Abstract
The murine immunoglobulin heavy-chain (Igh) locus provides an important model for understanding the replication of tissue-specific gene loci in mammalian cells. We have observed two DNA replication programs with dramatically different temporal replication patterns for the Igh locus in B-lineage cells. In pro- and pre-B-cell lines and in ex vivo-expanded pro-B cells, the entire locus is replicated early in S phase. In three cell lines that exhibit the early-replication pattern, we found that replication forks progress in both directions through the constant-region genes, which is consistent with the activation of multiple initiation sites. In contrast, in plasma cell lines, replication of the Igh locus occurs through a triphasic pattern similar to that previously detected in MEL cells. Sequences downstream of the Igh-Cα gene replicate early in S, while heavy-chain variable (Vh) gene sequences replicate late in S. An ∼500-kb transition region connecting sequences that replicate early and late is replicated progressively later in S. The formation of the transition region in different cell lines is independent of the sequences encompassed. In B-cell lines that exhibit a triphasic-replication pattern, replication forks progress in one direction through the examined constant-region genes. Timing data and the direction of replication fork movement indicate that replication of the transition region occurs by a single replication fork, as previously described for MEL cells. Associated with the contrasting replication programs are differences in the subnuclear locations of Igh loci. When the entire locus is replicated early in S, the Igh locus is located away from the nuclear periphery, but when Vh gene sequences replicate late and there is a temporal-transition region, the entire Igh locus is located near the nuclear periphery.
In Saccharomyces cerevisiae, the initiation of DNA replication is sequence specific (27; for a review, see reference 23). In mammalian cells, however, the mechanism underlying this process is still being defined. Although specific origins of replication have been mapped, initiation regions as large as 50 kb have also been described (for reviews, see references 20 and 22), making the potential role of sequence specificity less clear. In mammalian cells, the origins of DNA replication are activated in different intervals of S phase and the timing of the activation of these origins is precisely regulated. The regulation of the replication timing of a specific gene appears to reflect the transcriptional activity of surrounding sequences (for a review, see reference 58). Evidence that shows that the localization of some genes or even of an entire chromosome within the nucleus is nonrandom and is linked both to chromatin structure and replication timing has been presented (9, 16, 41; for a review, see reference 14). Thus, early- and late-replication foci tend to occupy different subnuclear domains in interphase cells (for a review, see reference 28). Recent observations suggest that nuclear structure might play an important role in defining when and where DNA replication initiates (for a review, see reference 21).
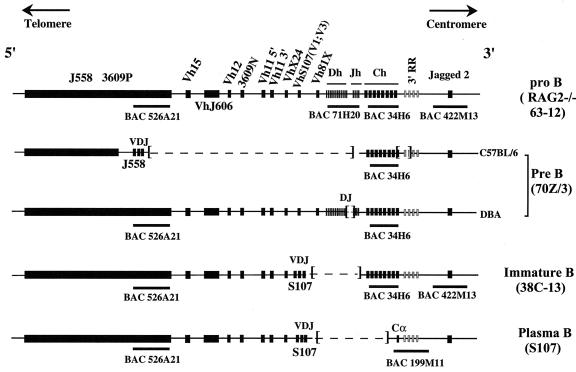
The immunoglobulin heavy-chain (Igh) locus (consisting of, in the 5′-to-3′ direction, 15 heavy-chain variable [Vh] gene families, 12 heavy-chain diversity [Dh] segments, 4 heavy-chain joining [Jh] segments, an intronic enhancer, 8 constant-region [Ch] genes, and the 3′ regulatory region) is an important model system for the study of mammalian DNA replication. The Igh locus shows successive changes in transcription and DNA rearrangements during B-cell development (Fig. 1). V(D)J joining occurs at the pro-B-cell stage, preceded by transcriptional activation of Vh genes. Class switching occurs later in B-cell development. These processes result in unique combinations of V, D, J, and C segments encoding and transcribing individual heavy-chain (and light-chain) genes in individual B cells (for a review, see reference 45). B-cell lines representing different stages of B-cell development are available for the study of the replication and nuclear localization of the Igh locus. The cell lines used in the present study are listed in Table 1. The mouse Igh locus is very large (approximately 3 Mb) but has been physically mapped and cloned in yeast artificial chromosome and bacterial artificial chromosome (BAC) arrays (13) and is being sequenced. The information derived from these arrays greatly facilitated the present study.
FIG. 1.
Rearrangements of the Igh locus during B-cell development.The germ line configuration of the Igh locus on murine chromosome 12 is shown (44, 57). In the 63-12 RAG2−/− pro-B-cell line, the entire locus is unrearranged and in germ line configuration, as is the case in MEL cells. The Igh locus is approximately 3 Mb in size and contains more than 100 Vh genes organized in 15 families, 12 Dh segments, 4 Jh segments, and 8 Ch genes. Only Vh gene families that were examined in the present study are indicated. The 3′ regulatory region (3′ RR) and the array of Ch, Dh, and Jh gene segments occupy ∼400 kb and are collectively referred to as the Igh-C locus. The Eμ enhancer is located between the Jh segments and Cμ gene (26, 29). Downstream of the Cα gene, four enhancers, represented by shaded rectangles, have been identified. They comprise the 3′ regulatory region (54). During the pre-B-cell stage of development, the locus undergoes VDJ rearrangement. As the result of VDJ rearrangement, one variable-region gene, one D segment, and one J segment are brought together, and the intervening sequences are deleted. Deleted regions are indicated by dashed lines. The two allelic chromosomes with different rearrangements in the 70Z/3 cell line are shown. Only the C57BL/6-derived chromosome is productively rearranged, resulting in the transcription of a Vh J558-Cμ gene (51, 53). This allele has a 34-kb deletion that removes Cα and two of the 3′ Igh enhancers (53). The DBA-derived chromosome has a DQ52-to-Jh2 rearrangement, which is transcribed as a sterile (Sμ) transcript (51). The 38C-13 B-lymphoma-cell line represents a B-cell line with a continuing Vκ rearrangement phenotype. The productively rearranged Vh gene of 38C-13 belongs to the S107 family (51). The S107 plasmacytoma cell line represents terminally differentiated B cells. Isotype switching to the Cα gene has occurred in the S107 cell line. The productively rearranged chromosome expresses IgA, and the Vh family used in this cell line is S107 V1. The positions of the BACs used in this study are shown by black rectangles under the maps.
TABLE 1.
Characteristics of B-lineage cell lines used in this study
| Cell line (reference) | Stage of B-cell development | VDJ recombination state(s) of Igh locus | Gene(s) at which transcription was detected |
|---|---|---|---|
| 63-12 (4) | RAG2−/− pro-B | Germ line configuration | VhJ558, Vh3609, Vh10 |
| 38B9 (1, 63) | Pre-B | VDJ recombined | VhJ558, Cμ |
| 22D6 (1, 32, 63) | Pre-B | VDJ recombined | VhJ558, Vh3609, VhS107 |
| 70Z/3 (51, 53) | Pre-B | VDJ rearranged on productive allele, DJ rearranged on nonproductive allele | VhJ558, Cμ |
| 18-81 (2, 61) | Pre-B | VDJ recombined | Vh81X, Vh81Y, Cμ, Cγ2b |
| 38C-13a (51) | Immature B | VDJ rearranged on productive allele, DJ rearranged on nonproductive allele | VhS107, Cμ |
| WEHI-231a (2) | Immature B | VDJ recombined | Cμ |
| S107 (15, 39) | Plasmacytoma | VDJ recombined and class switched to Cα gene on productive allele, the other allele translocated with c-myc | VhS107, Cα |
Although both 38C-13 and WEHI-231 cells are immature B cells, 38C-13 cells still express RAG protein and can undergo secondary light-chain recombination (42). Therefore, 38C-13 may resemble pre-B-cell lines.
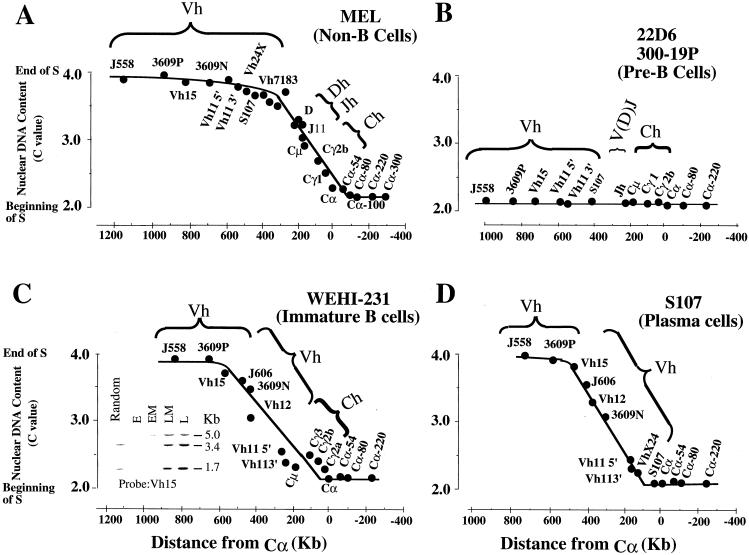
Previous studies using bromodeoxyuridine (BrdU) S-phase fractionation have shown that the replication timing of the Igh locus in non-B-cell lines, such as the murine erythroleukemia (MEL) cell line, is different from that in pre-B-cell lines (11, 34, 35). In MEL cells, an ∼400-kb sequence forms a transition region between an early-replication domain downstream of the 3′ regulatory region and a late-replication domain containing Vh genes (triphasic-replication pattern) (Fig. 2A). Sequences within this transition region replicate 3′ to 5′ progressively later during S. A previous study using neutral-alkaline two-dimensional (N/A 2D) gel electrophoresis has shown that the transition region is formed by a single replicon through which replication forks progress in the 3′-to-5′ direction (25). In contrast, the entire Igh locus is replicated within the first hour of S in two pre-B-cell lines. Previous studies also demonstrated that part of the Igh-C region replicates early in S in plasma cell lines (34).
FIG. 2.
