Abstract
Metazoan NXF1/p15 heterodimers promote export of bulk mRNA through nuclear pore complexes (NPC). NXF1 interacts with the NPC via two distinct structural domains, the UBA-like domain and the NTF2-like scaffold, which results from the heterodimerization of the NTF2-like domain of NXF1 with p15. Both domains feature a single nucleoporin-binding site, and they act synergistically to promote NPC translocation. Whether the NTF2-like scaffold (and thereby p15) contributes only to NXF1/NPC association or is also required for other functions, e.g., to impart directionality to the export process by regulating NXF1/NPC or NXF1/cargo interactions, remains unresolved. Here we show that a minimum of two nucleoporin-binding sites is required for NXF1-mediated export of cellular mRNA. These binding sites can be provided by an NTF2-like scaffold followed by a UBA-like domain (as in the wild-type protein) or by two NTF2-like scaffolds or two UBA-like domains in tandem. In the latter case, the export activity of NXF1 is independent of p15. Thus, as for the UBA-like domain, the function of the NTF2-like scaffold is confined to nucleoporin binding. More importantly, two copies of either of these domains are sufficient to promote directional transport of mRNA cargoes across the NPC.
Nucleocytoplasmic transport of macromolecules occurs through nuclear pore complexes (NPCs) and is mediated by saturable transport receptors. The receptors bind to nucleoporins, the components of the NPC, and to cargo molecules that need to be translocated across the pore. While the vast majority of transport receptors are members of a conserved family of proteins known as importins or exportins (or karyopherins), export of bulk mRNA is thought to be mediated by members of the conserved family of NXF proteins (reviewed in references 9, 26, and 27).
The yeast genome encodes a single NXF protein, Mex67p, but there are two NXFs in Caenorhabditis elegans and four in Drosophila melanogaster and Homo sapiens (14, 18, 33, 37, 41, 42). Of these, Saccharomyces cerevisiae Mex67p, C. elegans NXF1, and D. melanogaster NXF1 have been shown to be essential for the export of bulk polyadenylated RNA [poly(A)+ RNA] to the cytoplasm (15, 33, 37, 40). In Schizosaccharomyces pombe, Mex67p is implicated in but not essential for mRNA export (42).
Human NXF1 is also known as TAP and will be referred to as TAP. TAP is directly implicated in the nuclear export of simian type D retroviral RNAs bearing the constitutive transport element (CTE) (7, 12, 19). A role for TAP in the export of cellular mRNA is suggested by the observation that in Xenopus laevis oocytes, titration of TAP with an excess of CTE RNA leads to an mRNA export block (29, 32). Consistently, overexpression of TAP in human cultured cells or microinjection of the recombinant protein into Xenopus oocytes stimulates export of mRNAs that are otherwise exported inefficiently (8, 13, 14, 41).
TAP consists of two functional domains: an N-terminal cargo-binding domain and a C-terminal NPC-binding domain (Fig. 1). The cargo-binding domain comprises an N-terminal fragment for which no structural information is available, an RNP-type RNA-binding domain, and a leucine-rich repeat domain (24). The cargo-binding domain binds directly to the CTE RNA, exhibits a general affinity for RNA, and interacts with adaptor proteins that are thought to recruit TAP to cellular mRNA (7, 19, 21, 24; reviewed in reference 9).
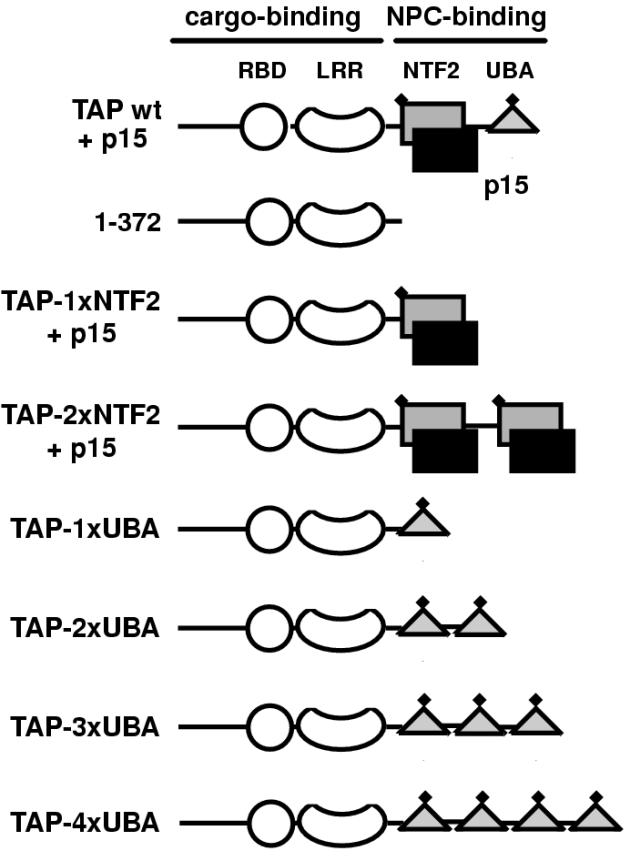
FIG. 1.
Structural and functional domains of human TAP bound to p15. The cargo-binding domain of TAP (residues 1 to 372) consists of an RNP-type RNA-binding domain (RBD, residues 119 to 198), a leucine-rich repeat domain (LRR, residues 203 to 362), and a less well conserved region N-terminal to the RBD for which no structural information is available (residues 1 to 119). The NPC-binding domain of TAP consists of the NTF2-like domain (grey square, residues 371 to 551) bound to p15 (black square) and a UBA-like domain (triangle, residues 563 to 619). Each of these domains features a nucleoporin-binding site (solid diamond).
