Abstract
Telomeres are essential for protecting the ends of chromosomes and preventing chromosome fusion. Telomere loss has been proposed to play an important role in the chromosomal rearrangements associated with tumorigenesis. To determine the relationship between telomere loss and chromosome instability in mammalian cells, we investigated the events resulting from the introduction of a double-strand break near a telomere with I-SceI endonuclease in mouse embryonic stem cells. The inactivation of a selectable marker gene adjacent to a telomere as a result of the I-SceI-induced double-strand break involved either the addition of a telomere at the site of the break or the formation of inverted repeats and large tandem duplications on the end of the chromosome. Nucleotide sequence analysis demonstrated large deletions and little or no complementarity at the recombination sites involved in the formation of the inverted repeats. The formation of inverted repeats was followed by a period of chromosome instability, characterized by amplification of the subtelomeric region, translocation of chromosomal fragments onto the end of the chromosome, and the formation of dicentric chromosomes. Despite this heterogeneity, the rearranged chromosomes eventually acquired telomeres and were stable in most of the cells in the population at the time of analysis. Our observations are consistent with a model in which broken chromosomes that do not regain a telomere undergo sister chromatid fusion involving nonhomologous end joining. Sister chromatid fusion is followed by chromosome instability resulting from breakage-fusion-bridge cycles involving the sister chromatids and rearrangements with other chromosomes. This process results in highly rearranged chromosomes that eventually become stable through the addition of a telomere onto the broken end. We have observed similar events after spontaneous telomere loss in a human tumor cell line, suggesting that chromosome instability resulting from telomere loss plays a role in chromosomal rearrangements associated with tumor cell progression.
Telomeres are nucleoprotein complexes at the ends of chromosomes that contain short DNA repeat sequences added on by telomerase (7, 45). Telomeres serve multiple functions, including protecting chromosome ends and preventing chromosome fusion (7, 13, 45). The loss of a telomere, either through improper maintenance or due to a double-strand break (DSB) occurring near the end of a chromosome, can have a variety of consequences. Chromosomes without telomeres can be “healed” by the addition of a new telomere, resulting in a terminal deletion. This chromosome healing can occur by a variety of mechanisms, including de novo addition by telomerase, as has been demonstrated in yeast (14) and tetrahymena (83). The addition of telomeres directly onto the end of a broken chromosome has been observed in human tumor cells (19) and mouse embryonic stem (ES) cells (74) and has been associated with human genetic diseases (17, 80, 82). However, the mechanism of direct telomere addition in mammalian cells is not known. Telomeres can also be obtained by the capture of preexisting telomeres, which in human tumor cells can result from cryptic translocation of terminal fragments from other chromosomes (47). Finally, telomeres can be obtained by replication of the ends of other chromosomes through break-induced replication (8), which has been demonstrated to occur in yeast by RAD51-independent homologous recombination (8, 25).
As first described by McClintock in maize (44), chromosomes that have lost their telomeres can fuse at their ends and become unstable as a result of repeated breakage and fusion during subsequent cell divisions, termed the breakage-fusion-bridge (B/F/B) cycle. Increased chromosome fusion and chromosome instability has been observed in cells from telomerase-deficient mice (27) and in mammalian cells with a variety of mutations that affect telomere function, including DNA-PKcs (4, 24), Ku (4, 31), and TRF2 (79). Chromosome fusion resulting from telomere loss appears to play an important role in chromosomal rearrangements associated with cancer. Telomere shortening in aging somatic cells has been suggested to lead to chromosomal fusions involved in the initiation of tumorigenesis (3). In addition, the high rate of telomere associations in many cancer cells (13) suggests that they commonly have a high spontaneous rate of telomere loss. Thus, cancer cells may often have fundamental defects in their ability to properly maintain telomeres. Consistent with this possibility, cancer cells commonly show a high rate of B/F/B events (22, 66), which have been associated with telomere dysfunction and chromosome instability (21).
The loss of a selectable marker gene located near the end of a chromosome has proven invaluable in the study of the types of events associated with telomere loss. In yeast that are deficient in DSB repair, the loss of telomeric marker genes after the introduction of a DSB with the HO endonuclease often involves chromosome healing (14, 33, 68). In contrast, telomerase-deficient yeast cells were found to have a high rate of gross chromosome rearrangements, resulting from chromosome fusions and nonreciprocal translocations that appeared to result from break-induced replication (25). Chromosome healing was not observed in this later study, as might be expected in telomerase-deficient cells. The spontaneous loss of a selectable marker gene located near the end of a chromosome was also found to be associated with gross chromosome rearrangements in yeast with mutations in genes involved in cellular response to DSBs (52, 53). These gross chromosome rearrangements resulted from both chromosome healing and nonreciprocal translocations with other chromosomes.
We have used selectable marker genes located adjacent to a telomere to investigate the consequences of telomere loss in mammalian cells. Studies in the human tumor cell line EJ-30 demonstrated a high spontaneous rate of rearrangements (10−4 event/cell/generation) involving a marker chromosome containing a herpes simplex virus thymidine kinase (HSV-tk) gene adjacent to a telomere. Most HSV-tk-deficient (HSV-tk−) subclones had nontelomeric DNA joined onto the end of the marker chromosome (19). In contrast, the spontaneous rate of rearrangement of the telomeric HSV-tk gene in a mouse ES cell clone was below the level of detection (<10−6 event/cell/generation) (74). However, telomere loss could be induced in mouse ES cell clones after the introduction of a DSB at a specific location near the HSV-tk gene with the I-SceI endonuclease, which has been widely used to study the influence of DSBs on both homologous and nonhomologous recombination in mammalian cells (32, 40, 60, 61, 65, 67). Two types of rearrangements were observed. Unlike previous studies with I-SceI-induced DSBs at interstitial sites, ∼90% of the mouse ES cell HSV-tk− subclones had telomeres added directly onto the site of the break (74). The remainder of the HSV-tk− subclones showed nontelomeric DNA joined onto the end of the chromosome, similar to the most common type of event observed in HSV-tk− subclones from the human EJ-30 tumor cell line. In the present study, we characterized the rearrangements in the mouse ES cell HSV-tk− subclones at both the DNA and chromosome levels. The generation of DSBs with the I-SceI endonuclease in these cells provides the opportunity to analyze the types of events resulting from telomere loss due to a DSB at a known location on individual chromosomes. The results demonstrate that DSBs occurring near telomeres can result in complex chromosome rearrangements and chromosome instability, which can be prevented or terminated by the addition of telomeric repeat sequences to the end of the broken chromosome.
MATERIALS AND METHODS
Cell lines and culture conditions.
The mouse ES cell line JM-1 was obtained from Roger Pedersen (University of California, San Francisco [UCSF]). JM-1 cells were grown on feeder layers consisting of STO cells that had been treated with 50 Gy of ionizing radiation as previously described (62). Leukemia inhibition factor (Gibco) was added to the medium at 1,000 U/ml. The pNPT-tel and pNPT2-tel plasmids were introduced into the ES cells by electroporation as previously described (74), and clones containing the integrated plasmid were selected in medium containing 300 μg of G418/ml. The EJ-30 cell line (obtained from William Dewey, UCSF) was subcloned from the EJ bladder cell carcinoma cell line, which is also named MGH-U1 (54). EJ-30 is deficient in p53 (11, 46) and expresses telomerase activity (unpublished observation). EJ-30 was grown in alpha minimal essential medium (UCSF Cell Culture Facility) supplemented with 5% fetal calf serum (Gibco), 5% newborn calf serum with iron (Gibco), 1 mM l-glutamine (Gibco), and gentamicin.
Plasmids.
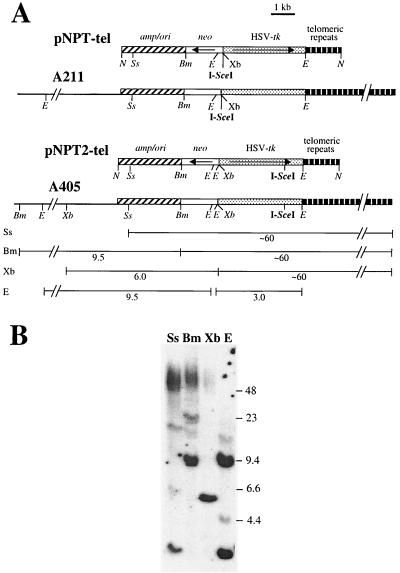
The plasmids used for analysis of terminal deletions, pNPT-tel and pNPT2-tel, were constructed from the pSXneo-1.6T2AG3 plasmid previously shown to seed new telomeres upon integration (28). The pSXneo-1.6T2AG3 plasmid contains an ampicillin resistance gene, a neomycin resistance (neo) gene, and 1.6 kb of telomeric repeat sequences. The neo gene has a promoter from the HSV-tk gene with a polyomavirus enhancer (76), while the HSV-tk gene has a phosphoglycerate kinase promoter for efficient expression in ES cells (72). Expression of the HSV-tk gene makes cells sensitive to ganciclovir, which has been used for analysis of mutations in the HSV-tk gene (9, 50). In pNPT-tel, an 18-bp I-SceI recognition site was introduced between the HSV-tk gene and the neo gene, while in pNPT2-tel, the I-SceI site was introduced in the BstEII site in the 3′-untranslated region of the HSV-tk gene near the telomeric repeat sequences (Fig. 1). Transfection was performed after linearization of the plasmids with the NotI restriction enzyme, which placed the telomeric repeat sequences in the proper orientation at one end.
FIG. 1.
Structure of the linearized pNPT-tel and pNPT2-tel plasmids before and after integration on the end of a chromosome in clones A211 and A405. (A) The pNPT-tel and pNPT2-tel plasmids were linearized with NotI prior to transfection so that the telomeric repeats would be in the proper orientation at one end to seed a new telomere. The location of the pSP73 vector (amp/ori), neo gene (neo), HSV-tk gene, and telomeric repeat sequences and the orientation of the neo and HSV-tk genes (arrows) are indicated. The structures of the integrated pNPT-tel plasmid in clone A211 and the pNPT2-tel plasmid in clone A405 are also shown. The location of the BamHI (Bm), EcoRI (E), NotI (N), SspI (Ss), and XbaI (Xb) restriction sites and the 18-bp recognition site for the I-SceI endonuclease (I-SceI) are shown. The sizes of fragments generated by the various restriction enzymes are indicated for clone A405. (B) Southern blot analysis of genomic DNA isolated from clone A405 digested with SspI (Ss), BamHI (Bm), XbaI (Xb), or EcoRI (E) and separated by pulsed-field gel electrophoresis prior to Southern blot anlaysis. Hybridization was performed with the pNTP-Δ plasmid (which is similar to pNPT-tel but lacks telomeric repeat sequences) as a probe. The locations of DNA size markers are shown. Terminal restriction fragments are long and heterogeneous in length due to variations in the lengths of the extensive telomeric repeat sequences found in different cells in the population.
Introduction of DSBs and isolation of HSV-tk− subclones.