Replication timing of the murine Igh locus in B-lineage cell lines. The time in S phase when a particular segment is replicated is expressed in terms of the C value (y axis) and is plotted versus its distance in kilobases from the Cα gene (x axis), and the results are shown as filled circles. The C value is the haploid DNA content of the cells at a particular time in S phase. At the G1/S boundary, the C value is 2.0; after DNA replication is completed, the C value is 4.0. All the timing data were obtained by the BrdU S-phase fractionation technique. BrdU-labeled DNA was prepared from cells at four different intervals of S phase (early, early middle, late middle, and late) and separated by centrifugal elutriation. The graphs shown represent the average times of replication of the two alleles. According to our timing data from the BrdU S-phase fractionation, the maximum differences in the replication times of the two alleles are not greater than 1.5 h for sequences replicating early and late in S phase and not greater than 2.5 h for sequences within the transition region. These estimates are based on the hybridization signals detected in BrdU-labeled DNA replicated in the four intervals of S phase. (A) Three temporal domains were detected in MEL cells. The early-replication (downstream of Cα) and late-replication regions (Vh genes) are connected by sequences (∼400 kb) that are replicated sequentially throughout S (11, 25, 36, 47). The times of replication of the Vh11, Vh15, Vh3609P, and J558 genes were obtained in this study. (B) Replication of the Igh locus was completed within the first hour of S phase in two pre-B-cell lines. The data presented here were obtained from the 22D6 cell line, but similar results were obtained for the 300-19P cell line. (C and D) The time in S when replication of the murine Igh locus occurs in the WEHI-231 B-cell line (C) and the S107 plasma B-cell line (D) resembles that in the MEL non-B-cell line, whose results are shown in panel A. Three temporal domains were observed. VDJ rearrangement and isotype switching result in deletion of the internal portion of the Igh locus. The lengths of the transition regions are similar to that in MEL cells. Also shown in panel C is the autoradiogram after hybridization (with a Vh15 probe) to DNA transfer of BrdU-labeled WEHI-231-cell DNA replicated during four intervals of S (E, early; EM, early middle; LM, late middle; L, late).
One explanation for the dramatically different replication programs detected in non-B cells and pre-B cells is that the DNA replication program is linked to the transcriptional status of the Igh locus. In MEL cells, the Igh locus is transcriptionally silent, while in pre-B-cell lines, this locus is actively transcribed. Since the pattern of transcription along the locus changes during B-cell development, we decided to examine the replication of the Igh locus in B-cell lines at different stages of B-cell development.
The experiments presented here reveal two distinct programs of replication for the Igh locus in B-lineage cells. In pro- and pre-B-cell lines, including those derived from RAG2−/− mice, the entire Igh locus is replicated early in S phase. The temporal pattern of Igh replication in other cell lines representing late stages of B-cell development (an immature-B-cell line and plasma cell lines) resembles that in non-B cells. The temporal-transition regions in the different cell lines are similar in length but encompass different sequences, reflecting individual DNA rearrangement events associated with VDJ and class switch recombination. The replication timing of the Igh locus in ex vivo-expanded pro-B cells and lipopolysaccharide (LPS)-stimulated spleen B cells is consistent with the data obtained from the cell lines.
In this study, we address the potential mechanism for this dramatic difference in replication programs. N/A 2D gel electrophoresis demonstrates that replication forks progress in both directions through the examined Igh-C genes in cell lines in which the entire locus replicates early in S. Our results suggest that the early replication of the Igh locus is achieved through the activation of one or more replication origins. In contrast, in B cells in which the Igh locus replicates through a triphasic pattern, replication forks progress in a single 3′-to-5′ direction (opposite to the transcriptional orientation) through all the constant-region genes we examined. Our observations that the directions of replication fork progression can differ in B-lineage cell lines raise the possibility that changes in Igh replication are involved in processes of Igh rearrangement and expression. We have found that, associated with the changes in replication, the entire Igh locus is located away from the nuclear periphery when it is replicated early in S phase but is located near the nuclear periphery when it is replicated in a triphasic pattern.
MATERIALS AND METHODS
Cell lines.
The 70Z/3 pre-B-cell line was adapted from a methylnitrosourea-induced tumor that arose in a thymectomized (DBA/2 × C57BL/6) mouse (51). 18-81 (2) and 22D6 (1) are Abelson virus-transformed pre-B-cell lines. A RAG2−/− fetal-liver-derived Abelson virus-transformed pro-B-cell line, 63-12, contains an unrearranged Igh locus, but transcription through Vh genes is activated (4). The 38B9 pre-B-cell line and ex vivo-expanded pro-B cells (obtained from a fetal [14.5-day-old] liver) were kindly provided by Steven Kosak in Harinder Singh’s laboratory (University of Chicago) (40). The 38C-13 B-cell line, which produces membrane and secreted forms of Cμ (51), was developed from a 7,12-dimethylbenzanthracene-induced tumor in a C3H/eB mouse. WEHI-231 is an IgM-producing B-cell line (2) kindly provided by P. Brodeur (Tufts University). The S107 (IgA-producing) murine myeloma cell line was originally obtained from M. Scharff (15). One of the Igh alleles has undergone reciprocal translocation with the c-myc gene (39).
All cell lines were maintained as exponential cultures at a density between 2 × 105 and 7 × 105 cells/ml. The doubling times were ∼12 h for the MEL, 70Z/3, 38C-13, 18-81, 22D6, 38B9, and RAG2−/− 63-12 cell lines and ∼16 h for WEHI-231 and S107. All cell lines were cultured at 37°C with 5% CO2 in RPMI 1640 medium, except S107 and MEL cells, which were grown in Dulbecco's modified Eagle's medium. The RPMI 1640 medium was supplemented with 10% fetal bovine serum (GIBCO BRL), penicillin-streptomycin (GIBCO BRL), 2 mM l-glutamate (GIBCO BRL), and 50 μM 2-mercaptoethanol (Sigma), but the Dulbecco's modified Eagle's medium was supplemented only with 10% fetal bovine serum and penicillin-streptomycin. Splenic B cells, depleted of red cells, T cells, and macrophages and treated with LPS (40 μg/ml) for 48 h, were kindly provided by Steven Gordon (Albert Einstein College of Medicine). After stimulation, 90% of the cells were B220-positive B cells.
Centrifugal elutriation, BrdU S-phase fractionation, and 2D gel electrophoresis.
Centrifugal elutriation and BrdU S-phase fractionation were carried out as previously described (11). BrdU-labeled DNA obtained from different intervals of S phase was transferred and chemically bonded to diazobenzyloxymethyl paper (3) before Southern hybridization so that the times of replication of many different segments could be compared on the same DNA transfer. N/A 2D (25, 50) and neutral-neutral (N/N) 2D (8) gel electrophoreses were carried out as previously described. We estimated the relative amounts of replication intermediates in matrix-associated DNA by densitometry of signals within the linear response range of the film. For each segment analyzed by N/A 2D gel electrophoresis, the ratio of the intensity of parental plus nascent molecules to the intensity of the signal produced by nonreplicating linear molecules (1N spot) was calculated. This ratio was calculated for six segments in MEL cells from five different centrifugal elutriations and for five segments in 70Z/3 cells from four different centrifugal elutriations. For this analysis, replication intermediates were recovered from cells in fractions in early or early-middle S phase. In addition, we analyzed two segments from asynchronous (nonelutriated) 38C-13 cells and compared them to the same segments from asynchronous MEL cells. We also made these calculations for a few segments analyzed by N/N 2D gel electrophoresis.
FISH.
Fluorescence in situ hybridization (FISH) was performed on flattered nuclei as described previously (55). For the nuclear localization and replication timing studies, BACs containing various segments of the Igh cluster were used. The BACs were from the CitbCJ7 library made from the 129S1/SvImJ mouse strain and were purchased from Research Genetics (now Invitrogen). The BACs used were CT7-526A21, which contains Ch-proximal J558 genes and flanking sequences, CT7-71H20, which contains ∼90 kb of the Dh and Jh segments, CT7-34H6, which contains Igh-C sequences, CT7-199M11, which contains sequences downstream of the Cα gene, and CT7-422M13, which contains Jagged 2 gene sequences located approximately 300 kb downstream of Cα. The BAC colonies were cultured in Luria broth supplemented with chloramphenicol (12.5 μg/ml). DNA was purified with QIAGEN-tip 100 according to the instructions of the manufacturer, except that the elution buffer was prewarmed at 65°C before use. The BACs were labeled with biotin-16-dUTP or digoxigenin-11-dUTP by use of a nick translation kit (Roche). The probe (∼100 ng/slide) was then coprecipitated with mouse Cot-1 competitive DNA (∼30 μg) and salmon sperm DNA (∼5 μg) with 100% ethanol.
The slides were hybridized to the probes in a humid chamber for at least 14 h at 37°C. Biotinylated probes were detected with fluorescein isothiocyanate-avidin (Oncor), and digoxigenin-labeled probes were detected with rhodamine-conjugated antidigoxigenin antibody (Oncor). The slides were counterstained with DAPI (4′,6′-diamidino-2-phenylindole) and then mounted in antifade solution. The slides were visualized by conventional fluorescence microscopy, and digital images were taken with a charge-coupled-device camera and combined by Adobe Photoshop.
The replication timing in some cell lines was estimated by FISH, as described previously (7, 55, 59). The data obtained by FISH have been shown to be consistent with data obtained by the BrdU S-phase fractionation technique (59). The number of doublets (indicating replication) for each allele in the nuclei of cells in log phase was calculated. All the cell lines subjected to FISH for the estimation of replication timing have similar doubling times, and the percentage of S-phase cells was about 50%. To determine the subnuclear localization of Igh sequences, the distance from the center of each nucleus to the individual signal and the radius of each nucleus were measured with the NIH Image program (Scion Image 1.62). The data were analyzed with Microsoft Excel. At least 50 nuclei were analyzed in each experiment.
Probes used for N/A 2D gel electrophoresis and timing blots.
