Abstract
Two Saccharomyces cerevisiae plasma membrane-spanning proteins, Sho1 and Sln1, function during increased osmolarity to activate a mitogen-activated protein (MAP) kinase cascade. One of these proteins, Sho1, utilizes the MAP kinase kinase kinase Ste11 to activate Pbs2. We previously used the FUS1 gene of the pheromone response pathway as a reporter to monitor cross talk in hog1 mutants. Cross talk requires the Sho1-Ste11 branch of the HOG pathway, but some residual signaling, which is STE11 dependent, still occurs in the absence of Sho1. These observations led us to propose the existence of another osmosensor upstream of Ste11. To identify such an osmosensor, we screened for mutants in which the residual signaling in a hog1 sho1 mutant was further reduced. We identified the MSB2 gene, which encodes a protein with a single membrane-spanning domain and a large presumptive extracellular domain. Assay of the FUS1-lacZ reporter (in a hog1 mutant background) showed that sho1 and msb2 mutations both reduced the expression of the reporter partially and that the hog1 sho1 msb2 mutant was severely defective in the expression of the reporter. The use of DNA microarrays to monitor gene expression revealed that Sho1 and Msb2 regulate identical gene sets in hog1 mutants. A role for MSB2 in HOG1 strains was also seen in strains defective in the two known branches that activate Pbs2: an ssk1 sho1 msb2 strain was more osmosensitive than an ssk1 sho1 MSB2 strain. These observations indicate that Msb2 is partially redundant with the Sho1 osmosensing branch for the activation of Ste11.
Adaptation to high-osmolarity environments is of universal importance to cells. For example, vertebrate kidney medullary cells and free-living microorganisms are constantly exposed to conditions of changing osmolarity. The biochemical mechanisms by which eukaryotic cells sense high extracellular osmolarity are not understood in detail. Exposure of cells to high extracellular osmolarity elicits a common response: accumulation of a compatible solute inside the cell. In eukaryotes from yeasts to humans, hyperosmotic shock activates a conserved mitogen-activated protein kinase (MAPK) cascade (17). Identification of all of the osmosensors in a genome is required to fully understand how cells sense and adapt to conditions of changing osmolarity.
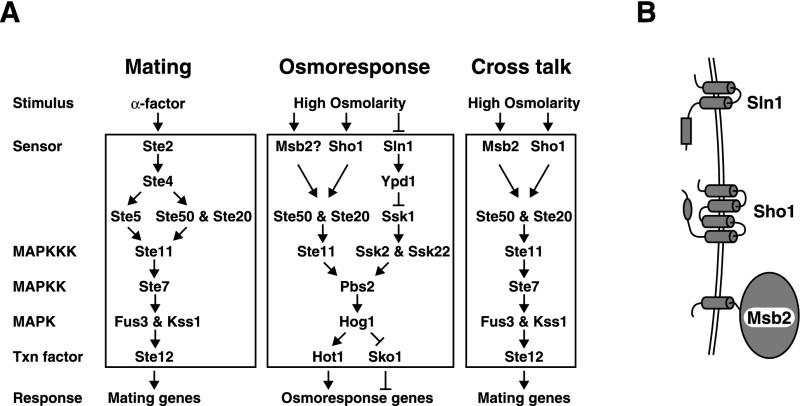
In Saccharomyces cerevisiae, two structurally unrelated membrane-spanning proteins function during conditions of high environmental osmolarity. Each regulates a unique branch of a pathway which converges on the MAPK kinase (MAPKK) Pbs2 (Fig. 1). The osmosensor Sln1 is a protein with two membrane-spanning domains and a cytoplasmic histidine kinase domain, which is homologous to sensor histidine kinases of bacteria, fungi, and plants. Sln1 is constitutively active in media of constant osmotic pressure, where it inhibits the activity of Ssk1 (31). Under conditions of high external osmolarity, Sln1 is inhibited, resulting in the activation of Ssk1 and hence of two redundant MAPKK kinases, Ssk2 and Ssk22 (24, 29), which then activate Pbs2. The second putative high-osmolarity sensor, Sho1, contains four transmembrane domains and an intracellular Src homology 3 (SH3) domain, which binds a proline-rich region of Pbs2 (23). Under conditions of high osmolarity, Sho1 utilizes Ste20 (a p21-activated protein kinase [PAK] homolog) and Ste50 to activate the MAPKK kinase Ste11 (10, 23, 27, 30), which then activates Pbs2. Pbs2 then phosphorylates the MAPK Hog1, resulting in the translocation of Hog1 into the nucleus and the transcriptional induction of a large number of genes, some of which are responsible for the production of glycerol, the compatible solute in yeast cells (1, 4, 12, 33).
FIG. 1.
Mating, osmoresponse, and cross talk pathways. (A) Components of each pathway are displayed. See the text for details. The cross talk pathway operates robustly in response to hyperosmotic shock in hog1 or pbs2 mutants. (B) Diagram of transmembrane proteins involved in the high-osmolarity-sensing pathway in yeast cells.
Prior studies on the HOG pathway identified a convenient and quantitative assay to examine upstream elements of the Sho1-Ste11 branch of the HOG pathway (27). In particular, it was observed that in hog1 or pbs2 mutants, high osmolarity causes the induction of the pheromone response pathway, as measured with a FUS1-lacZ reporter. We have shown that inappropriate activation of the pheromone response pathway (cross talk) utilizes the Sho1-Ste11 branch of the pathway and have identified Ste20 and Ste50 as additional components required for the HOG pathway (and for cross talk) (Fig. 1). Studies also indicate that the Sho1-Ste11 branch of the HOG pathway may provide input to the pseudohyphal growth pathway (21). It was observed that cross talk in hog1 mutants is completely abolished by mutation of STE11 or STE50 but only partially reduced by mutation of SHO1. We reasoned that the remaining cross talk in hog1 sho1 mutants was attributable to a third, unidentified osmosensor, as it was high osmolarity specific.
Here we describe our studies seeking to identify an additional osmosensor that feeds into Ste11. We have carried out a hunt for mutants defective in this osmosensor, which has led to the identification of MSB2 as the presumptive osmosensor. MSB2 was discovered as a multicopy suppressor of a temperature-sensitive mutation in the CDC24 gene (2), but its function has been completely obscure. We have found that Msb2 and Sho1 both provide inputs to the Ste11 branch of the HOG pathway in hog1 mutants and perhaps for the normal osmoresponse in HOG1 cells as well. Our data indicate that MSB2 encodes a weak but physiologically relevant osmosensing component in S. cerevisiae which functions in parallel to the Sho1 osmosensing branch.
MATERIALS AND METHODS
Strains, media, and genetic techniques.
Yeast strains are listed in Table 1. Yeast strains were grown in YEPD medium (1% yeast extract, 2% Bacto Peptone, 2% glucose) at 30°C. Synthetic complete minimal medium lacking nutrients (35) was used for maintaining plasmids and selecting gene replacements. d-Sorbitol and KCl (Sigma) were used at various final concentrations. Yeast transformations were done by the lithium acetate procedure (36). Yeast strains were derived from the EG123 strain background (trp1-1 leu2-3,112 ura3-52 his4 can1) or the W303 strain background (trp1 leu2 ura3 his3 ADE2 can1). Disruptions of S. cerevisiae genes with the Candida glabrata TRP1, LEU2, and HIS3 genes (16) replaced the entire open reading frames (ORFs) of the yeast genes with the C. glabrata genes as follows. The C. glabrata genes were amplified from plasmids with bifunctional oligonucleotide primers: the 46 5′ bases corresponded to sequences directly flanking the S. cerevisiae ORFs to be deleted, and the 20 3′ bases of the primers corresponded to universal plasmid sequences flanking the Candida genes. Ten microliters of the PCR product was used for yeast transformations. Gene disruptions constructed in this manner were confirmed by colony PCRs with gene-specific primers. For constructing the HOG1-GFP::HIS3MX6 fusion gene, we used a GFP::His3MX6 plasmid as described previously (22) and confirmed the integration event at the HOG1 locus by using gene-specific primers for colony PCRs. The HOG1-GFP fusion gene was functional, as assayed by growth on high-osmolarity medium. One HOG1-GFP integrant was crossed to various mutants to generate strains used in some of the experiments (see Fig. 5B).
