Abstract
A variety of cellular stresses activate the stress-responsive mitogen-activated protein (MAP) kinases p38 and JNK. In this study, we studied the activation mechanism of a human MAP kinase kinase kinase, MTK1 (also known as MEKK4), which mediates activation of both p38 and JNK. MTK1 has an extensive N-terminal noncatalytic domain composed of ∼1,300 amino acids. Full-length or near full-length MTK1 is catalytically inactive when expressed in Saccharomyces cerevisiae cells, as it is in mammalian cells. Deletion of a segment including positions 253 to 553 activates kinase, indicating that this segment contains the autoinhibitory domain. In the autoinhibited conformation, the MTK1 kinase domain cannot interact with its substrate, MKK6. By a functional complementation screening with yeast cells, GADD45 proteins (GADD45α, β, and γ) were identified as MTK1 activators. GADD45 proteins bind a site in MTK1 near the inhibitory domain and relieve autoinhibition. Mutants of full-length MTK1 were isolated that can interact with MKK6 in the absence of the activator GADD45 proteins. These MTK1 mutants are constitutively active, in both yeast and mammalian cells. A model of MTK1 autoinhibition by the N-terminal inhibitory domain and activation by GADD45 binding is presented.
In eukaryotes from yeasts to mammals, various cellular stresses generate intracellular signals that converge on the stress-responsive mitogen-activated protein kinases (MAPKs). In mammalian cells, two families of stress-responsive MAPKs, JNK and p38, are activated by stimuli such as UV radiation, ionizing radiation, hyperosmolarity, oxidative stress, and translation inhibitors, as well as cytokines such as interleukin 1, tumor necrosis factor alpha, and transforming growth factor β (7, 8). These MAPKs are activated by their cognate MAPK kinases (MAPKKs) through phosphorylation of conserved threonine and tyrosine residues in the kinase domain activation loop (15, 28). A MAPKK is activated by specific MAPKK kinases (MAPKKKs) through phosphorylation of conserved threonine and/or serine residues (1). An interacting set of MAPKKKs, MAPKKs, and MAPKs constitutes a specific MAPK cascade. Numerous MAPKKKs are now known to function upstream of the JNK and p38 MAPKs, undoubtedly reflecting the diverse stimuli that lead to JNK and p38 activation. While it is believed that different MAPKKKs are activated by disparate mechanisms, individual mechanisms are either unknown or only vaguely understood. Human MTK1 (and its mouse homolog MEKK4) is one of the MAPKKKs that act upstream of JNK and p38 (16, 36). In this report, we characterize the MTK1 activation mechanism by making use of our previous observation that MTK1 can be expressed functionally in Saccharomyces cerevisiae.
In yeast cells, hyperosmotic stress activates a stress-responsive MAPK pathway that is composed of three MAPKKKs (Ssk2, Ssk22, and Ste11), the MAPKK Pbs2, and the MAPK Hog1 (2, 3, 22, 23, 31). The activities of the Hog1 MAPK pathway are essential for yeast cells to survive in high-osmolarity environments. Thus, Δssk2 Δssk22 Δste11 triple mutants are osmosensitive because such mutants lack all three MAPKKKs (22, 31). The presence of any one of the three MAPKKKs, however, is sufficient for yeast cells to survive in high-osmolarity conditions. By functional complementation, we previously isolated a human homolog of the Ssk2 and Ssk22 MAPKKKs, namely MTK1 (36). The kinase domain of MTK1 is highly homologous to those of Ssk2 and Ssk22 (42 and 36% identities, respectively), and when expressed in yeast cells, it specifically activates Pbs2. Nonetheless, their N-terminal regulatory (noncatalytic) domains have only limited similarity, suggesting that the MTK1 regulatory mechanism is distinct from that of Ssk2/Ssk22. Indeed, yeast Ssk2/Ssk22 MAPKKKs are activated by a two-component response regulator whose homolog does not exist in mammalian cells (30, 32). Thus, it is unlikely that MTK1 is activated in yeast cells by the activation machinery for the Ssk2/Ssk22 MAPKKKs. Consistent with this reasoning, all of the MTK1 cDNA clones isolated in the functional complementation screening lacked the N-terminal regulatory region to various extents (36).
It is known that many protein kinases of the MAPKKK family, including Ssk2 and MTK1, can be constitutively activated by eliminating their N-terminal sequences (4, 31, 42). Thus, it is likely that their regulatory N-terminal regions contain autoinhibitory domains and that any activation mechanism of such kinases entails a dissociation of the autoinhibitory domain from the catalytic domain. A simple activation mechanism is a proteolytic removal of the N-terminal inhibitory sequence; for example, MEKK1 may be cleaved by an activated caspase (5, 11). Phosphorylation by an upstream kinase, such as Ste20 and P21-activated kinases (PAKs), is another proposed mechanism for activating MAPKKKs (18, 29, 45). Also, binding of an activator protein is a possible activation mechanism. It is well established, for example, that the Raf family kinases are activated by binding of the small G protein Ras (8) and that the apoptosis signal-regulating kinase 1 MAPKKK is activated by binding of TRAF2 or Daxx (6, 26). In an analogous manner, yeast Ssk2 is activated by binding of the Ssk1 response regulator protein (23, 30, 32). As for MTK1, it was previously reported that a family of GADD45-like proteins (GADD45α, GADD45β [MyD118], and GADD45γ [CR6]) bind tightly to an N-terminal region of MTK1 and coexpression of the full-length MTK1 and GADD45 resulted in strong activation of the MTK1 kinase (37). GADD45α is a p53-regulated small protein of 21 kDa and was originally identified as one of the genes that are induced by radiation and other cellular stresses (13). While the exact function of GADD45α is still unclear, it seems to serve as a key coordinator of stress response by regulating cell cycle, genomic stability, DNA repair, and other stress-related responses (35). GADD45β and GADD45γ are both highly homologous to GADD45α and may have overlapping but distinct cellular functions.
It was unclear, however, how GADD45 interaction leads to MTK1 activation. In this report, we demonstrate that the MTK1 N-terminal region inhibits the C-terminal kinase domain by preventing the kinase domain from interacting with its substrate, MKK6. Furthermore, we show that the GADD45 proteins can activate MTK1, in yeast as well as mammalian cells, by freeing the MTK1 kinase domain from the N-terminal autoinhibitory domain.
MATERIALS AND METHODS
Yeast strains.
The following yeast strains were used: FP75 (MATa ura3 leu2 trp1 his3 ssk2::LEU2 ssk22::LEU2 ste11::HIS3) and L40 (MATa leu2 trp1 his3 ade2 LYS2::lexAop-HIS3 URA3::lexAop-lacZ gal80)
Buffers and media.
Lysis buffer contains 20 mM Tris-HCl (pH 7.5), 1% Triton X-100, 10% glycerol, 137 mM NaCl, 2 mM EDTA, 50 mM β-glycerophosphate, 10 mM NaF, 1 mM sodium vanadate, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 10 μg of leupeptin/ml, and 10 μg of aprotinin/ml. Kinase buffer contains 25 mM Tris-HCl (pH 7.5), 20 mM MgCl2, 0.5 mM sodium vanadate, 25 mM β-glycerophosphate, 2 mM dithiothreitol, and 1 mM EGTA. Sodium dodecyl sulfate (SDS) loading buffer is 65 mM Tris-HCl (pH 6.8), 5% (vol/vol) 2-mercaptoethanol, 2% SDS, 0.1% bromophenol blue, and 10% glycerol. Yeast extract-peptone-dextrose (YPD) medium contains 10 g of yeast extract, 20 g of tryptone, and 20 g of dextrose per liter. YPGal medium contains 10 g of yeast extract, 20 g of tryptone, and 20 g of galactose per liter.
Yeast expression plasmids.
p414ADH, p414TEF, p415TEF, and p426GAL vectors have been described previously (24). pRS422-THL, which contains the TEF1 promoter, a hemagglutinin (HA) tag, a nuclear localization signal (NLS), a multicloning site (MCS), and the CYC1 terminator sequence, was created as follows. A sequence encoding an HA tag (YPYDVPDYA) and an NLS (PKKKRKVED) (17) was amplified by PCR and cloned into p415TEF, which contains the TEF1 promoter and the CYC1 terminator sequence. From this intermediate construct, the sequence containing the TEF1 promoter followed by the HA tag, NLS, MCS, and the CYC1 terminator was excised and subcloned into pRS422 (9) to generate pRS422-THL. The complete open reading frames (ORFs) of human GADD45α, β, and γ were individually cloned into pRS422-THL to create pRS422-THL-GADD45α, β, and γ. p414ADH-MTK1 was constructed by subcloning a fragment derived from pACT-MTK1 that contains the MTK1 ORF and an HA tag at its N terminus into p414ADH. The full-length MTK1-FL constructs actually start with the Met22 codon because we believe this is a more likely initiation codon than Met1.
