Abstract
Stimulation of transcriptional elongation by the human immunodeficiency virus type 1 Tat protein is mediated by CDK9, a kinase that phosphorylates the RNA polymerase II carboxyl-terminal domain (CTD). In order to obtain direct evidence that this phosphorylation event can alter RNA polymerase processivity, we prepared transcription elongation complexes that were arrested by the lac repressor. The CTD was then dephosphorylated by treatment with protein phosphatase 1. The dephosphorylated transcription complexes were able to resume the transcription elongation when IPTG (isopropyl-β-d-thiogalactopyranoside) and nucleotides were added to the reaction. Under these chase conditions, efficient rephosphorylation of the CTD was observed in complexes containing the Tat protein but not in transcription complexes prepared in the absence of Tat protein. Immunoblots and kinase assays with synthetic peptides showed that Tat activated CDK9 directly since the enzyme and its cyclin partner, cyclin T1, were present at equivalent levels in transcription complexes prepared in the presence or absence of Tat. Chase experiments with the dephosphorylated elongation transcription complexes were performed in the presence of the CDK9 kinase inhibitor DRB (5,6-dichloro-1-β-d-ribofuranosyl-benzimidazole). Under these conditions there was no rephosphorylation of the CTD during elongation, and transcription through either a stem-loop terminator or bent DNA arrest sequence was strongly inhibited. In experiments in which the CTD was phosphorylated prior to elongation, the amount of readthrough of the terminator sequences was proportional to the extent of the CTD modification. The change in processivity is due to CTD phosphorylation alone, since even after the removal of Spt5, the second substrate for CDK9, RNA polymerase elongation is enhanced by Tat-activated CDK9 activity. We conclude that phosphorylation of the RNA polymerase II CTD by CDK9 enhances transcription elongation directly.
Transcription in eukaryotic cells is regulated by a series of phosphorylation events involving the carboxyl-terminal domain (CTD) of the largest subunit of RNA polymerase II (Pol II) (for a review, see reference 55). Initially, Pol II with a hypophosphorylated CTD (Pol IIa) is recruited to the promoter. The hypophosphorylated CTD is able to form a tightly packed complex at the promoter, together with general initiation factors and the mediator complex. During initiation, the CTD becomes highly phosphorylated, creating a form of the enzyme called Pol IIo. The large conformational change induced in the enzyme during the transition from initiation to early elongation releases the phosphorylated CTD and permits it to interact with a variety of elongation factors, as well as with proteins involved in mRNA processing (28).
The CTD comprises 52 tandem repeats with the consensus sequence Tyr1-Ser2-Pro3-Thr4-Ser5-Pro6-Ser7. Three cyclin-dependent kinases—CDK7, CDK8, and CDK9—are known to phosphorylate the CTD during transcription (55). The three CTD kinases primarily phosphorylate the CTD at Ser5, although Ser2 can also be phosphorylated by CDK9. Each enzyme displays a distinct preference for individual heptads in the CTD repeat. The distinctive phosphorylation patterns produced by each kinase provide unique signals to the transcription machinery (53).
The activities of the CTD kinases are associated with specific stages of transcription. CDK7/cyclin H is a component of the general transcription factor TFIIH and is the kinase that phosphorylates the CTD after the preinitiation complex is formed (2). The CDK9/cyclin T1 (CycT1) kinase was originally identified as part of the positive transcription factor b (P-TEFb), a factor that is critical for the transition of Pol II into productive elongation (52). The role of CDK8/cyclin C is poorly understood, but it is known to be a component of the mammalian SRB/mediator complex (64), a regulatory complex targeted by viral transcription activators such as E1a and VP16 (1, 24).
CDK9 also plays a crucial role in the activation of transcription elongation by the human immunodeficiency virus type 1 (HIV-1) Tat protein (for reviews, see references 33, 52, and 60). In the absence of Tat, the RNA Pol II complexes engaged at the HIV promoter are dysfunctional and elongate poorly. Recruitment of Tat to elongating transcription complexes enhances processivity and permits synthesis of a high level of full-length transcripts (16, 32). Tat associates with the elongating RNA polymerase after binding to trans-activation-responsive region (TAR) RNA, an RNA stem-loop structure that is located at the 5′ end of HIV transcripts (34). The recognition sequence of Tat on TAR is restricted to the region surrounding a trinucleotide bulge located near the apex of the TAR RNA stem-loop structure. However, CycT1, the cyclin partner of CDK9, is able to increase the affinity of Tat for TAR RNA by extending the recognition sequence on TAR RNA to include the apical loop sequence (4, 21, 22, 59, 65, 71). The quaternary complex between CDK9, CycT1, Tat, and TAR RNA induces conformational changes in the enzyme that result in the activation of the CDK9 kinase (4, 8, 19, 22, 31, 39) and, subsequently, the release of TAR (34). The Tat-activated CDK9 is then able to extensively phosphorylate the CTD, creating a unique and highly phosphorylated form that we have designated Pol IIo* (27, 29, 41, 72, 74). In addition to phosphorylating the RNA polymerase CTD, CDK9 is also able to target Spt5, an elongation factor that enhances the stability of transcription complexes at terminator sequences (6, 30, 36, 66).
It has been surprisingly difficult to obtain experimental evidence that RNA polymerase processivity is regulated directly by CTD phosphorylation by CDK9. Tat activity can be blocked selectively by inhibitors that block CDK9 kinase activity (41) or interfere with CycT1 (7) or by immunodepletion of CDK9 from nuclear extracts (18). However, the use of these compounds or CDK9-depleted extracts does not permit isolation of transcription complexes that contain hypophosphorylated CTD since modification of CTD by CDK7 during initiation produces a highly phosphorylated polymerase. Here we have exploited the unique features of HIV transcriptional regulation to compare the processivity of hypophosphorylated elongation complexes to complexes that have been exclusively phosphorylated by CDK9. The results show that only the phosphorylated elongation complexes are able to read through terminator sequences efficiently.
MATERIALS AND METHODS
Templates.
Plasmids carrying the wild-type HIV-1 long terminal repeat (LTR) (pW1), the mGC mutation in the Tat-binding site of TAR (G26:C39 to C:G), or the mLG mutation of the apical loop of TAR (G32-34 to UUU) were described previously (12). pH3.3, which contains two tandem bent DNA arrest sequences (35, 57), was constructed by replacing the stem-loop terminator sequence of pW1 (located between the NheI and AccI sites) by the sequence 5′-CTAGCATTTTTAAAAGAGGGACGTTTTTTTCCCTTTTTTGGAGAGGCGGAAACTTGATGT-3′. The plasmid pΔTerm was created by replacing the terminator sequence by a DNA sequence of equivalent length from the corresponding region of the HIV genome (5′-CTAGCAGGAGAGAGATGGGTGCGAGAGCGTCGGTATTAAGCGGGGGAGAATTAGATAAGT-3′). Plasmid DNAs were linearized with XbaI and biotinylated at both the 5′ and 3′ ends by incorporation of biotin-16-dUTP (Roche) as described previously (34).
Preinitiation complexes.
In order to form preinitiation complexes, 0.5 μg of DNA was linearized, biotinylated, and immobilized on streptavidin-coated magnetic beads (Dynal). The beads were incubated in 40-μl reaction mixtures containing 20 μl of HeLa cell nuclear extract, 50 mM KCl, 4 mM MgCl2, 20 mM HEPES (pH 7.9), 11.25 μM ZnSO4, 3 mM dithiothreitol (DTT), 1 μg of poly(dI-dC), and 50 μM dATP (which was used as a phosphate donor) in the presence or absence of 20 ng of Tat for 10 min at 30°C. The HeLa nuclear extracts were preincubated with hexokinase (20 U/ml of extract) and 11 mM glucose for 5 min at 30°C in order to remove endogenous ATP and then treated with 100 μM DNA-PK inhibitor LY294002 (Sigma) for a further 5 min at 30°C.
To label the preinitiation complexes with [γ-32P]ATP, the complexes were first assembled on the immobilized DNA templates as described above in the absence of nucleotides. The complexes were washed once with EBCD buffer (50 mM Tris-HCl [pH 8.0], 120 mM NaCl, 0.5% [vol/vol] NP-40, 5 mM DTT, 4 mM MgCl2), resuspended in 40 μl of 20 mM HEPES (pH 7.9)-50 mM KCl-4 mM MgCl2-3 mM DTT-11.25 μM ZnSO4 and 10 μCi of [γ-32P]ATP as a phosphate donor, and incubated for another 10 min at 30°C. The preinitiation complexes were washed twice with EBCD wash buffer and analyzed by immunoblotting or protease digestion as described below.
Elongation complexes paused at residue 14.
Early-stage elongation complexes paused at the uridine residue at position 14 were obtained by adding 10 μCi of [α-32P]UTP, 50 μM GTP, 50 μM CTP, 2.5 μg of creatine kinase/ml, and 10 μM creatine phosphate to the preinitiation reaction mixtures, followed by incubation for a further 10 min at 30°C. Because of the low levels of UTP present during the labeling step, 125 μM UTP was added to the reaction for 2 min to chase the smaller transcripts to position 14.
Elongation complexes.
Streptavidin-coated magnetic beads carrying 0.5 μg of biotinylated template DNA were added to 40-μl reaction mixtures containing 20 μl of HeLa cell nuclear extract (preincubated for 5 min at 30°C with 100 μM DNA-PK inhibitor LY294002), 20 mM HEPES (pH 7.9), 50 mM KCl, 4 mM MgCl2, 11.25 μM ZnSO4, 3 mM DTT, 10 μM creatine phosphate, 2.5 μg of creatine kinase/ml, 50 μM GTP, 50 μM ATP, 50 μM CTP, 5 μM UTP, 10 μCi of [α-32P]UTP (400 Ci/μmol; Amersham), 1 μg of poly(dI-dC), and 100 ng of LacR. Where indicated, 20 ng of Tat protein was added to the reactions. Reaction mixtures were incubated for 20 min at 30°C with occasional mixing.
Dephosphorylation and 32P labeling of RNA polymerase.
Elongation complexes arrested by lac repressor were treated with 2.5 U of protein phosphatase 1 (PP1; New England Biolabs) for 1 h at 30°C with occasional mixing. The dephosphorylated complexes were washed with TMZ buffer (10 mM HEPES [pH 7.9], 2 mM DTT, 6.25 μM ZnSO4, 4 mM MgCl2) to remove PP1 for the following chase experiments. To label RNA polymerase with 32P, the dephosphorylated complexes were first washed with EBCD containing 0.1% (vol/vol) Sarkosyl to remove nonspecifically bound kinases from the elongation complexes. The washed complexes were labeled with 10 μCi of [γ-32P]ATP for 5 min, followed by the addition of 0.5 μM unlabeled ATP for a further 5 min.
To purify 32P-labeled RNA Pol II, transcription complexes bound on the magnetic beads were resuspended in 15 μl of 0.5% sodium dodecyl sulfate (SDS) and boiled for 5 min to dissolve the proteins. Pol II was immunoprecipitated from the SDS-protein solution by the addition of 15 μl of 0.5% NP-40, 2 μg of Pol II (N-20) antibody, 20 μl of phosphate-buffered saline, and 60 μl of 2× EBCD buffer. After incubation for 2 h at 4°C with roll mixing, 10 μl of protein A-Sepharose beads was added, and the samples were incubated for an additional hour. The beads were collected by brief centrifugation and washed three times with EBCD buffer prior to analysis.
Chase of elongation complexes arrested at LacR.
Although a high proportion of the arrested transcription complexes pause at the lac repressor, a number of polymerases can be found attached to the templates at more distal sites. To minimize this background in the chase experiments, the long transcripts were removed by treatment with RNase H (34). Transcripts extending beyond the LacR site were removed by RNase H treatment in the presence of RHX1 (CTAGCCTCCGCTAGTCAAAA, nucleotides 270 to 289), RHLAC (CGCCTCTTGCCGTGCGCGCC, nucleotides 220 to 239), 1 U of RNase H, and 40 U of RNasin as described previously (12).