TABLE 1.
Yeast strains used in this study
| Strain | Relevant genotype | Source or reference |
|---|---|---|
| IH1793 | MATα lys1 | Collection of I. Herskowitz |
| EG123 background | ||
| SO329 | MATaFUS1-lacZ::LEU2 | 27 |
| SO330 | MATahog1::hisG FUS1-lacZ::leu2 | 27 |
| SO1158 | MATahog1::hisG sho1::TRP1 FUS1-lacZ::leu2 | This study |
| SO567 | MATahog1::hisG msb2::LEU2cg FUS1-lacZ::leu2 | This study |
| SO552 | MATahog1::hisG sho1::TRP1 msb2::LEU2cg FUS1-lacZ::leu2 | This study |
| SO331 | MATahog1::hisG ste4::LEU2 FUS1-lacZ::leu2 | 27 |
| SO391 | MATahog1::hisG ste20::TRP1 FUS1-lacZ::LEU2 | 27 |
| SO373 | MATahog1::hisG ste50::hisG::URA3::hisG FUS1-lacZ::leu2 | 27 |
| SO333 | MATahog1::hisG ste11::URA3 FUS1-lacZ::LEU2 | 27 |
| SO1159 | MATahog1::hisG far1::TRP1cg FUS1-lacZ::leu2 | This study |
| SO1160 | MATahog1::hisG sln1::TRP1cg FUS1-lacZ::leu2 | This study |
| SO1161 | MATamsb2::LEU2cg FUS1-lacZ::leu2 | This study |
| W303 background (ADE+) | ||
| SO992 | MATa | Rob Nash and Andrew Murray |
| SO609 | MATapbs2::LEU2 | This study |
| SO1162 | MATahog1::hisG | This study |
| SO1163 | MATahog1::hisG sho1::TRP1 | This study |
| SO1164 | MATahog1::hisG msb2::LEU2cg | This study |
| SO1165 | MATahog1::hisG sho1::TRP1 msb2::LEU2cg | This study |
| SO1004 | MATamsb2::LEU2cg | This study |
| SO1000 | MATasho1::TRP1 | This study |
| SO1016 | MATasho1::TRP1 msb2::LEU2cg | This study |
| SO996 | MATassk1::HIS3cg | This study |
| SO1012 | MATassk1::HIS3cg msb2::LEU2cg | This study |
| SO1008 | MATassk1::HIS3cg sho1::TRP1cg | This study |
| SO1020 | MATassk1::HIS3cg sho1::TRP1cg msb2::LEU2cg | This study |
| SO1128 | MATassk1::HIS3cg ste11::TRP1cg | This study |
| SO579 | MATaHOG1-GFP::HIS3mx | This study |
| SO611 | MATaHOG1-GFP::HIS3mx pbs2::LEU2 | This study |
| SO589 | MATaHOG1-GFP::HIS3mx msb2::LEU2cg | This study |
| SO593 | MATaHOG1-GFP::HIS3mx msb2::LEU2cg sho1::TRP1 | This study |
| SO595 | MATaHOG1-GFP::HIS3mx ssk1::HIS3cg | This study |
| SO597 | MATaHOG1-GFP::HIS3mx ssk1::HIS3 msb2::LEU2cg | This study |
| SO599 | MATaHOG1-GFP::HIS3mx ssk1::HIS3cg sho1::TRP1 | This study |
| SO603 | MATaHOG1-GFP::HIS3mx ssk1::HIS3cg sho1::TRP1 msb2::LEU2cg | This study |
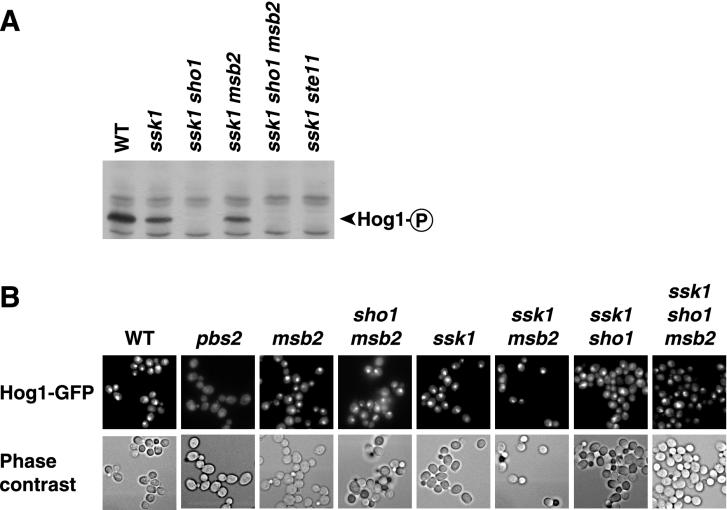
FIG. 5.
Msb2 does not appear to regulate Hog1 activity. (A) Detection of phosphorylated Hog1. Yeast cells were treated with 0.7 M NaCl for 5 min, and protein blots were probed with anti-phospho-p38 antibody as described previously (27). WT, wild type. (B) Hog1-GFP localization after 1.0 M KCl exposure. The indicated strains were treated with high-osmolarity medium for 10 min and assayed as described in Materials and Methods.
Yeast mutagenesis and cloning techniques.
A hog1 sho1 FUS1-lacZ MATa strain was mutagenized with ethyl methane sulfonate (Sigma) to 77% killing and plated on YEPD medium plates at a density of approximately 300 colonies per plate (18). After growth at 30°C for 3 to 7 days, colonies were patched onto YEPD medium plates at a density of 50 per plate. The plates were incubated at 30°C overnight and replica plated to two plates, a no. 3 Whatman filter on a YEPD medium-1 M sorbitol plate and a synthetic complete minimal medium plate spread with a mating tester strain. After growth overnight, β-galactosidase assays were performed with the filters, and the results were compared with those for mating assay plates. Patches that exhibited reduced β-galactosidase activity but still mated well were selected for further analysis. Such strains were purified and transformed separately with a control vector (pRS316) and a pRS316-SHO1 plasmid. Transformants were patched onto plates of synthetic complete minimal medium lacking uracil (SC−ura), replica plated to YEPD medium-1 M sorbitol, and assayed for FUS1-lacZ activity. Mutants in which cross talk (activation of FUS1-lacZ) was restored by the SHO1 plasmid were backcrossed twice to a hog1 sho1 strain, and strains with mutations which segregated as single gene traits were saved as putative osmosensor mutants.
For cloning the gene(s) defective in these mutants, strains were transformed with a yeast genomic library (see below) and plated on SC−ura at a density of approximately 500 to 2,000 transformants/plate. After growth for 3 days, colonies were replica plated to a no. 3 Whatman filter on YEPD medium-1 M sorbitol and incubated overnight at 30°C. Filter β-galactosidase assays were performed to identify lacZ-expressing strains. Such blue colonies were picked directly from the filters, streaked to SC−ura plates, and retested.
Construction of a yeast genomic library.
Yeast genomic DNA was prepared from strain SO329 as described previously (18). This DNA was then treated with Sau3A for various times, and 8- to 12-kb DNA fragments were selected on 0.8% agarose gels. After excision and purification (Qiaexgel extraction kit; Qiagen), the DNA was ligated to BamHI-digested pRS316 (38). Approximately 18,000 independent Escherichia coli transformants were recovered and used to prepare plasmid DNA (Qiagen plasmid kit). Ten of 10 independent transformants contained plasmids with inserts of the expected sizes.
Microscopy techniques.
