Abstract
The yeast Mcm1 protein is a member of the MADS box family of transcriptional regulatory factors, a class of DNA-binding proteins that control numerous cellular and developmental processes in yeast, Drosophila melanogaster, plants, and mammals. Although these proteins bind DNA on their own, they often combine with different cofactors to bind with increased affinity and specificity to their target sites. To understand how this class of proteins functions, we have made a series of alanine substitutions in the MADS box domain of Mcm1 and examined the effects of these mutations in combination with its cofactors that regulate mating in yeast. Our results indicate which residues of Mcm1 are essential for viability and transcriptional regulation with its cofactors in vivo. Most of the mutations in Mcm1 that are lethal affect DNA-binding affinity. Interestingly, the lethality of many of these mutations can be suppressed if the MCM1 gene is expressed from a high-copy-number plasmid. Although many of the alanine substitutions affect the ability of Mcm1 to activate transcription alone or in combination with the α1 and Ste12 cofactors, most mutations have little or no effect on Mcm1-mediated repression in combination with the α2 cofactor. Even nonconservative amino acid substitutions of residues in Mcm1 that directly contact α2 do not significantly affect repression. These results suggest that within the same region of the Mcm1 MADS box domain, there are different requirements for interaction with α2 than for interaction with either α1 or Ste12. Our results suggest how a small domain, the MADS box, interacts with multiple cofactors to achieve specificity in transcriptional regulation and how subtle differences in the sequences of different MADS box proteins can influence the interactions with specific cofactors while not affecting the interactions with common cofactors.
Mcm1 is an essential protein in yeast and a founding member of the MADS box family of transcriptional regulatory factors, a highly conserved group of proteins found in virtually all eukaryotic organisms (38, 40). Although members of this family share conserved DNA-binding and dimerization domains, they regulate a wide range of cellular functions, from basic metabolism to control of the cell cycle and determination of cell type. MADS box proteins bind with high affinity and specificity to their target genes in vitro; however, they often require accessory or cofactor proteins to specifically bind to their target sites in vivo. For example, in plants, MADS box proteins form heterodimers to regulate specific sets of genes required for flower development, and in mammals, the SRF protein interacts with several different cofactors to activate different sets of genes in response to serum stimulation (35, 37, 46). In many cases, the cofactor interactions of MADS box proteins determine which genes are regulated and if these genes are transcriptionally activated or repressed. Therefore, determination of how MADS box proteins interact with different cofactors to achieve target specificity and proper transcriptional regulation is essential for understanding the underlying mechanisms of regulation of many cellular and developmental processes.
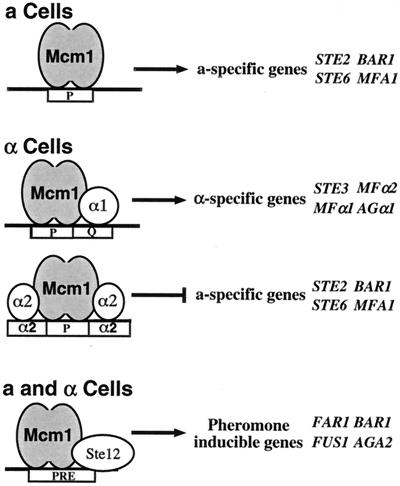
A simple example of a combinatorial network involving a MADS box protein is the transcriptional regulatory system that specifies cell mating type in the yeast Saccharomyces cerevisiae (16). Yeasts have two haploid cell types, a and α, that differ by their cell surface receptors and the pheromones they secrete. Exposure of an a or α cell to the pheromone of the opposite mating type triggers a signal transduction cascade, inducing genes that are required for mating and the formation of the diploid a/α cell type. In a cells, the MADS box protein Mcm1 binds to target sites upstream of the promoters of a-specific genes to activate their transcription (2) (Fig. 1). In α cells, Mcm1 combines with the α1 protein to activate transcription of α-specific genes (4, 20, 33, 43). Mcm1 also combines with the α2 protein in α cells to bind to the pheromone response element (PRE) to repress transcription of a-specific genes (23, 32). In addition to determining the expression of the cell type-specific genes, Mcm1 interacts with the Ste12 protein in haploid a and α cells to activate genes required for mating and cell fusion (10, 12, 13, 19, 31). The interaction of Mcm1 with its different cofactors, therefore, determines the target specificity of Mcm1 and, in the case of α2, also changes it from functioning as a transcriptional activator to functioning as a repressor.
FIG. 1.
Role of Mcm1 in a regulatory network for control of mating type functions. In a cells, Mcm1 activates a-specific gene expression. In α cells, Mcm1 activates α-specific gene expression in complex with α1 and represses a-specific genes in complex with α2. In the presence of a pheromone, Mcm1 combines with Ste12 in both haploid cell types to activate high-level expression of genes involved in cell cycle arrest and mating. Representative target genes in each class are shown.
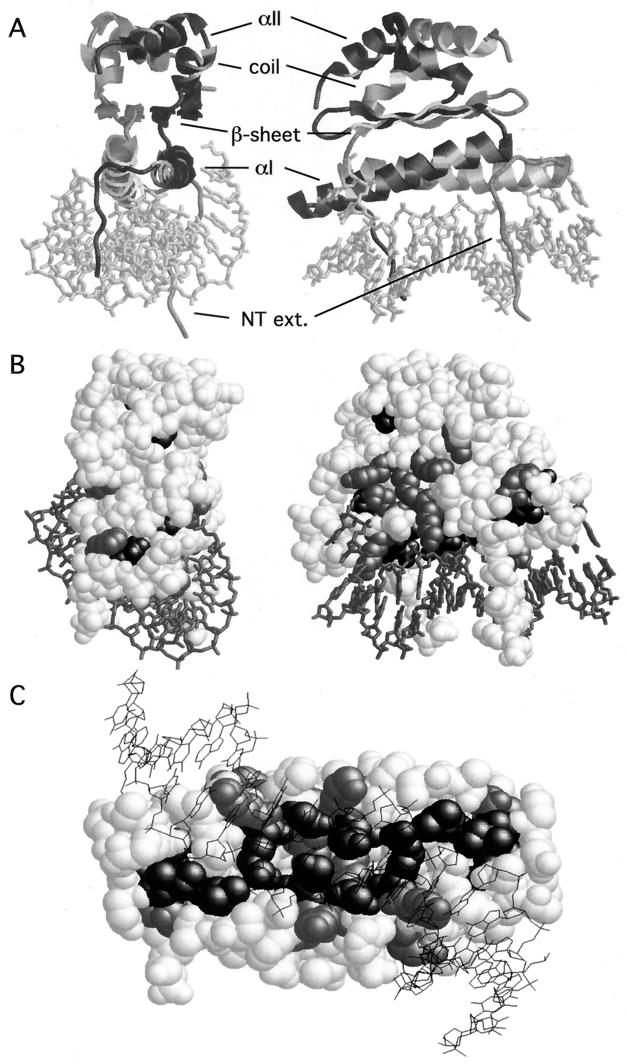
The crystal structure of the ternary complex of Mcm1 bound with α2 to DNA provides an excellent model for understanding how Mcm1 binds DNA and interacts with this cofactor (44). Comparison of the Mcm1 structure with the mammalian SRF and MEF2 proteins in complex with DNA revealed that the protein conformation and many of the DNA contacts are remarkably conserved between the yeast and human proteins (34, 39). The MADS box domain forms a protein dimer composed of three layers (Fig. 2A ). The first layer consists of antiparallel coiled-coil α helices, one from each monomer, positioned above the center of the recognition site. Residues in these helices make numerous phosphate- and base-specific contacts with the major groove. The middle layer consists of a hydrophobic four-stranded antiparallel β sheet, formed from two β strands of each monomer, and is aligned roughly parallel to the coiled-coil α helices. The β loop between the strands in each monomer makes phosphate contacts in the major groove at positions outside of the conserved CArG-box [CC(A/T)6GG] binding site. These contacts contribute to the observed DNA-bending activity of many of the MADS box proteins (1, 34, 44). The top layer of the protein consists of an extended coil region followed by an α helix of three turns. This α helix packs against the corresponding face of the other monomer, adding to the dimerization interface. There is also an N-terminal extension from the αI helix of each monomer that folds back toward the protein and makes several base-specific and phosphate backbone contacts with positions at the center of the recognition site. Deletion studies have shown that an 80-residue fragment of Mcm1 containing the MADS box domain is sufficient to bind DNA and mediate interactions with different cofactors (6, 9, 36, 48). In this study, we investigated how this small domain interacts with many different cofactors and how these interactions may influence the activity of the protein. Through extensive mutagenesis of the Mcm1 MADS box domain, we have identified residues that are required for transcriptional regulation in complex with the α1, α2, and Ste12 cofactors. Interaction of Mcm1 with its cofactors maps to the same region of the MADS box domain, but the requirements within this same interface are very different among the different cofactors. These results show how the same interface can specify interactions with different cofactors.
FIG. 2.
Model of Mcm1 bound to DNA and positions of mutations that produce a lethal phenotype. (A) The views of the Mcm1 dimer binding to its site are derived from the coordinates of the crystal structure of the α2-Mcm1-DNA ternary complex (44). The two views represent a 90° rotation around the vertical axis. A cartoon of the Mcm1 dimer is shown with the DNA as a stick figure at the bottom of the structure. One monomer is in gray, and the other is in black. The positions of the secondary structures that form the different layers of the dimer are shown. NT ext., N-terminal extension. (B and C) Positions of alanine substitutions that fail to complement an mcm1 null mutation. A space-filling model of the Mcm1 dimer is shown in which the relative orientation of the models is roughly the same as that in panel A. Positions at which alanine substitutions cause a lethal phenotype in low copy numbers but result in viability at high copy numbers are gray. Positions at which alanine substitutions cause a lethal phenotype at low and high copy numbers are black. Many of these residues are buried in the interior of the dimer and are not visible from the surface. Panel C shows the lethal alanine substitutions in Mcm1 with a view of the dimer from the bottom of the structure seen through the DNA. Residues in the N-terminal extension were removed from this view to show the lethal residues that contact the DNA.
