Abstract
Activator protein 1 (AP-1) is a group of dimeric transcription factors composed of Jun, Fos, and ATF family proteins. Both gain- and loss-of-function studies have revealed specific roles for individual AP-1 components in cell proliferation, differentiation, apoptosis, and other biological processes. However, little is known about the functions of specific AP-1 dimers. To test the importance of AP-1 composition in transcriptional activation, AP-1 monomers were joined via a flexible polypeptide tether to force specific pairing. The resultant single-chain AP-1 molecules showed DNA binding specificity and transcriptional activation potentials similar to those of untethered dimers, even in the presence of dominant-negative AP-1 monomers. c-Jun-containing dimers showed distinct promoter specificity in transient-transfection experiments, depending on the Fos, Fra, or ATF partner. When stably expressed in NIH 3T3 cells, c-Jun∼Fra2, but not c-Jun∼Fra1 and c-Jun∼cFos (the tilde indicates a tethered dimer), inhibited G0 arrest at confluency and under low-serum conditions and specifically activated cyclin A expression. These data suggest that the choice of dimerization partner defines the role of c-Jun in gene activation and cell cycle regulation and that single-chain AP-1 molecules provide a powerful tool for assessing the role of specific AP-1 dimers.
Activator protein 1 (AP-1) transcription factors play important roles in specifying and executing essential genetic programs. Proteins that form AP-1 belong to the basic leucine zipper (bZIP) family and bind DNA as obligate homo- or heterodimers (for review, see reference 33). The bZIP motif consists of a DNA-binding domain rich in basic amino acids and an adjacent protein-protein dimerization domain known as the leucine zipper structure, which is characterized by leucines at every seventh residue (21). c-Jun and c-Fos are the prototype AP-1 components. The related proteins, JunB, JunD, Fra1, Fra2, and FosB, show expression patterns and transactivation potentials distinct from those of c-Jun or c-Fos. While Jun proteins can dimerize with each other and with the Fos proteins, Fos proteins cannot form homodimers (26). The AP-1 complexes are not limited to Jun and Fos dimers, since certain Jun and Fos proteins have been shown to dimerize with other bZIP proteins, such as ATF2, MAF, and their related proteins (for review, see reference 5). Various AP-1 dimers differ in DNA binding affinity and specificity. For example, Jun-Fos heterodimers preferentially bind to the heptamer consensus, 5′-TGA(C/G)TCA-3′, known as the TPA response element (TRE), while Jun-ATF dimers prefer the octamer consensus 5′-TGACGTCA-3′ known as the cyclic AMP response element (CRE). These binding sites have been found in the regulatory regions of a wide range of genes, including transcription factors, matrix-degrading enzymes, cell adhesion molecules, cyclins, and cytokines. Jun-Fos dimers that have similar DNA binding specificities can differ in transcriptional activity due to nonconserved domains located outside the bZIP region that can be regulated by phosphorylation. It is therefore plausible that AP-1 dimers of different composition execute specific cellular programs.
AP-1 complexes in exponentially growing mouse NIH 3T3 fibroblasts are predominantly dimers containing c-Jun, JunD, and Fra2 (19, 20). In serum-starved fibroblasts, JunD accumulates while the level of the other members decreases. Mitogenic stimulation induces the synthesis of c-Jun, JunB, c-Fos, Fra1, Fra2, and FosB in a defined order (19, 20). In addition, we have recently shown that the abundance and phosphorylation of Jun proteins vary during the cell cycle (2).
The combinatorial character of the AP-1 transcriptional complex makes the interpretation of overexpression experiments difficult. Introduction of a new bZIP species into the cell can produce primary effects from homodimerization of itself or from heterodimerization with usually unidentified endogenous bZIP proteins, which cause potentially widespread effects by titrating out certain monomers and shifting the equilibrium within the AP-1 pool. Furthermore, the expression level and phosphorylation status of individual bZIP proteins in the recipient cell can be dramatically different depending on cell type and cell context. To overcome these difficulties, we developed a tethering strategy. Inspired by successful examples using other dimeric molecules, we joined two AP-1-forming monomers by a flexible polypeptide so that they formed an intramolecular dimer. In this study, we characterized the biochemical properties and biological activities of tethered Jun∼Fos proteins (the tilde indicates a tethered dimer). These single-chain AP-1 molecules revealed differences in the transcriptional regulation of specific promoters and showed that c-Jun in combination with Fra2 inhibits cell cycle arrest under both confluency and low-serum conditions for cultured NIH 3T3 mouse fibroblasts.
MATERIALS AND METHODS
Plasmids and retroviruses.
To construct the tethered Jun∼Fos molecules, several restriction sites and a FLAG epitope were introduced by high-fidelity PCR using specific oligonucleotide primers and cloned into pBluescript. For c-Jun, JunB, and JunD, a HindIII site was generated at the 3′ end of each coding sequence, removing the stop codon. For c-Fos, Fra1, and Fra2, an XhoI site was created in front of the initiation codon, while the FLAG sequence with a stop codon (5′-GAC TAC AAG GAC GAC GAC GAC AAG TGA-3′) was introduced at the end of the coding sequence followed by an EcoRI site. An XbaI-HindIII fragment from the pBluescript-cJun plasmid was ligated with a HindIII-XhoI double-stranded oligonucleotide linker encoding the polypeptide tether (5′-AAG CTT GGG GGA TCA GGC GGA GGT GGA GGA TCC GGT GGC GGT GGC TCG AGC-3′) and then inserted into XbaI-XhoI-linearized pBluescript-cFosFLAG vector. Other tethered molecules were constructed in the same way. Constructed coding regions were verified by sequencing. The c-Fos template used throughout our studies contained five amino acid substitutions present in v-Fos. Miller and colleagues have shown that these substitutions do not modify the properties of mouse c-Fos; in particular, they did not confer a transformed phenotype (23).
The mutated version of c-Jun∼Fra1 (c-Jun∗∼Fra1) was constructed by the replacement of an AccI-Psp 1406 fragment from pBluescript-cJun-∼Fra1 by the corresponding mutated fragment from RSVcJunDB-4 (a kind gift from D. Bohmann), where the critical Lys-Cys amino acid pair, located in the c-Jun DNA binding domain, was mutated to Ile-Asp (3).
To construct the Jun∼Fos expression vectors, the corresponding cDNA coding sequences were excised from pBluescript constructs by using the appropriate restriction sites and ligated into either a cytomegalovirus (CMV)-driven expression vector (pCG; a kind gift from F. Thierry) or the pBabe-puro vector (24). Retroviral supernatants were prepared as previously described (22) using transient transfection into the pNXeco packaging cell line. The ΔJun expression vector was described previously (13).
Culture conditions and generation of cell lines.
NIH 3T3 and 293 cells were cultured in Dulbecco minimal essential medium (DMEM) containing 7% (or 0.5%) fetal calf serum (FCS). To generate NIH 3T3/c-Jun∼Fos and NIH 3T3/puro control cells, NIH 3T3 cells were seeded at a density of 105 cells in a 6-cm-diameter dish and infected with approximately 106 CFU of each pBabe virus in 2 ml of DMEM containing 8 μg of Polybrene/ml for 3 h. The infection medium was subsequently diluted by addition of an equal volume of fresh medium, and cells were cultured for an additional 12 h. Cells were then rinsed and split in three 10-cm-diameter dishes, and 5 μg of puromycin/ml was added to the culture medium after 24 h. Puromycin-resistant colonies were then pooled.
Fuorescence microscopy.
Fluorescence microscopy was described previously (20). Cells were costained with DAPI (4′,6′-diamidono-2-phenylindole) (Sigma) for DNA and with a mouse monoclonal anti-FLAG antibody (Kodak) followed by fluorescein isothiocyanate (FITC)-coupled anti-mouse antibody (Amersham) for the expressed protein. Cells were observed under a Zeiss Axiophot epifluorescence microscope, and photographs were taken with Kodak films.
Flow cytometry and measurement of DNA synthesis.
For flow cytometry cell cycle analysis, cells were tripsinized and collected by centrifugation at 2,000 rpm for 5 min and resuspended in 0.1% Na citrate, 0.1% Triton X-100, 50 μg of propidium iodide/ml, and 50 μg of DNase-free RNase (HSS buffer)/ml. To discriminate sub-G1 cells, trypsinized cells were eventually fixed overnight in ice-cold 70% ethanol and rinsed with phosphate-buffered saline prior to resuspension in HSS buffer. Analysis was carried out on an Epics XL Flow Cytometer (Coulter), and cell cycle phases were estimated using the Multiplus Software (Phoenix Systems).
In vitro protein synthesis and EMSA.
Proteins were prepared for in vitro studies using the TNT rabbit reticulocyte coupled transcription-translation system (Promega). The mixture was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to nitrocellulose filters, and probed using the indicated antibodies or used for electromobility shift assays (EMSA).
