Abstract
The t(8;21)(q22;q22) translocation, which fuses the ETO gene on human chromosome 8 with the AML1 gene on chromosome 21 (AML1-ETO), is one of the most frequent cytogenetic abnormalities associated with acute myelogenous leukemia (AML). It is seen in approximately 12 to 15% of AML cases and is present in about 40% of AML cases with a French-American-British classified M2 phenotype. We have generated a murine model of the t(8;21) translocation by retroviral expression of AML1-ETO in purified hematopoietic stem cells (HSC). Animals reconstituted with AML1-ETO-expressing cells recapitulate the hematopoietic developmental abnormalities seen in the bone marrow of human patients with the t(8;21) translocation. Primitive myeloblasts were increased to approximately 10% of bone marrow by 10 months posttransplant. Consistent with this observation was a 50-fold increase in myeloid colony-forming cells in vitro. Accumulation of late-stage metamyelocytes was also observed in bone marrow along with an increase in immature eosinophilic myelocytes that showed abnormal basophilic granulation. HSC numbers in the bone marrow of 10-month-posttransplant animals were 29-fold greater than in transplant-matched control mice, suggesting that AML1-ETO expression overrides the normal genetic control of HSC pool size. In summary, AMLI-ETO-expressing animals recapitulate many (and perhaps all) of the developmental abnormalities seen in human patients with the t(8;21) translocation, although the animals do not develop leukemia or disseminated disease in peripheral tissues like the liver or spleen. This suggests that the principal contribution of AML1-ETO to acute myeloid leukemia is the inhibition of multiple developmental pathways.
The t(8;21)(q22;q22) translocation, which fuses the ETO gene on human chromosome 8 with the AML1 gene on chromosome 21, is seen in approximately 12 to 15% of acute myelogenous leukemia (AML) cases and in about 40% of AML cases with a French-American-British classified M2 phenotype (10, 27). AML1 (also known as Runx1) is a transcription factor with significant homology to the product of the Drosophila segmentation gene Runt (11, 23). It binds the enhancer core target sequence, TGT/cGGT, in association with a non-DNA-binding subunit, CBFβ (5, 20, 28, 40). Both proteins (together referred to as core-binding factor [CBF]) interact through the DNA-binding, Runt homology domain of AML1. The inversion (16) disrupts the CBFβ gene and is found in an additional 12% of AML cases (18). Null mutations in either CBF subunit in mice resulted in embryonic lethality that was associated with intracranial hemorrhaging and a complete absence of definitive hematopoiesis (30, 36, 38, 39).
The t(8;21) translocation fuses the N-terminal 177 amino acids of AML1, which includes the Runt homology domain that binds DNA and interacts with CBFβ, in frame with amino acids 30 to 604 of ETO. The fusion protein deletes the C-terminal activation domain of AML1. The ETO gene is homologous to the Drosophila gene nervy and can associate with transcriptional corepressor complexes that include mSin3, histone deacetylases, and nuclear hormone corepressors, which are involved in transcriptional repression (19). Gene knock-in experiments in mice have shown that AML1-ETO acts in a dominant-repressive manner to block AML1-dependent transcription (29, 42). Animals heterozygous for an AML1-ETO knock-in allele displayed a phenotype similar to that of AML1 or CBFβ knock-out mice in that they died early in embryonic life (embryonic day 13.5) and exhibited intracranial bleeding and a block in definitive hematopoiesis. One important difference between the knock-out and knock-in phenotypes was the presence of dysplastic hematopoietic progenitor cells within the fetal livers of the knock-in mice that could be readily established as immortalized cell lines in vitro (29).
The consequence of AML1-ETO expression on myeloid lineage development has been explored by using transformed myeloid cell lines that retain some capacity to terminally differentiate. Expression of AML1-ETO in the myeloid cell line 32D.3 inhibits C/EBPα-dependent transcription that correlates with a block in granulocytic differentiation in vitro (41). Inhibition of C/EBPα function in these experiments was related to the direct association of AML1-ETO with C/EBPα. Mice that develop in the absence of C/EBPα lack neutrophils and are blocked in granulocytic development at the myeloblast stage (44). Significant downregulation of C/EBPα has also been seen in patient samples bearing the t(8;21) translocation, thus establishing C/EBPα as a potentially critical target gene in AML1-ETO-associated leukemia (31, 32).
Recent efforts to establish animal models for the t(8;21) translocation have involved both transgenic and Cre-Lox-mediated interchromosomal translocation approaches. One study used the tetracycline-OFF (TET-OFF) system to conditionally activate expression of an AML1-ETO transgene in vivo (35). No abnormal hematopoiesis in peripheral blood or bone marrow was observed over a 2-year time period in AML1-ETO-expressing animals. In vitro colony-forming cell assays showed no difference in the number or type of colonies that were generated from AML1-ETO or control bone marrow samples. However, AML1-ETO-expressing cells did exhibit an enhanced replating potential during serial passage in methylcellulose. Cells isolated from these platings exhibited an immature myeloid morphology.
A second transgenic approach used a myeloid-specific promoter, MRP8, to drive expression of AML1-ETO specifically in the myeloid lineage (43). Again, no abnormal hematopoiesis was seen in the animals in the absence of additional mutations generated by N-ethyl-N-nitrosourea treatment of newborn mice. Finally, Buchholz et al. used the Cre recombinase system to conditionally activate an interchromosomal AML1-ETO translocation after the onset of definitive hematopoiesis (6). No characterization of hematopoiesis was made in this study.
Morphological and phenotypic analysis of bone marrow from t(8;21) AML patients has revealed a number of characteristic abnormalities in myeloid lineage cells. Large basophilic blasts with a prominent Golgi zone, abnormal granulation, cytoplasmic vacuoles, and a single Auer rod are common (1, 26, 37). Nuclear maturation in granulocytes is generally characterized by abnormal nuclear condensation at the metamyelocyte stage and a failure to segment properly (37). Some patient samples exhibit a marked marrow eosinophilia (>5%) with distinct basophilic granulation (37). In leukemic samples, it is not clear which of these abnormalities are attributable to the activity of AML1-ETO.
In our experiments, we transduced a purified population of hematopoietic stem cells (HSC) with an AML1-ETO-expressing retrovirus that coexpresses the green fluorescent protein (GFP) from an internal ribosome entry site (IRES). HSC that were transduced with the AML1-ETO retrovirus were resorted and then transplanted into lethally irradiated, congenic animals that differed at the Ly-5 locus. Our results showed a striking phenotype in both the stem cell compartment and the myeloid cell lineages of all reconstituted AML1-ETO animals. We observed a progressive increase in both the frequency and absolute number of HSC in the bone marrow of AML1-ETO-expressing animals that was 29-fold greater than stem cell numbers in GFP control animals by 10 months posttransplant. The expansion of AML1-ETO-expressing HSC no longer seemed to be restricted by the normal genetic control of HSC pool size (8, 25) but was exhaustible, based on serial transplantation experiments. At 10 months posttransplant, myeloid colony-forming progenitors were expanded approximately 50-fold, which was consistent with increases in the percentage of myeloblasts and promyelocytes to 5 to 14% of total bone marrow. Eosinophil development was also affected, in that we observed a significant increase in immature eosinophil myelocytes that exhibited abnormal basophilic granulation.
In summary, animals that express AML1-ETO in HSC recapitulate many (if not all) of the developmental abnormalities seen in human patients with the t(8;21) translocation but require additional secondary mutations for disease progression to acute myeloid leukemia.
MATERIALS AND METHODS
Generation of retrovirus.
AML1-ETO was cloned upstream of the IRES element into the EcoRI site of the parental murine stem cell virus (MSCV) IRES GFP vector (13). Retroviral constructs were transiently transfected into BOSC23 ecotropic packaging cells by calcium phosphate coprecipitation (15, 33). Viral supernatants were titrated using NIH 3T3 cells. Titers ranged between 3 × 106 and 1 × 107 IU/ml.
HSC isolation and retroviral transduction.
An enriched population of HSC of the surface phenotype Sca-1+c-Kit+ Lin− were isolated by fluorescence-activated cell sorting (FACS) and prestimulated in cytokines as previously described (15). Bone marrow cells from 5-fluorouracil-treated mice (isolated 4 days post-intraperitoneal injection of 150 mg of 5-fluorouracil/kg of body weight) were treated with ACK (0.15 M NH4Cl and 0.01 M KHCO3) for 5 min on ice to lyse red blood cells and then stimulated for 24 h. After stimulation, cells were cocultured on transiently transfected and irradiated (30 Gy) BOSC23 cells in the presence of 4 μg of Polybrene/ml for 48 h prior to transplantation.
Transplantation.