The probes used for the N/A 2D gel electrophoresis of the Igh-C locus were described previously (25), except for the probes for the Cα region (probe 24 and 25). Probe 24 is a 0.6-kb HindIII/EcoRI fragment from the 3′ end of the 4.5-kb EcoRI fragment (Cα, exons CH2 and CH3), and probe 25 is a 0.9-kb EcoRI/HindIII fragment from the 5′ end of the same fragment (30). For the timing studies, the probes for some of the Vh genes; for the Dh, Jh, and constant-region genes; and for downstream sequences have been described previously (25). The new probes used in these studies for the Vh region genes include the following: the VhX24 probe, a 500-bp EcoRI fragment containing a 300-bp VhX24 sequence and 200 bp of 3′-end-flanking sequences (33); the Vh11 probe, a 321-bp EcoRI/XbaI fragment that originated from PCR of a cDNA library and that can detect two Vh11 genes (52); the Vh12 probe, a 0.3-kb EcoRI/XbaI fragment that originated from PCR of a cDNA library and that can detect a single Vh12 gene (52); the VhJ606 probe, a 0.6-kb EcoRI/BamHI fragment that contains coding region sequences corresponding to amino acids 16 to 98 and 3′-end-flanking sequences and that can detect the murine VhJ606 gene family (62); the Vh15 probe, a 297-bp PCR product that contains Vh15 coding sequences and that can detect the mouse Vh15 gene family (44); the Vh3609P probe, a 1.6-kb EcoRI/BamHI fragment that contains coding sequences of the Vh3609P gene family (10); and the J558 probe, a 0.5-kb EcoRI fragment that contains part of the J558 coding sequences and 3′-end-flanking sequences and that can detect the J558 gene family (63).
RESULTS
Two dramatically different programs of DNA replication of the Igh locus occur in B-lineage cells.
It has previously been shown that in two pre-B-cell lines (22D6 and 300-19P), the VhS107, Dh, Jh, and Igh-C genes replicate early in S (11, 34). Here, we examined the replication timing of other Vh gene families and of sequences located downstream of Cα in 22D6 cells using the BrdU S-phase fractionation method. These experiments showed that the entire Igh locus replicates within the first hour of S (Fig. 2B). This result is in sharp contrast to the observed temporal organization of Igh gene replication in MEL and other non-B-cell lines (Fig. 2A) (11, 25). Here, we studied replication timing in other pre-B-cell lines as well as in primary cells by FISH. FISH can be applied to relatively few cells and, in particular, to primary cells; it is generally not possible to obtain a sufficient number of primary cells for the BrdU S-phase fractionation method (11).
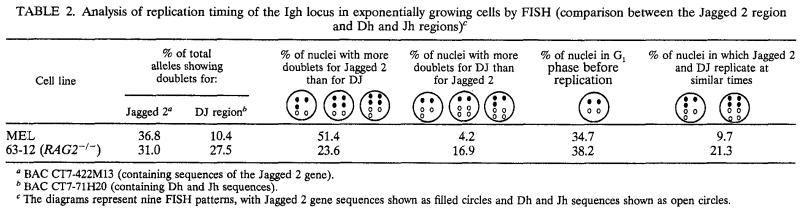
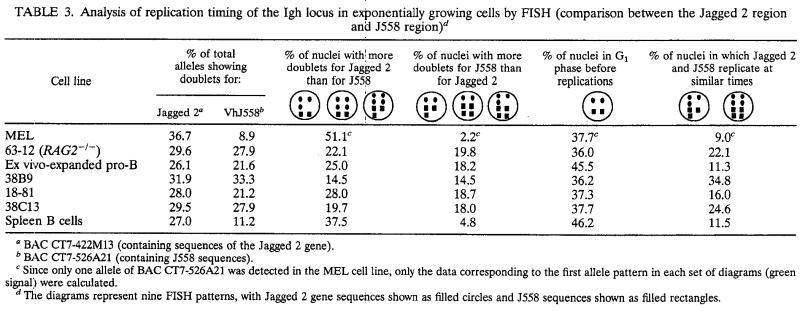
We first demonstrated that FISH is an accurate method for the measurement of the replication timing of the Igh locus by applying this technique to MEL cells for which we had previously measured the replication timing of the Igh locus using the BrdU method. The replication timing was determined by measuring the proportion of alleles displaying doublets (55, 59; for a review, see reference 7). A gene replicated at the start of S shows doublets at each of its alleles throughout S phase, while a gene replicated late in S shows doublets only briefly before division. Thus, a gene that replicates early in S phase can be detected by the presence of a large rather than a small number of unsynchronized cells displaying doublets for the contrasting genes. The Jagged 2 gene is located approximately 300 kb downstream of the Cα gene and replicates early in S in MEL cells, as detected by the BrdU method. In MEL cells, 36.8% of the total Jagged 2-hybridizing alleles displayed doublets. Consistent with the later replication of the Vh, Dh, and Jh segments, the percentages of total alleles showing doublets for these regions are less than that for the Jagged 2 gene: 8.9% for J558 genes and 10.4% for the Dh and Jh segments. The data shown in Tables 2 and 3 also indicate that the Jagged 2-hybridizing region replicates earlier than the upstream J558 genes and the Dh and Jh segments: 51.1% of the nuclei have more doublets for Jagged 2 than for J558 genes, and 51.4% have more doublets for Jagged 2 than for the Dh and Jh segments. Only a few nuclei show more doublets for J558 genes (2.2%) or for Dh and Jh segments (4.2%) than for Jagged 2. These data show that the replication timing determined for MEL cells by FISH (Tables 2 and 3) agrees with that previously determined by the BrdU method (Fig. 2A).
TABLE 2.
Analysis of replication timing of the Igh locus in exponentially growing cells by FISH (comparison between the Jagged 2 region and Dh and Jh regions)c
BAC CT7-422M13 (containing sequences of the Jagged 2 gene).
BAC CT7-71H20 (containing Dh and Jh sequences).
The diagrams represent nine FISH patterns, with Jagged 2 gene sequences shown as filled circles and Dh and Jh sequences shown as open circles.
TABLE 3.
Analysis of replication timing of the Igh locus in exponentially growing cells by FISH (comparison between the Jagged 2 region and J558 region)d
BAC CT7-422M13 (containing sequences of the Jagged 2 gene).
BAC CT7-526A21 (containing J558 sequences).
Since only one allele of BAC CT7-526A21 was detected in the MEL cell line, only the data corresponding to the first allele pattern in each set of diagrams (green signal) were calculated.
The diagrams represent nine FISH patterns, with Jagged 2 gene sequences shown as filled circles and J558 sequences shown as filled rectangles.
We therefore used FISH to examine the replication timing of the Igh locus in several pre-B-cell lines. Based on our observations that the percentages of total alleles showing doublets for the J558- and the Jagged 2-hybridizing regions are similar (21 to 33%) (Tables 2 and 3), we found that in the 38B9 and 18-81 cell lines the entire Igh locus replicates early in S. In agreement with this finding, the data in columns 3 and 4 of Tables 2 and 3 indicate that the probabilities that the J558 genes replicate earlier or later than the Jagged 2 gene are similar. Because most of the Dh and Jh regions were deleted in pre-B-cell lines as a result of VDJ joining, these regions were not examined. We have determined that sequences spanning the entire Igh locus examined here replicate early in S in four pre-B-cell lines. Based on these results, we propose that the Igh locus replicates early in cell lines representing an early stage of B-cell development.
During the pro- and pre-B-cell stages of development, the Igh locus is transcriptionally activated, after which VDJ joining events occur independently on each allele, coupled with DNA deletion of intervening segments. We wanted to determine whether VDJ joining is required for the activation of early replication of the Igh locus, perhaps through deletion of cis-acting elements that inhibit the activation of replication origins within the transition region early in S. We therefore used FISH to examine replication timing in a RAG2−/− pro-B-cell line, 63-12, which cannot undergo VDJ joining (4). As in the pre-B-cell lines studied here, sequences spanning the entire Igh locus replicate early in S phase in 63-12 cells (Tables 2 and 3). We have also found that the Igh locus replicates early in three additional RAG-deficient pro-B-cell lines (data not shown). Therefore, early replication of the Igh locus occurs in cell lines representing early stages of B-cell differentiation prior to VDJ joining.
These experiments extend our previous studies that distinguished two replication programs for the Igh locus. In MEL cells and other non-B-cell lines (11), Igh genes replicate throughout S, while in pro- and pre-B-cell lines, Igh genes replicate early in S. Since Igh genes are transcribed in pro- and pre-B-cell lines but not in MEL cells, these observations raise the possibility that replication of the Igh locus early in S reflects Igh transcriptional activity. We therefore examined the replication timing of the Igh locus in B-cell lines representing later stages of B-cell development. Examination of the 38C-13 immature-B-cell line by FISH showed that the entire locus replicates early in this cell line (Table 3). However, the temporal organization of the replication of the Igh locus in three other cell lines (WEHI-231 immature B cells [Fig. 2C], S107 plasma cells [Fig. 2D], and LP1.2 plasma cells [data not shown]), which were examined by BrdU S-phase fractionation, resembles that of non-B MEL cells rather than that of the pro- and pre-B-cell lines. Although the rearranged variable-region gene and the associated constant gene are transcribed and replicate early in S in these cell lines, these transcriptions do not result in early replication of the entire locus.
We examined ex vivo-expanded pro-B cells and LPS-stimulated splenic B cells (Table 3) to determine whether the two different replication programs for the Igh locus are also apparent in primary B cells at different stages of B-cell maturation. The J558 Vh gene family (furthest from the constant region) replicates as early as downstream region genes in ex vivo-expanded pro-B cells. In contrast, this same region replicates later in S phase than the downstream-region sequences in LPS-stimulated splenic B cells. This indicates that the temporal order of Igh DNA replication that we observed in cultured cells parallels that in primary cells.
A recent study has indicated that there is asynchrony in the times of replication of the two Igh alleles (48), with a difference in replication timing of <1.5 h (H. Cedar, personal communication). Only the μ heavy chain was examined by FISH in that recent study (48). While our FISH data are consistent with this relatively small degree of asynchrony, this asynchrony is not great enough to affect our conclusions about the dramatic changes in replication timing of the entire locus in different B-lineage cells. In pro- and pre-B-cell lines and in ex vivo-expanded pro-B cells, the entire locus on both alleles is replicated early in S phase, though one allele can replicate somewhat earlier than the other. In a B-cell line, plasma cell lines, and non-B-cell lines, the triphasic-replication pattern we detected is triphasic for both alleles (see the legend for Fig. 2).
