Abstract
During rRNA biogenesis, multiple RNA and protein substrates are modified and assembled through the coordinated activity of many factors. In Saccharomyces cerevisiae, the double-stranded RNA nuclease Rnt1p and the H/ACA snoRNA pseudouridylase complex participate in the transformation of the nascent pre-rRNA transcript into 35S pre-rRNA. Here we demonstrate the binding of a component of the H/ACA complex (Gar1p) to Rnt1p in vivo and in vitro in the absence of other factors. In vitro, Rnt1p binding to Gar1p is mutually exclusive of its RNA binding and cleavage activities. Mutations in Rnt1p that disrupt Gar1p binding do not inhibit RNA cleavage in vitro but slow RNA processing, prevent nucleolar localization of H/ACA snoRNA-associated proteins, and reduce pre-rRNA pseudouridylation in vivo. These results demonstrate colocalization of various components of the rRNA maturation complex and suggest a mechanism that links rRNA pseudouridylation and cleavage factors.
In eukaryotes, the 25S/28S, 18S, and 5.8S rRNAs are produced by RNA polymerase I as single RNA transcripts that are processed, modified, and assembled into ribosomes within the nucleolus (41). In Saccharomyces cerevisiae, trimming of the nascent pre-rRNA 3′ end and RNA modifications produce the first stable rRNA precursor (50). The 35S pre-rRNA is subjected to a series of further cleavages that remove external transcribed spacers 1 and 2 and internal transcribed spacers 1 and 2 to produce mature rRNA. Changes in the sequence of cleavage or modification sites within the pre-rRNA transcript usually alter the cleavage pattern at distal sites, indicating an interdependent processing pathway (3, 5, 36). However, the mechanistic basis for this coordination remains undefined.
At least six endonucleolytic cleavages are required for the production of mature rRNA in yeast cells, and two endonucleases are known to be involved in rRNA maturation (50). The first is RNase MRP, a ribonucleoprotein endonuclease related to the tRNA processing enzyme RNase P (43). MRP cleaves at site A3, one of two redundant sites that lead to the formation of mature 18S rRNA (46). The second endonuclease is the double-stranded RNA (dsRNA)-specific RNase (Rnt1p), which is the orthologue of the bacterial pre-rRNA processing enzyme RNase III (2, 40). Rnt1p performs the earliest cleavage event at the 3′ end of the pre-rRNA transcript immediately after transcription (2). Rnt1p is also required for the processing of several snRNAs (1, 11, 47, 49) and snoRNAs, including members of both C/D and H/ACA snoRNA families (12). Rnt1p accurately cleaves most of the snoRNA substrates in vitro in the absence of other factors, with the exception of U18, which requires the presence of the C/D small nucleolar ribonucleoprotein (snoRNP) Nop1p (17). Rnt1p is required for normal growth at 30°C and is essential for growth at 37°C (31). Deletion of the enzyme inhibits the cleavage of the 25S pre-rRNA and delays cleavage at site A0 upstream of the 18S rRNA (2, 26). Like MRP, Rnt1p cleaves model substrates at sites identical to those observed in vivo, indicating that no other factors are required for this cleavage reaction (2, 35, 38). Less is known about the other endonucleolytic events; however, many of those require the presence of snoRNPs. For example, cleavage at sites A1 and A2 requires the U3, U14, snR30, and snR10 snoRNPs (32, 37, 48). After endonucleolytic cleavage of the pre-rRNA, the immature termini are trimmed with 5′-3′ exonucleases (e.g., Rat1p and Xrn1p) and 3′-5′ exonucleases (the exosome) to produce mature rRNA (4, 16).
The rRNA undergoes extensive base modifications, including 2′ O-ribose methylation and pseudouridylation (50). Methylation is achieved by an RNP complex that contains most of the C/D box snoRNAs and their associated proteins, with the exception of U3 (25). This complex recognizes the methylation sites by pairing the snoRNAs with a complementary rRNA sequence to allow base methylation, possibly by Nop1p/fibrillarin (6). rRNA pseudouridylation is performed by a snoRNP complex that contains H/ACA snoRNAs and at least four proteins, Gar1p, Nhp2p, Nop10p, and Cbf5p, which is likely a pseudouridylase (39, 51). All four proteins are required for both pseudouridylation and normal cleavage of the 18S pre-rRNA. Depletion of Nhp2p, Cbf5p, or Nop10p results in rapid degradation of Gar1p and the H/ACA snoRNAs, including snR30 and snR10, which are needed for 18S pre-rRNA cleavage (22, 29). Gar1p appears to participate in rRNA cleavage and pseudouridylation through a different mechanism (9, 19). Depletion of Gar1p blocks rRNA pseudouridylation without affecting the accumulation of the snoRNAs or the protein components of the H/ACA snoRNP complex (9).
In vivo labeling experiments have revealed a temporal order for rRNA maturation, where rRNA modifications occur predominantly on the 35S pre-rRNA immediately after cleavage of the nascent 3′ end (50). To maintain this order, one would expect the presence of a coordinating signal recognized by the machinery for both rRNA cleavage and modification. It was thought that this signal is provided by a conserved sequence motif, but an extensive search within the pre-rRNA sequence has failed to identify such a signal in cis (5). This result suggests that the coordination of ribosome biogenesis may be activated instead by protein signals that function in trans. To understand how rRNA modification is regulated and to find potential signals that coordinate ribosome biogenesis, we have searched for proteins that connect pre-rRNA cleavage and modification. The endoribonuclease Rnt1p, which performs the first pre-rRNA cleavage, was used to screen yeast genomic libraries for interacting proteins by using a two-hybrid assay. The screen identified an interaction between Rnt1p and Gar1p, a component of the pseudouridylation snoRNP complex. This interaction was verified in vitro and shown to be required for the nucleolar localization of Gar1p and its associated proteins. This work reveals a mechanism that ensures the colocalization and hence the coordinated functions of components of the pre-rRNA cleavage and modification machinery.
MATERIALS AND METHODS
Strains and plasmids.
Yeast cells were grown and manipulated according to standard procedures (20, 44). The ΔRNT1 strain was described previously (13). Yeast stain PJ69-4A was described previously (24). Gar1p, Rnt1p, and Cbf5p were expressed by using pGBDU (24) and pACT2 (Clontech Labs, Inc., Palo Alto, Calif.). pGBDU plasmids that contain fragments of RNT1 were described previously (31). BD-dsRBD3 was constructed by removing an AvrII-BglII fragment from BD-dsRBD (DS1) (31). BD-dsRBD4 was cloned by inserting an Rnt1p C-terminal PstI fragment from BD-dsRBD (DS1) into pGBDU-C2. Gar1p was PCR amplified from yeast genomic DNA with primers 5′-TAATGAGTTTCAGAGG-3′ and 5′-CTAGCTAGATTATCTTC-3′ and cloned into the SmaI site of pUNI15 (33), resulting in pUNI-Gar1p. The glutathione S-transferase (GST)-Gar1p bacterial expression plasmid was generated by site-specific recombination with the pHB2-GST vector (34). AD-Gar1p/1-205 was cloned by insertion of an EcoRI-XhoI GAR1 fragment from pUNI-Gar1p into the EcoRI-XhoI sites of pACT2. pCu423-Gar1p was made by inserting a PCR fragment of GAR1 amplified from pUNI-Gar1p into the EcoRV site of pCu423 (27) with the 5′end primer 5′-TAATGAGTTTCAGAGG-3′ and the 3′ end primer 5′-CTAATGGTGGTGATGATGATGTCTTCTACCTCCTC-3′, which introduced a six-His tag at the 3′ end of GAR1. The pGBDU-Cbf5p and pGAD-Cbf5p plasmids were made by inserting an EcoRI-XhoI fragment originating from pCBF5-BFG (52) into the EcoRI-SalI sites of pGBDU-C3 and pGAD-C3, respectively. Rnt1p-ST was made by amplifying RNT1 by PCR with primers 5′-CAAGCTTTTGGATCCAATGGGCTC-3′ and 5′-GGCTTAAAAATCTAAC-3′. This amplification removed the native stop codon in Rnt1p, causing translation termination in the vector-encoded stop codon. Rnt1p-M was obtained by random PCR mutagenesis (10). Rnt1p-AA was constructed by using double PCR with an oligonucleotide that introduces a mutation (5′-CAAAAGAATGCGGCAAGAAAATT-3′) into Rnt1p and a primer complementary to the sequence of the vector pGBDU. Immunofluorescence studies were performed by using the ΔRNT1 strain transformed with pCu423-Gar1p, pCBF5-BFG, AD-Gar1p, BD-C6F5p, and either pGBDU, pGBDU-Rnt1p, or pGBDU-Rnt1p-AA.
