Abstract
Up-regulation of the c-jun gene is a critical event in the retinoic acid (RA)-mediated differentiation of embryonal carcinoma F9 cells. Activating transcription factor 2 (ATF-2) and p300 cooperate in the activation of transcription of the c-jun gene during the differentiation of F9 cells. We show here that the overexpression of Jun dimerization protein 2 (JDP2), a repressor of AP-1, inhibits the transactivation of the c-jun gene by ATF-2 and p300 by recruitment of the histone deacetylase 3 (HDAC3) complex, thereby repressing the RA-induced transcription of the c-jun gene and inhibiting the RA-mediated differentiation of F9 cells. Moreover, chromatin immunoprecipitation assays showed that the JDP2/HDAC3 complex, which binds to the differentiation response element within the c-jun promoter in undifferentiated F9 cells, was replaced by the p300 complex in response to RA, with an accompanying change in the histone acetylation status of the chromatin, the initiation of transcription of the c-jun gene, and the subsequent differentiation of F9 cells. These results suggest that JDP2 may be a key factor that controls the commitment of F9 cells to differentiation and shed new light on the mechanism by which an AP-1 repressor functions.
Murine F9 cells, a line of embryonal carcinoma cells derived from a teratocarcinoma (4), can be induced to differentiate to endoderm-like cells by exposure to retinoic acid (RA) (36). The RA-induced differentiation results in dramatic changes in gene expression, which include a rapid increase in the rate of transcription of the c-jun gene (9, 46). Moreover, constitutive expression of the c-jun gene results in the differentiation of various lines of embryonal carcinoma cells, such as F9 and P19 cells (10, 44, 46), suggesting that the induction of transcription of c-jun by RA may play an important role in the events that result in differentiation. It was reported previously that a sequence element in the c-jun promoter, the differentiation response element (DRE), is necessary and sufficient for the RA-induced expression of the c-jun gene (26). This element binds the differentiation regulatory factor (DRF) complex, one component of which is the adenovirus E1A-associated protein p300 (11, 28), which has histone acetyltransferase activity (3, 33, 45); another component is activating transcription factor 2 (ATF-2), which is a DNA-binding subunit of the DRF complex (23, 24). ATF-2 and p300 interact with each other in the DRF complex and cooperate in the control of transcription in response to differentiation-inducing signals, such as RA or E1A. However, it is likely that other factors in addition to ATF-2 and p300 are present in the DRF complex (23). In a previous attempt to identify additional proteins in the DRF complex during the differentiation of F9 cells, Jin et al. isolated Jun dimerization protein 2 (JDP2), a repressor of AP-1, as a candidate protein by using a yeast-two hybrid screening system with ATF-2 as the bait (20).
JDP2 was initially isolated on the basis of its ability to interact specifically with the AP-1 transcription factor c-Jun (2). JDP2 is a relatively small (163 amino acids), ubiquitously expressed bZIP protein that can form stable heterodimers with c-Jun, JunB, or JunD, and it acts as a repressor of c-Jun and the c-Jun/c-Fos heterodimer (2). JDP2 also interacts with ATF-2 both in vitro and in vivo via its bZIP domain and binds to the cyclic AMP response element to repress the cyclic AMP response element-dependent transcription that is mediated by ATF-2 (20). Thus, JDP2 may be a general repressor of transcription that is related to the Jun/Fos/ATF-2 family. The AP-1 transcription factor is a collection of dimers composed of Jun proteins (c-Jun, JunB, and JunD), Fos proteins (c-Fos, FosB, Fra1, and Fra2), or ATF proteins (ATF-1, ATF-2, ATF-2a, and ATF-3) (22); it is involved in a variety of transcriptional responses that are associated with the progression of the cell cycle, cell differentiation, apoptosis, and tumorigenesis (1, 5, 22, 35, 39, 47). It seems likely that JDP2 is involved in all such processes as an AP-1 repressor and, indeed, an inhibitory role for JDP2 in the regulation of UV-induced apoptosis via suppression of the expression of p53 was reported recently (34). However, details of the physiological role of JDP2 remain unknown, and the mechanisms by which JDP2 acts also remain to be clarified.
We demonstrate here that JDP2 is an inhibitory subunit of the DRF complex that can repress the RA-induced transcription of the c-jun gene by recruitment of the histone deacetylase 3 (HDAC3) complex to the DRE, thereby inhibiting the RA-mediated differentiation of F9 cells. We also provide evidence suggesting that JDP2 may be a key factor in the commitment of F9 cells to RA-dependent differentiation.
MATERIALS AND METHODS
Plasmids and antibodies.
pGEX-JDP2 was generated by ligating a PCR-amplified fragment of DNA that included the coding sequence (amino acids 1 to 163) of JDP2 into pGEX-4T-1 (Amersham Pharmacia Biotech, Uppsala, Sweden) in frame. pcDNA-JDP2 was prepared by subcloning the JDP2 coding region, which had been amplified by PCR, into pcDNA4/HisMax (Invitrogen BV, Groningen, The Netherlands). pGEX-ATF-2 and pECE-ATF-2 were gifts from M. Green. pcDNA-Flag-HDAC3 was provided by W.-M. Yang. pGEX-HDAC3 was constructed by inserting the HDAC3-encoding fragment from pcDNA-Flag-HDAC3 into the EcoRI site of pGEX-4T-1. 3× DRE/tk-CAT, −730/+874 c-jun-CAT, pACT-p300, pRSV-LacZ, and pBluescript-ATF-2 were prepared as described elsewhere (20, 28). 3× mDRE/tk-CAT was generated by site-directed mutagenesis, which changed the DRE site into the mutant DRE (mDRE) site (5′-TTACCTTTTCCGAAAGCCTT-3′, where underlining shows mutated nucleotides); three tandem repeats of this mDRE were inserted into the HindIII site of tk-CAT. pcDNA4/LacZ, a control plasmid that encoded β-galactosidase, was purchased from Invitrogen BV.
Monoclonal antibodies against JDP2 (immunoglobulin G1 [IgG1]) were prepared by standard procedures (18). Supernatants from cultures of hybridoma cells were screened by Western blotting for their ability to recognize purified glutathione S-transferase (GST)-JDP2 specifically and not GST alone. Rabbit antibodies against ATF-2 (N-96), mSin3 (AK-11), and HDAC3 (H-99) and goat antibodies against HDAC3 (N-19), HDAC4 (L-19), HDAC5 (P-16), HDAC6 (C-16), and β-actin were purchased from Santa Cruz Biotechnology Inc. (Santa Cruz, Calif.). Rabbit antibodies against HDAC1 (H-51), HDAC2 (H-54), acetyl-histone H4, and sodium butyrate were purchased from Upstate Biotechnology Co. (Lake Placid, N.Y.). Preparation of the p300-specific polyclonal antibodies was described elsewhere (28). Trichostatin A (TSA) was obtained from Wako Chemical Co. (Osaka, Japan).
EMSAs.
Details of the preparation of nuclear extracts, translation of proteins in vitro, and electrophoretic mobility shift assays (EMSAs) were described elsewhere (20). The DNA probe was obtained by annealing 5′-AGCTAGCATTACCTCATCCCGTGAGCCTT-3′ and 5′-GATCAAGGCTCACGGGATGAGGTAATGCT-3′. Competition experiments and supershift assays were performed by prior incubation of proteins with a nonradiolabeled double-stranded oligodeoxynucleotide competitor or with appropriate antibodies for 20 min.
Cell cultures, transfection, and assays of CAT activity.