Hog1-green fluorescent protein (GFP) and cell morphology were visualized by using a Nikon Microphot-SA microscope with a ×60 objective lens and a Princeton Instruments cooled charge-coupled device camera (RTE/CCD-1300-V).
Microarray analysis.
Protocols and material source information were obtained from http://microarrays.org/. DNA microarrays were fabricated at the University of California, San Francisco, by spotting full-length yeast ORFs (derived from PCR products) onto glass microscope slides. ORF DNA microarray analysis, total RNA isolation, and mRNA purification were performed as described previously (8), except that for each RNA sample to be assayed, 4 μg of mRNA was used for cDNA synthesis in the presence of amino-allyl dUTP. cDNA was then labeled with Cy3 or Cy5 (37).
Microarrays were scanned with GenePix 4000A or 4000B microarray scanners (Axon Instruments, Inc., Foster City, Calif.). Genes were assigned to the resulting microarray images with GenePix Pro software. After removal of data from gene spots that were damaged, data were uploaded to the database program AMAD (available at http://microarrays.org/software.html), which also normalized the data over the entire array. Red/green (r/g) expression ratios (the ratio of means) were used if the spot intensity was above 150 U for each channel; this strategy reduced variance due to a weak fluorescence signal. We further normalized the data by adjusting each of 16 sectors of the array to an average ratio of 1.0; this step decreased array position variation in the data set. Ultimately, expression data were manipulated by using FileMaker Pro, Microsoft Excel, Cluster, and TreeView. Cluster and TreeView (11) are available at http://rana.lbl.gov/EisenSoftware.htm.
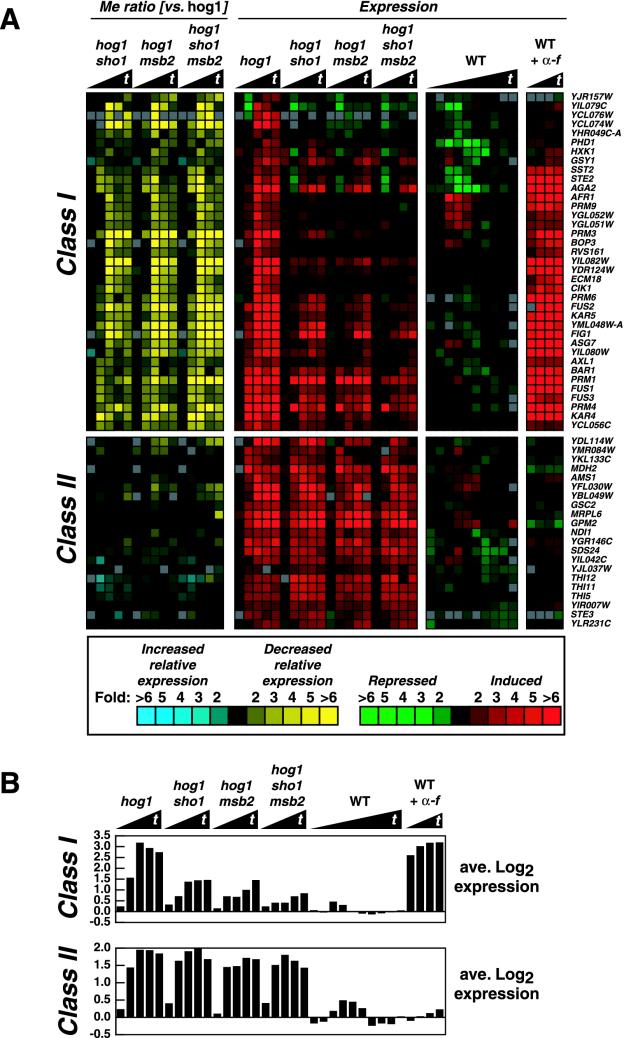
For genes displayed in Fig. 2 and 3, we considered genes which were induced or repressed by a factor of twofold at two or more time points (in the time series of any strain displayed in either figure). We then calculated mutant effect (Me) ratios, which relate the expression of the mRNA in one strain to that in another strain, by dividing the r/g expression ratios of the means (for each time in the time series). The Me ratio is a quotient, with “versus” representing a division sign. For example, in Fig. 3C, the Me ratio of sho1 versus sho1 msb2 strains represents the expression value of the sho1 strain divided by the expression value of the sho1 msb2 strain. For gene expression to be considered defective (compared to wild-type expression) in the various mutant strains, a gene had to have at least two Me ratios which were ≥3 or ≤0.333 in the time series. In Fig. 2, genes were also considered defective if two Me ratios (compared to those of a hog1 strain) were ≥2 or ≤0.5 in the time series. Figure 2A displays a subset of the cross talk genes shown in Fig. ii at our website at http://www.sacs.ucsf.edu/home/HerskowitzLab/MSB2/. Data can also be downloaded for each microarray figure or for the entire genome from our website.
FIG. 2.
Sho1 and Msb2 both control similar sets of genes whose expression is induced by cross talk. The wild-type (WT) strain (SO992) was treated with 1 M sorbitol for 0, 5, 10, 20, 30, 40, 60, 90, 120, and 180 min. In a different experiment, the wild-type strain was treated with α-factor (α-f) for 10, 20, 30, and 40 min. The hog1 mutant strains (SO1162 to SO1165) were treated with 1 M sorbitol for 0, 30, 60, 120, and 180 min. Cy5-labeled cDNA derived from mRNA from each strain and time point was hybridized to a DNA microarray along with Cy3-labeled cDNA derived from mRNA from an untreated wild-type strain. Scanning the microarrays yielded the r/g expression ratios. The black triangles beneath the strain designations denote increasing times of treatment (t). (A) Fifty-eight cross talk genes that exhibited altered regulation in the mutant strains were selected as described in Materials and Methods. The color scale on the bottom of the figure gives the level of relative mRNA abundance (expression: red or green) or the fold difference between strains (Me ratio: blue or yellow). The expression values were clustered along with the Me ratio values, and class I and class II genes are explained in the text. (B) The average r/g expression ratios (in log2 scale) for the genes in each class are displayed.
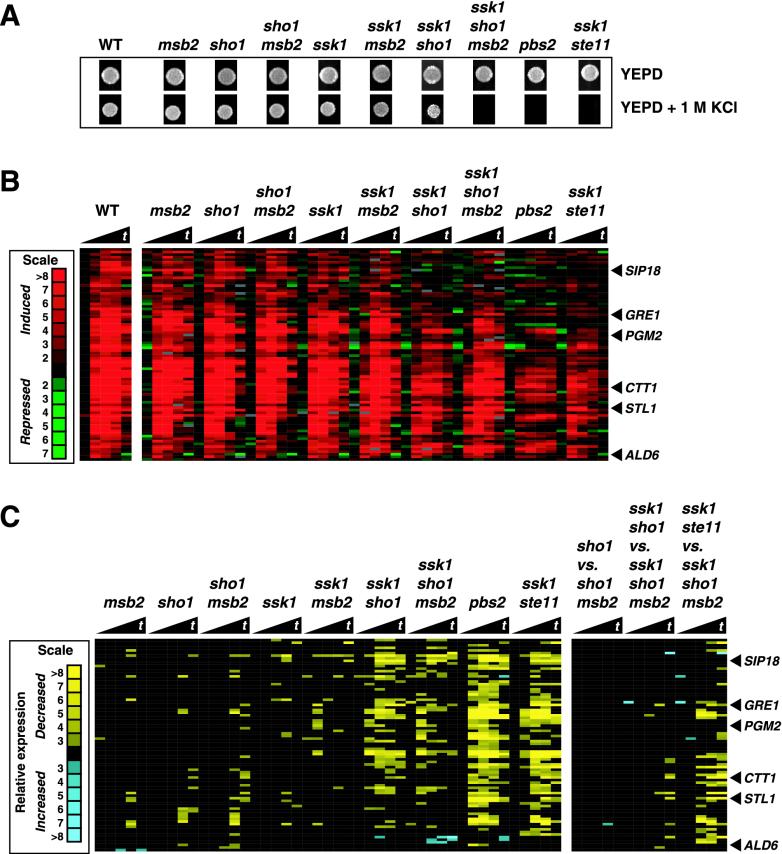
FIG. 3.
