Abstract
Neurabin I, a neuronal actin-binding protein, binds protein phosphatase 1 (PP1) and p70 ribosomal S6 protein kinase (p70S6K), both proteins implicated in cytoskeletal dynamics. We expressed wild-type and mutant neurabins fused to green fluorescent protein in Cos7, HEK293, and hippocampal neurons. Biochemical and cellular studies showed that an N-terminal F-actin-binding domain dictated neurabin I localization at actin cytoskeleton and promoted disassembly of stress fibers. Deletion of the C-terminal coiled-coil and sterile alpha motif domains abolished neurabin I dimerization and induced filopodium extension. Immune complex assays showed that neurabin I recruited an active PP1 via a PP1-docking sequence,457KIKF460. Mutation of the PP1-binding motif or PP1 inhibition by okadaic acid and calyculin A abolished filopodia and restored stress fibers in cells expressing neurabin I. In vitro and in vivo studies suggested that the actin-binding domain attenuated protein kinase A (PKA) phosphorylation of neurabin I. Modification of a major PKA site, serine-461, impaired PP1 binding. Finally, p70S6K was excluded from neurabin I/PP1 complexes and required the displacement of PP1 for recruitment to neurabin I. These studies provided new insights into the assembly and regulation of a neurabin I/PP1 complex that controls actin rearrangement to promote spine development in mammalian neurons.
Cross talk between protein kinases and phosphatases regulates synaptic strength and information processing in mammalian brain (33). Prior studies identified protein phosphatase 1 (PP1) as a key regulator of activity-dependent changes in synaptic function underlying the two major forms of plasticity known as long-term potentiation (3) and long-term depression (LTD) extensively studied for hippocampal neurons (19). LTD-inducing stimuli promoted distribution of PP1 to dendritic spines (18), where it associated with the actin-rich cytoskeletal structure known as the postsynaptic density or PSD (30). This localization was critical for effective dephosphorylation of PP1 substrates such as Ca2+/calmodulin-dependent protein kinase II (30) and dl-α-amino-3-hydroxy-5-methylisoxazolepropionic acid receptors (34) and down-regulation of synaptic function. This has focused attention on the cellular mechanisms that target PP1 to the neuronal actin cytoskeleton (32).
Neurabin I (NrbI), identified by F-actin binding (20), shares structural homology with spinophilin (1) or neurabin II (NrbII) (25), which was isolated as a PP1-binding protein. PP1 complexes containing both neurabins have been demonstrated in extracts from rat brain (16). In addition, the PDZ (PSD-95/Dlg/ZO-1 homology) domain in NrbI recruited p70 ribosomal S6 protein kinase (p70S6K) (4) and kalirin-7 (24), a GTP-exchange factor, molecules that have been implicated in the control of neuronal morphology. The C terminus of NrbI contained coiled-coil and SAM (sterile alpha motif) domains, shown to mediate homodimerization and heterodimerization of other proteins (14, 27), which may contribute to the in vitro actin-cross-linking or bundling activity of neurabins (20). The C terminus of NrbI also bound the trans-Golgi protein TGN38 (28) and suggested that NrbI was a multifunctional protein scaffold that regulated both membrane and cytoskeletal functions.
By immunohistochemistry, NrbII was localized to dendritic spines and thus called spinophilin (1). In contrast, NrbI was present in both spines and growth cones (20). Subcellular fractionation showed that both neurabins are present in highly purified preparations of PSD (25, 32) and growth cones (R. T. Terry-Lorenzo and S. Shenolikar, unpublished observations). Ectopic expression of NrbI (20) and NrbII (25) in cultured cells demonstrated that both proteins localized to actin cytoskeleton, consistent with their ability to bind polymerized F-actin. More recent studies showed that the cytoskeletal association of neurabins was highly dynamic and regulated by growth factors (29) and small GTPases (25). This suggested that, in response to physiological signals, neurabins delivered signaling proteins, such as PP1 and p70S6K, to the actin cytoskeleton to control cell morphology. However, to date the role of NrbI signaling in cells has not been investigated.
Disruption of the mouse spinophilin/NrbII gene resulted in significant deficits in PP1 signaling required for LTD. Changes in neuronal morphology, specifically an overabundance of spines (9), were noted in the hippocampus of young NrbII-null mice. In contrast, antisense depletion of NrbI inhibited neurite outgrowth (20) and suggested distinct roles for the two neurabin isoforms in neurons. We have undertaken a combined molecular and cell biological analysis of rat NrbI to define its role in signal transduction and cell morphology. Our studies examined the contribution of specific structural determinants in the assembly of cellular NrbI complexes and their impact on the rearrangement of actin cytoskeleton. Our data suggested that NrbI targets PP1 activity to actin-rich structures in cultured cells to promote filopodia and disassemble the stress fiber network. In this regard, our studies provided the first insights into the role of the NrbI/PP1 complex in cytoskeletal dynamics and spine development in mammalian neurons.
MATERIALS AND METHODS
Phosphorylase b was purchased from Calzyme. Actin Binding Kit BK001 was from Cytoskeleton. [γ-32P]ATP and [32P]orthophosphoric acid were purchased from NEN. Adenosine 3′,5′-cyclic monophosphorothiolate, Sp isomer-triethylammonium salt (Sp-cAMPS); adenosine 3′,5′-cyclic monophosphorothiolate, Rp isomer-triethylammonium salt (Rp-cAMPS); latrunculin B; okadaic acid; and calyculin A were from Calbiochem. Anti-p70S6K antibody was obtained from Santa Cruz Biotechnology, anti-green fluorescent protein (anti-GFP) was from Research Diagnostics, and anti-PP1 was from Transduction Laboratories. A polyclonal antibody was generated by injecting rabbits with the recombinant PDZ domain (amino acids 488 to 593) of rat NrbII. The expression plasmid pXS-HA-p70S6K was provided by R. T. Abraham (Burnham Institute).
Expression of NrbI in Escherichia coli.
His-NrbI (1-615), His-NrbI (103-615), His-NrbI (615-1095), and His-NrbI (103-1095) were generated by PCR with full-length rat NrbI cDNA as template and cloned into pRSET-B (Invitrogen). BL21(DE3)pLysS competent cells (Stratagene) were transformed with plasmid DNA and grown in Luria-Bertani broth at 37°C until the optical density at 600 nm was 0.6. Protein expression was induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at 25°C for 3 h. Cells were lysed by sonication following their incubation in phosphate-buffered saline (PBS) containing 10 mM imidazole, 0.5% Triton X-100, 1 mM phenylmethylsulfonyl fluoride (PMSF), 1 mM benzamidine, and 1 mg of lysozyme/ml. Extracts were clarified by centrifugation at 3,000 × g for 25 min, and the supernatant was incubated with Ni+-nitrilotriacetic acid (NTA) agarose (Qiagen). The Ni+-NTA column was washed three times with lysis buffer and once with the same buffer containing 25 mM imidazole prior to elution with buffer containing 350 mM imidazole.
NrbI expression in mammalian cells.
The cDNA encoding full-length NrbI (1-1095) was subcloned into BglII/SmaI sites in pEGFP-C1 (Clontech). GFP-NrbI (1-552) was generated with a BglII/BamHI fragment of the NrbI cDNA subcloned into pEGFP-C1. GFP-NrbI (286-1095) was generated by inserting the KpnI/SmaI fragment into pEGFP-C1. GFP-NrbI (553-1095) was generated by inserting the BamHI fragment into pEGFP-C2. GFP-NrbI (1-552, 4A) was generated with the QuikChange Site-Directed Mutagenesis Kit (Stratagene). The first reaction used the oligonucleotide GAAATCCCAGCAAATAGGGCAGCTAAGTTTAGTTGTGCTCCG to substitute alanines for lysine-457 and isoleucine-458. The second reaction used this product as template and the oligonucleotide GAAATCCCAGCAAATAGGGCAGCTGCGGCTAGTTGTGCTCCG to substitute alanines for lysine-459 and phenylalanine-460. GFP-NrbI (286-1095, S461A) was also generated by site-directed mutagenesis with the oligonucleotide GCAAATAGGAAAATTAAGTTTGCTTGTGCTCCGATTAAGGTTTTC to substitute an alanine for serine-461.
