Abstract
Rad23 contains a ubiquitin-like domain (UbLR23) that interacts with catalytically active proteasomes and two ubiquitin (Ub)-associated (UBA) sequences that bind Ub. The UBA domains can bind Ub in vitro, although the significance of this interaction in vivo is poorly understood. Rad23 can interfere with the assembly of multi-Ub chains in vitro, and high-level expression caused stabilization of proteolytic substrates in vivo. We report here that Rad23 interacts with ubiquitinated cellular proteins through the synergistic action of its UBA domains. Rad23 plays an overlapping role with Rpn10, a proteasome-associated multi-Ub chain binding protein. Mutations in the UBA domains prevent efficient interaction with ubiquitinated proteins and result in poor suppression of the growth and proteolytic defects of a rad23Δ rpn10Δ mutant. High-level expression of Rad23 revealed, for the first time, an interaction between ubiquitinated proteins and the proteasome. This increase was not observed in rpn10Δ mutants, suggesting that Rpn10 participates in the recognition of proteolytic substrates that are delivered by Rad23. Overexpression of UbLR23 caused stabilization of a model substrate, indicating that an unregulated UbLR23-proteasome interaction can interfere with the efficient delivery of proteolytic substrates by Rad23. Because the suppression of a rad23Δ rpn10Δ mutant phenotype required both UbLR23 and UBA domains, our findings support the hypothesis that Rad23 encodes a novel regulatory factor that translocates ubiquitinated substrates to the proteasome.
A family of proteins that contain amino-terminal ubiquitin (Ub)-like (UbL) domains has been shown to play a role in proteolysis (12, 13, 20, 24, 33, 35). Rad23 is the best-characterized protein in this class, and Rad23 proteins of Saccharomyces cerevisiae and humans are biochemically similar (17, 18, 30, 35, 37). Rad23 binds DNA repair factors that play essential roles in nucleotide excision repair (NER) (29, 32, 37) and specifically interacts with Rad4 to facilitate the assembly of a DNA repair complex on UV-damaged DNA (19, 22). Extensive characterization of Rad23 proteins from yeast and humans showed that they promoted the assembly of DNA repair complexes (19, 22) and stimulated the activity of Rad4/XPC (36). We reported previously that the UbL domains in both yeast and human Rad23 could interact with catalytically active proteasomes (35). This interaction is important for DNA repair because removal of UbLR23 caused sensitivity to UV light (47), although this mutant retained the ability to bind Rad4 (P. Tongaonkar and K. Madura, unpublished results). However, a well-defined biochemical function for Rad23 was unavailable.
The addition of purified Rad23 protein to a reconstituted ubiquitination reaction mixture effectively blocked the formation of multi-Ub chains on a test substrate histone, H2B (30, 41). In agreement with this result, we found that the overexpression of Rad23 inhibited the degradation of test substrates in vivo (30). Additionally, it has been shown that overexpression of Rad23 causes stabilization of Pds1 (8). The discovery that Rad23 can bind Ub (5, 7), multi-Ub chains (33, 48), and ubiquitinated proteins (30) provides a mechanistic basis for these findings.
Rad23 contains two Ub-associated (UBA) domains (21, 43) that play an important role in regulating the abundance of the cell cycle regulator Pds1 (5, 8). Recent studies showed that the UBA domains in Rad23, as well as Ddi1 and Dsk2, could interact with Ub and multi-Ub chains (5, 7, 13, 33, 48). Interestingly, Dsk2 and Ddi1 also contain UbL domains, and double mutants (rad23Δ dsk2Δ and rad23Δ ddi1Δ) display growth defects that are not apparent in the single-mutant strains (6, 8). Similar to results from our studies with Rad23 (30, 35), the Dsk2 protein was recently reported to bind ubiquitinated proteins and the proteasome in yeast cells (13, 33). Collectively, these findings suggest that proteins that contain both UbL and UBA domains represent a novel class of proteolytic regulators that have similar and partially redundant functions.
Rad23 has a role that overlaps with that of Rpn10 (26), a subunit in the proteasome that binds multi-Ub chains (44). Loss of both proteins (rad23Δ rpn10Δ) resulted in pleiotropic growth and proteolytic defects that included extreme sensitivity to cold temperatures and inability to grow in the presence of amino acid analog canavanine (26). In addition, ubiquitinated proteins accumulated in the double mutant, suggesting a failure in their delivery to the proteasome. In agreement with this conjecture, we found that ΔUbLrad23 (a mutant lacking the UbL domain) could bind ubiquitinated proteins but was unable to alleviate any of the defects of a rad23Δ rpn10Δ mutant, underscoring the importance of the Rad23-proteasome interaction.
We report here that Rad23 can bind ubiquitinated cellular proteins. Significantly, this interaction required the UBA domains, since specific mutants were deficient in binding ubiquitinated cellular proteins. Because Rad23 mutants that lacked either UbLR23 or functional UBA domains failed to suppress the defects of rad23Δ rpn10Δ mutants, we conclude that the interactions with both ubiquitinated proteins and the proteasome are required for Rad23 function. High-level expression of Rad23 resulted in a dramatic increase in the levels of ubiquitinated proteins in the proteasome. Furthermore, the increased association between ubiquitinated proteins and the proteasome required Rpn10. These intriguing results are to our knowledge the first evidence that reveals the interaction between ubiquitinated proteins and the proteasome. Collectively, our results provide compelling support for a “shuttle factor hypothesis,” wherein Rad23 and other proteins that contain UbL and UBA domains, such as Dsk2 (6) and Ddi1 (8), regulate the translocation of proteolytic substrates to the proteasome. As a further test of this idea, we found that overexpression of GST-UbLR23 resulted in significant stabilization of a test substrate, suggesting that unregulated interaction between UbLR23 and the proteasome can interfere with the efficient delivery of proteolytic substrates by Rad23.
MATERIALS AND METHODS
Yeast strains and plasmids.
A bar1Δ mutant was constructed with plasmid pJGSST1 in haploid yeast strain JD47-13C (MATa his3-Δ200 trp1-Δ63 lys2-801 ura3-52 leu2-112), which was kindly provided by J. Dohmen. The yeast RAD23 gene was deleted in JD47-13C by using plasmid pDG28, which was provided by R. D. Gietz. KMY1188 is an 5-fluoroorotic acid-resistant derivative of this rad23Δ strain. Plasmids expressing Flag-Rad23 and the set of glutathione S-transferase (GST)-Rad23 fusion proteins were described previously (35).
Protein and immunological methods.
Pulse-chase and immunoblotting experiments were performed essentially as described previously (2, 30). Briefly, exponential-stage yeast cells (∼30 ml; A600 = 1) were washed and suspended in labeling buffer containing 250 μCi of a [35S]Met-[35S]Cys (NEN/Dupont) combination and incubated for 5 min at 30°C. The incorporation of radioisotope was terminated by suspending the cells in medium containing 0.5 mg of cycloheximide/ml and excess unlabeled methionine and cysteine. Aliquots were withdrawn at intervals and frozen in liquid nitrogen. After aliquots from all the time points were collected, 0.5-mm acid-washed glass beads (Sigma Chemical Co., St. Louis, Mo.) were added and the cells were lysed by vortexing. The incorporation of 35S label into trichloroacetic acid (TCA)-insoluble material was determined by scintillation counting, and equal amounts of protein were adjusted to equal volume and incubated with specific antibodies. Immunoprecipitations were typically carried out for 3 h at 4°C with constant mixing. The bound proteins were washed, resuspended in sodium dodecyl sulfate (SDS)-sample buffer, boiled, and resolved in SDS-containing polyacrylamide gels. Experiments involving the characterization of β-galactosidase (β-Gal) test proteins were performed using 8% polyacrylamide gels, while 10% acrylamide gels were used in all other experiments. The separated proteins were transferred to nitrocellulose with a Hoefer semidry transfer apparatus and blocked with 5% milk powder. Antibodies against β-Gal were purchased from Promega Inc. (Madison, Wis.). Anti-Flag, anti-Ub, and anti-GST antibodies were purchased from Sigma Chemical Co. Antibodies against GST-Rad23 were prepared by Pocono Rabbit Co. For immunoblot detection, an enhanced chemiluminescence kit was purchased from NEN/Dupont. Protein A-Sepharose was purchased from Repligen Inc. (Cambridge, Mass.), and GST-Sepharose was purchased from Amersham/Pharmacia Biotech (Piscataway, N.J.). Protein interactions were quantitated with Kodak 1D-3.5 software.
Cell-cycle analysis.
The growth of yeast bar1-1 mutant cells (4910-3-3a; MATa cdc7-4 his7 ura1 bar1) was arrested by adding 10 ng of alpha-factor (Sigma)/ml. The efficiency of arrest was determined microscopically, and after greater than 95% of the cells had arrested in G1 they were resuspended in fresh medium lacking alpha-factor to resume mitotic growth. An aliquot of the culture was withdrawn immediately after resuspension and suspended for 5 min in labeling buffer containing 200 μCi of a [35S]Met-[35S]Cys combination. Further incorporation of 35S label was terminated by the addition of cycloheximide, and the cells were incubated for further 60 min (chase). Equal-volume aliquots were removed during the chase after 0, 10, and 60 min and frozen in liquid nitrogen. Similar pulse-chase experiments were performed at 30-min intervals five additional times during the cell cycle. Equal amounts of TCA-insoluble material were reacted with antibodies against native Rad23, and the precipitated proteins were separated by SDS-polyacrylamide gel electrophoresis (PAGE), transferred to nitrocellulose, and exposed to X-ray film. The same filter was subsequently incubated with antibodies against Ub, and the cross-reacting material was detected by enhanced chemiluminescence.
