Abstract
How a given Ras prreotein coordinates multiple signaling inputs and outputs is a fundamental issue of signaling specificity. Schizosaccharomyces pombe contains one Ras, Ras1, that has two distinct outputs. Ras1 activates Scd1, a presumptive guanine nucleotide exchange factor (GEF) for Cdc42, to control morphogenesis and chromosome segregation, and Byr2, a component of a mitogen-activated protein kinase cascade, to control mating. So far there is only one established Ras1 GEF, Ste6. Paradoxically, ste6 null (ste6Δ) mutants are sterile but normal in cell morphology. This suggests that Ste6 specifically activates the Ras1-Byr2 pathway and that there is another GEF capable of activating the Scd1 pathway. We thereby characterized a potential GEF, Efc25. Genetic data place Efc25 upstream of the Ras1-Scd1, but not the Ras1-Byr2, pathway. Like ras1Δ and scd1Δ, efc25Δ is synthetically lethal with a deletion in tea1, a critical element for cell polarity control. Using truncated proteins, we showed that the C-terminal GEF domain of Efc25 is essential for function and regulated by the N terminus. We conclude that Efc25 acts as a Ras1 GEF specific for the Scd1 pathway. While ste6 expression is induced during mating, efc25 expression is constitutive. Moreover, Efc25 overexpression renders cells hyperelongated and sterile; the latter can be rescued by activated Ras1. This suggests that Efc25 can recruit Ras1 to selectively activate Scd1 at the expense of Byr2. Reciprocally, Ste6 overexpression can block Scd1 activation. We propose that external signals can partly segregate two Ras1 pathways by modulating GEF expression and that GEFs can influence how Ras is coupled to specific effectors.
Ras G proteins act as molecular switches for signal transduction pathways that are important for cell proliferation, differentiation, cell death, and organization of the cytoskeleton (reviewed in reference 29). In humans, there are three RAS genes (H-, K-, and N-RAS) which encode four Ras proteins with more than 90% identity in amino acid sequence. The biochemical properties of these Ras proteins are very similar and straightforward. Ras can bind either GTP or GDP. In the resting state of the cell, Ras is primarily GDP bound and inactive. In response to signals, Ras switches to the active GTP-bound state, a process catalyzed by guanine nucleotide exchange factors (GEFs). Activated Ras stimulates effector proteins to turn on downstream pathways. How a given Ras protein functions in the cell, however, is anything but straightforward. By one count, there are at least three Ras effectors (Raf, phosphatidylinositol 3-kinase, and Ral GDS; reviewed in reference 29) and three families of GEFs containing at least five members (Sos1, Sos2, GRF1/Cdc25Mm, GRF2, and GRP [2]). Under in vitro conditions, most known Ras effectors and GEFs can frequently interact with more than one Ras protein, but how they actually match up with one another in the cell is poorly understood.
We use the fission yeast Schizosaccharomyces pombe as a genetic model organism to study Ras functions. S. pombe contains a single Ras homolog, Ras1, which interacts with two effectors that control two distinct functions. Ras1 activates the Byr2 protein kinase (a MEKK homolog) to mediate mating pheromone signaling (31). Inactivation of this pathway blocks sexual differentiation and results in sterility. The second Ras1 effector is Scd1 (4; also known as Ral1 [12]), a presumptive GEF for Cdc42. Inactivation of this pathway affects numerous functions, the most readily observable of which is a change of cell morphology from elongated to round. In addition, we have recently identified an additional function of this pathway—namely, the ability to interact with a conserved protein complex containing Yin6 and Moe1 to affect spindle formation and chromosome segregation (5, 18, 34). The scd1 null (scd1Δ) mutants are also sterile, but this sterility does not seem to result from abnormalities in mating pheromone signaling (4). Cells lacking scd1 can sporulate efficiently and induce the expression of mam2 (encoding a mating pheromone receptor), both of which require mating pheromones. It is possible that Scd1 may contribute to mating by affecting functions such as cell polarity and cytoskeleton organization.
Even though GTP-bound Ras1 can bind both Byr2 and Scd1 with high affinity, the molecular pathways controlled by Scd1 and Byr2 are not interchangeable in the cell. Deleting byr2 blocks mating pheromone signaling to cause sterility but does not affect cell morphology (31), chromosome segregation, or spindle formation (5, 18). Conversely, scd1Δ cells are round and defective in chromosome segregation and spindle formation (5, 18) but still respond to mating pheromones (4). Byr2 overexpression cannot rescue the abnormal cell shape of scd1Δ cells, and Scd1 overexpression cannot rescue the sterility of byr2Δ cells (4). Thus, the S. pombe Ras1 pathways are similar to the Ras pathways in the mammalian systems in that a given Ras protein must somehow coordinate interactions with multiple factors.
In S. pombe, the best characterized Ras1 GEF is Ste6 (14). Like other Ras GEFs from mammals (Sos and GRF) and the budding yeast (Cdc25), Ste6 contains a “classic” catalytic domain in its C terminus. ste6Δ cells, like byr2Δ cells, are sterile but have a normal cell shape. In addition, ste6 expression is barely detectable during vegetative growth but is induced by signals for mating, nutrient starvation, and mating pheromones (15). These intriguing phenomena suggest that Ste6 is specific for Ras1 function in mating pheromone signaling. Since Ras1 also controls the morphogenic pathway involving Scd1, we hypothesized that this function of Ras1 is regulated by another GEF.
A gene encoding a second potential Ras1 GEF, efc25 (term derived from “exchange factor cdc25-like”), was isolated unexpectedly in a study intended to isolate the gene encoding a subunit of the DNA polymerase (30). The Efc25 protein contains a C-terminal Cdc25-like catalytic domain, which shares over 30% identity in amino acid sequence with other Ras GEFs. The N terminus of Efc25 is not homologous to any known proteins. Although a detailed study of Efc25 had not been carried out, the phenotype of efc25Δ cells has been reported, round but fertile (30). These interesting observations collectively suggest that Efc25 may specifically regulate the Ras1-Scd1 pathway and that GEFs can play a very important role in establishing specificity for Ras signaling.
In this study, we investigated whether Efc25 indeed specifically activates the Ras1-Scd1 pathway and whether GEFs play key roles in allowing a single Ras protein to control two downstream pathways. Our data indicate that the functions of Efc25 and Ste6 are not interchangeable and that Efc25 specifically regulates the Ras1-Scd1 pathway. We reveal the influence of mating signals on the expression of GEFs. Furthermore, our data support a model in which GEFs play a role in directing Ras1 into a given downstream pathway.
MATERIALS AND METHODS
Parental strains and microbial manipulation.
