Abstract
Polyadenylation and splicing are highly coordinated on substrate RNAs capable of coupled polyadenylation and splicing. Individual elements of both splicing and polyadenylation signals are required for the in vitro coupling of the processing reactions. In order to understand more about the coupling mechanism, we examined specific protein-RNA complexes formed on RNA substrates, which undergo coupled splicing and polyadenylation. We hypothesized that formation of a coupling complex would be adversely affected by mutations of either splicing or polyadenylation elements known to be required for coupling. We defined three specific complexes (AC′, AC, and BC) that form rapidly on a coupled splicing and polyadenylation substrate, well before the appearance of spliced and/or polyadenylated products. The AC′ complex is formed by 30 s after mixing, the AC complex is formed between 1 and 2 min after mixing, and the BC complex is formed by 2 to 3 min after mixing. AC′ is a precursor of AC, and the AC′ and/or AC complex is a precursor of BC. Of the three complexes, BC appears to be a true coupling complex in that its formation was consistently diminished by mutations or experimental conditions known to disrupt coupling. The characteristics of the AC′ complex suggest that it is analogous to the spliceosomal A complex, which forms on splicing-only substrates. Formation of the AC′ complex is dependent on the polypyrimidine tract. The transition from AC′ to AC appears to require an intact 3′-splice site. Formation of the BC complex requires both splicing elements and the polyadenylation signal. A unique polyadenylation-specific complex formed rapidly on substrates containing only the polyadenylation signal. This complex, like the AC′ complex, formed very transiently on the coupled splicing and polyadenylation substrate; we suggest that these two complexes coordinate, resulting in the BC complex. We also suggest a model in which the coupling mechanism may act as a dominant checkpoint in which aberrant definition of one exon overrides the normal processing at surrounding wild-type sites.
In vitro RNA processing experiments with pre-mRNA substrates capable of polyadenylation (PA) and splicing have shown that the two processing reactions become interdependent. This is indicated by the observation that alterations in the efficiency of one processing reaction directly affect the efficiency of the other. Thus, the reactions are highly coordinated or coupled. In mammals, this coordination, or coupling, appears to involve the proper definition of the last exon of the mRNA. The most compelling model for this is the exon definition mechanism proposed by Berget and colleagues (4, 25, 29). This model proposes a means for the definition of all exons (first exons, interior exons, and last exons); however, much of the data that support the model come from studies of first and last exons.
First exons contain an m7GpppG 5′-cap structure instead of a 3′-splice site (3′-SS). The 5′-cap structure of a mammalian pre-mRNA is necessary for the efficient utilization of the adjacent 5′-SS for definition of the first exon and the removal of the first intron (16). Efficient recognition of the cap-proximal 5′-SS by U1 snRNP is facilitated by an interaction between the U1 snRNP and the nuclear cap binding complex bound to the cap (17). This interaction appears to define the exon.
Last exons contain a PA signal instead of a 5′-SS. Processing and removal of the last intron appear to involve interactions between splicing components at the 3′-SS of the last exon and components of the PA complex at the PA signal (10, 18, 23, 25, 26). Such interactions would define the exon. By using coupled in vitro splicing and PA substrates, it has been shown that mutations in the PA signal (the AAUAAA or downstream sequences), which eliminated PA, also caused decreased splicing, i.e., decreased removal of the last intron (10, 23). Likewise, mutations in the 3′-SS or the polypyrimidine tract (PPT) of the last exon, which eliminated splicing, caused inhibition of PA (10, 26, 40). In addition, a number of experiments have established that splicing and PA influence each other in vivo (8, 21, 22, 36, 39).
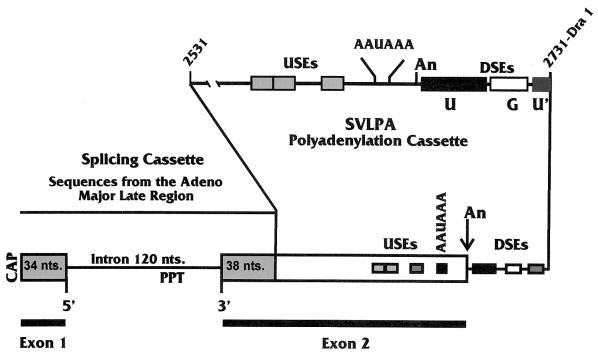
Most mammalian PA signals consist of the sequence AAUAAA and a GU- or U-rich element downstream of AAUAAA (the downstream element [DSE]). However, some signals, such as the PA signal for the late messages of simian virus 40 (the SV40 late PA signal [SVLPA]), are more complex, containing multiple DSEs as well as efficiency elements located upstream of the AAUAAA sequence (upstream elements [USEs]). In the SVLPA, three DSEs have been identified by mutagenesis. A U-rich element (U in Fig. 1), located between 14 and 40 nucleotides (nt) downstream of AAUAAA (9), appears similar to other DSEs in that it is the binding site for the 64-kDa component of the cleavage stimulatory factor (CStF) (38). A G-rich DSE (G in Fig. 1) between 45 and 58 nt downstream of the sequence AAUAAA has been shown to bind the 50-kDa protein hnRNP H (1, 2). The binding of hnRNP H may enhance the efficiency of PA (2, 27). Finally, a second U-rich DSE (U′ in Fig. 1) was identified by deletion analysis between 59 and 67 nt downstream of AAUAAA (32, 33). It can function as an hnRNP C protein binding site and also as a DSE in synthetic PA signals (43, 44).
FIG. 1.