The LacR-arrested complexes were chased in the presence or absence of DRB (5,6-dichloro-1-β-d-ribofuranosyl-benzimidazole) after dephosphorylation by PP1. The beads carrying the templates were resuspended in 40 μl of the chase solution containing 20 mM HEPES (pH 7.9), 75 mM KCl, 4 mM MgCl2, 11.25 μM ZnSO4, 3 mM DTT, 1 μg of poly(dI-dC), 10 μM creatine phosphate, 2.5 μg of creatine kinase/ml, 50 μM DNA-PK inhibitor LY294002 (Sigma), 250 μM GTP, 250 μM ATP, 250 μM CTP, 5 μM UTP, 1 μg of protein phosphatase inhibitor 2, and 25 mM IPTG (isopropyl-β-d-thiogalactopyranoside) and then incubated for various times up to 10 min at 30°C. Aliquots were removed at each step in the transcription reactions, washed once with EBCD buffer, and then analyzed in parallel for RNA and protein content.
Analysis of RNA transcripts.
To analyze RNA transcripts, immobilized transcription complexes on streptavidin magnetic beads were resuspended in 40 μl of TE buffer (10 mM Tris-HCl [pH 8.0], 1.0 mM EDTA), and 50 μl of stop solution (0.6 M sodium acetate, 2% SDS, 40 mM EDTA, 200 μg of tRNA/ml) was added to the sample. After phenol extraction and ethanol precipitation, the RNA was dissolved in 8 μl of RNA loading buffer (80% [vol/vol] formamide, 1 mM EDTA, 0.1% [wt/vol] bromophenol blue, 0.1% [wt/vol] xylene cyanol), denatured at 95°C for 5 min, and fractionated on 6% polyacrylamide gels containing 7 M urea.
Protease digestion.
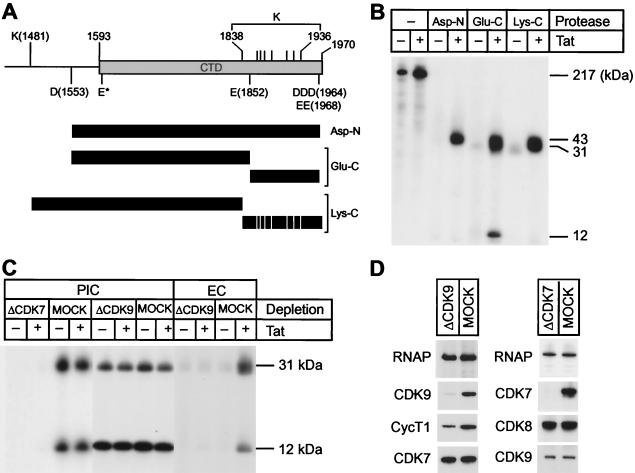
Immunoprecipitated RNA polymerase was digested by adding 200 ng of Glu-C, Asp-N, or Lys-C protease to reactions containing 100 mM Tris-HCl (pH 7.8 for Glu-C and pH 8.5 for Asp-N and Lys-C) and 0.01% SDS. After incubation for 16 h at 37°C, the digested peptide products were dissolved in SDS loading buffer and separated on 4 to 12% Bis-Tris-SDS gels. After transfer to nitrocellulose membranes, labeled proteins were detected by autoradiography. Densitometry (Molecular Dynamics) was used to determine the extent of phosphorylation on each peptide.
In vitro kinase assay.
P-TEFb (TAK) used for the kinase assay was immunoprecipitated from cleared HeLa nuclear extract. To clear the nuclear extracts, 5 μg of rabbit immunoglobulin G (IgG; Santa Cruz Biotechnology) was added to 100 μl of extract at 4°C for 30 min, followed by the addition of 50 μl of bovine serum albumin (BSA)-treated protein A-Sepharose beads (Amersham Pharmacia Biotech, Inc.) for a further 30 min. The beads were removed by brief centrifugation. The cleared extracts were then incubated with 5 μg of anti-CDK9 antibody (H-169; Santa Cruz Biotechnology) for 1 h at 4°C and further incubated with 100 μl of protein A-Sepharose beads for an hour at 4°C. The P-TEFb-bound beads were collected by brief centrifugation and washed three times with 1 ml of EBCD buffer and twice with 1 ml of TKB/Mg buffer (50 mM Tris-HCl [pH 7.4], 10 mM MgCl2, 5 mM DTT).
We used 2.5 μl of beads carrying the immunoprecipitated P-TEFb or P-TEFb present in the LacR-arrested and dephosphorylated elongation complexes obtained from a 40-μl transcription reaction for the in vitro kinase assays. P-TEFb was incubated in a solution containing TKB/Mg buffer, 20 μg of the CTD peptide containing five heptad repeat sequences (YSPTSPKYSPTSPTYSPTTPKYSPTSPTYSPTSPV), and 1 μCi of [γ-32P]ATP in the presence or absence of 2 μM unlabeled ATP and 2.5 mM MnCl2 for 1 h at 30°C. A total of 20 ng of Tat was included in the kinase reaction of immunoprecipitated CDK9 where indicated. The phosphorylated peptides were separated on 4 to 20% Bis-Tris-SDS gels (Invitrogen, Inc.) and detected by autoradiography. The extent of the reaction was measured quantitatively by densitometry (Molecular Dynamics).
Immunodepletion of nuclear extracts.
Protein A-Sepharose beads (100 μl) were saturated with BSA for 30 min, and the BSA-treated beads were incubated with 15 to 20 μg of specific antibodies for 1 h at 4°C. Rabbit polyclonal anti-CDK7 (C-19; Santa Cruz Biotechnology), rabbit polyclonal anti-CDK9 (H-169; Santa Cruz Biotechnology), or monoclonal anti-Spt5 (DRB sensitivity-inducing factor [DSIF]; BD Transduction Laboratories) was used for the depletion of specific proteins from extracts. The equivalent amount of mouse IgG or rabbit IgG (Santa Cruz Biotechnology) was used for mock depletion of the extracts. A total of 100 μl of antibody-bound beads was used to deplete 100 μl of nuclear extract. The beads were divided into three aliquots, and each aliquot was used for one round of depletion. The first round of depletion was carried out for 2 h, followed by two further rounds of depletions which were incubated for 1 h each by using fresh aliquots of the antibody-bound beads. After the third round, the nuclear extract was incubated for 20 min with 20 μl of BSA-saturated protein A-Sepharose beads to remove the remaining antibodies from the nuclear extract. All of the incubation steps were performed at 4°C with roll mixing.
Immunoblots.
The proteins were separated on 4% Tris glycine-SDS gels or 3 to 8% Tris acetate-SDS gels (Invitrogen, Inc.) for the analysis of the RNA Pol II and Spt5 or were separated on 10% or 4 to 12% Bis-Tris-SDS gels for the analysis of the other proteins and transferred onto a nitrocellulose membrane (Schleicher & Schuell). The RNA Pol II was detected by using either the polyclonal Pol II N-20 antibody (Santa Cruz Biotechnology) or the anti-CTD monoclonal antibody 8WG16 (BAbCO). Phosphoserine 2 (Ser2) or phosphoserine 5 (Ser5) modifications of the CTD were detected by using the H5 or H14 antibodies (BAbCO), respectively. Spt5 was detected with a polyclonal antibody (a gift from Richard Gaynor) directed against the C-terminal region (residues 852 to 1087) of the protein. Additional transcription factors were detected with the anti-CDK7 (C-19), anti-CDK8 (C-19), anti-CDK9 (H-169), anti-CycH (D-10), and anti-CycT1 (C-20) antibodies (Santa Cruz Biotechnology). The proteins were visualized by using the ECL Detection Kit.
RESULTS
Experimental design.
In order to examine the effect of the phosphorylation of the RNA polymerase CTD by CDK9 during transcription, we developed a technique for preparing unphosphorylated elongation complexes. Since initiation at the HIV promoter is completely blocked at kinase inhibitor concentrations that prevent formation of the Pol IIo form of the enzyme, we first prepared elongation complexes arrested by the lac repressor (LacR) by the method of Keen et al. (34). The phosphate groups present on the CTD arising from the combined action of CDK7 and CDK9 were then removed from the paused elongation complexes by treatment with PP1 (Fig. 1). The dephosphorylated transcription complexes (Pol IIa) were able to resume transcription elongation after the addition of IPTG and nucleotides. During the transcription reaction, CDK9 that is present in the Tat-activated transcription complexes was able to rephosphorylate the CTD and convert it into the Pol IIo* form. The rephosphorylation of the CTD by CDK9 can be blocked during elongation by the addition of the kinase inhibitor DRB.
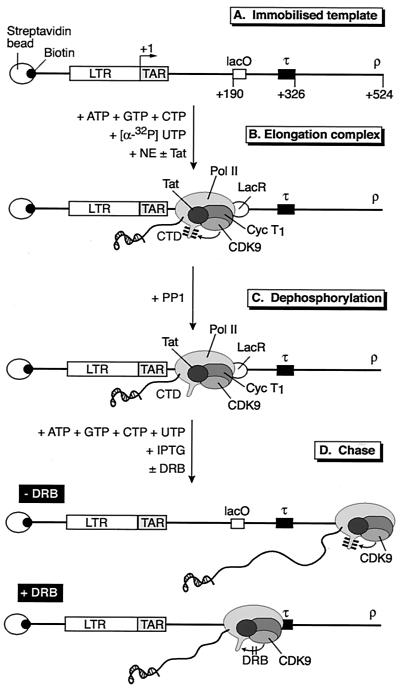
FIG. 1.
Strategy used for analyzing transcription elongation complexes. (A) Structure of HIV-LTR template. DNA templates containing the lac operator (lacO) binding site for the lac repressor protein (LacR) and a terminator (τ) sequence were biotinylated and bound to streptavidin beads. (B) Elongation complexes were trapped by the lac repressor (LacR) after incubation of the immobilized templates with HeLa nuclear extract (NE) in the presence of nucleotide triphosphates and LacR and in the absence or presence of Tat. The CTD of the RNA polymerase was phosphorylated during the elongation reaction due to the activity of CDK7 and CDK9. (C) Elongation complexes arrested by LacR were treated with PP1 to remove phosphate groups from the CTD. (D) The phosphatase-treated complexes can resume transcription elongation after the addition of nucleotides and IPTG. During the chase reaction the CTD became phosphorylated by CDK9. The addition of DRB blocked the rephosphorylation of the CTD and induced pausing of the transcription complex at the terminator sequences.
In the experiments described below, we used this experimental system to compare the processivity of hypophosphorylated RNA polymerase, prepared by phosphatase treatment of elongation complexes, to hyperphosphorylated polymerases prepared from elongation complexes exposed to Tat. In addition, we studied the pattern of CTD phosphorylation by CDK9 in elongation complexes and CDK7 in preinitiation complexes.
Tat-dependent CTD hyperphosphorylation in LacR-arrested transcription complexes.
Figure 2A shows the pattern of CTD phosphorylation in purified transcription elongation complexes before and after treatment with PP1. The different forms of the enzyme were detected by immunoblotting with an antibody (N-20) that recognizes the central domain of the largest subunit of Pol II. As expected, Pol IIa is the predominant form of the enzyme found in the extracts. This is converted to the slower-migrating Pol IIo form during transcription in the absence of Tat. In the presence of Tat, the Pol IIo* form is created due to additional phosphorylation of the CTD by CDK9 during elongation (29). Treatment with PP1 removes all of the phosphates from the CTD and increases the mobility of the polymerase in the gels to match that of the IIa form.
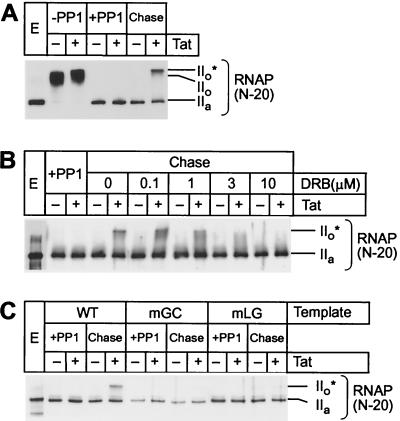
FIG. 2.
