Abstract
Expression of human T-cell leukemia virus type 1 (HTLV-1) is regulated by the viral transcriptional activator Tax. Tax activates viral transcription through interaction with the cellular transcription factor CREB and the coactivators CBP/p300. One key property of the coactivators is the presence of histone acetyltransferase (HAT) activity, which enables p300/CBP to modify nucleosome structure. The data presented in this manuscript demonstrate that full-length p300 and CBP facilitate transcription of a reconstituted chromatin template in the presence of Tax and CREB. The ability of p300 and CBP to activate transcription from the chromatin template is dependent upon the HAT activity. Moreover, the coactivator HAT activity must be tethered to the template by Tax and CREB, since a p300 mutant that fails to interact with Tax did not facilitate transcription or acetylate histones. p300 acetylates histones H3 and H4 within nucleosomes located in the promoter and 5′ proximal regions of the template. Nucleosome acetylation is accompanied by an increase in the level of binding of RNA polymerase II transcription factor TFIID and RNA polymerase II to the promoter. Interestingly, we found distinct transcriptional activities between CBP and p300. CBP, but not p300, possesses an N-terminal activation domain which directly activates Tax-mediated HTLV-1 transcription from a naked DNA template. Finally, using the chromatin immunoprecipitation assay, we provide the first direct experimental evidence that p300 and CBP are associated with the HTLV-1 long terminal repeat in vivo.
Human T-cell leukemia virus type 1 (HTLV-1) is a human lentivirus and the etiologic agent of adult T-cell leukemia and the neurological disorder tropical spastic paraparesis-HTLV-1-associated myelopathy (20, 37, 52, 54, 74). Studies have shown that the transactivator protein Tax, encoded by the pX region of HTLV-1, is a potent activator of the HTLV-1 long terminal repeat (LTR) (for reviews, see references 8, 11, 72, and 73). Tax has also been shown to activate transcription of a number of cellular genes, including interleukin-2 (IL-2) and IL-2Rα (5, 13, 16, 18, 23, 48). Tax does not bind to DNA directly but activates transcription by recruiting or modifying the activity of cellular transcription factors, including CREB (cyclic AMP-responsive element binding protein), serum responsive factor (SRF), and NF-κB (1, 6, 8, 17, 31, 34, 36, 41, 43, 46, 56, 62-64, 70, 71). Because Tax plays such an important role in gene expression and pathogenesis of HTLV-1, numerous studies have been directed toward the mechanism of Tax transactivation. It has been shown that Tax activates expression of viral genes via the LTR. Three highly conserved 21-bp repeat elements located within the LTR, termed the Tax-response elements (TRE), are critical to Tax-mediated transcriptional activation (10). Tax associates with the LTR through interaction with CREB (75). The formation of this Tax-CREB promoter complex serves as a high-affinity binding site for the recruitment of the multifunctional cellular coactivators CBP, p300, and PCAF (21, 25, 33, 42, 45). The direct interaction of Tax with CBP allows the binding of the coactivator in the absence of CREB phosphorylation, permitting specific activation of the viral LTR (42, 44).
CBP and p300 are often referred to as p300/CBP, since they exhibit strong sequence similarity, have similar biological functions, bind to common transcription factors, and are considered in many cases as functional homologues. p300 and CBP play key roles in cellular differentiation, growth control, and homeostasis (for reviews, see references 22 and 24). The importance of p300/CBP for normal cellular metabolism has been revealed by gene knockout studies. Gene knockouts in mice indicate that p300 and CBP are required for normal embryonic development and viability (40, 69). In humans, mutations in the CBP gene are associated with Rubinstein-Taybi syndrome, a haploinsufficiency disorder resulting in mental retardation and other developmental abnormalities (50). Chromosomal translocations resulting in the fusion of CBP with monocytic leukemia zinc finger protein or with mixed lineage leukemia protein have been found in subtypes of acute myeloid leukemia (9). p300 gene mutations have also been reported in epithelial cancers (19).
The transcriptional activity of p300/CBP is tightly linked to the intrinsic histone acetyltransferase (HAT) activity domain. Martinez-Balbas et al. demonstrated that a region of CBP encompassing the HAT domain could stimulate transcription when tethered to a promoter in vivo (49). Furthermore, Parekh and Maniatis demonstrated that virus-induced hyperacetylation of histones at the beta interferon promoter is correlated with the interaction between the activator and p300/CBP (53). Using an in vitro transcription system with a chromatin template, Kadonaga's group studied the mechanisms of transcriptional enhancement by the p300 coactivator (38). They demonstrated that p300 could mediate estrogen receptor-activated gene transcription initiation in vitro in the context of a chromatin template but not in that of a naked DNA template. Recently, using activator-dependent transcription on chromatin templates, Kundu et al. reported that p300-mediated transcription by GAL4-VP16 involved targeted histone acetylation by p300 (39). Most recently, Asahara et al. (4) reported that cyclic AMP-dependent recruitment of p300 to the somatostatin promoter stimulated acetylation of histone H4 and in vitro transcription from a chromatin template. Interestingly, transcription is activated through a chromatin-dependent mechanism in which recruitment of p300 facilitates interaction of CREB with RNA polymerase II (RNAP II) transcription factor TFIID.
Our laboratory previously reported that CBP stimulates Tax-mediated HTLV-1 LTR transcription initiation and reinitiation from a naked DNA template in vitro (35). We also demonstrated that p300 and PCAF function as coactivators for Tax-stimulated HTLV-1 LTR transcription (33). Harrod et al. subsequently reported that Tax could interact directly with both PCAF and p300/CBP in a coactivator-activator 21-bp complex (25). The existence of such a complex raised an interesting question as to which HAT (57) is required for Tax-mediated transcription. Moreover, the identity and nature of the exact molecular mechanism by which HAT increases transcription from the HTLV-1 LTR were unclear. To address these questions, we have developed a Tax-dependent in vitro transcription system using reconstituted HTLV-1 TRE chromatin templates. We show that Tax functions with p300 or CBP to activate transcription from the chromatin template. p300 mediates HTLV-1 LTR expression by targeted nucleosomal acetylation, which results in recruitment of RNAP II and TFIID onto the chromatin template. Finally, using a chromatin immunoprecipitation (ChIP) assay, we demonstrate that p300 associates with the integrated HTLV-1 LTR in vivo.
MATERIALS AND METHODS
Chromatin reconstitution.
Reconstitution was performed on a 2.0-kb NdeI-BsrFI fragment from the 4xTRE plasmid containing four copies of the 21-bp repeat I inserted upstream of the HTLV-1 TATA and promoter (−52 to −1) and G-free cassette. For a control promoter, the −52 to −1 region is cloned upstream of the G-free region (both plasmids [2] were a kind gift from M. Anderson of the Medical College of Georgia). The DNA was biotinylated by filling in the NdeI 3′ end using Klenow polymerase (New England Biolabs) and the nucleotides α-S-dTTP, α-S-dGTP, α-S-dCTP (Sigma), and biotin-dATP. Biotin-labeled DNA (20 μg) was immobilized on 2.5 mg of streptavidin beads (Dynabeads M-280; Dynal) using the KiloBase binder kit (Dynal). After washing, the bead-DNA mixtures were resuspended in embryo extract (EX) buffer [10 mM HEPES (pH 7.6), 10 mM KCl, 1.5 mM MgCl2, 0.5 mM EGTA, 10% glycerol, 10 mM glycerophosphate, 1 mM dithiothreitol (DTT), and 1 mM 4-(2-aminoethyl)-benzenesulfonyl fluoride hydrochloride (AEBSF) containing 0.05% NP-40] (EX-N), 0.001% thimerosal, 0.3 mg of bovine serum albumin (BSA) per ml, and 1 mM AEBSF to a DNA concentration of 0.1 mg/ml.
Late Drosophila embryo extracts were made according to the procedure of Becker et al. (7), and chromatin reconstitution was carried out as described previously (15). Specifically, chromatin was reconstituted by adding 2 μg of immobilized DNA, 1 mg of late Drosophila extracts, and 2 μg of histone octamers from mouse cells in a total volume of 200 μl. After rotation for 4 h at room temperature, the reconstitution extract was removed using magnetic beads. The chromatin was incubated at room temperature for 5 min with 0.05% Sarkosyl and 0.3 mg of BSA per ml in EX-N buffer to remove Drosophila remodeling and assembly complexes. The chromatin was then washed once each with cold 200 mM NaCl-EX-N buffer and EX-N buffer containing 0.3 mg of BSA per milliliter and stored in a solution containing EX-N buffer, 0.3 mg of BSA per ml, and protease inhibitors.
Chromatin reconstitution was also carried out on a supercoiled plasmid by using Drosophila embryo extract. Chromatin was reconstituted by adding 2 μg of DNA, 1 mg of Drosophila extract, and 2 μg of histone octamers from mouse cells in a total volume of 200 μl. After rotation for 4 h at room temperature, the chromatin was analyzed by micrococcal nuclease digestion and used for in vitro transcription assays.
Micrococcal nuclease digestion assay.
Chromatin was analyzed by digesting 250 ng (25 μl) at each time point with 3 U of micrococcal nuclease per microliter and 0.3 mM CaCl2 for 0, 0.5, 1, and 5 min at room temperature. Reactions were stopped with 6.3 μl of 2.5% Sarkosyl-0.1 M EDTA, and then the reaction mixtures were incubated for 1 h with 1 μl of 0.5-mg/ml RNase A. Proteins were then digested overnight at 37°C with 4 μl of 10-mg/ml proteinase K and 0.2% sodium dodecyl sulfate (SDS). Following an ethanol precipitation, DNA was analyzed on a 1% agarose gel.
Restriction enzyme accessibility assay.
