Abstract
hnRNPK and hnRNP E1/E2 mediate translational silencing of cellular and viral mRNAs in a differentiation-dependent way by binding to specific regulatory sequences. The translation of 15-lipoxygenase (LOX) mRNA in erythroid precursor cells and of the L2 mRNA of human papilloma virus type 16 (HPV-16) in squamous epithelial cells is silenced when either of these cells is immature and is activated in maturing cells by unknown mechanisms. Here we address the question of how the silenced mRNA can be translationally activated. We show that hnRNP K and the c-Src kinase specifically interact with each other, leading to c-Src activation and tyrosine phosphorylation of hnRNP K in vivo and in vitro. c-Src-mediated phosphorylation reversibly inhibits the binding of hnRNP K to the differentiation control element (DICE) of the LOX mRNA 3′ untranslated region in vitro and specifically derepresses the translation of DICE-bearing mRNAs in vivo. Our results establish a novel role of c-Src kinase in translational gene regulation and reveal a mechanism by which silenced mRNAs can be translationally activated.
hnRNP K and hnRNP E1/E2 regulate human papilloma virus type 16 (HPV-16) L2 capsid protein mRNA and reticulocyte 15-lipoxygenase (LOX) mRNA expression in the course of cellular differentiation. The expression of the virus capsid protein L2 is restricted to terminally differentiated epithelial cells in the superficial layers of the squamous epithelium by repression of L2 mRNA translation in the deeper layers (13). The underlying inhibitory mechanism employs hnRNPs K and E1/E2 interacting with a specific cis-acting element in the 3′ end of L2 mRNA (5). LOX is a key enzyme in erythroid cell differentiation. It can attack phospholipids of the mitochondrial membranes and participates in their breakdown in late reticulocytes (20, 24). LOX expression is temporally restricted by translational silencing of LOX mRNA in erythroid precursor cells in the bone marrow and in early reticulocytes (12). The differentiation control element (DICE) in the 3′ untranslated region (UTR) of LOX mRNA binds the KH domain proteins hnRNP K and hnRNP E1 to form translationally silenced mRNPs in immature erythroid cells (15, 16). The hnRNP K/E1-DICE complex blocks 80S ribosome assembly by inhibition of 60S ribosomal subunit joining (15). This erythroid silencing mechanism can be recapitulated in vitro and in HeLa cells transfected with DICE-regulated reporter mRNAs and hnRNP K alone or together with hnRNP E1 (16). Like LOX mRNA silencing, the HPV-16 L2 mRNA mechanism has been shown to operate in HeLa cells as well (5).
The C-terminal part of hnRNP K contains proline-rich domains, which enable hnRNP K to interact with the SH3 domains of members of the Src kinase family (3) such as c-Src itself (23, 25, 27), Fyn, and Lyn (27). c-Src and Lck, a further member of the Src kinase family, have been shown to be able to phosphorylate hnRNP K in vitro and to affect its binding to RNA (19). The functional significance of hnRNP K tyrosine phosphorylation by members of the Src family of kinases is as yet unknown.
Here we show that hnRNP K specifically binds and activates c-Src. c-Src mediates tyrosine phosphorylation of hnRNP K and inhibition of its DICE binding activity. Moreover, we demonstrate that c-Src kinase specifically regulates hnRNP K function as a translational silencer in vivo. Our results identify a specific role of c-Src in posttranscriptional regulation via hnRNP K, and suggest a mechanism for how the differentiation-dependent translation of cellular and viral RNA could be activated in maturing cells.
MATERIALS AND METHODS
Plasmids.
For luciferase (LUC) indicator constructs, the LUC cDNA from pGEM-LUC (Promega) was inserted into pSG5 (16). LUC-DICE and LUC-NR were made by insertion of the DICE or nonregulatory (NR) sequences of the LOX mRNA 3′ UTR into the EcoRV site of pSG5 before insertion of the LUC open reading frame (18). pSG5-His-hnRNP K was generated using pSG5-hnRNP K (16) by insertion of an oligonucleotide coding for 10 histidine residues between the SmaI and XhoI sites, N-terminal to hnRNP K. The tyrosine-to-phenylalanine mutants pSG5-His-hnRNP K(Y4F) (Y 230, 234, 236, 380 F) and pSG5-His-hnRNP K(Y6F) (Y 72, 225, 230, 234, 236, 380 F) were made with a site-directed mutagenesis kit (Stratagene) according to manufacturer's recommendations. pSG5-FLAG-hnRNP E1 was generated using pSG5-hnRNP E1 (16) by insertion of the FLAG peptide between the SmaI and XhoI sites, N terminal to hnRNP E1. The different Src mutants cloned into the simian virus 40 (SV40) expression vector pSGT, were described previously [pSGT-c-Src, pSGT-Src(KP), pSGT-Src(Y527F), and pSGT-Src(−SH3)] (7, 8). The plasmids SV40-beta-gal, pSGT-Src(Y416F), and pSGT-Src(−SH2) were kind gifts from Stefania Gonfloni and Jana Kretschmar.
Expression, in vitro phosphorylation, and dephosphorylation of recombinant protein.
The cDNAs coding for hnRNP K and hnRNP E1 were cloned into the pET-16b plasmid (Novagen) for transformation of Escherichia coli BL21(DE3). Expression and purification of His-hnRNP K and His-hnRNP E1 were carried out as described elsewhere (16). A 200-ng portion of each recombinant His-hnRNP K or E1 protein was tyrosine phosphorylated with c-Src (only His-hnRNP K), Src(KP), or Src(Y416F), immunopurified with the Src-217 antibody (8) from transiently transfected HeLa cells (300 μg of total protein) for 2 h at 30°C, and dephosphorylated with λ-phosphatase (New England Biolabs) for 2 h at 30°C in 50 mM Tris (pH 7.5), 20 mM MnCl2, 1 mM dithiothreitol (DTT), and 0.1 mM ATP.
