Abstract
Eukaryotic translation initiation factor 4GI (eIF4GI) is an essential protein that is the target for translational regulation in many cellular processes and viral systems. It has been shown to function in both cap-dependent and cap-independent translation initiation by recruiting the 40S ribosomal subunit to the mRNA cap structure or internal ribosome entry site (IRES) element, respectively. Interestingly eIF4GI mRNA itself has been reported to contain an IRES element in its 5′ end that facilitates eIF4GI protein synthesis via a cap-independent mechanism. In HeLa cells, eIF4GI exists as several isoforms that differ in their migration in sodium dodecyl sulfate (SDS) gels; however, the nature of these isoforms was unclear. Here, we report a new cDNA clone for eIF4GI that extends the 5′ sequence 340 nucleotides beyond the previously published sequence. The new extended sequence of eIF4GI is located on chromosome 3, within two additional exons immediately upstream of the previously published eIF4GI sequence. When mRNA transcribed from this cDNA clone was translated in vitro, five eIF4GI polypeptides were generated that comigrated in SDS-polyacrylamide gels with the five isoforms of native eIF4GI. Furthermore, translation of eIF4GI-enhanced green fluorescent protein fusion constructs in vitro or in vivo generated five isoforms of fusion polypeptides, suggesting that multiple isoforms of eIF4GI are generated by alternative translation initiation in vitro and in vivo. Mutation of two of the five in-frame AUG residues in the eIF4GI cDNA sequence resulted in loss of corresponding polypeptides after translation in vitro, confirming alternate use of AUGs as the source of the multiple polypeptides. The 5′ untranslated region of eIF4GI mRNA also contains an out-of-frame open reading frame (ORF) that may down-regulate expression of eIF4GI. Further, data are presented to suggest that a proposed IRES embedded in the eIF4GI ORF is able to catalyze synthesis of multiple eIF4GI isoforms as well. Our data suggest that expression of the eIF4GI isoforms is partly controlled by a complex translation strategy involving both cap-dependent and cap-independent mechanisms.
Eukaryotic translation initiation factor 4G (eIF4G) is a large modular protein that in association with eIF4E (cap-binding protein) and eIF4A (RNA helicase) forms the translation initiation complex eIF4F. eIF4G contains many domains that bind translation factors and mRNA simultaneously in preinitiation complexes. eIF4G can be roughly divided into three domains, N terminal, central, and C terminal. The N-terminal third of eIF4G contains binding sites for poly(A)-binding protein (PABP) and eIF4E as well as a large proline-rich segment of undefined function (25, 32, 38). The central region, whose structure was recently solved, is the most highly conserved region of eIF4G and contains binding domains for eIF3, eIF4A, and an mRNA recognition motif (32, 39). The C-terminal domain contains another binding domain for eIF4A and a binding domain for mnk-1 and mnk-2 kinases, which activate eIF4E via phosphorylation (30, 44, 47). A functional core region has been described that comprises the binding domains for eIF4E, eIF3, and eIF4A and is capable of minimally supporting translation initiation in Saccharomyces cerevisiae and mammalian systems (14, 42). The C-terminal domain has been proposed to enhance and regulate the activity of the core domain (14, 42).
Since eIF4G can bind eIF4E and eIF3 simultaneously, it is generally accepted that eIF4G forms a molecular bridge linking the 5′ end of the mRNA to the ribosomal subunit and thus is an essential factor in the recruitment of the 40S ribosomal subunits to the 5′ end of capped mRNA (reviewed in references 20 and 43). In contrast, eIF4G is also required in cap-independent translation initiation on picornavirus RNAs to recruit the 40S ribosomal subunit to mRNA (37). In this case eIF4G and other factors bind to a large secondary-structure element within the RNA that is termed the internal ribosome entry site (IRES); however, eIF4E is not required. Two independent isoforms of human eIF4G, now termed eIF4GI and eIF4GII, have been described previously (21). These isoforms are only 46% conserved overall but contain homologous protein binding domains and interact with the same factors (eIF4E, eIF4A, and eIF3) to form functional eIF4F complexes (21). eIF4GI is the predominant form of eIF4G in HeLa cells and is estimated to comprise about 85% of the total in eIF4F complexes, partly based on examination of unique cleavage products in silver-stained gels (5, 40).
Recently, 5′-3′ interactions in mRNA have also been shown to be important for translation initiation and regulation. Specifically, eIF4G is able to bridge 5′ and 3′ ends of mRNA by simultaneous binding of eIF4E (cap structure) and PABP [poly(A) tail], forming pseudocircular mRNA structures (50, 52). 5′-3′ interactions mediated by this complex stimulate translation initiation synergistically (4, 41a), play crucial roles in the rate of mRNA turnover, and may explain the location of translation enhancers in the 3′ nontranslated region of certain mRNAs (9, 15, 50).
The essential role of eIF4GI in translation initiation also makes this protein a central target for regulation of translation. During poliovirus or rhinovirus infection of HeLa cells, cap-dependent translation is rapidly shut off (16, 46). The mechanism involves proteolytic cleavage of eIF4GI between the eIF4E and eIF3 binding domains, thus severing the bridge between the 40S ribosomal subunit and the 5′ cap structure (16, 32, 34, 36, 54). The C-terminal segment of eIF4GI that is released by cleavage contains eIF3 and eIF4A binding domains and functions efficiently to initiate cap-independent translation of viral RNA via the viral IRES elements (3, 29). Additionally, eIF4GI and eIF4GII are cleaved by caspase 3 during apoptosis at times when translation is being down-regulated (11, 40, 41). Other viral proteins are also known to modify translation by binding to eIF4GI and interfering with its function. Influenza ns1 has recently been shown to bind to the N-terminal region of eIF4GI, which recruits eIF4GI specifically to viral mRNA at the expense of host translation (2). Further, adenovirus 100-kDa protein can reduce cellular cap-dependent translation by binding to eIF4GI, and evicting the mnk kinase (13). During heat shock translation is down-regulated at times when levels of functional eIF4GI are reduced, and it has been recently shown that hsp27 binds eIF4GI to sequester it in nonfunctional complexes (12, 33).
Interestingly, eIF4GI appears as a series of four to five polypeptide bands with apparent molecular masses of 200 to 220 kDa when resolved in sodium dodecyl sulfate (SDS)-polyacrylamide gels. Despite the enormous amount of information known about eIF4GI structure and function, no molecular explanation has emerged as to the nature of these isoforms. The original eIF4GI clone was obtained from a human brain cDNA library and was reported to have a long (450-nucleotide [-nt]) 5′ untranslated region (5′ UTR) (hereafter referred to as brain expression [BE] 5′ UTR) which contained an IRES element. This clone generates a single polypeptide of approximately 195 kDa when translated in vitro, and it has been proposed that posttranslational modifications or alternate splicing may generate the multiple isoforms observed in vivo (25, 53). eIF4GI has been shown to be extensively phosphorylated, but no correlation between phosphorylation and specific isoforms has been established (45). More recently, two groups independently reported finding additional cDNA sequences for eIF4GI that extended the 5′ end and expanded the originally reported eIF4GI open reading frame (ORF) by 156 amino acids (25, 27). The nature of the polypeptide(s) generated by the addition of these extended sequences has not been described. Interestingly, the extension segment has been proposed to contain a different IRES element, conferring the property of cap-independent expression to eIF4GI itself (27).
Here we report a new eIF4GI cDNA sequence that contains an additional 340-nt 5′-terminal extension beyond previous clones and produces full-length eIF4GI that, when translated in vitro, comigrates on gels with native eIF4GI. By fusing the new 5′ sequence of eIF4GI onto enhanced green fluorescent protein (EGFP) cDNA, we show that the multiple isoforms of eIF4GI result from alternative translation initiation at different in-frame AUG codons in vitro and in vivo. Further, we show that expression of eIF4GI is influenced by an upstream ORF and several isoforms can be translated from dicistronic RNAs. Thus, eIF4GI expression in vivo is likely mediated via a complex translation regulation scheme involving both cap-dependent and cap-independent mechanisms that generate multiple isoforms of eIF4GI.
MATERIALS AND METHODS
Cells and mammalian cell transfections.
HeLa S3 cells were grown in SMEM (Invitrogen) supplemented with 9% bovine calf serum-1% fetal calf serum, and penicillin-streptomycin (Invitrogen). HeLa CCL2 and 293T cells were maintained in Iscove's DMEM (Invitrogen) or high glucose DMEM (Invitrogen), respectively, with the same amounts of serum and antibiotics. In vivo labeling and processing of cell lysates were carried out as previously described (26). Transfections of HeLa cells were performed using the calcium phosphate method (10) with minor modifications. Sixteen hours posttransfection cells were washed once in phosphate-buffered saline (PBS) containing 10% dimethyl sulfoxide, and the medium was replaced. Following another 20 h of growth, cells were pelleted, washed twice with PBS, and resuspended in DMEM containing 10% dialyzed calf serum and lacking methionine. Transfected cells were labeled at 37°C for 1.5 h by the addition of Tran35S-label (10 μCi/ml; ICN). Labeled cells were washed with PBS and subjected to detergent lysis (20 mM HEPES, pH 7.6; 10 mM KCl; 1.2 mM magnesium acetate; 2.5 mM dithiothreitol; 1% NP-40) for 10 min at 37°C. Nuclei were pelleted, and supernatant lysates were collected. 293T cells were transfected with plasmid constructs using Lipofectamine (Invitrogen) according to the manufacturer's protocols. Cells were harvested 36 h posttransfection and subjected to detergent lysis as described above.
Immunoprecipitation.
