Abstract
Retinoids exhibit antineoplastic activities that may be linked to retinoid receptor-mediated transrepression of activating protein 1 (AP1), a heterodimeric transcription factor composed of fos- and jun-related proteins. Here we show that transcriptional activation of an AP1-regulated gene through the mitogen-activated protein kinase (MAPK)-extracellular signal-regulated kinase (ERK) pathway (MAPKERK) is characterized, in intact cells, by a switch from a fra2-junD dimer to a junD-fosB dimer loading on its promoter and by simultaneous recruitment of ERKs, CREB-binding protein (CBP), and RNA polymerase II. All-trans-retinoic acid (atRA) receptor (RAR) was tethered constitutively to the AP1 promoter. AP1 transrepression by retinoic acid was concomitant to glycogen synthase kinase 3 activation, negative regulation of junD hyperphosphorylation, and to decreased RNA polymerase II recruitment. Under these conditions, fra1 loading to the AP1 response element was strongly increased. Importantly, CBP and ERKs were excluded from the promoter in the presence of atRA. AP1 transrepression by retinoids was RAR and ligand dependent, but none of the functions required for RAR-mediated transactivation was necessary for AP1 transrepression. These results indicate that transrepressive effects of retinoids are mediated through a mechanism unrelated to transcriptional activation, involving the RAR-dependent control of transcription factors and cofactor assembly on AP1-regulated promoters.
AP1 response elements are cis-acting DNA sequences mediating transcriptional responses to many physiological and pharmacological stimuli, including phorbol esters, growth factors, mitogenic hormones, and UV and other cellular stresses. These DNA sequences, originally defined as 12-O-tetradecanoylphorbol-13-acetate (TPA) response elements, preferentially bind heterodimers of proteins belonging to the bZIP transcription factor family (3). These transcription factors include products of immediate-early genes, such as members of the fos (c-fos, fosB, fra1, and fra2) and jun (c-jun, junB, and junD) (21) families. The transcriptional activity of the AP1 complex is regulated through different modes. First, its activity is determined by the composition of the AP1 dimer, which may display a different affinity for a given response element (10, 33, 34). The level of expression of AP1 factors is thus one key step of AP1 regulation through modulation of the number of possible combinations of dimers. Second, posttranslational modifications exert a strong control on AP1 activity. Notably, phosphorylation of c-jun at Ser 63 and Ser 73 increases its affinity for the coactivator CREB-binding protein (CBP) without altering its DNA binding activity (4). This posttranslational modification is regulated mostly through the mitogen-activated protein kinase (MAPK)-jun N-terminal kinase (JNK) signaling module (MAPKJNK), which controls the activity of JNK. Thus, activation of the AP1 complex is regulated through a complex network of protein kinases, which renders this transcription factor highly reactive to extracellular stimuli (21). Indeed, 10 MAPK families have been identified in mammalian cells, two of them (ERK1 and -2) being preferentially activated through growth factor receptors and by phorbol esters such as TPA. Stress-activated protein kinases (SAPKs) are predominantly activated through stressful stimulis and proinflammatory cytokines (such as JNK and p38s), but cross-regulation between the MAPKERK, MAPKJNK, and MAPKp38 pathways has been reported (16, 28).
AP1 regulates the expression of several genes involved in oncogenic transformation and cellular proliferation such as those coding for metalloproteases, VEGF, and transforming growth factor β (TGF-β), and AP1 transactivation is required for tumor promotion in vivo (54). Therefore, there is considerable interest in identification of compounds able to downregulate AP1 activity and thereby oppose unregulated cell growth leading to benign or malignant hyperplasia and cancer. A number of ligands for nuclear receptors, including glucocorticoids, retinoids, and fatty acids display such AP1 repressive activity, which seems to be the basis for their beneficial therapeutic effects. All-trans-retinoic acid receptors (RARs) exert two types of action on gene activity. The first one is referred to as transactivation and is characterized by structural transitions occurring in the receptor structure upon agonist binding. This leads to the formation of protein interaction interface(s) allowing recruitment of nuclear coactivators to the retinoid X receptor (RXR)-RAR dimer and therefore to the promoter (reviewed in reference 52). Transrepression of AP1 is the second activity of RARs, for which several observations may provide a model for transcriptional interference with membrane receptor-controlled signaling pathways. They include c-jun DNA binding inhibition (45), competitive titration by RAR and AP1 of limiting amounts of CBP (20), or downregulation of JNK activity (7, 24). Molecular determinants governing the transrepressive activity of RAR are, however, likely to be distinct from those ruling its transactivation potential. This view is supported by the description of dissociated retinoids, which are unable to elicit a transactivating response by RARs, yet induce transrepression of AP1 (9, 17, 29, 36) as well as that of receptor mutants with altered transactivation, but wild-type transrepression properties (25).
In this study, we examined the transrepressive activity of retinoids on the ERK-regulated AP1 activity in HeLa cells. We observed that atRA treatment of target cells led to the alteration of AP1 complex composition bound to an AP1 site. We report here for the first time that ERKs, CBP, and RARα are associated with the AP1-responsive promoter in vivo under stimulating conditions. This complex underwent structural alterations under transrepressive conditions, which were correlated with the dissociation of ERKs and CBP from the promoter. Using synthetic retinoids and receptor mutants, we also demonstrate that AP1 inhibition by human RARα (hRARα) is independent of nuclear coactivator recruitment and other functions, such as dimerization with RXR and specific DNA binding.
MATERIALS AND METHODS
Cell line and culture conditions.
The HeLa cells used were from a subclone isolated from a HeLa Tet-On (Clontech) cell population. They were routinely maintained in Dulbecco's modified Eagle medium (Glutamax-1; Gibco-BRL, Cergy-Pontoise, France) supplemented with 10% fetal calf serum, 50 U (each) of penicillin and streptomycin per ml, and 100 μg of G418 per ml.
Retinoids and chemicals.
atRA was purchased from Sigma (St Quentin-Fallavier, France). 9-cis-Retinoic acid was obtained from Hoffman-Laroche. All CD compounds were kindly provided by U. Reichert (Galderma). CD3105 (AGN 192870) is an RAR antagonist (22); CD2409 {(4-[1-hydroxy-3-(5,5,8,8-tetramethyl-5,6,7,8-tetrahydronaphthalen-2-yl)-prop-2-ynyl]-benzoic acid} is a dissociated anti-AP1 ligand (50). PD98059 and all other kinase inhibitors were purchased from Calbiochem (France-Biochem, Meudon, France).
Transient transfections.
HeLa cells were transfected by the polyethyleneimine method (25), and the luciferase activity was assayed as previously described (35).
cDNAs, plasmids, and reporter genes.
pSG5-based expression vectors for hRARα, hRARβ, and hRARγ have been described previously (5). hRARα mutants have been described elsewhere (26, 35). pSG5 hRARα K380R-D383R was a kind gift from H. de Thé. The pSG5-hRARα K244-262A double mutant was generated by site-directed mutagenesis (QuickChange; Stratagene) with pSG5-hRARα K244A (35) as a template. The RXGRE-tk Luc reporter gene (designated DR5-tk Luc in this study) is a pGL3-based vector (Promega, Charbonnières, France) and has been described elsewhere (14). The AP1-tk Luc reporter gene is a pGL3-based vector in which the thymidine kinase (tk) promoter is hooked to four repeats of the consensus AP1 site TGAGTCA. pcDNA3-myc-His6-hRARα was constructed by inserting the RARα cDNA in frame within the pcDNA 3.1 myc/His vector (InVitrogen). pGEX-based expression vectors used for bacterial overexpression of glutathione S-transferase (GST)-fused coactivators have been described elsewhere (25). pSG5 c-jun, pBK c-fos, pRSV junB and pRSV junD, pRSV fra1, and the dominant-negative JNK expression vector (dnJNK) were gifts from B. Wasylik, I. Verma, M. Nemer, M. Yaniv, and J. M. Blanchard, respectively. Constitutively active ras, MEK1, wild-type MKK7, and JNK1 vectors were generous gifts from R. Davis.
Western blotting and antibodies.
Whole-cell extracts were prepared as follows. First, 5 × 106 cells were grown and treated with retinoids and/or TPA. Monolayers were scraped rapidly in ice-cold 1× phosphate-buffered saline (PBS), and cells were lysed in 1 volume of sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer and briefly sonicated. Western blotting was carried out as described previously (13, 35, 47). Anti-protein kinase C (anti-PKC), anti-JNKK1/MKK4, anti-ERK1 and -2, anti-MEK1 and -2, and anti-MKK1 antibodies were obtained from Transduction Laboratories (Lexington, Ky.). Anti-phospho ERK antibody was obtained from Promega. Antibodies recognizing JNK1, c-jun (N), junB (N), junD (329), c-fos (no. 4), fosB (102), fra1 (R-20), fra2 (L-15), and ATF2 were purchased from Santa Cruz Biotechnology (Santa Cruz, Calif.). Calbiochem and New England BioLabs were the sources of anti-c-jun and anti-phosphorylated c-jun antibodies. Anti-RNA polymerase II (pol II) antibodies were from Santa Cruz Biotechnology (total RNA pol II, sc 899) and Covance/BabCo (anti-RNA pol IIA, 8WG16), and the DRIP205 antiserum was a gift from C. Rachez and L. P. Freedman. Peroxidase-coupled antimouse, antigoat, or antirabbit immunoglobulin G was obtained from Sigma.