Two-hybrid screen and analysis.
The two-hybrid screen was conducted by using the Rnt1p gene fused to the Gal4 DNA binding domain (BD) (31). Yeast strain PJ69-4A harboring the pGBDU-Rnt1p plasmid was transformed with three different yeast DNA libraries fused to the Gal4 AD (24). The transformants were selected by using synthetic complete defined (SCD) medium without uracil, leucine, and lysine. Cells with interacting proteins were selected by replica plating on SCD medium lacking either adenine or histidine and supplemented with 20 mM 3-aminotriazole (24). The dependence of cell growth on the protein interaction was examined by testing bait and prey plasmid cosegregation. Cells were grown on a medium that selects only for the interaction (SCD without adenine) and then tested for growth on a medium that selects for the interacting plasmids (SCD without leucine and uracil). Colonies that kept both plasmids after growth on the interaction selection medium were tested for the Rnt1p-dependent interaction. Plasmids harboring Rnt1p were eliminated by growth on medium containing 5-fluoroorotic acid, and colonies were retested. Cells that did not grow without BD-Rnt1p were retransformed with pGBDU-C3 and retested for the interaction. Cells needing BD-Rnt1p and not BD to activate the test promoters were stored. The prey plasmids from 122 colonies were sequenced (24) and identified by using a yeast genome database (14). Two-hybrid assays with Gar1p or Cbf5p and with the various versions of Rnt1p were conducted as described previously (31).
Northern blot analysis.
Northern blot analysis was performed as described earlier (1) with oligonucleotides against snR43 (12) or snR10 (5′ CATGGGTCAAGAACGCCCCGGAGGGG-3′). Analysis of pre-rRNA processing was performed by using probes B and C, which were described earlier (2).
Cleavage and gel shift assays.
The radiolabeled RNAs used in the enzymatic assays were generated with T7 RNA polymerase in the presence of [α-32P]UTP. The RNA substrates were produced from a T7 promoter of the plasmid pRS316 rRNA 3′end (30). For in vitro cleavage, 24 pmol of RNA was incubated with 0.1 pmol of Rnt1p for 20 min at 30°C in 20 μl of reaction buffer. Stop buffer was added, and the samples were loaded onto an 8% polyacrylamide gel (31). RNA binding reactions were performed as described previously (30).
Protein purification.
Recombinant six-His-Rnt1p was purified as described previously (30). GST-Gar1p was produced in Escherichia coli BL21(DE3)/pLysS (Promega Corporation, Madison, Wis.) and purified on a 5-ml glutathione-Sepharose column (APB, Baie d'Urfé, Québec, Canada). The column was washed with 15 column volumes of phosphate-buffered saline (PBS) (pH 7.3) and then eluted with 5 column volumes of elution buffer (50 mM Tris [pH 8.0], 10 mM reduced glutathione). The protein fractions were dialyzed (50% glycerol, 0.5 M KCl, 30 mM Tris [pH 8.0], 0.1 mM dithiothreitol [DTT], 0.1 mM EDTA) and stored at −20°C.
Pull-down assays and Western blotting.
The protein quantities used in assays with purified components are described for each experiment. When assays were performed with extracts, 20 μg of purified Rnt1p was added to bacterial extracts expressing GST-Gar1p or GST alone in PBS (21). The extracts were incubated with 50 μl of glutathione-Sepharose 4B (APB) for 1 h with rotation at 4°C. The beads were washed four times with PBS, transferred to a new tube, washed twice with PBS, and resuspended in 1 volume of Laemmli buffer dye (28). The proteins were separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and visualized with Coomassie blue R (Sigma-Aldrich Canada Ltd., Oakville, Ontario, Canada) or by Western blot analysis (31). The effect of RNA on the protein interaction was tested by adding total yeast RNA, bacterial tRNA (Sigma-Aldrich Canada), or T7 RNA transcripts of the 25S pre-rRNA 3′ end (2) to the binding assay samples. Proteins were visualized with anti-GST antibodies (APB) or anti-Rnt1p antibodies. Western blot analysis of total proteins involved proteins extracted from yeast cells grown to logarithmic phase in the appropriate media as described previously (31). Antibodies against Rnt1p were produced by subcutaneous immunization of a New Zealand White rabbit (Cocalico Biologicals, Inc., Reamstown, Pa.). Crude sera containing anti-Rnt1p antibodies were used at a dilution of 1:15,000.
Pulse-labeling of cellular RNA and determination of pseudouridine content of rRNA.
Cells were grown in SCD medium to an optical density at 600 nm (OD600) of 0.6 at 26°C. The RNA was labeled in vivo by incubation with 1 mCi of [32P]orthophosphate for 15 or 90 min at 26°C. The RNA was extracted, run on a 1.0% agarose-formaldehyde gel, and visualized by autoradiography to observe rRNA processing. The ψ content in the 25S rRNA was determined as described previously (52) with modifications. The 25S rRNA was separated by agarose gel electrophoresis and eluted from total RNA that had been labeled by incubating the cells for 90 min in the presence of [32P]orthophosphate. The RNA was digested with RNase T2 (CLT, Burlington, Ontario, Canada), and 50,000 cpm of digested RNA was used for two-dimensional thin-layer chromatography (TLC) as described earlier (52). The ribonucleotides were visualized by autoradiography and quantified with InstantImager (Packard, Meriden, Conn.).
Glycerol gradient sedimentation analysis.
Glycerol gradient sedimentation analysis was performed as described earlier (8) with several modifications. Yeast cells (OD600, 100) were grown at 26°C in SCD medium lacking histidine, leucine, and uracil. Total proteins were extracted by using glass beads in lysis buffer (20 mM Tris-HCl [pH 8], 5 mM MgCl2, 150 mM NaCl, 0.2% Triton X-100, 1 mM DTT supplemented with 1 mM phenylmethylsulfonyl fluoride and protease inhibitors [156 μg of benzamidine/ml, 1 μg of aprotinin/ml, 1 μg of leupeptin/ml, 1 μg of pepstatin A/ml, and 1 μg of antipain/ml]). The proteins were clarified by two centrifugation steps at 10,000 × g and loaded onto 5 to 35% glycerol gradients prepared in lysis buffer (with the concentration of Triton X-100 reduced to 0.1% and that of DTT reduced to 0.5 mM). The gradients were centrifuged for 5 h at 150,000 × g. Fractions of 1 ml were collected and subjected to SDS-PAGE. The protein bands were visualized by Western blotting and the RNA bands were visualized by Northern blotting as described above.
Immunolocalization.
Yeast cells were grown to an OD600 of 0.5 at 26°C in liquid medium, fixed by the addition of 37% formaldehyde (Sigma-Aldrich Canada) to 5%, and resuspended in pH 6.5 potassium phosphate-buffered 5% formaldehyde. The fixation time varied from 30 to 60 min (42). The primary antibodies used were polyclonal anti-His (Santa Cruz Biotechnology, Santa Cruz, Calif.) at a dilution of 1:1,000 to visualize six-His-Gar1p, monoclonal anti-hemagglutinin (HA) (Roche Diagnostics, Laval, Québec, Canada) at a dilution of 1:2,000 to detect HA-Cbf5p, and polyclonal anti-Nhp2p (22) at a 1:1,000 dilution. Anti-Nop1p antibody was used at a dilution of 1:10,000 (gift from J. P. Aris, University of Florida). All antibodies were revealed by Oregon green 488-conjugated goat secondary antibodies specific for either mouse or rabbit immunoglobulin G (Molecular Probes, Eugene, Oreg.) and used at a 1:1,000 dilution. Antibodies were incubated at 25°C for 2 h in PBS containing 1 mg of bovine serum albumin/ml. Yeast DNA was stained with 1 μg of 4′,6′-diamidino-2-phenylindole (DAPI) (Sigma-Aldrich Canada)/ml. The cells were observed with an epifluorescence Olympus IX70 microscope at a magnification of ×100 (Carsen Group Inc., Markham, Ontario, Canada).
RESULTS
Yeast two-hybrid screen for protein partners of Rnt1p.