MATERIALS AND METHODS
Strains.
Cell viability and autonomous transcriptional activation by Mcm1 were measured in strain YY2052 [MATa Π(PAL)-lacZ::FUS1 leu2-3,112 trp1-1 ura3 his3-11,15 ade2-1 can1-100 mcm1::LEU2/pSL1574-CEN/ARS MCM1 URA3], which was provided by G. Sprague (7). α1-Mcm1-mediated activation and α2-Mcm1-mediated repression were assayed in strain JM01 (MATα leu2-3,112 trp1-1 ura3 his3-11,15 ade2-1 can1-100 mcm1::LEU2/pSL1574-CEN/ARS MCM1 URA3), a derivative of YY1888 (6). Transcriptional activation by Mcm1 in combination with Ste12 was assayed in strain JM02 (MATa leu2-3,112 trp1-1 ura3 his3-11,15 ade2-1 can1-100 mcm1::LEU2/pSL1574-CEN/ARS MCM1 URA3). Derivatives of plasmid pJM231 containing site-directed mutations in MCM1 were transformed into the appropriate strain, and loss of plasmid pSL1574, containing the wild-type gene, was selected on medium containing 5-fluoroorotic acid (5-FOA). These strains were then transformed with pJM300 or pJR018, CYC1-lacZ transcription reporter vectors in which the CYC1 upstream activating sequence elements have been replaced with the α1-Mcm1 binding site from the STE3 promoter or the Ste12-Mcm1 binding site from the STE2 promoter. α2-Mcm1-mediated repression was measured in strain JM01 transformed with pJM120, a CYC1-lacZ transcription reporter vector containing a consensus α2-Mcm1 binding site (50). β-Galactosidase assays were performed as described previously (22), and the activation or repression values are averages of three independent transformants for each mutant.
Plasmids.
Individual alanine substitutions at each position in the MADS box domain of Mcm1 were constructed by cloning double-stranded oligonucleotides containing the desired codon changes into pJM231, a derivative of the pRS313 vector that contains an engineered MCM1 gene expressed from its native promoter (1). To express Mcm1 mutant proteins from a high-copy-number plasmid, pJM231 derivatives (CEN HIS3) were digested with SphI to yield a 3.3-kb fragment containing the entire MCM1 gene. The SphI fragments were cloned into pAB1, a modified version of the pRS423 plasmid (2μm HIS3) in which the polylinker region has been removed by digestion with PvuII and replaced with an SphI site. pDA105, a derivative of pJM231 with three copies of the hemagglutinin tag inserted at the C terminus of MCM1, was used for Western analysis of the mcm1-encoded mutant proteins.
The α1-dependent reporter plasmids were constructed by digesting pTBA23, a CYC1-lacZ transcription reporter plasmid, with XhoI and BglII and cloning in double-stranded oligonucleotides containing the α1-Mcm1 sites from the STE3 gene (5′GATCTCTGTCATTGTGACACTAATTAGGAAAC), the AGA1 gene (5′GATCTCTTCCTAATTAGGTCATCAATGACCTC), the MFα2 gene (5′GATCTTTTCCTAATTAGTCCTTCAATAGAACC), and the MFα1 gene (5′GATCTCTTCCTAATTAGGCCATCAACGACAGC). The Ste12-Mcm1 site was taken from the STE2 gene (5′-GATCTACCATGTAAATTTCCTAATTGGGTAAGTACATGATGAAACACATATG). These sites have been used previously to monitor Mcm1-dependent transcription (3, 7). All constructs were verified by sequence analysis. The α2-dependent reporter vectors containing the α2-Mcm1 binding sites from the BAR1, STE2, and ASG7 promoters were described previously (49).
Protein purification.
The mutant Mcm1 proteins used in the in vitro DNA-binding assays were purified from Escherichia coli BL21 cells transformed with pHA227, a protein expression vector in which the sequences coding for residues 1 to 97 of Mcm1 were fused in frame to the gene coding for the maltose-binding protein. The protein was purified from 100-ml cultures of the expression strain as described previously (1). The Mcm1 proteins isolated by this procedure were >95% homogeneous and consisted of two nonnative N-terminal amino acids, Gly1 and Ser2, followed by native Mcm1 residues 1 to 97. The concentrations of the wild-type and mutant proteins were determined and normalized by Bradford assays and verified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Full-length α2 protein containing the six-His fusion was expressed in bacteria transformed with plasmid pJM163 and purified by Ni+2 affinity chromatography (28). The α1 protein was purified from bacteria containing the α1 expression vector pSL2187 as described previously (15).
Electrophoretic mobility shift assays.
The labeled fragment containing the STE6 α2-Mcm1 binding site that was used as a probe in electrophoretic mobility shift assays was generated by isolating the 86-bp fragment from an EcoRI-HindIII restriction digest of pCK1 (22) and filling the 5′ overhangs with 32P by using Klenow polymerase. A labeled fragment containing the α1-Mcm1 binding site from the STE3 promoter was generated by digesting pJM336 with KpnI and EcoRI, filling in the ends with 32P by using Klenow polymerase, and gel purification of the 84-bp fragment. DNA-binding assays were performed as described previously (47). Purified Mcm1 proteins were diluted in 50 mM Tris (pH 7.6)-500 mM NaCl-1 mM EDTA-10 mM 2-mercaptoethanol-1 mg of bovine serum albumin per ml and mixed with constant amounts of α1 or α2 diluted in the same buffer. The relative binding affinity of the mutant Mcm1 proteins was calculated by using best-fit analysis of the probe fraction bound at different protein concentrations and comparing it to that of the wild type.
Biological assays.
Mating pheromone production was monitored by measuring the sizes of the growth inhibition halos that formed around the strains when they were patched onto a dilute lawn of pheromone-sensitive strains RC634 (MATa sst1 ade2 his6 met1 ura1 rme1) and RC757 (MATα sst2-1 met1 his6 can1 cyh2) (8). Mcm1 protein levels were determined by Western blot analysis as described previously (21). Rat monoclonal antibody specific to hemagglutinin (Roche) was used at 1:5,000 as the primary antibody, and goat anti-rabbit antiserum conjugated to horseradish peroxidase (Bio-Rad) was used as the secondary antibody.
RESULTS
Residues in the Mcm1 MADS box domain are essential for viability.
Mcm1 is an essential protein in yeast, and deletion studies have shown that the MADS box domain of the protein is sufficient to support growth and cell type regulation (6, 9, 33). To identify which residues within this domain of Mcm1 are required for viability, we individually replaced nearly every residue in the MADS box domain with an alanine. The mutant plasmids were transformed into yeast strain YY2052, which contains a chromosomal mcm1::LEU2 null mutation and is maintained by plasmid pSL1574 containing wild-type MCM1 and URA3 (7). Cells that had lost the wild-type copy of MCM1 were selected by plating on medium containing the drug 5-FOA (5). Failure of transformants to grow after wild-type MCM1 has been lost indicates that the alanine substitution significantly affects Mcm1 activity and that the mutant is unable, at low copy numbers, to complement the mcm1 null mutation.
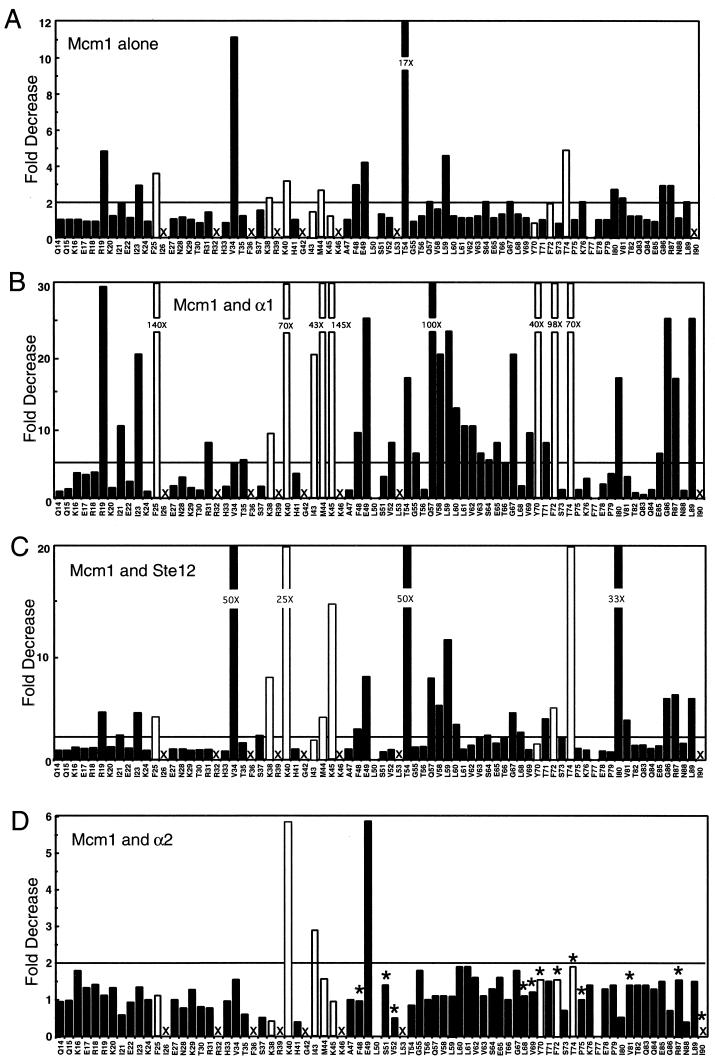
The alanine scanning mutagenesis indicated that 17 of the 75 side chains that were mutated in the Mcm1 MADS box domain are essential for cell growth (Fig. 2B and 3A ). Many of the lethal mutations were in side chains that contact the DNA (K38, R39, K40, G42, K45, and K46), and we have previously shown that these mutations cause significant reductions in the DNA-binding affinity of the protein in vitro (1). Substitutions at other conserved positions in the α helices that are not in contact with the DNA, such as R32, F36, I43, M44, and L53, also failed to complement the mcm1 null mutation and were lethal to the cell. These residues, along with another lethal mutation, I90A, are positioned along the dimer interface and are likely to be critical for stability of the dimer. In the C terminus of the MADS box domain of Mcm1, alanine substitutions at only four positions, Y70, F72, T74, and I90, were lethal. The Y70, F72, and T74 side chains lie adjacent to each other on the outer β strand. Two of these residues, Y70 and F72, have large hydrophobic side chains that extend away from the interior of the protein and are positioned above the α helix (αI) that mediates DNA binding (Fig. 2B). These side chains are partially solvent exposed and therefore could potentially be involved in interactions with essential cofactors. However, Y70, F72, and T74 may be required to stabilize the α helix so it makes proper DNA contacts. In support of this model, we found that these mutant proteins have significantly decreased DNA-binding affinities (data not shown).
