Abstract
Yeast mutants lacking telomerase are capable of maintaining telomeres by an alternate mechanism that depends on homologous recombination. We show here, by using Kluyveromyces lactis cells containing two types of telomeric repeats, that recombinational telomere elongation generates a repeating pattern common in most or all telomeres in survivors that retain both repeat types. We propose that these patterns arise from small circles of telomeric DNA being used as templates for rolling-circle gene conversion and that the sequence from the lengthened telomere is spread to other telomeres by additional, more typical gene conversion events. Consistent with this, artificially constructed circles of DNA containing telomeric repeats form long tandem arrays at telomeres when transformed into K. lactis cells. Mixing experiments done with two species of telomeric circles indicated that all of the integrated copies of the transforming sequence arise from a single original circular molecule.
Telomeres are the protective DNA-protein complexes at the ends of chromosomes (reviewed in references 35 and 57). The DNA of telomeres in most eukaryotic organisms is composed of tandem arrays of 5- to 8-bp direct repeats. These repeats in part serve as binding sites for specific proteins that cap the telomeres and prevent them from eliciting the repair responses that are normally activated by broken DNA ends. Loss of this function can lead to cell cycle arrest, telomere fusions, and high rates of recombination near telomeres (3, 6, 14, 29, 33, 34, 38, 39, 48). The functioning of telomeres is thus critical for proper cell growth and chromosome stability. Because DNA polymerases are unable to fully replicate ends, the DNA of even properly capped telomeres is intrinsically unstable and prone to progressive shortening with every cell division (26, 42). Most organisms avoid this problem because of the enzyme telomerase, which adds new telomeric repeats de novo onto telomeric ends (18, 36). Most human somatic cells have little or no telomerase activity, and their telomeres are subject to gradual shortening (19, 22, 49). Sufficient telomere shortening in human cells triggers the permanent growth arrest of replicative senescence (7, 27). This is thought to be an adaptation to reduce the rate of cancer formation, as almost all human cancers are found to have an active telomere maintenance pathway, most typically due to the presence of telomerase (22).
Although sequence addition by telomerase is the major mechanism of telomere elongation in eukaryotic cells, it is not the only mechanism. The ends of chromosomes in the fruit fly Drosophila melanogaster are maintained by occasional transposition of retrotransposons (4, 24, 53). Other organisms, including the midge Chironomus sp., the mosquito Anopheles gambiae, and the plant Alium cepa, have telomeres composed of complex repeat families that are thought to be maintained by recombination (5, 13, 43, 47).
Recombinational telomere elongation can also occur in some cells that normally utilize telomerase. Yeast cells without telomerase undergo gradual telomere shortening and growth senescence within 50 to 100 cell divisions (26, 32, 50). Cells that survive beyond the point of senescence have restored longer tracts of telomeric repeats to their chromosomal ends through recombination. In S. cerevisiae, there are two genetically distinguishable pathways of recombinational telomere maintenance, one which amplifies subtelomeric sequences while maintaining short telomeres (type 1) and the other that lengthens telomeric repeat arrays (type 2) (23, 25, 51). In Kluyveromyces lactis, which lacks the subtelomeric blocks of telomeric repeats found in Saccharomyces cerevisiae, only the equivalent of type 2 survivors are seen (31).
Some human cells are capable of maintaining telomeric repeat arrays at their chromosome ends in the absence of telomerase. This telomerase-independent pathway of telomere maintenance is called alternate lengthening of telomeres (ALT). ALT was first observed in certain in vitro immortalized cell lines (10, 37, 44). Subsequently, it has been found that 5 to 10% of human cancers also appear to maintain long telomeres despite an absence of telomerase (9). Recently, direct evidence for telomeric recombination in ALT cells has been found, suggesting that these cells maintain telomeres through homologous recombination (15). These data indicate that recombinational telomere maintenance can occur in human cells and is likely of significance to a number of human cancers.
How cells can utilize recombination to generate longer telomeric repeat tracts has remained perplexing. In K. lactis cells, short telomeres are subject to large increases in the rates of subtelomeric gene conversion and can lead to the frequent spreading of a marker gene from one telomere to most or all other telomeres in the cell (33). We show here, by marking K. lactis telomeres with two types of telomeric repeat, that recombinational telomere elongation generates a common repeating pattern within most telomeres in a given survivor. We further show that long repeating arrays are formed at chromosome ends when cells are transformed with DNA circles containing telomeric repeats. These results suggest that rolling-circle gene conversion may be a mechanism for recombinational telomere elongation.
MATERIALS AND METHODS
Strains.
The K. lactis ter1-Δ strain was derived from haploid, wild-type TER1 K. lactis 7B520 (Ura3− His3− Trp−) (56). TER1 was deleted using the plasmid “loop in, loop out” procedure (32). The ter1-Δ strain with wild-type telomeric repeats at the base and Bcl telomeric repeats at the tips was derived by transforming a senescing ter1-Δ strain with an integrative plasmid (pTER-BX:UA TER1-Bcl) containing TER1-Bcl, forming a ter1-Δ/TER1-Bcl heteroallele. Plating on 5-fluoroorotic acid (5-FOA) selected for cells that had looped out either the TER1-Bcl allele or the ter1-Δ allele. Clones containing only the ter1-Δ allele were identified by their early senescence rough colony phenotype. K. lactis transformation was done by electroporation as described for S. cerevisiae (2).
Telomere cloning.
Telomeres were cloned by plasmid rescue as follows. A pRS423-derived plasmid, with a HIS3 marker gene, containing a ∼590-bp EcoRI-XbaI fragment of K. lactis subtelomeric sequence was transformed into the ter1-Δ/TER1-BclIheteroallele strain where it integrated (looped in) next to a telomere. Transformants were selected on plates lacking histidine and then plated on 5-FOA to select for ter1-Δ cells and passaged by serial streaking on solid medium to produce senescent cells and, later, the formation of postsenescence survivors. Genomic DNA from His+ survivors was cleaved with XhoI, to detach the plasmid-telomere fragment from chromosomal DNA, treated with T4 polymerase to blunt ends, and ligated to circularize the fragment. After transformation into Escherichia coli, plasmids containing cloned telomeres could be recovered. As selection for a subtelomeric marker leads to the spread of that marker to multiple telomeres in senescing ter1-Δ cells (33), different cloned telomeres from a given survivor likely represent different chromosome ends.
Southern hybridizations and quantitation of URA3 copy number.
Southern blotting was done using Hybond N+ membrane. All hybridizations were done in 500 mM Na2HPO4 and 7% sodium dodecyl sulfate. The telomeric probe used in our hybridizations is the Klac1-25 oligonucleotide (32). This probe was used at 50°C. The subtelomeric probe was generated from the ∼590-bp EcoRI-XbaI. The URA3 probe used was a HindIII fragment from pMH3 containing the S. cerevisiae URA3 gene (20). The subtelomeric and URA3 probes were hybridized at 65°C. The number of copies of URA3 was determined by analysis with a PhosphorImager.
Isolation of circle S and circle P.
The two circles (circle S and circle P) differ at a single restriction enzyme site (for circle S, SalI; for circle P, PvuI). Circle S was constructed by circularizing a BamHI-BglII fragment from plasmid pMH3-1Tel, and circle P was constructed in a like manner after first filling in the SalI site to create a PvuI site. pMH3-1Tel was derived by integrating a DpnI fragment from pAK25 (33) containing ∼11.5 telomeric repeats and ∼120 bp of subtelomeric sequence into pMH3.