In vitro transcription and Northwestern analysis.
Uncapped 32P-labeled DICE RNA (specific activity, 7 × 107 cpm/μg) used in Northwestern blot assays was transcribed with T3 RNA polymerase (Stratagene) and purified as described previously (18). Recombinant proteins His-hnRNP K and E1 were in vitro phosphorylated and dephosphorylated as indicated (see Fig. 5), resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), blotted onto a nitrocellulose membrane (Schleicher and Schüll), denatured in 8 M urea, and slowly renatured (10 dilution steps of urea in Tris-buffered saline for 10 min each). Finally, the membrane was rinsed in Tris-buffered saline and incubated for 30 min in 25 mM NaCl, 10 mM MgCl2, 10 mM HEPES (pH 8.0), 0.1 mM EDTA, and 1 mM DTT. 32P-labeled DICE binding was carried out in 50 mM NaCl, 10 mM MgCl2, 10 mM HEPES (pH 8.0), 0.1 mM EDTA, and 1 mM DTT for 10 min; unbound probe was washed off afterwards with binding buffer. Binding of the DICE was analyzed by autoradiography.
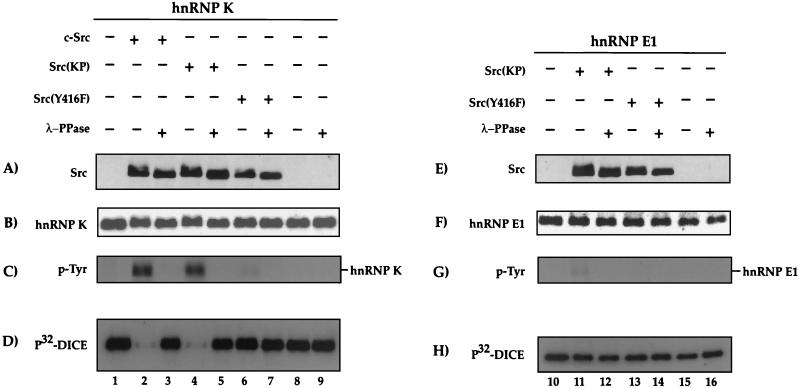
FIG. 5.
Tyr-phosphorylation of hnRNP K reversibly affects its DICE binding activity. His-hnRNP K (lanes 1 to 9) and His-hnRNP E1 (lanes 10 to 16) were in vitro phosphorylated with either c-Src (lanes 2 and 3), Src(KP) (lanes 4, 5, 11, and 12) or Src(Y416F) (lanes 6, 7, 13, and 14) immunopurified from transiently transfected or nontransfected (lanes 8, 9, 15, and 16) HeLa cells. Dephosphorylation was subsequently achieved by incubation with λ-phosphatase (λ-PPase) (lanes 3, 5, 7, 9, 12, 14, and 16). The reaction products were resolved by SDS-PAGE and analyzed in Western blot assays using either anti-Src antibody (A and E), anti-His antibody (hnRNP K and hnRNP E1) (B and F), or antiphosphotyrosine antibody (p-Tyr) (C and G). (D and H) DICE binding activity of hnRNPs K and E1 was examined with a 32P-labeled DICE probe by Northwestern blotting.
HeLa cell transfection and analysis.
HeLa cells were transiently transfected by the calcium phosphate method (9). For the experiments shown in Fig. 1, cDNAs coding for c-Src and different Src mutants (His-hnRNP K and FLAG-hnRNP E1) were cotransfected as indicated. For Fig. 3, 5 μg of pSG5-His-hnRNP K was cotransfected with 5 μg of either pSGT-c-Src, pSGT-Src(−SH3), or pSGT-Src(−SH2). For Fig. 4, 5 μg of pSG5-His-hnRNP K, pSG5-His-hnRNP K(Y4F), or pSG5-His-hnRNP K(Y6F) was cotransfected with 5 μg of pSGT vector, pSGT-c-Src, or pSGT-Src(Y416F). For Fig. 6, 5 μg of LUC-DICE or LUC-NR and, for normalization, 5 μg of SV40 β-galactosidase control plasmid, as well as 5 μg of pSG5-U1A or pSG5-His-hnRNP K, were cotransfected with 5 μg of pSGT vector or pSGT-c-Src, pSGT-Src(KP), or pSGT-Src(Y416F), respectively. The inhibitor of Src PP2 [4-amino-5-(4-chlorophenyl)-7-(t-butyl)pyrazolo(3,4-d)pyrimidine], or, as a negative control, PP3 [4-amino-7-phenylpyrazolo(3,4-d)pyrimidine] (Calbiochem) (11), was added where indicated.
FIG. 1.
hnRNP K, but not hnRNP E1, is a substrate and activator of c-Src. HeLa cells were transiently transfected with His-tagged hnRNP K (A to G) or FLAG-tagged hnRNP E1 (H to N) and c-Src, the activated mutants Src(KP) and Src(Y527F), or the inactive autophosphorylation site mutant Src(Y416F). HeLa cell lysate was resolved by SDS-PAGE and analyzed in Western blot assays (lanes 1 to 8) with either anti-His (A), anti-hnRNP E1 (H), anti-c-Src (B and I), antiphosphotyrosine (p-Tyr) (C and J), or anti-Src(416) (D and K) antibodies. His-tagged hnRNP K was immunoprecipitated with an anti-His antibody (E to G) and FLAG-tagged hnRNP E1 with an anti-FLAG antibody (L to N), resolved by SDS-PAGE, and analyzed by Western blot assays with antibodies against the His tag (E) or the FLAG tag (L) to assess the amounts of precipitated hnRNP K and hnRNP E1, p-Tyr to analyze the Tyr phosphorylation status of hnRNP K and hnRNP E1 (F and M), or Src antibody to visualize coprecipitated c-Src (G and N). The increased background in panels H to N results from the necessity of exposing the Western blots significantly longer than those in panels A to G to discern the specific signals. wt, wild type.
