Abstract
In the budding yeast Saccharomyces cerevisiae, null alleles of several DNA repair and recombination genes confer defects in recombination that grow more severe with decreasing sequence length, indicating that they are required for short-sequence recombination (SSR). RAD1 and RAD10, which encode the subunits of the structure-specific endonuclease Rad1/10, are critical for SSR. MRE11, RAD50, and XRS2, which encode the subunits of M/R/X, another complex with nuclease activity, are also crucially important. Genetic evidence suggests that Rad1/10 and M/R/X act on the same class of substrates during SSR. MSH2 and MSH3, which encode subunits of Msh2/3, a complex active during mismatch repair and recombination, are also important for SSR but play a more restricted role. Additional evidence suggests that SSR is distinct from nonhomologous end joining and is superimposed upon basal homologous recombination.
Eukaryotic genomes are characterized by an abundance of short, repetitive sequences (11). In the yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe, recombination between dispersed, short sequences, such as tRNA genes and δ elements, can give rise to genome rearrangements (56, 69). In humans, recombination between Alu elements create genome rearrangements that are involved in several diseases (15). These observations suggest that careful control of recombination between short sequences may be essential for the maintenance of eukaryotic genome stability.
In S. cerevisiae, homologous recombination occurs less frequently per unit length for sequences of less than 300 bp (1, 26, 65) and becomes extremely inefficient for sequences of less than 30 bp (36), suggesting that short-sequence recombination (SSR) is distinct from general recombination. Mutations in several DNA repair genes have been shown to selectively limit recombination between short sequences, demonstrating that SSR is under separate genetic control. Often, the effects of these mutations can be correlated with changes in the processing of recombination intermediates. For instance, certain mutations in genes encoding subunits of the nucleotide excision repair (NER) and transcription factor IIH (TFIIH) complexes increase SSR and block the degradation of the ends of broken DNA molecules (5, 6, 32, 35). Similarly, loss of RAD27, which encodes a structure-specific nuclease important in DNA replication and repair (18, 23, 27, 34, 51, 64), increases SSR and decreases the cleavage of single strands adjacent to heteroduplex DNA (44). However, the precise relationships between these DNA processing defects and the control of SSR remain unclear.
To better understand how SSR is restricted, we need to develop an understanding of how it is propagated. We have begun this process by identifying the factors that are required for SSR and determining how they are related to one another. We hypothesized that these factors are most likely to be previously identified components of the double-strand break (DSB) repair machinery that play as yet undescribed roles in recombination between short sequences. Several recombination genes were selected for analysis because their null alleles confer phenotypes that seemed likely to provide insight into the basis of SSR. RAD52 was selected because the rad52-null mutant of budding yeast is defective for most homologous recombination (47), which would be helpful in determining the involvement of the basal recombination machinery in SSR. RAD1 was chosen because the rad1- and rad10-null mutants are particularly defective for recombination between short sequences, suggesting that the NER and recombination endonuclease Rad1/10 (7, 17, 30, 53, 60, 61, 67, 70, 71, 76) is an important component of SSR (5). MSH2 and MSH3, which encode subunits of the mismatch repair apparatus (52), were picked because the msh2- and msh3-null mutants display recombination defects that are similar to those observed in rad1 and rad10 mutants (58, 66), suggesting that they could also be required for SSR. MRE11, RAD50, and XRS2, which encode the subunits of M/R/X, a complex that plays a role in many processes related to DNA recombination and repair (20), were chosen because their null alleles confer defective degradation of broken DNA molecules (25, 43, 73), suggesting that they could be involved in SSR. Finally, HDF1 (yKU70) and DNL4 were selected because they are required for the repair of DSBs by nonhomologous end joining (NHEJ) (38, 74, 77), which has been shown to rejoin broken DNA molecules by using very short (1- to 4-bp) homologous sequences (31, 59), and might, therefore, be part of the SSR apparatus.
The effects of null alleles of these genes on SSR, both singly and in various combinations, were used to determine their relevance to SSR and their relationships to each other. We measured SSR by assaying DNA fragment insertion into genomic target sequences. These assays revealed that SSR is composed of at least two pathways that are superimposed upon general homologous recombination controlled by the RAD52 gene. One pathway is controlled by the RAD1 and RAD10 genes, and the other is controlled by MRE11, RAD50, and XRS2. Our results indicate that these pathways compete for the same class of intermediates. The MSH2 and MSH3 genes are also important for SSR and act as members of both pathways, but their significance is more limited than that of the other genes. DNL4 and HDF1, however, play very small roles in SSR, suggesting that the apparatuses for NHEJ and SSR are essentially separate.
MATERIALS AND METHODS
Yeast plasmids and strains.
All of the plasmids used in this study were constructed, maintained, and amplified by standard techniques (57). Plasmids and DNA fragments were transformed into yeast by electroporation. All of the yeast strains used in our work are isogenic with W303-1A (70) and are listed in Table 1. Construction and maintenance of yeast strains were done by established protocols (12). Construction of the rad1::LEU2 (54), rad10::LEU2 (5), rad50::hisG (3, 4), rad52::TRP1 (63), msh2::hisG (52), hdf1::TRP1 (38), mre11::hisG (10), mre11-6 (75), and mre11-H125N (40) mutant alleles was done as previously described. The msh3::hisG strain was constructed by transforming yeast with a 5.8-kb SpeI/MscI fragment of the plasmid pLAY410 where the 3.8-kb hisG::URA3::hisG universal disruptor (2) replaces a 1.2-kb segment of the coding sequence of MSH3. The xrs2::TRP1 and dnl4::LEU2 alleles were constructed by transforming yeast with PCR fragments created with primers that had 45 bp of sequence upstream or downstream from the open reading frame to be replaced plus 15 to 26 bp of sequence flanking both sides of the TRP1 and LEU2 sequences in pUC-TRP1 and pUC-LEU2 (55), which were the templates for PCR. The sequences of the xrs2::TRP1 primers were 5′-CATTTGAAATCGGTATAGATACTAACAACGCCATCGTCATCACTAAACGCGTCAGCGGGTGTTGGCGGG-3′ and 5′-CAGTTAATGCCTCCAGCAACCTGTAGTGCTTCTTTAAACCGATCCGCACACCGCATAGGCAAGTGCAC-3′. The dnl4::LEU2primers were 5′-AAAATAAAAATCTAGAACTGAAGGAAATAGTAACGGATTATTTAGGTAGCGGGTGTTGGCGGGTGTCGGGGC-3′ and 5′-TAATGTACATATGTAGGATAGTATTAAATAAACTTCAAAAAAGGCGAATGGCGCGACGCGCCC-3′. Transformants were selected on medium lacking tryptophan or leucine. Trp+ and Leu+ transformants were screened by Southern blotting to determine in which clones gene replacement had successfully occurred (G. M. Manthey and A. M. Bailis, unpublished data). The sam2::his3::ura3::KAN-MX allele that was used as a target for DNA fragment insertion in our assays was constructed by transforming yeast with a 5-kb XhoI/NotI fragment from pLAY346 containing the sam2::his3::ura3::KAN-MX construct. Transformants were selected on medium containing 200 μg of G418 per ml and screened by Southern blotting (Manthey and Bailis, unpublished). The sam2::his3::ura3::KAN-MX construct contained a 1.5-kb BglII/EcoRI KAN-MX marker replacing a 60-bp StuI/ApaI fragment of the coding sequence in a 1.2-kb BamHI/BamHI URA3 clone, which, in turn, replaces a 60-bp BglII/BglII fragment of the coding sequence in a 1.3-kb BamHI/XhoI HIS3 clone that has been inserted into the SalI site in a 950-bp EcoRV/XhoI SAM2 fragment. The length of the HIS3 sequence incorporated into the sam2::his3::ura3::KAN-MX target encompasses all of the HIS3 sequences on the four his3::URA3 fragments used in our DNA fragment insertion experiments.
TABLE 1.