EMSA and GST pulldown assays.
Nuclear coactivators and dimerization assays with GST-tagged proteins as baits were performed as described previously (13, 14). Electrophoretic mobility shift assays (EMSA) were run as described in reference 41 with in vitro-translated receptors.
IP-kinase assays.
For immunoprecipitation (IP)-kinase assays, the activities of ERKs, JNK, and p38 were assayed with kits purchased from Cell Signaling Technologies (Beverly, Mass.) according to the manufacturer's instructions. MEK1 activity was assayed with a kit purchased from Upstate Biotechnology (Lake Placid, N.Y.), and glycogen synthase kinase 3 (GSK-3) activity was assayed similarly with an anti-GSK-3 antibody (Transduction Laboratories) by using a phospho-glycogen synthase peptide (Upstate Biotech) as a substrate.
DNA-IP assays.
For DNA-IP assays, 4 × 106 to 6 × 106 HeLa cells were transfected as described above with AP1-tk Luc and pSG5-hRARα vectors. Forty-eight hours after transfection, cells were treated for 1 h with atRA and/or phorbol ester. Proteins were cross-linked to DNA by addition of 1% formaldehyde to the medium for 15 min at 37°C. The reaction was quenched upon addition of 100 mM glycine for 15 min at room temperature, and cells were collected in ice-cold 1× PBS. Cells were lysed in 10 mM Tris-HCl (pH 7.4), 10 mM EDTA, 1% SDS, 100 μM Na2VO3, 10 mM sodium butyrate, 1% protease inhibitor cocktail (Sigma), and soluble DNA-protein complexes were prepared by sonication of cellular lysates. After cell debris had been spun down, soluble protein-DNA complexes were diluted 10-fold in IP buffer (10 mM Tris-HCl [pH 7.4], 10 mM EDTA, 20 mM NaCl,1% Triton X-100, 100 μM Na2VO3, 10 mM NaBu, 1% protease inhibitor cocktail) and incubated with 2 to 5 μg of antibody overnight at 4°C. Immune complexes were collected by addition of 50 μl of a 50% protein A-Sepharose slurry and incubation at room temperature for 2 h. Beads were successively washed with IP buffer, IP buffer plus 1.2 M NaCl, and a mixture containing 10 mM Tris-HCl (pH 7.4), 1% NP-40, 1% deoxycholate, 1 mM EDTA, 0.25 M LiCl, and 100 μM Na2VO3. After a final wash in a mixture of 10 mM Tris-HCl (pH 7.4) and 1 mM EDTA, immune complexes were eluted from beads in 250 μl of a mixture containing 0.1 M NaHCO3 and 1% SDS. Supernatants were heated for at least 4 h at 65°C in the presence of 1 mg of proteinase K per ml to reverse cross-links. DNA was purified with the QIAquick DNA purification kit (Qiagen). Target sequences were amplified by using RV3 and GLP2 primers from Promega (25 to 28 PCR cycles) and resolved on a 1.2% agarose gel.
Real-time PCR.
AP1 promoter sequences were amplified with the TaqMan PCR master mix (Applied Biosystems, Courtaboeuf, France). The FAM/TAMRA coupled-probe (+31/+52: CCCAGCGTCTTGTCATTGGGCGA), forward (−53/−31: GGTGCCAGAACATTTCTCTATCG), and backward (+81/+96: GACCTCGGACCGCGC) primers were designed to amplify a 140-bp DNA fragment encompassing the four repeats of the AP1 response element (TGACTCA) present in the reporter gene used in all experiments. Position +1 corresponds to the first base of the second AP1 response element. Reactions (40 cycles) and data analysis were carried out with an ABI Prism 770 (Applied Biosystems).
RESULTS
Retinoids repress the MEK1-dependent AP1 response in HeLa cells.
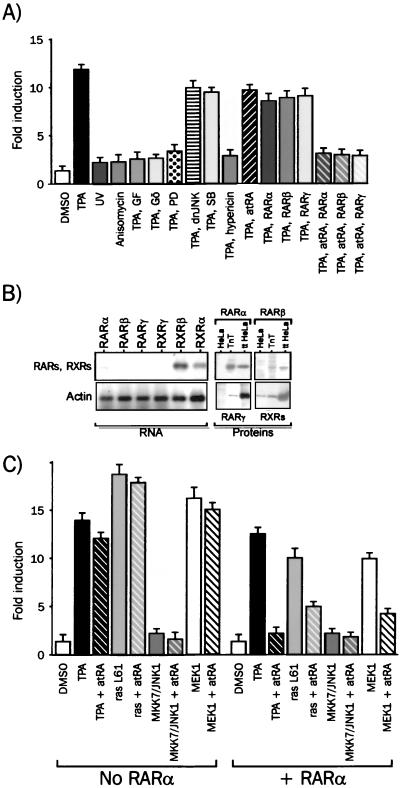
We first investigated which signaling module or modules mediate the TPA-induced AP1 activity by monitoring the activity of a transiently transfected AP1-responsive reporter gene in HeLa cells(Fig. 1A). The activity of the AP1-tk Luc reporter gene increased 10- to 15-fold when cells were treated for 6 h with 100 nM TPA. Specific activators of SAPKs, such as anisomycin and UV, were unable to activate the AP1-tk Luc reporter gene (Fig. 1A), as well as tumor necrosis factor alpha (TNF-α) (M. Benkoussa and P. Lefebvre, unpublished observations), suggesting that SAPK signaling modules are not fully functional in this cell line. The TPA-induced increase in luciferase activity was abrogated in the presence of GF109203X, a specific inhibitor of PKC-α, -β, -γ, -δ, and -ε isoforms (48) and of Gö6976, a specific inhibitor of PKC-α and -βI (32). Since PKC-β is not expressed in our cell line (M. Benkoussa and P. Lefebvre, unpublished), PKC-α is a likely molecular relay for TPA-mediated activation of AP1. PD98059 selectively inhibits MAPK kinases 1 and 2 (MEK1 and -2, respectively) (2), which act as an upstream activator of ERK1 and -2. The complete inhibition of the TPA-induced response by 10 μM PD98059 suggests that MEK1 and -2 is an important regulator of the TPA-induced AP1 response. Hypericin is known to inhibit PKCs and ERKs at 50% inhibitory concentrations of 3.3 μM and 4 nM, respectively. We thus used hypericin at 100 nM, a concentration affecting mostly ERK activity, and were able to block the TPA-induced AP1 response. Cotransfection of a dominant-negative mutant of JNK, dnJNK (40), led to a decrease of 15 to 20% of the TPA-induced luciferase activity, indicating that JNK activation plays a minor role in the transcriptional activation process of the promoter. The p38-specific inhibitor SB203580 was able to reduce the TPA-induced AP1 activity by about 20%. Thus, TPA triggers in HeLa cells a composite MEK1 and -2-dependent response involving mainly the MAPKERK signaling module. Both MAPKJNK and MAPKp38 also contributed to this response, albeit to a much lesser extent. Additionally, atRA was able to repress the TPA-induced AP1 activation in a receptor- and ligand-dependent manner. Indeed, the RAR-mediated repression, which ranged from 5 to 20% in the presence of endogenous RARα, was increased to 85 to 95% when RARα, RARβ, or RARγ was overexpressed (Fig. 1A and B).
FIG. 1.
Inhibition of the MAPKERK pathway by retinoids. (A) The MAPKERK signaling pathway activates AP1 in HeLa cells. HeLa cells were transfected and treated for 6 h with the following activators and/or inhibitors of kinases: 100 nM TPA, 500 nM GF109203X, and 500 nM Gö6976 (PKCs, GF, and Gö), 10 μM PD98059 (MEK1 and -2; PD), 100 nM SB203580 (p38α and -β; SB), 100 nM hypericin (ERKs). SAPK activators were used as follows: anisomycin, 50 ng/ml (subinhibitory concentration) (18); UV, 40 J/m2. Bar diagrams for this and the following experiments represent the average (± standard deviation) of at least five experiments carried out with six independent assays. The basal level of reporter gene expression was scaled up to 1, and all activities are expressed relative to the basal level, which did not vary significantly. dnJNK, dominant-negative mutant of JNK1. (B) Retinoid receptor expression in HeLa cells. mRNAs were analyzed by reverse transcription-PCR for RAR and RXR isoform transcripts. Actin was used as an external control. PCR products were resolved on an agarose gel and visualized by ethidium bromide staining (RNA panel). Fifty micrograms of whole-cell extract was analyzed by Western blotting with antibodies against RARα, RARβ, RARγ, and RXRs (HeLa). Controls were provided by RARs or RXRs extracted from cells transfected with expression vectors coding for each receptor (ttHeLa) or synthesized by coupled in vitro transcription and translation (TnT) (Proteins panel). (C) MAPK sensitivity to retinoid inhibition. HeLa cells were transfected with AP1-tk Luc and expression vectors coding for RARα, constitutively active ras (rasLG1), wild-type MKK7 and wild-type JNK1 or constituvely active MEK1. Cells were treated for 6 h with 1 μM atRA, and the resulting luciferase activities were plotted as in panel A.