Tat and TAR stimulate hyperphosphorylation of CTD in transcription elongation complexes. (A) Rephosphorylation by CDK9. Elongation complexes assembled on the pW1 templates were dephosphorylated by PP1 treatment (Pol IIa). Chase of the dephosphorylated complexes from the LacR site in the presence of all four nucleotides and IPTG permits rephosphorylation of the RNA polymerase CTD (Pol IIo*) in the presence (+) of 20 ng of Tat but not in the absence (−) of Tat. (B) Inhibition of CDK9 by DRB. Between 0 and 10 μM DRB was included in the chase reactions containing dephosphorylated elongation complexes. Transcription was performed on the pW1 DNA templates in the absence (−) and presence (+) of 20 ng of Tat. (C) Activation of CDK9 by Tat and TAR. Elongation complexes were assembled on templates carrying wild-type TAR (WT) or mutant TAR elements in the Tat-binding site (mGC) or in the CycT1-binding site (mLG) in the absence (−) or presence (+) of 20 ng of Tat. After dephosphorylation of RNA polymerase CTD, the complexes were chased as described above.
Chase experiments with phosphatase-treated transcription complexes prepared in the presence of Tat show that they are able to rephosphorylate the CTD. This produces an RNA polymerase species that migrates at the same position in the gel as the Pol IIo* form (Fig. 2A). Remarkably, there is no measurable phosphorylation of the RNA polymerase complexes prepared in the absence of Tat.
Inhibition profiles of the kinase reaction with the cyclin-dependent kinase inhibitors DRB (Fig. 2B) and H8 (data not shown) match those of purified P-TEFb (the TAK kinase, comprising CDK9 and CycT1) (29, 41, 74). Further evidence that CDK9 is the enzyme responsible for the production of the Pol IIo* form of RNA polymerase in this experiment comes from the observation that CTD phosphorylation also requires a functional TAR RNA element. As shown in Fig. 2C, when the dephosphorylation and rephosphorylation experiments were performed with templates carrying well-characterized point mutations in either the Tat-binding site (mGC) or in the CycT1-binding site (mLG), no Pol IIo* was detected.
CDK9 is able to phosphorylate Ser5 and Ser2 of the CTD heptad repeat during elongation.
The phosphorylation pattern of the RNA polymerase CTD induced by CDK7 in preinitiation complexes and by CDK9 in elongation complexes was compared by using a series of antibodies that recognize different epitopes on the RNA Pol II backbone and CTD region (Fig. 3). In these experiments, the N-20 and 8WG16 antibodies were used to detect both the unphosphorylated IIa and phosphorylated IIo forms of RNA Pol II. We also employed two antibodies that recognize specific epitopes on a small subset of the RNA polymerases carrying phosphorylated CTDs. The H5 antibody is generally believed to recognize CTDs carrying Ser2 phosphorylation sites, whereas the H14 antibody recognizes epitopes carrying Ser5 phosphorylation sites.
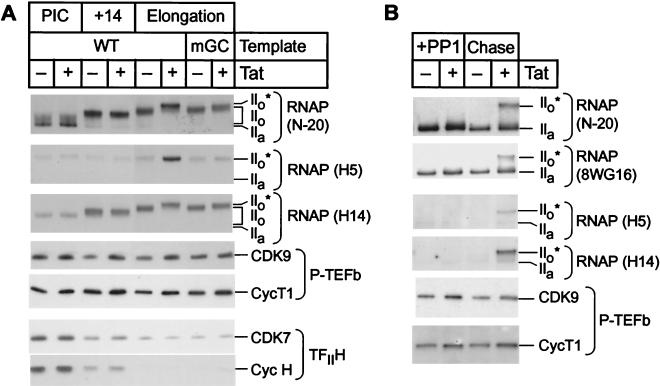
FIG. 3.
CDK9 phosphorylates Ser5 and Ser2 of the CTD in elongation complexes. (A) Transcription reactions. Preinitiation complexes (PIC) were assembled on immobilized wild-type template (WT) by using hexokinase/glucose-treated HeLa nuclear extract in the presence of 50 μM dATP and in the absence (−) or presence (+) of 20 ng of Tat. Transcription complexes paused at the uridine residue at position 14 were obtained after elongation of the preinitiation complexes in the absence of ATP. Standard transcription reactions were performed in parallel with templates carrying either the wild-type TAR element (WT) or a mutation in the Tat-binding site (mGC). Protein composition and phosphorylation of RNA Pol II CTD were analyzed from different transcription complexes by immunoblotting with the N-20, H5, and H14 antibodies directed against RNA Pol II (RNAP) and antibodies against CDK9, CycT1, CDK7, and CycH. (B) Rephosphorylation reactions. Transcription elongation complexes arrested by LacR were dephosphorylated by PP1, washed with EBCD buffer containing 0.1% Sarkosyl, and chased in the presence of 25 mM IPTG and all four nucleotide triphosphates. The proteins were detected by immunoblotting with the N-20, 8WG16, H5, and H14 antibodies directed against RNA Pol II and antibodies against CDK9 and CycT1.
In order to analyze the patterns of RNA polymerase phosphorylation in preinitiation complexes, complexes paused during early elongation, and complexes paused after elongation through TAR, we used our recently developed three-stage transcription system (6). In the first stage of the reaction, preinitiation complexes that contained only nonphosphorylated RNA Pol II were assembled by using nuclear extracts that were depleted of ATP by treatment with hexokinase and glucose (29). Phosphorylation of the RNA Pol II CTD is then initiated by adding dATP. The second stage of the reaction creates transcription complexes that are paused at position 14, a stage that is shortly after initiation but before complete clearance of the promoter. These complexes were obtained by chasing the preinitiation complexes in the presence of all of the ribonucleotides except ATP, with dATP as a phosphate donor for the phosphorylation of the RNA Pol II CTD (47, 50, 51). Because there is no ATP remaining in the extract, elongation stops at position 14, immediately before the first adenine on the template, which is at position 15. The addition of ATP in the reaction allows transcription to resume and permits analysis of late-stage elongation complexes.
As shown in Fig. 3A, conversion of preinitiation complexes to elongation complexes results in additional CTD phosphorylation events that are readily detected by the N-20 antibody. For example, during the chase from the preinitiation complexes to position 14 there is additional CTD phosphorylation that results in the formation of a heterogeneous and slowly migrating population of RNA Pol IIo forms. As expected, these early phosphorylation events are independent of Tat. Further phosphorylation of the polymerase takes place during late elongation, with the result that the molecular mass of the IIo population increases.
During Tat-activated elongation, there is additional CTD phosphorylation, resulting in the formation of the IIo* form of the polymerase, as detected by the N-20 antibody. As expected, there was no detectable IIo* form polymerase when templates carrying the mGC mutation in TAR were used; however, there was a small, but significant increase in the size of the IIo polymerase population.
In contrast, the H5 epitope, which is indicative of Ser2 phosphorylation, was at background levels in the preinitiation complexes and early elongation complexes (position 14) but readily detectable in the Tat-activated elongation complexes. As described below, the simplest explanation for this observation is that the H5 antibodies recognize CTDs that have been phosphorylated by CDK9 but not by CDK7. All of the RNA polymerases recognized by H5 migrate unusually slowly in the gels, at a position corresponding to the IIo* form of the RNA polymerase detected by the N-20 antibody. In elongation complexes that have been activated by Tat, there is more than 10-fold more polymerase that can be detected by this antibody.
A third distinctive pattern is exhibited by the H14 antibody, which binds exclusively to phosphorylated CTD repeats and is therefore unable to detect the IIa form found in the preinitiation complexes. The mobility of the phosphorylated species detected by H14 closely corresponds to the mobility of the phosphorylated forms of RNA polymerase detected by N-20, but the antibody does not recognize the IIo* form of the polymerase as well as the IIo form. Thus, although the H14 antibody is able to detect increased levels of phosphorylated polymerases after Tat activation of transcription from templates carrying the wild-type TAR element, the changes detected by this antibody are somewhat less pronounced than those detected by the N-20 antibody. Since the epitope recognized by the N-20 antibody is found outside of the CTD region, the antibody is able to bind with equivalent affinities to both the phosphorylated and the nonphosphorylated polymerases. As a result, the changes detected by this antibody provide the most accurate measurements of the polymerase population distribution.
To confirm that the CDK9 kinase is activated by Tat in late-stage elongation complexes, we also performed Western blotting assays on transcription complexes that had been arrested at the lac repressor, dephosphorylated by treatment with PP1, and then allowed to resume transcription (Fig. 3B). Because of the phosphatase step, all of the CTD modifications arising during early elongation are removed and the only events detected in this experiment are due to enzymatic activity during late-stage elongation. Analysis of Western blots containing rephosphorylated elongation complexes (Fig. 3B) confirms that CDK9 is only able to phosphorylate the CTD when Tat is present. Under these conditions, the rephosphorylated RNA polymerase is readily detected by the N-20, 8WG16, H5, and H14 antibodies. Thus, both the Ser2 (detected by H5) and the Ser5 (detected by H14) residues become phosphorylated during the chase reactions. Remarkably, the major polymerase species detected by each of these antibodies has a mobility corresponding to the IIo* form of the polymerase, and there are no intermediate products corresponding to the IIo form detected during this reaction. Once again, the N-20 antibody provides the best measure of the overall extent of CTD phosphorylation. The 8WG16 antibody gives a similar pattern to the N-20 antibody, but since it binds preferentially to unphosphorylated CTD repeats, the use of this antibody exaggerates the proportion of the unphosphorylated RNA Pol IIa species in the population.
It is important to note that control immunoblots in Fig. 3 demonstrate that both CDK9 and CycT1 are present at equivalent levels in elongation complexes prepared in the presence and absence of Tat. In contrast, CDK7 and CycH are only found in preinitiation complexes (Fig. 3A). These results are in agreement with previous results that demonstrate that CDK9 associates with both preinitiation complexes and elongation complexes, whereas CDK7 is only found in preinitiation complexes (29, 70, 72). Thus, CDK9, but not CDK7, remains associated with the elongation complexes and appears to be selectively activated only in the presence of Tat. Furthermore, CDK9 is likely to be the only CTD kinase that is capable of generating the H5 epitope, presumably by modifying the CTD heptad repeat at Ser2. Recent experiments by Zhou et al. (73) suggest that CDK9 is inhibited by the TFIIH found in the HIV-1 preinitiation complex. Since TFIIH is released after position 14 this mechanism provides a good explanation as to why Ser2 phosphorylation due to CDK9 activity is not observed in the preinitiation complexes or early-stage elongation complexes.
Our results differ somewhat from those of Zhou et al. (72), who reported that CDK9 is active in preinitiation complexes where it primarily phosphorylates Ser2. These authors suggested that Tat modifies the substrate specificity of CDK9 to permit additional phosphorylation of the CTD at Ser5. One possible experimental difference that could account for this discrepancy is that Zhou et al. (72) did not deplete endogenous ATP from nuclear extract prior to formation of the preinitiation complexes. As a result, they may have analyzed complexes that have started transcription and therefore activated CDK9.
Constitutive activation of CDK9 by Tat.
In the experiments described above, the activation of the CDK9 was measured in situ by using the CTD of the RNA polymerase as the substrate for the enzyme. The failure of CDK9 to phosphorylate the CTD in the absence of Tat could either be because the CDK9 kinase is inactive in the absence of Tat or because the CTD was inaccessible to the enzyme. To distinguish between these two possibilities, the activity of CDK9 present in elongation complexes was also measured by using short synthetic peptides as substrates (53).
Figure 4 shows the phosphorylation of a synthetic CTD peptide containing five repeat sequences (YSPTSPKYSPTSPTYSPTTPKYSPTSPTYSPTSPV) by the kinase present in the paused elongation complexes. To provide a standard for CDK9 kinase activity, P-TEFb was purified by immunoprecipitation from HeLa cell nuclear extracts with antibodies against CDK9. The immunoprecipitated enzyme was able to phosphorylate the CTD peptide in parallel reactions. In the experiment shown in Fig. 4, the peptides are labeled with 32P and then fractionated on SDS gels. As more sites on the CTD repeats become phosphorylated, the mobility of the peptide is reduced, giving rise to the three distinct bands seen in Fig. 4. The phosphorylation reactions were performed with 1 μCi of [γ-32P]ATP in the absence of unlabeled ATP to obtain products of maximal specific activity or in the presence of 2 μM unlabeled ATP in order to measure peptide phosphorylation at ATP concentrations near the Km of the kinase.