Three libraries representing yeast genomic DNAs fused in three reading frames to the Gal4 activation domain (AD) (24) were screened by using the Rnt1p sequence fused to the DNA BD of Gal4 (BD-Rnt1p). Plasmids carrying yeast genomic DNAs were transformed with BD-Rnt1p into a yeast strain harboring three marker genes (24). Interactions were scored by monitoring yeast growth on medium lacking adenine. From 3 × 106 transformants screened, 5,820 colonies grew under selection. About half of these colonies were eliminated because they grew in the absence of BD-Rnt1p, and the remaining 2,979 colonies were stored. From this stock, 122 different plasmids were retrieved and sequenced, revealing 55 known proteins and 23 unknown open reading frames. One of these plasmids, which showed a strong interaction with Rnt1p, contained a DNA fragment encoding the C-terminal 184 amino acids (aa) of the snoRNA binding protein Gar1p. This fragment of Gar1p includes the amino acid sequence required for the viability and nuclear localization of Gar1p (18). As we are interested in pre-rRNA processing, we decided to first analyze the Rnt1p/Gar1p interaction.
Gar1p specifically interacts with the C-terminal domain of Rnt1p in vivo.
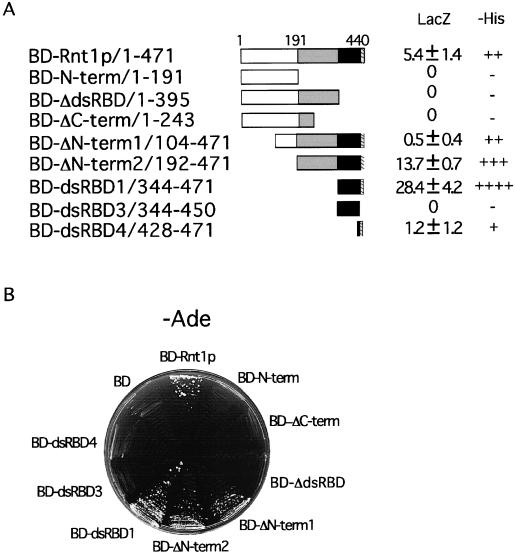
A full-length genomic copy of Gar1p was fused to the Gal4 AD (AD-Gar1p) and tested for an interaction with Rnt1p. As shown in Fig. 1, AD-Gar1p interacted with BD-Rnt1p, confirming the interaction identified by the two-hybrid screen. Rnt1p has an N-terminal domain required for correct protein conformation, a nuclease domain essential for RNA cleavage, and a dsRNA BD (dsRBD) needed for RNA binding (31). To map the Gar1p BD of Rnt1p, a series of Rnt1p deletion mutants were tested for interactions with Gar1p. Deletions of the dsRBD (BD-N-term/1-191, BD-ΔdsRBD/1-395, and BD-ΔC-term/1-243) disrupted the interaction with Gar1p, while deletions of the N-terminal domain (BD-ΔN-term1/104-471 and BD-ΔN-term2/192-471) or the nuclease domain (BD-dsRBD1/344-471) did not (Fig. 1). Deletion of 21 aa downstream of the dsRNA binding motif (BD-dsRBD3/344-450) abolished the interaction. The mutant with the last 44 aa (BD-dsRBD4/428-471), including 10 aa of the dsRBD, interacted weakly with Gar1p (Fig. 1). This result suggests that the amino acid residues downstream of the canonical dsRBM (Fig. 2A) form a primary Gar1p binding site. As shown in Fig. 2B, the introduction of a stop codon at position 441 of Rnt1p, which deletes the last 30 aa, blocked the interaction with Gar1p. Therefore, the C-terminal 30 aa of Rnt1p constitute the principal Gar1p binding site, but other elements in the dsRBD strengthen the interaction.
FIG. 1.
Gar1p interacts with the C-terminal domain of Rnt1p in vivo. (A) Schematic representation of the BD-Rnt1p fragments tested for interactions with AD-Gar1p. Rnt1p has three domains; the N-terminal domain (white box), the nuclease domain (gray box), and the dsRBD (black box). The hatched box at the end of the dsRBD represents the primary Gar1p binding site. Interaction was assessed by the ability of the plasmid to support cell growth on medium without histidine (−His) (interactions: ++++, very strong; +++, strong; ++, moderate; +, weak; −, none) (A) or adenine (−Ade) (B). The strength of the interaction was determined by using a liquid LacZ assay (A), and the expression level averaged from three experiments is shown in Miller units (mean and standard deviation) (36).
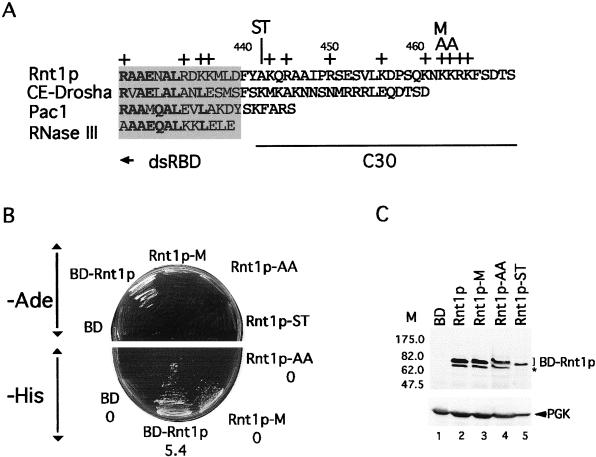
FIG. 2.
Point mutations at the end of the Rnt1p C-terminal domain disrupt its interaction with Gar1p. (A) Comparison of the C termini of different RNase III (40), fission yeast Pac1 (45), worm CE-Drosha (15), and budding yeast Rnt1p (2). The shaded area represents the terminal part of the canonical dsRBD. Conserved residues are shown in boldface. The C30 region of Rnt1p that interacts with Gar1p is underlined. The plus signs indicate positively charged amino acids. The Rnt1p-ST, Rnt1p-M, and Rnt1p-AA mutants, which disrupt the Gar1p interaction, are indicated at the top. Throughout this study, all proteins were tested as fusions with the Gal4 BD. (B) Interaction assay for Gar1p and Rnt1p proteins that carry mutations in the C30 region. Yeast cells carrying Gar1p fused to the Gal4 AD and different Rnt1p mutants fused to the Gal4 BD were tested on medium without adenine (upper panel) or without histidine (lower panel). The interaction strength (average of three experiments) in Miller units (36) is indicated below the name of each plasmid in the lower panel. (C) Western blot analysis of Rnt1p mutations. Proteins were extracted from ΔRNT1 cells expressing different mutations in Rnt1p or expressing the Gal4 BD. Proteins were separated by SDS-PAGE and examined with polyclonal antibodies against Rnt1p. Anti-3-phosphoglycerate kinase (anti-Pak) antibodies were used as an internal control. The molecular mass markers (M) are indicated on the left in kilodaltons. The asterisk indicates proteins that were formed due to a stop codon that was removed during the construction of Rnt1p-ST (see Material and Methods).
The region containing the C-terminal 30 aa (C30 region) has no apparent binding motif but has a number of positively charged amino acids that could support an ionic interaction with Gar1p (Fig. 2A). We systematically changed these residues into uncharged amino acids and retested them for interactions with Gar1p. As shown in Fig. 2B, substitution of methionine in place of lysine 463 (Rnt1p-M) weakened the interaction between Rnt1p and Gar1p, while substitution of alanine in place of lysines 463 and 464 (Rnt1p-AA) completely blocked the interaction. These results show that the Gar1p/Rnt1p interaction is mediated by selected amino acid residues downstream of the canonical dsRBM.
Rnt1p interacts with Gar1p in vitro in the absence of any other factor.
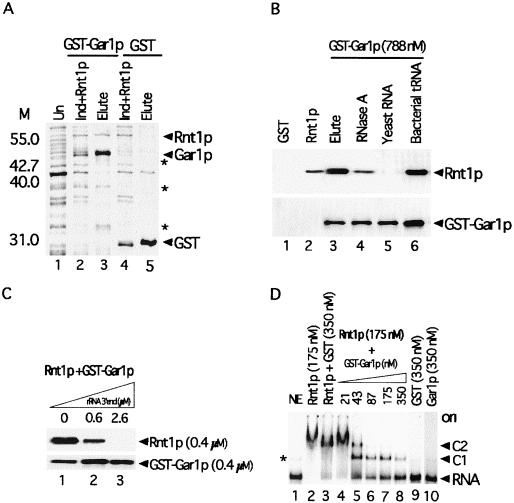
To determine whether Rnt1p interacts directly with Gar1p, we expressed both proteins in bacteria and assessed their interaction in vitro (Fig. 3). Pure six-His-Rnt1p was added to extracts of bacterial cells expressing either Gar1p fused to GST or GST alone, and the complexes were precipitated with glutathione-agarose beads. As shown in Fig. 3A, both GST-Gar1p and six-His-Rnt1p were present in the lysate (lane 2) and were coprecipitated together (lane 3). In contrast, Rnt1p was not precipitated when incubated with an extract expressing GST (Fig. 3A, lane 5). Therefore, Rnt1p specifically interacts with Gar1p in vitro in the absence of any other yeast protein or RNA.