F9 cells that had been stably transfected with the −730/+874 c-jun-CAT reporter (26) were transfected with 1 μg of pcDNA-JDP2 or pcDNA4/LacZ empty vector by using Lipofectamine 2000 reagent (Gibco-BRL, Rockville, Md.) or FuGENE6 transfection reagent (Roche Diagnostic Co., Tokyo, Japan) according to the protocol of the respective manufacturer. After selection of cells with 100 μg of Zeocin (Invitrogen BV)/ml for more than 3 weeks, we examined the expression of JDP2 and β-galactosidase by immunoblotting analysis with antibodies against JDP2 and by measurment of β-galactosidase activity, respectively. Stably transfected cells were analyzed for chloramphenical acetyltransferase (CAT) activity by Northern blotting and by a reverse transcription (RT)-PCR assay. For studies of transient expression, cells were transfected with plasmid DNA and harvested 24 h after transfection, and then CAT activity was measured by using extracts in amounts that had been normalized by reference to β-galactosidase activity as described elsewhere (28).
Immunoprecipitation, Western blotting analysis, and Northern blotting analysis.
Immunoprecipitation and Western blotting were performed as described elsewhere (28). Total RNA was prepared with TRIzol reagent (Gibco-BRL), and Northern blotting was performed according to the manufacturer's protocol.
Assays of HDAC activity.
Proteins in cell extracts were immunoprecipitated with appropriate antibodies, and immunoprecipitates were subjected to assays of HDAC activity with an HDAC assay kit (Upstate Biotechnology Co.) in accordance with the instructions of the manufacturer. The preparation of GST fusion proteins was described elsewhere (20).
GST pull-down assays.
The transcription and translation of pBluescript-ATF-2 and pcDNA-Flag-HDAC3 in vitro and the purification of GST and of GST fusion proteins were performed as described elsewhere (20). For protein-binding assays, 4 μg of GST or GST-protein was incubated with 35S-labeled ATF-2 or HDAC3 (5 × 106 cpm) in a final volume of 500 μl of binding buffer (50 mM Tris [pH 7.5], 5 mM MgCl2, 100 mM NaCl, 10% glycerol, 0.5 mg of bovine serum albumin per ml, 5 mM β-mercaptoethanol) at 4°C for 1 h, and then 30 μl of a suspension of glutathione-Sepharose beads (Amersham Pharmacia Biotech) was added. After incubation for 2 h at 4°C, samples were extensively washed three times with phosphate-buffered saline, and protein complexes were eluted with a buffer (50 mM Tris [pH 7.5], 5 mM MgCl2, 100 mM NaCl, 10% glycerol, 5 mM β-mercaptoethanol) that contained 10 mM glutathione for analysis by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) (10% acrylamide). Gels were dried prior to exposure to X-ray film.
Immunoprecipitation of chromatin (ChIP).
F9 cells were fixed in 1% formaldehyde at room temeprature for 5 min. Soluble chromatin was prepared as described elsewhere (6) and subjected to immunoprecipitation in lysis buffer (50 mM Tris [pH 8.0], 140 mM NaCl, 1% Triton X-100, 0.1% SDS, 1 mM EDTA, standard protease inhibitors) with 1 to 2 μg of antibody at 4°C for 5 h. Immunocomplexes were then recovered by the addition of protein A or G plus agarose beads (Sigma-Aldrich, St. Louis, Mo.) and incubation at 4°C for 2 h. After the beads were washed extensively, DNA was eluted, and cross-linking was reversed by the addition of 200 μl of elution buffer (1% SDS, 0.1 M NaHCO3) and incubation overnight at 65°C. DNA was extracted with phenol and chloroform (1:1, vol/vol), precipitated in ethanol, and then analyzed by PCR with primers that corresponded to the DRE, namely, 5′-AACTGTAGGAGCGCAGCGG-3′ (forward direction) and 5′-ATTCTTCTCTGGGCCCG-3′ (reverse direction).
Real-time RT-PCR.
The relative abundance of mRNAs was examined by a real-time fluorescence detection method (13) with TaqMan chemistry (PE Applied Biosystems, Foster City, Calif.). In brief, total RNA was treated with 1 U of DNase (Promega Co., Madison, Wis.) per 20 μg of RNA, and analysis by RT-PCR was performed with TaqMan one-step RT-PCR Master Mix reagents from PE Applied Biosystems. Twenty nanograms of total RNA in a total volume of 10 μl was used for RT-PCR analysis. Each mixture of 200 nM primers, 100 nM TaqMan probe, 1× Master Mix, 1× MultiScribe and RNase Inhibitor Mix, and RNA template was incubated at 48°C for 30 min and then at 95°C for 10 min. The PCR then was allowed to proceed for 40 cycles of incubation at 95°C for 15 s and at 60°C for 60 s. A standard curve was obtained by amplification of serially diluted samples with known concentrations of RNA from cells that had been stably transfected with the pcDNA4/LacZ empty vector. Real-time reactions and quantification were performed with a sequence detector (ABI 7700; PE Applied Biosystems). Glyceraldehyde-3-phosphate dehydrogenase transcripts were also quantified as an internal control. The following oligodeoxynucleotides were used for real-time RT-PCR: (i) laminin B1 primers, 5′-CTGCCCCAGTATACGGCATC-3′ (forward direction) and 5′-AGGGCATGAGAACAAGCGAGT-3′ (reverse direction); laminin B1 probe, 5′-TGACGTGGAGAGCCCTTACACGTTCA-3′; (ii) collagen 4α1 primers, 5′-GGCGGTACACAGTCAGACCAT-3′ (forward direction) and 5′-GGAATAGCCGATCCACAGTGA-3′ (reverse direction); collagen 4α1 probe, 5′-TTCCGCAGTGCCCTAACGGTTGGT-3′; and (iii) Hoxa-1 primers, 5′-CCCCTCTGACCATGGGATTAC-3′ (forward direction) and 5′-CCGCCGCAGCTGTTG-3′(reverse direction); Hoxa-1 probe, 5′-CTTTCCAATCCTGCGCGGTCAGTG-3′.
RESULTS
JDP2 is an inhibitory subunit of the DRF complex.
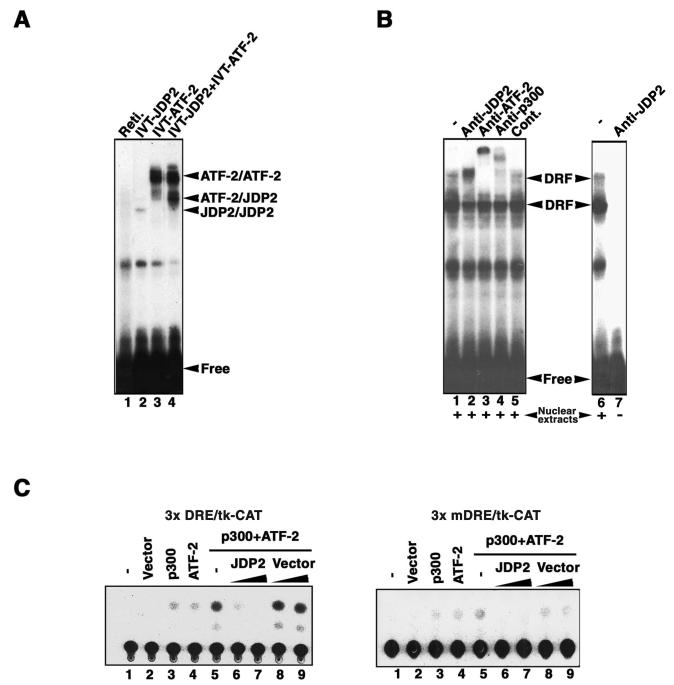
We performed gel EMSAs to determine whether JDP2 binds to DRE and is included in the DRF complex. As shown in Fig. 1A, homodimers of ATF-2 and of JDP2 that had been translated in vitro each bound to the DRE probe (lanes 2 and 3), while proteins in the reticulocyte lysate failed to bind to the DRE probe by themselves (lane 1). JDP2 also formed a heterodimer with ATF-2 in the presence of the DRE probe; the molecular mass of the heterodimer was different from those of homodimers of ATF-2 and of JDP2 (Fig. 1A, lane 4). To determine whether JDP2 was directly included in the DRF complex, we also performed EMSAs with DRE as the probe and antibodies against JDP2. As shown in Fig. 1B, the retarded bands that corresponded to DRF were shifted still further upon the addition of antibodies against JDP2 (lane 2). Antibodies specific for p300 and for ATF-2 also caused supershifting of the DRF complex, as reported previously (Fig. 1B, lanes 3 and 4) (23, 28). However, control antibodies had no effect on the mobility of the DRF complex (Fig. 1B, lane 5). In addition, antibodies against JDP2 alone failed to bind to the DRE probe (Fig. 1B, lane 7). These results suggested that JDP2 bound to DRE as a heterodimer with ATF-2 and that it was included in the DRF complex.