Therefore, two distinct DNA replication programs have been detected for the Igh locus in B-lineage cells. In pro- and pre-B-cell lines and ex vivo-expanded pro-B-cells, the entire Igh locus is replicated early in S phase. In contrast, in plasma cell lines, the locus is replicated through a triphasic replication program. Interestingly, in WEHI-231 B cells, the Igh genes were replicated through a triphasic program but in the 38C-13 B-cell line, the Igh genes replicated early in S. The 38C-13 B-cell line has attributes of B-lineage cells at early stages of development, since the RAG gene and surrogate light-chain genes are still actively transcribed and light-chain rearrangement is ongoing (12, 42) (see Discussion).
Early replication of the Igh locus is achieved by replication forks progressing in both directions through the Igh-C region.
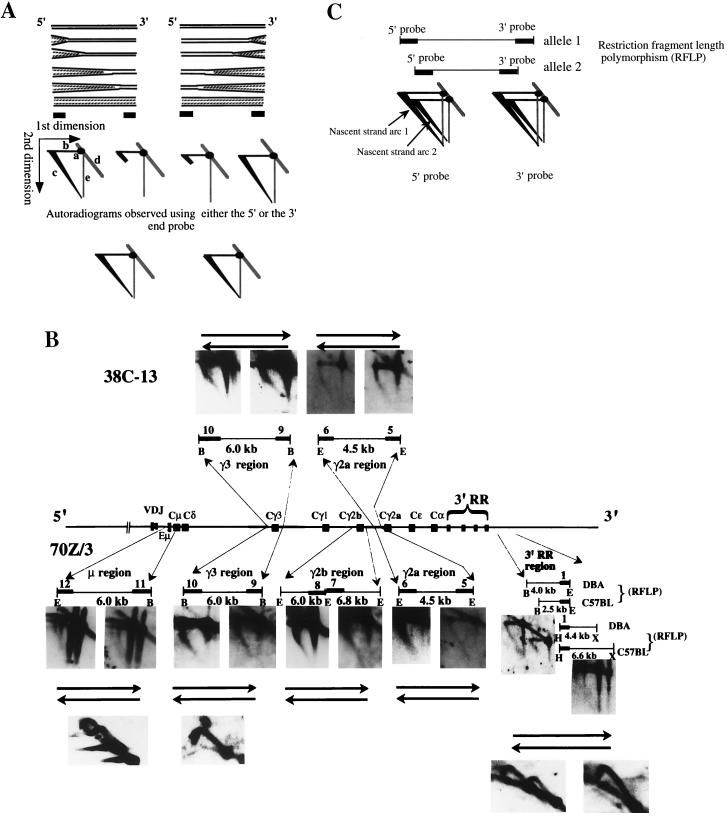
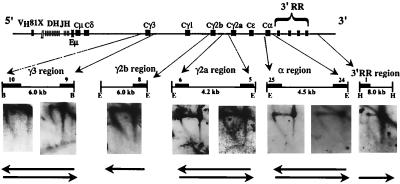
We identified an initiation region in MEL cells that is located downstream of the 3′ regulatory region: replication forks originating in this initiation region accomplish the replication of the transition region (25; J. Zhou, N. Ashonian, M. Delepine, F. Matsuda, C. Chevillard, R. Riblet, C. L. Schildkraut, and B. K. Birshtein, unpublished data). If the same initiation region were used to replicate Igh-C genes in cell lines in which the entire Igh locus replicates early, then forks would again move in a 3′-to-5′ direction through the Igh-C region. Alternatively, early replication of the Igh-C region may involve additional initiation sites that are activated early in S phase and that initiate forks progressing in both directions. To discriminate between these possibilities, we examined the direction of replication fork movement through the Igh-C cluster by N/A 2D gel electrophoresis. With the available unique probes, we were able to examine Igh-C sequences in five segments distributed over 250 kb. In the 70Z/3 pre-B-cell line, the 38C-13 B-cell line, and the RAG2−/−-derived 63-12 cell line, we found that replication forks progressed in both directions through all examined segments of the Igh-C cluster (Fig. 3B and 4). These data suggest that in addition to the 3′ initiation region, there must be at least one other origin upstream of the Igh-C cluster that is activated early in S in these cell lines. VDJ rearrangement is not essential for the progression of forks in both directions.
FIG. 3.
(A) Schematic representation of replication forks progressing in both directions through a segment, as detected by the N/A 2D gel electrophoresis technique. We considered two instances in which replication forks progress in both directions through a segment, as detected by either a 5′- or 3′-end probe: (i) replication forks progress towards each other within the examined segment and (ii) replication forks progress in one direction in some cells (one population) and in the opposite direction in other cells (the other population) within the examined segment. These two instances can be discriminated by N/N 2D gel electrophoresis. In the first case, a random or specific termination pattern is expected, while in the second case, no termination is observed. Since we did not detect termination within the segment by N/N 2D gel electrophoresis (Fig. 3B), only the second instance is shown in the diagram. Two identical segments in two different populations of cells are shown. The positions of the probes (filled rectangles) are shown below the segments. The signals that appeared on the film after hybridization to the specific probes are schematically shown below the specific probes and are labeled a to e as follows: a is the nonreplicated linear segments specifically detected by either the 3′- or 5′-end probe (called the 1N spot), b is the full-length parental strands from which the nascent strands are released in the second dimension at alkaline pH, c is the newly replicated nascent strands of multiple (both large and small) sizes, d is the cross-hybridization signals to the nonreplicating linear molecules, and e is the single-stranded molecules that result from breaks in the linear nonreplicating molecules (from the 1N spot). In the segment on the right, replication forks progress in the 3′-to-5′ direction. In this case, the probe at the 3′ end of the segment detects nascent strands of multiple sizes, while the probe at the 5′ end detects only large nascent strands. In the segment on the left, replication forks move from the 5′ end to the 3′ end of the segment. The probe at the 5′ end detects nascent strands of different sizes, and the 3′-end probe detects only large nascent strands. Since both of these populations of cells are present, both the 3′- and 5′-end probes detect nascent strands of multiple sizes (from different cells). The long diagonal nascent strand arcs detected by these probes are shown at the bottom of the figure. (B) Replication forks progress in both directions through examined segments of the Igh-C region in the early-replicating Igh locus. The examined restriction fragments and the positions of the probes (filled rectangles) are indicated. Both the 5′- and 3′-end probes detected nascent strands of all sizes, demonstrating that replication forks enter the segment from upstream and from downstream. Horizontal arrows below (70Z/3) or above (38C-13) the autoradiograms indicate the direction of replication fork movement. In two regions in the 70Z/3 cell line (μ region and 3′ regulatory region [RR]), the direction of replication fork progression on two allelic chromosomes was examined. Two separate arcs are visible for each allele since the probe detects two different restriction fragments (one from each allele). The difference in the sizes of the segments on the two allelic chromosomes for the 3′ regulatory region is much greater than that for the μ region. Thus, the two nascent strand arcs for the 3′ regulatory region are far apart while the two nascent strand arcs for the μ region are close to each other. Nascent strands of all sizes were detected in all segments after Southern hybridization to either the 5′- or 3′-end probe, indicating that replication forks progress in both directions on both alleles. Twelve fragments were examined by N/N 2D gel electrophoresis, which detects the shapes of the replicated molecules (examples are shown). Only a Y-shaped arc was detected in all segments examined, suggesting that replication forks progress through the fragments in either direction. Abbreviations: E, EcoRI; B, BamHI; H, HindIII; X, XbaI; RFLP, restriction fragment length polymorphism. (C) Schematic representation of replication forks progressing in both directions on two allelic chromosomes detected by N/A 2D gel electrophoresis. This scheme represents the ideal pattern of the autoradiogram one would detect if the replication forks move in both directions through the two allelic chromosomes, which can be distinguished by an RFLP. The two separated nascent strand arcs are indicated by arrows. For the Cμ segment, the smallest nascent strands originating from both chromosomes were not easily detectable, in contrast to what is shown in panel B. In the original autoradiogram, nascent strands smaller than 1.5 kb were weakly detected. This may be due to the fact that the intensities of the signals of the nascent strands from a single allele were weaker (by a factor of 2) than those of strands derived from both alleles simultaneously. In the 3′ regulatory region (B), nascent strands of all sizes originating from the DBA allele were easily detected with the 3′-end probe. However, on the C57BL allele, the smallest nascent strands (up to 800 bp) were not apparent on the autoradiogram since the conditions of N/A 2D gel electrophoresis were not ideal for both segments. Since these two segments (2.5 and 4.0 kb) differ in size by a factor of almost 2, the gel could not be run in a range optimal for both. The 5′-end probe for the HindIII/XbaI segment in the 3′ regulatory region detected nascent strands of all sizes from both alleles, which were apparent on the autoradiogram although they appear very weak in the gels shown in panel B. Since these allelic segments differ in size by only one-third, they can be distinguished by use of the same gel conditions.
FIG. 4.
VDJ rearrangement is not essential for activation of B-cell-specific origins. In the 63-12 cell line derived from RAG2−/− mice, the Igh-C locus is unrearranged, but germ line transcription through Vh genes is activated (4). The restriction fragments examined by N/A 2D gel electrophoresis are indicated by lines below the map. The arrows indicate the probes used. As shown on the autoradiogram, probes from the 5′ and 3′ ends detected nascent strands of all sizes, indicating that replication forks move in both directions through Igh-C. The autoradiograms for Cγ2b and the 3′ regulatory region (RR) were obtained with probes located at only one end of the segments. The 5′-end probe for the Cγ2b region did not detect replication intermediates, although several different membranes were hybridized multiple times. These membranes produced a very strong signal derived from replication intermediates when they were hybridized to other probes. In the 3′ regulatory region, the unique 3′-end probe for the HindIII fragment contained repeated sequences and thus could not be used to examine the direction of replication fork movement. E, EcoRI; H, HindIII.