The generation of HSV-tk− subclones from the mouse ES clones was accomplished by transient expression of I-SceI endonuclease to generate DSBs within the plasmid sequences. Transient expression was performed by electroporation of pCBASce, which contains the I-SceI gene with a chicken β-actin promoter and has been found to provide a high efficiency of cutting at I-SceI sites in mammalian cells (61). pCBASce was electroporated into the ES cells as previously described (74). After we waited 6 days for turnover of the existing HSV-tk, previously shown to be the required time for the generation of resistance to ganciclovir (9, 50), the cells were plated in medium containing 2 μM ganciclovir and 300 μg of G418/ml. Individual colonies were isolated by ring cloning after approximately 2 weeks. The maximum frequency of ganciclovir/G418-resistant colonies generated by I-SceI was ∼10−4. With clone A211, three of six experiments successfully generated cells that had I-SceI-induced DNA rearrangements, whereas in clone A405, one of six experiments was successful.
Cloning and analysis of DNA sequences.
Genomic DNA was purified as previously described (51) and digested with restriction enzymes according to the manufacturer's instructions. Genomic DNA was fractionated by agarose gel electrophoresis by either standard protocols or pulsed-field gel electrophoresis with 1% agarose gels in 0.5× TBE (0.045 M Tris-borate, 0.001 M EDTA), at 200 V and 10°C, and pulsed at 2.0- to 3.6-s intervals. For Southern blot analysis, DNA was depurinated by treatment with 0.25 M HCl for 30 min and transferred in 0.5 M NaOH onto a charged nylon Hybond-N+ membrane (Amersham) by using a vacuum transfer apparatus (Pharmacia). Prehybridization for 3 h and hybridization overnight were performed at 65°C in 5× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7]), 5× Denhardt's solution, 0.5% sodium dodecyl sulfate (SDS), and 0.25 mg of salmon sperm DNA/ml. Probes were labeled with [α-32P]dCTP (New England Nuclear) by using a High Prime Labeling Kit (Roche). Filters were washed three times in 2× SSPE with 0.1% SDS at room temperature, twice in 1× SSPE with 0.1% SDS at 65°C, and twice in 0.1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) with 0.1% SDS at 65°C.
The cellular DNA adjacent to the integration site in clone A211 was isolated by using long PCR of circularized genomic DNA and plasmid-specific primers in reverse orientations. Circularization of genomic DNA from clone A211 digested with BamHI was performed by diluting the DNA to 1 μg/ml in ligase buffer (20 mM Tris [pH 7.5], 10 mM MgCl2, 1 mM dithiothreitol, 1 mM ATP) containing 20 U of ligase (Gibco)/ml and incubation overnight at 14°C. DNA was then concentrated by using Ultrafree-MC 30,000 NMWL filtration units (Millipore). Long PCR was performed by using the Expand Template PCR Kit (Roche) according to the manufacturer's protocol. The primers were in reverse orientation within the origin of replication for the plasmid (5′-TATCCGGTAAGCGGCAGG-3′) and the ampicillin resistance gene (5′-ACCAATGCTTAATCAGTGAGGC-3′). PCR was performed with a hot start of 92°C for 2 min, followed by 10 cycles of 92°C for 10 s, 58°C for 30 s, and 68°C for 8 min and then 20 cycles of 92°C for 10 s, 58°C for 30 s, and 68°C for 8 min, with an extension of 20 s per cycle and an extension of 68°C for 10 min to complete the reaction. The PCR product was purified by using a QIAquick spin column (Qiagen), and the purified PCR product was cloned into pCR2.1 by using the TOPO-TA Cloning Kit (Invitrogen) for sequencing and further analysis.
In clone A211, analysis of the nucleotide sequence (GenBank accession no. AF442770) showed no homology to sequences in the mouse genome database. However, an informative polymorphic SINE/B2 repeat region was identified by PCR with the primers m13-pA2 (5′-GGGTCAGCTAGATGGCTCAG-3′) and m13-pB2 (5′-GTATGGTGTGCAGGTGATCG-3′) with an initial incubation at 94°C for 2 min; followed by 30 cycles of 94°C for 30 s, 58°C for 30 s, and 72°C for 45 s; and completed with an extension of 72°C for 10 min. The PCR produced 195- and 171-bp products from genomic DNA of mouse strains Mus musculus C57BL/6J and Mus spretus SPRET/Ei, respectively. This sequence-tagged site (STS), designated D18Rorl211, was used to map the genomic location of the integration site by using the Jackson Interspecific Backcross Panels (C57BL/6J × M. spretus)F1 × C57BL/6J (Jackson BSB panel) and the (C57BL/6JEi × SPRET/Ei)F1 × SPRET/Ei (Jackson BSS panel). D18Rorl211 was found to be closely linked to the loci D18Mit7 and D18Mit42, which are located at the end of chromosome 18 near the telomere (Mouse Genome Database accession no. J72310). A 140-kb BAC clone, 31E18, was identified by screening a mouse genomic DNA library (Mouse ES release I; Incyte Genomics) by using this D18Rorl211 STS.
The cloning of integrated plasmid sequences containing the recombination junctions from the HSV-tk− subclones of clone A211 was performed by PCR with one primer specific for the neo gene (5′-GAGCACAGCTGCGCAAGG-3′) and one primer specific for the ampicillin resistance gene (5′-GATGCTGAAGATCAGTTGG-3′). Both primers are in the same orientation (Fig. 1) and therefore will amplify inverted repeats. PCR was performed as previously described (59) by using an initial incubation for 2 min at 95°C; followed by 40 cycles of 95°C for 30 s, 60°C (for subclone FS-1) or 62°C (for subclone FS-2) for 30 s, and 72°C for 30 s; followed by one cycle of 95°C for 30 s, 60 oC (for subclone FS-1) or 62°C (for subclone FS-2) for 30 s, and 72°C for 2 min. The PCR products were cloned into the pCRII cloning vector (Invitrogen) by using the protocols provided by the manufacturer. Multiple clones of each of PCR product were analyzed to confirm that rearrangements had not occurred during cloning.
The cloning of the cellular DNA adjacent to the integration site in A405 and the recombination junctions in the HSV-tk− subclones of A405 was achieved by plasmid rescue. Plasmid rescue was performed by first digesting the DNA with BclI, followed by the circularization and concentration of the DNA as described above for clone A211. The DNA was then electroporated (2.5 kV, 200 Ω, 25 μF) by using 0.2-cm cuvettes (Bio-Rad) into electrocompetent DH12s bacteria (Gibco), and selection for the circularized plasmid and adjacent cellular DNA was performed with ampicillin. Sequence analysis of the genomic DNA adjacent to the integration site in clone A405 demonstrated an exact match to a BAC (RPCI23-169K7; GenBank accession no. AC021643) containing DNA from near the end of chromosome 15. The plasmid integrated into a region with a high degree of homology to the 5′ end of the promoter for the human heterogeneous ribonucleoprotein A1 gene (GenBank accession no. X73096) and adjacent to exon 1 of the heterochromatin protein 1α (GenBank accession no. AK008792)
Cytogenetic and FISH analysis.
Standard slides of 70% confluent mouse ES cell cultures were used to prepared metaphase spreads for cytogenetic and fluorescence in situ hybridization (FISH) analyses, as previously described (41). Briefly, aged slides were dehydrated in an ethanol series of 70, 90, and 100%, followed by denaturing with 70% formamide in 2× SSC. Analyses of subclones of A211 were performed with biotin-labeled chromosome 18-specific painting probe (Cambio), the digoxigenin-labeled subtelomeric BAC clone, 31E18, and fluorescein isothiocyanate (FITC)- or Cy3-labeled peptide nucleic acid (PNA) oligonucleotide of the telomeric repeat (5′-CCCTAACCCTAACCCTAA-3′). BAC DNA was prepared by using the MIDI plasmid preparation kit (Qiagen) according to the manufacturer's protocol for low-copy and large-insert plasmids and was labeled with digoxigenin by nick translation (Roche). Then, 200 to 500 ng of the BAC DNA was precipitated with 10-fold excess human Cot-I DNA (Roche) and 20-fold excess mouse Cot-I DNA (Gibco), and FISH was performed at high stringency in 50% formamide, 20% dextran sulfate, and 2× SSC at 37°C overnight after denaturation at 80°C for 5 min and preannealing at 37°C for 2 h to remove repeat sequences. Depending on the experiment, either the chromosome 18-specific painting or the PNA probe was mixed to the BAC probe before denaturation. Slides were then washed twice in 2× SSC at room temperature for 10 min before three washes at high stringency in 0.1× SSC at 61°C for 10 min. Detection of FISH signals was achieved by anti-digoxigenin-FITC (Roche) or avidin-rhodamine (Roche) according to the manufacturer's protocol. The slides were mounted on Vectasheild (Vector Laboratories) with 0.1 μg of DAPI (4′,6′-diamidino-2-phenylindole; Sigma)/ml as a counterstain.
For subclones of A405, FISH analysis was performed with the chromosome 15-specific painting probe (Cambio), the BAC RPCI23-169K7, the PAC RPCI21-561P12 (which has been mapped to chromosome 15 band E; Molecular Cytogenetic Resources [http://www.biologia.uniba.it/rmc]), and the PNA telomere probe under the conditions described above. FISH for analysis of the integrated plasmid sequences was performed as previously described (15, 36).
RESULTS
Establishment of clones with selectable marker genes integrated adjacent to a telomere.
Mouse ES cell clones containing selectable marker genes integrated immediately adjacent to a telomere were established by transfection with linearized plasmids containing telomeric repeat sequences on one end (Fig. 1A). The integration of the plasmid sequences on the ends of broken chromosomes results in new telomeres “seeded” from the telomeric repeat sequences within the plasmid (5, 16, 28). The seeded telomeres on these marker chromosomes are elongated in culture, and their length and dynamics become similar to the other telomeres in the cell (73, 75). The plasmids used in the present study contain a neo gene for positive selection with G418 and an HSV-tk gene for negative selection with ganciclovir. These plasmids also contain an I-SceI recognition site for the introduction of DSBs with the I-SceI endonuclease.
Two mouse ES cell clones, A211 and A405, which contain a plasmid integrated adjacent to a telomere have been isolated. Telomeric integration sites are identified by the heterogeneity in the length of the terminal restriction fragment (5, 19, 74), due to variation in the length of the telomere in different cells in the population. The terminal fragments average ∼60 kb in length, similar to other telomeres in mouse cells (84). Clone A211 contains a single telomeric copy of the pNPT-tel plasmid (74), which has an I-SceI recognition site located between the neo and HSV-tk genes (Fig. 1A). Clone A405 was obtained by transfection with the pNPT2-tel plasmid, which is similar to pNPT-tel except that the I-SceI recognition site is located at the end of the HSV-tk gene near the telomeric repeat sequences (Fig. 1A). Like clone A211, clone A405 also contains a single copy of the plasmid integrated adjacent to a telomere, as demonstrated by Southern blot analysis with a variety of restriction enzymes (Fig. 1B).
