Abstract
Trypanosome RNA editing is a unique U insertion and U deletion process that involves cycles of pre-mRNA cleavage, terminal U addition or U removal, and religation. This editing can occur at massive levels and is directed by base pairing of trans-acting guide RNAs. Both U insertion and U deletion cycles are catalyzed by a single protein complex that contains only seven major proteins, band I through band VII. However, little is known about their catalytic functions, except that band IV and band V are RNA ligases and genetic analysis indicates that the former is important in U deletion. Here we establish biochemical approaches to distinguish the individual roles of these ligases, based on their distinctive ATP and pyrophosphate utilization. These in vitro analyses revealed that both ligases serve in RNA editing. Band V is the RNA editing ligase that functions very selectively to seal in U insertion (IREL), while band IV is the RNA editing ligase needed to seal in U deletion (DREL). In combination with our earlier findings about the cleavage and the U-addition/U-removal steps of U deletion and U insertion, these results show that all three steps of these editing pathways exhibit major differences and suggest that the editing complex could have physically separate regions for U deletion and U insertion.
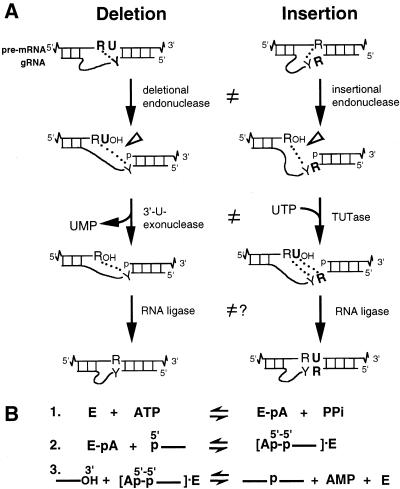
Trypanosomatids are early diverging protozoa that exhibit unique biological features, including RNA editing of their mitochondrial transcripts. This editing is posttranscriptional and involves numerous cycles of U insertion and less frequent U deletion. It creates start and stop codons as well as amino acid coding triplets that can constitute virtually the entire open reading frame (ORF) (reviewed in references 1, 3, 16, 35, 37, and 38). The information for this massive processing is encoded in small trans-acting guide RNAs (gRNAs) and is provided to the pre-mRNA through base pairing, using Watson-Crick and G-U interactions (4). These gRNAs consist of a 5′ anchor sequence that can duplex with cognate pre-mRNA, a central sequence that guides the U additions and U removals so that the mRNA becomes its complement, and a 3′ oligo(U) tail that tethers the upstream, very purine-rich pre-mRNA (Fig. 1A). Editing progresses 3′ to 5′ along the pre-mRNA, generally using multiple overlapping gRNAs.
FIG. 1.
Mechanism of RNA editing. (A) A cycle of U deletion or U insertion involves gRNA-directed cleavage of the pre-mRNA (open arrowhead), either 3′-U-exo or TUTase acting on the upstream fragment, and RNA ligase rejoining (4, 9, 22, 34). The two kinds of editing use distinct cleavage activities (10), and the 3′-U-exo is not a reverse TUTase reaction (9, 29), as indicated (≠). R = purine(s); Y = pyrimidine(s). The gRNA forms anchor and tether duplexes with pre-mRNA (indicated by base pairing), and its central guiding region becomes complementary upon accurate editing (4, 5). Dotted lines indicate that upon complete U removal or U addition, one or more base pairs could align the mRNA fragments, providing a “ligation bridge” (34). (B) The three steps in the action of an RNA ligase enzyme (E).
RNA editing at selected Trypanosoma brucei editing sites has been faithfully reproduced in vitro (22, 33). These reactions use in vitro-synthesized radiolabeled pre-mRNA and cognate gRNA corresponding to the 3′ portion of the ATPase subunit 6 (A6) transcript; totally unedited mRNA and gRNA specify U deletion at the first editing site (ES1), while RNAs that already pair at ES1 specify U insertion at the second editing site (ES2). These reactions are catalyzed by T. brucei mitochondrial extract (33) or various enriched preparations (2, 22, 24, 28, 29). The preparation with the simplest protein profile consists of a complex of only seven major proteins, called bands I through VII, which appear approximately equimolar based on silver staining (29). This purified complex catalyzes U deletion and U insertion very actively, the most efficient editing cycles yet reported when using optimized model gRNAs (10, 13; J. Cruz-Reyes et al., unpublished data).
Studies of this T. brucei in vitro editing and a related system from leishmania, for which detection of editing requires PCR amplification (6), have provided strong evidence that each editing cycle involves three sequential enzymatic activities: (i) endonuclease cleavage of the pre-mRNA at the 5′ end of the duplex with the gRNA's anchor region, (ii) terminal U removal or U addition by a 3′-U-specific exonuclease (3′-U-exo) or terminal-U-transferase (TUTase), respectively, and (iii) pre-mRNA sealing by RNA ligase (4, 9, 15, 22, 34; Fig. 1A). The anchor duplex can then extend to the next mismatch, where the subsequent editing cycle can begin. Throughout purification of the editing complex, as well as using various other fractionation protocols, we find these enzymatic activities copurify with each other and with the seven-protein complex (29; L. Rusché, C. Huang, and S. O'Hearn, unpublished observations).
Although fractions with this relatively simple set of proteins catalyze the editing reactions, the individual activities of these proteins in the U-deletion and U-insertion cycles have remained elusive. The best understood are band IV and band V of the minimal editing complex (29). Evidence that they are RNA ligases includes that they auto-adenylylate with ATP (31) and deadenylylate with pyrophosphate (PPi) or with ligatable RNA but that they do not deadenylylate with ligatable DNA or with nonligatable RNA (29). This fits well with the known mechanism of RNA ligase action (Fig. 1B). First, the ligase autoadenylylates (E + ATP ⇋ E-AMP + PPi), using ATP to form a covalent protein-AMP intermediate while releasing PPi. This reaction occurs in the absence of RNA and reverses with high concentrations of PPi. The AMP is then transferred to the 5′ phosphate of a donor RNA, generating a 5′-5′ pyrophosphate linkage, and finally the 3′ hydroxyl of the acceptor RNA displaces this 5′ AMP, forming the new phosphodiester bond. The adenylylation/deadenylylation reaction can conveniently be followed by using [α-32P]ATP.
