Abstract
Deficiency of liver arginase (AI) causes hyperargininemia (OMIM 207800), a disorder characterized by progressive mental impairment, growth retardation, and spasticity and punctuated by sometimes fatal episodes of hyperammonemia. We constructed a knockout mouse strain carrying a nonfunctional AI gene by homologous recombination. Arginase AI knockout mice completely lacked liver arginase (AI) activity, exhibited severe symptoms of hyperammonemia, and died between postnatal days 10 and 14. During hyperammonemic crisis, plasma ammonia levels of these mice increased >10-fold compared to those for normal animals. Livers of AI-deficient animals showed hepatocyte abnormalities, including cell swelling and inclusions. Plasma amino acid analysis showed the mean arginine level in knockouts to be approximately fourfold greater than that for the wild type and threefold greater than that for heterozygotes; the mean proline level was approximately one-third and the ornithine level was one-half of the proline and ornithine levels, respectively, for wild-type or heterozygote mice—understandable biochemical consequences of arginase deficiency. Glutamic acid, citrulline, and histidine levels were about 1.5-fold higher than those seen in the phenotypically normal animals. Concentrations of the branched-chain amino acids valine, isoleucine, and leucine were 0.4 to 0.5 times the concentrations seen in phenotypically normal animals. In summary, the AI-deficient mouse duplicates several pathobiological aspects of the human condition and should prove to be a useful model for further study of the disease mechanism(s) and to explore treatment options, such as pharmaceutical administration of sodium phenylbutyrate and/or ornithine and development of gene therapy protocols.
Arginase (EC 3.5.3.1) is the fifth and final enzyme of the urea cycle, the major pathway for the detoxification of ammonia in mammals. There are at least two forms of arginase in mammals, AI and AII, located in the cytoplasm and mitochondrion, respectively. The principal ureagenic enzyme activity (AI) is most abundant in normal mammalian liver and acts in coordination with the other enzymes of the urea cycle to sequester and eliminate excess nitrogen from the body (7). The second form (AII) is found in many organs, with the highest levels found in kidney and prostate and lower levels in macrophages, lactating mammary glands, and brain, often in the absence of the other urea cycle enzymes (7, 18). In humans, deficiency of the liver isoform (AI) causes hyperargininemia (OMIM 207800; Online Mendelian Inheritance in Man [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=OMIM]), a metabolic disorder characterized by neurological impairment, with deterioration of the cortex and pyramidal tracts and progressive dementia, spasticity, and growth retardation and punctuated by infrequently fatal episodes of hyperammonemia (7). Some of the symptoms of AI deficiency can be partially alleviated by dietary and pharmaceutical interventions, but no completely effective therapy or cure is currently available.
The human arginase AI locus is about 12 kb long, consists of eight exons and seven introns (14, 16), and is located at chromosome band 6q23 in the human genome (14). Comparison of the genomic structure of human and rodent arginase AI genes showed a high degree of conservation of both exon and intron numbers, sizes, and arrangement. The human (3, 6) (GenBank accession no. NM000045 and M14502 [http://www.ncbi.nlm.nih.gov/Entrez/]), mouse (GenBank accession no. NM007482 and AB047402), and rat (4, 11; GenBank accession no. NM017134 and J02720) AI cDNAs have all been cloned and encode proteins of very similar size (322 amino acids for human and 323 amino acids for mouse and rat). Comparison of amino acid sequences from human and rat and a number of lower organisms revealed extremely high amino acid homology in six conserved regions of all known arginases (7, 9). Three highly conserved histidine residues (at positions 101, 126, and 141), along with adjacent aspartates (a functional arrangement found in many hydrolytic enzymes), are found within these highly conserved areas and are critical for enzyme function. X-ray crystallographic analysis of the rat liver arginase at 2.1 Å resolution showed that the enzyme exists as a trimer (10). Each subunit requires and binds two Mn2+ residues for function, and each monomeric subunit is able to catalyze the hydrolytic reaction independently (12). The location and types of mutations found in human arginase deficiency have been extensively studied and catalogued (1, 17, 19). As might be expected, all natural missense mutations were found in a highly conserved residue and all but one was found in a conserved region of 5 or more amino acids. In contrast, natural chain termination or nonsense mutations appear to occur randomly in the molecule. Although higher arginine levels and absent enzyme activity occur with the more severe nonsense mutations, no clinically useful correlations between the specific mutation, the severity of disease, or the ability to control arginine levels have been found (7).
Unlike patients with severe deficiencies of the other enzymes of the urea cycle, AI-deficient patients have fewer hyperammonemic crises, the few cases of which are often less severe and more easily managed; AI deficiency is rarely if ever fatal in the perinatal or neonatal period. Also unlike the other deficiencies, ureagenesis is not completely abolished by arginase AI deficiency. A logical explanation for these phenomena is provided by the observation that kidney arginase (AII) activity is often significantly (3- to 34-fold) enhanced in AI-deficient patients during hyperammonemic episodes (5). That this increase in arginase activity is mediated by increased expression of the AII gene in response to high arginine levels is supported by the documented increases in arginase AII RNA, protein, and enzyme activity in human embryonic kidney cells grown in arginine-rich media (our unpublished data). The increased kidney arginase activity probably catalyzes a reduced level of ureagenesis, thus partially compensating for the lack of AI activity, and may provide a partial explanation for the longevity of these patients compared to those with other urea cycle disorders.
The stereotypical symptoms of AI deficiency and the differences in symptoms, progression, and outcome compared to those of the other urea cycle disorders suggest a unique pathogenic mechanism. Though the origin of the excess ammonia is apparent, as is the liver injury caused by this acute, toxic insult, the neurological symptoms are too subtle and slowly progressive to be caused solely by hyperammonemia. Little or nothing is known about the identity of the proximate neurotoxin or the actual mechanism of injury to the brain that causes the progressive neurological deterioration and mental retardation in AI deficiency. In order to study the pathophysiology of AI deficiency systematically and in the absence of any natural animal model of the disease, we constructed a knockout mouse strain carrying a nonfunctional AI gene. This model may serve to develop approaches to the therapy for this disorder, helped only incompletely by dietary treatment and ammonia diversion in the affected patients.