Diagram of the 392-nt splicing and PA RNA substrate (WT Sp/PA), also called MXSVL (23). It is composed of two separable cassettes, one for splicing and one for PA. Each cassette can function on its own in splicing and PA reactions, respectively. When together, they can function in coupled splicing and PA under the appropriate conditions (see text). The splicing cassette is derived from the adenovirus major late region; the 5′-SS (5′), 3′-SS (3′), and the PPT are shown. The PA cassette contains the PA signal from the SV40 late coding region, the SVLPA, SV40 nt 2531 to 2731. The structural features of the SVLPA are shown: these include the USEs, the AAUAAA, the cleavage and PA site (An), and the DSEs (see the text for details). When the substrate is spliced, polyadenylated exons 1 and 2 are joined.
Efficiency elements located upstream of AAUAAA have been characterized in a number of PA signals (5-7, 12, 13, 20, 30, 31, 34, 35, 41, 42). In the SVLPA signal, the USEs impart characteristics that increase efficiency or provide special levels of control to the PA signals (7, 35). No consensus or obvious similarity has been observed among the USEs that have been characterized. However, in the SVLPA (Fig. 1), the USE contains three discrete elements that have the consensus sequence AUUUGURA (35).
In previous studies of the in vitro coupling of splicing and PA (11), we utilized a pre-mRNA substrate called MXSVL (26) (Fig. 1), which contains an adenovirus splicing cassette attached to the SVLPA signal (26). In those studies, we showed that mutation of either the PPT, the 3′-SS, the AAUAAA, or the DSEs affected the coupling of splicing and PA. These data confirmed and expanded the work of Berget and colleagues (23, 26, 29) and support the model of exon definition. In order to understand more about the coupling mechanism that utilizes these elements, we examined specific protein-RNA complexes formed on RNA substrates that undergo coupled splicing and PA. Previous studies (24) have shown that a progression of protein-RNA complexes form on the MXSVL substrate in a time-dependent fashion. We have drawn from these initial biochemical and kinetic studies and hypothesized that formation of a coupling complex would be adversely affected by mutations of either splicing or PA elements known to be required for coupling. In the following studies, we define and characterize three specific complexes (AC′, AC, and BC) that are similar to those previously detected (24). These complexes form rapidly on a coupled splicing and PA (Sp/PA) substrate, well before the formation of spliced and/or polyadenylated products. The BC complex appears to be a true coupling complex, in that its formation was consistently diminished by mutations or experimental conditions known to disrupt coupling. We discuss a model in which the coupling mechanism may act as a dominant checkpoint at which aberrant definition of one exon overrides the normal processing at surrounding wild-type sites.
MATERIALS AND METHODS
Plasmids encoding precursor RNA substrates.
All plasmids were propagated in Escherichia coli strain DH5a or HB101 and prepared by standard procedures. Substrate RNAs for in vitro splicing and PA reactions were prepared by in vitro transcription from linearized plasmid templates. Figure 2A shows diagrams of the substrates discussed in the text. Plasmid pSP64-MXSVL (26) was used to generate the wild-type Sp/PA transcript (WT Sp/PA) RNA. We previously made various mutations of this plasmid, which produce substrate RNA transcripts bearing (i) a point mutation in the 3′-SS (−3′ Sp/PA), (ii) a mutation in the AAUAAA and upstream sequences (−AUA Sp/PA), (iii) a mutation in the PPT (−PPT Sp/PA), and (iv) linker substitution mutations (LSMs) across the downstream region (DM2, DM3, and DM4; Fig. 2A and B) (11).
FIG. 2.
(A) Wild-type and mutant Sp/PA substrates used in these studies. At the top is the diagram of the WT Sp/PA substrate detailed in Fig. 1; below are diagrams of various mutant substrates. These include the following: −3′-SS Sp/PA, which cannot splice and is debilitated in coupling, due to a point mutation in the 3′-SS; −AUA Sp/PA, which cannot polyadenylate and is debilitated in coupling, due to mutation of the AAUAAA sequence and linker substitution mutagenesis of the USEs; and −PPT Sp/PA, which cannot splice and is debilitated in coupling, due to mutation of the PPT. The bottom diagram shows three separate LSMs in the DSEs of the SVLPA, DM1, DM2, and DM3. The exact sequences mutated are shown in panel B. DM2 and DM4 debilitate PA and coupling; DM3 has little effect on either process and is included as a positive control. (B) The sequence of the downstream region of the SVLPA starting at AAUAAA and showing the cleavage and PA site (arrow), the first U-rich region (U; double underline), the G-rich region (G; single underline), and the second U-rich region (U′; single underline). The thick underlined areas show the sequence mutated by linker substitution in the three downstream mutations DM2, DM3, and DM4.
Preparation of precursor RNA substrates.
32P-labeled or nonlabeled RNA substrates for coupled Sp/PA reactions and complex analysis were made by in vitro transcription as previously described (11).
Preparation of nuclear extracts for in vitro splicing and PA reactions and complex formation analyses.
HeLa-S3 cells obtained from the National Cell Culture Center (Minneapolis, Minn.) were used to prepare nuclear extracts by the procedure of Moore and Sharp (19) with minor variation as previously described (11). In vitro splicing and PA reactions were done as previously described (11). Complex formation analyses were done with identical reaction mixtures, except the reactions were terminated by the addition of 5 μl of termination buffer (0.1% bromphenol blue, 0.1% xylene cyanol FF, 20 μg of heparin per ml, 5 mM EDTA) and immediate flash-freezing in liquid N2. In competition experiments, the reactions were identical, except for the addition of cold competitor transcripts as described in the text. Nondenaturing, native 3.5% polyacrylamide gels (60:1) in 75 mM Tris-glycine buffer (pH 8.8) were used to resolve RNA-protein complexes. Gels were allowed to polymerize for 2 h at room temperature and then cooled to 4°C in a cold room prior to electrophoresis; all subsequent electrophoretic steps were carried out at 4°C. Frozen samples were thawed on ice immediately prior to loading on the gel. Fifteen microliters of the sample was loaded onto the gel, and the remaining 15 μl was saved for analysis of RNA processing products. Electrophoresis was carried out in 75 mM Tris-glycine buffer prechilled to 4°C. Gels were run at 300 V (constant voltage) for approximately 3 h or until the xylene cyanol dye front reached the bottom edge of the gel. Gels were dried and exposed to autoradiography.