The NPC-binding domain of TAP (residues 371 to 619) consists of two distinct structural domains connected by a flexible linker: the NTF2-like domain, which is related to nuclear transport factor 2 (NTF2), and a C-terminal region related to ubiquitin-associated (UBA) domains (the so-called UBA-like domain) (Fig. 1) (10, 11, 36). The NTF2-like domain of TAP interacts with p15 to form a compact heterodimer with an overall structure similar to that of the NTF2 homodimer (Fig. 1) (10, 21, 36).
With the exception of D. melanogaster NXF4, the NTF2-like domain is conserved in all members of the NXF family, including S. cerevisiae Mex67p, even though p15 is absent in budding yeast (14, 15, 21, 36). The NTF2-like domain of S. cerevisiae Mex67p heterodimerizes instead with Mtr2p, a putative p15 analogue (21, 35). The UBA-like domain, in contrast, is less well conserved among members of the NXF family and is absent in D. melanogaster NXF4, C. elegans NXF2, H. sapiens NXF3, and H. sapiens NXF5 (14, 15, 18, 37, 41).
Although no structural information is available on the binding of nucleoporins to the UBA-like domain, a hydrophobic surface patch that affects NPC association of this domain was identified by site-directed mutagenesis (11). Single point mutations in this domain and a deletion of the entire domain have a similar effect on nucleoporin binding, suggesting that this domain provides a single nucleoporin-binding site (10, 11). The recently solved structure of the NTF2-like domain of TAP bound to p15 revealed that this heterodimeric NTF2-like scaffold also features a single nucleoporin-binding site located at the TAP side of the heterodimer (10). A clear implication of this structural study is that p15 contributes indirectly to nucleoporin binding by TAP by maintaining the proper folding of the NTF2-like domain (10).
Formation of TAP/p15 heterodimers is strictly required for TAP-mediated export of RNA cargoes that do not carry a CTE (8, 10, 13, 14, 39). CTE-bearing RNAs, in contrast, can be exported by TAP mutants that are impaired in p15 binding both in Xenopus oocytes and in cultured cells (1, 20). An essential role for metazoan p15 in the export of cellular mRNA was recently reported in Drosophila cells. In these cells, depletion of p15 by double-stranded RNA (dsRNA) interference inhibits growth and results in a strong nuclear accumulation of poly(A)+ RNA (15, 39). The growth arrest and the mRNA export block are observed in spite of the fact that D. melanogaster NXF1 is neither degraded nor mislocalized in cells depleted of p15 (15). Therefore, in the absence of p15, D. melanogaster NXF1 cannot perform its essential mRNA export function.
Despite the essential role of p15 and therefore of the NTF2-like domain of NXF1 in the export of cellular mRNA, it is still unclear whether this domain contributes only to the nucleoporin-binding activity of NXF1 or whether it would also have additional functions, such as being required for cargo binding and/or NXF1 recycling. Moreover, it has been proposed that p15 may impose a directionality on the export process by modulating the overall affinity of TAP/NXF1 for nucleoporins (22, 23, 39). In this study, we generated TAP derivatives that lack the NTF2-like domain but have two or more UBA-like domains. These proteins stimulate the export of a variety of RNA cargoes both in cultured cells and in Xenopus oocytes. This stimulation of export is independent of p15. A derivative of D. melanogaster NXF1 having two UBA-like domains but no NTF2-like domain partially restored mRNA export in Drosophila cells depleted of either endogenous p15 or NXF1. These results indicate that, as for the UBA-like domain, the role of the NTF2-like scaffold is confined to nucleoporin binding.
MATERIALS AND METHODS
Plasmids.
Most of the plasmids used in this study have been described before (8). TAP-1xUBA carries a deletion of amino acid residues 372 to 551, encompassing the NTF2-like domain. TAP-1xNTF2 carries a deletion of residues 567 to 613, encompassing the UBA-like domain. In previous studies, the equivalent constructs were referred to as TAPΔNTF2 and TAPΔUBA, respectively (8, 10).
To generate TAP-2xNTF2, first a PCR-amplified DNA fragment encoding TAP-1xNTF2 (amino acid residues 1 to 551) was cloned between the NcoI and KpnI sites of the pGEXCS vector. Second, a PCR-amplified DNA fragment encoding the NTF2-like domain of TAP (residues 372 to 551) was cloned at the 3′ end of TAP-1xNTF2 between the KpnI site and a BamHI site of pGEXCS. To generate TAP-2xUBA, first a PCR-amplified DNA fragment encoding TAP-1xUBA was cloned between the NcoI and KpnI sites of the pGEXCS vector. Second, a PCR-amplified DNA fragment encoding the UBA-like domain of TAP (residues 551 to 619) was cloned at the 3′ end of TAP-1xUBA between the KpnI site and a BamHI site of pGEXCS. TAP-3xUBA was generated by in-frame insertion of a PCR-generated DNA encoding one UBA at the unique KpnI site of pGEXCS-TAP-2xUBA. The 5′ PCR primer used to amplify the UBA-like domain also introduced an in-frame NotI restriction site to enable us to determine the orientation of the cDNA fragment following insertion. TAP-4xUBA was generated by in-frame insertion of a PCR-generated DNA encoding one UBA at the unique NotI site of pGEXCS-TAP-3xUBA.