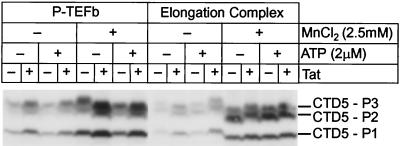
FIG. 4.
Activation of P-TEFb by Tat. In vitro kinase reactions were performed by using P-TEFb purified by immunoprecipitation from HeLa nuclear extract or CDK9 in transcription elongation complexes arrested by LacR. The kinase reactions were performed using a peptide carrying 5 heptad repeat sequences (CTD5) as the substrate. Reactions were performed in the presence of 1 μCi of [γ-32P]ATP and in the absence (−) or presence (+) of 2 μM cold ATP, 2.5 mM MnCl2, and 20 ng of Tat. The phosphorylated peptides were separated by SDS-4 to 20% PAGE, transferred onto nitrocellulose membranes, and detected by autoradiography.
As shown in Fig. 4, the CDK9 kinase was able to phosphorylate the synthetic peptides. Both the immunoprecipitated P-TEFb and the enzyme present in the elongation complexes were active in the absence of Tat. However, there was a 2.5-fold activation of the immunoprecipitated enzyme when Tat was added to the reaction. Similarly, transcription complexes that have incorporated Tat are 2.5-fold more active than complexes prepared in the absence of Tat.
Since the Western blotting experiments have shown that approximately equal amounts of CDK9 are present in elongation complexes prepared in the presence or absence of Tat, our observation that Tat increases the CDK9 kinase activity present in the elongation complexes is consistent with the idea that the enzyme is selectively activated by Tat. If this were the case, then enhanced levels of enzyme activity should also be detected under conditions where the kinase is activated by some other mechanism. To test this hypothesis, the kinase assays were also performed in buffers containing Mn2+. Under these conditions, CDK9 kinase activity is stimulated nonspecifically (53, 74). As shown in Fig. 4, the control experiments with the immunoprecipitated P-TEFb show that CDK9 kinase activity is activated fourfold when the kinase assays are performed in the presence of Mn2+ instead of in the presence of Mg2+. Similarly, elongation complexes assayed in the presence of Mn2+ prepared both in the presence and absence of Tat show strongly enhanced CDK9 activity. Surprisingly, under these conditions there is comparatively poor activation of the CDK9 kinase activity by Tat. For example, in the presence of Mn2+ the purified P-TEFb shows only a 1.6-fold increase in the presence of Tat, and there is only a 1.1-fold activation of the enzyme in the elongation complexes.
Thus, we have been able to obtain direct enzymatic evidence that CDK9 is present in elongation complexes prepared in the absence of Tat, but the enzyme is comparatively inactive unless Tat is present.
CDK9 phosphorylates both the N-terminal and C-terminal regions of the CTD.
The human RNA Pol II CTD is composed of 52 repeats of the heptapeptide sequence YSPTSPS. Standard approaches to mapping CTD phosphorylation sites are impractical since the repetitive nature of the amino acid sequence makes it difficult to sequence the CTD or isolate short peptides carrying phosphorylated amino acids.
In order to determine whether specific regions of the CTD were phosphorylated by CDK9 during elongation, we developed a novel mapping method based on our ability to prepare paused elongation complexes (Fig. 5). Arrested elongation complexes were first treated with PP1 and then labeled by incorporation of [γ-32P]ATP during the rephosphorylation reaction. The 32P-labeled RNA Pol II was then purified by immunoprecipitation with the N-20 antibody, and the CTD was cleaved with a variety of proteases. In contrast to Fig. 3A, which measures the accumulation of phosphorylated species during transcription, this method detects only the phosphorylation products that are produced during the rephosphorylation reaction.
FIG. 5.
Protease mapping of CTD phosphorylation sites in elongation complexes. (A) CTD domain. The diagram shows the position of protease cleavage sites for Asp-N, Glu-C, and Lys-C. Asp-N cleaves peptide bonds N terminally at aspartic acids (D). Glu-C cleaves peptide bonds at the carboxyl side of glutamic acids (E), as well as at the carboxyl side of aspartic acids (D). Cleavage at E* by Glu-C was prevented by the proline residue on the carboxyl side. Lys-C specifically cleaves peptide bonds on the carboxyl-terminal side of lysine (K) residues. Cleavage products of each protease are shown as shaded bars. (B) Elongation complexes. LacR-arrested and dephosphorylated elongation complexes were labeled with 10 μCi of [γ-32P]ATP as described in the text. The 32P-labeled RNA Pol II was immunoprecipitated with N-20 antibody and digested by the indicated proteases. The cleavage products were separated on SDS-4 to 12% PAGE gels, transferred to nitrocellulose membranes, and detected by autoradiography. (C) Glu-C digest. Preinitiation and/or elongation complexes were prepared with CDK7-, CDK9-, or mock-depleted HeLa nuclear extracts. RNA Pol II from the complexes was labeled with 10 μCi of [γ-32P]ATP, purified by immunoprecipitation, and digested with Glu-C. The labeled peptides were separated in SDS-4 to 12% PAGE gels, transferred to nitrocellulose membranes, and then detected by autoradiography. (D) Immunoblot analysis of CDK9- or mock-depleted extracts (left panel) and CDK7- or mock-depleted extracts (right panel). Note that CDK9 depletion did not affect the levels of CDK7 or RNA Pol II and that CDK7 depletion did not change the levels of CDK8, CDK9, or RNA Pol II.
As shown in Fig. 5A, the proteases Asp-N, Glu-C, and Lys-C are each able to cleave the CTD into fragments that can be readily resolved by SDS-polyacrylamide gel electrophoresis (PAGE). Asp-N cleaves peptide bonds N-terminally at aspartic acids (D) to produce an almost entire CTD fragment, together with an additional 40 residues on the amino-terminal side. The Glu-C protease cleaves peptide bonds at the carboxyl side of glutamic acid (E) and aspartic acid (D) residues. This produces a large fragment of 31 kDa containing approximately two-thirds of the CTD from the amino end of the CTD and a small fragment of 12 kDa derived from the C terminus. Lys-C cleaves peptide bonds on the C terminus of lysine (K) residues. This enzyme produces a large fragment of 36 kDa containing the N-terminal region of the CTD and numerous small fragments that are too small to be resolved on the gels used in these experiments.
Consistent with the immunoblotting results there are much lower levels of 32P labeling in the absence of Tat, confirming that CDK9 is mostly inactive in these complexes. As shown in Fig. 5B, the CTD of RNA polymerase becomes highly phosphorylated throughout its length by the Tat-activated kinase. For example, the Lys-C fragment, which lacks the C-terminal region, is labeled to approximately the same specific activity as the Asp-N fragment, which contains most of the C terminus of the CTD. Similarly, CDK9 labeled the 31- and 12-kDa peptides generated by Glu-C cleavage with a ratio of 1.2 to 1.0, after correction for the number of heptad repeats found in the two fragments.
To provide direct evidence that CDK9 is the kinase responsible for CTD phosphorylation in the elongation complexes, similar experiments were performed with immunodepleted extracts. As shown in Fig. 5D, the immunodepletion with antibodies to CDK9 removed the majority of the CDK9 from the extract but did not reduce the amounts of CDK7. In the absence of CDK9, CTD phosphorylation in elongation complexes was reduced to background levels. Furthermore, there was no activation of kinase activity by Tat in the CDK9-depleted extracts (Fig. 5C).
As an additional control, we also mapped the phosphorylation sites on the CTD recognized by CDK7 in preinitiation complexes (Fig. 5C). There is no detectable labeling of the CTD by [γ-32P]ATP in the preinitiation complexes prepared with CDK7-depleted extracts, even though both CDK9 and CDK8 are present in the complexes (Fig. 5D). However, as previously observed (29), CDK7 was able to induce extensive phosphorylation of the CTD in the absence of Tat.
Cleavage mapping of the CTD showed that, in contrast to CDK9, CDK7 preferentially phosphorylates the C-terminal Glu-C fragment. In the experiments shown in Fig. 5C, CDK9 labeled the 31- and 12-kDa Glu-C large fragments with a ratio of 1.5 to 1.0, after correction for the number of heptad repeats found in the two fragments. In contrast, CDK7 phosphorylated the C-terminal 12-kDa Glu-C fragment preferentially and gave ratios of between 0.13 and 0.52 to 1.0 when the large and small fragments were compared (Fig. 5C).
Phosphorylation of the CTD by CDK9 promotes readthrough of terminator and arrest sequences.
We have shown previously that Tat can promote readthrough of an artificial terminator sequence comprising an RNA stem-loop structure, followed by a poly(U) tract (25). In order to test whether Tat can help to overcome blocks imposed by other elongation blocks we also prepared templates carrying the arrest sequence present in the human histone H3.3 gene intron (Fig. 6A) (6). This sequence contains two tandem arrest sites that carry polypyrimidine tracts flanked by purines. The effect of these sequences is to bend the DNA template and force polymerase pausing (3, 35). As a control, we also included a template that carried only HIV sequences.
FIG. 6.
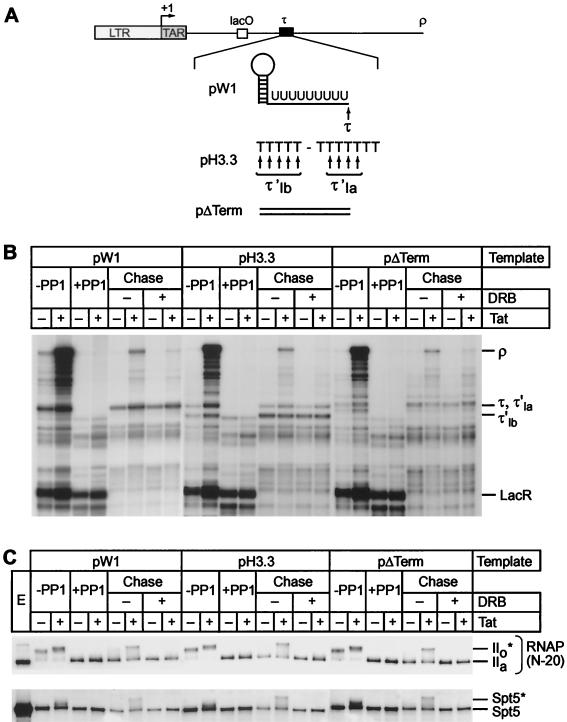
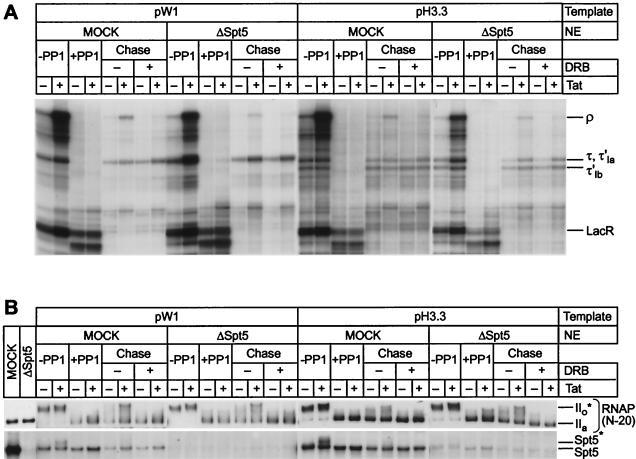
Phosphorylation of the CTD is required for efficient transcription through terminator sequences. (A) Structure of templates. The pW1 template carries a terminator formed by an RNA stem-loop, followed by nine uridines (τ). pH3.3 contains two tandem arrest sequences (τ′1a and τ′1b) that induce DNA bending. In pΔTerm, the terminator sequence was replaced by the original HIV sequence. (B) Transcription reactions were performed with three different immobilized DNA templates (pW1, pH3.3, or pΔTerm). Reactions contained 100 ng of LacR and were performed in the absence (−) or presence (+) of 20 ng of Tat. After labeling for 20 min with [α-32P]UTP (−PP1), the immobilized templates were purified and treated with RNase H in the presence of the RHX1 and RHLAC oligonucleotides to remove the labeled RNA transcripts from transcription complexes that had read through the LacR site. The arrested complexes were then dephosphorylated by PP1 treatment (+PP1). After the complexes were washed with TMZ buffer, the dephosphorylated complexes were chased by the addition of 250 μM ATP, GTP, and CTP; 5 μM UTP; and 25 mM IPTG in the absence (−) or presence (+) of 100 μM DRB. Positions of transcripts at the runoffs (ρ), terminator (τ), and lac repressor (LacR) in pW1 template are indicated. τ′1a and τ′1b indicate the stop sites from the pH3.3 template. (C) Immunoblot. Samples of the reactions shown in panel B were immunoblotted with the N-20 antibody against RNA Pol II and the anti-Spt5 antibody. Tat-dependent hyperphosphorylation of RNA polymerase CTD and Spt5 was observed during the chase of dephosphorylated complexes assembled on each template.