FIG. 1.
JDP2 is an inhibitory subunit of the DRF complex. (A) JDP2 binds to DRE as a heterodimer with ATF-2. An EMSA was performed with JDP2 and ATF-2 that had been translated in vitro (IVT) with 32P-labeled DRE as the DNA probe. Reti., reticulocyte lysate. The positions of the ATF-2/ATF-2 homodimer, the ATF-2/JDP2 heterodimer, and the JDP2/JDP2 homodimer are indicated. Free, free DNA probe. (B) JDP2 is a component of the DRF complex. Nuclear extracts from F9 cells were incubated without (lane 1) or with antibodies against (Anti-) JDP-2, ATF-2, and p300 and control (Cont.) antibodies (lanes 2 to 5, respectively). The mixtures were analyzed by an EMSA with DRE as the DNA probe. Results obtained for the DRE probe with nuclear extract (lane 6) and with JDP2-specific antibodies alone (lane 7) are also shown. The positions of DRF are indicated. (C) JDP2 represses DRE-dependent transactivation by ATF-2 and p300. F9 cells were cotransfected with 3× DRE/tk-CAT (0.2 μg; left panel) or with 3× mDRE/tk-CAT (0.2 μg; right panel) plus 0.5 μg of pRSV-LacZ, together with various combinations of empty vector (vector), pECE-ATF-2, and pACT-p300 (2.0 μg each) or increasing amounts of pcDNA-JDP2 (lanes 6 and 7) and empty vector (lanes 8 and 9) (0.5 and 1.0 μg, respectively). At 24 h after transfection, cells were collected and CAT activity was determined. The data represent the results from one of at least two independent experiments that gave similar results.
To investigate the role of JDP2 in the regulation of DRE-mediated transactivation, we cotransfected F9 cells with various combinations of a DRE/tk-CAT reporter construct and plasmids that encoded JDP2, ATF-2, and p300. Then we monitored DRE-mediated reporter activity. As anticipated, only cotransfection of plasmids that encoded ATF-2 and p300 significantly activated the transcription of the DRE/tk-CAT reporter gene (Fig. 1C, left panel, lanes 1 to 5). However, the expression of JDP2 but not of the empty vector suppressed the ability of ATF-2 and p300 to activate the DRE/tk-CAT reporter gene in a dose-dependent manner (Fig. 1C, left panel, lanes 6 to 9). In a control experiment with the mDRE/tk-CAT construct, in which 4 nucleotides within DRE had been mutated, we detected only very low CAT activity in every instance (Fig. 1C, right panel). These results demonstrated that JDP2 specifically repressed the DRE-dependent transcription of the reporter construct that was mediated by ATF-2 and p300. Considering all the results together, we concluded that JDP2 is an inhibitory component of the DRF complex.
JDP2 suppresses the RA-dependent transcription of the c-jun gene.
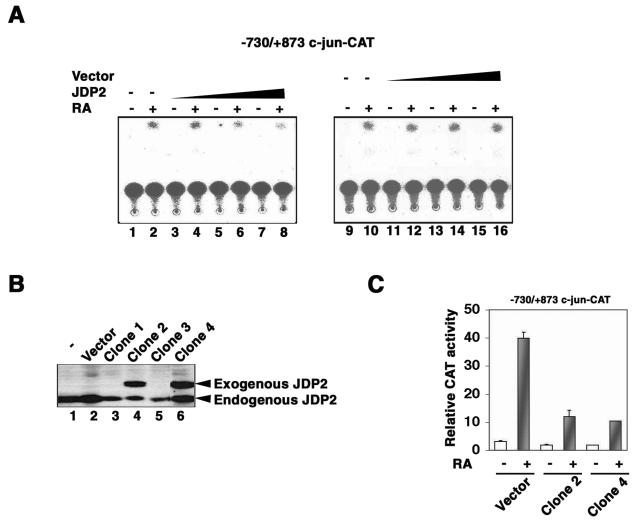
Since DRE is both necessary and sufficient for the RA-induced expression of the c-jun gene (26), the repression by JDP2 of the DRE-dependent transcription induced by ATF-2 and p300 implies a role for JDP2 in the regulation of the RA-mediated transcription of the c-jun gene. We examined the effect of the transient expression of JDP2 on the RA-induced activation of the c-jun promoter. F9 cells which had been stably transfected with the −730/+874 c-jun-CAT reporter construct were transfected with either pcDNA-JDP2 or empty vector pcDNA4/LacZ; 24 h later, they were treated with RA. Assays of CAT activity were performed after the cells were incubated with RA at 10−6 M for 72 h. As shown in Fig. 2A, transient expression of JDP2 but not of the empty vector repressed the CAT activity that was mediated by the c-jun promoter, and the effect of JDP2 was dose dependent. Next, we established F9-derived cell lines that stably coexpressed either JDP2 or the empty vector and the −730/+874 c-jun-CAT reporter construct. Figure 2B shows that two positive clones (clones 2 and 4) constitutively expressed JDP2. The viability of these cells was apparently unaffected by the enhanced expression of JDP2 (data not shown). As shown in Fig. 2C, the two positive clones (clones 2 and 4) and the control cell line that had been stably transfected with empty vector (vector) had very limited CAT activity in the absence of stimulation by RA. After treatment with RA at 10−6 M for 60 h, however, the extent of the enhancement of CAT activity in clones 2 and 4 was much lower (5- to 6-fold) than that (up to 13-fold) in the control cell line (vector). These results indicated that the expression of JDP2 repressed the RA-mediated activation of the c-jun promoter.
FIG. 2.
Inhibition of RA-induced activation of the c-jun promoter by JDP2. (A) Transient expression of JDP2 inhibits RA-mediated activation of a c-jun promoter construct, −730/+874 c-jun-CAT. F9 cells that had been stably transfected with −730/+874 c-jun-CAT were transfected with increasing amounts of pcDNA-JDP2 (JDP2) or the pcDNA4/LacZ empty vector (vector) (3.0, 6.0, or 9.0 μg each) plus pRSV-LacZ (1.0 μg). At 24 h after transfection, the cells were cultured without or with 1 μM RA for 72 h and assayed for CAT activity. The data represent the results from one of three independent experiments that gave similar results. (B) Stably transfected F9 cells harboring −730/+874 c-jun-CAT, together with pcDNA-JDP2 or pcDNA4/LacZ, were established as described in Materials and Methods. The expression of JDP2 was examined by immunoblotting analysis with antibodies against JDP2. Endogenous JDP2 and the exogenously expressed JDP2 fusion protein (clones 2 and 4) are indicated by arrowheads. (C) Stable expression of JDP2 inhibits RA-mediated activation of the c-jun promoter. pcDNA4/LacZ transfectants (vector) and clones 2 and 4 were incubated without or with 1 μM RA for 60 h. Samples were assayed for CAT activity as described in Materials and Methods. The values shown are the averages and standard deviations of the results from three independent experiments.