RESULTS
Repeating patterns are produced by recombinational telomere elongation.
To investigate the mechanism by which recombination generates elongated telomeres, we constructed yeast cells with a disrupted telomerase RNA gene (ter1-Δ) and containing telomeres composed of basal wild-type repeats and terminal mutant repeats (Bcl repeats) (Fig. 1A and Materials and Methods). We reasoned that the pattern of the two repeat species present in the telomeres that underwent recombinational elongation would provide clues about the mechanism that produced the elongation. The Bcl mutation present in the terminal repeats does not perturb telomere function and produces a BclI restriction site by changing one position within the 25-bp telomeric repeat (30, 33, 54).
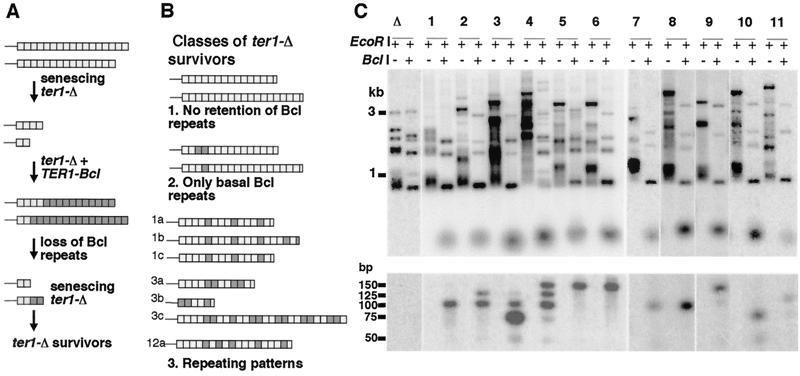
FIG. 1.
Repeating structure within telomeres of ter1-Δ survivors. (A) Strategy for construction of ter1-Δ strains with two types of telomeric repeats. The ter1-Δ strain with wild-type telomeric repeats at the base and Bcl telomeric repeats at the tips was derived by forming a ter1-Δ/TER1-Bcl heteroallele. Plating on 5-FOA selected for cells that had looped out either the TER1-Bcl allele or the ter1-Δ allele. Clones containing only the ter1-Δ allele were identified by their senescent rough colony phenotype. (B) Classes of outcomes observed in postsenescence survivors derived from ter1-Δ strains with wild-type and Bcl repeats. Light gray boxes indicate wild-type repeats, dark gray boxes indicate Bcl repeats, and white boxes indicate partial repeats not distinguishable as either wild-type or Bcl. Thin lines represent subtelomeric sequence. Telomere structures shown for outcomes 1 and 2 are approximate; based upon BclI restriction digestion and not sequencing. Telomere structures for outcome 3 are cloned and sequenced examples of recombinationally elongated telomeres from three independent postsenescence survivors. There is no sequence variation from wild-type telomeric repeats in the cloned telomeres except for the position that is expected to create a BclI restriction site. (C) Southern blot, hybridized with a telomeric probe, of a senescent ter1-Δ strain (lane Δ) and 11 postsenescence survivors derived from it that retained Bcl repeats (lanes 1 to 11). Pairs of lanes show DNA from individual survivors digested with EcoRI and EcoRI-BclI. Among the 12 telomeres, subtelomeric EcoRI sites are present ∼1 to 3.5 kb from telomeric ends. The double digest yields blocks of wild-type repeats. The lower panel shows these blocks resolved on a 4% NuSieve agarose gel. Size markers for both panels are indicated. Cloned telomeres in panel B (1a, 1b, and 1c and 3a, 3b, and 3c) came from clones 1 and 3, respectively, of panel C. Cloned telomere 12a of panel B was isolated from a clone not shown in panel C.
Digestion of genomic DNA from most postsenescence survivors indicated that elongated telomeres in these cells either lacked Bcl repeats entirely (Fig. 1B, outcome 1) or had them confined to the basal regions of one or more telomeres (Fig. 1B, outcome 2) (data not shown). These telomere structures are inconsistent with Bcl repeats having become amplified during the recombinational telomere elongation and survivors containing them and were not studied further. Other survivors (34 of 231), however, were more informative. BclI digestion of genomic DNA from these survivors, but not from pre-survivor cells, excised very small fragments that intensely hybridized to a telomeric probe (Fig. 1C). These were predicted to be blocks of wild-type repeats bordered on each side by half Bcl repeats. For most such survivors, all telomeres in the cell were cleaved by BclI and hence contained Bcl repeats. When the BclI-cleaved DNA was run on high-percentage agarose gels (lower panel of Fig. 1C and data not shown), it was found that the small telomeric fragments were the sizes expected for multiples of the K. lactis 25-bp telomeric repeat, with the great majority being between 50 and 150 bp (one to five wild-type repeats, correcting for the presence of a half Bcl repeat on each end). Within a given survivor clone, the excised small fragments were mostly a single size (most notably clones 1, 5, 6, 7, and 8). Different clones, however, displayed different sizes of BclI fragments. These data suggested that a common pattern of wild-type and Bcl repeats was often present in most or all elongated telomeres generated in a postsenescence survivor. The presence of a single predominant size class for blocks of wild-type repeats further suggested the possibility that the elongated telomeres often had a simple repeating pattern of wild-type and Bcl repeats. To address this, we cloned telomeres from three independent survivors containing interspersed wild-type and Bcl repeats. Sequencing telomeric DNA from these survivors confirmed the frequent presence of repeating patterns (Fig. 1B, outcome 3). The telomeres from survivors we examined commonly contained a repeating unit composed of four repeats, either three wild-type repeats and one Bcl repeat or two wild-type repeats and two Bcl repeats. The blocks of wild-type repeats observed in the sequenced clones were in agreement with our restriction digestion data (Fig. 1C and data not shown). Our results confirmed that recombinational telomere elongation in K. lactis commonly involved (i) a process capable of generating repeating patterns within telomeres and (ii) a process capable of spreading a common repeating pattern to multiple telomeres.
DNA circles containing telomeric repeats can greatly promote telomere elongation.
A possible way for a short telomere in a senescing telomerase deletion mutant to both become elongated and acquire a repeating pattern would be for it to initiate a gene conversion event utilizing a small circle of telomeric repeats as a template. A telomere extended by rolling-circle gene conversion would acquire a repeating pattern that matched the size and pattern of the circle that was copied.