Immunoprecipitation from in vitro translation reactions or transfected HeLa lysates was carried out in IP buffer (20 mM Tris-HCl, pH 8.0; 150 mM NaCl; 0.5% NP-40; 5 mM EDTA). Lysates were precleared by incubation with protein A-agarose beads for 60 min at 4°C prior to the addition of antibody. Antibody (as indicated in the figure legends) was incubated with lysates for 1 h at 4°C, followed by incubation together with protein A-agarose overnight at 4°C. Agarose beads were subjected to multiple washes with IP buffer. Immunoprecipitated proteins were resolved by SDS-polyacrylamide gel electrophoresis (PAGE), transferred to nitrocellulose membranes where indicated, and detected by autoradiography or immunoblotting.
In vitro translation reactions.
Nuclease-treated rabbit reticulocyte lysates (Promega) were programmed with capped or uncapped T7 RNA transcripts in the presence of [35S]methionine-cysteine (Tran35S-label) and incubated at 30°C for 90 min. Where indicated the translation mix was then incubated with 0.05 μg of purified coxsackievirus B3 (CVB3) 2A protease (2Apro) cotranslationally or for 15 min at 30°C before RNA addition. Translations were analyzed by SDS-PAGE-autoradiography as indicated. CVB3 2Apro was expressed and purified as previously described (26).
Plasmids.
pCR2.1-4Gext (27) was amplified by PCR using primers GCTCTAGACCATGgctagcatgactggtggacagcaaATGAACACGCCTCCTCAGCCCCGCC and CCCAAGCTTCAGACATGATCTCCTCTGTGATATCCTTTCC. The T7 tag epitope is underlined, boldface type indicates the initiation codon in this construct, and double underlining indicates the eIF4GI initiation codon based on the published sequence (27). The resulting PCR fragment spanning the coding region of 4Gext was cloned into pcDNA3.1 (Invitrogen) to generate pcT74Gext. pT74GI, containing the extended eIF4GI coding sequence, was constructed by cloning the EcoRI-EcoRI fragment, spanning the original eIF4GI coding region, from pAD-4G (28, 53) fused into pcT74Gext. pc4G-IRES was constructed by cloning the 5′ UTR of the original eIF4G clone (53) from pCMV-CAT/4GUTR/L (18, 19) into pcDNA3.1. The full-length eIF4GI cDNA clone DKFZp762O191Q3 (accession no. AL120751), referred to in this work as pSP4GI, was obtained from the German Cancer Research Center (DKFZ) and was cloned into pSPORT vector (Gibco). The full-length cDNA was subjected to further sequence analysis and resubmitted to GenBank as accession no. AY082886. p4G(395-821)-EGFP and pIRES-EGFP were constructed by cloning the EGFP gene from pEGFP-N2 (Clontech) into the pT74GI or pc4G-IRES, respectively. p4G(1-821)-EGFP was generated by cloning the 5′ end of eIF4GI cDNA from pSP4GI as a SalI/EcoRI fragment into p4G(395-821)-EGFP linearized with NheI/EcoRI. p4G(1-364)-EGFP was constructed by cleavage of p4G(1-821)-EGFP with SgrAI and AgeI, followed by ligation. p4G(1-274)-EGFP was constructed by isolation of a PCR fragment from pSP4GI, using primers 5′-CGGGATCCAAGCTTCTAGACCTGTGTGCCCCTGGGCCAGGCCC-3′ (forward) and 5′-GCGTGATCAAAGCTTTCATCTTCATTTGGTGCCACATCAGGGTC-3′ (reverse), that was digested with HindIII and inserted into vector pEGFP that contained both cytomegalovirus and T7 RNA polymerase promoters upstream of the EGFP ORF. p4G(5′uORF)-EGFP was constructed by digesting p4G(1-274)-EGFP with AgeI, followed by blunting the DNA overhangs with Klenow fragment and ligation.
pSP4GI ΔAUG275, ΔAUG395, and ΔAUG275,395 were generated by site-directed mutation of pSP4GI. In each case the indicated ATG was mutated to GTG using a QuikChange XL mutagenesis kit (Stratagene) and primers to pSP4GI that incorporated the base changes (31-nt complementary primer pairs [IDT]). p(5′uORF)-SP4GI was constructed using a QuikChange XL mutagenesis kit and primers to pSP4GI that incorporate a single adenosine insertion after nt 274 in the eIF4GI 5′ UTR coding region.
pRL-EGFP was generated by ligation of an NheI-Bsp120I fragment containing EGFP from pEGFP into pRL-HL (24) digested with XbaI and NotI, to remove the hepatitis C virus IRES and firefly luciferase gene. To construct pRL-4G(275-873)-EGFP, pSP4GI was subjected to PCR with primers 5′-CCTGTGTGCCCCTGGGCCAGGCCCG-3′ and 5′-GCCCCAGACATGATCTCCTCTGTGAT-3′ (IDT) and products were gel purified and ligated into pGEM-T Easy (Promega) to generate pGEM-TE 4G(275-873). pGEM-TE 4G(275-873) was digested with NotI and ligated into pRL-EGFP digested with Bsp120I. A thermodynamically stable hairpin previously described as blocking 43S ribosomal scanning (8) was amplified by PCR using primers 5′-CTAGCTAGCTTATAAAAGCAGTGGCTGCGG-3′ and 5′-GTCGCTAGCATTCTGTCTGTTTTGGGGGAT-3′ that added NheI sites to each end of the product DNA. Following digestion with NheI, the DNA was ligated into the NheI site of pRL-4G(275-873)-EGFP just downstream of the T7 promoter to generate phpRL-4G(275-873)-EGFP.
Antibodies.
Rabbit polyclonal antiserum specific to the N-terminal end of human eIF4GI (36) or antipeptide serum specific to a region in the N-terminal domain of eIF4GI (amino acids 231 to 251) (1) was used as previously described. Rabbit polyclonal anti-eIF4GI antiserum used for immunoprecipitation was produced by expression of the C-terminal 480 amino acid residues of eIF4GI as a His-tagged polypeptide in Escherichia coli. Protein was purified by metal chelate chromatography (Talon; Clontech) and injected in five boosts (50 μg each) over 3 months into rabbits to generate hyperimmune serum. Mouse monoclonal anti-green fluorescent protein (anti-GFP) antibody was obtained from Quantum, and rabbit polyclonal anti-EGFP antibody was obtained from Santa Cruz. Rabbit polyclonal antibody against T7 epitope tag was obtained from Invitrogen.
Northern blotting.
Transfected 293 cells were lysed directly into Trizol extraction reagent (Invitrogen). RNA samples were run on a denaturing formaldehyde gel and blotted to a nylon membrane (MSI) using the Northern Max kit (Ambion). Membranes were probed overnight at 68°C with a [32P]GTP-labeled antisense riboprobe directed to the EGFP ORF. Following four washes (described in the Ambion protocol) the membranes were exposed to film overnight at −70°C.
Nucleotide sequence accession number.
The full-length eIF4GI cDNA clone DKFZp762O191Q3 that was subjected to further sequence analysis in this study has been resubmitted to GenBank under accession no. AY082886.
RESULTS
Sequence of complete eIF4GI cDNA.
We obtained the 4Gext cDNA sequence reported by Johannes and Sarnow (27) that was then cloned onto the 5′ end of the original eIF4GI clone (pAD-4G) to produce plasmid pT74GI. We also searched GenBank, including the EST (expressed sequence tag) database, using the 4Gext region as the query sequence to find any additional 5′ extension sequences. One clone generated by the Molecular Genome Analysis Program at the DKFZ (accession no. AL120751) from a human melanoma cell line, was found to contain the 4Gext region and an additional sequence at the 5′ end (Fig. 1A). The published sequence listed only 637 nt in the clone; however, further sequence analysis of both ends and several internal regions of the insert and restriction analysis in our laboratory (data not shown) revealed that the DKFZp762O191Q3 clone contained the entire eIF4GI coding region with 5′ and 3′ UTRs (Fig. 1 and 2). This clone is referred to as pSP4GI.
FIG. 1.
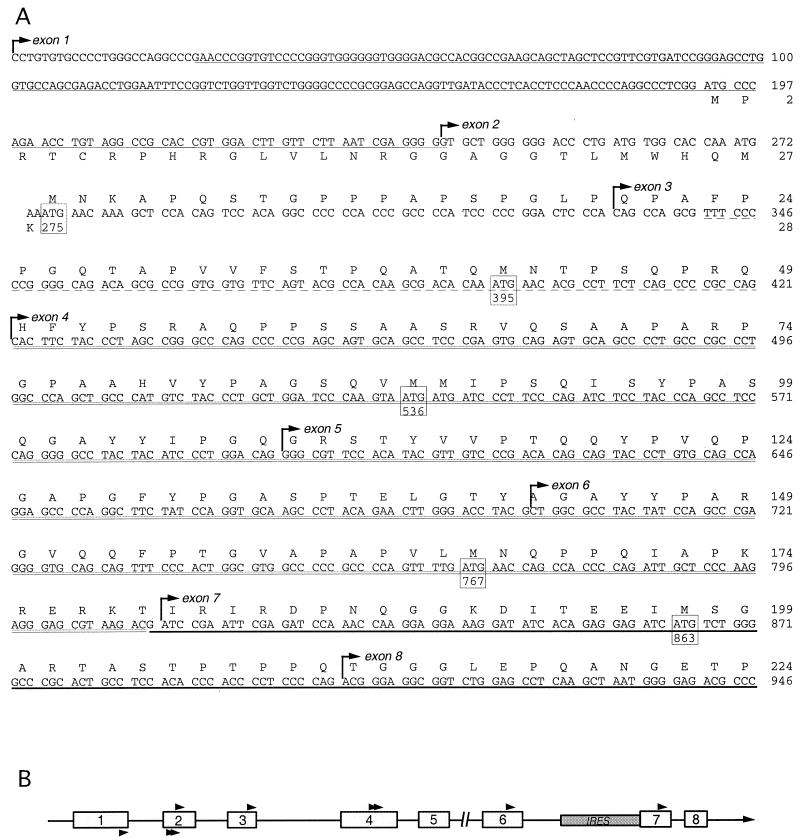
(A) 5′ terminal sequence of eIF4GI cDNA. The sequence begins with the first nucleotide of cDNA insert in pSP4GI. The amino acid sequence of eIF4GI is shown above the nucleotide sequence starting at nt 275. The amino acid sequence of the upstream ORF beginning at nt 192 is shown below the nucleotide sequence. In frame ATG codons within the eIF4GI ORF are boxed. Boldface underlining denotes nucleotides shared between the original eIF4GI clone, 4Gext sequence, and pSP4GI. Double underlining indicates nucleotide sequence shared between 4Gext reported by Gradi et al. (21), that reported by Johannes and Sarnow (27), and pSP4GI. Dotted underlining indicates sequence shared between the longer Johannes 4Gext segment (27) and pSP4GI. The single-underlined sequence is shared between an unmapped cDNA extension reported by Poncet and Cohen (accession no. AF002815.1) and pSP4GI. The complete eIF4GI cDNA sequence present in pSP4GI has been deposited in GenBank under accession no. AY082886. Exon junctions are indicated by arrows. (B) A linear map of the 5′ end of the eIF4GI genomic locus (located at 3q27). Exons are depicted as open boxes, and translation initiation codons are depicted as arrowheads. Arrowheads above the line are in frame with the main eIF4GI ORF, while arrowheads below the line indicate uORF initiator codons. The break in the line indicates the position of a gap in the contiguous sequence which is not yet defined. The shaded box indicates the location of the IRES described in the BE 5′ UTR of the original eIF4GI cDNA (18, 53).