Yeast strains used in this study
| Strain | Genotypea |
|---|---|
| ABX425-6B | MATaHIS3 sam2::his3::ura3::KAN-MX |
| ABX462-5B | MATα HIS3 sam2::his3::ura3::KAN-MX rad52::TRP1 |
| ABX441-26A | MATaHIS3 sam2::his3::ura3::KAN-MX rad1::LEU2 |
| ABX464-12A | MATaHIS3 sam2::his3::ura3::KAN-MX msh2::hisG |
| ABX545-12A | MATaHIS3 sam2::his3::ura3::KAN-MX msh3::hisG |
| ABX441-21A | MATα HIS3 sam2::his3::ura3::KAN-MX mre11::hisG |
| ABX480-1A | MATaHIS3 sam2::his3::ura3::KAN-MX mre11-H125N |
| ABX518-6C | MATaHIS3 sam2::his3::ura3::KAN-MX mre11-6 |
| ABX441-22D | MATaHIS3 sam2::his3::ura3::KAN-MX rad50::hisG |
| ABX441-64A | MATα HIS3 sam2::his3::ura3::KAN-MX xrs2::TRP1 |
| ABX449-7C | MATα HIS3 sam2::his3::ura3::KAN-MX hdf1::TRP1 |
| ABX448-4D | MATα HIS3 sam2::his3::ura3::KAN-MX dnl4::LEU2 |
| ABX441-39A | MATα HIS3 sam2::his3::ura3::KAN-MX rad1::LEU2 mre11::hisG |
| ABX441-20D | MATaHIS3 sam2::his3::ura3::KAN-MX rad1::LEU2 rad50::hisG |
| ABX441-25A | MATα HIS3 sam2::his3::ura3::KAN-MX rad1::LEU2 xrs2::TRP1 |
| ABX478-20C | MATaHIS3 sam2::his3::ura3::KAN-MX radI::LEU2 msh2::hisG |
| ABX479-9C | MATaHIS3 sam2::his3::ura3::KAN-MX mre11::hisG msh2::hisG |
| ABX526-56D | MATaHIS3 sam2::his3::ura3::KAN-MX rad1::LEU2 mre11::hisG msh2::hisG |
| ABX449-11A | MATaHIS3 sam2::his3::ura3::KAN-MX rad1::LEU2 hdf1::TRP1 |
| ABT151 | MATa::LEU2/pLAY97 (URA3 HOcs)/pGHOT (TRP1 GAL::HO) |
| ABT380 | MATa::LEU2 mre11::hisG/pLAY97/(URA3 HOcs)/pGHOT (TRP1 GAL1::HO) |
| ABT381 | MATa::LEU2 mre11-6/pLAY97 (URA3 HOcs)/pGHOT (TRP1 GAL1::HO) |
| ABT382 | MATa::LEU2 mre11-H125N/pLAY97 (URA3 HOcs)/pGHOT (TRP1 GAL1::HO) |
All strains are isogenic to W303-1A (MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 rad5-535) (70). Only deviations from this genotype are listed.
DNA fragment insertion assay.
The DNA fragment insertion assay used in this work is substantially similar to those previously described (44). his3::URA3 DNA fragments were prepared by restriction endonuclease digestion of pLAY144 (5), phenol extraction, and ethanol precipitation. Purified DNA fragments were used to transform yeast cells by electroporation, selecting for expression of the URA3 segment of the fragment. Efficiencies of transformation with the DNA fragments varied by twofold or less from strain to strain (data not shown). Insertion of the his3::URA3 fragments into alternative genomic targets was distinguished by replica plating to appropriate dropout media. The nature of the recombination events in selected, independent recombinants was confirmed by DNA sequencing. Insertion into the wild-type HIS3 allele at the HIS3 locus replaces 60 bp of the HIS3 coding sequence with the 1.2-kb URA3 marker and results in His− G418r Ura+ transformants. Insertion into the his3::ura3::KAN-MX allele at the SAM2 locus replaces a 1.5-kb KAN-MX marker with 60 bp of the URA3 coding sequence and results in His+ G418s Ura+ transformants. Recombination at the URA3 locus replaces a point mutation in the ura3-1 allele by gene conversion and results in His+ G418r Ura+ transformants.
Stability of HO-digested plasmid.
A more detailed description of the assay used to determine HO-digested plasmid stability was published previously (5). Single colonies of strains ABT151 (wild type), ABT380 (mre11::hisG), ABT381 (mre11-6), and ABT382 (mre11-H125N) were used to inoculate 300 ml of synthetic medium lacking uracil and tryptophan and containing 3% glycerol and 3% lactate. Uracil was omitted to select for the URA3 marker on pLAY97, a centromere-containing plasmid that also bears a 117-bp fragment of MATa containing the recognition site for HO endonuclease. Tryptophan was omitted to select for the TRP1 marker on pGHOT (46), another centromere plasmid containing the HO endonuclease gene linked to a galactose-inducible promoter. Glycerol and lactate were used as carbon sources to maintain the expression of the HO gene in neither an induced nor a repressed state. Cultures were grown at 30°C to a density of 5 × 106 cells/ml before a 50-ml aliquot was removed, and the cells were pelleted and frozen at −20°C. HO endonuclease expression was then induced by adding 25 ml of 20% galactose to the culture. After 30 min, the cells from another aliquot were collected and frozen. The remaining culture was filtered through sterile 0.2-μm nitrocellulose filters, and the cells were resuspended in prewarmed synthetic medium lacking tryptophan and containing 2% glucose. Uracil was provided because pLAY97 is frequently lost after HO cleavage, and glucose was used to repress HO gene expression. At regular intervals, aliquots were removed and the cells were collected as described above. DNA prepared from the frozen cells was digested with the NcoI restriction endonuclease and analyzed by Southern blot hybridization using 32P-labeled pBluescript (Stratagene, La Jolla, Calif.), the backbone of pLAY97. Hybridization patterns were visualized and quantitated by phosphorimaging (Molecular Dynamics). The stability of HO-digested pLAY97 was determined by comparing the levels of the 1.5- and 3.5-kb NcoI- and HO-digested fragments with that of the NcoI-digested 5.0-kb fragment. Stability of HO-digested DNA was plotted as the log of the percentage of HO-digested DNA remaining versus time. Curves were generated by least-squares analysis. Half-lives of broken DNA molecules were reported for two separate determinations.
RESULTS
DNA fragment insertion assay for SSR.
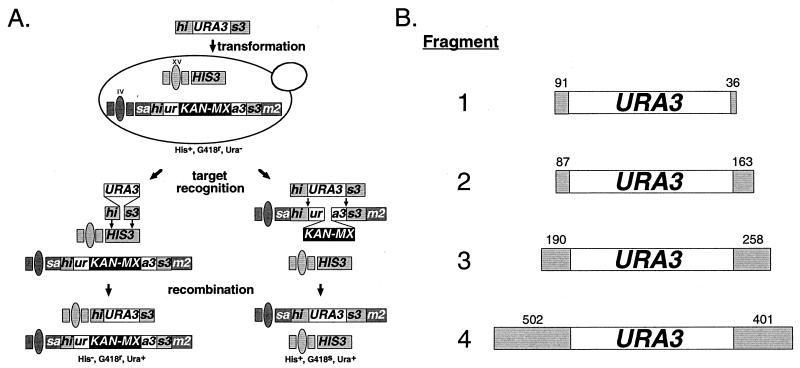
We previously used the insertion of DNA fragments of various lengths into alternative genomic targets to describe the effects of specific DNA repair mutations on SSR (44). The assay used in this study was very similar to the previously used assay but was adapted for use with haploid cells (Fig. 1). DNA fragments consisting of a URA3 marker flanked by different lengths of the HIS3 sequence (his3::URA3, Fig. 1B) were transformed into wild-type or mutant cells. Fragment insertion into the HIS3 locus on chromosome XV uses only the terminal HIS3 sequences, while insertion into the sam2::his3::ura3::KAN-MX allele at the SAM2 locus on chromosome IV uses the entire his3::URA3 sequence (Fig. 1A). Total homology to the HIS3 locus varied over sevenfold from the smallest to the largest his3::URA3 fragment, while homology to sam2::his3::ura3::KAN-MX varied by less than 65% (Fig. 1B). Therefore, changing the length of the HIS3 sequences on the his3::URA3 fragments should have a significant effect on their insertion into HIS3 but much less of an effect on overall fragment length and insertion into sam2::his3::ura3::KAN-MX. This makes the ratio of fragment insertions into HIS3 and sam2::his3::ura3::KAN-MX (H−/K−) an appropriate gauge of the cell's ability to use the terminal HIS3 sequences for homologous recombination.