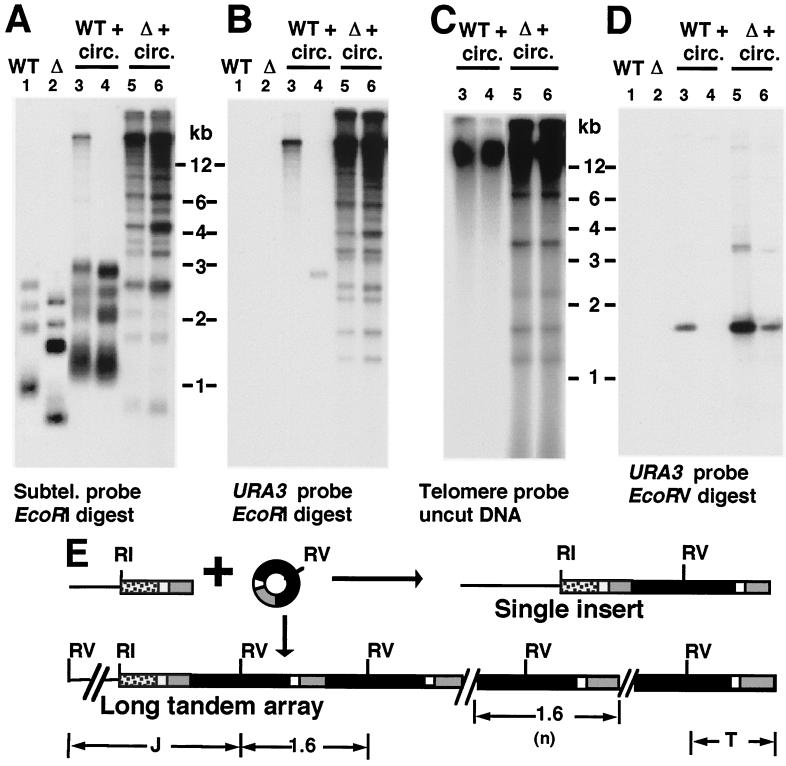
A rolling-circle model predicts that an exogenously provided DNA circle containing telomeric repeats could become incorporated at telomeres as a long tandem array. To test this, a DNA circle was constructed by ligating the ends of a restriction fragment containing URA3 and a K. lactis telomere. The resulting 1.6-kb monomeric circle was purified by isolation from a gel and then transformed into K. lactis ter1-Δ cells as well as cells containing a functional telomerase (TER1). As the circle lacked a replication origin, maintenance of sequences derived from it required incorporation into a chromosome. We analyzed some of the transformants that grew on uracil-lacking plates by Southern blotting and hybridization to determine their telomere structure. The terminal EcoRI fragments of the chromosomes were visualized through use of a subtelomeric probe that hybridizes to 11 of the 12 such fragments in the haploid K. lactis genome (Fig. 2A). Some TER1 transformants had structures consistent with integration of a single copy of the transforming circle at a telomere or at nontelomeric positions (Fig. 2A and B, lane 4, and data not shown). However, many TER1 transformants (32 of 48) and all ter1-Δ transformants (45 of 45) appeared to have one or more telomeric EcoRI fragments that were greatly elongated, running at or near limit mobility in the gel (>15 kb [Fig. 2A and B, lanes 3, 5, and 6]). Untransformed controls, in contrast, contained no telomeric fragments larger than 3.5 kb (including the single telomere not hybridizing to the subtelomeric probe). The elongated telomeres hybridized very intensely to URA3 (Fig. 2B) and telomeric probes (data not shown and Fig. 3). These data indicated that telomeres in the transformants had become elongated, and often greatly elongated, by incorporation of sequence from the transforming 1.6-kb circle. The extent of subtelomeric sequence present as elongated bands (Fig. 2A) is indicative of how many telomeres were elongated by addition of sequence derived from the transforming DNA circle and differed between TER1 and ter1-Δ transformants. TER1 transformants had one or sometimes 2 lengthened telomeres, while ter1-Δ transformants had many and often most of their 12 telomeres lengthened. Control experiments using linear DNA containing URA3, subtelomeric sequence, and telomeric repeats, but with ends incompatible with self-ligation, readily integrated by replacing one native telomere or more but never formed tandem arrays (33, 55); data not shown), indicating that formation of the highly elongated telomeres depended upon the transforming DNA having a circular structure.
FIG. 2.
Long tandem arrays formed at telomeres after transformation with a DNA circle containing URA3 and telomeric repeats. (A) Southern blot, hybridized with a subtelomeric probe, of EcoRI-digested DNA from wild-type TER1 (WT) and ter1-Δ (Δ) strains untransformed (lanes 1 and 2) and after transformation (lanes 3 to 6) with a 1.6-kb URA3-telomere circle. (B) Same filter as in panel A after stripping and reprobing with a URA3 probe. (C) Southern blot, hybridized to a telomeric probe, of uncut genomic DNA from TER1 (WT) and ter1-Δ (Δ) strains transformed with the 1.6-kb URA3-telomere circle. Transformants shown are the same as those shown in panels A and B. (D) Southern blot, hybridized with a URA3 probe, of EcoRV-digested DNA from wild-type TER1 (WT) and ter1-Δ (Δ) strains untransformed (lanes 1 and 2) and after transformation (lanes 3 to 6) with a 1.6-kb URA3-telomere circle. (E) Diagram of 1.6-kb URA3-telomere circle transformation and structures of single and multiple tandem inserts and a telomere. Gray boxes indicate blocks of telomeric repeats, black boxes indicate URA3, white boxes indicate a short subtelomeric sequence present on DNA circles, and stippled boxes indicate subtelomeric sequence used as a probe in panel A and not present on the circles. Subtelomeric EcoRI sites are 1 to 3.5 kb from telomeric ends in untransformed K. lactis cells. Positions of EcoRI sites(RI) and EcoRV (RV) sites are indicated. Abbreviations: J, centromere-proximal junction fragment; T, telomeric fragment.
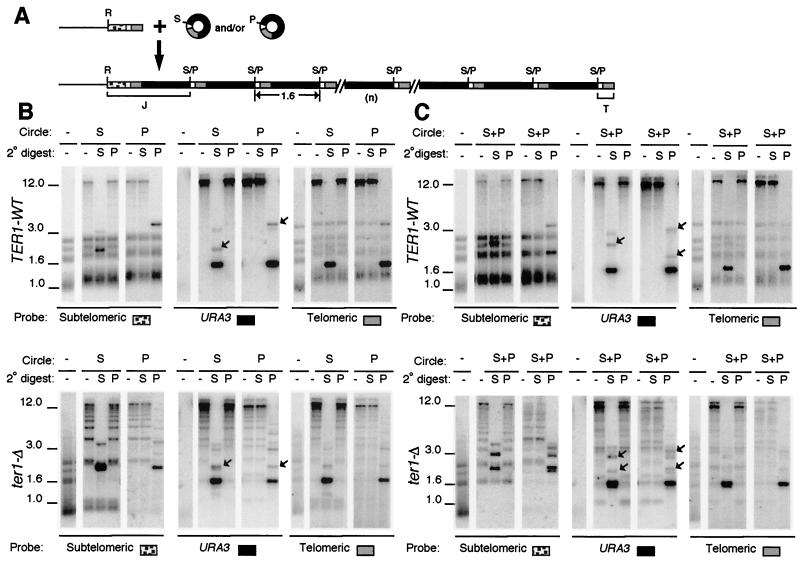
FIG. 3.