Expression of recombinant proteins.
TAP and TAP mutants were expressed as glutathione S-transferase (GST) fusions in the Escherichia coli BL21-codonPlus strain (Stratagene) with the corresponding pGEXCS plasmids. TAP and TAP-1xNTF2 were coexpressed with untagged p15-1 protein. Recombinant proteins were purified as described previously (12). For oocyte injections, recombinant proteins were dialyzed against 1.5× phosphate-buffered saline (PBS) supplemented with 10% glycerol. TAP and TAP mutants were expressed in human cultured cells as fusions with enhanced green fluorescent protein (EGFP). To this end, cDNA fragments encoding these proteins were excised from the corresponding pGEXCS constructs as NarI-BamHI fragments and cloned into the vector pEGFP-C1 (Clontech) between the AccI and BamHI restriction sites. p15-1 was expressed with an N-terminal tag consisting of two immunoglobulin-binding domains from protein A of Staphylococcus aureus (zz tag) as described before (plasmid pEGFP-N3-zzp15-1) (8).
DNA transfections, CAT assays, and RNase protection.
DNA transfections and chloramphenicol acetyltransferase (CAT) assays were performed as described before (8). Human 293 cells were transfected with Polyfect transfection reagent (Qiagen) according to the manufacturer's instructions. The transfected DNA mixture consisted of 0.25 μg of the CAT reporter plasmid pCMV128 (16), 0.5 μg of pEGFP-C1 plasmid encoding TAP or TAP mutants, and, when indicated, 0.5 μg of pEGFP-N3 plasmid, encoding zzp15. Transfection efficiency was determined by including 0.5 μg of pCH110 plasmid (Pharmacia) encoding β-galactosidase (β-Gal), as β-Gal expression from this vector is not affected by TAP overexpression (8). The total amount of plasmid DNA transfected in each sample was held constant by adding the appropriate amount of the corresponding parental plasmids without insert and was brought to a total of 2 μg by adding pBSSKII plasmid.
When Rev-M10 fusions were tested, the pEGFP-C1 plasmids were replaced by plasmids encoding RevM10 fusions of TAP or TAP mutants (pCMV-HA-RevM10). Plasmid pCMV-HA-RevM10 was used as a negative control. Cells were harvested 48 h after transfection. CAT and β-Gal activities were measured as described previously (8). In all experiments, the induced CAT enzyme expression levels were normalized to the activity of the β-Gal internal control. Protein expression levels were analyzed by Western blot with anti-GFP or antihemagglutinin (anti-HA) antibodies.
For RNase protection assays, total RNA was isolated from transfected cells with Trizol Reagent (Life Technologies). To isolate cytoplasmic RNA, cells were lysed for 30 s in 130 mM NaCl-5 mM KCl-25 mM Tris-0.2% NP-40-0.1% sodium deoxycholate-0.002% dextran sulfate. Following centrifugation for 1 min at 1,000 × g, the supernatant was extracted twice with phenol-chloroform, and the RNA was precipitated. Antisense riboprobes encompassing nucleotides 291 to 411 and 1 to 100 of the β-Gal and CAT open reading frames, respectively, were generated by in vitro transcription. Following DNase digestion, the probes were purified on denaturing 6% acrylamide-7 M urea gels, eluted, precipitated, and resuspended in 100% formamide at 300,000 cpm/μl.
The riboprobes (1 μl each) were mixed with 10 μg of cellular RNA and 40 μg of yeast total RNA in a final volume of 50 μl of hybridization buffer consisting of 40 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid), pH 6.4], 400 mM NaCl, 1 mM EDTA, and 80% formamide. The RNA mixtures were denatured by incubation at 85°C for 5 min. Hybridizations were carried out overnight at 50°C. The next day, 300 μl of RNase digestion buffer [10 mM Tris (pH 8.0), 5 mM EDTA, 300 mM NaCl] was added to each sample. RNase digestions were carried out for 45 min at 37°C with 2 U of Rnase T1 (Roche) and 12 μg of RNase A per sample. Digestions were stopped by adding proteinase K (50 μg per sample) and sodium dodecyl sulfate (SDS) to a final concentration of 0.5%. Samples were further incubated for 15 min at 37°C, extracted with phenol-chloroform, precipitated, resuspended in 80% formamide, and analyzed on a 6% acrylamide-7 M urea denaturing gel, followed by autoradiography.
To determine the subcellular localization of TAP derivatives, the corresponding pEGFP-C1 constructs were used. HeLa cells were transfected with FuGENE6 (Roche). Plasmid pEGFP-N3-zzp15-1 was cotransfected with wild-type TAP and TAP-2xNTF2. Approximately 20 h after transfection, cells were fixed in formaldehyde. Fluorescent in situ hybridization (FISH) was performed essentially as described before (15).
Xenopus oocyte microinjections.
Oocyte injections and analysis of microinjected RNA by denaturing gel electrophoresis and autoradiography were performed as described previously (32). Quantitation was done by FluorImager (Fuji FLA-2000). The concentration of recombinant proteins in the injected samples is indicated in the figure legends. All DNA templates for in vitro synthesis of labeled RNAs have been described (1, 12, 17, 25, 32).
Drosophila cell culture and RNA interference.