The rate of transcription of the c-jun gene increases dramatically during the RA-mediated differentiation of F9 cells. Our observation that the ectopic expression of JDP2 inhibited the activity of the c-jun promoter implied that JDP2 may act as a repressor of the RA-mediated transcription of the c-jun gene. To examine this possibility, we treated the above-mentioned cells with RA for different times and then monitored the level of transcription of the c-jun gene by Northern blotting. As shown in Fig. 3, the expression of JDP2 (in clones 2 and 4) delayed and decreased the extent of the RA-induced transcription of the c-jun gene compared to that in cells transfected with the empty vector alone. These results suggested that JDP2 inhibited the RA-induced expression of the c-jun gene that was mediated by DRE within the c-jun promoter.
FIG. 3.
Repression of RA-mediated transcription of the c-jun gene by JDP2. (A) Levels of RA-induced transcription of the c-jun gene. F9 cells that had been stably transfected with pcDNA-JDP2 (clones 2 and 4) or pcDNA4/LacZ (vector) were incubated with 1 μM RA for the indicated times. The levels of c-jun and β-actin transcripts were determined by Northern blotting. (B) The relative amounts of c-jun transcripts from panel A were quantified with a phosphorimager. Amounts were normalized by reference to the values for β-actin mRNA.
JDP2 functions in an HDAC-dependent manner.
HDACs act as repressors in the regulation of the expression of many genes (14, 15, 31). In an attempt to determine whether an HDAC may also be involved in the mechanism of repression by JDP2, we cotransfected F9 cells with a DRE/tk-CAT reporter construct and a variety of plasmids. Then we monitored DRE-mediated reporter activity in the presence and absence of TSA, an inhibitor of HDACs. As anticipated, the addition of pcDNA-JDP2 suppressed the promoter activity of the DRE/tk-CAT reporter gene that was mediated by either pACT-p300 or pECE-ATF-2 or the combination of those two plasmids in the absence of TSA (Fig. 4A, lanes 2, 4, and 6). This suppression of the transcription of the DRE/tk-CAT reporter gene by JDP2 was, however, clearly reversed upon incubation of the cells with 100 ng of TSA/ml (Fig. 4A, lanes 7 to 12). These data suggested that HDAC activity might be required for the transcriptional repression induced by JDP2.
FIG. 4.
JDP2 recruits an HDAC. (A) Treatment with TSA reverses repression by JDP2. F9 cells were transfected with 0.2 μg of 3× DRE/tk-CAT plus 0.5 μg of pRSV-LacZ in the presence or absence of pECE-ATF-2 or pACT-p300 (2.0 μg each) and pcDNA-JDP2 (1.0 μg). At 8 h after transfection, the cells were incubated without or with TSA (100 ng/ml) for 16 h and assayed for CAT activity; the results were normalized by reference to β-galactosidase activity (28). The results shown are from one of three independent experiments that gave similar results. (B) Recruitment by JDP2 of an HDAC. Proteins in extracts of F9 cells were immunoprecipitated with antibodies against JDP2 and assayed for HDAC activity in the presence or absence of sodium butyrate, an inhibitor of HDAC. Immunoprecipitates obtained with antibodies specific for mSin3 and mouse IgG were used as controls. GST and GST-fused JDP2 were also examined for HDAC activity. The values shown are the averages and standard deviations of the results from three independent experiments.
The association of JDP2 with HDAC activity implied that a protein complex that included JDP2 might have HDAC activity. We confirmed this possibility by measuring the HDAC activity of protein complexes that were recruited by JDP2. The HDAC activity of the JDP2-specific immunoprecipitate was as high as that of the mSin3-specific complex and more than five times higher than that of the control IgG complex (Fig. 4B). Moreover, treatment of immunoprecipitates that included JDP2 with sodium butyrate, an inhibitor of HDACs, abolished the HDAC activity (Fig. 4B). To exclude the possibility that JDP2 itself might have HDAC activity, we produced GST-fused JDP2 in Escherichia coli and examined its HDAC activity. As shown in Fig. 4B, neither GST alone nor GST-JDP2 had any such activity. Thus, our results indicated that JDP2 might function as a general repressor through the recruitment of an HDAC.
JDP2 is physically and functionally associated with HDAC3.
To identify the HDAC that is associated with the repressor activity of JDP2, we performed EMSAs by incubation of nuclear extracts from F9 cells and the DRE probe with a variety of antibodies specific for different types of HDACs. As shown in Fig. 5A, the formation of a DNA-protein complex (DRF) generated two major bands, and both bands disappeared only when antibodies specific for HDAC3 but not for HDAC1, HDAC2, HDAC4, HDAC5, and HDAC6 were included. Thus, it appeared that HDAC3 was part of the DRF complex that became associated with JDP2. We next performed immunoprecipitations and Western blotting analysis with nuclear extracts of F9 cells to determine whether JDP2 interacts with HDAC3 in vivo. Western blotting analysis revealed the presence of HDAC3 but not of HDAC1 or HDAC2 in immunoprecipitates of extracts prepared with antibodies specific for JDP2 (Fig. 5B). In a complementary experiment, JDP2 was coprecipitated with HDAC3 from nuclear extracts by antibodies against HDAC3 (Fig. 5B, lowest panel). These results suggested that JDP2 might interact specifically with HDAC3 in vivo. A very strong signal due to HDAC3 was detected in immunoprecipitates obtained with the JDP2-specific antibodies (Fig. 5B, uppermost panel), while relatively smaller amounts of JDP2 were pulled down by the HDAC3-specific antibodies (lowest panel). This difference might have been due to the nature of the antibodies used in the immunoprecipitation assay; the JDP2-specific antibodies were monoclonal, but the HDAC3-specific antibodies were polyclonal. Thus, the specifity and/or the affinity of the JDP2-specific antibodies might have been greater than those of the HDAC3-specific antibodies. Moreover, since the band of HDAC3 migrated in a manner very similar to that of the band of the heavy (H) chain of IgG, some of the signal due to the heavy chain might have overlapped the signal due to HDAC3. We also cannot altogether exclude the possibility that some other unidentified bZIP factor(s), in addition to JDP2 in the DRF complex, might interact with HDAC3.
FIG. 5.
JDP2 associates with HDAC3. (A) HDAC3 is included in the DRF complex. Nuclear extracts from F9 cells were incubated without (lane 1) or with (lanes 2 to 7) antibodies against HDAC1 through HDAC6 and then analyzed by an EMSA with a DRE probe. The bands of DNA-protein complexes (DRF) are indicated. Free, free DNA probe. (B) JDP2 specifically interacts with HDAC3 in vivo. Proteins in extracts of F9 cells were immunoprecipitated with antibodies against (Anti-) JDP2 (uppermost and middle two panels) or HDAC3 (lowest panel). Then, Western blotting analysis (WB) was performed with antibodies against (Anti-) HDAC3, HDAC1, HDAC2, and JDP2, respectively. In the uppermost panel, the positions of HDAC3, the heavy chain of IgG, a nonspecific band (NS), and the light chain of IgG are indicated by arrows. In input lanes, an aliquot of cell lysate (approximately 10%) was used directly for Western blotting. Mouse IgG was used as the negative control. The molecular masses of marker proteins are shown on the left. (C) Interaction between JDP2 and HDAC3 in vitro. (Left panel) Purified GST and GST fusion proteins (about 3.0 μg each) were analyzed by SDS-PAGE (10% polyacrylamide) and then stained with Coomassie brilliant blue. (Right panel) 35S-labeled HDAC3, obtained by translation in vitro (IVT-HDAC3), or ATF-2 (IVT-ATF-2) was separately incubated with GST and various GST fusion proteins. A suspension of glutathione-Sepharose beads was added. After extensive washing, bound proteins were eluted, resolved by SDS-PAGE (10% polyacrylamide), and autoradiographed. Input lanes contained 5% the radiolabeled protein used in binding reactions. (D) HDAC3 enhances the inhibitory activity of JDP2. F9 cells were cotransfected with 0.2 μg of 3× DRE/tk-CAT plus 0.5 μg of pRSV-LacZ, together with various combinations of pECE-ATF-2 or pACT-p300 (1.0 μg each) and pcDNA-JDP2 (0.1 μg) and increasing amounts of pcDNA-Flag-HDAC3 (0.25 μg in lane 4, 0.5 μg in lane 5, and 1.0 μg in lanes 6 and 7). At 8 h after transfection, the cells were incubated without (lanes 1 to 6) or with (lane 7) TSA for 16 h and assayed for CAT activity. The values shown are the averages and standard deviations of the results from three independent experiments.