FIG. 1.
DNA fragment insertion assay. (A) Description of the assay. DNA fragments electroporated into haploid yeast cells are inserted by recombination into the HIS3 locus on chromosome XV or the his3::ura3::KAN-MX marker at the SAM2 locus on chromosome IV. The fragments align with HIS3 by using the terminal HIS3 sequences, whereas the entire sequence is used to align the fragment with his3::ura3::KAN-MX. Insertion at HIS3results in a His− G418r (Kan+) Ura+ cell. Insertion at sam2::his3::ura3::KAN-MX results in a His+ G418s (Kan−) Ura+ cell. (B) DNA fragment substrates. The DNA fragments used in the experiments are depicted. HIS3 sequences are represented by shaded boxes with sequence lengths marked above in base pairs. The URA3 sequence is 1,071 bp long. The four fragments total 1,198, 1,321, 1,519, and 1,974 bp, respectively.
DNA fragment insertion requires the basal DSB repair machinery.
We investigated the effects of mutations in several DNA repair and recombination genes on his3::URA3 fragment insertion in an effort to define the factors required for SSR. Similar to previously reported results (50, 60), we found that the central recombination gene RAD52 was required to observe the insertion of DNA fragments into either of the targets discussed above, as well as gene conversion of the ura3-1 allele at the URA3 locus, as no Ura+ transformants were obtained with the rad52-null mutant. Therefore, all recombination between the DNA fragments and the genome is RAD52 dependent. This suggests that, in our assays, SSR requires general homologous recombination.
Genes encoding key subunits of several important DNA repair complexes are required for SSR.
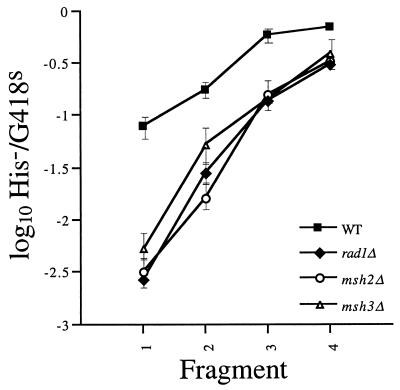
It was previously reported that his3::URA3 DNA fragments are inserted less efficiently into the HIS3 locus of rad1 and rad10 mutant cells than into that of wild-type cells (60, 61). This established that RAD1 and RAD10 play a role in the insertion of DNA sequences into the genome by recombination. We have also previously shown that decreasing HIS3 sequence length reduces fragment insertion into the HIS3 locus of rad1 and rad10 mutant cells more than that into wild-type cells (5). This indicates that the roles of RAD1 and RAD10 in recombination increase as sequence length decreases. Similarly, in the present study, we found that the rad1-null mutation had a much greater impact on the H−/K− ratio obtained with the shortest his3::URA3 fragment (127 bp of HIS3 homology), a 29-fold reduction from the wild type, than with the longest (903 bp of HIS3 homology), a 2.5-fold reduction (Fig. 2). Together, these results suggest that RAD1 and RAD10 play a critical role in the maintenance of SSR.
FIG. 2.
DNA fragment insertion in wild-type and mutant cells. The his3::URA3 fragments were electroporated into wild-type cells (ABX425-6B) and rad1::LEU2 (ABX441-26A), msh2::hisG (ABX464-12A), and msh3::hisG (ABX545-12A) mutant cells. Ura+ transformants were screened for DNA fragment insertion into HIS3 or sam2::his3::ura3::KAN-MX by replica plating to medium lacking histidine or containing 200 μg of G418 per ml. The mean ratios of His− G418r (Kan+) Ura+-to-His+ G418s (Kan−) Ura+ transformants (H−/K−) were determined from a minimum of five independent trials with each fragment in each strain. Log10 values of the mean ratios ± 2 standard errors were plotted. Each trial consisted of a minimum of 400 transformants.
Like RAD1 and RAD10, the mismatch repair genes MSH2 and MSH3 facilitate the removal of nonhomologous sequences from the 3′ ends of recombining molecules (66). However, this role is distinct from the role played by RAD1 and RAD10 because it is important only when the recombining sequences are 1 kb or less in length. In our analysis, the msh2- and msh3-null mutants displayed H−/K− ratios with all four his3::URA3 fragments that were not significantly different from those obtained with the rad1 mutant (Fig. 2), indicating that MSH2 and MSH3, like RAD1 and RAD10, play critical roles in propagating SSR.
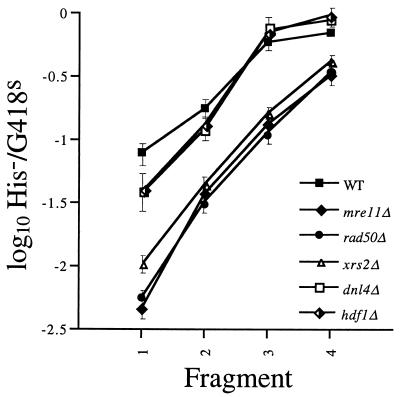
MRE11, RAD50, and XRS2 code for the subunits of another complex that has nuclease activity (19, 40, 72, 75) and plays multiple roles in the maintenance of genome stability (20). The H−/K− ratios obtained by inserting all four his3::URA3 fragments into the genomes of the mre11- and rad50-null mutants (Fig. 3) were very similar to the values obtained with the rad1-, msh2-, and msh3-null mutants (Fig. 2), suggesting that MRE11 and RAD50 are also required for SSR. Meanwhile, the ratio obtained by inserting the smallest fragment into xrs2-null mutant cells was slightly (two- to threefold) higher (Fig. 3). This suggests that XRS2 may play an independent and less critical role in SSR than do MRE11 and RAD50 or that the M/R/X complex plays a more peripheral role in the maintenance of SSR than a complex that includes only Mre11 and Rad50.
FIG. 3.
DNA fragment insertion into wild-type and mutant cells. The his3::URA3 fragments were electroporated into wild-type and mre11::hisG (ABX441-21A), rad50::hisG (ABX441-22D), xrs2::TRP1 (ABX441-64A), dnl4::LEU2 (ABX448-4D), and hdf1::TRP1 (ABX449-7C) mutant cells. Transformants were screened, and H−/K− ratios were determined and plotted as described in the legend to Fig. 2 and in Materials and Methods.
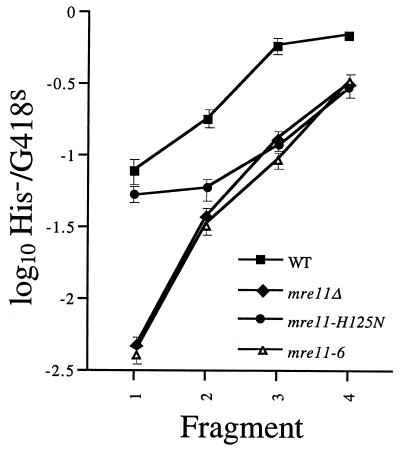
Several mre11 alleles have been isolated that encode proteins that lack one or more of the nuclease activities identified with Mre11 in vitro (19, 40, 75). We chose two of these alleles, mre11-6, which encodes a protein missing DNA binding site A (75), and mre11-H125N, which changes an amino acid in one of four conserved phosphoesterase motifs (10, 40), to investigate the potential role of Mre11 nuclease activity in SSR. Mre11-6 was previously found to lack single-stranded DNA (ssDNA) endonuclease, 3′ → 5′ ssDNA exonuclease, and 3′ → 5′ double-stranded DNA exonuclease activities (75), while Mre11-H125N, which was not assayed for the latter two activities, did not display ssDNA endonuclease activity (40). Both the mre11-6 and mre11-H125N mutations block meiotic DSB processing and confer a mild methyl methanesulfonate sensitivity. In addition, the mre11-H125N mutation was found not to change mitotic DSB processing and rejoining, NHEJ, or telomere maintenance (40). The mre11-null mutant is blocked for both mitotic and meiotic DSB processing, exhibits profound sensitivity to methyl methanesulfonate, and is defective for NHEJ and telomere maintenance (20). Therefore, both mutants share some characteristics with the mre11-null mutant but not others, suggesting that Mre11 performs both nuclease-dependent and -independent functions.