To demonstrate that forks move in both directions through Igh-C on both alleles, we took advantage of polymorphisms in 70Z/3 cells. N/A 2D gel electrophoresis was performed with probes specific for a polymorphic segment within the Cμ region as well as for a segment in the 3′ regulatory region within which there was deletion on one of the chromosomes (Fig. 1). Nascent strands of all sizes were detected in both the μ and the 3′-regulatory-region segments after hybridization to probes at either the 5′ or 3′ end (Fig. 3B), indicating that replication forks progress in both directions on both alleles. If replication forks progressed through a specific segment in both directions on a single allele within the same cell, then termination intermediates would be present either at specific or random sites. However, using N/N 2D gel electrophoresis to analyze the shapes of replication intermediates on the same segments we studied by N/A 2D gel electrophoresis, we found that none of the examined segments exhibited a detectable termination pattern (Fig. 3B). This finding is consistent with the fact that the majority of forks progress in one direction on a single allele in some cells and in the other direction through these segments in other cells. It is still possible that a few replication forks terminate within these segments, but such an occurrence would be at a frequency below the level detectable by N/N 2D gel electrophoresis. In addition to revealing fork progression in both directions, N/A 2D gel electrophoresis shows that similar numbers of forks progress from upstream origins and downstream origins for several segments throughout the Igh-C region (determined from the intensities of signals) (Fig. 3B and 4) (see Discussion). Our data demonstrate that each segment we examined is replicated by either an upstream or downstream origin.
Our previous data demonstrated that the 2-Mb Vh region replicates in a 1-h interval late in S phase in MEL cells (Fig. 2A), indicating that it must contain several initiation regions. In B-lineage cell lines in which the entire Igh locus replicates early in S phase, these initiation sites (or other sites within the Vh region) are activated early in S, because the entire Igh locus replicates during the first hour of S (Fig. 2B and Table 2) (11, 34). If only the Vh-associated origins or the origin region 3′ of the transition region were used to replicate a single allele, then forks would necessarily have to move much faster through this region than in MEL cells. Because there is no evidence for a higher rate of fork movement (see Discussion), the data shown in Fig. 2 to 4 and Table 2 suggest the activation of an additional origin or origins.
Replication forks progress in one direction through the constant-region genes in B-lineage cell lines that exhibit a temporal-transition region.
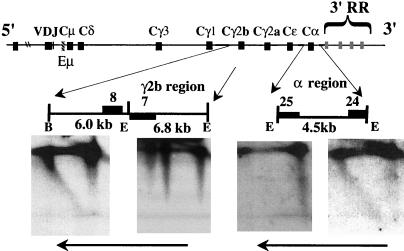
It has previously been reported that the replication of the temporal-transition region in MEL cells is achieved by a single fork progressing in a 3′-to-5′ direction (25). Our study shows that the temporal organization of replication in the Igh locus in WEHI-231 B cells and S107 plasma cells is similar to that in MEL cells. To determine whether the temporal-transition regions in the WEHI-231 B-cell and S107 plasma cell lines are also achieved by the same mechanism, we examined the direction of replication fork movement in these cell lines. Only constant-region genes were examined, because there was no strong unique probe available for variable-region genes. A probe for the 3′ end of the examined segments detected nascent strands of all sizes, while a probe for the 5′ end detected only long nascent strands for the two constant-region genes examined in WEHI-231 cells (Fig. 5) and for the Cα region in S107 cells (data not shown). These results, together with the timing data, provide evidence that replication forks also progress in the 3′-to-5′ direction through the transition region in these two cell lines, as they do in MEL cells. We could not examine the direction of replication fork movement in primary cells by using N/A 2D gel electrophoresis since this technique requires very large numbers of cells. However, the fork direction data from these cell lines provide models for hypotheses to be tested in primary cells.
FIG. 5.
Replication forks progress in one direction through the constant region in the WEHI-231 immature-B-cell line. The positions of the examined segments are shown under the map of the Igh locus. Only 3′-end probes detected nascent strands of all sizes, indicating that replication forks progress from 3′ to 5′ in two regions (Cγ2b, Cα) in WEHI-231. Unique probes were not available to analyze both ends of the two segments containing the γ2b gene. Instead, we used a probe at the 3′ end of one segment and at the 5′ end of the adjacent segment. Probe 7 detected only large nascent strands in the WEHI-231 cell line but detected nascent strands of all sizes in the 70Z/3 cell line (Fig. 3B). The vertical lines to the left of the 1N spot observed for the 6.8-kb EcoRI fragment in WEHI-231 indicate linear, nonreplicating molecules resulting from partial digestion, which can be distinguished from nascent strands that form diagonal arcs extending from the horizontal parental-strand signals (see the diagram in Fig. 3A). The numbered probes are indicated by filled rectangles, and the horizontal arrows indicate the direction of replication fork movement. RR, regulatory region; E, EcoRI; B, BamHI.
Subnuclear location of the Igh locus in B-lineage cells.
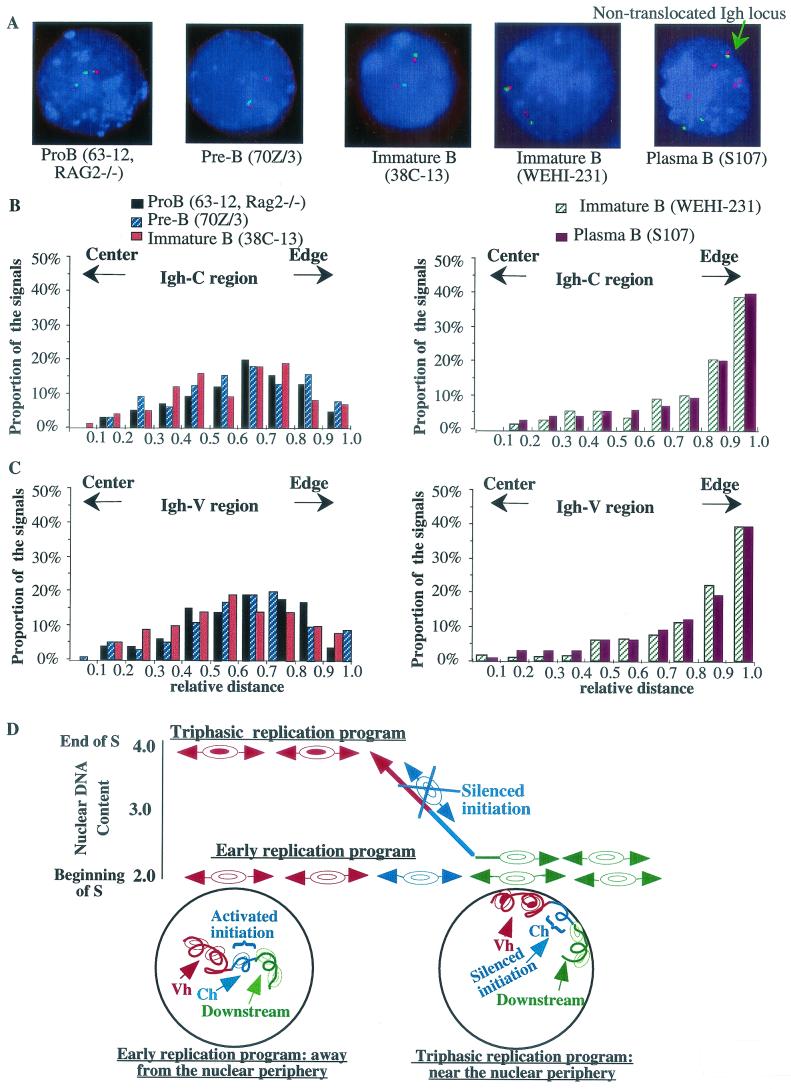
Replication at different times in S can occur in different subnuclear domains (for a review, see reference 28). To determine the subnuclear localization of Igh loci in B-lineage cell lines that exhibit different temporal-replication programs and fork movement directions, we used FISH on nuclei prepared by 2D gel electrophoresis with probes for Vh and Ch genes (Fig. 6A ). The relative position of the Igh locus was determined as the ratio of the distance from the signal to the center of the nucleus to the radius of the nucleus. Previous reports have shown that the data collected in this fashion are consistent with those collected from three-dimensionally preserved nuclei (16, 17).
FIG. 6.
The position of the Igh locus within the nuclei of B-cell lines is coordinated with the replication program of the Igh locus. (A) Interphase nuclei were hybridized to BAC 34H6 (containing the constant region) (red signals) and BAC 526A21 (containing J558 genes; Igh-V) (green signals). The nuclei were counterstained with DAPI (blue). In the 70Z/3 cell line, only the nonproductively rearranged allele was detected by BAC 526A21. BAC 199M11 (red signals) was used to examine the S107 cell line, because all the constant-region genes, except Cα, were deleted during isotype switching. The green arrow indicates the nontranslocated Igh locus in the S107 cell line. (B and C) Analyses of the subnuclear locations of Igh-C (B) and Igh-V (C) in B-lineage cell lines that showed two different replication programs. Each bar represents the frequency of the signal detected at a particular distance relative to the nuclear center and nuclear edge. The relative position of the signal in each nucleus was normalized to the radius of that nucleus. A value of 0 represents a location at the center of the nucleus, and a value of 1.0 represents a location at the edge of the nucleus. For example, bars between 0.4 and 0.5 show the proportion of the signals with relative positions between 0.4 and 0.5. The distributions of the Igh-V and Igh-C region genes are significantly different in different B-cell lines. In cell lines in which the entire Igh locus replicates early in S, both Igh-C and Igh-V signals are predominantly located away from the nuclear periphery. In B-cell lines that have triphasic-replication patterns (WEHI-231 and S107), Igh signals are preferentially associated with the nuclear periphery. (D) The top portion of the figure indicates the times in S when the different temporal domains of the Igh locus replicate. This is a diagram of the data shown in Fig. 2 (with the same values on the x and y axes), but the directions of replication fork movement and the replication initiation sites are also indicated. The replication initiation regions are indicated by pairs of concentric ellipses (at arbitrary sites). In the early-replication program, the entire Igh locus is replicated within the first hour of S phase and replication forks proceed in both directions through the constant-region genes (blue) and the downstream region (green). The locus is situated away from the nuclear periphery (diagram in the circle in the lower left portion of the figure). Our N/A 2D gel electrophoresis data are consistent with the idea that one or more B-cell-specific origins are activated in the region that serves as a temporal-transition region in MEL cells. In the triphasic-replication program, initiation at origins located in the Vh region (red) is delayed until the end of S. The origins located downstream (green) of the Igh locus are still activated early in these cell lines, and the leftward-moving replication fork from this region progresses toward the late-activation origins (red) in the Vh region, generating the temporal-transition region (∼500 kb in length) (the blue lines indicate constant-region genes, and the red lines indicate Vh region genes). Different sequences encompass the transition region in these two cell lines (the sequences of these transition regions are also different from those in the MEL cell line); therefore, the transition appears to be at least partially sequence independent. In both cell lines, any bidirectional initiation sites within the sequences of the transition regions, including Igh-C in WEHI-231 and the Vh region in WEHI-231 and S107, are silent. The locus is situated near the nuclear periphery (diagram in the circle in the lower right portion of the figure). We suggest that the temporal organization of the replication of the Igh locus is accomplished by (i) either the activation (early-replication program) or silencing (triphasic-replication program) of any initiation site(s) located within the transition region and (ii) the regulation of the firing times of initiation sites located within the Vh region. The silencing or activation of the initiation site(s) within the transition region and the regulation of the firing time of the Vh origins may be accomplished as a result of an alteration of the subnuclear position of the Igh locus.