The assays were carried out as previously described (15). Specifically, a 40-μl reaction mixture containing 0.04 μg of biotinylated chromatin and 10 U of restriction enzyme in EX buffer was incubated at 37°C for 15 min. Reactions were stopped by the addition of 10 μl of 2.5% Sarkosyl with 0.1 M EDTA. The supernatants were removed and protein elution buffer (10 mM Tris, 2 M NaCl, 0.1% SDS, 0.1% NP-40) was added, followed by incubation at 37°C for 30 min. Beads were washed with protein elution buffer followed by Tris-EDTA buffer and then resuspended in 30 μl of labeling solution containing [γ-32P]ATP and 1 μl of T4 polynucleotide kinase and incubated at 37°C for 1 h. Beads were then washed twice with Tris-EDTA, and the DNA fragment was released from the beads by digesting with EcoRI and analyzed by electrophoresis on a 1% agarose gel. Digested and undigested chromatin were quantitated using the ImageQuant program. Accessibility of a site to a specific enzyme was determined by the percentage of cleavage, calculated by dividing the amount of digested chromatin by the total amount of chromatin.
Protein purification.
The CREB protein is expressed from the pET-15b vector (kindly provided by R. Goodman's laboratory) under the T7 promoter. Transformed Escherichia coli BL21(DE3)(pLysS) was induced by 1 mM isopropyl-d-thiogalactopyranoside for 3 h. The protein was purified through Mono Q and heparin columns and dialyzed against buffer E (25 mM Tris-HCl [pH 7.5], 50 mM NaCl, 0.1% Triton X-100, 5% glycerol, 1 mM DTT). The Tax protein with a six-histidine tag at the C terminus (TaxH6) was purified from E. coli as previously described (33). p300 protein was expressed as Flag-tagged fusion proteins in baculovirus-infected cells and purified through an anti-Flag M2 column. Baculovirus CBP was a kind gift from D. Thanos of Columbia University. CBP was expressed as His-tagged protein in baculovirus-infected cells and purified as previously described (12). ATP-utilizing chromatin assembly and remodeling factor (ACF) was kindly provided by J. T. Kadonaga of the University of California, San Diego, and baculovirus nucleosome assembly protein 1 (NAP1) was a gift from T. Ito of the Saitama Medical School. The proteins were purified as described before (29).
Chromatin pulldown assay.
Different combinations of proteins (Tax, CREB, p300, and CBP) were incubated with 400 μg of chromatin template in 60 μl of acetylation buffer containing 10 mM Tris, 10 mM KCl, 5 mM MgCl2, 10% glycerol, 1 mM DTT, 1 mM AEBSF, 10 mM sodium butyrate, and 0.5 mM acetyl coenzyme A (acetyl-CoA) at 30°C for 30 min. Nuclear extract (200 μg) was diluted in 200 μl of buffer [50 mM Tris (pH 7.6), 50 mM NaCl, 0.5 mM EDTA, 1 mM DTT, 5 mM MgCl2, 0.1% Triton, 5% glycerol, 2.5 mg of BSA per ml, 10 μg of poly(dI-dC) per ml, and 1 mM sodium butyrate], and the reaction was continued for 30 min at 30°C. The beads were then washed four times with binding buffer without BSA and poly(dI-dC). The reaction mixture was then split into two aliquots. One portion was used to analyze the proteins bound to the chromatin. Proteins were eluted by SDS loading buffer and loaded onto a 4 to 20% Tris-glycine gel. Immunoblotting was performed using anti-Tax (hybridoma; AIDS Research and Reference Reagent Program), anti-TATA-binding protein (TBP) (Santa Cruz Biotech), and anti-RNAP II (Berkeley Antibody Company) antibodies. The other half of the chromatin sample was deproteinized, and DNA was released from the beads by digestion with EcoRI. Then DNA fragments were analyzed by electrophoresis on a 1% agarose gel and stained with ethidium bromide.
In vitro IP assay.
In vitro IP was carried out as described before (33). Tax (200 ng) was incubated with 200 ng of either the region comprising amino acids 1 to 670 of p300 [p300 (1-670)] or amino acids 1 to 682 of CBP [CBP (1-682)] protein in buffer A (50 mM Tris-HCl [pH 7.6], 50 mM NaCl, 0.5 mM EDTA, 1 mM DTT, 5 mM MgCl2, 0.1% Triton, 5% glycerol, 2.5 mg of BSA/ml) for 1 h at 4°C. One microliter of anti-Tax monoclonal antibody or a control mouse antibody was added and incubated for 2 h. Protein complexes were then incubated with 25 μl of protein A-protein G agarose beads (Calbiochem) overnight. The agarose beads were washed three times with buffer B (50 mM Tris-HCl [pH 7.6], 150 mM NaCl, 1% NP-40, 1 mM EDTA, 5% glycerol) and boiled in 1× SDS protein gel loading buffer. Proteins were separated by SDS-polyacrylamide gel electrophoresis (PAGE), and Western blotting was performed to detect p300 amino acids 1 to 670 (p300N) or the N-terminal domain of CBP (CBPN).
Histone acetylation assay.
Core histone acetylation assays were performed as described by Ogryzko et al. (51). A total of 0.2 μg of the BsrFI-NdeI fragment of 4xTRE was assembled into the chromatin template by using purified ACF and NAP1 as described by Ito et al. (29). The chromatin template was then mixed with 100 ng of p300 in the presence or absence of 50 ng of Tax and 50 ng of CREB. The reaction mixtures were incubated for 30 min at 30°C in 30-μl reaction volumes of acetylation buffer containing 10 mM HEPES, 10 mM KCl, 5 mM MgCl2, 10% glycerol, 1 mM DTT, 1 mM AEBSF, 10 mM sodium butyrate, and 1 μl of [14C]acetyl-CoA (55 mCi/mmol; Amersham Pharmacia Biotech). Reaction products were resolved by SDS-PAGE, dried, and exposed to a Molecular Dynamics PhosphorImager plate.
ChIP assay.
The ChIP assay was carried out using the acetylated histone H3 or H4 antibody ChIP assay kit (Upstate Biotechnology) by following the methods described by Kundu et al. (39). After the chromatin reconstitution and acetylation reactions, chromatin was digested with MNase and EDTA was added to stop the reaction. The nucleosomes were precleared with salmon-sperm DNA-protein A agarose beads. The supernatants were diluted 10-fold with ChIP dilution buffer and split into three equal parts for IP with (i) no antibody, (ii) anti-acetylated histone H3 antibody, or (iii) anti-acetylated histone H4 antibody. After overnight rotation at 4°C, the immune complexes were collected by addition of protein A agarose beads. DNA was purified by proteinase K digestion, phenol extraction, and ethanol precipitation and amplified by PCR using the indicated primers. The PCR products were analyzed by electrophoresis using 1% agarose gel and visualized with ethidium bromide staining.
The in vivo ChIP assay was performed by cross-linking protein to DNA in intact HTVL-I cells (MT-2 and HUT102) by using a Surelite SSP UV laser (Nagaich et al., unpublished data) (3). Samples were then processed as described by Shang et al. (60) with minor modifications. Cells were washed with ice-cold PBS and buffer I (0.25% Triton X-100, 10 mM EDTA, 0.5 mM EGTA, 10 mM HEPES [pH 6.5]). Cells were then suspended in 0.3 ml of lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris-HCl [pH 8.1], and 1× protease inhibitor cocktail [Roche Molecular Biochemicals]), sonicated three times for 10 s each, and cleared by centrifugation for 10 min at 12,000 × g. Supernatants were collected and diluted in IP buffer (1% Triton X-100, 2 mM EDTA, 150 mM NaCl, 20 mM Tris-HCl [pH 8.1]), followed by preclearing with 2 μg of sheared salmon sperm DNA and protein A-protein G beads (Oncogene Research) for 2 h at 4°C. IP was carried out using anti-p300 (Upstate), anti-CBP (Upstate Biotech), anti-p65 (Oncogene Research), anti-Tax (Tab172), or preimmune-immunoglobulin G. DNA was subjected to PCR analysis using specific primers for the HTLV-1 LTR or for the IL-15Rα as a control.
In vitro chromatin transcription assay.
Nuclear extracts of HeLa cells were made by the method of Dignam et al. (14) and were dialyzed against buffer containing 20 mM HEPES (pH 7.9), 50 mM KCl, 5 mM MgCl2, and 1 mM DTT. After chromatin reconstitution, 25 μl of chromatin reconstitute reaction mixture (containing 250 ng of DNA) was incubated with or without purified Tax/CREB (50 ng) and p300/CBP for 30 min at 30°C. The p300-selective HAT inhibitor Lys-CoA was used to examine specific HAT activity. Lys-CoA (50 μM) was incubated with p300 for 30 min on ice before addition to the chromatin template. After incubation, HeLa nuclear extract (3- to 4-mg/ml protein) and transcription buffer containing 20 mM HEPES (pH 7.9), 80 mM KCl, 10 mM MgCl2, and 1 mM DTT were added. After incubation at 30°C for 30 min, 4 μl of reaction mixture (ATP, CTP, 3′-O-methyl-GTP, RNase inhibitor, and [α-32P]UTP) was added and the incubation continued for 1 h at 30°C. Ten units of T1 RNase was added, and the incubation was continued for another 10 min. The transcription reaction was terminated by the addition of 200 μl of stop buffer (20 mM Tris-HCl, 1 mM EDTA, 100 mM NaCl, 1% SDS, and 10 μg of yeast tRNA; GIBCO catalogue no. 16051-039). RNA was phenol-chloroform extracted, ethanol precipitated, and separated by using 6% PAGE. The gel then was visualized and quantitated with a PhosphorImager.
RESULTS
Assembly of HTLV-1 LTR chromatin template.