FIG. 3.
In vivo transcription assays of mcm1 alanine mutations. Alanine substitutions were made at each position in Mcm1, and the mutants were assayed for expression of lacZ reporter constructs by autonomous activation at a P(PAL) site (A), in complex with α1 at an α1-Mcm1 binding site from the STE3 promoter (B), in complex with Ste12 at a PRE site from the STE2 promoter (C), and for repression in complex with α2 at an α2-Mcm1 binding site from the STE6 promoter (D). Data are expressed as fold decreases with respect to wild-type activity. A value of 1 indicates wild-type activity, whereas large bars indicate significant decreases in activity. The solid bars represent mutations resulting in viability at low copy numbers; open bars represent mutations that cause inviability at low copy numbers but allow viability at high copy numbers. The fold decreases in activity are indicated for mutations that are above the top axis. Values above the horizontal lines were considered significant. Each bar represents the average of three transformants with standard deviations that usually varied by less than 10%. The X's indicate those mutations that are lethal when expressed at low or high copy numbers. Positions L50 and F77 were not tested. The asterisks in panel D denote those residues in Mcm1 that directly contact α2 in the crystal structure of the α2-Mcm1-DNA ternary complex (44).
Lethality of many mcm1 mutations is suppressed by high levels of expression.
After screening the panel of mcm1 alanine mutations for viability, we learned that some of the lethal mutant proteins result in viability if expressed at higher levels (E. Dubois and F. Messenguy, personal communication). We therefore cloned the mutations that cause inviability onto a 2μm plasmid and performed the plasmid shuffle to determine if any of these mutations is rescued by higher levels of expression. Surprisingly, we found that only 8 of the 17 mutations that caused inviability at low copy numbers also failed to complement the null mutation when expressed from a 2μm plasmid (Fig. 2B and 3). Several of the mutations that caused inviability at high copy numbers are at residues in contact with the DNA (Fig. 2C). As predicted from the analysis of the crystal structure, substitution of alanine for residue G42 causes a lethal phenotype, likely because of steric interference with the DNA (44). The two other residues, R39 and K46, are conserved among all of the MADS box proteins and make nearly identical contacts in the MADS box cocrystal structures (34, 39, 44). Our data show that these residues are essential for the function of the protein. In contrast, substitution of alanine for three other residues that directly contact DNA, K38, K40, and K45, caused inviability at low copy numbers but not when expressed at high copy numbers. This result was unexpected because these are also highly conserved residues among MADS box proteins, and two of these mutations, K38A and K45A, produced a greater-than-1,000-fold decrease in DNA-binding affinity (1). The finding that these mutations permit viability at high copy numbers indicates that these proteins are expressed and are folded sufficiently to interact with the cofactors to express the Mcm1-dependent genes that are required for growth. These results also indicated that strong DNA-binding activity is not required to maintain viability if sufficient levels of the protein are present.
Interestingly, the I90A mutation allowed viability when on a high-copy-number plasmid in a MATa strain but not in a MATα strain. The levels of available Mcm1 protein may account for the difference in viability between the two mating types. In an α strain, a significant portion of the Mcm1 protein may be sequestered into a complex with α1 and α2, whereas in an a strain, there may be more Mcm1 protein available to interact with other cofactors, such as Fkh2, to activate essential genes (17, 24, 25). This result, along with the fact that higher levels of MCM1 expression can suppress the lethality of many of the mutations, suggest that Mcm1 levels may be limiting within the cell. In further support of this model, we found that some of the mutants grew very slowly and that only a few colonies were able to survive the plasmid swap selection on 5-FOA plates. In fact, when we rescreened the inviable mutants by plating a higher number of cells on 5-FOA plates, we found a few colonies with the T54A and L59A mutations, substitutions we had previously considered to cause inviability (1). These mutants did not grow well and, as will be discussed below, were defective in many Mcm1 functions. It is possible that a small increase in the expression or activity of the mutant protein allows these clones to survive without the wild-type plasmid. We also observed differences in the viability of some of the mutations in different strain backgrounds, further suggesting subtle differences in the expression of the mutant Mcm1 proteins may influence cell viability (E. Dubois and F. Messenguy, personal communication). We have examined a number of the mutant proteins by Western blot analysis and found a three- to fourfold increase in the level of expression at high copy numbers compared to low copy numbers, further indicating that relatively small differences in the levels of some of the mutant proteins are sufficient to restore viability.
Residues required for Mcm1-mediated transcriptional activation.
Although a few mutations failed to complement the mcm1 null mutation and were therefore lethal to the cell, a large number of the alanine substitutions resulted in viability when present at either low or high copy numbers. To determine if these substitutions affect Mcm1 activity, the mutant proteins resulting in viability were assayed for the ability to autonomously activate transcription in vivo. In this assay, strain YY2052, which contains an integrated lacZ reporter gene under the control of a P(PAL) Mcm1 binding site, was transformed with all of the mcm1 mutations that result in viability and β-galactosidase activity was measured (Fig. 3A and 4A) (7, 20). The majority of the mutations activated transcription of the P(PAL) reporter at nearly wild-type levels, suggesting that they do not dramatically alter the structure or function of the protein. There were, however, a number of mutations that produced a significant decrease in activation. As expected, several of these mutations, R19A, V34A, K38A, and K40A, are at residues that contact the DNA and, as shown previously, caused significant decreases in the DNA-binding affinity of the protein (1). Replacement of other residues, such as T54, L59, I80, and G86, that lie at the dimer interface, and T74, which forms an H bond between the β strands in each monomer, may slightly affect the folding of the protein so that it cannot efficiently bind DNA or interact with cofactors required for activation. Residue I23 in the N-terminal extension and residues F48 and E49 in the αI helix also showed reductions in activation when mutated to alanine. Residue E49 packs up against I23 and is likely important for positioning of the N-terminal extension, which is required for DNA binding (1). Interestingly, although the protein with the F48A mutation had a threefold decrease in transcriptional activation, it bound DNA slightly better than did wild-type Mcm1, suggesting that this residue plays a role in protein-protein interactions with transactivating factors (1). The V34A mutation had the second largest effect on Mcm1-mediated autonomous activation of any of the mutations in the MADS box domain. Although this residue makes two base-specific contacts, the mutation produced only a modest (2.7-fold) decrease in DNA-binding affinity (1). However, this mutation caused a large decrease in DNA bending by the protein, suggesting that DNA bending is important for Mcm1-mediated activation at a P(PAL) site.
FIG. 4.
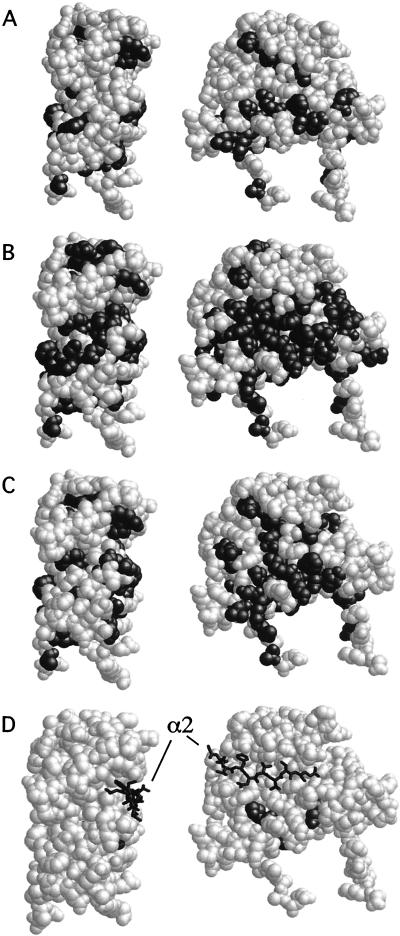
Position in Mcm1 of alanine substitutions that affect transcriptional regulation with its cofactors. The positions of alanine substitutions that affect autonomous activation (A), activation in complex with α1 (B), activation in complex with Ste12 (C), and repression in complex with α2 (D) are black. Shaded residues are those that lie above the line in the corresponding panels in Fig. 3. The views shown are in the same orientation as those in Fig 2A and B. The positions of the residues in the linker region of α2 that contact Mcm1 are shown as stick figures in panel D.