Msb2 is redundant with Sho1 in the Ste11-dependent branch of the high-osmolarity response pathway, as monitored by growth on high-osmolarity medium, but not for gene expression. Wild-type (WT) (SO992), msb2 (SO1004), sho1 (SO1000), sho1 msb2 (SO1016), ssk1 (SO996), ssk1 msb2 (SO1012), ssk1 sho1 (SO1008), ssk1 sho1 msb2 (SO1020), pbs2 (SO609), and ssk1 ste11 (SO1128) strains were tested for osmotolerance and gene expression. (A) Growth on YEPD medium (2 days at 30°C) and YEPD medium-1 M KCl (3 days at 30°C) plates. Equivalent numbers of cells from liquid cultures were spotted onto the indicated plates. (B) Expression of high-osmolarity-induced genes whose RNA levels were altered in any of the strains tested were clustered and are displayed. Red indicates increased mRNA abundance. Strains from panel A were treated with 0.5 M KCl for 0, 10, 20, 30, and 40 min prior to harvesting of the yeast cells for RNA preparation. The black triangles beneath the strain designations denote increasing times of treatment (t). (C) The Me ratios of the genes from panel B are displayed. The nine comparisons in the left panel were made by dividing the r/g expression values of the mutants by the wild-type r/g expression values. The three comparisons in the right panel are mutant-to-mutant Me ratios. Yellow indicates decreased relative mRNA expression.
RESULTS
Identification of a third putative osmosensor, Msb2.
To identify a hypothesized third osmosensor, we mutagenized a hog1 sho1 FUS1-lacZ strain with ethyl methanesulfonate and screened for mutants which displayed reduced levels of FUS1-lacZ activity in the presence of 1 M sorbitol. Because we expected to find numerous mutants defective in the pheromone response pathway (for example, affecting the STE11, STE7, and STE12 genes), we tested mutants with reduced FUS1-lacZ activity for mating competence. Of 2,600 mutagenized colonies screened, 52 strains were found that could mate and had reduced cross talk. If the mutations affected a new osmosensor and not downstream signaling components or the reporter gene, then the restoration of SHO1 function to the strain should restore FUS1-lacZ activity. We thus introduced a low-copy-number SHO1 plasmid into all of our remaining mutants and assayed for the recovery of cross talk. Thirty-seven mutants displayed increased cross talk after the introduction of an SHO1 plasmid but not of a control plasmid. Mutants were crossed to a hog1 sho1 strain twice to confirm single-gene segregation. These procedures yielded 21 strains with single-gene traits that were candidates for having a mutation affecting a third osmosensor gene.
We constructed a yeast genomic library in the low-copy-number plasmid pRS316 and isolated plasmids which restored expression to the FUS1-lacZ reporter. We attempted to clone the mutant gene(s) for 6 of the strains which displayed relatively little residual cross talk, and the remaining 15 strains were not studied further. Only SHO1- and FUS1-lacZ-containing plasmids were isolated for three of the mutants (both types of plasmids give an increased LacZ signal for trivial reasons). One of the remaining mutants was complemented by a STE7 plasmid and another mutant was complemented by a STE20 plasmid, indicating that these mutants may have reduced-function alleles in STE7 and STE20. These putative ste7 and ste20 strains were not studied further. The remaining mutant was complemented by a library plasmid containing the MSB2 gene and by a plasmid (3) that carries only the MSB2 gene. The mutation in this strain was allelic to an MSB2 deletion allele (as determined by linkage analysis), indicating that it contains a mutation at or near the MSB2 locus. Taken together, these data indicate that MSB2 is required for cross talk signaling in hog1 sho1 mutants.
Quantitative analysis of the role of MSB2 in cross talk signaling.
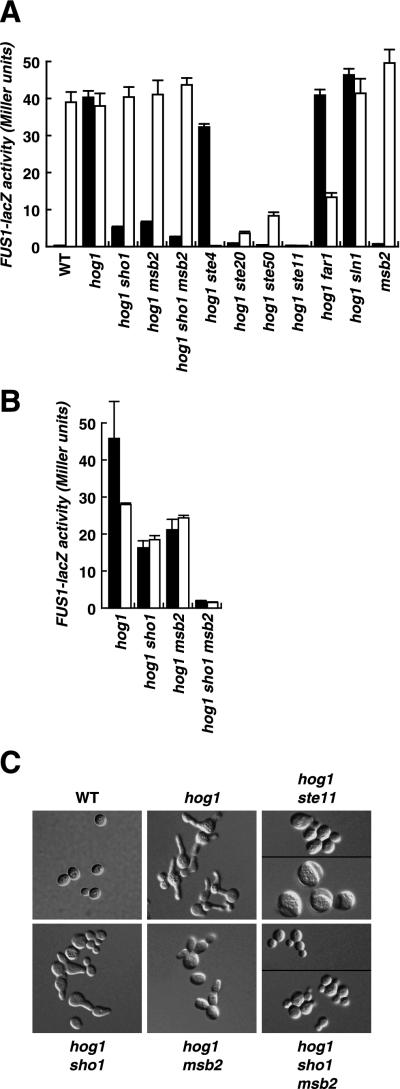
To characterize the role of MSB2 in signaling, we measured cross talk to FUS1-lacZ. As shown in Fig. 4A, a hog1 strain exhibits substantial activation of FUS1-lacZ in response to 1 M sorbitol (40 U of β-galactosidase activity after 5 h of induction). As observed previously (27), inactivation of STE20, STE50, or STE11 completely abolished cross talk in the hog1 strain (less than 1.0 U of activity). Mutation of SLN1 or STE4 still allowed robust cross talk, indicating that the osmosensor Sln1 and the pheromone receptor upstream of Ste4 (Ste2) are not responsible for providing inputs to this response pathway. In contrast, inactivation of SHO1 or MSB2 in the hog1 strain resulted in a marked reduction of cross talk signaling (Fig. 4A, black bars). Moreover, a hog1 sho1 msb2 strain exhibited even less cross talk (2.7 U) than the hog1 sho1 (5.2 U) or hog1 msb2 (6.4 U) strain at 5 h of induction with 1 M sorbitol. These results are consistent with the idea that Sho1 and Msb2 each independently provide input to Ste20 and Ste50. The remaining cross talk in the hog1 sho1 msb2 strain is apparently due to the activity of at least one additional osmosensor which stimulates Ste11. We tested this set of strains for the induction of FUS1-lacZ by α-factor as well (Fig. 4A, white bars). This analysis demonstrated that the msb2 and sho1 mutations did not affect signaling through the pheromone response pathway initiated from the α-factor receptor.
FIG. 4.
Msb2 promotes cross talk in hog1 mutants. (A) MSB2, SHO1, and a subset of pheromone response pathway genes are required for cross talk to activate FUS1-lacZ. Strains were grown to exponential phase in liquid YEPD medium, shifted to YEPD medium-1 M sorbitol (black bars) or YEPD medium-α-factor (white bars) and grown for 5 h, and harvested for quantitative β-galactosidase assays as described previously (27). Strains were wild type (WT) (SO329), hog1 (SO330), hog1 sho1 (SO1158), hog1 msb2 (SO567), hog1 sho1 msb2 (SO552), hog1 ste4 (SO331), hog1 ste20 (SO391), hog1 ste50 (SO73), hog1 ste11 (SO333), hog1 far1 (SO1159), hog1 sln1 (SO1160), and msb2 (SO1161). (B) Mutation of both SHO1 and MSB2 is required to completely block cross talk induction of FUS1-lacZ during long incubation in YEPD medium-1 M sorbitol. Strains were treated with 1 M sorbitol for 18 h (black bars) or 24 h (white bars), and β-galactosidase activity was assayed. Error bars in panels A and B indicate standard deviations. (C) Dependence of cross talk (as monitored by induction of a shmoo-like morphology) on Sho1 and Msb2. Strains from panel A were grown in liquid YEPD medium-1 M sorbitol for 24 h and assayed by differential interference contrast microscopy.