We next sought to identify at which step or steps atRA interferes with the signal transduction process. AP1 activity was monitored in the presence or absence of retinoids as described above in cells overexpressing RARα and constitutively active protein kinases belonging to MAPK signaling pathways (Fig. 1C). Constitutively active ras (ras L61) induced a strong AP1 activity, which was highly sensitive to atRA inhibition. Similarly, overexpression of MEK1 induced a potent AP1 response sensitive to atRA inhibition. Coexpression of MKK7 and JNK1, two potent upstream activators of the jun transcription factor family, poorly activated the AP1 response, and this response was not sensitive to atRA. Similar results were obtained when a constitutive activator of the MAPKJNK pathway, SEK1 (SEK1 ED) (44), was overexpressed (M. Benkoussa and P. Lefebvre, unpublished). Thus atRA exerts its transrepressive activity through regulation of a MEK1 and -2 and/or ERK1 and -2-controlled event.
The ability of retinoids to inhibit AP1 activation may potentially be ascribed to several mechanisms, one of which is downregulation of the expression of essential relays of MAPK or SAPK signaling cascades. Preliminary experiments allowed us to rule out such a hypothesis for PKCs, c-raf, MEK-1 and -2, JNKK, JNK1 and -2, and ERK1 and -2 (M. Benkoussa, M.-H. Delmotte, and P. Lefebvre, unpublished observations). Expression of other proteins potentially involved in AP1 modulation such as p21/Waf-1 (a retinoid-regulated inhibitor of cyclin-dependent kinase and a regulator of NF-κB activity) (30) and p53 (a regulator of p21/Waf-1 gene transcription) was not significantly altered in our system (M. Benkoussa and P. Lefebvre, unpublished).
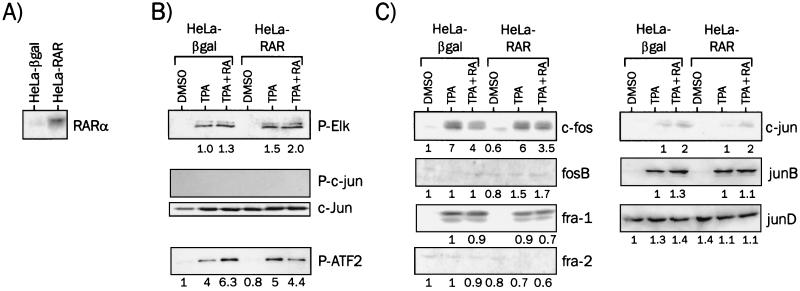
We then established HeLa-derived cell lines overexpressing hRARα or β-galactosidase as a control to further investigate the role of this receptor as a transrepressor. The level of expression of RARα in the HeLa-RAR subclone was at least 10- to 15-fold higher than that of the wild-type (C. Brand and P. Lefebvre, unpublished observations) or β-galactosidase-positive HeLa cells (Fig. 2A). Stimulation of HeLa cells by TPA led to a strong increase of ERK1 and -2 activity, as assayed by an IP-kinase assay (Fig. 2B). However, atRA was not able to counteract this upregulation in either the wild-type or RAR-overexpressing background. Similar assessments of JNK and p38 activities were carried out, and while JNK remained nondetectable in our system, p38 activity was slightly increased upon TPA treatment, with no significant effect of retinoic acid in both cellular backgrounds.
FIG. 2.
Components of MAPK signaling modules in HeLa cells. (A) Expression of hRARα in HeLa-βgal or HeLa-RAR cell lines. Whole-cell extracts (50 μg of protein) were analyzed by Western blotting with an anti-RARα antibody. (B) MAPK activities in the wild-type HeLa or HeLa-RAR background. Cells were treated for 6 h with the indicated compounds, and cell extracts were assayed for ERK1 and -2 (P-Elk), JNK1 and -2 (P-c-jun) and p38 (P-ATF2) activities by IP-kinase assays. Representative results are shown. The GST-c-jun fusion protein, used as a substrate in the JNK assay, was detected with a specific anti-c-jun antibody to ensure that the negative result was not due to aberrant loading of gels. For the experiment shown, the numbers indicate the fold induction over the reference set to 1. (C) Expression levels of c-fos, c-jun, and related transcription factors. Western blot analyses were carried out with specific antibodies for c-fos- and c-jun-related proteins. Results were quantified and expressed as in panel B.
Expression levels of AP1 components were examined by Western blot analysis of whole cell extracts (Fig. 2C). TPA treatment strongly increased c-fos expression levels (Fig. 2C), consistent with the reported upregulation of the c-fos promoter by ERK-phosphorylated Elk1 (31). atRA, however, did not significantly alter this effect in either in the wild-type or HeLa-RAR background. This is in contrast to a previous report showing that retinoids oppose TPA-induced upregulation of c-fos in pituitary cells (39). fra1 responded similarly to c-fos in both cases, whereas fosB and fra2 were barely detectable under these conditions. While c-jun was weakly expressed in our cellular model, the expression level of junD was high under all conditions and was not regulated by retinoids and/or TPA. junB turned out to be strongly induced by TPA, but not downregulated by retinoids
From these data, we conclude that AP1 inhibition is unlikely to proceed through modulation of levels of MAPK activity or AP1 factors, a conclusion strengthened by the observation that protein synthesis inhibitors did not prevent AP1 activation and retinoid-mediated AP1 repression (M. Benkoussa and P. Lefebvre, unpublished).
GSK-3 is involved in atRA-mediated inhibition of AP1.
The activation state of several protein kinases known to potentially regulate AP-1 was further evaluated as a function of time by an IP-kinase assay in response to TPA and/or retinoids. Both ERK1 and -2 and MEK1 were activated in response to TPA treatment, but atRA did not inhibit this activation process (Fig. 3A). GSK-3 has been identified as an ERK-regulated negative modulator of c-jun and junD DNA binding activity, acting by phosphorylation of serine residues in the DNA binding domain of these transcription factors (12, 38). The activation state of GSK-3 was found to be insensitive to MAPKERK pathway activation by TPA, but significantly enhanced after a 30-min treatment with atRA. Note that each kinase underwent a sustained activation in time, likely due to the metabolic stability of TPA. Finally, the phosphorylation state of junD at S100, reflecting its transcriptional activation, was assayed by Western blotting with a specific anti-phospho junD antibody. As expected, TPA promoted a strong hyperphosphorylation of this transcription factor, which was abrogated in the presence of atRA. To confirm these kinase assays, the activation state of ERK1 and -2, MEK1 and -2, and of GSK-3 was also assessed after a 6-h treatment with antiphosphokinase antibodies (Fig. 3B). As it could be predicted from the IP-kinase assays, ERK1 and -2 and MEK1 were hyperphosphorylated (activated) upon TPA treatment, and insensitive to atRA. GSK-3 exists in most cell lines as two isoforms, α and β, and phosphorylation of these kinases leads to the inactivation of their catalytic properties. In our system, both isoforms were phosphorylated (inactive) in control cells, but atRA treatment lowered specifically the phosphate content of the β isoform. This shows that GSK-3β activation is positively regulated by atRA, in agreement with the IP-kinase assay. Finally, junD was strongly phosphorylated in the N-terminal transactivation domain (Ser 100) upon TPA treatment, and Ser 100 phosphorylation was abrogated in the presence of atRA.
FIG. 3.
GSK-3 is a retinoid-dependent partial inhibitor of AP1 transcriptional activity. (A) Activity of protein kinases in response to phorbol ester and/or atRA. HeLa-RAR cells were treated for the indicated times with the indicated combination of retinoid and TPA and whole-cell extracts were prepared. One hundred micrograms of proteins was subjected to IP with anti-ERK, anti-MEK1, or GSK-3 antibodies (1 μg), and immunoprecipitates were incubated with Elk-1, ERK2, and MBP, and phospho-glycogen synthase peptide, respectively, as substrates. junD phosphorylation was assayed by Western blotting as below and quantified by densitometric analysis of fluorograms. Each assay was performed in duplicate. (B) Phosphorylation of protein kinases in response to TPA and/or atRA. HeLa cell extracts, prepared from cells treated for 6 h with the indicated combination of retinoid and TPA, were probed for their content in phosphorylated adducin (as a control), GSKα and -β, ERK1 and -2, MEK1 and -2 and junD. Representative fluorograms are shown. (B) Lithium partially inhibits atRA-induced AP1 repression. HeLa cells were transfected with the AP1-tk Luc reporter gene and pSG5-hRARα. Cells were treated 2 days after transfection with the indicated combinations of 1 μM atRA, 100 nM TPA, 10 mM NaCl, or 10 mM LiCl. The resulting luciferase activity was assayed 6 h later, and the results are expressed as described in the legend to Fig. 1.
Our observations suggested that atRA-mediated activation of GSK-3 could potentially inhibit AP1 through phosphorylation of junD and impaired DNA binding of AP1 complexes. GSK-3α and -β are related protein serine kinases having a critical role in the dorsoventral patterning of vertebrates (15, 19), and severe effects of lithium on embryonic development are probably due to its ability to inhibit specifically GSK-3 (23, 46). To determine whether GSK-3 activity affects the retinoid-mediated inhibition of AP1, we incubated HeLa cells transfected with the AP1 reporter gene and the RARα expression vector with 10 mM lithium chloride or 10 mM sodium chloride and challenged them with TPA and/or atRA (Fig. 3C). Neither lithium nor NaCl had a significant effect on AP1 activity. However, the transrepressive activity of atRA was partially blunted in the presence of lithium, suggesting a contribution of GSK-3 to retinoid-controlled AP1 inhibition. These data indicate that GSK-3 activity is required for full inhibition of AP1 activity by retinoids.