Construction of RAD23 mutations.
DNA oligonucleotides to delete the UBA1 domain or to generate mutations in UBA1 and UBA2 domains of yeast RAD23 were synthesized. Oligonucleotides complementary to the 5′ and 3′ ends of the RAD23 gene contained EcoRI and KpnI DNA restriction sites for cloning into a yeast expression vector that encoded a Flag epitope. However, for expression of the same RAD23 alleles in E. coli we amplified the DNA with oligonucleotides that contained NcoI and BamHI DNA restriction sites on the 5′ and 3′ ends and cloned the DNA into pET11d (Novogen, Inc.). The oligonucleotides used for generating the UBA mutations are shown below, and the mutated genes were subjected to DNA sequencing for confirmation. The primers used for generating a deletion of UBA1, described previously (30), were 461 (5′-GAATATGCACTGATGGGTATTCCAG-3′), 462 (5′-GAATACCCATCAGTGCATATTCCACCGC-3′), and 470 (5′-GCGGATCCTCAGTCGGCATGATCGCTGAATGCGATATTTGCTGCAGC-3′)
RESULTS
Rad23 binds ubiquitinated proteins.
In previous studies we and others showed that Rad23 could bind free Ub (5, 7), although it was probable that ubiquitinated proteins were its natural targets. High-level expression of Rad23 led to the accumulation of ubiquitinated proteins in yeast cells (Fig. 1A, lanes 3 and 4), while a control strain that did not overexpress Rad23 (lane 1) and one that overexpressed Rpn10 (lane 2) did not display this effect. We considered it less likely that Rad23 promoted the accumulation of unanchored multi-Ub chains because we found that high-level expression of Rad23 resulted in the stabilization of test substrates (30). Therefore, we believe that the accumulation of anti-Ub cross-reacting material in extracts prepared from cells that overexpressed Rad23 represents ubiquitinated proteins. Overexpression of Rad23 has also been reported to result in the stabilization of the cell cycle regulator Pds1 (8).
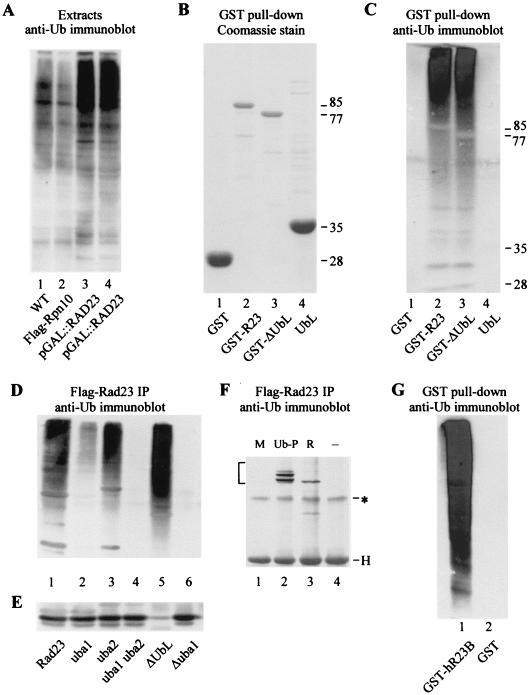
FIG. 1.
Rad23 binds ubiquitinated proteins through UBA domains. (A) Protein extracts were prepared from yeast cells that expressed Flag-Rpn10 from the copper-inducible CUP1 promoter (lane 2) or Rad23 from the galactose-inducible GAL1 promoter (lanes 3 and 4, independent preparations). Extracts were also prepared from a wild-type control strain (WT), which did not overexpress either protein (lane 1). Equal amounts of protein were resolved in an SDS-10% polyacrylamide gel, transferred to nitrocellulose (Bio-Rad), and incubated with anti-Ub antibodies (Sigma). The immunoblot (representing the entire SDS-polyacrylamide gel) was developed with enhanced chemiluminescence (NEN/Dupont). (B) GST (lane 1) and GST fusion proteins that expressed full-length Rad23 (lane 2), ΔUbLrad23 (lane 3), and the UbLR23 domain (lane 4) were purified from yeast extracts on glutathione-Sepharose (Amersham/Pharmacia Biotech) and are shown by Coomassie staining. The sizes of the various proteins are indicated at the right. The faint bands that are visible in lane 4 represent proteasome subunits that were coprecipitated with UbLR23. (C) An immunoblot that contained the purified GST fusion proteins in the order described for panel B was incubated with anti-Ub antibodies. The image represents the entire SDS-10% polyacrylamide gel, and the approximate positions of the fusion proteins are indicated. (D) Flag-Rad23 (lane 1) and derivatives that contained defective UBA domains were expressed in yeast and immunoprecipitated (IP) on Flag-agarose (Sigma). The proteins that were bound to the Rad23 proteins were separated by SDS-PAGE, transferred to nitrocellulose, and incubated with antibodies against Ub. Lanes 2 to 6, Flag-rad23uba1, Flag-rad23uba2, Flag-rad23uba1uba2, Flag-ΔUbLrad23, and Flag-rad23Δuba1, respectively. (E) The blot shown in panel D was stripped and incubated with antibodies against Cim5/Rpt1 to examine the interaction between the Rad23 derivatives and the proteasome. The faint band seen in lane 5 reflects a weak cross-reaction against the heavy chain of the anti-Cim5/Rpt1 antibodies, which comigrated with Cim5. (F) A yeast strain expressing Flag-Rad23 was transformed with plasmids expressing Met-β-Gal (M; lane 1), Ub-Pro-β-Gal (Ub-P; lane 2), or Arg-β-Gal (R; lane 3), and protein extracts were incubated with Flag-agarose. The bound proteins were resolved by SDS-PAGE and examined by immunoblotting with antibodies against β-Gal. Lane 4, yeast extract that contained Flag-Rad23, but none of the β-Gal proteins (control). Bracket, positions of the β-Gal proteins that were copurified with Flag-Rad23; ∗, nonspecific reaction against cellular protein; H, electrophoretic positions of the heavy chain of the anti-β-Gal antibodies. (G) Human hHR23-B was expressed in yeast cells as a fusion to GST and purified on glutathione-Sepharose (lane 1). Lane 2, extracts from a yeast strain that expressed GST. The bound proteins were resolved in a 10% polyacrylamide gel, and an immunoblot was incubated with antibodies against Ub. The image represents the entire SDS-10% polyacrylamide gel.
We had previously generated a set of GST-Rad23 fusion proteins from yeast cells (35) (Fig. 1B) to examine their interaction with the proteasome. We used these reagents here to examine the interaction between Rad23 and ubiquitinated proteins. Equal amounts of extract were incubated with glutathione-Sepharose, and the proteins that were bound to the GST fusion proteins were examined in an immunoblot with antibodies against Ub (Fig. 1C). Both full-length Rad23 (lane 2) and a mutant that lacked the N-terminal UbL domain (ΔUbLrad23; lane 3) formed a strong interaction with ubiquitinated proteins. In contrast, ubiquitinated proteins were not copurified with GST-UbLR23 (lane 4), although this domain forms a strong interaction with the proteasome, which recognizes and degrades ubiquitinated proteins. It is likely that ubiquitinated substrates are normally not detected in association with the proteasome because they are rapidly degraded. Similar to GST-UbLR23, ubiquitinated proteins were not purified with GST (lane 1).
UBA domains in Rad23 mediate the interaction with ubiquitinated proteins.