Mammalian expression plasmid.
pFlag-MTK1, pFlag-MTK1-K/R, and pHA-GADD45γ were described previously (37). The deletion constructs, such as pFlag-MTK1(1322-1607), were generated by a PCR-based method. pSRα-HA-MKK6-K/A (kinase-dead mutant) was constructed by subcloning the MKK6-K/A sequence amplified by PCR into the pSRα-HA vector (38).
Isolation of MTK1 activators by functional complementation in yeast cells.
Osmosensitive yeast mutant FP75 (Δssk2 Δssk22 Δste11) was transformed with a full-length MTK1 construct (p414ADH-MTK1-FL or p414TEF-HA-MTK1-FL). These transformants, which were still osmosensitive, were then supertransformed with a HeLa or Jurkat cDNA library constructed in the yeast expression vector pNV7 (25). Osmoresistant transformants were selected on plates containing 1.5 M sorbitol (and 2% galactose to induce the expression of cDNA in the pNV7 vector). pNV7-derived plasmids were recovered from osmoresistant transformants (33) and retransformed into FP75 to eliminate false-positive clones. cDNA inserts from reproducibly positive clones were subcloned into the pBluescript vector and sequenced.
Two-hybrid analyses.
Two-hybrid analysis was carried out essentially as described previously (12) with pACTII for GAL4 activator domain fusion proteins (20) and either pBTM116 (39) or its derivative pBTM118 for LexA DNA-binding domain (DB) fusion proteins. The following plasmids and their derivatives were used. pACT-MTK1-FL, which encodes a fusion protein between the GAL4 activator domain and the full-length MTK1 ORF, has been described previously (37). pACT-MTK1-FL-K/R was generated by replacing Lys1371 with an arginine codon by PCR-mediated mutagenesis. The pBTM118 vector was constructed by replacing the MCS of pBTM116 with that of pUC18 by using the unique EcoRI and PstI sites. pBTM-MKK6-K/A was constructed by subcloning the complete MKK6-K/A ORF (36) into pBTM118. The three-hybrid analysis was performed by using the pRS422-THL construct in addition to the two vectors used in the two-hybrid analysis. Yeast strain L40 was triply transformed with pACT (LEU2+), pBTM (TRP1+), and pRS422-THL (ADE2+) plasmids, plated on a selective medium lacking leucine, tryptophan, and adenine, and incubated for 3 days at 30°C. Each colony was streaked on both a selective medium lacking leucine, tryptophan, and adenine and a medium which also lacked histidine to examine histidine prototrophy. For each two-hybrid test, LacZ (β-galactosidase [β-Gal]) activity was also visualized in situ by using 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) as previously described (39).
Isolation of MTK1 mutants that constitutively interact with MKK6.
Yeast L40 cells doubly transformed with pBTM-MKK6-K/A and either pACT-MTK1-FL, pACT-MTK1-FL-K/R, or pACT-MTK1(253-1607)-K/R, were cultured in a selective medium lacking leucine and tryptophan. These transformants were then plated on plates of synthetic dextrose medium lacking leucine, tryptophan, and Listidine at ∼107 cells/plate. After 4 to 7 days at 30°C, 70 His+ colonies were randomly picked from 34 plates, and both histidine prototrophy and β-Gal activity were evaluated. pACT-based plasmids were recovered from His+ β-Gal-positive colonies and retransformed into L40 with pBTM-MKK6-K/A to eliminate false positives. Nucleotide sequencing of the entire MTK1 ORF from 11 MTK1 mutant plasmids revealed that each mutant plasmid had either one or two missense mutations. Finally, an individual mutation from each MTK1 mutant plasmid was introduced into the parent pACT-MTK1 vector by the swapping of a short restriction fragment so that all mutations could be compared in a common structure. This process also served to eliminate any possibility that a second mutation exists elsewhere in the original mutant plasmids. The reconstructed pACT-MTK1 mutant plasmids were individually cotransformed with pBTM-MKK6-K/A into L40, to verify both histidine prototrophy and β-Gal activity.
Immunoblotting analyses.
Immunoblotting analyses were carried out essentially as described previously (37, 38). The following antibodies were used in this study: mouse monoclonal antibody (mAb) M2 specific to the Flag epitope (Sigma), rat mAb 3F10 specific to the HA epitope (Roche), and rabbit polyclonal antiserum specific to phospho-MKK3/MKK6 (Cell Signaling Technology).
Tissue culture and in vitro kinase kinase assays.
COS-7 or HeLa cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, penicillin, and streptomycin. Cells grown in 35-mm-diameter dishes were transfected with the appropriate expression plasmids with either Effectene reagent (Qiagen) or Lipofectamine and PLUS reagents (Invitrogen). The total amount of plasmid DNA was adjusted to either 0.4 (see Fig. 8B) or 2 (see Fig. 8A and C) μg per plate with empty vector DNA. Thirty-six hours after transfection, cells were lysed in lysis buffer supplemented with 0.5% deoxycholate. Cell lysates were incubated with anti-Flag M2 mAb for 2 h at 4°C. Immune complexes were recovered with the aid of protein G-Sepharose beads (Pharmacia), washed twice with lysis buffer containing 500 mM NaCl, washed twice with lysis buffer, and washed two additional times with kinase buffer. Immunoprecipitates were resuspended in 30 μl of kinase buffer containing 3 μg of the specific substrate glutathione S-transferase (GST)-MKK6-K/A. The kinase reaction was initiated by the addition of 20 μCi of [γ-32P]ATP and 20 μM nonradioactive ATP. After a 15-min incubation at 30°C, reactions were terminated by adding 7.5 μl of 5× SDS loading buffer. Samples were boiled for 5 min, separated by SDS-polyacrylamide gel electrophoresis on 7.5% polyacrylamide gels, dried, and visualized by autoradiography.
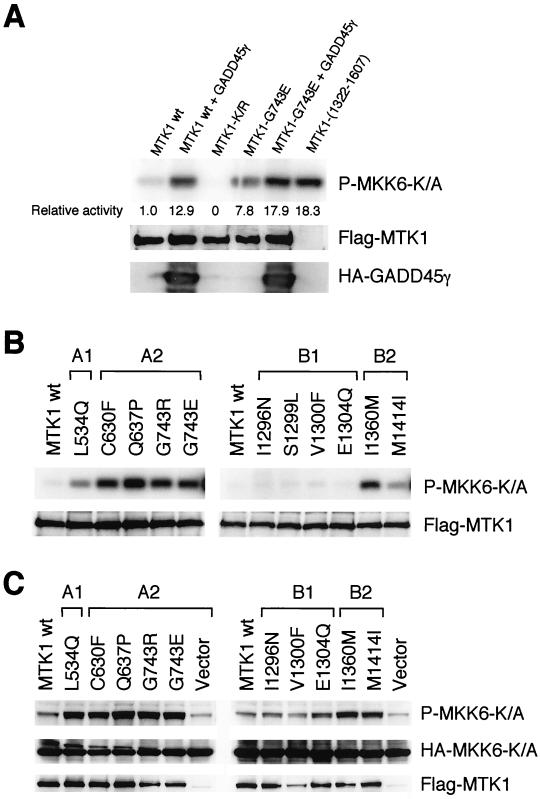
FIG. 8.