EMSA for AP-1 were performed as described previously (30). Briefly, equivalent amounts of in vitro-synthesized proteins or cell extracts were preincubated for 10 min in 10 mM HEPES (pH 7.0), 100 μg of bovine serum albumin (BSA)/ml, 20% glycerol, 2 mM dithiothreitol, 0.5 mM spermidine, 2 mM MgCl2, and 1 μg of poly(dI-dC). Then, 50 ng of double-stranded 32P-labeled oligonucleotide was added and the mixture was incubated on ice for 15 min. DNA-protein complexes were resolved on 8% native polyacrylamide-Tris-borate-EDTA (TBE) gels, dried, and visualized by autoradiography and phosphorimager (Molecular Dynamics).
The following oligonucleotides were used: MMP1, corresponding to the TRE element present in the collagenase promoter (17) (5′-GGC TAG CTG ACT CAG ATG TCC-3′); Jun1 and Jun2, corresponding to the two CRE-like element present in the human c-jun promoter (38) (5′-AGC TGG GGT GAC ATC ATG GG-3′ and 5′-AGC TAG CAT TAC CTC ATC CC-3′, respectively); CCND1-1 and CCND1-2, corresponding to the TRE and CRE elements presents in the human cyclin D1 promoter (15) (5′-TTA AAA TGA GTC AGA ATG GAG ATC ACT GT-3′ and 5′-CTT AAC AAC AGT AAC GTC ACA CGG ACT AC-3′, respectively); and CCNA2, corresponding to the CRE element present in the human cyclin A promoter (14) (5′-CGC CTT GAA TGA CGT CAA GGC CGC-3′).
Preparation of protein extracts and immunoblotting.
Whole-cell extracts were prepared in a mixture of 50 mM Tris (pH 7.6), 400 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% NP-40, 0.5 μg of of leupeptin/ml, 0.5 μg of aprotinin/ml, 0.5 μg of pepstatin A/ml, and 1 mM phenylmethylsulfonyl fluoride (all from Sigma). Western blotting procedures and c-Jun antibodies were described previously (20). Cyclin A-, cyclin D1-, cyclin E-, p21-, p27-, actin-, and tubulin-specific antibodies were purchased from Santa Cruz.
Reporter assays.
NIH 3T3 and 293 cells were transfected by calcium phosphate coprecipitation with Renilla luciferase expression vector and one of the following reporter plasmids: MMP1-luc (Coll517luc [17]) provided by P. Herrlich, CCND1-luc (Δ973CD1luc [15]) provided by R. Müller, Jun-Luc (Junluc [38]) provided by H. van Dam, and CCNA2-luc (pWTluc [14]) provided by J. Sobczac-Thepot. In the case of 293 cells, Jun, Fos, or Jun∼Fos, expression vectors were included. Luciferase assays were performed using the Promega dual luciferase kit by following the manufacturer's instructions.
Proliferation curves.
Triplicate cultures of NIH 3T3/puro (control) or NIH 3T3/c-Jun∼Fos cells were seeded in medium containing 7 or 0.5% serum. Adherent cells were trypsinized and counted daily using a Coulter ZI cell counter.
RESULTS
Design of tethered Jun-Fos proteins.
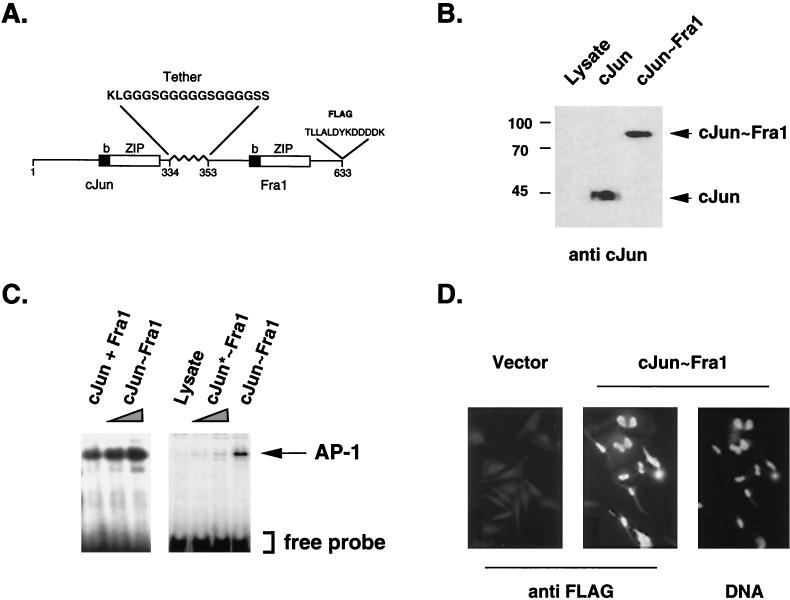
Coding regions of c-Jun and Fra1 cDNAs were fused in frame via a synthetic oligonucleotide which encodes a glycine-rich polypeptide tether of 18 amino acids (Fig. 1A). Glycine residues maximize flexibility, and interspersed serine residues increase hydrophilicity. Since the three-dimensional structures of intact Jun and Fos proteins are not known, the length of the tether was chosen based on estimated dimensions of the leucine zipper and basic-domain helices and on the postulated sizes and flexibilities of the different domains relative to each other (11). To facilitate subsequent analyses, a FLAG epitope tag was added at the C terminus of the fusion protein. To test whether the resulting molecule formed single-chain AP-1 by forced dimerization, coding sequences of c-Jun, Fra1, or c-Jun-Fra1 (the covalent dimer) were translated in the reticulocyte lysate system. c-Jun, Fra1, or the c-Jun∼Fra1 polyprotein were the predominant translation products, migrated at the expected size, and were recognized by the appropriate antibodies (Fig. 1B). Moreover, the tethered c-Jun∼Fra1 protein showed similar mobility on gel filtration columns as mixed, nondenaturated monomers (data not shown). EMSA showed that c-Jun∼Fra1 protein bound the consensus TRE element with a mobility shift similar to that of a mixture of c-Jun and Fra1 (Fig. 1C). These results strongly suggest that the single-chain molecules fold correctly to form genuine pseudodimers. Mutation of the critical Lys-Cys amino acid pair in the basic region of the c-Jun part (Fig. 1C) or excess of unlabeled AP-1 probe abolished formation of the complex between c-Jun∼Fra1 and the TRE (data not shown), further demonstrating the maintenance of specific DNA recognition after intramolecular dimerization. Moreover, the c-Jun∼Fra1 protein showed nuclear localization in transiently transfected HeLa cells, as visualized by indirect immunofluorescence using the anti-FLAG antibody (Fig. 1D) as well as anti-c-Jun and anti-Fra1 antibodies (data not shown). Similar constructs were generated for all combinations between Jun (c-Jun, JunB, and JunD) and Fos (c-Fos, Fra1, and Fra2) proteins and showed nuclear localization and DNA binding properties comparable to those of mixtures of the corresponding monomers (not shown).
FIG. 1.
In vitro analysis of forced AP-1 dimers. (A) Forced dimer design, example of c-Jun∼Fra1. The closed and open boxes denote the basic DNA binding domain (b) and the leucine zipper (ZIP), respectively, of both c-Jun and Fra1 connected via a flexible Glyn-Ser1 linker. A FLAG epitope tag was inserted at the C terminus of Fra1. (B) Western blot analysis of in vitro-synthesized c-Jun∼Fra1 protein. The expected size of c-Jun∼Fra1 determined by SDS-PAGE was recognized by a c-Jun-specific antibody. In vitro-synthesized c-Jun and an unrelated synthesis product (Lysate) were loaded as controls. (C) EMSA of the forced c-Jun∼Fra1 dimer. Equal amounts of in vitro-translated c-Jun∼Fra1 protein as well as cotranslated c-Jun plus Fra1 monomers were assayed for the ability to bind the TRE element of the collagenase promoter (left). A mutated c-Jun∼Fra1 dimer (c-Jun∗∼Fra1) was produced using a point mutant of c-Jun with impaired DNA binding activity (3). The arrow indicates the position of the AP-1-specific retarded band. (D) Nuclear localization of the c-Jun∼Fra1 forced dimer. HeLa cells were transiently transfected with a CMV-driven expression vector for c-Jun∼Fra1 (c-Jun∼Fra1) or the corresponding empty vector (Vector). Immunofluorescence staining of the cells was then performed using an anti-FLAG monoclonal antibody followed by an FITC-coupled secondary antibody (anti-FLAG). Nuclei were visualized using DAPI staining (DNA).
Tethered Jun∼Fos polyproteins are more resistant to interfering monomers.