FIG. 2.
3′-terminal sequence of eIF4GI cDNA clone. Shown is a comparison of the original eIF4GI cDNA clone reported by Yan et al. (53) and pSP4GI. The stop codon terminating the eIF4GI ORF is underlined, and boldface type indicates the nonhomologous residues in the two sequences.
Alignment of the 5′ end of pSP4GI and the 4Gext region indicated that the amino-terminal end of eIF4GI continues the previously predicted ORF, extending the ORF by an additional 40 amino acids (Fig. 1A). The resulting eIF4GI ORF now contains a total of 1,600 amino acids, encoding a polypeptide with a predicted molecular mass of 174.5 kDa. Unlike the earlier 4Gext sequence, which contained a continuous ORF through the most-5′ nucleotide in the sequence, the pSP4GI sequence contains several in-frame upstream stop codons. Two stop codons are located within 43 and 67 nt of the beginning of the eIF4GI ORF. This supports the hypothesis that the pSP4GI sequence represents a full-length eIF4GI coding sequence. All the numbering of amino acids and nucleotides of eIF4GI mRNA appearing in this work are now revised from earlier published numbering systems to reflect this additional sequence.
The 5′ UTR of pSP4GI cDNA is 274 nt long and contains a short upstream ORF (uORF) of 28 amino acids, overlapping with the large eIF4GI coding region by 1 nt (Fig. 1A). This uORF contains three in-frame AUG codons at positions 192, 258, and 270, none of which are in favorable Kozak consensus sequences (31). In contrast, the predicted eIF4GI initiation codon located at nt 275 to 277 is in a good sequence context to favor translation initiation. Interestingly, there are five in-frame AUG codons between nt 275 and nt 865, the spacing of which corresponds well with the observed differences in gel migration between the various native eIF4GI isoforms (Fig. 3C). Specifically, translation initiation at AUG codons beginning at nt 275, 395, 536, 767, and 863 could produce polypeptides differing by the appropriate molecular masses to generate the pattern of eIF4GI isoforms observed.
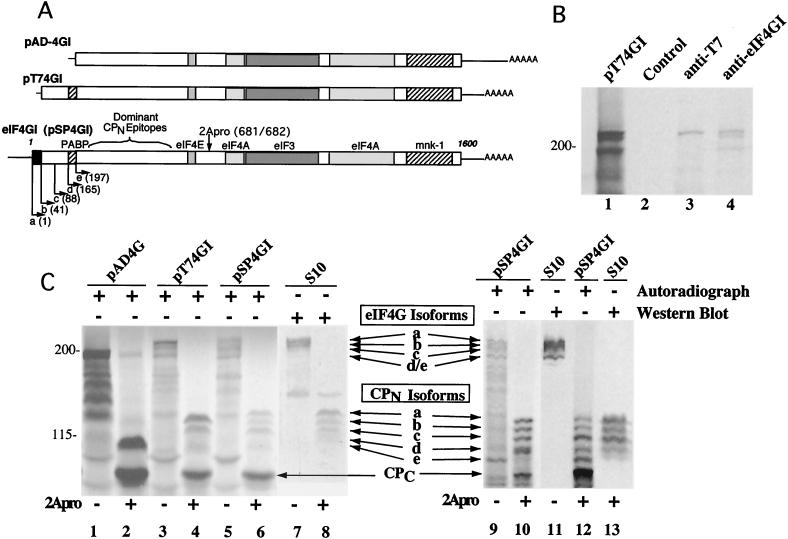
FIG. 3.
(A) Linear maps of eIF4GI cDNA clones. Boxed regions indicate the eIF4GI ORF, and the binding domains for other translation initiation factors are labeled. Arrows indicate proposed start points for translation. Letters on each arrow indicate the eIF4GI isoform produced, and the number in parentheses indicates the first amino acid of the isoform. The location of dominant amino-terminal immunogenic epitopes and the site of enterovirus 2Apro cleavage are also marked on pSP4GI. (B) In vitro expression of a T7-tagged eIF4GI cDNA. mRNA transcribed from pT74GI was translated in vitro and subjected to immunoprecipitation using normal rabbit serum (lane 2), rabbit polyclonal anti-T7 tag antibody (lane 3), or antiserum specific for the C-terminal domain of eIF4GI (lane 4). Starting nonprecipitated polypeptides (lane 1) and immunoprecipitated polypeptides were analyzed by SDS-PAGE-autoradiography. (C) In vitro expression of eIF4GI cDNAs. mRNAs transcribed from pAD4G, pT74GI, and pSP4GI were translated in vitro, and full-length polypeptides or 2Apro cleavage products of those polypeptides were analyzed by SDS-PAGE followed by transfer to nitrocellulose membranes and autoradiography. In both panels, S10 denotes Western analysis of HeLa cytoplasmic lysate using polyclonal antisera specific for eIF4GI residues 1 to 600 (left panel) (36) or an eIF4GI peptide located in residues 231 to 251 (right panel) (1). In the left panel, 10 μl of HeLa cytoplasmic lysate containing human eIF4GI was combined with 5 μl of reticulocyte translation reaction mixture before SDS-PAGE. Lanes 7 and 8 in the left panel are a direct Western blot overlay of the autoradiogram shown in lanes 5 and 6. Migration of intact eIF4G isoforms is noted as is migration of CPN and the CPC. The multiple smaller polypeptides in translation reactions which migrate faster than the band with the 195-kDa apparent molecular mass are aberrant translation products derived from improper initiation. The unmarked band appearing in the center of lanes 7 and 8 represents the p170 fragment of eIF3, which cross-reacts with this antiserum.
Apart from a few single-nucleotide alterations, the 3′ UTR of the new eIF4GI cDNA was identical to the previously published sequence through the first 234 nt, where it ended in a poly(A) sequence (Fig. 2). The original 3′ UTR cDNA sequence, derived from brain tissue, continues for an additional 319 nt before the poly(A) junction.
The full-length pSP4GI eIF4GI sequence was mapped onto the recently drafted genomic sequence for eIF4GI on human chromosome 3 to determine if the new predicted 5′ exons existed upstream of the known eIF4GI locus. Indeed, we found that the new sequence in pSP4GI constitutes two new upstream exons and that splice junctions joining these exons are normal. The first 8 of 32 eIF4GI exons are graphically depicted in Fig. 1B. To confirm the validity of this cDNA sequence we designed primers that would amplify nt 1 to 873 and performed reverse transcription-PCR of HeLa cytoplasmic RNA. The expected 0.87-kb DNA product was isolated, sequenced, and confirmed to be homologous with the 5′ sequence of pSP4GI (data not shown). While there is no canonical TATA box upstream of the new exon 1 of eIF4GI, the presence of several putative Sp1 binding sites and lack of an obvious splice acceptor site suggests that the new cDNA clone may encode a full-length eIF4GI mRNA. Interestingly, the initial eIF4GI cDNA clone derived from brain tissue contained an IRES element in the 5′ UTR (18). This IRES sequence is located in the eIF4GI gene in an intron between exons 6 and 7 (numbering reflects our full-length cDNA), as previously predicted (21). Analysis of the genomic sequence of the 3′-terminal region of the eIF4GI gene indicates that both the original published 3′ UTR and the new pSP4GI cDNA 3′ UTR sequences are derived from the same exon (exon 32). The differences observed in the 3′ UTR between the two cDNA clones probably result from the usage of alternative poly(A) cleavage sites.
Expression of extended eIF4GI cDNA clones.
The 4Gext region and the truncated polypeptide it encodes were previously tested for the ability to bind PABP in isolation from the bulk of the eIF4GI gene region and were not reconstructed on the genuine 3′ end of eIF4GI (25). This region was also reported to contain an IRES; however, the nature of polypeptides expressed using this 4Gext region was not examined (27). We constructed the plasmid pT74GI, which contains this extension fused to the original eIF4GI ORF derived from pAD-4GI. pAD-4GI contains the original eIF4GI clone with a unique 5′ UTR containing a different IRES element.