A plasmid allowing the expression of GFP-tagged D. melanogaster NXF1 in SL2 cells was generated by inserting the corresponding NXF1 cDNA into a pBS-based vector containing the Drosophila actin promoter upstream of the EGFP cDNA, followed by multiple cloning sites and the BgH1 terminator (pBSactEGFP). D. melanogaster NXF1-1xUBA was derived from plasmid pBSactEGFP-DmNXF1 by deleting the NTF2-like domain (amino acid residues 360 to 547) by the QuikChange site-directed mutagenesis kit (Stratagene). The PCR primers were designed to introduce in-frame NsiI and MfeI sites at position 360. DmNXF1-2xUBA was generated by in-frame insertion of a PCR-generated DNA fragment encoding the UBA-like domain of D. melanogaster NXF1 (residues 611 to 672) within the NsiI site of pBSactEGFP-NXF1-1xUBA. A BamHI site was introduced at the 3′ end of the UBA cDNA in order to determine the orientation of this fragment following insertion.
Stable Drosophila cell lines expressing GFP and GFP fusions of D. melanogaster NXF1, NXF1-1xUBA, and NXF1-2xUBA were established by cotransfecting the corresponding pBSactEGFP constructs with a plasmid encoding puromycin acetyltransferase at a ratio of 5:1. SL2 cells were transfected with Lipofectin (Invitrogen) according to the manufacturer's instructions and selected in medium containing 10 μg of puromycin/ml. Cells expressing the GFP fusions were enriched by fluorescence-activated cell sorting.
dsRNA interference was performed essentially as described before (15). NXF1 dsRNA corresponds to a fragment comprising the NTF2-like domain (amino acids 360 to 546). p15 dsRNA corresponds to the first 350 nucleotides of the D. melanogaster p15 coding region. FISH with an oligo(dT) probe was performed as described previously (15).
RESULTS
A minimum of two nucleoporin-binding sites is required for TAP-mediated nuclear RNA export.
Precise deletion of either of the nucleoporin-binding domains of TAP dramatically reduces its export activity (8, 10). To investigate whether the export activity of TAP mutants having only one nucleoporin-binding domain could be rescued by duplication of the same domain, we generated TAP derivatives having two UBA-like domains in tandem but no NTF2-like domain (TAP-2xUBA) or two NTF2-like domains in tandem but no UBA-like domain (TAP-2xNTF2) (Fig. 1). The export activity of these proteins in cultured cells was measured with the CAT reporter assay as described previously (8). In this assay, TAP expression vectors are cotransfected with the CAT gene encoded by plasmid pCMV128 (Fig. 2A) (16). As an internal control, a plasmid encoding β-Gal was cotransfected (see Materials and Methods).
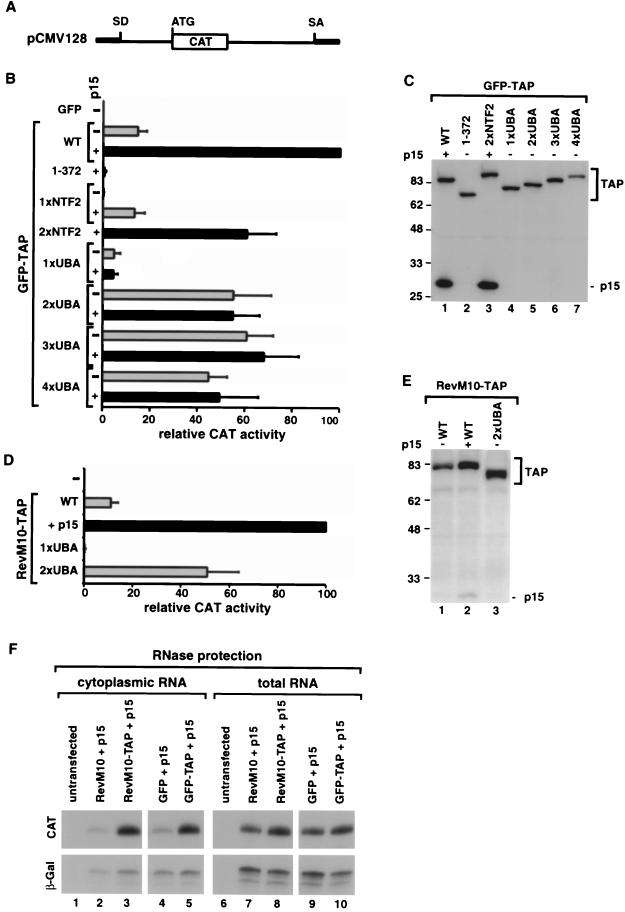
FIG. 2.