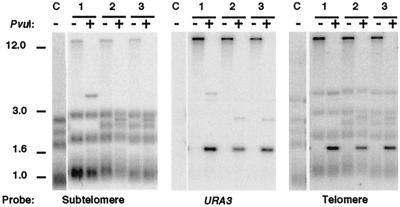
Transformation with two species of circle produces arrays derived from only one species. (A) Diagram of telomere structures before and after introduction of a mixture of two species of 1.6-kb URA3-telomere circles that differ only by a single restriction site. Gray and black boxes are blocks of telomeric repeats and URA3, respectively. White boxes are subtelomeric sequence present on DNA circles, and stippled boxes are subtelomeric sequence not present on the circles. Abbreviations: R, S, and P, sites for EcoRI, SalI, and PvuI, respectively; S/P, sites that will either be SalI or PvuI depending upon which circle the site is derived from; J and T, junction fragment with subtelomeric sequences and terminal telomeric fragment, respectively. (B) Southern blots of representative clones of TER1 (top) and ter1-Δ (bottom) transformed with either circle S or circle P are shown hybridized with subtelomere, URA3, or telomeric probe, as indicated. Untransformed control is shown digested with EcoRI, and transformants are shown digested with EcoRI (-), EcoRI-SalI (S), or EcoRI-PvuI (P). The type of transforming circle used is indicated on top. A faint band at 3.2 kb in URA3-probed lanes containing the dark 1.6-kb fragment are trace partials left over from cleaving the tandem arrays. Positions of molecular weight markers (in kilobase pairs) are indicated. (C) Southern blots of two representative clones each of TER1 (top) and ter1-Δ (bottom) transformed with both circle S and circle P are shown hybridized with subtelomere, URA3, or telomeric probe, as indicated. The two clones represent one example each of clones exhibiting tandem arrays of either an S version or a P version. Digests were done as for panel B. (B and C) Junction fragments (J) are marked with arrows in the URA3.
Cutting DNA from transformants with long telomeres (both TER1 and ter1-Δ) with restriction enzymes that cleaved the transforming circle at a single position generated a 1.6-kb fragment that hybridized intensely to URA3 (Fig. 2D) and telomeric probes and was often plainly visible on ethidium bromide-stained gels (data not shown). Such digests also produced one or more faint bands (visible with URA3 and subtelomeric probes) of sizes consistent with centromere-proximal junction fragments (Fig. 2E), and short diffuse terminal fragments (visible with URA3 and telomeric probes; Fig. 2E and data not shown). These results are fully consistent with the sequence from the circle having integrated at telomeres as long tandem arrays. The number of copies of the 1.6-kb unit present in transformants was estimated by measuring URA3 hybridization signal on a Southern blot using a strain with a single integrated copy of URA3 as a control. The copy number of the URA3-telomere insert was estimated to be 6 to 20 in TER1 transformants and 30 to 180 in ter1-Δ transformants (data not shown). The highest copy numbers are the equivalent of about 2% of total genomic DNA.
Unlike the elongated telomeres of TER1 transformants, those of ter1-Δ transformants ran as complex ladders of bands (Fig. 2B and 3B). Most of these bands hybridized with a subtelomeric probe and likely represent telomeres containing different numbers of the integrated 1.6-kb sequence (1 to 10 or more copies). However, not all bands in lanes from the ter1-Δ transformants that hybridized to the URA3 probe also hybridized to the subtelomeric probe (see for example, the lowest band visible in lanes 5 and 6 of Fig. 2B). This suggested that DNA present in these bands was not associated with telomeres. When uncut genomic DNAs from the ter1-Δ transformants were run on gels, several bands that hybridized to telomeric and URA3 probes were present that migrated well ahead of the bulk chromosomal DNA band (Fig. 2C and data not shown). These bands were not detected in wild-type transformants or in an untransformed ter1-Δ control. We conclude that extra-chromosomal derivatives of the 1.6-kb sequence were present in these cells. Although we have not characterized these DNA species, their positions in gels suggests that they could be circular and linear monomers and multimers of the 1.6-kb URA3-telomere sequence. Lacking an ARS element, such structures could not be stably maintained in cells but might instead be frequently produced by the high level of recombination experienced near telomeres in ter1-Δ cells (33). Our experiments have shown that none of the sequences of the 1.6-kb URA3-telomere circle are capable of driving autonomous replication in K. lactis (data not shown).
In addition to having one or two telomeres extended by tandem arrays from the transformed 1.6-kb circles, all other telomeres in TER1 transformants were slightly elongated, apparently by the addition of extra telomeric repeats (Fig. 2A and 3B). This may reflect an ability of the extra telomeric repeat arrays on the greatly extended telomere to bind and titrate a protein such as Rap1 that negatively regulates telomere length. A similar lengthening phenomenon has been observed in S. cerevisiae when extra copies of telomeric repeats were present on a multicopy plasmid (46).
All integrated copies of the 1.6-kb sequence are derived from a single transforming circular DNA molecule.
To test whether the tandem arrays that formed at telomeres in TER1 and ter1-Δ transformants were derived from a single original transforming DNA circle or from independent integrations of multiple circles, we performed a mixing experiment. A second form of the 1.6-kb circle was constructed by first filling in a unique SalI fragment to create a unique PvuI fragment in the precursor plasmid. The two forms of 1.6-kb circle (SalI [circle S] or PvuI [circle P]; Fig. 3A) were then generated, purified, and introduced singly or together (in similar amounts) into ter1-Δ and TER1 strains, and the telomeres from the resulting transformants were then examined (Fig. 3B and C). Figure 3A shows the expected general structure if sequence from circle S and/or circle P form tandem arrays at chromosome ends. The composition of these arrays was expected to differ if derived from a single transforming circle rather than being derived from many circles. If a single circular molecule was the ultimate source of all the tandem arrays in a transformant, then the arrays should be cleaved completely down to 1.6-kb units by the restriction enzyme specific to one form of circle and completely resistant to digestion with the enzyme specific for the other form of circle. In contrast, if arrays form from integration of multiple transforming circles, then telomeric DNA from any given transformant should contain both S and P versions of the 1.6-kb sequence and should be cleavable with both SalI and PvuI. Transformation rates of the two circles, transformed singly or together, were similar, indicating that transformation was not biased for one molecule or the other.
Figure 3B shows hybridization data from representative control clones of TER1-WT (top) and ter1-Δ (bottom) that had been individually transformed with either circle S or circle P. Filters of genomic DNA cleaved with EcoRI, EcoRI-SalI, and EcoRI-PvuI were sequentially probed with subtelomeric, URA3, and telomeric probes. As seen earlier with circle S alone (Fig. 2), tandem telomeric arrays were formed, and these were cleaved down to 1.6-kb units that hybridized to telomeric and URA3 probes when digested with the restriction enzyme specific for the transforming circle (SalI for circle S and PvuI for circle P). Arrays remained at limit mobility, and no 1.6-kb URA3-hybridizing bands were observed when samples were digested with the enzyme specific for the other form of circle. Subtelomeric junction fragments (Fig. 2A) that hybridized to subtelomeric and URA3 probes are indicated with arrows in the panels showing the URA3 hybridization. Very short and diffuse terminal fragments (Fig. 2A) were also observed with the telomeric probe (data not shown).
Figure 3C shows hybridization data from representative clones of TER1-WT (top) and ter1-Δ (bottom) that had been transformed with a mixture of comparable amounts of circle S and circle P. Digests and hybridizations shown are arranged as for Fig. 3B. The data show that transforming K. lactis cells with mixtures of S and P circles produced tandem telomeric arrays that, within a given transformant, contained sequences derived from only one form of circle. All 22 of 22 TER1 and 28 of 28 ter1-Δ contained tandem arrays of either the P version or the S version of the 1.6-kb sequence, but not both. Twelve of the 22 TER1 transformants had utilized circle S and the other 10 had utilized circle P to form tandem arrays. Eight of 28 ter1-Δ transformants had utilized circle S, and the other 20 had utilized circle P. These data indicate that even in cells with >100 integrated total copies spread among multiple telomeres, all copies can ultimately be derived from a single transforming molecule. The absence of clones containing sequences from both forms of circle may indicate that, in our experiments, only one circular molecule normally entered a cell about to be transformed.