The analysis of the nuclear localization of the Igh locus in different cell lines is shown in Fig. 6B and C. In the cell lines in which the entire locus replicates early in S (such as the 63-12 pro-B-cell line), in three pre-B-cell lines (70Z/3, 18-81, and 22D6 [data are shown only for the 70Z/3 cell line]), and in 38C-13 B cells, the Igh locus (both Vh and Ch genes) was located away from the periphery of the nucleus (>75% of the signals have a relative distance in the range from 0 to 0.8). In 70Z/3 cells, only the Ch region can be detected for the productively rearranged allele because Vh sequences that can be detected by the Vh probe are deleted as a result of VDJ joining. Both Vh and Ch signals can be detected for the nonproductively rearranged allele. Both alleles are transcribed, and there is no detectable difference in the location of the two alleles in this cell line. These data are in accord with the results of a previous study of the 38B9 pre-B-cell line, in which the Igh locus is located away from the nuclear periphery, as shown by 2D and three-dimensional experiments (40).
However, the nuclear location of the Igh locus was quite different in cell lines, such as MEL cells (data not shown), WEHI 231 B cells, and S107 plasma cells, in which the Igh locus is replicated by a triphasic pattern. In these cells, the Igh locus was preferentially positioned at the nuclear periphery (>60% of the signals have a relative position between 0.8 and 1.0). In the S107 plasma cell line, there are several chromosomes containing the Igh locus. One allele contains the productively rearranged and expressed Cα gene. A second allele has undergone reciprocal translocation with the c-myc locus on chromosome 15, as a result of which Vh and Cα genes are on different chromosomes. One set of closely spaced Vh and Ch signals represents the single nontranslocated allele (Fig. 6A), while Vh and Ch signals that are further apart likely detect products of the reciprocal translocation that are present in multiple copies. Both the nontranslocated allele (see Fig. 6B and C for statistical data of this allele) and the allele in which Vh is translocated to 5′ myc exon 1 are located near the nuclear periphery. However, the Cα allele that is separated from Vh sequences and associated with myc as a result of the myc-Igh translocation is located further from the nuclear periphery than sequences that we have detected on the nontranslocated chromosome. Thus, Vh sequences of the Igh locus may be important for targeting the locus to the nuclear periphery in later stages of development.
An examination of primary cells also revealed a developmental difference in the subnuclear localization of the Igh locus with respect to the nuclear periphery. In ex vivo-expanded pro-B cells, <12% of the signals have a relative position between 0.8 and 1.0, while in LPS-stimulated splenic B cells, ∼35 to ∼40% of the signals have a relative position between 0.8 and 1.0. A difference was found in the positions of the expressed and nonexpressed alleles with respect to gamma satellite heterochromatin in LPS-stimulated (for 4 days) splenic B cells (60). In our study, we examined the subnuclear location of the Igh locus with respect to the nuclear periphery, which had not been reported previously (60).
Thus, associated with the changes of the replication programs in Igh loci, there are changes in the subnuclear position of Igh genes in B-lineage cells. In cells in which the entire Igh locus replicates early, the locus was located away from the nuclear periphery. A more peripheral localization was observed in cells in which a temporal-transition region (triphasic replication) was present.
DISCUSSION
We have identified two distinct temporal patterns of Igh DNA replication. In pro- and pre-B-cell lines, all Igh sequences examined replicated early in S. In two plasma cell lines, as in non-B cells, Igh sequences exhibited a triphasic pattern of replication. Differences in the times of replication of the Igh locus were also detected in primary B cells. In ex vivo-expanded pro-B cells, all Igh sequences examined replicated early in S, which was also the case in pro- and pre-B-cell lines. In contrast, in LPS-stimulated splenic B cells, sequences downstream of the Igh locus replicated early in S while Vh sequences replicated late in S. Interestingly, different patterns were observed for the two different B-cell lines: 38C-13 showed an early-replication pattern, and WEHI-231 showed a triphasic-replication pattern. This difference might be related to the fact that the 38C-13 cell line exhibits features of pre-B cells. RAG genes are expressed in the 38C-13 cell line, which undergoes secondary light-chain rearrangements (42). We used reverse transcription PCR to confirm that the RAG2 gene is transcribed in the 38C-13 cell line studied here as well as in the 70Z/3 pre-B-cell line. RAG2 transcription was not detected in the WEHI-231 immature-B-cell line.
It is important to note that the Igh locus is maintained in germ line configuration in both MEL cells, which exhibit a triphasic pattern of Igh replication, and the 63-12 RAG−/− cell line, in which all Igh genes replicate early. Thus, replication origins within the Igh locus can be activated at different times during S phase in the same DNA sequence context, which can perhaps be accounted for by differences in chromatin structure.
Molecular mechanism for early replication of the Igh locus.
The most likely mechanism for accomplishing the early and rapid replication of Igh in pro- and pre-B cells is that one or more additional initiation sites are activated within the sequences that form the temporal-transition region in non-B cells. Such an initiation site could be present at a specific site, at several preferred sites, or at random sites. Notably, this would be the first example of developmentally regulated replication origin activation in mammalian cells.
Alternative mechanisms to account for the early replication of the Igh locus appear unlikely. One alternative is that on each chromosome, DNA replication starts from both of the two known origin-containing regions, i.e., the Vh region and downstream of the 3′ regulatory region (25; Zhou et al., unpublished data). If this were true, then sequences near one of the origins would almost always be replicated from that particular origin. For example, replication forks would be expected to progress predominantly 3′ to 5′ through the 3′ regulatory region, which is located 50 kb from the downstream initiation region. However, in 70Z/3 cells, we detected forks progressing through the 3′ regulatory region in both directions on both alleles. Moreover, by quantitating the ratios of the intensities of the nascent strands to those of the parental strands, we detected similar numbers of forks progressing in both directions through several other Igh-C segments in the three cell lines we examined (segments Cγ3 and Cγ2a for all three cell lines and segments Cμ and Cγ2b in 70Z/3). In fact, analysis of these segments by N/N 2D gel electrophoresis demonstrated that most replication forks do not terminate within these segments. These data suggest that a single allele replicates with forks progressing from upstream origins in some cells and from downstream origins in other cells.
Another alternative is that the Igh locus on an individual chromosome is replicated by either one or the other of the two origin-containing regions. To complete the replication of 400 kb of the locus in ∼1 h, the forks would have to proceed at a relatively high rate of ∼6.7 kb/min. We indirectly examined the rate of fork movement through the Igh locus in cell lines in which the Igh locus replicates early in S (70Z/3, 63-12, and 38C-13) by comparing these cell lines to MEL cells, in which the rate of fork movement through the transition region is 1.1 kb/min (replication of the 400-kb sequences through a 6-h interval of S). The cell cycle times of these lines are similar, but the Igh locus is replicated in 1 h in B cells when it replicates early in S versus in 6 h in MEL cells. If there were an increase in the rate of replication fork movement in the Igh locus in those B-cell lines, then the number of Igh replication forks (for any particular restriction segment) associated with the nuclear matrix at a particular time would be smaller than that in MEL cells. This would result in a significant decrease in the amount of nuclear-matrix-associated replication intermediates (DNA associated with the nuclear matrix is greatly enriched in replication intermediates) prepared from these cells compared to the amount in MEL cells. The ratio of the amount of Igh replication intermediates prepared from MEL cells to the amount of those from the B-cell lines in which the Igh locus replicates early in S was <1.6, implying that replication forks should not progress any faster than 1.8 kb/min (1.6 × 1.1 kb/min) in those B-cell lines. Although this analysis is dependent on several assumptions, such as similar recoveries of replication intermediates, we have found similar results from many different elutriations as well as from asynchronous cell populations (see Materials and Methods). Therefore, at this rate of replication fork progression, it seems unlikely that the two origin-containing regions are sufficient to replicate the Igh-C region early in S; instead, one or more additional initiation sites are likely to be activated within Igh-C sequences.
We note that an initiation site was previously detected at Eμ (5). However, we have not detected initiation at this or any other segment within the Igh-C locus by N/N 2D gel electrophoresis. Together with the data from N/A 2D gel electrophoresis, these observations suggest that activation of origin-containing segments within the Igh locus occurs inefficiently, probably at multiple sites.
Temporal-transition region.
The same kind of temporal-transition region was detected in the WEHI-231 and S107 cell lines, which have deletions of some DNA sequences within the Igh locus due to VDJ joining and isotype switching, as is the case in MEL cells, in which the Igh locus is in the germ line configuration. Thus, specific DNA sequences are not required for the formation of the temporal-transition region. For example, some Vh genes that replicate late in MEL cells are part of the transition region in WEHI-231 and S107 cells. However, the lengths of the transition regions (400 to 600 kb) are similar in all three cell lines.
N/A 2D gel electrophoresis showed that, as in MEL cells, replication forks progress in one direction through the examined constant-region genes (3′ to 5′, opposite to the transcriptional orientation) in the WEHI-231 and S107 cell lines. The replication timing data together with the results of our N/A 2D gel electrophoresis fork direction studies support the conclusion that the transition region is replicated by a single replicon, which indicates that any potential bidirectional initiation site(s) is silent within this ∼500-kb transition region. We speculate that when Vh transcription is limited to the recombined Vh gene, the initiation sites within the Igh locus are activated only later in S phase, perhaps reflecting a different chromatin structure. Therefore, the replication fork from the downstream region that is activated early progresses throughout S phase to replicate the transition region sequences until it meets the origins that are activated late; this results in a gradual temporal transition. It is interesting that while it has previously been demonstrated by autoradiography that the average distance between initiation sites is approximately 100 kb (24, 38), it has recently been pointed out that many replicons are much larger in size (6), and here we show that in some B-lineage cell lines the replicon size in the Igh locus is about 500 kb.