MATERIALS AND METHODS
Isolation of AI genomic clones.
A mouse ES129 genomic library in λFixII (Stratagene, La Jolla, Calif.) was probed with the AI cDNA by using standard techniques. Two clones spanning the entire genomic sequence of mouse AI were obtained and confirmed by Southern blotting (data not shown), as well as PCR to detect the presence of each of the eight exons of arginase AI (data not shown).
Construction of the gene-targeting vector.
AI genomic sequences constituting the 5′ and 3′ homologous arms of the AI gene-targeting vector were amplified by long-range, high-fidelity PCR with Pfu or Pfu Turbo DNA polymerase (Stratagene) by using specific primers obtained from Life Technologies (Bethesda, Md.). The primer pair comprised of MAI/E4/R/S (5′ AAACTCTTCAGATGCTTCCAACTGCCAG 3′) and MAI/E2/F/S (5′ AAACTCTTCATTCAGCCTCGAGGAGGGGTA 3′) was used to amplify a 4.5-kb AI genomic fragment from exon 2 to intron 3 (the 5′ arm of the targeting vector). Besides being complementary to AI genomic sequences, these primers also contained sequences needed to clone the products with the Seamless cloning kit (Stratagene), as well as the unique restriction sites, for excision of the fragment. Primers RAI/E4I4b/F (5′ AAAGGTAAAAGGCTCCTCAGAAC 3′) and MAIR1 (5′ GGGAATTCACAGTCATTTCTTAGGTGGTTT 3′) were used to amplify a 2.6-kb fragment encompassing an AI genomic sequence from intron 4 to exon 8, using Pfu DNA polymerase. PCR conditions were as recommended by the manufacturer (Stratagene). Both fragments were cloned into the plasmid pBluescriptIIKS(+) (Stratagene). The insert fragments were sequenced to confirm the integrity of each AI exon and intron-exon boundary regions, including splice site elements. Despite repeated attempts, we were unable to introduce the 5′ (AI exon 2-intron 3) and 3′ (AI intron 5-exon 8) arms together into the 5′ and 3′ sides of the neomycin resistance (NeoR) gene in the backbone vector pPNT, probably due to the presence of inverted repeat elements in the intronic sequences (data not shown). Hence, we introduced the NeoR and herpes simplex virus thymidine kinase (HSV-tk) genes from pPNT into the 5′ and 3′ sides, respectively, of the mouse AI intron 4-exon 8 fragment. An ∼7.0-kb fragment containing the NeoR gene, the 3′ AI arm, and the HSV-tk gene was cut out of this construct by using EcoRI and XhoI (New England Biolabs, Beverly, Mass.) and gel purified with the QIAEX gel purification kit (Qiagen, Chatsworth, Calif.). The 4.5-kb 5′ AI arm was cut out of pBluescriptIIKS(+) by using XhoI and EcoRI and purified by the same methodology. A four-way ligation of these fragments with the XhoI-cut long and short arms of the phage vector λFixII (Stratagene) was performed with T4 DNA ligase (New England Biolabs), and the ligation product was packaged with the GigaPack kit (also from Stratagene), all according to the manufacturer's recommendations. Plaques were obtained by infection of the Escherichia coli host strain, XLI-BLUEMRAP2, with the packaged product and plating on Luria-Bertani plates according to standard protocols. Over 400 isolated plaques were screened by a combination of plaque lifts and hybridization and by short- and long-range PCR with vector- and insert-specific primers to identify five correctly assembled clones constituting AI gene-targeting vector candidates (data not shown). The identities of these clones were further confirmed by direct sequencing of the insert, including AI sequences and cloning boundaries. The resultant AI gene replacement vector (see Fig. 1) contained a NeoR cassette flanked 5′ by 4.5 kb (exon 2-intron 3) and 3′ by 2.6 kb (intron 5-exon 8) of the homologous AI sequence. In this construct, the 163-bp-long AI exon 4, which contains several highly conserved residues, including His101, a residue implicated in Mn2+ binding, was replaced by the NeoR cassette. Hence, any polypeptide produced by alternative or cryptic splicing would lack arginase activity. The HSV-tk cassette for selection against nonhomologous recombinants is located 3′ of the 2.6-kb fragment.
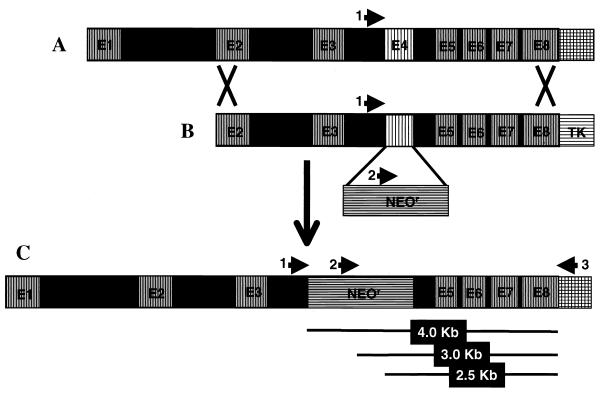
FIG. 1.