Congenic, C57BL/6-Ly-5.1 (Ly-5.1) mice (3 to 4 months of age) were used as transplant recipients. Prior to transplantation, Ly-5.1 mice were lethally irradiated with 10 Gy in a split dose separated by 3 h. Then 300 to 400 resorted GFP+/Ly-5.2+ HSC and a radioprotective dose of 2 × 105 Ly-5.1 bone marrow cells were transplanted into anesthetized mice by retro-orbital injection. A total of 4 × 106 bone marrow cells were used in serial transplant experiments and 1 × 106 to 6 × 106 bone marrow cells were used in 5-fluorouracil transplants. Mice were maintained for 2 to 3 weeks on acidified water containing neomycin sulfate (1.1 g/liter) and polymixin B sulfate (106 U/liter) or sulfamethoxazole (400 mg/liter).
Histology.
For cytospin preparation, 4 × 104 bone marrow cells in phosphate-buffered saline (PBS)-12% fetal calf serum or methylcellulose colonies in 150 μl of Iscove's modified Dulbecco’s medium (IMDM)-12% fetal calf serum were centrifuged onto glass slides and stained with Wright-Giemsa. Blood and bone marrow counts were determined manually.
Myeloid colony-forming assay.
A total of 1,000 each of AML1-ETO/GFP+ or GFP− myeloid scatter-gated cells isolated from the same mouse were sorted into Iscove's IMDM medium supplemented with 10% heat-inactivated fetal calf serum and then plated into MethoCult3434 medium (StemCell Technologies) supplemented with granulocyte-macrophage colony-stimulating factor (0.5 ng/ml; R & D Systems). Colonies were typed at day 10.
Western blot.
Approximately 3 × 106 myeloid scatter-gated cells were sorted as either AML1-ETO/GFP+ or GFP− from two AML1-ETO animals 3 months posttransplant. Cells were lysed in Laemmli buffer and run on a 10% polyacrylamide gel. AML-ETO was detected with a rabbit polyclonal antibody raised against a peptide encoding residues 32 to 50 of the human AML1 protein (11). The primary staining was visualized with a goat anti-rabbit immunoglobulin-horseradish peroxidase conjugate secondary antibody and enhanced chemiluminescence (Amersham Pharmacia).
Northern blot.
Total RNA from approximately 8 × 106 myeloid scatter-gated cells was isolated with RNA Stat-60 according to the manufacturer's instructions (Tel-test B, Inc., Friendswood, Tex.). Total RNA (7.5 μg) was run on a 1% agarose-0.6% formaldehyde gel, transferred to a Hybond-N (Amersham) membrane, and hybridized according to the supplier's protocol. A murine glyceraldehyde-3-phosphate dehydrogenase (Ambion) and C/EBPα probe (kindly provided by Dan Tenen, Harvard University) were used for detection.
RESULTS
Generation of a murine model of the t(8;21) translocation.
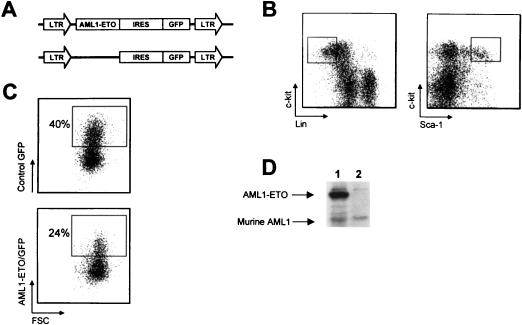
HSC of the phenotype c-Kit+Sca-1+Lin− (Fig. 1B) were double-sorted to a purity of >98% and then transduced with retroviral supernatant containing either the control or AML1-ETO vector (Fig. 1A). Each vector was derived from MSCV and contained an IRES to allow coexpression of GFP. Transduction efficiencies ranged from 20 to 28% for the AML1-ETO virus and 30 to 40% for the control virus (Fig. 1C). Transduced HSC isolated from C57BL/6-Ly-5.2 mice were resorted for GFP expression and then transplanted into lethally irradiated, congenic C57BL/6-Ly-5.1 animals at a dose of approximately 300 GFP+ cells per recipient. AML1-ETO-expressing animals were also generated by transplanting retrovirally transduced whole bone marrow cells isolated from 5-fluorouracil-treated animals (see Materials and Methods).
FIG. 1.
Retroviral transduction of murine hematopoietic stem cells. (A) Schematic diagram of MSCV retroviral constructs (control MSCV IRES GFP and MSCV AML1-ETO IRES GFP). (B) Gating used for sorting the HSC phenotype c-Kit+Sca-1+Lin− (where Lin represents a cocktail of antibodies to the mature blood cell antigens Mac-1, Gr-1, Ter119, B220, CD3, CD4, CD5, and CD8). (C) Flow cytometric analysis of HSC 24 h after retroviral transduction. Approximately 300 Ly-5.2+ HSC from control or AML1-ETO transductions were transplanted with a radioprotective dose of 2 × 105 Ly-5.1+ whole bone marrow cells into each Ly-5.1+ recipient animal. (D) Western blot analysis of GFP+ (lane 1) or GFP− (lane 2) myeloid scatter-gated cells FACS-sorted from the bone marrow of an 8-week-posttransplant AML1-ETO animal probed with a polyclonal anti-AML1 antibody.
Expression of AML1-ETO from the retroviral vector was confirmed by Western blot analysis with a polyclonal anti-AML1 antibody and GFP+ myeloid lineage cells sorted from the bone marrow of an 8-week-postreconstitution AML1-ETO animal (Fig. 1D). The anti-AML1 antibody was raised against a peptide encoding residues 32 to 50 of the human AML1 protein (11). The immunizing peptide has a three-amino-acid difference from the murine and human sequence, so a direct comparison between the levels of retrovirally expressed AML1-ETO and endogenous AML1 protein in myeloid lineage cells is not possible. Western blot analysis with lysates from 2.5 million FACS-sorted AML1-ETO-expressing myeloid cells and 2.5 million Kasumi-1 cells, which represent a human AML cell line that expresses the t(8;21) translocation, showed that Kasumi-1 cells expressed AML1-ETO at twofold-higher levels than retrovirally expressed AML1-ETO in primary cells (data not shown).
Abnormal myelopoiesis and decreased B lymphopoiesis in AML1-ETO/GFP+ peripheral blood cells.
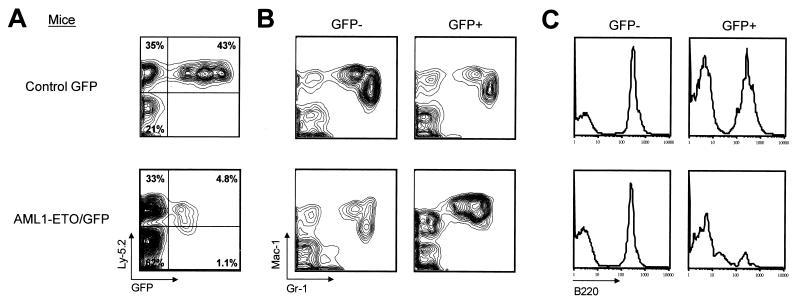
The effect of the AML1-ETO fusion protein on hematopoiesis was monitored in AML1-ETO-expressing and control GFP animals by FACS analysis of peripheral blood. All AML1-ETO (n = 29) and control GFP (n = 26) recipients were reconstituted with up to 85% of peripheral blood cells expressing the Ly-5.2 donor marker (Fig. 2A). Donor cells that silenced expression of the GFP marker were present in all reconstituted animals (15). Peripheral blood myeloid cells were analyzed by costaining with Mac-1 (CD11b) and Gr-1. AML1-ETO/GFP+ cells showed an abnormal Mac-1/Gr-1 phenotype in all AML1-ETO mice compared to control GFP mice or to non-AML1-ETO-expressing cells (GFP−) within the AML1-ETO mice (Fig. 2B).
FIG. 2.
Abnormal myelopoiesis and decreased B lymphopoiesis in AML1-ETO/GFP+ peripheral blood cells. (A) Flow cytometric analysis of peripheral blood cells from animals at 2.5 months posttransplantation stained with an antibody to the Ly-5.2 donor marker. Peripheral blood cells were gated as GFP− or GFP+ and analyzed for (B) simultaneous Mac-1 and Gr-1 or (C) B220 expression. Note the reduction in Mac-1loGr-1hi cells that represent mature neutrophils and an overrepresentation of Mac-1hiGr-1int cells in the AML1-ETO/GFP+ population compared to the GFP+ control. FACS plots are representative of all cocultured whole bone marrow transplants of control GFP (n = 21) and AML1-ETO-expressing (n = 26) mice and all purified HSC transplants of control GFP (n = 5) and AML1-ETO-expressing (n = 3) mice.