Figure 6B compares the efficiency of blocking by the stem-loop terminator and bent DNA arrest sequence in cell-free transcription reactions. In these reactions, late-stage elongation complexes prepared in the presence or absence of Tat and paused at the lac repressor were generated according to the method of Keen et al. (34). After dephosphorylation of the CTD by treatment with PP1, the paused complexes were then chased in the absence of new label. As shown in Fig. 6B, the phosphatase-treated transcription complexes paused at the lac repressor site were efficiently chased after the addition of a full complement of nucleotides. As expected, the stem-loop terminator sequence induced termination at position 290 (τ), whereas the histone H3.3 arrest site introduced two stop sites of the expected sizes (τ′1a and τ′1b). Comparable stop sites were absent from the control template. During the chase reaction, Tat promoted the synthesis of long transcripts and greatly enhanced readthrough of both terminator sequences. However, inhibition of CDK9 activity during the elongation reaction by DRB abolished the Tat response. Similar levels of interference with elongation were obtained when these templates were used in standard transcription reactions (6). Thus, Tat is able to promote efficient readthrough of elongation blocks acting at either the RNA or the DNA level. Readthrough of both types of block provides a sensitive assay for changes in RNA polymerase processivity.
Only complexes prepared in the presence of Tat were able to read through the two types of elongation blocks efficiently and produce significant amounts of full-length transcripts. During the chase reactions the RNA polymerase CTD was converted to the IIo* form whenever Tat was present (Fig. 6C). In addition to phosphorylating the RNA polymerase CTD, CDK9 is also able to phosphorylate the elongation factor Spt5 (6, 36, 51). As shown in Fig. 6C, Spt5 is phosphorylated in parallel to the CTD and converted into a slowly migrating species (Spt5*).
Blocking the phosphorylation reaction by the addition of DRB not only prevented phosphorylation of the CTD (Fig. 6C) but also blocked the ability of Tat to promote readthrough of the elongation blocks (Fig. 6B). Similarly, Tat-activated transcription was seen with templates lacking the terminator or arrest sequences. However, on these templates more transcription complexes were able to reach the end of the template in the absence of Tat (and/or presence of DRB) since the additional blocks to elongation were removed.
Thus, transcription elongation efficiency through terminator sequences is dependent upon CDK9-dependent phosphorylation events involving the RNA polymerase CTD and, possibly, Spt5.
Phosphorylation of RNA polymerase CTD stimulates RNA polymerase processivity.
In the preceding experiment, the distribution of RNA polymerases after a 10-min chase was evaluated, and clear differences between phosphorylated and nonphosphorylated transcription complexes were detected. However, this experiment does not demonstrate whether or not these changes arose because of changes in RNA polymerase processivity or other effects, such as changes in RNA polymerase stability on the template or changes in the proportion of transcription complexes that were able to resume elongation during the chase. In order to measure whether CTD phosphorylation by CDK9 stimulates the processivity of transcription elongation, the kinetics of elongation by the phosphatase-treated elongation complexes arrested by LacR were examined (Fig. 7). As described previously, DRB was added to one set of reactions in order to block rephosphorylation of the CTD by CDK9 during the chase.
FIG. 7.
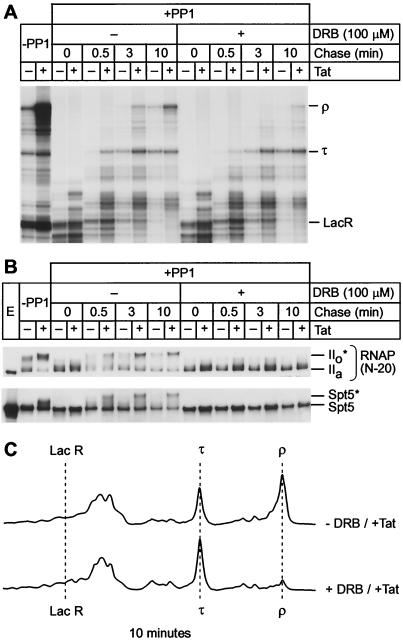
CTD phosphorylation by CDK9 enhances processivity of RNA Pol II. (A) Elongation complexes were assembled in the presence of LacR with HeLa nuclear extract on the pW1 DNA template and treated with RNase H and protein phosphatase. The complexes were then chased for the indicated times (0, 0.5, 3, and 10 min) in the absence (−) or presence (+) of DRB. RNA polymerase processivity is increased by the phosphorylation of CTD in the presence of Tat, but the addition of the CDK9 inhibitor DRB abolishes the activation of RNA Pol II processivity by Tat. (B) Immunoblots showing phosphorylation extent of RNA Pol II and Spt5 were performed with N-20 antibody against RNA Pol II or anti-Spt5 antibody. (C) Quantitative analysis of the gel shown in panel A by densitometry. The changes in the distributions of transcripts produced by the rephosphorylated transcription complexes (−DRB/+Tat) and unphosphorylated transcription complexes (+DRB/+Tat) during the chase reactions provide a direct measure of changes in RNA polymerase processivity.
As shown in Fig. 7A, virtually all of the arrested transcription complexes resume elongation within 0.5 min of adding nucleotides. After 3 min, most of the complexes chased in the absence of DRB have reached the terminator (τ) both in the presence and in the absence of Tat. However, at this early time, comparatively few complexes are able to reach the end of templates (ρ) even in the presence of Tat. By 10 min, the elongation reaction is essentially complete in the absence of DRB and there are sevenfold more transcripts present at the end of the template in the presence of Tat than in its absence. In contrast, addition of DRB to the chase reactions effectively blocked Tat stimulation of transcription and slowed the reaction kinetics.
This change in the distribution of RNA polymerases can also be measured quantitatively by comparing the densitometry traces shown in Fig. 7C. A useful measure of the efficiency of the elongation reaction is to compare the ratios of runoff transcripts to transcripts paused at the terminator (ρ/τ). For example, after the 10-min chase in the absence of DRB there is a ρ/τ ratio of 1.8-fold, whereas in the presence of DRB there is a ρ/τ ratio of only 0.15.
Immunoblots of the corresponding reactions (Fig. 7B) show that both the CTD of RNA Pol II and Spt5 were efficiently rephosphorylated when the chase reaction was performed in the presence of Tat, but there was only very weak rephosphorylation of the proteins in the absence of Tat. The rephosphorylation reaction is progressive, and intermediate phosphorylation products are observed after 0.5 min. However, the reaction reaches completion by 3.0 min. As expected, the addition of DRB completely prevented the rephosphorylation of RNA polymerase and Spt5.
Thus, the reaction kinetics show that hyperphosphorylation of elongation complexes by CDK9 directly correlates with an increase in the processivity of transcription elongation.
CTD phosphorylation prior to elongation can enhance polymerase processivity.
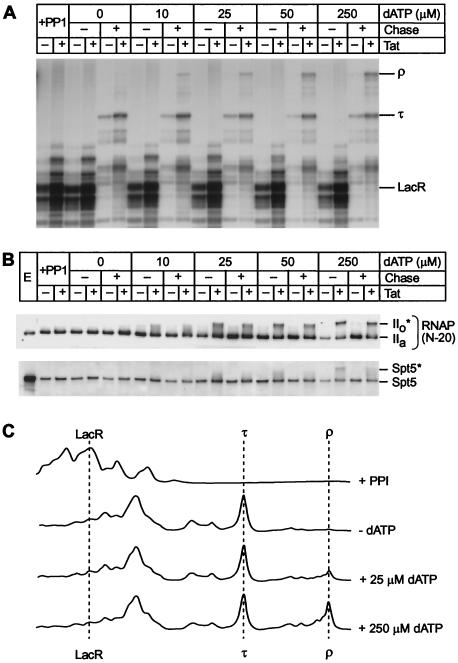
Is phosphorylation of the CTD by Tat-activated CDK9 kinase linked to transcription through pause sites, or does it have a general effect on RNA polymerase processivity? We addressed this question experimentally by performing separate phosphorylation and elongation reactions. As shown in Fig. 8B, the CDK9 kinase present in the Tat-activated transcription complexes is able to rephosphorylate the CTD and Spt5 in paused polymerases after the addition of dATP and none of the other ribonucleotide triphosphates. Under these conditions there is no additional elongation, and the paused elongation complexes remain bound to the template adjacent to the lac repressor. Conversion of the IIa form to the IIo* form is dependent upon the amount of dATP added until the reaction is saturated at 250 μM dATP. The phosphorylation of Spt5 proceeds in parallel in these reactions. After a brief wash of the phosphorylated transcription complexes to remove unincorporated dATP, 100 μM DRB was added to prevent further phosphorylation of RNA polymerase and Spt5. Transcription elongation was subsequently initiated by the addition of IPTG and the four ribonucleotide triphosphates. As shown in Fig. 8A and C, in the presence of Tat the extent of CTD and Spt5 phosphorylation was positively correlated with the efficiency of transcription through the termination signal (τ). As the dATP concentration is increased, progressively more transcription complexes become phosphorylated and reach the end of templates.
FIG. 8.
Activation of elongation complexes by phosphorylation prior to elongation. (A) Transcription. Elongation complexes were assembled by using the pW1 DNA template in the presence of lac repressor and dephosphorylated (+PP1) as described in the legend to Fig. 6. The complexes were rephosphorylated by adding between 0 and 250 μM dATP as a phosphate donor. After a brief wash to remove unincorporated dATP, 100 μM DRB was then added to prevent further phosphorylation of the complex during the chase reaction. The rephosphorylated complexes were chased by addition of all four ribonucleotide triphosphates and 25 mM IPTG. Positions of transcripts at the runoff (ρ), terminator (τ), and lac repressor (LacR) are shown. (B) Immunoblot. Samples from the reactions shown in panel A were immunoblotted with N-20 antibody against RNA Pol II and the anti-Spt5 antibody. (C) Quantitative analysis of the gel shown in panel A by densitometry. Note that there is a strong correlation between the extent of CTD phosphorylation and the amount of the runoff product (ρ).
We conclude that phosphorylation of the CTD results in a modified polymerase with enhanced processivity. Therefore, although the CTD must be phosphorylated for efficient elongation, this event can take place at any time during elongation, and there is no requirement to continuously phosphorylate the polymerase during its transit along the DNA template or when it encounters an elongation block.
CTD phosphorylation in the absence of Spt5 can enhance polymerase processivity.
Spt5 is a subunit of the DRB sensitivity-inducing factor (i.e., DSIF). In normal transcription, DSIF works in concert with the negative elongation factor (NELF) to induce polymerase pausing near promoter start sites (61-63, 67, 68). This pausing event can be reversed by a CDK9-dependent phosphorylation event. In addition to playing a role in early transcription, Spt5 also acts as a generalized elongation factor. In the first experiment to demonstrate a direct role for Spt5 in HIV transcription, Gaynor and coworkers (30, 66) showed that immunodepletion of Spt5 from transcription extracts strongly inhibited Tat-activated transcription but only had minimal effects on basal transcription. Similarly, our previous studies (6) have also shown that Spt5 plays a positive role in transcription elongation by enhancing the stability of transcription complexes at terminator sequences with the consequence that more transcription complexes are able to read through strong arrest sites.
