a Pax6 consensus sequence probe (P6Con) that binds Pax4 with high affinity. As expected, both Pax4 and Pax6 bound effectively to the P6Con probe, but only Pax6 bound to B8 (Fig. 7C). In addition, preincubation of either IVT Pax6 or βTC-3 nuclear extract with monoclonal Pax6 antibody specifically eliminated B8 activator complex formation (Fig. 7D).
FIG. 7.
Pax6 binds to B8 in vitro. (A) [35S]methionine was used to label IVT Pax6, Pax4, Nkx6.1, and Hb9. The positions of the full-length proteins are indicated by asterisks, and the molecular size markers are labeled. (B) Binding assays with B8 were performed in the presence (+) or absence (−) of IVT Pax6, Pax4, Nkx6.1, or Hb9 protein. The B8 binding complexes formed with βTC-3 nuclear extract are shown. The specific complex (arrow) was identified by competition with the wild-type (wt) and block mutant (mt) oligonucleotides. Nonspecific binding is indicated with asterisks. (C) Binding assays with B8 or P6Con (12) were performed with βTC-3 nuclear extract or IVT Pax4 or Pax6 in the presence or absence of 50-fold wild-type (wt) competitor. (D) Binding to B8 was conducted with βTC-3 nuclear extract (βTC-3 NE) or IVT Pax6 in the absence (−) or presence (+) of a monoclonal Pax6 or Nkx2.2 antibody. The arrow indicates the β-cell-enriched binding complex.
The mouse B8 (mB8) element is 90% identical to the consensus sequence for the bipartite Pax6 paired DNA-binding domain (Fig. 8A ). As an additional test for the role of Pax6 in area II reporter gene activation, a single-base-pair binding mutation was created in this context. The G-to-T mutation in the mouse sequence (A to T in the human sequence) was chosen because it had been shown to specifically reduce Pax6 binding to the mouse glucagon G1 element (2). This mutation also prevented mB8 from competing for Pax6 binding in gel shift assays (compare Tmt to B8, Fig. 8B). In addition, B8 Tmt decreased the activity of human area II by an amount similar to that caused by the B8 block mutation in transfection assays (Fig. 8C). These results suggest that the binding of Pax6 to B8 regulates area II-mediated transcription.
FIG. 8.
Pax6 binding contributes to area II activation. (A) The sequences of the mouse and human B8 and the B8 point mutant (T mt) are compared to a derived Pax6 binding oligonucleotide (P6Con [12]). The nucleotide similarity of the mouse and human sequences to the consensus Pax6 element is indicated with nonmatching nucleotides in lowercase. W is A or T; R is A or G; K is G or T; S is C or G; M is A or C. (B) Binding reactions were conducted with the B8-probeby using βTC-3 nuclear extracts (βTC-3 NE) or IVT Pax6. A 50-fold molar excess of unlabeled B8, B8 Tmt, and P6Con competitor was used. (C) Wild-type and mutant human area II:pTK constructs were transfected into βTC-3 and INS-1 cells. The ratio of mutant to wild-type area II:pTK activity is presented ± the standard error. Note that the B8 block mutant (B8mt) and point mutant activities (B8 Tmt) are reduced to a similar level relative to the wild type.
Pax6 and HNF3β bind to area II in vivo.
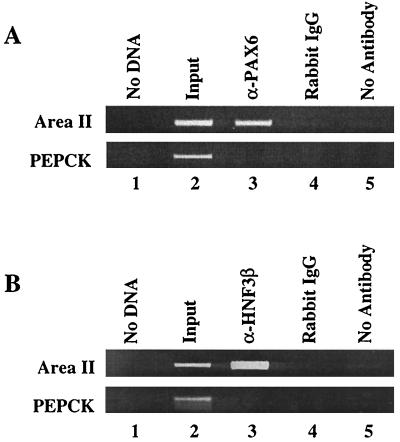
To determine whether Pax6 and HNF3β bind within the area II region of the endogenous pdx-1, ChIP assays were carried out. The antibodies to Pax6 and HNF3β immunoprecipitated area II sequences from βTC-3 cells, in contrast to the rabbit IgG or the no-antibody controls (Fig. 9). In addition, neither the Pax6 nor the HNF3β antibody immunoprecipitated the promoter sequences from the PEPCK gene, which is not transcribed in βTC-3 cells. The same results were obtained with chromatin from MIN6 β cells (data not shown). These results are consistent with both Pax6 and HNF3β playing a central role in regulating the β-cell expression of pdx-1 in vivo.