Notably absent in the AML1-ETO/GFP+ population was a subset of Mac-1loGr-1hi cells that represents an essentially pure population of mature neutrophils (17). In addition, there was an overrepresentation of a unique subset of cells that expressed high levels of Mac-1 and intermediate levels of Gr-1. This subset of cells was present in the peripheral blood of all AML1-ETO mice at all time points analyzed and did not increase in frequency between 2 and 10 months posttransplant (n = 3 for animals analyzed up to 10 months and n = 26 for animals analyzed between 1 and 6 months posttransplant). In all analyses, the GFP− cells within the AML1-ETO mice resembled the GFP− and GFP+ cell profiles from control animals.
Peripheral lymphoid cells in transplant recipients were analyzed by staining for B220 and CD3 expression on B and T cells, respectively. Analysis of the B220+ population in AML1-ETO and control GFP mice showed that B220 expression was significantly lower in AML1-ETO/GFP+ cells compared to controls (Fig. 2C). At this point, it is not clear whether the B220lo cells represent an immature B-cell population in the periphery or simply downregulation of B220 expression on mature B cells. The number of cells expressing CD3 was dramatically decreased in AML1-ETO/GFP+ cells, although this observation was also seen in some of the control GFP+ animals, making it difficult to draw definitive conclusions on the role of AML1-ETO in T-cell development at this point (data not shown).
Abnormal myelopoiesis in AML1-ETO-expressing bone marrow cells.
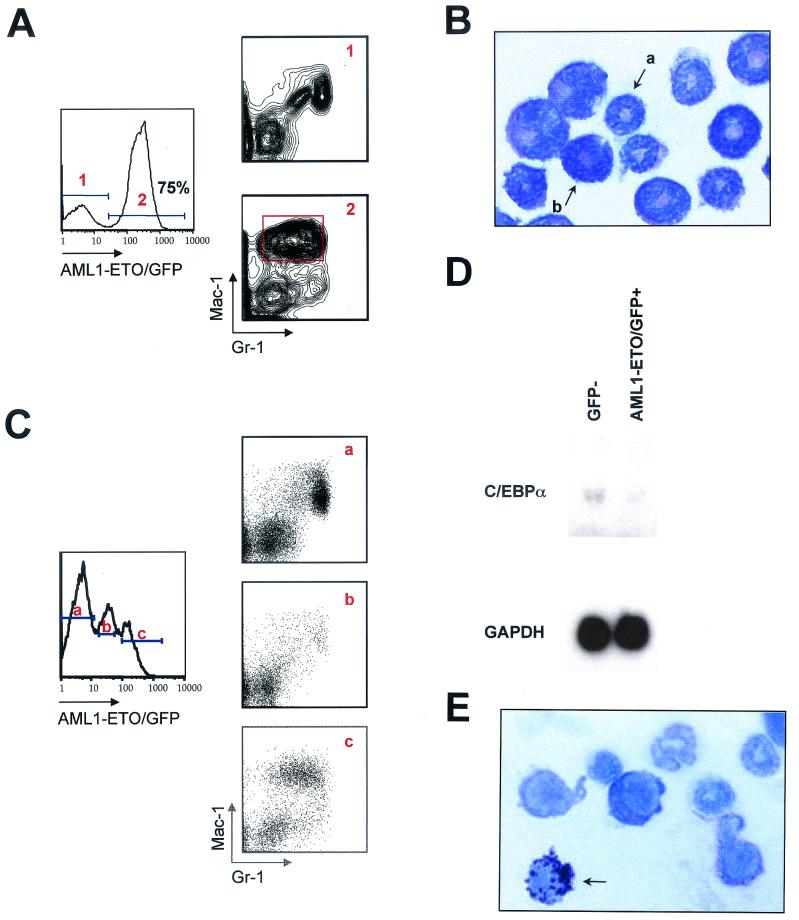
Given the abnormal myeloid phenotype in AML1-ETO/GFP+ peripheral blood cells, AML1-ETO-expressing mice were sacrificed to further investigate myeloid cell development in the bone marrow. AML1-ETO mice were sacrificed at 10 months posttransplant and analyzed for myeloid cell abnormalities by Mac-1/Gr-1 staining. All AML1-ETO mice (n = 3) exhibited the same Mac-1hiGr-1int population in the majority of AML1-ETO/GFP+ bone marrow cells compared to GFP− control myeloid cells analyzed from the same bone marrow (Fig. 3A). The appearance of this abnormal population in bone marrow was dependent on the level of AML1-ETO expression, as demonstrated by an AML1-ETO-expressing mouse that expressed both low and high levels of GFP (Fig. 3C). The dose-dependent phenotype in the myeloid lineage was not unexpected because AML1-ETO functions as a dominant inhibitor of normal AML1 activity (21, 29, 42).
FIG. 3.
Abnormal myelopoiesis in AML1-ETO-expressing bone marrow cells. Flow cytometric analysis of bone marrow from a 10-month-posttransplant AML1-ETO mouse. (A) Bone marrow cells were gated on (panel 1) GFP− and (panel 2) AML1-ETO/GFP+ bone marrow cells and analyzed for expression of Mac-1 and Gr-1. The data are representative of all AML1-ETO-transplanted animals between 2 and 10 months posttransplant. The Mac-1/Gr-1 profile in panel 1 is identical to what is seen in bone marrow from control GFP animals. (B) Wright-Giemsa-stained cytospin preparation of AML1-ETO/GFP+, Mac-1hiGr-1int cells gated as shown in panel A (×100 magnification). Arrows indicate (a) a banded neutrophil and (b) a metamyelocyte. (C) Graded levels of AML1-ETO expression show distinct Mac-1/Gr-1 phenotypes in bone marrow. (D) Northern blot analysis of RNA isolated from GFP− and AML1-ETO/GFP+ bone marrow cells from a 3-month-posttransplant AML1-ETO animal. The blot was probed with a 3′ fragment of the C/EBPα cDNA and a glyceraldehyde-3-phosphate dehydrogenase (GAPDH) probe. Quantitation of transcript levels was done on a phosphoimager. (E) Wright-Giemsa staining of an eosinophil myelocyte, showing abnormal basophilic granulation from the bone marrow of a 10-month-posttransplant AML1-ETO animal.
In order to determine the morphology and function of the cells residing in the Mac-1hiGr-1int population, we sorted these cells for Wright-Giemsa staining and for assays of myeloid colony-forming potential in methylcellulose. We observed no myeloid colony-forming activity in the Mac-1hiGr-1int population when 2,000 of these cells were plated in triplicate in methylcellulose over a 10-day in vitro culture period (data not shown). Plating 1,000 GFP− control myeloid lineage cells gave rise to 1 to 10 myeloid colonies (see below). Wright-Giemsa cytospin preparations indicated that 95% of the Mac-1hiGr-1int cells were metamyelocytes and immature band-form neutrophils (Fig. 3B), which is consistent with the lack of myeloid colony-forming activity in the population. In addition, there were no observed myeloblasts or promyelocytes in counts of 1,000 Mac-1hi Gr-1int cells from 10 independent microscope fields from two animals.
In the animal shown in Fig. 3A, 38% of the total marrow was comprised of this myeloid subset. The other animals analyzed had 8 and 14% Mac-1hiGr-1int cells in the bone marrow at 10 months posttransplant. Interestingly, morphological characterization of bone marrow from human patients with the t(8;21) translocation also showed abnormal nuclear condensation at the metamyelocyte stage (37).
Recent studies have demonstrated that AML1-ETO downregulates transcription of C/EBPα, a transcription factor necessary for granulocytic differentiation, in patients with t(8;21)-associated leukemia (31). To determine whether C/EBPα expression was affected in AML1-ETO/GFP+ cells, RNA was isolated from FACS-sorted myeloid AML1-ETO/GFP+ and myeloid GFP− cells from the same AML1-ETO-expressing animal. Northern analysis showed that the level of C/EBPα mRNA expression in AML1-ETO-expressing cells was 2.5-fold lower than in GFP− myeloid lineage cells (Fig. 3D). These results confirm that AML1-ETO expression causes a downregulation of C/EBPα levels in myeloid lineage cells.
Increased myeloid progenitors in the presence of AML1-ETO.