Formation of telomeric tandem arrays is reduced but not eliminated in a rad52 mutant.
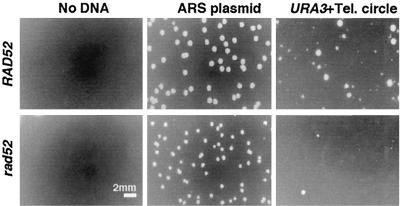
We next tested whether the formation of the long URA3-telomere arrays was dependent upon RAD52, a gene required for most forms of homologous recombination in S. cerevisiae. Because rad52 ter1-Δ mutants have extremely poor viability (31), this experiment was necessarily limited to a rad52 TER1 strain. When transformed with the 1.6-kb P circle, a rad52 TER1 strain showed a reduced ability to form Ura3+ transformants compared to a RAD52 TER1 strain (Fig. 4). A control ARS plasmid transformed both strains equally well. The precise degree to which the rad52 mutation reduced circle P transformation was difficult to determine because of the heterogeneous colony size observed on transformation plates (Fig. 4). However, we estimate that the degree of reduction is 75 to 95%. The colonies that did form in a rad52 background were examined to determine whether long telomeric arrays had formed. While the tiniest colonies of both rad52 and RAD52 strains did not contain detectable URA3 sequences (and were presumably abortive transformants), the small to medium transformant colonies did. As shown in Fig. 5, these transformants routinely (23 of 43) had the same characteristics as RAD52 TER1 circle P transformants, a novel telomere running at limit mobility in EcoRI digests which was cut down to a 1.6-kb fragment by PvuI and which hybridized intensely to URA3 and telomeric probes. The transformants were confirmed as being derived from the original rad52 deletion mutants through hybridization to a RAD52 gene probe. Our data indicate that absence of RAD52 function significantly reduces, but does not eliminate, the ability of a transforming circle to generate a long tandem array at a telomere. This result is consistent with either a single mechanism that is partially RAD52 dependent or with two mechanisms, the major of which is RAD52 dependent, for forming tandem arrays by copying the 1.6-kb circle.
FIG. 4.
URA3-telomere circle transformation into rad52 and RAD52 strains. Photographs show sections of plates with Ura+ transformants. Equal amounts of both the URA3-telomere (Tel.) circle and the autonomously replicating ARS plasmid control were used for both strains.
FIG. 5.
Long tandem arrays at telomeres can be formed in a TER1 rad52 strain transformed with a URA3-telomere circle. Shown is a Southern blot of untransformed control (C) and three rad52 clones transformed with a URA3-telomere circle. The untransformed control is shown digested with EcoRI, and the transformed clones are shown digested with EcoRI and EcoRI-PvuI, as indicated. The same filter is shown hybridized with subtelomeric, URA3, and telomeric probes. Faint bands at 1.6 kb in EcoRI-PvuI-digested samples hybridized with the subtelomeric sequence are residual signal from prior URA3 hybridization. Size markers (in kilobases) are shown at left.
DISCUSSION
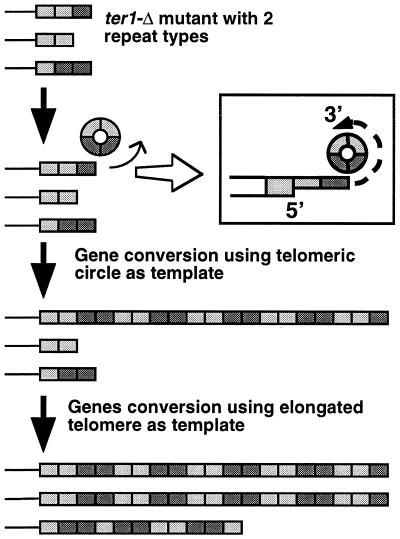
To explain recombinational telomere elongation in ter1-Δ survivors, we propose the “roll-and-spread” model shown in Fig. 6. It proposes that formation of long telomeres by recombinational telomere elongation is brought about by two distinct types of events. In one, a rolling-circle gene conversion utilizes a tiny circle of telomeric DNA as a template to generate a long telomere in a single step. If the circle contained two species of telomeric repeat, the resulting elongated telomere would have a repeating pattern based upon the unit structure of the circle being copied. In the second, more common type of event, other gene conversions using the elongated telomere as a template spread its sequence onto other telomeres in the cell.
FIG. 6.
Roll-and-spread model. The formation of one long telomere is postulated to occur via a rolling-circle gene conversion, copying either the 1.6-kb circle (circle transformants) or a very small telomeric circle (ter1-Δ survivors). The inset depicts a telomeric end processed to have a 3′ single strand overhang (shaded thin boxes) that strand invade a telomeric circle. In ter1-Δ cells, the very high rate of telomeric gene conversion can spread sequence from one long telomere onto many or all other telomeres of the cell under selective pressure for postsenescence survivors. The net result is that a common pattern is present in most or all of the elongated telomeres.
It was previously shown that short telomeres, even in the absence of growth senescence, caused gene conversion rates at subtelomeric positions to be highly elevated (33). In a ter1-Δ strain, the degree of increase was measured as ∼800 times higher than that seen in a TER1 control. This enormously elevated gene conversion frequency was shown to be readily capable of copying a marker gene initially next to a single telomere onto most or all other chromosome ends in the cell. Recent data indicate that recombination between telomeres themselves is also greatly enhanced in a strain lacking telomerase (Z. Topcu and M. McEachern, unpublished data).
Our findings reported here show that recombinationally elongated telomeres within a given postsenescence survivor typically contain a common pattern of telomeric repeats. This strongly supports the hypothesis that the generation of postsenescence survivors in ter1-Δ mutants is dependent upon spreading of a sequence from one elongated telomere to many, and sometimes all, other telomeres in the cell. As even a single short or missing telomere is likely to be enough to prevent normal growth (3, 48), there would be strong selective pressure on ter1-Δ cells to spread a long telomeric sequence to all other telomeres. The spreading of an elongated telomeric repeat array between different chromosome ends may not be a completely random process. Conceivably, by virtue of having many telomeric repeats, a long telomere would preferentially be a target of strand invasion by short recombinogenic telomeres and therefore preferentially be a donor of sequence information.
The results with transforming circular DNA molecules provide strong circumstantial support for rolling-circle gene conversion being a process that K. lactis cells can carry out. Our data clearly show that a single circular molecule is responsible for generating the long tandem arrays in TER1 and ter1-Δ transformants. The typical structure of TER1 transformants, where a single telomere has acquired all the copies of 1.6-kb sequence, argues that integration occurs through a concerted process. In principle, the 1.6-kb circle could integrate singly at a telomere and then expand into a tandem array through multiple unequal crossovers. This would have to be an extremely efficient process to account for long arrays being present as soon as transformants can be examined. At least two additional things argue against unequal recombination accounting for array formation. First, the expected intermediate structures (telomeres with only a few tandem copies of the 1.6-kb sequence) are not observed in TER1 transformants, even in rad52 cells which have a greatly reduced rate of homologous recombination. Second, we have shown directly (Fig. 2) that a TER1 transformant carrying a single copy insert (the transformant shown in lane 4 of Fig. 2A to D) will exist stably for at least 100 cell divisions without expanding into a long tandem array (data not shown).