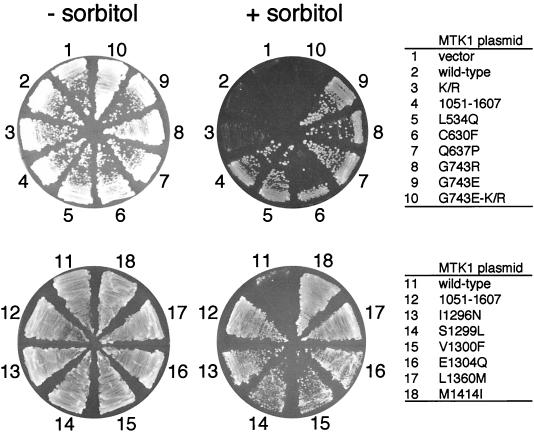
Kinase activities of MTK1 mutants. (A and B) In vitro kinase activities of MTK1 mutants. HeLa (A) or Cos-7 (B) cells were transiently transfected with Flag-tagged MTK1 (wild type or mutant) with or without HA-tagged GADD45γ. Flag-tagged MTK1 was immunoprecipitated from cell extracts, and its kinase activity was assayed in vitro with the substrate GST-MKK6-K/A in the presence of [γ-32P]ATP. Phosphorylated GST-MKK6-K/A was detected by autoradiography following SDS-polyacrylamide gel electrophoresis. Expression levels of Flag-MTK1 and HA-tagged GADD45γ were monitored by immunoblotting. In panel A, phosphorylated GST-MKK6-K/A was also detected by PhosphorImager (Molecular Dynamics) and quantitated. Kinase-dead MTK1-K/R was used as a negative control, and N-terminally truncated MTK1(1322-1607) was used as a positive control. The expression level of Flag-tagged MTK1(1322-1607) was comparable to that of the others, although it is not visible in this figure due to its smaller size. (C) In vivo kinase activities of MTK1 mutants. Cos-7 cells were transiently transfected with Flag-tagged MTK1 (wild type or mutant) together with HA-tagged MKK6-K/A. The phosphorylation status of transfected HA-MKK6-K/A was detected by immunoblotting with phospho-MKK6-specific antibody. Expression levels of HA-MKK6-K/A and Flag-MTK1 were also monitored by immunoblotting. wt, wild-type.
RESULTS
Inhibition of the MTK1 kinase domain by its N-terminal region.
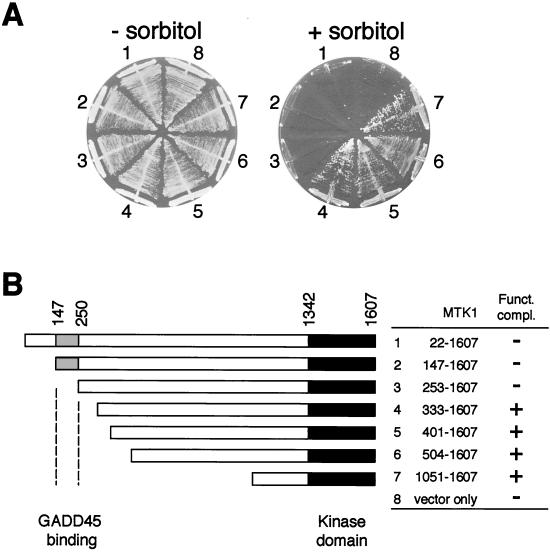
N-terminally truncated MTK1 mutants that lack most of the noncatalytic domain, being constitutively active, complement the osmosensitive defect of Δssk2 Δssk22 Δste11 triple mutants (36). In contrast, full-length MTK1 (MTK1-FL) failed to complement the same yeast mutants, suggesting the presence of an autoinhibitory domain within the MTK1 N-terminal region. In order to define the location of the autoinhibitory domain more precisely, especially in terms of its relationship to the GADD45 binding domain (residues 147 to 250; Fig. 1B), we constructed a series of MTK1 N-terminal deletion mutants and examined whether they can complement Δssk2 Δssk22 Δste11 triple mutants. As demonstrated by the results shown in Fig. 1A and summarized in Fig. 1B, deletion of 332 or more of the N-terminal amino acids resulted in successful complementation of Δssk2 Δssk22 Δste11, indicating that activation of the MTK1 kinase domain took place. Deletion of 252 or fewer of the N-terminal amino acids, however, did not activate the kinase domain. MTK1-FL was expressed equally well and as stably as other deletion constructs (data not shown). Thus, the autoinhibitory domain and the GADD45-binding domain seem contiguous to each other with relatively little overlap, if any.
FIG. 1.
MTK1 N-terminal region inhibits its kinase activity. (A) Yeast strain FP75 (Δssk2 Δssk22 Δste11) was transformed with either a centromeric vector plasmid (p414ADH) or one of its derivatives containing various MTK1 segments. Transformed cells were spread on YPD agar plates supplemented with (+) or without (−) 1.5 M sorbitol. FP75 cells grow on plates containing sorbitol only when the MTK1 kinase activity is expressed. The segment of MTK1 cDNA sequence included in each construct is indicated, both schematically and numerically, in panel B. (B) The topmost bar represents the full-length MTK1 cDNA (amino acids from positions 22 through the C terminus [position 1607]), and other bars represent deletion derivatives. The gray section indicates the GADD45-binding site, whereas the filled section indicates the kinase catalytic domain. Funct. compl., functional complementation; +, complementation; −, no complementation.
Isolation of MTK1 activators by using functional complementation in yeast mutants.
In mammalian cells, MTK1-FL is presumably activated by an upstream activator mechanism; however, a homologous activation mechanism does not seem to exist in the yeast cells, resulting in the failure of MTK1-FL to rescue the yeast Δssk2 Δssk22 Δste11 triple mutants. We thus reasoned that, if the mammalian activator of MTK1 is coexpressed with MTK1-FL, the latter would be activated even in yeast cells and that the consequent activation of MTK1-FL would be manifested by functional complementation of the Δssk2 Δssk22 Δste11 triple mutation. Based on this prediction, we screened for MTK1 activators. Yeast strain FP75 (Δssk2 Δssk22 Δste11) was transformed with either MTK1-FL or HA-tagged MTK1-FL. In either case, these cells are osmosensitive, indicating a failure to express the active MTK1 kinase. We then transformed these cells with two different human cDNA libraries made from mRNA of Jurkat T cells and HeLa cells. Transformants were then replicated on selective plates containing osmostressor (1.5 M sorbitol), and osmoresistant colonies were isolated. In all, we identified 31 human cDNA clones that complemented the osmosensitivity of FP75 carrying a full-length MTK1 plasmid from an estimated total of ∼10 million transformed yeast cells. Of the 31 cDNA clones, 5 encoded N-terminally truncated MTK1 and an additional 11 clones encoded the TAK1 MAPKKK. These clones complemented the Δssk2 Δssk22 Δste11 cells even in the absence of MTK1-FL, indicating that they directly activated the yeast Pbs2 MAPKK rather than activating MTK1-FL. TAK1 has been shown, under certain conditions, to activate another yeast MAPKK, Ste7 (43). These clones were not characterized any further.
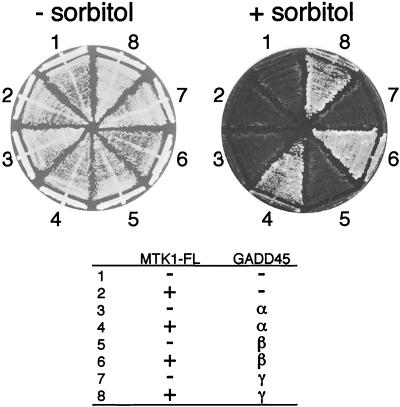
Of the remaining 15 clones, 1 clone encoded GADD45α and 14 clones encoded GADD45β. Although we did not find the third member of the GADD45 family, namely GADD45γ, in our screenings, a GADD45γ construct did also complement the osmosensitivity of FP75 plus MTK1-FL (Fig. 2). A GADD45 clone alone, in the absence of MTK1-FL, could not complement the osmosensitive defect of FP75. From these observations, it was concluded that coexpression of the GADD45 proteins releases MTK1-FL from kinase autoinhibition.
FIG. 2.
Activation of full-length MTK1 (MTK1-FL) by GADD45 proteins. Yeast strain FP75 was transformed with two expression plasmids (p414TEF for MTK1-FL and p426GAL for GADD45) with (+) or without (−) the indicated cDNA sequences. p426GAL is a multicopy plasmid with the galactose-inducible GAL1 promoter. Transformed cells were spread on YPGal agar plates supplemented with (+) or without (−) 1.5 M sorbitol.
It was previously shown by two-hybrid analyses that the GADD45 proteins bind to a site near the MTK1 N terminus (residues 147 to 250) (37). The functional interaction between GADD45 and MTK1-FL, as defined here by complementation of the Δssk2 Δssk22 Δste11 mutants, requires the same binding site: the short N-terminal deletion mutant MTK1(253-1607), which contains MTK1 amino acids from position 253 through the C terminus but lacks the GADD45 binding site, cannot be activated by GADD45 proteins under the conditions that activate MTK1-FL (see Fig. 4B) (37). Thus, the binding of GADD45 to the MTK1 GADD45-binding domain (between residues 147 and 250) appears to nullify the autoinhibitory effect of a nearby domain (including, but not limited to, the residues between 253 and 332), resulting in the activation of the C-terminal kinase domain.