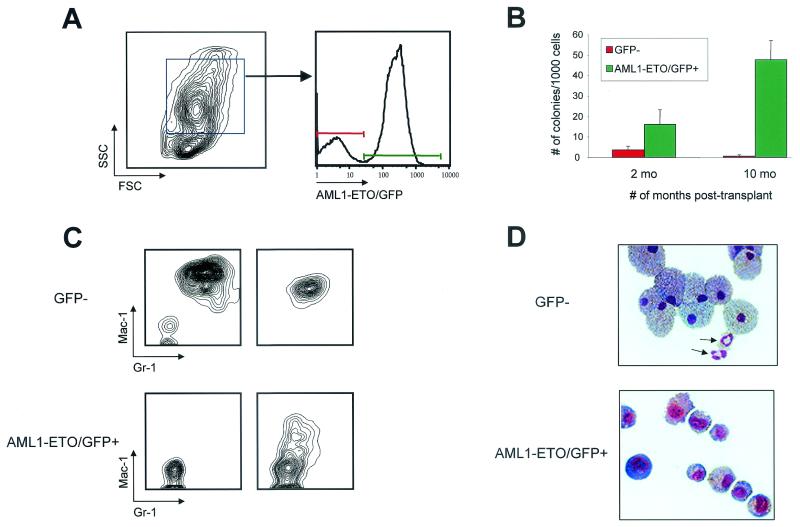
Changes in the number of myeloid progenitors in bone marrow were determined by in vitro colony-forming cell assays with GFP+ and GFP− cells isolated from AML1-ETO mice at 2 and 10 months posttransplant. One thousand myeloid scatter-gated AML1-ETO/GFP+ or GFP− cells from the same animal were sorted and then cultured in methylcellulose for 10 days (Fig. 4A). AML1-ETO/GFP+ cells isolated from 2-month-posttransplant animals (n = 3) gave rise to large colonies with an average of 16 myeloid colonies per 1,000 cells plated in triplicate, compared to somewhat smaller colonies observed in GFP− cell platings, which averaged 4 myeloid colonies per 1,000 cells plated (Fig. 4B). The fourfold increase in progenitor numbers compared to controls was statistically significant (P < 0.001). The expansion of myeloid progenitors was further increased in the bone marrow of 10-month-posttransplant animals (n = 3), where 1,000 AML1-ETO/GFP+ cells gave rise to an average of 48 myeloid colonies, compared to an average of 1 myeloid colony in GFP− control cells (Fig. 4B). The percentages of total myeloid cells in bone marrow (GFP+ and GFP−) were 58, 41, and 72% from the three AML1-ETO 10-month animals. The percentages of GFP+ myeloid cells in the same animals were 44, 46, and 91%, respectively. This indicates that there was not preferential expansion of GFP+ myeloid lineage cells in these animals (except in the latter case) even though the frequencies of specific myeloid subpopulations were significantly altered in cells that expressed AML1-ETO.
FIG. 4.
Increase in myeloid colony-forming cells in AML1-ETO animals. (A) Myeloid scatter-gated cells were sorted as GFP− or AML1-ETO/GFP+ from a 10-month-old AML1-ETO mouse, and 1,000 cells from each population were plated in triplicate into M3434 methylcellulose medium (10 ng of murine recombinant interleukin-3, 10 ng of human recombinant interleukin-6, and 50 ng of murine recombinant stem cell factor per ml) supplemented with 0.5 ng of granulocyte-macrophage colony-stimulating factor (R & D Systems) per ml. (B) Three independent AML1-ETO animals at 2 and 10 months posttransplant were used in the analysis. Colonies were enumerated (1 colony of >200 cells) and characterized 10 days after plating. (C) Representative FACS plots of individual methylcellulose colonies stained with Mac-1 and Gr-1. Two representative plots are shown for each sample. (D) Cytospin preparations of GFP− and AML1-ETO/GFP+ colonies stained with Wright-Giemsa. Arrows indicate mature, segmented neutrophils among the GFP− cells that were not seen in any AML1-ETO-expressing colonies.
Wright-Giemsa-stained cytospins of colonies derived from AML1-ETO/GFP+ cell platings showed a mixed lineage phenotype that included immature myeloid cells and mature macrophages (Fig. 4D). There were no segmented neutrophils present in AML1-ETO-expressing colonies. In contrast, cytospin preparations of GFP− colonies showed a number of mature segmented neutrophils (arrows in Fig. 4D). FACS analysis of individual colonies stained with Mac-1 and Gr-1 confirmed that GFP− colonies were almost completely differentiated (9 of 10 colonies were Mac-1+Gr-1+). In contrast, AML1-ETO/GFP+ colonies remained primarily undifferentiated, with negative or low-level expression of Mac-1 in only a fraction of the cells from a single colony (Fig. 4C).
To assess the percentages of myeloid cell types in the bone marrow of the three animals used for methylcellulose assays at 10 months posttransplant, myeloid-gated GFP+ and GFP− cells were cytospun and stained with Wright-Giemsa. The three AML1-ETO/GFP+ fractions of marrow were highly shifted in representation toward primitive myeloid cell types, with 17, 48, and 21% myeloblast/promyelocytes, compared to 1, 3, and 3%, respectively, of the same cell subsets in the GFP− controls (Table 1). Overall, the frequency of myeloblast/promyelocytes in bone marrow of the three AML1-ETO animals (after normalization for the total percentage of GFP+ myeloid cells) was 4.6, 9.5, and 14.0%. These results support the data from the in vitro colony-forming cell assays, indicating that a substantial increase in myeloid progenitor populations had occurred by 10 months posttransplant in the AML1-ETO animals.
TABLE 1.
Differential counts of sorted myeloid bone marrow cells from 10-month-posttransplant AML1-ETO animalsa
| GFP expression | % of >300 cells/sample
|
|||||
|---|---|---|---|---|---|---|
| Blasts + pro | Mye | Meta + band | Baso | Mature eosino | Eosino myelo | |
| Yes | 17 | 12 | 69 | <1 | <1 | 2 |
| 48 | 7 | 44 | <1 | <1 | 1 | |
| 21 | 6 | 73 | <1 | <1 | <1 | |
| No | 1 | 7 | 89 | <1 | 3 | <1 |
| 3 | 1 | 92 | <1 | 4 | <1 | |
| 3 | 8 | 89 | <1 | <1 | <1 | |
Blasts + pro, myeloblasts and promyelocytes; Mye, myelocytes; Meta + band, metamyelocytes and band nuclear granulocytes; Baso, basophils; Mature eosino, mature eosinophils; Eosino myelo, eosinophilic myelocytes. Statistical analysis (t test) showed statistically significant differences between GFP− and AML1-ETO/GFP+ cells in myeloblasts and promyelocytes (P ≤ 0.05), metamyelocytes and band nuclear granulocytes (P < 0.04), and mature eosinophils (P ≤ 0.01).
One criterion used in the characterization of AML in humans is the presence of greater than 20% myeloblasts in bone marrow (12). Although the percentage of myeloblasts/promyelocytes in the 10-month-posttransplant AML1-ETO animals was not 20%, the results clearly indicate that a highly abnormal condition exists in the myeloid lineage that becomes more pronounced over time. The lack of leukemia in the AML1-ETO animals was further supported by bone sections characterized at 4 months posttransplant, which did not show evidence of granulocytic foci. This was also true of the spleen and liver at this stage (data not shown). Finally, there was a significant increase in immature eosinophil myelocytes that exhibited abnormal basophilic granulation in AML1-ETO-expressing animals at both early and late times posttransplant (one at 2 months, one at 3 months, and three at 10 months; Fig. 3E and Table 1). A similar abnormality in eosinophil development is also seen in human patients with the t(8;21) translocation (37).
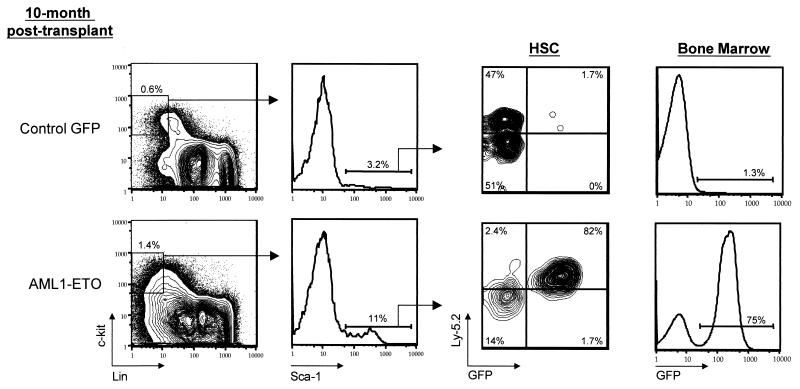
Expansion of HSC in AML1-ETO-expressing mice.
In order to characterize the HSC compartment in reconstituted animals, we performed five-color FACS analysis of bone marrow isolated from animals transplanted with cells expressing either the AML1-ETO or control GFP vector (Fig. 5). HSC in reconstituted animals have the same cell surface phenotype (c-Kit+Sca-1+Lin−) as HSC isolated from unmanipulated bone marrow (24). Bone marrow cells isolated from the tibias and femurs were quantitatively harvested and counted prior to staining to determine absolute HSC numbers. FACS analysis was performed at 2 and 10 months posttransplant of purified HSC and at 2.5 months posttransplant of transduced whole bone marrow cells isolated from 5-fluorouracil-treated animals (Table 2). The latter samples were analyzed to determine whether HSC expansion and absolute number would be influenced by the presence of approximately 106 bone marrow cells that were cotransduced and injected with HSC.