Several recent studies have focused on these two RNA ligases, reporting their encoding genes and that knock-out of the band IV gene is lethal and inhibits editing in vivo (19, 25, 28, 29, 32). However, we noted that when the band IV protein was genetically knocked out, the editing complex had reduced stability, and that could cause impaired function of other essential editing proteins (19). To analyze this ligase's functional role, distinct from a structural role, a more exacting genetic approach was then used: transgenic trypanosomes were constructed that could inducibly replace ∼2/3 of the band IV protein with a catalytically inactive version, and it was showed that this decreased joining in U deletion by ∼1/2, demonstrating that U deletion uses band IV ligase activity (19). However, that genetic approach did not discern whether U deletion could also use the band V ligase at lesser efficiency. Furthermore, that genetic study did not resolve which ligase seals in U insertion, which constitutes ∼90% of trypanosome editing events—whether it is the band V ligase or the remaining band IV ligase. Both scenarios seemed possible. The former scenario was earlier suggested, since there are two different RNA ligases in the editing complex and two forms of editing, so one ligase could serve U deletion and the other could serve U insertion (10, 11). However, one ligase could seal both forms of normal editing and the other serves a different function, such as sealing in the surprisingly frequent illegitimate editing (14, 19, 23) or in another process catalyzed by this multifunctional protein complex (17). Other possibilities are that both forms of editing could be served by one ligase in the insect host and by the other ligase in the mammalian host stage of the trypanosome life cycle or that the ligases could be partly redundant.
The aim of this study was to biochemically define the major roles of both RNA ligases in the in vitro editing cycles. Since band IV appears important in joining U deletion in vivo (19), we expected that would be the case also in vitro, but we wanted to determine whether band V showed any ability to join this minor form of editing. More critically, we wanted to address whether the major form of editing, U insertion, is sealed by band V ligase or instead preferentially uses band IV ligase. We approached this by first showing that the band IV and band V ligase proteins exhibit different adenylylation and deadenylylation properties, which enabled their biochemical distinction by ATP and PPi titration. These analyses revealed that band V can efficiently ligate the pre-mRNA in U insertion and showed that band IV but not band V serves to ligate in U deletion. The ligases constitute the first two subunits of the editing complex whose specific roles are defined. Furthermore, demonstration of this distinct ligase utilization, in conjunction with our earlier studies showing major differences also at the first two steps of the editing cycles (9, 10, 29), indicates that U deletion and U insertion differ at all three reaction steps.
Nomenclature.
Various laboratories have identified the two T. brucei RNA ligases as band IV and band V (29, 30), TbMP52 and TbMP48 (32) or p52 and p48 (25). The latter numbers indicate the sizes of the T. brucei preproteins, which are a few kilodaltons larger than the mature mitochondrial enzymes that are present in the editing complex. Additionally, in related organisms that also exhibit such editing, Leishmania tarentolae and Crithidia fasciculata, the corresponding proteins are somewhat different sizes (27; L. Rusché, unpublished observations). For a common nomenclature (7, 8), REL1 and REL2 (RNA editing ligase [32]) could suffice, except those designations are already used to identify other proteins in the NCBI database. Based on the specific roles of these ligases that we determine below and those previously published (19), we propose to designate the RNA editing ligase that is needed for U deletion as DREL (band IV/TbMP52/p52) and the RNA editing ligase that is specific for U insertion as IREL (band V/TbMP48/p48). This descriptive and unifying nomenclature will be used throughout the rest of this paper.
MATERIALS AND METHODS
In vitro synthesis of RNA.
Pre-edited A6 mRNA used for in vitro U deletion at ES1 or U insertion at ES2 was transcribed from PCR-amplified template (m[0,4] [10]) and was 3′ end labeled (9) or 5′ end labeled (see Fig. 6D). We similarly transcribed the gRNAs, and all RNAs were gel purified (13). The gRNA D32a and D30CC direct enhanced deletion of three U residues at ES1 while the gRNA I47G directs enhanced insertion of two U residues at ES2 (13, 19; Cruz-Reyes et al., unpublished data). These gRNAs are active using either purified editing complex or unfractionated mitochondrial extract and support sufficiently high levels of editing to readily assess partial ligase activation.
FIG. 6.
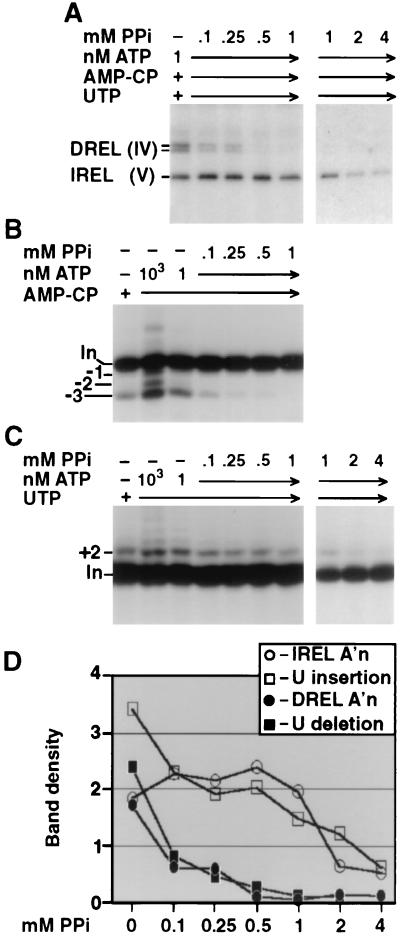
U deletion and U insertion at various PPi concentrations under common conditions. (A and B) U deletion and U insertion assays were as in Fig. 5B and C except that no ATP was added and reaction mixtures contained both UTP and AMP-CP. Because the purified editing complex contains partially adenylylated DREL and almost fully adenylylated IREL, it supports some U deletion and substantial U insertion in the absence of added ATP (10). The editing complexes should have active endonuclease, even when DREL is unadenylylated, so would not be expected to seal all the cleaved mRNA. Note also that the editing reactions used near-nanomolar amounts of purified editing complex, in which the IREL was largely adenylylated, so they could generate that much ATP. (C) Quantitation of the U deletion and U insertion products from the gels of panels A and B, as shown in Fig. 2E. (D) 3′-U-exo assay using 5′-end-labeled mRNA. Reactions were otherwise performed as described for Fig. 5B. This gel region shows the cleaved mRNA, with weak bands (45, 44, and 43 nt) representing removal of zero, one, and two U residues and the major band (42 nt) representing complete three-U removal (9, 34). Marker fragments are from nuclease P1 digestion of the same input mRNA under G-selective conditions (12, 36) and a hydroxide ladder. Band density is in units of 1,000 pixels (see Materials and Methods).
Editing and adenylylation analysis.
Mitochondrial extract (2 × 1010 cell equivalents/ml) was prepared from procyclic T. brucei strain TREU-667, and from this extract the editing complex was purified by Q-Sepharose and DNA-cellulose chromatography (29, 37). Editing was assayed (11) in 20-μl reaction mixtures with 10 mM KCl-MRB [25 mM Tris-HCl (pH 8.0), 10 mM Mg(OAc)2, 10 mM KCl, 1 mM EDTA, 0.5 mM dithiothreitol (DTT), and 5% glycerol], generally adding 50 μg of bovine serum albumin/ml. The reaction mixtures contained ATP and tetrapotassium pyrophosphate (PPi) as indicated. They also contained ∼30 fmol of mRNA and ∼1.2 pmol of gRNA that had been preannealed (11). U deletion reaction mixtures further contained 1 mM AMP-CP (Fluka) to support cleavage while minimizing ATP contamination (10), and U insertion reaction mixtures also contained 150 μM UTP (ATP-free; Pharmacia); common reaction conditions were used with both nucleotides. Since the ligases, especially IREL, become activated at very low ATP concentration, ATP sufficient to interfere with these studies is present in many commercial nucleotide preparations, especially those of UTP but also those of AMP-CP (only 10−5 or 10−6 parts contamination is already a problem). Reaction mixtures using purified editing complex (∼2 μl of peak fractions; ∼108 cell equivalents), assembled as described below, were incubated 1 h at 26°C. Reactions using mitochondrial extract (0.5 μl; ∼107 cell equivalents) were supplemented with 15 U of RNasin (Promega) and 10 mM DTT and were incubated 45 min at 26°C. RNA was recovered and analyzed on 1-m-long gels (10). The RNAs were sized as previously described (12, 36). Autoradiograms were scanned using a FluorChem 800 Advanced Fluorescence, Chemiluminescence and Visible Light Imaging System with AlphaEaseFC software, with band densities reported in units of 1,000 pixels.