RESULTS
Formation of specific complexes on the MXSVL substrate.
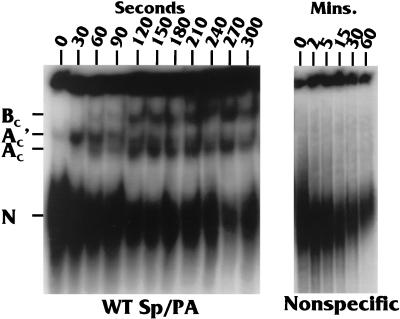
Figures 1 and 2A show the wild-type MXSVL Sp/PA substrate (WT Sp/PA) used in our studies of coupled splicing and PA (26). It contains a splicing cassette derived from the adenovirus major late region and a PA cassette composed of the SVLPA signal, which has been described in the introduction. In order to determine whether specific complexes form on the WT Sp/PA substrate, we incubated 32P-labeled substrate in HeLa cell extracts capable of coupled splicing and PA. After various times of incubation (0 to 300 s) at 30°C, the reactions were stopped, flash-frozen in liquid nitrogen, and kept frozen at −80°C until all of the samples could be analyzed on a native gel (see Materials and Methods). Figure 3 shows the time course of the complex formation. A complex of proteins, denoted N in Fig. 3, forms very rapidly. It can be seen at the zero time point, which is, in reality, the time it takes to mix the substrate and extract in the cold and quick-freeze the mixture (about 5 to 7 s). This complex may result from heterogeneous binding of hnRNPs in the extract, since a similar band formed on a nonspecific RNA transcript of equal size and contained no processing elements (Nonspecific in Fig. 3). However, by 30 s of incubation, specific complexes began to form only on the WT Sp/PA substrate. Three specific complexes (denoted AC′, AC, and BC) form over time. The subscript C (for coupled) is used to distinguish these complexes, which form on a coupled Sp/PA substrate, from the well-characterized splicesomal A and B complexes, which form on splicing-only substrates.
FIG. 3.
Time course studies of protein-RNA complex formation on the WT Sp/PA substrate and a nonspecific RNA substrate containing no processing elements. The complexes noted are a nonspecific complex (N), which forms rapidly on either substrate, and three specific complexes (AC, AC′, and BC), which form only on the WT Sp/PA complex. The subscript C (for “coupled”) is used to denote these complexes as forming on a coupled Sp/PA substrate and to differentiate them from spliceosomal complexes, which form on splicing-only substrates.
The AC′ complex appeared within 30 s of mixing at 30°C, and by 60 s, the AC complex appeared. Finally, the BC complex appeared by 120 s. The AC′ complex diminished with time, being nearly imperceptible by 120 s, suggesting that it may be a precursor to one or both of the other complexes. The AC and BC complexes remain relatively stable up to the last time point at 300 s. In some experiments in which the gel resolution was very good, the band representing the BC complex was a doublet (described below).
It should be noted that the intensity of the nonspecific complex (N) is significantly reduced over the 300-s time course with the WT Sp/PA substrate, whereas its level is more constant over a similar time course with the nonspecific substrate (compare the 0-s, 120-s or 2-min, and 300-s or 5-min time points). This indicates that the proteins in the N complex may either (i) be displaced by the proteins of the specific complexes or (ii) be included, with other factors, as part of more organized specific complexes.
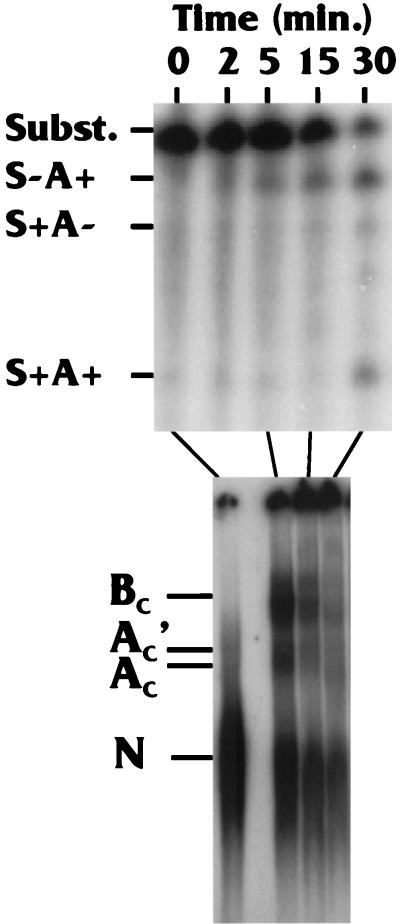
In longer time course studies with the WT Sp/PA substrate, we have seen the BC complex continuing to accumulate up to 15 min and then decreases significantly by 30 min (for example, see the bottom panel of Fig. 5). In the experiment shown in Fig. 4, the reaction mixtures were divided, with one-half used for complex analysis and the other half analyzed for RNA processing products, which are displayed in the top panel of Fig. 4. Three products are noted: polyadenylated but not spliced (S−A+); spliced but not polyadenylated (S+A−); and spliced and polyadenylated (S+A+), which represents the fully processed product. It should be noted that efficient production of the fully processed S+A+ product occurs by 30 min, well after the formation of complexes AC′, AC, and BC, as expected if the formation of these complexes is a prerequisite of coupled processing.
