Abstract
B-cell lineage-specific activator protein (BSAP), encoded by the Pax-5 gene, is critical for B-cell lineage commitment and B-cell development but is not expressed in terminally differentiated B cells. We demonstrate a direct connection between BSAP and B-lymphocyte-induced maturation protein 1 (Blimp-1), a transcriptional repressor that is sufficient to drive plasmacytic differentiation. Blimp-1 binds a site on the Pax-5 promoter in vitro and in vivo and represses the Pax-5 promoter in a binding-site-dependent manner. By ectopically expressing Blimp-1 or a competitive inhibitor of Blimp-1, we show that Blimp-1 is both necessary and sufficient to repress Pax-5 during plasmacytic differentiation of primary splenic B cells. Blimp-1-dependent repression of Pax-5 is sufficient to regulate BSAP targets CD19 and J chain and is necessary but not sufficient to induce XBP-1. We further show that repression of Pax-5 is required for Blimp-1 to drive differentiation of splenocytes to immunoglobulin M-secreting cells. Thus, repression of Pax-5 plays a critical role in the Blimp-1-dependent program of plasmacytic differentiation.
B-cell lineage-specific activator protein (BSAP), encoded by the Pax-5 gene, is critical for commitment to the B-lymphocyte lineage and early B-cell development (14, 30). Pax-5−/− B cells arrest at the pro-B stage (16, 30) and can differentiate into multiple hematopoietic lineages (14). BSAP is also important for proliferation and isotype switching in germinal center B cells (4, 11, 16, 20, 30). BSAP is expressed throughout B-cell development until the terminally differentiated plasma cell stage (1, 4). BSAP can either activate or repress transcription (33). Targets of BSAP activation include VpreB, λ5, CD19, and blk (B lymphoid kinase) (8, 15, 17, 22, 35). BSAP represses J chain, the immunoglobulin heavy-chain 3′Cα enhancer and XBP-1 (12, 25, 28).
B-lymphocyte-induced maturation protein 1 (Blimp-1, encoded by the prdm1 gene) is a critical regulator of plasma cell differentiation, induced during cytokine-dependent differentiation of a B-cell lymphoma line (BCL-1) (29) and after lipopolysaccharide (LPS) treatment of primary murine splenocytes (2). Blimp-1 is expressed in all plasma cells and in a subset of germinal center B cells with a partial plasma cell phenotype but not in memory B cells (3). Ectopic expression of Blimp-1 in BCL-1 cells and in primary splenic B cells is sufficient to cause terminal differentiation and immunoglobulin M (IgM) secretion (2, 19, 26, 29).
Blimp-1 is a transcriptional repressor. Its DNA-binding activity is conferred by five zinc-finger motifs (7), whereas association with histone deacetylases (34) and hGroucho (24) is required for transcriptional repression. One important target of Blimp-1 repression is c-myc (10). Although repression of c-myc is necessary for terminal differentiation of BCL-1 cells, it is not sufficient, suggesting the existence of additional Blimp-1 targets (9). Indeed, MHC2A, encoding CIITA, a coactivator for major histocompatibility class II (MHC-II) transcription, was recently identified as a Blimp-1 target, providing a mechanism for extinction of MHC-II expression during plasma cell differentiation (19). We demonstrate here that Blimp-1 represses Pax-5 and show that Blimp-1-dependent repression of Pax-5 is required for plasma cell differentiation.
MATERIALS AND METHODS
Cell culture.
BCL-1 (CW13.20-3B3, ATCC CRL 1669), P3X (P3X63Ag8), 18-81 Raji, and primary splenocytes were cultured in RPMI medium supplemented with 10% heat-inactivated fetal bovine serum (FBS; Gemini Bio-Products, Inc.), 20 μg of gentamicin (Gemini)/ml, and 50 μM β-mercaptoethanol. To induce differentiation of BCL-1, cells (5 × 105 cells/ml) were stimulated with interleukin-2 (IL-2) and IL-5, as described previously (29), for various times. 3T3 and Phoenix cells (G. Nolan, Stanford University) were cultured in Dulbecco modified Eagle medium supplemented with 10% FBS and 20 μg of gentamicin/ml. WI-L2 transfectants were cultured in the phenol red-free RPMI medium supplemented with 10% charcoal-dextran-treated FBS (HyClone) and penicillin-streptomycin (Gibco-BRL) and cultured in the presence of the selection antibiotic, hygromycin B (500 μg/ml; Gibco-BRL). 4-Hydroxytomaxifen was dissolved in 70% ethanol (1 μM) and CdSO4 (5 μM) from Sigma.
Plasmids.
To generate a Blimp-1 binding site mutated reporter, a wild-type luciferase reporter dependent on the Pax-5 promoter (BSAP-Luc) (18) was used as the template to PCR amplify two fragments by using two sets of the primers: set 1 (5′-GGTACCGGTCCCTCCCATTCAAAAGCT-3′ and 5′-GTCAGCTTGGAATCGCTCTCCGAGAGTGTT-3′) and set 2 (5′-GTCAGCTGCAAAACTGCATTGTCAGTGGC-3′ and 5′-CCGCGGGATCTGGGACCTGGTGGCTGA-3′). By religation of the two fragments with pGL-3B (Promega) at the KpnI and SacI sites, a reporter containing mutated Blimp-1 binding site (M BSAP-Luc) was generated and then confirmed by sequencing. The accession numbers for identifying the Blimp-1 binding site on the mouse and human Pax-5 promoter in this study are AF148961 and AF268279, respectively. To generate retroviruses carrying the cDNA encoding BSAP, a NotI fragment of BSAP cDNA from BSAP-pEEB (B. Birshtein, Albert Einstein College of Medicine) was cloned into the NotI site of pGC-IRES-YFP vector (G. Fathman, Stanford University). Retroviruses carrying TBlimp cDNA were constructed by blunt-ended ligation of TBlimp (34) into ClaI site of pGC-IRES-YFP vector. Vxy-BlimpFLAG-puro was cloned by ligation of cDNA encoding BlimpFLAG into the XhoI site of a retroviral vector, Vxy-puro (L. Staudt, National Cancer Institute). VxyPRDI-BF1-puro was cloned as described elsewhere (A. L. Shaffer et al., unpublished data).
Immunofluorescence.
Immunofluorescence was performed on human tonsils by incubating fixed sections with two primary antibodies, rabbit anti-Blimp-1 and mouse anti-BSAP (provided by G. Cattoretti), according to a previously described protocol (3).
Northern blots.
Northern blotting was performed as described previously (5). An XhoI/AvrII fragment of Blimp-1 cDNA from pSKBlimp-1 (5) and an NcoI fragment of BSAP cDNA, an XbaI fragment of J chain cDNA from JcX plasmid (provided by J. Wallin, University of California, Berkeley) and a PstI fragment of human GAPDH (glyceraldehyde-3-phosphate dehydrogenase) from pTRI-hGAPDH (Ambion) were used for making probes.
EMSA.