Schematic diagram of the AI gene-targeting event. (A) The genomic structure of mouse arginase I (AI) with eight exons and seven introns. (B) The AI gene-replacement vector, containing AI genomic sequences from exon 2 (E2) to exon 8 (E8): exon 4 (E4) is replaced by the neomycin resistance (NeoR) gene, and the HSV-tk (TK) gene provides strong selection against nonhomologous intergration events. (C) The product of homologous recombination of the AI gene-replacement vector into the mouse genome. The arrows represent the locations of primers used for genotyping the resultant ES cells and mice. 1, primer MAIF/I3; 2, primer NeoF(412); 3, primer MAIR3′UTR(∗+59). PCR A contains primers 1 and 2, and reaction B contains primers 2 and 3. The lines below segment C show the approximate sizes of the different products obtained. See Fig. 2 for an example of results obtained from wild-type, heterozygous, and AI-deficient mice.
Derivation of the AI-deficient mouse.
Embryonic stem (ES) cell and embryo manipulations for construction of the AI-deficient mice, including electroporation of the vector into ES cells, microinjection of three selected clones into blastocysts, and implantation of these chimeric embryos into pseudopregnant female mice, were performed in the University of California Los Angeles transgenic core facility by using now-standard methodologies. Briefly, for each of the three selected clones, 200 μg (larger amounts of DNA were needed due to the large size of the lambda backbone relative to that of the plasmid) of NotI-digested AI gene-targeting vector DNA was introduced into ES129 cells by electroporation. ES cell clones were grown in the presence of G418 antibiotic to select for clones carrying the neomycin resistance marker (i.e., presence of the gene replacement vector) and then treated with ganciclovir to select for loss of HSV-tk (an inevitable consequence of an homologous recombination event). Double-selected clones were amplified, and genomic DNA was extracted by using standard techniques. Clones containing a disrupted AI allele resulting from a single homologous recombination event were identified by short- and long-range PCR using primers located within the untargeted regions of the AI gene [primers MAIF/I31 (5′ AACCAGCACCTCTAAGGTCTATGG 3′) and MAI3′UTR(∗+59) (5′ TTGGGAGGAGAAGGCGTTTGC 3′)] in combination with ones located within the NeoR [Neo3′FL (5′ CTTCTGAGGGGATCGATCCGTCCTGTAAGT 3′) and Neo5′RL (5′ CACCCAAGCGGCCGGAGAACCTGCGTGCAA 3′)] and HSV-tk [TK5′R (5′ CCAAGCGGCCGGAGAACCTGCGTGCAA 3′)] genes. The identities of these clones were confirmed by sequencing of some of the resultant fragments (data not shown). Out of over 100 ES cell clones screened in this manner, 6 were confirmed to contain a disrupted AI allele, and 3 of these clones were used to obtain 12 male, AI-chimeric founder mice by introducing the ES cells into blastocysts by microinjection according to established protocols. Mice heterozygous for the AI gene were obtained by breeding three of these founders (from two different ES cell clones) with 6-week-old C57BL/6 females.
Genotyping of the mice.
Genomic DNA from tail or toe samples from 3-week-old progeny of these matings was extracted by standard protocols, and heterozygotes were identified by PCR using primers located in AI genomic segments common to both wild-type and disrupted AI alleles [primers MAIF/I31 and MAI3′UTR(∗+59)] in combination with one located within the NeoR gene [primer NeoF(412) (5′ GCCCATTCGACCACCAAG 3′)] (see Fig. 2 for location of primers and Fig. 3 for typical results obtained). Mice heterozygous for the disrupted AI allele (AI+/−) were mated to obtain animals homozygous for the disrupted AI allele (AI−/−). For genotyping the progeny of heterozygote matings, genomic DNA was extracted by standard techniques from tail or toe snip samples from 6- to 10-day-old pups and used as template for PCR as described above.
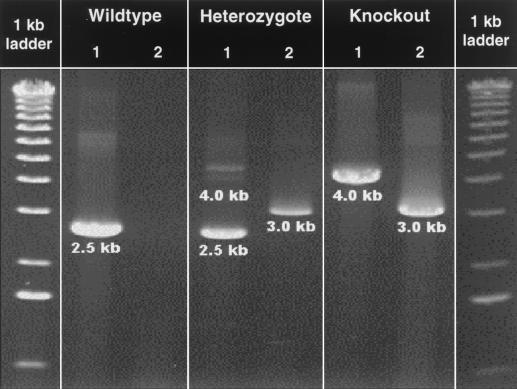
FIG. 2.
Genotyping of wild-type, heterozygous, and AI-deficient mice by long-range PCR. Reaction A (lanes 1) contains two AI-specific primers, one located within the targeted region (MAIF/I3) and one outside the targeted region of the AI gene [MAIR3′UTR(∗+59)]. Reaction B (lanes 2) contains a NeoR gene-specific primer [NeoF(412)] and an AI-specific primer located outside the targeted region [MAIR3′UTR(∗+59)]. For location of the primers on the wild-type and modified AI genes, see Fig. 1. As expected, wild-type mice exhibited a 2.5-kb band in reaction A but no product in reaction B; heterozygous mice exhibited a 2.5-kb and a 4.0-kb (often very weak or absent) band in reaction A and a 3.0-kb band in reaction B; and AI-deficient mice exhibited a strong 4.0-kb band in reaction A and a 3.0-kb band in reaction B.
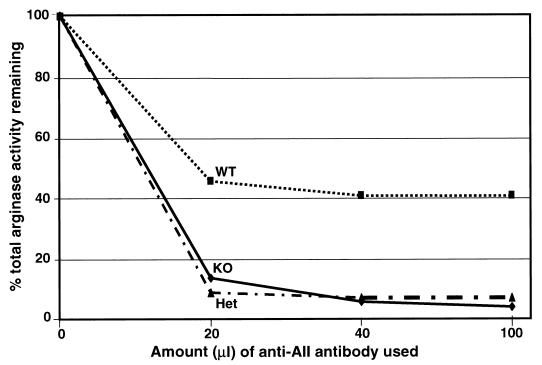
FIG. 3.