FIG. 5.
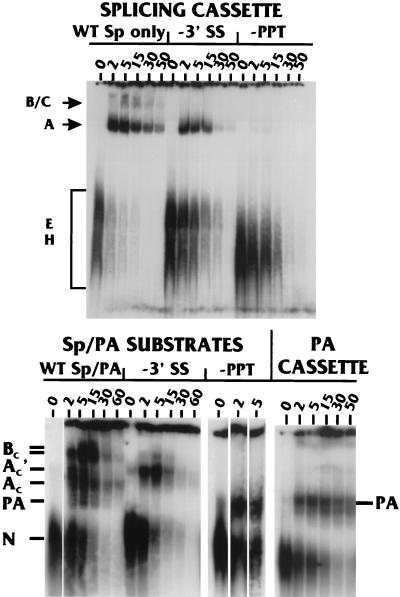
Comparison of splicing and PA complexes with known spliceosomal complexes and correlation of complexes with coupling. The top panel shows time course studies (in minutes) of complex formation on the splicing cassette (WT Sp only) from the WT Sp/PA substrate (See Fig. 1) as well as mutant splicing cassettes with a point mutation in the 3′-SS (−3′ SS) and LSM of the PPT (−PPT). The well-characterized spliceosomal complexes E, H, A, B, and C are noted. The bottom panel shows similar complex formation time courses (in minutes) with the WT Sp/PA substrate and mutant Sp/PA substrates (Fig. 2A) containing a point mutation in the 3′-SS (−3′ SS) and mutation of the PPT (−PPT). The last four lanes show complex formation on the isolated PA cassette from the WT Sp/PA substrate (Fig. 1).
FIG. 4.
Comparison of the rate of complex formation with the appearance of spliced and polyadenylated products. Reaction mixtures were split in half, with one-half being used for complex formation analysis and the other being analyzed for processing products: polyadenylated but not spliced (S−A+); spliced but not polyadenylated (S+A−); and spliced and polyadenylated (S+A+), which represents the fully processed product. See the text for details.
Comparison of Sp/PA complexes with characterized spliceosomal complexes.
A number of spliceosomal complexes formed on splicing substrates have been well characterized (28). These complexes, in temporal order of formation, are denoted E, A, B, and C. In addition, a nonspecific H complex has been described, which has characteristics similar to those of the N complex we detected on both specific and nonspecific substrates (37). To assess the similarity between the spliceosomal complexes and the complexes detected on the WT Sp/PA substrate, we compared complex formation on (i) the separate splicing cassette and PA cassette, which together make up MXSVL (Fig. 1); (ii) the separate splicing and PA cassettes containing specific mutations; and (iii) WT Sp/PA and mutant Sp/PA substrates. The top panel of Fig. 5 shows a time course of complex formation on the splicing cassette (WT Sp only); the spliceosomal complexes H, E, A, B, and C have been visually identified by comparison with published data (15). Point mutation of the 3′-SS (−3′-SS) allows elimination of formation of the B and C complexes, whereas the A, E, and nonspecific H complexes do form. In contrast, mutation of the PPT of the splicing cassette (−PPT) significantly inhibited all complex formation, except for that of the nonspecific H complex.
In the lower panel of Fig. 5, a repeat time course of the complex formation on the WT Sp/PA substrate (Fig. 1) is shown, along with similar time courses using substrates containing the point mutation in the 3′-SS or the mutation in the PPT (−3′-SS and −PPT, respectively; see also −3′-SS Sp/PA and −PPT Sp/PA in Fig. 2). With the WT Sp/PA substrate, the AC′ complex formed early and decreased significantly by 5 min followed by formation of the AC and BC complexes. However, by using the −3′-SS Sp/PA substrate, the AC′ complex formed and accumulated to high levels by 5 min, and the AC and BC complexes failed to form efficiently. Comparison of complex formation on the coupled −3′-SS Sp/PA substrate and −3′-SS splicing-only substrate suggests a relationship between the AC′ complex and the spliceosomal A complex. Specifically, (i) these are the only significant complexes formed on the mutant substrates, and (ii) they each appear to be precursors of other complexes. Thus, the AC′ complex may primarily contain spliceosomal factors.
A complex specific for the PA signal can also be discerned on several substrates; this is the band designated PA in the lower panel of Fig. 5. First, the PPT mutation (−PPT) severely inhibited all complex formation on the splicing-only substrate (Fig. 5, upper panel); however, a specific complex (PA) did form on the −PPT Sp/PA substrate, suggesting that this complex results from the presence of the PA signal in the substrate. A similarly migrating complex also formed on the PA cassette (Fig. 5, PA only), which contains the SVLPA signal and no splicing elements (Fig. 1). Furthermore, this complex did not form on a PA-only substrate containing a U-to-G point mutation in AAUAAA (data not shown). The PA signal-specific complex migrates faster than the AC′ and AC complexes and can be seen among the complexes formed on the WT Sp/PA substrate, where it forms rapidly and transiently (see the 2-, 5-, and 15-min time points with the WT Sp/PA substrate; Fig. 5, lower panel). The PA complex was not always detected, as will be noted from looking at all of the data with the WT Sp/PA substrate: we feel this is due to unknown experimental variation, which specifically affected the transient nature of this complex. Since the Ac′ and PA complexes both form very transiently on the WT Sp/PA complex, we suggest that they are the primary complexes (formed on the individual splicing elements and PA signal, respectively), which combine in the formation of the coupling complexes.
Correlation of complexes with coupling.