To clarify whether the interaction between JDP2 and HDAC3 is direct or indirect, we prepared various GST fusion proteins (Fig. 5C, left panel) and used them in GST pull-down assays (Fig. 5C, right panel). We incubated GST-JDP2 with 35S-labeled HDAC3 and immobilized the complex on glutathione-Sepharose beads. After extensive washing, the bound proteins were fractionated by SDS-PAGE. HDAC3 did not bind to GST-JDP2 directly in vitro (Fig. 5C, right panel, lane 3). The converse was also true (data not shown). Moreover, HDAC3 also failed to interact directly with ATF-2 in vitro (Fig. 5C, right panel, lanes 4 and 8). In contrast, in a control experiment, we detected the direct association of 35S-labeled ATF-2 with GST-JDP2 (Fig. 5C, right panel, lane 7) but not with GST alone (lane 6). Taken together, our results suggested that JDP2 might indirectly recruit HDAC3 to DRE via some factor(s) in the DRF complex other than ATF-2.
To determine the contribution of HDAC3 to the JDP2-induced repression of transcription, we cotransfected cells with the DRE/tk-CAT reporter construct and a variety of combinations of plasmids that encoded p300, ATF-2, JDP2, and HDAC3 or the empty vector. The combination of ATF-2 and p300 enhanced the transcription of the DRE/tk-CAT reporter gene (Fig. 5D, lane 2), and JDP2 suppressed the resultant reporter activity significantly (lane 3). Furthermore, the coexpression of HDAC3 with JDP2 increased the repressive activity of JDP2 in a dose-dependent manner as the level of HDAC3 was increased (Fig. 5D, lanes 4 to 6). The activity of the DRE/tk-CAT reporter gene was restored upon the addition of TSA (Fig. 5D, lane 7). Thus, it appeared that JDP2 was functionally associated with HDAC3.
JDP2 inhibits the RA-induced differentiation of F9 cells.
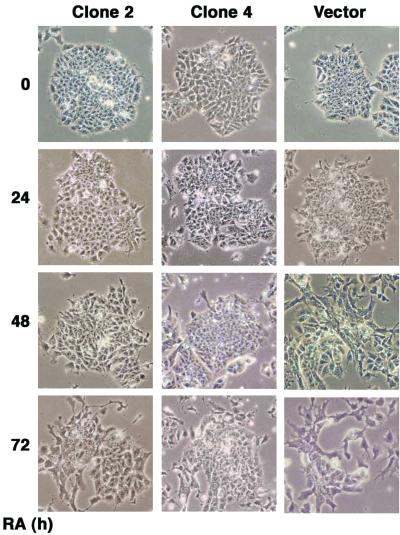
To investigate the effects of JDP2 on RA-mediated differentiation, we generated stable lines of F9 cells that expressed JDP2 or the empty vector alone as described above. The cells were treated for different times with 1 μM RA to induce differentiation, and morphological changes were monitored. Compared with morphological changes in cells that had been stably transfected with the empty vector, such changes in JDP2-expressing cells (clones 2 and 4) were significantly delayed or absent (Fig. 6).
FIG. 6.
Overexpression of JDP2 suppresses RA-mediated morphological changes in F9 cells that are associated with differentiation. F9 cells that had been stably transfected with pcDNA4/LacZ (vector) or pcDNA-JDP2 (clones 2 and 4) were incubated without or with 1 μM RA. The cells were then photographed at the indicated times after exposure to RA under a phase-contrast microscope (magnification, ×125).
We next compared the levels of expression of a number of RA-inducible genes and of genes whose expression is a marker of differentiation after the exposure of cells to RA (1 μM); we measured the levels of expression by real-time RT-PCR (Fig. 7A and B). The levels of the induced expression of genes for collagen type 4α1 and laminin B1, two markers of endodermal differentiation (36, 40), were fivefold lower in JDP2-expressing cells than in control cells after 72 h of treatment with RA (Fig. 7C and D). Similarly, the induction of another RA-responsive gene, the gene for Hoxa-1 (29), was 50% lower in JDP2-expressing cells than in control cells (Fig. 7E). These results indicated that the ectopic expression of JDP2 inhibited the RA-induced differentiation of F9 cells.
FIG. 7.
Stable expression of JDP2 inhibits RA-induced expression of marker genes for differentiation. (A and B) Generation of a calibration curve. For quantification of levels of mRNA by real-time RT-PCR, a calibration curve was generated by analysis of serially diluted reference samples. Amplification curves were generated first by plotting the normalized reporter signal (ΔRn) against the cycle number to illustrate the changes in the level of the product of RT-PCR with increasing number of cycles. Then, the standard curve was generated by plotting the threshold cycle (Ct) against the starting amount of sample with PRISM 7700 SDS software (PE Applied Biosystems). The linearity of the standard curve confirmed that the starting quantity was inversely proportional to its Ct. (C to E) Quantification of levels of transcripts of RA-inducible genes and of genes that are markers of cell differentiation. The relative levels of expression of genes for laminin B1 (C), collagen 4α1 (D), and Hoxa-1 (E) in F9 cells that had been stably transfected with pcDNA4/LacZ (empty vector) or pcDNA-JDP2 (JDP2), without or with treatment with RA, were obtained by determining Ct values from the standard curve. All experiments were performed in triplicate (results are means and standard deviations), and negative controls without template RNA were always included in each experiment. Furthermore, the results were normalized by reference to the values for glyceraldehyde-3-phosphate dehydrogenase mRNA.
Changes in the levels of JDP2 during the commitment of F9 cells to RA-dependent differentiation.
We next addressed the exact role of endogenous JDP2 in the regulation of the RA signaling pathway. One possibility that we considered was that JDP2 might be associated with the inhibition of the expression of the c-jun gene in undifferentiated F9 cells via the recruitment of HDAC3 to the promoter region and might thus maintain cells in an undifferentiated state. To investigate this hypothesis, we examined the expression of JDP2 and its DNA-binding and protein-protein interactions during the initiation of differentiation. We also monitored the DNA-binding activity of other components of the DRF complex, as well as the acetylation status of histones associated with DRE.
We first measured the levels of expression of JDP2 mRNA and JDP2 itself by Northern blotting and Western blotting analyses, respectively. Unexpectedly, the levels of both the mRNA and the protein appeared not to change in F9 cells that had been treated with 1 μM RA for 24 h, corresponding to the time during which cells commit to differentiation in response to RA (27) (data not shown). We next examined the DNA-binding activity of JDP2 in vivo by using a ChIP assay. We used antibodies specific for JDP2 to immunoprecipitate formadehyde-cross-linked, sonicated chromatin from F9 cells that had been incubated with or without RA. We examined the presence of DRE in immunoprecipitates of JDP2 by PCR. As shown in Fig. 8A, treatment of F9 cells with RA for 24 h resulted in a marked decrease in the extent of binding of JDP2 to DRE. We obtained a similar result with antibodies specific for HDAC3 (Fig. 8A). However, the DNA-binding activities of ATF-2 and p300, other components of the DRF complex, increased significantly after the incubation of cells with RA for 24 h. We next examined the interaction of JDP2 with HDAC3 in vivo in F9 cells that had been incubated with or without RA. Cell lysates from F9 cells, after incubation with or without RA, were immunoprecipitated with antibodies against JDP2 or control mouse IgG. HDAC3 was coimmunoprecipitated with antibodies specific for JDP2 but not with control IgG (Fig. 8B). However, the extent of binding of JDP2 to HDAC3 was significantly reduced after the treatment of cells with RA for 24 h (Fig. 8B). Similarly, the binding of JDP2 to ATF-2 diminished after such treatment with RA (Fig. 8B).