HEK293 and Cos7 cells were maintained in Dulbecco modified Eagle medium (DMEM) containing 10% fetal bovine serum and penicillin-streptomycin-amphotericin B (Fungizone; Gibco-BRL) at 37°C in 5% CO2. DNA transfections used 6 μl of Lipofectamine (Gibco-BRL) and 1 μg of plasmid DNA. For microscopy, cells were grown on slides coated with poly-d-lysine. After 24 h, cells were washed twice with ice-cold PBS; fixed for 10 min in 4% (wt/vol) paraformaldehyde; and, when needed, permeabilized for 5 min with 0.2% Triton X-100 in PBS, blocked for 1 h with 4% bovine serum albumin in PBS, and incubated with rhodamine-conjugated phalloidin (Molecular Probes) at a 1:200 dilution. Cells were analyzed with a Zeiss LSM410 laser scanning confocal microscope with a krypton-argon laser.
To obtain primary hippocampal neurons, the CA1 region was dissected from the brains of postnatal 2- to 3-day-old rats, dispersed, and grown at a density of approximately 105 neurons in a 22-mm-diameter dish (13). After being cultured for 7 days in vitro (DIV), the neurons were transfected with 1 μl of Lipofectamine 2000 (Gibco-BRL) and 1.5 μg of plasmid DNA. After 24 h or more, cells were fixed with 4% (wt/vol) paraformaldehyde and 4% (wt/vol) sucrose and analyzed with a Zeiss Axioscope microscope.
For time-lapse video microscopy, HEK293 cells were plated on fibronectin-coated coverslips and transfected with 1 μg of plasmid DNA by using 3 μl of Fugene 6 (Roche) in serum-free DMEM. After 24 h, a coverslip was placed on the heated stage of a Zeiss Axiovert 135T inverted fluorescence microscope in a chamber containing 1 ml of phenol-red-free DMEM supplemented with 10% fetal bovine serum and 1× antibiotic-antimycotic mixture at 37.3°C. Images were captured every 150 s for 1 h with a Hamamatsu Orca II cooled charge-coupled device camera controlled by Isee software (Inovision).
Immunoprecipitation of GFP-NrbI.
HEK293 cells (106) washed with ice-cold PBS were lysed in 500 μl of RIPA buffer (1% NP-40, 0.5% deoxycholic acid, 0.1% sodium dodecyl sulfate [SDS], 150 mM NaCl, 50 mm Tris-HCl [pH 8.0], 1 mM PMSF, 1 mM benzamidine) on ice for 15 min. Lysates were subjected to centrifugation at 16,000 × g for 5 min at 4°C, and the supernatants were incubated with anti-GFP for 1 h at 4°C, followed by protein G-Sepharose for 1 h at 4°C. Beads were washed three times with NETN (10 mM Tris-HCl [pH 8.0], 100 mM NaCl, 0.5% NP-40, 1 mM EDTA) and boiled in SDS sample buffer prior to SDS-polyacrylamide gel electrophoresis (PAGE). Gels were transferred to a polyvinylidene difluoride membrane for immunoblotting.
Immune complex phosphatase assays.
GFP-NrbI immunoprecipitates adsorbed to protein G-Sepharose were washed in NETN buffer as described above followed by 50 mM Tris-HCl (pH 7.5)-1 mM EDTA-0.1% (vol/vol) β-mercaptoethanol and assayed for protein phosphatase activity with 32P-labeled phosphorylase a at 37°C for 30 min (26).
Metabolic labeling of HEK293 cells.
HEK293 cells expressing GFP-NrbI were incubated in DMEM lacking sodium phosphate and sodium pyruvate but containing 12.5 μCi of [32P]orthophosphoric acid for 2 h. Cells were stimulated with 100 μM Sp-cAMPS or Rp-cAMPS for 15 min, washed twice with ice-cold PBS, and lysed in 500 μl of RIPA buffer containing 50 mM sodium fluoride for 15 min on ice. Lysates were clarified by centrifugation at 16,000 × g for 5 min at 4°C. Supernatants were precleared with protein G-Sepharose (1 h at 4°C), which was removed by centrifugation at 1,000 × g for 3 min. The supernatants were incubated with anti-GFP for 1 h at 4°C, followed by protein G-Sepharose (1 h at 4°C). These beads were washed three times with NETN-250 (10 mM Tris-HCl [pH 8.0], 250 mM NaCl, 0.5% NP-40, 1 mM EDTA) and twice with NETN prior to the addition of SDS sample buffer and subjected to SDS-7.5% (wt/vol) PAGE and autoradiography.
F-actin binding assays.
F-actin binding was analyzed with the Actin Binding Protein Biochem Kit BK001. Briefly, His-NrbI (7 μM) was incubated with F-actin, polymerized in a polymerization buffer, for 30 min at 24°C and centrifuged through a sucrose cushion at 150,000 × g for 1.5 h at 24°C. Supernatants and pellets were subjected to SDS-10% (wt/vol) PAGE, and the proteins were stained with Coomassie blue.
Pull-downs from brain extracts.
Rat brain cortex (Pel-Freez) was homogenized with 2.5 volumes of 50 mM Tris-HCl (pH 7.5) containing 5 mM EDTA, 5 mM EGTA, 10 mM NaCl, 1% deoxycholate, 1 mM PMSF, 1 mM benzamidine, 1 μg of leupeptin/ml, and 1 μg of aprotinin/ml. The homogenate was centrifuged at 100,000 × g for 1 h, and the supernatant was dialyzed against 50 mM Tris-HCl (pH 7.5) containing 1 mM EDTA, 1 mM EGTA, 10 mM NaCl, 0.1 mM PMSF, and 0.1 mM benzamidine. Lysates (4 mg of total protein) were precleared with Ni+-NTA agarose before they were incubated with 20 μg of His-tagged NrbI bound to Ni+-NTA agarose for 1 h at 4°C. Beads were washed four times with PBS containing 5 mM imidazole, and bound proteins were eluted with 50 μl of SDS buffer for SDS-10% (wt/vol) PAGE and Western immunoblotting.
Pull-downs of rat brain extracts with microcystin-LR-Sepharose were performed as previously described (6).
In vitro phosphorylation of neurabins.
His-NrbI proteins (10 μM) were incubated with protein kinase A (PKA) purified from bovine heart (2) in 50 μl of 10 mM Tris-HCl (pH 7.5) containing 300 μM dithiothreitol, 1 mM MgCl2, 100 μM ATP, and 0.03 μCi of [γ-32P]ATP at 37°C. Aliquots (4 μl) were removed at intervals, and 50 μl of bovine serum albumin (10 mg/ml) and 200 μl of 20% trichloroacetic acid were added. Samples incubated at 4°C for 5 min were centrifuged at 13,000 × g for 5 min, and 32P incorporation into the trichloroacetic acid-precipitated fraction was determined by Cerenkov counting.
RESULTS
N-terminal F-actin-binding domain localizes NrbI to actin cytoskeleton.
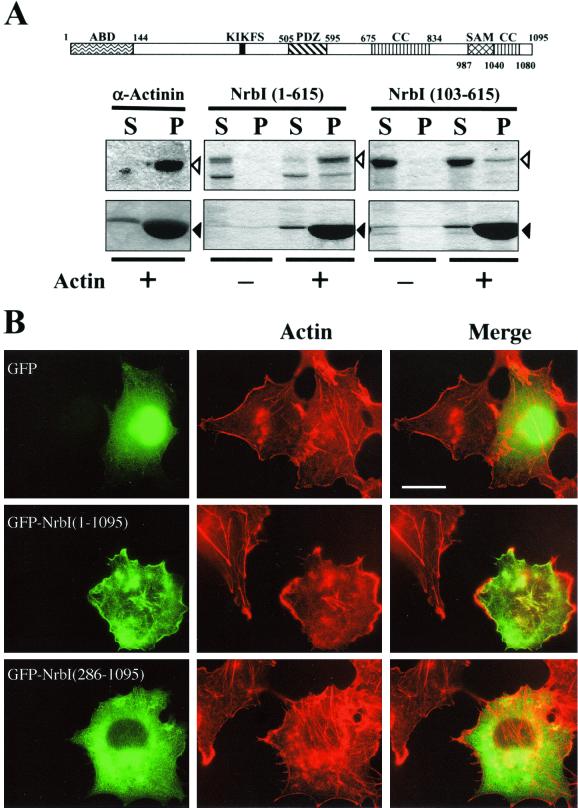
An N-terminal actin-binding region was identified in rat neurabins (20, 25) that was partly conserved in Xenopus laevis neurabin (accession no. AF363388) but absent from Caenorhabditis elegans (accession no. U80487) and Drosophila melanogaster neurabins (11). Thus, we analyzed hexahistidine-tagged recombinant rat NrbI polypeptides for in vitro F-actin binding (Fig. 1A). NrbI (1-615), like recombinant α-actinin, a known F-actin-binding protein, was completely sedimented with polymerized F-actin. In contrast, NrbI (103-615) was largely soluble, with a very small amount of NrbI (103-615) consistently bound to F-actin. Earlier work showed that an N-terminal fragment (144 amino acids) of rat NrbI bound F-actin in vitro (20). A longer NrbI (1-210) polypeptide, however, bound F-actin more effectively. Moreover, myc-tagged NrbI (145-1095) expressed in Cos7 cells was partially localized at the actin cytoskeleton (20). This suggested that more extended sequences may dictate NrbI localization with the cytoskeleton.