Identification of arginase isoforms in kidney extracts of wild-type (WT), heterozygous (Het), and AI-deficient (KO) mice by immunoprecipitation with an anti-rat AII antibody. Only ∼60% of the arginase activity from kidney extracts of wild-type mice was precipitated by the anti-AII antibody, in agreement with previous data obtained. Almost 100% of arginase activity in AI-deficient and heterozygous mice was precipitated by this antibody, thus proving that arginase AI is not expressed in any organ of the AI knockout mouse. The reason for the lack of arginase AI activity in the kidneys of heterozygous mice is unclear at this time.
Arginase activity assays.
Arginase activities of all samples were assayed by the method of Schimke (13), as modified by Spector et al. (15).
Identification of arginase isoforms by immunoprecipitation.
Approximately 200 mg of frozen tissue (brain, kidney, or liver) was thawed on ice and immediately homogenized in a mixture containing 0.5 ml of 20 mM Tris (pH 7.4), 100 mM KCl, and 10 mM MnCl2. Insoluble materials were pelleted by centrifugation, and aliquots containing about 0.25 to 0.5 U of arginase were mixed with increasing amounts of a rabbit anti-rat mammary gland arginase II antibody (8). Preimmune rabbit serum was added to normalize the amount of serum protein in each sample. Samples were incubated for 30 min at 37°C and then kept at 4°C overnight. Antigen-antibody complexes were removed by addition of IgGSorb (The Enzyme Center, Inc., Boston, Mass.) and centrifugation. The arginase activity remaining in the supernatant was measured as described previously (15).
Determination of plasma ammonia concentrations.
Blood was drawn by cardiac puncture from AI-deficient animals in the final stages of life, as well as from normal and heterozygous animals, which was sacrificed at the same time. The blood was collected in heparinized tubes, cells were removed by centrifugation, and plasma ammonia concentrations were assayed immediately with a plasma ammonia assay kit (Sigma Chemical Co., St. Louis, Mo.) according to the manufacturer's recommendations.
Plasma amino acid analysis.
Blood obtained by cardiac puncture (and collected in heparinized tubes) from four homozygous AI knockout, four wild-type, and two heterozygous animals was centrifuged to remove cells. The plasma was frozen on dry ice and shipped to the Clinical Biochemistry Laboratory, directed by William E. O'Brien, at the Baylor College of Medicine, Houston, Tex. After ninhydrin derivatization, amino acid concentrations were determined by high-performance liquid chromatography.
Gross and histopathological examination of mouse tissues.
For the purpose of postmortem examination, mice were sacrificed by CO2 asphyxiation. For light microscopy, tissues were immersion fixed in 4% formalin (except for the lungs, which were always inflated with fixative before immersion) for 48 h. The maximum time between death and tissue fixation was 10 min. After fixation, tissues were routinely processed and embedded in paraffin and 4-μm-thick sections were cut and stained with hematoxylin and eosin stain. Other special stains used as required were the periodic acid-Schiff, Picro Mallory trichrome, Alcian blue, and luxol fast blue-Crescil violet stains (2).
RESULTS
We used three primers, two located in the AI genomic regions and one in the NeoR gene (Fig. 1), in two combinations to genotype all our animals. As expected, wild-type mice gave a single strong ∼2.5-kb band in the reaction with AI-specific primers but no bands in that containing the AI plus NeoR-specific primers. Heterozygotes produced a 2.5-kb and a 4.0-kb band (often very faint or not visible) with the AI-specific primers and a strong ∼3.0-kb band with the AI plus NeoR-specific primers. The weakness or lack of the 4.0-kb band may be explained by the fact that the smaller of two possible products is often favored during PCR. Finally, homozygous knockout animals produced a 4.0-kb band with the AI-specific primers and a 3.0-kb band with the AI plus NeoR-specific primers (Fig. 2). Genotypic analysis of 402 offspring of AI heterozygote matings by this method revealed a non-Mendelian distribution of genotypes (39.1% wild type, 39.6% heterozygotes, 21.4% knockout), with a significant deficit of the knockout allele in the population (Table 1). However, the fact that the genotypes were in Hardy-Weinberg equilibrium weakens the possibility that the deficit was due to a loss of individuals carrying the disrupted alleles in utero and suggests the possibility that sperm lacking the capability to produce arginase AI may be less “fit” and may participate in fertilization less often than those carrying the wild-type allele.
TABLE 1.
Distribution of genotypes among 402 progeny of AI heterozygote matings
| Genotype | No. of phenotypes
|
(Observed − expected)2/ expected | P value | |
|---|---|---|---|---|
| Observed | Expected | |||
| AI+/+ | 157 | 100.5 | 31.763 | ≫0.001 |
| AI+/− | 159 | 20.1 | 8.776 | |
| AI−/− | 86 | 100.5 | 2.092 | |
| Total | 402 | 402 | 42.631 | |
AI-deficient mice completely lacked liver arginase (AI) activity (Table 2), exhibited severe symptoms of hyperammonemia, and died at postnatal days 10 to 14 (usually between days 10 and 12). Unlike human patients, in whom kidney arginase activity ascribed to the AII isoform is induced 3- to 34-fold during hyperammonemic episodes, no increase in total arginase activity was found in the kidneys of knockout mice; arginase activities in wild-type, heterozygous, and knockout brain lysates were also similar. Immunoprecipitation with a polyclonal rat anti-AII antibody brought down the arginase activity in normal kidneys by only 50 to 60%. In contrast to wild-type mice, the same antibody precipitated nearly all of the arginase activity from the kidneys of AI-deficient mice, confirming the lack of AI expression in the knockout animals (Fig. 3). Surprisingly, the anti-AII antibody also precipitated almost all of the activity from heterozygous mouse kidneys, suggesting that AI expression was also very low in the kidneys of these mice. Comparison of the arginase activity (Table 2) and immunoprecipitation data (Fig. 3) suggests that AII activity is induced twofold in the kidneys of knockout and heterozygous mice compared to that of wild-type animals.