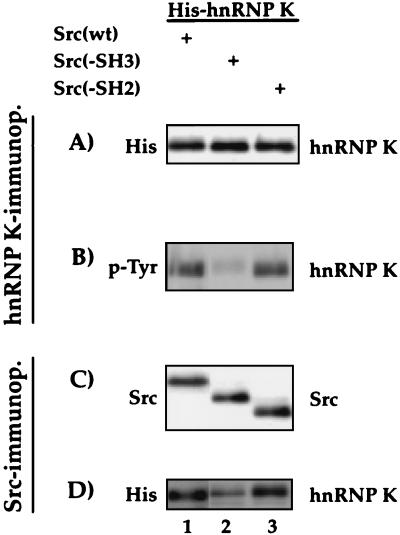
FIG. 3.
The SH3 domain of c-Src is important for the interaction with and phosphorylation of hnRNP K. HeLa cells were transfected with His-hnRNP K and c-Src, Src(−SH3), or Src(−SH2) expression vectors. The products of a His immunoprecipitation (A and B) or a Src immunoprecipitation (C and D) were analyzed after SDS-PAGE in Western blot assays using antibodies against the His-tag to detect the precipitated His-hnRNP K (A) and p-Tyr to examine Tyr phosphorylation of hnRNP K (B). After Src immunoprecipitation, c-Src expression was assessed with an anti-c-Src antibody (C) and the coprecipitated His-hnRNP K was detected with an anti-His antibody (D).
FIG. 4.
Tyr-phosphorylation of Y4F and Y6F His-hnRNP K mutants. (Top) Amino acid sequence of hnRNP K with the Tyr residues that are mutated to Phe residues in the His-hnRNP K mutants Y4F and Y6F underlined and numbered; the other 11 Tyr residues are underlined. (Bottom) HeLa cells were transfected with cDNAs coding for wild-type, Y4F, or Y6F His-hnRNP K, together with control plasmid, c-Src, or Src(Y416F). After immunoprecipitation with anti-His antibody, SDS-PAGE, and Western blotting, the levels of hnRNP K were determined with an anti-His antibody (A), the Tyr phosphorylation of hnRNP K was assayed with an antiphosphotyrosine antibody (p-Tyr) (B), and its interaction with Src was detected with an anti-Src antibody (C).
FIG. 6.
c-Src activates the translation of LUC-DICE mRNA silenced by hnRNP K. (A) HeLa cells were transiently cotransfected with LUC-DICE or LUC-NR expression vectors and expression plasmids coding for U1A (as a specificity control) or His-hnRNP K and for c-Src, the activated mutant Src(KP), or the inactive form Src(Y416F). SV40-beta-gal was also cotransfected, and β-galactosidase activity was used to correct for differences in transfection efficiency. Extracts were used to analyze the enzymatic activity of the expressed LUC protein. (B) LUC-DICE and His-hnRNP K expression vectors were cotransfected with or without c-Src, Src(KP), or Src(Y416F). After 16 h of transfection, the medium was changed to a low fetal bovine serum concentration (0.2%) for 24 h. Following serum starvation, fresh medium containing 10% fetal bovine serum was added for 4 h. Where indicated, 10 μM PP2 or PP3 was added for 2 h before addition of fresh medium including the indicated substances. From the HeLa cell lysate, His-hnRNP K was immunoprecipitated with an anti-His antibody. (Top) Proteins were resolved by SDS-PAGE and analyzed in Western blot assays using antibodies against phosphotyrosine (p-Tyr) and His tag. (Bottom) Extracts were analyzed in an enzymatic LUC activity assay.
Luciferase activity assays (see Fig. 6) were carried out as described in reference 2.
HeLa cell lysate preparation and immunoprecipitation was performed as described previously (16) with anti-His (H5; Qiagen) or anti-c-Src (Src-217) (8). Western blotting onto polyvinylidene difluoride membranes was performed according to standard protocols. Antibodies against His, FLAG (M2; Sigma), hnRNP E1 (D. H. Ostareck, unpublished data), c-Src (Src-217), p-Tyr (PY99; Santa Cruz Biotechnology), and Src(416) (New England Biolabs) were used to identify the corresponding proteins as indicated in the figure legends.
In vitro Src kinase assay.
c-Src and Src(Y527F) expressed in human HEK-293 cells were immunopurified with a Src-specific monoclonal antibody (Src-217) prebound to protein G-Sepharose (Amersham Pharmacia) from 85 μg of total protein. Equal amounts of the immunopurified kinases were incubated in the presence or absence of 125 or 375 ng of purified recombinant hnRNP K or E1 in a kinase assay buffer in the presence of 80 μM ATP, 2 μCi of [γ-32P]ATP (3,000 Ci/mmol; Amersham), and 3 μM acid-denatured enolase as an exogenous substrate at 30°C for 10 min (6). The specific inhibitor of Src, PP2, and the inactive control PP3, both at 1 μM, were added to the kinase assay where indicated. After SDS-PAGE separation, the proteins were Coomassie stained. Phosphorylation of enolase and hnRNPs K and E1 was analyzed by autoradiography. The bands corresponding to enolase and hnRNPs K and E1 were excised, and incorporated radioactivity was quantified by scintillation counting (Cerenkov; Beckman LS6000-SC).