FIG. 8.
Levels of each component of DRF and of the acetylation of histone H4 during the commitment to differentiation. (A) Alterations in binding to DNA of components of DRF complexes. Soluble chromatin was prepared from F9 cells that had been incubated without (0 h) or with (24 h) RA and then was immunoprecipitated (IP) with various antibodies (Anti-). The same extracts were incubated with mouse IgG to determine the nonspecific precipitation of chromatin. Precipitated genomic DNA was analyzed by PCR with primers that corresponded to the sequence that spanned DRE. The uppermost panel shows the level of DRE that was amplified prior to immunoprecipitation, confirming that equivalent amounts of DNA were present in the samples. (B) Interaction of JDP2 with HDAC3 or ATF-2. Proteins in extracts of F9 cells that had been incubated without (0 h) or with (24 h) RA were immunoprecipitated with antibodies against JDP2. Immunoprecipitated proteins were subjected to Western blotting analysis with antibodies specific for ATF-2, HDAC3, and β-actin. In input lanes, an aliquot of cell lysate (approximately 10%) was used directly for Western blotting. Mouse IgG was used as a negative control. (C) Immunoprecipitation of chromatin with antibodies against acetylated histone H4. The ChIP assay was performed as described for panel A, except that the antibodies were specific for acetylated histone H4 (Anti-AcH4). Input DNA obtained from an aliquot of soluble chromatin (20%) was purified in parallel with the immunoprecipitated samples.
The results described above suggested that alterations in histone acetylation status might be involved in the JDP2-dependent control of the transcription of the c-jun gene. To examine this possibility, we immunoprecipitated isolated chromatin with antibodies specific for acetylated histone H4. Subsequent changes in the products of PCR should reflect changes in acetylated histone H4 bound to the DRE sequence. As shown in Fig. 8C, no products of PCR associated with acetylated histone H4 were detected in untreated F9 cells. However, incubation of the cells with RA resulted in a remarkable increase in the amount of acetylated histone H4 associated with DRE at 24 h. These data strongly supported the hypothesis that JDP2 is associated with the maintenance of the hypoacetylation of histones associated with the c-jun promoter through the recruitment of HDAC3 to DRE in undifferentiated F9 cells, thereby inhibiting the transcription of the c-jun gene. When F9 cells are treated with RA, the JDP2/HDAC3 complex may be replaced by p300, with the resultant hyperacetylation of the histones associated with the c-jun promoter and, as a consequence, the rapid initiation of transcription of the c-jun gene and triggering of the commitment of F9 cells to RA-induced differentiation.
DISCUSSION
Possible role of JDP2 in the RA-dependent regulation of the transcription of the c-jun gene.
The c-Jun protein, a major component of the AP-1 transcription factor, is expressed at a very low level in undifferentiated F9 cells. Levels of endogenous c-jun mRNA begin to rise dramatically within 18 to 24 h of the start of exposure to RA and reach a steady state after 60 to 72 h (26). The mechanisms of transactivation of the c-jun gene in stimulated F9 cells are relatively well understood, but it is unclear how the expression of the gene can be inhibited in nonstimulated F9 cells but can respond rapidly to RA. In this study, we identified JDP2 as an inhibitory DNA-binding subunit of DRF on the basis of the following observations. First, JDP2 bound directly to DRE as a homodimer or as a heterodimer with ATF-2 (Fig. 1A). Second, in EMSAs, bands that corresponded to DRF were shifted still further upon the addition of antibodies against JDP2 (Fig. 1B). Third, JDP2 interacted directly with ATF-2 in vivo and in vitro, as well as in a yeast two-hybrid system (20). Finally, the overexpression of JDP2 not only repressed DRE-mediated transactivation by ATF-2 and p300 but also inhibited the RA-induced activation of the promoter of c-jun and the transcription of the c-jun gene (Fig. 1C, 2, and 3). It is possible that JDP2, acting as a repressive component of DRF, may have properties consistent with a role in inhibiting the expression of c-jun in nonstimulated F9 cells and in the rapid response to stimulation by RA during the commitment of F9 cells to differentiation. This hypothesis is also supported by the observation that, while the level of expression of JDP2 was apparently unchanged, the binding of JDP2 to DRE and the association of JDP2 with HDAC3 and with ATF-2 decreased markedly 24 h after the start of treatment of F9 cells with RA (Fig. 8A and B). In contrast, the binding to DRE of p300 and ATF-2 was significantly enhanced after treatment of cells with RA for 24 h (Fig. 8B). Furthermore, the replacement of the ATF-2/JDP2/HDAC3 combination by the ATF-2/p300 combination in the DRF complex was accompanied by a change in the acetylation status of histones associated with DRE, from hypo- to hyperacetylation, during the commitment of F9 cells to differentiation in response to RA (Fig. 8C). TSA alone can induce the differentiation of F9 cells (data not shown), indicating that the deacetylation status of histones is closely associated with the maintenance of the undifferentiated state of F9 cells. The levels of recruitment of the ATF-2/JDP2/HDAC3 complex and the ATF-2/p300 complex may be critical for the determination of the choice between the undifferentiated state and the commitment of cells to differentiation in response to RA. It is possible, moreover, that JDP2 is a key factor in the regulation of the transcription of the c-jun gene in F9 cells in response to RA.
JDP2 blocks the RA-induced differentiation of F9 cells.
The c-Jun protein plays an important role in the differentiation of F9 cells. It is expressed in many organs during murine development, as well as in the adult mouse (1, 43). Mouse embryos with null mutations in the c-jun gene die at midgestation, a phenomenon that suggests an essential function for the product of this gene in mouse development (19, 21). In F9 cells, RA and E1A stimulate the activity of the c-jun promoter and rapidly increase the level of expression of c-jun (25, 46). Moreover, constitutive expression of c-jun results in the differentiation of F9 cells, suggesting a significant role for c-jun in the regulation of cell differentiation (44, 46). The observed inhibition of transcription of the c-jun gene by JDP2 led us to investigate the role of JDP2 in the RA-induced differentiation of F9 cells. As we expected, the overexpression of JDP2 repressed RA-mediated differentiation, with both a delay in and a decrease in the extent of morphological changes associated with differentiation (Fig. 6), as well as repression of the transcription of RA-inducible genes and genes for markers of differentiation (Fig. 7C to E). The inhibitory effect of JDP2 on differentiation may involve the regulation of the transcription of c-jun, in view of the key role of c-jun in differentiation and the activity of JDP2 as a negative regulator of the expression of this gene. It is also possible that JDP2 inhibits differentiation not only by suppression of the transcription of c-jun but also by direct repression of the expression of other RA-inducible genes and/or genes for markers of differentiation.
The expression of c-jun is also involved in RA-induced apoptosis and the arrest of cell growth. Thus, JDP2, as a repressor of c-Jun, may also be associated with these phenomena, in addition to its role in the regulation of differentiation. These issues remain to be investigated. Confirmation of a role for JDP2 in the regulation of the differentiation of F9 cells would provide novel insight both into RA signaling and into mechanisms of differentiation.
Molecular mechanism of the repression of the expression of c-jun by JDP2.