FIG. 9.
Pax6 binds to area II in vivo. Cross-linked chromatin from βTC-3 cells was incubated with antibodies raised to a C-terminal epitope of mouse Pax6 or HNF3β. The anti-Pax6 (A) and the anti-HNF3β (B) immunoprecipitated DNA were analyzed by PCR with area II and PEPCK promoter-specific primers (lane 3). As controls, PCRs were run with no DNA (lane1), on input chromatin (1:100 dilution, lane 2), and with DNA obtained after precipitation with rabbit IgG (lane 4) or no antibody (lane 5).
DISCUSSION
PDX-1 is expressed in the progenitor cells of the endocrine and exocrine pancreas, primarily in the islet β cells of the pancreas in adults. Cell-type-specific expression of PDX-1 is required for normal pancreas development and glucose homeostasis in mammals. In the present study, we examined the basis for β-cell-selective transcription of the pdx-1 gene. Previous analysis of 5′-flanking pdx-1 sequences suggested that cis-acting elements involved in tissue-specific transcriptional control were contained within conserved area I and area II sequences, since both of these domains are located within the PBhsplacZ transgene that is expressed primarily in islet β cells in neonates (60). The significance of area II was further demonstrated here. Area II and not area I independently targeted expression to islet β cells in vivo; thus, this PstBst subdomain acts as a tissue-specific regulator. Mutation of the conserved HNF3 binding site in area II compromised islet cell expression of PBhsplacZ, indicating that HNF3α, -β, or -γ had an essential role in the regulation of pdx-1 transcription. To localize additional regulatory elements within area II, blocks of conserved sequence within this subdomain were systematically mutated, and their effects on activation were determined in transfection assays. Several potential positive- and negative-acting control sites were identified, and Pax6, a factor involved in endodermal development and islet hormone gene expression, was shown to be associated with area II activation in vitro. Most significantly, our ability to detect Pax6 and HNF3β binding within the area II region of the endogenous pdx-1 gene provides additional support for a direct role in regulation.
Tissue-specific gene expression appears to be controlled by regulatory factors functioning in a cooperative manner. For example, specific positive- and negative-acting gene sequences control the elevation in α-fetoprotein expression in the fetal liver, the low levels in adults, and reexpression during liver regeneration and cancer (45, 52, 58). Similar to α-fetoprotein, pdx-1 is expressed in embryos throughout the pancreatic buds but is later downregulated in most exocrine cells and is primarily expressed in β cells in the adult endocrine pancreas (32, 47). In addition, expression is increased in response to partial pancreatectomy (62). Thus, pdx-1 is expressed in a developmental and tissue-specific pattern that is also likely mediated by both positive- and negative-acting sequences. Specific positive-acting sequences have been demonstrated that drive transient β-cell-specific activity in the embryo and neonate but not in the adult (XhoBgl [13]), whereas the islet cell-specific sequence is active at all ages (PstBst [13, 60]). This independently acting, islet-specific regulatory segment was first identified in HSS1. Three subregions (i.e., areas I, II, and III) were identified within HSS1 for further study. Because the PstBst fragment spanning areas I and II drives tissue-specific transgene expression, we developed area IhsplacZ and area IIhsplacZ transgenic mice to determine their role in this activity. In a parallel study, area III conserved sequences were also tested by transgenic analysis (13). Strikingly, only area II, but not area I (this report) or III (13), was capable of directing islet cell-specific transgene expression in neonatal mice. This implies that area II represents the principal islet control domain of PstBst and, presumably, also HSS1.
Among the species studied, area II is only found within the mammalian pdx-1 genes (e.g., human, mouse, and rat [15; K. Gerrish and R. Stein, unpublished data]), implying that β-cell expression in unrelated species is driven by other non-area II sequences. Furthermore, the limited expression of the area II transgene to a fraction of islet cells indicates that functional interactions with non-area II transcription factors are critical for expression throughout the entire islet β-cell population. The factors that contribute to control include HNF3β in areas I (15, 29) (Fig. 9B) and II (15, 60), PDX-1 (16, 29, 30) and HNF1α (16) in area I, and, as reported here, Pax6 in area II.