We found that mre11-null and mre11-6 mutant cells presented nearly identical, reduced H−/K− ratios for all four DNA fragments, but the mre11-H125N mutant displayed a significant variation (Fig. 4). The mre11-H125N ratios observed with the three largest DNA fragments were very close to those of the other mre11 mutants; however, the ratio obtained with the smallest fragment was more than 12-fold higher and was only 2-fold lower than the wild-type ratio. This indicates that the three mre11 alleles reduce recombination with the longer sequences equally but that mre11-H125N has much less of an effect on recombination with the shortest sequence. This demonstrates that MRE11 plays genetically distinct roles in recombination with the three longer sequences and the shortest sequence, which we operationally define as SSR.
FIG. 4.
DNA fragment insertion into wild-type and mutant cells. The his3::URA3 fragments were electroporated into wild-type and mre11::hisG mre11-H125N (ABX480-1A) and mre11-6 (ABX518-6C) mutant cells. Transformants were screened, and H−/K− ratios were determined and plotted as described in the legend to Fig. 2 and in Materials and Methods.
One possible explanation for the role of MRE11, RAD50, and XRS2 in SSR could be that these recombination events resemble MRE11-, RAD50-, and XRS2-dependent NHEJ. Other laboratories have shown that DNA fragments and plasmids can be inserted into the genome by using little, if any, homology (31, 39, 59). DNA sequence analysis of recombination junctions revealed that most insertion events involve 1 to 4 bp of homology between the ends of the fragment and the genome, similar to a predominant class of NHEJ events. In addition, these events are dramatically reduced in rad50 -null mutants (62), indicating that RAD50, and perhaps MRE11 and XRS2, is required for insertion of DNA molecules into the genome by using very short lengths of homology.
In order to ascertain whether NHEJ is involved in SSR, we performed DNA fragment insertion assays with NHEJ-defective hdf1- and dnl4-null mutant strains. The H−/K− ratios obtained with the mutants were similar to the wild-type ratio and nearly indistinguishable from each other (Fig. 3). The mutant ratios were slightly (two- to threefold) reduced with the smallest fragment, suggesting that NHEJ makes, at most, a very small contribution to our SSR events. Therefore, the roles played by MRE11, RAD50, and XRS2 in SSR may not be related to their roles in NHEJ. Interestingly, the H−/K− ratios obtained with the largest fragment were slightly (twofold) elevated in the hdf1 and dnl4 strains, indicating that NHEJ subtly inhibits recombination between longer sequences in our assay.
Complex interactions between important factors control SSR in budding yeast cells.
A previous analysis indicated that rad1 and rad10 are epistatic with respect to SSR, as are mre11, rad50, and xrs2 (Manthey and Bailis, unpublished). This suggests that RAD1 and RAD10 work together, as do MRE11, RAD50, and XRS2. We conducted an epistasis analysis to explore the relationship between these two groups of genes and others with respect to SSR.
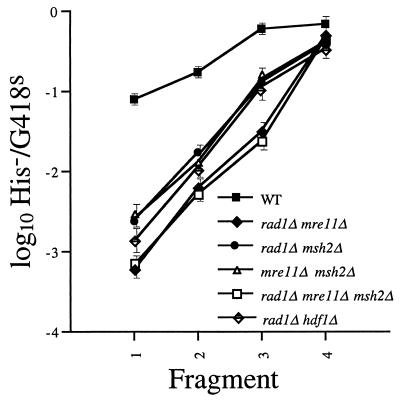
DNA fragment insertions into various single and double mutants were compared. For example, the combination of the rad1-null mutation with the mre11-null mutation led to lower H−/K− ratios than either single mutation (Fig. 5). The extent of the decreases, 130-fold for the shortest fragment in the double mutant versus 18- to 29-fold for the single mutants, indicates that combining the mutations has synergistic effects. Very similar results were obtained with the rad1 rad50 and rad1 xrs2 double mutants (data not shown).
FIG. 5.
DNA fragment insertion into wild-type and mutant cells. The his3::URA3 fragments were electroporated into wild-type; rad1::LEU2 mre11::hisG (ABX441-39A), rad1::LEU2 msh2::hisG (ABX478-20C), and mre11::hisG msh2::hisG (ABX479-9C) double-mutant; and rad1::LEU2 mre11::hisG msh2::hisG (ABX526-56D) and rad1::LEU2 hdf1::TRP1 (ABX449-11A) triple-mutant cells. Transformants were screened, and H−/K− ratios were determined and plotted as described in the legend to Fig. 2 and in Materials and Methods.
Other laboratories have previously shown that null alleles of MSH2, MSH3, RAD1, and RAD10 are epistatic to one another with respect to recombination, indicating that they are members of the same recombination pathway (29, 58, 66). Similarly, our data indicate that null alleles of MSH2 and RAD1 are epistatic to one another with respect to SSR because the H−/K− ratios from the rad1 msh2 double mutant (Fig. 5) were not significantly different from the single-mutant ratios (Fig. 2). Intriguingly, the msh2-null allele also had an epistatic relationship with the mre11-null allele (Fig. 5). We further explored the relationships among MSH2, RAD1, and MRE11 by assaying a rad1 mre11 msh2 triple mutant and found that the H−/K− ratios were not significantly different from the rad1 mre11 double-mutant ratios (Fig. 5), indicating that the msh2-null allele has no effect on SSR when RAD1, MRE11, or both are mutated.
Epistasis analysis revealed that the H−/K− ratios from the rad1 hdf1 double mutant were slightly (twofold) lower for the two shortest fragments than those from the rad1 single mutants (Fig. 5). Since the fold effect of the hdf1 allele relative to the wild type is slightly greater (Fig. 3), the relationship between the rad1 and hdf1 alleles with respect to SSR is most likely synergistic, indicating that the function of RAD1 is in a separate, competitive pathway from HDF1. Unfortunately, the extreme genome instability of the rad50 hdf1 double mutant (unpublished results) prevented our analysis of the strain and determination of the relationship between NHEJ and MRE11-, RAD50-, and XRS2-dependent SSR.
Defective processing of broken DNA molecules correlates with SSR deficiency in mre11 mutant strains.
Several laboratories have demonstrated that the mre11-, rad50-, and xrs2-null mutants all exhibit a reduced rate of exonucleolytic degradation of broken DNA molecules (24, 25, 65, 73). In contrast, we previously showed that the rad1-null mutant displays no such change (5). Interestingly, we also found that combining the rad1-null allele with the rad3-G595R allele, which slows degradation, leads to a synergistic decrease in SSR (5). We concluded that blocking the degradation of the ends of DNA molecules reduced SSR in cells that lack Rad1/10 nuclease activity. This suggests that the decrease in SSR observed in the rad1 mre11 double mutant may, in part, be due to a similar combination of defects.
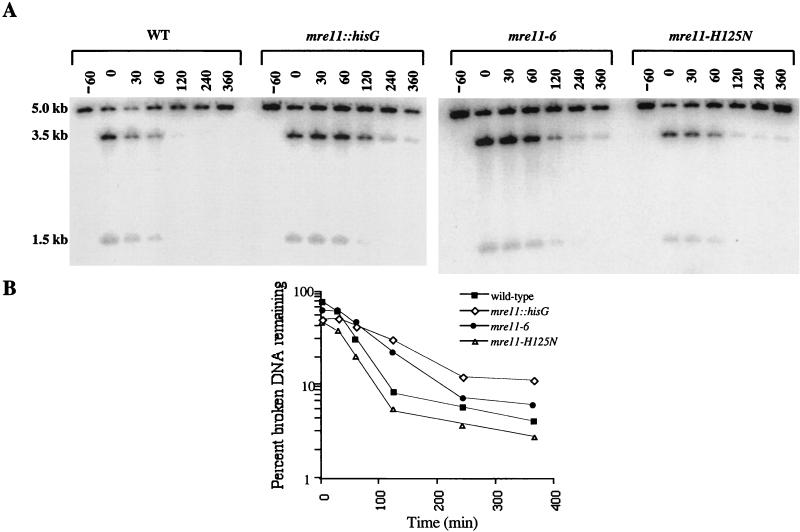
We investigated the link between mitotic DSB stability and SSR in the three mre11 mutants described above by using an assay for measuring the degradation of a single-copy plasmid following digestion in vivo with HO endonuclease (5). Similar to the results presented by others (73) the mre11-null mutant strain displayed a markedly decreased rate of decay of broken plasmid molecules (Fig. 6). The half-life of an HO-digested plasmid was nearly threefold longer in mre11-null mutant cells (82 to 87 min) than in wild-type cells (30 to 34 min). Its half-life in mre11-6 mutant cells was similarly elevated (68 to 77 min). Similar to the results previously described by Moreau et al. (40), the stability of HO-digested DNA molecules in mre11-H125N mutant cells was closer to that of the wild type (37 to 44 min). The different half-lives in these strains could not be accounted for by differences in the repair of the broken molecules by NHEJ (Manthey and Bailis, unpublished). Therefore, the mre11-null and mre11-6 mutants that presented definitive SSR defects as described above (Fig. 4) were also found to possess a defect in the exonucleolytic processing of broken DNA molecules, while the mre11-H125N mutant exhibited significantly less severe defects.