FIG. 5.
Expansion of hematopoietic stem cells in AML1-ETO mice. HSC analysis from a 10-month-posttransplant AML1-ETO mouse. Bone marrow cells were stained with c-Kit, lineage marker antibodies (see text), Sca-1, and the Ly-5.2 donor marker. The percentages of cells in individual gated populations are indicated.
TABLE 2.
Absolute number and frequency of hematopoietic stem cells in transplanted animalsa
| Time posttransplant (mo) | AML1-ETO/GFP mice
|
Control GFP mice
|
Average expansion (fold) | ||
|---|---|---|---|---|---|
| Absolute HSC no. | Frequency of HSC (%) | Absolute HSC no. | Frequency of HSC (%) | ||
| 2 | 7,755 | 0.017 | 1,509 | 0.004 | |
| 10,802 | 0.018 | 2,326 | 0.007 | 3 | |
| 5,895 | 0.015 | ||||
| 10 | 93,960 | 0.116 | 6,162 | 0.011 | |
| 118,556 | 0.163 | 10,200 | 0.020 | 29 | |
| 505,200 | 0.800 | ||||
| 2.5b | 52,100 | 0.146 | 10,980 | 0.015 | |
| 16,480 | 0.032 | 10,890 | 0.015 | 9 | |
| 259,980 | 0.619 | 13,530 | 0.022 | ||
Hematopoietic stem cells were derived from the femurs and tibias of transplanted mice. Average expansion is a multiple of the average number of HSC in AML1-ETO-transplanted animals over the average number in control GFP-transplanted animals at a given time point.
Animals from whole bone marrow transduction.
Figure 5 shows a representative analysis and gating of one AML1-ETO and one control GFP animal analyzed at 10 months posttransplant. Table 2 summarizes the results from eight AML1-ETO and eight control animals analyzed at the indicated time points. There was a modest expansion (threefold) in the absolute number of phenotypically defined HSC in AML1-ETO-expressing animals at 2 months posttransplant and a dramatic expansion (29-fold) by 10 months. One animal at 10 months had more than 50 times the expected number of c-Kit+Sca-1+Lin− cells. HSC from AML1-ETO animals transplanted with cocultured whole bone marrow cells were expanded 9.3-fold compared to control GFP animals at 2.5 months posttransplant. At every time point analyzed, the lowest number of HSC in an AML1-ETO animal was higher than the highest HSC number in any of the control GFP animals (Table 2).
The absolute number and frequency of HSC in control GFP animals were highly consistent in all animals, which suggests that the genetic control of hematopoietic stem cell pool size was maintained in primary transplant recipients expressing the control vector (8, 25). In contrast, AML1-ETO-expressing HSC no longer seemed to be restricted by the regulatory mechanisms that influence homeostasis within the stem cell compartment. Consistent with this speculation was the observation that the increase in HSC number in the AML1-ETO animals was due to an expansion of AML1-ETO/GFP+ HSC within the HSC compartment. The percentages of AML1-ETO/GFP+ HSC in the total HSC compartment ranged from 72 to 99% in seven of eight AML1-ETO animals (one AML1-ETO animal had 44% GFP+ HSC), with a mean percentage of GFP+ HSC of 82% (n = 8). This was in contrast to control GFP animals, in which the mean percentage of GFP+ HSC was 15% (n = 8). GFP− donor (Ly-5.2+) and recipient (Ly-5.2−)-type HSC were present in all animals.
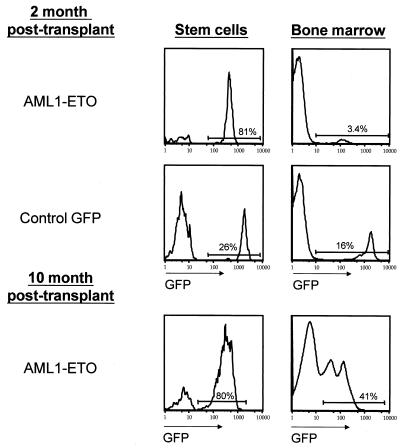
Delayed differentiation in AML1-ETO-expressing hematopoietic stem cells.
Despite the high percentage of AML1-ETO/GFP+ HSC at 2 months posttransplant (75 and 81%, n = 2), the percentage of AML1-ETO/GFP+ cells in the bone marrow was only 3.5 and 3.4%, respectively (Fig. 6). In the control GFP animals, the percentage of GFP+ HSC more closely approximated the GFP percentage in the bone marrow. The delayed appearance of more differentiated GFP+ cells in bone marrow was consistent with a delay in the appearance of GFP+ peripheral blood cells in animals transplanted with AML1-ETO-transduced HSC (n = 5 for AML1-ETO; data not shown). In addition, AML1-ETO-expressing HSC were unable to radioprotect lethally irradiated recipient animals at a dose of 600 cells (n = 6), whereas the same dose of control HSC radioprotected and reconstituted four of five animals (data not shown). This supports the notion that AML1-ETO-expressing HSC show a reduced ability to differentiate and an enhanced tendency to undergo cell division events that favor self-renewal. In spite of an apparent partial block in differentiation at 2 months posttransplant, the percentage of GFP+ cells in older AML1-ETO-expressing animals increased to proportions seen in controls (Fig. 6), which was largely due to an accumulation of GFP+ myeloid lineage cells.
FIG. 6.
Delayed differentiation in AML1-ETO-expressing stem cells. The percentage of AML1-ETO-expressing (GFP+) cells in the stem cell population and in whole bone marrow was contrasted at early (2 months, n = 3) and late (10 months, n = 3) times postreconstitution. The ratio of GFP+ cells in the stem cell compartment and in the bone marrow of control GFP animals was similar to the ratio seen in older AML1-ETO animals.
Maintenance of abnormal myelopoiesis is dependent on sustained expression of AML1-ETO in HSC.
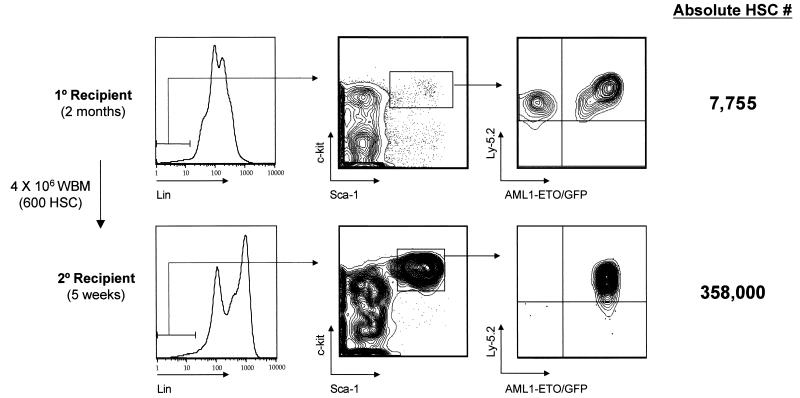
The lack of leukemia in AML1-ETO-expressing animals by 10 months posttransplant suggests that secondary mutations or additional time is necessary for disease progression. In an attempt to accelerate a disease phenotype, 4 × 106 bone marrow cells from primary transplant recipients at either 2 or 10 months posttransplant were serially transplanted into multiple secondary recipient animals. Interestingly, only one of four secondary recipients were reconstituted in bone marrow with AML1-ETO/GFP+ cells at 5 weeks posttransplant with marrow isolated from a 2-month primary donor, even though the bone marrow inoculum would have contained approximately 600 GFP+ HSC and about 114,000 GFP+ myeloid lineage cells (Fig. 7 and Table 3 ). Of the 600 GFP+ HSC, 60 would be expected to rehome to the bone marrow and approximately 12 would rehome to the tibias and femurs, which represent about 20% of the total marrow cellularity.
FIG. 7.
AML1-ETO expression in stem cells is required for maintenance of abnormal myelopoiesis. Bone marrow from one primary recipient AML1-ETO animal was serially transplanted at a dose of 4 × 106 cells into each of four lethally irradiated secondary mice. Flow cytometric analysis of HSC in one of four secondary animals is shown at 5 weeks posttransplant. All secondary transplant animals received 114,000 AML1-ETO-expressing myeloid cells along with approximately 600 AML1-ETO/GFP+ HSC in the bone marrow inoculum. WBM, whole bone marrow.
TABLE 3.