As seen in Fig. 4, transformants of the 1.6-kb circle that grew on plates lacking uracil were heterogeneous in size compared to ARS plasmid controls. Some colony size heterogeneity is characteristic of integrative transformation in general for K. lactis. In some organisms, telomeres are known to be able to transcriptionally silence nearby genes (1, 17, 41), and we cannot rule out the possibly that some degree of telomeric silencing might occur in our 1.6-kb circle transformants. However, the 1.6-kb circle transformants remain Ura+ over at least 100 generations of nonselective growth (data not shown), suggesting that silencing is either uncommon or insufficient to render cells Ura−. Also, telomeric silencing failed to be observed when a single copy of the same URA3 gene fragment used in the 1.6-kb circles was placed within ∼120 bp of a K. lactis telomere (33).
Although both TER1 and ter1-Δ cells have the ability to greatly extend their telomeres when transformed with a DNA circle containing telomeric repeats, there are notable differences in the behavior of the two strains that likely reflect the differences between normal telomeres and dysfunctional short telomeres. While TER1 circle transformants have only one or two telomeres with tandem arrays of the URA3-telomere sequence, ter1-Δ transformants typically have multiple telomeres with tandem arrays. This is almost certainly due to the highly recombinogenic nature of short telomeres and their propensity to spread a sequence from one telomere to other telomeres. Consistent with this, atypical ter1-Δ transformants, initially containing few telomeres with tandem arrays, have a much greater proportion of their telomeres with arrays after an additional passaging of ∼125 cell divisions, even in the absence of selection for URA3 (data not shown). A second difference between TER1 and ter1-Δ circle transformants is that only the latter contain detectable extrachromosomal species derived from the URA3-telomere sequence. The mechanism(s) by which these form is not known and could potentially be different from that responsible for the formation of tiny telomeric circles. A third difference concerns the size heterogeneity of the tandem arrays at telomeres. While arrays in TER1 cells typically exist as a single band running at limit mobility in our gels, arrays in ter1-Δ are highly heterogeneous (1 to >10 copies of the 1.6-kb sequence). This, coupled with the abundance of extrachromosomal species derived from the 1.6-kb sequence in their cells, suggests that the tandem arrays of ter1-Δ transformants are prone to recombining at high rates. This would not be surprising, as the terminal group of telomeric repeats in an array, which is likely responsible for all telomere function, remains both relatively short (data not shown) and is still subject to gradual sequence loss. We suggest that the telomeres of our circle transformants reach an equilibrium where recombination events that lengthen and spread the arrays are balanced by recombination events and perhaps other processes that shorten or delete them.
Rolling-circle gene conversion could readily account for the patterns of repeats we observed in the telomeres of ter1-Δ survivors. If a small circle of DNA containing both wild-type and Bcl repeats were used as a template for rolling-circle gene conversion, it would produce a repeating pattern of those two repeat types in the resulting elongated telomere, as observed in most of those survivors that retained any Bcl repeats. Although the major class of survivors we observed lacked Bcl repeats, this does not suggest the existence of a second mechanism of recombinational elongation. Rather, these survivors likely arose from copying circles of DNA composed solely of wild-type repeats. As the senescing cells contained only wild-type repeats in the more basal part of the telomere (as diagrammed in Fig. 1A), it is not surprising that Bcl repeats could be completely lost in most cells prior to the formation of survivors with elongated telomeres.
The simplest form of our hypothesis would be for a short telomere to undergo elongation by direct strand invasion into a small circle of telomeric DNA followed by rolling-circle DNA synthesis (Fig. 6). However, it is conceivable that rolling-circle synthesis and integration at a telomere could occur as separate steps. Rolling-circle replication might be initiated extrachromosomally on a telomeric circle if the 3′ end from a gap or broken telomeric fragment were available as a primer. Once an extrachromosomal tandem array was formed, strand invasion by a telomere followed by gene conversion could lead to the array becoming incorporated at a chromosome end.
Our data would suggest that circles as small as 100 bp can serve as templates for rolling-circle DNA synthesis. Although we have thus far been unable to identify small telomeric circles in K. lactis ter1-Δ cells (unpublished data), there is precedent for DNA circles composed of telomeric repeats being present in some mammalian cells (45), and circles as small as ∼100 bp are formed in vivo from the unusual mitochondrial telomeres of the yeast Candida salmanticensis (52). There is also precedent in vitro for circles as small as 34 nucleotides serving as templates for rolling-circle DNA synthesis (16). How tiny telomeric circles might form in senescing ter1-Δ cells is not clear, but they could easily be imagined to be an occasional by-product of the very high rates of subtelomeric and telomeric recombination that occur when telomeres become very short (33). Recombination between repeats of a single telomere or annealing between broken single-stranded fragments of telomeric DNA would each, in principle, be able to form circles containing whole numbers of telomeric repeats. Shortening of telomeres through intratelomeric recombination has been documented to sometimes occur in S. cerevisiae (11). If a small circle of telomeric repeats was produced by this process in a ter1-Δ strain, it conceivably could immediately be utilized by that telomere for rolling-circle gene conversion. We suggest that a limiting factor for recombinational telomere elongation may be the formation of telomeric circles and not the ability to utilize circles for telomeric elongation. Our results with transforming artificially created circles indicate that both TER1 and ter1-Δ cells can effectively utilize DNA circles to lengthen their telomeres.
We cannot rule out the possibility that the telomere elongation in some or all ter1-Δ postsenescence survivors arises through unequal recombination between telomeres rather than rolling-circle gene conversion. However, it is more difficult to imagine how unequal recombination could generate the long telomere sizes or the repeating patterns common to most telomeres that are seen in survivors. Irregularities in the patterns of some recombinationally elongated telomeres (Fig. 1B, clones 1b, 3c, and 12a) indicate that telomeric repeat arrays at chromosome ends can be altered by recombination events other than those that copy DNA circles.
A notable difference between the URA3-telomere circle transformants and ter1-Δ postsenescence survivors is the much lower degree of telomere elongation in the latter (typically hundreds of base pairs and not more than a few thousand base pairs). This might reflect a markedly lower degree of processivity for a DNA polymerase copying a very small circle. It could easily be imagined that a 100-bp circle would not be large enough for the proper assembly of a fully processive DNA polymerase holoenzyme.
Postsenescence survivors in S. cerevisiae are of two types. Type 2 survivors have elongated telomeric arrays very similar to the survivors in K. lactis (51). Type 1 survivors, in contrast, have long alternating arrays of telomeric repeats and subtelomeric Y′ elements (26), a pattern very reminiscent of the arrays of URA3 and telomeric repeats seen in K. lactis cells transformed with the 1.6-kb circle. The different structures of type 1 and type 2 survivors, combined with differences in the genes required for them (12, 23), indicate that there are mechanistic differences in how they arise. It will be of interest to determine whether the K. lactis circle transformants resemble type 1 transformants in other ways.