In order to determine how the AC′, AC, and BC complexes are associated with coupling, we performed complex formation time course studies with MXSVL substrates that were mutated in various splicing or PA elements. We hypothesized that the formation of a complex specifically associated with coupling would be affected by mutations in either splicing elements or PA elements, which are known to be involved in coupling. In this regard, it has previously been established that a point mutation of the 3′-SS (−3′SS Sp/PA, Fig. 2A), an LSM in the PPT (−PPT Sp/PA, Fig. 2A), and an LSM in the U-rich downstream element of the SVLPA signal (Fig. 2A and B) will each disrupt coupling of splicing and PA in vitro (11, 14). Thus, complex formation was performed on the MXSVL-based substrates containing these mutations.
The complex formation analyses comparing the WT Sp/PA, −3′-SS Sp/PA substrate (−3′-SS), and the −PPT Sp/PA (−PPT) in Fig. 5 (upper panel) have been discussed above. The data show that mutation of the 3′-SS inhibits the formation of the AC and BC complexes, and mutation of the PPT inhibits formation of the AC′, AC, and BC complexes. These data suggest that all three complexes (AC′, AC, and BC) may be important in coupling. Thus, we next examined complex formation on substrates with PA element mutations. The mutations we used are the linker substitution mutants DM2, DM3, and DM4 (Fig. 2A) within the downstream region of the SVLPA (14). Figure 2B shows the position of the DM2, DM3, and DM4 mutations within the sequence of the downstream region beginning with the sequence AAUAAA and showing the position of the cleavage site, the first U-rich region, the G-rich region, and the second U-rich region. DM2, DM3, and DM4 have previously been tested for their effects on PA and coupling (11). DM3 mutates a region between the U-rich and G-rich DSEs and has little effect on PA efficiency or coupling; it was used here as a positive control. DM2 and DM4 mutate the U-rich and G-rich downstream regions, respectively; each has been shown to significantly inhibit both PA and the coupling of splicing and PA (11, 14).
Figure 6 shows a time course of complex formation on WT Sp/PA, DM2, DM3, and DM4. Complex formation of the DM3 mutation is very similar to that on the WT Sp/PA substrate, as anticipated, since this mutation does not affect PA or coupling. In contrast, the DM4 mutation was deleterious to the formation of all complexes, especially complex BC. The separate lanes in Fig. 6 show a similar experiment with DM2, which also significantly affected the formation of complex BC.
FIG. 6.
Correlation of complexes with coupling: effects of DSE mutations on complex formation. The panel shows time course studies (in minutes) of complex formation on Sp/PA substrates, including the wild-type substrate (WT Sp/PA) and substrates with LSMs in the downstream region of the SVLPA: DM2, DM3, and DM4 (Fig. 1 and 2 and the text).
The data from Fig. 5 and 6 suggest that mutations in either splicing elements or elements of the PA signal affect the formation of the various complexes. However, the consistent loss of complex BC strongly correlates this complex with coupling.
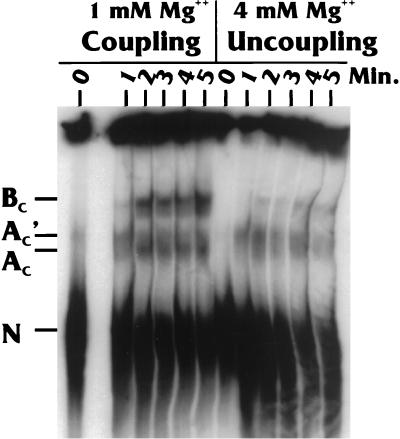
Effects of magnesium concentrations on complex formation.
It has previously been established that coupling is very dependent on the magnesium concentration in the in vitro reaction (11, 26). In our extracts, the optimal Mg2+ concentration is 1 to 1.5 mM; all of the experiments described above were done within this concentration range. However, at 4 mM Mg2+ and higher, coupling is significantly reduced (11). We previously demonstrated that the reduction in coupling at 4 mM Mg2+ is not due to the inhibition of one of the processing reactions; in fact, the levels of total PA and total splicing are very similar at both 1 and 4 mM Mg2+. The difference is that splicing and PA occur less frequently on the same RNA molecules, since the reactions are uncoupled (11). Thus, if complex AC′, AC, or BC is associated with coupling, we would predict less-efficient coupling complex formation at the suboptimal (4 mM) Mg2+ concentrations.
The data shown in Fig. 7 indicate that the formation of complex AC′ (and possibly the complex AC) is very similar at both Mg2+ concentrations. However, at the higher Mg2+ concentration, the formation of complex BC is significantly inhibited. The Mg2+ concentration-dependent formation of the BC complex supports the conclusion that it is a coupling complex. In addition, little BC complex forms when there is zero Mg2+ (data not shown), a condition in which the system is also uncoupled (11)
FIG. 7.
Effects of Mg2+ concentration on complex formation on the WT Sp/PA substrate. The time course reactions (in minutes) were identical, except in terms of the magnesium concentrations (1 or 4 mM).
Competition analysis of complex formation.