FIG. 6.
Stability of a single-copy plasmid following cutting by HO endonuclease in wild-type (WT) and mre11 mutant cells. (A) Autoradiogram of plasmid DNA linearized in vivo. DNA was prepared from the wild-type (ABT151) strain and the mre11::hisG (ABT380), mre11-H125N (ABT381), and mre11-6 (ABT382) mutant strains carrying plasmids pLAY97 and pGHOT before and at intervals after a brief (30-min) period of HO endonuclease expression. The DNA was digested with NcoI prior to agarose gel electrophoresis and blotting to a nylon membrane. pLAY97 bands were revealed by hybridization with 32P-labeled pBluescript. Digestion of pLAY97 with NcoI before cutting with HO endonuclease yields a 5.0-kb signal. This becomes 3.5- and 1.5-kb signals after cutting with HO. Degradation of HO-cleaved pLAY97 was indicated by changes in the intensity of the 3.5- and 1.5-kb fragments relative to that of the 5.0-kb fragment. (B) Stability of HO-digested plasmid DNA. The stability of HO-digested pLAY97 in wild-type, mre11Æ, mre11-H125N, and mre11-6 cells was determined by dividing the sum of the 3.5- and 1.5-kb signals by the sum of the 5.0-, 3.5-, and 1.5-kb signals, as determined by phosphorimaging. The resulting values were plotted versus time.
DISCUSSION
This report is a partial description of the apparatus required for SSR in budding yeast. In order to characterize the SSR machinery, we used a DNA fragment insertion assay to measure recombination in response to changes in DNA sequence length in a variety of DNA repair and recombination mutant strains. This assay was chosen because of the simplicity of changing DNA sequence length and the similarity of its mechanism to the rescue of broken chromosomes by break-induced recombination (42, 45, 47). Similar to results from other laboratories (50, 60), we found that DNA fragment insertion is completely dependent on RAD52, which is required for most recombination in budding yeast cells (47). This suggests that the pathways that mediate recombination between short sequences are superimposed upon the general recombination apparatus.
Our previous work showed that rad1- and rad10-null mutants display recombination defects that worsen progressively with decreasing DNA sequence length (5). The results from our current experiments confirm this, demonstrating that RAD1 is required for SSR in budding yeast (Fig. 2). Previously, we postulated that Rad1/10 is required for cleavage of the chromosomal target strand displaced by the invading DNA fragment, which creates the opportunity to form a covalent joint between the DNA fragment and genomic target by ligation (5). Symington and colleagues also suggest that Rad1/10 may cleave D loops during plasmid gap repair (68). We further postulated that the requirement for Rad1/10-facilitated covalent joint formation is related to the length of the homologous sequences because the shorter the length of homology, the less stable the heteroduplex connecting the fragment to the target. This instability might necessitate the formation of covalent joints that tether the fragment to the genomic target before the heteroduplex can dissolve, aborting recombination.
The results of the epistasis analysis presented here (Fig. 5) also indicate that another multiprotein complex, M/R/X, whose subunits are encoded by the MRE11, RAD50, and XRS2 genes, occupies a pathway for SSR that acts upon the same class of intermediates processed by Rad1/10 (22), although the possibility that they act sequentially cannot be rigidly excluded (41). M/R/X plays a role in many cellular processes, including mitotic and meiotic recombination, telomere maintenance, NHEJ, and DNA damage signaling (9, 20), and has been shown to possess several nuclease activities in vitro (19, 40, 72, 75). However, Mre11 is the only subunit with intrinsic nuclease activity and can be thought of as the core of a modular nuclease whose activity can be modified by adding or subtracting additional subunits (48, 72). It is unclear which, if any, of the nuclease activities of M/R/X are required for mitotic recombination in budding yeast. Nuclease-defective mre11 alleles, including the mre11-H125N allele that conferred little effect on SSR in our studies (Fig. 4), were previously found to have wild-type levels of mitotic recombination (40, 68), suggesting that Mre11 nuclease activity is not generally required. Similarly, genetic evidence suggests that Mre11 nuclease activity may not play a role in the exonucleolytic digestion of the 5′ strands of DSBs, since several nuclease-defective mre11 alleles, including mre11-H125N, minimally affect DSB processing (Fig. 6) (40). In contrast, every nuclease-defective mre11 mutant that has been tested is also defective for DNA damage signaling (14), suggesting that some sort of nucleolytic digestion of broken DNA molecules may be required to elicit the DNA damage signal in mitotic cells (33, 49). This indicates that the functions of Mre11 in DNA damage signaling and SSR are genetically separate.
Regardless of whether M/R/X plays a direct or an indirect role in the degradation of broken DNA molecules, we have observed a correlation between the effects of three different mre11 alleles on the exonucleolytic degradation of DNA ends and their effects on SSR. We found that both SSR and the stability of DNA ends were significantly altered in mre11-null and mre11-6 mutant cells, while neither was greatly altered in the mre11-H125N mutant (Fig. 4 and 6). This might suggest that Mre11-mediated degradation of DNA ends plays a role in the propagation of SSR. If this is true, however, it is unlikely to be the sole function of Mre11 in SSR, as several TFIIH/NER mutants defective for the degradation of DNA ends exhibit increased SSR (5, 6, 32, 35). We favor an alternative scenario in which the decreased degradation in the mre11 mutants may be indicative of the failure of a whole suite of proteins to act at the DNA ends, including those that propagate SSR. In support of this notion, recent evidence suggests that RAD50 may be required to load Msh2 onto certain recombination intermediates at the ends of DNA molecules (16).
Interestingly, the mre11-H125N mutant displayed a defect in recombination with longer fragments that was very similar to the defects observed in the mre11-null and mre11-6 mutants, while recombination with the shortest fragment was far more defective in the mre11-null and mre11-6 mutants (Fig. 4). These results suggest that MRE11 controls both general recombination and SSR and that these controls are separate. Further, it suggests that the recombination phenotypes of the mre11-null and mre11-6 mutants are due to defects in both controls.
The msh2 and msh3 single mutants had profound SSR defects, equivalent to those of the rad1, mre11, and rad50 mutants (Fig. 2 and 3), indicating that MSH2 and MSH3 play an important role in SSR. This suggests that the Msh2/3 complex may play a role in SSR that is similar to the role proposed in other studies of recombination (58, 66). Consistent with these studies, we observed that the msh2 and rad1 mutations were epistatic to one another with respect to SSR (Fig. 5), suggesting that the Rad1/10 and Msh2/3 complexes occupy the same pathway for SSR (22). Unexpectedly, we found that the msh2- and mre11-null alleles were also epistatic (Fig. 5). Since the evidence suggests that Rad1/10 and M/R/X are in separate pathways and loss of MSH2 had no impact on this relationship in a rad1 mre11 msh2 triple mutant (Fig. 5), we speculate that our SSR assays may involve two processes, a dominant process that is dependent on Rad1/10, M/R/X, and Msh2/3 and a subsidiary process dependent only on Rad1/10 and M/R/X. It will be of further interest to discover whether MSH6 is involved in this control, as the Msh2/6 heterodimer has been shown to bind to recombination intermediates (37). It will also be important to determine if recruitment of additional factors by either of the heterodimers is required for SSR (8).