The absence of area II and PstBst mutant transgene activity within some β cells strongly suggests that non-area II control sequences and HNF3β help define the islet expression pattern of the pdx-1 gene in vivo. The presence of a mutationally sensitive HNF3β binding site within each of the pdx-1 gene subdomains that stimulate β-cell-selective reporter gene expression (15, 29, 39, 60) is indicative of HNF3β's role in pdx-1 regulation. ChIP analysis clearly indicates that HNF3β binds within the area II region in vivo (Fig. 9B). Importantly, pdx-1 mRNA expression is decreased 50 to 60% in β cells upon conditional inactivation of the HNF3β gene by insulin-driven Cre recombinase expression in vivo (K. Kaestner, unpublished data). Together, these data indicate that HNF3β is an important regulator of pdx-1 transcription in the β cell. Although it is unclear how HNF3β mediates transcriptional control, both the chromatin remodeling (17) and transcriptional activation potential (8) of this factor may be of significance.
Since HNF3β (15, 60) and PDX-1 (18, 34) are essentially expressed within all islet β cells, one expected outcome of mutating the area II HNF3β binding site would be a reduction in PBhsplacZ activity in all cells. In previous studies, the reduction in pdx-1 transcriptional activity in HNF3β binding site mutants in transfection assays and HNF3β-null embryoid bodies was reported as a general reduction in expression (15). These experiments evaluated the population of cells en masse, and any restriction to a subpopulation would have appeared as a reduction in expression. Thus, the apparent discrepancy with our results reflects the different assays used in these experiments. In the current study, the area II HNF3β binding site mutation did not appear to significantly reduce transgene activity within expressing cells as expected but instead limited activity to a fraction of the cells relative to wild-type PBhsplacZ (Fig. 2 and 3). Area IIhsplacZ expression was similarly limited to a fraction of the β cells (Fig. 1 and 3). It is unlikely that the sites of chromosomal integration or copy number influence these transgene expression patterns since very similar results were found in three independent PBhsplacZ HNF3β site mutant lines and in seven independent area IIhsplacZ lines.
The limited activity of the PBhsplacZ HNF3β site mutant and area IIhsplacZ may reflect heterogeneity in expression and/or activity of cooperating transcriptional activators of the pdx-1 gene within the islet β-cell population. Evidence for functional heterogeneity among normal and transformed β cells has been shown to exist at the metabolic level (4, 24, 36, 41). The varied sensitivity to area II mutations seen in the HIT-T15, βTC-3, and INS-1 cell lines (Fig. 4) may, in addition, reflect heterogeneity in the transcriptional regulatory capacity among islet β cells both in vitro and in vivo. Whether the restricted expression of the area II or PstBst mutant transgene denotes islet β cells that also have different metabolic and/or neogenic potential is under investigation.
To begin to identify other factors that are necessary for area II activity, we prepared a comprehensive series of block mutations within the conserved sequences. Our transfection analyses demonstrated that no single mutation profoundly affected area II-mediated activation. Only 3 (B8, B7, and B13) of the 17 block mutants affected area II-mediated activation by ∼50% or greater in our β-cell lines, with the mutation of B8 repressing activity and the mutation of B7 and B13 stimulating activity (Fig. 4). The B8 and B13 mutations in PstBst recapitulated their effects in area II alone.
Our biochemical analysis focused on identifying the B8 activator. Pax6, a member of the paired-box, homeodomain transcription factor family was shown to specifically bind to B8 element sequences in vitro, and more significantly, to bind to area II sequences in vivo. Pax6 also appears to be involved in islet insulin, somatostatin, and glucagon gene activation (37). These observations indicate that Pax6 is one component of a complex regulatory network involved in defining pdx-1 promoter activity in the pancreas.
Pax6 and Pax4 represent the only known members of the paired-box, homeodomain transcription factor family involved in pancreatic islet development (10). Both of these proteins have similar in vitro DNA-binding properties (27). Furthermore, Pax4 can inhibit insulin- and glucagon-driven reporter expression in transfected cells by competing for and reducing Pax6-mediated activation (35, 42). As a consequence, competition between Pax4, which is expressed in the pancreas only during embryogenesis (42), and Pax6 may be involved in regulating gene expression in islet progenitor cells. Pax6 and Pax4 gene ablation studies indicate that these factors also have distinct and nonoverlapping gene targets during islet cell development (44, 50). The B8 element is different from many other Pax6 binding sites in not binding Pax4, implying a distinct role for Pax6 in pdx-1 transcription (Fig. 7). Even though PDX-1 levels appear normal in Pax6−/− embryos (37, 50), these animals die at birth (44, 50) due to central nervous system problems. Therefore, Pax6 may be required for normal pdx-1 expression and islet function at later points. Several mouse models of islet neogenesis also support a function for Pax6 in pdx-1 expression (28, 43, 61).