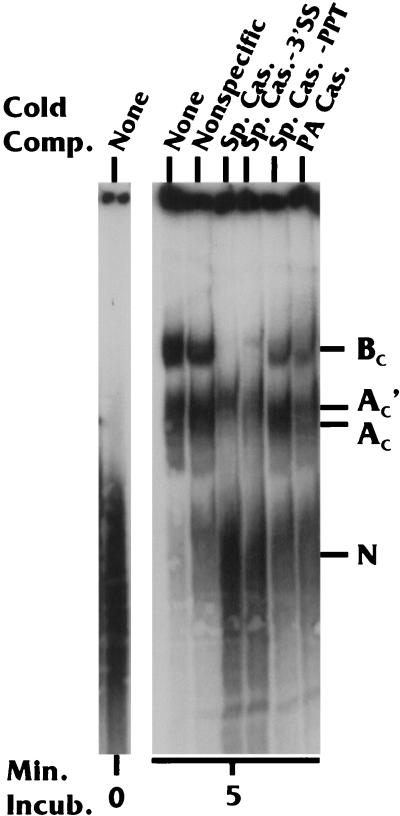
In competition analysis experiments, complex formation on the WT Sp/PA substrate was challenged with excess cold RNA competitors consisting of WT and mutant splicing and PA cassettes. Nonspecific competitor had only a modest effect on complex formation compared to the sample with no RNA competitor (Fig. 8). When the splicing cassette (Fig. 6) was used as the competitor, formation of the BC complex was abolished, and the migration of the AC and AC′ complexes may have been altered. To determine which elements of the splicing cassette were significant in the inhibition of complex formation, we introduced splicing cassette competitors containing the point mutation in the 3′-SS or the LSM in the PPT. The −3′-SS competitor remained very effective in complex inhibition; however, the −PPT competitor was not as effective, since it only moderately inhibited the formation of the BC complexes. These data suggest that while both the PPT and the 3′-SS are important elements involved in coupling complex formation, the PPT appears to be more influential, since its mutation formed a much less effective competitor. The competitor containing the PA cassette (Fig. 1) primarily competed the formation of the BC complexes. In fact, complete inhibition of the BC complexes could be obtained with increased amounts of the PA cassette inhibitor (data not shown).
FIG. 8.
Complex formation on the WT Sp/PA substrate in the presence of specific cold competitor (Comp.) RNAs. None, no competitor; Nonspecific, a nonspecific RNA containing no processing elements; Sp. Cas., wild-type splicing cassette; Sp. Cas. −3′SS, splicing cassette with a point mutation in the 3′-SS; Sp. Cas. −PPT, splicing cassette with an LSM in the PPT; PA Cas., PA cassette.
The cumulative data from Fig. 8 show that the coupling complexes, especially the BC complexes, can be inhibited by competition with either the splicing cassettes or the PA cassettes. This agrees with the results presented above and further supports the conclusion that the BC complex is a coupling complex.
DISCUSSION
In previous studies, our laboratory (11) and others (3, 4, 26, 29, 40) have examined the roles of individual elements of splicing and PA signals in the coupling of PA and splicing. These in vitro experiments have shown that the major effectors of coupling are the PPT, the 3′-SS, the AAUAAA sequence, and the DSE of the PA signal. In order to understand more about the coupling functions of these elements, we examined specific protein-RNA complexes that form on RNA substrates capable of coupled splicing and PA. We hypothesized that formation of a coupling complex would be adversely affected by mutation of either splicing or PA elements known to be necessary for coupling. We have identified at least three specific complexes—AC′, AC, and BC—which form rapidly, well before the appearance of processing products (Fig. 4). This agrees with previous data suggesting that coupling is a very early step in processing (25, 40). The AC′ complex forms by 30 s after mixing, the AC complex forms between 1 and 2 min of mixing, and BC forms by 2 to 3 min after mixing. The data suggest that AC′ is a precursor to AC. This is indicated by the following: (i) the kinetics of appearance and disappearance of the AC′ complex compared with the appearance of the AC complex (Fig. 1); (ii) the analysis of complex formation on the −3′-SS Sp/PA substrate (Fig. 5), where the AC′ complex formed and accumulated to high levels over time, although there was no formation of either the AC or BC complexes. The data also suggest that the AC′ and AC complexes are precursors of the BC complex. This is indicated by the kinetics of formation of the BC complex compared with those of the AC′ and AC complexes. In addition, some experiments have shown that the AC′ and AC complexes accumulated under conditions in which the BC complex does not form (see, for example, the DM4 mutation in Fig. 6).
The AC′ complex has characteristics that suggest that it is primarily splicesomal in nature, analogous to the spliceosomal A complex, which forms on a splicing-only substrate. In addition, a PA-specific complex (PA complex) forms relatively stably on PA-only substrates (Fig. 5). Our data (i) show that both the AC′ and PA complexes form very transiently on a WT Sp/PA substrate and (ii) suggest that they are the primary complexes formed, respectively, on the splicing elements and PA signal. We suggest that the AC′ and PA complexes combine to form coupling-specific complexes on the Sp/PA substrate. The cumulative complex formation data suggest that the BC complex is the most definitive coupling-specific complex, since its formation is strongly affected by mutations in both splicing elements (e.g., the 3′-SS and PPT) and PA signal elements (e.g., AAUAAA and DSE). In addition, the formation of the BC complex is significantly diminished under Mg2+ conditions known to uncouple splicing and PA.
The competition data (Fig. 8) support the present and previous observations (11, 23, 26, 40) that coupling complexes depend on the PPT, the 3′-SS, the AAUAAA sequence, and the DSEs. The contribution of the PPT was particularly striking in the competition analysis. In addition, the observation that only a PA-specific complex formed on the coupled substrate mutated in the PPT (Fig. 5) supports a pivotal role of the PPT in coupling. These data support previous findings suggesting that the binding of U2AF to the PPT is involved in the coupling of splicing and PA through interaction with poly(A) polymerase (40).
We suggest that formation of the AC′ complex is dependent on the PPT. This is based on the competition data and the observation that the AC′ complex forms when either the 3′-SS or the PA signal is mutated, but not when the PPT is mutated (Fig. 5 and 6). The transition from AC′ to AC appears to require an intact 3′-SS, since AC does not appear when the 3′-SS is mutated, and its formation is not significantly affected by mutations in the PA signal. Finally, as previously mentioned, the formation of the BC complex requires both splicing elements and the PA signal and may represent a complex containing both the AC′ and PA complexes.