Mcm1 mutations showed defects in activation in combination with α1.
While mutations at relatively few positions decreased activation by Mcm1 alone, a large number of the mutations significantly affected the ability of Mcm1 to activate transcription with α1 (Fig. 3B and 4B). Many of the mutations that had moderate (2- to 4-fold) effects on autonomous activation by Mcm1 caused large (10- to 100-fold) decreases in combination with α1. The majority of these residues are clustered and buried in the inner and outer β strands. However, a few of these residues, Q57, Y70, and F72, are partially solvent exposed and, along with other residues that affect activation with α1, such as K40, M44, and K45, form a solvent-exposed surface in the middle layer of the protein (Fig. 4B). These residues may directly interact with α1 or, alternatively, are important for the alternative conformation when Mcm1 binds with α1 (45). The fact that so many of the mutations showed significant differences in the level of activation in combination with α1 than when Mcm1 activates transcription autonomously or in combination with Ste12 also indicates differences in the way that Mcm1 mediates transcriptional activation with these different cofactors. In further support of these differences, the mutation that produced the second greatest decrease in Mcm1-mediated activation alone, V34A, produced a relatively minor decrease in activation in combination with αl.
Since many of the alanine substitutions caused large decreases in activation in complex with α1 but had relatively little effect on autonomous activation, we were concerned that the α1-Mcm1 site from the STE3 promoter may be more sensitive to mutations in Mcm1 than are sites from other α-specific genes. To compare the sensitivities of the different α1-Mcm1 binding sites, we constructed lacZ reporter promoters containing α1-Mcm1 binding sites from the promoters of the MFα1 AGAα1, and MFα2 α-specific genes and assayed these reporters in combination with a set of mcm1 mutations (Table 1). Although there were differences in strength between these α1-Mcm1 activator sites, we found the relative activities produced by the mcm1 mutations to be very similar at each of the α1-Mcm1 sites. These results suggested that the defects in activation caused by many of the mcm1 alanine mutations with the STE3 reporter are consistent with other α1-Mcm1 sites.
TABLE 1.
Activation by Mcm1 mutant proteins at different α1-Mcm1 sitesa
| Mutation | STE3 (8.6 U) | MFα1A (5.3 U) | AGA1 (43 U) | MFα1B (36 U) |
|---|---|---|---|---|
| None (wild type) | 100 | 100 | 100 | 100 |
| R19A | 3 | 6 | 2 | 3 |
| I21A | 10 | 8 | 6 | 7 |
| I23A | 5 | 4 | 2 | 1 |
| E49A | 4 | 6 | 4 | 4 |
| G67A | 5 | 19 | 6 | 12 |
| S73A | 89 | 77 | 101 | 110 |
| P75A | 54 | 83 | 60 | 82 |
| G86A | 4 | 11 | 3 | 1 |
The values shown are percentages of wild-type transcriptional activation activity by α1-Mcm1 sites from the indicated genes assayed in a heterologous promoter. The values in parentheses are levels of expression in β-galactosidase units of the reporter in a wild-type strain. A reporter lacking a site gives 0.5 U of activity in the same strain.
Many of the alanine substitutions in the N-terminal region of the Mcm1 MADS box domain (residues 14 to 54) strongly decreased the DNA-binding affinity of the protein (1). Although not in direct contact with the DNA, alanine substitutions at residues in the C-terminal region of the MADS box domain may also have a large effect on DNA binding by Mcm1 alone and in combination with its cofactors. A number of the mutant proteins were therefore expressed in bacteria, purified, and assayed for DNA binding in combination with α1 (Fig. 5). In general, there was a good correlation between the level of cooperative binding with α1 in vitro and transcriptional activation in vivo. For example, proteins with the V34A and F48A mutations cooperatively bound with α1 at levels comparable to that of the wild-type protein and these mutant proteins showed relatively minor reductions in activation of the reporter in vivo (Fig. 3B). In contrast, mutant proteins such as those with the K40A and Y70A mutations, which showed large reductions in activation, also showed reduced cooperative binding with α1. These proteins bound to the STE3 sites with reasonable affinity on their own but failed to bind cooperatively with α1, indicating that these substitutions likely affect interactions with α1. However, some of the mutations in the C-terminal region of the domain, such as V69A, F72A, T74A, and L89A, caused significant decreases in binding to the site in the absence of α1 (data not shown). However, many of these mutant proteins were able to interact with α1, although with significantly reduced binding affinity, suggesting that the reduced activation by these mutant proteins is due to the reduced binding affinity of the complex.
FIG. 5.
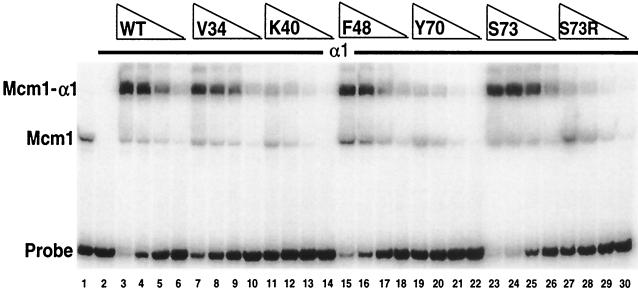
DNA binding of Mcm1 alanine mutant proteins in combination with α1. Shown is the affinity of binding to the STE3 α1-Mcm1 site of wild-type (WT) Mcm1 (lanes 3 to 6) and mutant Mcm1 proteins with the V34A (lanes 7 to 10), K40A (lanes 11 to 14), F48A (lanes 15 to 18), Y70A (lanes 19 to 22), S73A (lanes 23 to 26), and S73R (lanes 27 to 30) mutations in the presence of α1. All of the Mcm1 proteins contain the entire MADS box domain (residues 1 to 97) and are titrated as fivefold dilutions from a concentration of 2 × 10−9 M (lanes 3, 7, 11, 15, 19, 23, and 27). Lane 1 contains wild-type Mcm1 in the absence of α1. Lanes 2 to 30 contain 100 ng of partially purified α1. The positions of Mcm1 and the Mcm1-α1 complex are indicated.
It was previously shown that an S73R substitution in Mcm1 caused a large decrease in α1-Mcm1-mediated transcriptional activation and DNA binding (7). In contrast, the S73A substitution produced wild-type activity in our assay system. We also observed this difference in the activity of the mutant proteins in vitro (Fig. 5). This result suggests that while the Ser side chain may not be critical for the specificity of the interaction between the proteins, the α1 protein is likely to be in close proximity to this side chain because the Arg side chain interferes with binding.
Mcm1 residues required for activation in combination with Ste12.
To monitor Ste12-Mcm1-dependent activation, we used a lacZ reporter promoter that contains the Mcm1 binding site from the STE2 promoter. Even in the absence of a mating pheromone, this construct had levels of lacZ expression that are dependent on Ste12 and that are 100-fold greater than those of a reporter promoter lacking the site (19) (data not shown). Given the high level of Ste12-dependent activation, we assayed activation of the mcm1 mutations in combination with Ste12 in the absence of a mating pheromone (Fig. 3C and 4C). Although the magnitudes differ, many of the substitutions that affect Mcm1-dependent activation of the P(PAL) site alone also decreased activation by STE2 PRE in combination with Ste12. A number of these residues are involved in DNA binding or position the N-terminal arm. Interestingly, in contrast to its effect on activation with α1, the V34A substitution caused a large decrease in transcriptional activation with Ste12, suggesting that DNA bending plays an important role in activation by this complex. As with α1, residues Q57 and T74 are important for the interaction of Mcm1 with Ste12. These residues form a solvent-exposed cleft near the ends of the β strands.
Although we observed Ste12-dependent activation of the reporter promoter in the absence of a pheromone, in the presence of α pheromone, there was a further twofold increase in the level of lacZ expression in our reporter system. Upon induction of the mating signal transduction pathway with a mating pheromone, Ste12 is phosphorylated (18, 41). It is possible that this phosphorylation alters the conformation of Ste12 and its interactions with Mcm1. If this were the case, we might expect to see a different profile of the phenotypes produced by the alanine mutations that affect activation with Ste12 in the presence of α mating factor. However, we tested a subset of the mcm1 mutations for Ste12-dependent activation in the presence of α pheromone and although there was an increase in the levels of expression of the reporter promoter, the relative effects of each of the mcm1 mutations were roughly the same in the presence and absence of α pheromone (data not shown).
Alanine mutations of residues in Mcm1 that contact α2 in the crystal structure had little effect on repression.