Serial transplantation of AML1-ETO bone marrowa
| Time posttransplant (mo) | Primary recpt | Secondary recpt | % GFP+ HSC | Absolute no. of GFP+ HSC | % GFP+ WBM |
|---|---|---|---|---|---|
| 2 | A | 75.0 | 5,816 | 3.4 | |
| 1 | A1 | 0 | 0 | 0 | |
| 1 | A2 | 0 | 0 | 0 | |
| 1 | A3 | 97.2 | 347,976 | 1.8 | |
| 1 | A4 | 0 | 0 | 0 | |
| 10 | E | 97.3 | 491,559 | 75.4 | |
| 4 | E1 | ND | ND | 35.0 | |
| 4 | E2 | ND | ND | 18.1 | |
| 6 | E3 | 69.8 | 5,641 | 11.8 | |
| 2 | E4 | 86.9 | 21,134 | 37.2 |
recpt, recipient. Secondary recipients each received 4 × 106 whole bone marrow cells (WBM) from the primary recipient. ND, not determined.
The three negative animals all showed high donor reconstitution and no GFP+ HSC, suggesting that donor GFP− HSC may have outcompeted GFP+ HSC during engraftment or that GFP+ HSC homed less efficiently to marrow than GFP− HSC. The one animal that was donor reconstituted with AML1-ETO/GFP+ cells showed an enormous expansion of the HSC phenotype (from a predicted 12 HSC to 358,000 GFP+ HSC in both tibias and femurs in 5 weeks; Fig. 7). Approximately 33% of the total GFP+ cells in the marrow of this secondary recipient were c-Kit+Sca-1+Lin−, supporting the observation that AML1-ETO-expressing HSC are partially blocked in their ability to differentiate. Of note was the lack of abnormal myelopoiesis in the absence of AML1-ETO/GFP+ HSC in the three negative secondary animals. This suggests that the 114,000 coinjected AML1-ETO/GFP+ myeloid lineage cells do not expand extensively and retain a relatively short half-life in vivo.
Four secondary recipients derived from injection of 4 × 106 bone marrow cells from a 10-month primary transplant animal were all highly reconstituted with AML1-ETO/GFP+ cells in peripheral blood for up to 6 months posttransplant (Table 3). One animal that was sacrificed at 2 months posttransplant had 21,134 total HSC, which represented a modest 33-fold expansion in HSC number over the 2-month reconstitution period. This was in contrast to the 30,000-fold expansion in 5 weeks seen in secondary recipient A3 (Table 3). The observation that four of four animals were highly reconstituted with AML1-ETO/GFP+ cells from a 10-month primary donor and only one of four secondary animals was reconstituted with the same number of bone marrow cells isolated from a 2-month donor may be related to the predicted number of GFP+ HSC in the inocula. The GFP+ HSC number from the 10-month donor was approximately 32,000 cells, which was in contrast to the 600 GFP+ HSC from the 2-month primary donor.
The total expansion of AML1-ETO/GFP+ HSC in vivo also seemed to be limited by some uncharacterized mechanism. This conclusion is based on the observation that HSC expansion was more severely limited with bone marrow from primary animals that already displayed substantial HSC expansion (Table 3). This may indicate that the genetic mechanisms regulating the replicative life span of HSC are distinct from those that control the steady-state number of stem cells in vivo.
DISCUSSION
Myeloid developmental abnormalities associated with AML1-ETO expression.
We have generated a murine model of the developmental defects observed in human patients with t(8;21)-associated AML by retroviral expression of the AML1-ETO fusion protein in HSC. The developmental abnormalities seen in AML1-ETO-expressing myeloid lineage cells were evident at three levels. First, more differentiated metamyelocytes and band-form neutrophils exhibiting a Mac-1hiGr-1int phenotype accumulate in bone marrow and become the predominant cell population by 10 months posttransplant (Fig. 3). These cells are also evident in the periphery and are diagnostic for the presence of an 8;21 translocation in bone marrow long before the marrow becomes highly abnormal. However, the Mac-1hiGr-1int cells in the periphery do not accumulate to the same extent as in bone marrow, suggesting that some developmental cue necessary for emigration of these cells from the marrow may be absent or that these cells turn over rapidly in the periphery. It remains possible that this maturational cue may involve a nuclear condensation abnormality in metamyelocytes, which is seen in t(8;21) AML.
A second developmental abnormality in the myeloid lineage was seen in bone marrow eosinophils. We observed a unique population of immature eosinophil myelocytes in three of three AML1-ETO animals at 10 months posttransplant that were not observed in non-AML1-ETO-expressing myeloid cells (Table 1). Immature eosinophils that exhibited basophilic granules were also detected at early times posttransplant (one at 2 months and one at 3 months), which indicates that AML1-ETO interferes with normal eosinophil development. Marrow eosinophilia is frequently observed in human AML M2 patients with the t(8;21) translocation, where the eosinophils also have developmental abnormalities associated with abnormal nuclear maturation and basophilic granulation (37).
Finally, early myeloid progenitor cells that give rise to myeloid colonies in methylcellulose expand approximately 50-fold over non-AML1-ETO-expressing myeloid progenitors by 10 months posttransplant. These cells have a reduced ability to differentiate, as evidenced by the immature morphology and cell surface antigen profile of myeloid colonies in vitro (Fig. 4), which may contribute to their accumulation in bone marrow. In AML1-ETO mice, myeloblasts and promyelocytes increased to between 5 and 14% of total marrow at 10 months posttransplant, which indicates that myelopoiesis within the bone marrow has become highly abnormal but not leukemic at this stage. Consistent with this interpretation was the absence of Auer rods in the myeloblasts, which are typically seen in human leukemic blasts.
Hematopoietic stem cell expansion in AML1-ETO-expressing animals.
The substantial increase in the absolute number and frequency of c-Kit+Sca-1+Lin− cells in AML1-ETO-expressing animals (Table 2) suggests that AML1-ETO can override the normal genetic control of HSC pool size in mice (8, 25). Interestingly, the absolute expansion of AML1-ETO-expressing HSC was exhaustible in vivo, based on serial transplantation experiments. We observed no AML1-ETO/GFP+ cells in the bone marrow of seven tertiary transplant mice reconstituted with bone marrow from the highly reconstituted secondary recipient animal shown in Fig. 7 (data not shown). These results suggest that AML1-ETO may decrease the proliferative capacity or homing efficiency of HSC in the context of a transplant assay. Consistent with this interpretation is the observation that AML1 regulates expression of certain integrin genes that may be inhibited by AML1-ETO (34).
In addition, AML1-ETO expression in myeloid cell lines was associated with decreased proliferation and a block in myeloid development (7). The increase in HSC numbers in AML1-ETO animals was gradual (Table 2, 2 months) and was associated with an apparent partial block in their ability to differentiate (Fig. 6). This again suggests that proliferation kinetics in the HSC population was not increased by AML1-ETO expression and that HSC accumulation may be related to an enhanced self-renewal potential in the presence of AML1-ETO. It also remains possible that AML1-ETO may enhance HSC survival and thereby increase HSC numbers in vivo. Studies by Domen et al. have shown that expression of the antiapoptosis gene bcl-2 in HSC resulted in a 2.4-fold increase in HSC numbers in vivo compared with nontransgenic control littermates (9). However, from this result, it would seem unlikely that blocking cell death would be the only mechanism responsible for the magnitude of HSC increase that we observed in the AML1-ETO-expressing animals. This issue is currently being explored. In any case, the large expansion in the HSC pool size would presumably allow a much larger population of cells that could acquire additional mutations, leading to a more aggressive phenotype resembling AML.
Secondary mutations are required for disease progression associated with t(8;21).
The large increase in the frequency of myeloid progenitor cells within the bone marrow of AML1-ETO animals occurred over a 10-month period (Fig. 3), which indicates that disease progression associated with an 8;21 translocation is rather slow. Secondary mutations that have been associated with AML with an M2 phenotype and the t(8;21) translocation include activating mutations in the tyrosine kinase receptors c-KIT and FLT3 and in the RAS proto-oncogene (2, 3, 16). Activating mutations in FLT3 associated with an internal tandem duplication of the juxtamembrane domain are seen in about 25% of AML cases and 9% of AML cases with an M2 phenotype and t(8;21) (16). Approximately 20 to 30% of AML cases have mutations in the N-ras or K-ras gene, which are not usually seen in association with the FLT3 mutations (14). Interestingly, the principal secondary mutations associated with the t(8;21) translocation would all provide a potent mitogenic stimulus that could rapidly expand the abnormal developmental stages that we observed in the AML1-ETO animals.
Animal models of the 8;21 translocation.