The generic wild-type strain is SP870 (h90 ade6-210 leu1-32 ura4-D18). The following strains are all derived from SP870: SPSCD1U (scd1::ura4), as described in reference 4, SPBU (byr2::ura4), SPRU (ras1::ura4), and SPRN (ras1::ura4::pUC), as described in reference 31. All moe1Δ and yin6Δ cells were described in references 5 and 34, respectively. Strain MOE1N is essentially the same as MOE1U (moe1::ura4), except that its ura4 was disrupted by homologous recombination (5). ste6Δ (ste6::ura4) cells were as described previously (15) and were named STE6U for this study. The rich medium was YEAU (5), and the minimal medium was MM supplemented with the appropriate supplements (1). A nitrogen (N)-free MM was prepared by eliminating NH4Cl. To induce sexual differentiation, homothallic (h90) cells were pregrown to log phase (2× 106 to 5 × 106 cells/ml) at 30°C in the MM. They were then washed twice with the N-free MM and finally resuspended in an equal volume of the N-free MM at 30°C. Time points were taken after the shift, and aliquots were centrifuged and frozen at −70°C for further Northern and Western analysis.
Plasmid constructions.
PCR was performed to modify the open reading frame (ORF) for byr2 such that it can be cloned into pARTCM (4) at the SalI site. The resulting vector was named pARTCMBYR2. An EcoRI-BamHI fragment containing the ORF of efc25 was cloned into pBluescript SK(−) to create pBSEFC25EB. The HindIII site of pBSEFC25EB was changed into a BamHI site by a linker to create pBSEFC25BB, which allows efc25 to be released from pBSEFC25BB as a BamHI fragment. This fragment was cloned into pARTCM to create pARTCMEFC25. A 2.2-kb region upstream of the efc25 ORF, containing the presumptive efc25 promoter, was amplified by PCR and swapped with the adh1 promoter in pARTCMEFC25 to create pEPCMEFC25. A 1.6-kb BamHI-XbaI from pARTCMEFC25 was cloned into the BamHI-XbaI sites of pBluescript SK(−) to create pBSEFC25N. A BamHI-SacI fragment from pBSEFC25N was cloned into the BamHI-SacI sites of pARTCM to create pARTCMEFC25N. An XbaI-BamHI fragment containing the last 1.3 kb of the efc25 from pBSEFC25EB was cloned into the XbaI-BamHI sites of pBluescript SK(−). The XbaI site was changed to BamHI by a linker to create pBSEFC25CBB, and the BamHI fragment from pBSEFC25CBB was cloned into the BamHI site of pARTCM to create pARTCMEFC25C. A blunt-ended SacI fragment of ade6 was cloned into the EcoRV site of pARTCMEFC25C to create pARTCMEFC25CA. The ste6 ORF was amplified by PCR to contain BamHI and SacI sites for cloning into pARTCM to create pARTCMSTE6. To create pREP1STE6, pARTCMSTE6 was digested with SacI, blunt ended, and cut with BamHI. The resulting fragment was cloned into the BamHI-SmaI sites of pREP1 (21). To generate plasmids for deleting efc25, efc25 genomic DNA was amplified using primers to contain a BamHI site and SacI site for cloning into pBluescript SK(−) to create pBSEFC25. pBSEFC25 was digested by EcoNI and MunI and was blunt ended. The resulting vector was ligated with blunt-ended ura4 and LEU2 to create pBSEFC25::ura4 and pBSEFC25::LEU2. A BamHI-SacI fragment containing the ORF of ras1 was created by PCR and cloned into pARTCM to produce pARTCMR1G. A 1.1-kb SphI-PstI fragment, containing the presumptive ste6 promoter, was created by PCR. This fragment was swapped with the adh1 promoter in pARTCMSTE6 to create pSPCMSTE6. To create pARTCMA, a blunt-ended fragment of ADE2 was cloned into the EcoRV site of pARTCM.
Strain constructions.
To create ras1Δ cells (strain SPR1L), SP870 was transformed with ras▿Hd/pUC7 (22). Strains EFC25U (efc25::ura4) and EFC25L (efc25::LEU2) were created by transforming strain SP870 with a BamHI-SacI fragment from pBSEFC25::ura4 and pBSEFC25::LEU2, respectively. The resulting efc25Δ cells are phenotypically indistinguishable from the efc25Δ strains as described before (30). A tea1Δ strain, TEA1U (tea1::ura4), was created using plasmid pSISTea1Δ as described (20). All double mutants were created by protoplast fusion of single deletion mutants followed by tetrad analysis. Tagging Efc25 at the C terminus abolishes its function (data not shown). To tag efc25 at the N terminus with the coding sequence of c-Myc, we created strain MYCEFC25 by transforming strain EFC25U with a MluI fragment of pEPCMEFC25 (linearized in ars, term derived from “autonomous replicating sequence”), which presumably allows the DNA cassette containing efc25 promoter-c-MYC-efc25 to integrate into ars. The resulting strain MYCEFC25 is phenotypically indistinguishable from the wild-type strain (SP870).
Western and Northern blot analysis.
The preparation of cell lysate and Western blotting were performed as described earlier (4). Antibodies 9E10 (4) and TAT1 (5) were used to detect c-Myc-tagged proteins and tubulins, respectively. For Northern blotting, total RNA was extracted as described earlier (28). A 951-bp XbaI fragment of efc25 and a 927-bp BclI fragment of ste6 (15) were used as templates for the preparation of radiolabeled probes. The probes were prepared by using the Prime-It Random Primer labeling kit (Stratagene), and the hybridization was carried out with the ExpressHyb solution (Clontech). A part of the scd1 and byr2 ORFs (+609 to +1265 and +901 to +1979, respectively) was amplified by PCR as templates for probe preparation. Nonradioactive probes and hybridization were performed according to the manufacturer's protocol (Roche).
Microscopy.
Cell morphology was documented under differential interference contrast microscopy. F-actin staining was performed using rhodamine-conjugated phalloidin, and DNA was visualized by 4′,6′-diamidino-2-phenylinole (DAPI) (1). Microtubules were visualized by immunostaining using the TAT1 antibody against tubulin (34).
RESULTS
Efc25 acts upstream of Ras1.