The UBA domains in Rad23 (43) and other proteins bind Ub (5, 7, 13, 33, 48). In addition, ubiquitinated proteins could be coprecipitated with Rad23 and Dsk2 from yeast cell extracts (13, 30). To further characterize the interaction between the UBA domains in Rad23 and Ub, we generated a set of mutants that were based on studies described previously (5). We deleted UBA1 in Rad23 and also generated mutants that contained single amino acid substitutions in each UBA domain to prevent their interaction with Ub. Each derivative was expressed with an amino-terminal Flag epitope to facilitate rapid purification from yeast cells. (Flag-Rad23 can functionally replace the native protein.) In agreement with the results in Fig. 1C, ubiquitinated proteins were efficiently copurified with Flag-Rad23 (Fig. 1D, lane 1). However, a single amino acid substitution in UBA1 (L183A; rad23uba1) caused an 80% reduction in the amount of ubiquitinated protein that was recovered (lane 2). In contrast, a similar mutation in UBA2 (L392A; rad23uba2) decreased the binding by only 20% (lane 3), consistent with a weaker interaction with Ub, which was previously described (5, 7). A Rad23 mutant that contained mutations in both UBA domains (rad23uba1uba2) was completely defective (lane 4), and the interaction with ubiquitinated proteins was ∼1% of that detected with the wild-type protein. In contrast to the single amino acid substitution in UBA1 (rad23uba1), deletion of the entire domain (rad23Δuba1; removal of residues 141 to 190) caused a very severe defect (lane 6), and the interaction with ubiquitinated proteins was ∼1% of the level seen with the wild-type protein, despite the presence of intact UBA2. Since the UBA domains in Rad23 have been reported to participate in the formation of homodimers (4, 5), it is possible that they contribute to the conformational integrity of the protein, and a deletion could cause significant structural perturbations. Similarly, dimerization between Rad23 and other UBA-containing proteins (33) might also contribute to its stabilization. Removal of UbLR23 (ΔUbLrad23), which is believed to form an independently folded domain (35; our unpublished studies), did not affect the interaction with ubiquitinated proteins (lane 5). The same filter was also incubated with antibodies against Cim5/Rpt1 (35), one of the six ATPases present in the 19S regulatory particle of the proteasome (16). With the exception of ΔUbLrad23 (Fig. 1E, lane 5), all the Rad23 derivatives contained UbLR23 and were able to bind the proteasome (Fig. 1E). The ability of the ΔUbLrad23 mutant to bind ubiquitinated proteins but not the proteasome is consistent with the idea that shuttle factors deliver proteolytic substrates to the proteasome. We examined the interaction between Flag-Rad23 and well-characterized proteolytic substrates to establish that the anti-Ub cross-reacting material that coprecipitated with Flag-Rad23 included proteolytic substrates (and not just unanchored multi-Ub chains). Plasmids that expressed the test substrates Ub-Pro-β-Gal and Arg-β-Gal and the control protein Met-β-Gal were transformed into a yeast strain that harbored the Flag-Rad23-expressing construct. Yeast extracts were prepared and applied to Flag-agarose beads, and the interaction with the β-Gal substrates was determined by immunoblotting (Fig. 1F). As expected, we found that well-characterized proteolytic substrates Ub-Pro-β-Gal and Arg-β-Gal (2, 45) coprecipitated with Flag-Rad23 (lanes 2 and 3) but not with Met-β-Gal, which is not targeted for degradation (lane 1). The human counterparts of Rad23 (hHR23-A and hHR23-B) can also bind the proteasome (35) and block the assembly of substrate-linked multi-Ub chains (30). To further assess their similarity with the yeast protein, we expressed GST-hHR23-B in yeast and found that ubiquitinated proteins were copurified (Fig. 1G, lane 1). Since the sequence and structure of Ub are highly conserved between yeast and humans, these results suggest that the UBA domains in Rad23 proteins play an evolutionarily conserved role in binding ubiquitinated proteins.
UBA mutants fail to stabilize substrates in vivo.
We next investigated if mutations in the UBA domains had an effect on the biochemical properties of Rad23. We first measured the binding between the mutant Rad23 proteins and proteolytic substrates Arg-β-Gal and Ub-Pro-β-Gal. Flag-Rad23 and Flag-rad23uba2 could efficiently coimmunoprecipitate ubiquitinated Ub-Pro-β-Gal (Fig. 2A, lanes 2 and 11) and Arg-β-Gal (lanes 3 and 12) but not the control substrate Met-β-Gal (lanes 1 and 10). In contrast, Rad23 derivatives that contained mutations in UBA1 were significantly more impaired in binding these substrates (Fig. 2A, lanes 4 to 9 and 13 to 15) in proportion to their ability to interact with ubiquitinated cellular proteins (Fig. 1D). In contrast to Flag-rad23uba2, Flag-rad23Δuba1 and Flag-rad23uba1uba2 interacted poorly with Arg-β-Gal and Ub-Pro-β-Gal. In the experiment shown in Fig. 2A, the cellular levels of Flag-rad23uba1, Flag-rad23uba2, and Flag-rad23uba1uba2 mutant proteins were approximately threefold higher than the level of Flag-Rad23, accounting for the higher levels of bound substrates. (Figure 2B is a Ponceau-S-stained filter that was subsequently immunoblotted in Fig. 2A.) For reasons that are not clear, the single amino acid substitution in rad23uba2 resulted in a noticeable decrease in electrophoretic mobility (Fig. 2B, lanes 10 to 15). However, it should be noted that native Rad23 also displays aberrant mobility in SDS-polyacrylamide gels, as it is typically detected as a 58- to 60-kDa protein despite a predicted size of 42 kDa. (A cross-reacting band that comigrated with Met-β-Gal was detected by immunoblotting, but not following pulse-chase analysis and immunoprecipitation, as shown in Fig. 2C.)
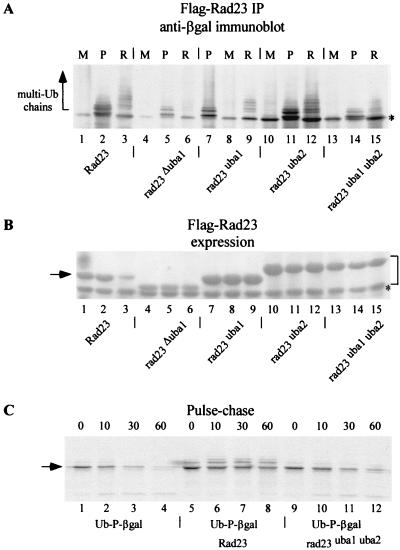
FIG. 2.
The UBA domains in Rad23 are required for binding substrates and inhibiting degradation. (A) Yeast strains expressing Met-β-Gal (M), Ub-Pro-β-Gal (P), and Arg-β-Gal (R) were transformed with plasmids encoding Flag-Rad23, Flag-rad23Δuba1, Flag-rad23uba1, Flag-rad23uba2, and Flag-rad23uba1uba2. All the Rad23 proteins were expressed well in yeast cells, and the levels of Flag-rad23uba1, Flag-rad23uba2, and Flag-rad23uba1uba2 were approximately threefold higher than that of Flag-Rad23 (see panel B). Equal amounts of protein extracts were incubated with Flag-agarose, and the bound material was examined in an immunoblot with antibodies against β-Gal (Promega). Both Ub-Pro-β-Gal and Arg-β-Gal interacted with Flag-Rad23, while the UBA mutants were impaired to various degrees in their interaction with these proteolytic substrates. Weak cross-reaction against a nonspecific protein that comigrated with Met-β-Gal was detected by immunoblotting (asterisk) but not by immunoprecipitation (see panel C). Arrow, positions of multiubiquitinated proteolytic substrates. (B) Expression of Flag-Rad23 and the various UBA mutants in yeast cells. The filter shown in panel A was initially stained with Ponceau-S, and the image revealed that Flag-rad23uba1, Flag-rad23uba2, and Flag-rad23uba1uba2 were expressed well in yeast cells and were present at higher levels than Flag-Rad23 (lanes 1 to 3; arrow). Also, the mutation in UBA2 caused an aberrant decrease in the electrophoretic mobility of Rad23 mutant proteins (bracket), and the reason for this is not known. (C) Pulse-chase measurements were performed to measure the stability of Ub-Pro-β-Gal in strains that expressed either Flag-Rad23 or Flag-rad23uba1uba2. The first set of pulse-chase measurements (lanes 1 to 4) represents the stability of Ub-Pro-β-Gal in a wild-type strain (arrow). The expression of high levels of Rad23 (from the PCUP1 promoter) resulted in stabilization of Ub-Pro-β-Gal (lanes 5 to 8), which was ligated to Ub. In contrast, expression of Flag-rad23uba1uba2 had no effect on Ub-Pro-β-Gal stability (lanes 9 to 12), consistent with its inability to bind this substrate efficiently.
The overexpression of Rad23 causes stabilization of proteolytic substrates in yeast cells. We therefore measured protein stability by pulse-chase methods to confirm that the failure of Flag-rad23uba1uba2 to bind ubiquitinated proteins would result in constitutive degradation of Ub-Pro-β-Gal. As expected, Ub-Pro-β-Gal was efficiently ubiquitinated and degraded in a wild-type cell (half-life [t1/2], ∼7 min; Fig. 2C, lanes 1 to 4), while overexpression of Flag-Rad23 prevented the expansion of multi-Ub chains on Ub-Pro-β-Gal (lanes 5 to 8), resulting in strong stabilization (t1/2, >100 min). In contrast to Flag-Rad23, Flag-rad23uba1uba2 did not inhibit the multiubiquitination and degradation of Ub-Pro-β-Gal (t1/2, ∼10 min; lanes 9 to 12), demonstrating that the interaction between UBA domains in Rad23 and Ub moieties on a substrate is important for controlling protein stability.
Rad23 forms transient interactions with ubiquitinated proteins.