FIG. 3.
Rnt1p specifically interacts with Gar1p in vitro in the absence of other factors. (A) Coprecipitation of the GST-Gar1p fusion with six-His-Rnt1p in bacterial cell extracts. Shown is a stained gel of total proteins from uninduced cells (lane 1), total proteins from cells expressing GST-Gar1p supplemented with six-His-Rnt1p (lane 2), proteins eluted from glutathione-agarose beads incubated with an extract containing GST-Gar1p and six-His-Rnt1p (lane 3), total proteins from cells expressing GST and six-His-Rnt1p (lane 4), and proteins eluted from beads incubated with an extract containing GST and six-His-Rnt1p (lane 5). The protein positions are indicated on the right. The asterisks indicate degradation products of GST-Gar1p. The molecular weight markers (M) are indicated on the left. (B) Gar1p interacts with Rnt1p in the absence but not in the presence of yeast RNA. Rnt1p was incubated with GST (lane 1), GST-Gar1p (lane 3), GST-Gar1p pretreated with 10 μg of RNase A (lane 4), GST-Gar1p and 10 μg of yeast total RNA (lane 5), or GST-Gar1p and 10 μg of bacterial tRNA (lane 6). The proteins were visualized by using antibodies against Rnt1p (top panel) or GST (bottom panel). The input Rnt1p was loaded directly onto the gel as a control (lane 2). (C) An Rnt1p substrate disrupts its interaction with Gar1p. Coprecipitation of GST-Gar1p and six-His-Rnt1p was tested in the presence of increasing amounts of in vitro transcribed 25S pre-rRNA 3′ end (2). (D) Gar1p competes with RNA for binding to Rnt1p in a gel shift assay. RNA was incubated with no protein (lane 1), six-His-Rnt1p (lane 2), six-His-Rnt1p and GST (lane 3), or six-His-Rnt1p and increasing amounts of GST-Gar1p (lanes 4 to 8). GST (lane 9) or Gar1p (lane 10) alone was incubated with RNA as a negative control. The positions of shifted RNAs are indicated on the right. C1 and C2 represent the complexes that Rnt1p forms when binding with the 25S pre-rRNA 3′ end (30). The asterisk indicates a minor form of naked RNA that runs faster than the protein-RNA complex. ori, origin.
Purified recombinant Rnt1p and Gar1p were treated with RNases and tested for an interaction in a GST pull-down assay. As shown in Fig. 3B, the two proteins were coprecipitated in the absence of bacterial proteins (lane 3). Treatment with 10 μg of RNase A (Fig. 3B, lane 4) slightly decreased the coprecipitation of the Rnt1p/Gar1p complex. Higher concentrations of RNase A did not further affect the coprecipitation of the complex (data not shown), suggesting that Rnt1p can interact with Gar1p in the absence of RNA.
The Rnt1p dsRBD appears to overlap the Gar1p binding site (Fig. 1), and we wished to test whether the presence of RNA could alter the Rnt1p interaction with Gar1p. As shown in Fig. 3B, the addition of bacterial tRNA to a Gar1p binding reaction mixture did not affect complex formation (lane 6), whereas an equal amount of yeast total RNA abolished the Rnt1p interaction with Gar1p (lane 5). These results indicate that components of yeast RNA, possibly Rnt1p substrates, block the Rnt1p interaction with Gar1p.
To more specifically determine whether Rnt1p substrates can disrupt the interaction with Gar1p, we incubated the two proteins with increasing amounts of a model rRNA 3′ end (2) and observed the effects on complex formation. In the absence of RNA, both proteins were coprecipitated (Fig. 3C, lane 1), while the addition of substrate concentrations equal to or greater than that of Rnt1p inhibited complex precipitation (lanes 2 and 3). These results show that Rnt1p substrates disrupt the interaction with Gar1p. In addition, we tested the effect of Gar1p on the capacity of Rnt1p to bind to RNA. As shown in Fig. 3D, lanes 6 to 8, the addition of increasing amounts of Gar1p reduces RNA binding to Rnt1p. Rnt1p alone or together with GST completely bound to and shifted the substrate (Fig. 3D, lanes 2 and 3). Gar1p did not interact with the 25S rRNA 3′end alone (Fig. 3D, lane 10), and no new complexes that could correspond to an Rnt1p/Gar1p/RNA tertiary complex were observed (Fig. 3D). We conclude that Rnt1p cannot simultaneously bind to Gar1p and its RNA substrate.
Mutations that disrupt the Gar1p/Rnt1p complex slow rRNA processing in vivo without affecting Rnt1p catalytic activity in vitro.
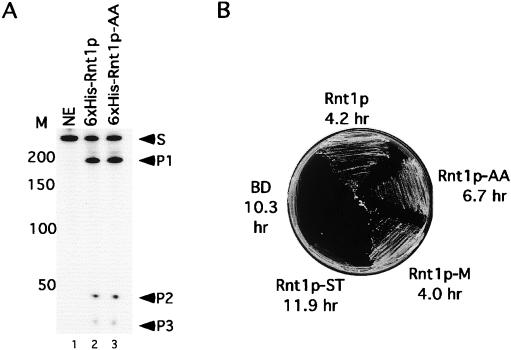
The Rnt1p-ST and Rnt1p-AA mutants, which disrupted the interaction with Gar1p, were expressed in bacteria, and their catalytic activities were tested in vitro. Rnt1p-ST was inactive (data not shown), while Rnt1p-AA efficiently bound to and cleaved its RNA substrate (Fig. 4A). We conclude that Rnt1p interaction and catalytic activities are distinct.
FIG. 4.
Mutations that disrupt the Rnt1p interaction with Gar1p slow yeast growth without affecting RNA cleavage in vitro. (A) Six-His-Rnt1p-AA efficiently cleaves RNA in vitro. A model 25S pre-rRNA 3′end substrate was incubated alone (lane 1) or with equal amounts of either six-His-Rnt1p (lane 2) or six-His-Rnt1p-AA (lane 3). The positions of the substrate (S) and the cleavage products (P1, P2, and P3) are indicated on the right. The DNA markers (M) are indicated on the left. (B) Mutations within the C30 region reduce cell growth. Cells lacking RNT1 were transformed with plasmids carrying different RNT1alleles and tested for growth at 37°C on plates. The doubling time at 26°C in liquid medium was also measured, and the average of three experiments is indicated below the name of each plasmid.
Mutations that disrupted the Rnt1p/Gar1p interaction were transformed into ΔRNT1 cells, and their effects on cell growth and RNA cleavage were observed. Because the fusion of Rnt1p to the Gal4 BD does not affect Rnt1p function (31), the mutants were maintained as BD fusions to allow direct comparisons of Gar1p binding efficiency and Rnt1p activity in vivo. Rnt1p complemented the RNT1 deletion and enabled yeast cells to grow at both permissive (26°C) and restrictive (37°C) temperatures (Fig. 4B). Cells expressing the BD or Rnt1p-ST with the BD failed to grow at 37°C and grew slowly at 26°C. These results are consistent with the inability of Rnt1p-ST to cleave RNA in vitro (data not shown) and suggest an essential role for the C30 region in Rnt1p folding or activity. The Rnt1p-M mutant, which weakened the interaction with Gar1p (Fig. 2B), grew efficiently at both 26 and 37°C, while the Rnt1p-AA mutant, which disrupted the Rnt1p/Gar1p interaction (Fig. 2B), showed partial growth at both 26 and 37°C (Fig. 4B). We conclude that the Rnt1p interaction with Gar1p is not essential but is required for optimum cell growth.
The effect of Rnt1p-AA on rRNA processing in vivo was examined by Northern blot analysis. A probe specific for sequences downstream of the Rnt1p cleavage site (Fig. 5A) or a probe specific for the 5.8S rRNA (Fig. 5B) hybridized to RNA extracted from cells lacking Rnt1p (lane 2) and revealed an accumulation of 3′-extended 25S rRNA, 35S pre-rRNA, and 27S pre-rRNA, as described previously (2, 26). However, RNA extracted from cells expressing Rnt1p-AA (Fig. 5A and B, lane 3) was similar to that extracted from wild-type cells (lane 1), revealing no accumulation of unprocessed rRNA. Similarly, the processing of other known Rnt1p snRNA and snoRNA substrates continued in cells expressing Rnt1p-AA, as judged by Northern blot analysis (data not shown). These results suggest that mutations disrupting the Rnt1p/Gar1p complex do not affect the accumulation of mature rRNA.