DNA ends lacking telomeric repeats in yeast are processed to generate 3′ overhangs which can then strand invade homologous duplex DNA to initiate a localized gene conversion or establish a replication fork that can sometimes copy sequence all the way to the end of the donor chromosome (8). The latter process, termed break-induced replication, could be the mechanism by which our 1.6-kb circles are copied to produce highly elongated telomeres. In S. cerevisiae, break-induced replication appears to be highly RAD52 dependent (28). It is noteworthy then that telomere elongation promoted by DNA circles in K. lactis is partially RAD52 independent. This could mean it occurs by a different mechanism, or it may simply be due to differences in the experimental systems. Disruption of RAD52 function is known to greatly reduce but not completely eliminate telomerase-independent telomere elongation in K. lactis ter1-Δ cells (31).
Our experiments have shown that DNA circles containing telomeric repeats are potent triggers of recombinational telomere elongation. The use of endogenously formed circular DNA molecules as templates to extend telomeric ends could potentially explain other examples of telomerase-independent telomere elongation. As mentioned, type II survivors of S. cerevisiae cells lacking telomerase elongate telomeres in a manner essentially identical to that seen in K. lactis ter1-Δ survivors, and type I survivors amplify alternating Y′ elements and telomeric sequences. DNA circles composed of Y′ and telomeric repeats have been reported to exist in S. cerevisiae (21). Mammalian cells that are ALT+ (and believed to maintain telomeres by recombination) often build very long telomeres in the absence of telomerase (10). In one reported example, the amplified DNA appeared to be a tandem array of telomeric and nontelomeric sequences (40). It will be of great interest to determine whether such telomeric circles underlie the ALT phenomenon of mammalian cells.
Acknowledgments
We are very grateful to Elizabeth Blackburn for her support in the early stages of this work and to the reviewers and members of the McEachern laboratory for critical reading of the manuscript. We thank Tracy Fulton for construction of the ter1-Δ/TER1-BclI heteroallele strain.
This work was supported by grants from the American Cancer Society (RPG GMC-99746) and from the NIH (GM61645-01).
REFERENCES
- 1.Baur, J. A., Y. Zou, J. W. Shay, and W. E. Wright. 2001. Telomere position effect in human cells. Science 292:2075-2077. [DOI] [PubMed] [Google Scholar]
- 2.Becker, D. M., J. D. Fikes, and L. Guarente. 1991. A cDNA encoding a human CCAAT-binding protein cloned by functional complementation in yeast. Proc. Natl. Acad. Sci. USA 88:1968-1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bennett, C. B., A. L. Lewis, K. K. Baldwin, and M. A. Resnick. 1993. Lethality induced by a single site-specific double-strand break in a dispensable yeast plasmid. Proc. Natl. Acad. Sci. USA 90:5613-5617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biessmann, H., S. B. Carter, and J. M. Mason. 1990. Chromosome ends in Drosophila without telomeric DNA sequences. Proc. Natl. Acad. Sci. USA 87:1758-1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biessmann, H., J. Donath, and M. F. Walter. 1996. Molecular characterization of the Anopheles gambiae 2L telomeric region via an integrated transgene. Insect Mol. Biol. 5:11-20. [DOI] [PubMed] [Google Scholar]
- 6.Blasco, M. A., H. W. Lee, M. P. Hande, E. Samper, P. M. Lansdorp, R. A. DePinho, and C. W. Greider. 1997. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell 91:25-34. [DOI] [PubMed] [Google Scholar]
- 7.Bodnar, A. G., M. Ouellette, M. Frolkis, S. E. Holt, C. P. Chiu, G. B. Morin, C. B. Harley, J. W. Shay, S. Lichtsteiner, and W. E. Wright. 1998. Extension of life-span by introduction of telomerase into normal human cells. Science 279:349-352. [DOI] [PubMed] [Google Scholar]
- 8.Bosco, G., and J. E. Haber. 1998. Chromosome break-induced DNA replication leads to nonreciprocal translocations and telomere capture. Genetics 150:1037-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bryan, T. M., A. Englezou, L. Dalla-Pozza, M. A. Dunham, and R. R. Reddel. 1997. Evidence for an alternative mechanism for maintaining telomere length in human tumors and tumor-derived cell lines. Nat. Med. 3:1271-1274. [DOI] [PubMed] [Google Scholar]
- 10.Bryan, T. M., A. Englezou, J. Gupta, S. Bacchetti, and R. R. Reddel. 1995. Telomere elongation in immortal human cells without detectable telomerase activity. EMBO J 14:4240-4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bucholc, M., Y. Park, and A. J. Lustig. 2001. Intrachromatid excision of telomeric DNA as a mechanism for telomere size control in Saccharomyces cerevisiae. Mol. Cell. Biol. 21:6559-6573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen, Q., A. Ijpma, and C. W. Greider. 2001. Two survivor pathways that allow growth in the absence of telomerase are generated by distinct telomere recombination events. Mol. Cell. Biol. 21:1819-1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cohn, M., and J. E. Edstrom. 1992. Telomere-associated repeats in Chironomus form discrete subfamilies generated by gene conversion. J. Mol. Evol 35:114-122. [DOI] [PubMed] [Google Scholar]
- 14.de Lange, T. 1998. Length control of human telomeres. Cancer J. Sci. Am. 4(Suppl. 1):S22-S25. [PubMed] [Google Scholar]
- 15.Dunham, M. A., A. A. Neumann, C. L. Fasching, and R. R. Reddel. 2000. Telomere maintenance by recombination in human cells. Nat. Genet. 26:447-450. [DOI] [PubMed] [Google Scholar]
- 16.Fire, A., and S. Q. Xu. 1995. Rolling replication of short DNA circles. Proc. Natl. Acad. Sci. USA 92:4641-4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gottschling, D. E., O. M. Aparicio, B. L. Billington, and V. A. Zakian. 1990. Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell 63:751-762. [DOI] [PubMed] [Google Scholar]
- 18.Greider, C. W., and E. H. Blackburn. 1989. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature 337:331-337. [DOI] [PubMed] [Google Scholar]
- 19.Harley, C. B., N. W. Kim, K. R. Prowse, S. L. Weinrich, K. S. Hirsch, M. D. West, S. Bacchetti, H. W. Hirte, C. M. Counter, C. W. Greider, et al. 1994. Telomerase, cell immortality, and cancer. Cold Spring Harb. Symp. Quant. Biol. 59:307-315. [DOI] [PubMed] [Google Scholar]
- 20.Hollingsworth, N. M., and B. Byers. 1989. HOP1: a yeast meiotic pairing gene. Genetics 121:445-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Horowitz, H., and J. E. Haber. 1985. Identification of autonomously replicating circular subtelomeric Y′ elements in Saccharomyces cerevisiae. Mol. Cell. Biol. 5:2369-2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim, N. W., M. A. Piatyszek, K. R. Prowse, C. B. Harley, M. D. West, P. L. Ho, G. M. Coviello, W. E. Wright, S. L. Weinrich, and J. W. Shay. 1994. Specific association of human telomerase activity with immortal cells and cancer. Science 266:2011-2015. [DOI] [PubMed] [Google Scholar]