It was therefore important to determine whether the enhanced processivity we observed after CDK9-dependent phosphorylation of elongation complexes was a direct consequence of the phosphorylation of the CTD rather than due to an effect of DSIF that was mediated by Spt5 phosphorylation. For example, CDK9 could be acting primarily to relieve a block during late elongation that was imposed by DSIF that is analogous to the blocks imposed by DSIF during early elongation. Alternatively, the phosphorylated form of Spt5 could be enhancing RNA polymerase processivity.
Chase experiments were therefore performed with extracts that had been selectively depleted of Spt5 by antibodies (6). As shown in Fig. 9B, phosphorylation of the RNA polymerase CTD proceeds efficiently in the absence of Spt5. The Spt5-depleted transcription complexes were also able to resume elongation during the chase experiments (Fig. 9A), but only complexes that had been exposed to Tat, and therefore carried phosphorylated CTD sequences, were able to efficiently read through the elongation blocks. In the absence of Spt5, the amount of transcripts reaching the end of the templates (ρ) is somewhat reduced. As described previously, this is due to the ability of Spt5 to block premature dissociation of the nascent RNA chains from the transcription complexes during elongation (6).
FIG. 9.
Spt5 is not required for Tat-dependent activation of elongation and RNA polymerase phosphorylation. (A) Transcription. Elongation complexes were assembled in the presence of lac repressor (−PP1), dephosphorylated (+PP1), and then chased with either mock-depleted or Spt5-depleted HeLa nuclear extracts. Reactions were performed with either the pW1 template or the pH3.3 template as described in the legend to Fig. 6. Positions of transcripts at the runoff (ρ), terminator (τ, τ′1a, and τ′1b), and lac repressor (LacR) are shown. (B) Immunoblot. Samples from the reactions shown in panel A were immunoblotted with N-20 antibody against RNA Pol II and the anti-Spt5 antibody. Note that hyperphosphorylated polymerases can elongate efficiently through both the terminator site and the arrest sites even in the absence of Spt5.
Since Tat is able to enhance transcription elongation even in the absence of Spt5, we conclude that phosphorylation of the RNA polymerase CTD by Tat-activated CDK9 directly enhances the processivity of the polymerase.
DISCUSSION
Phosphorylation of the RNA polymerase CTD by CDK9 enhances polymerase processivity.
It is now well established that HIV-1 Tat stimulates hyperphosphorylation of RNA polymerase CTD during transcription elongation (29, 41, 72, 74). However, it has been a significant experimental challenge to measure the effects of CDK9 on transcriptional elongation because many kinases, including CDK7, CDK8, and DNA-PK, are also able to phosphorylate the CTD. We therefore developed a method that allowed us to analyze the phosphorylation of RNA polymerase during elongation and in the absence of new initiation events. Our technique was based on the observation that phosphorylated RNA polymerases (IIo or IIo*) that have been arrested by LacR can be converted to the IIa form by protein phosphatase treatment and still retain their elongation capacity. During subsequent chases, the paused polymerases are able to resume elongation, and in the presence of Tat, the CTD becomes phosphorylated.
Here we provide strong evidence that the enzyme responsible for CTD phosphorylation during elongation is CDK9. First, the phosphorylation reaction is both Tat and TAR dependent. Second, Tat-activated phosphorylation showed inhibition profiles of DRB and H8 (data not shown) on phosphorylation of RNA polymerase CTD similar to those of CDK9 as reported previously (29, 74). Finally, immunodepletion of CDK9 prevented CTD phosphorylation during elongation.
Under our experimental conditions only a fraction of RNA polymerases are converted to the IIo* form during Tat-activated phosphorylation, and a similar fraction of the purified transcription complexes are activated by Tat. As shown in Fig. 8, the proportion of active and inactive enzyme can be varied by controlling the extent of the phosphorylation reaction by using different dATP concentrations. It therefore seems likely that the IIo* form represents the entire fraction of the RNA polymerase population that becomes activated by Tat.
The chase experiments reported here demonstrate that phosphorylation of RNA transcription complexes by CDK9 alters the processivity of the enzyme and allows it to overcome transcription at two distinct elongation blocks. The first block we studied was an RNA stem-loop structure followed by nine uridine residues. This artificial sequence, which closely resembles bacterial termination signals, causes RNA polymerase to stop elongation and to release transcripts in cell-free transcription systems. The second block is a sequence normally found in the histone H3.3 gene carrying structural elements that bend DNA. This sequence causes transcription arrest, but does not induce transcript release, in cell-free systems and in vivo (3, 35). Bent DNA sequences are also potent arrest sites in yeast (11, 38). We also showed in the present study that phosphorylation of RNA polymerase by CDK9 enhances readthrough of both types of block, indicating a generalized role of hyperphosphorylation of CTD in processivity of RNA Pol II.
The terminator and arrest site sequences that we used in these experiments provide effective tools for measuring small changes in RNA polymerase processivity. However, the HIV genome does not carry any specific termination signals. Instead, there are numerous sites where pausing can occur. The cumulative effect of each of these pauses is to prevent the RNA polymerase from transcribing the entire HIV genome unless Tat is present.
The Tat-activated CDK9 is also able to phosphorylate the transcription elongation factor Spt5 during elongation. Since Spt5 is one of the subunits of DSIF, which can act as an inhibitor of early elongation events, it is conceivable that the effects we observed were due to reversal of a block imposed by DSIF and its cofactor NELF (61-63, 67, 68). However, as shown here, equivalent chase efficiencies and Tat activation can be achieved in the absence of Spt5. Consistent with these results, recent experiments by Bourgeois et al. (6) have shown that Spt5 acts as a complementary factor for Tat-activated transcription by inhibiting RNA release from paused RNA polymerases. In addition to Spt5, other elongation factors such as TFIIS, TFIIF, and Tat-SF1 may cooperate with Tat to promote polymerase transcription through various blocks. However, none of these factors appears to be a substrate for CDK9.
Thus, we conclude that it is only the phosphorylation of the CTD that has a direct effect on polymerase processivity.
Constitutive activation of the CDK9 kinase by Tat.
Little is known about how CDK9 becomes activated by Tat. Although Tat, CDK9, and CycT1 can form stable complexes with TAR RNA during elongation (65), TAR appears to be only used as a loading site for Tat and does not remain tightly associated with the elongation complex (34). It therefore seems likely that protein-protein interactions between Tat, CycT1, and CDK9 mediate conformational changes that permit the enzyme to become constitutively active. Identifying the sequences and structures that regulate these events will be critical for defining the mechanism of action of Tat.
Using our cell-free system, we demonstrated for the first time that CDK9 is tightly regulated during transcriptional elongation and activated only in the presence of Tat. In the absence of Tat, or after transcription through a defective TAR element, the CDK9 present in elongation complexes cannot phosphorylate RNA polymerase CTD, even though the enzyme is present at the same level as in complexes that have not incorporated Tat.
The inability of CDK9 to phosphorylate the CTD in the absence of Tat could either be because the enzyme is inactive and/or because the CTD is inaccessible to the enzyme. Several lines of evidence suggest that CDK9 is activated by Tat. First, we have found with synthetic peptide substrates that CDK9 in transcription complexes shows very low activity in the absence of Tat. Second, in agreement with a variety of other studies, we found that isolated P-TEFb kinase, which contains the CDK9/CycT1 complex, can also be activated by the addition of recombinant Tat (20, 72).
Activation of cyclin-dependent kinases usually occurs in two steps. First, binding of the kinases to their respective cyclin partners induces conformational changes in the enzyme. Second, in order to form a stable binary complex with its activating partner, cyclin H, CDK7 must be phosphorylated on a conserved threonine (Thr170) in its activation segment, or T-loop (40, 44). It seems likely that activation of CDK9 by CycT1 involves phosphorylation at an analogous site. However, this event is not sufficient to explain the tight regulation of CDK9 by Tat since CycT1 is always present in the elongation complexes.
One possible mechanism for Tat regulation of CDK9 is that it could alter the conformation of regulatory sequences found on the C terminus of CDK9 and the C terminus of CycT1. This could lead to an activation of the enzyme by preventing them from occupying the active site of the enzyme. This hypothesis is supported by the observation that these regions of the proteins can be phosphorylated by CDK9. Furthermore, autophosphorylation of CDK9 is known to enhance the binding of the CDK9/CycT1 complex to TAR (18, 20). Similarly, Zhou et al. (73) have suggested that TFIIH is able to inhibit CDK9 activity in preinitiation complexes and early-stage elongation complexes by blocking CDK9 autophosphorylation.
Certain tightly regulated cyclin-dependent kinases are also under the control of two classes of inhibitors: CIPs and INKs (reviewed by Pavletich [49]). The CIP family typically bind the cyclin-dependent kinase/cyclin complex, whereas the INK4 family tend to target the free cyclin-dependent kinase. Together, these inhibitors exert a tight on-off control on many cyclin-dependent kinase proteins within the cell cycle. It is conceivable that similar inhibitors are able to switch transcription on or off via interactions with CDK7 and CDK9. For example, one recently discovered feature of CDK9 regulation is that more than half of the enzyme in HeLa cells is found in an inactive complex with 7SK RNA (45, 69). 7SK is a powerful inhibitor of HIV-1 Tat activation since it both inhibits the kinase activity of CDK9 and prevents recruitment of P-TEFb to the HIV promoter (45, 69). Although 7SK RNA itself does not participate in the control of CDK9 during elongation, it is conceivable that other RNA sequences, such as TAR, could interact with P-TEFb to inhibit its activity in the absence of Tat.
Phosphorylation of the CTD is also regulated by CTD phosphatases during elongation (10, 37, 42, 43, 56). For example, Marshall et al. (42, 43) have reported that CTD phosphatases are strongly inhibited by Tat. Under our conditions, a low level of protein phosphatase activity was detected in the paused elongation complexes under conditions where CDK9 was inactive, i.e., after prolonged incubation in the presence of DRB or in the absence of ATP. However, we were unable to detect large differences in phosphatase activity in the presence or absence of Tat (data not shown). These observations imply that it is the activation of the CDK9 kinase activity, rather than inhibition of the CTD phosphatase, that is primarily responsible for the enhanced phosphorylation of the RNA polymerase induced by Tat that we have observed.
Sequential phosphorylation of the CTD by CDK7 and CDK9.
The CTD of RNA polymerase is extensively phosphorylated during transcription initiation and during transition to early elongation. It has been proposed that Tat may play a role during these stages of transcription by activating TFIIH (14, 46) or CDK9 (27). However, we have been unable to detect by sensitive immunoblotting and 32P-incorporation assays any increase in either CDK7 or CDK9 activity when Tat is added to preinitiation complexes. This suggests that both enzymes become inaccessible to Tat because of the structure of the preinitiation complex. Support for this idea comes from the observation that immunodepletion of CDK7 completely abolished phosphorylation of RNA polymerase CTD in preinitiation complexes, whereas immunodepletion of CDK9 had no effect on CTD phosphorylation in preinitiation complexes. Similarly, it has recently been reported that the CAK component of TFIIH is not required for Tat-activated transcription (9).
In order to compare the patterns of phosphorylation induced by CDK7 and CDK9 (53, 72) during three distinct stages of transcription, we used antibodies that recognize epitopes associated with the presence of phosphorylated Ser2 (H5) and Ser5 (H14) in the heptapeptide repeat of the CTD (48). In the preinitiation complexes and early elongation complexes, the primary modification of the CTD is due to Ser5 phosphorylation by CDK7.
CDK7 dissociates from the transcription complexes shortly after transcription initiation (29, 70, 72). However, there is a significant increase in both Ser5 and Ser2 phosphorylation by CDK9 during Tat-dependent elongation. These new phosphorylation signals appear to be distinct from those produced by CDK7. Ramanathan et al. (53) compared the substrate specificity of CDK7, CDK8, and CDK9 by using synthetic peptide substrates. Their data suggest that each kinase displays differential preferences according to where a specific heptad repeat is located in the CTD. Additional evidence that Ser2 phosphorylation of RNA Pol II occurs only during elongation comes from chromatin immunoprecipitation in Saccharomyces cerevisiae. In these experiments, Ser5 phosphorylation was detected primarily in polymerases located at promoter regions and Ser2 phosphorylation was found only in polymerases located in coding regions (37).