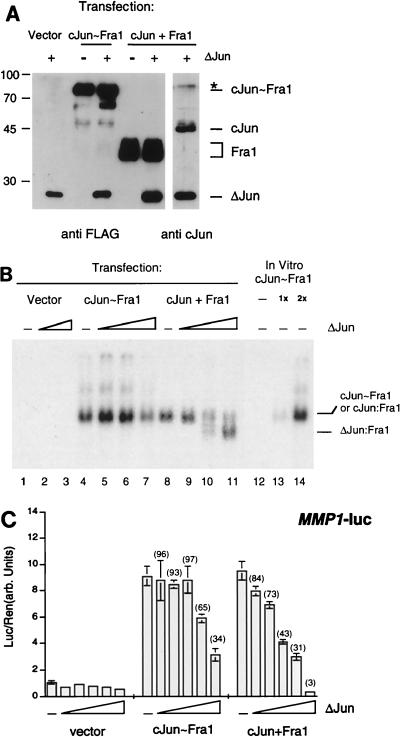
We next investigated whether tethered Jun∼Fos molecules bind DNA predominantly as intramolecular pseudodimers or promiscuously as intermolecular dimers with other single chains or with free monomers in the cell. To distinguish between these two possibilities, sensitivity to intervening monomers was compared between c-Jun∼Fra1 and a mixture of c-Jun and Fra1 monomers. We used ΔJun, a dominant-negative form of c-Jun lacking amino acids 1 to 168, as the intervening monomer. Since it retains the bZIP domain of c-Jun, ΔJun dimerizes with Jun and Fos proteins and binds to the TRE but shows reduced transactivation capacity (16). If tethered c-Jun∼Fra1 folds into a stable intramolecular dimer, it should be less sensitive to excess ΔJun than intermolecular dimers. A c-Jun∼Fra1 vector or a mixture of c-Jun- and FLAG-tagged Fra1-expressing vectors (c-Jun plus Fra1) were cotransfected with an expression vector encoding FLAG-tagged ΔJun in 293 cells, and proteins of the expected sizes were visualized by anti-FLAG and anti-c-Jun antibodies (Fig. 2A). In the absence of ΔJun, nuclear extracts from cells transfected with either c-Jun∼Fra1 or c-Jun plus Fra1 shifted the TRE oligonucleotide similarly to in vitro-translated c-Jun∼Fra1 (Fig. 2B, lanes 4, 8, and 14). When ΔJun was cotransfected, the c-Jun-plus-Fra1 monomer mixture was efficiently disrupted as judged by the quasi disappearance of the major retarded band and the concomitant appearance of a faster migrating species corresponding to ΔJun-containing dimers (Fig. 2B, lanes 9 to 11). In contrast, DNA binding of the tethered c-Jun∼Fra1 molecule was far more resistant to the addition of ΔJun (Fig. 2B, lanes 4 to 7), and a decrease of DNA binding was observed only at the highest ΔJun level tested (Fig. 2B, lane 7). This suggests that some interaction between the tethered protein and ΔJun can occur when the latter is present in vast excess. Transfected ΔJun seems to preferentially associate with free Fra1 monomer since homodimerization with itself (lanes 2 and 3) or with transfected c-Jun (lanes 9 to 11) was not detected under the experimental conditions used.
FIG. 2.
DNA binding and ΔJun sensitivity of c-Jun∼Fra1. (A) Western blot analysis of transfected-cell extracts. 293 cells were transiently transfected with c-Jun∼Fra1, c-Jun plus Fra1, and ΔJun expression vectors driven by the CMV promoter. Except for c-Jun, all proteins contained a FLAG epitope and were detected using the FLAG antibody (left), while c-Jun and ΔJun were detected using an antibody directed against the C-terminal region of c-Jun (right). The endogenous Jun and Fra1 proteins remained undetectable in the conditions of the experiment. ∗, a cross-reactive band recognized by the c-Jun polyclonal antibody (right). For transfection, 0.5 μg of empty vector, ΔJun, and c-Jun∼Fra1 and 0.25 μg of each c-Jun and Fra1 vectors were used. (B) EMSA of transfected-cell extracts for their ability to bind collagenase AP-1 site. In vitro-translated c-Jun∼Fra1 was also included (lanes 12 to 14). The postulated identity of the retarded bands specific for c-Jun∼Fra1, c-Jun-Fra1, and ΔJun-Fra1 are indicated. All lanes are from the same gel and can be compared directly. For the assay, 0 μg (lanes 1, 4, and 8), 2.5 μg (lanes 2, 5, and 9), 5.5 μg (lanes 3, 6, and 10), and 9.5 μg (lanes 7 and 11) of ΔJun was cotransfected together with the empty expression vector (0.5 μg), c-Jun plus Fra1 (0.25 μg of each), or c-Jun∼Fra1 (0.5 μg) plasmids. (C) Reporter assay. 293 cells were transiently transfected with c-Jun∼Fra1 (0.5 μg) or mixtures of c-Jun (0.25 μg) plus Fra1 (0.25 μg) and increasing amounts (0, 0.2, 0.5, 1, 2, and 4 μg, respectively) of ΔJun expression vector. All transfections contained 1 μg of the MMP1-luc reporter plasmid, and each amount of total DNA transfected was made the same by using the empty expression vector. Luciferase activity was normalized to a Renilla internal control. Numbers in parentheses are the percentages of activity compared to the zero ΔJun data point.
The effect of ΔJun on c-Jun∼Fra1 was further examined using a luciferase reporter construct driven by TREs within the 517-bp promoter fragment of the human collagenase gene (MMP1-luc [17]). As shown in Fig. 2C, both c-Jun∼Fra1 and c-Jun plus Fra1 activated the collagenase promoter to a similar extent in the absence of ΔJun. When the ΔJun expression vector was cotransfected, the forced dimer was substantially resistant to added ΔJun, whereas low doses of ΔJun efficiently inhibited transcriptional activation by c-Jun plus Fra1 (Fig. 2C). Only at higher levels of ΔJun was forced dimer activation progressively reduced, but to a lesser extent. This could have been due to competitive occupation of the AP-1 target sites by ΔJun dimers or to interactions between ΔJun and c-Jun∼Fra1. Similar results were obtained using the tethered c-Jun∼cFos molecule (not shown). These data support the view that tethering Jun to Fos molecules strongly reduces their direct interaction with endogenous bZIP monomers and can be used as a tool to study molecular and phenotypic effects of individual AP-1 dimers.
Transcriptional specificity of single-chain AP-1 molecules.
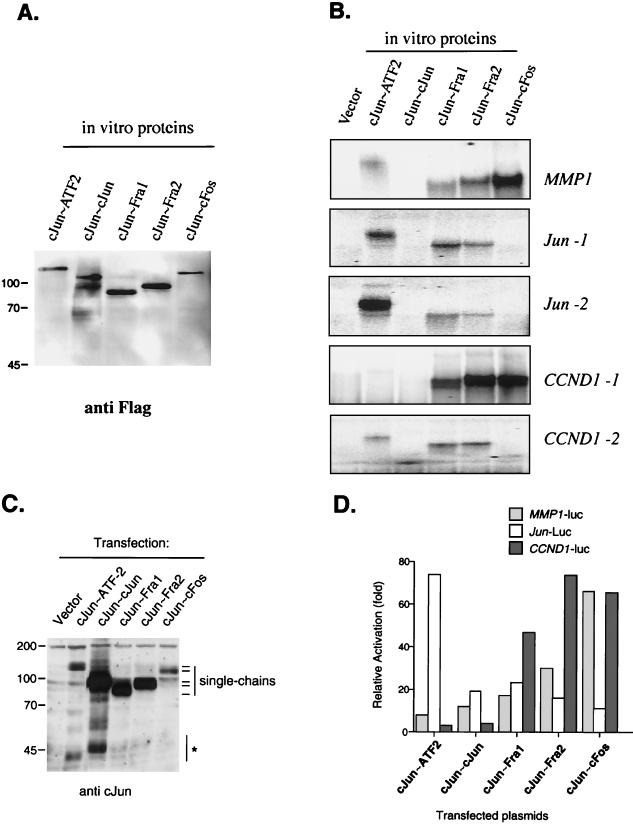
A major unsolved issue concerns the differences in transcriptional activation and target gene specificity of different AP-1 heterodimers. We therefore examined the binding and transactivation selectivities of five c-Jun-containing tethered AP-1 proteins by using gel retardation and reporter assays. c-Jun∼ATF2, c-Jun∼c-Jun, c-Jun∼Fra1, c-Jun∼Fra2, and c-Jun∼cFos were produced in reticulocyte lysate and assayed for binding to five oligonucleotides derived from three different promoters: a consensus TRE element present in the human collagenase promoter (MMP1) actively bound by Jun-Fos dimers, two CRE-like elements (Jun-1 and Jun-2) present in the human c-jun promoter that have been shown to bind efficiently Jun-ATF2 dimers and not Jun-Fos dimers (38), and two elements (CCND1-1 and CCND-2) present in the human cyclin D1 promoter that have been proposed to be targets for Jun-containing dimers (15). The relative amounts of synthesized proteins were controlled by anti-Flag Western blotting (Fig. 3A), and the results of a representative gel retardation experiment is shown in Fig. 3B: noticeably, c-Jun∼cFos dimers showed an exclusive binding to TRE elements (MMP1 and CCND1-1), compared with CRE elements. The two other Jun∼Fos tethered dimers showed a milder site selectivity since c-Jun∼Fra1 and c-Jun∼Fra2 could bind the CRE elements present in Jun-1, Jun-2, and CCND1-2. On the other hand, and in agreement with the previously published studies (37, 38), the c-Jun∼ATF2 dimer showed a clear preference for CRE-type elements, especially Jun-1. Finally, we could not detect any binding of the c-Jun∼c-Jun tethered homodimer in the binding conditions we used.