To activate as well as to specifically radiolabel the ligases, adenylylation reactions (31) were carried out for 5 min on ice in an editing reaction mixture containing the purified editing complex and the indicated amounts of ATP but lacking the RNAs. For labeling, the ATP was [α-32P]ATP (3,000 Ci/mmol [ICN], except 30 Ci/mmol in Fig. 2A). For analysis, these reaction mixtures were precipitated overnight at −20°C with 10 μg of bovine serum albumin plus 3 volumes of acetone, recovered by 10 min of centrifugation at 17,500 × g, and analyzed by sodium dodecyl sulfate (SDS)-10% polyacrylamide gel electrophoresis (PAGE). For ATP titrations shown in Fig. 2 to 4, the ligases of the purified complex were first fully deadenylylated with 10 mM PPi (29) by preincubating for 5 min on ice a mixture containing 2 μl of the editing complex plus 1 μl of 30 mM PPi in 10 mM KCl-MRB for each reaction. Diluting 3 μl of this mixture in the final 20 μl of editing reaction mixture reduced the PPi concentration to 1.5 mM. (A higher PPi significantly inhibits the endonuclease of U deletion.) PPi and pyrophosphatase (PPiase) treatment for Fig. 4 was as previously described (29); control reactions showed that the PPiase level was saturating. We only use PPiase treatment for adenylylation analysis because it can interfere with other steps of editing (data not shown). For PPi titrations shown in Fig. 5 to 7, the otherwise complete reaction mixtures were preincubated for 5 min on ice before the RNA (in 2 μl) was added. To score deadenylylation in Fig. 5, before the PPi incubation, the ligases were partially tagged by preincubating the editing complex for 5 min on ice with 1 nM ATP (for Fig. 5A, it was [α-32P]ATP).
FIG. 2.
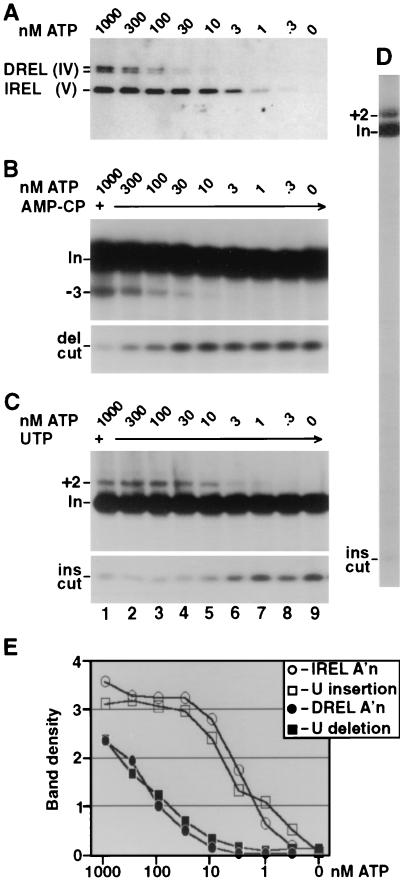
Adenylylation, U deletion, and U insertion directed by the editing complex at various ATP concentrations. The purified complex was first fully decharged with 10 mM PPi, then diluted to 1.5 mM PPi, and supplemented with the indicated ATP. (A) Adenylylation assay. Autoradiogram of protein gel assessing DREL and IREL activation using [α-32P]ATP. Mixtures that were incubated longer or that contained the editing RNAs yielded similar results, and higher ATP concentrations had no additional effect (not shown). The DREL doublet corresponds to two isoforms (29, 30). (B and C) U deletion and U insertion assays. Electrophoretic analyses show RNA products generated by incubating the treated complex with the indicated amounts of unlabeled ATP and 3′-end-labeled A6 pre-mRNA, preincubated with gRNA D32a (B) or I47G (C). Also, U deletion reaction mixtures contained 1 mM AMP-CP (B), while the U insertion reaction mixtures contained 150 μM UTP (C). The upper panels show the gel region containing input mRNA (In) and the guided (−3) U deletion product or (+2) U insertion product; the lower panels show the gel region containing the fragment remaining from the gRNA-directed cleavage at the U deletion site (del cut) or at the U insertion site (ins cut). (D) A larger region of a gel assessing U insertion, as in panel C, using 1 μM ATP. (E) Quantitation of autoradiograms showing adenylylation of the two ligase proteins in panel A and the U deletion and U insertion products from different exposures of the gels of panels B and C. Band density is in units of 1,000 pixels (see Materials and Methods).
FIG. 4.
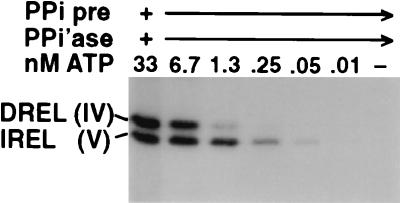
Two ligases have different apparent affinities for ATP. Adenylylation reactions using decharged editing complex were carried out as described for Fig. 2A, except the PPi was degraded with excess PPiase prior to [α-32P]ATP addition. The reactions also used 1 mM AMP-CP and an editing complex isolated at a different time than the reactions for the other figures (and used ∼0.1 nM rather than ∼1 nM). Reaction mixtures were analyzed on minigels, so the DREL isoforms barely resolved. The conclusion of this experiment was also verified by titrating ATP without any PPi addition; then, the IREL band is very light, since it was mostly precharged, but that which became adenylylated again required significantly less ATP than did DREL adenylylation (data not shown).
FIG. 5.
Adenylylation, U deletion, and U insertion at various PPi concentrations. The purified editing complex was first charged with the indicated amount of [α-32P]ATP (labeled [A] and unlabeled [B and C]) prior to treatment with indicated amounts of PPi plus the other indicated components. (A) Adenylylation assay performed as described for Fig. 2. The similar intensity labeling of DREL and IREL arose because 1 nM ATP charged a small fraction of DREL molecules and a large fraction of available IREL molecules, but most IREL molecules in the purified editing complex were preadenylylated (29). The same result was obtained in the absence of added AMP-CP and UTP (data not shown). (B and C) U deletion and U insertion assays performed as described for Fig. 2. (B) The ratio of partial-to-complete U deletion is similar in all lanes. Unlike the cleavage of U insertion, cleavage of U deletion diminishes at >1 mM PPi (not shown). (D) Quantitation of the deadenylylation of the two ligase proteins shown in panel A and the U deletion and U insertion products from the gels of panels B and C, as in Fig. 2E. Band density is in units of 1,000 pixels (see Materials and Methods).