FIG. 4.
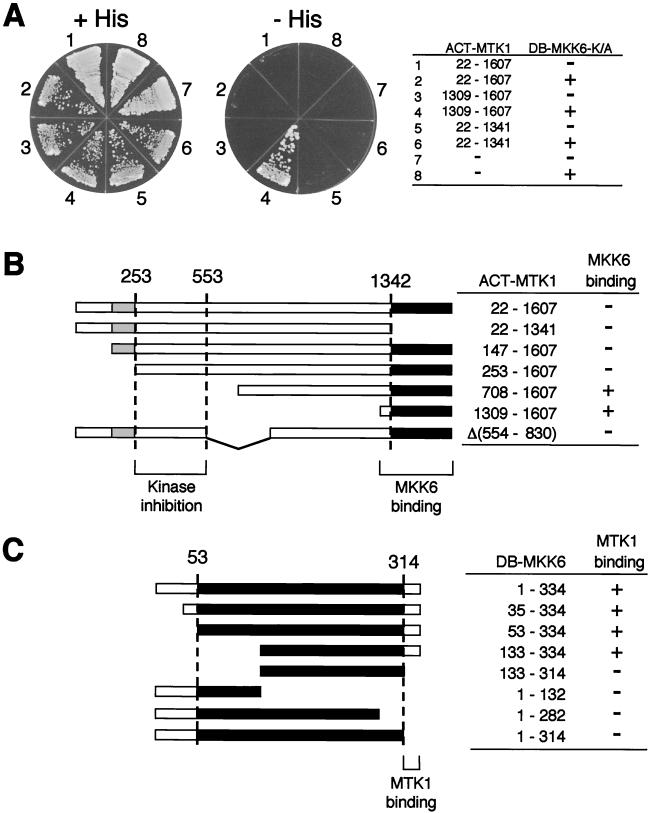
Three-hybrid interaction analysis. Yeast L40 cells were transformed with three plasmids (ACT-MTK1, DB-MKK6-K/A, and GADD45 constructs) as indicated. A minus sign indicates the corresponding vector (pACTII, pBTM118, or pRS422-THL) without an insert; a plus sign indicates the vector with an insert. Transformed cells were spread on the appropriate synthetic agar plates supplemented with (+His) or without (−His) histidine.
Interaction between the MTK1 kinase domain and MKK6.
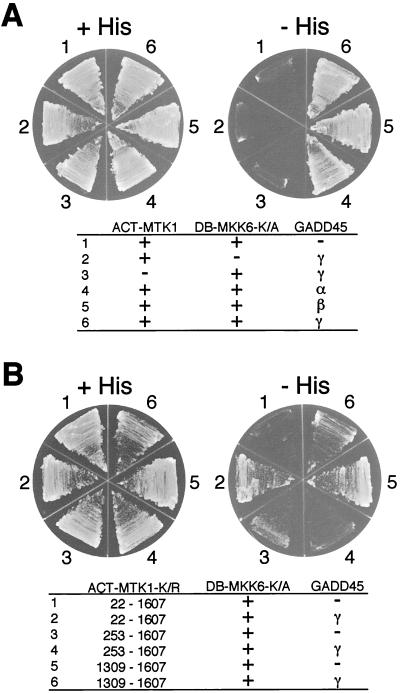
MTK1 activates the MKK6 MAPKK by phosphorylating the conserved threonine and serine residues (Ser207 and Thr211) in the kinase domain activation loop (36). This activation process necessarily involves a direct contact between the MTK1 kinase domain and the MKK6 kinase. One possible mechanism of MTK1 autoinhibition is, therefore, to obstruct such an interaction. We thus examined the interaction between the MTK1 and MKK6 kinase domains by using the yeast two-hybrid method. For this purpose, we constructed a LexA DB-MKK6-K/A fusion protein and a series of Gal4 transcription activation domain (ACT)-MTK1 fusion proteins. DB-MKK6-K/A contains the full-length MKK6 with the Lys82→Ala substitution (kinase dead) mutation because coexpression of the wild-type MKK6 with an active MTK1 construct was toxic to yeast cells. Yeast strain L40 was cotransformed with combinations of ACT-MTK1 and DB-MKK6-K/A constructs; when there is successful protein-protein interaction, cotransfected yeast cells will grow on histidine-deficient media.
As shown in Fig. 3A, ACT-MTK1(1309-1607), which contains only the MTK1 kinase domain, indeed interacted with DB-MKK6-K/A. In contrast, ACT-MTK1(1-1341), which contains only the MTK1 N-terminal regulatory region, did not bind DB-MKK6-K/A. Perhaps more important, the full-length MTK1 (ACT-MTK1-FL) also failed to interact with DB-MKK6-K/A, even though it contains the kinase domain. These results are consistent with our model that the MTK1 autoinhibitory domain masks its kinase domain from interacting with MKK6 (see Fig. 10).
FIG. 3.
Two-hybrid interaction analysis between MTK1 and MKK6. (A) Interaction of various MTK1 segments fused to the GAL4 activation domain (ACT-MTK1 constructs) with MKK6-K/A fused to the LexA DB (DB-MKK6-K/A) was tested. Vectors used in these experiments were pACTII and pBTM118. Segments of MTK1 included in the ACT-MTK1 constructs are indicated. A minus sign in the ACT-MTK1 column indicates the vector without an insert. Yeast L40 cells were transformed with two plasmids (ACT-MTK1 and DB-MKK6-K/A constructs) as indicated. −, vector without an insert; +, vector with an insert. Transformed L40 cells were spread on the appropriate synthetic agar plates supplemented with (+His) or without (−His) histidine. A kinase-defective MKK6-K/A mutant was used to reduce the toxic effect of MTK1 expression. MTK1(1309-1607) encodes the kinase catalytic domain, whereas MTK1(22-1341) encodes the N-terminal noncatalytic domain. In situ β-Gal assays gave essentially identical results (not shown). (B) Summary of the experiments in panel A and additional two-hybrid tests. The MTK1 coding sequence is represented by the horizontal bar. Gray and filled boxes are the GADD45-binding domain and the kinase catalytic domain, respectively. The segment between amino acid positions 253 and 553 is required for autoinhibition of the MTK1-MKK6 interaction. +, growth on −His plates; −, no growth on −His plates. (C) Summary of two-hybrid analyses for the MTK1 binding site in MKK6. Binding of full-length (positions 1 to 334) and various deletion constructs of DB-MKK6-K/A was tested with ACT-MTK1(1309-1607) as a bait. The MKK6 coding sequence is represented by the horizontal bar. Filled boxes represent the kinase catalytic domain. The C-terminal noncatalytic region of MKK6 is necessary to interact with the MTK1 kinase domain. +, growth on −His plates; −, no growth on −His plates.
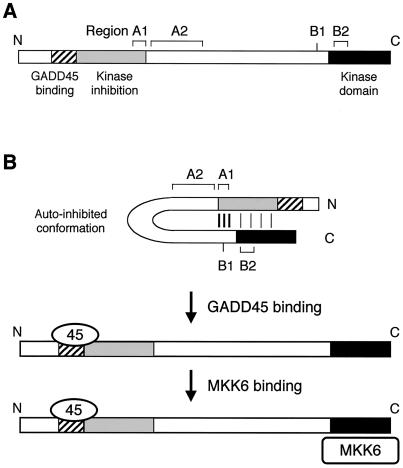
FIG. 10.
A speculative model of MTK1 autoinhibition by its N-terminal region and activation by GADD45 binding. (A) Schematic representation of MTK1 structure. The GADD45 binding site, kinase inhibitory domain, and kinase catalytic domain are shown. Also shown are the locations of four regions (A1, A2, B1, and B2) defined by constitutively active MTK1 mutations. (B) Model of MTK1 activation mechanism. In the autoinhibited conformation, the N-terminal inhibitory domain interacts with the C-terminal kinase domain so that substrates (such as MKK6) cannot interact. It is likely that there are multiple interactions between the N- and C-terminal regions. The A1 and B1 regions may interact strongly and serve as a clasp. Weaker interactions involving the kinase domain itself must also exist. Binding of GADD45 to a site near the inhibitory domain sterically interferes with the N-C interaction, making the kinase domain accessible to its substrates.