FIG. 3.
Site and transactivation specificity of c-Jun-containing dimers. (A) Western blot analysis of in vitro-synthesized proteins. Tethered c-Jun dimers (c-Jun∼ATF2, c-Jun∼c-Jun, c-Jun∼Fra1, c-Jun∼Fra2, and c-Jun∼c-Fos) of the expected size were visualized with a Flag monoclonal antibody. (B) EMSA of in vitro-synthesized proteins for their ability to bind collagenase AP-1 site (MMP1), c-jun CRE elements (Jun-1 and Jun-2), and cyclin D1 TRE and CRE elements (CCND1-1 and CCND1-2). Comparable amounts of lysate, corresponding to those in panel A, were assayed for binding. (C) Western blot analysis of transiently expressed proteins. Equal amounts of total-cell extracts from transiently transfected 293 cells were separated by SDS-PAGE and visualized with an anti-c-Jun polyclonal antibody. The positions of the different single-chain proteins are indicated. The same amount of DNA (3 μg) was used for all points. Note that the c-Jun∼c-Jun single chain contains twofold-more-reactive epitopes than the other proteins. ∗, c-Jun immunoreactive bands that could correspond to endogenous c-Jun or degradation products. (D) Activation of three different reporter constructs by the c-Jun forced dimers. Cells were transfected with 0.5 μg of the different CMV-driven expression vectors plus 1 μg of the indicated reporter construct (MMP1-luc, Jun-luc, and CCND1-luc). Luciferase activity was normalized to the Renilla internal control. Each point corresponds to the mean of two different experiments done in duplicate. Activation is relative to the values obtained with cells transfected with the corresponding reporter and the empty expression vector. Note that only values obtained from the same promoter can be compared to each other.
We then assayed the transactivation specificity of these five c-Jun-containing dimers by using transient cotransfection of the corresponding expression vectors together with reporter constructs. Three different promoter-luciferase constructs were used: the human collagenase promoter (MMP1) as a bona fide Jun-Fos-controlled promoter, a 1,090-bp human c-jun promoter fragment (JUN) containing both Jun-1 and Jun-2 elements, and a 970-bp human cyclin D1 promoter fragment (CCND1) containing both CCND1-1 and CCND1-2 elements. Expression of the tethered proteins was checked by Western blotting and the different single-chain proteins migrated as expected, with c-Jun∼ATF2 and c-Jun∼c-Fos being less abundant (Fig. 3C). Similar results were obtained using either HeLa (not shown) or 293 (Fig. 3D) cells. For the MMP1 reporter, and in agreement with the gel retardation results, c-Jun∼c-Fos was the strongest activator, followed by c-Jun∼Fra2, c-Jun∼Fra1, and the two non-Fos dimers. Since c-Jun∼c-Fos is present at lower levels than c-Jun∼Fra1 or c-Jun∼Fra2 (Fig. 3C), the difference among these three constructs may be even greater. In contrast, c-Jun∼ATF2, which was a poor activator of the MMP1 promoter, was the strongest activator for the Jun promoter, and c-Jun∼c-Fos was the least potent activator of this promoter. Finally, three constructs containing Fos proteins, c-Jun∼Fra1, c-Jun∼Fra2, and c-Jun∼cFos, activated the CCND1 promoter, while transactivation by c-Jun∼ATF2 and c-Jun∼c-Jun was only marginal. The molecular basis for the observed decreased selectivity of the CCND1 promoter towards the three different Fos proteins might be due to the lower binding selectivity of the CCND1-1 TRE element present in this promoter toward the three Fos dimers, as well as to the contribution of the CCND1-2 CRE element (Fig. 3B).
Taken together, these data indicate that c-Jun∼Fos dimers bind and efficiently activate TRE-type motifs while Jun∼ATF2 prefers the atypical CRE-like motifs that are found in the Jun promoter. Moreover, the different Jun∼Fos dimers show subtle differences in their relative binding and activation of AP-1-controlled promoters. c-Jun∼Fra1 and c-Jun∼Fra2 dimers bind and activate the Jun promoter more efficiently that c-Jun∼Fos-forced dimers.
Phenotypes of cultured cells stably expressing tethered Jun-Fos proteins.
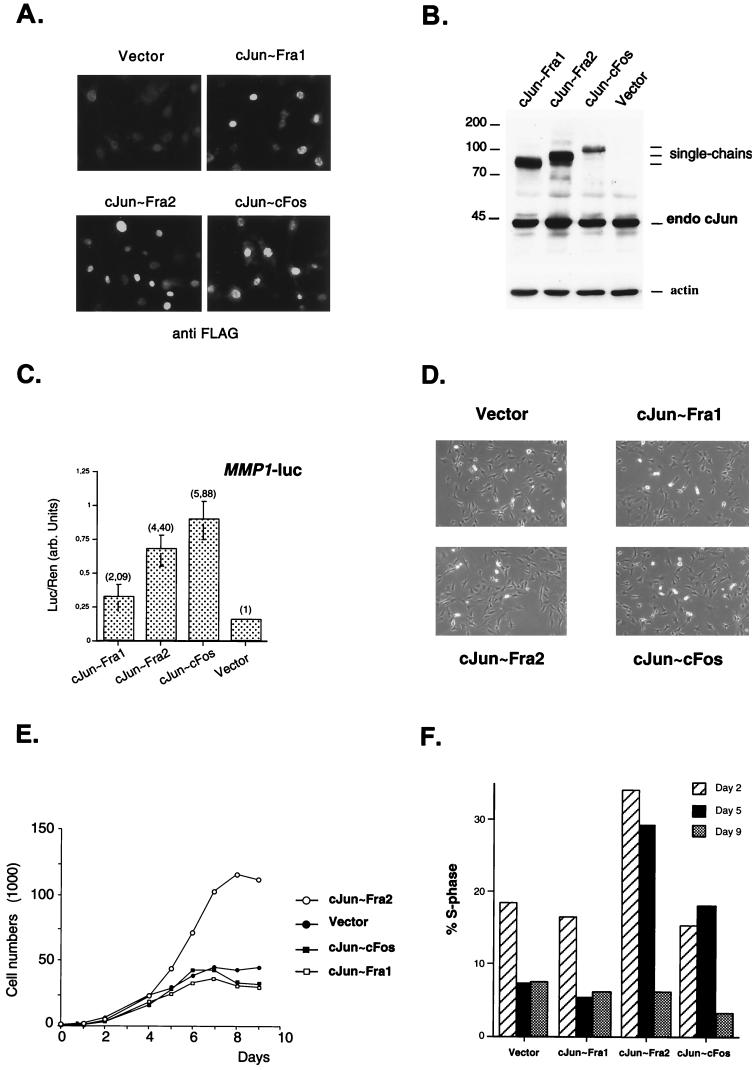
To analyze cellular phenotypes associated with specific AP-1 composition, we established NIH 3T3-derived cell lines stably expressing tethered Jun∼Fos proteins. Coding sequences for c-Jun∼Fra1, c-Jun∼Fra2, and c-Jun∼c-Fos were cloned into the pBabe-puro retroviral vector (24). After retroviral gene transfer and puromycin selection, pools of infected cells were analyzed by immunofluorescence using an anti-FLAG antibody, and the transduced proteins showed the expected nuclear localization (Fig. 4A). The expression level was also analyzed by Western blotting using either anti-c-Jun antibody (Fig. 4B) or anti-FLAG antibody (not shown). Compared to the amount of endogenous c-Jun expressed in these cells, exogenous proteins were estimated to be expressed at levels of one- to threefold relative to the endogenous c-Jun (Fig. 4B). In spite of its low expression level, c-Jun∼c-Fos activated a transiently transfected MMP1-luc reporter up to sixfold, while c-Jun∼Fra1 and c-Jun∼Fra2 activated the same reporter only two- and fourfold, respectively (Fig. 4C). The strong activation by c-Jun∼c-Fos is consistent with the data obtained using transient cotransfection in 293 cells (Fig. 3D), even though these pools of infected cells are likely to express less tethered proteins per cell than transiently transfected cells.
FIG. 4.