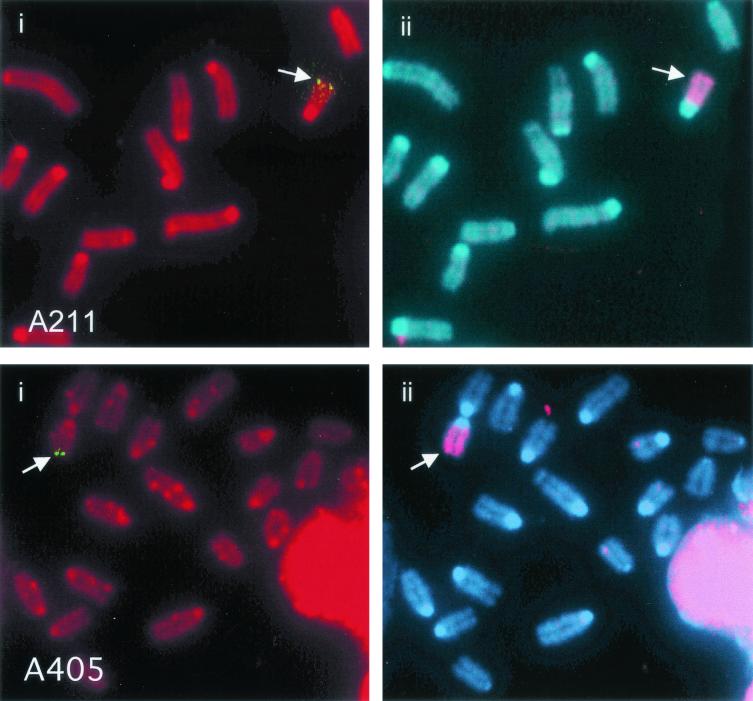
The pNPT-tel plasmid in clone A211 was integrated near the end of chromosome 18, as determined from the Jackson Laboratory Interspecific Backcross Mapping Panels by using an STS marker (D18Rorl211) identified within the cloned cellular DNA (see Materials and Methods). The integration site was confirmed by FISH analysis with the plasmid without telomeric repeat sequences as a probe (Fig. 2). Nucleotide sequence analysis of DNA cloned from the integration site in clone A405 demonstrated that the plasmid was located near the end of chromosome 15 (see Materials and Methods), which was also confirmed by FISH analysis (Fig. 2).
FIG. 2.
FISH analysis of telomeric plasmid integration sites in clones A211 and A405. (i) The location of the integration site (arrow) in A211 and A405 is demonstrated by hybridization with the pNTP-Δ plasmid (green). Chromosomes were counterstained with propidium iodide. (ii) The identity of the chromosome containing the integration sites was determined by hybridization with a chromosome 18-specific painting probe for clone A211 or a chromosome 15-specific painting probe for clone A405 (red). Chromosomes were counterstained with DAPI.
Inverted repeats involving the plasmid sequences in HSV-tk− subclones.
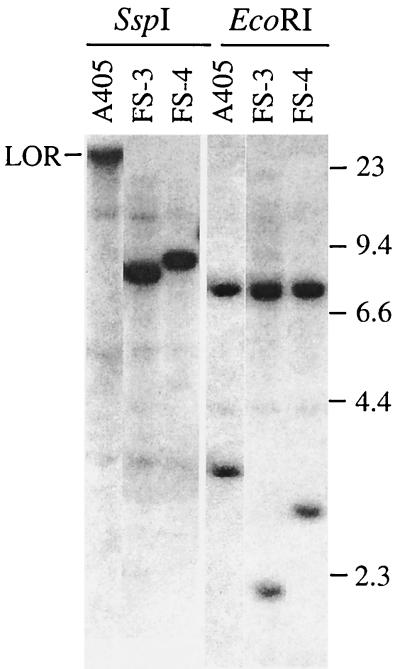
I-SceI endonuclease was used to introduce a DSB at a specific location in the plasmid sequences in clones A211 and A405. Transient expression of I-SceI was accomplished by electroporation of the pCBASce expression vector containing the I-SceI gene, which has previously been shown to efficiently generate DSBs in mouse ES cells (61). Subclones of A211 and A405 that have an inactivated HSV-tk gene were isolated by coselection in medium containing both G418 and ganciclovir to select for G418- and ganciclovir-resistant (G418r/Ganr) subclones. Coselection with G418 was necessary to eliminate HSV-tk− subclones that showed no change in the plasmid sequences, as a result of silencing or point mutations of the HSV-tk gene. The frequency of generating G418r/Ganr colonies by transient transfection with the I-SceI gene within A211 and A405 is significantly less (≤10−4) than the frequency of mutations generated at interstitial sites in other studies (32, 40, 60, 61, 65, 67). Whether this difference is due to the difficulty in generating DSBs near the telomere or due to the type of selection system used is not clear. As previously reported, the inactivation of the HSV-tk gene involves the addition of telomeric repeat sequences at the site of the break in more than 90% of the I-SceI-induced G418r/Ganr subclones (74). However, in some G418r/Ganr subclones, Southern blot analyses demonstrated the loss of the heterogeneous band containing the telomere and the appearance of new discrete bands, indicating the addition of nontelomeric DNA onto the end of the chromosome. We have now further analyzed two of the G418r/Ganr subclones of A211, FS-1, and FS-2 (74) and two additional G418r/Ganr subclones of A405, FS-3, and FS-4 (Fig. 3) that appeared to have nontelomeric DNA joined to the end of the chromosome.
FIG. 3.
Southern blot analysis of genomic DNA from clone A405 and its G418r/Ganr subclones FS-3 and FS-4. Genomic DNA was digested with either SspI or EcoRI and separated by standard agarose gel electrophoresis. Hybridization was performed with the pNTP-Δ plasmid as a probe. The lighter bands that are the same size in all three cell lines are due to cross-hybridization of the probe with the mouse endogenous pgk promoter. The location of DNA size markers and the limit of resolution (LOR) are shown.
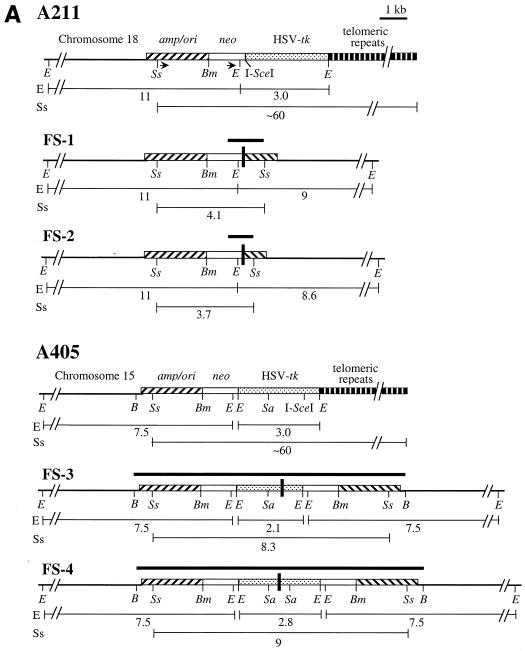
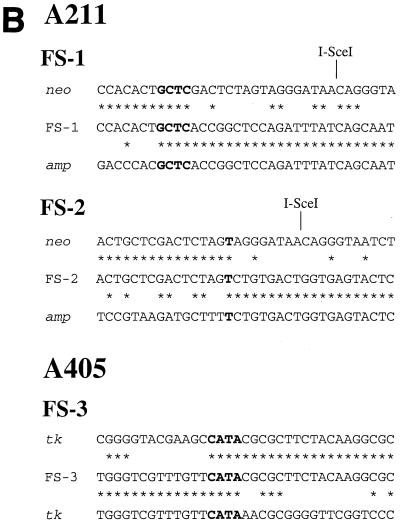
The cloning of the plasmid sequences containing the sites of recombination in the FS-1 and FS-2 subclones was accomplished by PCR, with oligonucleotide primers specific for the neo and ampicillin resistance genes. These primers are in the same orientation in the original pNPT-tel plasmid (Fig. 4A) and therefore do not produce a PCR product with genomic DNA from the parental clone A211. However, these primers did produce 1.2- and 0.8-kb PCR products with genomic DNA from the FS-1 and FS-2 subclones, respectively (data not shown). Nucleotide sequence analysis of multiple clones of these PCR products demonstrated the presence of inverted repeats in both FS-1 and FS-2 subclones (Fig. 4A). In FS-1, recombination occurred between plasmid sequences that were 17 bp away from the I-SceI site in one repeat and 2.6 kb away from the I-SceI site in the other repeat, with 4 bp of complementarity at the recombination junction (Fig. 4B). The sequence of the cloned PCR product is consistent with Southern blot analysis of genomic DNA from subclone FS-1, which demonstrated new 4.1-kb SspI and 9-kb EcoRI bands containing the novel junction fragments (see Fig. 3, lane 10, in reference 74). In subclone FS-2, recombination occurred between plasmid sequences that were 8 bp away from the I-SceI site in one repeat and 3.0 kb away from the I-SceI site in the other repeat (Fig. 4A), with 1 bp of complementarity at the junction (Fig. 4B). The structure of the cloned PCR fragment is consistent with Southern blot analysis of genomic DNA from subclones FS-2, which demonstrated new 3.7-kb SspI and 8.6-kb EcoRI bands containing the novel junction fragment (see Fig. 3, lane 15, in reference 74).
FIG. 4.
Cloning and analysis of inverted repeats in the G418r/Ganr subclones isolated from the mouse ES cell clones. (A) The structure of the integrated pNPT-tel plasmid in the parental A211 is compared with the inverted repeats in the FS-1 and FS-2 subclones. Similarly, the structure of the integrated pNPT2-tel plasmid in the parental A405 clone is compared with the inverted repeats in the FS-3 and FS-4 subclones. The fragments cloned by PCR for A211 or by plasmid rescue for A405 are indicated (heavy horizontal lines), as are the sizes of the restriction fragments consistent with Southern blot analysis of genomic DNA (light horizontal lines). The recognition site for I-SceI is indicated, as are some of the restriction enzymes used for mapping and Southern blot analysis, including BclI (B), BamHI (Bm), EcoRI (E), SacI (Sa), and SspI (Ss). The locations of the adjacent cellular DNA (chromosome 18 for A211 and chromosome 15 for A405), pSP73 vector sequences (amp/ori), neo gene (neo), HSV-tk gene, and telomeric repeat sequences are shown. The location of the oligonucleotide primers (arrows) used to amplify the fragments from subclones FS-1 and FS-2 and the locations of sites of recombination (heavy vertical bars) are shown. (B) Comparison of nucleotide sequences at the sites of recombination in the G418r/Ganr mouse ES cell subclones. The nucleotide sequences at the site of recombination in subclones FS-1 and FS-2 are compared with the sequence of the neo (neo) and ampicillin resistance (amp) genes. The nucleotide sequence at the site of recombination in subclone FS-3 is compared with sequences of the HSV-tk gene (tk) in opposite orientations. Nucleotides that are identical between sequences (asterisks) or show complementarity between the sequences at the site of recombination (boldface) are indicated.