TABLE 2.
Arginase activity in tissue extracts from wild-type, heterozygous, and AI-deficient animals
| Genotype | Arginase activity (U/mg of total protein)a
|
||
|---|---|---|---|
| Liver | Kidney | Brain | |
| AI+/+ | 100.76 ± 49.26 | 0.84 ± 0.25 | 0.28 ± 0.07 |
| AI+/− | 59.17 ± 23.22 | 0.84 ± 0.46 | 0.24 ± 0.09 |
| AI−/− | 1.57 ± 2.25 | 0.81 ± 0.36 | 0.27 ± 0.14 |
Values are given in micromoles of arginine cleaved per 30 min ± standard deviations.
Livers of AI-deficient animals, harvested during these hyperammonemic episodes, were grossly abnormal and exhibited several histopathologic features similar to those seen in the livers of two arginase-deficient patients who died exhibiting symptoms of hyperammonemia (Fig. 4). The livers were often enlarged and pale in appearance compared to those from normal (AI+/+) or heterozygotic (AI+/−) mice. Upon microscopic examination of Giemsa-stained sections, we observed many enlarged hepatocytes, with both nuclei and cytoplasm enlarged two- to three times. This caused a generalized masking of the normal architecture, with undefined plates and collapse of sinusoids, when compared to those in AI+/+ and AI+/− mice. There were also occasional cells or small groups of cells that appeared shrunken, with a homogeneous, densely stained eosinophilic cytoplasm. In addition, there were scattered cells showing lytic necrosis, with fragmentation of the nuclei. The hepatocytes contained a variety of intracytoplasmic inclusions, including large, well-defined, round or oval eosinophilic inclusions suggestive of giant mitochondria. In addition, we observed extralysosomal regions of nonstaining (or pale-blue staining) material with well-delineated but irregular contours suggestive of dilated endoplasmic reticulum (smooth and/or rough), as well as finely granular, intensely eosinophilic areas. Many nuclei were also pale with finely granulated chromatin and occasionally contained diabetic-type inclusions. One (and only one) mouse also showed mild periportal fibrosis and bile duct proliferation. A normal feature seen in all the mice was scattered small foci of hematopoiesis. Nonhepatocytes (Kupffer, Ito, and endothelial cells) were unremarkable. No significant aberrations were seen in the brains of AI knockout mice compared to those of wild-type or heterozygous mice. This is in contrast to the situation in humans, in whom astrocyte defects have been reported. However, this might be the result of the rapidity with which death occurs after the onset of symptoms (≤24 h).
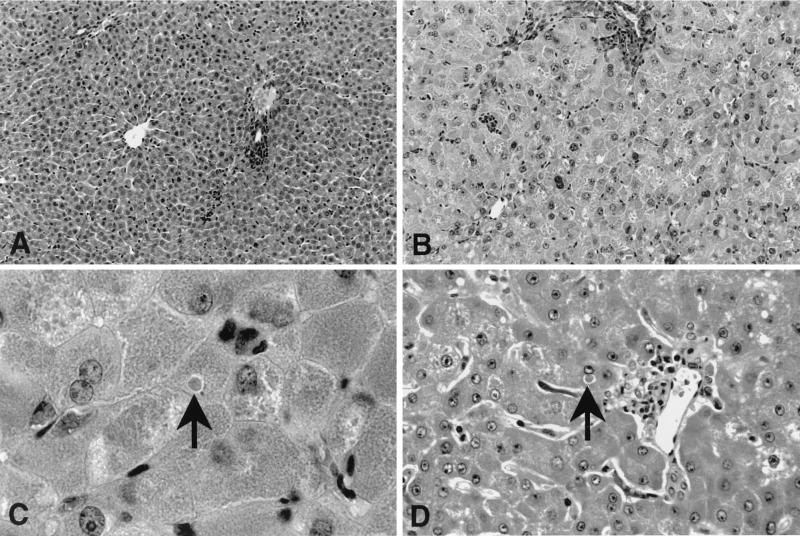
FIG. 4.
Histopathologic analysis of AI wild-type and AI-deficient mouse livers (hematoxylin-and-eosin-stained sections) harvested during hyperammonemic episodes. Typical fields (magnification, ×200) from wild-type (A) and knockout (B) mouse livers are shown. AI-deficient mice exhibited a two- to threefold enlargement of hepatocytes, with several different types of inclusions also present (see Results for a detailed description). High magnification (×1,000) views of arginase-deficient mouse (C) and human (D) liver sections are shown. Hepatocytes in the mouse model shared many histological features with those of human patients (see Results for a complete description), including dense, eosinophilic intracytoplasmic inclusion bodies (arrows).