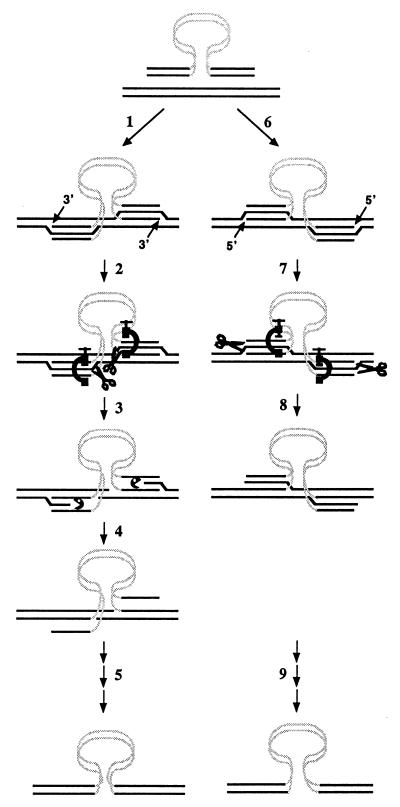
We offer the following model for the control of SSR based on the conclusions from current and previous experiments (Fig. 7). We suggest that SSR can begin by either 3′ or 5′ strand invasion of the genomic target sequences by the DNA fragment, generating two different types of substrates. We believe that 5′ strand invasion may occur because our previous studies of SSR in rad27 mutant strains strongly suggest that undegraded 5′ ends frequently participate in heteroduplex formation (44). Further, our observations that several TFIIH/NER mutations that retard 5′ strand resection significantly increase SSR suggest that 5′ strand invasion may be important (32, 35). Following strand invasion, Rad1/10 acts to cleave the displaced genomic target strand, as discussed previously (5). Recent in vitro experiments by the Sung laboratory have shown that the Mre11-Rad50 heterodimer can endonucleolytically cleave a stem-loop structure on the 5′ side of the loop (72), which is the same polarity with which Rad1/10 cleaves displaced DNA strands (7). We hypothesize that M/R/X might cleave genomic target strands displaced by the invading DNA fragment in a manner similar to that of Rad1/10, explaining the synergism between the rad1-null and mre11-null mutations. Alternatively, M/R/X might facilitate the action of another, as yet unidentified, nuclease. M/R/X has been shown to facilitate Dnl4-catalyzed end joining in vitro, suggesting that it may act as a platform for various enzymatic activities at the ends of DNA molecules (13). Cleavage of the target strand potentiates ligation of the fragment strand to the target either directly or subsequent to 5′ strand resection. The formation of a covalent joint between the fragment and target should stabilize their relationship and may be the rate-limiting step in SSR.
FIG. 7.
Model for Rad1/10, M/R/X, and Msh2/3 action during SSR. The his3::URA3 fragments (black lines, HIS3 sequences; shaded lines, URA3 sequences) align with the HIS3 genomic target (solid black lines). Either the 3′ or the 5′ end can be used for Rad52-dependent strand invasion. (Step 1) Invasion by the 3′ end displaces the target strand of like polarity and forms a short, unstable heteroduplex. (Step 2) Msh2/3 (C clamp) binds at the junction between homologous (HIS3) and nonhomologous (URA3) sequences and stabilizes the D loop. Rad1/10, M/R/X, or another, M/R/X-dependent, nuclease (scissors) cleaves on the stabilized 5′ side of the D loop, creating a free 5′ end that is susceptible to exonucleolytic degradation. (Step 3) Exonucleolytic degradation (pacman) brings the 5′ end of the target strand in register with the 3′ end of the invading 3′ strand of the fragment. (Step 4) Ligation of the 3′ end of the fragment creates a covalent link between the DNA fragment and the target that resists unraveling and termination of recombination. (Step 5) Strand invasion, nuclease cleavage, and ligation tether the fragment ends to the target sequences, completing fragment insertion. (Step 6) Invasion by the 5′ end displaces the target strand of like polarity and forms a short, unstable heteroduplex. (Step 7) Msh2/3 (C clamp) binds at the junction between homologous and nonhomologous sequences. However, Rad1/10, M/R/X, or an M/R/X-dependent nuclease (scissors) cleaves on the opposite, 5′, side of the D loop, where Msh2/3 may not have any effect. Endonucleolytic cleavage creates a ligatable end. (Step 8) Ligation of the 5′ end of the fragment to the genomic target creates a covalent link that should prevent loss of the recombination event by heteroduplex unraveling. (Step 9) Subsequent nuclease cleavage and ligation or another round of strand invasion, nuclease cleavage, and ligation ties the remaining fragment ends to the target sequences, completing fragment insertion.
The epistasis results suggest that Msh2/3 plays a more limited role in SSR than do Rad1/10 and M/R/X but one that overlaps those of both. We suggest that this may be due to an interaction between Msh2/3 and Rad1/10, M/R/X, or a M/R/X-associated nuclease following 3′ strand invasion but not 5′ strand invasion (Fig. 7). When the 3′ strand invades, we have proposed that the nucleases would cleave the displaced target strand near the position homologous to the junction between the HIS3 and URA3 segments on the incoming fragment. We further suggest that Msh2/3 bound at the junction between homologous and nonhomologous sequences, as suggested by Alani and colleagues (16), may stabilize the proximal arm of the D loop and facilitate endonucleolytic cleavage. When the 5′ strand invades, however, the nucleases would cleave the distal arm of the D loop, opposite from where Msh2/3 is bound. At that position, Msh2/3 may not play a role in determining the efficiency of endonucleolytic cleavage.
The presence of an apparatus committed to the maintenance of recombination between short sequences but not long sequences suggests that SSR may play an important role in S. cerevisiae. We suggest that SSR may be ideally suited for rescuing DSBs created in the wake of replication fork failure by sister chromatid recombination. Not requiring extensive heteroduplex formation may increase the rate of these local interactions and minimize the likelihood of potentially deleterious interactions with duplicate sequences elsewhere in the genome. Interestingly, a number of studies have linked the RAD1 and RAD50 genes to sister chromatid recombination, supporting a potential link with SSR (21, 28, 39). Future studies will focus on determining which interactions among the components of the SSR apparatus are the most critical in budding yeast in order to develop a better understanding of how eukaryotic cells maintain genome stability.
Acknowledgments
This work was supported by U.S. Public Health Service grant GM57484 to A.M.B., as well as funds from the Beckman Research Institute of the City of Hope and the City of Hope National Medical Center.
We thank J. McDonald, E. Alani, N. Kleckner, R. D. Kolodner, K. Shannon, D. T. Weaver, S. Moreau, L. Symington, D. Bressan, J. Petrini, J. Nickoloff, and B. Jensen for strains and plasmids. We also thank P. Sung, J. Petrini, J. Haber, and the members of the Bailis laboratory for stimulating discussions. Finally, we thank D. Bishop and several anonymous reviewers for insightful comments on the manuscript.