Nuclear extracts from P3X and 18-81 cells were isolated, and 10 μg of nuclear protein was used for electrophoretic mobility shift assay (EMSA) as described previously (10). Double-stranded probes were BSAP (GCAGTTTTGGAAAGTGAATCGCTCTCCGA and its complementary strand) or PRF (CGCGTACAGAAAGGGAAAGGACTAGCGCG and its complementary strand). The accession numbers for the regions containing Blimp-1 binding sites on the mouse Pax-5 promoter and mouse c-myc promoter in this study are AF148961 and M12345, respectively. For the competition or supershift analysis, nuclear extracts were incubated with competitor, preimmune serum, or polyclonal Blimp-1 antiserum for 15 min before the addition of radiolabeled probe.
Retrovirus preparation and infection.
Retroviruses were generated as described previously (19). Splenic B cells were purified by negative selection with Thy1.2 magnetic beads (Dynal) and cultured as described previously (19). Splenic B cells (106/ml) were stimulated with LPS (10 μg/ml) or/and anti-F(ab′)2 (5 μg/ml; Southern Biotechnology) overnight before the retrovirus infection at a multiplicity of infection of 0.2 to 2 in the presence of 5 μg of Polybrene/ml. At 3 days postinfection, cells were sorted by flow cytometry for expression of yellow fluorescent protein (YFP). For infection of Vxy-puro, Vxy-Blimp-1(PRDIBF-1)-puro, and VxyBlimpFLAG-puro in 18-81 cells, cells (2 × 105/ml) were infected in the presence of 5 μg of Polybrene/ml. At 2 days postinfection, cells were selected with puromycin (puro; 6 μg/ml) for 48 h, and surviving cells were purified with Histopaque according to the manufacturer's suggested protocol (Sigma).
ChIP assay.
A total of 5 × 106 18-81 cells and P3X cells were harvested as described earlier (34). To determine the acetylation levels of histone H3 in retrovirus-tranduced 18-81 cells, cells were infected with virus expressing human Blimp-1 (PRDIBF-1) and puro or control virus, expressing puro alone. Selected cells (2 × 106) were subjected to chromatin immunoprecipitation (ChIP) analysis as described previously (34). To assess binding of Blimp-1 to the endogenous Pax-5 promoter, 18-81 cells were infected with either virus expressing BlimpFLAG and puro or control virus expressing puro alone. The subsequent virus infection, puro selection, and purification procedures were performed as described above. A total of 2 × 106 cells were cross-linked by the addition of formaldehyde into a final concentration of 1% and incubated at 4°C for 20 min. Subsequent sonication and precipitation steps were performed essentially as described previously (34), except with protein G-agarose beads (Santa Cruz) and 5 μg of anti-FLAG antibody (M2) (Sigma). The PCR primers used were c-myc (5′-ACGTGCGTGACGCGGTCCA-3′ and 5′-GGTGTAAACAGTAATAGCG-3′), BSAP (5′-CGAAAATCTGCTCCAGTGAAT-3′ and 5′-GAGAAGCCTATAGAGCCCCGT-3′), and amylase (5′-CAGCTGTGCACATCATTG-3′ and 5′-TTCCTTGGCAATATCAACC-3′). A total of 107 WI-L2 transfectants were harvested 20 h after induction of the expression of FLAG-tagged protein. A ChIP assay to determine the binding of Blimp-1 to Pax-5 promoter in human WI-L2 transfectants was performed essentially as described above in the system of acute infection of BlimpFLAG expressing retrovirus in 18-81 cells. The PCR primers were CIITA (5′-GGTTCCATTGTGATCATCA-3′ and 5′-AAACTCTCCCTGCAAGGTG-3′), BSAP (5′-TTTCCTCGAGCTCTAGGCTCT-3′ and 5′-TAGCCACTGAAAGCCTGTGGG-3′), and CSF-1 (5′-CTCTTCCTCCTGATAGCTCCATGA-3′ and 5′-CACTATGTTAGCCAGGATGGTCTC-3′) and were designed to amplify regions of 230 to 280 bp. All of the PCRs were annealed at 55°C and amplified for 30 cycles. PCR products were then transferred onto nylon membranes and hybridized with either γ-32P-labeled probes that recognize internal sequences or α-32P-labeled random primed probe from the PCR-amplified genomic template.
Transfection.
A total of 2 × 105 3T3 cells were seeded in six-well plates 1 day before the experiment. Then, 2 μg of luciferase reporter driven by the Pax-5 promoter (BSAP-Luc) (18), mutated Pax-5 promoter (M BSAP-Luc), or CIITA promoter III (CIITA-Luc) was cotransfected with various amounts of Blimp-1 expression vector (pBDP1-F) or control vector carrying the reverse sequence of Blimp-1 cDNA (pBDP1-B), as indicated in Fig. 2D (29), into 3T3 cells by the calcium phosphate method. At 44 to 48 h posttransfection, cells were harvested to determine the luciferase content as described previously (34). A total of 3 × 106 Raji cells were transfected by electroporation at 200 V and 960 μF. At 44 to 48 h posttransfection, cells were harvested for luciferase assay. A total of 0.5 μg of Renilla luciferase reporter driven by the thymidine kinase promoter was used in each cotransfection experiment to normalize the transfection efficiency. Three preparations of plasmid were used.
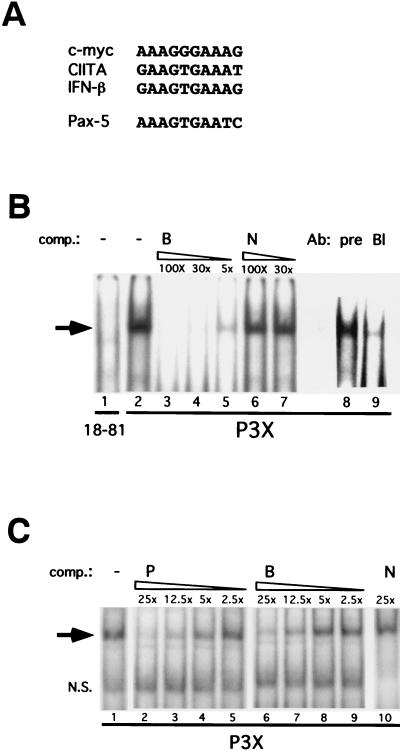
FIG. 2.