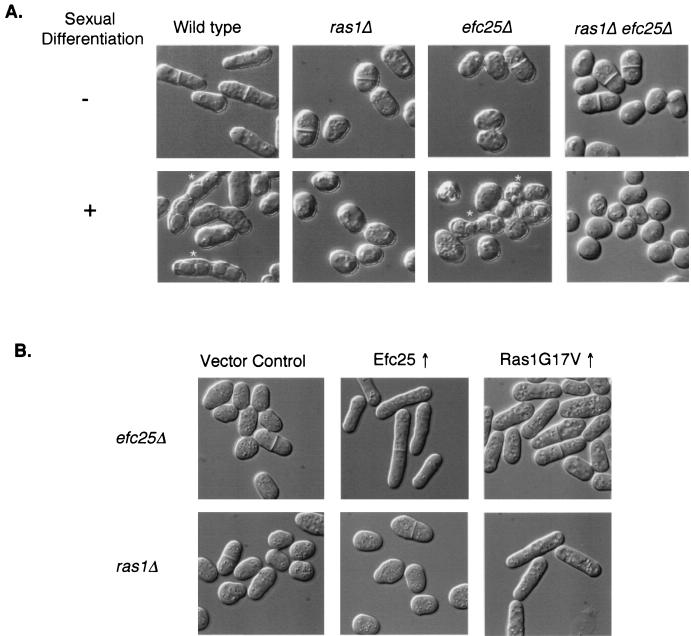
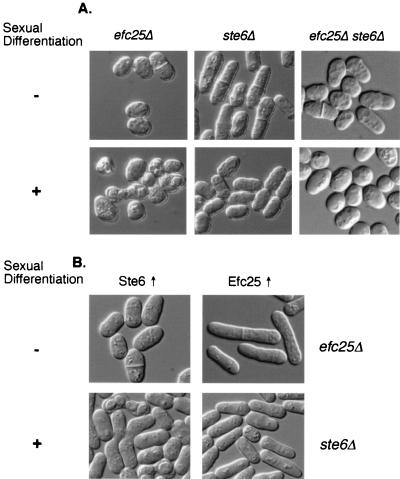
Efc25 was hypothesized as a Ras1 GEF, based on analyses of the null mutant phenotype and protein sequence. To characterize more vigorously the relationship between Efc25 and Ras1, we carried out a series of genetic experiments to determine whether Efc25 acts upstream of Ras1. First, we created a null strain lacking both ras1 and efc25 (ras1Δ efc25Δ). This strain is phenotypically indistinguishable from ras1Δ cells (round and sterile; Fig. 1A). Moreover, there is no obvious “novel” phenotype in the double mutant cells, and these cells do not show any detectable growth defects at 20 and 37°C. These results support the idea that Efc25 and Ras1 act on a linear pathway. Next, we determined in what order they act. As shown in Fig. 1B, the roundness of efc25Δ cells can be very efficiently rescued by the presence of a mutant allele of ras1, ras1G17V, which encodes a Ras1 protein that is constitutively active. Overexpression of the wild-type ras1 only weakly rescues the abnormal cell morphology of efc25Δ cells (data not shown). In contrast, overexpression of efc25 cannot rescue the abnormal morphology of ras1Δ cells. These results indicate that Efc25 acts upstream of Ras1 and is important for Ras1 activation.
FIG. 1.
Efc25 is upstream of Ras1. (A) The relevant genotype of the tested strains is indicated on top of each panel. Cells were plated on either rich medium and grown to log phase (−) or on MM plates for 3 days to induce sexual differentiation (+) and were then visualized under differential interference contrast microscopy. Asterisks mark asci, evidence for mating and sporulation. Strains used were SP870 (wild type), SPR1L (ras1Δ), and EFC25U (efc25Δ), and efc25Δ ras1Δ cells were derived from a fusion between strains SPR1L and EFC25U. (B) Strain EFC25U (efc25Δ) or SPRN (ras1Δ) was transformed with pARTCM (vector control), pARTCMEFC25 (Efc25↑), or pALRV (Ras1G17V↑ [22]), and the morphology of the cells in log phase was recorded.
Efc25 activates specifically the Ras1-Scd1 pathway but not the Ras1-Byr2 pathway.
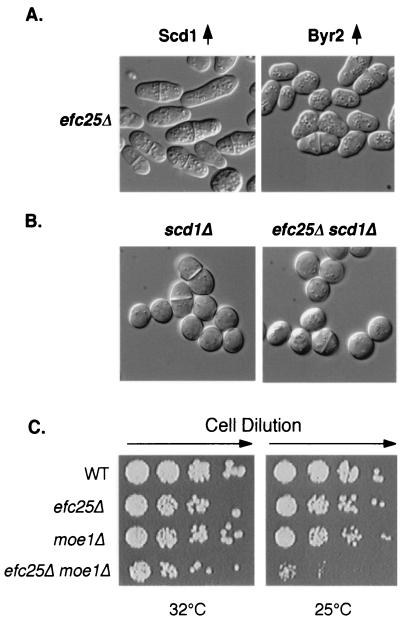
Since efc25Δ cells are round and fertile, it is possible that Efc25 can specifically regulate the Ras1-Scd1 pathway. To test this, we used genetics to ascertain whether Efc25 is upstream of Scd1. As shown in Fig. 2A, overexpression of scd1 but not of byr2 modestly rescues the roundness of efc25Δ cells, while efc25 overexpression does not rescue the defects of scd1Δ and byr2Δ cells (data not shown). In addition, efc25Δ scd1Δ cells are phenotypically indistinguishable from scd1Δ cells (Fig. 2B).
FIG. 2.
Efc25 is upstream of Scd1 but not of Byr2. (A) Strain EFC25U (efc25Δ) was transformed with pALASCD1 (Scd1↑ [4]) or pARTCMBYR2 (Byr2↑), and the cell morphology was recorded. See Fig. 1 for efc25Δ cells transformed with a vector control. (B) An efc25Δ scd1Δ strain was derived from fusing strains SPSCD1U (scd1Δ) and EFC25L, and cells in log phase were photographed. Note that scd1Δ cells are nearly spherical even in log phase (4). (C) Various cells were spotted in 1:5 serial dilutions on rich medium at 32 and 25°C and were allowed to grow for 2 and 4 days, respectively. We note that moe1Δ cells are cold sensitive for growth (5). At 20°C, these cells are viable but grow slowly. The wild-type (WT), moe1Δ, and efc25Δ strains used were SP870, MOE1L, and EFC25U. The efc25Δ moe1Δ strain was derived after a fusion between strains MOE1L and EFC25U.
We have uncovered several components that interact specifically with the Ras1-Scd1 pathway but not with the Ras1-Byr2 pathway. If Efc25 indeed specifically regulates the Scd1 pathway, Efc25 would similarly interact with these components. To test this, we first examined whether Efc25 can interact with Moe1, which has been shown to cooperate with the Ras1-Scd1 pathway to affect spindle formation and chromosome segregation. The deletion of moe1 together with mutations inactivating the Ras1-Scd1 pathway is synthetically lethal (reference 5 and Table 1). Consistent with the hypothesis that Efc25 is upstream of the Ras1-Scd1 pathway, we found that efc25Δ substantially worsens the growth defect of moe1Δ cells. At 25°C, while the single mutants are viable, efc25Δ moe1Δ cells can barely grow (Fig. 2C and Table 1).
TABLE 1.
efc25Δ worsens phenotype of moe1Δ and tea1Δ cells
| Relevant genotype of various double mutantsa | Cell growthb | Reference or source |
|---|---|---|
| ras1Δ tea1Δ | − | This study |
| efc25Δ tea1Δ | − | This study |
| byr2Δ tea1Δ | ++ | This study |
| scd1Δ tea1Δ | − | This study |
| ras1Δ moe1Δ | − | 5 |
| efc25Δ moe1Δ | −+ | This study |
| byr2Δ moe1Δ | ++ | This study |
| scd1Δ moe1Δ | − | 5 |
All double null mutants were derived from the same parental wild-type strain SP870 (Materials and Methods). All respective single null mutants are viable; see text for details.