Characterization of AP1 complexes bound to the TPA response element in intact cells.
We then attempted to characterize the composition of AP1 complexes binding to DNA by EMSA. Although informative, EMSAs did not reflect the alteration of the expression of AP1 components, as revealed by Western blot analysis of whole-cell extracts. We thus used an immunoprecipitation assay to quantify the biochemical composition of AP1 complexes bound to the AP1-responsive reporter gene promoter in vivo.
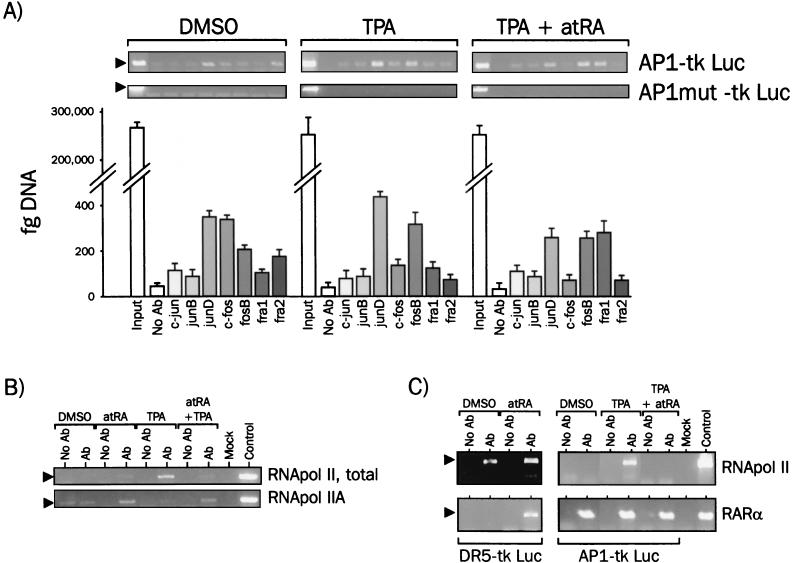
Using antibodies directed against each potential component of the AP1 complex, immunoprecipitation from untreated cells with each antibody followed by real-time PCR detection (DNA-IP) of the promoter region (including the AP1 site and the thymidine kinase [tk] promoter) showed that junD, c-fos, and fra2 were prominently bound to this promoter (Fig. 4A). After treatment for 1 h with 100 nM TPA, the composition of the AP1 complex was shifted towards a junD-fosB dimer. Surprisingly, c-fos was not found to be a major component of the AP1 complex under these conditions. When cells were challenged with TPA plus 1 μM atRA, junD, fosB, and fra1 were associated with the promoter, consistent with the reported inhibitory effect of fra1 on AP1-mediated transcription (33, 51). In contrast to untreated cells, junD binding was weaker, strengthening the hypothesis that atRA-regulated GSK-3 activation may play a role in AP1 transrepression. As a negative control, a reporter gene (AP1 mut) containing a mutated AP1 site that does not confer responsiveness to TPA was used, showing that loading of AP1 factors is strictly dependent on the AP1 site integrity.
FIG. 4.
Transcription factor assembly on the AP1 consensus response element in intact cells. (A) AP1 protein binding to the AP1 response element. HeLa cells were transfected with the AP1-tk Luc reporter gene together with the pcDNA myc-His hRARα expression vector. DNA-IP assays were performed after a 1-h treatment with TPA and/or atRA with antibodies (Ab) against AP1 proteins. As a control, a reporter gene containing an inactivated AP1 site and displaying no activity upon TPA treatment was used (AP1mut-tk Luc). Immunoprecipitated DNA was analyzed by semiquantitative PCR (top panels, 28 PCR cycles) and quantified by real-time PCR. Results are expressed as femtograms of DNA and displayed in the bar graph (bottom panel). The input lanes show that equivalent amounts of plasmid DNA were used for each DNA-IP. (B) atRA prevents RNA pol II loading on the AP1-responsive promoter. DNA-IP assays were performed as in panel A with either a nonphosphorylated RNA pol II CTD monoclonal antibody (RNA pol IIA, clone 8WG16, BaBCo) or a polyclonal antibody against total (phosphorylated and nonphosphorylated) RNA pol II (RNA pol II, total, sc899; Santa Cruz Biotechnology). (C) RARα is constitutively tethered to the AP1 binding site. DNA-IP assays were performed after transfection of cells with either the AP1-tk Luc or the DR5-tk Luc reporter genes together with RAR or RAR and RXR expression vectors, respectively. Antibodies directed against total RNA pol II or the RARα C terminus were used, and immunoprecipitates were analyzed for the presence of RARE or AP1 response element sequences.
By using a similar approach, we then assessed RNA pol II recruitment to the AP1-tk Luc promoter (Fig. 4B). We first used an antibody directed against the N terminus of the rpb1-encoded subunit, which recognizes this polypeptide irrespective of posttranslational modifications (Fig. 4B, RNA pol II total). In nonstimulated cells, little association of RNA pol II was detected on the promoter, as well as under atRA stimulation. As expected, a very strong increase in RNA pol II recruitment was observed after a 1-h stimulation with TPA. Surprisingly, atRA strongly inhibited TPA-induced RNA pol II loading to basal levels. The TFIIE-dependent, TFIIH-mediated phosphorylation of the C-terminal domain (CTD) of pol II is in many instances correlated to transcriptional activation of genes (8, 53). Conversely, hypophosphorylation is associated to the DNA-bound, elongation-incompetent form of pol II (RNA pol IIA). In nonstimulated cells, DNA-IP with an antibody recognizing the nonphosphorylated form of RNA pol II did not reveal any association of this enzyme with the promoter (RNA pol IIA; Fig. 4B). As expected, this transcriptionally inactive form was absent in TPA-treated cells. However, atRA was able to promote, alone or in the presence of TPA, a detectable interaction of RNA pol IIA with the promoter. Thus, atRA is likely to repress transcriptional activation of the AP1-responsive reporter gene by decreasing the overall level of RNA pol II recruitment, as well as by favoring the stalling of a small fraction of RNA pol II as a nonphosphorylated enzyme.
To determine whether RARα could be recruited to the AP1 response element, we performed DNA-IP assays with an antibody against the C terminus of RARα. The ability of RARα to bind to a DR5 retinoic acid response element (RARE) was evaluated in parallel by transfecting cells with RAR and RXR expression vectors together with a reporter gene (DR5-tk Luc), the sequence of which is strictly identical to that of the AP1 reporter gene (AP1-tk Luc), except for the response element (Fig. 4C). In naive cells, no RNA pol II was bound to the AP1 site, but RARα was found to be associated with the promoter, whereas under conditions of stimulation by TPA, both RNA pol II and RARα were associated with DNA. Under conditions of transrepression, only RARα was detected. This differential loading was response element specific, since atRA-induced recruitment of RARα to the DR5 RARE, which was concomitant to a strongly increased RNA pol II loading. Thus, transrepression by atRA may be attributed to decreased RNA pol II recruitment at the AP1-responsive promoter. RARα is also associated to this promoter under any condition, suggesting that ligand binding triggers an event leading to AP1 transcriptional inactivation. This unexpected finding prompted us to characterize more extensively the binding of RARα and other putative components of the transcriptional machinery to the promoter.
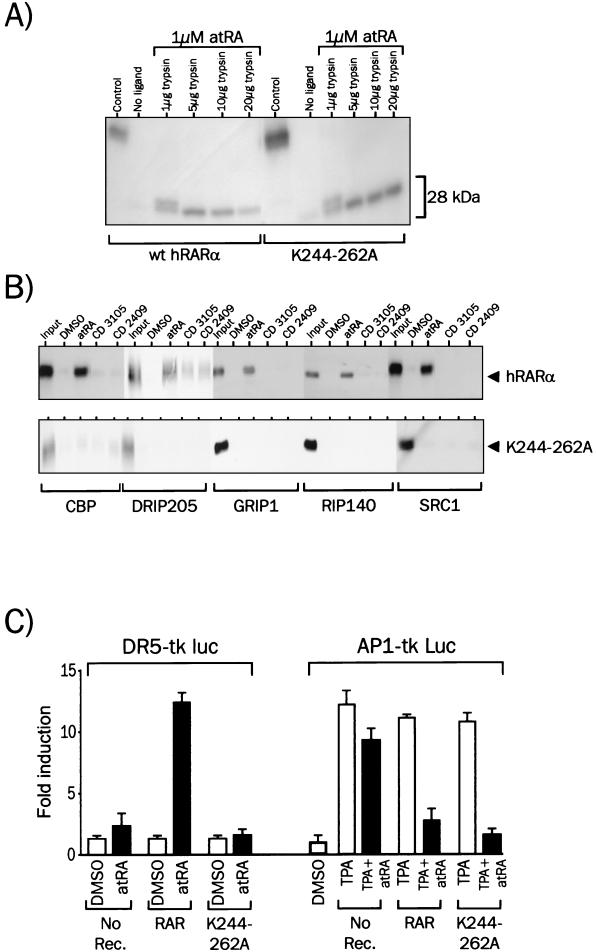
We therefore constructed a N-terminal fusion protein between the c-myc epitope and RARα, which displays wild-type transrepressive activity (M. Benkoussa and P. Lefebvre, unpublished) and performed the DNA-IP assay as described above. Antibodies directed against the C terminus of RARα, the c-myc epitope located at the N terminus of RARα, the NR2 box of DRIP205, the CREB binding domain of CBP, and the C terminus of ERK1 and -2 were thus used in the DNA-IP assay (Fig. 5). As described above, RARα was consistently detected in all conditions when an antibody directed against its C terminus was used. More surprisingly, the accessibility of the RARα N terminus was decreased to basal levels under transrepressive conditions. This decreased accessibility was also observed in the presence of a retinoid antagonist (CD3105), which displays an anti-AP1 activity similar to that of atRA (Fig. 6). This result demonstrates that the RARα N-terminus region is either masked or undergoes structural transitions when AP1 is transcriptionally active. ERKs have been shown to phosphorylate c-jun at Ser 63 and Ser 73 and to regulate its transcriptional activity (28). junD is, by homology, a likely substrate for ERK, and we asked whether this kinase could be detected at the AP1-regulated promoter. Very interestingly, ERK was detected under basal conditions, and this recruitment was strongly increased in TPA-stimulated cells. Both a retinoid agonist and a retinoid antagonist were able to downregulate ERK recruitment, and this behavior paralleled exactly that of CBP, a coactivator for AP1 (20). DRIP205, a nuclear receptor coactivator (42), was recruited to the AP1 promoter only in the presence of atRA and not CD3105, suggesting that this interaction occurs through RARα. Thus, incorporation of ERK and CBP at the AP1 promoter is affected negatively by retinoids, irrespective of their activity in transactivation assays.