Several studies have shown that p300 and CBP act as coactivators of Tax to activate transcription in vitro and in vivo. Unfortunately, none of these studies were performed on a chromatin template such that the HAT target and mechanism of transactivation could be analyzed. For the present study, we developed an in vitro reconstituted HTLV-1 LTR chromatin template by using a plasmid containing four copies of the 21-bp repeat (4xTRE) cloned upstream of the basal −52 to −1 HTLV-1 promoter (2). For a control promoter, the −52 to −1 promoter region containing no 21-bp repeats was cloned upstream of the G-free cassette. Drosophila embryo extracts were used to provide a method for reconstituting physiologically spaced nucleosomal arrays in vitro (67). Chromatin assembly was carried out using a 2.1-kb BsrFI-NdeI fragment of DNA from the 4xTRE plasmid immobilized on streptavidin beads. The immobilized chromatin facilitates purification and analysis of the protein composition of preinitiation and elongation transcription complexes (15).
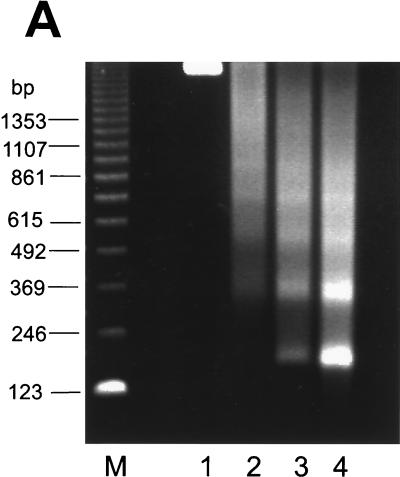
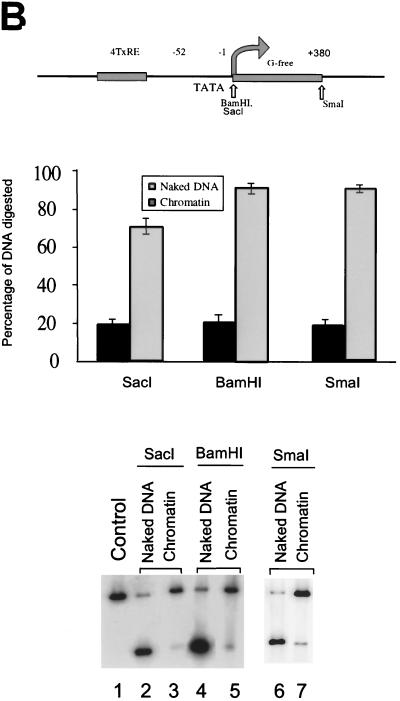
The reconstituted chromatin was initially analyzed by micrococcal nuclease digestion. As shown in Fig. 1A, a typical nucleosome ladder pattern was observed, suggesting that the nucleosomes are regularly spaced on the DNA. The nucleosomal DNA fragments had a periodicity of approximately 180 bp. We also used the restriction enzyme accessibility assay to analyze the reconstituted chromatin structure (Fig. 1B). The positions of the restriction enzyme sites are indicated in the top panel. The 4xTRE chromatin was more resistant to restriction enzyme digestion than the naked DNA. A total of 70 to 90% of naked DNA was cleaved by all three restriction enzymes. In contrast, less than 20% of the reconstituted chromatin DNA was cleaved.
FIG. 1.
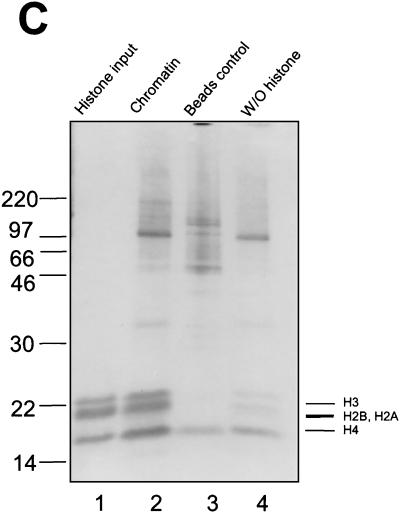
Characterization of reconstituted 4xTRE chromatin template. (A) Micrococcal digestion of 4xTRE chromatin template. Chromatin was digested with micrococcal nuclease for 0 min (lane 1), 0.5 min (lane 2), 1 min (lane 3), or 5 min (lane 4). DNA was purified and analyzed on a 1% agarose gel. (B) Restriction enzyme accessibility assay of reconstituted chromatin. The restriction enzyme sites are indicated in the schematic of the chromatin template. Restriction enzyme digestion was performed as described in Materials and Methods. The levels of cleaved DNA are indicated as percentages of total DNA. The middle panel represents the quantitation of the bands from the bottom panel. (C) Coomassie-stained SDS-16% PAGE showing the histone input (lane 1) and histones eluted from reconstituted chromatin template (lane 2). Lanes 3 and 4 are experimental controls which include beads with no DNA (lane 3) and reconstituted chromatin in the absence of exogenous histones (lane 4). The protein molecular size standards are indicated in kilodaltons.
We also analyzed the protein composition of the chromatin template (Fig. 1C). As expected, the major proteins associated with the chromatin are the histones H2A, H2B, H3, and H4, which are present at approximately equimolar amounts (Fig. 1C, lane 2). The ratio of histone proteins looks similar to that of the input histone sample (Fig. 1C, lane 1). Several other minor higher-molecular-weight bands between 46 and 220 kDa were also observed. These proteins come from the Drosophila embryo extract and also appear in the bead control lane (Fig. 1C, lane 3), suggesting that they represent a nonspecific interaction with the beads. Also present in the bead control lane was a nonspecific protein of approximately 14 kDa, which overlaps the histone H4 band.
Transcription activation of the chromatin template by Tax/CREB and p300 or CBP.
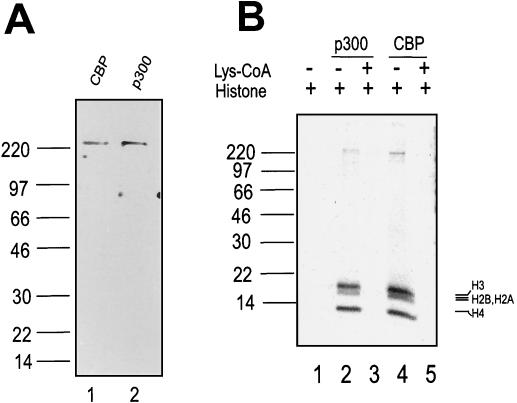
In the experiments described below, we tested the transcriptional activity of purified baculovirus p300 and CBP on naked DNA and the reconstituted chromatin template. The purity of these proteins is demonstrated by GelCode staining (Fig. 2A). Each protein gave a single band on an SDS gel and was judged to be more than 95% pure. The biological activity of the purified proteins was determined by a free histone acetylation assay (Fig. 2B). The results demonstrate that p300 and CBP acetylate the histone substrate (Fig. 2B, lanes 2 and 4). The HAT activity can be inhibited by specific HAT inhibitor Lys-CoA (43) (55), which inhibits p300 and CBP HAT activity (Fig. 2B, lane 3).
FIG. 2.
Purified p300 and CBP proteins. (A) GelCodeBlue-stained SDS-4 to 20% PAGE results showing p300 and CBP. (B) The activity of proteins was determined by free histone acetylation assay. Lys-CoA (50 μM) is a specific HAT inhibitor of p300 and CBP.
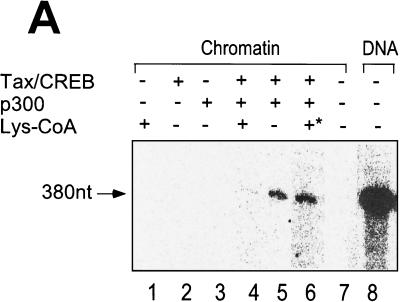
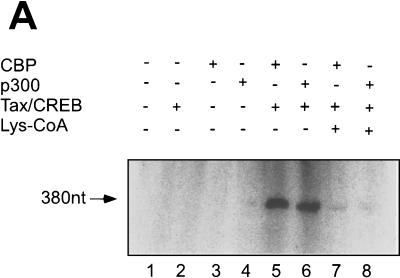
We next performed an in vitro transcription assay using the 4xTRE chromatin template and HeLa cell nuclear extract. Purified Tax/CREB and p300 were added (as described in Materials and Methods) after chromatin reconstitution and incubated with the template for 30 min at 30°C prior to in vitro transcription. Consistent with analyses of transcription from chromatin templates in other laboratories, we found that the chromatin structure significantly reduced the level of transcription from the DNA template (Fig. 3A, lanes 7 and 8). The addition of Tax/CREB or p300 alone to the in vitro transcription reaction mixture failed to activate transcription (Fig. 3A, lanes 2 and 3). The addition of Tax/CREB and p300 together, however, significantly increased the level of transcription (Fig. 3A, lane 5). The ability of p300 to activate transcription in the presence of Tax/CREB was dependent upon HAT activity, since the addition of Lys-CoA abolished the transactivation (Fig. 3A, lane 4). The addition of Lys-CoA to the in vitro transcription reaction mixture after the preincubation step did not inhibit transcription (Fig. 3A, lane 6), suggesting that the p300 HAT function is important for modification of the chromatin structure before transcription.
FIG. 3.