To determine which positions in Mcm1 affect cooperative binding with α2 and repression of a-specific genes, we assayed the mcm1 mutations in combination with α2. Previously, we performed alanine scanning mutagenesis of the residues in the linker region of α2 and identified a small patch of hydrophobic residues that are required for interaction with Mcm1 (28). The crystal structure of the α2-Mcm1-DNA ternary complex verified that these residues in α2 contact a hydrophobic surface in Mcm1 (44) (Fig. 4D). We therefore wanted to determine if alanine substitutions for these residues in Mcm1 also cause significant reductions in cooperative interaction with α2 and repression of a-specific genes. Surprisingly, we found that replacement of only three residues, K40, I43, and E49, with alanine produced significant (greater-than-twofold) decreases in repression (Fig. 3D and 4D). Replacement of residues that directly contact α2 in the crystal structure of the ternary complex, such as S51, V69, and R87, with alanine had a less-than-twofold effect on repression. The K40, I43, and E49 residues do not contact α2 in the crystal structure, but all of the proteins with mutations at these sites showed reductions in binding affinity of 50-fold or greater. However, the decreased repression by these mutant proteins is not simply due to a reduction in DNA-binding affinity because mutant proteins with even greater losses of DNA-binding affinity, such as those with the F25A, K38A, M44A, K45A, and T54A mutations, had nearly wild-type activity. The I43 and E49 residues are buried in the core of the protein, and it is possible that their replacement lowers the level of the protein so that there is not sufficient Mcm1 for full repression. The K40A mutation not only caused a decrease in the DNA-binding affinity of the protein but also caused a significant reduction in DNA bending (1). Although it is possible that bending by this residue may be required for cooperative binding with α2, other mutations that affect DNA bending (but to a lesser degree), such as V34A and S37A, have wild-type repression activity in combination with α2.
The α2-Mcm1 binding site used to assay the different mutations in Mcm1 is a consensus site derived by alignment of all of the natural binding sites found in the promoter of a-specific genes (50). This site functions as a very strong repressor site in the context of the heterologous promoter. It is therefore possible that the strong nature of this site makes it insensitive to partial decreases in Mcm1 activity, possibly explaining why replacement of residues contacted by α2 had little effect on repression. We therefore tested the repression of several mcm1 mutations with transcription reporters containing α2-Mcm1 binding sites from the BAR1, STE2, and ASG7 promoters (49). All of the mcm1 mutations tested produced levels of repression through these sites roughly similar to those achieved with the consensus α2-Mcm1 site (Table 2). This indicates that the failure to observe derepression by the mcm1 mutations is not due to the specific α2-Mcm1 binding site used in our assay system.
TABLE 2.
Repression by Mcm1 mutant proteins at different α2-Mcm1 sitesa
| Mutation | AMCS | BAR1 | STE2 | ASG7 |
|---|---|---|---|---|
| None (wild type) | 100 | 100 | 100 | 100 |
| R19A | 89 | 280 | 163 | 62 |
| I21A | 170 | 390 | 387 | 139 |
| I23A | 74 | 229 | 221 | 58 |
| E49A | 25 | 25 | 27 | 14 |
| S51A | 70 | 91 | 95 | 58 |
| V69A | 83 | 135 | 85 | 44 |
| V81A | 70 | 175 | 147 | 30 |
| R87A | 28 | 85 | 86 | 24 |
| L89A | 65 | 210 | 31 | 84 |
The values shown are percentages of wild-type transcriptional repression activity by the α2-Mcm1 consensus site (AMCS) and sites from the BAR1, STE2, and ASG7 promoters assayed in a heterologous promoter (50).
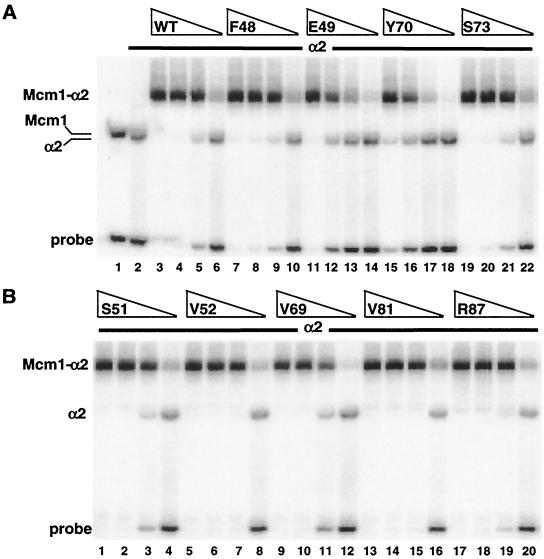
We next determined if these mutations affect cooperative binding with α2 to the α2-Mcm1 binding site from the STE6 promoter (Fig. 6). Although some of the mutations, such as E49A, Y70A, and I90A, caused a decrease in the formation of the cooperative α2-Mcm1 complex, they also produced a similar reduction in the DNA-binding affinity of the protein alone and did not affect the cooperative interactions with α2. Mutant proteins with substitutions of residues that directly contact α2, such as S51A, V52A, V69A, V81A, and R87A, showed roughly wild-type DNA-binding affinity alone (data not shown) and in complex with α2. These results are in agreement with the in vivo transcription assay and indicate that these mutations do not cause a significant decrease in cooperative interactions with α2.
FIG. 6.
DNA binding of mcm1-encoded alanine mutant proteins in combination with α2. Shown is the affinity of binding to the STE6 α2-Mcm1 site of wild-type (WT) Mcm1 (lanes 3 to 6) and mutant proteins with the F48A (lanes 7 to 10), E49A (lanes 11 to 14), Y70A (lanes 15 to 18), and S73A (lanes 19 to 22) mutations (A) and those with the S51A (lanes 1 to 4), V52A (lanes 5 to 8), V69A (lanes 9 to 12), V81A (lanes 13 to 16), and R87A (lanes 17 to 20) mutations (B) in the presence of α2. The Mcm1 proteins were diluted by fivefold dilutions from a concentration of 4 × 10−10 M (lanes 1, 3, 7, 11, 15, and 19) in the presence of 4.7 × 10−7 M α2. Lane 1 in panel A shows binding by wild-type Mcm1 in the absence of α2, and lane 2 shows binding by α2 in the absence of Mcm1. The positions of α2, Mcm1, and the Mcm1-α2 complex are indicated.
Radical mutations at residues in Mcm1 that directly contact α2 do not affect repression.
The alanine scanning experiments showed that removal of individual side chains in the Mcm1 MADS box domain, even in residues that directly contact α2 in the crystal structure of the ternary complex, do not affect repression of a-specific genes or cooperative binding with α2. To determine if this interaction can be disrupted, we made radical substitutions with larger or charged amino acids at the residues in Mcm1 that directly contact α2 (Table 3). A few of the radical mutations were lethal to the cell and therefore could not be assayed in vivo. Although some of the single mutations caused significant reductions in transcriptional activation through the P(PAL) site, with the exception of the S51F mutation, none had a more-than-twofold decrease in repression with α2. We therefore constructed a series of double-mutant proteins to determine if removal or disruption of multiple contacts would affect the interaction. Removal of two side chains that form the hydrophobic pocket in Mcm1 that accommodates the α2 F116 residue, such as R87A/V69A or R87A/V81A, had little effect on repression. However, removal of a side chain that contacts a different residue in α2, such as S51A or T71A, in addition to the mutation in the pocket (R87A), reduced repression. Finally, radical double substitutions, such as R87F/S51F and R87F/V69F, caused significant decreases in the level of repression at levels similar to those that we observed with mutations in the α2 linker region (28).
TABLE 3.
Effects of radical and double mutations at residues in Mcm1 that contact α2
| Mutation(s) | Viabilitya
|
% Activationb | % Repressionc | |
|---|---|---|---|---|
| CEN | 2 μM | |||
| None (wild type) | + | 100 | 100 | |
| S51F | + | 42 | 42 | |
| S51K | − | − | ||
| V69F | S | 10 | 70 | |
| Y70F | + | 78 | 125 | |
| Y70I | − | − | ||
| T71N | + | 78 | 60 | |
| F72Y | + | 79 | 106 | |
| S73R | + | 37 | 59 | |
| P75L | + | 74 | 90 | |
| V81H | + | 59 | 90 | |
| V81F | + | 30 | 91 | |
| R87E | S | 10 | 74 | |
| R87F | + | 26 | 82 | |
| R87A/S51A | + | 37 | 30 | |
| R87A/V69A | − | + | 58 | 175 |
| R87A/T71A | + | 22 | 56 | |
| R87A/S73A | + | 70 | 103 | |
| R87A/V81A | − | + | 110 | |
| R87F/S51F | + | 10 | 23 | |
| R87F/V69F | − | + | 37 | 10 |
| R87F/S73R | + | 40 | ||
+, normal growth after swapping out the wild-type copy of MCM1; −, no growth after 5 days; S, slow-growth phenotype.
Level of activation through the P(PAL) site based upon lacZ expression in comparison to that of the wild type.
Level of repression based upon lacZ expression in comparison to that of the wild type.
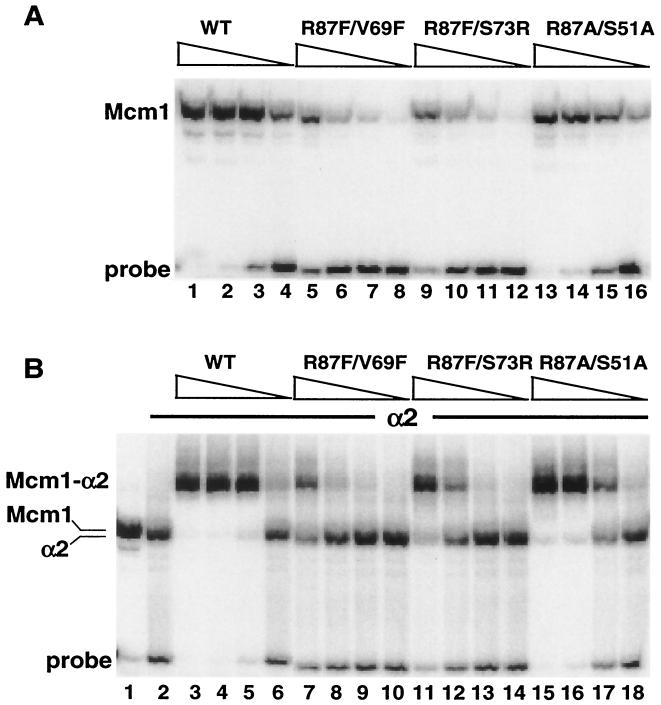
To test if the effects of the radical mutations on α2-Mcm1-mediated repression were due to decreased binding by the complex, we purified the mutant proteins and assayed their ability to bind DNA alone and in complex with α2 (Fig. 7). Even though some of the substitutions are at solvent-exposed residues away from the DNA-binding surface, some of the mutations caused a decrease in DNA-binding affinity in the absence of α2. It is possible that these changes affect the ability of the protein to fold properly and dimerize. These mutant proteins also showed significant decreases in cooperative binding with α2, indicating why these substitutions cause decreases in repression in vivo.