FIG. 5.
Transcription factors and cofactors loading at the AP1 response element in vivo. HeLa cells were transfected as described in the legend to Fig. 4. DNA-IP assays were carried out with anti-c-myc (RARα N terminus), anti-RARα AF2 (RARα C terminus), anti-pan-ERK, anti-DRIP205, and anti-CBP antibodies. atRA and CD3105 were both used at a final concentration of 1 μM, and TPA was used at a final concentration of 100 nM.
FIG. 6.
Transactivation and transrepression by retinoids. (A) Transactivation by retinoids. The activity of the DR5 RARE-driven reporter gene was monitored by assaying luciferase activity in HeLa cells transiently transfected with RGXR alone (left panel) or RGXR and hRARα. Cells were treated with the indicated ligand for 6 h. (B) Transrepression by retinoids. HeLa cells were transfected with receptor expression vectors as described above and the AP1-tk Luc reporter gene. TPA was used as the AP1 inducer (100 nM). The basal level of reporter gene expression was scaled up to 1, and all activities are expressed relative to the basal level, which did not vary significantly. The following concentrations were used in both assays: 50 nM atRA, 1 μM CD3105, 10 μM CD2409, and 100 nM TPA.
AP1 transrepression by retinoids does not correlate with their transactivating properties.
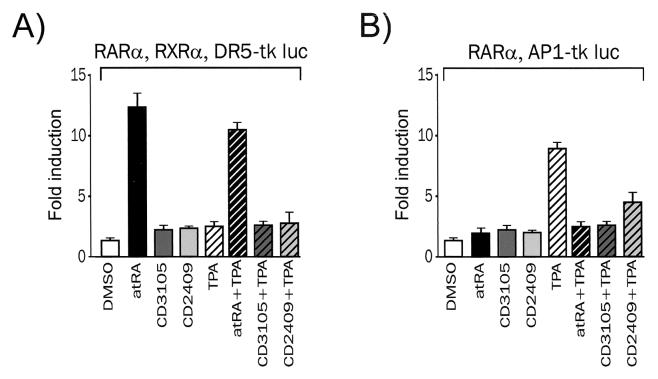
Several pieces of evidence suggest that RAR-mediated transrepression and transactivation are distinct and unrelated phenomena (see the introduction). We thus assayed the transactivating and transrepressing activities of hRARα by using retinoids with different chemical structures and distinct biological properties. atRA is used as the reference compound, whereas CD3105 is an RAR antagonist (22) and CD2409 is a dissociated, anti-AP1 retinoid (50).
We first tested these three retinoids in a standard transactivation assay by using the DR5-tk Luc reporter gene and expression vectors coding for RARα and RXRα. This assay confirmed the biological properties of these three representative retinoids in our system. atRA elicited a strong response (12- to 15-fold induction), whereas neither the antagonist CD3105 nor the dissociated anti-AP1 CD2409 retinoid stimulated luciferase activity after a 6-h treatment (Fig. 6A). HeLa cells were treated simultaneously with TPA and retinoids to assess whether activation of the PKC-MAPK pathway could affect hRARα function. A modest decrease of the responsiveness to atRA was observed in the presence of overexpressed hRARα under these conditions, in agreement with our previous results obtained in COS cells (47).
The ability of these three retinoids to promote RARα-mediated AP1 transrepression was assessed. In the absence of overexpressed hRARα, TPA strongly activated AP1 (10- to 15-fold induction). Retinoids alone were inactive, and a very weak inhibition of the TPA-induced luciferase activity was observed, suggesting a minor contribution of endogenous RARα. A moderate constitutive repression was observed in the presence of overexpressed RARα (Fig. 6A) and for hRARβ and hRARγ in this cell line and others (M. Benkoussa and P. Lefebvre, unpublished). Retinoids abrogated the TPA-induced luciferase activity, irrespective of their ability (or inability) to transactivate the DR5-driven promoter (Fig. 6B). Transcriptional interference is thus unlikely to require a function or functions necessary for receptor-mediated transcriptional activation
Coactivator recruitment is not a prerequisite for AP1 repression by hRARα.
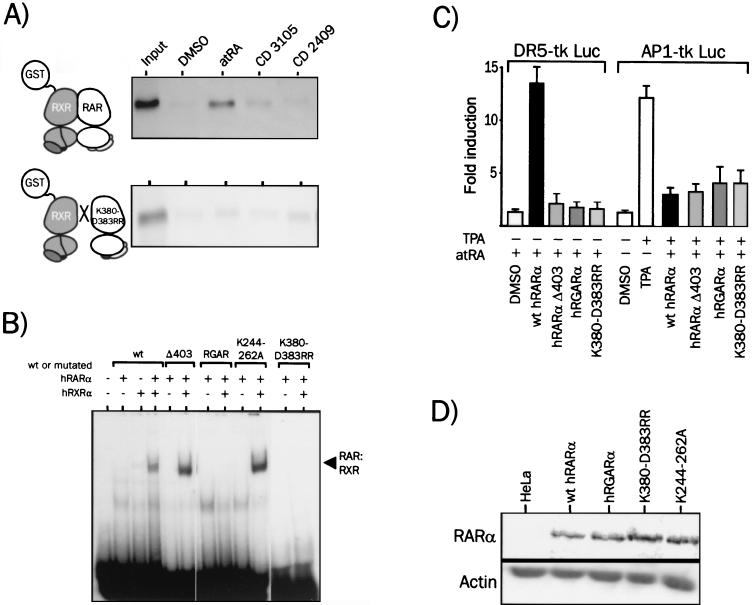
We showed recently that mutation of K244 or K262 in hRARα partially abolished nuclear receptor coactivator (NCoA) recruitment by hRARα and strongly impaired its transactivating activity (35). We therefore introduced a double K244-to-A and K262-to-A mutation in wild-type hRARα to generate a transcriptionally inactive receptor. The ligand-binding and DNA-binding activities of K244-262A were not altered, as judged from the limited proteolysis assay (Fig. 7A) and EMSA, respectively (Fig. 8B). The ability of K244-262A to interact with NCoAs in vitro was assessed by an in vitro protein-protein interaction assay (Fig. 7B). As expected, monomeric wild-type hRARα was able, when challenged with atRA, to interact with members of the p160 class of NCoAs, such as GRIP1, RIP140, and SRC1, and others, such as DRIP205 and CBP. On the contrary, ligands unable to elicit transactivation by hRARα (CD3105 and CD2409) were unable to promote a detectable interaction of hRARα with these coactivators (Fig. 7B). When the K244-262A mutant was used in a similar assay, none of the ligands used above promoted interaction of hRARα with any of these coactivators (Fig. 7B). Structural transitions leading to NCoA recruitment are thus completely abrogated by the K244-262A mutation, as well as corepressor binding (M. Benkoussa and P. Lefebvre, unpublished). Thus, the K244-262A mutation selectively inactivated NCoA and NCoR recruitment without compromising ligand and DNA binding activities of the receptor. Predictably, the K244-262A mutant did not activate transcription from a DR5-driven promoter (Fig. 7C), yet acted as a transrepressor in the presence of agonists, antagonists, and dissociated retinoids. Thus, NCoA recruitment is not required for AP1 transrepression. This conclusion is further strengthened by results obtained with hRARαΔ403, a C-terminally-truncated RARα mutant. This truncation removes both the F domain and the AF2-AD region of hRARα and yields a receptor exhibiting wild-type ligand binding activity (26), unaltered DNA binding activity (Fig. 8B), and impaired NCoA recruitment and is transcriptionally inactive (25) However, hRARαΔ403 displays full anti-AP1 activity (Fig. 8C).
FIG. 7.