Identification of phosphorylated Tyr residues.
hnRNP K (5 μg) was phosphorylated in vivo by cotransfection of c-Src (5 μg) into HeLa cells. The lysates were prepared and immunoprecipitation was performed as described above. After SDS-PAGE and Coomassie staining, the corresponding protein bands were isolated. The protein bands were excised, washed, reduced, and alkylated, followed by tryptic digestion overnight, as previously described (21). The resulting peptide mixture was desalted and fractionated by three elution steps using an OligoR3 microcolumn. Each fraction was then analyzed individually by nanoelectrospray mass spectrometry as previously described (14). For phosphopeptide detection, all experiments were carried out on an API III triple quadrupole mass spectrometer (Applied Biosystems, Sciex, Concord, Ontario, Canada) equipped with a nanoelectrospray ion source. Parent ion scans of m/z 79 for the detection of phosphopeptides in negative ion mode were acquired at a collision energy setting of 50 V (difference between R0 and R2 potentials on the API III instrument). Argon was used as the collision gas at a thickness of approximately 3 × 1014 molecules/cm2. Once the phosphopeptides were detected, the nanospray capillary with the analyte solution was transferred to a quadrupole time-of-flight instrument (Micromass, Manchester, United Kingdom), and the phosphopeptides were fragmented by collision with nitrogen in the collision cell. The collision energy was adjusted for each peptide individually. The resulting product ion spectra allowed the partial sequence determination of the peptides and therefore the localization of the phosphorylation sites.
RESULTS
hnRNP K specifically binds and activates Src kinase in vivo and in vitro.
Since activated c-Src and Lck can phosphorylate hnRNP K in vitro (19), we wanted to examine whether hnRNP K can be phosphorylated by active forms of Src kinase in transfected cells. HeLa cells were cotransfected with expression vectors for wild-type c-Src, constitutively active or inactive c-Src mutants, and histidine-tagged hnRNP K. Protein expression was monitored by Western blotting of total cell lysates using antibodies directed against the His tag (Fig. 1A), c-Src (Fig. 1B), phosphotyrosine (Fig. 1C), and c-Src phosphorylated at Tyr-416 (Fig. 1D), which indicates Src kinase activity. Surprisingly, transfection of wild-type c-Src with His-hnRNP K (Fig. 1A, compare lanes 1 and 4) causes an increase in tyrosine phosphorylation of cellular proteins (Fig. 1C, lanes 1 and 4). This increase in tyrosine phosphorylation reflects posttranslational Src kinase activation over the basal activity (Fig. 1D, compare lanes 1 and 2 with lane 4), because the levels of c-Src protein are similar (Fig. 1B, lanes 1, 2, and 4). As a positive control for the activated state of c-Src, we transfected 5 μg of His-hnRNP K together with the constitutively active mutant Src-KP or Src-Y527F (8) (Fig. 1B, lanes 5 and 6). The overall tyrosine phosphorylation of cellular proteins with 5 μg of His-hnRNP K and wild-type c-Src is at least as high as the levels observed with the activated mutants (Fig. 1C, compare lanes 4, 5, and 6). By contrast, the inactive autophosphorylation site mutant Src-Y416F fails to be activated by hnRNP K and does not increase cellular phosphotyrosine levels following the cotransfection of hnRNP K (Fig. 1C and D, lane 7). These findings suggest that hnRNP K is able to activate the tyrosine kinase activity of c-Src in vivo.
To test for a direct interaction of hnRNP K and different forms of Src in vivo, His-hnRNP K was precipitated with a His antibody. The immunoprecipitates were subsequently analyzed by Western blotting (Fig. 1E to G). As is evident from Fig. 1F and G, both the tyrosine-phosphorylated and the nonphosphorylated forms of hnRNP K interact with c-Src and the Src mutants (lanes 4 to 7). Thus, following cotransfection of hnRNP K and the wild type c-Src kinase into HeLa cells, the two proteins interact with each other, c-Src kinase is activated, and hnRNP K and other proteins become tyrosine phosphorylated. As is evident from the inactive Src(Y416F) mutant (Fig. 1F and G, lane 7), Src activity and hnRNP K phosphorylation are not required for the interaction.
We next assessed whether the ability to interact with and activate c-Src was shared by hnRNP E1, another KH domain RNA binding protein which together with hnRNP K binds to the DICE and regulates LOX expression (16). hnRNP E1 can bind only minor quantities of c-Src (Fig. 1N), is very weakly phosphorylated (Fig. 1M), and completely fails to activate c-Src in vivo (Fig. 1J and K, compare lanes 1 and 2 with lane 4). We conclude that the ability to interact with and activate c-Src is a specific feature of hnRNP K. We also found that the tyrosine kinase c-Abl, the cellular homologue of the transforming gene of Abelson murine leukemia virus, fails to associate with and be specifically activated by hnRNP K (data not shown).
hnRNP K is an RNA-binding protein without any domains that resemble protein kinases or protein phosphatases that could help explain the Src activation. To distinguish whether the activation of c-Src by hnRNP K is direct or indirect, we next tested whether recombinant hnRNP K could activate purified c-Src in vitro. Wild-type c-Src and the constitutively active Y527F mutant as a positive control were immunopurified from transfected human HEK-293 cells and incubated with the Src substrate enolase in the presence of [γ-32P]ATP. The basal phosphorylation of enolase (Fig. 2, lanes 2) is stimulated approximately fivefold by replacement of wild-type c-Src with the active Y527F mutant (lanes 1). Addition of recombinant hnRNP K to wild-type c-Src stimulates enolase phosphorylation in a dose-dependent manner up to sixfold (Fig. 2, lanes 3 and 4). The activation of c-Src by hnRNP K is inhibited by the selective inhibitor of Src kinase PP2 (Fig. 2, lanes 5) but not by PP3 (lanes 6), which was included as a negative control. By contrast to hnRNP K and in keeping with the in vivo data, recombinant hnRNP E1 (Fig. 2, lanes 7 and 8) fails to activate c-Src. We conclude that hnRNP K specifically activates c-Src in vivo and in vitro and that this activation is likely to be direct. c-Src, activated by hnRNP K, can phosphorylate both hnRNP K and an external substrate, enolase.