The symbols represent the following: ++, no apparent growth inhibition tested at 25, 32, and 37°C; −/+, growth inhibition at 25°C but not at 32°C; and −, nearly inviable, see text for more details.
Another unique function of the Ras1-Scd1 pathway is to influence cell polarity to maintain the elongation of cells. One of the key elements in cell polarity control is Tea1, a microtubule binding protein (20). We investigated whether the Ras1-Scd1 pathway could interact with Tea1 for cell polarity control and, if so, whether Efc25 could similarly interact with Tea1.
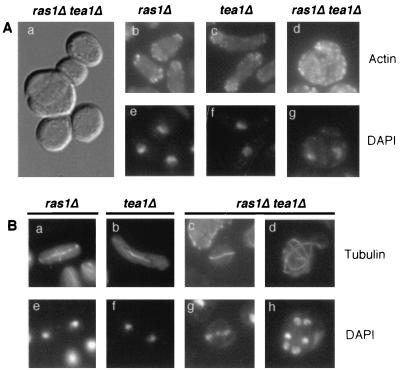
After analyzing tetrads from tea1Δ+ ras1Δ+ diploid strains (Table 1), we determined that more than 60% of tea1Δ ras1Δ spores either did not divide or divided only a few times, while the remaining 30% could only form a microcolony (cell colony diameter < 10% of that of the wild-type cells). We examined phenotypes of those ras1Δ tea1Δ cells in the microcolonies and found that nearly half of them appear to remain connected after septation and thus form large cell masses (Fig. 3A, micrograph a). Frequently, within these cell masses, more than half of the cells are multinucleated (Fig. 3A, micrographs d and g, and 3B, micrographs d and h), suggesting that Ras1 and Tea1 are important for cytokinesis and/or septation. In keeping with the idea that Tea1 can cooperate with the Ras1 pathway for cell polarity control, ras1Δ tea1Δ cells are nearly spherical (Fig. 3A, micrograph a), while ras1Δ cells in log phase are pear shaped, not completely round (Fig. 3A, micrograph b). Consistent with our previous finding that the Ras1-Scd1 pathway plays a role in mediating proper chromosome segregation, ras1Δ tea1Δ cells with lagging chromosomes can be detected (approximately 3% of the mitotic cells; Fig. 3B, micrographs c and g), an anomaly not readily detectable in the single mutants (Fig. 3B, micrographs a, b, e, and f). Additionally, we germinated all tea1Δ ras1Δ cells from tea1Δ/+ ras1Δ/+ diploids (by selective nutrient supplement) and found that these cells displayed the same abnormalities as shown in Fig. 3, indicating that these abnormalities are not unique to cells in microcolonies. Finally, tea1Δ is also synthetically lethal with efc25Δ and scd1Δ but not with byr2Δ (Table 1), and the double null mutants showed the same set of phenotypes as described above (not shown). We conclude that inactivation of both Tea1 and the Ras1-Scd1 pathway leads to a global disruption of cytoskeleton function and that Efc25 is upstream specifically of Scd1.
FIG. 3.
Phenotypes of tea1Δ ras1Δ cells. (A) Micrograph a, morphology of tea1Δ ras1Δ cells in a microcolony. Micrographs b to d, F-actin staining of strains carrying indicated null mutations grown in rich medium. The same cells were counterstained with DAPI to view DNA (micrographs e to g). (B) Tubulin staining of strains carrying indicated null mutations is shown in micrographs a to d; DAPI counterstaining of the same cell is shown in micrographs e to h. The ras1Δ and tea1Δ strains used were SPR1L and TEA1U. The ras1Δ tea1Δ strain was derived from a fusion of SPR1L and TEA1U strains.
The GEF domain is essential for Efc25 function and is regulated by the N terminus of Efc25.
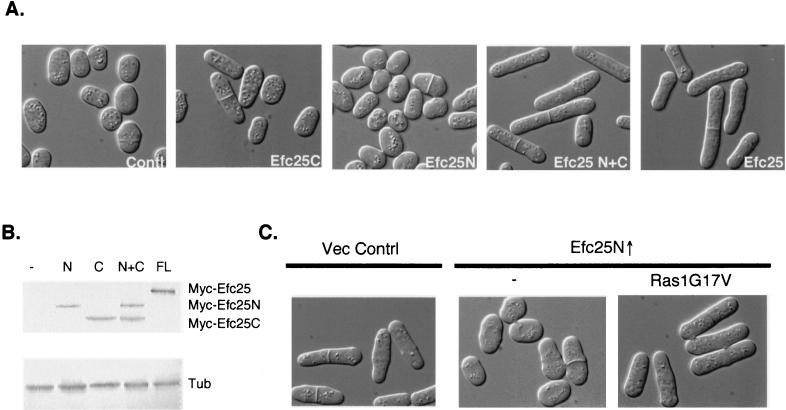
Our data illustrate that Efc25 acts upstream of Ras1 and plays a role in Ras1 activation. We further examined whether Efc25 acts as a Ras1 GEF by determining whether the GEF domain of Efc25 is important for its function. Two truncated forms of Efc25, Efc25N and Efc25C, were created. Efc25C contains the C terminus of Efc25 (amino acid residues 550 to 987), where the GEF domain is located, while Efc25N contains the N terminus of Efc25 (amino acid residues 1 to 550). As shown in Fig. 4A, Efc25C can modestly rescue the roundness of efc25Δ cells, but Efc25N cannot. This result supports the hypothesis that Efc25 acts as a Ras1 GEF.
FIG. 4.
The C-terminal GEF domain of Efc25 is functional and positively regulated by the N terminus. (A) The plasmids used to overexpress Efc25C, Efc25N, and full-length Efc25 in strain EFC25U (efc25Δ) were pARTCMEFC25CA (Efc25C), pARTCMEFC25N (Efc25N), and pARTCMEFC25 (Efc25), respectively. pARTCMEFC25N and pARTCMEFC25CA were cotransformed to express both Efc25N and Efc25C (Efc25N+C). The vector controls are pARTCM and pARTCMA. (B) Protein extracts from cells shown in panel A were analyzed by Western blots using an anti-c-Myc antibody (top), which detects various forms of c-Myc-tagged Efc25 proteins, as indicated on the right. Tubulin (Tub) levels were examined as a loading control (bottom). The overexpressed c-Myc-Efc25 proteins in various samples are marked on top of each lane as follows: none (−), Efc25N (N), Efc25C (C), and Efc25N and Efc25C (N+C), and full-length Efc25 (FL). (C) SP870 (wild-type) cells were transformed with pARTCM and pSLF173 (Vec Contrl [11]), pARTCMEFC25N (Efc25N↑) and pSLF173 (−), or pARTCMEFC25N (Efc25N↑) and pAURV (Ras1G17V [22]).