Two nucleoporin-binding domains are required for TAP-mediated translocation of RNP cargoes across the NPC. (A) Schematic representation of the cat reporter gene encoded by plasmid pCMV128 (16). SD and SA indicate the splice donor and splice acceptor sites of the intron, respectively. (B) Human 293 cells were transfected with a mixture of plasmids encoding β-Gal (pCH110), CAT (pCMV128), and either GFP alone or GFP fused N-terminally to TAP or a TAP mutant, as indicated on the left. When indicated, a pGFP-N3 derivative encoding zzp15 was cotransfected (+p15, black bars). Cells were collected 48 h after transfection, and β-Gal and CAT activities were determined. CAT activities were normalized to the activities of the β-Gal internal control. The stimulation of cat gene expression by TAP mutants measured in three independent experiments is expressed as a percentage of the activity of wild-type TAP. The values are means ± standard deviations. (C) Protein expression levels were analyzed by Western blot with anti-GFP antibodies. The amount of cell extract loaded per lane was corrected for minor differences in transfection efficiencies, as revealed by the β-Gal internal control. The positions of TAP derivatives fused to GFP or of zzp15 are shown on the right. (D) Human 293 cells were transfected with the reporter plasmids pCMV128-RRE and pCH110 and plasmids encoding RevM10 fusions of TAP, TAP-1xUBA, or TAP-2xUBA. When indicated, a plasmid encoding zzp15 (+p15) was cotransfected (black bar). Thereporter plasmid pCMV128-RRE is identical to pCMV128 (panel A) but carries the HIV RRE inserted in the intron, downstream of the cat gene. The stimulation of cat gene expression by TAP mutants measured in three independent experiments is expressed as a percentage of the activity of RevM10-TAP coexpressed with p15. The values are means ± standard deviations. (E) Protein expression levels were analyzed by Western blot with anti-HA antibodies. The amount of cell extract loaded per lane was normalized to the activity of the β-Gal internal control. The positions of TAP derivatives fused to RevM10 are shown on the right. (F) RNase protection analysis was performed with cytoplasmic or total RNA fractions isolated from 293 cells cotransfected with pCMV128 and plasmid pCH110 encoding β-Gal. In lanes 3, 5, 8, and 10, plasmids encoding p15 and RevM10 or GFP fusions of TAP were cotransfected. As controls, the reporter plasmids were cotransfected with empty vectors expressing RevM10 or GFP alone together with the plasmid expressing p15 (lanes 2, 4, 7, and 9).
In pCMV128, the CAT coding sequence is inserted into an inefficiently spliced intron, so cells transfected with this plasmid retain the unspliced pre-mRNA in the nucleus, yielding only trace levels of CAT activity (16). Overexpression of TAP/p15 heterodimers bypasses nuclear retention and promotes export of the inefficiently spliced pre-mRNA, resulting in about a 40-fold increase in CAT activity (Fig. 2B) (8). In all experiments described below, the induced CAT enzyme activity was normalized to the activity of the β-Gal internal control.
Deletion of both the NTF2-like and the UBA-like domains (TAP 1 to 372) abolishes export, but a protein in which only the NTF2-like domain has been deleted (TAP-1xUBA) exhibits 5% of the export activity of wild-type TAP (Fig. 2B; Table 1) (8, 10). When a second UBA-like domain was inserted in TAP-1xUBA to generate TAP-2xUBA, export was rescued to about 55% of the activity observed with wild-type TAP (Fig. 2B). Similarly, deletion of the UBA-like domain of TAP (TAP-1xNTF2) resulted in 12% export activity, but a protein having two NTF2-like domains in tandem (TAP-2xNTF2) retained 61% of the export activity of wild-type TAP when coexpressed with p15 (Fig. 2B and Table 1).
TABLE 1.
Relative export activity of TAP derivativesa
| TAP derivative | Mean stimulation (% of control) ± SD
|
|
|---|---|---|
| Xenopus oocytes | Human 293 cells | |
| TAP + p15 | 100 | 100 |
| TAP 1-372 | 0 | 0 |
| TAP-1xNTF2 + p15 | <10 | 12 ± 4 |
| TAP-2xNTF2 + p15 | n.d. | 61 ± 12 |
| TAP-1xUBA | <12 | 5 ± 3 |
| TAP-1xUBA + p15 | n.d. | 4 ± 2 |
| TAP-2xUBA | 61 ± 10 | 53 ± 20 |
| TAP-2xUBA + p15 | n.d. | 55 ± 11 |
| TAP-3xUBA | 45 ± 9 | 61 ± 11 |
| TAP-3xUBA + p15 | n.d. | 68 ± 14 |
| TAP-4xUBA | n.d. | 45 ± 8 |
| TAP-4xUBA + p15 | n.d. | 49 ± 18 |
The stimulation of Ad mRNA export in Xenopus oocytes or of cat gene expression in human 293 cells by TAP derivatives measured in three independent experiments is expressed as a percentage of the activity of full-length TAP. The concentration of the protein samples microinjected in Xenopus oocytes was 3 μM. n.d., not determined.
Western blot assays indicated that the expression levels of TAP mutants were comparable to that of TAP (Fig. 2C) when the amount of cell lysates analyzed was normalized to the activity of the β-Gal internal control. Thus, TAP-2xNTF2 is as active as TAP-2xUBA, but its export activity depends on p15 coexpression, while TAP-2xUBA exhibits the same activity regardless of the presence of exogenous p15 (Table 1 and Fig. 2B, compare grey and black bars).
It has been demonstrated that TAP can mediate the nuclear export of an RNA if tethered to that RNA via the RNA-binding domain of either the MS2 coat protein or the export-defective human immunodeficiency virus (HIV) Rev protein RevM10 (8, 13, 41). In this context, stimulation of RNA export by TAP is independent of its ability to bind RNA or RNA-associated proteins. To investigate whether the reduced export activity of TAP-2xUBA resulted from an impaired ability to interact with the cargo, we determined its export activity by tethering the protein directly to the pCMV128 pre-mRNA. To this end, wild-type TAP, TAP-1xUBA, and TAP-2xUBA were fused to the C terminus of RevM10. Although RevM10 cannot promote export of RNAs bearing the HIV Rev response element (RRE), it can target the fusion protein to a pCMV128 pre-mRNA carrying the RRE inserted in the intron (8, 13, 16).