FIG. 5.
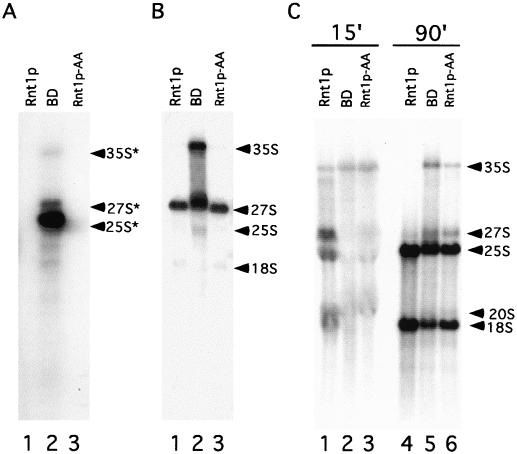
Mutations that disrupt the Rnt1p/Gar1p interaction delay pre-rRNA processing. Total RNA was extracted from cells expressing Rnt1p, BD, or Rnt1p-AA, separated on a denaturing agrose gel, and subjected to Northern blot analysis with a probe against the precursor mature 3′ end of the 25S (A) or 5.8S (B) rRNA to reveal rRNA precursors. (C) Pulse-labeling of rRNA. Cells expressing BD-Rnt1p, BD, or Rnt1p-AA were grown at 26°C on minimal medium lacking inorganic phosphate to mid-log phase and labeled by the addition of [32P]orthophosphate for 15 or 90 min before the RNA was extracted and separated on a 1.2% agarose-formaldehyde gel. The position of each RNA species is indicated on the right. The asterisks indicate rRNA precursors that extended beyond the Rnt1p processing site.
To determine whether the mutations K463/A and K464/A affect the rate of rRNA processing, we observed the kinetics of rRNA processing in vivo in cells that lack Rnt1p, express Rnt1p, or express Rnt1p-AA. The RNA was pulse-labeled in vivo for 15 or 90 min, and the resulting labeled RNA species were separated on an agarose gel. As shown in Fig. 5C, RNA extracted from cells expressing Rnt1p after 15 min of labeling contained pre-rRNA precursors and mature rRNA (lane 1), while RNA extracted from cells labeled for 90 min contained mainly mature rRNA (lane 4). As expected, BD-expressing cells that lacked Rnt1p showed a long delay in rRNA processing. For ΔRNT1 cells, 15 min of labeling resulted in the appearance of the 35S rRNA precursor as the most visible band, and very small amounts of processing intermediates or mature RNA were detected (lane 2). After 90 min of labeling, the 35S and 27S rRNA precursors remained visible (lane 5), while mature rRNA started to accumulate. These results suggest that deletion of Rnt1p not only blocks the processing of the 25S rRNA 3′ end, as previously described, but also delays all rRNA processing steps. Surprisingly, mutations that disrupted the Rnt1p/Gar1p complex but did not affect the steady-state accumulation of rRNA showed very long delays in rRNA processing, similar to that seen with a full RNT1 deletion. Furthermore, after 15 min, the pre-rRNA remained mainly in the form of the 35S rRNA precursor (Fig. 5C, lane 3), and significant amounts of this pre-rRNA could be seen after 90 min of labeling (lane 6). Interestingly, all pre-rRNAs in these mutants appeared to eventually mature, as revealed by analysis of the steady-state RNA population (Fig. 5A and B). We conclude that mutations disrupting the Rnt1p/Gar1p complex significantly slow the processing of rRNA primary transcripts, possibly due to a delay in both Rnt1p and Gar1p processing steps.
Disruption of the Rnt1p/Gar1p interaction destabilizes the H/ACA snoRNP complex and reduces rRNA pseudouridylation in vivo.
Gar1p is an integral component of the pseudouridylation complex, and its depletion blocks rRNA pseudouridylation (9). To determine the effects of disrupting the Rnt1p/Gar1p interaction on the stability of the Gar1p-associated pseudouridylation snoRNP complex, we examined the sedimentation patterns of Rnt1p and components of the Gar1p-associated snoRNP complex in cells expressing Rnt1p or Rnt1p-AA. Extracts of cells expressing Rnt1p or Rnt1p-AA were overlaid on a glycerol gradient, and the fractions were analyzed by Western blotting with antibodies against Rnt1p or tagged versions of Cbf5p and Gar1p. The pattern of the Gar1p-associated H/ACA snoRNA snR10 was detected by Northern blotting. As shown in the left panel of Fig. 6A, in cell extracts expressing Rnt1p, fractions 6 to 8 contained Gar1p, Cbf5p, Nhp2p, Rnt1p, and snR10. The putative snoRNP complex migrated between the 40S and 60S ribosomal subunits. The migration pattern of Rnt1p closely followed that of Gar1p. All the fractions with detectable Gar1p also contained Rnt1p, Cbf5p, Nhp2p, and snR10, suggesting that Rnt1p interacts with snoRNP-bound Gar1p and not a distinct form of Gar1p. In cell extracts expressing Rnt1p-AA, which does not interact with Gar1p, Rnt1p was no longer fractionated with the other proteins and was detected closer to the top of the gradient (Fig. 6A, right panel). Nhp2p appeared to be spread randomly across the gradient, while Cbf5p was again found close to the top of the gradient. Peculiarly, both Gar1p and snR10 migrated closer to the bottom of the gradient, probably due to aggregation or abnormal interaction with a larger complex. These results confirm that mutations K463/A and K464/A (Fig. 2A) disrupt the interaction with Gar1p in vivo and reveal a relationship between Rnt1p and the sedimentation pattern of members of the pseudouridylation snoRNA complex. We conclude that the Rnt1p association with Gar1p is required for the stability of the pseudouridylation snoRNP complex.
FIG. 6.
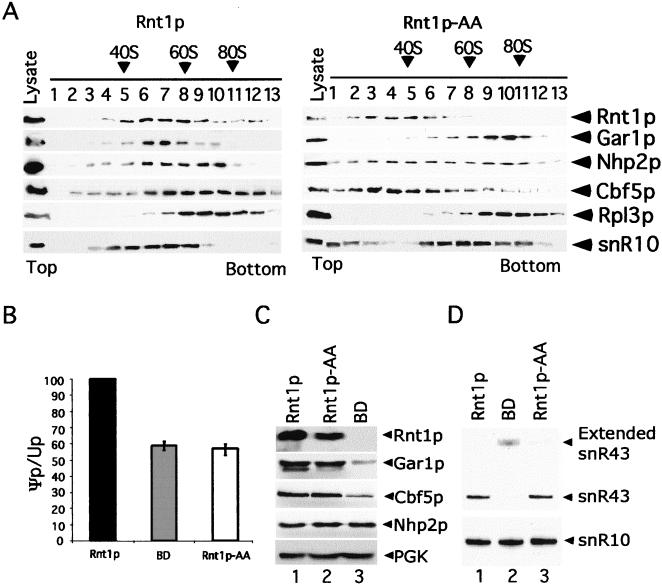
Inhibition of the Rnt1p/Gar1p interaction disrupts the H/ACA snoRNP complex and reduces rRNA pseudouridylation. (A) Sedimentation analysis of the Rnt1p/Gar1p complex. Cell extracts were separated on a 5 to 35% glycerol gradient, the fractions were loaded onto an SDS-polyacrylamide gel, and the bands corresponding to the different proteins were visualized by Western blot analysis. Antibodies against the large-subunit ribosomal protein Rpl3p were used as a marker. The position of the small subunit was determined by using rRNA as a marker (data not shown). The sedimentation pattern of H/ACA snoRNA was analyzed by Northern blotting with a probe against snR10. The positions of the different proteins and RNA are indicated on the right, while the fraction numbers are indicated at the top. (B) Quantitative analysis of 25S rRNA pseudouridylation. In vivo labeled 25S rRNA was digested with RNase T2 and subjected to two-dimensional TLC. The spots were quantified, and Ψp/Up ratios were determined and indicated as a percentage of the wild-type ratio. The results are averages of three different experiments, and error bars are indicated. (C) Effect of Rnt1p on the levels of expression of Gar1p, Cbf5p, and Nhp2p. Proteins from ΔRNT1 cells expressing tagged Gar1p and Cbf5p along with Rnt1p, Rnt1p-AA, or BD were analyzed by Western blotting. PGK was included as a loading control. (D) Northern blot analysis of H/ACA box snoRNA in cells expressing Rnt1p, Rnt1p-AA, or BD. RNA was extracted from different strains and detected by Northern blot analysis with radiolabeled probes against snR10 and snR43. The positions of the mature and extended RNA forms are indicated on the right.