In previous studies of coupling, we have suggested that the RNA-protein complexes formed to define an exon may become inhibitory to processing if definition of a subsequent exon fails (10, 11). This finding suggests a mechanism to monitor the successful definition of exons in a precursor RNA and to allow only those successfully defined precursors to be processed efficiently. Such a monitoring mechanism is again suggested by the present data. The separate splicing and PA cassettes (Fig. 1) can be processed in vitro very efficiently; however, as the present data show, when the cassettes are attached, a specific mutation in one cassette will decrease the processing of the other, wild-type, cassette. Thus, a functional processing complex formed on the wild-type cassette is inhibited by the abrogated complex formation on the other cassette. This model implies that the coupling mechanism is dominant over individual processing reactions. Thus, it may act as a dominant checkpoint at which aberrant processing at one site will override the normal processing of the surrounding, otherwise wild-type, sites.
Acknowledgments
We thank all of the members of the Alwine Laboratory for helpful discussion and support. We also thank Sherri Adams for critical reading of the manuscript.
This work was supported by Public Health Service grants R01 GM45773 and P01 CA72765.
REFERENCES
- 1.Bagga, P. S., G. K. Arhin, and J. Wilusz. 1998. DSEF-1 is a member of the hnRNP H family of RNA-binding proteins and stimulates pre-mRNA cleavage and polyadenylation in vitro. Nucleic Acids Res. 26:5343-5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bagga, P. S., L. P. Ford, F. Chen, and J. Wilusz. 1995. The G-rich auxiliary downstream element has distinct sequence and position requirements and mediates efficient 3′ end pre-mRNA processing through a trans-factor. Nucleic Acids Res. 23:1625-1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berger, S. L., and W. R. Folk. 1985. Differential activation of RNA polymerase III-transcribed genes by the polyoma virus enhancer and adenovirus E1A gene products. Nucleic Acids Res. 13:1413-1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berget, S. M. 1995. Exon recognition in vertebrate splicing. J. Biol. Chem. 270:2411-2414. [DOI] [PubMed] [Google Scholar]
- 5.Brackenridge, S., H. L. Ashe, M. Giacca, and N. J. Proudfoot. 1997. Transcription and polyadenylation in a short human intergenic region. Nucleic Acids Res. 25:2326-2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown, P. H., L. S. Tiley, and B. R. Cullen. 1991. Efficient polyadenylation with the human immunodeficiency virus type 1 long terminal repeat requires flanking U3-specific sequences. J. Virol. 65:3340-3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carswell, S., and J. C. Alwine. 1989. Efficiency of utilization of the simian virus 40 late polyadenylation site: effects of upstream sequences. Mol. Cell. Biol. 9:4248-4258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chiou, H. C., C. Dabrowski, and J. C. Alwine. 1991. Simian virus 40 late mRNA leader sequence involved in augmenting mRNA accumulation via multiple mechanisms, including increased polyadenylation efficiency. J. Virol. 65:6677-6685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conway, L., and M. Wickens. 1985. A sequence downstream of AAUAAA is required for formation of simian virus 40 late mRNA 3′ termini in frog oocytes. Proc. Natl. Acad. Sci. USA 82:3949-3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cooke, C., and J. C. Alwine. 1996. The cap and 3′-splice site similarly affect polyadenylation efficiency. Mol. Cell. Biol. 16:2579-2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cooke, C., H. Hans, and J. C. Alwine. 1999. Utilization of splicing elements and polyadenylation signal elements in the coupling of splicing and last-intron removal. Mol. Cell. Biol. 19:4971-4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeZazzo, J. D., and M. J. Imperiale. 1989. Sequences upstream of AAUAAA influence poly(A) site selection in a complex transcription unit. Mol. Cell. Biol. 9:4951-4961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeZazzo, J. D., J. E. Kilpatrick, and M. J. Imperiale. 1991. Involvement of long terminal repeat U3 sequences overlapping the transcription control region in human immunodeficiency virus type I mRNA 3′ end formation. Mol. Cell. Biol. 11:1624-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hans, H., and J. C. Alwine. 2000. Functionally significant secondary structure of the simian virus 40 late polyadenylation signal. Mol. Cell. Biol. 20:2926-2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hong, W., M. Bennett, Y. Xiao, R. Feld-Kramer, C. Wang, and R. Reed. 1997. Association of U2 snRNP with the spliceosomal complex E. Nucleic Acids Res. 25:354-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Izaurralde, E., J. Lewis, C. McGuigan, M. Jankowska, E. Darzynkiewicz, and I. W. Mattaj. 1994. A nuclear cap binding complex involved in pre-mRNA splicing. Cell 78:657-668. [DOI] [PubMed] [Google Scholar]
- 17.Lewis, J. D., E. Izaurralde, A. Jarmolowski, C. McGuigan, and I. W. Mattaj. 1996. A nuclear cap-binding complex facilitates association of U1 snRNP with the cap-proximal 5′ splice site. Genes Dev. 10:1683-1698. [DOI] [PubMed] [Google Scholar]
- 18.Lutz, C. S., K. G. Murthy, N. Schek, J. L. Manley, and J. C. Alwine. 1996. Interaction between the U1snRNP-A protein and the 160 kD subunit of cleavage-polyadenylation specificity factor increases polyadenylation efficiency in vitro. Genes Dev. 10:325-337. [DOI] [PubMed] [Google Scholar]
- 19.Moore, C. L., and P. A. Sharp. 1984. Site-specific polyadenylation in a cell-free reaction. Cell 36:581-591. [DOI] [PubMed] [Google Scholar]