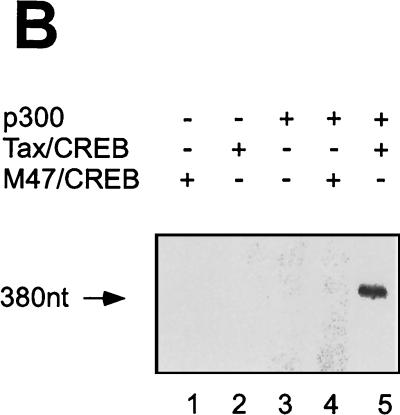
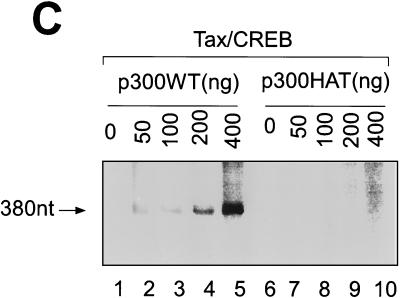
p300 mediates Tax/CREB-dependent transcription from a chromatin template through its HAT activity in vitro. The chromatin template was reconstituted as described in Materials and Methods. The chromatin template was incubated with Tax/CREB and p300 as indicated at 30°C for 30 min and then subjected to in vitro transcription using HeLa cell nuclear extract as described in Materials and Methods. [32P]RNA was purified and analyzed by electrophoresis on a denaturing polyacrylamide gel. (A) Transcription of 4xTRE chromatin with Tax/CREB ± p300. Lys-CoA (50 μM) was either preincubated with p300 (lane 4) or added after incubation of p300 with the chromatin template (lane 6). 380nt, nucleotide 380. (B) Tax mutant M47 fails to activate p300-dependent transcription in vitro. In vitro chromatin transcription reactions were performed as described in Materials and Methods. Tax, CREB, p300, and Tax mutant M47 were added as indicated. (C) A p300 HAT fails to activate transcription.
To address the specificity of Tax transactivation on the reconstituted chromatin template, we utilized Tax mutant M47 (61), which has a double mutation at amino acids 319 and 320. M47 lacks the ability to activate HTLV-1 LTR transcription either in vivo or in vitro (35, 61). Consistent with the results of previous studies, M47 did not cooperate with p300 to activate 4xTRE chromatin transcription (Fig. 3B, lane 4). The M47 Tax protein was active, as demonstrated by protein electroporation of Jurkat lymphocytes with M47 and a NF-κB reporter plasmid (data not shown).
We next wanted to determine if p300 must be tethered to Tax on the promoter. To address this point, we utilized a p300 mutant (p300 HAT) (51), which lacks the Tax/CREB binding site but still possesses HAT activity (amino acids 1195 to 1673) (see Fig. 5A, lane 8). In contrast to the significant transactivation observed with wild-type p300 (Fig. 3C, lanes 2 to 5), no transactivation was observed with p300 HAT (Fig. 3C, lanes 7 to 10). The results suggest that while the p300 HAT activity is critical for Tax-mediated transcription on the 4xTRE, p300 must be tethered to the chromatin template through Tax/CREB to activate transcription.
FIG. 5.
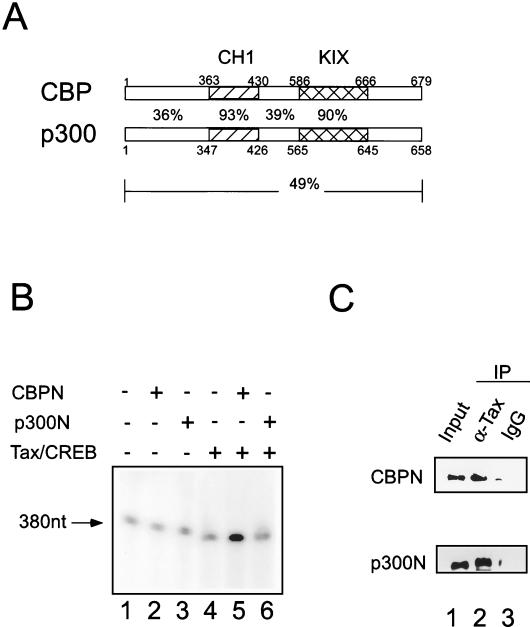
Comparison of CBPN and p300N during Tax-mediated HTLV-1 transcription of a naked DNA template. (A) Schematic of CBPN and p300N. Sequence identity is indicated. (B) CBPN, but not p300N, activates transcription from a naked DNA template. (C) Both CBPN and p300N interact with Tax. CBPN or p300N was incubated with Tax and subsequently immunoprecipitated with an anti-Tax monoclonal antibody. Immunoprecipitates were analyzed by SDS-PAGE and Western blot with either an anti-CBP or anti-p300 antibodies. Lane 1 represents input of CBPN or p300N. Tax interacts with CBPN or p300 (lane 2). IgG, immunoglobulin G.
CBP activates Tax-mediated transcription from chromatin in a HAT-dependent manner.
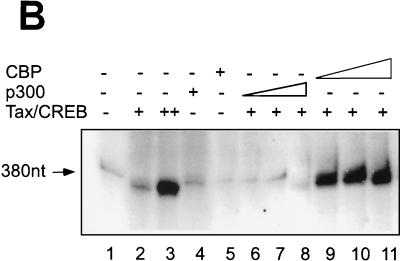
Given the homology between p300 and CBP, we were interested in determining whether CBP could activate transcription from the chromatin template in vitro. Purified recombinant CBP protein was added to the in vitro transcription system as described above for the preincubation and transcription reaction step. As shown in Fig. 4A, CBP, like p300, when added with Tax/CREB, significantly increases transcription from the chromatin template (Fig. 4A, lane 5). The magnitude of transcription activation by CBP is comparable to that by p300 (Fig. 4A, lanes 5 and 6). Like p300, CBP activation depends on the HAT activity, since preincubation of CBP with HAT inhibitor Lys-CoA inhibited transcriptional activation by CBP (Fig. 4A, lane 7).
FIG. 4.
CBP activates Tax-mediated HTLV-1 transcription in vitro from both chromatin and naked DNA templates. (A) CBP activates Tax-mediated chromatin transcription in a HAT-dependent manner. (B) CBP, but not p300, activates transcription from a naked DNA template. Increasing amounts of p300 (lanes 6 to 8, 100, 200, and 400 ng, respectively) and increasing amounts of CBP (lanes 9 to 11, 100, 200, and 400 ng, respectively) were added. (C) CBP activates naked DNA transcription in a HAT-independent manner. Preincubation of CBP with Lys-CoA (50 μM) did not eliminate transcription stimulation by CBP.
CBP, but not p300, activates transcription from a naked DNA template.
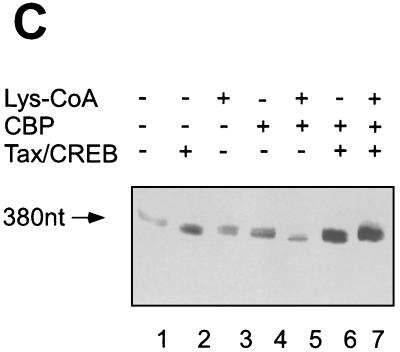
We next compared the abilities of CBP and p300 to activate transcription from a naked DNA template. In vitro transcription assays were carried out by adding Tax/CREB and p300 or CBP with HeLa cell nuclear extract without the addition of mock-reconstituted chromatin template. On the naked DNA template, Tax and CREB are able to activate transcription (Fig. 4B, lane 3). While the addition of CBP or p300 alone did not activate transcription (Fig. 4B, lanes 4 and 5), the addition of increasing concentrations of CBP to the Tax/CREB transcription reaction mixture significantly induced transcription (Fig. 4B; compare lanes 2 and 9 to lane 11). In contrast, the addition of p300 to the transcription assay did not stimulate transcription (Fig. 4B, lanes 6 to 8). Stimulation of Tax/CREB-mediated transcription by CBP is independent of its HAT activity, since when CBP was preincubated with Lys-CoA, it still activated transcription (Fig. 4C, lane 7).
Researchers have previously shown that CBPN, comprising amino acids 1 to 682, facilitates Tax transactivation in a HAT-independent manner, suggesting that CBP contains an N-terminal activation domain (35). To determine whether the N-terminal domain of p300 also contains a transactivation domain, we directly analyzed the transcriptional activity of the N-terminal domain of p300. In contrast to the results obtained with CBPN (Fig. 5B, lane 5), in vitro transcription studies with p300N failed to activate Tax-dependent transcription (Fig. 5B, lane 6). To examine the possibility that the lack of p300N transactivation was due to lack of interaction with Tax, we analyzed the ability of CBPN and p300N to interact with Tax. Purified Tax protein was incubated with p300N or CBPN and then immunoprecipitated with an anti-Tax monoclonal antibody. The presence of CBP or p300 was determined by Western blot analysis with either an anti-CBP antibody or an anti-p300 antibody. Both purified CBP (1-682) and p300 (1-670) proteins were found to interact with Tax protein (Fig. 5C). We searched a protein domain database, the Conserved Domain Database (http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml), to look for potential functional domains. While no distinct domains were identified, some unique regions were found. After alignment of the two sequences, we noticed that although there is high sequence identity within the CH1 and KIX regions of CBP and p300, there is relatively low sequence identity for the regions spanning amino acids 1 to 363 of CBP and 1 to 347 of p300 (Fig. 5A). The sequence differences between CBP and p300 in these regions may contribute to the different transactivation properties of CBP and p300.
p300 acetylates nucleosomal histones on the reconstituted template.