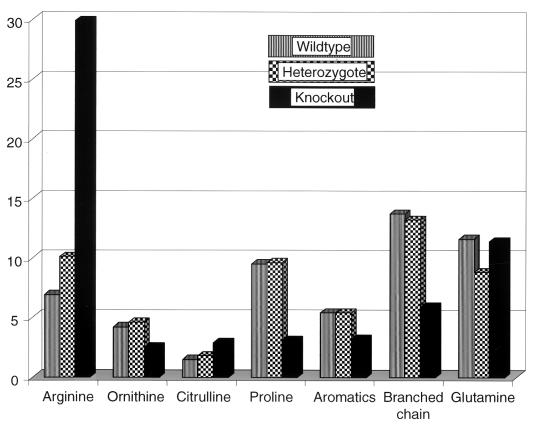
Compared to those of normal animals, plasma ammonia levels of AI-deficient animals, in the throes of metabolic crisis, were increased ≥10-fold (Table 3) to a level that is quite sufficient to explain the liver dysmorphology described above. Glucose and electrolyte (Ca2+, Mg2+, and K+) levels were normal (data not shown). The levels in plasma of several amino acids were also significantly perturbed in the AI-deficient versus the wild-type or heterozygous animals (Fig. 5). Knockout animals, assayed after symptoms appeared, had significantly elevated plasma arginine levels (836.43, 562.05, 572.60, and 299.56 μM in four animals) compared to levels found in the wild type (142.65, 126.79, 182.39, and 182.92 μM in four animals) or heterozygotes (281.57 and 154.03 μM in two animals). Proline levels were significantly depleted in AI-deficient animals (74.00, 28.00, 52.00, and 83.00 μM) versus those in the wild type (230.60, 201.32, 238.00, and 198.00 μM) or heterozygotes (192.00 and 222.00 μM). Ornithine levels were reduced in two of four AI-deficient mice (20.84 and 26.97 μM) but were in the normal (or close to normal) range for the other two (66.64 and 76.85 μM). Analysis of normalized mean values for plasma amino acid concentrations of mice from the three genotypes (Fig. 5) revealed the following. The mean arginine level in knockouts was ∼4-fold greater than that for the wild type and threefold greater than that for heterozygotes, whereas the mean proline level was approximately one-third and ornithine level was one-half of the respective proline and ornithine levels for the wild type or heterozygotes. Glutamic acid, citrulline, and histidine levels were elevated to about 1.5 times that seen in the phenotypically normal set. No significant changes in plasma glutamine concentrations were observed. Concentrations of the branched-chain amino acids valine, isoleucine, and leucine were reduced to 0.4 to 0.5 times concentrations found in the phenotypically normal mice.
TABLE 3.
Plasma ammonia levels of wild-type, heterozygous, and arginase AI-deficient mice
| Genotype | Phenotype | Mean ammonia concn (μmol/liter)a |
|---|---|---|
| AI+/+ | Normal | 554.10 ± 124.22 |
| AI+/− | Normal | 441.23 ± 95.88 |
| AI−/− | Liver arginase-deficient (terminal crisis) | 4,792.60 ± 1,021.02 |
Plasma ammonia values reported for phenotypically normal animals are the means measured in three animals each, whereas those for the AI-deficient animals are the averages of eight measurements. Normal plasma ammonia levels in humans are 12 to 47 μmol/liter. In comparison, normal ammonia levels in mice were much higher (20), at 412 μmol/liter, similar to the values we obtained for our phenotypically normal mice.
FIG. 5.
Perturbation of amino acid concentrations in plasma of AI-deficient mice in hyperammonemic crisis compared to those of wild-type and heterozygous mice. Normalized mean values obtained from four mice each (wild-type and knockout) or two mice (heterozygotic) were plotted (for raw data, see Results). Compared to wild-type animals, AI-deficient mice exhibited hyperargininemia (approximately fourfold increase), hypoornithinemia (one-half normal levels), and significantly reduced proline levels (one-third wild-type levels). Glutamic acid, histidine (not shown), and citrulline levels were elevated to about 1.5 times those seen in phenotypically normal animals. No significant changes in plasma glutamine concentrations were observed. Concentrations of the branched-chain amino acids (valine, isoleucine, and leucine) were reduced to about 50% of those for phenotypically normal animals, and the aromatics were reduced to ∼60% of those for phenotypically normal animals.
DISCUSSION
Using standard gene replacement techniques, we have introduced a null arginase AI allele into the mouse genome. Genotypic analysis of the progeny from matings between AI heterozygotes (AI+/−) by a rapid and accurate PCR method suggests a statistically significant deficit of the null allele in these populations. The distribution of genotypes was non-Mendelian but in Hardy-Weinberg equilibrium (Table 1), strongly suggesting the possibility that haploid germ cells carrying the disrupted AI allele are less fit and participate in the formation of zygotes less frequently than those spermatozoa that express the wild-type allele. Mice homozygous for the null allele (AI−/−) completely lack the activity of arginase AI. This cytoplasmic, predominantly hepatic enzyme is required for the function of the urea cycle, the primary mechanism for the excretion of excess nitrogen from the mammalian body. Predictably, the AI knockout mice had virtually no hepatic arginase activity and died at postnatal days 10 to 14, exhibiting symptoms of severe hyperammonemia, including decerebrate posture, lethargy, and a high-frequency tremor of the extremities, particularly the tail. In contrast to the severe phenotype seen in the AI knockout mice, hepatic arginase deficiency in humans represents the mildest of all the urea cycle disorders identified in humans. A plausible explanation for this discrepancy is provided by the observation that, both in vivo and in vitro, human kidney exhibits an often dramatic (3- to 34-fold) enhancement of expression and activity of the second, extrahepatic isoform of arginase (AII) in response to exposure to high concentrations of arginine (5). Kidney lysates prepared from AI knockout mice in the throes of hyperammonemic crisis had levels of arginase activity very similar to those seen in the kidneys of normal and heterozygous siblings (Table 2), which were harvested at the same time. This was a surprising finding, since approximately half of the relatively low (compared to that in the liver) arginase activity in normal mouse kidney has been ascribed to AI expression. As we expected (Fig. 3), immunoprecipitation with an anti-AII antibody brought down the arginase activity by only about half of that found in normal kidneys, whereas the same antibody precipitated nearly all of the arginase activity from kidneys of AI-deficient and heterozygous mice. Since total kidney arginase activities were very similar in normal, heterozygous, and knockout mice, these data suggest that there is an approximately twofold induction of AII activity in the kidneys of the heterozygous and knockout mice. However, unlike in AI-deficient humans, this relatively low level of induction of arginase AII activity, at least at this age, does not seem to be sufficient to compensate for the loss of liver arginase activity and hence may account for the more rapid and severe clinical course of disease seen in our knockout mice. Levels of arginase activity in the brains of knockout mice were also similar to those seen in the brains of wild-type and heterozygous mice. However, since both the AI and AII isoforms of arginase are expressed in the mouse brain in approximately equal amounts (21; H. Yu, R. K. Iyer, W. W. Grody, and S. D. Cederbaum, unpublished data), it is likely that there is some induction of AII activity in the brains of knockout mice as well.