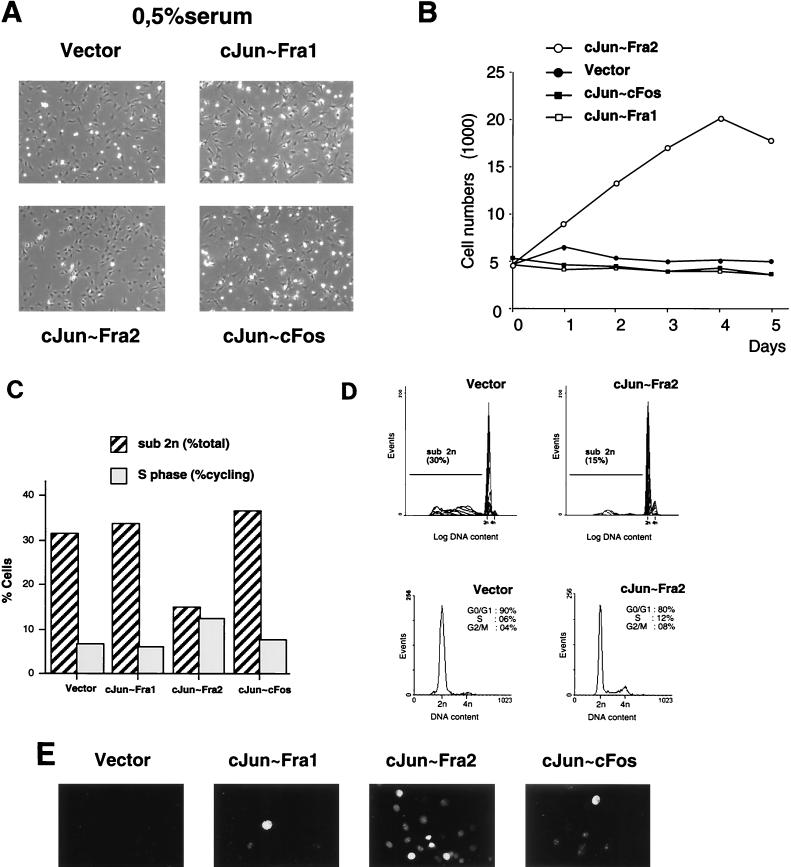
Analysis of c-Jun∼Fos forced dimers in stably transfected NIH 3T3 cells. (A) Immunofluorescence microscopy of the different cell lines. Staining of the cells was performed using anti-FLAG monoclonal antibody followed by FITC-coupled secondary antibody. (B) Western blot analysis. Equal amounts of total-cell extracts from the different stable cell lines were probed with an anti-c-Jun antibody. The positions of the different single chains and of the endogenous c-Jun protein are indicated. Vector denotes the puromycin-resistant control cell line, infected with the empty virus. The membrane was reprobed with an antiactin antibody (actin) as loading control. Molecular size markers (in kilodaltons) are on the left. (C) Reporter assay. The forced c-Jun-containing dimers activated transcription of a transiently transfected MMP1-luc reporter plasmid (2 μg). The values and standard errors were based on two independent experiments with duplicate points and normalized for transfection efficiency by using a Renilla luciferase internal control. The levels of activation, relative to the values obtained with the control (vector) cell line, are the numbers in parentheses. (D) Cell morphology in normal culture conditions. Control cells (vector) and three c-Jun∼Fos expressing clones were seeded at low density in DMEM containing 7% FCS. Photographs were taken under a phase contrast microscope after 2 days of culture. (E) Proliferation curve. Duplicate cultures of the control (vector) and the three c-Jun∼Fos cell lines were maintained in DMEM containing 7% FCS, and cells were counted at daily intervals. The mean values of duplicates are plotted against time. (F) Percentages of S-phase cells in the different cell lines after 2, 5, and 9 days of culture. Cell cycle analysis using conventional flow cytometry was performed on a fraction of the cells used for growth curves at the indicated time points.
No obvious morphological differences were observed between cell populations expressing the different single-chain AP-1 molecules (Fig. 4D). However, we noticed that c-Jun∼Fra2-expressing cells reached higher saturation density than the others. To investigate this further, cell cultures expressing vector, c-Jun∼Fra1, c-Jun∼c-Fos, and c-Jun∼Fra2 were seeded at low density in medium containing 7% serum and adherent cells were counted daily. During the first 4 days of culture, all growth curves were virtually identical (Fig. 4E). However, at later time points, when cell numbers for control, c-Jun∼Fra1, and c-Jun∼c-Fos became constant, the c-Jun∼Fra2 cells continued growing for another 2 days and reached a substantially higher saturation density (Fig. 4E). Moreover, estimated doubling times were shorter for c-Jun∼Fra2 cells (approximately 19 h) than for the three other cell lines (22 to 24 h). To explore the molecular basis for the different sensitivity to confluency, cell cycle parameters were determined by flow cytometry on days 2, 5, and 9. Control and c-Jun∼Fra1 cells were already growth arrested after 5 days of culture, as judged by the low percentage of cells in S phase at days 5 and 9. In contrast, c-Jun∼Fra2 and c-Jun∼c-Fos cells were still actively synthesizing DNA at day 5, based on higher S-phase cells than at day 9 (Fig. 4F). The higher S-phase cell population of cells expressing c-Jun∼Fra2 (on days 2 and 5) was associated with higher percentage of G2/M cells and a lower percentage of G0/G1 cells (data not shown). These data suggest that c-Jun∼Fra2 promotes cell proliferation at otherwise saturation density. The shortened doubling time was correlated with an increase in S, G2, and M compartments of the cell cycle. In other words, cells expressing c-Jun∼Fra2, but not other tethered dimers, are less sensitive to contact inhibition.
An alternative to confluency, deprivation of serum or growth factors is widely used to render NIH 3T3 cells quiescent. We therefore cultured the control and the three c-Jun∼Fos cells in medium containing 0.5% serum. After 48 h, c-Jun∼Fra2 cultures showed fewer floating, shrunken cells than control, c-Jun-Fra1 or c-Jun∼c-Fos cultures (Fig. 5A). While the population of adherent control, c-Jun∼Fra1 or c-Jun∼c-Fos cells did not increase in serum with a low concentration, c-Jun-Fra2 cells substantially increased in number (Fig. 5B). In addition, flow cytometry analyses indicated that this increase correlated with an increase in S and G2/M cells (Fig. 5C and D) and with a substantial decrease in cells with less than diploid DNA content, representative of dead cells (Fig. 5C and D). To visualize replicating cells, cultures were pulse labeled with BrdU for 2 h after 48 h of serum deprivation. While nuclear labeling was undetectable in the control cells and low in c-Jun∼Fra1 and c-Jun∼c-Fos cultures, numerous BrdU-positive cells were detected in c-Jun∼Fra2 cultures, confirming that these cells actively synthesize DNA in low-serum culture conditions (Fig. 5E). These results indicate that c-Jun∼Fra2 expressing cells are able to maintain cell cycle progression in both high-density and low-serum culture conditions and to better resist cell death induced by growth factor deprivation.
FIG. 5.
Effect of ectopic expression of c-Jun∼Fos forced dimers in low-serum-level conditions. (A) Culture morphology in low-serum-level conditions. Control (vector) cells and c-Jun∼Fos expressing clones were seeded in DMEM containing 0.5% FCS. Photographs were taken under phase-contrast microscope after 2 days of culture. (B) Proliferation curve. Control cells (vector) and the three c-Jun∼Fos cell lines were cultured in duplicate in DMEM containing 0.5% FCS, and cells were counted daily. The mean values of duplicates are shown plotted against time. (C) Cell cycle distribution of the different cell lines. Cell cycle analysis and quantification of a sub-G1 fraction was performed using flow cytometry after 2 days of culture in DMEM containing 0.5% FCS. (D) Representative cell death (top) and cell cycle (bottom) profiles for c-Jun∼Fra2 and control (vector) cell lines. (E) BrdU labeling of S phase cells. The control (vector) and c-Jun∼Fos cell lines cultures were cultured in DMEM containing 0.5% FCS. BrdU-positive nuclei appear white.
Molecular basis for proliferation defects in c-Jun∼Fra2 cells.