FIG. 7.
U deletion and U insertion directed by unfractionated mitochondrial extract at various PPi concentrations. (A and B) U deletion and U insertion assays were performed as described for Fig. 6A and B except for using whole extract. As standardly observed for U insertion reactions performed with whole mitochondrial extract, some RNA degradation products (x) and chimeric RNA (chi) were also obtained. (C) Quantitation of the U deletion and U insertion products from the gels of panels A and B, as described for Fig. 2E. Band density is in units of 1,000 pixels (see Materials and Methods).
RESULTS
Differences in DREL and IREL adenylylation.
We wanted to biochemically distinguish the two RNA ligases of the purified editing complex (29) that catalyzes U deletion and U insertion (10). We earlier noted that both ligases are largely unadenylylated in mitochondrial extract, but during the initial step of purification, the smaller ligase (band V; IREL) became largely adenylylated while the larger ligase (band IV; DREL) did not (29; these proteins remained in ∼1:1 abundance throughout purification). This differential adenylylation suggests that these ligases might have differences in their charging with ATP and/or decharging with PPi which we could use to distinguish their roles in RNA editing in vitro.
Differential ATP titrations of two ligases and two forms of editing.
Purified, fully deadenylylated editing complex was incubated in adenylylation reaction mixtures with increasing concentrations of radiolabeled ATP. AMP transfer to the smaller ligase protein was observed at <1/10 the ATP concentration needed for the larger ligase protein. This biochemical distinction of the two ligases prompted us to assess the joining step of U deletion and U insertion using this same range of ATP concentrations and similarly pretreated editing complex. The substrate was 3′-end-labeled A6 pre-mRNA annealed to either gRNA D32a to direct deletion of three U's at ES1 (Fig. 2B) or to gRNA I47G to direct insertion of two U's at ES2 (Fig. 2C). The figures show only small regions of the gels, but the entire lane assessing U deletion (9, 13) or U insertion (Fig. 2D) generally showed only minor unexplained bands.
Notably, the two forms of editing responded differently to the ATP titrations, suggesting that they were sealed by different ligases. Significantly more ATP was required for U deletion than for U insertion, with activation occurring within the 30 to 300 nM range (Fig. 2B) and the 1 to 30 nM range (Fig. 2C), respectively. Critically, the ATP amount required for U deletion coincides with that needed for adenylylation of the larger ligase protein, therefore called DREL (Fig. 2A, B, and E), while the ATP amount required for U insertion agrees with that needed for adenylylation of the smaller ligase protein, therefore called IREL (Figs. 2A, C, and E).
Importantly, the step of U deletion and of U insertion being affected by the ATP titration is mRNA ligation. This is strongly implicated because ATP is not required for the initial endonuclease cleavage of either U deletion or U insertion (10; Fig. 2B and C, lower panels), nor is it important for the 3′-U-exo or TUTase activities that support the U removal and U addition steps, respectively (29). Since neither of the first two steps of U deletion and U insertion are affected by ATP while the remaining step of the editing pathways, mRNA ligation, is dependent on ATP and coincidentally requires the same amounts of ATP for activation as are needed for the full cycle editing reactions, there is little doubt that the ATP effect on editing reflects the ligase active in that pathway.
Verifying the deduction that ligation is the step that defines the ATP responses of the editing cycles, the amount of cleaved mRNA that remained unligated in the editing reactions (Fig. 2B and C, lower panels, del cut and ins cut bands) is inversely proportional to the amount of edited product in that reaction (upper panels). Accordingly, the amount of this unligated cleaved mRNA is also inversely proportional to the level of activation of the respective ligase by adenylylation (Fig. 2A). Thus, the amount of remaining cleaved mRNA can provide a second discriminator to correlate the ligases with the editing pathways.
These data provide strong evidence that U deletion is sealed by DREL, consistent with in vivo observations (19) and new information that U insertion is sealed by IREL. Additionally, because IREL is activated with less ATP than is needed for U deletion and DREL, the data also indicate that this U deletion is not detectably sealed by IREL.
Verification of ATP titration analysis.
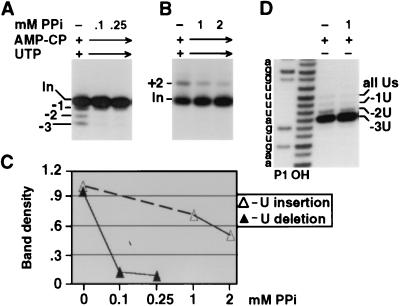
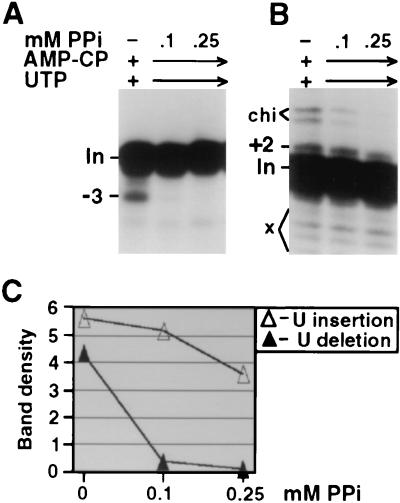
One potential caveat is that U deletion and U insertion are optimized and typically assayed under different reaction conditions (10, 11, 22, 34). In particular, U deletion requires >1 mM adenosine ribonucleotide to maximize its endonucleolytic cleavage, while high adenosine ribonucleotide concentrations inhibit the cleavage step of U insertion (10). Furthermore, U insertion requires UTP for TUTase action (22), while UTP addition to U deletion reactions elicits more partial (−1 and −2) and less of the complete (−3) editing product (11; Fig. 3A). The optimized reaction conditions in Fig. 2B and C differed only in that U deletion used 1 mM AMP-CP (10) and U insertion used 150 μM UTP; both nucleotide preparations had been selected for their negligible ATP contamination (data not shown). To nonetheless confirm that reagent differences were not biasing our analyses, we therefore performed the ATP titrations under common reaction conditions using both 1 mM AMP-CP and 150 μM UTP (Fig. 3A and B). Again, more ATP was required for U deletion than for U insertion (Fig. 3C) and ATP titration of adenylylation was virtually identical to that in Fig. 2A (data not shown), further supporting that these reagents provide negligible amounts of ATP. Therefore, even when reactions with peak fractions of purified editing complex were carried out under identical conditions, DREL also correlated with U deletion and IREL correlated with U insertion. Furthermore, joining of the partial and complete U deletion products required the same ATP concentration (Fig. 3A), indicating that they both use DREL.
FIG. 3.