The cloning of the sites of recombination in the FS-3 and FS-4 subclones of A405 was accomplished by rescue of the plasmid sequences after the digestion of genomic DNA with the BclI restriction enzyme. Mapping with restriction enzymes and nucleotide sequence analysis of multiple clones again demonstrated the presence of inverted repeats, except that the recombination junctions involved sequences within the HSV-tk gene. In subclone FS-3, recombination occurred at sites that were originally 1.3 kb apart in the HSV-tk gene, occurring 0.5 and 1.8 kb away from the I-SceI site (Fig. 4A), with 4 bp of complementarity at the recombination junction (Fig. 4B). The structure of the rescued fragment is consistent with Southern blot analysis of the genomic DNA from subclone FS-3, which demonstrated new 8.3-kb SspI and 2.1-kb EcoRI bands containing the novel junction fragments. In subclone FS-4, identical sequences were observed on both sides of the 600-bp SacI fragment, with a gap of ∼50 bp in the center that could not be sequenced. The inability to sequence this junction is apparently due to the presence of a nearly perfect inverted repeat, which can form hairpin structures that interfere with the sequencing reaction. Thus, the recombination site occurred at nearly identical locations in the two HSV-tk genes 1.0 kb away from the I-SceI site (Fig. 4A). The structure of the rescued fragment is consistent with Southern blot analysis of genomic DNA from subclone FS-4, which demonstrated new 9-kb SspI and 2.8-kb EcoRI bands containing the novel junction fragments. The presence of inverted repeats in FS-3 and FS-4 is also consistent with Southern blot analysis, which showed that the upper EcoRI band is increased in intensity relative to the lower band compared with the parental A405, indicating the duplication of the larger fragment containing the neo and ampicillin resistance genes (Fig. 4A).
Tandem duplications and chromosome instability following DSBs near a telomere.
Sister chromatid fusion is a known mechanism by which inverted repeats can be formed at the end of a chromosome after the loss of a telomere (44). Chromosomes that have undergone sister chromatid fusion are thought to demonstrate a variety of characteristic rearrangements, including large tandem duplications at the end of the chromosome and amplification of the subtelomeric regions adjacent to the break site (44). FISH and cytogenetic analyses were performed on three of the G418r/Ganr subclones to determine whether they contained these characteristic chromosome rearrangements.
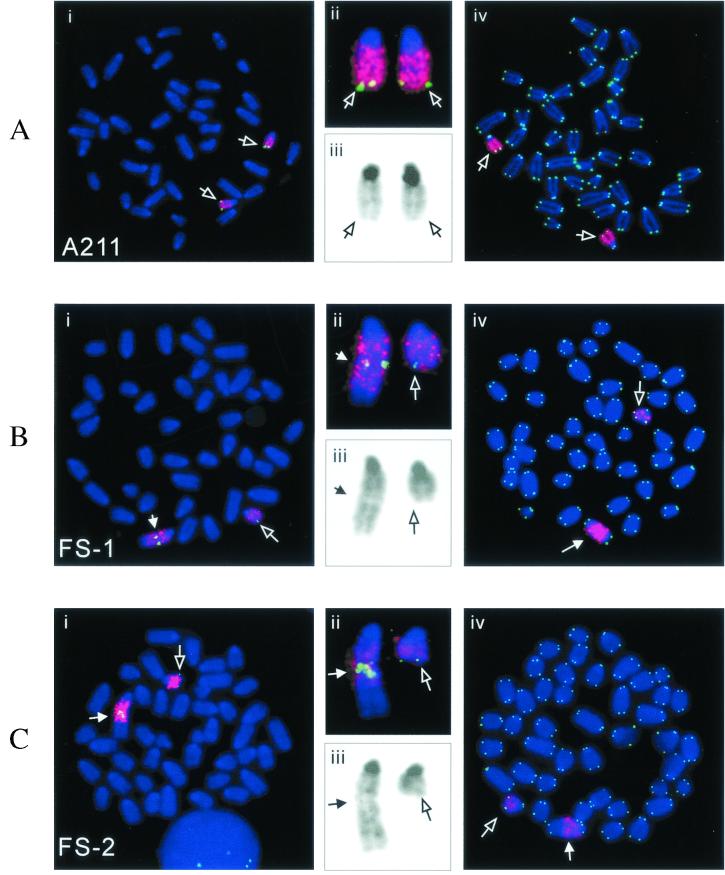
FISH analysis of metaphase chromosomes from clone A211 with both a chromosome 18-specific painting probe and the 31E18 BAC clone containing sequences subtelomeric to the integration site demonstrated two indistinguishable chromosome 18 homologues (Fig. 5A). In contrast to the parental A211 clone, both subclones FS-1 (Fig. 5B) and FS-2 (Fig. 5C) contain one homologue of chromosome 18 that is rearranged. In subclone FS-1, there is a large tandem duplication of chromosome 18, with hybridization of the BAC clone near the center of the chromosome. In addition, an unidentified chromosome fragment that does not appear to be pericentromeric heterochromatin has been joined on to the end of the tandem duplication. Despite being rearranged, the marker chromosome was relatively stable at the time of cloning, since it was the same in every metaphase spread examined (n = 50). FISH analysis with a telomere-specific probe demonstrated that the rearranged chromosome had telomeres on both ends in all of the cells examined (Fig. 5Biv). However, no telomeric repeat sequences were observed within the rearranged chromosome, indicating that telomere loss preceded chromosome fusion. Despite the stability of the rearranged chromosome in subclone FS-1, the hybridization signal for the 31E18 BAC clone is consistently enhanced in the rearranged chromosome (∼5-fold) relative to the normal homologue in the same metaphase spread (Fig. 5Bii), demonstrating amplification of the subtelomeric region. This amplification of the subtelomeric DNA suggests a transient period of instability in the rearranged chromosome during the 6-day period between treatment with I-SceI and selection with ganciclovir, which is required to allow for turnover of the existing HSV-tk.
FIG. 5.
Chromosome rearrangements in the FS-1 and FS-2 subclones isolated from clone A211. Metaphase spreads from clone A211 (A) and its subclones FS-1 (B) and FS-2 (C) were hybridized with both chromosome 18-specific painting (red) and subtelomeric BAC 31E18 (green) probes (i). An enlarged view of the chromosome 18 homologues (ii) and reverse video of the blue channel showing the DAPI GC banding (iii) are also shown. A different metaphase spread hybridized with both chromosome-18-specific (red) and telomere-specific (green) probes is shown for clone A211 and its subclones, FS-1 and FS-2 (iv). The open arrows point to the normal chromosome-18 homologues, while the closed arrow points to the rearranged chromosome 18.
FISH analysis on subclone FS-2 demonstrated that it also contains one normal-looking chromosome 18 and one chromosome 18 that contains a large tandem duplication, with hybridization of the BAC clone near the center (Fig. 5C). In addition, like subclone FS-1, subclone FS-2 consistently demonstrated increased hybridization of the BAC probe in the rearranged chromosome 18 compared to the normal homologue in the same metaphase spread (Fig. 5Cii), indicating amplification of the subtelomeric BAC sequences. Subclone FS-2 also had an unidentified chromosome fragment translocated onto the end of the rearranged chromosome, although this fragment is longer than that observed in subclone FS-1 (Fig. 5Cii). As in subclone FS-1, the rearranged chromosome was identical in all of the cells examined and always had telomeres on both ends (Fig. 5Civ), and therefore it was relatively stable at the time of isolation of this subclone. Southern blot analysis did not show a significant increase in hybridization with the plasmid probe in either subclone FS-1 or FS-2 compared to A211 (74). However, although the formation of the inverted repeat would duplicate the sequences in the subtelomeric BAC clone, it would cause a net loss in plasmid sequences (7 kb in the original plasmid versus 5.0 kb in FS-1 and 4.6 kb in FS-2) (Fig. 4A). An additional duplication of the region would result in a 4-fold increase in the BAC sequences over that found in A211 but only a 1.4-fold increase in plasmid sequences in FS-1 and a 1.3-fold increase in FS-2 (7 kb to 10 and 9.2 kb, respectively).
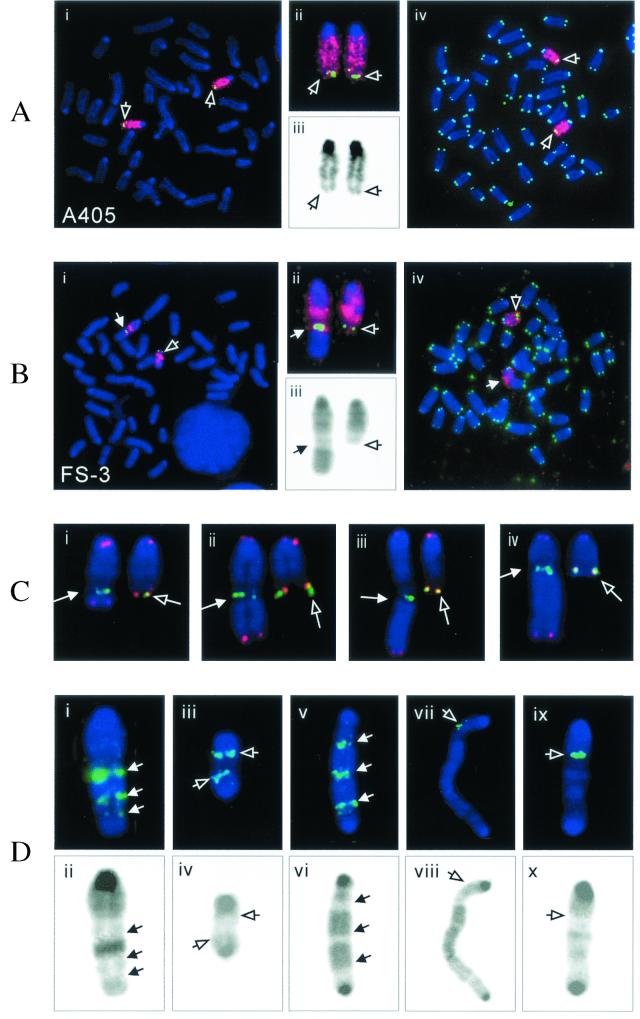
FISH analysis was also performed on metaphase chromosomes from clone A405 and its G418r/Ganr subclone FS-3, with either a chromosome 15-specific painting probe or the 169K7 BAC clone that contains sequences subtelomeric to the plasmid integration site. The results demonstrated that clone A405 contains two indistinguishable homologues of chromosome 15 (Fig. 6A). However, one of the chromosome 15 homologues in subclone FS-3 showed considerable heterogeneity in different cells in the population (Fig. 6B to D). This heterogeneity in chromosome structure must have occurred after the isolation of this subclone, because Southern blot analysis demonstrated that all of the cells in the population contained the same inverted repeat. Thus, the marker chromosome was still unstable at the time of isolation of subclone FS-3, despite the fact that this subclone was isolated 6 days after the treatment with I-SceI. In the majority of metaphase spreads in subclone FS-3, there was no apparent duplication of chromosome 15 distal to the subtelomeric BAC probe (Fig. 6B). However, there was amplification of subtelomeric sequences and large duplications on the end of the chromosome in some metaphase spreads (Fig. 6B, C, and Di to vi). Amplification of the subtelomeric sequences in some cells in the population was consistent with a small but significant increase in hybridization with the plasmid probe by Southern blot analysis in subclones FS-3 and FS-4 (Fig. 3).
FIG. 6.