FIG. 1.
N-terminal domain localizes GFP-NrbI to actin cytoskeleton. (A) Domain structure of rat NrbI. The protein interaction domains highlighted include an actin-binding domain (ABD), the PSD95/Dlg/ZO-1 domain (PDZ), the coiled-coil domain (CC), and the SAM domain. The PP1-binding domain (black box) is characterized by a canonical KIKF sequence. Also shown is the association of His-tagged rat NrbI (1-615) and rat NrbI (103-615) in vitro with polymerized F-actin. α-Actinin, a known F-actin-binding protein, was used as a control. Proteins were incubated in the presence (+) or absence (−) of F-actin in the actin polymerization buffer (cytoskeleton). Samples were centrifuged at 150,000 × g through a sucrose cushion, and particulate (P) and supernatant (S) fractions were analyzed by SDS-10% (wt/vol) PAGE and stained with Coomassie blue. Open arrowheads indicate the test proteins, and closed arrowheads indicate actin. (B) Fluorescence micrographs of Cos7 cells expressing GFP, GFP-NrbI (1-1095), or GFP-NrbI (286-1095). Cells were fixed with paraformaldehyde following transfection, permeabilized, and stained with rhodamine-phalloidin. Bar, 30 μm.
We expressed GFP, GFP-NrbI (1-1095), and GFP-NrbI (286-1095) in Cos7 cells. GFP was widely distributed throughout the cell (Fig. 1B), and little overlap of GFP, seen as yellow, with F-actin-containing stress fibers or cortical actin cytoskeleton, stained by rhodamine-conjugated phalloidin, was seen. In contrast, full-length GFP-NrbI (1-1095) localized predominantly with actin-based structures, highlighted by the merged images. Interestingly, stress fibers in cells expressing GFP-NrbI (1-1095) were largely collapsed, as emphasized by actin staining in adjacent nontransfected cells. Treatment of cells with latrunculin B, an actin-depolymerizing agent, resulted in the redistribution of GFP-NrbI (1-1095) to diffuse intracellular sites, indicating that NrbI localization was dictated by the polymerized actin (data not shown). Similar results were obtained for HEK293 cells in which both GFP-NrbI (1-1095) and NrbI (1-1095)-GFP with GFP fused to the C terminus of NrbI induced a rounded morphology concomitant with the loss of stress fibers (data not shown). GFP-NrbI (286-1095), which lacked F-actin binding, was thus largely distributed to intracellular sites (Fig. 1B), and the cells retained their network of stress fibers. Neither GFP-NrbI (1-1095) nor GFP-NrbI (286-1095) entered the cell nucleus. Association with cytoskeleton was also confirmed by lysing cells in buffers containing 0.75% Triton X-100, which maintains an intact cytoskeleton (21), and centrifugation at 150,000 × g on a layer of 6% sucrose. Immunoblotting these fractions showed that GFP-NrbI (1-1095) bound exclusively to actin cytoskeleton, while both GFP and GFP-NrbI (286-1095) were soluble (data not shown). This established that F-actin binding was essential for both NrbI localization with the cytoskeleton and disassembly of stress fibers.
C-terminal sequences in NrbI mediate dimerization.
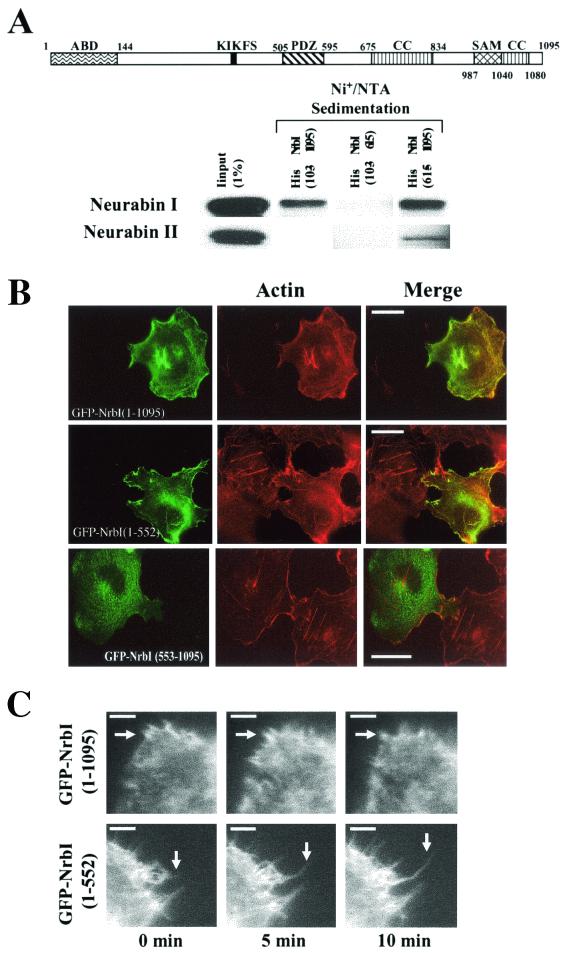
Gel filtration of recombinant neurabins (20, 25) and immunoprecipitation of individual neurabins from extracts (16) suggested that NrbI formed homodimers and heterodimers with NrbII. However, the structural basis for neurabin dimerization which potentially contributes to its in vitro actin-bundling activity (20) remained undefined. Studies of other proteins containing coiled-coil (14) and SAM (27) domains suggested these domains as excellent candidates for NrbI dimerization. Thus, we undertook pull-downs from rat brain extracts with His-tagged NrbI as bait (Fig. 2A). To ensure direct association of the recombinant proteins with brain neurabins, we deleted the N-terminal 103 amino acids representing the actin-binding domain (Fig. 1A). Both His-NrbI (103-1095) and His-NrbI (615-1095) bound a 190-kDa polypeptide representing rat NrbI and, to a lesser extent, also bound NrbII. His-NrbI (1-615) failed to sediment either rat neurabin, providing the first direct evidence that C-terminal sequences in NrbI mediated homodimerization and heterodimerization.
FIG. 2.
C-terminal sequences mediate dimerization of NrbI. The NrbI structure is schematically shown as in Fig. 1. (A) Proteins sedimented from rat brain deoxycholate extracts by 20 μg of His-NrbI (103-1095), His-NrbI (103-615), and His-NrbI (615-1095) bound to Ni+-NTA agarose were subjected to SDS-10% (wt/vol) PAGE and immunoblotted with anti-NrbI and anti-NrbII antibodies. (B) Fluorescence micrographs of Cos7 cells expressing GFP-NrbI (1-1095), GFP-NrbI (1-552), or GFP-NrbI (553-1095). Cells were fixed with paraformaldehyde following transfection, permeabilized, and stained with rhodamine-phalloidin. Bars, 30 μm. (C) Time-lapse video microscopy was undertaken for HEK293 cells expressing GFP-NrbI (1-1095) or GFP-NrbI (1-552) grown on fibronectin-coated coverslips. Images are shown at 5-min intervals with a white arrow highlighting an extending filopodium. Bars, 10 μm.