The C-terminal catalytic domain of budding yeast Cdc25 appears as active as full-length Cdc25 (17). By contrast, the catalytic domain of Efc25 (Efc25C) does not function as efficiently as full-length Efc25. The N termini of most known GEFs are highly diverse in primary sequence and many of these have been shown to play regulatory roles (6, 13; see Discussion). Therefore, we investigated whether the N terminus of Efc25 can similarly regulate Efc25 functions. Indeed, Fig. 4A shows that, while overexpression of Efc25N alone has no effect on the shape of efc25Δ cells, overexpression of Efc25C together with Efc25N efficiently restores the abnormal cell morphology of efc25Δ cells. The expression levels of Efc25C and Efc25N, whether expressed together or singularly, and of full-length Efc25 are similar, as determined by the Western blot analysis (Fig. 4B). These data support the hypothesis that the N terminus of Efc25 can positively regulate the GEF activity of Efc25.
If the N terminus of Efc25 positively regulates Efc25, overexpressing Efc25N in wild-type cells may be dominant negative. Consistent with this hypothesis, Efc25N overexpression in wild-type cells causes them to become round but does not affect mating (Fig. 4C) and, importantly, the cell roundness induced by Efc25N overexpression can be rescued by Ras1G17V (Fig. 4C). Evidently, Efc25N overexpression can interfere with Ras1 activation, which leads to inactivation of the Scd1 pathway.
The functions of Efc25 and Ste6 are not interchangeable.
Previous studies and our results presented above suggest that Efc25 activates the Ras1-Scd1 pathway, while Ste6 regulates the Ras1-Byr2 pathway. We examined whether the functions of Efc25 and Ste6 are interchangeable. An efc25Δ ste6Δ strain was created, and it is phenotypically indistinguishable from ras1Δ strains (Fig. 5A). This observation confirms that Efc25 and Ste6 are each on a separate Ras1 pathway and argues strongly that no other GEFs are necessary for Ras1 functions. Moreover, Ste6 expressed from various plasmids (containing the ste6 genomic promoter, the adh1 promoter, or the strongest nmt1 promoter), all of which fully rescue the sterility of ste6Δ cells, does not rescue the roundness of efc25Δ cells; reciprocally, Efc25 overexpression does not rescue the sterility of ste6Δ cells (Fig. 5B). These experiments indicate that the two Ras1 pathways are regulated by two GEFs whose functions are not interchangeable.
FIG. 5.
Efc25 and Ste6 are not functionally redundant. (A) efc25Δ (strain EFC25L), ste6Δ (strain STE6U), and efc25Δ ste6Δ (made from a fusion between strains EFC25L and STE6U) were plated either on rich medium and grown to log phase (−) or on MM and grown for 3 days to induce sexual differentiation (+). (B) Strains EFC25U (efc25Δ) and STE6U (ste6Δ) were transformed with pARTCMEFC25 (Efc25↑) or pARTCMSTE6 (Ste6↑), and cell morphology was determined under log phase (−) or starvation (+) conditions.
Regulation of the Ras1 pathways: GEF expression is mediated by different signals.
We went on to investigate the mechanisms by which a single Ras1 protein can regulate two pathways and what roles GEFs play in this process. Cell mating is induced by external signals, such as mating pheromones and nutrient starvation, which have been shown to induce ste6 expression transcriptionally (15). Therefore, we asked whether the expression of GEFs is a key to the regulation of Ras1 pathways.
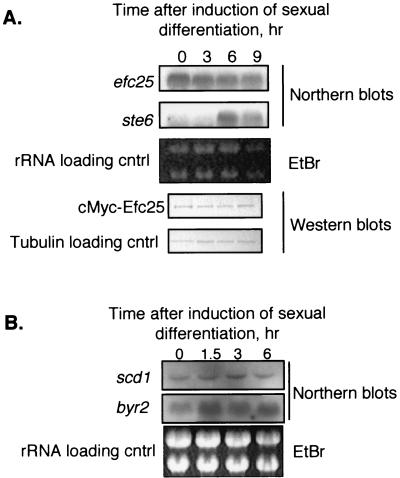
As shown in Fig. 6A, ste6 mRNA levels are weakly detectable before the induction for sexual differentiation but can be increased almost 10-fold after 6 h of induction as reported elsewhere (15). In contrast, efc25 seems to be constitutively expressed. The efc25 mRNA levels remain unchanged before and after the induction for sexual differentiation for up to 14 h (Fig. 6A and data not shown). To determine whether the accumulation of Efc25 protein follows the same pattern as that of the mRNA, we performed Western blots on cells in which the endogenous efc25 was tagged at the 5′ end with the coding sequence for the c-Myc epitope. The expression of Efc25 is thus controlled by its own promoter, and we showed that the tagged protein appears as functional as wild-type protein (Materials and Methods). As shown in Fig. 6A, Efc25 protein levels follow the same profile as those of the mRNA. These results reveal that expression of Ras1 GEFs can be differentially influenced by external signals, and that, as such, a particular Ras1 pathway can be selectively turned on in a timely fashion.
FIG. 6.
Expression of Ras1 GEFs and effectors in response to mating signals. (A) Homothallic cells capable of switching mating types were pregrown to log phase. These cells were then transferred to nitrogen-free medium, which induces secretion of mating pheromones and the onset of sexual differentiation. Time points were taken after the transfer as indicated. Strains SP870 (wild type) and MYCEFC25, containing a c-Myc-tagged Efc25, were used for Northern and Western blots, respectively. EtBr, ethidium bromide. (B) mRNAs were analyzed in wild-type strain SP870 as described for panel A.
Expression of ras1 is not dependent on mating signals; it increases approximately twofold during sexual differentiation (15). We performed Northern blot analyses to investigate whether the expressions of Ras1 effectors are coordinately regulated with that of the GEFs. As shown in Fig. 6B, like efc25, scd1 appears constitutively expressed. byr2 expression is not as extensively dependent on mating signals as is ste6 expression, and it can be increased two- to threefold during sexual differentiation.
Regulation of the Ras1 pathways: GEFs directs Ras1 to specific downstream effectors.
During the course of examining Efc25 overexpression, we noted that Efc25 overexpression in fact caused cells to elongate more than normal. The cells as shown in Fig. 1B and 4A are approximately 30% longer than normal. As mentioned earlier, Efc25 overexpression does not induce cell hyperelongation in ras1Δ and scd1Δ cells (Fig. 2B and data not shown), but it can do so in byr2Δ and byr1Δ cells (data not shown). These results suggest that the cell hyperelongation is caused by activation of the Scd1 pathway, not by a Byr2-dependent hypersexual effect (31).