To determine if pSP4GI or pT74GI represents a full-length eIF4GI cDNA clone and what types of polypeptides would be synthesized from mRNAs containing extension segments, RNA from plasmids was transcribed and translated in vitro. The polypeptide products were directly compared to those from endogenous eIF4GI by mixing samples prior to SDS-PAGE analysis (Fig. 3C, left panel). As shown, pSP4GI encodes a protein with an apparent molecular mass of about 220 kDa and generates several polypeptides that comigrate with all eIF4GI polypeptides detected in HeLa extract. However, in vitro translation of pAD4GI and pT74GI did not produce all isoforms of native eIF4GI detected on Western blots. Specifically, pT74GI did not produce the largest, slowest-migrating isoform of eIF4GI; however, this isoform was produced by pSP4GI. Migration of eIF4GI polypeptides is easier to discern after truncation by CVB3 2Apro, which cuts eIF4GI between amino acids 681 and 682 (new numbering based on pSP4GI clone sequence), generating a set of N-terminal cleavage products (CPN) and a single C-terminal cleavage product (CPC). In this case, it is clear that only CPN produced by pSP4GI RNA comigrates with the largest native eIF4GI CPN (Fig. 3C), and products generated by pT74GI RNA are missing this large CPN. This suggests that pT74GI contains an incomplete eIF4GI sequence.
Based on inspection of numerous gels, we determined that five isoforms of eIF4GI can be produced from pSP4GI RNA, and five isoforms of native eIF4GI can sometimes be detected by high-resolution SDS-PAGE, particularly in analysis of CPN (Fig. 3C, right panel). We have denoted these isoforms eIF4GI-a through eIF4GI-e, with eIF4GI-a denoting the slowest-migrating form. The eIF4GI-d and eIF4GI-e isoforms appear with various intensity in translation reactions and in Western blots with different antisera, for reasons that remain unknown. Since multiple eIF4GI polypeptide isoforms are produced from translation of a single pT74GI or pSP4GI RNA, these data strongly suggest that alternative translation initiation is capable of producing multiple isoforms of eIF4GI.
pT74GI was constructed with a T7 epitope tag fused to the eIF4GI start codon (AUG 395). To substantiate which eIF4GI polypeptide was derived from initiation at the first AUG codon in the pT74GI RNA, in vitro-translated pT74GI polypeptides were immunoprecipitated with anti-T7 antibody. Results showed that only the largest isoform reacted with the anti-T7 antibody (Fig. 3B), further suggesting that the multiple forms of eIF4GI generated from this construct contained different amino ends and do not arise from posttranslational modification(s).
Initiation at alternative AUG codons generates multiple forms of eIF4GI fusion proteins.
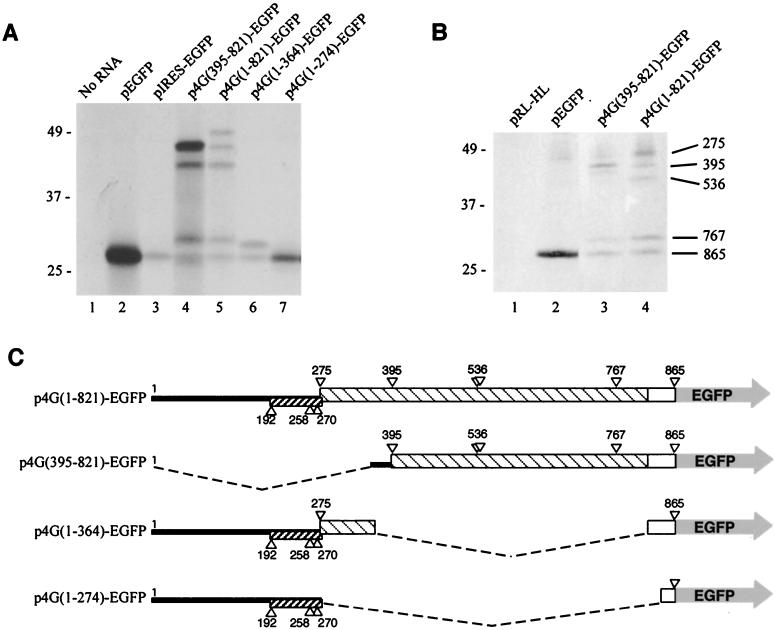
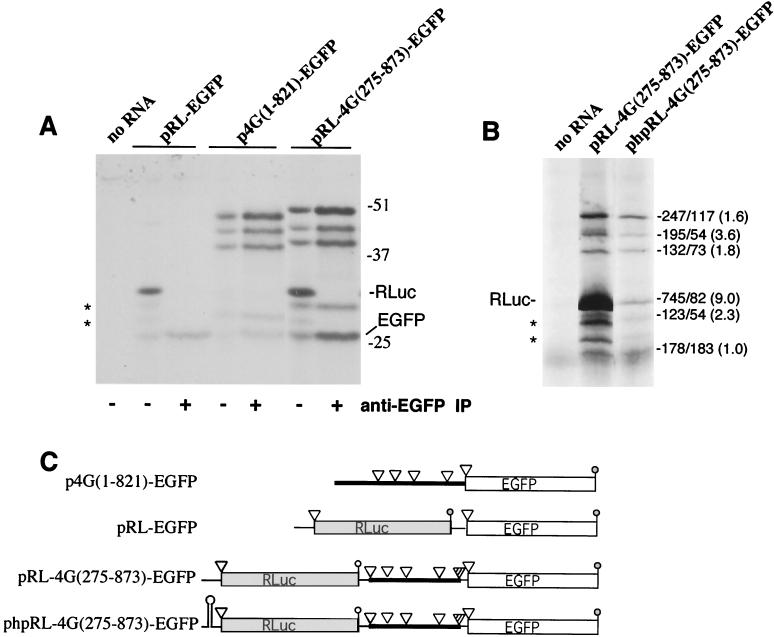
To test the hypothesis that alternative translation initiation occurs on the eIF4GI mRNA, constructs were made that contained regions of the eIF4GI 5′ end fused in frame with EGFP (the schematic map of these clones is depicted in Fig. 4C). All plasmids were used to generate capped mRNAs in vitro, which were determined to be intact by denaturing gel electrophoresis. Equimolar amounts of mRNAs were translated in vitro in rabbit reticulocyte lysates and protein products analyzed by SDS-PAGE-autoradiography (Fig. 4A). Translation of pEGFP RNA alone generated a large amount of a single EGFP polypeptide that migrated at the expected apparent molecular mass of 27 kDa (Fig. 4A, lane 2). When the original 5′ UTR/IRES described by Gan and Rhoads (18) was inserted upstream of the EGFP ORF, a single polypeptide corresponding in size to EGFP was made, though the amount of translation product was markedly reduced. However, when RNA containing the 4Gext segment fused in frame to EGFP was translated [p4G(395-821)-EGFP] four polypeptides were efficiently produced (Fig. 4A, lane 4). The fastest-migrating product comigrated with EGFP, whereas three products with molecular masses greater than that of EGFP were also produced. These have apparent molecular masses of 44, 39, and 31 kDa, which matches well with deduced molecular masses of proteins that would be produced by initiation of translation at eIF4GI AUG codons 395, 536, and 767, respectively. The largest polypeptide was translated much more efficiently than other fusion protein isoforms, suggesting that cap-dependent scanning may largely drive initiation on this RNA.
FIG. 4.
Expression of eIF4GI-EGFP fusion constructs. (A) In vitro expression of eIF4GI-EGFP fusion constructs. Capped mRNAs transcribed in vitro from plasmids were translated in vitro and analyzed by SDS-PAGE-autoradiography. Translation reactions were programmed with 0.25 pmol of each RNA. pIRES-EGFP mRNA contains the original eIF4G (BE) IRES cloned upstream of the EGFP initiator codon. Each EGFP fusion product was confirmed by immunoprecipitation with EGFP-specific antiserum (data not shown). (B) In vivo expression of eIF4GI-EGFP fusion constructs. Indicated plasmids were transfected into HeLa cells. Cells were radiolabeled at 36 h posttransfection, and EGFP fusion polypeptides were immunoprecipitated and analyzed by SDS-PAGE-autoradiography as described in Materials and Methods. Three times more cell lysate was used for the immunoprecipitation shown in lane 4 than for those shown in lanes 1 to 3. Numbers on the right denote assignment of the eIF4GI AUG serving as an initiator codon in the fusion polypeptide. (C) EGFP fusion constructs containing portions of eIF4GI extension sequences. The nucleotide numbering is based on the complete sequence in plasmid pSP4GI. Boxes indicate ORF segments, hatched regions denote eIF4GI coding sequence, and open boxes represent additional amino acids derived from multiple cloning sites on plasmid constructs. Triangles represent location of all AUG codons. Boxed areas below the line indicate the out-of-frame uORF.
When RNA containing the full eIF4GI leader sequence [p4G(1-821)] fused to EGFP was translated, five polypeptides were synthesized, four of which comigrated with products produced by p4G(395-821)-EGFP RNA (Fig. 4A, lane 5). The largest polypeptide migrated with an apparent molecular mass of 49 kDa, and migration of the whole set of fusion proteins was consistent with initiation occurring at AUGs 275, 395, 536, and 767 and the authentic EGFP initiator codon (nt 865). Thus, transfer of the first 821 nt of eIF4GI mRNA to a heterologous ORF promotes alternate selection of initiation codons. The initiation codons that were utilized from the p4G(1-821)-EGFP RNA seem to be the same as those utilized by the full-length eIF4GI mRNA. Unlike products translated from p4G(395-821)-EGFP RNA, translation of the largest polypeptide was not enhanced relative to the other fusion protein isoforms.