ATP titrations of U deletion and U insertion under common conditions. (A and B) U deletion and U insertion reactions using decharged editing complex were performed as described for Fig. 2 except for containing both UTP and AMP-CP, as indicated. (C) Quantitation of the U deletion and U insertion products from the gels of panels A and B, as in Fig. 2E. Band density is in units of 1,000 pixels (see Materials and Methods).
The ATP titrations of Fig. 2 and 3 contained both ATP and PPi, the PPi remaining from initially decharging the ligases which had become preadenylylated during purification of the editing complex (see Materials and Methods). Because adenylylation is a reversible reaction (E + ATP ⇋ E-AMP + PPi), the differences observed for the two ligases could reflect a difference in their apparent affinities for ATP (forward reaction), PPi (reverse reaction), or both. To examine their apparent affinities for ATP, we repeated an adenylylation experiment as described for Fig. 2A, except that after the initial deadenylylation, the PPi was depleted by treatment with excess PPiase before the addition of the ATP (Fig. 4). Adenylylation of IREL was again observed at ∼1/10 the ATP concentration needed for DREL adenylylation (Fig. 4). Note also that both ligases required less ATP to adenylylate when PPi was absent (Fig. 4) than when present (Fig. 2A), as expected for this reversible reaction. We conclude that IREL indeed has a higher apparent affinity for ATP than does DREL and that this difference is in large part responsible for the differing ligation efficiencies of U deletion and U insertion seen upon ATP titration in Fig. 2 and 3.
In summary, the results of Fig. 2 to 4 show that the roles of the two ligases in editing can be determined by analyzing ATP titrations. The conclusion that DREL is needed to seal in U deletion and that IREL serves to seal in U insertion is established by our correlation of the ATP levels required for both ligase activation and full-round editing, since the previous two steps of the editing cycles are ATP independent, and is further corroborated by comparing with the amount of residual cleaved mRNA that remains unligated. Further, analysis under conditions that yield partial U deletion products shows that they are also sealed by DREL, while analysis in the absence of PPi attests that these observed differences are largely due to different apparent affinities of the two ligases for ATP.
Deadenylylation analysis also shows distinct roles for two RNA ligases in editing.
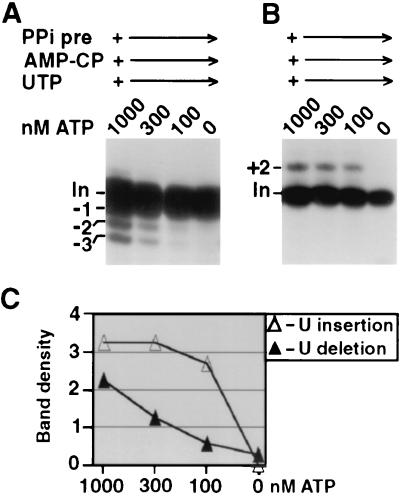
Wanting a second discriminator to verify the functional assignments of DREL and IREL based on their ATP responses, we examined whether these ligases also differed in their sensitivities to PPi. To initially monitor deadenylylation, the ligases were partially prelabeled using a very low level of [α-32P]ATP (Fig. 5A). Upon PPi titration, DREL readily became deadenylylated, whereas IREL was relatively resistant (Fig. 5A). We then titrated PPi in U deletion and U insertion reaction mixtures under these reaction conditions. U deletion was greatly suppressed by 0.1 mM PPi (Fig. 5B) and parallels DREL decharging (Fig. 5A and D), suggesting that DREL and not IREL seals in U deletion. In contrast, U insertion remained at 10-fold more PPi (Fig. 5C), more closely paralleling IREL decharging (Fig. 5A and D), suggesting that IREL can efficiently seal in U insertion. These correlations between ligase and form of editing are consistent with those obtained in the ATP titrations of Fig. 2 to 4.
Because the reactions of Fig. 5 contained a small amount of added ATP, they may reflect a combination of the ligases' sensitivity to PPi and ATP. To specifically score effects of PPi, we repeated the PPi titrations of editing without any added ATP (Fig. 6). This analysis was possible because much IREL and some DREL of the purified editing complex is preadenylylated (29), enabling U insertion and some U deletion in the absence of exogenous ATP (10; Fig. 5B and C, lanes 1 and 2). Titrating PPi without any added ATP using the purified editing complex, U deletion was again largely suppressed by 0.1 mM PPi (Fig. 6A) while U insertion remained at 10-fold more PPi (Fig. 6B). These results are much as were observed in Fig. 5 (compare Fig. 5D and 6C) and indicate that the different sensitivities of U deletion and U insertion to PPi titration seen in that experiment arise independently of the small amount of ATP added to those reaction mixtures. We conclude that the correlations of U deletion with DREL and of U insertion with IREL seen upon PPi titration (Fig. 5 and 6) are independent of those seen upon ATP titration (Fig. 2 to 4) and therefore provide two confirmatory lines of evidence.
It is important to note that the differential effects of PPi on U deletion versus on U insertion are not due to effects on the first two steps of the editing pathways. This is shown in Fig. 6D where the mRNA cleavage step and the 3′-U-exo step (scored together using 5′-end-labeled pre-mRNA) are not inhibited by up to ∼1.5 mM PPi. Thus, the different PPi sensitivities of U deletion and U insertion (Fig. 5 and 6) reflect the third step of the editing pathway, RNA ligation. Figure 6 further demonstrates that the partial (−1 U and −2 U) deletion products observed in the presence of UTP (11; Fig. 3A) are suppressed by PPi in synchrony with the complete −3 U deletion product (Fig. 6A), indicating that DREL also ligates these partial U deletion products.
In conclusion, data from PPi titration (Fig. 5 and 6) and ATP titration (Fig. 2 to 4) provide two independent lines of evidence that the ligation step of this complete and partial U deletion cycle requires DREL and does not function with IREL, while the ligation step of this U insertion cycle is actively supported by IREL.
Analysis of unfractionated mitochondrial extract.
Finally, to determine whether the differential ligase utilization in U deletion and U insertion seen in Fig. 2 to 6 could somehow arise from purification of the editing complex, we repeated a PPi titration (much as in Fig. 6A and B) but with whole mitochondrial extract (Fig. 7). Again, U deletion was effectively inhibited by 0.1 mM PPi while U insertion was maintained at higher PPi concentrations (Fig. 7A and B; compare with Fig. 5 and 6), indicating that also in whole extract, DREL is needed for U deletion and IREL serves U insertion. Other experiments with whole extract confirm that more PPi is needed to deadenylylate IREL than DREL (not shown), that less ATP is needed to adenylylate IREL than DREL (not shown), and that in U deletion both the cleavage and 3′-U-exo steps are less sensitive to PPi than is the ligation step (reference 9 and data not shown), as with the purified complex (Fig. 2 to 6). The finding that DREL is needed to seal U deletion and IREL serves to seal U insertion also in mitochondrial extract suggests that ligase utilization in editing is not affected by interactions of the minimal enzymatic complex with possible auxiliary components.
DISCUSSION
Biochemical distinction of DREL and IREL reveals their roles in U deletion and U insertion.