Inactivation of nuclear coactivator recruitment by RARα does not prevent retinoid-mediated AP1 transrepression. (A) Ligand-receptor interaction assay. Wild-type hRARα and hRARα K244-262A were translated in vitro and incubated with 1 μM atRA for 2 h at 4°C. Complexes were then incubated with trypsin, and products were resolved by SDS-10% PAGE. Representative autoradiographs are shown for each receptor. The 28-kDa band represents the trypsin-resistant receptor domain, which encompass most of the ligand binding domain. (B) Interaction of wild-type hRARα and hRARα K244-262A with nuclear coactivators. 35S-labeled hRARα was incubated in the presence of the indicated GST fusion proteins for 2 h and with retinoids. Coactivator-receptor complexes were isolated by adsorption of the GST moiety on agarose beads coupled to glutathione. After washing of the beads, complexes were analyzed by SDS-8% PAGE, and bound receptor was assayed by autoradiography. The affinity of the mutant receptor hRARα K244-262A for NCoAs was assayed as for wild-type RARα. (C) Transactivation and transrepression by the hRARα K244-262A mutant. The activity of the DR5-driven reporter gene was monitored by assaying luciferase activity in HeLa cells transiently transfected with RGXRα and hRARα. Cells were treated with 1 μM atRA for 6 h (left panel). HeLa cells were transfected with RAR expression vectors and the AP1-tk Luc reporter gene. TPA was used as the AP1 inducer at 100 nM, and atRA was used at 1 μM (right panel).
FIG. 8.
Dimerization of hRARα with RXR and DNA-specific recognition are not required for RAR-mediated AP1 inhibition. (A) Dimerization of hRARα with hRXRα in the presence of an agonist, an antagonist, or a dissociated retinoid. 35S-labeled hRARα was produced by in vitro coupled transcription-translation, incubated for 2 h with DMSO or 1 μM retinoids, and adsorbed on a Sepharose-GST-hRXRα affinity matrix. Bound receptors were visualized by autoradiography of the dried SDS-PAGE (8% polyacrylamide) gel. (B) DNA binding activities of hRARα mutants. hRARα or derivatives and hRXRα were obtained by in vitro translation. Receptors were then incubated for 30 min with the labeled DR5 probe, and complexes were resolved by nondenaturing 5% PAGE. Receptor-DNA complexes were then visualized by autoradiography of dried gels. The specific nature of the binding was assessed with a 50-fold excess of cold DR5 or SP1 oligonucleotide in the binding reaction mixture (M. Benkoussa and P. Lefebvre, unpublished). Receptor concentrations were adjusted in order to obtain a 1:1 stoichiometric ratio of RAR to RXR and to avoid significant binding of hRARα homodimers to the DR5 probe. (C) Transrepression and transactivation properties of hRARα mutants. The transactivation (RGXRE tk Luc) and transrepression (AP1-tk Luc) activities of hRGARα (mutated in the P box) of the dimerization-defective RARα (K380-D388RR) and of the truncated hRARα Δ403 were compared. (D) Expression levels of RARα and RARα mutants in transfected HeLa cells. Whole-cell extracts (25 μg of protein) were analyzed by Western blot analysis for their content in overexpressed receptor. Note that RARα Δ403 was not detectable because the antibody is directed against the F domain of RARα.
Dimerization of hRARα with hRXRα and specific DNA binding are not required for AP1 inhibition.
AP1 transrepression by hRARα does not require coexpression of RXR, leading to the prediction that RAR might act as a monomer. We thus wanted to test this hypothesis by first comparing the capacity of representative retinoids to promote hRARα interaction with hRXRα, independently of DNA binding (Fig. 8A). A GST pulldown assay was used, allowing the detection of DNA-independent, ligand-induced heterodimer formation (14). As previously reported, atRA stabilized efficiently hRARα interaction with hRXRα under these conditions. On the contrary, both CD3105 and CD2409 were unable to trigger heterodimer formation, both in vitro, as shown by GST-pulldown assays (Fig. 8A), and in vivo, as shown by a two-hybrid assay in mammalian cells (14). The RXR-RAR dimerization interface is defined by 25 conserved amino acids located in helices 7 to 11 (6, 41). Ten out of 25 RARα amino acid side chains, including those of K380 and D383, are engaging into salt bridges and contribute to 35% of the 967-Å2 RXR-RAR dimerization interface (6). The double mutation yields a receptor mutant that is unable to form heterodimers in solution (Fig. 8A). Consequently, hRARα K380R-D383R does not bind DNA (Fig. 8B) and displays no transactivation potential when tested on a DR5 RARE-driven reporter gene (Fig. 8C) and DR2- and TREpal-driven reporters (P. Lefebvre, unpublished data). However, this mutant fully repressed AP1 activity in a ligand-dependent manner (Fig. 8C). Taken together, these results strongly suggest that dimerization with RXR is not mandatory for transrepression.
We then wished to test whether the ability of the receptor to bind a RARE is required for transrepression. The P box of hRARα, which contains three amino acids essential for specific DNA recognition, was thus mutated to confer specific binding of hRARα to a GRE. As expected, the RAR mutant RGAR did not bind to a DR5 response element in the presence of hRXRα (Fig. 8B) and was inactive in the transactivation assay (Fig. 8C). However, this mutant was fully active in the transrepression assay (Fig. 8C), demonstrating that the ability to bind specifically to cognate response elements is not required for AP1 transrepression.
DISCUSSION
One of the most fascinating question regarding nuclear receptor functions is how a single polypeptide can exhibit distinct transcriptional activities and how the ligand may selectively dictate these activities. Here we report the functional analysis of RARα as an AP1 inhibitor and the characterization of AP1 activation and transrepression processes in a defined cellular background. We also observed that AP1-responsive reporter genes driven by distinct AP1 response elements (from the collagenase A promoter and the atrial natriuretic factor [ANF] promoter) were equally sensitive to retinoid inhibition (M. Benkoussa and P. Lefebvre, unpublished). JNK inhibition has been proposed to be a key step in AP1 repression by glucocorticoids and retinoids (7). In our model, however, the MEK1 and -2-ERK pathway is the main signaling module regulating AP1 activation and sensitive to inhibition by atRA, while the MAPKJNK pathway is poorly active and not sensitive to retinoids. Our results also suggest that transrepression is not a reciprocal phenomenon. This conclusion is consistent with our earlier findings demonstrating that okadaic acid, a protein phosphatase inhibitor, strongly potentiated retinoid-induced transcription (27), whereas it is a known activator of MAPKs, including ERK1 and -2 and JNK (43).
These observations therefore hinted at a new target or targets for retinoids within the MAPKERK signaling module. RARα remains nuclear upon TPA treatment (M. Benkoussa and P. Lefebvre, unpublished observations), excluding a possible direct interaction of RAR with cytoplasmic components of signaling cascades. Moreover, neither the intracellular concentration, activation status, nor subcellular localization of key components of MAPK signaling modules was affected by retinoids (M. Benkoussa and P. Lefebvre, unpublished), suggesting that transcriptional interference is likely to take place in the nuclear compartment.
Activation of the MAPKERK pathway led to preferential loading of junD and fosB on the AP1 response element and to recruitment of RNA pol II, whereas fra2-junD are dimers assembled on the AP1 response element in nonstimulated cells. Activation of the AP1-responsive promoter thus results from displacement of fra1 and preferential binding of fosB to DNA in intact cells. Treatment by atRA led to a favored recruitment of fra1 to the promoter, thus appearing as a competitor of fosB for the access to the AP1 response element, in agreement with the reported transcriptional inactivity of fra1, which, unlike and c-fos and fosB, does not harbor a C-terminal transactivation domain (34, 51).
In vitro protein-protein interaction assays suggested that RAR-c-jun interaction might be responsible for transrepression (45). Similarly, c-jun interacts physically with VDR and represses the granulocye-macrophage colony-stimulating factor gene (49). junD, highly homologous to c-jun, is a common component of AP1 complexes in our system, as well as RARα. Thus, a direct interaction between junD and RARα may occur in intact cells. However, neither supershift experiments nor in vitro protein-protein interaction assays showed an interaction between AP1 components and RARα (M. Benkoussa and P. Lefebvre, unpublished observations), in agreement with a previous report (37). The possibility that weak interactions are not detected by standard in vitro assays is likely; alternatively, a stable RAR-AP1 interaction might occur only in the presence of each component of the complex. AP1, RARα, and ERK1 and -2 are indeed likely to form a multiprotein complex the activity of which is regulated through RAR. Most notably, the transition from an apo RAR-containing complex to a holo RAR-containing complex was accompanied by ERK and CBP displacement. This is an unprecedented observation, suggesting that ERK regulates AP1 activity by phosphorylating either fosB or junD when bound to DNA. Leppä and colleagues (28) have elegantly demonstrated that c-jun is a downstream target for ERKs in both PC12 and NIH 3T3 cells and that activation of these kinases resulted in c-jun phosphorylation at Ser 63 and 73. While this possibility remains to be formally established in our model, this would suggest that retinoids are able to modify junD transcriptional activity by regulating ERK access to its substrate (Ser 100 of junD) in a receptor- and ligand-dependent manner. This retinoid-controlled hypophosphorylation of junD could lead, in analogy with c-jun (4), to a decreased affinity for CBP and therefore to transcriptional inhibition. This hypothesis is supported by our data showing that CBP is, like ERKs, excluded from the AP1-regulated promoter under repressive conditions. In addition, CBP tethering to the promoter is unlikely to result from CBP-RAR interaction, since the RARα K244-262A mutant, unable to recruit various coactivators in vitro, also displays full transrepressive activity.