- 20.Moreira, A., M. Wollerton, J. Monks, and N. J. Proudfoot. 1995. Upstream sequence elements enhance poly(A) site efficiency of the C2 complement gene and are phylogenetically conserved. EMBO J. 14:3809-3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nesic, D., J. Cheng, and L. E. Maquat. 1993. Sequences within the last intron function in RNA 3′-end formation in cultured cells. Mol. Cell. Biol. 13:3359-3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nesic, D., and L. E. Maquat. 1994. Upstream introns influence the efficiency of final intron removal and RNA 3′-end formation. Genes Dev. 8:363-375. [DOI] [PubMed] [Google Scholar]
- 23.Niwa, M., and S. M. Berget. 1991. Mutation of the AAUAAA polyadenylation signal depresses in vitro splicing of proximal but not distal introns. Genes Dev. 5:2086-2095. [DOI] [PubMed] [Google Scholar]
- 24.Niwa, M., and S. M. Berget. 1991. Polyadenylation precedes splicing in vitro. Gene Expr. 1:5-14. [PMC free article] [PubMed] [Google Scholar]
- 25.Niwa, M., C. C. MacDonald, and S. M. Berget. 1990. Are vertebrate exons scanned during splice site selection? Nature (London) 360:277-280. [DOI] [PubMed] [Google Scholar]
- 26.Niwa, M., S. D. Rose, and S. M. Berget. 1990. In vitro polyadenylation is stimulated by the presence of an upstream intron. Genes Dev. 4:1552-1559. [DOI] [PubMed] [Google Scholar]
- 27.Qian, Z., and J. Wilusz. 1991. An RNA-binding protein specifically interacts with a functionally important domain of the downstream element of the simian virus 40 late polyadenylation signal. Mol. Cell. Biol. 11:5312-5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reed, R. 2000. Mechanisms of fidelity in pre-mRNA splicing. Curr. Opin. Cell Biol. 12:340-345. [DOI] [PubMed] [Google Scholar]
- 29.Robberson, B. L., G. J. Cote, and S. M. Berget. 1990. Exon definition may facilitate splice site selection in RNAs with multiple exons. Mol. Cell. Biol. 10:84-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Russnak, R. 1991. Regulation of polyadenylation in hepatitis B viruses: stimulation by the upstream activating signal PS1 is orientation-dependent, distance-dependent, and additive. Nucleic Acids Res. 19:6449-6456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Russnak, R., and D. Ganem. 1990. Sequences 5′ to the polyadenylation signal mediate differential poly(A) site use in hepatitis B viruses. Genes Dev. 4:764-776. [DOI] [PubMed] [Google Scholar]
- 32.Sadofsky, M., and J. C. Alwine. 1984. Sequences on the 3′ side of hexanucleotide AAUAAA affect efficiency of cleavage at the polyadenylation site. Mol. Cell. Biol. 4:1460-1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sadofsky, M., S. Connelly, J. L. Manley, and J. C. Alwine. 1985. Identification of a sequence element on the 3′ side of AAUAAA which is necessary for simian virus 40 late mRNA 3′-end processing. Mol. Cell. Biol. 5:2713-2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sanfacon, H., P. Brodmann, and T. Hohn. 1991. A dissection of the cauliflower mosaic virus polyadenylation signal. Genes Dev. 5:141-149. [DOI] [PubMed] [Google Scholar]
- 35.Schek, N., C. Cooke, and J. C. Alwine. 1992. Definition of the upstream efficiency element of the simian virus 40 late polyadenylation signal by using in vitro analyses. Mol. Cell. Biol. 12:5386-5393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scott, J. M., and M. J. Imperiale. 1996. Reciprocal effects of splicing and polyadenylation on human immunodeficiency virus type 1 pre-mRNA processing. Virology 224:498-509. [DOI] [PubMed] [Google Scholar]
- 37.Staknis, D., and R. Reed. 1994. SR proteins promote the first specific recognition of pre-mRNA and are present together with the U1 small nuclear ribonucleoprotein particle in a general splicing enhancer complex. Mol. Cell. Biol. 14:7670-7682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takagaki, Y., and J. L. Manley. 1997. RNA recognition by the human polyadenylation factor CstF. Mol. Cell. Biol. 17:3907-3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsai, T. F., M. J. Wu, and T. S. Su. 1998. Usage of cryptic splice sites in citrullinemia fibroblasts suggests role of polyadenylation in splice-site selection during terminal exon definition. DNA Cell Biol. 17:717-725. [DOI] [PubMed] [Google Scholar]
- 40.Vagner, S., C. Vagner, and I. W. Mattaj. 2000. The carboxyl terminus of vertebrate poly(A) polymerase interacts with U2AF 65 to couple 3′-end processing and splicing. Genes Dev. 14:403-413. [PMC free article] [PubMed] [Google Scholar]
- 41.Valsamakis, A., N. Schek, and J. C. Alwine. 1992. Elements upstream of the AAUAAA within the human immunodeficiency virus polyadenylation signal are required for efficient polyadenylation in vitro. Mol. Cell. Biol. 12:3699-3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Valsamakis, A., S. Zeichner, S. Carswell, and J. C. Alwine. 1991. The human immunodeficiency virus type 1 polyadenylation signal: a 3′-LTR element upstream of the AAUAAA necessary for efficient polyadenylation. Proc. Natl. Acad. Sci. USA 88:2108-2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wilusz, J., D. I. Feig, and T. Shenk. 1988. The C proteins of heterogeneous nuclear ribonucleoprotein complexes interact with RNA sequences downstream of polyadenylation cleavage sites. Mol. Cell. Biol. 8:4477-4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wilusz, J., and T. Shenk. 1990. A uridylate tract mediates efficient heterogeneous nuclear ribonucleoprotein C protein-RNA cross-linking and functionally substitutes for the downstream element of the polyadenylation signal. Mol. Cell. Biol. 10:6397-6407. [DOI] [PMC free article] [PubMed] [Google Scholar]