Translation efficiency of p4G(1-821)-EGFP RNA was consistently much lower than that of p4G(395-821)-EGFP RNA, suggesting that the additional upstream sequence (nt 1 to 394) may regulate translation. In order to test this hypothesis, EGFP expression constructs containing eIF4GI nt 1 to 364 or nt 1 to 274 were also made (Fig. 4C). Both of these constructs translated at a lower efficiency than did pEGFP RNA (Fig. 4A, compare lane 2 to lanes 6 and 7). Translation efficiency was more impaired in p4G(1-364)-EGFP RNA than in p4G(1-274)-EGFP RNA, which contains only the 5′ UTR. This difference could be explained in p4G(1-364)-EGFP RNA by the maintenance of a portion of the main eIF4GI ORF in frame with EGFP and of the overlap of stop and start codons at the uORF/eIF4GI ORF junction.
It was also important to determine whether alternative translation initiation from these clones could be reproduced in vivo. To address this, HeLa cells were transfected with p4G(1-821)-EGFP, p4G(395-821)-EGFP, or pEGFP. Cells were pulse-labeled with [35S]methionine 36 h posttransfection, and radiolabeled proteins were immunoprecipitated with monoclonal anti-EGFP antibodies and detected by autoradiography. HeLa cells transfected with p4G(395-821)-EGFP generated four polypeptides that could be immunoprecipitated with anti-GFP antibodies (Fig. 4B). Though all four polypeptides were consistently observed, the fusion protein initiating at AUG 395 was consistently predominant and the fusion protein initiating at AUG 536 generated only a very weak band. When p4G(1-821)-EGFP was transfected into cells, five polypeptides were immunoprecipitated, suggesting that all five initiator codons were utilized in vivo. Interestingly, the dominant polypeptide precipitated was the largest, suggesting that in vivo more ribosomes are directed to AUG 275 than to the downstream initiator codons. This relative pattern of expression of the five polypeptides was not duplicated in vitro in reticulocyte lysates (compare Fig. 4A, lane 5) in which the middle-sized polypeptide initiating at AUG 536 was usually more dominant. However, consistent with in vitro data, the overall level of expression from the full-length p4G(1-821) leader sequence was consistently much lower than that of the cognate construct lacking the eIF4GI 5′ UTR [p4G(395-821)-EGFP], requiring immunoprecipitation from three times the number of transfected cells to easily visualize translation products. Taken together, these data indicate that alternative translation initiation associated with the eIF4GI RNA 5′ region is not merely an artifact of the in vitro translation system but rather a specific mechanism for generation of alternative protein isoforms.
Translation initiation at in-frame AUGs in eIF4GI mRNA.
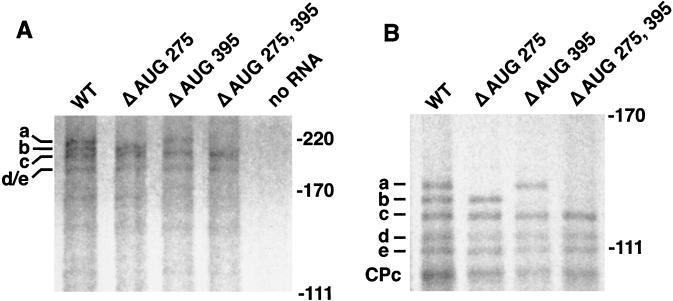
To further substantiate the hypothesis that alternate translation initiation produces eIF4GI isoforms from a single RNA species, we introduced point mutations into the full-length eIF4GI RNA. In these constructs AUG 275, AUG 395, or both AUGs were mutated to a GUG codon that does not normally mediate translation initiation. Analysis of in vitro translation products generated by these mutated mRNA species demonstrated that AUG 275 and AUG 395 initiate synthesis of the a and b isoforms of eIF4GI, respectively, as illustrated by the disappearance of either isoform or both when corresponding RNAs were translated (Fig. 5A). To better visualize the elimination of isoform synthesis in the mutants, each translation lysate was also cleaved with CVB3 2Apro prior to analysis (Fig. 5B).
FIG. 5.
Mutation of AUG codons interrupts expression of eIF4GI isoforms. (A) Wild-type pSP4GI cDNA (WT) was mutated to replace AUG with GUG at nt 275 (Δ AUG 275), nt 395 (Δ AUG 395), or both (Δ AUG 275, 395), and then mRNA was transcribed, translated in vitro, and analyzed as described in Materials and Methods. (B) Same as panel A, except eIF4GI translation products were cleaved by inclusion of 2Apro in translation lysates. eIF4GI isoforms are identified on the left, and migration of marker proteins in kilodaltons is denoted on the right of panels.
The eIF4GI 5′ uORF alters eIF4GI translation.
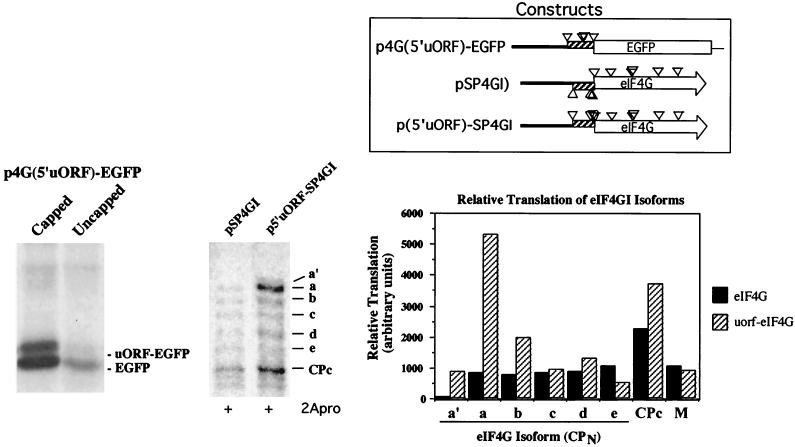
We observed that the eIF4GI 5′ UTR was able to reduce translation efficiency of EGFP fusion protein (Fig. 4A, lanes 5 and 6). The 5′ UTR contains a small upstream ORF that may down-regulate eIF4GI initiation, particularly since the uORF stop codon partly overlaps the AUG codon initiating synthesis of the eIF4GI-a isoform (AUG 275). This uORF contains three in-frame AUG codons; however, all three are in poor Kozak consensus sequences. To test whether the uORF is able to recruit ribosomes, we made an eIF4GI-EGFP fusion construct in which the uORF was in frame with the EGFP ORF [p4G(5′uORF)-EGFP (Fig. 6)]. In vitro translation of capped and uncapped RNAs derived from this construct generated two polypeptides (Fig. 6, left panel). The faster-migrating polypeptide comigrated with EGFP, and the slower-migrating polypeptide displayed a mobility consistent with initiation at AUG 192, i.e., the beginning of the uORF. This suggests that the uORF is functional and can recruit ribosomes in vitro. Also, expression of capped versus uncapped RNA resulted in differential translation of the two polypeptides. With capped RNA, significantly more ribosomes initiated at the downstream AUG than the upstream uORF AUG, consistent with leaky scanning. When uncapped mRNA was tested, overall translation levels dropped as expected; however, the initiation from the upstream uORF AUG dropped much more than initiation from the EGFP AUG. Thus, initiation at AUG 192 may proceed by a cap-dependent mechanism.
FIG. 6.
In vitro expression of eIF4GI uORF. Constructs which fuse the eIF4GI 5′ uORF in frame with either EGFP or the main ORF of eIF4GI were generated by PCR and cloning or by site-directed mutagenesis as indicated in Materials and Methods. (Left panel) Equal amounts of capped or uncapped T7 transcripts from p4G(5′uORF)-EGFP were translated in nuclease-treated rabbit reticulocyte lysates for 1 h prior to SDS-PAGE-autoradiography. (Middle panel) pSP4GI (wild type)- or p5′uORF-SP4GI-capped T7 transcripts were translated in vitro and subsequently treated with CVB3 2Apro, for 1 h each, prior to SDS-PAGE-autoradiography. (Right panel) Quantitation of translation of pSP4GI and p5′uORF-SP4GI polypeptides by scanning densitometry. M, vector-specific background band used to show equal loading.
To determine if the uORF was functional in the context of full-length eIF4GI mRNA, an additional construct was made in which the uORF was shifted 1 nt to bring it in frame with the eIF4GI ORF (Fig. 6). Translation products derived from this construct (p5′uORF-SP4GI) were compared to eIF4GI translation products after cleavage with CVB3 2Apro, which aids separation and visualization of isoforms in gels. Interestingly, the intensity of the eIF4GI-a CPN isoform was sharply increased when the frame-shifted RNA was translated. Quantitation of polypeptides produced from this mutant mRNA compared to native eIF4GI mRNA showed increased production of both eIF4GI-a and eIF4GI-b CPN isoforms as well as a corresponding increase in CPC (Fig. 6, right panel). The data also revealed a new CPN polypeptide was formed that migrated more slowly than eIF4GI-a CPN, termed eIF4GI-a′. This was consistent with weak initiation at AUG 192, at the uORF 5′ AUG. These results suggest that the uORF recruits ribosomes; however, the poor Kozak consensus surrounding AUG 192 enables many ribosomes to scan further downstream to AUG codons in a better context. In the case of the native eIF4GI mRNA, the juxtaposition of the uORF stop codon over AUG 275 results in termination and loss of some ribosomes, potentially down-regulating overall eIF4GI expression.
Production of multiple eIF4GI isoforms from dicistronic RNA.