To define the segment of MTK1 that is required to inhibit the interaction between MTK1 and MKK6, we tested additional deletion derivatives of the ACT-MTK1-K/R. In these experiments, we used the kinase-dead MTK1-K/R mutant because expression of catalytically active and N-terminally truncated MTK1 is somewhat toxic to yeast cell growth, presumably by overactivating the endogenous Pbs2 MAPKK (36). As summarized in Fig. 3B, deletion of the first 252 residues or deletion of the sequence between residues 554 and 830 did not induce MTK1-MKK6 interaction. In contrast, deletion of the first 708 amino acids did cause the MTK1-MKK6 interaction. Thus, it can be concluded that the presence of the sequence between residues 253 and 553 is required for the inhibition of the MTK1-MKK6 interaction. It should be noted that deletion of the GADD45-binding site (residues 147 to 250) did not induce the MTK1-MKK6 interaction, indicating that the autoinhibitory domain and the GADD45-binding domain are separable.
To define the segment of MKK6 that is required for its interaction with MTK1, we examined the capacity of various DB-MKK6-K/A deletion constructs to interact with ACT-MTK1(1309-1607). As summarized in Fig. 3C, deletion of the most C-terminal 20 amino acids that lie outside the catalytic domain abolished MTK1 binding completely, whereas the N-terminal region is dispensable. Thus, the observed MTK1-MKK6 interaction required the C-terminal region of MKK6. It seems likely that the binding of the MKK6 C terminus to MTK1 is a necessary recruitment step for an efficient phosphorylation of MKK6 by MTK1.
GADD45 proteins stimulate the interaction between full-length MTK1 and MKK6.
We showed above that coexpression of GADD45 and MTK1-FL in yeast cells activates the intrinsic catalytic activity of the MTK1 kinase domain. It is likely that this activation is mediated by an MTK1 conformational change, resulting in a dissociation of the N-terminal inhibitory domain from the MTK1 kinase domain (see Fig. 10). If so, coexpression with GADD45 should allow the otherwise inert MTK1-FL to productively interact with MKK6. This prediction was borne out by a three-hybrid analysis: expression of either GADD45α, β, or γ induced interaction between ACT-MTK1-FL and DB-MKK6-K/A (Fig. 4). GADD45 proteins alone did not induce the reporter gene (HIS3) expression (Fig. 4A), indicating that GADD45 stimulates the interaction between ACT-MTK1-FL and DB-MKK6-K/A. As might be further predicted, GADD45 proteins did not have any effect on the ACT-MTK1(253-1607)-K/R construct that lacks the GADD45-binding domain (Fig. 4B). By further two-hybrid analyses, we have excluded the possibility that GADD45 proteins interact with MKK6 (as well as with MTK1), forming a bridge between the two molecules (data not shown). We thus conclude that GADD45 proteins, by binding to MTK1, induce a conformational change in MTK1, and allow the MTK1 kinase domain to interact with MKK6.
Isolation of MTK1-FL mutants that are capable of interacting with MKK6.
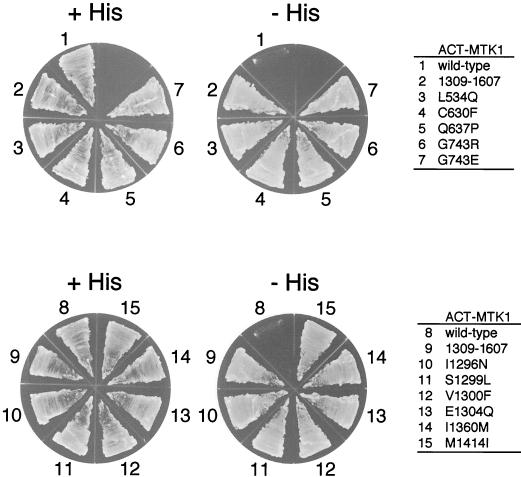
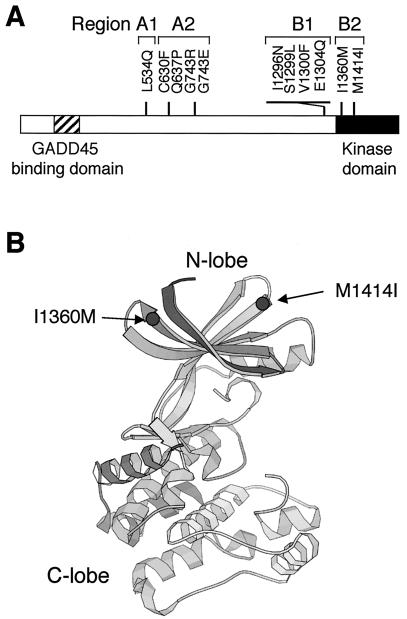
Yeast two-hybrid tester strain L40 does not grow on histidine-deficient media when cotransformed with the ACT-MTK1-FL and DB-MKK6-K/A constructs because these molecules do not interact. The same cells, however, gave rise to occasional His+ colonies. These His+ cells, we reasoned, potentially contained MTK1 mutants that were no longer autoinhibitory. We recovered ACT-MTK1-FL plasmids from spontaneously arisen His+ cells and tested their ability to interact with DB-MKK6-K/A by retransformation into the parental L40 cell with DB-MKK6-K/A. In this manner, we isolated 11 MTK1-FL mutants that became capable of interacting with MKK6-K/A (Fig. 5). Sequencing of the entire coding regions of the 11 MTK1-FL mutants revealed that 9 mutants had only one mutation each. Two were double mutants, but segregation of the two mutations showed that only one was entirely responsible for the mutant phenotype in each case. These mutations are represented by a one-letter amino acid code. L534Q, for example, stands for a mutation of leucine at the 534th amino acid position to glutamine. The 11 mutations are clustered in two regions: region A near the middle of the noncatalytic domain and region B near the junction between the noncatalytic domain and the kinase domain (Fig. 6A). These regions are further subdivided into A1, A2, B1, and B2, as shown in Fig. 6A, for reasons which are explained later. Although the three-dimensional structure of the MTK1 kinase domain is unknown, a structure is available for related protein kinases, such as PAK1 (19). A comparison of the PAK1 and MTK1 sequences, with reference to the PAK1 crystallographic structure, reveals that MTK1 Ile1360 should be located at the end of the β2 strand, near the surface loop between β2 and β3 (Fig. 6B). Similarly, Met1414 is expected to be located at the beginning of the β5 strand, near the β4-β5 surface loop. When placed in the consensus protein kinase folding, these sites are both exposed on the external surface of the kinase N lobe.
FIG. 5.
Mutants of full-length MTK1 that can interact with MKK6 in the absence of GADD45. Yeast L40 cells were transformed with an ACT-MTK1 construct (pACT-MTK1-FL wild type or one of its derivatives, as indicated on the figure) and a common DB-MKK6-K/A construct (pBTM-MKK6-K/A). Transformed yeast cells were spread on the appropriate synthetic agar plates supplemented with (+His) or without (−His) histidine. Mutations are designated by a one-letter amino acid code: L534Q, for example, stands for a mutation of leucine at the 534th amino acid position to glutamine. ACT-MTK1(1309-1607), which contains only the MTK1 kinase catalytic domain, was used as a positive control.
FIG. 6.
Locations of the constitutively active MTK1 mutations. (A) The positions of the 11 MTK1 mutations are shown along a bar representing the MTK1 coding sequence. Four regions (A1, A2, B1, and B2) are indicated. The GADD45-binding domain and the kinase catalytic domain are also shown. (B) The predicted three-dimensional locations of the two mutations in region B2 (I1360M and M1414I) are shown by using the crystallographic structure of the PAK1 kinase domain (19).
If these mutations abolish or weaken the autoinhibition by the N-terminal region, it will be predicted that these MTK1 mutants are constitutively active. In order to test this prediction, these MTK1 mutants were transformed into the yeast strain FP75 (Δssk2 Δssk22 Δste11). The wild-type MTK1-FL and MTK1(1051-1607) served as negative and positive controls, respectively. As shown in Fig. 7, all 11 mutants could rescue the osmosensitive defect of FP75, although the region B1 mutants grew more slowly (it is not clearly visible in Fig. 7). Thus, at least in yeast cells, these mutants are constitutively active.
FIG. 7.
MTK1 mutants that can interact with MKK6 in the absence of GADD45 are constitutively active in yeast cells. The osmosensitive yeast strain FP75 was transformed with p414ADH-MTK1-FL or one of its derivatives, as indicated in the cDNA sequences. K/R is the catalytically inactive K1371R mutation in the MTK1 kinase domain and was used as a negative control. MTK1(1051-1607) is a constitutively active N-terminal truncation mutant used here as a positive control. Transformed cells were spread on YPD agar plates supplemented with (+) or without (−) 1.5 M sorbitol.