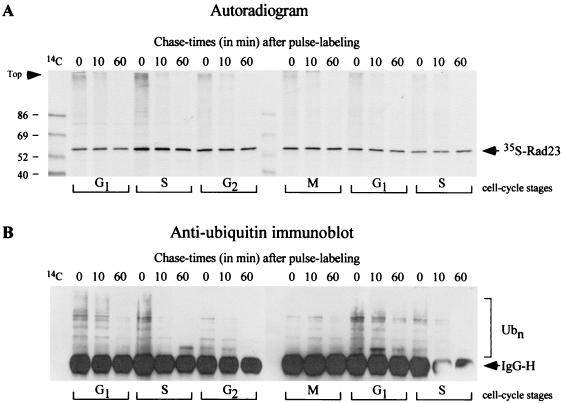
In previous studies we reported that Rad23 and another DNA repair protein, Rad4, could be cofractionated with the proteasome (35). Based on this result we speculated that Rad23 might translocate proteolytic substrates to the proteasome, although it is not known if Rad4 is a substrate for degradation. We also observed that high-level expression of Rad23 resulted in the stabilization of well-characterized proteolytic substrates in vivo (30). One interpretation of this result is that, at high-level expression, free Rad23 (representing the fraction that is not bound to substrate) binds the proteasome and interferes with the efficient delivery of proteolytic substrates, resulting in their stabilization. However, at physiological levels of expression, Rad23 might form transient interactions with substrates to facilitate their delivery to the proteasome. This conjecture is supported by the ability of Rad23 to bind Ub (5, 7) and multi-Ub chains (33, 48) and its overlapping functions with Rpn10 (26). To test this idea, we measured the interaction between Rad23 and ubiquitinated proteins in a time course assay. The growth of yeast cells was inhibited in G1 phase by the addition of alpha-mating factor to the medium (27). The culture was released from G1 arrest and allowed to grow synchronously for approximately two generations, during which time we conducted pulse-chase analysis at 30-min intervals to measure the interaction between ubiquitinated proteins and native Rad23 protein. We found that Rad23 formed an association with high-molecular-weight, 35S-labeled material throughout the cell cycle (Fig. 3A, zero time point lanes). As noted previously, native Rad23 is stable (47), despite its interaction with the proteasome (35). When the same filter was incubated with anti-Ub antibodies, we detected a strong reaction against high-molecular-weight species (Fig. 3B), consistent with the hypothesis that Rad23 forms transient interactions with cellular proteins that are ubiquitinated. Significantly, the interaction between Rad23 and ubiquitinated proteins decreased rapidly during the chase (compare intensities in the 0- and 60-min lanes). We speculate that this decrease in anti-Ub cross-reacting material might reflect proteasome-mediated degradation of the proteins that are bound to Rad23. However, we cannot discount the possibility that the decrease results from the dissociation of the bound ubiquitinated proteins or the action of de-ubiquitinating enzymes.
FIG. 3.
Rad23 binds transiently to ubiquitinated proteins during the mitotic cell cycle. (A) A yeast bar1-1 mutant was incubated with alpha-factor, and growth was arrested in G1 (27). The cells were washed extensively and resuspended in fresh medium, and aliquots were withdrawn at 0, 30, 60, 90, 120, and 150 min, corresponding approximately to the cell cycle stages indicated at the bottom. Each aliquot was subjected to pulse-chase analysis with 35S label, and native Rad23 was immunoprecipitated from equal amounts of labeled protein by using polyclonal antibodies. The precipitated proteins were separated by SDS-PAGE and transferred to nitrocellulose, and the filter was exposed to X-ray film. The position of 35S-labeled Rad23 is shown. The presence of high-molecular-weight 35S-labeled material, which was bound to Rad23 during the cell cycle, is particularly noticeable in the 0-min sample of each pulse-chase. (B) To confirm that the high-molecular-weight material represented ubiquitinated proteins, the same filter was incubated with antibodies against Ub and detection was by enhanced chemiluminescence. The polyclonal anti-Ub antibodies reacted strongly against the heavy chain of the polyclonal anti-Rad23 antibodies (arrow). IgG-H, immunoglobulin heavy chain. The bracket on the right indicates high-molecular-weight Ub-cross-reacting material. 14C molecular weight protein standards are present in the left lane and are detected by autoradiography (A).
The interaction between Rad23 and ubiquitinated proteins increased following DNA damage.
The removal of UbLR23 from Rad23 prevents interaction with the proteasome (35) and causes sensitivity to DNA damage (47), suggesting that Rad23 has a proteolytic role in its diverse biological activities, including DNA repair. Additionally, genetic results showed that Rad23 plays an role overlapping with that of Rpn10 (26), a proteasome-associated multi-Ub chain-binding factor (44), and physical interactions between Rad23 and subunits in the catalytic particle of the proteasome have been reported (42). However, recent studies have indicated that the UBA domains are not required for the NER-specific functions of Rad23 (5). It was also proposed that during NER the interaction between Rad23 and the proteasome does not involve proteolysis (14, 34). The emerging evidence, which suggests that Rad23 interacts with ubiquitinated cellular proteins (5, 7, 33, 48), is consistent with a role in proteolysis, although this conjecture will have to be confirmed with a physiological substrate. It is relevant in this regard that Rad23 was recently reported to interact specifically with multi-Ub chains (33, 48) that have been shown to promote substrate degradation by the proteasome (23, 25). If Rad23 binds the distal end of a multi-Ub chain, it would likely prevent further conjugation of Ub, consistent with our previous studies (30). The interaction between Rad23 and ubiquitinated proteins (Fig. 1C) might prevent deubiquitination, although subsequent translocation to the proteasome by Rad23 is expected to promote substrate degradation. These contrasting activities of Rad23 could allow a constitutively degraded protein to be transiently stabilized to set in motion specific biochemical functions. This form of regulation might be particularly beneficial for proteins that cause cellular toxicity or for other deleterious effects.
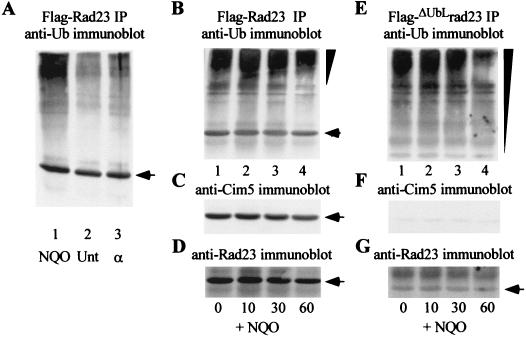
To explore the link between NER and proteolysis, we examined Rad23 interaction with ubiquitinated proteins following DNA damage. An actively growing culture of yeast cells was exposed to the UV mimetic 4-nitrosoquinoline oxide (4-NQO) or to alpha-mating factor. Protein extracts were prepared, and Flag-Rad23 was precipitated. The proteins were separated by SDS-PAGE, transferred to nitrocellulose, and incubated with antibodies against Ub. Equivalent amounts of ubiquitinated proteins were bound to Flag-Rad23 in untreated and alpha-factor-treated cells (Fig. 4A, lanes 2 and 3). In contrast, the interaction between Rad23 and ubiquitinated proteins increased approximately twofold after treatment with 4-NQO (lane 1). (The quantitation of these results was adjusted for the amount of Flag-Rad23 that was precipitated.) The relatively modest increase in interaction with ubiquitinated proteins following DNA damage may reflect the fact that Rad23 participates in diverse pathways including stress response, cell cycle control, and DNA repair. Therefore, a twofold increase may be quite significant if it arises exclusively from increased levels of DNA repair-specific targets. To further characterize the effect of DNA damage, we compared the binding between ubiquitinated proteins and either Flag-Rad23 or Flag-ΔUbLrad23, a mutant that causes a defect in DNA repair (47). Yeast cells that expressed either Flag-Rad23 or Flag-ΔUbLrad23 were exposed to 4-NQO for 60 min and then resuspended in medium lacking this DNA-damaging agent. Aliquots of the culture were withdrawn after 0, 10, 30, and 60 min, and protein extracts were prepared. Equal amounts of protein were incubated with Flag-agarose, and the ubiquitinated proteins that were bound to Rad23 were detected by immunoblotting (Fig. 4B and E). We found that the overall amount of ubiquitinated proteins that was bound to Rad23 decreased over the 60-min duration. Intriguingly, we observed that the ubiquitinated species that were bound to Flag-Rad23 were redistributed toward higher-molecular-weight forms during the course of the incubation (Fig. 4B). The interaction between Rad23 and the proteasome was unaffected, as revealed by the precipitation of Cim5/Rpt1 (Fig. 4C). In contrast to these results, the ubiquitinated species that were bound to Flag-ΔUbLrad23 were more heterogeneous in size (Fig. 4E), and we did not observe a significant redistribution to higher-molecular-weight forms (Fig. 4E). As expected, Flag-ΔUbLrad23 did not show an appreciable interaction with the proteasome (Fig. 4F). The filters used for Fig. 4D and G (which are the same as those used for Fig. 4B and E) were probed with anti-Rad23 antibodies, and the positions of Flag-Rad23 and Flag-ΔUbLrad23 are indicated. Since substrate-ubiquitinating factors are present in the proteasome (40, 46, 49), these findings raise the possibility that the complete ubiquitination of some proteolytic substrates might occur after their translocation to the proteasome by Rad23.
FIG. 4.