Blimp-1 binds to the Pax-5 promoter and represses Pax-5 promoter activity. (A) The alignment of the putative Blimp-1 binding site in the murine Pax-5 promoter with known sites in the murine c-myc promoter, human CIITA promoter III, and human IFN-β promoter. (B) EMSA with nuclear extracts from P3X plasmacytoma and 18-81 pre-B cells and a 30-bp probe derived from the putative Blimp-1 site in the murine Pax-5 promoter. Oligonucleotide B contains the Blimp-1 site in the murine Pax-5 promoter. Oligonucleotide N is a nonspecific oligonucleotide. pre, preimmune serum; Bl, Blimp-1-specific polyclonal antiserum. The molar excess of cold competitors is indicated. The arrow indicates the specific binding complex. (C) Comparison of the binding affinity of Blimp-1 to sites in the mouse c-myc promoter and mouse Pax-5 promoter. EMSA was performed with nuclear extracts from P3X and a probe containing the Blimp-1 binding site in the mouse c-myc promoter. Oligonucleotide P contains the Blimp-1 site in the mouse c-myc promoter. Oligonucleotide B contains the Blimp-1 site in the mouse Pax-5 promoter. N is a nonspecific oligonucleotide. The molar excess of cold competitors is indicated. N.S., nonspecific binding complex. (D) 3T3 cells were cotransfected with different amounts of Blimp-1 expression vectors, as indicated in the figure, and 2 μg of luciferase reporter driven by a 1.8-kb region of the mouse Pax-5 promoter (BSAP-Luc), the Pax-5 promoter with a mutated Blimp-1 binding site (M BSAP-Luc), or CIITA promoter III (CIITA-Luc). The relative luciferase activity represents the normalized luciferase activity derived from cells transfected with the Blimp-1 expression vector (pBDP1-F) divided by the normal-ized luciferase activity derived fromcells transfected with a control vector (pBDP1-B). The luciferase light units from one representative experiment were BSAP-Luc plus 2 μg of control vector 700137 and MBSAP-Luc plus 2 μg of control vector 548658. Averages ± the standard deviations from four independent experiments are shown. Diagrams of BSAP-Luc and M BSAP-Luc with the Blimp-1 binding sequence and mutated sequence and their relative distance to the transcription start sites in the Pax-5 promoter are indicated. The hatched box represents the Pax-5 gene sequence, and the arrow indicates the transcription start site. (E) Raji cells were cotransfected with 5 μg of Blimp-1 expression plasmid and 10 μg of luciferase reporter driven by the mouse Pax-5 promoter (WT) or with a mutated Blimp-1 site (M). The relative luciferase activity was determined as described in panel D. The luciferase light units from one representative experiment were the wild type plus the control expression vector 36840 and the mutant plus the control expression vector 34650.
RT-PCR.
RNA was isolated by the Trizol method (Gibco-BRL). A total of 250 ng of RNA was used for DNase (Promega) treatment, and the subsequent reverse transcription (RT) procedure as described previously (19). The primers used for PCR analysis were GAPDH (5′-TTAGCACCCCTGGCCAAGG-3′ and 5′-CTTACTCCTTGGAGGCCATG-3), BSAP (5′-GCGTGTTTGAGAGACAGCACT-3′ and 5′-AAGAATACTGAGGGTGGCTGT-3′), J chain (5′-CTAGGATCATCCCTTCCAC-3′ and 5′-TGATACCTAAGTGGGACCA-3′), CD19 (5′-ACAGGACTGGAAGAAGAAG-3′ and 5′-ACTGAATTGAGTGGAGCTG-3′), Blimp-1 (5′-GCCAACCAGGAACTTCTTGTGT-3′ and 5′-AGGATAAACCACCCGAGGGT-3′), and XBP-1 (5′-GCTGCGGAGGAAACTGAAA-3′ and 5′-GGGAGGCTGGTAAGGAACT-3′). All of the reactions were annealed at 55°C and amplified for 35 cycles, except the reactions with GAPDH and XBP-1 (30 cycles for each). The PCR products were analyzed by electrophoresis in 1.5% agarose gels.
Enzyme-linked immunospot (ELISPOT) assay.
Dilutions (from 5,000 to 50) of YFP+ splenic B cells were seeded into a 96-well plate and incubated on nitrocellulose membranes (Bio-Rad) coated with mouse antibody [Ig(H+L); Southern Biotechnology] at 37°C for 2.5 h. Cells attached to the membranes were lysed by phosphate-buffered saline (PBS)-EDTA for 15 min, followed by two washes of the membrane in PBS with 0.1% Tween 20 and one wash in PBS. The membrane was incubated at room temperature for 30 min with PBS with 1% bovine serum albumin. The membranes were then incubated with alkaline phosphatase-conjugated goat anti-mouse IgM (Southern Biotechnology) at a dilution of 1:1,000 in PBS plus 1% bovine serum albumin at room temperature for 1 h. IgM-secreting cells were then detected and counted by the addition of enzyme substrate.
RESULTS
Expression of BSAP and Blimp-1 in B cells.
Double immunofluorescence staining was used to analyze BSAP and Blimp-1 in human tonsils. Blimp-1 was detected in cells in the extrafollicular area of the tonsils where plasma cells are found, in a subset of germinal center B cells, and in squamous epithelium, as previously described (3) (Fig. 1Aa and c). BSAP+ B cells are located in the germinal centers (CD10+; data not shown) and the mantle zone (Fig. 1Ab and d). Superimposition of Blimp-1 and BSAP stains (Fig. 1Aa and b and Ac and d) revealed a nearly mutually exclusive pattern of expression for these proteins. In particular, in the germinal center, most B cells expressed BSAP, but the subset expressing Blimp-1 did not express BSAP.
FIG. 1.
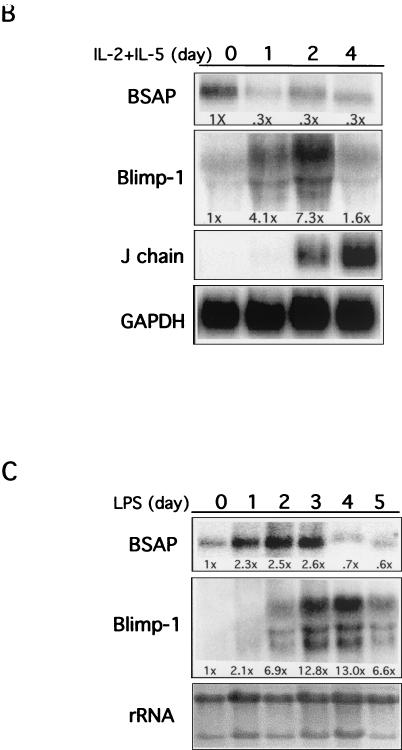
Expression of Blimp-1 and BSAP. (A) Blimp-1 and BSAP in tonsillar B cells. Fluorescent nuclear staining for Blimp-1 in green (a and c) and for BSAP in red (b and d), with merged images in the center. Germinal centers (GC), extrafollicular areas (EFA), and the mantle zone (MZ) are indicated. Magnifications: a and b, ×10; c and d, ×40. A cluster of Blimp-1+ plasma cells (PC) (left, boxed) and Blimp-1+ squamous epithelial cells (SE) (upper right) are indicated. (B) A total of 107 BCL-1 cells, growing in log phase, were stimulated with the cytokines IL-2 and IL-5. At the indicated times, cells were harvested, and the total RNA was prepared and analyzed by Northern blotting. GAPDH was used as the internal control. The fold changes in BSAP and Blimp-1 RNA, normalized to GAPDH, are shown under each lane. The fold change in J chain was not quantitated since no mRNA was measurable prior to treatment. (C) Splenocytes were stimulated with LPS (10 μg/ml) for 0 to 5 days, and RNA was analyzed by Northern blotting at the indicated times. 28S and 18S rRNA stained with ethidium bromide were used as loading controls. The fold changes, normalized to 28S rRNA, are shown below each lane.