Chromosome rearrangements in the FS-3 subclone isolated from clone A405. Metaphase spreads of the parental clone A405 (A) and subclone FS-3 (B) were hybridized with both chromosome-15-specific painting (red) and subtelomeric BAC 169K7 (green) probes (i). An enlarged view of the chromosome-15 homologues (ii) and reverse video of the blue channel showing the DAPI GC banding (iii) are also shown. A different metaphase spread hybridized with both chromosome 15-specific (red) and telomere-specific (green) probes (iv) is also shown for clone A405 and subclone FS-3. The open arrows point to the normal chromosome 15 homologues, while the closed arrows point to the rearranged chromosome 15. (C) Heterogeneity of the rearranged chromosomes in the different second-generation subclones of FS-3 (i to iv) as indicated by the variable lengths of the translocated fragments from other chromosomes. The junction of the rearranged chromosome (arrow) is indicated by the BAC 169K7 subtelomeric signal (green). Hybridization with the telomere-specific probe (red) is observed at both ends of these rearranged chromosomes, but no telomere-specific hybridization was observed within the rearranged chromosome. The normal (open arrow) and rearranged (closed arrow) homologues from the same metaphase spreads are shown for comparison. (D) Complex rearrangements involving the marker chromosome. A variety of large duplications (i and ii) and dicentric chromosomes (iii to x) are shown. The upper panel shows hybridization with subtelomeric BAC 169K7 (green, arrow in panels i and v) or the PAC 561P12 originally located in the center of chromosome 15 (green, open arrows in panels iii, vii, and ix), while the lower panel shows the corresponding DAPI GC banding by reverse video of the blue channel. Dicentric chromosomes were identified by the presence of pericentric heterochromatin (darkly stained regions) on both ends in the metaphase spreads stained with DAPI.
Similar to subclones FS-1 and FS-2, fragments of unidentified chromosomes were also translocated onto the rearranged end of the chromosome in subclone FS-3. However, unlike subclones FS-1 and FS-2, there was extensive variability in the lengths of these translocated fragments in different cells in the population (Fig. 6B and C), demonstrating that extensive rearrangement of the chromosome occurred after cloning. Nonetheless, the presence of telomeres on the end of the rearranged chromosome 15 in all of the metaphases examined (Fig. 6Biv and Ci to iv) suggests that the rearranged chromosome 15 was stable in most cells at the time of analysis. Consistent with this conclusion, the structure of the rearranged chromosome was the same in all metaphases examined within each of the 20 second-generation subclones examined, although there was extensive heterogeneity in the structure of the rearranged chromosome among the different second-generation subclones. Nonetheless, there was some degree of instability in the marker chromosome in the FS-3 subclone, since dicentrics involving the marker chromosome were observed in 4 of the 100 metaphases analyzed (Fig. 6Div, vi, viii, and x). Hybridization with the 561P12 PAC clone that contains sequences near the center of chromosome 15 demonstrated that one of these dicentric chromosomes was composed of two copies of chromosome 15 (Fig. 6Diii), while others involved fusion of chromosome 15 with other chromosomes (Fig. 6Dvii and ix). No hybridization with the telomeric repeat sequence probe was apparent at internal sites within the rearranged chromosomes, demonstrating that the translocations and dicentric chromosomes involving the marker chromosome and other chromosomes involved the loss of the ends of the chromosomes containing the telomere.
Isolation and DNA sequence analysis of inverted repeats resulting from spontaneous telomere loss in a human tumor cell line.
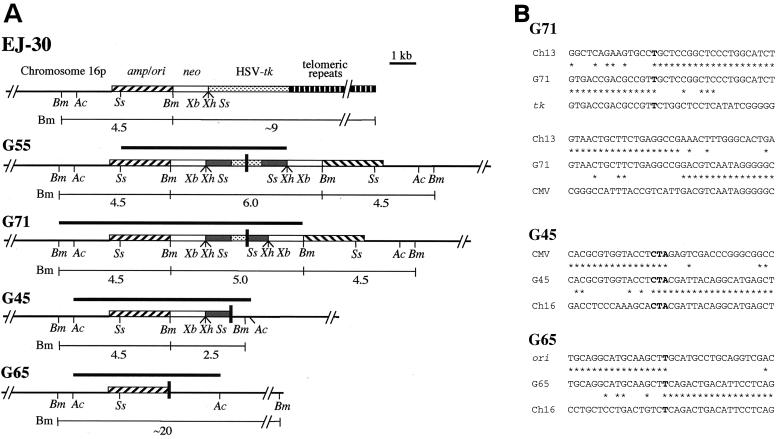
Inverted repeats similar to those found in the mouse ES cell subclones were also observed on the end of a marker chromosome in G418r/Ganr subclones isolated from the EJ-30 human tumor cell line after spontaneous telomere loss. Multiple clones of EJ-30 that have a single copy of the pNCT-tel plasmid integrated at a telomere were isolated (19). Like pNPT-tel, the pNCT-tel plasmid contains an HSV-tk gene for negative selection in ganciclovir (Fig. 7A). However, unlike the mouse ES cell clones, the clones of the EJ-30 cell line demonstrated a high rate of spontaneous inactivation of the HSV-tk gene. The analysis of G418r/Ganr subclones showed that most of them had nontelomeric DNA joined on the end of the marker chromosome. We previously reported that two of these G418r/Ganr subclones contained inverted repeats on the end of the chromosome (19). In subclone G55, the recombination site occurred within the HSV-tk gene at nearly the same location in both plasmids (Fig. 7A). However, like subclone FS-4, the site of recombination could not be sequenced, apparently due to the presence of the nearly perfect inverted repeat. In subclone G71, the inverted repeats resulted from recombination between sequences that were originally 1.3 kb apart and included the insertion of a small fragment of cellular DNA (Fig. 7A). There was 1 bp of complementarity on one end of the insertion, and no complementarity on the other end (Fig. 7B). In both subclones, the structures of the rescued fragments are consistent with the size of the fragments detected in genomic DNA by Southern blot analysis.
FIG. 7.
Cloning and analysis of inverted repeats in G418r/Ganr subclones isolated from the human EJ-30 tumor cell line. (A) The structure of the integrated pNCT-tel plasmid in the parental EJ-30 clone is compared with inverted repeats in the G55, G71, G45, and G65 subclones. The fragments cloned by plasmid rescue are indicated (heavy horizontal lines), as are the sizes of the restriction fragments that are consistent with bands observed by Southern blot analysis of genomic DNA (light horizontal lines). The recognition sites are shown for some of the restriction enzymes used for mapping and Southern blot analysis, including AccI (Ac), BamHI (Bm), SspI (Ss), XbaI (Xb), and XhoI (Xh). The locations of the adjacent cellular DNA (chromosome 16p), pSP73 vector sequences (amp/ori), neo gene (neo), HSV-tk gene, telomeric repeat sequences, and sites of recombination (black vertical bars) are shown. (B) Nucleotide sequences at the sites of recombination in the G418r/Ganr subclones from EJ-30. The nucleotide sequences at the sites of recombination at either end of the187-bp insert in subclone G71 are compared with human DNA sequence from chromosome 13 (Ch13), a sequence from the HSV-tk gene (tk), and a sequence from the cytomegalovirus (CMV) promoter. The nucleotide sequence at the site of recombination in subclone G45 is compared with the sequence in the HSV-tk gene (tk) and the sequence 4.9 kb from the integration site in chromosome 16 (contig no. NT_000669) in the opposite orientation (Ch16). The nucleotide sequence at the site of recombination in subclone G65 is compared with sequences in the CMV promoter on the HSV-tk gene (CMV) and the sequence 6.3 kb from the integration site in chromosome 16 in the opposite orientation (Ch16). Nucleotides that are identical between sequences (asterisks) or show complementarity between the sequences at the site of recombination (boldface) are indicated.
We have now rescued the plasmid sequences from two additional G418r/Ganr subclones of EJ-30: G45 and G65. These two subclones were selected because, unlike subclones G55 and G71, they did not appear to have amplified the plasmid sequences (see Fig. 2B in Fouladi et al. [19]). The plasmid sequences and adjacent cellular DNAs from G45 and G65 were rescued after digestion of genomic DNA with AccI. Nucleotide sequence analysis demonstrated that these subclones also contain inverted repeats on the end of the marker chromosome (Fig. 7A). However, unlike subclones G55 and G71, they contain plasmid sequences on one end of the inverted repeat and cellular sequences proximal to the integration site on chromosome 16 on the other end. The plasmid and cellular sequences at the site of recombination in subclones G45 and G65 were originally 9.8 and 8.7 kb apart, respectively. There are 3 bp of complementarity at the recombination site in subclone G45 and 1 bp of complementarity at the recombination site in subclone G65 (Fig. 7B). The structure of the cloned fragments was consistent with Southern blot analysis, which demonstrated 2.5- and 4.5-kb BamHI bands in subclone G45 and a single large BamHI band in subclone G65 (see Fig. 2B in Fouladi et al. [19]). Thus, all four G418r/Ganr subclones analyzed from the EJ-30 cell line also demonstrated inverted repeats similar to those found in the G418r/Ganr mouse ES cell subclones.
DISCUSSION
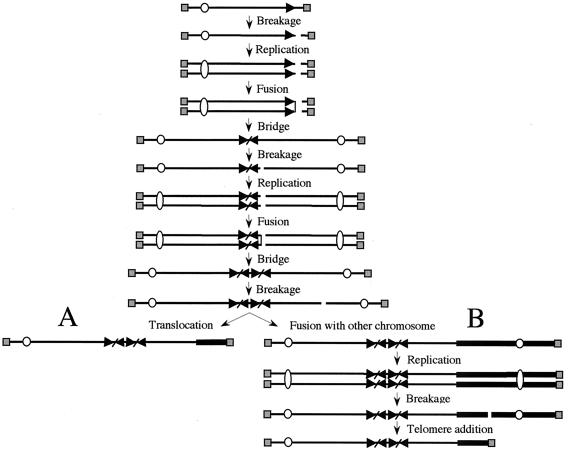
The inverted repeats and large duplications on the end of the marker chromosomes in the G418r/Ganr mouse ES cell (Fig. 4, 5, and 6) and human EJ-30 tumor cell (Fig. 7) (19) subclones are consistent with a mechanism involving sister chromatid fusion. Sister chromatid fusion would result in a dicentric chromosome that would break during cell division, generating an inverted repeat on the end of the chromosome in one daughter cell and a terminal deletion in the other daughter cell (Fig. 8). We have previously reported that the most common event observed in the mouse ES cell G418r/Ganr subclones is the addition of a new telomere to the site of the break (74). The results presented here suggest that sister chromatid fusion is the most common event in cells that fail to add a telomere to the end of the broken chromosome. Similar results were obtained in the EJ-30 human tumor cell line (Fig. 7) (19), although in these cells sister chromatid fusion occurred much more frequently than telomere addition. Earlier cytogenetic studies have concluded that sister chromatid fusions occur after the loss of a telomere in maize (44) and that they are the initial event in gene amplification in hamster cells (43, 71, 77). Sister chromatid fusion has also been proposed as a mechanism for the creation of the inverted duplications on the ends of chromosomes in human genetic disease (18, 30) and for gene amplification in human cancer (29, 69).
FIG. 8.