Other animal models of the t(8;21) translocation have been generated as previously described (see the introduction). Our results are entirely consistent with the results from other studies with the exception that we observed a striking phenotype in the bone marrow of AML1-ETO-expressing animals in the absence of any secondary mutations induced by chemical mutagenesis. From our studies, we can make four observations that may help to resolve the apparent discrepancies in the animal models. First, it is clear that the level of AML1-ETO expression must be high enough to titrate out the activity of both wild-type AML1 alleles in order to see any phenotype in transgenic or retroviral models (Fig. 3C). Second, it is important that bone marrow characterization be done at early and late times postreconstitution, given the slow nature of disease progression that we observed both in the HSC compartment and within the myeloid lineages (Fig. 4B and Table 2). Third, AML1-ETO must be expressed in the HSC compartment in order to sustain abnormal myelopoiesis in the bone marrow based on serial transplantation experiments (4) (Fig. 7 and Table 3). Finally, disease progression may be accelerated in the retroviral model due to an increased number of HSC that express the t(8;21) translocation at the beginning of the experiment. If very few HSC express AML1-ETO, as might be the case in the inducible transgenic models, we would expect a longer incubation period before highly abnormal conditions exist in the bone marrow.
Currently, all models (with the exception of Cre-Lox-mediated interchromosomal translocation [6]) suffer from the limitations that expression levels of AML1-ETO may not parallel those seen in human t(8;21) patients, and the temporal pattern of expression in hematopoietic cells may also differ. With respect to the latter point, we did observe AML1 expression in primitive HSC, suggesting that an 8;21 translocation in this population would be expressed (22; C. de Guzman and C. Klug, unpublished data). One distinct advantage of the retroviral system over other approaches has been the ability to identify AML1-ETO-expressing cells with GFP as a surrogate marker for AML1-ETO. This has allowed us to study the abnormalities associated with AML1-ETO expression in animals that exhibit very low percentages of cells that are AML1-ETO/GFP+.
The generation of a murine model of the t(8;21) translocation is significant in that the unique contribution of AML1-ETO to leukemia and developmental dysfunction can be studied at great depth at both the cellular and molecular levels. The animals will also be valuable as tools to explore therapies that specifically target cells that express AML1-ETO and provide a means to address the secondary mutations that are required for disease progression.
Acknowledgments
We thank John Kearney for help with the fluorescent microscope; Dan Tenen for the C/EBPα probe; Raymond Davidson for animal care; and Hyung Kim, C. Scott Swindle, and Claudiu Cotta for helping in retroviral transduction and transplantation. We also thank Tom Ryan, Trent Schoeb, Max Cooper, and the Division of Developmental and Clinical Immunology for valuable discussions and support.
This work was supported by a Howard Hughes faculty development award to C.A.K. (53000281) and a Molecular and Viral Oncology Predoctoral Fellowship grant (5T32CA09467) to C.D.G. A.J.W. holds an MRC Senior Clinical Fellowship through the Department of Haematology, University of Cambridge.
REFERENCES
- 1.Andrieu, V., I. Radford-Weiss, X. Troussard, C. Chane, F. Valensi, M. Guesnu, E. Haddad, F. Viguier, F. Dreyfus, B. Varet, G. Flandrin, and E. Macintyre. 1996. Molecular detection of t(8;21)/AML1-ETO in AML M1/M2: correlation with cytogenetics, morphology and immunophenotype. Br. J. Haematol. 92:855-865. [DOI] [PubMed] [Google Scholar]
- 2.Bartram, C. R., W. D. Ludwig, W. Hiddemann, J. Lyons, M. Buschle, J. Ritter, J. Harbott, A. Frohlich, and J. W. Janssen. 1989. Acute myeloid leukemia: analysis of ras gene mutations and clonality defined by polymorphic X-linked loci. Leukemia 3:247-256. [PubMed] [Google Scholar]
- 3.Beghini, A., P. Peterlongo, C. B. Ripamonti, L. Larizza, R. Cairoli, E. Morra, and C. Mecucci. 2000. c-kit mutations in core binding factor leukemias. Blood 95:726-727. [PubMed] [Google Scholar]
- 4.Bonnet, D., and J. E. Dick. 1997. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat. Med. 3:730-737. [DOI] [PubMed] [Google Scholar]
- 5.Bravo, J., Z. Li, N. A. Speck, and A. J. Warren. 2001. The leukemia-associated AML1 (Runx1)-CBF beta complex functions as a DNA-induced molecular clamp. Nat. Struct. Biol. 8:371-378. [DOI] [PubMed] [Google Scholar]
- 6.Buchholz, F., Y. Refaeli, A. Trumpp, and J. M. Bishop. 2000. Inducible chromosomal translocation of AML1 and ETO genes through Cre/loxP-mediated recombination in the mouse. EMBO Rep. 1:133-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burel, S. A., N. Harakawa, L. Zhou, T. Pabst, D. G. Tenen, and D. E. Zhang. 2001. Dichotomy of AML1-ETO functions: growth arrest versus block of differentiation. Mol. Cell. Biol. 21:5577-5590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Haan, G., and G. Van Zant. 1997. Intrinsic and extrinsic control of hemopoietic stem cell numbers: mapping of a stem cell gene. J. Exp. Med. 186:529-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Domen, J., S. H. Cheshier, and I. L. Weissman. 2000. The role of apoptosis in the regulation of hematopoietic stem cells: overexpression of Bcl-2 increases both their number and repopulation potential. J. Exp. Med. 191:253-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Downing, J. R. 1999. The AML1-ETO chimaeric transcription factor in acute myeloid leukaemia: biology and clinical significance. Br. J. Haematol. 106:296-308. [DOI] [PubMed] [Google Scholar]
- 11.Erickson, P., J. Gao, K. S. Chang, T. Look, E. Whisenant, S. Raimondi, R. Lasher, J. Trujillo, J. Rowley, and H. Drabkin. 1992. Identification of breakpoints in t(8;21) acute myelogenous leukemia and isolation of a fusion transcript, AML1/ETO, with similarity to Drosophila segmentation gene, runt. Blood 80:1825-1831. [PubMed] [Google Scholar]
- 12.Harris, N. L., E. S. Jaffe, J. Diebold, G. Flandrin, H. K. Muller-Hermelink, J. Vardiman, T. A. Lister, and C. D. Bloomfield. 1999. World Health Organization classification of neoplastic diseases of the hematopoietic and lymphoid tissues: report of the Clinical Advisory Committee meeting-Airlie House, Virginia, November 1997. J. Clin. Oncol. 17:3835-3849. [DOI] [PubMed] [Google Scholar]
- 13.Hawley, R. G., F. H. Lieu, A. Z. Fong, and T. S. Hawley. 1994. Versatile retroviral vectors for potential use in gene therapy. Gene Ther. 1:136-138. [PubMed] [Google Scholar]
- 14.Kiyoi, H., T. Naoe, Y. Nakano, S. Yokota, S. Minami, S. Miyawaki, N. Asou, K. Kuriyama, I. Jinnai, C. Shimazaki, H. Akiyama, K. Saito, H. Oh, T. Motoji, E. Omoto, H. Saito, R. Ohno, and R. Ueda. 1999. Prognostic implication of FLT3 and N-RAS gene mutations in acute myeloid leukemia. Blood 93:3074-3080. [PubMed] [Google Scholar]
- 15.Klug, C. A., S. Cheshier, and I. L. Weissman. 2000. Inactivation of a GFP retrovirus occurs at multiple levels in long-term repopulating stem cells and their differentiated progeny. Blood 96:894-901. [PubMed] [Google Scholar]
- 16.Kottaridis, P. D., R. E. Gale, M. E. Frew, G. Harrison, S. E. Langabeer, A. A. Belton, H. Walker, K. Wheatley, D. T. Bowen, A. K. Burnett, A. H. Goldstone, and D. C. Linch. 2001. The presence of a FLT3 internal tandem duplication in patients with acute myeloid leukemia (AML) adds important prognostic information to cytogenetic risk group and response to the first cycle of chemotherapy: analysis of 854 patients from the United Kingdom Medical Research Council AML 10 and 12 trials. Blood 98:1752-1759. [DOI] [PubMed] [Google Scholar]