The murine B-cell lymphoma line BCL-1 provides a good model for studying plasma cell differentiation since it acquires a plasma cell phenotype, including induction of surface Syndecan-1, IgM secretion, and cessation of cell cycle, in response to treatment with IL-2 plus IL-5 (9, 10, 29). Indeed, Blimp-1 was first identified in cytokine-treated BCL-1 cells and is sufficient to cause differentiation of BCL-1 cells in the absence of cytokines (10, 29). In order to monitor the expression of BSAP mRNA during induction of Blimp-1 in BCL-1 cells, Northern blot analyses were performed after cytokine treatment. Blimp-1 mRNA was induced approximately four- and sevenfold after 1 and 2 days of cytokine treatment, and levels fell by day 4 (Fig. 1B). BSAP mRNA declined threefold after 1 day of treatment and remained low thereafter (Fig. 1B). In addition, transcripts of J chain, a gene previously shown to be repressed by BSAP (25), were significantly induced after 2 days of cytokine stimulation (Fig. 1B). Thus, induction of Blimp-1 mRNA correlates with reduction of BSAP mRNA and derepression of the BSAP target, J chain, during differentiation of BCL-1 cells.
We also monitored Blimp-1 and BSAP mRNAs during differentiation of primary B cells in response to LPS. Murine splenocytes were cultured with LPS and levels of Blimp-1 and BSAP mRNA were monitored. In response to LPS, splenic B cells differentiate into plasma cells, a process characterized by the induction of Blimp-1 mRNA, Syndecan-1, IgM secretion, and a decrease in c-Myc (2). Since the initial population of cells was developmentally heterogeneous, the patterns are more complicated. Blimp-1 was undetectable initially, present at day 2, and more fully induced to approximately 13-fold on days 3 and 4. BSAP mRNA initially increased and then decreased on day 4 when the Blimp-1 levels were high (Fig. 1C). Thus, inverse expression patterns between Blimp-1 and BSAP mRNAs were observed during plasmacytic differentiation of primary splenic B cells and BCL-1 cells. Together with the expression pattern observed for Blimp-1 and BSAP protein in tonsillar B cells (Fig. 1A), these data show that expression of BSAP decreases as Blimp-1 is induced, a finding consistent with the possibility that Blimp-1 represses Pax-5 transcription.
Blimp-1 repression of the Pax-5 promoter.
To test whether Pax-5 is a direct target of Blimp-1, we analyzed a 1.8-kb region of the murine Pax-5 promoter which has previously been shown to have basal activity (18). Alignment of known Blimp-1 binding sites in murine c-myc (10), human beta interferon (IFN-β) (6), and human CIITA promoter III (19) with sequences in a 1.8-kb region of the mouse Pax-5 promoter identified a putative Blimp-1 site, AAAGTGAATC, at −926 to −917 bp (Fig. 2A). Nuclear extract from P3X, a plasmacytoma expressing Blimp-1 (10), was used in EMSAs with a 30-bp probe encompassing the site from bp −926 to −917 from the Pax-5 promoter (Fig. 2B). A complex was observed with P3X nuclear extract (lanes 2 and 8) that was absent in reactions containing nuclear extract from 18-81 pre-B cells lacking Blimp-1 (lane 1). The specificity of the complex was demonstrated by competition with cold probe (lanes 3 to 5) but not with nonspecific DNA (lanes 6 and 7). The complex was ablated by antiserum against Blimp-1 but not by preimmune serum (lanes 8 and 9), demonstrating that it contained Blimp-1.
To compare the affinity of Blimp-1 binding sites in the Pax-5 and c-myc genes, P3X nuclear extracts were incubated with a probe containing the Blimp-1 binding site from the murine c-myc promoter and cold competitors corresponding to the c-myc and Pax-5 sites. Blimp-1 binds to the Pax-5 site with an affinity that is ∼2-fold lower than that for the c-myc site (Fig. 2C).
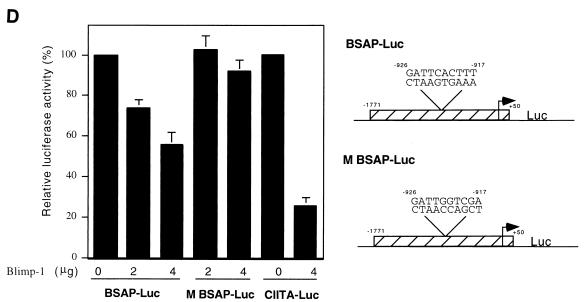
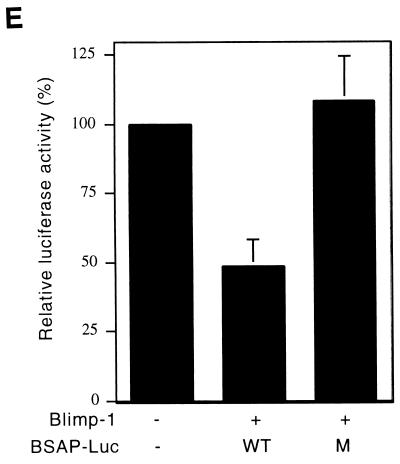
A luciferase reporter dependent on 1.8 kb of the Pax-5 promoter (BSAP-Luc) was used to determine whether Blimp-1 represses Pax-5 promoter activity. Although this portion of the Pax-5 gene contains basal promoter activity, elements which confer B-cell specificity have not been identified (18) and are not present in our reporter. Cotransfection of BSAP-Luc with a Blimp-1 expressing plasmid in 3T3 cells showed that the transcriptional activity of the Pax-5 promoter was repressed by Blimp-1 in a dose-dependent manner to ca. 50% (Fig. 2D). Blimp-1-dependent repression of the CIITA promoter was used as a positive control (19). Similar repression by Blimp-1 on the Pax-5 promoter was observed in Raji cells (Fig. 2E). A reporter with a mutated Blimp-1 binding site (M BSAP-Luc) was not subject to Blimp-1 repression in either 3T3 or Raji cells (Fig. 2D and E), establishing the binding site specificity of the repression.
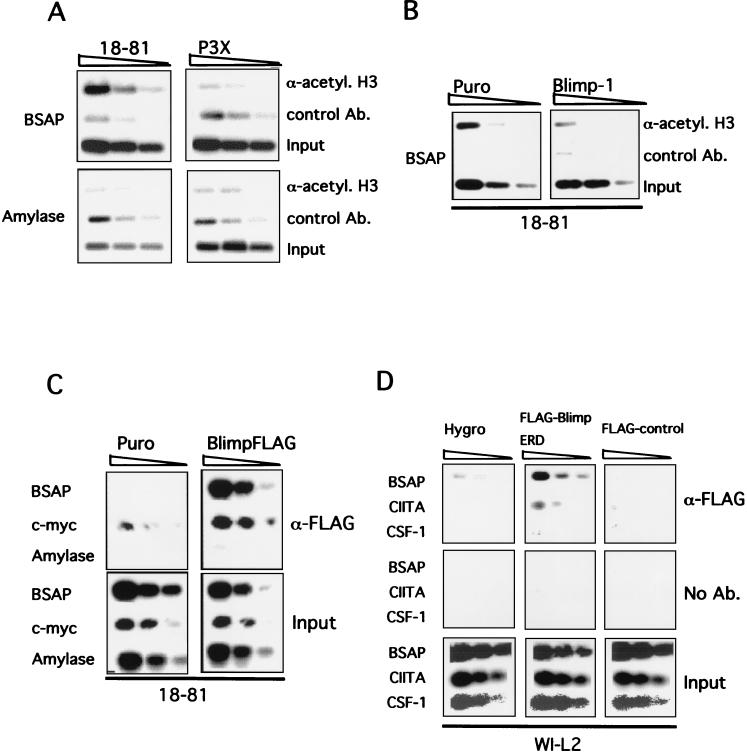
ChIP was used to investigate Blimp-1 action on the endogenous Pax-5 promoter in vivo. Since association of Blimp-1 with histone deacetylases is required for transcriptional repression (34), we investigated the acetylation status of histone H3 at the Blimp-1 site in the Pax-5 promoter by using antibody specific to acetylated histone H3 to immunoprecipitate chromatin. (Antibody specific to CDK2 was used as a control in this experiment.) Histone H3 was significantly more acetylated at the Blimp-1 binding site in the Pax-5 promoter in the pre-B line 18-81, lacking Blimp-1, compared to the plasmacytoma P3X expressing endogenous Blimp-1 (Fig. 3A). This difference was specific to Pax-5 since no difference in histone H3 acetylation between the two cell lines was observed in the control α-amylase-2 gene (Fig. 3A). To determine whether the deacetylation of histone H3 associated with the Pax-5 promoter in plasmacytoma cells was due to the action of Blimp-1, bicistronic retroviruses expressing Blimp-1 and puro resistance or a control virus expressing puro resistance were used to transduce 18-81 pre-B cells. After 2 days of puro selection, ChIP was again used to determine the acetylation status of histone H3. Histone H3 at the Blimp-1 site in the Pax-5 promoter was deacetylated in cells expressing Blimp-1 relative to control cells (Fig. 3B). Thus, the presence of either endogenous or ectopically expressed Blimp-1 correlates with the deacetylation of histone H3 near the Blimp-1 site of the Pax-5 promoter, a finding consistent with Blimp-1-dependent repression involving histone deacetylation.