Analysis of FUS1-lacZ activity after longer periods of induction (18 and 24 h) further demonstrated that both SHO1 and MSB2 contribute to the activation of the cross talk pathway (Fig. 4B and Fig. i at our website). At 18 h after induction, the hog1 sho1 and hog1 msb2 strains exhibited 35 and 46%, respectively, the induction observed for the hog1 SHO1 MSB2 strain. Strikingly, the inactivation of both SHO1 and MSB2 in the hog1 strain caused a reduction in signaling to only 4% the level in the hog1 SHO1 MSB2 strain. Assaying the induction of FUS1-lacZ at 24 h yielded similar conclusions: inactivation of SHO1 or MSB2 led to 66 or 87% signaling activity, respectively, whereas inactivation of both SHO1 and MSB2 led to only 5% signaling compared to that seen with the hog1 SHO1 MSB2 strain. These observations show that Msb2 and Sho1 play partially redundant roles in the signaling to FUS1-lacZ that is observed in this Hog1-deficient strain background.
Msb2 promotes cell elongation in hog1 mutants.
During mating, yeast cells polarize their cytoskeleton and grow toward the mating partner to form a pear shape or shmoo. In addition to activating the FUS1-lacZ reporter, cross talk in hog1 mutants also induces cells to exhibit a shmoo-like morphology (Fig. 4C) (27). This morphology is largely dependent on Sho1 but not on upstream elements of the pheromone response pathway, such as Ste4 (27). However, some residual cell elongation remains in hog1 sho1 mutants, especially after long incubation in the presence of 1 M sorbitol (24 h) (Fig. 4C). We hypothesized that MSB2 might also contribute to this response and thus examined the effect of the deletion of MSB2 on cell elongation. Indeed, mutation of MSB2 reduced cell elongation and did so to a greater extent than mutation of SHO1 (Fig. 4C), although some cells still formed shmoo-like structures. Strikingly, a hog1 strain deficient in both SHO1 and MSB2 exhibited little, if any, morphological change in response to high osmolarity. In fact, the morphology of hog1 sho1 msb2 cells was similar to that of hog1 ste11 cells, which exhibit no cross talk, with the exception that the hog1 ste11 strain frequently produced abnormally large cells. These observations on high-osmolarity-induced morphological changes reinforce the prior conclusion that Msb2 and Sho1 are partially redundant for providing inputs to Ste11 in a Hog1-deficient strain background.
Msb2 and Sho1 control the expression of a subset of genes in hog1 mutants.
To learn more about the role of Msb2 in the cross talk pathway of signal transduction, we monitored gene expression with DNA microarrays. We sought genes with expression patterns that differed between wild-type and hog1 strains or between hog1 and hog1 sho1, hog1 msb2, or hog1 sho1 msb2 strains. We examined RNA levels over a relatively long time period, after 0, 30, 60, 120, and 180 min of 1 M sorbitol treatment, and with additional intermediate time points for the wild type. To focus on genes with altered regulation in a multiple-mutant strain relative to the regulation in a hog1 strain, we calculated Me ratios (see Materials and Methods), which relate the RNA level in one strain to that in another. There were 462 genes in six major classes with altered expression in hog1 mutants compared to the wild-type or between hog1 mutants (Fig. ii at our website). The different patterns of gene expression in hog1 mutants will be more fully described elsewhere (S. M. O'Rourke and I. Herskowitz, unpublished data). However, two classes included cross talk-induced genes (genes induced in hog1 mutants but not in the wild type) in response to increased osmolarity. Class I was dependent on Sho1 and Msb2 for induction, while class II was not, as revealed by mutation of SHO1 and MSB2. Figure 2A displays the response profiles for 58 class I and class II genes. On average, class I genes were induced 9-fold in hog1 mutants and class II genes were induced 3.8-fold in hog1 mutants after 1 h of sorbitol treatment (Fig. 2B). The average expression of class I and class II genes in the wild type was near basal levels after 1 h (0.92- and 0.85-fold, respectively).
The induction of class I genes was reduced when SHO1, MSB2, or SHO1 and MSB2 were deleted. Indeed, this class contains the native FUS1 gene, which displays regulation similar to that seen with the FUS1-lacZ reporter gene, as described for Fig. 4A. Strikingly, all of the class I genes were also induced by α-factor. Genes involved in pheromone reception and signaling (STE2, SST2, BAR1, and FUS3) as well as genes involved in cell-cell adherence, cell fusion, and nuclear fusion (AGA2, FUS2, PRM1, KAR4, and KAR5) are likewise found in class I. The effect of a SHO1 mutation on these genes is clear, but significant residual induction is still present in a hog1 sho1 strain (Fig. 2B). For example, the average induction of the class I genes at 180 min in a hog1 strain is 6.6-fold, but in a hog1 sho1 strain, the induction is 2.7-fold. Mutation of MSB2 has a similar effect at 180 min: the average induction in a hog1 msb2 strain is 2.7-fold. Finally, a hog1 sho1 msb2 strain induces this set of genes only 1.8-fold. Thus, the induction of a variety of α-factor-induced cross talk genes is similar to the results obtained for the FUS1-lacZ reporter gene: Msb2 and Sho1 provide partially redundant inputs to the cross talk pathway. An interesting observation is that mutation of SHO1 or MSB2 altered the expression of identical gene sets. This observation suggests that these two transmembrane proteins indeed activate the same pathway.
Despite the fact that class II genes are cross talk induced, they do not depend on SHO1 or MSB2 for their induction (Fig. 2A, Me ratios). These genes show an average induction of three- to fourfold in any strain where hog1 is deleted (Fig. 2B). In addition, they are not generally α-factor inducible. Many of these genes code for enzymes involved in metabolic processes. Two are involved in cell wall metabolism (AMS1 and GSC2), three are involved in vitamin B1 and pyrimidine biosynthesis (THI5, THI11, and THI12), and several function in energy production (GPM2, NDI1, and MDH2). Many of these genes are induced during nitrogen starvation and stationary phase (13). It is possible that these genes are induced in response to secondary stress damage due to a deficient osmotic response or that a different cross talk circuit is activated which is unrelated to the pheromone response pathway. At any rate, these genes clearly form an SHO1- and MSB2-insensitive class of cross talk genes.
Msb2 controls osmosensitivity in HOG1 strains.
The studies above indicate that Msb2 functions in parallel to Sho1 in a signaling pathway that is operative in a hog1 mutant background, the cross talk pathway (Fig. 1). To determine whether Msb2 functions in a HOG1 strain, we examined the contribution of MSB2 to growth under conditions of high osmolarity. It was shown previously that inactivation of the two inputs to Pbs2 and Hog1 by mutations in both SSK1 and STE11 leads to osmosensitivity, whereas mutation in either SSK1 or STE11 alone does not (23, 30) (Fig. 3A). Strains defective in both SSK1 and SHO1 also exhibit greater osmosensitivity than strains defective in only SSK1, but an ssk1 sho1 strain is not as osmosensitive as an ssk1 ste11 strain (Fig. 3A). We therefore tested whether MSB2 contributes to the osmoresistant phenotype of the ssk1 sho1 strain by examining the properties of an ssk1 sho1 msb2 strain; the triply defective strain is more osmosensitive than the ssk1 sho1 MSB2 strain and is almost as sensitive to high osmolarity as the ssk1 ste11 strain (Fig. 3A) (unpublished data). We interpret these observations to indicate that both Sho1 and Msb2 may activate Ste11 and consequently Hog1 and thus promote the osmoresistance of wild-type strains. However, the ssk1 msb2 strain is not osmosensitive (unlike the ssk1 sho1 strain) (Fig. 3A), suggesting that Msb2 provides a relatively smaller contribution to osmotolerance and presumably to Ste11 activity.