Vectors expressing RevM10 fusions were cotransfected into 293 cells with the CAT reporter plasmid pCMV128-RRE and a plasmid encoding β-Gal as an internal control. As reported (8), expression of the RevM10-TAP fusion moderately stimulated CAT activity. In contrast, its coexpression with p15 increased CAT expression dramatically (by 200- to 250-fold) (Fig. 2D). TAP-1xUBA was used as a negative control because deletion of either the NTF2-like or the UBA-like domain abolished the export activity of RevM10-TAP (Fig. 2D) (8). In contrast, TAP-2xUBA fused to RevM10 retained 50% of the export activity of wild-type TAP. This export activity was observed in the absence of exogenous p15 (Fig. 2D). Western blot assays indicated that the expression level of TAP-2xUBA was comparable to that of TAP (Fig. 2E).
To confirm that the stimulation of CAT protein expression observed in the assays shown in Fig. 2B and 2D reflects an increased export of the unspliced cat mRNA encoded by pCMV128, we performed an RNase protection assay with cytoplasmic and total RNA fractions derived from transfected cells. Unspliced cat mRNA was detected at low levels in the cytoplasm of cells cotransfected with pCMV128 and the control vectors encoding p15 and either RevM10 or GFP alone (Fig. 2F, lanes 2 and 4). In contrast, the cat mRNA was exported to the cytoplasm of cells coexpressing p15 and TAP fused either to RevM10 or GFP (Fig. 2F, lanes 3 and 5). Therefore, TAP stimulates the nuclear export of the reporter RNA either by binding directly to that RNA via a heterologous RNA-binding domain (Fig. 2D) or by being recruited to that RNA, most probably via protein-protein interactions (Fig. 2B).
Altogether, these results indicate that a minimum of two nucleoporin-binding sites is required for TAP-mediated export of the pCMV128 reporter RNA. These binding sites can be provided by an NTF2-like domain followed by a UBA-like domain (as in the wild-type protein) or by two NTF2-like or two UBA-like domains in tandem. In the latter case, the export activity of TAP is independent of p15. Because a TAP derivative containing two UBA-like domains but no NTF2-like domain exhibited similar export activity regardless of whether this protein was tethered directly to the cargo, we conclude that the NTF2-like domain of TAP and hence p15 are unlikely to play a crucial role in cargo binding.
The export activity of TAP does not increase linearly with the number of nucleoporin-binding sites.
The proteins having two UBA-like domains or two NTF2-like scaffolds in tandem exhibited about 50 to 60% of the export activity of wild-type TAP, suggesting that the two nucleoporin-binding sites in these constructs may not be accessible in the appropriate conformation for optimal binding to nucleoporins. We therefore investigated whether full restoration of export could be achieved by adding an increasing number of NPC-binding sites. TAP derivatives having three or four UBA-like domains in tandem had export activities similar to that with TAP-2xUBA (Fig. 2B and Table 1). Thus, once a minimum of two UBA-like domains is present, increasing the number of nucleoporin-binding sites does not have an additive effect on export efficiency.
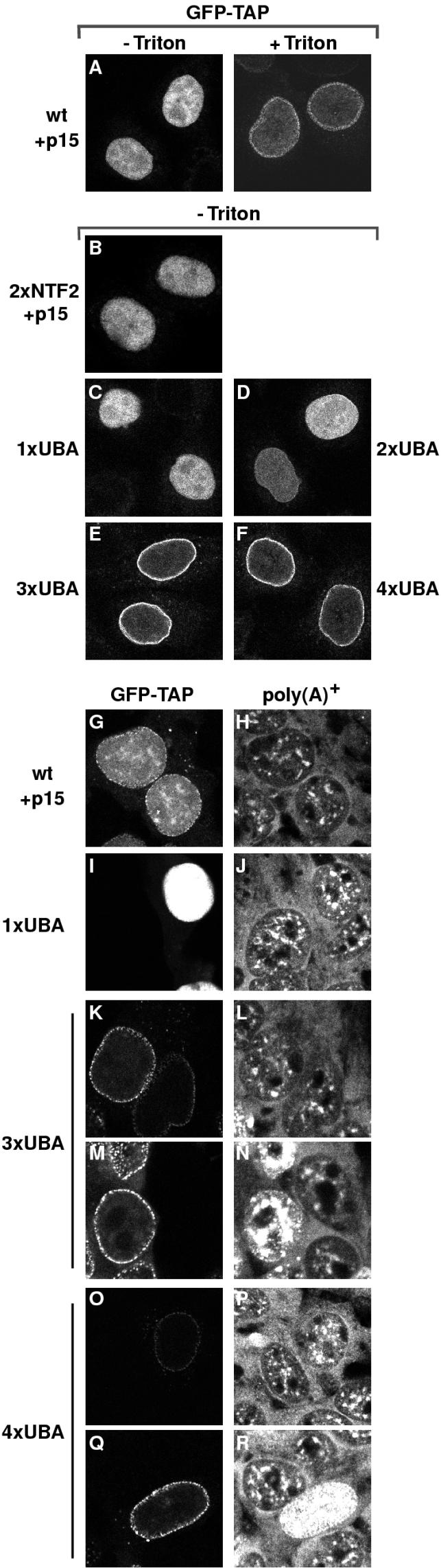
To determine whether mislocalization of the proteins could account for these effects, the subcellular localization of TAP constructs fused to GFP was analyzed in transfected HeLa and 293 cells (Fig. 3). At equilibrium, TAP localizes mainly in the nucleoplasm (Fig. 3A) (1, 4, 21). The pool of TAP associated with the nuclear envelope could be visualized in cells extracted with Triton X-100 before fixation (Fig. 3A [+Triton]) (1). Under these conditions, most of the nucleoplasmic pool of the protein was solubilized, but the fraction of TAP localized at the nuclear periphery was resistant to detergent extraction. Strikingly, TAP-3xUBA and TAP-4xUBA were exclusively visualized at the nuclear envelope without Triton X-100 preextraction (Fig. 3E, F, K, M, O, and Q). In contrast, under these conditions, TAP-1xUBA, TAP-2xUBA, and TAP-2xNTF2 were predominantly nuclear, like wild-type TAP (Fig. 3B, C, and D).