The ability of area II, but not of area I, to direct β-cell-specific transgene expression indicates that the transcription factors important for its activation are also essential in making the pdx-1 gene locus competent for transcription. In general, little is known about the mechanisms utilized to convert a silent gene to a transcriptionally active one. With analogy to the manner in which HNF3β functions in albumin gene activation (7), we presume that the earliest regulators bind within area II to initiate opening of the chromatin and transcription complex assembly prior to pdx-1 gene activation. The limited islet β-cell expression pattern observed for the PBhsplacZ area II HNF3β binding site mutant supports the proposal that HNF3β may serve in this capacity for pdx-1. However, detection of pdx-1 expression in HNF3β−/− embryoid bodies (15) indicates that other factors are also important in nucleating transcription complex assembly. Initiation of Pax6 expression occurs later than that of pdx-1, suggesting that Pax6 represents a factor that is recruited to transcriptionally active chromatin to modulate activity. Our area II mutational studies and DNase I footprinting have defined potential binding sites for factors that may act in conjunction with Pax6 and HNF3β to mediate tissue-specific control of the pdx-1 gene. We believe that identifying these regulatory factors will not only provide insight into the mechanisms involved in regulating pdx-1 transcription and pancreas development but also may help to identify heritable defects that cause insulin deficiency and diabetes.
Acknowledgments
We thank Eva Henderson and Min Guo for technical support and Klaus Kaestner at the University of Pennsylvania for allowing us to cite his data on the β-cell-specific knockout of the HNF3β gene prior to publication. The monoclonal Pax6 and Nkx2.2 antibodies were developed by Atsushi Kawakami and Thomas Jessell, respectively, and were obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by The University of Iowa, Department of Biological Sciences, Iowa City, Iowa. The Human Islet Core Lab at the Diabetes Research and Training Center, Washington University, St. Louis, Mo., provided the human islets.
This work was supported by grants from the NIH (RO1 DK50203 to R.S. and PO1 DK42502 to C.V.E.W.), a training grant in Molecular Endocrinology (5T32DK07563 to M.G.), and the Juvenile Diabetes Foundation (JDF 398212 to S.E.S.). The transgenic mice were developed with the assistance of the Transgenic/ES cell Shared Resource (NCIP30 CA68485 and NIH DK20593). Partial support was also provided from the Vanderbilt University Diabetes Research and Training Center Molecular Biology Core Laboratory (Public Health Service grant P60 DK20593 from the NIH).
REFERENCES
- 1.Ahlgren, U., J. Jonsson, L. Jonsson, K. Simu, and H. Edlund. 1998. Beta-cell-specific inactivation of the mouse Ipf1/Pdx1 gene results in loss of the β-cell phenotype and maturity onset diabetes. Genes Dev. 12:1763-1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersen, F. G., R. S. Heller, H. V. Petersen, J. Jensen, O. D. Madsen, and P. Serup. 1999. Pax6 and Cdx2/3 form a functional complex on the rat glucagon gene promoter G1-element. FEBS Lett. 445:306-310. [DOI] [PubMed] [Google Scholar]
- 3.Ang, S. L., and J. Rossant. 1994. HNF-3β is essential for node and notochord formation in mouse development. Cell 78:561-574. [DOI] [PubMed] [Google Scholar]
- 4.Bennett, B. D., T. L. Jetton, G. Ying, M. A. Magnuson, and D. W. Piston. 1996. Quantitative subcellular imaging of glucose metabolism within intact pancreatic islets. J. Biol. Chem. 271:3647-3651. [DOI] [PubMed] [Google Scholar]