- 23.Le, S., J. K. Moore, J. E. Haber, and C. W. Greider. 1999. RAD50 and RAD51 define two pathways that collaborate to maintain telomeres in the absence of telomerase. Genetics 152:143-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Levis, R. W., R. Ganesan, K. Houtchens, L. A. Tolar, and F. M. Sheen. 1993. Transposons in place of telomeric repeats at a Drosophila telomere. Cell 75:1083-1093. [DOI] [PubMed] [Google Scholar]
- 25.Lundblad, V., and E. H. Blackburn. 1993. An alternative pathway for yeast telomere maintenance rescues est1- senescence. Cell 73:347-360. [DOI] [PubMed] [Google Scholar]
- 26.Lundblad, V., and J. W. Szostak. 1989. A mutant with a defect in telomere elongation leads to senescence in yeast. Cell 57:633-643. [DOI] [PubMed] [Google Scholar]
- 27.Lustig, A. J. 1999. Crisis intervention: the role of telomerase. Proc. Natl. Acad. Sci. USA 96:3339-3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malkova, A., E. L. Ivanov, and J. E. Haber. 1996. Double-strand break repair in the absence of RAD51 in yeast: a possible role for break-induced DNA replication. Proc. Natl. Acad. Sci. USA 93:7131-7136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McClintock, B. 1942. The fusion of broken ends of chromosomes following nuclear fusion. Proc. Natl. Acad. Sci. USA 28:458-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McEachern, M. J., D. H. Underwood, and E. H. Blackburn. 2002. Dynamics of telomeric DNA turnover in yeast. Genetics 160:63-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McEachern, M. J., and E. H. Blackburn. 1996. Cap-prevented recombination between terminal telomeric repeat arrays (telomere CPR) maintains telomeres in Kluyveromyces lactis lacking telomerase. Genes Dev. 10:1822-1834. [DOI] [PubMed] [Google Scholar]
- 32.McEachern, M. J., and E. H. Blackburn. 1995. Runaway telomere elongation caused by telomerase RNA gene mutations. Nature 376:403-409. [DOI] [PubMed] [Google Scholar]
- 33.McEachern, M. J., and S. Iyer. 2001. Short telomeres in yeast are highly recombinogenic. Mol. Cell 7:695-704. [DOI] [PubMed] [Google Scholar]
- 34.McEachern, M. J., S. Iyer, T. B. Fulton, and E. H. Blackburn. 2000. Telomere fusions caused by mutating the terminal region of telomeric DNA. Proc. Natl. Acad. Sci. USA 97:11409-11414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McEachern, M. J., A. Krauskopf, and E. H. Blackburn. 2000. Telomeres and their control. Annu. Rev. Genet. 34:331-358. [DOI] [PubMed] [Google Scholar]
- 36.Morin, G. B. 1989. The human telomere terminal transferase enzyme is a ribonucleoprotein that synthesizes TTAGGG repeats. Cell 59:521-529. [DOI] [PubMed] [Google Scholar]
- 37.Murnane, J. P., L. Sabatier, B. A. Marder, and W. F. Morgan. 1994. Telomere dynamics in an immortal human cell line. EMBO J. 13:4953-4962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Naito, T., A. Matsuura, and F. Ishikawa. 1998. Circular chromosome formation in a fission yeast mutant defective in two ATM homologues. Nat. Genet. 20:203-206. [DOI] [PubMed] [Google Scholar]
- 39.Nakamura, T. M., J. P. Cooper, and T. R. Cech. 1998. Two modes of survival of fission yeast without telomerase. Science 282:493-496. [DOI] [PubMed] [Google Scholar]
- 40.Niida, H., Y. Shinkai, M. P. Hande, T. Matsumoto, S. Takehara, M. Tachibana, M. Oshimura, P. M. Lansdorp, and Y. Furuichi. 2000. Telomere maintenance in telomerase-deficient mouse embryonic stem cells: characterization of an amplified telomeric DNA. Mol. Cell. Biol. 20:4115-4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nimmo, E. R., A. L. Pidoux, P. E. Perry, and R. C. Allshire. 1998. Defective meiosis in telomere-silencing mutants of Schizosaccharomyces pombe. Nature 392:825-828. [DOI] [PubMed] [Google Scholar]
- 42.Olovnikov, A. M. 1973. A theory of marginotomy. J. Theor. Biol. 41:181-190. [DOI] [PubMed] [Google Scholar]
- 43.Pich, U., and I. Schubert. 1998. Terminal heterochromatin and alternative telomeric sequences in Allium cepa. Chromosome Res. 6:315-321. [DOI] [PubMed] [Google Scholar]
- 44.Reddel, R. R., T. M. Bryan, L. M. Colgin, K. T. Perrem, and T. R. Yeager. 2001. Alternative lengthening of telomeres in human cells. Radiat. Res. 155:194-200. [DOI] [PubMed] [Google Scholar]
- 45.Regev, A., S. Cohen, E. Cohen, I. Bar-Am, and S. Lavi. 1998. Telomeric repeats on small polydisperse circular DNA (spcDNA) and genomic instability. Oncogene 17:3455-3461. [DOI] [PubMed] [Google Scholar]
- 46.Runge, K. W., and V. A. Zakian. 1989. Introduction of extra telomeric DNA sequences into Saccharomyces cerevisiae results in telomere elongation. Mol. Cell. Biol. 9:1488-1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saiga, H., and J. E. Edstrom. 1985. Long tandem arrays of complex repeat units in Chironomus telomeres. EMBO J. 4:799-804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sandell, L. L., and V. A. Zakian. 1993. Loss of a yeast telomere: arrest, recovery, and chromosome loss. Cell 75:729-739. [DOI] [PubMed] [Google Scholar]
- 49.Shay, J. W., and S. Bacchetti. 1997. A survey of telomerase activity in human cancer. Eur. J. Cancer 33:787-791. [DOI] [PubMed] [Google Scholar]
- 50.Singer, M. S., and D. E. Gottschling. 1994. TLC1: template RNA component of Saccharomyces cerevisiae telomerase. Science 266:404-409. [DOI] [PubMed] [Google Scholar]
- 51.Teng, S. C., and V. A. Zakian. 1999. Telomere-telomere recombination is an efficient bypass pathway for telomere maintenance in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:8083-8093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tomaska, L., J. Nosek, A. M. Makhov, A. Pastorakova, and J. D. Griffith. 2000. Extragenomic double-stranded DNA circles in yeast with linear mitochondrial genomes: potential involvement in telomere maintenance. Nucleic Acids Res. 28:4479-4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Traverse, K. L., and M. L. Pardue. 1988. A spontaneously opened ring chromosome of Drosophila melanogaster has acquired He-T DNA sequences at both new telomeres. Proc. Natl. Acad. Sci. USA 85:8116-8120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tzfati, Y., T. B. Fulton, J. Roy, and E. H. Blackburn. 2000. Template boundary in a yeast telomerase specified by RNA structure. Science 288:863-867. [DOI] [PubMed] [Google Scholar]
- 55.Underwood, D. H., and M. J. McEachern. 2001. Totally mutant telomeres: single-step mutagenesis of tandem repeat DNA sequences. BioTechniques 30:934-935, 938. [DOI] [PubMed] [Google Scholar]
- 56.Wray, L. V., M. M. Witte, R. C. Dickson, and M. I. Riley. 1987. Characterization of a positive regulatory gene, LAC9, that controls induction of the lactose-galactose regulon of Kluyveromyces lactis: structural and functional relationships to GAL4 of Saccharomyces cerevisiae. Mol. Cell. Biol. 7:1111-1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zakian, V. A. 1996. Structure, function, and replication of Saccharomyces cerevisiae telomeres. Annu. Rev. Genet. 30:141-172. [DOI] [PubMed] [Google Scholar]