DNA damage results in increased interaction between Rad23 and ubiquitinated proteins. (A) A bar1Δ mutant was exposed to either 4-NQO (2 μg/ml) or alpha-factor (50 ng/ml) for 2 h, and protein extracts were prepared. Equal amounts of protein were incubated with Flag-agarose to precipitate Flag-Rad23, and the amount of ubiquitinated protein that was recovered was estimated by immunoblotting. Lane 1, extracts prepared from 4-NQO-treated cells; lanes 2 and 3, untreated and alpha-factor-treated cells, respectively. Arrows indicate the position of Flag-Rad23 (A to D) and Flag-ΔUbLrad23 (G). We used the KODAK-1D imaging system to quantitate the levels of Flag-Rad23 to estimate the interaction with ubiquitinated proteins following DNA damage. (B) A wild-type yeast strain expressing Flag-Rad23 was incubated with 4-NQO for 1 h and then washed and resuspended in fresh medium. Aliquots were withdrawn from this culture after 0, 10, 30, and 60 min, extracts were prepared, and Flag-Rad23 was precipitated to examine its interaction with ubiquitinated proteins. Ub-cross-reacting material was detected, and its distribution showed a noticeable shift toward higher-molecular-weight species during the 60-min incubation (indicated by the inverted triangle). The same filter was subsequently incubated with antibodies against Cim5/Rpt1 (C) and Rad23 (D). (E) A strain that expressed Flag-ΔUbLrad23 was examined as indicated for panels B to D. In contrast to the result in panel (B), the ubiquitinated proteins that coprecipitated with Flag-ΔUbLrad23 were more heterogeneous in size distribution (inverted triangle) and the shift toward higher-molecular-weight derivatives was reduced. The same filter was incubated with antibodies against Cim5/Rpt1 (F) and Rad23 (G).
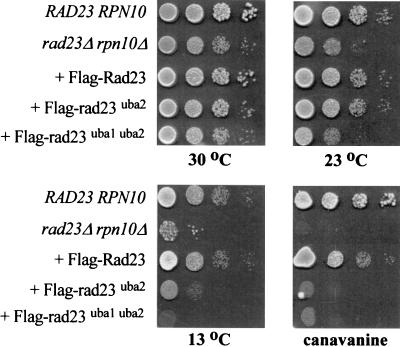
UBA mutants fail to suppress the growth and proteolytic defects of a rad23Δ rpn10Δ double mutant.
The rad23Δ rpn10Δ double mutant is unable to grow in the presence of amino acid analog canavanine or at low temperature (26). The rad23Δ rpn10Δ mutant also displays proteolytic defects and a delay during the G2 phase of the cell cycle. These pleiotropic phenotypes are completely suppressed following transformation with a plasmid that encodes either Rad23 or Rpn10 (26). To determine if the Rad23-proteasome interaction played a role in this suppression, we previously expressed ΔUbLrad23 in the rad23Δ rpn10Δ mutant and found that it was unable to alleviate any of the defects (26), underscoring the importance of proteasome interaction for Rad23 function. Here we investigated if the interaction between Rad23 and ubiquitinated proteins was also required for suppressing the defects of the rad23Δ rpn10Δ mutant. We expressed Flag-rad23uba1uba2 in the rad23Δ rpn10Δ mutant and found that it was unable to restore growth at low temperature or in the presence of amino acid analog canavanine (Fig. 5). A mutation in UBA2 enabled Flag-rad23uba2 to grow well at 30 and 23°C but not at 13°C, consistent with its moderate interaction with ubiquitinated proteins (Fig. 1D). In contrast, Flag-rad23uba1uba2 was as growth impaired as the rad23Δ rpn10Δ mutant at 30, 23, and 13°C, reflecting its reduced interaction with ubiquitinated cellular proteins. Interestingly, both mutant proteins failed to permit growth in medium containing canavanine, indicating that the generation of damaged proteins by amino acid analogs requires a fully functional Rad23 protein. Taken together, our genetic and biochemical results reveal a correspondence between the biological activity of Rad23 and the efficiency of its interaction with ubiquitinated proteins.
FIG. 5.
Suppression of the pleiotropic defects of the rad23Δ rpn10Δ mutant by Rad23 requires an interaction with ubiquitinated proteins. The ability of Flag-Rad23, Flag-rad23uba2, and Flag-rad23uba1uba2 to suppress the growth defect of the rad23Δ rpn10Δ mutant at low temperature and in the presence of amino acid analog canavanine was examined. Yeast cultures were grown to exponential phase, and 10-fold dilutions were spotted on synthetic medium and incubated at the temperatures indicated. A similar plate that contained 1 μg of canavanine/ml was incubated at 30°C. Growth was examined after 3 to 10 days of incubation, depending on the temperature.
Rad23 stimulates the interaction between ubiquitinated proteins and the proteasome.
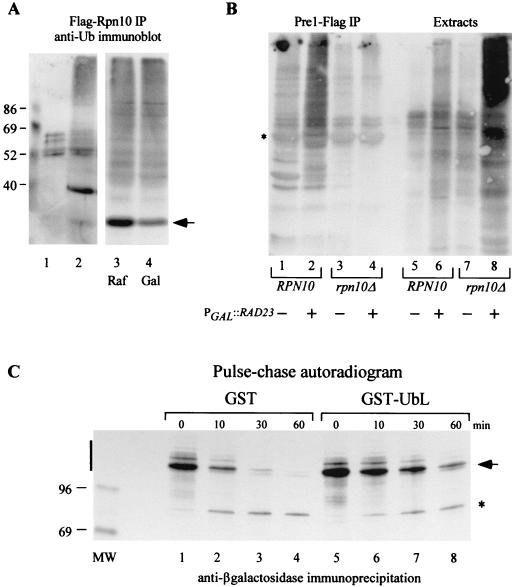
Rad23 and Rpn10 can bind ubiquitinated proteins through distinct interacting domains (5, 7, 11, 48), suggesting that they recognize structurally different features in a multi-Ub chain. In support of this argument, we note that a proteolytic substrate that contains only one or two Ub moieties is a very poor target for Rpn10 (31, 39), although it could be coimmunoprecipitated with Rad23 (30) (Fig. 2A). Furthermore, Rpn10 does not show appreciable interaction with mono-Ub, unlike Rad23 (5, 7). Consequently, it is possible that Rad23 and Rpn10 can simultaneously bind substrate-linked multi-Ub chains by recognizing different determinants within the chain. We therefore investigated if Rpn10 could bind ubiquitinated proteins, since this protein and its human counterpart S5a have been shown to bind unlinked multi-Ub chains in vitro (10, 44). In agreement with the in vitro results, we found that Flag-Rpn10 could interact with ubiquitinated proteins in yeast cell extracts (Fig. 6A).
FIG. 6.
Rad23 and Rpn10 promote the recognition of ubiquitinated proteins by the proteasome. (A) Yeast extracts containing Flag-Rpn10 were incubated with Flag-agarose beads, and the bound proteins were examined by immunoblotting with anti-Ub antibodies (lane 2). A control extract was also prepared from a yeast strain that did not express Flag-Rpn10 (lane 1). To examine the effect of Rad23 on the interaction between Rpn10 and ubiquitinated proteins, we prepared protein extracts from yeast strains that expressed both Flag-Rpn10 and galactose-inducible Rad23 (PGAL1::RAD23). Lanes 3 and 4, extracts prepared from raffinose (uninduced) and galactose (inducing) medium. The interaction between Flag-Rpn10 and cellular ubiquitinated proteins was not affected significantly by high-level expression of Rad23. Flag-Rpn10 (arrow) is visible in lane 2 (∼40 kDa). Lanes 1 to 2 and 3 to 4 are from different immunoblots. (B) Wild-type and rpn10Δ yeast strains expressing epitope-tagged proteasome subunit Pre1-Flag were transformed with a plasmid containing PGAL::RAD23. The cultures were grown in medium containing either glucose or galactose to regulate Rad23 expression. Equal amounts of protein extract were incubated with Flag-agarose to immunoprecipitate the proteasome, and the precipitated proteins were examined by immunoblotting using antibodies against Ub. Lanes 1 to 4, Pre1-Flag immunoprecipitation reactions from wild-type (lanes 1 and 2) and rpn10Δ (lanes 3 and 4) cells that were grown in either glucose- (lanes 1 and 3) or galactose-containing medium (lanes 2 and 4). Following incubation with antibodies against Ub, we found that high-level expression of Rad23 resulted in increased amounts of ubiquitinated proteins in the Pre1-Flag immunoprecipitate (compare lanes 1 and 2). However, this effect was not observed in a strain that lacked Rpn10 (compare lanes 2 and 4). Lanes 5 to 8, samples representing equal amounts of extract from the various cultures and corresponding to the samples examined in lanes 1 to 4. Note the significantly elevated levels of cellular ubiquitinated proteins in rpn10Δ cells that overexpressed Rad23 (lane 8). (The image represents the entire 10% polyacrylamide gel.) ∗, position of the heavy chain of the anti-Flag antibodies. (C) A strain that expressed Ub-Pro-β-Gal was transformed with plasmids expressing either GST or GST-UbLR23. Actively growing cultures were metabolically labeled with [35S]Met-[35S]Cys, and translation was terminated by the addition of cycloheximide and unlabeled methionine and cysteine. To examine the stability of Ub-Pro-β-Gal, equal amounts of TCA-insoluble material were incubated with antibodies against β-Gal, and bound proteins were separated in an SDS-8% polyacrylamide gel and visualized by autoradiography. Vertical bar, region of the autoradiogram that was quantitated to determine the stability of Ub-Pro-β-Gal; asterisk, appearance of a stable ∼90-kDa degradation product, described previously.