Most of the differences revealed by comparison of normalized mean amino acid levels in the plasma of AI knockout mice to those of phenotypically normal littermates (Fig. 5) can be explained by the expected perturbations of arginine and ornithine metabolism caused by loss of arginase AI activity. Compared to their wild-type and heterozygous compatriots, knockout mice exhibited significant alterations in the concentrations in plasma of several amino acids intimately involved in arginine metabolism. These include the expected, albeit modest, increases in arginine and decreased levels of proline and ornithine. Ornithine deficiency may inhibit the normal functioning of the early steps of the urea cycle and cause the hyperammonemia. In contrast, the elevated arginine could augment nitric oxide (NO) synthesis and lead to oxidative damage and, paradoxically, a compartmentalized increase in arginase AII activity could cause augmented synthesis of glutamate and/or γ-aminobutyric acid (GABA) and affect brain function. This model is ideal for studying these hypotheses if the animals can be protected from hyperammonemia. The AI-deficient animals also exhibited other common consequences of liver damage, including elevation of levels of glutamate, citrulline, and histidine in plasma and reduction of the branched-chain amino acids valine, isoleucine, and leucine. Knockout animals also exhibited a general reduction in the concentrations in plasma of most amino acids, possibly because they feed less as symptoms progress. In general, there was much more intersample variability within the knockout set than in the phenotypically normal animals. These observed changes could also be due to liver failure and the consequent perturbation of homeostasis.
Unlike normal adult mice, which obtain ornithine by removal of urea from arginine by arginase, neonatal mice obtain ornithine from glutamate by the action of ornithine aminotransferase (OAT), an enzyme that is highly expressed in the small intestines of newborn mice. OAT knockout mice die within 2 days of birth, exhibiting ornithine deficiency and severe hyperammonemia, but can be rescued by the intraperitoneal administration of arginine, which allows the sufficient production of ornithine (20). Gastrointestinal tract levels of OAT begin to drop at about 10 days after birth and reach very low levels at about 2 weeks postnatal. At this time, hepatic arginase becomes the primary provider of ornithine, and in close concordance with this switch, OAT knockout mice no longer require arginine administration for survival. Our AI-deficient mice died at about the same time (postnatal days 10 to 14) that intestinal OAT levels dropped, exhibiting severe hyperammonemia and symptoms virtually identical to those described for the OAT mice, and a plasma ornithine deficit was observed. From these observations, we postulated that these mice may be neonatal ornithine auxotrophs and might be rescued by ornithine supplementation.
At this time, we have a good model for acute hyperammonemia, but our goal is to study the effects of prolonged hyperargininemia on somatic growth and neurological development. Clearly the symptoms exhibited by the AI-deficient mice are consistent with perturbation of the urea cycle. However, because of the early death (before weaning) of these mice, it is currently not possible to study the mechanisms of brain injury in this model. As discussed previously, the slow but progressive neurological deterioration seen in human AI-deficient patients cannot be explained solely by the acute toxic insult represented by sporadic exposure to high levels of ammonia and the identity of the proximate toxin is hence currently unknown. Candidates for this role include arginine itself, as well as some products of alternate (i.e., not mediated by arginase) pathways of arginine metabolism such as NO, glutamate, and GABA. NO plays a prominent role in the process of inflammation and injury in many tissues, including the brain, and glutamate excitotoxicity is a well-studied mechanism for neuronal death. Comparative in situ and immunohistochemical studies of wild-type and AI-knockout mice in our laboratory (21; Yu et al., unpublished data) have already revealed that AI is much more widely distributed and expressed in the brain than previously suspected and suggest an important role for this isoform in brain function. Further studies in situ, as well as with cultured brain cells from wild-type and knockout animals, should help to further elucidate the role of this isoform in the brain.
Among several options for the prevention of early hyperammonemia is treatment with sodium phenylacetate (or phenylbutyrate), a diversion therapy used successfully in AI-deficient humans to reduce ammonia and often arginine levels. In addition, to test our hypothesis (see above) that these mice are suffering from the adverse consequences of hypo-ornithinemia, we will also administer ornithine, either alone or in combination with phenylacetate. Since many of the symptoms of human arginase deficiency are reduced or alleviated by overexpression of the second isoform AII, it may be possible to cure AI deficiency by inducing expression of arginase AII in liver or even in extra-hepatic tissues by using novel pharmaceutical agents (including [possibly] as-yet-undiscovered arginine analogs that induce AII; to be useful these agents should preferably not contribute to the worsening of the existing hyperargininemia), and indeed expression may be naturally induced in older mice. This mouse model for AI deficiency may also be useful for the development and testing of gene therapy vectors and protocols for treatment of the human condition. Once we are able to keep the AI knockout mice alive past weaning, we should be able to analyze any adverse effects of protracted hyperargininemia on brain function as well as somatic growth.
Acknowledgments
This work was supported in part by the Mental Retardation Research Center at UCLA and by USPHS grants HD-06576, HD-36415, and HD-04612.
We thank Anthony Butch, of the Department of Pathology and Laboratory Medicine at UCLA, for the analysis of blood electrolyte and glucose levels.
REFERENCES
- 1.Ash, D. E., L. R. Scolnick, Z. F. Kanyo, J. G. Vockley, S. D. Cederbaum, and D. W. Christianson. 1998. Molecular basis of hyperargininemia: structure-function consequences of mutations in human liver arginase. Mol. Genet. Metab. 64:243-249. [DOI] [PubMed] [Google Scholar]
- 2.Bancroft, D., and A. Stevens (ed.). 1996. Theory and practice of histological techniques. Churchill Livingstone, Ltd., Edinburgh, Scotland.