- 5.Bretherton-Watt, D., N. Gore, and D. S. Boam. 1996. Insulin upstream factor 1 and a novel ubiquitous factor bind to the human islet amyloid polypeptide/amylin gene promoter. Biochem. J. 313:495-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carty, M. D., J. S. Lillquist, M. Peshavaria, R. Stein, and W. C. Soeller. 1997. Identification of cis- and trans-active factors regulating human islet amyloid polypeptide gene expression in pancreatic β cells. J. Biol. Chem. 272:11986-11993. [DOI] [PubMed] [Google Scholar]
- 7.Cirillo, L. A., and K. S. Zaret. 1999. An early developmental transcription factor complex that is more stable on nucleosome core particles than on free DNA. Mol. Cell 4:961-969. [DOI] [PubMed] [Google Scholar]
- 8.Denson, L. A., M. H. McClure, C. W. Bogue, S. J. Karpen, and H. C. Jacobs. 2000. HNF3β and GATA-4 transactivate the liver-enriched homeobox gene, Hex. Gene 246:311-320. [DOI] [PubMed] [Google Scholar]
- 9.de Wet, J. R., K. V. Wood, M. DeLuca, D. R. Helinski, and S. Subramani. 1987. Firefly luciferase gene: structure and expression in mammalian cells. Mol. Cell. Biol. 7:725-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dohrmann, C., P. Gruss, and L. Lemaire. 2000. Pax genes and the differentiation of hormone-producing endocrine cells in the pancreas. Mech. Dev. 92:47-54. [DOI] [PubMed] [Google Scholar]
- 11.Dutta, S., S. Bonner-Weir, M. Montminy, and C. Wright. 1998. Regulatory factor linked to late-onset diabetes? Nature 392:560. [DOI] [PubMed] [Google Scholar]
- 12.Epstein, J., J. Cai, T. Glaser, L. Jepeal, and R. Maas. 1994. Identification of a Pax paired domain recognition sequence and evidence for DNA-dependent conformational changes. J. Biol. Chem. 269:8355-8361. [PubMed] [Google Scholar]
- 13.Gannon, M., L. W. Gamer, and C. V. E. Wright. 2001. Regulatory regions driving developmental and tissue-specific expression of the essential pancreatic gene pdx1. Dev. Biol. 238:185-201. [DOI] [PubMed] [Google Scholar]
- 14.Gannon, M., M. K. Ray, K. Van Zee, F. Rausa, R. H. Costa, and C. V. Wright. 2000. Persistent expression of HNF6 in islet endocrine cells causes disrupted islet architecture and loss of β cell function. Development 127:2883-2895. [DOI] [PubMed] [Google Scholar]
- 15.Gerrish, K., M. Gannon, D. Shih, E. Henderson, M. Stoffel, C. V. Wright, and R. Stein. 2000. Pancreatic β cell-specific transcription of the pdx-1 gene: the role of conserved upstream control regions and their hepatic nuclear factor 3β sites. J. Biol. Chem. 275:3485-3492. [DOI] [PubMed] [Google Scholar]
- 16.Gerrish, K. E., M. A. Cissell, and R. Stein. 2001. The role of hepatic nuclear factor 1α and PDX-1 in transcriptional regulation of the pdx-1 gene. J. Biol. Chem. 276:22071-22076. [DOI] [PubMed] [Google Scholar]
- 17.Gualdi, R., P. Bossard, M. Zheng, Y. Hamada, J. R. Coleman, and K. S. Zaret. 1996. Hepatic specification of the gut endoderm in vitro: cell signaling and transcriptional control. Genes Dev. 10:1670-1682. [DOI] [PubMed] [Google Scholar]
- 18.Guz, Y., M. R. Montminy, R. Stein, J. Leonard, L. W. Gamer, C. V. Wright, and G. Teitelman. 1995. Expression of murine STF-1, a putative insulin gene transcription factor, in β cells of pancreas, duodenal epithelium and pancreatic exocrine and endocrine progenitors during ontogeny. Development 121:11-18. [DOI] [PubMed] [Google Scholar]
- 19.Harrison, K. A., J. Thaler, S. L. Pfaff, H. Gu, and J. H. Kehrl. 1999. Pancreas dorsal lobe agenesis and abnormal islets of Langerhans in Hlxb9-deficient mice. Nat. Genet. 23:71-75. [DOI] [PubMed] [Google Scholar]
- 20.Heinemeyer, T., E. Wingender, I. Reuter, H. Hermjakob, A. E. Kel, O. V. Kel, E. V. Ignatieva, E. A. Ananko, O. A. Podkolodnaya, F. A. Kolpakov, N. L. Podkolodny, and N. A. Kolchanov. 1998. Databases on transcriptional regulation: TRANSFAC, TRRD and COMPEL. Nucleic Acids Res. 26:362-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hogan, B., R. Beddington, F. Constantini, and E. Lacy. 1994. Manipulating the mouse embryo: a laboratory manual, 2nd ed. Cold Spring Harbor Press, Cold Spring Harbor, N.Y.