Although the function of Rad23 remains to be clearly elucidated, we speculated that it might regulate the stability of proteolytic substrates by controlling their delivery to the proteasome (26, 30, 35). Based on the genetic interaction between Rad23 and Rpn10, it was likely that both proteins participated in this activity. An attractive model for Rad23 function posits that it binds ubiquitinated proteins through the two UBA domains, while a subsequent interaction with the proteasome (involving the UbLR23 domain), would permit the transfer of the proteolytic substrate to Rpn10. One might predict that the expansion of a multi-Ub chain would be inhibited while the substrate was bound to Rad23, consistent with both in vitro and in vivo results (30). The presence of two UBA domains in Rad23 provides a mechanistic basis for the interaction with proteolytic substrates that are ligated to Ub.
Our previous genetic studies showed that Rad23 and Rpn10 are both required for efficient stress tolerance and intracellular proteolysis (26). These results are in agreement with biochemical studies, which showed that the UbL domain in human Rad23 can bind S5a (20), the counterpart of yeast Rpn10. However, because Rad23 can interact with the proteasome in rpn10Δ cells, it is likely that it can bind multiple subunits in the proteasome. We investigated if Rpn10 contributed to the recognition of ubiquitinated substrates by the proteasome because of the inability of Rad23 mutants to suppress the defects of the rad23Δ rpn10Δ mutant. We expressed an epitope-tagged proteasome subunit (Pre1-Flag) in wild-type and rpn10Δ cells, as well as a plasmid that expressed RAD23 at high levels from the galactose-inducible GAL1 promoter. Proteasomes were immunopurified on Flag-agarose, and the proteins were separated by SDS-PAGE, transferred to nitrocellulose, and incubated with anti-Ub antibodies. Low levels of ubiquitinated proteins were associated with proteasomes that were immunoprecipitated from wild-type cells (Fig. 6B, lane 1). In contrast, much lower levels of ubiquitinated cellular proteins were bound to proteasomes that were purified from rpn10Δ cells (lane 3), which may be due to the absence of this multi-Ub chain-binding protein. Alternatively, proteasomes have been reported to be unstable in rpn10Δ cells (15), which could result in poor recovery of intact proteasomes and any bound ubiquitinated substrates. Significantly larger amounts of ubiquitinated proteins were precipitated with the proteasome when Rad23 was overexpressed in a wild-type cell (lane 2). Remarkably, a similar increase was not detected when Rad23 was overexpressed in a strain that lacked Rpn10 (rpn10Δ strain; lane 4). This result is all the more striking given that the high expression of Rad23 in rpn10Δ cells resulted in dramatically increased amounts of intracellular ubiquitinated proteins (lane 8). Collectively, our results support the hypothesis that Rad23 binds ubiquitinated substrates and regulates their delivery to Rpn10 in the proteasome.
We expressed high levels of either GST or GST-UbLR23 (Fig. 1B, lanes 1 and 4) in a strain that also expressed Ub-Pro-β-Gal to further explore the idea that Rad23 translocates proteolytic substrates to the proteasome. Based on the assumption that the interaction between UbLR23 and the proteasome might block the delivery of substrates, we performed a pulse-chase analysis to measure Ub-Pro-β-Gal stability in the presence of high levels of UbLR23. Actively growing yeast cells were metabolically labeled with [35S]Met-[35S]Cys for 5 min, and translation was terminated by the addition of cycloheximide and unlabeled methionine and cysteine. Equal amounts of labeled cellular protein were incubated with anti-β-Gal antibodies, and the precipitated material was resolved by SDS-PAGE and visualized by autoradiography. We found that the stability of Ub-Pro-β-Gal was slightly less than 7 min in a strain that expressed GST, consistent with the results in Fig. 2C. In contrast, the stability of this substrate was increased dramatically (to ∼41 min) in cells that contained high levels of UbLR23, supporting the idea that unregulated UbLR23-proteasome interaction can interfere with the delivery of proteolytic substrates to the proteasome.
DISCUSSION
Rad23 was initially characterized as a DNA repair protein whose interaction with the repair factor Rad4 (XPC in humans) (18, 43) was reported to promote assembly of repair proteins at sites of DNA lesions (19, 22). The discovery of a UbL domain at the amino terminus of Rad23 suggested a potential proteolytic function, especially since full functionality was achieved when UbLR23 was replaced by Ub (47). We reported previously that Rad23 interacted with catalytically active proteasomes through its UbLR23 domain (35). Proteasome interaction is important during DNA repair because loss of UbLR23 in yeast Rad23 resulted in sensitivity to DNA damage. However, it is unclear if the Rad23-proteasome interaction mediates a proteolytic event during NER, and a potential nonproteolytic function has been suggested (14, 34). However, while it is conceivable that the Rad23-proteasome interaction does not involve proteolysis, several lines of evidence are strongly supportive of a proteolytic role. Our studies, as well as other reports, showed that Rad23 can bind Ub (5, 7), regulate the assembly of substrate-linked multi-Ub chains (30), and control the stability of proteins in vivo (8, 30). Additionally, Rad23 was recently reported to bind multi-Ub chains in vitro (33, 48). Another study proposed that Rad23 might control the stability of Rad4 (1), which is intriguing because we showed previously that Rad4-hemagglutinin (HA) could be copurified with Rad23 and the 26S proteasome after several chromatography steps (35). Rad23 interacts with Rad4 (and XPC in humans) through a motif that is unrelated to the UBA domain (28), and it is therefore not surprising that Flag-rad23uba1uba2 can bind Rad4-HA efficiently (L. Chen and K. Madura, unpublished results), despite its inability to bind ubiquitinated proteins. Because Rad23 may not require UBA domains to control Rad4 stability (28), it is not clear if UBA mutations alone would be expected to confer a significant defect in DNA repair.
The expression of high levels of Rad23 resulted in the accumulation of ubiquitinated proteins in yeast cells. The UBA domains in Rad23 are required for the interaction with ubiquitinated cellular proteins, and mutants that were unable to bind ubiquitinated proteins displayed biochemical and physiological defects. The interaction between Rad23 and ubiquitinated proteins increased transiently after DNA damage, and these ubiquitinated proteins were converted to higher-molecular-weight derivatives over time. In contrast, a Rad23 mutant that was unable to bind the proteasome (ΔUbLrad23) formed a persistent interaction with ubiquitinated proteins, with no significant increase in the formation of higher-molecular-weight conjugates. One interpretation of this result is that, for some substrates, complete assembly of a multi-Ub chain might require an interaction with the proteasome (40).
The role of Rad23 overlaps with that of Rpn10 (26), a subunit in the proteasome that binds multi-Ub chains (44). Rad23 mutants that are unable to bind ubiquitinated proteins failed to stabilize test substrates in vivo or suppress the proteolytic and growth defects of a rad23Δ rpn10Δ mutant. Consistent with previous in vitro studies, we discovered that ubiquitinated proteins in yeast protein extracts could be copurified with Flag-Rpn10. Rad23 and Rpn10 contain different Ub-binding domains and might interact with structurally distinct features within a multi-Ub chain. Although we previously showed that Rad23 could bind short, substrate-linked multi-Ub chains, the results shown here also suggest that Rad23 might bind substrates that contain longer multi-Ub chains. We note that the distal Ub moieties in a long multi-Ub chain are not tightly packed and may resemble a short, substrate-linked multi-Ub chain (9). In contrast, Rpn10 recognizes a hydrophobic patch that is formed by the compaction of Ub moieties in a long lysine-48-linked multi-Ub chain (3, 39). The interaction between Rad23 and a multi-Ub chain might prevent dismantling by Ub-specific proteases, resulting in transient stabilization of the substrate, consistent with our in vitro and in vivo data (30). Subsequent delivery of the ubiquitinated substrate by Rad23 to Rpn10 in the proteasome could initiate degradation. We note that a recent study demonstrated that Rad23 specifically interacted with multi-Ub chains in vivo and displayed specificity only for chains that promote the degradation of proteins by the proteasome (33).
To examine the role of Rad23 and Rpn10 in the targeting of proteolytic substrates, we immunoprecipitated proteasomes from rpn10Δ and wild-type cells using an epitope-tagged subunit in the 20S core particle (Pre1-Flag). A low level of ubiquitinated proteins was coprecipitated with proteasomes from a wild-type strain. In contrast, much lower levels of ubiquitinated proteins were detected in proteasomes that were isolated from the rpn10Δ mutant, consistent with the well-characterized interaction of Rpn10 with multi-Ub chains. The levels of proteasome-associated ubiquitinated proteins increased dramatically when Rad23 was overexpressed, supporting the idea that it might deliver substrates to the proteasome. Significantly, overexpression of Rad23 in rpn10Δ cells resulted in the accumulation of very high levels of intracellular ubiquitinated proteins, although these ubiquitinated proteins could not be copurified with the proteasome. A plausible interpretation of these results is that the high levels of ubiquitinated proteins that arise from overexpression of Rad23 are not efficiently delivered to the proteasome in rpn10Δ cells. Under normal conditions, the Rad23-proteasome interaction may allow substrates to be transferred to Rpn10 to initiate further ubiquitination by proteasome-associated E3 (49) and E2 (40) factors. A failure to efficiently deliver ubiquitinated substrates to the proteasome could underlie the pleiotropic growth and proteolytic defects of the rad23Δ rpn10Δ mutant.