FIG. 2.
hnRNP K but not hnRNP E1 activates Src catalytic activity in vitro. Wild-type Src (wt) and the constitutively active Src(Y527F) mutant as a positive control were immunopurified from transfected human 293 HEK cells, and equal amounts were assayed with an excess of the substrate enolase in the presence of [γ-32P]ATP, incubated with increasing amounts (125 [+] and 375 [++] ng) of purified hnRNP K (lanes 3 and 4) or hnRNP E1 (lanes 7 and 8). The Src inhibitor PP2 (lanes 5 and 9) and the control PP3 (lanes 6 and 10) were added at a concentration of 1 μM. Phosphorylation of enolase and hnRNP K was analyzed by autoradiography (an example is shown in panel B). (A) The bars show the sum of the hnRNP K and enolase phosphorylation signals with the standard deviation observed in three repeat experiments.
The SH3 domain of c-Src is important for interaction with hnRNP K.
A peptide corresponding to the SH3 domain of c-Src has been shown to interact with the proline-rich domain of hnRNP K in vitro (23, 25, 27). To evaluate the importance of this region for the binding of c-Src to hnRNP K in HeLa cells, we compared His-hnRNP K binding to c-Src with Src mutants lacking either its SH3 or its SH2 domain, Src(−SH3) and Src(−SH2), respectively, by coimmunoprecipitation with antihistidine (Fig. 3A and B) or anti-Src (Fig. 3C and D) antibodies. In contrast to c-Src and Src(−SH2), the Src(−SH3) mutant (Fig. 3, lanes 2) displays a much reduced interaction with hnRNP K (Fig. 3D) and yields barely detectable levels of tyrosine phosphorylation of hnRNP K (Fig. 3B). Hence, the SH3 domain of Src is important for both the binding and the phosphorylation of hnRNP K, but a weak interaction between the two proteins can also be established in the absence of the SH3 domain.
Multiple tyrosine residues in hnRNP K are targets of Src.
Earlier analyses of hnRNP K predicted four tyrosine residues as targets of Src (Tyr 230, 234, 236, and 380) (22), and Tyr 230, 234, and 236 have been shown to contribute to hnRNP K phosphorylation by Src in vitro (19). In initial experiments in HeLa cells, we found that the hnRNP K mutant in which the tyrosine residues 230, 234, and 236 were changed to phenylalanine (called Y3F) was still phosphorylated by c-Src (data not shown). A Y4F hnRNP K mutant (with the sites 230, 234, 236, and 380 changed) can bind both wild-type c-Src and the inactive Src mutant Src(Y416F) (Fig. 4C, lanes 1 to 6). Y4F is also still phosphorylated by c-Src (Fig. 4B, lane 5), although at a lower level (81%) than wild-type hnRNP K (compare lane 2). In order to identify the tyrosine phosphorylation sites of hnRNP K targeted by c-Src in vivo, phosphopeptide mapping by mass spectrometry was carried out. By using parent ion scans, which provide a selective detection method for phosphopeptides (14), 6 phosphorylated tyrosines out of the 17 tyrosine residues present in hnRNP K (Fig. 4, top) were determined. We confirmed that Tyr 230, 234, 236, and 380 are phosphorylated and identified two additional targets of c-Src, Tyr 72 and Tyr 225 (data not shown).
Based on this result, the appropriate Y6F hnRNP K mutant was generated and tested. Tyrosine phosphorylation of hnRNP K (Y6F) was further reduced compared to that of both hnRNP K wild-type (32%) and hnRNP K (Y4F) (41%) in HeLa cells cotransfected with c-Src (Fig. 4B, lanes 2, 5, and 8). No hnRNP K phosphorylation was observed following cotransfection with the negative control Src(Y416F) (lanes 3, 6, and 9). Nonetheless, even the Y6F mutant still interacted with and was phosphorylated by c-Src (Fig. 4B and C, lanes 8). These findings indicate that Y 72, 225, 230, 234, 236, and 380 contribute to phosphorylation by c-Src but that additional tyrosine residues of hnRNP K are targeted by the kinase.
Attempts to map additional tyrosine residues in the hnRNP K Y6F mutant phosphorylated by c-Src by mass spectrometry were challenged by insufficiently low signal-to-noise ratios.
Tyrosine phosphorylation of hnRNP K by c-Src reversibly inhibits DICE binding.
To directly examine the effect of Src-mediated phosphorylation on the binding of hnRNP K to the DICE, His-tagged recombinant human hnRNP K was incubated with immunopurified c-Src, the activated mutant Src (KP), the inactive mutant Src(Y416F), or phosphorylation buffer without the kinase (Fig. 5). Subsequently, successful and specific tyrosine phosphorylation of hnRNP K was confirmed with an antiphosphotyrosine antibody (Fig. 5C). The phosphorylation of hnRNP K by c-Src in vitro causes a small reduction of its electrophoretic mobility compared to the nonphosphorylated protein (Fig. 5B, compare lanes 2 and 4 with lanes 1 and 3). Importantly, a Northwestern blot with a radioactively labeled DICE shows that the binding of hnRNP K to the DICE is drastically reduced after the phosphorylation by c-Src kinase (Fig. 5D). Subsequent treatment with λ-phosphatase reverses both the tyrosine phosphorylation and the inability of hnRNP K to bind to the DICE probe (Fig. 5C and D, lanes 3 and 5). As predicted by the data in Fig. 1 and 2, Src does not affect the ability of hnRNP E1 to bind to the DICE (Fig. 1E to H). These results show that the tyrosine phosphorylation of hnRNP K by c-Src permits the specific and reversible regulation of the binding of hnRNP K to its physiological binding site, the DICE.
c-Src rescues translation inhibited by hnRNP K in vivo.