FIG. 3.
TAP-3xUBA and TAP-4xUBA localize to the nuclear rim. (A) HeLa cells were transfected with a plasmid expressing GFP-TAP. When cells were directly fixed in formaldehyde, TAP was predominantly detected throughout the nucleoplasm, excluding nucleoli (−Triton). When cells were extracted with Triton X-100 prior to fixation, a punctate labeling pattern was visible at the nuclear periphery (+Triton). (B to F) HeLa cells were transfected with plasmids expressing GFP fusions of TAP derivatives as indicated. Approximately 20 h after transfection, cells were fixed in formaldehyde and directly observed with a confocal microscope. TAP-2xNTF2, TAP-1xUBA, and TAP-2xUBA were detected throughout the nucleoplasm (B, C, and D). An additional punctate labeling pattern was visible at the nuclear rim for TAP-2xUBA. TAP-3xUBA and TAP-4xUBA localized predominantly at the nuclear rim (E, F). (G to R) Human 293 cells were transfected with plasmids encoding GFP fusions of TAP derivatives as indicated. Approximately 20 h after transfection, bulk poly(A)+ RNA was detected by FISH with an indocarbocyanine-labeled oligo(dT) probe. All images were taken with the same settings so the GFP signals could be compared directly. In panels K, M, O, and Q, examples of cells expressing TAP-3xUBA or TAP-4xUBA at different levels are shown. In about 10 to 20% of transfected cells, TAP-3xUBA and TAP-4xUBA were expressed at higher levels (M and Q), resulting in a strong accumulation of the poly(A)+ signal within the nucleus (N and R). wt, wild type.
The predominant localization of TAP-3xUBA and TAP-4xUBA at the nuclear rim suggests that these proteins have a higher affinity for nucleoporins and may interfere with the export process. We therefore analyzed the distribution of bulk poly(A)+ RNA in 293 cells expressing these TAP constructs by FISH with an oligo(dT) probe. Transfected cells expressing TAP-3xUBA and TAP-4xUBA at very high levels (as judged by the intensity of the GFP signal) accumulated poly(A)+ RNA within the nucleus (Fig. 3M, N, Q, and R). This inhibition of bulk mRNA export was observed in about 10 and 20% of cells expressing TAP-3xUBA and TAP-4xUBA, respectively, but not in cells in which TAP-1xUBA was expressed at similar or higher levels (Fig. 3I and J). Thus, it is possible that the reduced stimulation of CAT expression observed with TAP-4xUBA (45%) relative to TAP-3xUBA (61%) is in part due to an increased inhibitory effect of this protein.
Together, these observations agree with studies showing that high-affinity interactions between nucleoporins and transport receptors are dispensable for NPC passage (30). Indeed, a high rate of transport would be impossible if the individual interactions of the receptor with nucleoporins were of high affinity (30). Consistently, by increasing the number of UBA-like domains from two to four, the affinity for nucleoporins was increased (as judged by the localization of the constructs at the nuclear rim) without a corresponding increase in the export activity. Rather, these derivatives interfered with multiple export pathways (see also below), probably by saturating binding sites at the NPC.
TAP derivatives that lack the NTF2-like scaffold but have two or three UBA-like domains stimulate mRNA export in Xenopus oocytes.
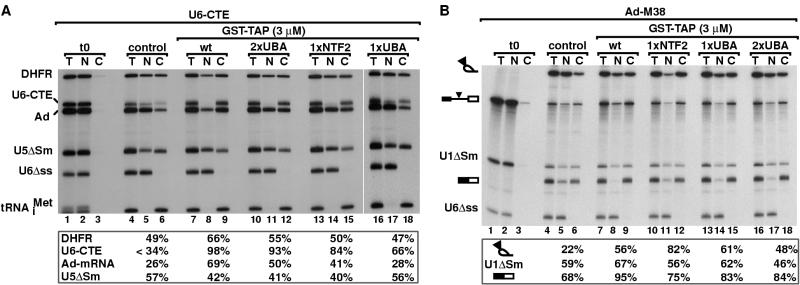
The ability of TAP-2xUBA and TAP-3xUBA to stimulate mRNA export directly was confirmed in X. laevis oocytes. Oocyte nuclei were injected with a mixture of labeled RNAs and purified TAP proteins expressed in E. coli as GST fusions (Fig. 4C). Wild-type TAP and TAP-1xNTF2 were coexpressed with p15 to ensure the proper folding of the NTF2-like domain. TAP-2xNTF2 could not be expressed at high levels in E. coli. The RNA mixture consisted of U5ΔSm and U6Δss snRNAs, the human initiator methionyl tRNA, and two mRNAs differing in their export efficiencies (8, 25). These were fushi tarazu (Ftz) mRNA and an mRNA derived from the adenovirus major late region (Ad mRNA). U6Δss RNA is not exported from the nucleus and serves as an internal control for nuclear injection (38). U5 snRNA and tRNAs are exported by CRM1 and exportin-t, respectively (reviewed in reference 26).