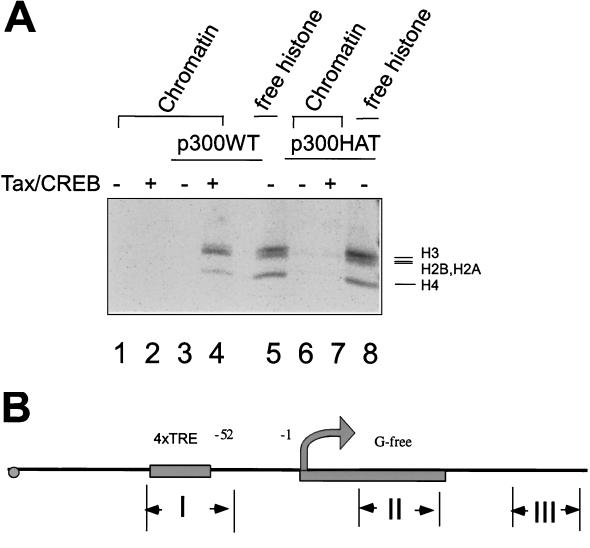
Since our studies indicate that p300 coactivates Tax function through its HAT activity, we explored the mechanism by which p300 may increase Tax-dependent transcription. In particular, we asked if Tax recruits p300 to acetylate nucleosomal histones within the promoter. For this purpose, we reconstituted the HTLV-1 LTR into chromatin by using the recombinant chromatin assembly system as described by Kadonaga and colleagues (28, 30). The fragment of 4xTRE DNA obtained using BsrFI and NdeI was incubated with histone octamers in the presence of NAP1 and ACF proteins in the reconstitution buffer at 27°C for 4 h. The quality of 4xTRE chromatin assembled in this purified system was assayed by micrococcal nuclease digestion, with results similar to those shown in Fig. 1A (data not shown). After assembly of 4xTRE chromatin, the templates were incubated with Tax/CREB and/or p300 in the presence of [14C]acetyl-CoA. The reactions were then terminated, and the proteins were denatured and analyzed by SDS-PAGE and PhosphorImager analysis. The results of these experiments demonstrate that p300, when tethered to the chromatin template via Tax and CREB, acetylates all four of histones 2A, 2B, H3, and H4 on the chromatin template (Fig. 6A, lanes 3 and 4). Consistent with the transcription results presented above, the p300 mutant (p300 HAT), which lacks the interaction site for Tax, failed to acetylate histones on the chromatin template (Fig. 6A, lane 7).
FIG. 6.
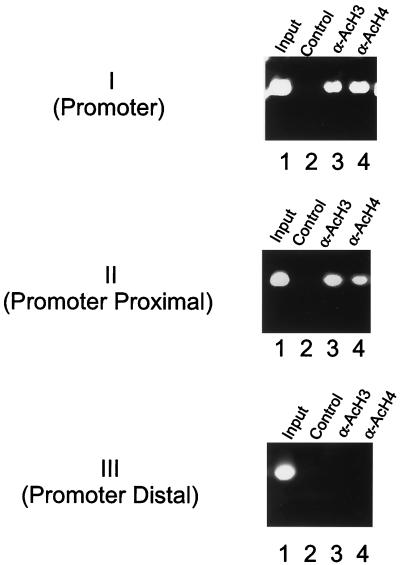
p300 targets chromatin acetylation. (A) Chromatin HAT assay. 4xTRE assembled into chromatin with purified ACF and NAP1 were incubated with Tax/CREB ± p300 in the presence of [14C]acetyl-CoA. Acetylation reaction products were analyzed by SDS-PAGE, and labeled proteins were detected by PhosphorImager. (B) ChIP assays. Chromatin templates were incubated with Tax/CREB + p300 as described for panel A and digested with micrococcal nuclease prior to IP with antibodies against acetylated histone H3 and H4. Recovered DNA was used as template in PCR amplification with primers against different regions of 4xTRE as indicated in the top panel. Input DNA (5%) and protein A agarose controls were included as control reaction ingredients.
p300 acetylates histones H3 and H4 in nucleosomes with the promoter region.
We next performed in vitro ChIP assays. After reconstitution, the chromatin template was incubated with Tax/CREB and p300 in the presence of acetyl-CoA. The template was then digested with micrococcal nuclease into mono- and dinucleosomes as described previously (27). The fragmented chromatin DNA was immunoprecipitated using anti-acetyl-H4 and anti-acetyl-H3 antibodies. Following IP, the DNA was deproteinized and subjected to PCR analysis using primers in the promoter region, G-free region, and downstream region sequences (Fig. 6B). Nucleosomes within the promoter region, which contains the TRE binding site for Tax/CREB/p300, as well as those within the promoter proximal downstream region, were specifically precipitated with the anti-acetyl-H3 and H4 antibodies but not the control antibody (Fig. 6B, top and middle IP assay panels). In contrast, the anti-acetyl-H3 and -H4 antibodies did not precipitate promoter distal nucleosomes located at the 3′ end of the template DNA (Fig. 6B, bottom IP assay panel). The results of this experiment suggest that p300 acetylates histones H3 and H4 within nucleosomes that are located in the promoter region and immediately downstream of the RNA initiation site.
Acetylation by p300 enhanced recruitment of TFIID and RNAP II onto the HTLV-1 LTR chromatin template.
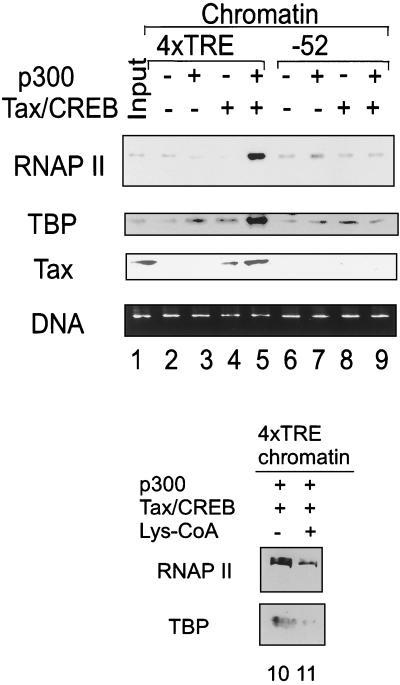
Acetylation of nucleosomes is thought to alter DNA-histone interaction by modifying histone tails. Given the location of the acetylated nucleosomes in the promoter region, it was of interest to determine whether recruitment of basal transcription factors, including RNAP II and TFIID, increases in the presence of p300. The reconstituted immobilized chromatin template was preincubated with Tax/CREB and p300 and then incubated with HeLa nuclear extract in the absence of ribonucleotides to allow formation of the preinitiation complex. The immobilized preinitiation complexes were precipitated, washed gently, and split into two aliquots. One portion was used to analyze the proteins bound to the chromatin template by SDS-PAGE and Western blot analysis with anti-Tax, anti-TBP, and anti-RNAP II antibodies. The other half of the chromatin sample was deproteinized, and the DNA was analyzed by 1% agarose gel electrophoresis. Tax was found to be specifically bound to 4xTRE chromatin but not to the control −52 chromatin (Fig. 7). The level of Tax binding was increased in the presence of p300 (Fig. 7, lanes 4 and 5), suggesting that p300 stabilizes the complex. We saw similar loading of TFIID and RNAP II when either Tax/CREB or p300 was incubated with chromatin alone (Fig. 7, lanes 2 to 4). In contrast, the amount of template-bound RNAP II and TFIID was increased in reactions containing both Tax/CREB and p300 (Fig. 7, lane 5). The increase in TFIID and RNAP II binding was not observed with the control −52 chromatin template, which lacks the Tax-responsive 21-bp repeat elements (Fig. 7, lane 9). Ethidium bromide staining of the agarose gel demonstrated that equivalent amounts of template DNA were recovered in each of the samples (Fig. 7). These results, along with those from the transcription and nucleosome acetylation assays, provide strong evidence that the Tax/CREB/p300 complex targets acetylation of promoter proximal nucleosomes, allowing enhanced binding of TFIID and RNAP II. Consistent with this interpretation, preincubation of p300 with Lys-CoA in the chromatin template binding assay decreased RNAP II and TBP binding (lanes 10 and 11).
FIG. 7.
p300 facilitates recruitment of TFIID and RNAP II to the HTLV-1 chromatin template. Biotinylated LTR chromatin templates were incubated with Tax/CREB ± p300, followed by incubation with HeLa cell nuclear extract as described in Materials and Methods. After incubation and washing, proteins were eluted from beads and analyzed by SDS-PAGE and Western blot using antibodies against Tax, TBP, and RNAP II. Also, DNA fragments were recovered from the beads, analyzed by 1% agarose gel electrophoresis, and stained with ethidium bromide. Input HeLa cell nuclear extract (5%) (lane 1) was included.
In parallel studies, we compared the loading of TFIID and RNAP II on naked DNA. In contrast to the results obtained with the chromatin template, on the naked DNA template there is a higher basal level but no enhanced recruitment of either TFIID or RNAP II in the presence of Tax/CREB and p300 (data not shown).
In vivo, Tax and p300 bind to the HTLV-1 LTR.
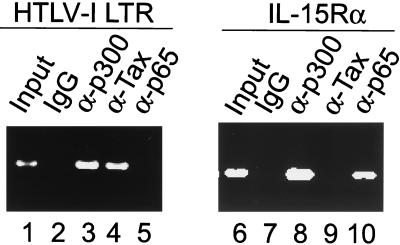
Although extensive studies on the physical and functional interaction between Tax and CBP/p300 have been reported (26, 42, 58, 68), there is no direct evidence that Tax and its coactivators interact with the HTLV-1 LTR chromatin in vivo. To address whether Tax and p300 bind to the integrated HTLV-1 LTR, we performed ChIP assays using MT2 cells, a HTLV-1-transformed cell line. Proteins were cross-linked to the DNA, extracts were prepared, and the DNA was fragmented as described in Materials and Methods. IPs were carried out using control, anti-Tax, anti-p300, and anti-p65 antibodies. Following IP, the DNA was amplified by PCR using probes specific for the HTLV-1 LTR or the cellular IL-15Rα promoter. The IL-15Rα promoter was included as a control in the assay because it is a known Tax-responsive cellular gene that is activated through the Tax/NF-κB pathway (47). The results of the HTLV-1 LTR PCR amplification demonstrate that Tax and p300 bind specifically to the promoter (Fig. 8, left panel). In contrast, the LTR is not immunoprecipitated with NF-κB anti-p65 antibody or control immunoglobulin.