It is known that early- and late-replication factories are physically separated in the nucleus (38, 49). An apparent requirement for a gradual transition between early- and late-replication domains in the Igh locus may account for the formation of a transition region, which, if replicated by a single replication fork progressing at a rate of ∼1.1 kb/min, would require most of S phase to be completely replicated (11, 25). This might be accomplished by a single replication factory that replicates sequences throughout S (6).
Replication and subnuclear localization of Igh genes in B-lineage cells.
The correlations between the subnuclear localization and replication timing of the Igh locus are displayed in Fig. 6D. In cell lines in which the entire Igh locus is replicated early in S, the locus is maintained away from the nuclear periphery and origins within the Vh region are activated early in S. One or more new replication origins are activated within the region that previously was a temporal-transition region in non-B cells. Activation of germ line transcription of Vh region genes and VDJ recombination of the heavy-chain locus occurs during these early stages of B-cell development, including in the cell lines studied here (4, 32, 63). In later stages of B-cell development, germ line transcription is off (32) and the entire Igh locus no longer replicates early in S. Correspondingly, the DNase I sensitivity of Vh and Jh segments is increased in pro-B and pre-B cells compared to that in non-B cells and plasma cells (32, 43). In addition, in both the T-cell receptor (46) and Igh (43) loci, changes in histone acetylation appear to occur prior to VDJ joining. In addition to the constellation of associated changes in chromatin structure, germ line transcription, and VDJ joining, an early-replication pattern and a movement of the locus away from the nuclear periphery were discovered. It seems likely that these changes are mechanistically linked in the process of Igh activation and rearrangement.
In other cell lines that showed a triphasic-replication pattern, including one B-cell line and two plasma cell lines, origins within the Vh region are either silenced or activated late in S phase, while the downstream origin(s) is still activated early in S phase. The Igh locus is located near the nuclear periphery. This perinuclear location of the Igh locus may be required to ensure that Vh origins can be activated only at the end of S phase. Our data also suggest that perinuclear localization alone is not sufficient to ensure that origins fire late, because the downstream origins still fire early in this perinuclear region. It is possible that only Vh origins are flanked by late-determination elements, as has been reported for late-firing origins in yeast (37, 56), and that when they are targeted to the perinuclear region, their activation time is late in S.
Although not described here in detail, the kappa chain locus also shows changes in replication timing. For example, most Vκ segments replicate earlier in the 22D6 pre-B-cell line than in plasma cell lines (36). In addition, the κ locus was situated away from the nuclear periphery in pro-B cells and at the periphery of the nucleus in T cells (40). Thus, both the Igh and kappa loci show differences in replication in different B-lineage cell lines.
Potential role of replication fork direction in B-cell development.
Recent studies have shown that the timing of DNA replication of immunoglobulin loci plays an important role in allelic exclusion (48), which emphasizes the potentially important role of replication in B-cell development. Our studies demonstrate that dramatic differences in times of replication and the directions of replication fork movement in pro- and pre-B-cell lines from those in plasma cells and MEL cells appear to be due to the activation of a new replication origin in the pro- and pre-B-cell lines. While most of our research has been carried out with several cultured cell lines, we show here that the replication timing of ex vivo-expanded pro-B cells differs from that of splenic B cells in a way that parallels what we observe in cultured cells. Therefore, it is likely that the changes in the direction of replication fork movement also occur in primary B-lineage cells, although the 2D gel electrophoresis technique is not suitable for the determination of fork direction in these cells.
For the Igh locus, we observed a change in the direction of replication fork progression as the result of an activation of a new initiation site(s). This has not been reported for other mammalian loci, including three housekeeping gene loci (lamin, c-myc, and dihydrofolate reductase gene loci) and the tissue-specific β globin gene locus. We speculate that activation of a new initiation site(s) in the Igh cluster may be important for some specific aspects of B-cell development, possibly by producing a change in the direction of replication fork movement. It has been shown that replication fork direction plays an important role in mating type switching in Schizosaccharomyces pombe (18), and it has been speculated that fork direction may also play a role in the Igh locus (19). It has been pointed out that replication fork direction may affect the repair of double-strand breaks and therefore may have an impact on somatic hypermutation (31). The present study documents a developmentally regulated change in replication fork direction. Further studies of this change should shed light on its significance for differentiation at the Igh locus.
Acknowledgments
J.Z. and O.V.E. contributed equally to this work.
This work was supported by NIH grants GM45751 (C.L.S.), AI13509 (B.K.B.), and AI23548 (R.R.). Support was also provided by Cancer Center Core Grant NIH/NCI P30CA13330.
We thank R. Little and L. Nguyen for their contribution to the early stages of this work, P. Norio for many helpful discussions, and J. Huberman, D. Gilbert, P. Dijkwel, H, Singh, S. Kosak, M. Scharff, A. Sepulveda, F. Garrett, E. Bouhassira, and J. Greally for critical reading of the manuscript. We thank Q. Yang and S. Shenoy for help with FISH. Microscopy was performed at the Analytical Imaging Facility, and the DNA content of separated cell fractions was determined at the fluorescence-activated cell sorter facility of the Albert Einstein College of Medicine.
REFERENCES
- 1.Alt, F., N. Rosenberg, S. Lewis, E. Thomas, and D. Baltimore. 1981. Organization and reorganization of immunoglobulin genes in A-MULV-transformed cells: rearrangement of heavy but not light chain genes. Cell 27:381-390. [DOI] [PubMed] [Google Scholar]
- 2.Alt, F. W., N. Rosenberg, V. Enea, E. Siden, and D. Baltimore. 1982. Multiple immunoglobulin heavy-chain gene transcripts in Abelson murine leukemia virus-transformed lymphoid cell lines. Mol. Cell. Biol. 2:386-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alwine, J. C. 1981. Hybridization analysis of specific RNAs by electrophoretic separation in agarose gels and transfer to diazobenzyloxymethyl paper. Gene Amplif. Anal. 2:565-572. [PubMed] [Google Scholar]
- 4.Angelin-Duclos, C., and K. Calame. 1998. Evidence that immunoglobulin VH-DJ recombination does not require germ line transcription of the recombining variable gene segment. Mol. Cell. Biol. 18:6253-6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ariizumi, K., Z. Wang, and P. W. Tucker. 1993. Immunoglobulin heavy chain enhancer is located near or in an initiation zone of chromosomal DNA replication. Proc. Natl. Acad. Sci. USA 90:3695-3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berezney, R., D. D. Dubey, and J. A. Huberman. 2000. Heterogeneity of eukaryotic replicons, replicon clusters, and replication foci. Chromosoma 108:471-484. [DOI] [PubMed] [Google Scholar]
- 7.Boggs, B. A., and A. C. Chinault. 1997. Analysis of DNA replication by fluorescence in situ hybridization. Methods 13:259-270. [DOI] [PubMed] [Google Scholar]
- 8.Brewer, B. J., and W. L. Fangman. 1987. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell 51:463-475. [DOI] [PubMed] [Google Scholar]
- 9.Bridger, J. M., S. Boyle, I. R. Kill, and W. A. Bickmore. 2000. Re-modelling of nuclear architecture in quiescent and senescent human fibroblasts. Curr. Biol. 10:149-152. [DOI] [PubMed] [Google Scholar]
- 10.Brodeur, P. H., M. A. Thompson, and R. J. Riblet. 1984. The content and organization of mouse Igh-V families. UCLA Symp. Mol. Cell. Biol. 18:445-453. [Google Scholar]
- 11.Brown, E. H., M. A. Iqbal, S. Stuart, K. S. Hatton, J. Valinsky, and C. L. Schildkraut. 1987. Rate of replication of the murine immunoglobulin heavy-chain locus: evidence that the region is part of a single replicon. Mol. Cell. Biol. 7:450-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caspi, Y., M. Taya, N. Hollander, and J. Haimovich. 1995. Light chain loss and reexpression leads to idiotype switch. Surrogate light chains are probably responsible for this process. Curr. Top. Microbiol. Immunol. 194:179-186. [DOI] [PubMed] [Google Scholar]
- 13.Chevillard, C., J. Ozaki, C. Herring, and R. Riblet. 2002. A 3 megabase yeast artificial chromosome contig spanning the C57BL mouse Igh locus. J. Immunol. 168:5659-5666. [DOI] [PubMed] [Google Scholar]
- 14.Cockell, M., and S. M. Gasser. 1999. Nuclear compartments and gene regulation. Curr. Opin. Genet. Dev. 9:119-205. [DOI] [PubMed] [Google Scholar]
- 15.Cohn, M., G. Notani, and S. A. Rice. 1969. Characterization of the antibody to the C-carbohydrate produced by a transplantable mouse plasmacytoma. Immunochemistry 6:111-123. [DOI] [PubMed] [Google Scholar]
- 16.Croft, J. A., J. M. Bridger, S. Boyle, P. Perry, P. Teague, and W. A. Bickmore. 1999. Differences in the localization and morphology of chromosomes in the human nucleus. J. Cell Biol. 145:1119-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Csink, A. K., and S. Henikoff. 1998. Large-scale chromosomal movements during interphase progression in Drosophila. J. Cell Biol. 143:13-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dalgaard, J. Z., and A. J. Klar. 1999. Orientation of DNA replication establishes mating-type switching pattern in S. pombe. Nature 400:181-184. [DOI] [PubMed] [Google Scholar]