The DREL (band IV) and IREL (band V) RNA ligases that constitute two of the seven major proteins of the T. brucei editing complex have different apparent affinities for ATP and PPi in their activating adenylylation reaction (E + ATP ⇌ E-AMP + PPi), enabling their individual roles in RNA editing to be distinguished. DREL adenylylation and U deletion exhibit similar activation profiles with ATP and similar inactivation profiles with PPi, while IREL adenylylation and U insertion become activated with an order of magnitude less ATP and remain active with an order of magnitude more PPi (Fig. 2 to 6). Additional studies indicate that the step of the U deletion and U insertion cycle affected by ATP or PPi concentration within the examined range is indeed ligation, not cleavage or the U-exo/U-transferase step. These combined results provide separate corroborative evidence for a functional correlation of U insertion with band V, called IREL (RNA editing ligase specific for U insertion) and a correlation of U deletion with band IV, called DREL (RNA editing ligase needed for U deletion). Therefore, the two RNA ligases of the editing complex are demonstrated to exhibit distinct roles in editing. These are the first two proteins of the basic enzymatic complex whose roles in editing have been discerned.
The role of DREL (band IV) was initially approached using genetic knock-out analysis, which showed this ligase to be essential for trypanosome viability and for in vivo editing (30, 32). However, the finding that this protein is important in stabilizing the editing complex (19) made unclear whether its requirement was primarily to maintain other editing components in an intact editing complex or also to catalyze ligation in editing. A catalytic requirement for DREL was then demonstrated genetically (19), using this protein's autoregulation (30) to inducibly replace ∼2/3 of the wild-type protein with a version bearing a single charge-conservative mutation that destroys catalytic activity but maintains its structural features. This reduced U deletion by about one-half without apparently affecting U insertion, indicating that U deletion uses at least largely DREL (band IV; 19). However, by such methods it would have been extremely challenging to address whether IREL partly served U deletion. Moreover, that genetic approach would require a completely different set of constructs to ascertain whether U insertion was sealed specifically by IREL or efficiently by DREL. U insertion constitutes ∼90% of the editing events in vivo, so this major form of editing remained to be analyzed. We submit that the in vitro approach reported here enables determinations in a convenient system that allows the ligases to be selectively activated and scored.
Distinction of U deletion and U insertion cycles at all three catalytic steps.
Models for U deletion and U insertion have ranged from these editing cycles sharing all catalytic activities (a common endonuclease, a TUTase that runs in reverse to provide 3′-U-exo, and a common ligase [18, 38]) to their using distinct activities at all three editing steps (10, 11). The latter notion has been supported by the observed differences between U deletion and U insertion (11), including that at their first and second enzymatic steps. Specifically, the endonuclease cleavages have different cofactor requirements (10) and the TUTase and 3′-U-exo steps use different catalytic centers (the former consuming UTP and the latter generating UMP [9, 29]). Moreover, U deletion and U insertion also exhibit different requirements for various gRNA features (13) which affect the cleavage and the RNA ligation steps (Cruz-Reyes et al., unpublished data). We have now directly examined the third step of the editing cycles, RNA ligation, and found that the two RNA ligases of the editing complex are utilized differently: DREL and not IREL serves in U deletion and IREL serves in U insertion. Thus, all three reaction steps of U deletion and U insertion show major differences.
Even though U deletion and U insertion differ at all three reaction steps, the existence of a common enzymatic complex and the observation that UTP affects U deletion (9, 11, 29) raises the possibility of functional cross talk between activities of the U deletion and U insertion pathways. In our titrations of ATP and PPi, the distinguishing intermediate concentrations of both reagents activate IREL and not DREL. The finding that U deletion occurs only at the high but not at the intermediate concentrations of ATP and only at the low but not at the intermediate concentrations of PPi makes clear that only DREL and not IREL supports this form of editing. Furthermore, the activity of U insertion at these intermediate concentrations makes clear that IREL supports that form of editing. However, our analyses, which generally used very active preparations of editing complex, are not diagnostic for whether DREL could also support U insertion. This is because IREL seals the vast majority of the RNA cleaved at the U insertion site, leaving minimal substrate on which DREL could act. It therefore remained unclear from these data whether at physiological ATP concentrations, orders of magnitude higher than we used in these studies, U insertion is sealed only by IREL or by a combination of IREL and DREL. Because DREL appears to have rather relaxed structural requirements (see below), it seems possible that DREL could serve in U insertion, at least if IREL is insufficient. Indeed, when using somewhat less active aliquots of editing complex, there is a suggestion that DREL may seal in U insertion in vitro (Fig. 5). If DREL could replace IREL in U insertion in vivo, then IREL might not be an essential protein. Intriguingly, current studies using RNA interference (RNAi) and genetic knock-outs indicate that IREL is not needed for trypanosome viability (S. O'Hearn, C. Huang, and B. Sollner-Webb, unpublished data, and reference 39). This suggests that in vivo, DREL could act in U insertion, at least when IREL is not available.
The distinctive features of IREL and DREL enable additional understandings.
Our identification of the individual roles for the two ligases of the editing complex explains and extends a number of interesting observations in the literature concerning the ligases of editing. (i) The first observation concerns the ligases' requirements for the gRNA to precisely base pair with the two mRNA ends and align them for ligation, called a ligation bridge (26). U deletion is not dependent on such a potential ligation bridge (13), and since DREL seals U deletion (Fig. 2 to 6), we conclude that a precise ligation bridge is not critical for DREL activity. The activity of DREL at sealing partial U deletions (Figs. 3A and 6A) and at sealing RNA cleaved for U insertion but not extended by TUTase (19), which also could not form a ligation bridge, further supports this conclusion. Conversely, U insertion has appeared dependent on the gRNA forming a ligation bridge (Cruz-Reyes et al., unpublished data, and reference 21). Since IREL serves specifically in U insertion (Fig. 2 to 6), this implies that IREL is dependent on the gRNA forming such a paired structure. It further indicates that those U insertion reactions used mainly IREL and not DREL.
(ii) IREL (band V) has been observed to become selectively adenylylated upon initial fractionation of the editing complex (29). This likely arises because IREL is adenylylated at ∼1 nM ATP (Fig. 4), which is <1/10 the concentration needed for DREL and an amount small enough to potentially become available in crude extract.
(iii) Igo et al. (20) reported a “pre-cleaved U insertion assay” that uses a purified editing complex, UTP, and three base-paired RNA oligonucleotides to mimic a U insertion site after the mRNA cleavage step. Incubation without exogenous ATP favored ligation of correctly edited product, while incubation with ATP also favored joining of the input fragments without added U residues (20). These authors inferred that joining of the U insertion product used preadenylylated ligase and that joining of the input fragments represents some aberrant form of ligation (20). We submit that their data are well-explained by our understanding of two ligases (IREL and DREL) in the purified editing complex. IREL specifically serves in U insertion (Fig. 2 to 6) and is substantially preadenylylated (29) (Fig. 5C), consistent with this enzyme selectively sealing Igo's edited fragments without added ATP, as they observed (20; see also reference 10). DREL, on the other hand, is largely unadenylylated (29) (Fig. 5B) and so requires ATP addition for maximal activity. It joins RNAs not held by a precise ligation bridge (13) (Fig. 2 to 7), like a cleaved U insertion site (19), which Igo's input fragments mimic. Thus, Igo's input RNAs should be joined specifically by DREL when ATP is added, explaining the observed joining characteristics (20).