The mechanism of transcriptional repression by JDP2 remains to be fully clarified. Aronheim et al. (2) proposed that JDP2 may repress the transcriptional activity of c-jun by forming a stable c-Jun/JDP2 heterodimer and that JDP2 may compete with activators for binding to c-Jun. Alternatively, JDP2 may enhance the binding of c-Jun to the promoter, thereby providing further inhibition via competition with active heterodimers for binding to the same elements. However, another mechanism seems plausible, as in the case of the repression by JDP2 of ATF-2-mediated transactivation. In this study, we demonstrated that the repression of the transcription of the c-jun gene by JDP2 depended on an HDAC with, most probably, recruitment of HDAC3 to DRE of the c-jun promoter. Our conclusion is supported by the observation that the suppression by JDP2 of DRE-mediated transactivation by ATF-2 and p300 was reversed when cells were treated with TSA, an HDAC-specific inhibitor (Fig. 4A), and by the observation that immunoprecipitates obtained with antibodies specific for JDP2 had HDAC activity (Fig. 4B). In addition, support for the association of JDP2 with HDAC3 was provided by the demonstration of their specific interaction in vivo (Fig. 5B) and by their coexistence in a DRF complex, as determined by an EMSA (Fig. 5A) and the ChIP assay (Fig. 8B). Furthermore, augmentation of the repressive activity of JDP2 by the enhanced expression of HDAC3 also suggested a functional relationship between JDP2 and HDAC3 (Fig. 5D). Unexpectedly, we failed to detect any direct interaction between JDP2 and HDAC3 (Fig. 5C) (data not shown). Our results suggest that JDP2 may recruit HDAC3 indirectly via some other unidentified component(s) of DRF. We have, in fact, identified some other components of DRF by an EMSA with a DRE probe, and these components include corepressors, subunits of chromatin-remodeling factors, and other proteins (data not shown). It is possible that JDP2 recruits HDAC3 to DRE through binding to some of these factors in DRF.
Involvement of HDAC3 in cell differentiation.
The activities of histone deacetylases have been linked to transcription, progression of the cell cycle, gene silencing, cell differentiation, regulation of the neuronal phenotype, DNA replication, and the response to DNA damage (14, 32). Several proteins that associate with HDAC3 were recently identified, including some corepressors, such as N-CoR, SMRT, and KAP-1; a component of the SWI/SNF complex; receptor-interacting protein 140; and transduction beta-like protein 1 (16, 17, 30, 37, 38, 41, 42). However, the biological functions of HDAC3, including its role in cell differentiation, remain to be characterized. In the present study, we found that HDAC3 associated with JDP2 to form a novel repressor complex and that JDP2 suppressed the RA-induced transcription of c-jun through the recruitment of HDAC3 to the promoter region. We also obtained evidence suggesting that the association of JDP2 with HDAC3 may be involved in the maintenance of the hypoacetylation status of histones and may be a key phenomenon in undifferentiated embryonal carcinoma cells. Our results suggest a potential role for HDAC3 in the regulation of cell differentiation. Among the class I HDACs, HDAC1 and HDAC2 are more closely related to each other than to HDAC3 (8, 12), suggesting a possible unique role of HDAC3. Indeed, we detected only HDAC3 and not HDAC1 or HDAC2 in the DRF complex (Fig. 5A), and we also found that HDAC3 and not HDAC1 or HDAC2 specifically associated with JDP2 in vivo (Fig. 5B). The observation that HDAC1 or HDAC2 and HDAC3 exist in different N-CoR complexes (37) and the observation that some mutations in retinoblastoma protein (Rb) inhibit the binding of Rb to HDAC1 and HDAC2 but have no effect on the binding of Rb to HDAC3 (7), in conjunction with our present results, support the hypothesis that it is HDAC3 and not HDAC1 or HDAC2 that is likely to be important in the regulation of the commitment of F9 cells to differentiation in response to RA.
Acknowledgments
We thank K. Itakura, G. Gachelin, M. Green, W.-M. Yang, and R. Eckner for reagents and for many helpful discussions about the manuscript.
This work was supported by grants from the BioResource Research Project of RIKEN, the Uehara Memorial Foundation, and the Ministry of Education, Culture, Sports, Science and Technology of Japan (to K.K.Y.).
REFERENCES
- 1.Angel, P., and M. Karin. 1991. The role of Jun, Fos and AP-1 complex in cell proliferation and transformation. Biochim. Biophys. Acta 1072:129-159. [DOI] [PubMed] [Google Scholar]
- 2.Aronheim, A., E. Zandi, H. Hennemann, S. J. Elledge, and M. Karin. 1997. Isolation of an AP-1 repressor by a novel method for detecting protein-protein interactions. Mol. Cell. Biol. 17:3094-3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bannister, A. J., and T. Kouzarides. 1996. The CBP co-activator is a histone acetyltransferase. Nature 384:641-643. [DOI] [PubMed] [Google Scholar]
- 4.Bersteine, E. G., M. L. Hooper, S. Grandchamp, and B. Ephrussi. 1973. Alkaline phosphatase activity in the differentiation of mouse teratoma. Proc. Natl. Acad. Sci. USA 70:3899-3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bossy-Wetzel, E., and M. Yaniv. 1997. Induction of apoptosis by the transcription factor c-Jun. EMBO J. 16:1695-1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braunstein, M., A. B. Rose, S. G. Holmes, C. D. Allis, and J. R. Broach. 1993. Transcriptional silencing in yeast is associated with reduced nucleosome acetylation. Genes Dev. 7:592-604. [DOI] [PubMed] [Google Scholar]
- 7.Dahiya, A., M. R. Gavin, R. X. Luo, and D. C. Dean. 2000. Role of the LXCXE binding site in Rb function. Mol. Cell. Biol. 20:6799-6805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dangond, F., D. A. Hafler, J. K. Tong, J. Randall, N. Utku, and S. R. Gullans. 1998. Differential display cloning of a novel human histone deacetylase (HDAC3) cDNA from PHA-activated immune cells. Biochem. Biophys. Res. Commun. 26:648-652. [DOI] [PubMed] [Google Scholar]
- 9.de Groot, R. P., J. Schoorlemmer, S. T. van Genesen, and W. Kruijer. 1990. Differential expression of jun and fos genes during differentiation of mouse P19 embryonal carcinoma cells. Nucleic Acids Res. 18:3195-3202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Groot, R. P., F. A. E. Kruyt, S. T. van Genesen, and W. Kruijer. 1990. Ectopic expression of c-jun leads to differentiation of P19 embryonal carcinoma cells. EMBO J. 9:1831-1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eckner, R., T.-S. Yao, E. Oldread, and D. M. Livingston. 1994. Molecular cloning and functional analysis of the adenovirus E1A-associated p300-kDa protein (p300) reveal a protein with properties of a transcriptional adapter. Genes Dev. 8:869-884. [DOI] [PubMed] [Google Scholar]
- 12.Emiliani, S., W. Fischle, C. van Lint, Y. Al-Abed, and E. Verdin. 1998. Characterization of a human RPD3 ortholog, HDAC3. Proc. Natl. Acad. Sci. USA 95:2795-2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gibson, U. E., C. A. Heid, and P. M. Williams. 1996. A novel method for real-time quantitative RT-PCR. Genome Res. 6:995-1001. [DOI] [PubMed] [Google Scholar]
- 14.Gray, S. G., and T. J. Ekström. 2001. The human histone deacetylase family. Exp. Cell Res. 262:75-83. [DOI] [PubMed] [Google Scholar]