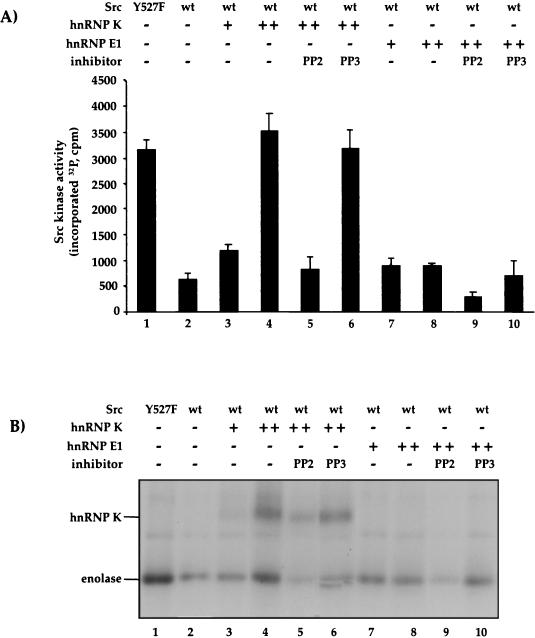
Cotransfection of hnRNP K with a LUC reporter mRNA bearing a DICE in its 3′ UTR (LUC-DICE) recapitulates LOX silencing and causes a specific, DICE-dependent reduction of LUC activity in transfected HeLa cells. The stability of the LUC reporter mRNA is not affected by coexpressed hnRNP K (16) (data not shown). For use as a negative control mRNA that is not silenced by hnRNP K, the DICE was replaced in LUC-NR by NR sequences of the LOX mRNA 3′ UTR (16). To investigate whether hnRNP K phosphorylation by c-Src controls its activity as a repressor of LUC-DICE mRNA translation in vivo, HeLa cells were transfected with LUC-DICE or LUC-NR reporter constructs (Fig. 6). These LUC indicator constructs were cotransfected with an hnRNP K expression vector (or an expression vector for the RNA binding protein U1A as a specificity control) and different forms of c-Src (Fig. 6A). As expected, LUC activity from LUC-DICE, but not LUC-NR, is inhibited by hnRNP K. This inhibition is relieved by coexpression of wild type c-Src or of the constitutively active Src-KP (K249E, P250E) mutant. By contrast, cotransfection of the inactive mutant Src-Y416F fails to stimulate LUC activity. This shows that a Src mutant that binds hnRNP K (Fig. 1) but fails to phosphorylate it is insufficient to activate LUC-DICE translation. LUC-NR expression is not affected by hnRNP K alone or in combination with any of the Src mutants, demonstrating the specificity of the observed effects for the hnRNP K/DICE interaction.
To further evaluate the role of c-Src in the activation of LUC-DICE expression, we used the potent and selective inhibitor of Src kinase PP2, or PP3 as a negative control (11). Treatment of transfected HeLa cells with PP2, but not PP3, abolishes the stimulatory effect of c-Src or Src-KP on LUC-DICE expression (Fig. 6B). This pharmacological effect of PP2 is associated with a lack of hnRNP K phosphorylation but not with a failure to express the hnRNP K protein (Fig. 6B). These data demonstrate that tyrosine phosphorylation of hnRNP K by c-Src can regulate the function of the hnRNP K/DICE mRNA silencing system in vivo.
DISCUSSION
In this report, we describe a novel biochemical and functional interplay between the c-Src tyrosine kinase and hnRNP K. Our findings primarily shed light on two aspects of this interplay: first, the unexpected activation of c-Src by its substrate hnRNP K, and second, a signal-dependent mechanism for the translational activation of a silenced mRNA.
c-Src-hnRNP K interaction.
Earlier work described an interaction of hnRNP K with the SH3 domain of Src (23, 25, 27). It was shown that hnRNP K can be phosphorylated by c-Src and Lck in vitro and that the binding of hnRNP K to polyribo(C) was reduced as a consequence of phosphorylation by Lck (19). For a different member of the KH domain protein family, Sam68, a reduction of its poly(U) binding activity following tyrosine phosphorylation by the Src kinase family member Fyn has been reported (26).
We have found that hnRNP K and c-Src specifically interact with each other in vivo (Fig. 1). To our surprise, we observed that cells which were cotransfected with hnRNP K and the wild-type form of c-Src display increased Src kinase activity (Fig. 1D). As a consequence, hnRNP K is phosphorylated on tyrosine residues by wild-type c-Src (Fig. 1C and F). hnRNP E1 shares none of these features of hnRNP K (Fig. 1J, K, and M), demonstrating the specificity of the functional interaction between hnRNP K and c-Src. Moreover, hnRNP K but not hnRNP E1 activates c-Src in vitro (Fig. 2A and B). These findings show that hnRNP K can directly activate c-Src (Fig. 1A to D). Thus, not only is hnRNP K a substrate of the Src kinase, but it can also function as its activator.
Binding of hnRNP K to and phosphorylation by Src kinase involves the SH3 domain of Src in vivo (Fig. 3). hnRNP K contains three proline-rich sequence motifs that can bind to the isolated SH3 domain of Src in vitro, with the third domain (residues 303 to 318) showing the highest binding activity (25). Like hnRNP K, the proteins Sin (1) and Cas (4) can bind c-Src and activate it. The activation of c-Src by hnRNP K is strong. Studying the principal mechanism and structure-function relationships of the activation of c-Src by hnRNP K should therefore be of particular interest. Conceivably, hnRNP K could alter the conformation of c-Src to trigger autophosphorylation and activation.