To investigate the importance of NrbI dimerization, we expressed GFP-NrbI (1-552) and GFP-NrbI (553-1095) in Cos7 cells. As noted above, full-length GFP-NrbI (1-1095) localized with the actin cytoskeleton and resulted in collapse of stress fibers. GFP-NrbI (1-552) also localized with the actin cytoskeleton, and its distribution at the cell periphery was even more prominent (Fig. 2B). This was confirmed by biochemical fractionation as described above (data not shown). Cells expressing GFP-NrbI (1-552) not only demonstrated the loss of stress fibers but also projected extensive filopodia. GFP-NrbI (553-1095), representing the isolated dimerization domain, largely failed to localize with the cytoskeleton and was distributed in the cytoplasm. Cells expressing GFP-NrbI (553-1095) retained an intact stress fiber network.
GFP-NrbI (1-552) induced even more striking filopodia in HEK293 cells (Fig. 2C). Time-lapse video microscopy of living cells focused on the GFP-NrbI-containing filopodia seen in cells expressing GFP-NrbI (1-1095) and GFP-NrbI (1-552). Expression of wild-type (WT) GFP-NrbI (1-1095) resulted in the projection of short, highly dynamic filopodia that failed to extend further over the 10-min time frame. In contrast, not only did GFP-NrbI (1-552)-induced filopodia continue to extend over this time period (arrows in Fig. 2C) but also many of these projections remained stable for 60 min. These studies suggested that the dynamics of NrbI dimerization dictated its effects on cell morphology, promoting dissolution of stress fibers and filopodium extension.
PP1 recruitment required for NrbI-induced filopodia and stress fiber disassembly.
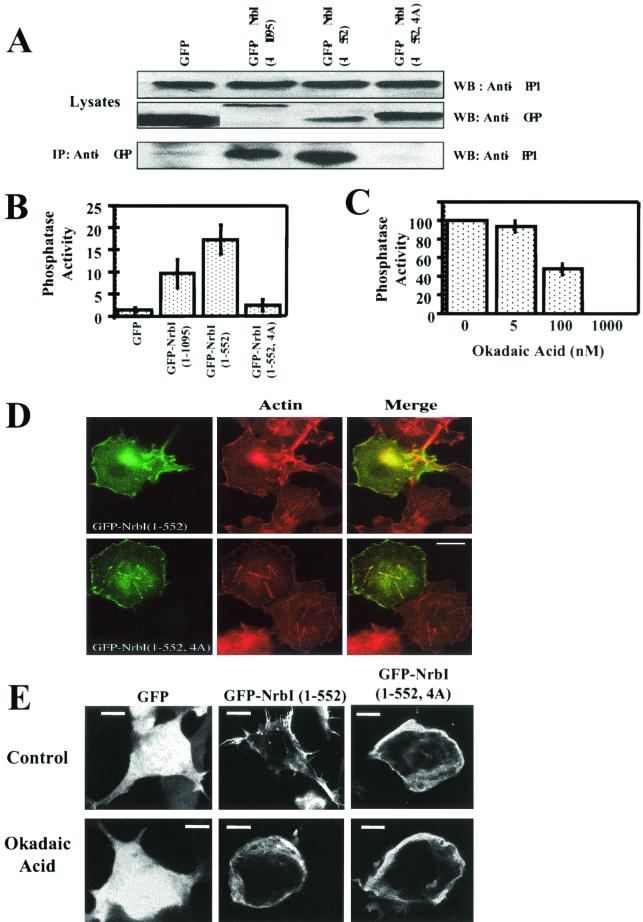
GFP, GFP-NrbI (1-1095), and GFP-NrbI (1-552) expression in HEK293 cells had no effect on overall PP1 levels (Fig. 3A). Immunoprecipitation with anti-GFP monoclonal antibody showed that, despite its much higher expression, GFP alone bound only a trace of PP1 (Fig. 3A). In contrast, immunoprecipitates containing GFP-NrbI (1-1095) and GFP-NrbI (1-552) contained readily detectable PP1. Quantitative analysis of the immunoblots suggested that GFP-NrbI (1-552) bound PP1 more effectively than did WT GFP-NrbI (1-1095) (data not shown). Substituting alanines for 457KIKF460, a proposed PP1-binding motif, eliminated PP1 association with GFP-NrbI (1-552, 4A).
FIG. 3.
PP1 is required for NrbI (1-552)-induced cytoskeletal rearrangement. (A) Lysates (approximately 5% of input used in immunoprecipitations) of HEK293 cells expressing GFP, GFP-NrbI (1-1095), GFP-NrbI (1-552), or GFP-NrbI (1-552, 4A) were immunoblotted with anti-PP1 and anti-GFP. Cells (106) were lysed in RIPA buffer and clarified, and GFP-NrbI was immunoprecipitated with anti-GFP. The immunoprecipitates were subjected to SDS-10% (wt/vol) PAGE and analyzed by immunoblotting for PP1. IP, immunoprecipitation; WB, Western blot. (B) Phosphorylase a phosphatase activity associated with the washed GFP-NrbI immunoprecipitates. Phosphatase activity is shown as nanomoles of [32P]phosphate released by equivalent amounts of immunoprecipitates, quantified by immunoblotting with anti-GFP. The data represent averages of three independent experiments performed in duplicate (shown with standard error bars). (C) Sensitivity of the immunoprecipitated GFP-NrbI (1-552)-bound phosphatase activity to okadaic acid, represented as a percentage of that for the control without okadaic acid. The data represent the averages of three independent experiments and are shown with standard error bars. (D) Fluorescence micrographs of Cos7 cells expressing either GFP-NrbI (1-552) or GFP-NrbI (1-552, 4A). Cells were fixed with paraformaldehyde following transfection, permeabilized, and stained with rhodamine-phalloidin. Bar, 20 μm. (E) HEK293 cells expressing GFP, GFP-NrbI (1-552), or GFP-NrbI (1-552, 4A) treated with or without okadaic acid (1 μM) for 15 min prior to fixation. Bars, 25 μm.
Anti-GFP immunoprecipitates were also assayed for PP1 activity with the standard substrate, phosphorylase a. Extensive washes of anti-GFP immunoprecipitates with either NETN or assay buffer failed to release PP1 (data not shown), and the immune complex assays established for the first time that the NrbI-bound PP1 is an active protein phosphatase. When corrected for equivalent amounts of protein, immunoprecipitates containing GFP-NrbI (1-552) consistently contained 50 to 60% more phosphatase activity than did GFP-NrbI (1-1095) (Fig. 3B). As anticipated, immunoprecipitates of GFP-NrbI (1-552, 4A) possessed little phosphatase activity, similar to GFP alone. These data showed that PP1 utilized the 457KIKF460 sequence for NrbI binding and bound GFP-NrbI (1-552) more effectively than GFP-NrbI (1-1095).
To confirm the identity of the NrbI-bound phosphatase, immune complex assays were performed with GFP-NrbI (1-552) in the presence of increasing concentrations of okadaic acid (Fig. 3C). Okadaic acid (5 nM), sufficient to inhibit most PP2A-like phosphatases, had no effect on GFP-NrbI (1-552)-bound phosphatase, and concentrations exceeding 100 nM, up to 1 μM, were needed to fully inhibit the enzyme, consistent with its being exclusively PP1 (5).
As indicated above, GFP-NrbI (1-552) expression collapsed stress fibers and induced filopodia in Cos7 cells (Fig. 3D). In contrast, the non-PP1-binding GFP-NrbI (1-552, 4A), while still localized to the actin cytoskeleton, failed to induce filopodia, and stress fibers remained intact in these cells. Similarly, in HEK293 cells, GFP-NrbI (1-552) elicited striking filopodia (Fig. 3E), which were completely eliminated in GFP-NrbI (1-552, 4A)-expressing cells, which possessed a rounded morphology similar to cells expressing WT GFP-NrbI (1-1095) (data not shown). Treatment of GFP-NrbI (1-552)-expressing HEK293 cells with the phosphatase inhibitor okadaic acid (1 μM) (Fig. 3E) or calyculin A (10 nM) rapidly eliminated the filopodia. The brief (15-min) treatment with okadaic acid or calyculin A had no effect on the morphology of cells expressing GFP or GFP-NrbI (1-552, 4A). This indicated that the loss of PP1 binding or inhibition of phosphatase activity had very similar effects on cell morphology and established the importance of PP1 recruitment for NrbI-induced changes in morphology in both Cos7 and HEK293 cells.
Neurabin is a phosphoprotein in cells.