- 19.Dalgaard, J. Z., and A. J. Klar. 2001. Does S. pombe exploit the intrinsic asymmetry of DNA synthesis to imprint daughter cells for mating-type switching? Trends Genet. 17:153-157. [DOI] [PubMed] [Google Scholar]
- 20.DePamphilis, M. L. 1999. Replication origins in metazoan chromosomes: fact or fiction? Bioessays 21:5-16. [DOI] [PubMed] [Google Scholar]
- 21.DePamphilis, M. L. 2000. Review: nuclear structure and DNA replication. J. Struct. Biol. 129:186-197. [DOI] [PubMed] [Google Scholar]
- 22.Dijkwel, P. A., and J. L. Hamlin. 1999. Physical and genetic mapping of mammalian replication origins. Methods 18:418-431. [DOI] [PubMed] [Google Scholar]
- 23.Dutta, A., and S. P. Bell. 1997. Initiation of DNA replication in eukaryotic cells. Annu. Rev. Cell Dev. Biol. 13:293-332. [DOI] [PubMed] [Google Scholar]
- 24.Edenberg, H. J., and J. A. Huberman. 1975. Eukaryotic chromosome replication. Annu. Rev. Genet. 9:245-284. [DOI] [PubMed] [Google Scholar]
- 25.Ermakova, O. V., L. Nguyen, R. D. Little, N. Ashouian, R. Riblet, B. K. Birshtein, and C. L. Schildkraut. 1999. Evidence that a single replication fork proceeds from early to late replicating domains in the Igh locus in a non B cell line. Mol. Cell 3:321-330. [DOI] [PubMed] [Google Scholar]
- 26.Friedman, K. L., and B. J. Brewer. 1995. Analysis of replication intermediates by two-dimensional agarose gel electrophoresis. Methods Enzymol. 262:613-627. [DOI] [PubMed] [Google Scholar]
- 27.Friedman, K. L., J. D. Diller, B. M. Ferguson, S. V. Nyland, B. J. Brewer, and W. L. Fangman. 1996. Multiple determinants controlling activation of yeast replication origins late in S phase. Genes Dev. 10:1595-1607. [DOI] [PubMed] [Google Scholar]
- 28.Gilbert, D. M. 2001. Nuclear position leaves its mark on replication timing. J. Cell Biol. 152:F11-F16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gillies, S. D., S. L. Morrison, V. T. Oi, and S. Tonegawa. 1983. A tissue-specific transcription enhancer element is located in the major intron of a rearranged immunoglobulin heavy chain gene. Cell 33:717-728. [DOI] [PubMed] [Google Scholar]
- 30.Gregor, P. D., and S. L. Morrison. 1986. Myeloma mutant with a novel 3′ flanking region: loss of normal sequence and insertion of repetitive elements leads to decreased transcription but normal processing of the alpha heavy-chain gene products. Mol. Biol. Cell 6:1903-1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haber, J. E. 2001. Hypermutation: give us a break. Nat. Immunol. 2:902-903. [DOI] [PubMed] [Google Scholar]
- 32.Haines, B. B., and P. H. Brodeur. 1998. Accessibility changes across the mouse Igh-V locus during B cell development. Eur. J. Immunol. 28:4228-4235. [DOI] [PubMed] [Google Scholar]
- 33.Hartman, A. B., and S. Rudikoff. 1984. VH genes encoding the immune response to beta-(1,6)-galactan: somatic mutation in IgM molecules. EMBO J. 3:3023-3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hatton, K. S., V. Dhar, E. H. Brown, M. A. Iqbal, S. Stuart, V. T. Didamo, and C. L. Schildkraut. 1988. Replication program of active and inactive multigene families in mammalian cells. Mol. Cell. Biol. 8:2149-2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hatton, K. S., V. Dhar, T. A. Gahn, E. H. Brown, D. Mager, and C. L. Schildkraut. 1988. Temporal order of replication of multigene families reflects chromosomal location and transcriptional activity. Cancer Cells 6:335-340. [Google Scholar]
- 36.Hatton, K. S., and C. L. Schildkraut. 1990. The mouse immunoglobulin kappa light chain genes are located in early and late replicating regions of chromosome 6. Mol. Cell. Biol. 10:4314-4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heun, P., T. Laroche, M. K. Raghuraman, and S. M. Gasser. 2001. The positioning and dynamics of origins of replication in the budding yeast nucleus. J. Cell Biol. 152:385-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jackson, D. A., and A. Pombo. 1998. Replicon clusters are stable units of chromosome structure: evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J. Cell Biol. 140:1285-1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kirsch, I. R., J. V. Ravetch, S. P. Kwan, E. E. Max, R. L. Ney, and P. Leder. 1981. Multiple immunoglobulin switch region homologies outside the heavy chain constant region locus. Nature 293:585-587. [DOI] [PubMed] [Google Scholar]
- 40.Kosak, S. T., J. Skok, M. M. Le Beau, R. Riblet, A. G. Fisher, and H. Singh. 2002. Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science 296:158-162. [DOI] [PubMed] [Google Scholar]
- 41.Li, F., J. Chen, M. Izumi, M. C. Butler, S. M. Keezer, and D. M. Gilbert. 2001. The replication timing program of the Chinese hamster beta-globin locus is established coincident with its repositioning near peripheral heterochromatin in early G1 phase. J. Cell Biol. 154:283-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maes, J., Y. Caspi, F. Rougeon, J. Haimovich, and M. Goodhardt. 2000. Secondary V(D)J rearrangements and B cell receptor-mediated down-regulation of recombination activating gene-2 expression in a murine B cell line. J. Immunol. 165:703-709. [DOI] [PubMed] [Google Scholar]
- 43.Maes, J., L. P. O'Neill, P. Cavelier, B. M. Turner, F. Rougeon, and M. Goodhardt. 2001. Chromatin remodeling at the Ig loci prior to V(D)J recombination. J. Immunol. 167:866-874. [DOI] [PubMed] [Google Scholar]
- 44.Mainville, C. A., K. M. Sheehan, L. D. Klaman, C. A. Giorgetti, J. L. Press, and P. H. Brodeur. 1996. Deletional mapping of fifteen mouse VH gene families reveals a common organization for three Igh haplotypes. J. Immunol. 156:1038-1046. [PubMed] [Google Scholar]
- 45.Max, E. E. 1999. Immunoglobulins: molecular genetics, p. 111-182. In W. E. Paul (ed.), Fundamental immunology, 4th ed. Lippincott-Raven Press, New York, N.Y.
- 46.McMurry, M. T., and M. S. Krangel. 2000. A role for histone acetylation in the developmental regulation of VDJ recombination. Science 287:495-498. [DOI] [PubMed] [Google Scholar]
- 47.Michaelson, J. S., O. Ermakova, B. K. Birshtein, N. Ashouian, C. Chevillard, R. Riblet, and C. L. Schildkraut. 1997. Regulation of the replication of the murine immunoglobulin heavy chain gene locus: evaluation of the role of the 3′ regulatory region. Mol. Cell. Biol. 17:6167-6174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mostoslavsky, R., N. Singh, T. Tenzen, M. Goldmit, C. Gabay, S. Elizur, P. Qi, B. E. Reubinoff, A. Chess, H. Cedar, and Y. Bergman. 2001. Asynchronous replication and allelic exclusion in the immune system. Nature 414:221-225. [DOI] [PubMed] [Google Scholar]
- 49.Nakayasu, H., and R. Berezney. 1989. Mapping replicational sites in the eucaryotic cell nucleus. J. Cell Biol. 108:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nawotka, K. A., and J. A. Huberman. 1988. Two-dimensional gel electrophoretic method for mapping DNA replicons. Mol. Cell. Biol. 8:1408-1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nelson, K. J., J. Haimovich, and R. P. Perry. 1983. Characterization of productive and sterile transcripts from the immunoglobulin heavy-chain locus: processing of μm and μs mRNA. Mol. Cell. Biol. 3:1317-1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pennell, C. A., K. M. Sheehan, P. H. Brodeur, and S. H. Clarke. 1989. Organization and expression of VH gene families preferentially expressed by Ly-1+ (CD5) B cells. Eur. J. Immunol. 19:2115-2121. [DOI] [PubMed] [Google Scholar]
- 53.Saleque, S., M. Singh, and B. K. Birshtein. 1999. Ig heavy chain expression and class switching in vitro from an allele lacking the 3′ enhancers DNase I-hypersensitive hs3A and hs1,2. J. Immunol. 162:2791-2803. [PubMed] [Google Scholar]
- 54.Saleque, S., M. Singh, R. D. Little, S. L. Giannini, J. S. Michaelson, and B. K. Birshtein. 1997. Dyad symmetry within the mouse 3′ IgH regulatory region includes two virtually identical enhancers (C alpha3′E and hs3). J. Immunol. 158:4780-4787. [PubMed] [Google Scholar]
- 55.Selig, S., K. Okumura, D. C. Ward, and H. Cedar. 1992. Delineation of DNA replication time zones by fluorescence in situ hybridization. EMBO J. 11:1217-1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sharma, K., M. Weinberger, and J. A. Huberman. 2001. Roles for internal and flanking sequences in regulating the activity of mating-type-silencer associated replication origins in Saccharomyces cerevisiae. Genetics 159:35-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shimizu, A., N. Takahashi, Y. Yaoita, and T. Honjo. 1982. Organization of the constant-region gene family of the mouse immunoglobulin heavy chain. Cell 28:499-506. [DOI] [PubMed] [Google Scholar]
- 58.Simon, I., and H. Cedar. 1996. Temporal order of DNA replication, p. 387-408. In M. L. DePamphilis (ed.), DNA replication in eukaryotic cells. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 59.Simon, I., T. Tenzen, B. E. Reubinoff, D. Hillman, J. R. McCarrey, and H. Cedar. 1999. Asynchronous replication of imprinted genes is established in the gametes and maintained during development. Nature 401:929-932. [DOI] [PubMed] [Google Scholar]
- 60.Skok, J. A., K. E. Brown, V. Azuara, M. L. Caparros, J. Baxter, K. Takacs, N. Dillon, D. Gray, R. P. Perry, M. Merkenschlager, and A. G. Fisher. 2001. Nonequivalent nuclear location of immunoglobulin alleles in B lymphocytes. Nat. Immunol. 2:848-854. [DOI] [PubMed] [Google Scholar]
- 61.Wahl, M. R., G. B. Beck-Engeser, and P. D. Burrows. 1984. Allelic inclusion in the pre-B-cell line 18-81. Proc. Natl. Acad. Sci. USA 81:867-870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang, X. F., and K. Calame. 1985. The endogenous immunoglobulin heavy chain enhancer can activate tandem VH promoters separated by a large distance. Cell 43:659-665. [DOI] [PubMed] [Google Scholar]
- 63.Yancopoulos, G. D., and F. W. Alt. 1985. Developmentally controlled and tissue-specific expression of unrearranged VH gene segments. Cell 40:271-281. [DOI] [PubMed] [Google Scholar]