FIG. 8.
Binding of Tax and p300 to HTLV-1 LTR in vivo. ChIP (see Materials and Methods for details) demonstrated promoter occupancy by Tax and p300 from HTLV-1-transformed MT-2 cells. As a control, IL-15Rα promoter was occupied by p65 and p300.
The results of the IL-15Rα promoter PCR amplification demonstrate that NF-κB p65 and p300 are associated with the activated cellular promoter, but Tax is not. The absence of Tax association with the IL-15Rα promoter is not surprising, since the primary mechanism of Tax activation of NF-κB operates through an increase in IκBα phosphorylation and degradation, releasing NF-κB to translocate to the nucleus and activate transcription. In addition to MT-2 cells, another HTLV-1-transformed cell, HUT102, was used for the in vivo ChIP assay. Identical results were obtained in these assays (data not shown).
The presence of endogenous p300 and CBP (data not shown) on the HTLV-1 LTR in vivo suggests that both coactivators are involved in the stimulation of Tax-mediated HTLV-1 transcription. These studies provide the first direct evidence that p300 and CBP associate with the HTLV-1 LTR in vivo.
DISCUSSION
The data presented in this manuscript demonstrate that p300 and CBP facilitate transcription of a Tax-responsive chromatin template in the presence of Tax and CREB, which provide the docking site for the coactivator. The ability of p300 and CBP to activate transcription from the chromatin template is dependent upon HAT activity. Our results suggest that the primary substrates of p300 HAT activity are histones 2A, 2B, H3, and H4 within the nucleosome. These acetylation patterns are consistent with the results of Ito et al., who found that p300 acetylated all four histones in the context of a nucleosomal array (29). In contrast, Kundu et al. (39) demonstrated that interaction of p300 with GAL4-VP16 resulted in the targeted acetylation of histones H3 and H4 within a reconstituted chromatin template. Hyperacetylation of histones H3 and H4 in the beta interferon promoter have also been correlated with transcriptional activation (53). The similarity of these observations to those regarding the estrogen-responsive pS2 promoter, in which acetylation of histones facilitates the binding of TBP, suggests that this may be a common mechanism of activation of many RNAP II promoters (59). It will be of interest to determine whether the observed differences in histone acetylation reflect subtle changes in nucleosome conformation or accessibility on different promoters.
Histone modifications can generate synergistic or antagonistic interaction of chromatin-associated proteins which dictate transition between transcriptionally active and inactive chromatin states (32). Using an in vitro ChIP assay, we observed that acetylation of nucleosomes occurs in the promoter and promoter proximal downstream regions but not in the far downstream region of the template. Furthermore, in our analysis of RNAP II and TFIID, we demonstrated that as a consequence of acetylation of nucleosomal histones, the recruitment of TFIID and RNAP II was enhanced. These results are consistent with the finding that the acetylation of nucleosomal histone tails increases accessibility and allows the transcriptional machinery to be associated with promoter region of chromatin. Indeed, recent in vivo studies of estrogen receptor (ER)-dependent transcription demonstrated that acetylation of nucleosomal histone could enhance TBP recruitment (59). In a separate study, Hassan et al. (27) analyzed the distribution, function, and retention of the yeast SWI/SNF complex by a sequence-specific transcription activator. These studies demonstrated that activator recruitment of SWI/SNF bound the complex to promoter proximal nucleosomes and led to localized disruption of chromatin structure. Interestingly, the retention of SWI/SNF to the promoter required the continued presence of the transcription activator or acetylation of histones. It will be of interest to analyze the interaction of chromatin remodeling factors, such as SWI/SNF, with the HTLV-1 chromatin template.
Our results provide interesting functional differences between CBP and p300. Although CBP and p300 are often referred to as homologues, gene knockout studies provide evidence that they function differently in vivo (69). Yao et al. observed that despite the presence of normal levels of CBP, p300−/− fibroblasts were defective in retinoic acid-dependent transcription (69). Interestingly, the activity of CREB was normal, indicating that the presence of CBP, but not that of p300, is sufficient to activate the CREB pathways (69). Studies in this laboratory have shown that CBP has a transcription activation domain that is independent of its HAT activity (35). In contrast, in vitro transcription studies with p300N failed to activate Tax-dependent transcription (Fig. 5B). Analysis of CBP and p300 amino acid sequences in the N-terminal region revealed only 34% sequence identity in the region encompassing amino acids 1 to 363. Swope et al. (65) previously reported that CBP (1-460) contains a transcriptional activation domain. Thus, it will be of interest to further characterize the N-terminal transactivation domain of CBP in activating Tax-mediated transcription. We have also analyzed the function of full-length CBP and p300 in in vitro transcription systems. However, CBP and p300 may also contain overlapping functions. Both proteins stimulate transcription from a chromatin template in a HAT-dependent manner. Further analysis of the direct targets for p300 and CBP will be of importance. In vivo studies of the ER gene demonstrated that there is a sequential recruitment of coactivators onto the promoter in response to stimuli. p300 is recruited to the promoter first, followed by CBP and PCAF. The time course of p300 and CBP association with the ER promoter suggested a link between p300 and preinitiation complex formation. CBP function was associated with transcription reinitiation (35).
Since CBP stimulates transcription from both chromatin and naked DNA templates, we would expect to observe greater levels of stimulation of chromatin transcription by CBP than p300. However, in our studies CBP activates chromatin transcription to a level similar to that seen with p300. We suspect that additional chromatin remodeling activities, such as ATP-dependent chromatin remodeling activities, are necessary to fully relieve chromatin repression. Indeed, Hassan et al. (27) reported that HAT complexes could stabilize SWI/SNF binding to the promoter region. Thus, CBP may have different functions in a multistep activation pathway during stimulation of HTLV-1 chromatin transcription and certain of these steps may be limiting in the in vitro transcription assay. Experiments to test this hypothesis are under way.
Although extensive studies on the physical and functional interaction between Tax and CBP/p300 have been reported, there is no direct evidence that Tax and these coactivators interact with the HTLV-1 LTR in vivo. It has been shown in vitro that Tax interacts with the HTLV-1 LTR through protein-protein interactions with the CREB protein (17). Several studies have shown that Tax binds to the KIX and C/H1 domains of CBP/p300 and facilitates binding of the coactivator to the Tax-responsive 21-bp repeats (26, 66). Co-IP experiments using either HTLV-1-transformed or -transfected cells have demonstrated that Tax interacts with CBP and p300 in vivo. To extend these studies and demonstrate that Tax and p300 bind to the integrated HTLV-1 LTR, we performed an in vivo ChIP assay using MT2 cells, an HTLV-1-transformed cell line. Our results demonstrate that, in fact, Tax and p300/CBP are bound to the integrated HTLV-1 LTR chromatin template. The presence of endogenous p300 and CBP on the HTLV-1 LTR in vivo suggests both coactivators are involved in the Tax-dependent regulation of HTLV-1 transcription. It will be of importance to analyze the interaction of CBP and p300 with the HTLV-1 promoter at various stages of transcriptional activation and determine the distinct role each coactivator plays in Tax-mediated transcription.
REFERENCES
- 1.Adya, N., L. J. Zhao, W. Huang, I. Boros, and C. Z. Giam. 1994. Expansion of CREB's DNA recognition specificity by Tax results from interaction with Ala-Ala-Arg at positions 282-284 near the conserved DNA-binding domain of CREB. Proc. Natl. Acad. Sci. USA 91:5642-5646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, M. G., K. E. Scoggin, C. M. Simbulan-Rosenthal, and J. A. Steadman. 2000. Identification of poly(ADP-ribose) polymerase as a transcriptional coactivator of the human T-cell leukemia virus type 1 Tax protein. J. Virol. 74:2169-2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Angelov, D., S. Khochbin, and S. Dimitrov. 1999. UV laser footprinting and protein-DNA crosslinking. Application to chromatin. Methods Mol. Biol. 119:481-495. [DOI] [PubMed] [Google Scholar]
- 4.Asahara, H., B. Santoso, E. Guzman, K. Du, P. A. Cole, I. Davidson, and M. Montminy. 2001. Chromatin-dependent cooperativity between constitutive and inducible activation domains in CREB. Mol. Cell. Biol. 21:7892-7900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ballard, D. W., E. Bohnlein, J. W. Lowenthal, Y. Wano, B. R. Franza, and W. C. Greene. 1988. HTLV-1 tax induces cellular proteins that activate the kappa B element in the IL-2 receptor alpha gene. Science 241:1652-1655. [DOI] [PubMed] [Google Scholar]
- 6.Baranger, A. M., C. R. Palmer, M. K. Hamm, H. A. Giebler, A. Brauweiler, J. K. Nyborg, and A. Schepartz. 1995. Mechanism of DNA-binding enhancement by the human T-cell leukaemia virus transactivator Tax. Nature 376:606-608. [DOI] [PubMed] [Google Scholar]
- 7.Becker, P. B., T. Tsukiyama, and C. Wu. 1994. Chromatin assembly extracts from Drosophila embryos. Methods Cell. Biol. 44:207-223. [DOI] [PubMed] [Google Scholar]
- 8.Bex, F., and R. B. Gaynor. 1998. Regulation of gene expression by HTLV-1 Tax protein. Methods 16:83-94. [DOI] [PubMed] [Google Scholar]
- 9.Borrow, J., V. P. J. Stanton, J. M. Andresen, R. Becher, F. G. Behm, R. S. Chaganti, C. I. Civin, C. Disteche, I. Dube, A. M. Frischauf, D. Horsman, F. Mitelman, S. Volinia, A. E. Watmore, and D. E. Housman. 1996. The translocation t(8;16)(p11;p13) of acute myeloid leukaemia fuses a putative acetyltransferase to the CREB-binding protein. Nat. Genet. 14:33-41. [DOI] [PubMed] [Google Scholar]
- 10.Brady, J., K. T. Jeang, J. Duvall, and G. Khoury. 1987. Identification of p40x-responsive regulatory sequences within the human T-cell leukemia virus type 1 long terminal repeat. J. Virol. 61:2175-2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brady, J. N. 1996. Biology of HTLV-1: host cell interactions, p. 79-112. In P. Hollsberg and D. A. Hafl (ed.), Human T-cell lymphotropic virus type 1. John Wiley and Sons Ltd., Chichester, England.