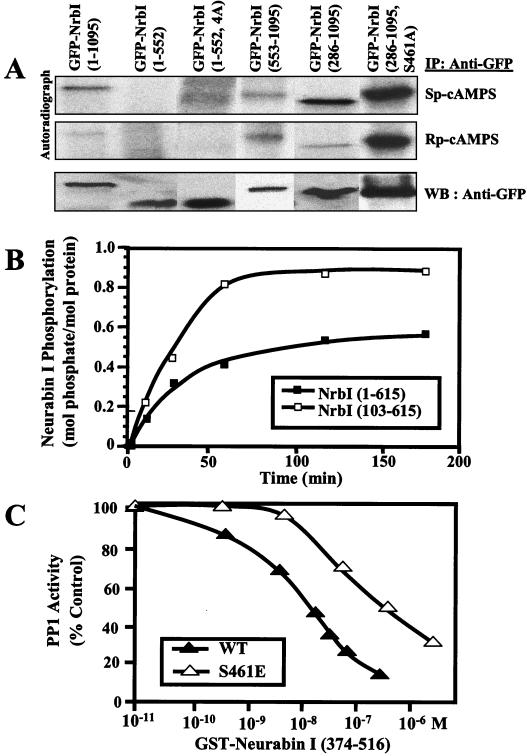
While in vitro studies showed PKA phosphorylation of NrbI (17), the phosphorylation of NrbI in vivo in response to PKA agonists had not been investigated. HEK293 cells expressing GFP-NrbI proteins were metabolically labeled with [32P]orthophosphate, and cells were treated with the PKA agonist Sp-cAMPS or the antagonist Rp-cAMPS (Fig. 4A). Immunoprecipitation and autoradiography established that GFP-NrbI (1-1095) was a phosphoprotein in vivo and that its phosphorylation was enhanced by activation of PKA. By comparison, GFP-NrbI (1-552) was essentially unphosphorylated (Fig. 4A). PP1 binding did not dictate NrbI dephosphorylation, as GFP-NrbI (1-552, 4A), which fails to bind PP1, was also unphosphorylated in the presence of either a PKA agonist or an antagonist. Treatment of cells with 1 μM okadaic acid to inhibit PP1 activity increased cellular protein phosphorylation but failed to promote the phosphorylation of GFP-NrbI (1-552) or GFP-NrbI (1-552, 4A) (data not shown). The inability to phosphorylate GFP-NrbI (1-552) was not due to elimination of the major PKA sites in NrbI, as phosphorylation of GFP-NrbI (553-1095), which showed some basal incorporation of [32P]phosphate (in the presence of Rp-cAMPS), was not elevated following PKA activation (Fig. 4A). Remarkably, GFP-NrbI (286-1095), which lacked the N-terminal actin-binding domain, was more highly phosphorylated than was WT GFP-NrbI (1-1095) in the presence of Sp-cAMPS (Fig. 4A) and mutation of serine-461, a major PKA site identified in vitro (17), to alanine abolished PKA-induced phosphorylation of GFP-NrbI (286-1095, S461A) in HEK293 cells. This suggested that the actin-binding domain and/or NrbI localization at the cytoskeleton suppressed NrbI phosphorylation by PKA on serine-461.
FIG. 4.
Phosphorylation of NrbI in HEK293 cells. (A) Cells expressing GFP-NrbI (1-1095), GFP-NrbI (1-552), GFP-NrbI (1-552, 4A), GFP-NrbI (553-1095), GFP-NrbI (286-1095), or GFP-NrbI (286-1095, S461A) were metabolically labeled with [32P]orthophosphoric acid for 120 min at 37°C and subsequently treated for 15 min with a PKA agonist, Sp-cAMPS (100 μM), or an antagonist, Rp-cAMPS (100 μM). Cells were lysed in RIPA buffer and clarified, and GFP-NrbI was immunoprecipitated with anti-GFP. Immunoprecipitates were subjected to SDS-7.5% (wt/vol) PAGE, and the gels were dried and autoradiographed. The bottom panel shows relative levels of each GFP-NrbI protein in the immunoprecipitates by immunoblotting with anti-GFP. IP, immunoprecipitation; WB, Western blot. (B) In vitro phosphorylation of His-NrbI (1-615) and His-NrbI (103-615) by PKA and [γ-32P]ATP. The data are representative of three independent experiments. (C) Inhibition of PP1 activity with phosphorylase a as substrate in the presence of increasing concentrations of WT GST-NrbI (374-516) and the mutant S461E. The data are representative of three independent experiments. PP1 activity is shown as a percentage of that for the control without GST-NrbI.
To elucidate the direct role of the actin-binding domain in PKA phosphorylation of NrbI, we analyzed in vitro phosphorylation of recombinant NrbI (1-615) and NrbI (103-615) lacking in vitro actin binding (Fig. 1A) by purified PKA. NrbI (1-615) was phosphorylated in a time-dependent manner and incorporated approximately 0.5 mol of phosphate per mol of protein (Fig. 4B). Deletion of the actin-binding domain enhanced PKA phosphorylation, and NrbI (103-615) incorporated approximately twofold-more [32P]phosphate. This demonstrated that the actin-binding domain suppressed NrbI phosphorylation by PKA. Mutation of serine-461 to alanine confirmed that this was a major PKA site in vitro (R. T. Terry-Lorenzo and S. Shenolikar, unpublished observation).
Serine-461 is located immediately adjacent to 457KIKF460, the core PP1-docking site, and as seen with other PP1 regulators, its phosphorylation modified PP1 binding (17). In the absence of stoichiometric phosphorylation of NrbI in vivo, we were unable to demonstrate reduced PP1 binding to either GFP-NrbI (1-1095) or GFP-NrbI (286-1095) following PKA activation. In vitro analysis was also confounded by the dephosphorylation of PKA-phosphorylated NrbI by PP1. Thus, we mutated serine-461 to glutamic acid to mimic PKA phosphorylation in recombinant His-tagged NrbI (374-516). NrbI (374-516) inhibited PP1 activity in a dose-dependent manner with a 50% inhibitory concentration of approximately 10 nM. Mutation of serine-461 to glutamate attenuated the ability of NrbI (374-516, S461E) to inhibit PP1, right-shifting the dose-response curve by approximately 10-fold (Fig. 4C). This confirmed that NrbI phosphorylation at serine-461 modified PP1 binding and/or regulation (17).
PP1 precludes p70S6K binding to NrbI.
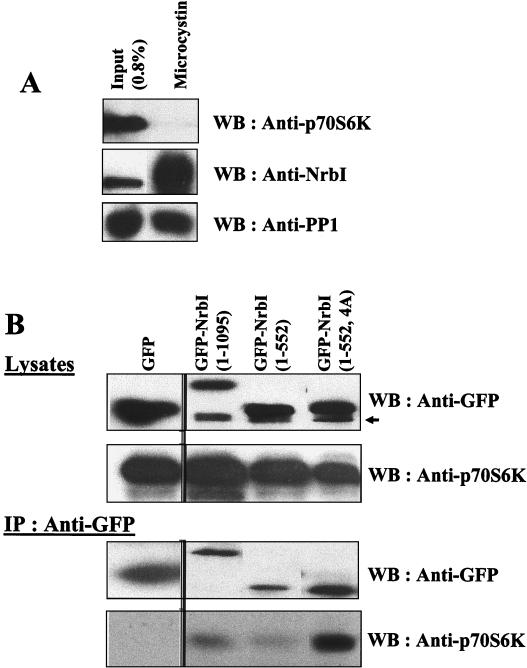
In addition to PP1, NrbI, via an adjacent PDZ domain, binds p70S6K (4), which may oppose PP1 in regulating neurite outgrowth (23) and cytokinesis (31). However, affinity isolation of PP1 complexes from rat brain extracts by using the immobilized PP1 inhibitor microcystin-LR quantitatively extracted NrbI, but the complexes contained no detectable p70S6K (Fig. 5A). We also coexpressed hemagglutinin (HA)-tagged p70S6K with GFP-NrbI proteins in HEK293 cells. Anti-GFP immunoprecipitates showed PP1 binding to both GFP-NrbI (1-1095) and GFP-NrbI (1-552) (Fig. 3A), but no significant p70S6K association was observed with either protein (Fig. 5B). Similarly, anti-PP1 immunoprecipitates contained the two GFP-NrbI proteins (data not shown), but anti-p70S6K or anti-HA antibodies that effectively sedimented HA-tagged p70S6K from the transfected cells failed to show any association with either NrbI or PP1 (data not shown). This suggested that the majority of cellular NrbI complexes in tissue, as well as the cultured cells, contained PP1 and excluded p70S6K. Evidence for potential competition between PP1 and p70S6K for NrbI binding came from the immunoprecipitation of GFP-NrbI (1-552, 4A), which failed to bind PP1. The non-PP1-binding mutant NrbI (1-552, 4A) sedimented significant amounts of p70S6K, readily detectable by immunoblotting with anti-HA or anti-p70S6K antibodies (Fig. 5B). Neither p70S6K overexpression nor treatment of cells with rapamycin to inhibit p70S6K activation had any effect on cell morphology (data not shown). Thus, the function of the NrbI/p70S6K complex is currently unknown, but our data suggest that PP1 displacement by mechanisms such as PKA phosphorylation at serine-461 may promote the formation of NrbI/p70S6K complexes and that the competition between PP1 and p70S6K for NrbI binding could play a role in cytoskeletal dynamics.