Using a combination of immunoprecipitation and protease digestion of RNA polymerase engaged in preinitiation or elongation, we were also able to analyze the phosphorylation pattern of RNA polymerase CTD in preinitiation complexes and elongation complexes. Analysis of the 32P-labeled Glu-C digestion products showed that distinct regions of the CTD are phosphorylated by CDK7 and CDK9. The amino-terminal part of the CTD is preferentially phosphorylated by CDK9 kinase in the elongation complexes. In contrast, in preinitiation complexes the carboxyl-terminal parts of the CTD are phosphorylated preferentially by CDK7. Similarly, Ramanathan et al. (53) reported that fragments of the CTD that corresponded to the carboxyl-terminal region of CTD were preferred as substrates by purified CDK7, whereas the amino-terminal region was preferred by CDK9.
Role of CDK9 during basal transcription from the HIV LTR.
It has been difficult to determine the precise role played by CDK9 in promoting transcription from the HIV LTR in the absence of Tat. Recent experiments by Renner et al. (54) have shown that addition of recombinant DSIF and NELF to isolated RNA Pol II elongation complexes increases the time that the polymerase spends at pause sites. P-TEFb is able to reverse this negative effect due to its ability to phosphorylate the Spt5 subunit of DSIF. A key feature of this system is that the negative factors begin to exert their influence very quickly, and independently of transcript length, whereas there is pronounced kinetic delay before P-TEFb can overcome these elongation blocks. These results strongly suggest that during basal transcription from the HIV LTR P-TEFb is required to promote early-stage elongation and permit transcription complexes to reach TAR. In agreement with this model, our Western blotting results show that CTD phosphorylation at Ser5 is increased in late-stage elongation complexes compared to early-stage elongation complexes even in the absence of Tat. However, in the absence of Tat, there is no increase in Ser2 phosphorylation, a finding consistent with the idea that the substrate specificity of CDK9 might be influenced by Tat (72).
Structural basis for the regulation of polymerase processivity by CTD phosphorylation.
Much of the recent work on the RNA polymerase CTD has focused on its role in providing a platform for RNA splicing and processing factors (for a review, see reference 28). Different regions of the repeat appear to act as binding sites for different sets of processing factors. For example, the CTD carboxyl terminus spanning heptads 27 to 52 can support capping, splicing, and 3′-end processing, whereas the amino terminus only supports capping (17). However, in addition to providing a mechanism that couples transcription and mRNA processing, the CTD also plays an important role in regulating elongation.
Recent cross-linking (15) and X-ray crystallographic (13, 23) studies have provided important clues as to how phosphorylation of the CTD could be used to increase RNA polymerase processivity. In preinitiation complexes the CTD is buried close to the DNA template and surrounded by basal transcription factors, including TFIIH, and proteins of the mediator complex. Cross-linking studies have shown that in preinitiation complexes the polymerase is orientated with CTD making contacts with the DNA template at around position 16, downstream of the transcription start site (15). The repeat unit of each heptapeptide in the CTD contains two SPXX motifs (SPTS and SPSY) that allow the CTD to fold into a compact beta-turn stabilized by a side chain-main chain interaction (5). Peptides with this structure can bind DNA by bis-intercalation (26, 58). Phosphorylation of the CTD would be expected to disrupt these interactions by introducing negative charges that are repelled by the phosphates on the DNA helix. Thus, phosphorylation of the CTD during initiation can destabilize extensive downstream contacts between the polymerase and the DNA template.
Similar principles probably apply during elongation. In this case, regions near the N terminus of the CTD that are hypophosphorylated because of limited activity by CDK7, or CTD phosphatase activity, would be free to interact with the DNA template. This would be expected to stabilize the transcription complex at pause sites but also limit processivity. Phosphorylation of the N-terminal region of the RNA polymerase CTD by CDK9 could then be used to overcome these blocks to elongation.
In conclusion, we have demonstrated that phosphorylation of the RNA polymerase CTD at novel sites by CDK9 can directly alter its processivity. This novel biochemical mechanism explains how Tat is able to activate transcription elongation during HIV replication.
Acknowledgments
We thank our colleagues at the Laboratory of Molecular Biology, M. J. West and M. Bailey for helpful discussions, and A. D. Lowe for the preparation of nuclear extracts used in these experiments.
Y.K.K. is a postdoctoral fellow of the Korea Science and Engineering Foundation.
REFERENCES
- 1.Akoulitchev, S., S. Chuikov, and D. Reinberg. 2000. TFIIH is negatively regulated by cdk8-containing mediator complexes. Nature 407:102-106. [DOI] [PubMed] [Google Scholar]
- 2.Akoulitchev, S., T. P. Mäkelä, R. A. Weinberg, and D. Reinberg. 1995. Requirement for TFIIH kinase activity in transcription by RNA polymerase II. Nature 377:557-560. [DOI] [PubMed] [Google Scholar]
- 3.Awrey, D. E., R. G. Weilbaecher, S. A. Hemming, S. M. Orlicky, C. M. Kane, and A. M. Edwards. 1997. Transcription elongation through DNA arrest sites: a multistep process involving both RNA polymerase II subunit RPB9 and TFIIS. J. Biol. Chem. 272:14747-14754. [DOI] [PubMed] [Google Scholar]
- 4.Bieniasz, P. D., T. A. Grdina, H. P. Bogerd, and B. R. Cullen. 1998. Recruitment of a protein complex containing Tat and cyclin T1 to TAR governs the species specificity of HIV-1 Tat. EMBO J. 17:7056-7065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bienkiewicz, E. A., A.-Y. M. Woody, and R. W. Woody. 2000. Conformation of the RNA polymerase II C-terminal domain: circular dichroism of long and short fragments. J. Mol. Biol. 297:119-133. [DOI] [PubMed] [Google Scholar]
- 6.Bourgeois, C. F., Y. K. Kim, M. J. Churcher, M. J. West, and J. Karn. 2002. Spt-5 cooperates with Tat by preventing premature RNA release at terminator sequences. Mol. Cell. Biol. 22:1079-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chao, S.-H., K. Fujinaga, J. E. Marion, R. Taube, E. A. Sausville, A. M. Senderowicz, B. M. Peterlin, and D. H. Price. 2000. Flavopiridol inhibits p-TEFb and blocks HIV-1 replication. J. Biol. Chem. 275:28345-28348. [DOI] [PubMed] [Google Scholar]
- 8.Chen, D., Y. Fong, and Q. Zhou. 1999. Specific interaction of Tat with the human but not rodent pTEFb complex mediates the species-specific Tat activation of HIV-1 transcription. Proc. Natl. Acad. Sci. USA 96:2728-2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, D., and Q. Zhou. 1999. Tat activates human immunodeficiency virus type 1 transcriptional elongation independent of TFIIH kinase. Mol. Cell. Biol. 19:2863-2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cho, H., T.-K. Kim, H. Mancebo, W. S. Lane, O. Flores, and D. Reinberg. 1999. A protein phosphatase functions to recycle RNA polymerase II. Genes Dev. 13:1540-1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Christie, K. R., D. E. Awrey, A. M. Edwards, and C. M. Kane. 1994. Purified yeast RNA polymerase II reads through intrinsic blocks to elongation in response to the yeast TFIIS analogue, p37. J. Biol. Chem. 269:936-943. [PubMed] [Google Scholar]
- 12.Churcher, M. J., A. D. Lowe, M. J. Gait, and J. Karn. 1995. The RNA element encoded by the trans-activation-responsive region of human immunodeficiency virus type 1 is functional when displaced downstream of the start of transcription. Proc. Natl. Acad. Sci. USA 92:2408-2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cramer, P., D. A. Bushnell, and R. D. Kornberg. 2001. Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution. Science 292:1863-1867. [DOI] [PubMed] [Google Scholar]
- 14.Cujec, T. P., H. Okamoto, K. Fujinaga, J. Meyer, H. Chamberlin, D. O. Morgan, and B. M. Peterlin. 1997. The HIV trans-activator Tat binds to the CDK-activating kinase and activates the phosphorylation of the carboxy-terminal domain of RNA polymerase II. Genes Dev. 11:2645-2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Douziech, M., D. Forget, J. Greenblatt, and B. Coulombe. 1999. Topological localization of the carboxyl-terminal domain of RNA polymerase II in the initiation complex. J. Biol. Chem. 274:19868-19873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feinberg, M. B., D. Baltimore, and A. D. Frankel. 1991. The role of Tat in the human immunodeficiency virus life cycle indicates a primary effect on transcriptional elongation. Proc. Natl. Acad. Sci. USA 88:4045-4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fong, N., and D. L. Bentley. 2001. Capping, splicing, and 3′ processing are independently stimulated by RNA polymerase II: different functions for different segments of the CTD. Genes Dev. 15:1783-1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fong, Y. W., and Q. Zhou. 2000. Relief of two built-in autoinhibitory mechanisms in p-TEFb is required for the assembly of a multicomponent transcription elongation complex at the HIV-1 promoter. Mol. Cell. Biol. 20:5897-5907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fujinaga, K., R. Taube, J. Wimmer, T. P. Cujec, and B. M. Peterlin. 1999. Interactions between human cyclin T, Tat, and the transactivation response element (TAR) are disrupted by a cysteine to tyrosine substitution found in mouse cyclin T. Proc. Natl. Acad. Sci. USA 96:1285-1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garber, M. E., T. P. Mayall, E. M. Suess, J. Meisenhelder, N. E. Thompson, and K. A. Jones. 2000. CDK9 autophosphorylation regulates high-affinity binding of the human immunodeficiency virus type 1 Tat-P-TEFb complex to TAR RNA. Mol. Cell. Biol. 20:6958-6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garber, M. E., P. Wei, and K. A. Jones. 1998. HIV-1 Tat interacts with cyclin T1 to direct P-TEFb CTD kinase complex to TAR RNA. Cold Spring Harbor Symp. Quant. Biol. 63:371-380. [DOI] [PubMed] [Google Scholar]
- 22.Garber, M. E., P. Wei, V. N. Kewel-Ramani, T. P. Mayall, C. H. Herrmann, A. P. Rice, D. R. Littman, and K. A. Jones. 1998. The interaction between HIV-1 Tat and human cyclin T1 requires zinc and a critical cysteine residue that is not conserved in the murine CycT1 protein. Genes Dev. 12:3512-3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gnatt, A. L., P. Cramer, J. Fu, D. A. Bushnell, and R. D. Kornberg. 2001. Structural basis of transcription: an RNA polymerase II elongation complex at 3.3 Å resolution. Science 292:1876-1882. [DOI] [PubMed] [Google Scholar]
- 24.Gold, M. O., J.-P. Tassan, E. A. Nigg, A. P. Rice, and C. H. Herrmann. 1996. Viral transactivators E1A and VP16 interact with a large complex that is associated with CTD kinase activity and contains CDK8. Nucleic Acids Res. 24:3771-3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Graeble, M. A., M. J. Churcher, A. D. Lowe, M. J. Gait, and J. Karn. 1993. Human immunodeficiency virus type 1 trans-activator protein Tat, stimulates transcriptional read-through of distal terminator sequences in vitro. Proc. Natl. Acad. Sci. USA 90:6184-6188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harding, M. M., G. Y. Krippner, C. J. Shelton, A. Rodger, K. J. Sanders, J. P. Mackay, and A. S. Prakash. 1997. DNA-binding studies of XSPTSPSZ, derivatives of the intercalating heptad repeat of RNA polymerase II. Biopolymers 42:387-398. [DOI] [PubMed] [Google Scholar]