Using mutagenesis and mass spectrometry, we found that an hnRNP K mutant (Y4F) in which four previously implicated tyrosine residues had been changed is still tyrosine phosphorylated by c-Src, indicating that further tyrosine residues might be targets of c-Src in vivo (Fig. 4). Mass spectrometry analysis of hnRNP K phosphorylated in vivo by c-Src revealed two additional phosphorylated residues (Tyr 72 and 225) (data not shown). Even the corresponding six-tyrosine mutant (hnRNP K Y6F) is still tyrosine phosphorylated by c-Src (Fig. 4). These findings show that c-Src can phosphorylate multiple tyrosine residues in hnRNP K and that further sites can become phosphorylated when c-Src and hnRNP K interact and the primary target residues are mutated.
Translational activation by c-Src.
Phosphorylation of hnRNP K by c-Src inhibits the binding of hnRNP K to the DICE in vitro (Fig. 5) and activates the translation of DICE/hnRNP K-silenced mRNAs in vivo (Fig. 6). When hnRNP K lacks a stretch of 97 amino acids (amino acids 240 to 337) bearing the proline-rich regions between the second and third KH domains, LUC-DICE translation is fully silenced, but the translational activation by Src is abolished (data not shown). This indicates that translational repression and translational activation by c-Src are separable functions of hnRNP K. Our findings directly implicate c-Src in translational regulation via hnRNP K. They also raise the intriguing possibility of c-Src-mediated activation of LOX translation in erythroid differentiation. One can also envisage a phosphorylation-dependent activation of the HPV-16 L2 mRNA translation in the course of terminal differentiation of squamous epithelial cells. In addition to hnRNP K, hnRNP E1 contributes to LOX mRNA silencing (15, 16), and its role in erythroid differentiation also needs to be considered. Since hnRNP E1 is not a direct target of Src, it will be interesting to determine whether the dimerization between hnRNPs K and E1 (17) may indirectly mediate Src effects on hnRNP E1.
In summary, we have shown that tyrosine phosphorylation of hnRNP K by c-Src inactivates cytoplasmic hnRNP K as a translational repressor and thus mediates the specific translational activation of a DICE-silenced mRNA. In conjunction with the recent finding that serine phosphorylation of hnRNP K by ERK drives the cytoplasmic accumulation of hnRNP K and promotes the subsequent inhibition of translation (10), these findings reveal that hnRNP K is a specific posttranscriptional regulatory factor which is subject to both positive and negative regulation by different phosphorylation pathways.
Acknowledgments
We thank S. Gonfloni and J. Kretzschmar for providing plasmids. We thank J. Cooper for discussions and experimental suggestions.
This work was supported by a fellowship of the Deutsche Akademie der Naturforscher Leopoldina to A.O.-L. C.C. is a FEBS fellowship recipient. K.B. holds grants from the NIH (GM45134 and DK45978). Funds from the Gottfried Wilhelm Leibniz Prize were awarded by the Deutsche Forschungsgemeinschaft to M.W.H.
REFERENCES
- 1.Alexandropoulos, K., and D. Baltimore. 1996. Coordinate activation of c-Src by SH3- and SH2-binding sites on a novel, p130 Cas-related protein, Sin. Genes Dev. 10:1341-1355. [DOI] [PubMed] [Google Scholar]
- 2.Brasier, A. R., J. E. Tate, and J. F. Habener. 1989. Optimized use of the firefly luciferase assay as a reporter gene in mammalian cell lines. BioTechniques 7:1116-1122. [PubMed] [Google Scholar]
- 3.Brown, M. T., and J. A. Cooper. 1996. Regulation, substrates and functions of src. Biochim. Biophys. Acta 1287:121-149. [DOI] [PubMed] [Google Scholar]
- 4.Burnham, M. R., P. J. Bruce-Staskal, M. T. Harte, C. L. Weidow, A. Ma, S. A. Weed, and A. H. Bouton. 2000. Regulation of c-Src activity and function by the adapter protein Cas. Mol. Cell. Biol. 20:5865-5878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collier, B., L. Goobar-Larsson, M. Sokolowski, and S. Schwartz. 1998. Translational inhibition in vitro of human papillomavirus type 16 L2 mRNA mediated through interaction with heterogenous ribonucleoprotein K and poly(rC)-binding protein 1 and 2. J. Biol. Chem. 273:22648-22656. [DOI] [PubMed] [Google Scholar]
- 6.Gonfloni, S., F. Frischknecht, M. Way, and G. Superti-Furga. 1999. Leucine 255 of Src couples intramolecular interactions to inhibition of catalysis. Nat. Struct. Biol. 6:760-764. [DOI] [PubMed] [Google Scholar]
- 7.Gonfloni, S., A. Weijland, J. Kretzschmar, and G. Superti-Furga. 2000. Crosstalk between the catalytic and regulatory domains allows bidirectional regulation of Src. Nat. Struct. Biol. 7:281-286. [DOI] [PubMed] [Google Scholar]