FIG. 5.
PP1 precludes NrbI binding to p70S6K. (A) Immunoblots of proteins from rat brain deoxycholate extracts (input) bound to microcystin-LR-Sepharose. The bound proteins were subjected to SDS-10% (wt/vol) PAGE and immunoblotted with anti-NrbI and anti-PP1 antibodies. (B) HEK293 cells expressing HA-tagged p70S6K and either GFP, GFP-NrbI (1-1095), GFP-NrbI (1-552), or GFP-NrbI (1-552, 4A) were lysed in RIPA buffer, lysates were clarified by centrifugation, and GFP-NrbI was immunoprecipitated with anti-GFP. Immunoprecipitates were subjected to SDS-10% (wt/vol) PAGE and immunoblotted with anti-GFP and anti-p70S6K antibodies. Cell lysates (5% of input) were also blotted with anti-GFP and anti-p70S6K. The arrow indicates a nonspecific immunoreactive band. IP, immunoprecipitation; WB, Western blot.
NrbI overexpression induces filopodia and dendritic spines in cultured hippocampal neurons.
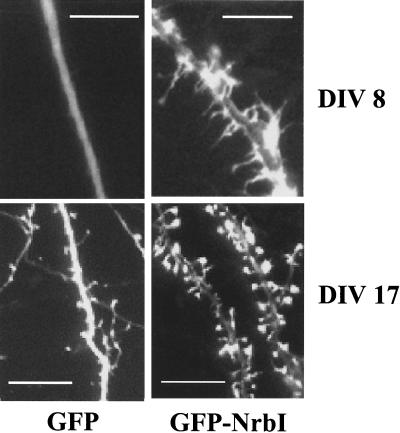
To assess the relevance of NrbI in dictating neuronal morphology, we overexpressed GFP-NrbI (1-1095) in 7-DIV-cultured hippocampal neurons, which express both neurabin isoforms. Compared to that of GFP alone, GFP-NrbI (1-1095) expression induced numerous filopodia by 8 DIV (Fig. 6). Unlike GFP, which was distributed throughout the cell, GFP-NrbI (1-1095) was localized preferentially within the dendritic projections. Following 17 DIV, GFP-NrbI (1-1095)-expressing neurons developed numerous large spines with GFP-NrbI (1-1095) concentrated within these structures. These data suggested that neuronal NrbI complexes containing PP1 played a role in the development of filopodia and their subsequent maturation into dendritic spines in neurons.
FIG. 6.
Role for NrbI in spine development. Cultured rat hippocampal neurons were cultured for 7 DIV prior to transfection with the plasmid expressing either GFP or WT GFP-NrbI (1-1095). The following day (DIV 8) extensive filopodia were induced only in the NrbI-expressing neurons, with GFP-NrbI localized within these structures. Following DIV 17, spine development was observed in all neurons with GFP distributed throughout the cell. In contrast, NrbI overexpression resulted in the development of numerous and much larger spines, with the GFP-NrbI being highly concentrated in these structures (bars, 5 μm).
DISCUSSION
Rapid growth of filopodia is observed in dendrites during the development of the nervous system and following increased synaptic activity. While some filopodia subsequently retract, others proceed to form new spines and synapses. In adult neurons, even mature spines undergo dynamic changes in shape that are mediated by actin rearrangement, and pharmacological agents that promote actin depolymerization result in serious deficits in synaptic efficacy (15). We have analyzed NrbI-bound PP1 as a potential convergence point for signals that regulate both synaptic plasticity and spine morphology. As in vitro studies of NrbI are limited by the inability to express full-length NrbI (4, 17, 20), a key goal of our studies was to establish a cell-based assay that exploited in vitro structure-function analyses of NrbI to yield new insights into the physiological role of the NrbI/PP1 complex in the mammalian nervous system.
WT GFP-NrbI (1-1095) bound polymerized F-actin both in vitro and in vivo and modified the morphology of HEK293 and Cos7 cells to reorganize the stress fiber network. GFP-NrbI (286-1095), which lacked actin binding, was cytosolic and had little effect on cell morphology, thereby establishing that actin binding was critical for cytoskeletal reorganization by NrbI. Previous studies showed that NrbI promoted in vitro bundling or cross-linking of actin fibers (20). As ectopically expressed NrbI and NrbII formed homodimers and heterodimers (16), this raised the possibility that the presentation of two adjacent actin-binding domains in a dimeric NrbI may promote actin bundling. Thus, we deleted the C-terminal coiled-coil and SAM domains, shown elsewhere for other proteins to self-associate (14, 27), and by using pull-downs of WT NrbI and NrbII from rat brain extracts established that homodimerization and heterodimerization of neurabins are mediated via their C termini. The monomeric GFP-NrbI (1-552) induced dramatic changes in cell morphology, disrupting the stress fiber network and promoting highly extended filopodia. Live video microscopy showed the rapid growth of filopodia in the GFP-NrbI (1-552)-expressing cells that extended significantly further than those in cells expressing WT GFP-NrbI (1-1095). The NrbI C terminus (amino acids 553 to 1095), while sufficient to bind NrbI (and NrbII), did not localize with the actin cytoskeleton or modify cell morphology. Cell fractionation and immunoprecipitation studies suggested that GFP-NrbI (1-552) bound both F-actin and PP1 more effectively than did WT GFP-NrbI (1-1095), hinting that NrbI dimerization may attenuate its actin- and PP1-binding properties.
The importance of PP1 recruitment for NrbI-induced morphology was established by mutating the core PP1-binding sequence, KIKF, in NrbI to alanines. This not only abolished PP1 binding in vitro (data not shown) and in vivo, but unlike GFP-NrbI (1-552), GFP-NrbI (1-552, 4A) failed to induce filopodia in HEK293 and Cos7 cells or disrupt stress fibers in Cos7 cells. Pharmacological inhibition of PP1 activity by okadaic acid or calyculin A rapidly collapsed filopodia induced by GFP-NrbI (1-552), showing that, in addition to binding F-actin, NrbI must recruit an active PP1 to modify cell morphology. Immune complex assays confirmed that the NrbI-bound PP1 was an active protein phosphatase. This was unexpected, as in vitro studies (26, 27) (Fig. 4C) showed that PP1-binding NrbI peptides inhibit its activity against phosphorylase a. The molecular basis for the difference in the NrbI/PP1 complexes assembled in vitro and in vivo is unclear, but the difference may reflect the presence of additional proteins or covalent modifications of NrbI in cells. In any case, our data supported an active role for the NrbI/PP1 complex in cytoskeletal reorganization.