REFERENCES
- 1.Ahn, B. Y., K. J. Dornfeld, T. J. Fagrelius, and D. M. Livingston. 1988. Effect of limited homology on gene conversion in a Saccharomyces cerevisiae plasmid recombination system. Mol. Cell. Biol. 8:2442-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alani, E., L. Cao, and N. Kleckner. 1987. A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics 116:541-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alani, E., R. Padmore, and N. Kleckner. 1990. Analysis of wild-type and rad50 mutants of yeast suggests an intimate relationship between meiotic chromosome synapsis and recombination. Cell 61:419-436. [DOI] [PubMed] [Google Scholar]
- 4.Alani, E., S. Subbiah, and N. Kleckner. 1989. The yeast RAD50 gene encodes a predicted 153-kD protein containing a purine nucleotide-binding domain and two large heptad-repeat regions. Genetics 122:47-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bailis, A. M., and S. Maines. 1996. Nucleotide excision repair gene function in short-sequence recombination. J. Bacteriol. 178:2136-2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bailis, A. M., S. Maines, and M. C. Negritto. 1995. The essential helicase gene RAD3 suppresses short-sequence recombination in Saccharomyces cerevisiae. Mol. Cell. Biol. 15:3998-4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bardwell, A. J., L. Bardwell, A. E. Tomkinson, and E. C. Friedberg. 1994. Specific cleavage of model recombination and repair intermediates by the yeast Rad1-Rad10 DNA endonuclease. Science 265:2082-2085. [DOI] [PubMed] [Google Scholar]
- 8.Bowers, J., P. T. Tran, R. M. Liskay, and E. Alani. 2000. Analysis of yeast MSH2-MSH6 suggests that the initiation of mismatch repair can be separated into discrete steps. J. Mol. Biol. 302:327-338. [DOI] [PubMed] [Google Scholar]
- 9.Bressan, D. A., B. K. Baxter, and J. H. J. Petrini. 1999. The Mre11-Rad50 Xrs2 protein complex facilitates homologous recombination-based double-strand break repair in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:7681-7687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bressan, D. A., H. A. Olivares, B. E. Nelms, and J. H. J. Petrini. 1998. Alteration of N-terminal phosphoesterase signature motifs inactivates Saccharomyces cerevisiae Mre11. Genetics 150:591-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Britten, R. J., and D. E. Kohne. 1968. Repeated sequences in DNA: hundreds of thousands of copies of DNA sequences have been incorporated into the genomes of higher organisms. Science 161:529-540. [DOI] [PubMed] [Google Scholar]
- 12.Burke, D., D. Dawson, and T. Stearns (ed.). 2000. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 13.Chen, L., K. Trujillo, W. Ramos, P. Sung, and A. E. Tomkinson. 2001. Promotion of Dnl4-catalyzed DNA end-joining by the Rad50/Mre11/Xrs2 and Hdf1 and Hdf2 complexes. Cell 8:1105-1115. [DOI] [PubMed] [Google Scholar]
- 14.D'Amours, D., and S. P. Jackson. 2001. The yeast Xrs2 complex functions in S phase checkpoint regulation. Genes Dev. 15:2238-2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deininger, P. L., and M. A. Batzer. 1999. Alu repeats and human disease. Mol. Genet. Metab. 67:183-193. [DOI] [PubMed] [Google Scholar]
- 16.Evans, E., N. Sugawara, J. E. Haber, and E. Alani. 2000. The Saccharomyces cerevisiae Msh2 mismatch repair protein localizes to recombination intermediates in vivo. Mol. Cell 5:789-799. [DOI] [PubMed] [Google Scholar]
- 17.Fishman-Lobell, J., and J. E. Haber. 1992. Removal of nonhomologous DNA ends in double-strand break recombination: the role of the yeast ultraviolet repair gene RAD1. Science 258:480-484. [DOI] [PubMed] [Google Scholar]
- 18.Frank, G., J. Qiu, M. Y. Somsouk, Y. Weng, L. Somsouk, J. P. Nolan, and B. Shen. 1998. Partial functional deficiency of E160D flap endonuclease-1 mutant in vitro and in vivo is due to defective cleavage of DNA substrates. J. Biol. Chem. 273:33064-33072. [DOI] [PubMed] [Google Scholar]
- 19.Furuse, M., Y. Nagase, M. Lopes, C. Lucca, M. Ferrari, G. Liberi, M. Muzi Falconi, and P. Plevani. 2000. Distinct roles of two separable in vitro activities of yeast Mre11 in mitotic and meiotic recombination. EMBO J. 17:6412-6425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haber, J. E. 1998. The many interfaces of Mre11. Cell 95:583-586. [DOI] [PubMed] [Google Scholar]
- 21.Hartsuiker, E., E. Vaessen, A. M. Carr, and J. Kohli. 2001. Fission yeast Rad50 stimulates sister chromatid recombination and links cohesion with repair. EMBO J. 20:6660-6671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haynes, R. H., and B. A. Kunz. 1981. DNA repair and mutagenesis in yeast, p. 371-414. In J. N. Strathern, E. W. Jones, and J.R. Broach (ed.), The molecular biology of the yeast Saccharomyces: life cycle and inheritance. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 23.Holmes, A., and J. E. Haber. 1999. Double-strand break repair requires both leading and lagging strand DNA polymerases. Cell 96:415-424. [DOI] [PubMed] [Google Scholar]
- 24.Ivanov, E. L., V. G. Korolev, and F. Fabre. 1992. XRS2, a DNA repair gene of Saccharomyces cerevisiae, is needed for meiotic recombination. Genetics 132:651-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ivanov, E. L., N. Sugawara, C. I. White, F. Fabre, and J. E. Haber. 1994. Mutations in XRS2 and RAD50 delay but do not prevent mating-type switching in Saccharomyces cerevisiae. Mol. Cell. Biol. 14:3414-3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jinks-Robertson, S., M. Michelitch, and S. Ramcharan. 1993. Substrate length requirements for efficient mitotic recombination in Saccharomyces cerevisiae. Mol. Cell. Biol. 13:3937-3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson, R. E., G. K. Kovvali, L. Prakash, and S. Prakash. 1998. Requirement of the yeast Rth1 5′ to 3′ exonuclease for the stability of simple repetitive DNA. Science 269:238-239. [DOI] [PubMed] [Google Scholar]
- 28.Kadyk, L. C., and L. H. Hartwell. 1993. Replication-dependent sister chromatid recombination in rad1 mutants of Saccharomyces cerevisiae. Genetics 133:469-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kirkpatrick, D. T., and T. D. Petes. 1997. Repair of DNA loops involves DNA-mismatch and nucleotide-excision repair proteins. Nature 387:929-931. [DOI] [PubMed] [Google Scholar]
- 30.Klein, H. 1989. Different types of recombination events are controlled by the RAD1 and RAD52 genes of Saccharomyces cerevisiae. Genetics 120:367-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kramer, K. M., J. A. Brock, K. Bloom, J. K. Moore, and J. E. Haber. 1994. Two different types of double-strand breaks in Saccharomyces cerevisiae are repaired by similar RAD52-independent, nonhomologous recombination events. Mol. Cell. Biol. 14:1293-1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee, B.-S., L. Bi, D. J. Garfinkel, and A. M. Bailis. 2000. Nucleotide excision repair/TFIIH helicases Rad3 and Ssl2 inhibit short-sequence recombination and Ty1 retrotransposition by similar mechanisms. Mol. Cell. Biol. 20:2436-2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee, S. E., J. K. Moore, A. Holmes, K. Umezu, R. D. Kolodner, and J. E. Haber. 1998. Saccharomyces Ku70, Mre11/Rad50, and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell 94:399-409. [DOI] [PubMed] [Google Scholar]
- 34.Lieber, M. 1997. The FEN-1 family of structure-specific nucleases in eukaryotic DNA replication, recombination and repair. Bioessays 19:233-240. [DOI] [PubMed] [Google Scholar]
- 35.Maines, S., M. C. Negritto, X. Wu, G. M. Manthey, and A. M. Bailis. 1998. Novel mutations in the RAD3 and SSL1 genes perturb genome stability by stimulating recombination between short repeats in Saccharomyces cerevisiae. Genetics 150:963-976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Manivasakam, P., S. C. Weber, J. McElver, and R. H. Schiestl. 1995. Micro-homology mediated PCR targeting in Saccharomyces cerevisiae. Nucleic Acids Res. 23:2799-2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marsischky, G. T., S. Lee, J. Griffith, and R. D. Kolodner. 1999. Saccharomyces cerevisiae MSH2/6 complex interacts with Holliday junctions and facilitates their cleavage by phage resolution enzymes. J. Biol. Chem. 274:7200-7206. [DOI] [PubMed] [Google Scholar]