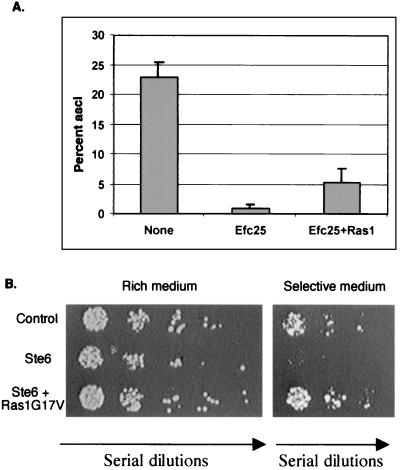
Interestingly, these hyperelongated cells are also sterile (Fig. 7A). Is it possible then that Efc25 can preferentially recruit Ras1 to activate Scd1 and thus render Ras1 unavailable for Byr2? To test this, we examined whether Ras1 overexpression could rescue the sterility resulting from Efc25 overexpression. As shown in Fig. 7A, Ras1 overexpression increases cell mating by sevenfold (P < 0.05, Student t test). Moreover, overexpression of Byr2 or Scd1 cannot efficiently rescue sterility (data not shown). This result is in keeping with the fact that Byr2 overexpression is a poor suppressor for mating for ras1Δ cells (31) and thus suggests that Efc25 overexpression blocks Byr2's access to nearly all the Ras1. We conclude from these data that overexpression of Efc25 can preferentially recruit Ras1 to perform a “morphogenic” function and thus block Byr2's access to Ras1.
FIG. 7.
Efc25 overexpression can block the Ras1-Byr2 pathway, while Ste6 overexpression can block the Ras1-Scd1 pathway. (A) To overexpress Efc25 and Ras1 in strain SP870 (wild type), pSLFEFC25 and pARTCMR1G were used. The control vectors were pARTCM and pSLF173. The percentage of asci in approximately 1,000 cells per transformed colony was counted after 3 days of growth on MM. Bars represent standard deviations from the results of three different colonies of each transformation. (B) MOE1N (moe1Δ) cells were transformed with pARTCM and pSLF173 (control), pARTCMSTE6 (Ste6) and pSLF173, or pARTCMSTE6 and pAURV (Ste6 + Ras1G17V). The resulting cells were spotted on MM or nonselective rich medium (as a control for the amount of cells spotted) and grown at 32°C.
Next, we performed reciprocal experiments to investigate whether Ste6 overexpression can titrate out Ras1 for the Scd1 pathway. moe1Δ cells are very sensitive to the loss of function in the Ras1-Scd1 pathway; therefore, we tested the effects of Ste6 overexpression on the Ras1 pathway in these cells. Our data show that Ste6 overexpression severely worsens the growth defect of both moe1Δ (Fig. 7B) and yin6Δ (not shown) cells and that the presence of Ras1G17V can effectively rescue these abnormalities.
DISCUSSION
S. pombe Ras1 is an excellent model for studying how a given Ras protein coordinates various inputs and outputs during signal transduction. Through a detailed genetic study of Efc25, we conclude that Ras1 GEFs play a critical role in directing Ras1 to downstream pathways. Our data illustrate that the functions of Efc25 and Ste6 are not interchangeable. Efc25 appears to act as a GEF to activate Ras1 specifically for the Scd1pathway, while Ste6 seems to activate Ras1 specifically to turn on the Byr2 pathway. We show further that the expression of GEFs (and thus the activation of Ras1) can be differentially influenced by mating signals and that the presence of a given GEF seems capable of recruiting Ras1 to activate a cognate downstream pathway.
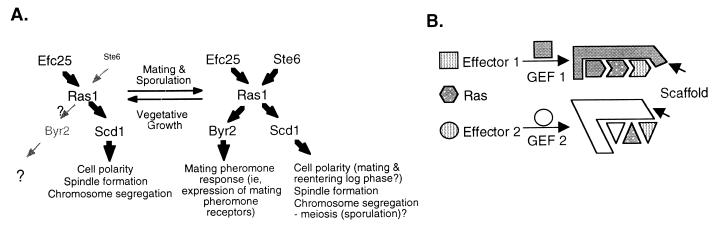
The presumptive dynamic interaction between GEFs, Ras1, and Ras1 effectors is depicted in Fig. 8A. It seems evident that, during vegetative growth, the dominant Ras1 pathway is that of Efc25-Ras1-Scd1. This is likely because the presence of Efc25 seems to efficiently and selectively recruit Ras1 and Scd1 and because very little Ste6 is present. As cells undergo sexual differentiation, Ste6 protein levels rise substantially, and Ste6 is predicted to recruit some of the Ras1 to eventually activate Byr2.
FIG. 8.
Model. (A) The two Ras1 pathways are partly regulated at the transcriptional levels by signals of mating (see text for details). Ste6 is shown with a smaller font for log-phase cells because it is not as highly expressed as those cells entering sexual differentiation. (B) GEFs can mediate Ras1 binding to a given effector, and this binding can also be mediated by scaffold proteins.
The functions of the Efc25-Ras1-Scd1 pathway during vegetative growth are quite apparent, controlling polarized cell extension and mitotic fidelity. The Ste6-Ras1-Byr2 pathway has been shown to regulate expression of genes encoding the mating pheromone receptors (33), an activity that is clearly important for sexual differentiation. It is rather surprising to learn that Efc25 and Scd1 are also expressed during sexual differentiation. What functions are likely to be controlled by them?
It has been shown that, after prolonged starvation, ras1Δ cells display disorganized “birth scars,” which are orderly deposits of cell wall materials left behind from previous cell divisions (25). These cells lag behind wild-type cells in reentry into vegetative growth upon addition of fresh nutrients, and the delay correlates with the time needed to reorganize their birth scars. This observation supports a hypothesis that Ras1 is required for maintaining cell polarity during prolonged starvation, the loss of which may delay cell division upon reentry into the cell cycle. It is also possible that cell polarity and cytoskeleton organization may be important for mating (23, 24). This partly explains why ras1Δ and scd1Δ cells cannot mate, even as the mating pheromone pathway is activated by Byr2 overexpression (4, 31). There is one caveat to this: unlike scd1Δ cells, efc25Δ cells mate nearly as efficiently as wild-type cells. We surmise that, in the absence of efc25, Ras1 and Scd1 (and proteins downstream of Scd1) may be partially active and capable of promoting mating.
The Ras1-Scd1 pathway is also important for spindle formation and chromosome segregation, which are conceivably important not only for mitosis but also for meiosis. In keeping with this, we observed that diploid cells defective in the Yin6-Moe1 complex (34) and in Scd1 (Y.-C. Li and E. C. Chang, unpublished results) frequently sporulate to produce abnormal asci with fewer than the normal four spores, indicative of meiotic chromosome missegregation (7).