FIG. 3.
Blimp-1 causes deacetylation of histone H3 at the Blimp-1 site of the Pax-5 promoter and binds the promoter in vivo. (A) Acetylation of histone H3 associated with the Blimp-1 binding site at Pax-5 promoter in 18-81 pre-B cells and P3X plasmacytoma cells. 18-81 and P3X cells were harvested, and chromatin was cross-linked prior to immunoprecipitation with anti-acetylated histone H3 antibody or control antibody. Immunoprecipitated chromatin was subjected to PCR analysis with primers specific for the Blimp-1 binding site at the Pax-5 promoter. Fourfold dilutions of immunoprecipitated chromatin or input chromatin were used in this experiment. (B) Ectopic expression of Blimp-1 causes the deacetylation of histone H3 associated with the Blimp-1 site in the Pax-5 promoter. 18-81 cells were infected with retrovirus, VxyPRDI-BF1-puro (Blimp-1), expressing human Blimp-1 and puro resistance or control virus (Puro), Vxy-puro, expressing puro resistance alone. Infected cells were selected by puro and purified for the ChIP assay. Fourfold serial dilutions of immunoprecipitated chromatin or input chromatin were used for PCR analysis. (C) Blimp-1 binds to Pax-5 promoter in vivo. 18-81 cells were infected with Vxy-puro (Puro) or VxyBlimpFLAG-puro (BlimpFLAG) viruses; puro-resistant cells were used for ChIP assay. Fourfold dilutions of immunoprecipitated chromatin or input chromatin were analyzed by PCR. (D) WI-L2 stable transfectants were induced to express the indicated FLAG-tagged protein. After 20 h of induction, cells were harvested for ChIP assay to assess the binding of Blimp-1 on the Pax-5 promoter in vivo. Twofold dilutions of immunoprecipitated chromatin or input chromatin were analyzed by PCR.
To investigate binding of Blimp-1 to the endogenous Pax-5 promoter, 18-81 cells were infected with a bicistronic retrovirus expressing FLAG-tagged Blimp-1 (BlimpFLAG) and puro resistance or with a control expressing puro resistance. (This approach was used because antibody capable of immunoprecipitating Blimp-1 is not currently available.) The expression of BlimpFLAG and its ability to bind to DNA were verified by the EMSA by using a probe for the Blimp-1 site on the c-myc promoter (data not shown). After puro selection, chromatin was prepared and then immunoprecipitated with anti-FLAG antibody, and DNA was then analyzed by PCR. The Blimp-1 sites on the Pax-5 promoter, the c-myc promoter, and a control site on the α-amylase-2 gene were amplified. BlimpFLAG bound to both the c-myc and the Pax-5 sites but not to the α-amylase site (Fig. 3C), showing that Blimp-1 binds to its cognate site on the Pax-5 promoter in vivo. We performed the same experiment with a WI-L2 stable transfectant, where the expression of a FLAG-tagged BlimpERD fusion protein is inducible by cadmium and 4-hydroxytomaxifen (Shaffer et al., unpublished). The expression of FLAG-tagged protein after 20 h of induction was confirmed by immunoblotting with anti-FLAG antibody (not shown). A transfectant carrying the empty plasmid with the hygromycin B resistance gene alone (Hygro) and a transfected line expressing another FLAG-tagged protein (FLAG control) were used as negative controls for the immunoprecipitation. The Blimp-1 site in CIITA promoter III was used as a positive control, and a region in the CSF-1 (colony-stimulating factor 1) gene was used as a negative control. FLAG-BlimpERD bound specifically to both the Pax-5 and CIITA promoters but not to the CSF-1 gene (Fig. 3D). In summary, the ChIP data provide strong support for the idea that Blimp-1 binds and represses the endogenous Pax-5 promoter in vivo.
Roles of BSAP and Blimp-1 during plasmacytic differentiation of primary cells.
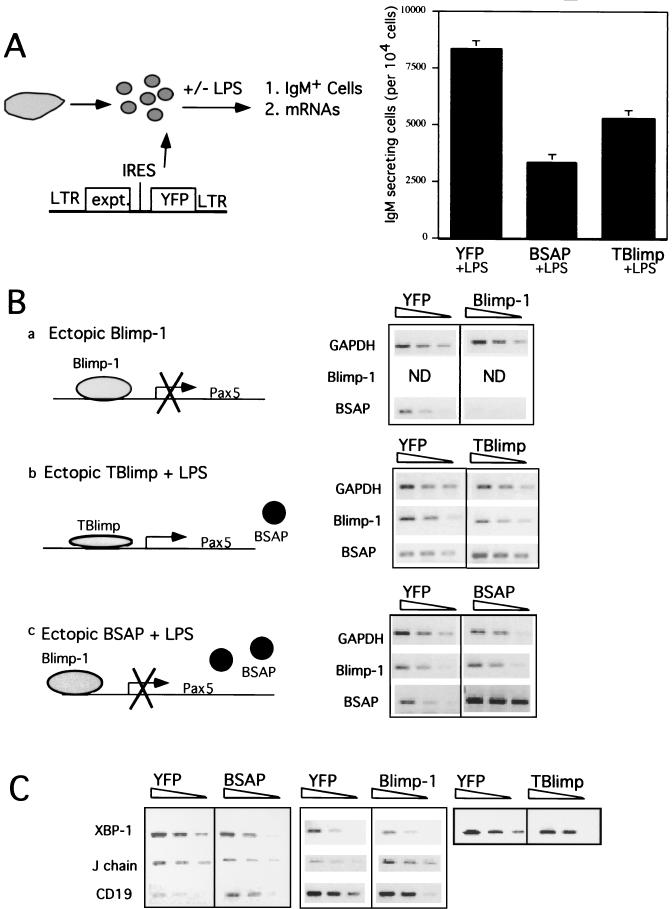
LPS treatment of murine splenic B cells induces Blimp-1 and drives plasmacytic differentiation (Fig. 1C) (2); furthermore, ectopic expression of Blimp-1 is sufficient to cause terminal differentiation of splenic B cells to IgM-secreting cells (19, 26; Shaffer et al., unpublished). We chose this model of plasmacytic differentiation of primary B cells to investigate the roles of Blimp-1 and BSAP. Bicistronic retroviruses expressing YFP and Blimp-1, TBlimp, or BSAP were used to infect primary murine splenic B cells, and their effects on differentiation and gene expression were observed. Splenic B cells were purified, activated, infected with retrovirus and, as indicated, treated with LPS as described previously (19). By using IgM secretion as an indication of plasmacytic differentiation, we confirmed previous results (19) showing that ectopic expression of Blimp-1 is sufficient to induce differentiation (data not shown). TBlimp is a truncated form of Blimp-1 that competes with full-length Blimp-1 for binding sites and blocks endogenous Blimp-1 activity (5, 34). TBlimp inhibited the formation of IgM-secreting cells after LPS treatment, confirming that endogenous Blimp-1 is required for plasmacytic differentiation in this system (Fig. 4A). Enforced expression of BSAP also inhibited the formation of IgM-secreting cells after LPS treatment (Fig. 4A). Thus, repression of Pax-5 is necessary for plasmacytic differentiation.