- 22.Inoue, H., J. Nomiyama, K. Nakai, A. Matsutani, Y. Tanizawa, and Y. Oka. 1998. Isolation of full-length cDNA of mouse PAX4 gene and identification of its human homologue. Biochem. Biophys. Res. Commun. 243:628-633. [DOI] [PubMed] [Google Scholar]
- 23.Jacoby, D. B., N. D. Zilz, and H. C. Towle. 1989. Sequences within the 5′-flanking region of the S14 gene confer responsiveness to glucose in primary hepatocytes. J. Biol. Chem. 264:17623-17626. [PubMed] [Google Scholar]
- 24.Jonkers, F. C., and J. C. Henquin. 2001. Measurements of cytoplasmic Ca2+ in islet cell clusters show that glucose rapidly recruits β cells and gradually increases the individual cell response. Diabetes 50:540-550. [DOI] [PubMed] [Google Scholar]
- 25.Jonsson, J., L. Carlsson, T. Edlund, and H. Edlund. 1994. Insulin-promoter-factor 1 is required for pancreas development in mice. Nature 371:606-609. [DOI] [PubMed] [Google Scholar]
- 26.Jorgensen, M. C., H. Vestergard-Petersen, J. Ericson, O. D. Madsen, and P. Serup. 1999. Cloning and DNA-binding properties of the rat pancreatic β-cell-specific factor Nkx6.1. FEBS Lett. 461:287-294. [DOI] [PubMed] [Google Scholar]
- 27.Kalousova, A., V. Benes, J. Paces, V. Paces, and Z. Kozmik. 1999. DNA binding and transactivating properties of the paired and homeobox protein Pax4. Biochem. Biophys. Res. Commun. 259:510-518. [DOI] [PubMed] [Google Scholar]
- 28.Kritzik, M. R., E. Jones, Z. Chen, M. Krakowski, T. Krahl, A. Good, C. Wright, H. Fox, and N. Sarvetnick. 1999. PDX-1 and Msx-2 expression in the regenerating and developing pancreas. J. Endocrinol. 163:523-530. [DOI] [PubMed] [Google Scholar]
- 29.Marshak, S., E. Benshushan, M. Shoshkes, L. Havin, E. Cerasi, and D. Melloul. 2000. Functional conservation of regulatory elements in the pdx-1 gene: PDX-1 and hepatocyte nuclear factor 3β transcription factors mediate β-cell-specific expression. Mol. Cell. Biol. 20:7583-7590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marshak, S., E. Ben-Shushan, M. Shoshkes, L. Havin, E. Cerasi, and D. Melloul. 2001. Regulatory elements involved in human pdx-1 gene expression. Diabetes 50(Suppl. 1):S37-S38. [DOI] [PubMed] [Google Scholar]
- 31.Nordeen, S. K., P. P. D. Green, and D. M. Fowlkes. 1987. A rapid, sensitive, and inexpensive assay for chloramphenicol acetyltransferase. DNA 6:173-178. [DOI] [PubMed] [Google Scholar]
- 32.Offield, M. F., T. L. Jetton, P. A. Labosky, M. Ray, R. W. Stein, M. A. Magnuson, B. L. Hogan, and C. V. Wright. 1996. PDX-1 is required for pancreatic outgrowth and differentiation of the rostral duodenum. Development 122:983-995. [DOI] [PubMed] [Google Scholar]
- 33.Ohlsson, H., K. Karlsson, and T. Edlund. 1993. IPF1, a homeodomain-containing transactivator of the insulin gene. EMBO J. 12:4251-4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peshavaria, M., L. Gamer, E. Henderson, G. Teitelman, C. V. Wright, and R. Stein. 1994. XIHbox 8, an endoderm-specific Xenopus homeodomain protein, is closely related to a mammalian insulin gene transcription factor. Mol. Endocrinol. 8:806-816. [DOI] [PubMed] [Google Scholar]
- 35.Petersen, H. V., M. C. Jorgensen, F. G. Andersen, J. Jensen, T. F.-Nielsen, R. Jorgensen, O. D. Madsen, and P. Serup. 2000. Pax4 represses pancreatic glucagon gene expression. Mol. Cell. Biol. Res. Commun. 3:249-254. [DOI] [PubMed] [Google Scholar]
- 36.Sanchez-Soto, M. C., M. E. Larrieta, R. Vidaltamayo, and M. Hiriart. 1999. Heterogeneity in glutamic acid decarboxylase expression among single rat pancreatic β cells. Diabetologia 42:1086-1092. [DOI] [PubMed] [Google Scholar]