(iv) Further, IREL (band V) became selectively adenylylated upon initial fractionation of the editing complex (29), likely because IREL is adenylylated at ∼1 nM ATP (Fig. 4), <1/10 the concentration needed for DREL and an amount small enough to potentially become available in crude extract.
(v) Finally, while DREL and IREL achieve half adenylylation (E + ATP ⇌ E-AMP + PPi) at nanomolar ATP and millimolar PPi concentrations (Fig. 2 to 6), mitochondria and cells contain many orders of magnitude more ATP and orders of magnitude less PPi. Thus, while manipulation of adenylylation has proven convenient in vitro, both ligases should be fully activated in vivo. Therefore, their charging should not limit cellular RNA editing. This is unlike the endonuclease step of editing which is responsive to ATP at physiological levels (10), nearly a million-fold higher than needed for ligase charging.
ATP and PPi titrations provide confirmatory ligase assignments.
Our initial ATP and PPi titrations contained both ATP (including [α-32P]ATP for labeling) and PPi (diluted from prior deadenylylation in Fig. 2 and 3, or added for titration in Fig. 5), so in those experiments the ligases' differential adenylylation states could not be unequivocally attributed to different apparent affinities for either component. We therefore also performed reactions that added these reagents individually. Adenylylation without PPi used editing complex that had been decharged and then fully PPiase treated (Fig. 4) or that had not been decharged (Fig. 4 legend), and both these complementary analyses indicated that IREL has ∼10-fold-higher apparent affinity for ATP than does DREL. PPi titrations were performed without exogenous ATP (Fig. 6), using precharged ligase molecules (most of the IREL and some DREL), and showed that U deletion by DREL is inactivated at <1/10 the PPi needed to inactivate U insertion by IREL. Thus, differences in DREL and IREL charging were observed in titrations where only ATP or only PPi were added.
Our studies further demonstrate that the differential effects of ATP and PPi in U deletion and U insertion are not due to a possible auxiliary component that was lost upon editing complex purification (Fig. 7), an ATP or ATPase contaminant in specific reaction buffers (Fig. 3 and 6), or effects on the previous two steps of editing (9) (Fig. 2B, bottom, and Fig. 6C). We therefore conclude that one ligase (IREL) of the editing complex is specialized for the major form of editing (U insertion) while the other ligase (DREL) is needed for the minor form of editing (U deletion).
The finding that the two forms of editing use the two ligases differently, as a reviewer pointed out, “reveals that nature does not always believe in minimalism.” Rather, the presence of two ligases in the simple editing complex and their differential roles in U deletion and U insertion likely reflects that different ligase features are advantageous for the two forms of editing. However, the whole process of RNA editing—requiring posttranscriptional editing to form 80% of the codons of certain transcripts—provides a striking demonstration that our ideas of simplicity need not reflect what survives natural selection.
Acknowledgments
We thank Laura Rusché for providing the purified editing complex, Sean O'Hearn for letting us mention provocative unpublished results, Michele Klingbeil, Paul Englund, Pierre Coulombe, Randy Bryant, and members of our lab for helpful discussions, Marilyn Parsons, Al Mildvan, and Pete Peterson for advice on nomenclature, and Vadim S. Alatortsev for computer and software expertise in the preparation of figures.
We thank the NIH (grant GM34231) for support.
The first two authors contributed equally to this work.
REFERENCES
- 1.Alfonzo, J., O. Thiemann, and L. Simpson. 1997. The mechanism of U-insertion/deletion RNA editing in kinetoplastid mitochondria. Nucleic Acids Res. 25:3751-3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allen, T., S. Heidmann, R. Reed, P. Myler, H. U. Göringer, and K. Stuart. 1998. Association of guide RNA binding protein gBP21 with active RNA editing complexes in Trypanosoma brucei. Mol. Cell. Biol. 18:6014-6022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benne, R., and D. Speijer. 1998. RNA editing types and characteristics, p. 551-554. In H. Grosjean and R. Benne (ed.), Modification and editing of RNA. ASM Press, Washington, D.C.
- 4.Blum, B., N. Bakalara, and L. Simpson. 1990. A model for RNA editing in kinetoplastid mitochondria: “guide” RNA molecules transcribed from maxicircle DNA provide the edited information. Cell 60:189-198. [DOI] [PubMed] [Google Scholar]
- 5.Blum, B., and L. Simpson. 1990. Guide RNAs in kinetoplastid mitochondria have a nonencoded 3′ oligo(U) tail involved in recognition of the preedited region. Cell 62:391-397. [DOI] [PubMed] [Google Scholar]
- 6.Byrne, E., G. Connell, and L. Simpson. 1996. Guide RNA-directed uridine-insertion RNA editing in vitro. EMBO J. 15:6758-6765. [PMC free article] [PubMed] [Google Scholar]
- 7.Clayton, C. 2000. Why do we need standard genetic nomenclature for parasite genes and gene products? Acta Trop. 75:119-122. [DOI] [PubMed] [Google Scholar]
- 8.Clayton, C., M. Adams, R. Almeida, T. Baltz, M. Barrett, P. Bastien, S. Belli, S. Beverley, N. Biteau, J. Blackwell, C. Blaineau, M. Boshart, F. Bringaud, G. Cross, A. Cruz, W. Degrave, J. Donelson, N. El-Sayed, G. Fu, K. Ersfeld, W. Gibson, K. Gull, A. Ivens, J. Kelly, L. Vanhamme, et al. 1998. Genetic nomenclature for Trypanosoma and Leishmania. Mol. Biochem. Parasitol. 97:221-224. [DOI] [PubMed] [Google Scholar]
- 9.Cruz-Reyes, J., and B. Sollner-Webb. 1996. Trypanosome U deletional RNA editing involves gRNA-directed endonuclease cleavage, terminal U-exonuclease and RNA ligase activities. Proc. Natl. Acad. Sci. USA 93:8901-8906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cruz-Reyes, J., L. Rusché, K. Piller, and B. Sollner-Webb. 1998. Trypanosoma brucei RNA editing: adenosine nucleotides inversely affect U deletion and U insertion reactions at mRNA cleavage. Mol. Cell 1:401-409. [DOI] [PubMed] [Google Scholar]