FIG. 4.
Effects of ectopic expression of Blimp-1, BSAP, and TBlimp on splenic B-cell terminal differentiation. (A) ELISPOT assay showing that ectopic expression of BSAP and TBlimp blocks the differentiation of murine splenocytes in response to LPS. Purified murine splenic B cells were infected with bicistronic retrovirus, pGC-BSAP-YFP, pGC-TBlimp-YFP, or control virus (pGC-YFP) and subsequently treated with LPS. Three days after virus infection, YFP+ cells were sorted and used for an ELISPOT assay. Error bars represent the standard deviations from two experiments each performed in triplicate. (B) Blimp-1 is necessary and sufficient for repression of Pax-5 during plasma cell differentiation. Purified splenic B cells were infected with retrovirus carrying Blimp-1 (pGC-Blimp-1-YFP) or control virus (pGC-YFP). Additionally, splenic B cells were cultured and infected with other retroviruses, pGC-BSAP-YFP (BSAP), pGC-TBlimp-YFP (TBlimp), and their control virus, pGC-YFP (YFP), as described in Materials and Methods. Three days after infection, YFP+ cells were sorted and used for semiquantitative RT-PCR analysis with primers specific for the indicated genes. GAPDH was used to control for equal amounts of cDNA. Fourfold dilutions of cDNA were used for PCR. Results are shown for ethidium bromide-stained PCR products separated by electrophoresis on a 1.5% agarose gel. Diagrams a to c show the rationale of the experimental strategies. (C) Blimp-1-dependent repression of Pax-5 alters mRNAs for BSAP downstream targets. cDNAs from panel B were used for PCR amplification with primers specific for CD19, J chain, and XBP-1 genes. GAPDH levels for the internal control in panel C were the same as in panel B.
Semiquantitative RT-PCR with RNA prepared from sorted YFP+ cells was used to determine how Blimp-1 and BSAP affected one another in these experiments (Fig. 4B). We reasoned that ectopic expression of Blimp-1 in splenic B cells would repress Pax-5 and other genes, revealing Blimp-1-dependent events (Fig. 4Ba). Indeed, consistent with our cotransfection studies, ectopic expression of Blimp-1 caused a significant (∼16-fold) decrease in BSAP mRNA in splenic B cells (Fig. 4B). Splenocytes treated with LPS but expressing TBlimp should reveal changes that require endogenous Blimp-1 (Fig. 4Bb). Consistent with repression of Pax-5 by ectopic Blimp-1, TBlimp blocked BSAP downregulation, showing that endogenous Blimp-1 is required for downregulation of BSAP in response to LPS. Finally, splenocytes treated with LPS but with forced expression of BSAP should reveal changes that require downregulation of BSAP (Fig. 4Bc). Importantly, ectopic expression of BSAP did not alter the induction of Blimp-1 mRNA, suggesting that prdm1 is not subject to feedback regulation by BSAP in the context of splenic B-cell differentiation (Fig. 4B). Since IgM secretion was inhibited by ectopically expressed BSAP (Fig. 4A) even though Blimp-1 was induced normally (Fig. 4B), these data show that plasmacytic differentiation cannot proceed in the absence of Pax-5 repression in this setting.
We also investigated the regulation of known BSAP target genes in this system (Fig. 4C). Ectopic expression of BSAP confirmed previous studies showing that BSAP activates CD19 and represses J chain and XBP-1 (8, 22, 25). In Blimp-1-expressing cells, where BSAP mRNA is downregulated (Fig. 4B), levels of mRNA for J chain are high and for CD19 are low, suggesting that Blimp-1-dependent repression of Pax-5 is sufficient to regulate BSAP targets CD19 and J chain. However, XBP-1 mRNA was not induced by ectopic expression of Blimp-1, suggesting that Blimp-1-dependent repression of Pax-5 is not sufficient to induce XBP-1 in this setting (Fig. 4C). However, treatment of murine splenocytes with LPS is known to induce XBP-1 (21) as well as Blimp-1. Therefore, we analyzed XBP-1 mRNA levels in splenocytes treated with LPS and expressing TBlimp to determine whether endogenous Blimp-1 is required for XBP-1 induction. XBP-1 mRNA levels were lower in TBlimp cells relative to controls (Fig. 4C), suggesting that endogenous Blimp-1 is required for XBP-1 induction. Thus, these results show that Blimp-1-dependent repression of Pax-5 is necessary for plasmacytic differentiation and plays a role in the regulation of BSAP target genes.
DISCUSSION
Repression of Pax-5 by Blimp-1.
The inverse expression pattern of BSAP and Blimp-1 in both human tonsillar B cells (Fig. 1A) and murine B cells induced to differentiate in vitro (Fig. 1B) supports the notion that Blimp-1 represses Pax-5. We have provided evidence that Blimp-1 binds from bp −926 to −917 on the mouse Pax-5 promoter in vitro (Fig. 2B and C) and in vivo (Fig. 3C and D). A probable Blimp-1 binding sequence, AGAGTGAATC, is present at a similar location in the human Pax-5 promoter at 992 to 983 bp 5′ to the transcription start site, suggesting conservation of Blimp-1-dependent repression between mice and humans. In addition, Pax-5 promoter activity is repressed by Blimp-1 in a site-dependent manner in cotransfections (Fig. 2D and E) and histones associated with the site are deacetylated in the presence of Blimp-1 in vivo. Thus, Pax-5 appears to be a direct target of Blimp-1. However, the element(s) responsible for B-cell-specific expression of Pax-5 is not known and may be distant from the transcription start site. Since Pax-5 regulatory elements are not fully characterized, the existence of an additional, currently unidentified, Blimp-1 response element(s) is a possibility.