Collectively, our findings provide compelling evidence that the proteasome recognizes multiubiquitinated substrates. Although a variety of genetic and biochemical approaches have shown that multiubiquitinated proteins are targeted to the proteasome, it has not been possible to clearly demonstrate the interaction between proteolytic substrates and the proteasome. Our findings predict important roles for both the UbLR23 and UBA domains of Rad23 in the translocation of proteolytic substrates to the proteasome. Since the role of Rad23 overlaps with those of other proteins that contain both UbL and UBA domains (Ddi1 [8] and Dsk2 [6, 33]), we speculate that this class of proteins may encode novel regulators that deliver substrates to the proteasome. Our current understanding of Rad23 function can be represented by a simple model in which Rad23 (and other proteins that contain both UbL and UBA domains) are shuttle factors that translocate proteolytic substrates to the proteasome (Fig. 7). In agreement with the shuttle factor model, we note that Rad23 can be cofractionated with Rad4 and the 26S proteasome (35) and can also promote the translocation of Png1 to the proteasome (38). A recent study has also reported that Rad23 controls the abundance of cell cycle regulator Pds1 (5, 8). The failure to control Pds1 levels may account, in part, for the G2-to-M phase transition defect of the rad23Δ rpn10Δ mutant.
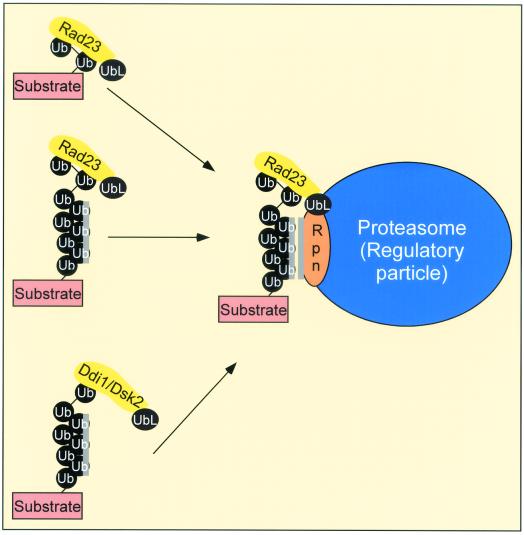
FIG. 7.
Model. Rad23 translocates ubiquitinated proteins to the proteasome. We provide evidence that Rad23 can interact with substrates that are ligated to multi-Ub chains (black circles). The interaction with ubiquitinated substrates could transiently inhibit further multi-Ub chain assembly and thereby stabilize the protein. However, subsequent delivery of the ubiquitinated substrate to specific proteasome-associated multi-Ub chain binding factors, such as Rpn10 (Rpn), could initiate degradation. Rad23 and Rpn10 may recognize different determinants in a multi-Ub chain, and consequently both proteins could interact simultaneously with substrate-linked multi-Ub chains. The interaction between Rpn10 and the multi-Ub chain is mediated by hydrophobic interactions, indicated by the stripe next to the multi-Ub chain and Rpn10. In contrast, Rad23 might interact with the distal Ub moieties in a multi-Ub chain. We speculate that proteasome-associated E2 and E3 factors could further ubiquitinate the substrate to promote efficient degradation. This scheme also anticipates that other UBA- and UbL-containing proteins, including Ddi1 and Dsk2, perform similar roles in the delivery of proteolytic substrates and regulators to the proteasome.
ADDENDUM IN PROOF
A recent paper has shown that the Dsk2 homolog from Schizosaccharomyces pombe also interacts with multiubiquitinated proteins via its UBA domains (R. T. Elder, X.-Q. Song, M. Chen, K. M. Hopkins, H. B. Lieberman, and Y. Zhao, Nucleic Acids Res. 30:581-591, 2002).
Acknowledgments
We thank Cherylene Schauber, Ellen Doss-Pepe, and Tatiana G. Ortolan for preliminary studies that led to the results shown in Fig. 1C. We thank J. Dohmen for strains and plasmids. Members of the laboratory are thanked for critical review of the manuscript.
This study was supported by Public Health Service grant CA-83875 from the National Cancer Institute.
REFERENCES
- 1.Araki, M., C. Masutani, M. Takemura, A. Uchida, K. Sugawawa, J. Kondoh, Y. Okhuma, and F. Hanaoka. 2001. Centrosome protein centrin 2/caltractin 1 is part of the xeroderma pigmentosum group C complex that initiates global genome nucleotide excision repair. J. Biol. Chem. 276:18665-18672. [DOI] [PubMed] [Google Scholar]
- 2.Bachmair, A., D. Finley, and A. Varshavsky. 1986. In vivo half-life of a protein is a function of its amino-terminal residue. Science 234:179-186. [DOI] [PubMed] [Google Scholar]
- 3.Beal, R. E., D. Toscano-Cantaffa, P. Young, M. Rechsteiner, and C. M. Pickart. 1998. The hydrophobic effect contributes to polyubiquitin chain recognition. Biochemistry 37:2925-2934. [DOI] [PubMed] [Google Scholar]
- 4.Bertolaet, B. L., D. J. Clarke, M. Wolff, M. H. Watson, M. Henze, G. Divita, and S. I. Reed. 2001. UBA domains mediate protein-protein interactions between two DNA damage-inducible proteins. J. Mol. Biol. 313:955-963. [DOI] [PubMed] [Google Scholar]
- 5.Bertolaet, B. L., D. J. Clarke, M. Wolff, M. H. Watson, M. Henze, G. Divita, and S. I. Reed. 2001. UBA domains of DNA damage-inducible proteins interact with ubiquitin. Nat. Struct. Biol. 8:417-422. [DOI] [PubMed] [Google Scholar]
- 6.Biggins, S., I. Ivanovska, and R. D. Rose. 1996. Yeast ubiquitin-like genes are involved in duplication of the microtubule organizing center. J. Cell Biol. 133:1331-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, L., U. Shinde, T. G. Ortolan, and K. Madura. 2001. Ubiquitin-associated (UBA) domains in Rad23 bind ubiquitin and promote inhibition of multi-ubiquitin chain assembly. EMBO Rep. 2:933-938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clarke, D. J., G. Mondesert, M. Segal, B. L. Bertolaet, S. Jensen, M. Wolff, M. Henze, and S. I. Reed. 2001. Dosage suppressors of pds1 implicate ubiquitin-associated domains in checkpoint control. Mol. Cell. Biol. 21:1997-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cook, W. J., L. C. Jeffrey, M. Carson, Z. Chen, and C. M. Pickart. 1992. Structure of a diubiquitin conjugate and a model for interaction with ubiquitin conjugating enzyme (E2). J. Biol. Chem. 267:16467-16471. [DOI] [PubMed] [Google Scholar]
- 10.Deveraux, Q., V. Ustrell, C. Pickart, and M. Rechsteiner. 1994. A 26S protease subunit that binds ubiquitin conjugates. J. Biol. Chem. 269:7059-7061. [PubMed] [Google Scholar]
- 11.Fu, H., S. Sadis, D. M. Rubin, M. Glickman, S. van Nocker, D. Finley, and R. D. Vierstra. 1998. Multiubiquitin chain binding and protein degradation are mediated by distinct domains within the 26S proteasome subunit Mcb1. J. Biol. Chem. 273:1970-1981. [DOI] [PubMed] [Google Scholar]
- 12.Funakoshi, M., S. Geley, T. Hunt, T. Nishimoto, and H. Kobayashi. 1999. Identification of XDRP1; a Xenopus protein related to yeast Dsk2p binds to the N-terminus of cyclin A and inhibits its degradation. EMBO J. 18:5009-5018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Funakoshi, M., T. Sasaki, T. Nishimoto, and H. Kobayashi. 2002. Budding yeast Dsk2p is a polyubiquitin-binding protein that can interact with the proteasome. Proc. Natl. Acad. Sci. USA 99:745-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gillette, T. G., W. Huang, S. J. Russell, S. H. Reed, S. A. Johnston, and E. C. Friedberg. 2001. The 19S complex of the proteasome regulates nucleotide excision repair in yeast. Genes Dev. 15:1528-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Glickman, M. H., D. M. Rubin, O. Coux, I. Wefes, G. Pfeifer, Z. Cjeka, W. Baumeister, V. A. Fried, and D. Finley. 1998. A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9-signalosome and eIF3. Cell 94:615-624. [DOI] [PubMed] [Google Scholar]
- 16.Glickman, M. H., D. M. Rubin, V. A. Fried, and D. Finley. 1998. The regulatory particle of the Saccharomyces cerevisiae proteasome. Mol. Cell. Biol. 18:3149-3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guzder, S. N., V. Bailly, P. Sung, L. Prakash, and S. Prakash. 1995. Yeast DNA repair protein RAD23 promotes complex formation between transcription factor TFIIH and DNA damage recognition factor RAD14. J. Biol. Chem. 270:8385-8388. [DOI] [PubMed] [Google Scholar]
- 18.Guzder, S. N., Y. Habraken, P. Sung, L. Prakash, and S. Prakash. 1995. Reconstitution of yeast nucleotide excision repair with purified Rad proteins, replication protein A, and transcription factor TFIIH. J. Biol. Chem. 270:12973-12976. [DOI] [PubMed] [Google Scholar]