Activities of MTK1 mutants.
We then investigated whether these constitutively active MTK1 mutants do indeed have higher kinase activities when expressed in mammalian cells, both by measuring the kinase activities of immunoprecipitated MTK1 in vitro and by monitoring the phosphorylation state of an MTK1 substrate in vivo. To these ends, Flag-tagged MTK1 constructs (wild type and a randomly chosen mutant, G743E) were constructed by using a mammalian expression vector. HeLa cells were transfected with the Flag-MTK1 expression constructs alone or together with an HA-tagged GADD45γ expression construct. As positive and negative controls, N-terminal truncation [Flag-MTK1(1322-1607)] and kinase-dead (Flag-MTK1-K/R) mutants were transfected, respectively. Forty-eight hours after transfection, cell lysates were prepared and the kinase activity of immunoprecipitated Flag-MTK1 was assayed in vitro by using a specific substrate, GST-MKK6-K/A. The immunoprecipitated full-length MTK1 (wild type) had only a weak kinase activity, indicating that it is in an autoinhibited conformation (Fig. 8A). Cotransfection of full-length MTK1 with GADD45γ resulted in a 13-fold increase in MTK1 kinase activity. In contrast, the MTK1-G743E mutant protein had significantly higher kinase activity even in the absence of GADD45 coexpression, indicating that the G743E mutation indeed activates the MTK1 kinase domain. Coexpression with GADD45γ further increased the kinase activity of MTK1-G743E; the effect seems additive, but its significance is unknown at present. By the same assay method, we found that all of the mutants in regions A1, A2, and B2 were also constitutively active (Fig. 8B). The region B1 mutants had, however, no higher kinase activities than the wild-type MTK1. It is possible that the yeast cell test was more sensitive than the in vitro kinase assay.
To test whether the elevated in vitro kinase activities of the MTK1 mutants actually reflect their in vivo catalytic activities, the phosphorylation state of intracellular MKK6 was monitored in MTK1-transfected cells. For this purpose, COS-7 cells were cotransfected with an HA-MKK6-K/A expression plasmid and a Flag-MTK1 (wild type or mutant) expression plasmid, and the phosphorylation status of HA-MKK6-K/A was monitored by anti-phospho-MKK6 antibody. Consistent with the results from the in vitro kinase assays, HA-MKK6-K/A was highly phosphorylated when coexpressed with any of the region A1, A2, or B2 MTK1 mutants but not when coexpressed with the region B1 mutants (Fig. 8C). Thus, it was concluded that the mutants in regions A1, A2, and B2 are indeed constitutively active when expressed in mammalian cells. The region B1 mutants do not appear to have significantly higher activities than the wild-type MTK1. It is possible that these mutants also have activities higher than those of the wild type but lower than the experimental sensitivity.
Interaction between the MTK1 N and C termini.
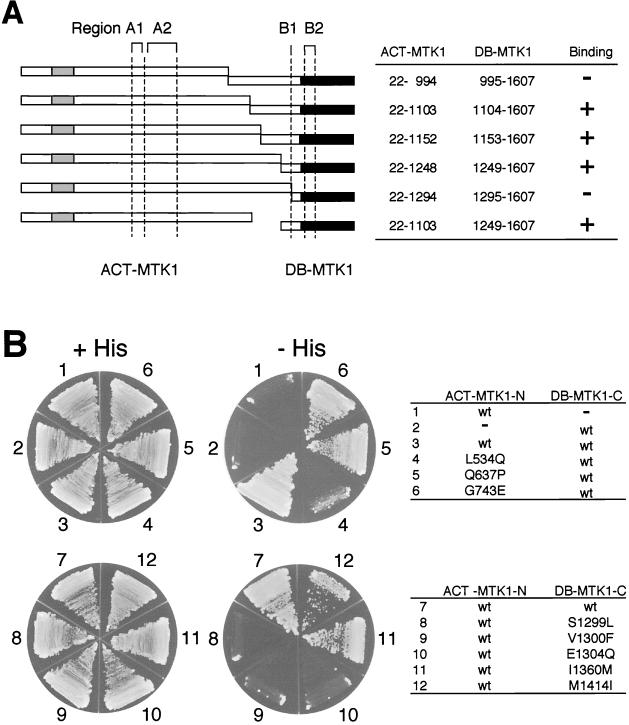
If the N-terminal region of MTK1 inhibits the C-terminal kinase domain by direct interaction, it might be possible to detect such physical interaction between the two regions. Our attempts to demonstrate the anticipated interaction by coprecipitation assays, however, were frustrated by the apparent presence of multiple interaction sites. In contrast, two-hybrid analyses gave more readily interpretable results. Figure 9A summarizes the results of our initial two-hybrid analysis between the N- and C-terminal regions of MTK1. As seen here, the N-terminal fragment (positions 22 to 1103) could interact with the nonoverlapping C-terminal fragment (positions 1249 to 1607). Of some interest is the observation that a shorter C-terminal fragment (positions 1295 to 1607) cannot interact with the N-terminal fragments. Thus, the C-terminal fragment must contain about 100 amino acids (positions 1249 to 1341) preceding the kinase catalytic domain. Significantly, this region overlaps with region B1 as defined by the mutant screening.
FIG. 9.
Two-hybrid interaction analysis between N- and C-terminal MTK1 segments. (A) Sets of ACT-MTK1 N-terminal constructs and DB-MTK1 C-terminal constructs, as shown schematically by horizontal bars and numerically in the table, were transformed into L40 tester cells. The presence and absence of the N-C interaction, as deduced from growth on histidine-deficient plates and β-Gal expression, are indicated by plus and minus signs, respectively. The GADD45 binding site (gray), kinase catalytic domain (black), and four regions (A1, A2, B1, and B2) are also indicated. (B) Some, but not all, constitutively active MTK1 mutants are defective in the N-C interaction, as detected by two-hybrid analysis. The ACT-MTK1-N construct contains the MTK1(22-1103) segment, and the DB-MTK1-C construct contains the MTK1(1104-1607) segment. Either a wild-type (wt) or a mutant sequence, as indicated in the figure, was tested in each pair. A minus sign indicates the corresponding vector (pACTII or pBTM118) without an insert. +His, with histidine; −His, without histidine.
We then examined, by using an interacting pair of MTK1 N and C fragments [MTK1(22-1103) and MTK1(1104-1607), respectively], whether activating the mutations we isolated above had any influence on their interaction. As shown in Fig. 9B, several mutations did abolish the N-C interaction. On the N fragment, the mutation in region A1 (L534Q) strongly suppressed the interaction. On the C fragment, mutations in region B1 (S1299L, V1300F, and E1304Q) strongly inhibited the interaction. These results, together with the deletion analysis shown in Fig. 9A, indicate that region A1 in the N fragment around position 534 and region B1 in the C fragment around position 1300 are important for the binding between the N and C fragments, possibly for their interaction with each other. In contrast, mutations in regions A2 and B2 did not interfere in the N-C interaction significantly, despite the fact that these mutants exhibited strong catalytic activities in vivo and in vitro. It is possible that mutations in the A2 and B2 regions affect autoinhibition indirectly by altering conformation of the protein. In fact, deletion of region A2 did not activate the MTK1 kinase domain (Fig. 3), indicating that region A2 is not directly involved in autoinhibition.
DISCUSSION
In this paper, we characterized the regulatory mechanism of the MTK1 MAPKKK. The main conclusions from this study can be summarized as follows: (i) the MTK1 N-terminal region contains an autoinhibitory domain that interacts with the C-terminal kinase domain, inhibits kinase activity, and prevents interaction with its substrate, MKK6, (ii) GADD45 proteins are MTK1 activators, and (iii) the binding of GADD45 to the N-terminal region of MTK1 eliminates inhibition of the kinase domain by the autoinhibitory domain. The isolation of constitutively active MTK1 mutants, many of which are located in the noncatalytic domain, further supports these conclusions. A speculative model of MTK1 activation by the GADD45 protein, based on these observations, is depicted in Fig. 10.