Johannes et al. proposed that eIF4GI RNA contained an IRES element in the region between nt 395 and 863; however, the polypeptide products produced by translation initiation from this element were not determined. This putative IRES is embedded in the eIF4GI ORF; thus, we wished to determine which of the multiple eIF4GI isoforms could be initiated via a cap-independent process. The eIF4GI sequence from nt 275 to 873 (lacking the 5′ UTR) was inserted into the dicistronic construct pRL-EGFP (Fig. 7C). Capped RNA was transcribed from dicistronic constructs and analyzed by denaturing gel electrophoresis to ensure that RNAs were not nicked and were of unit length (data not shown). Translation of capped RNA from the control pRL-EGFP construct revealed efficient translation of Renilla luciferase (Fig. 7A) and a small amount of leaky reinitiation at the downstream EGFP ORF, probably due to the close spacing of stop and start codons in this construct (42 nt apart). When the eIF4GI sequence was inserted in the intercistronic region of pRL-EGFP a significant enhancement in translation of EGFP and EGFP fusion proteins (12.6-fold, total fusion proteins) was observed. In addition, monocistronic and dicistronic RNAs generated eIF4GI-EGFP fusion proteins with nearly equivalent efficiency, suggesting that cap-independent translation is relatively robust in this context. The most-intriguing observation was formation of multiple eIF4GI-EGFP fusion isoforms from the dicistronic RNA, particularly since the putative IRES element must be located downstream of the AUG that generates the largest fusion polypeptide. All fusion polypeptide products generated by the dicistronic RNA migrated slightly more slowly than products from monocistronic RNA due to the insertion of a short linker sequence (Fig. 7C). Interestingly, translation appears not to initiate from one downstream AUG (AUG 863) in this context (Fig. 7C) for reasons that are unknown, but this could be due to altered RNA secondary structure in this construct.
FIG. 7.
(A) Multiple eIF4GI-EGFP fusion proteins translate from a dicistronic construct in vitro. In vitro translation was performed with RNA transcripts from a control dicistronic construct (pRL-EGFP), a monocistronic eIF4GI-EGFP fusion construct [p4G(1-821)-EGFP], and a dicistronic eIF4GI-EGFP fusion construct [pRL-4G(275-873)-EGFP]. Where indicated, EGFP and EGFP fusion polypeptides were immunoprecipitated (IP) prior to SDS-PAGE to isolate genuine fusion proteins from the two vector-specific background bands (*) that migrate between RLuc and EGFP. Numbers on the right indicate migration of molecular mass markers. (B) Second-cistron translation is independent of that of the first cistron. In vitro translation was preformed with equal amounts of RNA transcribed from the dicistronic eIF4GI-EGFP fusion construct [pRL-4G(275-873)-EGFP] and an analogous construct that contains a stable hairpin upstream of RLuc [phpRL-4G(275-873)-EGFP]. Translations were subjected to SDS-PAGE and autoradiography, and bands were quantitated by densitometry with NIH Image software (version 1.62). Quantitation of polypeptides is indicated on the right (no hairpin-hairpin constructs). Fold inhibition of expression of isoforms in the hairpin construct is given in parentheses. (C) Schematic showing the RNAs translated in this figure. Open triangles indicate translation initiation codons, and striped triangles are those which do not seem to function in a dicistronic construct.
To further test whether the second cistron translation occurs independently of the first cistron, we placed a thermodynamically stable hairpin between the T7 promoter and the Renilla luciferase gene. Quantitation by densitometry (Fig. 7B) and luciferase assay (data not shown) showed that addition of this hairpin reduced translation from the upstream Renilla luciferase cistron ninefold, whereas translation of the downstream cistron originating from the five eIF4GI AUG codons was decreased only about twofold on average (Fig. 7B). This suggests that translations of the upstream and downstream cistrons are largely independent. Other preliminary experiments suggest that expression of the fusion proteins from the dicistronic construct is not sensitive to blockage of cap-dependent translation by addition of coxsackievirus 2Apro to translation lysates (data not shown).
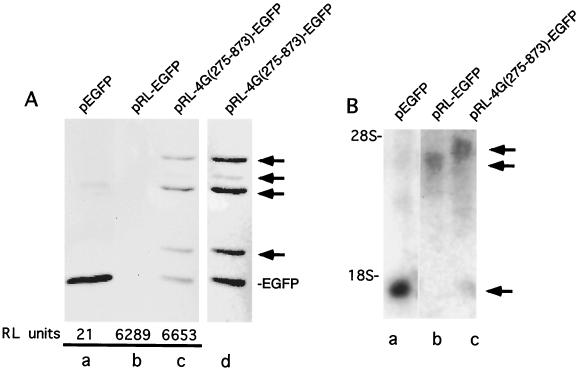
In order to determine which of these polypeptides could be produced from a dicistronic construct in vivo, we transfected plasmids pEGFP, pRL-EGFP, and pRL-4G(275-873)-EGFP, which express the indicated RNAs under the control of a cytomegalovirus promoter, into 293T cells. Thirty-six hours posttransfection cells were examined under an inverted fluorescence microscope and harvested. 293T cells expressing EGFP alone exhibited strong fluorescence, while cells transfected with pRL-4G(275-873)-EGFP plasmid DNA fluoresced only weakly (data not shown). No fluorescence was observed in cells transfected with pRL-EGFP. Analysis of cytoplasmic extracts from cells transfected with pRL-EGFP revealed high RLuc expression but that EGFP was undetectable (Fig. 8A, lane b), suggesting that very little readthrough of the RLuc stop codon occurred in vivo. In contrast, pRL-4G(275-873)-EGFP expressed relatively high levels of EGFP and eIF4GI-EGFP fusion proteins from the second cistron (Fig. 8A, lanes c and d). As was observed in vitro, multiple eIF4GI-EGFP fusion proteins were produced from the second cistron, though in this case translation initiation at AUG 395 was much weaker than that on the other AUGs and required further exposure to visualize (Fig. 8A, lane d). Northern blots of lysates from transfected cells indicate that only one anti-EGFP reactive RNA species was present in each set of transfections at the correct relative size, thus ruling out specific RNA cleavage or splicing as a significant source of second cistron translation initiation (Fig. 8B). These experiments demonstrate that multiple eIF4GI isoforms can be translated from a dicistronic construct both in vitro and in vivo.
FIG. 8.
(A) Multiple eIF4GI-EGFP fusion proteins are translated from a dicistronic construct in vivo. 293T cells were transfected with the indicated DNA constructs as described in Materials and Methods. Lysates were immunoprecipitated with a monoclonal anti-EGFP antibody, subjected to SDS-PAGE, and transferred to nitrocellulose. Western blotting was performed with polyclonal anti-EGFP antibodies. Lane a contains lysate from 2 × 106 transfected cells, while lanes b and c contain lysate from 107 cells. Relative luciferase activity from cell lysates is shown below the lane for the given transfected plasmid. (B) Northern blot analysis of transfected cells. Cells transfected with the indicated constructs were lysed in Trizol reagent, and RNA was isolated and run on a 1% agarose formaldehyde gel. RNA was then transferred to a nylon membrane and probed with a [32P]GTP-labeled antisense EGFP riboprobe. Migration of marker RNAs is shown on the left.
DISCUSSION
The human eIF4GI gene was initially cloned from a human brain cDNA library and was reported to have a long (450-nt) 5′ UTR (BE 5′ UTR) followed by an ORF coding for a single polypeptide 1,404 amino acids long (53). The BE 5′ UTR was reported to function as an IRES element in a dicistronic construct (18, 19); however, the single eIF4GI polypeptide synthesized from the original clone in vitro migrated faster than several native eIF4GI isoforms (28). Based on this observation, it was proposed that posttranslational modification of eIF4GI polypeptide might account for the multiple protein isoforms observed by Western blotting of cell lysates (28). More recently the coding region for eIF4GI was found to contain a 156-amino-acid extension (termed 4Gext) at the amino end, upstream of the proposed initiation site in the original published sequence (21, 25, 27) (Fig. 1A). The 4Gext region contains a different putative IRES (25, 27) and encodes a binding site for PABP that facilitates interactions between the 5′ and 3′ ends of mRNA (35, 49, 51). The 4Gext region, however, did not contain the long 5′ UTR of the original cDNA clone, and alignments between these clones sharply diverged at nt 810. This difference, and the presence of a splice acceptor site at the point of divergence led to proposals that the BE 5′ UTR sequence is part of an unspliced intron (21).
Here we describe a new cDNA clone of eIF4GI that extends the known ORF of eIF4GI by 40 amino acid residues, from 1,560 to 1,600 amino acids, and also contains a new 5′ UTR of 274 nt. The sequence of this clone overlaps both previous cDNA clones and, combined with mapping the cDNA onto the genomic DNA sequence, has significantly clarified the divergent cDNA sequence data obtained by other investigators. When expressed in rabbit reticulocyte lysate and compared to HeLa eIF4GI, the new cDNA clone was able to express a protein that comigrated with the largest isoform (isoform a) of endogenous eIF4GI (Fig. 3C). Further, when cleaved by 2Apro, the CPN of the largest native isoform and largest translated isoform comigrated in gels. Coupled with the presence of upstream stop codons and an overlapping out-of-frame uORF, the new clone likely represents a complete cDNA sequence of eIF4G mRNA. Examination of the human genomic sequence revealed that the entire 5′ UTR and the additional coding region of eIF4GI reported here were located in two additional upstream exons.
Since its first description in 1982 (17, 23), the nature of the multiple forms of eIF4GI observed in Western blots has remained unknown, though they had been postulated to result from posttranslational modifications (28). eIF4GI is a phosphoprotein; however, phosphorylation does not easily explain the size and discrete nature of the band shifts between eIF4GI isoforms. Also, the majority of phosphorylation of eIF4GI occurs in the C-terminal domain, which appears as a single band in gels after cleavage by 2Apro (Fig. 3C) (45). Immunoblot analysis using antibodies specific for N- or C-terminal epitopes has shown that all the modifications associated with isoforms are exclusively associated with the N terminus of eIF4GI (32, 34, 54).