- 12.Chen, C. J., Z. Deng, A. Y. Kim, G. A. Blobel, and P. M. Lieberman. 2001. Stimulation of CREB binding protein nucleosomal histone acetyltransferase activity by a class of transcriptional activators. Mol. Cell. Biol. 21:476-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Curtiss, V. E., R. Smilde, and K. L. McGuire. 1996. Requirements for interleukin 2 promoter transactivation by the Tax protein of human T-cell leukemia virus type 1. Mol. Cell. Biol. 16:3567-3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dignam, J. D., R. M. Lebovitz, and R. G. Roeder. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res 11:1475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fletcher, T. M., B. W. Ryu, C. T. Baumann, B. S. Warren, G. Fragoso, S. John, and G. L. Hager. 2000. Structure and dynamic properties of a glucocorticoid receptor-induced chromatin transition. Mol. Cell. Biol. 20:6466-6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Franchini, G. 1995. Molecular mechanisms of human T-cell leukemia/lymphotropic virus type 1 infection. Blood 86:3619-3639. [PubMed] [Google Scholar]
- 17.Franklin, A. A., M. F. Kubik, M. N. Uittenbogaard, A. Brauweiler, P. Utaisincharoen, M. A. Matthews, W. S. Dynan, J. P. Hoeffler, and J. K. Nyborg. 1993. Transactivation by the human T-cell leukemia virus Tax protein is mediated through enhanced binding of activating transcription factor-2 (ATF-2) ATF-2 response and cAMP element-binding protein (CREB). J. Biol. Chem. 268:21225-21231. [PubMed] [Google Scholar]
- 18.Franklin, A. A., and J. K. Nyborg. 1995. Mechanisms of Tax regulation of human T cell leukemia virus type 1 gene expression. J. Biomed. Sci. 2:17-29. [DOI] [PubMed] [Google Scholar]
- 19.Gayther, S. A., S. J. Batley, L. Linger, A. Bannister, K. Thorpe, S. F. Chin, Y. Daigo, P. Russell, A. Wilson, H. M. Sowter, J. D. Delhanty, B. A. Ponder, T. Kouzarides, and C. Caldas. 2000. Mutations truncating the EP300 acetylase in human cancers. Nat. Genet. 24:300-303. [DOI] [PubMed] [Google Scholar]
- 20.Gessain, A., F. Barin, J. C. Vernant, O. Gout, L. Maurs, A. Calender, and G. de The. 1985. Antibodies to human T-lymphotropic virus type-1 in patients with tropical spastic paraparesis. Lancet 2:407-410. [DOI] [PubMed] [Google Scholar]
- 21.Giebler, H. A., J. E. Loring, K. van Orden, M. A. Colgin, J. E. Garrus, K. W. Escudero, A. Brauweiler, and J. K. Nyborg. 1997. Anchoring of CREB binding protein to the human T-cell leukemia virus type 1 promoter: a molecular mechanism of Tax transactivation. Mol. Cell. Biol. 17:5156-5164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Giordano, A., and M. L. Avantaggiati. 1999. p300 and CBP: partners for life and death. J. Cell. Physiol. 181:218-230. [DOI] [PubMed] [Google Scholar]
- 23.Good, L., S. B. Maggirwar, and S. C. Sun. 1996. Activation of the IL-2 gene promoter by HTLV-1 tax involves induction of NF-AT complexes bound to the CD28-responsive element. EMBO J. 15:3744-3750. [PMC free article] [PubMed] [Google Scholar]
- 24.Goodman, R. H., and S. Smolik. 2000. CBP/p300 in cell growth, transformation, and development. Genes Dev. 14:1553-1577. [PubMed] [Google Scholar]
- 25.Harrod, R., Y. L. Kuo, Y. Tang, Y. Yao, A. Vassilev, Y. Nakatani, and C. Z. Giam. 2000. p300 and p300/cAMP-responsive element-binding protein associated factor interact with human T-cell lymphotropic virus type-1 Tax in a multi-histone acetyltransferase/activator-enhancer complex. J. Biol. Chem. 275:11852-11857. [DOI] [PubMed] [Google Scholar]
- 26.Harrod, R., Y. Tang, C. Nicot, H. S. Lu, A. Vassilev, Y. Nakatani, and C. Z. Giam. 1998. An exposed KID-like domain in human T-cell lymphotropic virus type 1 Tax is responsible for the recruitment of coactivators CBP/p300. Mol. Cell. Biol. 18:5052-5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hassan, A. H., K. E. Neely, and J. L. Workman. 2001. Histone acetyltransferase complexes stabilize swi/snf binding to promoter nucleosomes. Cell 104:817-827. [DOI] [PubMed] [Google Scholar]
- 28.Ito, T., M. Bulger, M. J. Pazin, R. Kobayashi, and J. T. Kadonaga. 1997. ACF, an ISWI-containing and ATP-utilizing chromatin assembly and remodeling factor. Cell 90:145-155. [DOI] [PubMed] [Google Scholar]
- 29.Ito, T., T. Ikehara, T. Nakagawa, W. L. Kraus, and M. Muramatsu. 2000. p300-mediated acetylation facilitates the transfer of histone H2A-H2B dimers from nucleosomes to a histone chaperone. Genes Dev. 14:1899-1907. [PMC free article] [PubMed] [Google Scholar]
- 30.Ito, T., M. E. Levenstein, D. V. Fyodorov, A. K. Kutach, R. Kobayashi, and J. T. Kadonaga. 1999. ACF consists of two subunits, Acf1 and ISWI, that function cooperatively in the ATP-dependent catalysis of chromatin assembly. Genes Dev. 13:1529-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jeang, K. T. 2001. Functional activities of the human T-cell leukemia virus type 1 Tax oncoprotein: cellular signaling through NF-kappa B. Cytokine Growth Factor Rev. 12:207-217. [DOI] [PubMed] [Google Scholar]
- 32.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 293:1074-1080. [DOI] [PubMed] [Google Scholar]
- 33.Jiang, H., H. Lu, R. L. Schiltz, C. A. Pise-Masison, V. V. Ogryzko, Y. Nakatani, and J. N. Brady. 1999. PCAF interacts with tax and stimulates tax transactivation in a histone acetyltransferase-independent manner. Mol. Cell. Biol. 19:8136-8145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jin, D. Y., V. Giordano, K. V. Kibler, H. Nakano, and K. T. Jeang. 1999. Role of adapter function in oncoprotein-mediated activation of NF-κB. Human T-cell leukemia virus type 1 Tax interacts directly with IκB kinase gamma. J. Biol. Chem. 274:17402-17405. [DOI] [PubMed] [Google Scholar]
- 35.Kashanchi, F., J. F. Duvall, R. P. Kwok, J. R. Lundblad, R. H. Goodman, and J. N. Brady. 1998. The coactivator CBP stimulates human T-cell lymphotrophic virus type 1 Tax transactivation in vitro. J. Biol. Chem. 273:34646-34652. [DOI] [PubMed] [Google Scholar]
- 36.Kibler, K. V., and K. T. Jeang. 2001. CREB/ATF-dependent repression of cyclin a by human T-cell leukemia virus type 1 Tax protein. J. Virol. 75:2161-2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kira, J., Y. Itoyama, Y. Koyanagi, J. Tateishi, M. Kishikawa, S. Akizuki, I. Kobayashi, N. Toki, K. Sueishi, and H. Sato. 1992. Presence of HTLV-1 proviral DNA in central nervous system of patients with HTLV-1-associated myelopathy. Ann. Neurol. 31:39-45. [DOI] [PubMed] [Google Scholar]
- 38.Kraus, W. L., and J. T. Kadonaga. 1998. p300 and estrogen receptor cooperatively activate transcription via differential enhancement of initiation and reinitiation. Genes Dev. 12:331-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kundu, T. K., V. B. Palhan, Z. Wang, W. An, P. A. Cole, and R. G. Roeder. 2000. Activator-dependent transcription from chromatin in vitro involving targeted histone acetylation by p300. Mol. Cell 6:551-561. [DOI] [PubMed] [Google Scholar]
- 40.Kung, A. L., V. I. Rebel, R. T. Bronson, L. E. Ch'ng, C. A. Sieff, D. M. Livingston, and T. P. Yao. 2000. Gene dose-dependent control of hematopoiesis and hematologic tumor suppression by CBP. Genes Dev. 14:272-277. [PMC free article] [PubMed] [Google Scholar]
- 41.Kuo, Y. L., Y. Tang, R. Harrod, P. Cai, and C. Z. Giam. 2000. Kinase-inducible domain-like region of HTLV type 1 tax is important for NF-κB activation. AIDS Res. Hum. Retrovir. 16:1607-1612. [DOI] [PubMed] [Google Scholar]
- 42.Kwok, R. P., M. E. Laurance, J. R. Lundblad, P. S. Goldman, H. Shih, L. M. Connor, S. J. Marriott, and R. H. Goodman. 1996. Control of cAMP-regulated enhancers by the viral transactivator Tax through CREB and the co-activator CBP. Nature 380:642-646. [DOI] [PubMed] [Google Scholar]
- 43.Lau, O. D., T. K. Kundu, R. E. Soccio, S. A. Ait, E. M. Khalil, A. Vassilev, A. P. Wolffe, Y. Nakatani, R. G. Roeder, and P. A. Cole. 2000. HATs off: selective synthetic inhibitors of the histone acetyltransferases p300 and PCAF. Mol. Cell 5:589-595. [DOI] [PubMed] [Google Scholar]