- 38.Milne, G. T., S. Jin, K. B. Shannon, and D. T. Weaver. 1996. Mutations in two Ku homologs define a DNA end-joining repair pathway in Saccharomyces cerevisiae. Mol. Cell. Biol. 16:4189-4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moore, J. K., and J. E. Haber. 1996. Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Mol. Cell. Biol. 16:2164-2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moreau, S., J. R. Ferguson, and L. S. Symington. 1999. The nuclease activity of Mre11 is required for meiosis but not mating type switching, end joining, or telomere maintenance. Mol. Cell. Biol. 19:556-566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morrison, A., A. L. Johnson, L. H. Johnston, and A. Sugino. 1993. Pathway correcting DNA replication errors in Saccharomyces cerevisiae. EMBO J. 12:1467-1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morrow, D. M., C. Connelly, and P. Hieter. 1997. “Break-copy” duplication: a model for chromosome fragment formation in Saccharomyces cerevisiae. Genetics 147:371-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nairz, K., and F. Klein. 1997. Mre11-S, a yeast mutation that blocks double-strand-break processing and permits nonhomologous synapsis in meiosis. Genes Dev. 11:2272-2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Negritto, M. T., J. Qiu, D. O. Ratay, B. Shen, and A. M. Bailis. 2001. Novel function of Rad27 (FEN-1) in short-sequence recombination. Mol. Cell. Biol. 21:2349-2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Negritto, M. T., X. Wu, T. Kuo, S. Chu, and A. M. Bailis. 1997. Influence of DNA sequence identity on efficiency of targeted gene replacement. Mol. Cell. Biol. 17:278-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nickoloff, J. A., E. Y. Chen, and F. Heffron. 1986. A 24-base-pair DNA sequence from the MAT locus stimulates intergenic recombination in yeast. Proc. Natl. Acad. Sci. USA 83:7831-7835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paques, F., and J. E. Haber. 1999. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63:349-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Paull, T. T., and M. Gellert. 1999. Nbs1 potentiates ATP-driven DNA unwinding and endonuclease cleavage by the Mre11/Rad50 complex. Genes Dev. 13:1276-1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Petrini, J. H. J. 2000. The Mre11 complex and ATM: collaborating to navigate S phase. Curr. Opin. Cell Biol. 12:293-296. [DOI] [PubMed] [Google Scholar]
- 50.Orr-Weaver, T. L., J. W. Szostak, and R. J. Rothstein. 1981. Yeast transformation: a model system for the study of recombination. Proc. Natl. Acad. Sci. USA 78:6354-6358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reagan, M. S., C. Pittenger, W. Siede, and E. C. Friedberg. 1995. Characterization of a mutant strain of Saccharomyces cerevisiae with a deletion of the RAD27 gene, a structural homolog of the RAD2 nucleotide excision repair gene. J. Bacteriol. 177:364-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reenan, R. A. G., and R. D. Kolodner. 1992. Isolation and characterization of two Saccharomyces cerevisiae genes encoding homologs of the bacterial HexA and MutS mismatch repair proteins. Genetics 132:963-973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reynolds, R. J., and E. C. Friedberg. 1981. Molecular mechanisms of pyrimidine dimer excision in Saccharomyces cerevisiae: incision of ultraviolet irradiated deoxyribonucleic acid in vivo. J. Bacteriol. 146:692-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ronne, H., and R. Rothstein. 1988. Mitotic sectored colonies: evidence of heteroduplex DNA formation during direct repeat recombination. Proc. Natl. Acad. Sci. USA 85:2696-2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rothstein, R. 1991. Targeting, disruption, replacement, and allele rescue: integrative DNA transformation in yeast. Methods Enzymol. 194:281-301. [DOI] [PubMed] [Google Scholar]
- 56.Rothstein, R., C. Helms, and N. Rosenberg. 1987. Concerted deletions and inversions are caused by mitotic recombination between delta sequences in Saccharomyces cerevisiae. Mol. Cell. Biol. 7:1198-1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 58.Saparbaev, M., L. Prakash, and S. Prakash. 1996. Requirement of mismatch repair genes MSH2 and MSH3 in the RAD1-RAD10 pathway of mitotic recombination in Saccharomyces cerevisiae. Genetics 142:727-736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schiestl, R. H., and T. D. Petes. 1991. Integration of DNA fragments by illegitimate recombination in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 88:7585-7589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schiestl, R. H., and S. Prakash. 1988. RAD1, an excision repair gene of Saccharomyces cerevisiae, is also involved in recombination. Mol. Cell. Biol. 8:3619-3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schiestl, R. H., and S. Prakash. 1990. RAD10, an excision repair gene of Saccharomyces cerevisiae, is involved in the RAD1 pathway of mitotic recombination. Mol. Cell. Biol. 10:2485-2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schiestl, R. H., J. Zhu, and T. D. Petes. 1994. Effect of mutations in genes affecting homologous recombination on restriction enzyme-mediated and illegitimate recombination in Saccharomyces cerevisiae. Mol. Cell. Biol. 14:4493-4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schild, D., B. Konforti, C. Perez, W. Gish, and R. K. Mortimer. 1983. Isolation and characterization of yeast DNA repair genes. I. Cloning of the RAD52 gene. Curr. Genet. 7:85-92. [DOI] [PubMed] [Google Scholar]
- 64.Sommers, C. H., E. J. Miller, B. Dujon, S. Prakash, and L. Prakash. 1995. Conditional lethality of null mutations in RTH1 that encodes the yeast counterpart of a mammalian 5′ to 3′ exonuclease required for lagging strand synthesis in reconstituted systems. J. Biol. Chem. 270:4193-4196. [DOI] [PubMed] [Google Scholar]
- 65.Sugawara, N., and J. E. Haber. 1992. Characterization of double-strand break-induced recombination: homology requirements and single-stranded DNA formation. Mol. Cell. Biol. 12:563-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sugawara, N., F. Paques, M. Colaiacovo, and J. E. Haber. 1997. Role of Saccharomyces cerevisiae Msh2 and Msh3 repair proteins in double-strand break-induced recombination. Proc. Natl. Acad. Sci. USA 94:9214-9219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sung, P., P. Reynolds, L. Prakash, and S. Prakash. 1993. Purification and characterization of the Saccharomyces cerevisiae Rad1/Rad10 endonuclease. J. Biol. Chem. 26:23691-23699. [PubMed] [Google Scholar]
- 68.Symington, L. S., L. E. Kang, and S. Moreau. 2000. Alteration of gene conversion tract length and associated crossing over during plasmid gap repair in a nuclease-deficient strain of Saccharomyces cerevisiae. Nucleic Acids Res. 28:4649-4656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Szankasi, P., C. Gysler, U. Zehntner, U. Leupold, J. Kohli, and P. Munz. 1986. Mitotic recombination between dispersed but related tRNA genes of S. pombe generates a reciprocal translocation. Mol. Gen. Genet. 202:394-402. [Google Scholar]
- 70.Thomas, B. J., and R. Rothstein. 1989. The genetic control of direct-repeat recombination in Saccharomyces: the effect of rad52 and rad1 on mitotic recombination of a GAL10 transcriptionally regulated gene. Genetics 123:725-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tomkinson, A. E., A. J. Bardwell, L. Bardwell, N. J. Tappe, and E. C. Friedberg. 1993. Yeast DNA repair and recombination proteins Rad1 and Rad10 constitute a single-stranded-DNA endonuclease. Nature 362:860-862. [DOI] [PubMed] [Google Scholar]
- 72.Trujillo, K. M., and P. Sung. 2001. DNA structure-specific nuclease activities in the Saccharomyces cerevisiae Rad50-Mre11 complex. J. Biol. Chem. 276:35458-35464. [DOI] [PubMed] [Google Scholar]
- 73.Tsubouchi, H., and H. Ogawa. 1998. A novel mre11 mutation impairs processing of double-strand breaks of DNA during both mitosis and meiosis. Mol. Cell. Biol. 18:260-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tsukamoto, Y., J. Kato, and H. Ikeda. 1996. Hdf1, a yeast Ku-protein homologue, is involved in illegitimate recombination, but not in homologous recombination. Nucleic Acids Res. 24:2067-2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Usui, T., T. Ohta, H. Oshiumi, J. Tomizawa, H. Ogawa, and T. Ogawa. 1998. Complex formation and functional versatility of Mre11 of budding yeast in recombination. Cell 95:705-716. [DOI] [PubMed] [Google Scholar]
- 76.Wilcox, D. R., and L. Prakash. 1981. Incision and postincision step of pyrimidine dimer removal in excision-defective mutants of Saccharomyces cerevisiae. J. Bacteriol. 148:618-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wilson, T. E., U. Grawunder, and M. R. Lieber. 1997. Yeast DNA ligase IV mediates non-homologous DNA end joining. Nature 388:495-498. [DOI] [PubMed] [Google Scholar]