PP1 binds both substrates (8, 10) and regulators (22) that undergo reversible phosphorylation. Covalent modification of PP1 regulators modulates PP1 binding (7) and activity (12). Analysis of metabolically labeled HEK293 cells established for the first time that GFP-NrbI (1-1095) was a phosphoprotein and that its phosphorylation in vivo was stimulated by the PKA agonist Sp-cAMPS. Surprisingly, GFP-NrbI (1-552) was essentially unphosphorylated in HEK293 cells. This was not due to elimination of PKA sites, as GFP-NrbI (553-1095) also showed low basal phosphorylation that was not further stimulated by Sp-cAMPS. Deletion of PP1 binding or inhibition of phosphatase activity failed to enhance GFP-NrbI (1-552) phosphorylation, suggesting that PP1 binding did not dictate the phosphorylation state of NrbI. Compared to WT GFP-NrbI (1-1095), soluble GFP-NrbI (286-1095) incorporated much higher levels of [32P]phosphate that was greatly enhanced by PKA activation. This raised the possibility that the actin-binding domain and/or association with the cytoskeleton inhibited NrbI phosphorylation by PKA. This was confirmed by in vitro studies that showed that NrbI (103-615) lacking N-terminal actin-binding sequences incorporated twofold-higher levels of phosphate than did NrbI (1-615). This did not, however, fully explain the absence of phosphate in GFP-NrbI (1-552), leading us to speculate that F-actin association may further impair NrbI phosphorylation in cells. Mutation of serine-461 to alanine in GFP-NrbI (286-1095) abolished PKA-stimulated phosphorylation and identified the serine immediately adjacent to the PP1-binding motif as a major in vivo PKA phosphorylation site. Analysis of recombinant NrbI (374-516, S461E), in which glutamic acid was introduced to mimic serine-461 phosphorylation, confirmed an earlier observation (17) that serine-461 modification diminished PP1 association with NrbI and suggested that assembly-disassembly of the NrbI/PP1 complex may be regulated by hormones that elevate intracellular cyclic AMP.
The PDZ domain adjacent to the core PP1-binding site recruits p70S6K, which has effects opposite of those of PP1 on actin dynamics (23). Affinity isolation of NrbI/PP1 complexes from rat brain or NrbI immunoprecipitation from HEK293 cells overexpressing p70S6K failed to show significant association of p70S6K with NrbI. This demonstrated that NrbI had a significant preference for PP1 over p70S6K and predicted that disruption of PP1 binding would facilitate p70S6K recruitment. Indeed, GFP-NrbI (1-552, 4A) that failed to bind PP1 showed enhanced p70S6K binding. These data can be summarized in the following model for the potential role of NrbI in transducing signals that regulate actin cytoskeleton (Fig. 7). Our data suggest that the majority of cellular NrbI is bound to PP1 and is associated with the actin cytoskeleton. This promotes the disassembly of stress fibers. While both dimeric and monomeric forms of NrbI induce filopodia in cultured cells, the enhanced ability by cells expressing monomeric NrbI to extend surface projections leads us to speculate that the dynamics of NrbI dimerization may also play a role in dictating the cell morphology. Physiological signals that activate Rac1 GTPase (29) promote NrbI shuttling to the cytoskeleton and may target PP1 to the actin cytoskeleton. In contrast, the cytosolic NrbI, by virtue of being an improved substrate for PKA (at serine-461), may display decreased PP1 binding and recruit p70S6K and other targets to the NrbI PDZ domain. This mechanism may also account for the antagonism between PP1 and p70S6K that regulates neuronal morphology. As neither p70S6K overexpression nor inhibition of p70S6K activity by rapamycin had any effect on HEK293 cell morphology, we were unable to establish a role for the NrbI-bound p70S6K in heterologous cells.
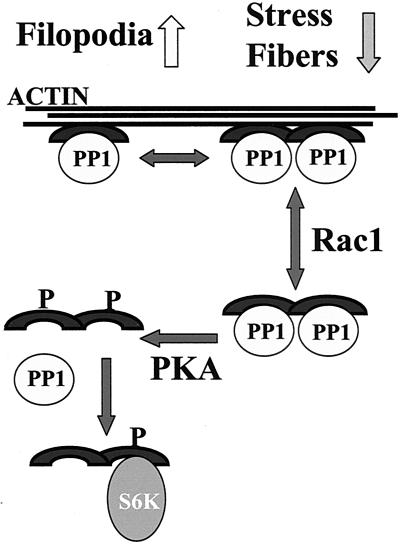
FIG. 7.
Model for the regulation of cell morphology by the NrbI/PP1 complex. Our experiments propose the following model for the regulation of the neuronal cytoskeleton by the NrbI/PP1 complex. NrbI function may be regulated at many steps including reversible dimerization of the actin- and PP1-bound NrbI, which may control the efficacy of filopodium extension and stress fiber disassembly. The NrbI/PP1 complex may also shuttle on and off the cytoskeleton in response to growth factor signals that activate Rac1 (29). We speculate that the enhanced phosphorylation of soluble NrbI by PKA at serine-461 displaces PP1, allowing recruitment of p70S6K and other targets of the NrbI PDZ domain to oppose PP1 in regulating actin rearrangement and cell morphology.
NrbI is expressed exclusively in neurons, while NrbII is more widely expressed. Both neurabins bind an overlapping set of targets that include PP1. Thus, disruption of the mouse NrbII/spinophilin gene that results in defective PP1 signaling and increases spine formation in the hippocampus of young NrbII-null mice (9) may represent not only the loss of NrbII/PP1 complexes but also the uncontested activity of NrbI/PP1. By overexpressing WT GFP-NrbI (1-1095), we artificially tipped the balance in favor of NrbI/PP1 complexes in cultured hippocampal neurons and induced filopodia, which subsequently matured into spines with GFP-NrbI (1-1095) localized within these structures. This suggested an active role of NrbI/PP1 in spine formation in developing neurons and may explain the abundance of spines seen in the NrbII-null mouse (9). In conclusion, our work has highlighted several key structural determinants and protein interactions required for the regulation of actin cytoskeleton by a NrbI/PP1 complex and thus set the stage for future studies that will elucidate the physiological role of the NrbI/PP1 complex in neurons.
Acknowledgments
We thank M. D. Ehlers and T. A. Blanpied (Duke University) for their help in culturing rat hippocampal neurons and Kathy Reaves for technical assistance.
The work was supported by NIH grants DK52054 and NS41063 (to S.S.), NS37508 (to R.J.C.), and CA40042 and GM56362 (to D.L.B.). C.J.O. is supported by a predoctoral fellowship from the Department of Defense Breast Cancer Program (DAMD17-18-1-8075), and R.T.T.-L. is supported by a predoctoral fellowship from the National Science Foundation.