To determine the effects of disrupting the Rnt1p/Gar1p interaction on Gar1p-dependent pseudouridylation, we examined the effects of mutations in Rnt1p on the level of 25S rRNA pseudouridylation. In vivo labeled 25S rRNA was gel purified and digested with RNase T2, and the ratio of ψp to Up nucleotides was examined by using TLC. Spot quantification showed that Rnt1p deletion or expression of Rnt1p-AA reduced 25S rRNA pseudouridylation by about 40% compared to the wild-type level (Fig. 6B). The levels of expression of Gar1p, Rnt1p, Nhp2p, and Cbf5p were normal in cells expressing Rnt1p-AA (Fig. 6C). The levels of expression of H/ACA snoRNAs snR10 and snR43 were also normal in cells expressing Rnt1p-AA (Fig. 6D) (data not shown). These results indicate that disruption of the Rnt1p/Gar1p complex reduces rRNA pseudouridylation without depleting the protein or RNA components of the pseudouridylation snoRNP complex. In contrast, deletion of Rnt1p reduces Gar1p and Cbf5p expression, possibly due to a general inhibition of snoRNA and rRNA processing (Fig. 6C) (data not shown). In ΔRNT1 cells, the depletion of Cbf5p was not associated with the depletion of snR10, as previously reported (29), because the remaining fraction of Cbf5p was sufficient to stabilize the reduced amount of mature H/ACA snoRNA in ΔRNT1 cells.
The interaction between Rnt1p and Gar1p is required for the nuclear localization of the pseudouridylation complex.
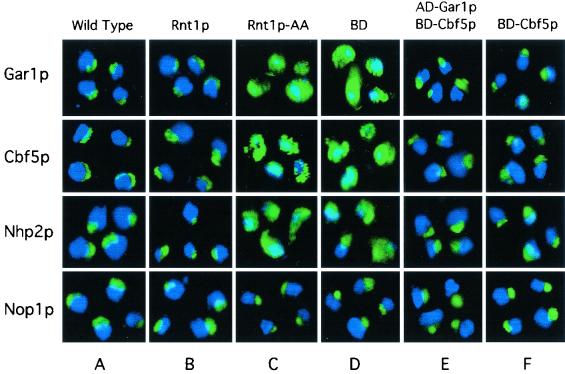
The localization patterns for Cbf5p, Nhp2p, and Gar1p were examined in the absence of Rnt1p or in the presence of Rnt1p-AA. The proteins were visualized in situ by using antibodies against Rnt1p, Nhp2p, HA-Cbf5p, and six-His-Gar1p. Wild-type and Rnt1p cells (Fig. 7A and B) showed a similar crescent-shaped staining of Cbf5p, Nhp2p, and Gar1p, corresponding to the nucleolus. Rnt1p-AA cells showed a staining pattern ranging from cytoplasmic staining to nearly normal nucleolar staining for all three proteins (Fig. 7C). Examination of 300 Rnt1p-AA cells revealed that 40% exhibited normal nucleolar staining for Nhp2p and 30% exhibited normal staining for either Gar1p or Cbf5p. In BD-expressing cells that lacked Rnt1p, most of the staining, including that for Nhp2p, was either cytoplasmic or cytoplasmic with overlapping nuclear staining (Fig. 7D). Patterns of immunostaining with antibodies against the C/D snoRNA-associated protein Nop1p were similar in wild-type, BD, and Rnt1p-AA cells (Fig. 7). These results indicate that the misslocalization of Gar1p, Nhp2p, and Cbf5p is not due to a general disruption of the nucleolus. We also examined the localization pattern of Rnt1p-AA and found it similar to that of wild-type Rnt1p (data not shown). This result suggests that Gar1p, Cbf5p, and Nhp2p nuclear localization requires an interaction between Rnt1p and Gar1p. To directly test this possibility, we expressed Gar1p and Cbf5p in fusions with the Gal4 BD or AD linked to the simian virus 40 (SV40) nuclear localization signal (NLS) in ΔRNT1 cells and observed their localization patterns. The SV40 NLS is a strong NLS that is routinely used in two-hybrid assays to import proteins into the nucleus (24). As shown in Fig. 7E, both AD-Gar1p and BD-Cbf5p correctly localized in the nucleolus, suggesting that Gar1p and Cbf5p need Rnt1p to enter the nucleus and not associate with the nucleolus. Interestingly, Gar1p- and Cbf5p-NLS fusions also restored the localization pattern of Nhp2p, suggesting that all three proteins interact and enter the nucleus together with a single NLS provided by Rnt1p. As shown in Fig. 7F, the expression of BD-Cbf5p alone in ΔRNT1 cells restored the nucleolar localization of Cbf5p, Nhp2p, and Gar1p. We conclude that the Rnt1p/Gar1p interaction is required for the efficient localization of Gar1p and its associated proteins.
FIG. 7.
Disruption of the Gar1p/Rnt1p interaction impairs the nucleolar localization of Gar1p, Cbf5p, and Nhp2p. In situ immunofluorescence analysis was conducted with wild-type cells (A) or ΔRNT1 cells expressing Rnt1p (B), Rnt1p-AA (C), BD (D), AD-Gar1p and BD-Cbf5p (E), or BD-Cbf5p alone (F). Blue represents DAPI staining of the nucleoplasm, while green represents antibodies against Gar1p, Cbf5p, Nhp2p, or Nop1p visualized by Oregon green-conjugated goat secondary antibodies. Costaining appears in blue-green.
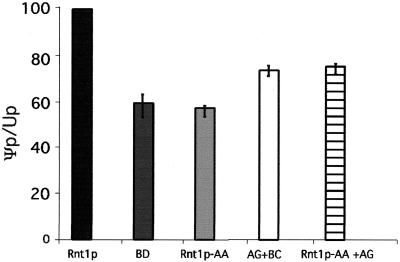
To determine whether Gar1p nucleolar localization is sufficient for rRNA pseudouridylation, we examined rRNA pseudouridylation in ΔRNT1 and Rnt1p-AA cells expressing AD-Gar1p or both AD-Gar1p and BD-Cbf5p carrying the SV40 NLS. As shown in Fig. 8, the expression of both AD-Gar1p and BD-Cbf5p restored rRNA pseudouridylation to about 75% the wild-type level, indicating that Gar1p is partially responsible for the pseudouridylation defect caused by the deletion of Rnt1p. This result is not surprising, since the deletion of Rnt1p blocks the processing of many H/ACA snoRNAs needed for rRNA pseudouridylation. The expression of both AD-Gar1p and Rnt1p-AA in ΔRNT1 cells did not completely restore 25S rRNA pseudouridylation. This result suggests that AD-Gar1p nucleolar localization remains less efficient than Rnt1p-dependent localization or that the Gar1p/Rnt1p interaction is also required for efficient rRNA processing.
FIG. 8.
Rnt1p-independent nucleolar localization of Gar1p partially restores rRNA pseudouridylation. RNA was extracted from ΔRNT1 cells expressing BD, Rnt1p, and Rnt1p-AA as described in the legend to Fig. 6. In addition, RNA was extracted from cells transformed with three different plasmids expressing BD, AD-Gar1p plus BD-Cbf5p (AG+BC), and Rnt1p-AA plus Gar1p (Rnt1p-AA+AG). Total RNA was separated on a denaturing gel, and 25S rRNA was extracted. The purified 25S rRNA was digested with RNase T2 and subjected to two-dimensional TLC. The spots were quantified, and Ψp/Up ratios were determined and indicated as a percentage of the wild-type ratio. The results are averages of three different experiments, and error bars are indicated.
DISCUSSION
In this study, we showed binding between the rRNA cleavage enzyme Rnt1p and Gar1p, a component of the pseudouridylation snoRNP complex. Mutations that disrupted the Rnt1p/Gar1p interaction did not inhibit the expression of Gar1p or its associated snoRNPs but slowed rRNA maturation and reduced pseudouridylation of 25S rRNA (Fig. 5 and 6). In situ immunofluorescence analysis showed that disrupting the Rnt1p/Gar1p interaction impairs the nucleolar localization of Gar1p, Nhp2p, and Cbf5p (Fig. 7). Fusion of an NLS to Gar1p or Cbf5p restored the nucleolar localization of Gar1p, Cbf5p, and Nhp2p independently of Rnt1p (Fig. 7). These results demonstrate a link between Rnt1p and the H/ACA snoRNP complex and suggest coordinated transport and function of the rRNA cleavage and pseudouridylation machineries.
Physical link between Rnt1p and the 35S pre-rRNA maturation complex.