- 8.Gonfloni, S., J. C. Williams, K. Hattula, A. Weijland, R. K. Wierenga, and G. Superti-Furga. 1997. The role of the linker between the SH2 domain and catalytic domain in the regulation and function of Src. EMBO J. 16:7261-7271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Graham, F. L., and A. J. Van der Eb. 1973. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology 52:456-464. [DOI] [PubMed] [Google Scholar]
- 10.Habelhah, H., K. Shah, L. Huang, A. Ostareck-Lederer, A. L. Burlingame, K. M. Shokat, M. W. Hentze, and Z. Ronai. 2001. ERK phosphorylation drives cytoplasmic accumulation of hnRNP K and subsequent inhibition of mRNA translation. Nat. Cell Biol. 3:325-330. [DOI] [PubMed] [Google Scholar]
- 11.Hanke, J. H., J. P. Gardner, R. L. Dow, P. S. Changelian, W. H. Brissette, E. J. Weringer, B. A. Pollok, and P. A. Connelly. 1996. Discovery of a novel, potent, and Src family-selective tyrosine kinase inhibitor. Study of Lck- and FynT-dependent T cell activation. J. Biol. Chem. 271:695-701. [DOI] [PubMed] [Google Scholar]
- 12.Höhne, M., B. J. Thiele, S. Prehn, E. Giessmann, B. Nack, and S. M. Rapoport. 1988. Activation of translationally inactive lipoxygenase mRNP particles from rabbit reticulocytes. Biomed. Biochim. Acta 47:75-78. [PubMed] [Google Scholar]
- 13.Howley, P. M. 1996. Papillomavirinae: the viruses and their replication, p. 2045-2076. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed., vol. 2. Lippincott-Raven, Philadelphia, Pa. [Google Scholar]
- 14.Neubauer, G., and M. Mann. 1999. Mapping of phosphorylation sites of gel isolated proteins by nanoelectrospray mass spectrometry: potentials and limitations. Anal. Chem. 71:235-242. [DOI] [PubMed] [Google Scholar]
- 15.Ostareck, D. H., A. Ostareck-Lederer, I. N. Shatsky, and M. W. Hentze. 2001. Lipoxygenase mRNA silencing in erythroid differentiation: the 3′UTR regulatory complex controls 60S ribosomal subunit joining. Cell 104:281-290. [DOI] [PubMed] [Google Scholar]
- 16.Ostareck, D. H., A. Ostareck-Lederer, M. Wilm, B. J. Thiele, M. Mann, and M. W. Hentze. 1997. mRNA silencing in erythroid differentiation: hnRNP K and hnRNP E1 regulate 15-lipoxygenase translation from the 3′ end. Cell 89:597-606. [DOI] [PubMed] [Google Scholar]
- 17.Ostareck-Lederer, A., D. H. Ostareck, and M. W. Hentze. 1998. Cytoplasmic regulatory functions of the KH-domain proteins hnRNPs K and E1/E2. Trends Biochem. Sci. 23:409-411. [DOI] [PubMed] [Google Scholar]
- 18.Ostareck-Lederer, A., D. H. Ostareck, N. Standart, and B. J. Thiele. 1994. Translation of 15-lipoxygenase mRNA is inhibited by a protein that binds to a repeated sequence in the 3′ untranslated region. EMBO J. 13:1476-1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ostrowski, J., D. S. Schullery, O. N. Denisenko, Y. Higaki, J. Watts, R. Aebersold, L. Stempka, M. Gschwendt, and K. Bomsztyk. 2000. Role of tyrosine phosphorylation in the regulation of the interaction of heterogenous nuclear ribonucleoprotein K protein with its protein and RNA partners. J. Biol. Chem. 275:3619-3628. [DOI] [PubMed] [Google Scholar]
- 20.Rapoport, S. M., and T. Schewe. 1986. The maturational breakdown of mitochondria in reticulocytes. Biochim. Biophys. Acta 864:471-495. [DOI] [PubMed] [Google Scholar]
- 21.Shevchenko, A., M. Wilm, O. Vorm, and M. Mann. 1996. Mass spectrometric sequencing of proteins from silver stained polyacrylamide gels. Anal. Chem. 68:850-858. [DOI] [PubMed] [Google Scholar]
- 22.Songyang, Z., S. E. Shoelson, M. Chaudhuri, G. Gish, T. Pawson, W. G. Haser, F. King, T. Roberts, S. Ratnofsky, R. J. Lechleider, B. G. Neel, R. B. Birge, J. E. Fajardo, M. M. Chou, H. Hanafusa, B. Schaffhausen, and L. C. Cantley. 1993. SH2 domains recognize specific phosphopeptide sequences. Cell 72:767-778. [DOI] [PubMed] [Google Scholar]
- 23.Taylor, S. J., and D. Shalloway. 1994. An RNA-binding protein associated with Src through its SH2 and SH3 domains in mitosis. Nature 368:867-871. [DOI] [PubMed] [Google Scholar]
- 24.van Leyen, K., R. M. Duvoisin, H. Engelhardt, and M. Wiedmann. 1998. A function for lipoxygenase in programmed organelle degradation. Nature 395:392-395. [DOI] [PubMed] [Google Scholar]
- 25.Van Seuningen, I., J. Ostrowski, X. R. Bustelo, P. R. Sleath, and K. Bomsztyk. 1995. The K protein domain that recruits the interleukin 1-responsive K protein kinase lies adjacent to a cluster of c-Src and Vav SH3-binding sites. Implications that K protein acts as a docking platform. J. Biol. Chem. 270:26976-26985. [DOI] [PubMed] [Google Scholar]
- 26.Wang, L. L., S. Richard, and A. S. Shaw. 1995. P62 association with RNA is regulated by tyrosine phosphorylation. J. Biol. Chem. 270:2010-2013. [DOI] [PubMed] [Google Scholar]
- 27.Weng, Z., S. M. Thomas, R. J. Rickles, J. A. Taylor, A. W. Brauer, C. Seidel-Dugan, W. M. Michael, G. Dreyfuss, and J. S. Brugge. 1994. Identification of Src, Fyn, and Lyn SH3-binding proteins: implications for a function of SH3 domains. Mol. Cell. Biol. 14:4509-4521. [DOI] [PMC free article] [PubMed] [Google Scholar]