- 44.Laurance, M. E., R. P. Kwok, M. S. Huang, J. P. Richards, J. R. Lundblad, and R. H. Goodman. 1997. Differential activation of viral and cellular promoters by human T-cell lymphotropic virus-1 tax and cAMP-responsive element modulator isoforms. J. Biol. Chem. 272:2646-2651. [DOI] [PubMed] [Google Scholar]
- 45.Lenzmeier, B. A., H. A. Giebler, and J. K. Nyborg. 1998. Human T-cell leukemia virus type 1 Tax requires direct access to DNA for recruitment of CREB binding protein to the viral promoter. Mol. Cell. Biol. 18:721-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li, X. H., K. M. Murphy, K. T. Palka, R. M. Surabhi, and R. B. Gaynor. 1999. The human T-cell leukemia virus type-1 Tax protein regulates the activity of the IκB kinase complex. J. Biol. Chem. 274:34417-34424. [DOI] [PubMed] [Google Scholar]
- 47.Mariner, J. M., V. Lantz, T. A. Waldmann, and N. Azimi. 2001. Human T cell lymphotropic virus type 1 Tax activates IL-15R alpha gene expression through an NF-kappa B site. J. Immunol. 166:2602-2609. [DOI] [PubMed] [Google Scholar]
- 48.Marriott, S. J., D. Trinh, and J. N. Brady. 1992. Activation of interleukin-2 receptor alpha expression by extracellular HTLV-1 Tax1 protein: a potential role in HTLV-1 pathogenesis. Oncogene 7:1749-1755. [PubMed] [Google Scholar]
- 49.Martinez-Balbas, M. A., A. J. Bannister, K. Martin, P. Haus-Seuffert, M. Meisterernst, and T. Kouzarides. 1998. The acetyltransferase activity of CBP stimulates transcription. EMBO J. 17:2886-2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Murata, T., R. Kurokawa, A. Krones, K. Tatsumi, M. Ishii, T. Taki, M. Masuno, H. Ohashi, M. Yanagisawa, M. G. Rosenfeld, C. K. Glass, and Y. Hayashi. 2001. Defect of histone acetyltransferase activity of the nuclear transcriptional coactivator CBP in Rubinstein-Taybi syndrome. Hum. Mol. Genet. 10:1071-1076. [DOI] [PubMed] [Google Scholar]
- 51.Ogryzko, V. V., R. L. Schiltz, V. Russanova, B. H. Howard, and Y. Nakatani. 1996. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell 87:953-959. [DOI] [PubMed] [Google Scholar]
- 52.Osame, M., K. Usuku, S. Izumo, N. Ijichi, H. Amitani, A. Igata, M. Matsumoto, and M. Tara. 1986. HTLV-1 associated myelopathy, a new clinical entity. Lancet i:1031-1032. [DOI] [PubMed] [Google Scholar]
- 53.Parekh, B. S., and T. Maniatis. 1999. Virus infection leads to localized hyperacetylation of histones H3 and H4 at the IFN-beta promoter. Mol. Cell 3:125-129. [DOI] [PubMed] [Google Scholar]
- 54.Poiesz, B. J., F. W. Ruscetti, A. F. Gazdar, P. A. Bunn, J. D. Minna, and R. C. Gallo. 1980. Detection and isolation of type C retrovirus particles from fresh and cultured lymphocytes of a patient with cutaneous T-cell lymphoma. Proc. Natl. Acad. Sci. USA 77:7415-7419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Polesskaya, A., I. Naguibneva, L. Fritsch, A. Duquet, S. A. Ait, P. Robin, A. Vervisch, L. L. Pritchard, P. Cole, and A. Harel-Bellan. 2001. CBP/p300 and muscle differentiation: no HAT, no muscle. EMBO J. 20:6816-6825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Portis, T., J. C. Harding, and L. Ratner. 2001. The contribution of NF-kappa B activity to spontaneous proliferation and resistance to apoptosis in human T-cell leukemia virus type 1 Tax-induced tumors. Blood 98:1200-1208. [DOI] [PubMed] [Google Scholar]
- 57.Schiltz, R. L., C. A. Mizzen, A. Vassilev, R. G. Cook, C. D. Allis, and Y. Nakatani. 1999. Overlapping but distinct patterns of histone acetylation by the human coactivators p300 and PCAF within nucleosomal substrates. J. Biol. Chem. 274:1189-1192. [DOI] [PubMed] [Google Scholar]
- 58.Scoggin, K. E., A. Ulloa, and J. K. Nyborg. 2001. The oncoprotein Tax binds the SRC-1-interacting domain of CBP/p300 to mediate transcriptional activation. Mol. Cell. Biol. 21:5520-5530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sewack, G. F., T. W. Ellis, and U. Hansen. 2001. Binding of TATA binding protein to a naturally positioned nucleosome is facilitated by histone acetylation. Mol. Cell. Biol. 21:1404-1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shang, Y., X. Hu, J. DiRenzo, M. A. Lazar, and M. Brown. 2000. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell 103:843-852. [DOI] [PubMed] [Google Scholar]
- 61.Smith, M. R., and W. C. Greene. 1990. Identification of HTLV-1 tax trans-activator mutants exhibiting novel transcriptional phenotypes. Genes Dev. 4:1875-1885. [DOI] [PubMed] [Google Scholar]
- 62.Sun, S. C., and D. W. Ballard. 1999. Persistent activation of NF-κB by the tax transforming protein of HTLV-1: hijacking cellular IκB kinases. Oncogene 18:6948-6958. [DOI] [PubMed] [Google Scholar]
- 63.Sun, S. C., E. W. Harhaj, G. Xiao, and L. Good. 2000. Activation of I-κB kinase by the HTLV type 1 Tax protein: mechanistic insights into the adaptor function of IKKγ. AIDS Res. Hum. Retrovir. 16:1591-1596. [DOI] [PubMed] [Google Scholar]
- 64.Suzuki, T., H. Hirai, J. Fujisawa, T. Fujita, and M. Yoshida. 1993. A trans-activator Tax of human T-cell leukemia virus type 1 binds to NF-kappa B p50 and serum response factor (SRF) and associates with enhancer DNAs of the NF-kappa B site and CArG box. Oncogene 8:2391-2397. [PubMed] [Google Scholar]
- 65.Swope, D. L., C. L. Mueller, and J. C. Chrivia. 1996. CREB-binding protein activates transcription through multiple domains. J. Biol. Chem. 271:28138-28145. [DOI] [PubMed] [Google Scholar]
- 66.van Orden, K., J. P. Yan, A. Ulloa, and J. K. Nyborg. 1999. Binding of the human T-cell leukemia virus Tax protein to the coactivator CBP interferes with CBP-mediated transcriptional control. Oncogene 18:3766-3772. [DOI] [PubMed] [Google Scholar]
- 67.Wu, C., T. Tsukiyama, D. Gdula, P. Georgel, M. Martinez-Balbas, G. Mizuguchi, V. Ossipow, R. Sandaltzopoulos, and H. M. Wang. 1998. ATP-dependent remodeling of chromatin. Cold Spring Harbor Symp. Quant. Biol. 63:525-534. [DOI] [PubMed] [Google Scholar]
- 68.Yan, J. P., J. E. Garrus, H. A. Giebler, L. A. Stargell, and J. K. Nyborg. 1998. Molecular interactions between the coactivator CBP and the human T-cell leukemia virus Tax protein. J. Mol. Biol. 281:395-400. [DOI] [PubMed] [Google Scholar]
- 69.Yao, T. P., S. P. Oh, M. Fuchs, N. D. Zhou, L. E. Ch'ng, D. Newsome, R. T. Bronson, E. Li, D. M. Livingston, and R. Eckner. 1998. Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell 93:361-372. [DOI] [PubMed] [Google Scholar]
- 70.Yin, M. J., and R. B. Gaynor. 1996. Complex formation between CREB and Tax enhances the binding affinity of CREB for the human T-cell leukemia virus type 1 21-base-pair repeats. Mol. Cell. Biol. 16:3156-3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yin, M. J., E. J. Paulssen, J. S. Seeler, and R. B. Gaynor. 1995. Protein domains involved in both in vivo and in vitro interactions between human T-cell leukemia virus type 1 tax and CREB. J. Virol. 69:3420-3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yoshida, M. 1995. HTLV-1 oncoprotein Tax deregulates transcription of cellular genes through multiple mechanisms. J. Cancer Res. Clin. Oncol. 121:521-528. [DOI] [PubMed] [Google Scholar]
- 73.Yoshida, M., and M. Seiki. 1987. Recent advances in the molecular biology of HTLV-1: trans-activation of viral and cellular genes. Annu. Rev. Immunol. 5:541-559. [DOI] [PubMed] [Google Scholar]
- 74.Yoshida, M., M. Seiki, K. Yamaguchi, and K. Takatsuki. 1984. Monoclonal integration of human T-cell leukemia provirus in all primary tumors of adult T-cell leukemia suggests causative role of human T-cell leukemia virus in the disease. Proc. Natl. Acad. Sci. USA 81:2534-2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhao, L. J., and C. Z. Giam. 1992. Human T-cell lymphotropic virus type 1 (HTLV-1) transcriptional activator, Tax, enhances CREB binding to HTLV-1 21-base-pair repeats by protein-protein interaction. Proc. Natl. Acad. Sci. USA 89:7070-7074. [DOI] [PMC free article] [PubMed] [Google Scholar]