FIG. 7.
DNA binding of radical double-mutation mcm1-encoded proteins in combination with α2. (A) Binding affinity of wild-type (WT) Mcm1 (lanes 1 to 4) and mutant proteins with the R87F/V69F (lanes 5 to 8), R87F/S73R (lanes 9 to 12), and R87A/S51A (lanes 13 to 16) mutations to the STE6 α2-Mcm1 site. The proteins were diluted by fivefold dilutions from a concentration of 4 × 10−10 M (lanes 1, 5, 9, and 13). (B) Binding affinity of wild-type Mcm1 (lanes 3 to 6) and mutant proteins with the R87F/V69F (lanes 7 to 10), R87F/S73R (lanes 11 to 14), and R87A/S51A (lanes 15 to 18) mutations in the presence of 4.7 × 10−7 M α2. The proteins were diluted by fivefold dilutions from a concentration of 4 × 10−10 M (lanes 3, 7, 11, and 15). In panel B, lane 1 shows the position of wild-type Mcm1 binding in the absence of α2 and lane 2 shows the position of α2 binding in the absence of Mcm1.
Effects of Mcm1 mutant proteins on the expression of endogenous genes.
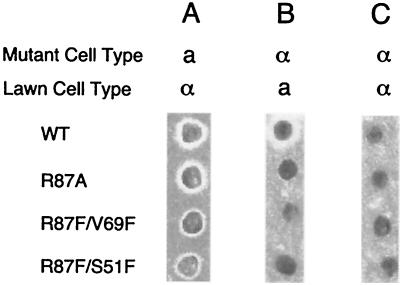
We have measured the effects of mcm1-encoded mutant proteins with heterologous transcription reporter promoters containing isolated Mcm1-binding elements. We next determined if these mutant proteins have a similar effect on the expression of the endogenous genes that are required for cell type establishment and mating. Each of the viable mutant proteins was assayed for mating pheromone production in the appropriate cell types with a mating pheromone halo assay (42). In this assay, mcm1 mutant strains in a and α mating type backgrounds were tested for pheromone production by patching of colonies onto lawns of strains of the opposite mating type. Pheromone secretion from the cell being tested causes cell cycle arrest in the lawn of cells of the opposite mating type, thereby forming a halo of nongrowth around the strain being tested. All of the mutants in the a cell type background produced a halo when patched onto a lawn of α cells, indicating that they retained sufficient activity to activate the endogenous a-specific genes in the cell (Fig. 8A). However, for some mutants, such as those with the R87F/V69F and R87F/S51F mutations, the size of the halo was decreased, indicating that lower pheromone levels were produced by these strains. These results correlate well with the reduced activation by these mutants in the lacZ reporter assays (Table 3). In α cells, the relative size of the halo correlates with the level of activation by the α1-Mcm1 complex of the MFα1 and MFα2 genes coding for the α pheromone. In agreement with the transcription reporter assays, mutants of the α cell type that were defective in activating transcription of the α1-Mcm1-dependent lacZ reporter produced little or no halo when patched onto a lawn of a cells (Fig. 8B). None of the mcm1 mutants of the α strain produced a halo when patched onto a lawn of α cells (Fig. 8C). This result indicates that although some of the mutants showed decreased repression of the lacZ reporter in combination with α2, they still retained sufficient activity to repress transcription of the endogenous a-specific genes.
FIG. 8.
Mating pheromone halo assays of the wild-type (WT) and mcm1 mutant strains. The wild-type strain and the indicated mcm1 mutant strains in a MATa background (A) or a MATα background (B and C) were patched onto lawns of MATα (A and C) and MATa (B) pheromone-sensitive strains. The presence of a zone of growth inhibition around the patched cells indicates that they produce a pheromone of the mating type opposite to that of the cells on the lawn.
DISCUSSION
MADS box protein Mcm1 is a global transcriptional regulator involved in the control of genes that specify diverse cellular processes that include mating type specificity, cell cycle regulation, minichromosome maintenance, arginine metabolism, osmotic regulation, and cell wall and membrane structure (11, 26, 27, 33). The N-terminal third of Mcm1 (residues 1 to 98) performs all of the essential Mcm1-dependent functions, including DNA binding, maintenance of cell viability, and interaction with all of its known cofactors (6, 9, 36). To determine how this domain carries out these different functions, we have scanned the entire MADS box domain of Mcm1 by making individual alanine substitutions to identify residues that are required for cell viability, transcriptional regulation, and interaction with the α1, α2, and Ste12 cofactors.
On the basis of their effects on viability, the mcm1 alanine mutations can be divided into three classes: (i) those that cause inviability at low or high copy numbers, (ii) those that cause inviability at low copy numbers but allow viability at high copy numbers, and (iii) those that allow viability at low copy numbers. Eight mutant proteins, those with the I26A, R32A, F36A, R39A, G42A, K46A, L53A, and I90A mutations, are in the first class of mutant proteins. Six of these eight have amino acid substitutions in the coiled-coil αI helix. Residues R39, G42, and K46 make direct contacts with the DNA, and alanine substitutions at these positions completely destroy the ability of the proteins to bind DNA (1). Residues I26, R32, F36, L53, and I90 are mostly buried and are likely to be essential for dimerization. With the exception of I90, all of the residues in this class are virtually invariant throughout all of the MADS box proteins and cluster at the dimer interface on either end of the long αI helix (40, 44). The high conservation of these residues, along with the strong phenotype of the alanine substitutions at these positions, indicates that these residues are likely to be essential for the function of most MADS box proteins. It is possible that these residues are important for binding by the ends of the two αI helices at the dimer interface. In contrast, buried residues at the dimer interface in the center of the αI helix, such as I43, or in the inner βI strand, such as V58 to V63, are more accommodating to removal of the side chain. These residues are not as well conserved among the MADS box proteins.
We were surprised by the number and positions of the mutations in the second class, which are inviable at low copy numbers but viable at high copy numbers. Many of the mutations in this class are at residues that are strongly conserved among the different MADS box proteins. For example, two of the residues in this class, K38 and K45, are invariant in all of the MADS box proteins and these side chains directly contact the DNA in the MADS box crystal structures (34, 39, 44). Residue K38 makes several base-specific contacts in the α2-Mcm1-DNA crystal structure, and the K38A mutant protein fails to bind DNA (1). The K45A mutant protein binds DNA with 1,000-fold decreased affinity compared to that of wild-type Mcm1. The K40 residue also directly contacts DNA in the crystal structures, and an alanine substitution at this position not only causes a decrease in the DNA-binding affinity of the protein but also causes a large reduction in the degree of DNA bending. Even though the other residues in this class of mutations do not directly contact DNA, alanine substitutions at these positions cause significant decreases in DNA-binding affinity. Although these residues are critical for DNA binding by Mcm1, the lethality of removal of these side chains can be suppressed by high-level expression of the mutant proteins. This suggests that these mutant proteins are expressed and are able to fold into a structure that, at high copy numbers, is sufficient to overcome the decreased activity caused by the mutation.
It is not clear why the lethal effects of the mutant proteins in the second class can be suppressed if the protein is present at high copy numbers while mutant proteins in the first class that show similar decreases in DNA-binding affinity are not viable at high copy numbers. Interestingly, the R39A, G42A, and K46A mutant proteins in the first class all contact the phosphate backbone, suggesting that these contacts are more important than the specific base pair contacts or DNA bending by other residues. It is possible that the cofactors that bind with Mcm1 to activate expression of essential genes provide a large portion of the DNA-binding specificity of the complex, so the base-specific contacts made by these residues are not essential. On the other hand, the phosphate backbone contacts made by R39, G42, and K46 may be essential for proper docking of the protein to the DNA.
Almost all of the mutations that cause a lethal phenotype at low or high copy numbers either directly contact the DNA or are buried and likely to be involved in folding or dimerization. In theory, there should also be solvent-exposed side chains that interact with other DNA-binding proteins to bind to target sites upstream of essential genes or that interact with transacting cofactors to recruit the basic transcription machinery. It is possible that the surface of Mcm1 that interacts with these putative cofactors is extensive, such that any single-alanine point mutation has little or no effect on the interaction. This would be similar to the effects of single-amino-acid substitutions on interaction with α2 that we observed. Alternatively, Mcm1 may serve as an architectural protein that binds to the promoters of essential genes and alters the chromatin structure by bending the DNA, allowing other transcription factors to bind to the promoter and thereby serving to indirectly activate transcription.