- 37.Sander, M., A. Neubuser, J. Kalamaras, H. C. Ee, G. R. Martin, and M. S. German. 1997. Genetic analysis reveals that PAX6 is required for normal transcription of pancreatic hormone genes and islet development. Genes Dev. 11:1662-1673. [DOI] [PubMed] [Google Scholar]
- 38.Schreiber, E., P. Matthias, M. M. Muller, and W. Schaffner. 1989. Rapid detection of octamer binding proteins with “mini-extracts,” prepared from a small number of cells. Nucleic Acids Res. 17:6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sharma, S., U. S. Jhala, T. Johnson, K. Ferreri, J. Leonard, and M. Montminy. 1997. Hormonal regulation of an islet-specific enhancer in the pancreatic homeobox gene STF-1. Mol. Cell. Biol. 17:2598-2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sharma, S., J. Leonard, S. Lee, H. D. Chapman, E. H. Leiter, and M. R. Montminy. 1996. Pancreatic islet expression of the homeobox factor STF-1 relies on an E-box motif that binds USF. J. Biol. Chem. 271:2294-2299. [DOI] [PubMed] [Google Scholar]
- 41.Smith, P., P. J. Millard, C. M. Fewtrell, and F. M. Ashcroft. 1997. Heterogeneity of β-cell Ca2+ responses to glucose. Adv. Exp. Med. Biol. 426:253-257. [DOI] [PubMed] [Google Scholar]
- 42.Smith, S. B., H. C. Ee, J. R. Conners, and M. S. German. 1999. Paired-homeodomain transcription factor PAX4 acts as a transcriptional repressor in early pancreatic development. Mol. Cell. Biol. 19:8272-8280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Song, S. Y., M. Gannon, M. K. Washington, C. R. Scoggins, I. M. Meszoely, J. R. Goldenring, C. R. Marino, E. P. Sandgren, R. J. Coffey, Jr., C. V. Wright, and S. D. Leach. 1999. Expansion of Pdx1-expressing pancreatic epithelium and islet neogenesis in transgenic mice overexpressing transforming growth factor alpha. Gastroenterology 117:1416-1426. [DOI] [PubMed] [Google Scholar]
- 44.Sosa-Pineda, B., K. Chowdhury, M. Torres, G. Oliver, and P. Gruss. 1997. The Pax4 gene is essential for differentiation of insulin-producing β cells in the mammalian pancreas. Nature 386:399-402. [DOI] [PubMed] [Google Scholar]
- 45.Spear, B. T. 1999. Alpha-fetoprotein gene regulation: lessons from transgenic mice. Semin. Cancer Biol. 9:109-116. [DOI] [PubMed] [Google Scholar]
- 46.Stoffers, D. A., J. Ferrer, W. L. Clarke, and J. F. Habener. 1997. Early-onset type-II diabetes mellitus (MODY4) linked to IPF1. Nat. Genet. 17:138-139. [DOI] [PubMed] [Google Scholar]
- 47.Stoffers, D. A., R. S. Heller, C. P. Miller, and J. F. Habener. 1999. Developmental expression of the homeodomain protein IDX-1 in mice transgenic for an IDX-1 promoter/lacZ transcriptional reporter. Endocrinology 140:5374-5381. [DOI] [PubMed] [Google Scholar]
- 48.Stoffers, D. A., V. Stanojevic, and J. F. Habener. 1998. Insulin promoter factor-1 gene mutation linked to early-onset type 2 diabetes mellitus directs expression of a dominant-negative isoprotein. J. Clin. Investig. 102:232-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stoffers, D. A., N. T. Zinkin, V. Stanojevic, W. L. Clarke, and J. F. Habener. 1997. Pancreatic agenesis attributable to a single nucleotide deletion in the human IPF1 gene coding sequence. Nat. Genet. 15:106-110. [DOI] [PubMed] [Google Scholar]
- 50.St-Onge, L., B. Sosa-Pineda, K. Chowdhury, A. Mansouri, and P. Gruss. 1997. Pax6 is required for differentiation of glucagon-producing α cells in mouse pancreas. Nature 387:406-409. [DOI] [PubMed] [Google Scholar]