- 27.Herrmann, C. H., and A. P. Rice. 1995. Lentivirus Tat proteins specifically associate with a cellular protein kinase, TAK, that hyperphosphorylates the carboxyl-terminal domain of the large subunit of RNA polymerase II: candidate for a Tat cofactor. J. Virol. 69:1612-1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hirose, Y., and J. L. Manley. 2000. RNA polymerase II and the integration of nuclear events. Genes Dev. 14:1415-1429. [PubMed] [Google Scholar]
- 29.Isel, C., and J. Karn. 1999. Direct evidence that HIV-1 Tat activates the Tat-associated kinase (TAK) during transcriptional elongation. J. Mol. Biol. 290:929-941. [DOI] [PubMed] [Google Scholar]
- 30.Ivanov, D., Y. T. Kwak, J. Guo, and R. B. Gaynor. 2000. Domains in the SPT5 protein that modulate its transcriptional regulatory properties. Mol. Cell. Biol. 20:2970-2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ivanov, D., Y. T. Kwak, E. Nee, J. Guo, L. F. Garcia-Martinez, and R. B. Gaynor. 1999. Cyclin T domains involved in complex formation with Tat and TAR RNA are critical for Tat-activation. J. Mol. Biol. 288:41-56. [DOI] [PubMed] [Google Scholar]
- 32.Kao, S.-Y., A. F. Calman, P. A. Luciw, and B. M. Peterlin. 1987. Anti-termination of transcription within the long terminal repeat of HIV-1 by Tat gene product. Nature 330:489-493. [DOI] [PubMed] [Google Scholar]
- 33.Karn, J. 1999. Tackling Tat. J. Mol. Biol. 293:235-254. [DOI] [PubMed] [Google Scholar]
- 34.Keen, N. J., M. J. Churcher, and J. Karn. 1997. Transfer of Tat and release of TAR RNA during the activation of the human immunodeficiency virus type-1 transcription elongation complex. EMBO J. 16:5260-5272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kerppola, T. K., and C. M. Kane. 1990. Analysis of signals for transcription termination by purified RNA polymerase II. Biochemistry 29:269-278. [DOI] [PubMed] [Google Scholar]
- 36.Kim, J. B., and P. A. Sharp. 2001. Positive transcription elongation factor b phosphorylates hSPT5 and RNA polymerase II carboxyl-terminal domain independently of cyclin-dependent kinase-activating kinase. J. Biol. Chem. 276:12317-12323. [DOI] [PubMed] [Google Scholar]
- 37.Komarnitsky, P., E.-J. Cho, and S. Buratowski. 2000. Different phosphorylated forms of RNA polymerase II, and associated mRNA processing factors during transcription. Genes Dev. 14:2452-2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kulish, D., and K. Struhl. 2001. TFIIS enhances transcriptional elongation through an artificial arrest site in vivo. Mol. Cell. Biol. 21:4162-4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kwak, Y. T., D. Ivanov, J. Guo, E. Nee, and R. B. Gaynor. 1999. Role of the human and murine cyclin T proteins in regulating HIV-1 Tat-activation. J. Mol. Biol. 288:57-69. [DOI] [PubMed] [Google Scholar]
- 40.Larochelle, S., J. Chen, R. Knights, J. Pandur, P. Morcillo, H. Erdjument-Bromage, P. Tempst, B. Suter, and R. P. Fisher. 2001. T-loop phosphorylation stabilizes the CDK7-cyclin H-MAT 1 complex in vivo and regulates its CTD kinase activity. EMBO J. 20:3749-3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mancebo, H. S. Y., G. Lee, J. Flygare, J. Tomassini, P. Luu, Y. Zhu, J. Peng, C. Blau, D. Hazuda, D. Price, and O. Flores. 1997. p-TEFb kinase is required for HIV Tat transcriptional activation in vivo and in vitro. Genes Dev. 11:2633-2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marshall, N. F., G. K. Dahmus, and M. E. Dahmus. 1998. Regulation of carboxyl-terminal domain phosphatase by HIV-1 Tat protein. J. Biol. Chem. 273:31726-31730. [DOI] [PubMed] [Google Scholar]
- 43.Marshall, N. F., and M. E. Dahmus. 2000. C-terminal domain phosphatase sensitivity of RNA polymerase II in early elongation complexes on the HIV-1 and adenovirus 2 major late templates. J. Biol. Chem. 275:32430-32437. [DOI] [PubMed] [Google Scholar]
- 44.Morgan, D. O. 1995. Principles of CDK regulation. Nature 374:131-134. [DOI] [PubMed] [Google Scholar]
- 45.Nguyen, V. T., T. Kiss, A. A. Michels, and O. Bensaude. 2001. 7SK small nuclear RNA binds to and inhibits the activity of CDK9/cyclin T complexes. Nature 414:322-325. [DOI] [PubMed] [Google Scholar]
- 46.Parada, C. A., and R. G. Roeder. 1996. Enhanced processivity of RNA polymerase II triggered by Tat-induced phosphorylation of its carboxy-terminal domain. Nature 384:375-378. [DOI] [PubMed] [Google Scholar]
- 47.Parada, C. A., J.-B. Yoon, and R. G. Roeder. 1995. A novel LBP-1-mediated restriction of HIV-1 transcription at the level of elongation in vitro. J. Biol. Chem. 270:2274-2283. [DOI] [PubMed] [Google Scholar]
- 48.Patturajan, M., N. K. Conrad, D. B. Bregman, and J. L. Corden. 1999. Yeast carboxyl-terminal domain kinase I positively and negatively regulates RNA polymerase II carboxyl-terminal domain phosphorylation. J. Biol. Chem. 274:27823-27828. [DOI] [PubMed] [Google Scholar]
- 49.Pavletich, N. P. 1999. Mechanisms of cyclin-dependent kinase regulation: structures of cdks, their cyclin activators, and Cip and INK4 inhibitors. J. Mol. Biol. 287:821-828. [DOI] [PubMed] [Google Scholar]
- 50.Ping, Y.-H., and T. M. Rana. 1999. Tat-associated kinase (P-TEF-b): a component of transcription preinitiation and elongation complexes. J. Biol. Chem. 274:7399-7404. [DOI] [PubMed] [Google Scholar]
- 51.Ping, Y. H., and T. M. Rana. 2001. DSIF and NELF interact with RNA polymerase II elongation complex and HIV-1 Tat stimulates P-TEFb-mediated phosphorylation of RNA polymerase II and DSIF during transcription elongation. J. Biol. Chem. 276:12951-12958. [DOI] [PubMed] [Google Scholar]
- 52.Price, D. H. 2000. P-TEFb, a cyclin-dependent kinase controlling elongation by RNA polymerase II. Mol. Cell. Biol. 20:2629-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ramanathan, Y., S. M. Rajpara, S. M. Reza, E. Lees, S. Shuman, M. B. Mathews, and T. Pe'ery. 2001. Three RNA polymerase II CTD kinases display distinct substrate preferences. J. Biol. Chem. 276:10913-10920. [DOI] [PubMed] [Google Scholar]
- 54.Renner, D. B., Y. Yamaguchi, T. Wada, H. Handa, and D. H. Price. 2001. A highly purified RNA polymerase II elongation control system. J. Biol. Chem. 276:42601-42609. [DOI] [PubMed] [Google Scholar]
- 55.Riedl, T., and J.-M. Egly. 2000. Phosphorylation in transcription: the CTD and more. Gene Express. 9:3-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schroeder, S. C., B. Schwer, S. Shuman, and D. Bentley. 2000. Dynamic assocation of capping enzymes with transcribing RNA polymerase II. Genes Dev. 14:2435-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Siva-Raman, L., D. Reines, and C. M. Kane. 1990. Purified elongation factor SII is sufficient to promote read-through by purified RNA polymerase II at specific termination sites in the human histone H3.3 gene. J. Biol. Chem. 265:14554-14560. [PubMed] [Google Scholar]
- 58.Suzuki, M. 1990. The heptad repeat in the largest subunit of RNA polymerase II binds by intercalating into DNA. Nature 344:562-565. [DOI] [PubMed] [Google Scholar]
- 59.Taube, R., K. Fujinaga, D. Irwin, J. Wimmer, M. Geyer, and B. M. Peterlin. 2000. Interactions between equine cyclin T1, Tat, and TAR are disrupted by a leucine-to-valine substitution found in human cyclin T1. J. Virol. 74:892-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Taube, R., K. Fujinaga, J. Wimmer, M. Barboric, and B. M. Peterlin. 1999. Tat transactivation: a model for the regulation of eukaryotic transcriptional elongation. Virology 264:245-253. [DOI] [PubMed] [Google Scholar]
- 61.Wada, T., G. Orphanides, J. Hasegawa, D.-K. Kim, D. Shima, Y. Yamaguchi, A. Fukada, K. Hisatake, O. Sangtaek, D. Reinberg, and H. Handa. 2000. FACT relieves DSIF/NELF-mediated inhibition of transcriptional elongation and reveals functional differences between p-TEFb and TFIIH. Mol. Cell 5:1067-1072. [DOI] [PubMed] [Google Scholar]
- 62.Wada, T., T. Takagai, Y. Yamaguchi, D. Watanabe, and H. Handa. 1998. Evidence that P-TEFb alleviates the negative effect of DSIF on RNA polymerase II-dependent transcription in vitro. EMBO J. 17:7395-7403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wada, T., T. Takagi, Y. Yamaguchi, A. Ferdous, T. Imai, S. Hirose, S. Sugimoto, K. Yano, G. A. Hartzog, F. Winston, S. Buratowski, and H. Handa. 1998. DSIF, a novel transcription elongation factor that regulates RNA polymerse II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev. 12:343-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang, G., G. T. Cantin, J. L. Stevens, and A. J. Berk. 2001. Characterization of mediator complexes from HeLa cell nuclear extract. Mol. Cell. Biol. 21:4604-4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wei, P., M. E. Garber, S.-M. Fang, W. H. Fischer, and K. A. Jones. 1998. A novel cdk9-associated c-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop specific binding to TAR RNA. Cell 92:451-462. [DOI] [PubMed] [Google Scholar]
- 66.Wu-Baer, F., W. S. Lane, and R. B. Gaynor. 1998. Role of the human homolog of the yeast transcription factor SPT5 in HIV-1 Tat-activation. J. Mol. Biol. 277:179-197. [DOI] [PubMed] [Google Scholar]
- 67.Yamaguchi, Y., T. Takagi, T. Wada, K. Yano, A. Furuya, S. Sugimoto, J. Hasegawa, and H. Handa. 1999. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell 97:41-51. [DOI] [PubMed] [Google Scholar]
- 68.Yamaguchi, Y., T. Wada, D. Watanabe, T. Takagi, J. Hasegawa, and H. Handa. 1999. Structure and function of the human transcription elongation factor DSIF. J. Biol. Chem. 274:8085-8092. [DOI] [PubMed] [Google Scholar]
- 69.Yang, Z., Q. Zhu, K. Luo, and Q. Zhou. 2001. The 7SK small nuclear RNA inhibits CDK9/cyclin T1 kinase to control transcription. Nature 414:317-322. [DOI] [PubMed] [Google Scholar]
- 70.Zawel, L., P. Kumar, and D. Reinberg. 1995. Recycling of the general transcription factors during RNA polymerase II transcription. Genes Dev. 9:1479-1490. [DOI] [PubMed] [Google Scholar]
- 71.Zhang, J., N. Tamilarasu, S. Hwang, M. E. Garber, I. Huq, K. A. Jones, and T. M. Rana. 2000. HIV-1 TAR RNA enhances the interaction between Tat and cyclin T1. J. Biol. Chem. 275:34314-34319. [DOI] [PubMed] [Google Scholar]
- 72.Zhou, M., M. A. Halanski, M. F. Radonovich, F. Kashanchi, J. Peng, D. H. Price, and J. N. Brady. 2000. Tat modifies the activity of CDK9 to phosphorylate serine 5 of the RNA polymerase II carboxyl-terminal domain during human immunodeficiency virus type 1 transcription. Mol. Cell. Biol. 20:5077-5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhou, M., S. Nekhai, D. C. Bharucha, A. Kumar, H. Ge, D. H. Price, J.-M. Egly, and J. N. Brady. 2001. TFIIH inhibits CDK9 phosphorylation during human immunodeficiency virus type 1 transcription. J. Biol. Chem. 276:44633-44640. [DOI] [PubMed] [Google Scholar]
- 74.Zhu, Y., T. Pe'ery, J. Peng, Y. Ramanathan, N. Marshall, T. Marshall, B. Amendt, M. B. Mathews, and D. H. Price. 1997. Transcription elongation factor p-TEFb is required for HIV-1 Tat trans-activation in vitro. Genes Dev. 11:2622-2632. [DOI] [PMC free article] [PubMed] [Google Scholar]