The majority of mcm1 alanine substitutions (58 of 75) fall into the third class, those that allow viability at low copy numbers. These mutations were tested for their effects on autonomous transcriptional activation through the P(PAL) site and activation with the α1 and Ste12 cofactors. Although most of the mutations caused greater effects on activation with α1, a comparison of the mutant proteins alone and in combination with α1 and Ste12 revealed some features that they have in common and are required for activation by the protein. In the C terminus of the MADS box domain, alanine substitutions at residues G67, T71, G86, R87, and L89 caused decreases in the level of activation by sites from the STE3 and STE2 promoters, while the same mutations had only small effects with activation on a P(PAL) site. This result suggests that there may be a region of Mcm1 that is involved in interaction with both α1 and Ste12. Alternatively, these residues may be involved in the interaction with a transcription cofactor that is recruited by both the α1-Mcm1 and Ste12-Mcm1 complexes and not by Mcm1 on its own. Of these residues, G67, T71, and R87 are partially solvent exposed and map to the hydrophobic pocket and groove on one surface of Mcm1. This is also the region that interacts with α2 in the crystal structure of the α2-Mcm1-DNA ternary complex (44). These results are in good agreement with previous findings that identified residues 69 to 81 of Mcm1 as important for both α1 and Ste12 interactions (7). Many of the mutations that affect the interactions with the different cofactors could alter the folding and dimerization of the protein. However, since most of these mutant proteins allow viability and show wild-type levels of repression and DNA binding with α2, these changes in folding are likely to be subtle.
Although many of the mutations in the Mcm1 MADS box domain have similar effects on activation with α1 and Ste12, there are some significant differences in the effects of mutations at the N terminus of the domain. For example, residues R31, T35, I43, V52, G55, L60 to T66, Y70, and F72 appear to be important for α1-mediated activation, while these residues have little effect on activation by Mcm1 alone or in complex with Ste12. Many of these mutations are at buried residues, and replacement of some of them may cause minor structural alterations. Although these changes do not appear to affect the essential role of the protein or strongly affect its interaction with Ste12 or α2, they do appear to affect interaction with α1. There are several explanations for this difference. In contrast to α2, the interaction of Mcm1 with α1 may be very specific, such that it cannot tolerate these small changes. It is also possible that the interaction between these proteins is relatively weak so that loss of any single contact is enough to significantly reduce the affinity and therefore activation by the complex. Another explanation is based on the differences in the mechanisms by which these complexes bind DNA. Even in the absence of the Ste12 and α2 cofactors, Mcm1 is bound to sites in the promoters of pheromone-responsive and a-specific genes, respectively. The Ste12 and α2 cofactors may therefore recognize contacts on the DNA, as well as binding surfaces on the Mcm1 protein. In contrast, the Mcm1 protein does not bind to sites in the α-specific genes in the absence of α1. The two proteins may therefore have to first form a complex in solution before binding to their target site. The formation of this complex may be more stringent and sensitive to mutations than interactions of Mcm1 with its other cofactors when bound to DNA.
The most striking difference between Mcm1 activation with α1 and that with Ste12 is shown by the protein with the V34A mutation. This mutant protein showed a greater-than-50-fold decrease in activation with Ste12 but, in complex with α1, activated α-specific genes nearly as well as the wild-type protein (Fig. 3). Residue V34 is critical for production of the large Mcm1-dependent bend in DNA (1). The fact that the V34A mutation has such profound effects on Ste12-dependent activation but has little effect on α1-mediated activation suggests that the mechanisms by which Mcm1 binds DNA when it is in complex with these two cofactors are likely very different. It is possible that DNA bending is required for activation by Mcm1 on its own or in complex with Ste12 but is not required when activating in complex with α1. Therefore, even in the absence of the V34 contact with the DNA, the α1-Mcm1 complex may produce a bend in the DNA that is similar to that produced by Mcm1 on its own. Alternatively, it is possible that DNA bending by Mcm1 produces a conformational change in the protein that is similar to the change induced by interaction with α1 (43).
Many of the mutations we have tested affect autonomous transcriptional activation or activation in complex with α1 or Ste12. We were therefore surprised to find that only a few mutations affect repression in complex with α2. Even residues within the hydrophobic pocket and along the hydrophobic groove that make direct contacts with α2 in the crystal structure have only minimal effects on repression when replaced with alanine. The observation that these mutations have similar effects on different α2-Mcm1 reporter promoters, do not alter cooperative binding with α2 in vitro, or affect repression of endogenous a-specific genes in vivo further supports our findings on the effects of these mutations. Even more surprising is the finding that most of the radical replacements of residues in Mcm1 that contact α2 did not significantly affect repression or cooperative binding with α2. It is possible that α2 interacts with Mcm1 in a manner different from that observed in the crystal structure. However, this is unlikely because, despite significant structural differences in residues in the linker region that do not contact Mcm1, the contacts by residues that do interact with Mcm1 are virtually identical in both of the α2 monomers (44). Furthermore, the effects of mutations in residues in the α2 linker region agree very well with predictions based on the crystal structure (28). The fact that most of the Mcm1 mutations do not affect repression indicates that the roles of the α2 and Mcm1 proteins in formation of the protein complex are very different. The region of interaction on α2 is relatively small, so that any single substitution changes a large percentage of the total contacts made by the protein. In comparison, Mcm1 appears to provide a large hydrophobic surface, in which multiple side chains, along with groups in the peptide backbone, contact each amino acid in the α2 linker. This model is supported by the fact that mutation of any single residue in Mcm1 has a minor effect on the interaction, while substitutions of residues in the linker region of α2 have a large effect on repression (28). Only when multiple contacts are disrupted by substitutions in Mcm1 with large amino acids is there an effect on repression. These results suggest that the overall structure of the Mcm1 interface, as opposed to individual contacts, plays an important role in the interaction with α2. The type of interaction between Mcm1 and α2 is therefore significantly different from the interaction between Mcm1 and α1 or Ste12, in which our mutational analysis has shown that individual side chains have important roles in the activity of the complex.
The differences in the types of interactions that we describe for Mcm1 with its mating cofactors may apply to the cofactor interactions of MADS box proteins from other organisms. As shown with α1 and Ste12 single mutations in Mcm1 affect transcriptional activation and DNA binding, indicating that subtle differences in the amino acid sequences of two MADS box proteins may provide determinants by which to distinguish which specific cofactors interact with each protein. However, as was shown by the effects of mutations in Mcm1 on the interaction with α2, the differences in sequence may not affect interactions with other cofactors that are common to both MADS box proteins. As an example, both the SRF and MEF2 MADS box proteins form a heterodimeric complex with myogenic cofactors such as the basic helix-loop-helix E12/MyoD heterodimer while also maintaining interactions with their own set of specific cofactors (14, 29, 30). By having different requirements within the same interface, as we have described for Mcm1 and its cofactors, MADS box proteins may be able to interact with the same set of cofactors, as well as with cofactors that only interact with specific MADS box proteins. This may explain how MADS box proteins with apparently similar DNA-binding specificities and the same cofactors are also able to interact with specific cofactors to differentially regulate distinct sets of genes.
Acknowledgments
We thank Stan Fields and George Sprague for providing strains and plasmids, Jonathan Mathias for constructing pJR018, and Deepu Abraham for pDA105. We thank Evelyne Dubois and Francine Messenguy for communication of unpublished data and the suggestion of cloning the mcm1 mutant proteins into a high-copy-number plasmid.
M.G and A.S. are recipients of a Howard Hughes Medical Institute Undergraduate Summer Research Fellowship and Rutgers Undergraduate Research Fellowships. M.G. is a recipient of a New Jersey Cancer Commission Undergraduate Fellowship. This research was supported by a grant from the National Institutes of Health (GM49265) to A.K.V.