Recent microarray studies have revealed that Blimp-1 represses a wide array of genes necessary for the identity and function of B cells. Consistent with the data reported here, Pax-5 was downregulated by Blimp-1 in those studies (Shaffer et al., unpublished). However, the microarray analyses also revealed that Blimp-1 downregulates early B-cell factor (EBF) and E2A. These transcription factors act together in early B-cell differentiation, and EBF directly activates Pax-5 transcription (18). Combining those results with the findings reported here, we suggest that Blimp-1 initiates a complex program of gene regulation that results in both direct repression of Pax-5 via binding at the bp site from −926 to −917 and indirect repression of Pax-5 due to the repression of EBF. This model is consistent with our observation that repression of Pax-5 by ectopic expression of Blimp-1 in vivo (Fig. 4A) was more robust than direct repression measured by cotransfection (Fig. 2D and E). It also emphasizes the importance of Pax-5 repression, which is consistent with our demonstration that downregulating BSAP is critical for the Blimp-1 program of plasmacytic differentiation (Fig. 4).
Pax-5 repression and plasma cell development.
The defect in repression of Pax-5 in splenic B cells expressing TBlimp, a blocking form of Blimp-1 (5, 34) (Fig. 4D), provides direct evidence that Blimp-1 is required for the repression of Pax-5 during LPS-dependent differentiation of splenic B cells. The importance of this repression was demonstrated by showing that enforced expression of BSAP blocks LPS-induced IgM secretion by splenic B cells (Fig. 4A). This observation is consistent with a previous study showing that overexpression of BSAP in the lymphoma line CH12.LX.A2 caused decreased immunoglobulin secretion (31). Furthermore, the data show that induction of Blimp-1 in the absence of Pax-5 repression is not sufficient to drive differentiation to IgM-secreting cells (Fig. 4C). Thus, our studies show that repression of Pax-5 is a critical component of the Blimp-1 program of plasma cell differentiation.
A combination of several mechanisms probably explains the requirement for Pax-5 repression in Blimp-1-dependent plasma cell differentiation. First, the ATF/CREB family protein XBP-1 has recently been shown to be required for plasma cell development and antibody secretion (21). BSAP represses XBP-1 (22), and our data (Fig. 4C) show that Blimp-1-dependent repression of Pax-5 is required, although it is not sufficient, for induction of XBP-1 in splenic B cells (Fig. 4B). Pax-5 also represses J chain transcription and immunoglobulin heavy-chain transcription via the 3′Cα enhancer; thus, it must be repressed to achieve abundant immunoglobulin expression and IgM secretion. Finally, Pax-5 promotes B-cell proliferation (32), and we have previously shown that enforced proliferation blocks plasma cell differentiation (9).
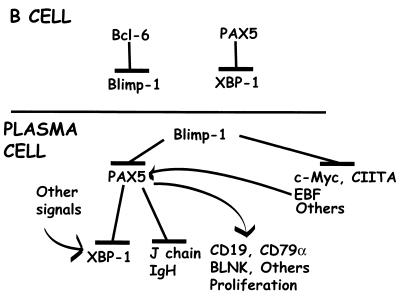
Combining our data with that of others, a putative regulatory cascade guiding development from B-cell to plasma cell can be formulated which illustrates a key role for BSAP (Fig. 5). In germinal center B cells, BCL-6 represses prdm1 (23, 27), thus inhibiting terminal differentiation while the germinal center reactions proceed. In addition, BSAP represses XBP-1 (22), providing additional inhibition of plasmacytic differentiation. When BCL-6 is downregulated (13), prdm1 repression is relieved and Blimp-1 drives commitment to a plasma cell fate (3). Blimp-1 represses multiple genes required for proliferation and B-cell identity and function (9, 10, 19; Shaffer et al., unpublished). One important target of Blimp-1 repression is Pax-5; Pax-5 is repressed both directly and indirectly via repression of EBF. Repression of Pax-5 allows induction of the XBP-1 (21, 22), immunoglobulin heavy-chain (28), and J-chain (25) genes required for IgM secretion. Decreased BSAP also decreases CD19 and signaling via BCR (15) and cell proliferation (32). Thus, repression of Pax-5 is both a critical component of Blimp-1's activity and a critical aspect of plasma cell differentiation.
FIG. 5.
Model for the regulation and activity of Blimp-1 and BSAP during plasmacytic differentiation.
Acknowledgments
We thank G. Siu and J. Piskurich for critically reading the manuscript and G. Cattoretti, B. Bishtein, G. Fathman, R. Grosschedl, and J. Wallin for providing reagents. We thank A. L. Shaffer for the WI-L2 stable transfectants, J. Liao and J. Weller for excellent technical assistance, and members of the Calame lab for helpful discussions.
This work was supported by the grants GM29361 and AI43576. K.-I. Lin is a fellow of the Leukemia and Lymphoma Society (5332-00).
REFERENCES
- 1.Adams, B., P. Dorfler, A. Aguzzi, Z. Kozmik, P. Urbanek, I. Maurer-Fogy, and M. Busslinger. 1992. Pax-5 encodes the transcription factor BSAP and is expressed in B lymphocytes, the developing CNS, and the adult testis. Genes Dev. 6:1589-1607. [DOI] [PubMed] [Google Scholar]
- 2.Angelin-Duclos, C., G. Cattoretti, D. Chang, K. Lin, Y. Lin, J. Yu, and K. Calame. 1999. The role of B lymphocyte-induced maturation protein-1 (BLIMP-1) in terminal differentiation of B cells and other cell lineages. Cold Spring Harbor Symp. Quant. Biol. 64:61-70. [DOI] [PubMed] [Google Scholar]
- 3.Angelin-Duclos, C., G. Cattoretti, K.-I. Lin, and K. Calame. 2000. Commitment of B lymphocytes to a plasma cell fate is associated with blimp-1 expression in vivo. J. Immunol. 165:5462-5471. [DOI] [PubMed] [Google Scholar]
- 4.Barberis, A., K. Widenhorn, L. Vitelli, and M. Busslinger. 1990. A novel B-cell lineage-specific transcription factor present at early but not late stages of differentiation. Genes Dev. 4:849-859. [DOI] [PubMed] [Google Scholar]
- 5.Chang, D., C. Angelin-Duclos, and K. Calame. 2000. BLIMP-1: trigger for differentiation of myeloid lineage. Nat. Immunol. 1:169-176. [DOI] [PubMed] [Google Scholar]
- 6.Keller, A. D., and T. Maniatis. 1991. Identification and characterization of a novel repressor of beta-interferon gene expression. Genes Dev. 5:868-879. [DOI] [PubMed] [Google Scholar]