Possible mechanisms for the generation of inverted repeats and rearrangement of the marker chromosome. A DSB in a chromosome before or during DNA replication or DSBs in both sister chromatids after DNA replication results in sister chromatid fusion. Due to the presence of two centromeres, the fused sister chromatids break during anaphase, resulting in the transfer of an inverted repeat to the end of one of the chromosomes. After cell division, the absence of a telomere on the broken chromosome leads to additional fusions, either between sister chromatids or with other chromosomes. (A) Due to the absence of a telomere, after replication the sister chromatids again fuse and break in the next cell cycle, resulting in further amplification of the subtelomeric DNA sequences and a large duplication on the end of the chromosome. The acquisition of a telomere by translocation from another chromosome then stabilizes the broken chromosome. Alternatively, if a new telomere is not acquired, the chromosome can undergo further rounds of sister chromatid fusion or fuse with other chromosomes. (B) The broken chromosome can also fuse with another chromosome. The dicentric chromosome breaks during anaphase when the two centromeres are pulled to opposite daughter cells, resulting in a translocation of the other chromosome to the end of the marker chromosome. The acquisition of a telomere then stabilizes the broken chromosome. Alternatively, if a new telomere is not acquired, the chromosome can undergo sister chromatid fusion or again fuse with another chromosome. The telomeres (░⃞), centromeres (○), and subtelomeric sequences (▸) on the marker chromosome are indicated.
The presence of inverted repeats in our study contrasts with the types of recombination events associated with I-SceI-induced DSBs at interstitial sites in mammalian cells. The introduction of DSBs at specific locations within the I-SceI endonuclease has been used to study both homologous recombination and nonhomologous end joining (NHEJ) at interstitial sites in mammalian cells (32, 40, 60, 61, 65, 67). The repair of I-SceI-induced DSBs by homologous recombination can involve nonreciprocal recombination (32, 61) or single-strand annealing (57) with few detectable chromosome rearrangements. The repair of I-SceI-induced DSBs in a single stably integrated selectable marker gene by NHEJ most often involves very small deletions of less than 100 bp, although some larger deletions have also been occasionally observed (39, 40, 65, 67). Complex chromosome rearrangements resulting from single I-SceI-induced breaks at interstitial sites are relatively uncommon. In one study of 253 I-SceI-induced mutations in an HSV-tk gene in mouse fibroblasts, 12.6% were found to have complex rearrangements, although the actual recombination events were not characterized (40). Amplification of a dihydrofolate reductase gene proximal to an I-SceI site in Chinese hamster fibroblasts appeared to result from B/F/B cycles; however, this type of event was rare compared to deletions (57). In contrast, chromosome translocations are a relatively common event (9 of 43) after I-SceI-induced DSBs within homologous sequences on two different chromosomes (60).
There are several possible explanations for the different spectrum of recombination events observed at interstitial and telomeric sites. One explanation is that the model systems used in the various studies select for different types of events. In the present study, the I-SceI sites were not located within the coding sequence of the selectable marker gene and therefore would not have been able to pick up small deletions. The failure to observe telomere addition or fusions at interstitial sites could also result from the fact that the loss of the end of the chromosome would often be lethal due to the loss of essential genes. As a result, both chromosome healing and chromosome fusion would be selected against at interstitial sites. Finally, the different types of events observed at interstitial and telomeric sites could reflect the differences in chromatin structure (35, 78) or diminished DNA repair capabilities (34, 55) within telomeric regions. Regardless of the mechanism, it is clear from our observations that DSBs occurring near the ends of chromosomes can result in extensive chromosome rearrangement and prolonged periods of chromosome instability.
Nucleotide sequence analysis of the sites of recombination suggests that NHEJ is the mechanism involved in the formation of inverted repeats in both the mouse ES cell and human EJ-30 tumor cell lines. The recombination junctions in the mouse ES cell subclones contained 1 to 4 bp of complementarity, while the recombination junctions in the EJ-30 subclones had 0 to 3 bp of complementarity (Table 1). In one EJ-30 subclone, a short fragment of human DNA was found inserted between the inverted plasmid sequences at the recombination junction. Similar junctions with little or no complementarity (40, 56, 64, 65, 67) and insertions of short fragments of cellular DNA (23, 38, 40, 48, 56, 63, 67) have been previously observed during repair of DSBs by NHEJ in mammalian cells.
TABLE 1.
Extent of deletions and complementarity at the recombination sites in G418r/Ganr subclones
| Cell line | Subclone | Deletiona | Complem entarity (bp)b |
|---|---|---|---|
| A211 | FS-1 | 17 bp/3.0 kb | 4 |
| FS-2 | 8 bp/2.6 kb | 1 | |
| A405 | FS-3 | 0.5 kb/1.8 kb | 4 |
| FS-4 | 1.0 kb/1.0 kb | ND | |
| EJ-30 | G55 | ≥0 bp | ND |
| G71 | ≥1.3 kb | 1/0c | |
| G45 | ≥9.8 kb | 3 | |
| G65 | ≥8.7 kb | 1 |
The distance from the recombination site to the I-SceI site in the A211 and A405 subclones and/or the distance between the sequences at the recombination site in the EJ-30 subclones is indicated. If we assume a single event initiated recombination in the EJ-30 subclones, the difference in the location in the plasmids would represent the minimum amount of degradation.
Complementarity at the recombination site. The recombination site in some subclones could not be sequenced (ND = not determined), apparently due to the presence of a perfect inverted repeat.
Complementarity on either end of a 187-bp insert of human DNA.
Because the DSBs were generated at a known location with the I-SceI endonuclease in the mouse ES cell subclones, the size of the deletions occurring on the ends at the site of the break could be precisely determined (Table 1). In clone A211, the I-SceI site is located immediately distal to the neo gene (Fig. 1A). Therefore, because the FS-1 and FS-2 subclones were coselected with G418, at least one of the plasmid sequences had to retain the neo gene. As expected, one of the DNA ends had recombined very close to the I-SceI site. However, deletions of 2.6 and 3.0 kb occurred on the other DNA end (Fig. 4). In the two A405 subclones, where the I-SceI site is located further away from the neo gene, deletions were observed in all four of the DNA ends generated by I-SceI. These deletions ranged from 0.5 to 1.8 kb, although it cannot be ruled out that deletions in A405 are required to inactivate the HSV-tk gene which is proximal to the I-SceI site.
Unlike the mouse ES cell studies, the size of the deletions in the G418r/Ganr subclones isolated from the EJ-30 tumor cell line could not be determined because the location of the initiating event was not known. In view of the close proximity of the sites of recombination, it is likely that a single event that occurred prior to or during DNA replication was involved. Thus, the differences in the site of recombination in the two sequences involved in the inverted repeat would be due to differences in the amount of degradation from the two ends. The plasmid and cellular sequences at the site of recombination in subclones G45 and G65 were originally 9.8 and 8.7 kb apart (Table 1), respectively, indicating that the deletions in the human EJ-30 tumor cell line are often larger than those occurring in the mouse ES cell line.
Consistent with the results for the mouse ES cell and human EJ-30 tumor cell lines, gross chromosome rearrangements resulting from telomere loss in yeast cells were also found to commonly involve large deletions (25). Although there is no previously published report on the extent of DNA degradation during chromosome fusion in mammalian cells, deletions have been observed in a number of studies involving repair of DSBs generated by I-SceI at interstitial sites. In one study, deletions of several kilobases were observed after intrachromosomal NHEJ at a DSB induced by I-SceI in the adenosine phosphoribosyltransferase gene in hamster cells, although the majority of mutants had deletions of less than 34 bp (67). Similarly, a study of I-SceI-induced DSB in the HSV-tk gene in mouse fibroblasts found that most deletions were less than 90 bp (40). However, larger deletions of up to 2.4 kb were observed at sites of NHEJ in translocations induced by two I-SceI sites on different chromosomes in mouse ES cells, suggesting that large deletions may be associated with interchromosomal recombination events (60).
Large inverted repeats have been demonstrated to be hotspots for recombination in Escherichia coli (10, 49), Saccharomyces cerevisiae (42), and mammals (1, 20, 81). Secondary recombination events might therefore have been responsible for the deletions on the two ends involved in the formation of the inverted repeat, since an absence of symmetry can increase the stability of inverted repeats (1, 42). However, deletions within the inverted repeats did not appear to occur during PCR or cloning in bacteria. Multiple DNA clones were rescued from each G418r/Ganr subclone, and all had structures that were consistent with plasmid sequences found within the genomic DNA as shown by Southern blot analysis. Thus, the inverted repeats were relatively stable in the bacterial strains used in our study, which contain mutations that increase the stability of inverted repeats. Although it is not possible to rule out secondary recombination events within the ES or EJ-30 subclones, Southern blot analysis did not reveal any polymorphism in the structure of the apparently perfect inverted repeats in the mouse ES cell subclone FS-4 (Fig. 3) or human EJ-30 subclone G55 (19).
The results presented here for the mouse ES cell subclones, like those obtained previously with the human EJ-30 tumor cell subclones (19), demonstrate that the loss of a telomere can promote chromosome instability. Although stable at the time of selection, the marked chromosome in subclones FS-1 and FS-2 had an approximately fivefold amplification of the subtelomeric BAC sequences and translocations from other chromosomes, indicating that multiple recombination events had occurred during the 6-day period between I-SceI treatment and selection. In addition, the marked chromosome in subclone FS-3 was highly unstable at the time of selection, as demonstrated by (i) the amplification of the subtelomeric sequences in some cells, (ii) the heterogeneity in the length of the duplications on the end of the chromosome, (iii) the variability in the lengths of the fragments of other chromosomes joined to the end, and (iv) the presence of dicentric chromosomes involving the marker chromosome and other chromosomes in some cells. The rearrangements that are observed in all three mouse ES cell subclones are consistent with B/F/B cycles (Fig. 8) and are similar in many respects to the types of rearrangements we previously observed in the EJ-30 subclones containing inverted repeats on the end of the marker chromosome (19). B/F/B cycles have previously been associated with sister chromatid fusion in maize (44) and in gene amplification in hamster cells (12, 43, 71, 77). Furthermore, B/F/B cycles resulting from telomere loss have been associated with chromosome instability in early-passage human tumor cell cultures (21). High-copy gene amplification in human tumor cells does not usually occur through B/F/B cycles (6, 26). However, B/F/B cycles have been found to be involved in low-copy gene amplification and have been proposed to serve as an early event in some high-copy gene amplification in human cancer (29, 69, 70). Telomere loss may also contribute to other types of chromosome rearrangements, such as nonreciprocal translocations commonly observed in tumor cells (58). Secondary events involving other chromosomes have been observed during gene amplification in hamster cells (77) and after telomere loss in cells from telomerase-deficient mice (3). The results presented here and by others (3, 66) also demonstrate that nonreciprocal translocations are a common event after telomere loss. Finally, the large inverted repeats generated by the sister chromatid fusions could also generate additional chromosome rearrangements, since it has been demonstrated that the hairpin structures formed by inverted repeats are hotspots for recombination (1, 10, 20, 42, 49, 81).