REFERENCES
- 1.Acton, T. B., J. Mead, A. M. Steiner, and A. K. Vershon. 2000. Scanning mutagenesis of Mcm1: residues required for DNA binding, DNA bending, and transcriptional activation by a MADS-box protein. Mol. Cell. Biol. 20:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ammerer, G. 1990. Identification, purification, and cloning of a polypeptide (PRTF/GRM) that binds to mating-specific promoter elements in yeast. Genes Dev. 4:299-312. [DOI] [PubMed] [Google Scholar]
- 3.Baur, M., R. K. Esch, and B. Errede. 1997. Cooperative binding interactions required for function of the Ty1 sterile responsive element. Mol. Cell. Biol. 17:4330-4337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bender, A., and G. F. Sprague, Jr. 1987. MATα1 protein, a yeast transcription activator, binds synergistically with a second protein to a set of cell-type-specific genes. Cell 50:681-691. [DOI] [PubMed] [Google Scholar]
- 5.Boeke, J. D., J. Trueheart, G. Natsoulis, and G. R. Fink. 1987. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 154:164-175. [DOI] [PubMed] [Google Scholar]
- 6.Bruhn, L., J.-J. Hwang-Shum, and G. F. Sprague, Jr. 1992. The N-terminal 96 residues of MCM1, a regulator of cell type-specific genes in Saccharomyces cerevisiae, are sufficient for DNA binding, transcription activation, and interaction with α1 Mol. Cell. Biol. 12:3563-3572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bruhn, L., and G. F. Sprague, Jr. 1994. MCM1 point mutants deficient in expression of α-specific genes: residues important for interaction with α1. Mol. Cell. Biol. 14:2534-2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan, R. K., and C. A. Otte. 1982. Isolation and genetic analysis of Saccharomyces cerevisiae mutants supersensitive to G1 arrest by a factor and α factor pheromones. Mol. Cell. Biol. 2:11-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christ, C., and B. K. Tye. 1991. Functional domains of the yeast transcription/replication factor MCM1. Genes Dev. 5:751-763. [DOI] [PubMed] [Google Scholar]
- 10.Dolan, J. W., C. Kirkman, and S. Fields. 1989. The yeast STE12 protein binds to the DNA sequence mediating pheromone induction. Proc. Natl. Acad. Sci. USA 86:5703-5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dubois, E., and F. Messenguy. 1991. In vitro studies of the binding of the ARGR proteins to the ARG5,6 promoter. Mol. Cell. Biol. 11:2162-2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Errede, B., and G. Ammerer. 1989. STE12, a protein involved in cell-type-specific transcription and signal transduction in yeast, is part of protein-DNA complexes. Genes Dev. 3:1349-1361. [DOI] [PubMed] [Google Scholar]
- 13.Fields, S., and I. Herskowitz. 1985. The yeast STE12 product is required for expression of two sets of cell-type specific genes. Cell 42:923-930. [DOI] [PubMed] [Google Scholar]
- 14.Groisman, R., H. Masutani, M. P. Leibovitch, P. Robin, I. Soudant, D. Trouche, and A. Harel-Bellan. 1996. Physical interaction between the mitogen-responsive serum response factor and myogenic basic-helix-loop-helix proteins. J. Biol. Chem. 271:5258-5264. [DOI] [PubMed] [Google Scholar]
- 15.Hagen, D. C., L. Bruhn, C. A. Westby, and G. F. Sprague, Jr. 1993. Transcription of α-specific genes in Saccharomyces cerevisiae: DNA sequence requirements for activity of the coregulator α1. Mol. Cell. Biol. 13:6866-6875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herskowitz, I., J. Rine, and J. Strathern. 1992. Mating-type determination and mating-type interconversion in Saccharomyces cerevisiae, vol. 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 17.Hollenhorst, P. C., M. E. Bose, M. R. Mielke, U. Muller, and C. A. Fox. 2000. Forkhead genes in transcriptional silencing, cell morphology and the cell cycle. Overlapping and distinct functions for FKH1 and FKH2 in Saccharomyces cerevisiae. Genetics 154:1533-1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hung, W., K. A. Olson, A. Breitkreutz, and I. Sadowski. 1997. Characterization of the basal and pheromone-stimulated phosphorylation states of Ste12p. Eur. J. Biochem. 245:241-251. [DOI] [PubMed] [Google Scholar]
- 19.Hwang-Shum, J. J., D. C. Hagen, E. E. Jarvis, C. A. Westby, and G. F. Sprague, Jr. 1991. Relative contributions of MCM1 and STE12 to transcriptional activation of a- and α-specific genes from Saccharomyces cerevisiae. Mol. Gen. Genet. 227:197-204. [DOI] [PubMed] [Google Scholar]
- 20.Jarvis, E. E., K. L. Clark, and G. F. Sprague. 1989. The yeast transcription activator PRTF, a homolog of the mammalian serum response factor, is encoded by the MCM1 gene. Genes Dev. 3:936-945. [DOI] [PubMed] [Google Scholar]
- 21.Justice, M. C., B. P. Hogan, and A. K. Vershon. 1997. Homeodomain-DNA interactions of the Pho2 protein are promoter-dependent. Nucleic Acids Res. 25:4730-4739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keleher, C. A., C. Goutte, and A. D. Johnson. 1988. The yeast cell-type-specific repressor α2 acts cooperatively with a non-cell-type-specific protein. Cell 53:927-936. [DOI] [PubMed] [Google Scholar]
- 23.Keleher, C. A., S. Passmore, and A. D. Johnson. 1989. Yeast repressor α2 binds to its operator cooperatively with yeast protein Mcm1. Mol. Cell. Biol. 9:5228-5230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koranda, M., A. Schleiffer, L. Endler, and G. Ammerer. 2000. Forkhead-like transcription factors recruit Ndd1 to the chromatin of G2/M-specific promoters. Nature 406:94-98. [DOI] [PubMed] [Google Scholar]
- 25.Kumar, R., D. M. Reynolds, A. Shevchenko, S. D. Goldstone, and S. Dalton. 2000. Forkhead transcription factors, Fkh1p and Fkh2p, collaborate with Mcm1p to control transcription required for M-phase. Curr. Biol. 10:896-906. [DOI] [PubMed] [Google Scholar]
- 26.Kuo, M. H., E. T. Nadeau, and E. J. Grayhack. 1997. Multiple phosphorylated forms of the Saccharomyces cerevisiae Mcm1 protein include an isoform induced in response to high salt concentrations. Mol. Cell. Biol. 17:819-832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lydall, D., G. Ammerer, and K. Nasmyth. 1991. A new role for MCM1 in yeast: cell cycle regulation of SW15 transcription. Genes Dev. 5:2405-2419. [DOI] [PubMed] [Google Scholar]
- 28.Mead, J., H. Zhong, T. B. Acton, and A. K. Vershon. 1996. The yeast α2 and Mcm1 proteins interact through a region similar to a motif found in homeodomain proteins of higher eukaryotes. Mol. Cell. Biol. 16:2135-2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Molkentin, J. D., B. L. Black, J. F. Martin, and E. N. Olson. 1995. Cooperative activation of muscle gene expression by MEF2 and myogenic bHLH proteins. Cell 83:1125-1136. [DOI] [PubMed] [Google Scholar]
- 30.Molkentin, J. D., and E. N. Olson. 1996. Combinatorial control of muscle development by basic helix-loop-helix and MADS-box transcription factors. Proc. Natl. Acad. Sci. USA 93:9366-9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oehlen, L. J., J. D. McKinney, and F. R. Cross. 1996. Ste12 and Mcm1 regulate cell cycle-dependent transcription of FAR1. Mol. Cell. Biol. 16:2830-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Passmore, S., R. Elble, and B. K. Tye. 1989. A protein involved in minichromosome maintenance in yeast binds a transcriptional enhancer conserved in eukaryotes. Genes Dev. 3:921-935. [DOI] [PubMed] [Google Scholar]
- 33.Passmore, S., G. T. Maine, R. Elble, C. Christ, and B. K. Tye. 1988. Saccharomyces cerevisiae protein involved in plasmid maintenance is necessary for mating of MATα cells. J. Mol. Biol. 204:593-606. [DOI] [PubMed] [Google Scholar]
- 34.Pellegrini, L., S. Tan, and T. J. Richmond. 1995. Structure of serum response factor core bound to DNA. Nature 376:490-498. [DOI] [PubMed] [Google Scholar]
- 35.Price, M. A., A. E. Rogers, and R. Treisman. 1995. Comparative analysis of the ternary complex factors Elk-1, SAP-1a and SAP-2 (ERP/NET). EMBO J. 14:2589-2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Primig, M., H. Winkler, and G. Ammerer. 1991. The DNA binding and oligomerization domain of MCM1 is sufficient for its interaction with other regulatory proteins. EMBO J. 10:4209-4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Riechmann, J. L., B. A. Krizek, and E. M. Meyerowitz. 1996. Dimerization specificity of Arabidopsis MADS domain homeotic proteins APETALA1, APETALA3, PISTILLATA, and AGAMOUS. Proc. Natl. Acad. Sci. USA 93:4793-4798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Riechmann, J. L., and E. M. Meyerowitz. 1997. MADS domain proteins in plant development. Biol. Chem. 378:1079-1101. [PubMed] [Google Scholar]
- 39.Santelli, E., and T. J. Richmond. 2000. Crystal structure of MEF2A core bound to DNA at 1.5 Å resolution. J. Mol. Biol. 297:437-449. [DOI] [PubMed] [Google Scholar]
- 40.Shore, P., and A. D. Sharrocks. 1995. The MADS-box family of transcription factors. Eur. J. Biochem. 229:1-13. [DOI] [PubMed] [Google Scholar]
- 41.Song, D., J. W. Dolan, Y. L. Yuan, and S. Fields. 1991. Pheromone-dependent phosphorylation of the yeast STE12 protein correlates with transcriptional activation. Genes Dev. 5:741-750. [DOI] [PubMed] [Google Scholar]
- 42.Sprague, G. F., Jr. 1991. Assay of yeast mating reaction. Methods Enzymol. 194:77-93. [DOI] [PubMed] [Google Scholar]
- 43.Tan, S., G. Ammerer, and T. J. Richmond. 1988. Interactions of purified transcription factors: binding of yeast MATα1 and PRTF to cell type-specific, upstream activating sequences. EMBO J. 7:4255-4264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tan, S., and T. J. Richmond. 1998. Crystal structure of the yeast MATα2/MCM1/DNA ternary complex. Nature 391:660-666. [DOI] [PubMed] [Google Scholar]
- 45.Tan, S., and T. J. Richmond. 1990. DNA binding-induced conformational change of the yeast transcriptional activator PRTF. Cell 62:367-377. [DOI] [PubMed] [Google Scholar]
- 46.Treisman, R., R. Marais, and J. Wynne. 1992. Spatial flexibility in ternary complexes between SRF and its accessory proteins. EMBO J. 11:4631-4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vershon, A. K., Y. Jin, and A. D. Johnson. 1995. A homeo domain protein lacking specific side chains of helix 3 can still bind DNA and direct transcriptional repression. Genes Dev. 9:182-192. [DOI] [PubMed] [Google Scholar]
- 48.Vershon, A. K., and A. D. Johnson. 1993. A short, disordered protein region mediates interactions between the homeodomain of the yeast α2 protein and the MCM1 protein. Cell 72:105-112. [DOI] [PubMed] [Google Scholar]
- 49.Zhong, H., R. McCord, and A. K. Vershon. 1999. Identification of target sites of the α2-Mcm1 repressor complex in the yeast genome. Genome Res. 9:1040-1047. [DOI] [PubMed] [Google Scholar]
- 50.Zhong, H., and A. K. Vershon. 1997. The yeast homeodomain protein MATα2 shows extended DNA binding specificity in complex with Mcm1. J. Biol. Chem. 272:8402-8409. [DOI] [PubMed] [Google Scholar]