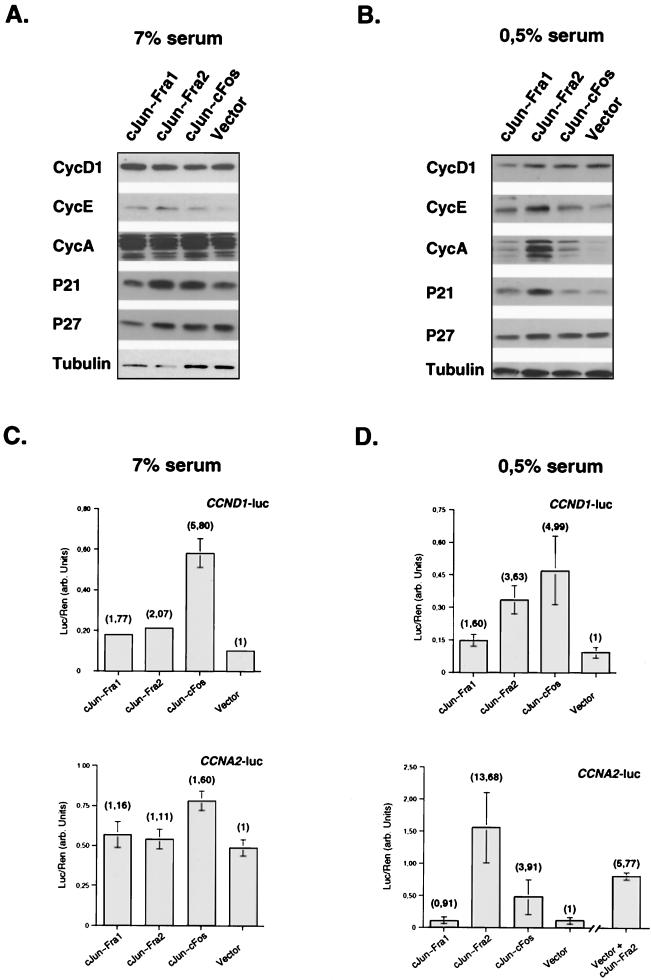
The cell cycle is regulated by cyclins, CDKs, and CDK inhibitors. Cyclins D and E regulate G1 progression, and cyclin A is required for both G1/S and G2/M transitions. Expression of cyclins declines when mitogens are withdrawn, leading to a relative excess of CDK inhibitors, which blocks S phase progression in cells deprived of growth factors (for review, see reference 34). To obtain further insights into the molecular basis for the resistance to growth arrest of the c-Jun∼Fra2 cells, we performed immunoblot analyses for cyclins and CDK inhibitors involved in G1 and G1/S progression. We prepared extracts from exponentially growing cells and serum-deprived cells. Protein levels of cyclin D1, cyclin A, cyclin E, p21, and p27 were rather similar between the different cells when they were exponentially growing in normal medium (Fig. 6A). In a low level of serum, however, cyclin E, cyclin A, and p21 protein levels were higher in the c-Jun∼Fra2 cells (Fig. 6B). These results suggest that the reduced sensitivity to growth factor deprivation of c-Jun∼Fra2 expressing cells is correlated with a failure to modulate cyclin expression.
FIG. 6.
Analysis of cell cycle genes in c-Jun∼Fos cell lines. (A and B) Western blotting analysis of control (Vector) and c-Jun∼Fos cell lines grown for 48 h under conditions of high (A) and low (B) serum levels. Equivalent amounts of total-cell extracts were visualized with the indicated antibodies (Santa Cruz). Tubulin expression was used as a loading control. (C and D) Activation of cyclin D1 (CCND1-luc) and cyclin A (CCNA2-luc) reporter constructs in the control and c-Jun∼Fos cell lines under conditions of high (C) and low (D) serum levels. Cells were transfected with 2 μg of the indicated reporter constructs. Luciferase activity (arbitrary units) was normalized to a Renilla luciferase internal control, and each point corresponds to the mean of two independent experiments done in duplicate. Activation relative to that of the control cell line is in parentheses. Vector + cJun∼Fra2, control (vector) cell line cotransfected with a c-Jun∼Fra2 expression vector and the CCNA2 reporter.
To investigate whether c-Jun∼Fra2 is directly involved in transcriptional regulation of cyclins, we analyzed the activation of transfected reporter promoters constructs for cyclin D1 (CCND1-luc) and cyclin A (CCNA2-luc) in exponentially growing and serum-deprived cells. Both constructs contain human promoter fragments (14, 15), which show similar regulation as their murine counterparts (8, 29). The CCND1 promoter is activated most efficiently by c-Jun∼c-Fos in normal serum conditions while activation of the CCNA2 promoter is only marginal (Fig. 6C). In a low serum concentration, c-Jun∼Fra2 activated the CCND1 promoter almost as efficiently as c-Jun∼c-Fos. Furthermore, c-Jun∼Fra2 substantially and reproducibly activated the CCNA2 reporter significantly better than the other dimers (Fig. 6D). Consistently, transient introduction of c-Jun∼Fra2 in the vector-infected control cells also activated the CCNA2 reporter (Fig. 6D, vector + c-Jun∼Fra2). These data suggest that c-Jun∼Fra2 is directly responsible for high cyclin A protein levels in serum-deprived c-Jun∼Fra2 cells. We also performed parallel experiments using a cyclin A reporter construct mutated in the CRE site located at −76 bp in the promoter. This site has been shown to be important for cyclin A regulation at the G1/S transition (7) and required for the downregulation of cyclin A transcription upon contact inhibition (41). The mutated reporter construct did not respond to c-Jun∼Fra2, in both low- and high-serum-level conditions (data not shown), suggesting that c-Jun∼Fra2 activates the cyclin A promoter through binding to the CRE element. Moreover, we showed, using in vitro-synthesized proteins and gel retardation assays, that c-Jun∼Fra2 can indeed bind an oligonucleotide derived from the cyclin A promoter and containing an intact CRE element (data not shown). Taken together, these expression studies indicate that c-Jun∼Fra2 can specifically affect, probably through the regulation of cyclin A expression, the cell cycle progression of mouse fibroblasts.
DISCUSSION
The AP-1 transcription complex plays an important role in specifying cell fate decision in several biological settings. The well-studied Jun/Fos families include positively and negatively acting monomers that can dimerize with each other in various combinations. An important implication is that changes in the levels of just one monomeric component could radiate to yield significant qualitative and quantitative shifts in many or even all members of the active multimer pool. This would, in turn, give rise to different regulatory outputs. Moreover, changes in concentration could be quite subtle and still have significant effects. For example, we and others have shown that cyclin D1 expression in fibroblasts is sensitive to a threefold difference in the relative levels of Jun proteins (2, 27, 40). Learning how these AP-1 shifts ultimately lead to major changes in cellular phenotype requires dealing with their combinatorial character. In this study, we have begun to test a new strategy for dissecting the regulatory capacity of specific AP-1 dimers.
Design and characterization of tethered Jun∼Fos molecules.
To reduce the complexity of the AP-1 network, we attempted to construct forced Jun∼Jun, Jun∼Fos, and Jun∼ATF dimers that cannot exchange with free monomers. Following the successful examples in the literature, using the MyoD and E47 bHLH transcription factors (25, 35), we designed synthetic molecules in which two complete AP-1 dimerization partners are placed in close proximity so that they preferentially form an intramolecular forced pseudodimer. One could suspect that linking one monomer to another might impair proper protein folding. However, since entire monomer units remain intact, tethering two monomers may be no more disruptive than routine domain shuffles that combine DNA-binding domains from one factor with a transactivation domain from another. Nor is it more disruptive than dimerization domain swapping or mutagenesis in order to restrict dimerization to one or more partners (1, 4, 31, 37).
It is possible that two linked molecules might associate with each other through intermolecular bZIP-bZIP interactions to generate tetrameric (or even higher-order) forms. Alternatively, linker proteolysis could produce free constituent monomers from the tethered construct. However, the DNA-binding properties of our single-chain proteins suggest that tethered Jun∼Fos constructs fold into active pseudodimers rather than forming tetramers or dimers of proteolytic products. In support of this, we observed a similar mobility on gel filtration columns for tethered AP-1 dimers as their unlinked dimeric counterparts (result not shown) and demonstrated that the major TRE-binding activity in c-Jun∼Fra1 expressing cells were resistant to ΔJun, except when in large molar excess. Resistance to the effect of dominant-negative c-Jun was also observed at the transcriptional level, since activation of the collagenase promoter by the c-Jun∼Fra1 protein was far less sensitive to ΔJun inhibition than activation by the corresponding monomer mixture. Resistance to ΔJun challenge implies that once introduced into cells, such tethered molecules are insulated from endogenous bZIP proteins. Conversely, resistance of the tethered form to ΔJun can be used to discriminate between direct activation by forced AP-1 dimers or secondary activation through induced expression of endogenous Jun and Fos proteins.
Our studies are in line with previous work using tethered MyoD∼E47 proteins that showed that intramolecular folding of this dimeric transcription factor could be achieved as soon as the tether was of sufficient length and flexibility and that such linked molecules behave like natural dimers (25). Moreover, structural studies performed with a tethered MASH-1 homodimer further supported the hypothesis that such constructs form intramolecular dimers (35).
Sequence and promoter selectivity.
In the course of this study, we have generated single-chain dimers between c-Jun and c-Fos, Fra1, Fra2, and ATF2 as well as c-Jun homodimers and confirmed that the different tethered proteins retain selectivity toward DNA target sites in a series of gel retardation and reporter assays. Using in vitro-synthesized proteins, we showed that tethered Jun∼Fos dimers prefer TRE elements while c-Jun∼ATF binds better to CRE sequences. We confirmed these observations by using transient-expression and reporter assays. In transient transfection, Jun∼Fos dimers activated the previously characterized Jun-Fos-controlled collagenase promoter more efficiently than c-Jun∼c-Jun or c-Jun∼ATF2 dimers, while the latter dimers were more potent activators of the Jun-ATF-controlled c-Jun promoter. Furthermore, mutation of the previously characterized AP-1 responsive elements on these reporter constructs abolished activation by the tethered dimers (not shown). These experiments showed that the tethering manipulations did not produce regulators with decreased selectivity when compared to free dimers, at least for these previously defined DNA target sites.