The extent of chromosome instability after telomere loss can be influenced by a variety of factors. The status of cell cycle checkpoints, which are commonly defective in cancer cells, could prevent B/F/B cycles through the elimination of cells containing dicentric or broken chromosomes (37). In this regard, both the EJ-30 human tumor cell line (11, 46) and mouse ES cells have been demonstrated to lack p53-dependent cell cycle checkpoints (2). As first proposed by McClintock, the addition of telomeres onto the ends of broken chromosomes can also influence both the initiation and duration of B/F/B cycles (44). The mouse ES cell line would appear to be relatively efficient in the addition of telomeres onto the ends of broken chromosomes compared to the EJ-30 human tumor cell line. Most of the G418r/Ganr subclones isolated from the mouse ES cell line have telomeres added onto the ends of the broken chromosomes (74), while most G418r/Ganr subclones isolated from the EJ-30 human tumor cell line have nontelomeric DNA joined to the end (19). In addition, the marker chromosome in the mouse ES cell subclone FS-3 always had telomeres and was stable in second-generation subclones, whereas the marker chromosomes in the EJ-30 subclone G71 often lacked telomeres and were unstable in many second-generation subclones. Thus, the high degree of chromosome instability in the human EJ-30 tumor cell line may result not only from an increased rate of spontaneous telomere loss but also from a reduced ability to add telomeres onto the broken ends and thereby prevent or terminate B/F/B cycles.
Acknowledgments
We thank Mary Barter (The Jackson Laboratory) for assistance in the interspecific backcross analysis and Luis Martins for helpful technical assistance.
The work in the J.P.M. laboratory was supported by National Institute of Environmental Health Science grant RO1 ES008427, and National Cancer Institute grant RO1 CA69044. The work in the L.S. laboratory was supported by contract number FIGH-CT-199-00002 from the CEC.
A.W.I.L and C.N.S. contributed equally to this study.
REFERENCES
- 1.Akgun, E., J. Zahn, S. Baumes, G. Brown, F. Liang, P. J. Romanienko, S. Lewis, and M. Jasin. 1997. Palindrome resolution and recombination in the mammalian germ line. Mol. Cell. Biol. 17:5559-5570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aladjem, M. I., B. T. Spike, L. W. Rodewald, T. J. Hope, M. Klemm, R. Jaenisch, and G. M. Wahl. 1998. ES cells do not activate p53-dependent stress responses and undergo p53-independent apoptosis in response to DNA damage. Curr. Biol. 8:145-155. [DOI] [PubMed] [Google Scholar]
- 3.Artandi, S. E., S. Chang, S.-L. Lee, S. Alson, G. J. Gottlieb, L. Chin, and R. A. DePinho. 2000. Telomere dysfunction promotes non-reciprocal translocations and epithelial cancers in mice. Nature 406:641-645. [DOI] [PubMed] [Google Scholar]
- 4.Bailey, S. M., J. Meyne, D. J. Chen, A. Kurimasa, G. C. Li, B. E. Lehnert, and E. H. Goodwin. 1999. DNA double-strand break repair proteins are required to cap the ends of mammalian chromosomes. Proc. Natl. Acad. Sci. USA 96:14899-14904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barnett, M. A., J. Buckle, E. P. Evans, A. C. G. Porter, D. Rout, A. G. Smith, and W. R. A. Brown. 1993. Telomere-directed fragmentation of mammalian chromosomes. Nucleic Acids Res. 21:27-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benner, S. E., G. M. Wahl, and D. D. Von Hoff. 1991. Double minute chromosomes and homogenously staining regions in tumors taken directly from patients verses in human tumor cell lines. Anticancer Drugs 2:11-25. [DOI] [PubMed] [Google Scholar]
- 7.Blackburn, E. H. 2001. Switching and signaling at the telomere. Cell 106:661-673. [DOI] [PubMed] [Google Scholar]
- 8.Bosco, G., and J. E. Haber. 1998. Chromosome break-induced replication leads to nonreciprocal translocations and telomere capture. Genetics 150:1037-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brisebois, J. J., and M. S. DuBow. 1993. Selection for spontaneous null mutations in a chromosomally integrated HSV-1 thymidine kinase gene yields deletions and a mutation caused by intragenic illegitimate recombination. Mutat. Res. 287:191-205. [DOI] [PubMed] [Google Scholar]
- 10.Collins, J. 1981. Instability of palindromic DNA in Escherichia coli. Cold Spring Harbor Symp. Quant. Biol. 45:409-416. [DOI] [PubMed] [Google Scholar]
- 11.Cooper, M. J., J. J. Haluschak, D. Johnson, S. Schwartz, L. J. Morrison, M. Lippa, G. Hatzivassiliou, and J. Tan. 1994. p53 mutations in bladder carcinoma cell lines. Oncol. Res. 6:569-579. [PubMed] [Google Scholar]
- 12.Coquelle, A., E. Pipiras, F. Toledo, G. Buttin, and M. Debatisse. 1997. Expression of fragile sites triggers intrachromosomal mammalian gene amplification and sets bounderies to early amplification. Cell 89:215-225. [DOI] [PubMed] [Google Scholar]
- 13.de Lange, T. 1995. Telomere dynamics and genome instability in human cancer, p. 265-293. In E. H. Blackburn and C. W. Greider (ed.), Telomeres. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 14.Diede, S. J., and D. E. Gottschling. 1999. Telomerase-mediated telomere addition in vivo requires DNA primase and DNA polymerase α and δ. Cell 99:723-733. [DOI] [PubMed] [Google Scholar]
- 15.Dutrillaux, B., and J. Couturier. 1981. La pratique de l'analysee chromosomique. Masson, Paris, France.
- 16.Farr, C., J. Fantes, P. Goodfellow, and H. Cooke. 1991. Functional reintroduction of human telomeres into mammalian cells. Proc. Natl. Acad. Sci. USA 88:7006-7010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flint, J., K. Thomas, G. Micklem, H. Raynham, K. Clark, N. A. Doggett, A. King, and D. R. Higgs. 1997. The relationship between chromosome structure and function at a human telomeric region. Nat. Genet. 15:252-257. [DOI] [PubMed] [Google Scholar]
- 18.Floridia, G., M. Piantanida, A. Minelli, C. Dellavecchia, C. Bonaglia, E. Rossi, G. Gimelli, G. Croci, F. Franchi, S. Gilgenkrantz, P. Grammatico, L. Dalpra, S. Wood, C. Danesino, and O. Zuffardi. 1996. The same molecular mechanism at the maternal meiosis I produces mono- and dicentric 8p duplications. Am. J. Hum. Genet. 58:785-796. [PMC free article] [PubMed] [Google Scholar]
- 19.Fouladi, B., D. Miller, L. Sabatier, and J. P. Murnane. 2000. The relationship between spontaneous telomere loss and chromosome instability in a human tumor cell line. Neoplasia 2:540-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gebow, D., N. Miselis, and H. L. Liber. 2000. Homologous and nonhomologous recombination resulting in deletion: effects of p53 status, microhomology, and repetitive DNA length and orientation. Mol. Cell. Biol. 20:4028-4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gisselsson, D., T. Jonson, A. Petersen, B. Strombeck, P. Dal Cin, M. Hoglund, F. Mitelman, F. Mertens, and N. Mandahl. 2001. Telomere dysfunction triggers extensive DNA fragmentation and evolution of complex chromosome abnormalities in human malignant tumors. Proc. Natl. Acad. Sci. USA 98:12683-12688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gisselsson, D., L. Pettersson, M. Hoglund, M. Heidenblad, L. Gorunova, J. Wiegant, F. Mertens, P. Dal Cin, F. Mitelman, and N. Mandahl. 2000. Chromosomal breakage-fusion-bridge events cause genetic intratumor heterogeneity. Proc. Natl. Acad. Sci. USA 97:5357-5362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Godwin, A. R., R. J. Bollag, D.-M. Christie, and R. M. Liskay. 1994. Spontaneous and restriction enzyme-induced chromosomal recombination in mammalian cells. Proc. Natl. Acad. Sci. USA 91:12554-12558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goytisolo, F. A., E. Samper, S. Edmonson, G. E. Taccioli, and M. A. Blasco. 2001. The absence of the DNA-dependent protein kinase catalytic subunit in mice results in anaphase bridges and increased telomeric fusions with normal telomere length and G-strand overhang. Mol. Cell. Biol. 21:3642-3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hackett, J. A., D. M. Feldser, and C. W. Greider. 2001. Telomere dysfunction increases mutation rates and genomic instability. Cell 106:275-286. [DOI] [PubMed] [Google Scholar]
- 26.Hahn, P. J. 1993. Molecular biology of double-minute chromosomes. Bioessays 15:477-484. [DOI] [PubMed] [Google Scholar]
- 27.Hande, P., E. Samper, P. Lansdorp, and M. A. Blasco. 1999. Telomere length dynamics and chromosomal instability in cells derived from telomerase null mice. J. Cell Biol. 144:589-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hanish, J. P., J. L. Yanowitz, and T. De Lange. 1994. Stringent sequence requirements for the formation of human telomeres. Proc. Natl. Acad. Sci. USA 91:8861-8865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hellman, A., E. Ziotorynski, S. W. Scherer, J. Cheung, J. B. Vincent, D. I. Smith, L. Trakhtenbrot, and B. Kerem. 2002. A role for common fragile site induction in amplification of human oncogenes. Cancer Cell 1:89-97. [DOI] [PubMed] [Google Scholar]
- 30.Hoo, J. J., M. Chao, K. Szego, M. Rauer, S. C. Echiverri, and C. Harris. 1995. Four new cases of inverted terminal duplications. Am. J. Med. Genet. 58:299-304. [DOI] [PubMed] [Google Scholar]
- 31.Hsu, H.-L., D. Gilley, S. A. Galande, M. P. Hande, B. Allen, S.-H. Kim, G. C. Li, J. Campisi, T. Kohwi-Shigematsu, and D. J. Chen. 2000. Ku acts in a unique way at the mammalian telomere to prevent end joining. Genes Dev. 14:2807-2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson, R. D., and M. Jasin. 2000. Sister chromatid gene conversion is a prominent double-strand break repair pathway in mammalian cells. EMBO J. 19:3398-3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kramer, K. M., and J. E. Haber. 1993. New telomeres in yeast are initiated with a highly selected subset of TG1-3 repeats. Genes Dev. 7:2345-2356. [DOI] [PubMed] [Google Scholar]
- 34.Kruk, P. A., N. J. Rampino, and V. A. Bohr. 1995. DNA damage and repair in telomeres: relation to aging. Proc. Natl. Acad. Sci. USA 92:258-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lejnine, S., V. L. Makarov, and J. P. Langmore. 1995. Conserved nucleoprotein structure at the ends of vertebrate and invertebrate chromosomes. Proc. Natl. Acad. Sci. USA 92:2393-2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lemieux, N., B. Dutrillaux, and E. Viegas-Pequignot. 1992. A simple method for simultaneous R- or G-banding and fluorescence in situ hybridization of small single-copy genes. Cytogenet. Cell Genet. 59:311-312. [DOI] [PubMed] [Google Scholar]
- 37.Lengauer, C., K. W. Kinzler, and B. Vogelstein. 1998. Genetic instabilities in human cancers. Nature 396:643-649. [DOI] [PubMed] [Google Scholar]
- 38.Liang, F., M. Han, P. J. Romanienko, and M. Jasin. 1998. Homology-directed repair is a major double-strand break repair pathway in mammalian cells. Proc. Natl. Acad. Sci. USA 95:5172-5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin, Y., T. Lukacsovich, and A. S. Waldman. 1999. Multiple pathways for repair of DNA double-strand breaks in mammalian cells. Mol. Cell. Biol. 19:8353-8360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin, Y., and A. S. Waldman. 2001. Capture of DNA sequences at double-strand breaks in mammalian cells. Genetics 158:1665-1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lo, A. W., G. C. Liao, M. Rocchi, and K. H. Choo. 1999. Extreme reduction of chromosome-specific alpha-sattellite array is unusually common in human chromosome 21. Genome Res. 9:895-908. [DOI] [PubMed] [Google Scholar]
- 42.Lobachev, K. S., D. A. Gordenin, and M. A. Resnick. 2002. The Mre11 complex is required for repair of hairpin-capped double-strand breaks and prevention of chromosome rearrangements. Cell 108:183-193. [DOI] [PubMed] [Google Scholar]
- 43.Ma, C., S. Martin, B. Trask, and J. L. Hamlin. 1993. Sister chromatid fusion initiates amplification of the dihydrofolate reductase gene in Chinese hamster cells. Genes Dev. 7:605-620. [DOI] [PubMed] [Google Scholar]
- 44.McClintock, B. 1941. The stability of broken ends of chromosomes in Zea mays. Genetics 41:234-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McEachern, M. J., A. Krauskopf, and E. H. Blackburn. 2000. Telomeres and their control. Annu. Rev. Genet. 34:331-358. [DOI] [PubMed] [Google Scholar]
- 46.McIlwrath, A. J., P. A. Vasey, G. M. Ross, and R. Brown. 1994. Cell cycle arrests and radiosensitivity of human tumor cell lines: dependence on wild-type p53 for radiosensitivity. Cancer Res. 54:3718-3722. [PubMed] [Google Scholar]
- 47.Meltzer, P. S., X.-Y. Guan, and J. M. Trent. 1993. Telomere capture stabilizes chromosome breakage. Nat. Genet. 4:252-255. [DOI] [PubMed] [Google Scholar]
- 48.Meuth, M. 1989. Illegitimate recombination in mammalian cells, p. 833-860. In D. E. Berg and M. M. Howe (ed.), Mobile DNA. American Society for Microbiology, Washington, D.C.