- 51.Turque, N., S. Plaza, F. Radvanyi, C. Carriere, and S. Saule. 1994. Pax-QNR/Pax-6, a paired box- and homeobox-containing gene expressed in neurons, is also expressed in pancreatic endocrine cells. Mol. Endocrinol. 8:929-938. [DOI] [PubMed] [Google Scholar]
- 52.Vacher, J., and S. M. Tilghman. 1990. Dominant negative regulation of the mouse α-fetoprotein gene in adult liver. Science 250:1732-1735. [DOI] [PubMed] [Google Scholar]
- 53.Vaisse, C., J. Kim, R. Espinosa III, M. M. Le Beau, and M. Stoffel. 1997. Pancreatic islet expression studies and polymorphic DNA markers in the genes encoding hepatocyte nuclear factor-3α, -3β, -3γ, -4γ, and -6. Diabetes 46:1364-1367. [DOI] [PubMed] [Google Scholar]
- 54.Waeber, G., N. Thompson, P. Nicod, and C. Bonny. 1996. Transcriptional activation of the GLUT2 gene by the IPF-1/STF-1/IDX-1 homeobox factor. Mol. Endocrinol. 10:1327-1334. [DOI] [PubMed] [Google Scholar]
- 55.Watada, H., Y. Kajimoto, H. Kaneto, T. Matsuoka, Y. Fujitani, J. Miyazaki, and Y. Yamasaki. 1996. Involvement of the homeodomain-containing transcription factor PDX-1 in islet amyloid polypeptide gene transcription. Biochem. Biophys. Res. Commun. 229:746-751. [DOI] [PubMed] [Google Scholar]
- 56.Watada, H., Y. Kajimoto, Y. Umayahara, T. Matsuoka, H. Kaneto, Y. Fujitani, T. Kamada, R. Kawamori, and Y. Yamasaki. 1996. The human glucokinase gene β-cell-type promoter: an essential role of insulin promoter factor 1/PDX-1 in its activation in HIT-T15 cells. Diabetes 45:1478-1488. [DOI] [PubMed] [Google Scholar]
- 57.Weinstein, D. C., A. Ruiz i Altaba, W. S. Chen, P. Hoodless, V. R. Prezioso, T. M. Jessell, and J. E. Darnell, Jr. 1994. The winged-helix transcription factor HNF-3β is required for notochord development in the mouse embryo. Cell 78:575-588. [DOI] [PubMed] [Google Scholar]
- 58.Wen, P., E. R. Groupp, G. Buzard, N. Crawford, and J. Locker. 1991. Enhancer, repressor, and promoter specificities combine to regulate the rat α-fetoprotein gene. DNA Cell Biol. 10:525-536. [DOI] [PubMed] [Google Scholar]
- 59.Whelan, J., D. Poon, P. A. Weil, and R. Stein. 1989. Pancreatic β-cell-type-specific expression of the rat insulin II gene is controlled by positive and negative cellular transcriptional elements. Mol. Cell. Biol. 9:3253-3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu, K. L., M. Gannon, M. Peshavaria, M. F. Offield, E. Henderson, M. Ray, A. Marks, L. W. Gamer, C. V. Wright, and R. Stein. 1997. Hepatocyte nuclear factor 3β is involved in pancreatic β-cell-specific transcription of the pdx-1 gene. Mol. Cell. Biol. 17:6002-6013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yamaoka, T., M. Yano, T. Yamada, T. Matsushita, M. Moritani, S. Ii, K. Yoshimoto, J. Hata, and M. Itakura. 2000. Diabetes and pancreatic tumours in transgenic mice expressing Pax6. Diabetologia 43:332-339. [DOI] [PubMed] [Google Scholar]
- 62.Zangen, D. H., S. Bonner-Weir, C. H. Lee, J. B. Latimer, C. P. Miller, J. F. Habener, and G. C. Weir. 1997. Reduced insulin, GLUT2, and IDX-1 in β cells after partial pancreatectomy. Diabetes 46:258-264. [DOI] [PubMed] [Google Scholar]
- 63.Zaret, K. 1999. Developmental competence of the gut endoderm: genetic potentiation by GATA and HNF3/fork head proteins. Dev. Biol. 209:1-10. [DOI] [PubMed] [Google Scholar]
- 64.Zhao, L., M. A. Cissell, E. Henderson, R. Colbran, and R. Stein. 2000. The RIPE3b1 activator of the insulin gene is composed of a protein(s) of approximately 43 kDa, whose DNA binding activity is inhibited by protein phosphatase treatment. J. Biol. Chem. 275:10532-10537. [DOI] [PubMed] [Google Scholar]