- 17.Lagasse, E., and I. L. Weissman. 1996. Flow cytometric identification of murine neutrophils and monocytes. J. Immunol. Methods 197:139-150. [DOI] [PubMed] [Google Scholar]
- 18.Liu, P., S. A. Tarle, A. Hajra, D. F. Claxton, P. Marlton, M. Freedman, M. J. Siciliano, and F. S. Collins. 1993. Fusion between transcription factor CBF beta/PEBP2 beta and a myosin heavy chain in acute myeloid leukemia. Science 261:1041-1044. [DOI] [PubMed] [Google Scholar]
- 19.Lutterbach, B., J. J. Westendorf, B. Linggi, A. Patten, M. Moniwa, J. R. Davie, K. D. Huynh, V. J. Bardwell, R. M. Lavinsky, M. G. Rosenfeld, C. Glass, E. Seto, and S. W. Hiebert. 1998. ETO, a target of t(8;21) in acute leukemia, interacts with the N-CoR and mSin3 corepressors. Mol. Cell. Biol. 18:7176-7184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meyers, S., J. R. Downing, and S. W. Hiebert. 1993. Identification of AML-1 and the (8;21) translocation protein (AML-1/ETO) as sequence-specific DNA-binding proteins: the runt homology domain is required for DNA binding and protein-protein interactions. Mol. Cell. Biol. 13:6336-6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meyers, S., N. Lenny, and S. W. Hiebert. 1995. The t(8;21) fusion protein interferes with AML-1B-dependent transcriptional activation. Mol. Cell. Biol. 15:1974-1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miyamoto, T., I. L. Weissman, and K. Akashi. 2000. AML1/ETO-expressing nonleukemic stem cells in acute myelogenous leukemia with 8;21 chromosomal translocation. Proc. Natl. Acad. Sci. USA 97:7521-7526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miyoshi, H., K. Shimizu, T. Kozu, N. Maseki, Y. Kaneko, and M. Ohki. 1991. t(8;21) breakpoints on chromosome 21 in acute myeloid leukemia are clustered within a limited region of a single gene, AML1. Proc. Natl. Acad. Sci. USA 88:10431-10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morrison, S. J., A. M. Wandycz, H. D. Hemmati, D. E. Wright, and I. L. Weissman. 1997. Identification of a lineage of multipotent hematopoietic progenitors. Development 124:1929-1939. [DOI] [PubMed] [Google Scholar]
- 25.Muller-Sieburg, C. E., and R. Riblet. 1996. Genetic control of the frequency of hematopoietic stem cells in mice: mapping of a candidate locus to chromosome 1. J. Exp. Med. 183:1141-1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nucifora, G., J. I. Dickstein, V. Torbenson, D. Roulston, J. D. Rowley, and J. W. Vardiman. 1994. Correlation between cell morphology and expression of the AML1/ETO chimeric transcript in patients with acute myeloid leukemia without the t(8;21) translocation. Leukemia 8:1533-1538. [PubMed] [Google Scholar]
- 27.Nucifora, G., and J. D. Rowley. 1995. AML1 and the 8;21 and 3;21 translocations in acute and chronic myeloid leukemia. Blood 86:1-14. [PubMed] [Google Scholar]
- 28.Ogawa, E., M. Maruyama, H. Kagoshima, M. Inuzuka, J. Lu, M. Satake, K. Shigesada, and Y. Ito. 1993. PEBP2/PEA2 represents a family of transcription factors homologous to the products of the Drosophila runt gene and the human AML1 gene. Proc. Natl. Acad. Sci. USA 90:6859-6863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Okuda, T., Z. Cai, S. Yang, N. Lenny, C. J. Lyu, J. M. van Deursen, H. Harada, and J. R. Downing. 1998. Expression of a knocked-in AML1-ETO leukemia gene inhibits the establishment of normal definitive hematopoiesis and directly generates dysplastic hematopoietic progenitors. Blood 91:3134-3143. [PubMed] [Google Scholar]
- 30.Okuda, T., J. van Deursen, S. W. Hiebert, G. Grosveld, and J. R. Downing. 1996. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell 84:321-330. [DOI] [PubMed] [Google Scholar]
- 31.Pabst, T., B. U. Mueller, N. Harakawa, C. Schoch, T. Haferlach, G. Behre, W. Hiddemann, D. E. Zhang, and D. G. Tenen. 2001. AML1-ETO downregulates the granulocytic differentiation factor C/EBPα in t(8;21) myeloid leukemia. Nat. Med. 7:444-451. [DOI] [PubMed] [Google Scholar]
- 32.Pabst, T., B. U. Mueller, P. Zhang, H. S. Radomska, S. Narravula, S. Schnittger, G. Behre, W. Hiddemann, and D. G. Tenen. 2001. Dominant-negative mutations of CEBPA, encoding CCAAT/enhancer binding protein-alpha (C/EBPα), in acute myeloid leukemia. Nat. Genet. 27:263-270. [DOI] [PubMed] [Google Scholar]
- 33.Pear, W. S., G. P. Nolan, M. L. Scott, and D. Baltimore. 1993. Production of high-titer helper-free retroviruses by transient transfection. Proc. Natl. Acad. Sci. USA 90:8392-8396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Puig-Kroger, A., C. Lopez-Rodriguez, M. Relloso, T. Sanchez-Elsner, A. Nueda, E. Munoz, C. Bernabeu, and A. L. Corbi. 2000. Polyomavirus enhancer-binding protein 2/core binding factor/acute myeloid leukemia factors contribute to the cell type-specific activity of the CD11a integrin gene promoter. J. Biol. Chem. 275:28507-28512. [DOI] [PubMed] [Google Scholar]
- 35.Rhoades, K. L., C. J. Hetherington, N. Harakawa, D. A. Yergeau, L. Zhou, L. Q. Liu, M. T. Little, D. G. Tenen, and D. E. Zhang. 2000. Analysis of the role of AML1-ETO in leukemogenesis, with an inducible transgenic mouse model. Blood 96:2108-2115. [PubMed] [Google Scholar]
- 36.Sasaki, K., H. Yagi, R. T. Bronson, K. Tominaga, T. Matsunashi, K. Deguchi, Y. Tani, T. Kishimoto, and T. Komori. 1996. Absence of fetal liver hematopoiesis in mice deficient in transcriptional coactivator core binding factor beta. Proc. Natl. Acad. Sci. USA 93:12359-12363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Swirsky, D. M., Y. S. Li, J. G. Matthews, R. J. Flemans, J. K. Rees, and F. G. Hayhoe. 1984. 8;21 translocation in acute granulocytic leukaemia: cytological, cytochemical and clinical features. Br. J. Haematol. 56:199-213. [DOI] [PubMed] [Google Scholar]
- 38.Wang, Q., T. Stacy, M. Binder, M. Marin-Padilla, A. H. Sharpe, and N. A. Speck. 1996. Disruption of the Cbfa2 gene causes necrosis and hemorrhaging in the central nervous system and blocks definitive hematopoiesis. Proc. Natl. Acad. Sci. USA 93:3444-3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang, Q., T. Stacy, J. D. Miller, A. F. Lewis, T. L. Gu, X. Huang, J. H. Bushweller, J. C. Bories, F. W. Alt, G. Ryan, P. P. Liu, A. Wynshaw-Boris, M. Binder, M. Marin-Padilla, A. H. Sharpe, and N. A. Speck. 1996. The CBFbeta subunit is essential for CBFα2 (AML1) function in vivo. Cell 87:697-708. [DOI] [PubMed] [Google Scholar]
- 40.Wang, S., Q. Wang, B. E. Crute, I. N. Melnikova, S. R. Keller, and N. A. Speck. 1993. Cloning and characterization of subunits of the T-cell receptor and murine leukemia virus enhancer core-binding factor. Mol. Cell. Biol. 13:3324-3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Westendorf, J. J., C. M. Yamamoto, N. Lenny, J. R. Downing, M. E. Selsted, and S. W. Hiebert. 1998. The t(8;21) fusion product, AML-1-ETO, associates with C/EBP-α, inhibits C/EBP-α-dependent transcription, and blocks granulocytic differentiation. Mol. Cell. Biol. 18:322-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yergeau, D. A., C. J. Hetherington, Q. Wang, P. Zhang, A. H. Sharpe, M. Binder, M. Marin-Padilla, D. G. Tenen, N. A. Speck, and D. E. Zhang. 1997. Embryonic lethality and impairment of haematopoiesis in mice heterozygous for an AML1-ETO fusion gene. Nat. Genet. 15:303-306. [DOI] [PubMed] [Google Scholar]
- 43.Yuan, Y., L. Zhou, T. Miyamoto, H. Iwasaki, N. Harakawa, C. J. Hetherington, S. A. Burel, E. Lagasse, I. L. Weissman, K. Akashi, and D. E. Zhang. 2001. AML1-ETO expression is directly involved in the development of acute myeloid leukemia in the presence of additional mutations. Proc. Natl. Acad. Sci. USA 98:10398-10403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang, D. E., P. Zhang, N. D. Wang, C. J. Hetherington, G. J. Darlington, and D. G. Tenen. 1997. Absence of granulocyte colony-stimulating factor signaling and neutrophil development in CCAAT enhancer binding protein alpha-deficient mice. Proc. Natl. Acad. Sci. USA 94:569-574. [DOI] [PMC free article] [PubMed] [Google Scholar]