In yeast cells, pre-rRNA processing starts immediately after transcription with Rnt1p cleavage downstream of the 25S rRNA to form 35S pre-rRNA (2, 26). This cleavage event was thought to be physically and functionally distinct from 35S pre-rRNA maturation. Here, we provide evidence that Rnt1p physically interacts with Gar1p, a factor required for the pseudouridylation and cleavage of 35S pre-rRNA (Fig. 1 and 2). This interaction links factors required for cleavage at the 3′ end of the 25S pre-rRNA with those needed for cleavage at the pre-rRNA 5′ end and rRNA pseudouridylation (9). The connection between processing of the nascent pre-rRNA transcript and the 35S pre-rRNA would ensure ordered pre-rRNA processing and prevent processing of incomplete or erroneous transcripts. Indeed, the disruption of the Gar1p/Rnt1p interaction slows cleavage at both the 5′ and the 3′ ends of the pre-rRNA (Fig. 5C) and reduces 25S rRNA pseudouridylation (Fig. 6B). The fact that rRNA cleavage and pseudouridylation are not completely blocked upon loss of the Rnt1/Gar1p interaction suggests that the interaction plays a coordinating rather than a housekeeping role in rRNA processing. The observed delay in rRNA processing and reduced pseudouridylation are unlikely to be caused by a defect in snoRNA maturation, as the maturation of H/ACA and C/D snoRNAs was not significantly affected by disruption of the Rnt1p/Gar1p interaction (data not shown). Furthermore, rRNA pseudouridylation was reduced to similar levels both when the Rnt1p/Gar1p complex was disrupted and when Rnt1p was deleted, despite the tremendous difference in the amounts of mature snoRNA observed.
It is not clear if the Gar1p/Rnt1p complex is the only direct connection between Rnt1p and the 35S rRNA processing machinery or whether other interactions reinforce this link. Recently, an interaction between Rnt1p and the C/D snoRNA-associated protein Nop1p was shown to play a role in the processing of U18 snoRNA, but the effect of this interaction on rRNA processing was not tested (17). In our screen, Gar1p was the only known factor related to pre-rRNA maturation. However, we have sequenced only 4% of the plasmids recovered, and more pre-rRNA maturation factors may exist in the pool. In any case, the connection between Rnt1p and the pseudouridylation complex appears to be mediated primarily by Gar1p. Disruption of the Gar1p interaction with Rnt1p alters the localization pattern not only for Gar1p but also for Nhp2p and Cbf5p (Fig. 7). In addition, Rnt1p does not interact directly with Cbf5p in a two-hybrid system, nor does Rnt1p interact with H/ACA snoRNAs in vivo or in vitro (Y. Yam, B. Lamontagne, S. Larose, and S. Abou Elela, unpublished observations).
Gar1p is required for coupled transport of Rnt1p and protein components of the pseudouridylation snoRNP complex.
The contribution of Gar1p to the pseudouridylation reaction is not known. Gar1p is not required for the production or stability of the pseudouridylation snoRNP complex, nor does it possess homology with pseudouridylases (7, 9). However, depletion of Cbf5p or Nhp2p results in the rapid degradation of Gar1p, while depletion of Gar1p does not affect the stability of any of the H/ACA snoRNP components but appears to affect the association of the snoRNP complex with rRNA (9, 23, 51). These findings suggest that Gar1p is not a core component of the pseudouridylation machinery and suggest a possible role for Gar1p as a regulator or coordinator of rRNA pseudouridylation. The role of Gar1p as a coordinator is further supported by the results shown in this study. Disruption of the Rnt1p/Gar1p interaction blocks not only the nuclear import of Gar1p but also the import of Cbf5p and Nhp2p. Therefore, the three proteins appear to enter the nucleus by a single NLS provided by Rnt1p, with Gar1p providing a structural bridge. This coordinated transport would ensure the simultaneous and equimolar import of the H/ACA snoRNP components and link it to the import of the rRNA cleavage enzyme Rnt1p.
In addition to its role in ensuring the coimport of the H/ACA snoRNP components, Gar1p may also help coordinate rRNA cleavage and pseudouridylation. The fact that Gar1p needs to interact with Rnt1p to reach the nucleus (Fig. 7) suggests that the two proteins enter the nucleus as a complex. However, since Rnt1p cannot simultaneously bind Gar1p and its RNA substrate (Fig. 3C), it is likely that the complex is disrupted upon encountering Rnt1p substrates in the nucleolus. This means that Rnt1p and Gar1p reach their site of activity and probably function in a coordinated manner. On the other hand, we cannot rule out the possibility that the binding of Rnt1p to Gar1p is required for the initiation of rRNA pseudouridylation. In this scenario, Rnt1p binding to Gar1p is blocked by Rnt1p substrates until processing is complete and RNA pseudouridylation is needed. It is clear that the interaction of Rnt1p and Gar1p physically links components of the rRNA processing and pseudouridylation machineries, and this observation underscores the possibilities for coordinated rRNA processing.
Acknowledgments
We thank J. Carbon, S. Fournier, J. Warner, J. Aris, J. P. Gélugne, P. James, S. Labbé, D. Thiele, and S. J. Elledge for sending plasmids, strains, and antibodies. We are indebted to B. Chabot, R. Wellinger, and all the members of the laboratory of Sherif Abou Elela for critical reading of the manuscript.
This work was supported by grant 216854 from the Natural Sciences and Engineering Research Council of Canada (NSERC) and grant MOP-14305 from the Canadian Institute for Health Research (CIHR). Support for the RNA group core was provided by CIHR. S.A.E. is Chercheur-Boursier Junior II of the Fonds de la Recherche en Santé du Québec.
REFERENCES
- 1.Abou Elela, S., and M. Ares, Jr. 1998. Depletion of yeast RNase III blocks correct U2 3′ end formation and results in polyadenylated but functional U2 snRNA. EMBO J. 17:3738-3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abou Elela, S., H. Igel, and M. Ares, Jr. 1996. RNase III cleaves eukaryotic preribosomal RNA at a U3 snoRNP-dependent site. Cell 85:115-124. [DOI] [PubMed] [Google Scholar]
- 3.Allmang, C., Y. Henry, J. P. Morrissey, H. Wood, E. Petfalski, and D. Tollervey. 1996. Processing of the yeast pre-rRNA at sites A(2) and A(3) is linked. RNA 2:63-73. [PMC free article] [PubMed] [Google Scholar]
- 4.Allmang, C., J. Kufel, G. Chanfreau, P. Mitchell, E. Petfalski, and D. Tollervey. 1999. Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J. 18:5399-5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Allmang, C., and D. Tollervey. 1998. The role of the 3′ external transcribed spacer in yeast pre-rRNA processing. J. Mol. Biol. 278:67-78. [DOI] [PubMed] [Google Scholar]
- 6.Bachellerie, J. P., and J. Cavaille. 1997. Guiding ribose methylation of rRNA. Trends Biochem. Sci. 22:257-261. [DOI] [PubMed] [Google Scholar]
- 7.Bagni, C., and B. Lapeyre. 1998. Gar1p binds to the small nucleolar RNAs snR10 and snR30 in vitro through a nontypical RNA binding element. J. Biol. Chem. 273:10868-10873. [DOI] [PubMed] [Google Scholar]
- 8.Billy, E., T. Wegierski, F. Nasr, and W. Filipowicz. 2000. Rcl1p, the yeast protein similar to the RNA 3′-phosphate cyclase, associates with U3 snoRNP and is required for 18S rRNA biogenesis. EMBO J. 19:2115-2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bousquet-Antonelli, C., Y. Henry, P. G'Elugne, J., M. Caizergues-Ferrer, and T. Kiss. 1997. A small nucleolar RNP protein is required for pseudouridylation of eukaryotic ribosomal RNAs. EMBO J. 16:4770-4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cadwell, R. C., and G. F. Joyce. 1992. Randomization of genes by PCR mutagenesis. PCR Methods Appl. 2:28-33. [DOI] [PubMed] [Google Scholar]
- 11.Chanfreau, G., S. A. Elela, M. Ares, Jr., and C. Guthrie. 1997. Alternative 3′-end processing of U5 snRNA by RNase III. Genes Dev. 11:2741-2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chanfreau, G., P. Legrain, and A. Jacquier. 1998. Yeast RNase III as a key processing enzyme in small nucleolar RNAs metabolism. J. Mol. Biol. 284:975-988. [DOI] [PubMed] [Google Scholar]
- 13.Chanfreau, G., G. Rotondo, P. Legrain, and A. Jacquier. 1998. Processing of a dicistronic small nucleolar RNA precursor by the RNA endonuclease Rnt1. EMBO J. 17:3726-3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cherry, J. M., C. Ball, S. Weng, G. Juvik, R. Schmidt, C. Adler, B. Dunn, S. Dwight, L. Riles, R. K. Mortimer, and D. Botstein. 1997. Genetic and physical maps of Saccharomyces cerevisiae. Nature 387:67-73. [PMC free article] [PubMed] [Google Scholar]
- 15.Filippov, V., V. Solovyev, M. Filippova, and S. S. Gill. 2000. A novel type of RNase III family proteins in eukaryotes. Gene 245:213-221. [DOI] [PubMed] [Google Scholar]
- 16.Geerlings, T. H., J. C. Vos, and H. A. Raue. 2000. The final step in the formation of 25S rRNA in Saccharomyces cerevisiae is performed by 5′→3′ exonucleases. RNA 6:1698-1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giorgi, C., A. Fatica, R. Nagel, and I. Bozzoni. 2001. Release of U18 snoRNA from its host intron requires interaction of Nop1p with the Rnt1p endonuclease. EMBO J. 20:6856-6865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Girard, J. P., C. Bagni, M. Caizergues-Ferrer, F. Amalric, and B. Lapeyre. 1994. Identification of a segment of the small nucleolar ribonucleoprotein-associated protein GAR1 that is sufficient for nucleolar accumulation. J. Biol. Chem. 269:18499-18506. [PubMed] [Google Scholar]
- 19.Girard, J. P., H. Lehtonen, M. Caizergues-Ferrer, F. Amalric, D. Tollervey, and B. Lapeyre. 1992. GAR1 is an essential small nucleolar RNP protein required for pre-rRNA processing in yeast. EMBO J. 11:673-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guthrie, C., and G. R. Fink. 1991. Guide to yeast genetics and molecular biology. Academic Press, Inc., San Diego, Calif.