The data presented here provide strong evidence that alternate use of in-frame AUG codons occurs in vitro and in vivo, which generates isoforms of eIF4GI that comigrate with the native eIF4GI isoforms in HeLa cell extracts. Since the same pattern of multiple translation products was observed when the eIF4GI N-terminal RNA sequence was appended to EGFP RNA, this region of RNA promotes the use of alternate AUG codons. From AUG mutagenesis data combined with deletion analyses, we assign AUGs 275, 395, 536, 767, and 863 as initiator codons for eIF4GI isoforms a through e, respectively. Indeed, during the revision of this work other researchers independently confirmed by mass spectroscopy that endogenous eIF4GI consists of multiple polypeptides with N termini corresponding to initiation at methionines 1, 41, 89, and 197 as we have predicted here (6). In several experiments, the relative abundance of eIF4GI-d and eIF4GI-e isoforms produced by in vitro translation of monocistronic RNA was variable. The reasons for this are unknown, but similar variability in these isoforms has been noted in immunoblots of HeLa eIF4GI and may reflect some degree of translation regulation or altered protein degradation.
Even though alternate use of translation initiation codons is occurring with eIF4GI-EGFP RNA in vivo, this does not exclude the possibility that alternative splicing may also play a role in the generation of multiple isoforms of eIF4GI. The original cDNA clone of eIF4GI containing the truncated ORF was appended to an IRES element that is not present in the pSP4GI clone of eIF4GI. This IRES element is located in an intron between exons 6 and 7 of the eIF4GI genomic sequence. The two sequences (pSP4GI and pAD-4GI) are homologous in the coding region downstream of nt 810 and sharply deviate at the 5′ exon 7 splice junction (Fig. 1A) (22).
Several lines of evidence in this work suggest that expression of eIF4GI is translationally regulated, possibly by multiple mechanisms. One attractive hypothesis is that a central regulator of translation (eIF4G) would itself be subjected to translational regulation. This would provide the ability for eIF4GI to sense changes in translational conditions in the cell and quickly respond by altering levels and types of eIF4GI isoforms produced. The presence of an upstream ORF in the 5′ UTR of eIF4GI mRNA provides one mechanism for translational regulation of expression of eIF4GI isoforms. Our data indicate that when the uORF is fused in frame to EGFP or the eIF4GI ORF, a larger fusion polypeptide was formed. Thus, the uORF was recognized by ribosomes even though a poor Kozak consensus sequence exists for all its initiators. All EGFP fusion mRNAs that contained this upstream element produced lower levels of EGFP fusion proteins than those in which this element was not present, suggesting that the upstream ORF may serve to down-regulate cap-dependent initiation of eIF4GI translation. We also noted that translation from the upstream AUG was highly cap dependent when the uORF was fused in frame to EGFP, yet translation from the downstream AUG was not, suggesting differential regulation.
The region of the eIF4GI mRNA between nt 340 and 810 contains a putative IRES element (27), providing a route for production of eIF4GI at times when cap-dependent translation is restricted (e.g., cell stress or mitosis). The synthesis of eIF4GI under the control of an IRES element fits well with the observed recovery of this protein following conditions where cap-dependent translation had been shut off, i.e., heat shock or hypoxia (48). Surprisingly, when this region of eIF4GI was expressed in a dicistronic RNA, multiple eIF4GI initiation codons were still utilized. This could result if an IRES element recruited ribosomes to bind upstream of its core in a fashion similar to that recently proposed for the IRES embedded in the human immunodeficiency virus gag ORF (7). After cap-independent initiation at an upstream AUG codon, leaky scanning could account for initiation at downstream AUG codons (i.e., codons 395, 536, and 767). A detailed investigation will be required to distinguish which of the isoforms are produced via leaky scanning as opposed to cap-independent initiation or both.
Finally, we do not know if individual isoforms of eIF4GI function equivalently in translation. Given that eIF4GI is a scaffolding protein with multiple protein binding partners, unique N-terminal amino acid sequences could provide binding sites for new ancillary translation factors which allow eIF4G to promote expression of specific mRNAs or regulate translation in other ways. Additionally, the eIF4GI-e isoform does not contain the PABP-binding domain, and in preliminary experiments we confirmed that eIF4GI isoforms a, b, and c bind PABP yet isoform e does not (data not shown). Further experiments are required to elucidate the functional differences between the eIF4GI isoforms.
Acknowledgments
We are indebted to R. E. Rhoads for furnishing plasmids pAD4G and pSKHFCI. We also thank P. Sarnow and G. Johannes for furnishing 4Gext cDNA, S. Lemon for furnishing pRL-HL, and A.-B. Shyu for providing us with the stable hairpin construct. We are grateful to M. Van Eden for critical review of the manuscript.
This work was supported by NIH grant GM 59803.
M.P.B. and M.Z. contributed equally to this work.
REFERENCES
- 1.Aldabe, R., E. Feduchi, I. Novoa, and L. Carrasco. 1995. Efficient cleavage of p220 by poliovirus 2A(pro) expression in mammalian cells: effects on vaccinia virus. Biochem. Biophys. Res. Commun. 215:928-936. [DOI] [PubMed] [Google Scholar]
- 2.Aragon, T., S. de La Luna, I. Novoa, L. Carrasco, J. Ortin, and A. Nieto. 2000. Eukaryotic translation initiation factor 4GI is a cellular target for NS1 protein, a translational activator of influenza virus. Mol. Cell. Biol. 20:6259-6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borman, A. M., R. Kirchweger, E. Ziegler, R. E. Rhoads, T. Skern, and K. M. Kean. 1997. elF4G and its proteolytic cleavage products: effect on initiation of protein synthesis from capped, uncapped, and IRES-containing mRNAs. RNA 3:186-196. [PMC free article] [PubMed] [Google Scholar]
- 4.Borman, A. M., Y. M. Michel, and K. M. Kean. 2000. Biochemical characterisation of cap-poly (A) synergy in rabbit reticulocyte lysates: the eIF4G-PABP interaction increases the functional affinity of eIF4E for the capped mRNA 5′ end. Nucleic Acids Res. 28:4068-4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bovee, M. L., B. Lamphear, R. E. Rhoads, and R. E. Lloyd. 1998. Direct cleavage of eIF4G by poliovirus 2A protease is inefficient in vitro. Virology 245:241-249. [DOI] [PubMed] [Google Scholar]
- 6.Bradley, C. A., J. C. Padovan, T. L. Thompson, C. A. Benoit, B. T. Chait, and R. E. Rhoads. 2002. Mass spectrometric analysis of the N terminus of translational initiation factor eIF4G-1 reveals novel isoforms. J. Biol. Chem. 277:12559-12571. [DOI] [PubMed] [Google Scholar]
- 7.Buck, C., X. Shen, M. Egan, T. Pierson, C. Walker, and R. Siliciano. 2001. The human immunodeficiency virus type 1 gag gene encodes an internal ribosome entry site. J. Virol. 75:181-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, C.-Y. A., N. Xu, and A.-B. Shyu. 1995. mRNA decay mediated by two distinct AU-rich elements from c-fos and granulocyte-macrophage colony-stimulating factor transcripts: different deadenylation kinetics and uncoupling from translation. Mol. Cell. Biol. 15:5777-5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chizhikov, V., and J. T. Patton. 2000. A four-nucleotide translation enhancer in the 3′-terminal consensus sequence of the nonpolyadenylated mRNAs of rotavirus. RNA 6:814-825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chu, G., and P. Sharp. 1981. SV40 DNA transfection of cells in suspension: analysis of efficiency of transcription and translation of T-antigen. Gene 13:197-202. [DOI] [PubMed] [Google Scholar]
- 11.Clemens, M. J., M. Bushell, and S. J. Morley. 1998. Degradation of eukaryotic polypeptide chain initiation factor (eIF) 4G in response to induction of apoptosis in human lymphoma cell lines. Oncogene 17:2921-2931. [DOI] [PubMed] [Google Scholar]
- 12.Cuesta, R., G. Laroia, and R. J. Schneider. 2000. Chaperone hsp27 inhibits translation during heat shock by binding eIF4G and facilitating dissociation of cap-initiation complexes. Genes Dev. 14:1460-1470. [PMC free article] [PubMed] [Google Scholar]
- 13.Cuesta, R., Q. Xi, and R. J. Schneider. 2000. Adenovirus-specific translation by displacement of kinase Mnk1 from cap-initiation complex eIF4F. EMBO J. 19:3465-3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Gregorio, E., T. Preiss, and M. W. Hentze. 1999. Translation driven by an eIF4G core domain in vivo. EMBO J. 18:4865-4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edskes, H. K., J. M. Kiernan, and R. J. Shepherd. 1996. Efficient translation of distal cistrons of a polycistronic mRNA of a plant pararetrovirus requires a compatible interaction between the mRNA 3′ end and the proteinaceous trans-activator. Virology 224:564-567. [DOI] [PubMed] [Google Scholar]
- 16.Etchison, D., and S. Fout. 1985. Human rhinovirus 14 infection of HeLa cells results in the proteolytic cleavage of the p220 cap-binding complex subunit and inactivates globin mRNA translation in vitro. J. Virol. 54:634-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Etchison, D., S. C. Milburn, I. Edery, N. Sonenberg, and J. W. B. Hershey. 1982. Inhibition of HeLa cell protein synthesis following poliovirus infection correlates with the proteolysis of a 220,000-dalton polypeptide associated with eukaryotic initiation factor 3 and a cap binding protein complex. J. Biol. Chem. 257:14806-14810. [PubMed] [Google Scholar]