- 49.Mizuuchi, K., M. Mizuuchi, and M. Gellert. 1982. Cruciform structures in palindromic DNA are favored by DNA supercoiling. J. Mol. Biol. 156:229-243. [DOI] [PubMed] [Google Scholar]
- 50.Murata, S., T. Matsuzaki, S. Takai, H. Yaoita, and M. Noda. 1995. A new retroviral vector for detecting mutations and chromosomal instability in mammalian cells. Mutat. Res. 334:375-383. [DOI] [PubMed] [Google Scholar]
- 51.Murnane, J. P., L. F. Fuller, and R. B. Painter. 1985. Establishment and characterization of a permanent pSVori−-transformed ataxia-telangiectasia cell line. Exp. Cell Res. 158:119-126. [DOI] [PubMed] [Google Scholar]
- 52.Myung, K., C. Chen, and R. D. Kolodner. 2001. Multiple pathways cooperate in the suppression of genome instability in Saccharomyces cerevisiae. Nature 411:1073-1076. [DOI] [PubMed] [Google Scholar]
- 53.Myung, K., A. Datta, and R. D. Kolodner. 2001. Suppression of spontaneous chromosomal rearrangements by S phase checkpoint functions in Saccharomyces cerevisiae. Cell 104:397-408. [DOI] [PubMed] [Google Scholar]
- 54.O'Toole, C. M., S. Povey, P. Hepburn, and L. M. Franks. 1983. Identity of some human bladder cancer cell lines. Nature 301:429-430. [DOI] [PubMed] [Google Scholar]
- 55.Petersen, S., G. Saretzki, and T. von Zglinicki. 1998. Preferential accumulation of single-stranded regions in telomeres of human fibroblasts. Exp. Cell Res. 239:152-160. [DOI] [PubMed] [Google Scholar]
- 56.Phillips, J. W., and W. F. Morgan. 1994. Illegitimate recombination induced by DNA double-strand breaks in a mammalian chromosome. Mol. Cell. Biol. 14:5794-5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pipiras, E., A. Coquelle, A. Bieth, and M. Debatisse. 1998. Interstitial deletions and intrachromosomal amplification initiated from a double-strand break targeted to a mammalian chromosome. EMBO J. 17:325-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Riboni, R., A. Casati, T. Nardo, E. Zaccaro, L. Ferretti, F. Nuzzo, and C. Mondello. 1997. Telomeric fusions in cultured fibroblasts as a source of genomic instability. Cancer Genet. Cytogenet. 95:130-136. [DOI] [PubMed] [Google Scholar]
- 59.Richard, C. W., III, D. A. Withers, T. C. Meeker, S. Maurer, G. A. Evans, R. M. Myers, and D. R. Cox. 1991. A radiation hybrid map of the proximal long arm of human chromosome 11 containing the multiple endocrine neoplasia type 1 (MEN-1) and bcl-1 disease loci. Am. J. Hum. Genet. 49:1189-1196. [PMC free article] [PubMed] [Google Scholar]
- 60.Richardson, C., and M. Jasin. 2000. Frequent chromosomal translocations induced by DNA double-strand breaks. Nature 405:697-700. [DOI] [PubMed] [Google Scholar]
- 61.Richardson, C., M. E. Moynahan, and M. Jasin. 1998. Double-strand break repair by interchromosomal recombination: suppression of chromosomal translocations. Genes Dev. 12:3831-3842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Robertson, E. J. 1987. Embryo-derived stem cells, p. 71-112. In E. J. Robertson (ed.), Teratocarcinomas and embryonic stem cells: a practical approach. Oxford University Press, Oxford, England.
- 63.Roth, D. B., X.-B. Chang, and J. H. Wilson. 1989. Comparison of filler DNA at immune, nonimmune, and oncogenic rearrangements suggests multiple mechanisms of formation. Mol. Cell. Biol. 9:3049-3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Roth, D. B., and J. H. Wilson. 1986. Nonhomologous recombination in mammalian cells: role for short sequence homologies in the joining reaction. Mol. Cell. Biol. 6:4295-4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rouet, P., F. Smih, and M. Jasin. 1994. Introduction of double-strand breaks into the genome of mouse cells by expression of a rare-cutting endonuclease. Mol. Cell. Biol. 14:8096-8106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rudolph, K. L., M. Millard, M. W. Bosenberg, and R. A. DePinho. 2001. Telomere dysfunction and evolution of intestinal carcinoma in mice and humans. Nat. Genet. 28:155-159. [DOI] [PubMed] [Google Scholar]
- 67.Sargent, R. G., M. A. Brenneman, and J. H. Wilson. 1997. Repair of site-specific double-strand breaks in a mammalian chromosome by homologous and illegitimate recombination. Mol. Cell. Biol. 17:267-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schulz, V. P., and V. A. Zakian. 1994. The Saccharomyces PIF1 DNA helicase inhibits telomere elongation and de novo telomere formation. Cell 76:145-155. [DOI] [PubMed] [Google Scholar]
- 69.Shuster, M. I., L. Han, M. M. Le Beau, E. Davis, M. Sawicki, C. M. Lese, N. H. Park, J. Colicelli, and S. M. Gollin. 2000. A consistent pattern of RIN1 rearrangements in oral squamous cell carcinoma cell lines supports a breakage-fusion-bridge cycle model for 11q23 amplification. Genes Chromosomes Cancer 28:153-163. [PubMed] [Google Scholar]
- 70.Singer, M. J., L. D. Mesner, C. L. Friedman, B. J. Trask, and J. L. Hamlin. 2000. Amplification of the human dihydrofolate reductase gene via double minutes is initiated by chromosome breaks. Proc. Natl. Acad. Sci. USA 97:7921-7926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Smith, K. A., M. B. Stark, P. A. Gorman, and G. R. Stark. 1992. Fusions near telomeres occur very early in the amplification of CAD genes in Syrian hamster cells. Proc. Natl. Acad. Sci. USA 89:5427-5431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Soriano, P., C. Montgomery, R. Geske, and A. Bradley. 1991. Targeted disruption of the c-src proto-oncogene leads to osteoporosis in mice. Cell 64:693-702. [DOI] [PubMed] [Google Scholar]
- 73.Sprung, C. N., G. Afshar, E. A. Chavez, P. Lansdorp, L. Sabatier, and J. P. Murnane. 1999. Telomere instability in a human cancer cell line. Mutat. Res. 429:209-223. [DOI] [PubMed] [Google Scholar]
- 74.Sprung, C. N., G. E. Reynolds, M. Jasin, and J. P. Murnane. 1999. Chromosome healing in mouse embryonic stem cells. Proc. Natl. Acad. Sci. USA 96:6781-6786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sprung, C. N., L. Sabatier, and J. P. Murnane. 1999. Telomere dynamics in human cancer cell line. Exp. Cell Res. 247:29-37. [DOI] [PubMed] [Google Scholar]
- 76.Thomas, K. R., and M. R. Capecchi. 1987. Site-directed mutagenesis by gene targeting in mouse embryo-derived stem cells. Cell 51:503-512. [DOI] [PubMed] [Google Scholar]
- 77.Toledo, F., G. Buttin, and M. Debatisse. 1993. The origin of chromosome rearrangements at early stages of AMPD2 gene amplification in Chinese hamster cells. Curr. Biol. 3:255-264. [DOI] [PubMed] [Google Scholar]
- 78.Tommerup, H., A. Dousmanis, and T. de Lange. 1994. Unusual chromatin in human telomeres. Mol. Cell. Biol. 14:5777-5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.van Steensel, B., A. Smogorzewska, and T. de Lange. 1998. TRF2 protects human telomeres from end-to-end fusions. Cell 92:401-413. [DOI] [PubMed] [Google Scholar]
- 80.Varley, H., S. Di, S. W. Scherer, and N. J. Royle. 2000. Characterization of terminal deletions at 7q32 and 22q13.3 healed by de novo telomere addition. Am. J. Hum. Genet. 67:610-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Waldman, A. S., H. Tran, E. C. Goldsmith, and M. A. Resnick. 1999. Long inverted repeats are an at-risk motif for recombination in mammalian cells. Genetics 153:1873-1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wong, A. C., Y. Ning, J. Flint, K. Clark, J. P. Dumanski, D. H. Ledbetter, and H. E. McDermid. 1997. Molecular characterization of a 130-kb terminal microdeletion at 22q in a child with mild mental retardation. Am. J. Hum. Genet. 60:113-120. [PMC free article] [PubMed] [Google Scholar]
- 83.Yu, G., and E. H. Blackburn. 1991. Developmentally programmed healing of chromosomes by telomerase in Tetrahymena. Cell 67:823-832. [DOI] [PubMed] [Google Scholar]
- 84.Zijlmans, J. M., U. M. Martens, S. S. S. Poon, A. K. Raap, H. J. Tanke, R. K. Ward, and P. M. Lansdorp. 1997. Telomeres in the mouse have large inter-chromosomal variations in the number of T2AG3 repeats. Proc. Natl. Acad. Sci. USA 94:7423-7428. [DOI] [PMC free article] [PubMed] [Google Scholar]