The contribution of Msb2 to resistance to high osmolarity in HOG1 strains suggests that Msb2 activates Hog1. Prior studies showed that Hog1 is rapidly phosphorylated by Pbs2 after a shift to high-osmolarity medium (4) and that Hog1 protein rapidly translocates to the nucleus after exposure of cells to high osmolarity (12, 33). To investigate whether Msb2 functions in osmoregulation by activating Hog1, we examined whether Msb2 governs the phosphorylation and nuclear localization of the Hog1 protein. In one set of experiments, we assayed tyrosine phosphorylation of Hog1 in wild-type, ssk1, ssk1 sho1, ssk1 msb2, and ssk1 sho1 msb2 strains (Fig. 5A). Although Hog1 was phosphorylated in wild-type, ssk1, and ssk1 msb2 strains in response to a variety of high-osmolarity conditions, we could not detect phosphorylation of Hog1 in ssk1 sho1 and ssk1 sho1 msb2 strains. In another set of experiments, we used a Hog1-GFP fusion (shown to have Hog1 activity; see Materials and Methods) to monitor Hog1 localization. As reported previously, the Hog1 protein localized to the nucleus in response to increased osmolarity in the wild-type strain. Some Hog1 was localized to the nucleus in the ssk1 sho1 strain, and similar localization was seen in the ssk1 sho1 msb2 strain (Fig. 5B). These observations indicate that the activity of Msb2 for stimulating the bona fide osmotic response pathway may be too weak to assay by Hog1 phosphorylation or nuclear localization (under the conditions that we tested). Alternatively, Msb2 may stimulate tolerance to high osmolarity in a manner that does not depend on Hog1.
A search for genes regulated by MSB2 in HOG1 strains.
In Fig. 2 we identified genes whose expression was regulated by MSB2 in hog1 strains. These were a subset of genes in the pheromone response pathway that were induced by the cross talk pathway. We next sought to determine whether Msb2 controls the expression of any osmoregulated genes in a HOG1 strain. We carried out two sets of experiments. The first set compares genes induced by 0.5 M KCl in strains in which the SLN1-SSK1 branch is operative; the second set compares genes induced by 0.5 M KCl in strains in which SSK1 is inactive. In the SSK1 set, we compared the wild type with msb2, sho1, and msb2 sho1 mutants (Fig. 3B). The Me plots in Fig. 3C show that the inactivation of these genes had little effect on the expression of 82 genes whose expression is induced by the HOG pathway. We have noted that a significant induction of HOG pathway-dependent genes still occurs in mutants which block the HOG pathway (O'Rourke and Herskowitz, unpublished). Furthermore, an Me plot comparing sho1 versus sho1 msb2 strains demonstrated that Msb2 plays little, if any, role in the sho1 strain (Fig. 3C, right panel). In the analysis of ssk1 strains, we analyzed ssk1, ssk1 msb2, ssk1 sho1, and ssk1 msb2 sho1 mutants (Fig. 3B). We observed a set of genes whose expression was dependent on SHO1 (that is, whose expression decreased in the ssk1 sho1 strain). However, mutation of MSB2 had little effect on the expression of these genes in either an ssk1 or an ssk1 sho1 background. A direct comparison of ssk1 sho1 with ssk1 sho1 msb2 demonstrated that the patterns were essentially identical, indicating that Msb2 did not contribute to the expression of any of these genes (Fig. 3C, right panel).
We have argued that both Sho1 and Msb2 provide inputs to Ste11. If they provide the only inputs to Ste11, then the ssk1 sho1 msb2 strain should exhibit a pattern similar to that of the ssk1 ste11 strain. A comparison of these two strains reveals that there are several genes whose RNA levels are reduced more in the ste11 ssk1 strain than in the ssk1 sho1 msb2 strain (Fig. 3B and C, right panel). This observation suggests that Ste11 has osmoregulatory inputs in addition to Sho1 and Msb2.
DISCUSSION
Earlier studies on the response to high osmolarity demonstrated that two branches provide inputs to the MEK and MAPK components of the Hog1 MAPK cascade. These branches are characterized by two integral membrane proteins, Sho1 and Sln1, which are presumed to be the osmosensors for these two branches. These two proteins are structurally different (having different sequences and numbers of membrane-spanning regions) and have great functional differences (Fig. 1B). In particular, Sho1 utilizes the PAK-like kinase Ste20, whereas Sln1 is part of a phosphorelay system. Prior studies focused on the Sho1-Ste20-Ste11 branch of the pathway under conditions in which signaling to the pheromone response pathway MAPK cascade (cross talk) was enhanced. We observed in hog1 and pbs2 mutants that FUS1-lacZ expression was completely dependent on STE11 but only partially dependent on SHO1, a finding which led us to propose the existence of an additional input to Ste11. We carried out a mutant hunt to identify components responsible for this residual cross talk and identified Msb2, a protein which has one presumptive membrane-spanning region and which we propose to be a third osmosensor. Physiological studies indicated that Msb2 functions in the cross talk pathway (that is, in the absence of Hog1) in a partially redundant manner with Sho1 to provide inputs to Ste20 and Ste11. The identification of Msb2 as a component of a third osmosensing branch may also explain the observation that ssk1 ste11 HOG1 strains are more osmosensitive than ssk1 sho1 HOG1 strains. This observation suggests that Msb2 is functionally redundant with Sho1 for activating Hog1 to promote osmotolerance. Enigmatically, we cannot find any other evidence that Msb2 activates Hog1 in HOG1 cells. The discovery of Msb2 raises a variety of questions, including the mechanism by which it may sense osmolarity and its possible role in providing input to the pseudohyphal and invasive growth pathways.
Msb2 and Sho1 provide partially redundant inputs to Ste20-Ste50-Ste11.
Several observations indicate that Msb2 and Sho1 are partially redundant and provide inputs to Ste11. Mutation of either MSB2 or SHO1 reduces the osmotically induced expression of FUS1-lacZ and other genes (in class I) (Fig. 2A) and the induction of the shmoo-like morphology in hog1 mutants; inactivation of both virtually eliminates FUS1-lacZ expression and the morphological response. Msb2 contributes to the residual tolerance of high osmolarity of an ssk1 sho1 strain (Fig. 3A). Thus, deletion of both SHO1 and MSB2 results in a phenotype like that resulting from an STE11 deletion for both cross talk signaling and osmoresistance. An interesting feature of Sho1 and Msb2 in signal transduction is that mutations in either gene reduce cross talk signaling (Fig. 4A and B). However, mutations in both genes together reduce cross talk even further. These results are puzzling but may indicate that Sho1 and Msb2 have a partially interdependent relationship in responding to increased osmolarity. For example, the Sho1 and Msb2 proteins may interact with each other for maximal activity. Alternatively, these two proteins may constitute a linear pathway with (as yet) unidentified and redundant components at both the Msb2 and the Sho1 steps in the pathway. Although we propose that Msb2 is an osmosensor, we cannot exclude the possibility that it is required for the activity, localization, or trafficking of another protein that is the true osmosensor. In fact, biochemical evidence for Sln1 or Sho1 directly sensing increased osmolarity is also lacking.
Structure of Msb2 and possible functional features.