- 11.Cruz-Reyes, J., L. Rusché, and B. Sollner-Webb. 1998. Trypanosoma brucei U insertion and U deletion activities co-purify with an enzymatic editing complex but are differentially optimized. Nucleic Acids Res. 26:3634-3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cruz-Reyes, J., K. Piller, L. Rusché, M. Mukherjee, and B. Sollner-Webb. 1998. Unexpected electrophoretic migration of RNA with different 3′ termini causes a RNA sizing ambiguity that can be resolved using nuclease P1. Biochemistry 37:6059-6064. [DOI] [PubMed] [Google Scholar]
- 13.Cruz-Reyes, J., A. Zhelonkina, and B. Sollner-Webb. 2001. Trypanosome RNA editing: simple guide RNA features enhance U deletion 100-fold. Mol. Cell. Biol. 21:884-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Decker, C., and B. Sollner-Webb. 1990. RNA editing involves indiscriminate U-changes throughout precisely defined editing domains. Cell 61:1001-1011. [DOI] [PubMed] [Google Scholar]
- 15.Frech, G., and L. Simpson. 1996. Uridine insertion into pre-edited mRNA by a mitochondrial extract from Leishmania tarentolae: stereochemical evidence for the enzyme cascade model. Mol. Cell. Biol. 16:4584-4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gott, J., and R. Emeson. 2000. Functions and mechanisms of RNA editing. Annu. Rev. Genet. 34:499-531. [DOI] [PubMed] [Google Scholar]
- 17.Grams, J., M. McManus, and S. Hajduk. 2000. Processing of polycistronic guide RNAs is associated with RNA editing complexes in Trypanosoma brucei. EMBO J. 19:5525-5532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hajduk, S. 1997. Defining the editing “reaction.” Trends Microbiol. 5:1-2. [DOI] [PubMed] [Google Scholar]
- 19.Huang, C., J. Cruz-Reyes, A. Zhelonkina, S. O'Hearn, E. Wirtz, and B. Sollner-Webb. 2001. Roles for the ligases in the RNA editing complex of T. brucei: band IV is needed for U deletion and RNA repair. EMBO J. 20:4694-4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Igo, R., Jr., S. Palazzo, M. Burgess, A. Panigrahi, and K. Stuart. 2000. Uridylate addition and RNA ligation contribute to the specificity of kinetoplast insertion RNA editing. Mol. Cell. Biol. 20:8447-8457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Igo, R., Jr., S. Lawson, and K. Stuart. 2002. RNA sequence and base pairing effects on insertion editing in Trypanosoma brucei. Mol. Cell. Biol. 22:1567-1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kable, M., S. Seiwert, S. Heidmann, and K. Stuart. 1996. RNA editing: a mechanism for gRNA-specified uridylate insertion into precursor mRNA. Science 273:1189-1195. [DOI] [PubMed] [Google Scholar]
- 23.Koslowsky, D., G. Bhat, L. Read, and K. Stuart. 1991. Cycles of progressive realignment of gRNA with mRNA in RNA editing. Cell 67:537-546. [DOI] [PubMed] [Google Scholar]
- 24.Madison-Antenucci, S., R. Sabatini, V. Pollard, and S. Hajduk. 1998. Kinetoplast RNA editing associated protein 1 (REAP-1): a novel editing complex protein with repetitive domains. EMBO J. 17:6368-6376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McManus, M., M. Shimamura, J. Grams, and S. Hajduk. 2001. Identification of candidate mitochondrial RNA ligases from Trypanosoma brucei. RNA 7:167-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moore, M., and P. Sharp. 1992. Site-specific modification of pre-mRNA: the 2′ hydroxyl group at the splice site. Science 256:992-997. [DOI] [PubMed] [Google Scholar]
- 27.Peris, M., A. Simpson, J. Grunstein, J. Liliental, G. Frech, L. Simpson. 1997. Native gel analysis of ribonucleoprotein complexes from a Leishmania tarentolae mitochondrial extract. Mol. Biochem. Para. 85:9-24. [DOI] [PubMed] [Google Scholar]
- 28.Panigrahi, A., S. Gygi, N. Ernst, R. Igo, S. Palazzo, A. Schnaufer, D. Weston, N. Carmean, R. Salavati, R. Aebersold, and K. Stuart. 2001. Association of two novel proteins, TbMP52 and TbMP48, with the Trypanosoma brucei RNA editing complex. Mol. Cell. Biol. 21:380-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rusché, L., J. Cruz-Reyes, K. Piller, and B. Sollner-Webb. 1997. Purification of a functional enzymatic editing complex from Trypanosoma brucei mitochondria. EMBO J. 16:4069-4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rusché, L., C. Huang, K. Piller, M. Hemann, E. Wirtz, and B. Sollner-Webb. 2001. The two RNA ligases of the Trypanosoma brucei RNA editing complex: cloning the essential band IV gene and identifying the band V gene. Mol. Cell. Biol. 21:979-989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sabatini, R., and S. Hajduk. 1995. RNA ligase and its involvement in guide RNA/mRNA chimera formation. Evidence for a cleavage-ligation mechanism of Trypanosoma brucei mRNA editing. J. Biol. Chem. 270:7233-7240. [DOI] [PubMed] [Google Scholar]
- 32.Schnaufer, A., A. Panigrahi, B. Panicucci, R. Igo, R. Salavati, and K. Stuart. 2001. An RNA ligase essential for RNA editing and survival of the bloodstream form of Trypanosoma brucei. Science 291:2159-2162. [DOI] [PubMed] [Google Scholar]
- 33.Seiwert, S., and K. Stuart. 1994. RNA editing: transfer of genetic information from gRNA to precursor mRNA in vitro. Science 266:114-117. [DOI] [PubMed] [Google Scholar]
- 34.Seiwert, S., S. Heidmann, and K. Stuart. 1996. Direct visualization of uridylate deletion in vitro suggests a mechanism for kinetoplastid RNA editing. Cell 84:831-841. [DOI] [PubMed] [Google Scholar]
- 35.Sollner-Webb, B. 1996. Trypanosoma RNA editing: resolved. Science 273:1182-1183. [DOI] [PubMed] [Google Scholar]
- 36.Sollner-Webb, B., J. Cruz-Reyes, and L. Rusché. 2001. Direct sizing of RNA fragments using RNase-generated standards, p. 378-383. In A. Nicholson (ed.), Methods in enzymology, vol. 342. Ribonucleases, part B. Academic Press, New York, N.Y. [DOI] [PubMed]
- 37.Sollner-Webb, B., L. Rusché, and J. Cruz-Reyes. 2001. Ribonuclease activities of Trypanosome RNA editing complex directed to cleave specifically at a chosen site, p. 154-174. In A. Nicholson (ed.), Methods in enzymology, vol. 341. Ribonucleases, part A. Academic Press, New York, N.Y. [DOI] [PubMed]
- 38.Stuart, K., T. Allen, S. Heidmann, and S. Seiwert, S. 1997. RNA editing in kinetoplastid protozoa. Microbiol. Mol. Biol. Rev. 61:105-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stuart, K., A. Panigrahi, A. Schnaufer, M. Drozdz, C. Clayton, and R. Salavati. 2002. Composition of the editing complex of Trypanosoma brucei. Philos. Trans. R. Soc. Lond. 357:71-79. [DOI] [PMC free article] [PubMed] [Google Scholar]