GADD45 proteins (GADD45α, β, and γ) were previously identified as MTK1 activators by using the two-hybrid screening strategy (37). It was also shown that GADD45 proteins bound and activated the full-length MTK1 in transfected mammalian cells as well as in an in vitro assay system (37). In this study, we added another line of evidence that GADD45 proteins are specific activators of MTK1. The function-based screening strategy employed in this study is entirely different, and more stringent, than the two-hybrid screening used in the earlier study; the functional screening method requires not only the protein-protein interaction but also the activation of the MTK1 kinase. The fact that we did not find any genes other than GADD45 in our functional screening may attest to its specificity. These results, however, do not preclude the presence of an alternative MTK1 activation mechanism(s). It is possible, for example, that GADD45 may simply be the most abundant activator in HeLa and Jurkat cells.
GADD45α was initially identified as a gene whose mRNA is rapidly induced by agents that cause DNA damage (UV, N-acetoxy-2-acetylaminofluorene, methyl methanesulfonate, and H2O2) (13, 14, 27). GADD45β and GADD45γ mRNA expression is also induced by various stress stimuli as well as by cytokines (37, 44). The role of the GADD45 proteins in the p38 and JNK pathways, however, was disputed by others based on several lines of evidence: unimpaired activation of p38 and JNK kinases in GADD45α-deficient mice, different time courses between p38/JNK activation and GADD45 expression, and the absence of an effect of the protein synthesis inhibitor cycloheximide (34, 40). Beside the fact that these studies used mutant mice with only the GADD45α gene deleted, their objections seem to reflect a tacit assumption that there should be only one mechanism to activate JNK/p38 for each external stimulus. Considering the fact that there are more than 10 different MAPKKKs that act upstream of the p38 and JNK kinases, it is more likely that several MAPKKKs, activated by different molecular mechanisms, contribute to JNK/p38 responses. Furthermore, there is now more direct evidence that GADD45 proteins, especially GADD45β and GADD45γ, serve crucial roles in p38 activation in T lymphocytes. It was found, by using gene knockout mice, that gamma interferon production in T helper type 1 (Th1) cells is dependent on the induction of the GADD45β or GADD45γ gene and on the resulting activation of the p38 MAPK (21, 44). Clearly, specific roles of individual MAPKKKs, as well as those of their activators, need to be studied further for a better understanding of their physiological significance.
Using a novel screening procedure, we isolated 11 MTK1 mutants that are constitutively active. These mutations are clustered in four regions, which are designated A1, A2, B1, and B2 (Fig. 6). Since no structural information is available for the N-terminal region of MTK1 or its homologs, it is premature to speculate on the structural consequences of the mutations in the regions A1, A2, and B1. Nonetheless, the presence of mutations in the N-terminal noncatalytic domain that influence the interactive properties of the C-terminal kinase domain compels us to conclude that a direct interaction between the N- and C-terminal domains must take place. In fact, our two-hybrid analyses (Fig. 9) indicated that an interaction between the MTK1 N- and C-terminal regions does occur. Interestingly, the mutations in regions A1 and B1 significantly weakened such an interaction, indicating that these regions have dominant roles in the N-C interaction. It is even possible that the A1 and B1 regions interact with each other, although there is no direct evidence. These regions, however, may not be directly involved in the kinase inhibition because mutants in these regions have relatively weak kinase activities (Fig. 8B). As graphically suggested in Fig. 10B, the strong A1-B1 interaction may aid the weaker, but inhibitory, interaction between the N-terminal autoinhibitory domain and the C-terminal kinase domain.
Because the two mutations in region B2, I1360M and M1414I, are within the conserved kinase domain, their locations can be predicted by using the crystallographic structures of similar protein kinases. Thus, it is predicted that these sites are both exposed on the external surface of the kinase N lobe (Fig. 6B). It is of interest that MTK1 Ile1360 is at the position corresponding to CDK4 Arg24, where mutations are known to exist that inhibit the binding of its inhibitor protein, p16INK4a (10, 41). By analogy, it is possible that the region B2 sites directly interact with the N-terminal autoinhibitory domain. A binding of the GADD45 protein to the GADD45-binding site near the autoinhibitory domain will interfere in the autoinhibitory interaction between the MTK1 N and C termini, leading to MKK6 binding and its activation by phosphorylation (Fig. 10B).
Acknowledgments
We thank Quint Medley, Vladimír Reiser, and Neil Krueger for valuable comments on the manuscript.
This work was supported by NIH grants GM-50909, GM-53415, and GM-56699 (to H.S.) and by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (to H.S. and M.T.). J.T. received the Japanese Heart Foundation & Bayer Yakuhin Research Grant Abroad.
REFERENCES
- 1.Alessi, D. R., Y. Saito, D. G. Campbell, P. Cohen, G. Sithanandam, U. Rapp, A. Ashworth, C. J. Marshall, and S. Cowley. 1994. Identification of the sites in MAP kinase kinase-1 phosphorylation by p74raf-1. EMBO J. 13:1610-1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boguslawski, G. 1992. PBS2, a yeast gene encoding a putative protein kinase, interacts with the RAS2 pathway and affects osmotic sensitivity of Saccharomyces cerevisiae. J. Gen. Microbiol. 138:2425-2432. [DOI] [PubMed] [Google Scholar]
- 3.Brewster, J. L., T. de Valoir, N. D. Dwyer, E. Winter, and M. C. Gustin. 1993. An osmosensing signal transduction pathway in yeast. Science 259:1760-1763. [DOI] [PubMed] [Google Scholar]
- 4.Cairns, B. R., S. W. Ramer, and R. D. Kornberg. 1992. Order of action of components in the yeast pheromone response pathway revealed with a dominant allele of the STE11 kinase and the multiple phosphorylation of the STE7 kinase. Genes Dev. 6:1305-1318. [DOI] [PubMed] [Google Scholar]
- 5.Cardone, M. H., G. S. Salvesen, C. Widmann, G. Johnson, and S. M. Frisch. 1997. The regulation of anoikis: MEKK-1 activation requires cleavage by caspases. Cell 90:315-323. [DOI] [PubMed] [Google Scholar]
- 6.Chang, H. Y., H. Nishitoh, X. Yang, H. Ichijo, and D. Baltimore. 1998. Activation of apoptosis signal-regulating kinase 1 (ASK1) by the adapter protein Daxx. Science 281:1860-1863. [DOI] [PubMed] [Google Scholar]
- 7.Chang, L., and M. Karin. 2001. Mammalian MAP kinase signalling cascades. Nature 410:37-40. [DOI] [PubMed] [Google Scholar]
- 8.Chen, Z., T. B. Gibson, F. Robinson, L. Silvestro, G. Pearson, B. Xu, A. Wright, C. Vanderbilt, and M. H. Cobb. 2001. MAP kinases. Chem. Rev. 101:2449-2476. [DOI] [PubMed] [Google Scholar]
- 9.Christianson, T. W., R. S. Sikorski, M. Dante, J. H. Shero, and P. Hieter. 1992. Multifunctional yeast high-copy-number shuttle vectors. Gene 110:119-122. [DOI] [PubMed] [Google Scholar]
- 10.Coleman, K. G., B. S. Wautlet, D. Morrissey, J. Mulheron, S. A. Sedman, P. Brinkley, S. Price, and K. R. Webster. 1997. Identification of CDK4 sequences involved in cyclin D1 and p16 binding. J. Biol. Chem. 272:18869-18874. [DOI] [PubMed] [Google Scholar]
- 11.Deak, J. C., J. V. Cross, M. Lewis, Y. Qian, L. A. Parrott, C. W. Distelhorst, and D. J. Templeton. 1998. Fas-induced proteolytic activation and intracellular redistribution of the stress-signaling kinase MEKK1. Proc. Natl. Acad. Sci. USA 95:5595-5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Durfee, T., K. Becherer, P.-L. Chen, S.-H. Yeh, Y. Yang, A. E. Kilburn, W.-H. Lee, and S. J. Elledge. 1993. The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev. 7:555-569. [DOI] [PubMed] [Google Scholar]