- 15.Grunstein, M. 1997. Histone acetylation in chromatin structure and transcription. Nature 389:349-353. [DOI] [PubMed] [Google Scholar]
- 16.Guenther, M. G., W. S. Lane, W. Fischle, E. Verdin, M. A. Lazar, and R. Shiekhattar. 2000. A core SMRT corepressor complex containing HDAC3 and TBL1, a WD40-repeat protein linked to deafness. Genes Dev. 14:1048-1057. [PMC free article] [PubMed] [Google Scholar]
- 17.Guenther, M. G., O. Barak, and M. A. Lazar. 2001. The SMRT and N-CoR corepressors are activating cofactors for histone deacetylase 3. Mol. Cell. Biol. 21:6091-6101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harlow, E., and D. Lane. 1988. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 19.Hilberg, F., A. Aguzzi, N. Howells, and E. F. Wagner. 1993. c-Jun is essential for normal mouse development and hepatogenesis. Nature 365:179-181. [DOI] [PubMed] [Google Scholar]
- 20.Jin, C., H. Ugai, J. Song, T. Murata, F. Nili, K. Sun, M. Horikoshi, and K. K. Yokoyama. 2001. Identification of mouse Jun dimerization protein 2 as a novel repressor of ATF-2. FEBS Lett. 489:34-41. [DOI] [PubMed] [Google Scholar]
- 21.Johnson, R. S., B. van Lingen, V. E. Papaioannou, and B. M. Spiegelman. 1993. A null mutation of the c-jun locus causes embryonic lethality and retarded cell growth in culture. Genes Dev. 7:1309-1317. [DOI] [PubMed] [Google Scholar]
- 22.Karin, M., Z. G. Liu, and E. Zandi. 1997. AP-1 function and regulation. Curr. Opin. Cell Biol. 9:240-246. [DOI] [PubMed] [Google Scholar]
- 23.Kawasaki, H., J. Song, R. Eckner, H. Ugai, R. Chiu, K. Taira, Y. Shi, N. Jones, and K. K. Yokoyama. 1998. p300 and ATF-2 are components of the DRF complex, which regulates retinoic acid- and E1A-mediated transcription of the c-jun gene in F9 cells. Genes Dev. 12:233-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kawasaki, H., R. Eckner, T.-P. Yao, K. Taira, R. Chiu, D. M. Livingston, and K. Yokoyama. 1998. Distinct roles of the co-activators p300 and CBP in retinoic acid-induced F9 cell differentiation. Nature 393:284-289. [DOI] [PubMed] [Google Scholar]
- 25.Kitabayashi, I., R. Chiu, G. Gachelin, and K. Yokoyama. 1991. E1A-dependent up-regulation of c-jun/AP-1 activity. Nucleic Acids Res. 19:649-655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kitabayashi, I., Z. Kawakami, R. Chiu, K. Ozawa, T. Matsuoka, S. Toyoshima, K. Umesono, R. M. Evans, G. Gachelin, and K. Yokoyama. 1992. Transcriptional regulation of the c-jun gene by retinoic acid and E1A during differentiation of F9 cells. EMBO J. 11:167-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kitabayashi, I., R. Chiu, K. Umezono, G. Gachelin, R. M. Evans, and K. K. Yokoyama. 1994. A novel pathway for RA-induced differentiation of F9 cells that is independent of receptor-mediated transactivation. In Vitro Cell. Dev. Biol. 30A:761-768. [DOI] [PubMed] [Google Scholar]
- 28.Kitabayashi, I., R. Eckner, Z. Arany, R. Chiu, G. Gachelin, D. M. Livingston, and K. K. Yokoyama. 1995. Phosphorylation of the adenovirus E1A-associated 300-kDa protein in response to retinoic acid and E1A during the differentiation of F9 cells. EMBO J. 14:3496-3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Langston, A. W., and L. J. Gudas. 1992. Identification of a retinoic acid-responsive enhancer 3′ of the murine homeobox gene Hox-1.6. Mech. Dev. 38:217-228. [DOI] [PubMed] [Google Scholar]
- 30.Li, J., J. Wang, Z. Nawaz, J. M. Liu, J. Qin, and J. Wang. 2000. Both corepressor proteins SMRT and N-CoR exist in large protein complexes containing HDAC3. EMBO J. 19:4342-4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nagy, L., H. Y. Kao, D. Chakravarti, R. J. Lin, C. A. Hassig, D. E. Ayer, S. L. Schreiber, and R. M. Evans. 1997. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell 89:373-380. [DOI] [PubMed] [Google Scholar]
- 32.Ng, H. H., and A. Bird. 2001. Histone deacetylases silences for hire. Trends Biochem. Sci. 25:121-126. [DOI] [PubMed] [Google Scholar]
- 33.Ogryzko, V. W., R. L. Schiltz, V. Russanova, R. H. Howard, and Y. Nakatani. 1996. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell 87:953-959. [DOI] [PubMed] [Google Scholar]
- 34.Piu, F., A. Aronheim, S. Katz, and M. Karin. 2001. AP-1 repressor protein JDP-2: inhibition of UV-mediated apoptosis through p53 down-regulation. Mol. Cell. Biol. 21:3012-3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schreiber, M., A. Kolbus, F. Piu, A. Szabowski, U. Möhle-Steinlein, J. Tian, M. Karin, P. Angel, and E. F. Wagner. 1999. Control of cell cycle progression by c-Jun is p53-dependent. Genes Dev. 13:607-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Strickland, S., and V. Mahdavi. 1978. The induction of differentiation in teratocarcinoma stem cells by retinoic acid. Cell 15:393-403. [DOI] [PubMed] [Google Scholar]
- 37.Underhill, C., M. S. Qutob, S. P. Yee, and J. Torchia. 2000. A novel nuclear receptor corepressor complex, N-CoR, contains components of the mammalian SWI/SNF complex and the corepressor KAP-1. J. Biol. Chem. 275:40463-40470. [DOI] [PubMed] [Google Scholar]
- 38.Urnov, F. D., J. Yee, L. Sachs, T. N. Collingwood, A. Bauer, H. Beug, Y. B. Shi, and A. P. Wolffe. 2000. Targeting of N-CoR and histone deacetylase 3 by the oncoprotein v-erbA yields a chromatin infrastructure-dependent transcriptional repression pathway. EMBO J. 19:4074-4090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vogt, T. P., and J. Bos. 1990. Jun: oncogene and transcriptional factor. Adv. Cancer Res. 55:1-35. [DOI] [PubMed] [Google Scholar]
- 40.Wang, S.-Y., and L. J. Gudas. 1983. Isolation of cDNA clones specific for collagen IV and laminin from mouse teratocarcinoma cells. Proc. Natl. Acad. Sci. USA 80:5880-5884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wei, L. N., X. Hu, D. Chandra, E. Seto, and M. Farooqui. 2000. Receptor-interacting protein 140 directly recruits histone deacetylases for gene silencing. J. Biol. Chem. 275:40782-40787. [DOI] [PubMed] [Google Scholar]
- 42.Wen, Y. D., V. Perissi, L. M. Staszewski, W. M. Yang, A. Krones, C. K. Glass, M. G. Rosenfeld, and E. Seto. 2000. The histone deacetylase-3 complex contains nuclear receptor corepressors. Proc. Natl. Acad. Sci. USA 97:7202-7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wilkinson, D. G., S. Bhatt, R. P. Ryseck, and R. Bravo. 1989. Tissue-specific expression of c-jun and JunB during organogenesis in the mouse. Development 106:465-471. [DOI] [PubMed] [Google Scholar]
- 44.Yamaguchi-Iwai, Y., M. Satake, Y. Murakami, M. Sakai, M. Muramatsu, and Y. Ito. 1990. Differentiation of F9 embryonal carcinoma cells induced by the c-jun and activated c-Ha-ras oncogenes. Proc. Natl. Acad. Sci. USA 87:8670-8674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang, X. J., V. V. Ogryzko, J. Nishikawa, B. H. Howard, and Y. Nakatani. 1996. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature 382:319-324. [DOI] [PubMed] [Google Scholar]
- 46.Yang-Yen, H. F., R. Chiu, and M. Karin. 1990. Elevation of AP-1 activity during F9 cell differentiation is due to increased c-jun transcription. Nat. New Biol. 2:351-361. [PubMed] [Google Scholar]
- 47.Yong, M. R., J.-J. Li, M. Rincon, R. A. Flavell, B. K. Sashyanarayana, R. Hunziker, and N. Colburn. 1999. Transgenic mice demonstrate AP-1 (activator protein-1) transactivation is required for tumor promotion. Proc. Natl. Acad. Sci. USA 96:9827-9832. [DOI] [PMC free article] [PubMed] [Google Scholar]