- 7.Keller, A. D., and T. Maniatis. 1992. Only two of the five zinc fingers of the eukaryotic transcriptional repressor PRDI-BF1 are required for sequence-specific DNA binding. Mol. Cell. Biol. 12:1940-1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kozmik, Z., S. Wang, P. Dorfler, B. Adams, and M. Busslinger. 1992. The promoter of the CD19 gene is a target for the B-cell-specific transcription factor BSAP. Mol. Cell. Biol. 12:2662-2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin, K.-I., Y. Lin, and K. Calame. 2000. Repression of c-myc is necessary but not sufficient for terminal differentiation of B lymphocytes in vitro. Mol. Cell. Biol. 20:8684-8695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin, Y., K. Wong, and K. Calame. 1997. Repression of c-myc transcription by Blimp-1, an inducer of terminal B cell differentiation. Science 276:596-599. [DOI] [PubMed] [Google Scholar]
- 11.Max, E. E., Y. Wakatsuki, M. F. Neurath, and W. Strober. 1995. The role of BSAP in immunoglobulin isotype switching and B-cell proliferation. Curr. Top. Microbiol. Immunol. 194:449-458. [DOI] [PubMed] [Google Scholar]
- 12.Neurath, M. F., W. Strober, and Y. Wakatsuki. 1994. The murine Ig 3′ alpha enhancer is a target site with repressor function for the B cell lineage-specific transcription factor BSAP (NF-HB, S alpha-BP). J. Immunol. 153:730-742. [PubMed] [Google Scholar]
- 13.Niu, H., B. H. Ye, and R. Dalla-Favera. 1998. Antigen receptor signaling induces MAP kinase-mediated phosphorylation and degradation of the BCL-6 transcription factor. Genes Dev. 12:1953-1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nutt, S. L., B. Heavey, A. G. Rolink, and M. Busslinger. 1999. Commitment to the B-lymphoid lineage depends on the transcription factor Pax5. Nature 401:556-562. [DOI] [PubMed] [Google Scholar]
- 15.Nutt, S. L., A. M. Morrison, P. Dorfler, A. Rolink, and M. Busslinger. 1998. Identification of BSAP (Pax-5) target genes in early B-cell development by loss- and gain-of-function experiments. EMBO J. 17:2319-2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nutt, S. L., P. Urbanek, A. Rolink, and M. Busslinger. 1997. Essential functions of Pax5 (BSAP) in pro-B cell development: difference between fetal and adult B lymphopoiesis and reduced V-to-DJ recombination at the IgH locus. Genes Dev. 11:476-491. [DOI] [PubMed] [Google Scholar]
- 17.Okabe, T., S. R. Bauer, and A. Kudo. 1992. Pre-B lymphocyte-specific transcriptional control of the mouse VpreB gene. Eur. J. Immunol. 22:31-36. [DOI] [PubMed] [Google Scholar]
- 18.O'Riordan, M., and R. Grosschedl. 1999. Coordinate regulation of B cell differentiation by the transcription factors EBF and E2A. Immunity 11:21-31. [DOI] [PubMed] [Google Scholar]
- 19.Piskurich, J. F., K. I. Lin, Y. Lin, Y. Wang, J. P. Ting, and K. Calame. 2000. BLIMP-I mediates extinction of major histocompatibility class II transactivator expression in plasma cells. Nat. Immunol. 1:526-532. [DOI] [PubMed] [Google Scholar]
- 20.Qiu, G., and J. Stavnezer. 1998. Overexpression of BSAP/Pax-5 inhibits switching to IgA and enhances switching to IgE in the I.29 mu B cell line. J. Immunol. 161:2906-2918. [PubMed] [Google Scholar]
- 21.Reimold, A. M., N. N. Iwakoshi, J. Manis, P. Vallabhajosyula, E. Szomolanyi-Tsuda, E. M. Gravallese, D. Friend, M. J. Grusby, F. Alt, and L. H. Glimcher. 2001. Plasma cell differentiation requires the transcription factor XBP-1. Nature 412:300-307. [DOI] [PubMed] [Google Scholar]
- 22.Reimold, A. M., P. D. Ponath, Y. S. Li, R. R. Hardy, C. S. David, J. L. Strominger, and L. H. Glimcher. 1996. Transcription factor B cell lineage-specific activator protein regulates the gene for human X-box binding protein 1. J. Exp. Med. 183:393-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reljic, R., S. D. Wagner, L. J. Peakman, and D. T. Fearon. 2000. Suppression of signal transducer and activator of transcription 3-dependent B lymphocyte terminal differentiation by BCL-6. J. Exp. Med. 192:1841-1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ren, B., K. J. Chee, T. H. Kim, and T. Maniatis. 1999. PRDI-BF1/Blimp-1 repression is mediated by corepressors of the Groucho family of proteins. Genes Dev. 13:125-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rinkenberger, J. L., J. J. Wallin, K. W. Johnson, and M. E. Koshland. 1996. An interleukin-2 signal relieves BSAP (Pax5)-mediated repression of the immunoglobulin J chain gene. Immunity 5:377-386. [DOI] [PubMed] [Google Scholar]
- 26.Schliephake, D. E., and A. Schimpl. 1996. Blimp-1 overcomes the block in IgM secretion in lipopolysaccharide/anti-mu F(ab′)2-costimulated B lymphocytes. Eur. J. Immunol. 26:268-271. [DOI] [PubMed] [Google Scholar]
- 27.Shaffer, A. L., X. Yu, Y. He, J. Boldrick, E. P. Chan, and L. M. Staudt. 2000. BCL-6 represses genes that function in lymphocyte differentiation, inflammation, and cell cycle control. Immunity 13:199-212. [DOI] [PubMed] [Google Scholar]
- 28.Singh, M., and B. K. Birshtein. 1993. NF-HB (BSAP) is a repressor of the murine immunoglobulin heavy-chain 3′ alpha enhancer at early stages of B-cell differentiation. Mol. Cell. Biol. 13:3611-3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Turner, C. A., Jr., D. H. Mack, and M. M. Davis. 1994. Blimp-1, a novel zinc finger-containing protein that can drive the maturation of B lymphocytes into immunoglobulin-secreting cells. Cell 77:297-306. [DOI] [PubMed] [Google Scholar]
- 30.Urbanek, P., Z. Q. Wang, I. Fetka, E. F. Wagner, and M. Busslinger. 1994. Complete block of early B cell differentiation and altered patterning of the posterior midbrain in mice lacking Pax5/BSAP. Cell 79:901-912. [DOI] [PubMed] [Google Scholar]
- 31.Usui, T., Y. Wakatsuki, Y. Matsunaga, S. Kaneko, H. Kosek, and T. Kita. 1997. Overexpression of B cell-specific activator protein (BSAP/Pax-5) in a late B cell is sufficient to suppress differentiation to an Ig high producer cell with plasma cell phenotype. J. Immunol. 158:3197-3204. [PubMed] [Google Scholar]
- 32.Wakatsuki, Y., M. F. Neurath, E. E. Max, and W. Strober. 1994. The B cell-specific transcription factor BSAP regulates B cell proliferation. J. Exp. Med. 179:1099-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wallin, J. J., E. R. Gackstetter, and M. E. Koshland. 1998. Dependence of BSAP repressor and activator functions on BSAP concentration. Science 279:1961-1964. [DOI] [PubMed] [Google Scholar]
- 34.Yu, J., C. Angelin-Duclos, J. Greenwood, J. Liao, and K. Calame. 2000. Transcriptional repression by blimp-1 (PRDI-BF1) involves recruitment of histone deacetylase. Mol. Cell. Biol. 20:2592-2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zwollo, P., and S. Desiderio. 1994. Specific recognition of the blk promoter by the B-lymphoid transcription factor B-cell-specific activator protein. J. Biol. Chem. 269:15310-15317. [PubMed] [Google Scholar]