Can we rule out the possibility that Efc25 can in fact regulate another Ras-like protein? We believe that this is highly unlikely, based on the S. pombe genome sequencing data (32). S. pombe has a total of 18 Ras-like proteins. In addition to Ras1, there are eight Rab-like and six Rho-like proteins, a single Spg1/Tem1 protein, and a single Spi1/Ran1 protein. These proteins are structurally substantially different from Ras and are thus regulated by unique GEFs. S. pombe does not have any Rap proteins, which are structurally very similar to Ras. The remaining Ras-like protein belongs to the Rheb subfamily (reviewed in reference 26). Phylogeny studies place Rheb distant from all other members in the Ras superfamily. In particular, there is a change from Gly to Arg at a position corresponding to the Gly-12 in human Ras proteins. This alteration is predicted to render Rheb constitutively GTP bound; thus, Rheb activation may not need any GEFs. The deletion of the S. pombe gene encoding Rheb (also known as rhb1) leads to a cell cycle arrest similar to that induced by nutrient starvation, and there was no genetic interaction with Ras1 (19). Consistent with the latter observation, we found that Rheb overexpression did not rescue the abnormal cell shape of efc25Δ cells (unpublished results).
Both Ras1 pathways are present during sexual differentiation, and this prompts us to speculate how these two pathways are coordinated. We made a surprising discovery in this study that supports a hypothesis that GEFs play a role in influencing the connection between Ras1 and its downstream pathways. We show that overexpression of Efc25 can selectively activate the Scd1 pathway at the expense of the Byr2 pathway; reciprocally, overexpression of Ste6 sequesters Ras1 for Scd1. There are at least two models that can explain this. In the first, we propose that the presence of a given GEF can induce a conformational change in Ras1 that favors the binding of a given effector and that these interactions can be further mediated by scaffold proteins (Fig. 8B). In previous studies of the Ras1-Scd1 pathway (3, 4), it was shown that components in this pathway interact with each other in a cooperative fashion—the presence of one component can strengthen the binding between other components in the same protein complex and identified at least one scaffold protein, Scd2. As an alternative, signaling specificity can be achieved if components of the two pathways are spatially segregated in the cell. These two mechanisms are not mutually exclusive and may both be operative to achieve maximal specificity.
The Ras pathways in mammalian cells are far more complex than those in S. pombe. Nevertheless, there is evidence that mammalian GEFs also play important roles in affecting the specificity of Ras signaling. GRF has been shown to efficiently activate H-Ras in NIH 3T3 cells without significantly activating N- or K-Ras (16). A Ca2+-calmodulin complex can bind the IQ domain in GRF1 (10), and GRP has binding sites for both Ca2+ and DAG (9). Thus, Ca2+ may directly or indirectly influence the activity of these GEFs to allow Ras to modulate Ca2+ signaling. Moreover, since Sos can bind growth factor receptors (27), while GRF1 and GRP can each bind Ca2+ and DAG, respectively, these GEFs may assemble unique Ras pathways in various parts of the cell where the growth factor receptor, Ca2+, or DAG is concentrated. The GEFs for Rho-like proteins have also been shown to affect the specificity of signaling (8, 35). Boriack-Sjodin et al. (2) have recently revealed the three-dimensional structure of a complex containing Ras and the catalytic domain of Sos (C-Sos). Intriguingly, they found that the binding of C-Sos causes a dramatic conformational change in the Ras Switch I region, which also encompasses the effector loop. It will be of interest to determine whether such conformational change can take place in the cell and whether it could play a role in modulating effector binding.
Acknowledgments
We thank Paul Nurse, Masayuki Yamamoto, and Keith Gull for providing reagents, Ying-chun Li for discussion, Gino Castriota and Julia Neugebauer for technical assistance, and Stephen Small for critically reading the manuscript. We particularly appreciate Giuseppe Baldacci for his kind and generous help.
This study was supported by grants from the American Cancer Society (RPG-97-137-01-MGO) and National Institutes of Health (CA90464) and the Whitehead Fellowship from New York University.
REFERENCES
- 1.Alfa, C., P. Fantes, J. Hyams, M. McLeod, and E. Warbrick. 1993. Experiments with fission yeast. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 2.Boriack-Sjodin, P. A., S. M. Margarit, D. Bar-Sagi, and J. Kuriyan. 1998. The structural basis of the activation of Ras by Sos. Nature 394:337-343. [DOI] [PubMed] [Google Scholar]
- 3.Chang, E., G. Bartholomeusz, R. Pimental, J. Chen, H. Lai, L.-H. L. Wang, P. Yang, and S. Marcus. 1999. Direct binding and in vivo regulation of the fission yeast p21-activated kinase Shk1 by the SH3 domain protein Scd2. Mol. Cell. Biol. 19:8066-8074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang, E. C., M. Barr, Y. Wang, V. Jung, H. Xu, and H. M. Wigler. 1994. Cooperative interaction of S. pombe proteins required for mating and morphogenesis. Cell 79:131-141. [DOI] [PubMed] [Google Scholar]
- 5.Chen, C.-R., Y.-C. Li, J. Chen, M. C. Hou, P. Papadaki, and E. C. Chang. 1999. Moe1, a novel conserved protein in Schizosaccharomyces pombe, interacts with a Ras effector, Scd1, to affect proper spindle formation. Proc. Natl. Acad. Sci. USA 96:517-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, R. A., T. Michaeli, L. Van Aelst, and R. Ballester. 2000. A role for the noncatalytic N terminus in the function of Cdc25, a Saccharomyces cerevisiae Ras-guanine nucleotide exchange factor. Genetics 154:1473-1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cooper, J. P., Y. Watanabe, and P. Nurse. 1998. Fission yeast Taz1 protein is required for meiotic telomere clustering and recombination. Nature 392:828-831. [DOI] [PubMed] [Google Scholar]
- 8.Daniels, R. H., F. T. Zenke, and G. M. Bokoch. 1999. alphaPix stimulates p21-activated kinase activity through exchange factor-dependent and -independent mechanisms. J. Biol. Chem. 274:6047-6050. [DOI] [PubMed] [Google Scholar]
- 9.Ebinu, J. O., D. A. Bottorff, E. Y. Chan, S. L. Stang, R. J. Dunn, and J. C. Stone. 1998. RasGRP, a Ras guanyl nucleotide-releasing protein with calcium- and diacylglycerol-binding motifs. Science 280:1082-1086. [DOI] [PubMed] [Google Scholar]