- 18.Gan, W., and R. E. Rhoads. 1996. Internal initiation of translation directed by the 5′-untranslated region of the mRNA for eIF4G, a factor involved in the picornavirus-induced switch from cap-dependent to internal initiation. J. Biol. Chem. 271:623-626. [DOI] [PubMed] [Google Scholar]
- 19.Gan, W. N., M. Lacelle, and R. E. Rhoads. 1998. Functional characterization of the internal ribosome entry site of eIF4G mRNA. J. Biol. Chem. 273:5006-5012. [DOI] [PubMed] [Google Scholar]
- 20.Gingras, A. C., B. Raught, and N. Sonenberg. 1999. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 68:913-963. [DOI] [PubMed] [Google Scholar]
- 21.Gradi, A., H. Imataka, Y. V. Svitkin, E. Rom, B. Raught, S. Morino, and N. Sonenberg. 1998. A novel functional human eukaryotic translation initiation factor 4G. Mol. Cell. Biol. 18:334-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gradi, A., Y. V. Svitkin, H. Imataka, and N. Sonenberg. 1998. Proteolysis of human eukaryotic translation initiation factor eIF4GII, but not eIF4GI, coincides with the shutoff of host protein synthesis after poliovirus infection. Proc. Natl. Acad. Sci. USA 95:11089-11094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grifo, J. A., S. A. Tahara, M. A. Morgan, A. J. Shatkin, and W. C. Merrick. 1983. New initiation factor activity required for globin mRNA translation. J. Biol. Chem. 258:5804-5810. [PubMed] [Google Scholar]
- 24.Honda, M., S. Kaneko, E. Matsushita, K. Kobayashi, G. Abell, and S. Lemon. 2000. Cell cycle regulation of hepatitis C virus internal ribosomal entry site-directed translation. Gastroenterology 118:152-162. [DOI] [PubMed] [Google Scholar]
- 25.Imataka, H., A. Gradi, and N. Sonenberg. 1998. A newly identified N-terminal amino acid sequence of human eIF4G binds poly(A)-binding protein and functions in poly(A)-dependent translation. EMBO J. 17:7480-7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Joachims, M., P. C. van Breugel, and R. E. Lloyd. 1999. Cleavage of poly(A)-binding protein by enterovirus proteases concurrent with inhibition of translation in vitro. J. Virol. 73:718-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johannes, G., and P. Sarnow. 1998. Cap-independent polysomal association of natural mRNAs encoding c-myc, BiP and eIF4G conferred by internal ribosome entry sites. RNA 4:1500-1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Joshi, B., R. Q. Yan, and R. E. Rhoads. 1994. In vitro synthesis of human protein synthesis initiation factor-4 gamma and its localization on 43-S and 48-S initiation complexes. J. Biol. Chem. 269:2048-2055. [PubMed] [Google Scholar]
- 29.Kolupaeva, V. G., T. V. Pestova, C. U. Hellen, and I. N. Shatsky. 1998. Translation eukaryotic initiation factor 4G recognizes a specific structural element within the internal ribosome entry site of encephalomyocarditis virus RNA. J. Biol. Chem. 273:18599-18604. [DOI] [PubMed] [Google Scholar]
- 30.Korneeva, N. L., B. J. Lamphear, F. L. C. Hennigan, W. C. Merrick, and R. E. Rhoads. 2001. Characterization of the two eIF4A-binding sites on human eIF4G-1. J. Biol. Chem. 276:2872-2879. [DOI] [PubMed] [Google Scholar]
- 31.Kozak, M. 1992. Regulation of translation in eukaryotic systems. Annu. Rev. Cell Biol. 8:197-225. [DOI] [PubMed] [Google Scholar]
- 32.Lamphear, B. J., R. Kirchweger, T. Skern, and R. E. Rhoads. 1995. Mapping of functional domains in eukaryotic protein synthesis initiation factor 4G (eIF4G) with picornaviral proteases: implications for cap-dependent and cap-independent translational initiation. J. Biol. Chem. 270:21975-21983. [DOI] [PubMed] [Google Scholar]
- 33.Lamphear, B. J., and R. Panniers. 1991. Heat shock impairs the interaction of cap-binding protein complex with 5′ messenger RNA cap. J. Biol. Chem. 266:2789-2794. [PubMed] [Google Scholar]
- 34.Lamphear, B. J., R. Q. Yan, F. Yang, D. Waters, H. D. Liebig, H. Klump, E. Kuechler, T. Skern, and R. E. Rhoads. 1993. Mapping the cleavage site in protein synthesis initiation factor-eIF-4γ of the 2A proteases from human coxsackievirus and rhinovirus. J. Biol. Chem. 268:19200-19203. [PubMed] [Google Scholar]
- 35.Le, H., R. L. Tanguay, M. L. Balasta, C.-C. Wei, K. S. Browning, A. M. Metz, D. J. Goss, and D. R. Gallie. 1997. Translation initiation factors eIF-iso4G and eIF-4B interact with the poly(A)-binding protein and increase its RNA binding activity. J. Biol. Chem. 272:16247-16255. [DOI] [PubMed] [Google Scholar]
- 36.Lloyd, R. E., H. G. Jense, and E. Ehrenfeld. 1987. Restriction of translation of capped mRNA in vitro as a model for poliovirus-induced inhibition of host cell protein synthesis: relationship to p220 cleavage. J. Virol. 61:2480-2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lomakin, I. B., C. U. Hellen, and T. V. Pestova. 2000. Physical association of eukaryotic initiation factor 4G (eIF4G) with eIF4A strongly enhances binding of eIF4G to the internal ribosomal entry site of encephalomyocarditis virus and is required for internal initiation of translation. Mol. Cell. Biol. 20:6019-6029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mader, S., H. Lee, A. Pause, and N. Sonenberg. 1995. The translation initiation factor eIF-4E binds to a common motif shared by the translation factor eIF-4γ and the translational repressors 4E-binding proteins. Mol. Cell. Biol. 15:4990-4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marcotrigiano, J., I. B. Lomakin, N. Sonenberg, T. V. Pestova, C. U. Hellen, and S. K. Burley. 2001. A conserved HEAT domain within eIF4G directs assembly of the translation initiation machinery. Mol. Cell 7:193-203. [DOI] [PubMed] [Google Scholar]
- 40.Marissen, W. E., A. Gradi, N. Sonenberg, and R. E. Lloyd. 2000. Cleavage of eukaryotic translation initiation factor 4GII correlates with translation inhibition during apoptosis. Cell Death Differ. 7:1234-1243. [DOI] [PubMed] [Google Scholar]
- 41.Marissen, W. E., and R. E. Lloyd. 1998. Eukaryotic translation initiation factor 4G is targeted for proteolytic cleavage by caspase 3 during inhibition of translation in apoptotic cells. Mol. Cell. Biol. 18:7565-7574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41a.Michel, Y. M., D. Poncet, M. Piron, K. M. Kean, and A. M. Borman. 2000. Cap-poly(A) synergy in mammalian cell-free extracts. J. Biol. Chem. 275:32268-32276. [DOI] [PubMed] [Google Scholar]
- 42.Morino, S., H. Imataka, Y. V. Svitkin, T. V. Pestova, and N. Sonenberg. 2000. Eukaryotic translation initiation factor 4E (eIF4E) site and the middle one-third of eIF4GI constitute the core domain for cap-dependent translation, and the C-terminal one-third functions as modulatory region. Mol. Cell. Biol. 20:468-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morley, S. J., P. S. Curtis, and V. M. Pain. 1997. eIF4G: translation's mystery factor begins to yield its secrets. RNA 3:1085-1104. [PMC free article] [PubMed] [Google Scholar]
- 44.Pyronnet, S., H. Imataka, A. C. Gingras, R. Fukunaga, T. Hunter, and N. Sonenberg. 1999. Human eukaryotic translation initiation factor 4G (eIF4G) recruits mnk1 to phosphorylate eIF4E. EMBO J. 18:270-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Raught, B., A.-C. Gingras, S. P. Gygi, H. Imataka, S. Morino, A. Gradi, R. Aebersold, and N. Sonenberg. 2000. Serum-stimulated, rapamycin-sensitive phosphorylation sites in the eukaryotic translation initiation factor 4GI. EMBO J. 19:434-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rose, J. K., H. Trachsel, K. Leong, and D. Baltimore. 1978. Inhibition of translation by poliovirus: inactivation of a specific initiation factor. Proc. Natl. Acad. Sci. USA 75:2732-2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scheper, G. C., N. A. Morrice, M. Kleijn, and C. G. Proud. 2001. The mitogen-activated protein kinase signal-integrating kinase Mnk2 is a eukaryotic initiation factor 4E kinase with high levels of basal activity in mammalian cells. Mol. Cell. Biol. 21:743-754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stein, I., A. Itin, P. Einat, R. Skaliter, Z. Grossman, and E. Keshet. 1998. Translation of vascular endothelial growth factor mRNA by internal ribosome entry: implications for translation under hypoxia. Mol. Cell. Biol. 18:3112-3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tarun, S. Z., and A. B. Sachs. 1996. Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J. 15:7168-7177. [PMC free article] [PubMed] [Google Scholar]
- 50.Tarun, S. Z., and A. B. Sachs. 1995. A common function for mRNA 5′ and 3′ ends in translation initiation in yeast. Genes Dev. 9:2997-3007. [DOI] [PubMed] [Google Scholar]
- 51.Wei, C. C., M. L. Balasta, J. H. Ren, and D. J. Goss. 1998. Wheat germ poly(a) binding protein enhances the binding affinity of eukaryotic initiation factor 4f and (iso)4f for cap analogues. Biochemistry 37:1910-1916. [DOI] [PubMed] [Google Scholar]
- 52.Wells, S. E., P. E. Hillner, R. D. Vale, and A. B. Sachs. 1998. Circularization of mRNA by eukaryotic translation initiation factors. Mol. Cell 2:135-140. [DOI] [PubMed] [Google Scholar]
- 53.Yan, R., W. Rychlik, D. Etchison, and R. E. Rhoads. 1992. Amino acid sequence of the human protein synthesis initiation factor eIF-4γ. J. Biol. Chem. 267:23226-23231. [PubMed] [Google Scholar]
- 54.Zamora, M., W. E. Marissen, and R. E. Lloyd. 2002. Multiple eIF4GI-specific protease activities present in uninfected and poliovirus-infected cells. J. Virol. 76:165-177. [DOI] [PMC free article] [PubMed] [Google Scholar]