- 3.Dizikes, G. J., W. W. Grody, R. M. Kern, and S. D. Cederbaum. 1986. Isolation of human liver arginase cDNA and demonstration of non-homology between the two human arginase genes. Biochem. Biophys. Res. Commun. 141:53-59. [DOI] [PubMed] [Google Scholar]
- 4.Dizikes, G. J., E. B. Spector, and S. D. Cederbaum. 1986. Cloning of rat liver arginase cDNA and elucidation of regulation of arginase gene expression in H4 rat hepatoma cells. Somat. Cell Mol. Genet. 12:375-384. [DOI] [PubMed] [Google Scholar]
- 5.Grody, W. W., R. M. Kern, D. Klein, A. E. Dodson, P. B. Wissman, S. H. Barsky, and S. D. Cederbaum. 1993. Arginase deficiency manifesting delayed clinical sequelae and induction of a kidney arginase isozyme. Hum. Genet. 91:1-5. [DOI] [PubMed] [Google Scholar]
- 6.Haraguchi, Y., M. Takiguchi, Y. Amaya, S. Kawamoto, I. Matsuda, and M. Mori. 1987. Molecular cloning and nucleotide sequence of cDNA for human liver arginase. Proc. Natl. Acad. Sci. USA 84:412-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iyer, R. K., C. P. Jenkinson, J. G. Vockley, R. M. Kern, W. W. Grody, and S. D. Cederbaum. 1998. The human arginases and arginase deficiency. J. Inherit. Metab. Dis. 1(Suppl. 21):86-100. [DOI] [PubMed] [Google Scholar]
- 8.Jenkinson, C. P., and M. R. Grigor. 1994. Rat mammary arginase: isolation and characterization. Biochem. Med. Metab. Biol. 51:156-165. [DOI] [PubMed] [Google Scholar]
- 9.Jenkinson, C. P., W. W. Grody, and S. D. Cederbaum. 1996. Comparative properties of arginases. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 114:107-132. [DOI] [PubMed] [Google Scholar]
- 10.Kanyo, B. F., L. R. Scolnick, D. E. Ash, and D. W. Christianson. 1996. Structure of a unique binuclear manganese cluster in arginase. Nature 383:554-557. [DOI] [PubMed] [Google Scholar]
- 11.Kawamoto, S., Y. Amaya, K. Murakami, F. Tokunaga, S. Iwanaga, K. Kobayashi, T. Saheki, S. Kimura, and M. Mori. 1987. Complete nucleotide sequence of cDNA and deduced amino acid sequence of rat liver arginase. J. Biol. Chem. 262:6280-6283. [PubMed] [Google Scholar]
- 12.Lavulo, L. T., T. M. Sossong, Jr., M. R. Brigham-Burke, M. L. Doyle, J. D. Cox, D. W. Christianson, and D. E. Ash. 2001. Subunit-subunit interactions in trimeric arginase. Generation of active monomers by mutation of a single amino acid. J. Biol. Chem. 276:14242-14248. [DOI] [PubMed] [Google Scholar]
- 13.Schimke, R. T. 1963. Studies on factors affecting the levels of urea cycle enzymes in rat liver. J. Biol. Chem. 238:1012-1018. [PubMed] [Google Scholar]
- 14.Sparkes, R. S., G. J. Dizikes, I., Klisak, W. W. Grody, T. Mohandas, C. Heinzmann, S. Zollman, A. J. Lusis, and S. D. Cederbaum. 1986. The gene for human liver arginase (ARG1) is assigned to chromosome band 6q23. Am. J. Hum. Genet. 39:186-193. [PMC free article] [PubMed] [Google Scholar]
- 15.Spector, E. B., M. Keirnan, B. Bernard, and S. D. Cederbaum. 1980. Comparative properties of fetal and adult red blood cell arginase: a possible prenatal diagnostic test for arginase deficiency. Am. J. Hum. Genet. 32:79-87. [PMC free article] [PubMed] [Google Scholar]
- 16.Takiguchi, M., Y. Haraguchi, and M. Mori. 1988. Human liver-type arginase gene: structure of the gene and analysis of the promoter region. Nucleic Acids Res. 16:8789-8802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vockley, J. G., D. E. Tabor, R. M. Kern, B. K. Goodman, P. B. Wissman, D. S. Kang, W. W. Grody, and S. D. Cederbaum. 1994. Identification of mutations (D128G, H141L) in the liver arginase gene of patients with hyperargininemia. Hum. Mutat. 4:150-154. [DOI] [PubMed] [Google Scholar]
- 18.Vockley, J. G., C. P. Jenkinson, H. Shukla, R. M. Kern, W. W. Grody, and S. D. Cederbaum. 1996. Cloning and characterization of the mouse and rat type II arginase genes. Genomics 38:118-123. [DOI] [PubMed] [Google Scholar]
- 19.Vockley, J. G., B. K. Goodman, D. E. Tabor, R. M. Kern, C. P. Jenkinson, W. W. Grody, and S. D. Cederbaum. 1996. Loss of function mutations in conserved regions of the human arginase I gene. Biochem. Mol. Med. 59:44-51. [DOI] [PubMed] [Google Scholar]
- 20.Wang, T., A. M. Lawler, G. Steel, I. Sipila, A. H. Milam, and D. Valle. 1995. Mice lacking ornithine-delta-aminotransferase have paradoxical neonatal hypoornithinaemia and retinal degeneration. Nat. Genet. 11:185-190. [DOI] [PubMed] [Google Scholar]
- 21.Yu, H., R. K. Iyer, R. M. Kern, W. I. Rodriguez, W. W. Grody, and S. D. Cederbaum. 2001. Expression of arginase isozymes in mouse brain. J. Neurosci. Res. 66:406-422. [DOI] [PubMed] [Google Scholar]