- 10.Farnsworth, C. L., N. W. Freshney, L. B. Rosen, A. Ghosh, M. E. Greenberg, and L. A. Feig. 1995. Calcium activation of Ras mediated by neuronal exchange factor Ras-GRF. Nature 376:524-527. [DOI] [PubMed] [Google Scholar]
- 11.Forsburg, S. L., and D. A. Sherman. 1997. General purpose tagging vectors for fission yeast. Gene 191:191-195. [DOI] [PubMed] [Google Scholar]
- 12.Fukui, Y., and M. Yamamoto. 1988. Isolation and characterization of Schizosaccharomyces pombe mutants phenotypically similar to ras1−. Mol. Gen. Genet. 215:26-31. [DOI] [PubMed] [Google Scholar]
- 13.Gross, A., S. Winograd, I. Marbach, and A. Levitzki. 1999. The N-terminal half of Cdc25 is essential for processing glucose signaling in Saccharomyces cerevisiae. Biochemistry 38:13252-13262. [DOI] [PubMed] [Google Scholar]
- 14.Hughes, D. A., Y. Fukui, and M. Yamamoto. 1990. Homologous activators of ras in fission yeast and budding yeast. Nature 344:355-357. [DOI] [PubMed] [Google Scholar]
- 15.Hughes, D. A., N. Yabana, and M. Yamamoto. 1994. Transcriptional regulation of a Ras nucleotide-exchange factor gene by extracellular signals in fission yeast. J. Cell Sci. 107:3635-3642. [DOI] [PubMed] [Google Scholar]
- 16.Jones, M. K., and J. H. Jackson. 1998. Ras-GRF activates Ha-Ras, but not N-Ras or K-Ras 4B, protein in vivo. J. Biol. Chem. 273:1782-1787. [DOI] [PubMed] [Google Scholar]
- 17.Lai, C. C., M. Boguski, D. Broek, and S. Powers. 1993. Influence of guanine nucleotides on complex formation between Ras and CDC25 proteins. Mol. Cell. Biol. 13:1345-1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li, Y.-C., C.-R. Chen, and E. C. Chang. 2000. Fission yeast Ras1 effector Scd1 interacts with the spindle and affects its proper formation. Genetics 156:995-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mach, K. E., K. A. Furge, and C. F. Albright. 2000. Loss of Rhb1, a Rheb-related GTPase in fission yeast, causes growth arrest with a terminal phenotype similar to that caused by nitrogen starvation. Genetics 155:611-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mata, J., and P. Nurse. 1997. tea1 and the microtubular cytoskeleton are important for generating global spatial order within the fission yeast cell. Cell 89:939-949. [DOI] [PubMed] [Google Scholar]
- 21.Maundrell, K. 1993. Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene 123:127-130. [DOI] [PubMed] [Google Scholar]
- 22.Nadin-Davis, S. A., A. Nasim, and D. Beach. 1986. Involvement of ras in sexual differentiation but not in growth control in fission yeast. EMBO J. 5:2963-2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Petersen, J., M. J. Heitz, and I. M. Hagan. 1998. Conjugation in S. pombe: identification of a microtubule-organising centre, a requirement for microtubules and a role for Mad2. Curr. Biol. 8:963-966. [DOI] [PubMed] [Google Scholar]
- 24.Petersen, J., O. Nielsen, R. Egel, and I. M. Hagan. 1998. F-actin distribution and function during sexual differentiation in Schizosaccharomyces pombe. J. Cell Sci. 111:867-876. [DOI] [PubMed] [Google Scholar]
- 25.Pichová, A., and E. Streiblová. 1992. Features of the cell periphery in the deformed ras1− cells of Schizosaccharomyces pombe. Exp. Mycol. 16:178-187. [Google Scholar]
- 26.Reuther, G. W., and C. J. Der. 2000. The Ras branch of small GTPases: Ras family members don't fall far from the tree. Curr. Opin. Cell Biol. 12:157-165. [DOI] [PubMed] [Google Scholar]
- 27.Schlessinger, J. 1994. How receptor tyrosine kinases activate Ras. Trends Biochem. Sci. 18:273-275. [DOI] [PubMed] [Google Scholar]
- 28.Schmitt, M. E., T. A. Brown, and B. L. Trumpower. 1990. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 18:3091-3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shields, J. M., K. Pruitt, A. McFall, A. Shaub, and C. J. Der. 2000. Understanding Ras: ‘it ain't over 'til it's over'. Trends Cell Biol. 10:147-154. [DOI] [PubMed] [Google Scholar]
- 30.Tratner, I., A. Fourticq-Esqueöute, J. Tillit, and G. Baldacci. 1997. Cloning and characterization of the S. pombe gene efc25+, a new putative guanine nucleotide exchange factor. Gene 193:203-210. [DOI] [PubMed] [Google Scholar]
- 31.Wang, Y., H.-P. Xu, M. Riggs, L. Rodgers, and M. Wigler. 1991. byr2, a Schizosaccharomyces pombe gene encoding a protein kinase capable of partial suppression of the ras1 mutant phenotype. Mol. Cell. Biol. 11:3554-3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wood, V., R. Gwilliam, M. A. Rajandream, M. Lyne, R. Lyne, A. Stewart, J. Sgouros, N. Peat, J. Hayles, S. Baker, D. Basham, S. Bowman, K. Brooks, D. Brown, S. Brown, T. Chillingworth, C. Churcher, M. Collins, R. Connor, A. Cronin, P. Davis, T. Feltwell, A. Fraser, S. Gentles, A. Goble, N. Hamlin, D. Harris, J. Hidalgo, G. Hodgson, S. Holroyd, T. Hornsby, S. Howarth, E. J. Huckle, S. Hunt, K. Jagels, K. James, L. Jones, M. Jones, S. Leather, S. McDonald, J. McLean, P. Mooney, S. Moule, K. Mungall, L. Murphy, D. Niblett, C. Odell, K. Oliver, S. O'Neil, D. Pearson, M. A. Quail, E. Rabbinowitsch, K. Rutherford, S. Rutter, D. Saunders, K. Seeger, S. Sharp, J. Skelton, M. Simmonds, R. Squares, S. Squares, K. Stevens, K. Taylor, R. G. Taylor, A. Tivey, S. Walsh, T. Warren, S. Whitehead, J. Woodward, G. Volckaert, R. Aert, J. Robben, B. Grymonprez, I. Weltjens, E. Vanstreels, M. Rieger, M. Schafer, S. Muller-Auer, C. Gabel, M. Fuchs, C. Fritzc, E. Holzer, D. Moestl, H. Hilbert, K. Borzym, I. Langer, A. Beck, H. Lehrach, R. Reinhardt, T. M. Pohl, P. Eger, W. Zimmermann, H. Wedler, R. Wambutt, B. Purnelle, A. Goffeau, E. Cadieu, S. Dreano, S. Gloux, V. Lelaure, et al. 2002. The genome sequence of Schizosaccharomyces pombe. Nature 415:871-880. [DOI] [PubMed] [Google Scholar]
- 33.Xu, H. P., M. White, S. Marcus, and M. Wigler. 1994. Concerted action of RAS and G proteins in the sexual response pathways of Schizosaccharomyces pombe. Mol. Cell. Biol. 14:50-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yen, H.-C. S., and E. C. Chang. 2000. Yin6, a fission yeast Int6 homolog, complexes with Moe1 and plays a role in chromosome segregation. Proc. Natl. Acad. Sci. USA 97:14370-14375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou, K., Y. Wang, J. L. Gorski, N. Nomura, J. Collard, and G. M. Bokoch. 1998. Guanine nucleotide exchange factors regulate specificity of downstream signaling from Rac and Cdc42. J. Biol. Chem. 273:16782-16786. [DOI] [PubMed] [Google Scholar]