- 19.Guzder, S. N., P. Sung, L. Prakash, and S. Prakash. 1998. Affinity of yeast nucleotide excision repair factor 2, consisting of the Rad4 and Rad23 proteins, for ultraviolet damaged DNA. J. Biol. Chem. 273:31541-31546. [DOI] [PubMed] [Google Scholar]
- 20.Hiyama, H., M. Yokoi, C. Masutani, K. Sugasawa, T. Maekawa, K. Tanaka, J. H. J. Hoeijmakers, and F. Hanaoka. 1999. Interaction of hHR23 with S5a. The ubiquitin-like domain of hHR23 mediates interaction with S5a subunit of 26S proteasome. J. Biol. Chem. 274:28019-28025. [DOI] [PubMed] [Google Scholar]
- 21.Hofman, K., and P. Bucher. 1996. The UBA domain: a sequence motif present in multiple enzyme classes of the ubiquitination pathway. Trends Biochem. Sci. 21:172-173. [PubMed] [Google Scholar]
- 22.Jansen, L. E. T., R. A. Verhage, and J. Brouwer. 1998. Preferential binding of yeast Rad4-Rad23 complex to damaged DNA. J. Biol. Chem. 273:33111-33114. [DOI] [PubMed] [Google Scholar]
- 23.Johnson, E. S., P. C. M. Ma, I. M. Ota, and A. Varshavsky. 1995. A proteolytic pathway that recognizes ubiquitin as a degradation signal. J. Biol. Chem. 270:17442-17456. [DOI] [PubMed] [Google Scholar]
- 24.Kleijnen, M. F., A. H. Shih, P. Zhou, S. Kumar, R. E. Soccio, N. L. Kedersha, G. Gill, and P. M. Howley. 2000. The hPLIC proteins may provide a link between the ubiquitination machinery and the proteasome. Mol. Cell 6:409-419. [DOI] [PubMed] [Google Scholar]
- 25.Koegl, M., T. Hoppe, S. Schlenker, H. D. Ulrich, T. U. Mayer, and S. Jentsch. 1999. A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell 96:635-644. [DOI] [PubMed] [Google Scholar]
- 26.Lambertson, D., L. Chen, and K. Madura. 1999. Pleiotropic growth and proteolytic defects caused by the loss of the proteasome-interacting factors Rad23 and Rpn10 of Saccharomyces cerevisiae. Genetics 153:69-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Madura, K., and S. Prakash. 1990. Transcript levels of the Saccharomyces cerevisiae DNA repair gene RAD23 increase in response to UV light and in meiosis but remain constant in the mitotic cell cycle. Nucleic Acids Res. 18:4737-4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Masutani, C., M. Araki, K. Sugasawa, P. J. van der Spek, A. Yamada, A. Uchida, T. Maekawa, D. Bootsma, J. H. J. Hoeijmakers, and F. Hanaoka. 1997. Identification and characterization of XPC-binding domain of hHR23B. Mol. Cell. Biol. 17:6915-6923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mu, D., D. S. Hsu, and A. Sancar. 1996. Reaction mechanism of human DNA repair excision nuclease. J. Biol. Chem. 271:8285-8294. [DOI] [PubMed] [Google Scholar]
- 30.Ortolan, T. G., P. Tongaonkar, D. Lambertson, L. Chen, and K. Madura. 2000. The Rad23 DNA repair protein is a negative regulator of substrate-linked multi-ubiquitin chain assembly. Nat. Cell Biol. 2:601-608. [DOI] [PubMed] [Google Scholar]
- 31.Piotrowski, J., R. Beal, L. Hoffman, K. D. Wilkinson, R. E. Cohen, and C. M. Pickart. 1997. Inhibition of the 26S proteasome by polyubiquitin chains synthesized to have defined lengths. J. Biol. Chem. 272:23712-23721. [DOI] [PubMed] [Google Scholar]
- 32.Prakash, S., and L. Prakash. 2000. Nucleotide excision repair in yeast. Mutat. Res. 30:13-24. [DOI] [PubMed] [Google Scholar]
- 33.Rao, H., and A. Sastry. 2002. Recognition of specific ubiquitin conjugates is important for the proteolytic functions of the ubiquitin-associated domain proteins Dsk2 and Rad23. J. Biol. Chem. 277:11691-11695. [DOI] [PubMed] [Google Scholar]
- 34.Russell, S. J., S. H. Reed, W. Huang, E. C. Friedberg, and S. A. Johnston. 1999. The 19S regulatory complex of the proteasome functions independently of proteolysis in nucleotide excision repair. Mol. Cell 3:687-695. [DOI] [PubMed] [Google Scholar]
- 35.Schauber, C., L. Chen, P. Tongaonkar, I. Vega, D. Lambertson, W. Potts, and K. Madura. 1998. Rad23 links DNA repair to the ubiquitin/proteasome pathway. Nature 391:715-718. [DOI] [PubMed] [Google Scholar]
- 36.Sugasawa, K., C. Masutani, A. Uchida, T. Maekawa, P. J. van der Spek, D. Bootsma, J. H. J. Hoeijmakers, and F. Hanaoka. 1996. HHR23B, a human Rad23 homolog, stimulates XPC protein in nucleotide excision repair in vitro. Mol. Cell. Biol. 16:4852-4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sugasawa, K., J. M. Y. Ng, C. Masutani, T. Maekawa, A. Uchida, P. J. van der Spek, A. M. P. Eker, S. Rademakers, C. Visser, A. Aboussekhra, R. D. Wood, F. Hanaoka, D. Bootsma, and J. H. J. Hoeijmakers. 1997. Two human homologs of Rad23 are functionally interchangeable in complex formation and stimulation of XPC repair activity. Mol. Cell. Biol. 17:6924-6931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suzuki, T., H. Park, M. A. Kwofie, and W. J. Lennarz. 2001. Rad23 provides a link between the Png1 deglycosylating enzyme and the 26S proteasome in yeast. J. Biol. Chem. 276:21601-21607. [DOI] [PubMed] [Google Scholar]
- 39.Thrower, J. S., L. Hoffman, M. Rechsteiner, and C. M. Pickart. 2000. Recognition of the polyubiquitin proteolytic signal. EMBO J. 19:94-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tongaonkar, P., L. Chen, D. Lambertson, B. Ko, and K. Madura. 2000. Evidence for an interaction between ubiquitin-conjugating enzymes and the 26S proteasome. Mol. Cell. Biol. 20:4691-4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tongaonkar, P., and K. Madura. 1998. Reconstituting ubiquitination reactions with affinity-purified components and 32P-ubiquitin. Anal. Biochem. 260:307-319. [DOI] [PubMed] [Google Scholar]
- 42.Uetz, P., L. Giot, G. Cagney, T. A. Mansfield, R. S. Judson, J. R. Knight, D. Lockshon, V. Narayan, M. Srinivasan, P. Pochart, A. Qureshi-Emili, Y. Li, B. Godwin, D. Conover, T. Kalbfleisch, G. Vijayadamodar, M. Yang, M. Johnston, S. Fields, and J. M. Rothberg. 2000. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403:623-631. [DOI] [PubMed] [Google Scholar]
- 43.van der Spek, P. J., C. C. Visser, F. Hanaoka, B. Smit, A. Hagemeijer, D. Bootsma, and J. H. J. Hoeijmakers. 1996. Cloning, comparative mapping, and RNA expression of the mouse homologues of the Saccharomyces cerevisiae nucleotide excision repair gene RAD23. Genomics 31:20-27. [DOI] [PubMed] [Google Scholar]
- 44.van Nocker, S., S. Sadis, D. M. Rubin, M. Glickman, H. Fu, O. Coux, I. Wefes, D. Finley, and R. D. Vierstra. 1996. The multiubiquitin-chain-binding protein Mcb1 is a component of the 26S proteasome in Saccharomyces cerevisiae and plays a nonessential, substrate-specific role in protein turnover. Mol. Cell. Biol. 16:6020-6028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Varshavsky, A. 1997. The ubiquitin system. Trends Biochem. Sci. 22:383-387. [DOI] [PubMed] [Google Scholar]
- 46.Verma, R., S. Chen, R. Feldman, D. Schieltz, J. Yates, J. Dohmen, and R. J. Deshaies. 2000. Proteasomal proteomics: identification of nucleotide-sensitive proteasome-interacting proteins by mass spectrometric analysis of affinity-purified proteasomes. Mol. Biol. Cell 11:3425-3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Watkins, J. F., P. Sung, L. Prakash, and S. Prakash. 1993. The Saccharomyces cerevisiae DNA repair gene RAD23 encodes a nuclear protein containing a ubiquitin-like domain required for biological function. Mol. Cell. Biol. 13:7757-7765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wilkinson, C. R. M., M. Seeger, R. Petersen-Hartmann, M. Stone, M. Wallace, C. Semple, and C. Gordon. 2001. Proteins containing the UBA domain are able to bind to multi-ubiquitin chains. Nat. Cell Biol. 3:939-943. [DOI] [PubMed] [Google Scholar]
- 49.Xie, Y., and A. Varshavsky. 2000. Physical association of ubiquitin ligases and the 26S proteasome. Proc. Natl. Acad. Sci. USA 97:2497-2502. [DOI] [PMC free article] [PubMed] [Google Scholar]