The structure of Msb2 differs considerably from those of Sho1 and Sln1 (Fig. 1B). It is a putative integral membrane protein with a single putative membrane-spanning domain (positions 1186 to 1208) and an overall serine-threonine content of 43%. It is proposed to have a large extracellular domain of 1,166 amino acids and a short intracellular tail of 98 amino acids but to lack any clear functional domains. In contrast, Sho1 has four proposed transmembrane segments and an intracellular SH3 domain which is used to bind downstream signaling proteins, notably Pbs2; Sln1 has two proposed transmembrane segments and a histidine kinase domain which is used for the regulation of Ypd1 and Ssk1. Msb2 functions in the Ste11 branch of the HOG pathway and thus should contain binding sites for some or all of Ste20, Ste11, and Ste50 and perhaps for Pbs2 as well. Msb2 lacks similarity to Sho1 and also lacks a recognizable SH3 domain, by which Sho1 associates with Pbs2 (23). One notable feature of the Msb2 protein is that its presumed extracellular domain contains seven 17-amino-acid repeated sequences, which are serine and threonine rich. The yeast protein with the greatest similarity to Msb2 (26.7% identity over its entire length) is Hkr1, which is also a type I membrane protein and has 1,802 amino acids. Hkr1 has a transmembrane domain at positions 1486 to 1506; its large presumptive extracellular domain also contains 13 repeats of a 26-amino-acid serine- and threonine-rich sequence. Hkr1 is a cell surface protein whose overexpression confers resistance to the killer toxin of Hansenula mrakii (15) and controls β-glucan synthesis and the axial budding pattern (39). Hkr1 and Msb2 thus may have similar localizations. Whether Hkr1 plays a role in osmoregulation remains to be determined. We suggest that Msb2 may link the plasma membrane and the cell wall through an interaction of the large extracellular domain, possibly the repeat sequences, with components of the cell wall. Such a linkage could be used to monitor mechanical stress between the plasma membrane and the cell wall.
MSB2 was discovered because its overexpression partially suppresses a cdc24-ts mutation in the presence of 1 M sorbitol (2, 3), but its function has otherwise been obscure. Cdc24 is a guanine nucleotide exchange factor for the small G protein Cdc42, which plays a role in the activation of Hog1 and Pbs2 through the Sho1 branch of the HOG pathway (32, 34). In particular, it is thought that Cdc42 regulates the PAK-like kinase Ste20, which subsequently activates Ste11. Moreover, Cdc42 is partially responsible for recruiting Pbs2 to sites of polarized growth during osmotic stress (34). Cdc24 is known to have a variety of binding partners, including Bud1/Rsr1 and Far1 (5, 25, 26, 28), which link the localization of Cdc24 to internal and external polarity signals. If Msb2 also regulates Cdc24, an osmolarity-sensitive input to the Cdc24-Cdc42 module would be created. Interestingly, βPix (a human guanine nucleotide exchange factor which regulates Cdc42) activates the p38 MAPK cascade via PAK1 (19), and the activities of both Cdc42 and PAK1 are stimulated by increased osmolarity (6, 20). Another potential link between Msb2 and elements of the HOG pathway is the recent finding (9) that Msb2 interacts in a two-hybrid assay with Cla4, a PAK-like kinase that is related to Ste20 and that also functions in the HOG pathway (32). Thus, Msb2 can potentially provide input to upstream elements of the HOG pathway via multiple mechanisms.
Role of Msb2 in HOG1 strains.
Msb2 plays a clear role in regulating gene expression through the cross talk pathway, that is, in a hog1 background. We have also observed that MSB2 contributes to osmoresistance in a HOG1 background: ssk1 sho1 MSB2 strains are more tolerant to high osmolarity than are ssk1 sho1 msb2 strains. The simplest interpretation of this result is that either Msb2 or Sho1 can signal to Pbs2 and Hog1. Our attempts to observe the regulation of Hog1 directly, by tyrosine phosphorylation or by nuclear localization of Hog1 protein, however, yielded no positive results. In addition, HOG1-dependent gene expression was not influenced by mutation of MSB2. We can propose two different explanations for this behavior of MSB2. Perhaps the assays used to detect Msb2 function in a HOG1 strain were not sufficiently sensitive. Another possibility is that Msb2 does not stimulate Hog1 activity. According to this explanation, Msb2 activates Ste11 in a way that does not lead to Pbs2 activation but that does activate Ste7 (note that Msb2 lacks an SH3 domain for binding to Pbs2). Thus, Msb2 may play a role in the weak cross talk activation of the pheromone response pathway that occurs in HOG1 cells (14) or in the activation of the pseudohyphal growth pathway. Why then would ssk1 sho1 msb2 strains be more sensitive to high osmolarity than ssk1 sho1 MSB2 strains? We suggest that perhaps weak expression of genes in the pheromone response pathway and/or the pseudohyphal growth pathway under the control of Msb2 contributes to tolerance to high osmolarity. Indeed, Cullen et al. found that the Sho1 protein signals to pheromone response pathway components for the monitoring of cell wall conditions (7). Thus, Msb2 may also utilize the pheromone response pathway for physiologically relevant processes.
Acknowledgments
We thank Joe Horecka, Linda Huang, Kenji Irie, Doug Jeffery, members of the laboratory of Wendell Lim, and Peter Pryciak for insightful discussions and Joseph DeRisi, Sang-Hyun Park, and Brian Pulliam for computer assistance. We also thank Holly Bennett and Joseph DeRisi for excellent training on the use of DNA microarrays and data analysis and Erin O'Shea for help in the preparation of the manuscript. We are grateful to John Pringle and Alan Bender for providing MSB2 plasmids.
This work was supported by National Institutes of Health (NIH) grant GM59466 (to I.H.). S.M.O. was supported by an NIH training grant, the Markey Program in Biological Sciences, the Herbert W. Boyer Fund, and a UCSF Chancellor's Fellowship.
REFERENCES
- 1.Albertyn, J., S. Hohmann, J. M. Thevelein, and B. A. Prior. 1994. GPD1, which encodes glycerol-3-phosphate dehydrogenase, is essential for growth under osmotic stress in Saccharomyces cerevisiae, and its expression is regulated by the high-osmolarity glycerol response pathway. Mol. Cell. Biol. 14:4135-4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bender, A., and J. R. Pringle. 1989. Multicopy suppression of the cdc24 budding defect in yeast by CDC42 and three newly identified genes including the ras-related gene RSR1. Proc. Natl. Acad. Sci. USA 86:9976-9980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bender, A., and J. R. Pringle. 1992. A Ser/Thr-rich multicopy suppressor of a cdc24 bud emergence defect. Yeast 8:315-323. [DOI] [PubMed] [Google Scholar]
- 4.Brewster, J. L., T. de Valoir, N. D. Dwyer, E. Winter, and M. C. Gustin. 1993. An osmosensing signal transduction pathway in yeast. Science 259:1760-1763. [DOI] [PubMed] [Google Scholar]
- 5.Butty, A. C., P. M. Pryciak, L. S. Huang, I. Herskowitz, and M. Peter. 1998. The role of Far1p in linking the heterotrimeric G protein to polarity establishment proteins during yeast mating. Science 282:1511-1516. [DOI] [PubMed] [Google Scholar]
- 6.Clerk, A., and P. H. Sugden. 1997. Activation of p21-activated protein kinase α (α PAK) by hyperosmotic shock in neonatal ventricular myocytes. FEBS Lett. 403:23-25. [DOI] [PubMed] [Google Scholar]