- 13.Fornace, A. J., Jr. 1992. Mammalian genes induced by radiation; activation of genes associated with growth control. Annu. Rev. Genet. 26:507-526. [DOI] [PubMed] [Google Scholar]
- 14.Fornace, A. J., Jr., I. Alamo, Jr., and M. C. Hollander. 1988. DNA damage-inducible transcripts in mammalian cells. Proc. Natl. Acad. Sci. USA 85:8800-8804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gartner, A., K. Nasmyth, and G. Ammerer. 1992. Signal transduction in Saccharomyces cerevisiae requires tyrosine and threonine phosphorylation of FUS3 and KSS1. Genes Dev. 6:1280-1292. [DOI] [PubMed] [Google Scholar]
- 16.Gerwins, P., J. L. Blank, and G. L. Johnson. 1997. Cloning of a novel mitogen-activated protein kinase kinase kinase, MEKK4, that selectively regulates the c-Jun amino terminal kinase pathway. J. Biol. Chem. 272:8288-8295. [DOI] [PubMed] [Google Scholar]
- 17.Goldfarb, D. S., J. Gariepy, G. Schoolnik, and R. D. Kornberg. 1986. Synthetic peptides as nuclear localization signals. Nature 322:641-644. [DOI] [PubMed] [Google Scholar]
- 18.King, A. J., H. Sun, B. Diaz, D. Barnard, W. Miao, S. Bagrodia, and M. S. Marshall. 1998. The protein kinase Pak3 positively regulates Raf-1 activity through phosphorylation of serine 338. Nature 396:180-183. [DOI] [PubMed] [Google Scholar]
- 19.Lei, M., W. Lu, W. Meng, M.-C. Parrini, M. J. Eck, B. J. Mayer, and S. C. Harrison. 2000. Structure of PAK1 in an autoinhibited conformation reveals a multistage activation switch. Cell 102:387-397. [DOI] [PubMed] [Google Scholar]
- 20.Li, L., S. J. Elledge, C. A. Peterson, E. S. Bales, and R. J. Legerski. 1994. Specific association between the human DNA repair proteins XPA and ERCC1. Proc. Natl. Acad. Sci. USA 91:5012-5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu, B., H. Yu, C. Chow, B. Li, W. Zheng, R. J. Davis, and R. A. Flavell. 2001. GADD45γ mediates the activation of the p38 and JNK MAP kinase pathways and cytokine production in effector Th1 cells. Immunity 14:583-590. [DOI] [PubMed] [Google Scholar]
- 22.Maeda, T., M. Takekawa, and H. Saito. 1995. Activation of yeast PBS2 MAPKK by MAPKKKs or by binding of an SH3-containing osmosensor. Science 269:554-558. [DOI] [PubMed] [Google Scholar]
- 23.Maeda, T., S. M. Wurgler-Murphy, and H. Saito. 1994. A two-component system that regulates an osmosensing MAP kinase cascade in yeast. Nature 369:242-245. [DOI] [PubMed] [Google Scholar]
- 24.Mumberg, D., R. Müller, and M. Funk. 1995. Yeast vectors for the controlled expression of heterologous proteins in different genetic background. Gene 156:119-122. [DOI] [PubMed] [Google Scholar]
- 25.Ninomiya-Tsuji, J., S. Nomoto, H. Yasuda, S. I. Reed, and K. Matsumoto. 1991. Cloning of a human cDNA encoding a CDC2-related kinase by complementation of a budding yeast cdc28 mutation. Proc. Natl. Acad. Sci. USA 88:9006-9010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nishitoh, H., M. Saitoh, Y. Mochida, K. Takeda, H. Nakano, M. Rothe, K. Miyazono, and H. Ichijo. 1998. ASK1 is essential for JNK/SAPK activation by TRAF2. Mol. Cell 2:389-395. [DOI] [PubMed] [Google Scholar]
- 27.Papathanasiou, M. A., N. C. Kerr, J. H. Robbins, O. W. McBride, I. Alamo, Jr., S. F. Barrett, I. D. Hickson, and A. J. Fornace, Jr. 1991. Induction by ionizing radiation of the gadd45 gene in cultured human cells: lack of mediation by protein kinase C. Mol. Cell. Biol. 11:1009-1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Payne, D. M., A. J. Rossomando, P. Martino, A. K. Erickson, J.-H. Her, J. Shabanowitz, D. F. Hunt, M. J. Weber, and T. W. Sturgill. 1991. Identification of the regulatory phosphorylation sites in pp42/mitogen-activated protein kinase (MAP kinase). EMBO J. 10:885-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Polverino, A., J. Frost, P. Yang, M. Hutchison, A. M. Neiman, M. H. Cobb, and S. Marcus. 1995. Activation of mitogen-activated protein kinase cascades by p21-activated protein kinases in cell-free extracts of Xenopus oocytes. J. Biol. Chem. 270:26067-26070. [DOI] [PubMed] [Google Scholar]
- 30.Posas, F., and H. Saito. 1998. Activation of the yeast SSK2 MAP kinase kinase kinase by the SSK1 two-component response regulator. EMBO J. 17:1385-1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Posas, F., and H. Saito. 1997. Osmotic activation of the HOG MAPK pathway via Ste11p MAPKKK: scaffold role of Pbs2p MAPKK. Science 276:1702-1705. [DOI] [PubMed] [Google Scholar]
- 32.Posas, F., S. M. Wurgler-Murphy, T. Maeda, E. A. Witten, T. C. Thai, and H. Saito. 1996. Yeast HOG1 MAP kinase cascade is regulated by a multistep phosphorelay mechanism in the SLN1-YPD1-SSK1 “two-component” osmosensor. Cell 86:865-875. [DOI] [PubMed] [Google Scholar]
- 33.Rose, M. D., and J. R. Broach. 1991. Cloning genes by complementation in yeast. Methods Enzymol. 194:195-230. [DOI] [PubMed] [Google Scholar]
- 34.Shaulian, E., and M. Karin. 1999. Stress-induced JNK activation is independent of Gadd45 induction. J. Biol. Chem. 274:29595-29598. [DOI] [PubMed] [Google Scholar]
- 35.Sheikh, M. S., M. C. Hollander, and A. J. Fornace, Jr. 2000. Role of Gadd45 in apoptosis. Biochem. Pharmacol. 59:43-45. [DOI] [PubMed] [Google Scholar]
- 36.Takekawa, M., F. Posas, and H. Saito. 1997. A human homolog of the yeast Ssk2/Ssk22 MAP kinase kinase kinases, MTK1, mediates stress-induced activation of the p38 and JNK pathways. EMBO J. 16:4973-4982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takekawa, M., and H. Saito. 1998. A family of stress-inducible GADD45-like proteins mediate activation of the stress-responsive MTK1/MEKK4 MAPKKK. Cell 95:521-530. [DOI] [PubMed] [Google Scholar]
- 38.Takekawa, M., and H. Saito. 1998. Protein phosphatase 2Cα inhibits the human stress-responsive p38 and JNK MAPK pathways. EMBO J. 17:4744-4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vojtek, A. B., S. M. Hollenberg, and J. A. Cooper. 1993. Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell 74:205-214. [DOI] [PubMed] [Google Scholar]
- 40.Wang, X., M. Gorospe, and N. J. Holbrook. 1999. Gadd45 is not required for activation of c-Jun N-terminal kinase or p38 during acute stress. J. Biol. Chem. 274:29599-29602. [DOI] [PubMed] [Google Scholar]
- 41.Wölfel, T., M. Hauer, J. Schneider, M. Serrano, C. Wölfel, E. Klehmann-Hieb, E. De Plaen, T. Hankeln, K.-H. M. zum Büschenfelde, and D. Beach. 1995. A p16INK4a-insensitive CDK4 mutant targeted by cytolytic T lymphocytes in a human melanoma. Science 269:1281-1284. [DOI] [PubMed] [Google Scholar]
- 42.Xu, S., D. Robbins, J. Frost, A. Dang, C. Lange-Carter, and M. H. Cobb. 1995. MEKK1 phosphorylates MEK1 and MEK2 but does not cause activation of mitogen-activated protein kinase. Proc. Natl. Acad. Sci. USA 92:6808-6812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yamaguchi, K., K. Shirakabe, H. Shibuya, K. Irie, I. Oishi, N. Ueno, T. Taniguchi, E. Nishida, and K. Matsumoto. 1995. Identification of a member of the MAPKKK family as a potential mediator of TGF-β signal transduction. Science 270:2008-2011. [DOI] [PubMed] [Google Scholar]
- 44.Yang, J., H. Zhu, T. L. Murphy, W. Ouyang, and K. M. Murphy. 2001. IL-18-stimulated GADD45β required in cytokine-induced, but not TCR-induced, IFN-γ production. Nat. Immunol. 2:157-164. [DOI] [PubMed] [Google Scholar]
- 45.Zhang, S., J. Han, M. A. Sells, J. Chernoff, U. G. Knaus, R. J. Ulevitch, and G. M. Bokoch. 1995. Rho family GTPases regulate p38 mitogen-activated protein kinase through the downstream mediator Pak1. J. Biol. Chem. 270:23934-23936. [DOI] [PubMed] [Google Scholar]