- 21.Harlow, E., and D. Lane. 1988. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 22.Henras, A., Y. Henry, C. Bousquet-Antonelli, J. Noaillac-Depeyre, J. P. Gelugne, and M. Caizergues-Ferrer. 1998. Nhp2p and Nop10p are essential for the function of H/ACA snoRNPs. EMBO J. 17:7078-7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Henry, Y., H. Wood, J. P. Morrissey, E. Petfalski, S. Kearsey, and D. Tollervey. 1994. The 5′ end of yeast 5.8S rRNA is generated by exonucleases from an upstream cleavage site. EMBO J. 13:2452-2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.James, P., J. Halladay, and E. A. Craig. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144:1425-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kiss-Laszlo, Z., Y. Henry, J. P. Bachellerie, M. Caizergues-Ferrer, and T. Kiss. 1996. Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell 85:1077-1088. [DOI] [PubMed] [Google Scholar]
- 26.Kufel, J., B. Dichtl, and D. Tollervey. 1999. Yeast Rnt1p is required for cleavage of the pre-ribosomal RNA in the 3′ ETS but not the 5′ ETS. RNA 5:909-917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Labbé, S., and D. J. Thiele. 1999. Copper ion inducible and repressible promoter systems in yeast. Methods Enzymol. 306:145-153. [DOI] [PubMed] [Google Scholar]
- 28.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 29.Lafontaine, D. L. J., C. Bousquet-Antonelli, Y. Henry, M. Caizergues-Ferrer, and D. Tollervey. 1998. The box H + ACA snoRNAs carry Cbf5p, the putative rRNA pseudouridine synthase. Genes Dev. 12:527-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lamontagne, B., and S. Abou Elela. 2001. Purification and characterization of Saccharomyces cerevisiae Rnt1p nuclease. Methods Enzymol. 342:159-167. [DOI] [PubMed] [Google Scholar]
- 31.Lamontagne, B., A. Tremblay, and S. Abou Elela. 2000. The N-terminal domain that distinguishes yeast from bacterial RNase III contains a dimerization signal required for efficient double-stranded RNA cleavage. Mol. Cell. Biol. 20:1104-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li, H. D., J. Zagorski, and M. J. Fournier. 1990. Depletion of U14 small nuclear RNA (snR128) disrupts production of 18S rRNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 10:1145-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu, Q., M. Z. Li, D. Leibham, D. Cortez, and S. J. Elledge. 1998. The univector plasmid-fusion system, a method for rapid construction of recombinant DNA without restriction enzymes. Curr. Biol. 8:1300-1309. [DOI] [PubMed] [Google Scholar]
- 34.Liu, Q., M. Z. Li, D. Liu, and S. J. Elledge. 2000. Rapid construction of recombinant DNA by the univector plasmid-fusion system. Methods Enzymol. 328:530-549. [DOI] [PubMed] [Google Scholar]
- 35.Lygerou, Z., C. Allmang, D. Tollervey, and B. Seraphin. 1996. Accurate processing of a eukaryotic precursor ribosomal RNA by ribonuclease MRP in vitro. Science 272:268-270. [DOI] [PubMed] [Google Scholar]
- 36.Melekhovets, Y. F., L. Good, S. A. Elela, and R. N. Nazar. 1994. Intragenic processing in yeast rRNA is dependent on the 3′ external transcribed spacer. J. Mol. Biol. 239:170-180. [DOI] [PubMed] [Google Scholar]
- 36a.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 37.Morrissey, J. P., and D. Tollervey. 1993. Yeast snR30 is a small nucleolar RNA required for 18S rRNA synthesis. Mol. Cell. Biol. 13:2469-2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nagel, R., and M. Ares, Jr. 2000. Substrate recognition by a eukaryotic RNase III: the double-stranded RNA-binding domain of Rnt1p selectively binds RNA containing a 5′-AGNN-3′ tetraloop. RNA 6:1142-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ni, J., A. L. Tien, and M. J. Fournier. 1997. Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell 89:565-573. [DOI] [PubMed] [Google Scholar]
- 40.Nicholson, A. W. 1999. Function, mechanism and regulation of bacterial ribonucleases. FEMS Microbiol. Rev. 23:371-390. [DOI] [PubMed] [Google Scholar]
- 41.Olson, M. O., M. Dundr, and A. Szebeni. 2000. The nucleolus: an old factory with unexpected capabilities. Trends Cell Biol. 10:189-196. [DOI] [PubMed] [Google Scholar]
- 42.Pringle, J. R., R. A. Preston, A. E. Adams, T. Stearns, D. G. Drubin, B. K. Haarer, and E. W. Jones. 1989. Fluorescence microscopy methods for yeast. Methods Cell Biol. 31:357-435. [DOI] [PubMed] [Google Scholar]
- 43.Reddy, R., and S. Shimba. 1995. Structural and functional similarities between MRP and RNase P. Mol. Biol. Rep. 22:81-85. [DOI] [PubMed] [Google Scholar]
- 44.Rose, M. D., F. Winston, and P. Hieter. 1990. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 45.Rotondo, G., and D. Frendewey. 1996. Purification and characterization of the Pac1 ribonuclease of Schizosaccharomyces pombe. Nucleic Acids Res. 24:2377-2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schmitt, M. E., and D. A. Clayton. 1993. Nuclear RNase MRP is required for correct processing of pre-5.8S rRNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 13:7935-7941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seipelt, R. L., B. Zheng, A. Asuru, and B. C. Rymond. 1999. U1 snRNA is cleaved by RNase III and processed through an Sm site-dependent pathway. Nucleic Acids Res. 27:587-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tollervey, D. 1987. A yeast small nuclear RNA is required for normal processing of pre-ribosomal RNA. EMBO J. 6:4169-4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van Hoof, A., P. Lennertz, and R. Parker. 2000. Yeast exosome mutants accumulate 3′-extended polyadenylated forms of U4 small nuclear RNA and small nucleolar RNAs. Mol. Cell. Biol. 20:441-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Venema, J., and D. Tollervey. 1999. Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet. 33:261-311. [DOI] [PubMed] [Google Scholar]
- 51.Watkins, N. J., A. Gottschalk, G. Neubauer, B. Kastner, P. Fabrizio, M. Mann, and R. Luhrmann. 1998. Cbf5p, a potential pseudouridine synthase, and Nhp2p, a putative RNA-binding protein, are present together with Gar1p in all H BOX/ACA-motif snoRNPs and constitute a common bipartite structure. RNA 4:1549-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zebarjadian, Y., T. King, M. J. Fournier, L. Clarke, and J. Carbon. 1999. Point mutations in yeast CBF5 can abolish in vivo pseudouridylation of rRNA. Mol. Cell. Biol. 19:7461-7472. [DOI] [PMC free article] [PubMed] [Google Scholar]