It is noteworthy that the activation of the cyclin D1 reporter by c-Jun-Fos dimers shows less sensitivity to the nature of the Fos partner than the collagenase reporter (Fig. 3). Moreover, the binding selectivity of Jun-Fos dimers toward TRE elements depends on the identity of the Fos partner since c-Jun∼Fra1 and c-Jun∼Fra2 dimers bind CRE elements better than c-Jun∼c-Fos. This could constitute the first indication of a reduced selectivity for Fos proteins of the AP-1 responsive sites on the cyclin D1 promoter together with the first indication of a different binding and transactivation activity among Jun-Fos dimers. Additional careful analyses, including comparison with corresponding mixtures of monomers, need to be performed, but this type of study could be extended to other promoters and will help in understanding the site specificity and transactivation properties of individual AP-1 dimers.
Phenotypic effects of stable Jun∼Fos expression in mouse fibroblasts.
We further characterized the biological properties of the different tethered c-Jun∼Fos dimers by establishing pools of fibroblast cells expressing tethered c-Jun∼Fra1, c-Jun∼Fra2, and c-Jun∼c-Fos proteins. The levels of overexpressed tethered proteins were modest compared to that of the endogenous c-Jun but sufficient to activate the collagenase reporter construct. Noticeably, the expression of the tethered c-Jun-c-Fos dimer was rather low, consistent with transient-transfection experiments. Repeated observation that c-Jun∼c-Fos is expressed much less than other c-Jun-forced dimers seem to reflect inherent instability of c-Fos messages and proteins (12, 36).
NIH 3T3/c-Jun∼Fra2 cultures reached higher saturation densities than control, c-Jun∼Fra1, and c-Jun∼cFos cells and were more resistant to serum withdrawal. Analysis of cell cycle distribution by flow cytometry indicated that under conditions where control cells stopped proliferating and accumulated in the G0/G1 compartment of the cell cycle, c-Jun∼Fra2-expressing cells were still able to progress through S and G2/M phases and further divide. This appeared to be the case both when cells were cultured to confluency and when they were deprived from growth factors by serum withdrawal. Nevertheless, sensitivity to cell-to-cell contacts was delayed rather than completely lost, since the c-Jun∼Fra2 cells cultures arrested at later time points (Fig. 4), remained in monolayers, and did not grow in soft agar (not shown).
Upon serum withdrawal, c-Jun∼Fra2 cultures showed a reproducible increase in cell numbers compared to the other cultures, together with a decrease in the G0/G1 phase, indicating that the c-Jun∼Fra2-expressing cells are indeed able to proliferate in a low level of serum. Moreover, we observed a substantial decrease in the sub-G1 compartment, representative of dead cells, in c-Jun∼Fra2 cultures relative to the control cell cultures. In conclusion, the increase in c-Jun∼Fra2 cell numbers in low-serum-level culture conditions can be due to a combination of increased proliferation and decreased cell death.
The two other cell populations showed milder and different phenotypes when results were analyzed more carefully. For example, even though the proliferation curve of the c-Jun∼c-Fos cells was identical to the control in both culture conditions, the percentage of S phase cells in the c-Jun∼c-Fos cultures was always slightly higher than in the control cultures. This could point to a possible role of this particular dimer in cell cycle progression. On the other hand, both c-Jun∼c-Fos and c-Jun∼Fra1 cells seemed to be slightly more sensitive to serum withdrawal than control cell lines, as judged by both morphology and the sub-G1 proportion (Fig. 5). It is possible that certain Jun∼Fos dimers alter cell cycle distribution, which was not apparent from their proliferation curves due to increased cell death. This has already been observed in other situations; for example, upon overexpression of the c-myc transcription factor (9). An interesting issue for future experiments would be to compare the resistance of these cell lines to death-inducing stimuli, as it has been proposed that both c-Jun and c-Fos are important mediators of the cellular response to DNA damage-induced apoptosis (18, 32, 40).
It is noteworthy that while Fra2 has been described as a less potent transactivator than c-Fos (for a review, see reference 10), overexpression of c-Jun∼Fra2 and not tethered c-Jun∼c-Fos dimers is sufficient to override, at least partially, the growth arrest signals induced by both increased cell density and serum withdrawal. We have previously shown that in NIH 3T3 cells, Fra2 is the predominant Fos monomer expressed when cells are growing exponentially and the other Fos proteins can be detected only when cells are subjected to extracellular stimulation, such as addition of serum (20). Since Fra2 is the major Fos species in nonstimulated exponentially growing NIH 3T3 fibroblasts, a restricted set of AP-1 dimers therefore seems sufficient to allow cell cycle progression. In other 3T3 cell lines, however, the set of possible AP-1 dimers can be larger since Fra1 has been shown to be expressed together with Fra2 in exponentially growing cells (19). The three Jun proteins are expressed in fibroblasts (19, 20), and numerous reports have suggested essential and opposite roles for Jun proteins in cell cycle progression; JunD and JunB monomers having a negative, antiproliferative role while c-Jun is the positive counterpart (2, 6, 27, 28, 39). We have previously shown that, when NIH 3T3 fibroblasts are deprived of serum or are grown to saturating densities, the relative expression levels of Jun and Fos proteins are modified since only JunD remains detectable, while c-Jun and Fra2 levels drop significantly (20). In NIH 3T3 mouse fibroblasts, down-regulation of c-Jun and Fra2 upon serum deprivation or cell-to-cell contact could therefore be a prerequisite for growth arrest. The phenotype of our c-Jun∼Fra2-expressing cells support this model. Consistent with the idea that full transformation requires several interacting Jun and Fos monomers, overexpression of the particular c-Jun∼Fra2 dimer in NIH 3T3 cells seems sufficient to initiate some aspects of transformation, like growth factor and contact-inhibition independence, but is not sufficient to completely bypass negative-growth regulators nor it is enough for full transformation. A combination of several Jun-Fos dimers might be necessary to achieve transformation. Alternatively, this partial effect may reflect the need for additional positive regulators other than AP-1.
We have analyzed the molecular basis of c-Jun∼Fra2-mediated growth factor independence. The results suggest that c-Jun∼Fra2-expressing cells continue to express cyclin A protein in a low-serum-level conditions. Moreover, a cyclin A promoter-reporter construct displayed a substantially higher activity when transfected in c-Jun∼Fra2 serum-deprived cells than in other cell lines and ectopic expression of c-Jun∼Fra2 increased cyclin A reporter activity. Therefore, the ability of this specific tethered dimer to override growth arrest signals and activate division-promoting pathways might be mediated, at least in part, through maintained expression of cyclin A in these cells. The molecular mechanisms leading to the modulation of cyclin D1, cyclin E, and p21 protein levels in the different c-Jun∼Fos-expressing cells remain to be further clarified but will certainly provide more insights into the role of individual AP-1 dimers in the control of cell cycle progression.
Acknowledgments
We are grateful to J.-P. Abastado for advice in the design of the peptide linker and to D. Lallemand for the gift of antibodies. We thank D. Bohmann for the gift of the RSVcJunDB-4 mutant construct and R. Müller, H. van Dam, P. Herrlich, and J. Sobczak-Thepot for different reporter constructs. We thank J. Weitzman and A. Szremska for critical reading of the manuscript and C. Coffinier, B. Arcangioli, and M. Pontoglio for fruitful discussions.
This work was supported by grants from the EEC Biomed and Training and Mobility Programs. L.B. was the recipient of awards from the Pasteur Weitzmann Council, the Fondation des Treilles, and the EMBO Short Term Fellowship. M.W. was the recipient of an initial award from the French Foreign Ministry and was supported by the Association for International Cancer Research (AICR).