- 7.Cullen, P. J., J. Schultz, J. Horecka, B. J. Stevenson, Y. Jigami, and G. F. Sprague, Jr. 2000. Defects in protein glycosylation cause SHO1-dependent activation of a STE12 signaling pathway in yeast. Genetics 155:1005-1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeRisi, J. L., V. R. Iyer, and P. O. Brown. 1997. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278:680-686. [DOI] [PubMed] [Google Scholar]
- 9.Drees, B. L., B. Sundin, E. Brazeau, J. P. Caviston, G. C. Chen, W. Guo, K. G. Kozminski, M. W. Lau, J. J. Moskow, A. Tong, L. R. Schenkman, A. McKenzie, I. I. I., P. Brennwald, M. Longtine, E. Bi, C. Chan, P. Novick, C. Boone, J. R. Pringle, T. N. Davis, S. Fields, and D. G. Drubin. 2001. A protein interaction map for cell polarity development. J. Cell Biol. 154:549-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Drogen, F., S. M. O'Rourke, V. M. Stucke, M. Jaquenoud, A. M. Neiman, and M. Peter. 2000. Phosphorylation of the MEKK Ste11p by the PAK-like kinase Ste20p is required for MAP kinase signaling in vivo. Curr. Biol. 10:630-639. [DOI] [PubMed] [Google Scholar]
- 11.Eisen, M. B., P. T. Spellman, P. O. Brown, and D. Botstein. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95:14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferrigno, P., F. Posas, D. Koepp, H. Saito, and P. A. Silver. 1998. Regulated nucleo/cytoplasmic exchange of HOG1 MAPK requires the importin beta homologs NMD5 and XPO1. EMBO J. 17:5606-5614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gasch, A. P., P. T. Spellman, C. M. Kao, O. Carmel-Harel, M. B. Eisen, G. Storz, D. Botstein, and P. O. Brown. 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11:4241-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hall, J. P., V. Cherkasova, E. Elion, M. C. Gustin, and E. Winter. 1996. The osmoregulatory pathway represses mating pathway activity in Saccharomyces cerevisiae: isolation of a FUS3 mutant that is insensitive to the repression mechanism. Mol. Cell. Biol. 16:6715-6723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kasahara, S., H. Yamada, T. Mio, Y. Shiratori, C. Miyamoto, T. Yabe, T. Nakajima, E. Ichishima, and Y. Furuichi. 1994. Cloning of the Saccharomyces cerevisiae gene whose overexpression overcomes the effects of HM-1 killer toxin, which inhibits β-glucan synthesis. J. Bacteriol. 176:1488-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kitada, K., E. Yamaguchi, and M. Arisawa. 1995. Cloning of the Candida glabrata TRP1 and HIS3 genes, and construction of their disruptant strains by sequential integrative transformation. Gene 165:203-206. [DOI] [PubMed] [Google Scholar]
- 17.Kultz, D., and M. Burg. 1998. Evolution of osmotic stress signaling via MAP kinase cascades. J. Exp. Biol. 201:3015-3021. [DOI] [PubMed] [Google Scholar]
- 18.Lawrence, C. W. 1991. Classical mutagenesis techniques, p. 273-281. In C. Guthrie and G. R. Fink (ed.), Guide to yeast genetics and molecular biology, vol. 194. Academic Press, Inc., San Diego, Calif. [Google Scholar]
- 19.Lee, S. H., M. Eom, S. J. Lee, S. Kim, H. J. Park, and D. Park. 2001. βPix-enhanced p38 activation by Cdc42/Rac/PAK/MKK3/6-mediated pathway. Implication in the regulation of membrane ruffling. J. Biol. Chem. 276:25066-25072. [DOI] [PubMed] [Google Scholar]
- 20.Lewis, A., C. Di Ciano, O. D. Rotstein, and A. Kapus. 2002. Osmotic stress activates Rac and Cdc42 in neutrophils: role in hypertonicity-induced actin polymerization. Am. J. Physiol. Cell Physiol. 282:C271-C279. [DOI] [PubMed] [Google Scholar]
- 21.Liu, H., C. A. Styles, and G. R. Fink. 1993. Elements of the yeast pheromone response pathway required for filamentous growth of diploids. Science 262:1741-1744. [DOI] [PubMed] [Google Scholar]
- 22.Longtine, M. S., A. McKenzie, I. I. I., D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 23.Maeda, T., M. Takekawa, and H. Saito. 1995. Activation of yeast PBS2 MAPKK by MAPKKKs or by binding of an SH3-containing osmosensor. Science 269:554-558. [DOI] [PubMed] [Google Scholar]
- 24.Maeda, T., S. M. Wurgler-Murphy, and H. Saito. 1994. A two-component system that regulates an osmosensing MAP kinase cascade in yeast. Nature 369:242-245. [DOI] [PubMed] [Google Scholar]
- 25.Nern, A., and R. A. Arkowitz. 1999. A Cdc24p-Far1p-Gβγ protein complex required for yeast orientation during mating. J. Cell Biol. 144:1187-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nern, A., and R. A. Arkowitz. 1998. A GTP-exchange factor required for cell orientation. Nature 391:195-198. [DOI] [PubMed] [Google Scholar]
- 27.O'Rourke, S. M., and I. Herskowitz. 1998. The Hog1 MAPK prevents cross talk between the HOG and pheromone response MAPK pathways in Saccharomyces cerevisiae. Genes Dev. 12:2874-2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park, H. O., E. Bi, J. R. Pringle, and I. Herskowitz. 1997. Two active states of the Ras-related Bud1/Rsr1 protein bind to different effectors to determine yeast cell polarity. Proc. Natl. Acad. Sci. USA 94:4463-4468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Posas, F., and H. Saito. 1998. Activation of the yeast SSK2 MAP kinase kinase kinase by the SSK1 two-component response regulator. EMBO J. 17:1385-1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Posas, F., and H. Saito. 1997. Osmotic activation of the HOG MAPK pathway via Ste11p MAPKKK: scaffold role of Pbs2p MAPKK. Science 276:1702-1705. [DOI] [PubMed] [Google Scholar]
- 31.Posas, F., S. M. Wurgler-Murphy, T. Maeda, E. A. Witten, T. C. Thai, and H. Saito. 1996. Yeast HOG1 MAP kinase cascade is regulated by a multistep phosphorelay mechanism in the SLN1-YPD1-SSK1 “two-component” osmosensor. Cell 86:865-875. [DOI] [PubMed] [Google Scholar]
- 32.Raitt, D. C., F. Posas, and H. Saito. 2000. Yeast Cdc42 GTPase and Ste20 PAK-like kinase regulate Sho1-dependent activation of the Hog1 MAPK pathway. EMBO J. 19:4623-4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reiser, V., H. Ruis, and G. Ammerer. 1999. Kinase activity-dependent nuclear export opposes stress-induced nuclear accumulation and retention of Hog1 mitogen-activated protein kinase in the budding yeast Saccharomyces cerevisiae. Mol. Biol. Cell 10:1147-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reiser, V., S. M. Salah, and G. Ammerer. 2000. Polarized localization of yeast Pbs2 depends on osmostress, the membrane protein Sho1 and Cdc42. Nat. Cell Biol. 2:620-627. [DOI] [PubMed] [Google Scholar]
- 35.Rose, M. D., F. Winston, and P. Hieter. 1990. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 36.Schiestl, R. H., and R. D. Gietz. 1989. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr. Genet. 16:339-346. [DOI] [PubMed] [Google Scholar]
- 37.Shoemaker, D. D., E. E. Schadt, C. D. Armour, Y. D. He, P. Garrett-Engele, P. D. McDonagh, P. M. Loerch, A. Leonardson, P. Y. Lum, G. Cavet, L. F. Wu, S. J. Altschuler, S. Edwards, J. King, J. S. Tsang, G. Schimmack, J. M. Schelter, J. Koch, M. Ziman, M. J. Marton, B. Li, P. Cundiff, T. Ward, J. Castle, M. Krolewski, M. R. Meyer, M. Mao, J. Burchard, M. J. Kidd, H. Dai, J. W. Phillips, P. S. Linsley, R. Stoughton, S. Scherer, and M. S. Boguski. 2001. Experimental annotation of the human genome using microarray technology. Nature 409:922-927. [DOI] [PubMed] [Google Scholar]
- 38.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yabe, T., T. Yamada-Okabe, S. Kasahara, Y. Furuichi, T. Nakajima, E. Ichishima, M. Arisawa, and H. Yamada-Okabe. 1996. HKR1 encodes a cell surface protein that regulates both cell wall β-glucan synthesis and budding pattern in the yeast Saccharomyces cerevisiae. J. Bacteriol. 178:477-483. [DOI] [PMC free article] [PubMed] [Google Scholar]