Functional domains necessary for RARα transrepressive activity are yet to be characterized. However, our results show that heterodimerization with RXR and specific DNA binding are not required. Importantly, coactivator recruitment to RARα is also dispensable as shown by the wild-type anti-AP1 activity of RARα K244-262A and the ability of a retinoid antagonist to fully elicit AP1 transrepression. We cannot, however, formally rule out, at this stage, that an yet unknown coactivator may be implicated in this process. We, however, do not favor this hypothesis in light of the high conservation of the receptor-coactivator interaction motif and of recent results reported for the glucocorticoid receptor (GR) (11). Preliminary results showed that isolated RARα AF1 or AF2 domains are weakly active in the transrepression assay (M. Benkoussa and P. Lefebvre, unpublished), suggesting that RARα structural integrity has to be maintained for full AP1 inhibition.
Our results thus provide strong support for the proposal that retinoids are able to trigger qualitative differences of AP1 complexes assembled onto an AP1 response element. More importantly, RARα appeared as a regulator of ERK access to its substrate bound to DNA, thereby altering AP1 transcriptional properties by regulating CBP association to the AP1 complex. In this respect, we note that p38, another MAPK, was recently described as part of the transcriptional machinery in yeast (1). Thus, kinase incorporation into the transcriptional machinery may be a general property of these enzymes. Furthermore, our data identify a new paradigm for transcriptional interference mediated by nuclear receptors, which act as monomeric entities and independently of their well-characterized transactivation activity.
Acknowledgments
We thank R. M. Evans (Salk Institute), J. D. Chen (University of Massachussetts), V. Cavailles (INSERM U148, Montpellier, France), D. D. Moore and B. W. O'Malley (Baylor College of Medicine), S. Michel and U. Reichert (CIRD-Galderma), L. P. Freedman and C. Rachez (MSKCC, New York, N.Y.), M. Yaniv (Institut Pasteur, Paris, France), J. M. Blanchard (I.G.M., C.N.R.S. Montpellier), Jim Woodgett (Ontario Cancer Institute), R. G. Davis (H.H.M.I., University of Massachusetts Medical School), and M. Nemer (I.R.C.M, Canada) for providing us with DNAs, retinoids, and antibodies. We thank C. Rachez and P. Sacchetti for careful reading of the manuscript and B. Masselot for technical help.
INSERM U459 is part of IFR 22 (INSERM, C.H. and U. de Lille, C.O.L. and University of Lille 2). This work was supported by grants from INSERM, Association pour la Recherche sur le Cancer and Ligue Nationale contre le Cancer (Comité du Nord-Pas de Calais).
REFERENCES
- 1.Alepuz, P. M., A. Jovanovic, V. Reiser, and G. Ammerer. 2001. Stress-induced MAP kinase hog1 is part of transcription activation complexes. Mol. Cell 7:767-777. [DOI] [PubMed] [Google Scholar]
- 2.Alessi, D. R., A. Cuenda, P. Cohen, D. T. Dudley, and A. R. Saltiel. 1995. PD 098059 is a specific inhibitor of the activation of mitogen-activated protein kinase kinase in vitro and in vivo. J. Biol. Chem. 270:27489-27494. [DOI] [PubMed] [Google Scholar]
- 3.Angel, P., M. Imagawa, R. Chiu, B. Stein, C. Jonat, and M. Karin. 1987. Phorbol-ester inducible genes contain a common cis-element recognized by a TPA-modulated trans-acting factor. Cell 49:729-739. [DOI] [PubMed] [Google Scholar]
- 4.Bannister, A. J., T. Oehler, D. Wilhelm, P. Angel, and T. Kouzarides. 1995. Stimulation of c-Jun activity by CBP: c-Jun residues Ser63/73 are required for CBP induced stimulation in vivo and CBP binding in vitro. Oncogene 11:2509-2514. [PubMed] [Google Scholar]
- 5.Benkoussa, M., B. Nomine, A. Mouchon, B. Lefebvre, J. M. Bernardon, P. Formstecher, and P. Lefebvre. 1997. Limited proteolysis for assaying ligand binding affinities of nuclear receptors. Recept. Signal. Transduct. 7:257-267. [PubMed] [Google Scholar]
- 6.Bourguet, W., V. Vivat, J. M. Wurtz, P. Chambon, H. Gronemeyer, and D. Moras. 2000. Crystal structure of a heterodimeric complex of RAR and RXR ligand-binding domains. Mol. Cell 5:289-298. [DOI] [PubMed] [Google Scholar]
- 7.Caelles, C., J. M. Gonzalez-Sancho, and A. Munoz. 1997. Nuclear hormone receptor antagonism with AP-1 by inhibition of the JNK pathway. Genes Dev. 11:3351-3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Castano, E., P. Gross, Z. X. Wang, R. G. Roeder, and T. Oelgeschlager. 2000. The C-terminal domain-phosphorylated IIO form of RNA polymerase II is associated with the transcription repressor NC2 (Dr1/DRAP1) and is required for transcription activation in human nuclear extracts. Proc. Natl. Acad. Sci. USA 97:7184-7189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, J. Y., S. Penco, J. Ostrowski, P. Balaguer, M. Pons, J. E. Starrett, P. Reczek, P. Chambon, and H. Gronemeyer. 1995. RAR-specific agonist/antagonists which dissociate transactivation and AP1 transrepression inhibit anchorage-independent cell proliferation. EMBO J. 14:1187-1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cook, S. J., N. Aziz, and M. McMahon. 1999. The repertoire of Fos and Jun proteins expressed during the G1 phase of the cell cycle is determined by the duration of mitogen-activated protein kinase activation. Mol. Cell. Biol. 19:330-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeBosscher, K., W. V. Berghe, L. Vermeulen, S. Plaisance, E. Boone, and G. Haegeman. 2000. Glucocorticoids repress NF-kappa B-driven genes by disturbing the interaction of p65 with the basal transcription machinery, irrespective of coactivator levels in the cell. Proc. Natl. Acad. Sci. USA 97:3919-3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Groot, R. P., J. Auwerx, M. Bourouis, and P. Sassone-Corsi. 1993. Negative regulation of Jun/AP-1: conserved function of glycogen synthase kinase 3 and the Drosophila kinase shaggy. Oncogene 8:841-847. [PubMed] [Google Scholar]
- 13.Delmotte, M. H., A. Tahayato, P. Formstecher, and P. Lefebvre. 1999. Serine 157, a retinoic acid receptor alpha residue phosphorylated by protein kinase C in vitro, is involved in RXR.RARalpha heterodimerization and transcriptional activity. J. Biol. Chem. 274:38225-38231. [DOI] [PubMed] [Google Scholar]
- 14.Depoix, C., M. H. Delmotte, P. Formstecher, and P. Lefebvre. 2001. Control of retinoic acid receptor heterodimerization by ligand-induced structural transitions. J. Biol. Chem. 276:9452-9459. [DOI] [PubMed] [Google Scholar]
- 15.Dominguez, I., K. Itoh, and S. Y. Sokol. 1995. Role of glycogen synthase kinase 3 beta as a negative regulator of dorsoventral axis formation in Xenopus embryos. Proc. Natl. Acad. Sci. USA 92:8498-8502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Efimova, T., P. LaCelle, J. F. Welter, and R. L. Eckert. 1998. Regulation of human involucrin promoter activity by a protein kinase C, Ras, MEKK1, MEK3, p38/RK, AP1 signal transduction pathway. J. Biol. Chem. 273:24387-24395. [DOI] [PubMed] [Google Scholar]
- 17.Fanjul, A., M. I. Dawson, P. D. Hobbs, L. Jong, J. F. Cameron, E. Harlev, G. Graupner, X. P. Lu, and M. Pfahl. 1994. A new class of retinoids with selective inhibition of AP-1 inhibits proliferation. Nature 372:107-111. [DOI] [PubMed] [Google Scholar]
- 18.Hazzalin, C. A., R. Le Panse, E. Cano, and L. C. Mahadevan. 1998. Anisomycin selectively desensitizes signalling components involved in stress kinase activation and fos and jun induction. Mol. Cell. Biol. 18:1844-1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He, X., J. P. Saint-Jeannet, J. R. Woodgett, H. E. Varmus, and I. B. Dawid. 1995. Glycogen synthase kinase-3 and dorsoventral patterning in Xenopus embryos. Nature 374:617-622. [DOI] [PubMed] [Google Scholar]
- 20.Kamei, Y., L. Xu, T. Heinzel, J. Torchia, R. Kurokawa, B. Gloss, S. C. Lin, R. A. Heyman, D. W. Rose, C. K. Glass, and M. G. Rosenfeld. 1996. A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell 85:403-414. [DOI] [PubMed] [Google Scholar]
- 21.Karin, M. 1996. The regulation of AP-1 activity by mitogen-activated protein kinases. Philos. Trans. R. Soc. Lond. Biol. 351:127-134. [DOI] [PubMed] [Google Scholar]
- 22.Klein, E. S., M. E. Pino, A. T. Johnson, P. J. Davies, S. Nagpal, S. M. Thacher, G. Krasinski, and R. A. Chandraratna. 1996. Identification and functional separation of retinoic acid receptor neutral antagonists and inverse agonists. J. Biol. Chem. 271:22692-22696. [DOI] [PubMed] [Google Scholar]