REFERENCES
- 1.Amati, B., M. W. Brooks, N. Levy, T. D. Littlewood, G. I. Evan, and H. Land. 1993. Oncogenic activity of the c-Myc protein requires dimerization with Max. Cell 72:233-245. [DOI] [PubMed] [Google Scholar]
- 2.Bakiri, L., D. Lallemand, E. Bossy-Wetzel, and M. Yaniv. 2000. Cell cycle-dependent variations in c-Jun and JunB phosphorylation: a role in the control of cyclin D1 expression. EMBO J. 19:2056-2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bohmann, D., and R. Tjian. 1989. Biochemical analysis of transcriptional activation by Jun: differential activity of c- and v-Jun. Cell 59:709-717. [DOI] [PubMed] [Google Scholar]
- 4.Castellazzi, M., L. Loiseau, F. Piu, and A. Sergeant. 1993. Chimeric c-Jun containing an heterologous homodimerization domain transforms primary chick embryo fibroblasts. Oncogene 8:1149-1160. [PubMed] [Google Scholar]
- 5.Chinenov, Y., and T. K. Kerppola. 2001. Close encounters of many kinds: Fos-Jun interactions that mediate transcription regulatory specificity. Oncogene 20:2438-2452. [DOI] [PubMed] [Google Scholar]
- 6.Chiu, R., P. Angel, and M. Karin. 1989. Jun-B differs in its biological properties from, and is a negative regulator of, c-Jun. Cell 59:979-986. [DOI] [PubMed] [Google Scholar]
- 7.Desdouets, C., C. Ory, G. Matesic, T. Soussi, C. Brechot, and J. Sobczak-Thepot. 1996. ATF/CREB site mediated transcriptional activation and p53 dependent repression of the cyclin A promoter. FEBS Lett. 385:34-38. [DOI] [PubMed] [Google Scholar]
- 8.Eto, I. 2000. Molecular cloning and sequence analysis of the promoter region of mouse cyclin D1 gene: implication in phorbol ester-induced tumour promotion. Cell Prolif. 33:167-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Evan, G. I., A. H. Wyllie, C. S. Gilbert, T. D. Littlewood, H. Land, M. Brooks, C. M. Waters, L. Z. Penn, and D. C. Hancock. 1992. Induction of apoptosis in fibroblasts by c-myc protein. Cell 69:119-128. [DOI] [PubMed] [Google Scholar]
- 10.Foletta, V. C. 1996. Transcription factor AP-1, and the role of Fra-2. Immunol. Cell Biol. 74:121-133. [DOI] [PubMed] [Google Scholar]
- 11.Glover, J. N., and S. C. Harrison. 1995. Crystal structure of the heterodimeric bZIP transcription factor c-Fos-c-Jun bound to DNA. Nature 373:257-261. [DOI] [PubMed] [Google Scholar]
- 12.Grosset, C., C. Y. Chen, N. Xu, N. Sonenberg, H. Jacquemin-Sablon, and A. B. Shyu. 2000. A mechanism for translationally coupled mRNA turnover: interaction between the poly(A) tail and a c-fos RNA coding determinant via a protein complex. Cell 103:29-40. [DOI] [PubMed] [Google Scholar]
- 13.Ham, J., C. Babij, J. Whitfield, C. M. Pfarr, D. Lallemand, M. Yaniv, and L. L. Rubin. 1995. A c-Jun dominant negative mutant protects sympathetic neurons against programmed cell death. Neuron 14:927-939. [DOI] [PubMed] [Google Scholar]
- 14.Henglein, B., X. Chenivesse, J. Wang, D. Eick, and C. Brechot. 1994. Structure and cell cycle-regulated transcription of the human cyclin A gene. Proc. Natl. Acad. Sci. USA 91:5490-5494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herber, B., M. Truss, M. Beato, and R. Müller. 1994. Inducible regulatory elements in the human cyclin D1 promoter. Oncogene 9:1295-1304. [PubMed] [Google Scholar]
- 16.Hirai, S., B. Bourachot, and M. Yaniv. 1990. Both Jun and Fos contribute to transcription activation by the heterodimer. Oncogene 5:39-46. [PubMed] [Google Scholar]
- 17.Jonat, C., B. Stein, H. Ponta, P. Herrlich, and H. J. Rahmsdorf. 1992. Positive and negative regulation of collagenase gene expression. Matrix Suppl. 1:145-155. [PubMed] [Google Scholar]
- 18.Kolbus, A., I. Herr, M. Schreiber, K. M. Debatin, E. F. Wagner, and P. Angel. 2000. c-Jun-dependent CD95-L expression is a rate-limiting step in the induction of apoptosis by alkylating agents. Mol. Cell. Biol. 20:575-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kovary, K., and R. Bravo. 1992. Existence of different Fos/Jun complexes during the G0-to-G1 transition and during exponential growth in mouse fibroblasts: differential role of Fos proteins. Mol. Cell. Biol. 12:5015-5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lallemand, D., G. Spyrou, M. Yaniv, and C. M. Pfarr. 1997. Variations in Jun and Fos protein expression and AP-1 activity in cycling, resting and stimulated fibroblasts. Oncogene 14:819-830. [DOI] [PubMed] [Google Scholar]
- 21.Landschulz, W. H., P. F. Johnson, and S. L. McKnight. 1988. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science 240:1759-1764. [DOI] [PubMed] [Google Scholar]
- 22.Matsuo, K., J. M. Owens, M. Tonko, C. Elliott, T. J. Chambers, and E. F. Wagner. 2000. Fosl1 is a transcriptional target of c-Fos during osteoclast differentiation. Nat. Genet. 24:184-187. [DOI] [PubMed] [Google Scholar]
- 23.Miller, A. D., T. Curran, and I. M. Verma. 1984. c-fos protein can induce cellular transformation: a novel mechanism of activation of a cellular oncogene. Cell 36:51-60. [DOI] [PubMed] [Google Scholar]
- 24.Morgenstern, J. P., and H. Land. 1990. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 18:3587-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neuhold, L. A., and B. Wold. 1993. HLH forced dimers: tethering MyoD to E47 generates a dominant positive myogenic factor insulated from negative regulation by Id. Cell 74:1033-1042. [DOI] [PubMed] [Google Scholar]
- 26.O'Shea, E. K., R. Rutkowski, and P. S. Kim. 1992. Mechanism of specificity in the Fos-Jun oncoprotein heterodimer. Cell 68:699-708. [DOI] [PubMed] [Google Scholar]
- 27.Passegué, E., and E. F. Wagner. 2000. JunB suppresses cell proliferation by transcriptional activation of p16(INK4a) expression. EMBO J. 19:2969-2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pfarr, C. M., F. Mechta, G. Spyrou, D. Lallemand, S. Carillo, and M. Yaniv. 1994. Mouse JunD negatively regulates fibroblast growth and antagonizes transformation by ras. Cell 76:747-760. [DOI] [PubMed] [Google Scholar]
- 29.Philips, A., S. Chambeyron, N. Lamb, A. Vie, and J. M. Blanchard. 1999. CHF: a novel factor binding to cyclin A CHR corepressor element. Oncogene 18:6222-6232. [DOI] [PubMed] [Google Scholar]
- 30.Piette, J., S. Hirai, and M. Yaniv. 1988. Constitutive synthesis of activator protein 1 transcription factor after viral transformation of mouse fibroblasts. Proc. Natl. Acad. Sci. USA 85:3401-3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pomerantz, J. L., P. A. Sharp, and C. O. Pabo. 1995. Structure-based design of transcription factors. Science 267:93-96. [DOI] [PubMed] [Google Scholar]
- 32.Schreiber, M., B. Baumann, M. Cotten, P. Angel, and E. F. Wagner. 1995. Fos is an essential component of the mammalian UV response. EMBO J. 14:5338-5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shaulian, E., and M. Karin. 2001. AP-1 in cell proliferation and survival. Oncogene 20:2390-2400. [DOI] [PubMed] [Google Scholar]
- 34.Sherr, C. J. 1996. Cancer cell cycles. Science 274:1672-1677. [DOI] [PubMed] [Google Scholar]
- 35.Sieber, M., and R. K. Allemann. 2000. Thermodynamics of DNA binding of MM17, a “single chain dimer” of transcription factor MASH-1. Nucleic Acids Res. 28:2122-2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsurumi, C., N. Ishida, T. Tamura, A. Kakizuka, E. Nishida, E. Okumura, T. Kishimoto, M. Inagaki, K. Okazaki, and N. Sagata. 1995. Degradation of c-Fos by the 26S proteasome is accelerated by c-Jun and multiple protein kinases. Mol. Cell. Biol. 15:5682-5687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Dam, H., S. Huguier, K. Kooistra, J. Baguet, E. Vial, A. J. van der Eb, P. Herrlich, P. Angel, and M. Castellazzi. 1998. Autocrine growth and anchorage independence: two complementing Jun-controlled genetic programs of cellular transformation. Genes Dev. 12:1227-1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Dam, H., D. Wilhelm, I. Herr, A. Steffen, P. Herrlich, and P. Angel. 1995. ATF-2 is preferentially activated by stress-activated protein kinases to mediate c-jun induction in response to genotoxic agents. EMBO J. 14:1798-1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weitzman, J. B., L. Fiette, K. Matsuo, and M. Yaniv. 2000. JunD protects cells from p53-dependent senescence and apoptosis. Mol. Cell 6:1109-1119. [DOI] [PubMed] [Google Scholar]
- 40.Wisdom, R., R. S. Johnson, and C. Moore. 1999. c-Jun regulates cell cycle progression and apoptosis by distinct mechanisms. EMBO J. 18:188-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yoshizumi, M., C. M. Hsieh, F. Zhou, J. C. Tsai, C. Patterson, M. A. Perrella, and M. E. Lee. 1995. The ATF site mediates downregulation of the cyclin A gene during contact inhibition in vascular endothelial cells. Mol. Cell. Biol. 15:3266-3272. [DOI] [PMC free article] [PubMed] [Google Scholar]