REFERENCES
- 1.Allen, P. B., C. C. Ouimet, and P. Greengard. 1997. Spinophilin, a novel protein phosphatase 1 binding protein localized to dendritic spines. Proc. Natl. Acad. Sci. USA 94:9956-9961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beavo, J. A., P. J. Bechtel, and E. G. Krebs. 1974. Preparation of homogeneous cyclic AMP-dependent protein kinase(s) and its subunits from rabbit skeletal muscle. Methods Enzymol. 38:299-308. [DOI] [PubMed] [Google Scholar]
- 3.Blitzer, R. D., J. H. Connor, G. P. Brown, T. Wong, S. Shenolikar, R. Iyengar, and E. M. Landau. 1998. Gating of CaMKII by cAMP-regulated protein phosphatase activity during LTP. Science 280:1940-1942. [DOI] [PubMed] [Google Scholar]
- 4.Burnett, P. E., S. Blackshaw, M. M. Lai, I. A. Qureshi, A. F. Burnett, D. M. Sabatini, and S. H. Snyder. 1998. Neurabin is a synaptic protein linking p70 S6 kinase and the neuronal cytoskeleton. Proc. Natl. Acad. Sci. USA 95:8351-8356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cohen, P., C. F. Holmes, and Y. Tsukitani. 1990. Okadaic acid: a new probe for the study of cellular regulation. Trends Biochem. Sci. 15:98-102. [DOI] [PubMed] [Google Scholar]
- 6.Connor, J. H., D. C. Weiser, S. Li, J. M. Hallenbeck, and S. Shenolikar. 2001. Growth arrest and DNA damage-inducible protein GADD34 assembles a novel signaling complex containing protein phosphatase 1 and inhibitor 1. Mol. Cell. Biol. 21:6841-6850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dent, P., D. G. Campbell, F. B. Caudwell, and P. Cohen. 1990. Identification of three in vivo phosphorylation sites on the glycogen-binding subunit of protein phosphatase 1 from rabbit skeletal muscle, and their response to adrenaline. FEBS Lett. 259:281-285. [DOI] [PubMed] [Google Scholar]
- 8.Durfee, T., K. Becherer, P. L. Chen, S. H. Yeh, Y. Yang, A. E. Kilburn, W. H. Lee, and S. J. Elledge. 1993. The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev. 7:555-569. [DOI] [PubMed] [Google Scholar]
- 9.Feng, J., Z. Yan, A. Ferreira, K. Tomizawa, J. A. Liauw, M. Zhuo, P. B. Allen, C. C. Ouimet, and P. Greengard. 2000. Spinophilin regulates the formation and function of dendritic spines. Proc. Natl. Acad. Sci. USA 97:9287-9292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Helps, N. R., X. Luo, H. M. Barker, and P. T. Cohen. 2000. NIMA-related kinase 2 (Nek2), a cell-cycle-regulated protein kinase localized to centrosomes, is complexed to protein phosphatase 1. Biochem. J. 349:509-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keegan, J., M. Schmerer, B. Ring, and D. Garza. 2001. The 62E early-late puff of Drosophila contains D-spinophilin, an ecdysone-inducible PDZ-domain protein dynamically expressed during metamorphosis. Genet. Res. 77:27-39. [DOI] [PubMed] [Google Scholar]
- 12.Kimura, K., M. Ito, M. Amano, K. Chihara, Y. Fukata, M. Nakafuku, B. Yamamori, J. Feng, T. Nakano, K. Okawa, A. Iwamatsu, and K. Kaibuchi. 1996. Regulation of myosin phosphatase by Rho and Rho-associated kinase (Rho-kinase). Science 273:245-248. [DOI] [PubMed] [Google Scholar]
- 13.Liao, D., X. Zhang, R. O'Brien, M. D. Ehlers, and R. L. Huganir. 1999. Regulation of morphological postsynaptic silent synapses in developing hippocampal neurons. Nat. Neurosci. 2:37-43. [DOI] [PubMed] [Google Scholar]
- 14.Lupas, A. 1996. Coiled coils: new structures and new functions. Trends Biochem. Sci. 21:375-382. [PubMed] [Google Scholar]
- 15.Luscher, C., R. A. Nicoll, R. C. Malenka, and D. Muller. 2000. Synaptic plasticity and dynamic modulation of the postsynaptic membrane. Nat. Neurosci. 3:545-550. [DOI] [PubMed] [Google Scholar]
- 16.MacMillan, L. B., M. A. Bass, N. Cheng, E. F. Howard, M. Tamura, S. Strack, B. E. Wadzinski, and R. J. Colbran. 1999. Brain actin-associated protein phosphatase 1 holoenzymes containing spinophilin, neurabin, and selected catalytic subunit isoforms. J. Biol. Chem. 274:35845-35854. [DOI] [PubMed] [Google Scholar]
- 17.McAvoy, T., P. B. Allen, H. Obaishi, H. Nakanishi, Y. Takai, P. Greengard, A. C. Nairn, and H. C. Hemmings, Jr. 1999. Regulation of neurabin I interaction with protein phosphatase 1 by phosphorylation. Biochemistry 38:12943-12949. [DOI] [PubMed] [Google Scholar]
- 18.Morishita, W., J. H. Connor, H. Xia, E. M. Quinlan, S. Shenolikar, and R. C. Malenka. 2001. Regulation of synaptic strength by protein phosphatase 1. Neuron 32:1133-1148. [DOI] [PubMed] [Google Scholar]
- 19.Mulkey, R. M., S. Endo, S. Shenolikar, and R. C. Malenka. 1994. Involvement of a calcineurin/inhibitor-1 phosphatase cascade in hippocampal long-term depression. Nature 369:486-488. [DOI] [PubMed] [Google Scholar]
- 20.Nakanishi, H., H. Obaishi, A. Satoh, M. Wada, K. Mandai, K. Satoh, H. Nishioka, Y. Matsuura, A. Mizoguchi, and Y. Takai. 1997. Neurabin: a novel neural tissue-specific actin filament-binding protein involved in neurite formation. J. Cell Biol. 139:951-961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nixon, J. B., and L. C. McPhail. 1999. Protein kinase C (PKC) isoforms translocate to Triton-insoluble fractions in stimulated human neutrophils: correlation of conventional PKC with activation of NADPH oxidase. J. Immunol. 163:4574-4582. [PubMed] [Google Scholar]
- 22.Park, I. K., P. Roach, J. Bondor, S. P. Fox, and A. A. DePaoli-Roach. 1994. Molecular mechanism of the synergistic phosphorylation of phosphatase inhibitor-2. Cloning, expression, and site-directed mutagenesis of inhibitor-2. J. Biol. Chem. 269:944-954. [PubMed] [Google Scholar]
- 23.Parker, E. M., A. Monopoli, E. Ongini, G. Lozza, and C. M. Babij. 2000. Rapamycin, but not FK506 and GPI-1046, increases neurite outgrowth in PC12 cells by inhibiting cell cycle progression. Neuropharmacology 39:1913-1919. [DOI] [PubMed] [Google Scholar]
- 24.Penzes, P., R. C. Johnson, R. Sattler, X. Zhang, R. L. Huganir, V. Kambampati, R. E. Mains, and B. A. Eipper. 2001. The neuronal Rho-GEF kalirin-7 interacts with PDZ domain-containing proteins and regulates dendritic morphogenesis. Neuron 29:229-242. [DOI] [PubMed] [Google Scholar]
- 25.Satoh, A., H. Nakanishi, H. Obaishi, M. Wada, K. Takahashi, K. Satoh, K. Hirao, H. Nishioka, Y. Hata, A. Mizoguchi, and Y. Takai. 1998. Neurabin-II/spinophilin. An actin filament-binding protein with one PDZ domain localized at cadherin-based cell-cell adhesion sites. J. Biol. Chem. 273:3470-3475. [DOI] [PubMed] [Google Scholar]
- 26.Shenolikar, S., and T. S. Ingebritsen. 1984. Protein (serine and threonine) phosphate phosphatases. Methods Enzymol. 107:102-129. [DOI] [PubMed] [Google Scholar]
- 27.Stapleton, D., I. Balan, T. Pawson, and F. Sicheri. 1999. The crystal structure of an Eph receptor SAM domain reveals a mechanism for modular dimerization. Nat. Struct. Biol. 6:44-49. [DOI] [PubMed] [Google Scholar]
- 28.Stephens, D. J., and G. Banting. 1999. Direct interaction of the trans-Golgi network membrane protein, TGN38, with the F-actin binding protein, neurabin. J. Biol. Chem. 274:30080-30086. [DOI] [PubMed] [Google Scholar]
- 29.Stephens, D. J., and G. Banting. 2000. In vivo dynamics of the F-actin-binding protein neurabin-II. Biochem. J. 345:185-194. [PMC free article] [PubMed] [Google Scholar]
- 30.Strack, S., M. A. Barban, B. E. Wadzinski, and R. J. Colbran. 1997. Differential inactivation of postsynaptic density-associated and soluble Ca2+/calmodulin-dependent protein kinase II by protein phosphatases 1 and 2A. J. Neurochem. 68:2119-2128. [DOI] [PubMed] [Google Scholar]
- 31.Sugiyama, H., P. Papst, M. Fujita, E. W. Gelfand, and N. Terada. 1997. Overexpression of wild type p70 S6 kinase interferes with cytokinesis. Oncogene 15:443-452. [DOI] [PubMed] [Google Scholar]
- 32.Terry-Lorenzo, R. T., M. Inoue, J. H. Connor, T. A. Haystead, B. N. Armbruster, R. P. Gupta, C. J. Oliver, and S. Shenolikar. 2000. Neurofilament-L is a protein phosphatase-1-binding protein associated with neuronal plasma membrane and post-synaptic density. J. Biol. Chem. 275:2439-2446. [DOI] [PubMed] [Google Scholar]
- 33.Winder, D. G., and J. D. Sweatt. 2001. Roles of serine/threonine phosphatases in hippocampal synaptic plasticity. Nat. Rev. Neurosci. 2:461-474. [DOI] [PubMed] [Google Scholar]
- 34.Yan, Z., L. Hsieh-Wilson, J. Feng, K. Tomizawa, P. B. Allen, A. A. Fienberg, A. C. Nairn, and P. Greengard. 1999. Protein phosphatase 1 modulation of neostriatal AMPA channels: regulation by DARPP-32 and spinophilin. Nat. Neurosci. 2:13-17. [DOI] [PubMed] [Google Scholar]