- 23.Klein, P. S., and D. A. Melton. 1996. A molecular mechanism for the effect of lithium on development. Proc. Natl. Acad. Sci. USA 93:8455-8459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee, H.-Y., N. Sueoka, W.-K. Hong, D. J. Mangelsdorf, F. X. Claret, and J. M. Kurie. 1999. All-trans-retinoic acid inhibits Jun N-terminal kinase by increasing dual-specificity phosphatase activity. Mol. Cell Biol. 19:1973-1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lefebvre, B., A. Mouchon, P. Formstecher, and P. Lefebvre. 1998. H11-H12 loop retinoic acid receptor mutants exhibit distinct trans-activating and trans-repressing activities in the presence of natural or synthetic retinoids. Biochemistry 37:9240-9249. [DOI] [PubMed] [Google Scholar]
- 26.Lefebvre, B., C. Rachez, P. Formstecher, and P. Lefebvre. 1995. Structural determinants of the ligand-binding site of the human retinoic acid receptor alpha. Biochemistry 34:5477-5485. [DOI] [PubMed] [Google Scholar]
- 27.Lefebvre, P., M. P. Gaub, A. Tahayato, C. Rochette-Egly, and P. Formstecher. 1995. Protein phosphatases 1 and 2A regulate the transcriptional and DNA binding activities of retinoic acid receptors. J. Biol. Chem. 270:10806-10816. [DOI] [PubMed] [Google Scholar]
- 28.Leppa, S., R. Saffrich, W. Ansorge, and D. Bohmann. 1998. Differential regulation of c-Jun by ERK and JNK during PC12 cell differentiation. EMBO J. 17:4404-4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li, Y., Y. Hashimoto, A. Agadir, H. Kagechika, and X. Zhang. 1999. Identification of a novel class of retinoic acid receptor beta-selective retinoid antagonists and their inhibitory effects on AP-1 activity and retinoic acid-induced apoptosis in human breast cancer cells. J. Biol. Chem. 274:15360-15366. [DOI] [PubMed] [Google Scholar]
- 30.Liu, M., A. Iavarone, and L. P. Freedman. 1996. Transcriptional activation of the human p21(WAF1/CIP1) gene by retinoic acid receptor—correlation with retinoid induction of U937 cell differentiation. J. Biol. Chem. 271:31723-31728. [DOI] [PubMed] [Google Scholar]
- 31.Marais, R., J. Wynne, and R. Treisman. 1993. The SRF accessory protein Elk-1 contains a growth factor-regulated transcriptional activation domain. Cell 73:381-393. [DOI] [PubMed] [Google Scholar]
- 32.Martiny-Baron, G., M. G. Kazanietz, H. Misschak, P. M. Blumberg, G. Kochs, G. Hug, D. Marme, and C. Schachtele. 1993. Selective inhibition of protein kinase C isozymes by the indolocarbazole Go 6976. Proc. Natl. Acad. Sci. USA 268:9194-9197. [PubMed] [Google Scholar]
- 33.McBride, K., and M. Nemer. 1998. The C-terminal domain of c-fos is required for activation of an AP-1 site specific for jun-fos heterodimers. Mol. Cell. Biol. 18:5073-5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Metz, R., T. Kouzarides, and R. Bravo. 1994. A C-terminal domain in FosB, absent in FosB/SF and Fra-1, which is able to interact with the TATA binding protein, is required for altered cell growth. EMBO J. 13:3832-3842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mouchon, A., M.-H. Delmotte, P. Formstecher, and P. Lefebvre. 1999. Allosteric regulation of the discriminative responsiveness of retinoic acid receptor to natural and synthetic ligands by retinoid X receptor and DNA. Mol. Cell. Biol. 19:3073-3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nagpal, S., J. Athanikar, and R. A. S. Chandraratna. 1995. Separation of transactivation and AP1 antagonism functions of retinoic acid receptor alpha. J. Biol. Chem. 270:923-927. [DOI] [PubMed] [Google Scholar]
- 37.Nicholson, R. C., S. Mader, S. Nagpal, M. Leid, C. Rochette-Egly, and P. Chambon. 1990. Negative regulation of the rat stromelysin gene promoter by retinoic acid is mediated by an AP1 binding site. EMBO J. 9:4443-4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nikolakaki, E., P. J. Coffer, R. Hemelsoet, J. R. Woodgett, and L. H. K. Defize. 1993. Glycogen synthase kinase-3 phosphorylates jun family members in vitro and negatively regulates their transactivating potential in intact cells. Oncogene 8:833-840. [PubMed] [Google Scholar]
- 39.Perez, P., A. Schonthal, and A. Aranda. 1993. Repression of c-fos gene expression by thyroid hormone and retinoic acid receptors. J. Biol. Chem. 268:23538-23543. [PubMed] [Google Scholar]
- 40.Philips, A., P. Roux, V. Coulon, J. M. Bellanger, A. Vie, M. L. Vignais, and J. M. Blanchard. 2000. Differential effect of Rac and Cdc42 on p38 kinase activity and cell cycle progression of nonadherent primary mouse fibroblasts. J. Biol. Chem. 275:5911-5917. [DOI] [PubMed] [Google Scholar]
- 41.Rachez, C., P. Sautiere, P. Formstecher, and P. Lefebvre. 1996. Identification of amino acids critical for the DNA binding and dimerization properties of the human retinoic acid receptor alpha. Importance of lysine 360, lysine 365, and valine 361. J. Biol. Chem. 271:17996-18006. [DOI] [PubMed] [Google Scholar]
- 42.Rachez, C., Z. Suldan, J. Ward, C. P. B. Chang, D. Burakov, H. Erdjument-Bromage, P. Tempst, and L. P. Freedman. 1998. A novel protein complex that interacts with the vitamin D3 receptor in a ligand-dependent manner and enhances VDR transactivation in a cell-free system. Genes Dev. 12:1787-1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rosenberger, S. F., J. S. Finch, A. Gupta, and G. T. Bowden. 1999. Extracellular signal-regulated kinase 1/2-mediated phosphorylation of JunD and FosB is required for okadaic acid-induced activator protein 1 activation. J. Biol. Chem. 274:1124-1130. [DOI] [PubMed] [Google Scholar]
- 44.Sanchez, I., R. T. Hughes, B. J. Mayer, K. Yee, J. R. Woodgett, J. Avruch, J. M. Kyriakis, and L. I. Zon. 1994. Role of SAPK/ERK kinase-1 in the stress-activated pathway regulating transcription factor c-Jun. Nature 372:794-798. [DOI] [PubMed] [Google Scholar]
- 45.Schule, R., P. Rangarajan, N. Yang, S. Kliewer, L. J. Ransone, J. Bolado, I. M. Verma, and R. M. Evans. 1991. Retinoic acid is a negative regulator of AP-1-responsive genes. Proc. Natl. Acad. Sci. USA 88:6092-6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stambolic, V., L. Ruel, and J. R. Woodgett. 1996. Lithium inhibits glycogen synthase kinase-3 activity and mimics wingless signalling in intact cells. Curr. Biol. 6:1664-1668. [DOI] [PubMed] [Google Scholar]
- 47.Tahayato, A., P. Lefebvre, P. Formstecher, and M. Dautrevaux. 1993. A protein kinase C-dependent activity modulates retinoic acid-induced transcription. Mol. Endocrinol. 7:1642-1653. [DOI] [PubMed] [Google Scholar]
- 48.Toullec, D., P. Pianetti, H. Coste, P. Bellevergue, T. Grand-Peret, M. Ajakane, V. Baudet, P. Boissin, E. Boursier, F. Loriolle, L. Duhamel, D. Charon, and J. Kirilovski. 1991. The bisindolylmaleimide GF 109203X is a potent and selective inhibitor of protein kinase C. J. Biol. Chem. 266:15771-15781. [PubMed] [Google Scholar]
- 49.Towers, T. L., T. P. Staeva, and L. P. Freedman. 1999. A two-hit mechanism for vitamin D3-mediated transcriptional repression of the granulocyte-macrophage colony-stimulating factor gene: vitamin D receptor competes for DNA binding with NFAT1 and stabilizes c-Jun. Mol. Cell. Biol. 19:4191-4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vega-Diaz, B., M. C. Lenoir, A. Ladoux, C. Frelin, M. Démarchez, and S. Michel. 2000. Regulation of vascular endothelial growth factor expression in human keratinocytes by retinoids. J. Biol. Chem. 275:642-650. [DOI] [PubMed] [Google Scholar]
- 51.Wisdon, R., and I. M. Verma. 1993. Transformation by Fos proteins requires a C-terminal transactivation domain. Mol. Cell. Biol. 13:7429-7438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu, L., C. K. Glass, and M. G. Rosenfeld. 1999. Coactivator and corepressor complexes in nuclear receptor function. Curr. Opin. Genet. Dev. 9:140-147. [DOI] [PubMed] [Google Scholar]
- 53.Yamamoto, S., Y. Watanabe, P. J. van der Spek, T. Watanabe, H. Fujimoto, F. Hanaoka, and Y. Ohkuma. 2001. Studies of nematode TFIIE function reveal a link between Ser-5 phosphorylation of RNA polymerase II and the transition from transcription initiation to elongation. Mol. Cell. Biol. 21:1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Young, M. R., J. J. Li, M. Rincon, R. A. Flavell, B. K. Sathyanarayana, R. Hunziker, and N. Colburn. 1999. Transgenic mice demonstrate AP-1 (activator protein-1) transactivation is required for tumor promotion. Proc. Natl. Acad. Sci. USA 96:9827-9832. [DOI] [PMC free article] [PubMed] [Google Scholar]