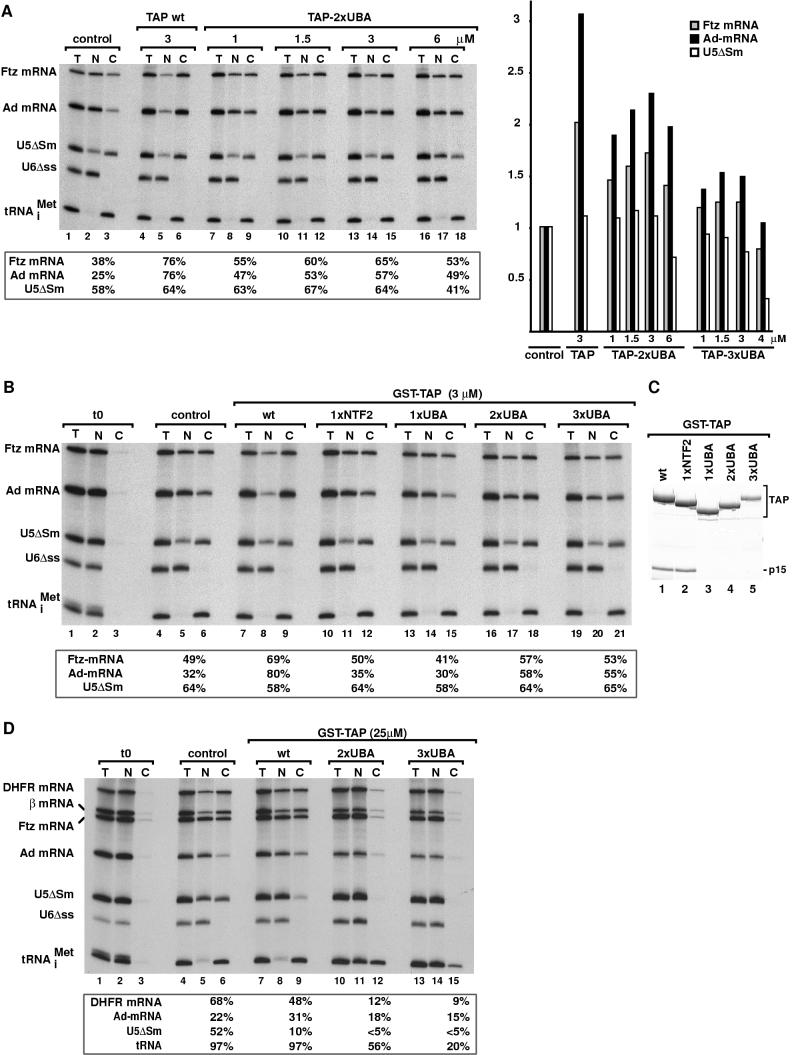
FIG. 4.
TAP-2xUBA and TAP-3xUBA stimulate mRNA export in Xenopus oocytes. (A and B) Xenopus oocyte nuclei were injected with mixtures of 32P-labeled RNAs and purified recombinant proteins as indicated. RNA samples from total oocytes (T) and nuclear (N) andcytoplasmic (C) fractions were collected 120 min after injection or immediately after injection (t0; lanes 1 to 3 in panel B). RNA samples were analyzed on 8% acrylamide-7 M urea denaturing gels. One oocyte equivalent of RNA, from a pool of 10 oocytes, was loaded per lane. The numbers above the lanes in panel A indicate the concentration (micromolar) of recombinant proteins in the injected samples. The numbers in boxes below the panels represent the percentage of a given RNA in the cytoplasm 120 min after injection in the presence of the proteins indicated above the lanes. Because the nuclear localization of U6Δss RNA and the export of tRNA (>94%) were not affected by any of the injected proteins, the respective values are not included. On the right of panel A, the effects of increasing concentrations of TAP-2xUBA or TAP-3xUBA on the export of U5ΔSm RNA and Ftz and Ad mRNAs are compared to those of TAP/p15 heterodimers. For the purpose of comparison, all values were normalized to the export observed in control oocytes. (C) TAP derivatives were expressed in E. coli as GST fusions, purified on glutathione-agarose beads, and visualized by Coomassie staining following SDS-polyacrylamide gel electrophoresis. (D) Xenopus oocyte nuclei were injected with mixtures of 32P-labeled RNAs and purified recombinant proteins as indicated. RNA samples from total oocytes (T) and nuclear (N) and cytoplasmic (C) fractions were collected 120 min after injection or immediately after injection (t0; lanes 1 to 3) and analyzed as in panels A and B. DHFR, β-globin, and Ftz mRNAs exhibited similar behavior, so only the quantitation for DHFR mRNA is shown. The nuclear localization of U6Δss RNA was not affected by any of the injected proteins.
In preliminary experiments, the concentration of TAP-2xUBA and TAP-3xUBA required to stimulate mRNA export specifically was determined. A representative example of one of these experiments is shown in Fig. 4A. Oocyte nuclei were coinjected with increasing concentrations of TAP-2xUBA or TAP-3xUBA and the RNA mixture described above. The stimulation of mRNA export in the presence of these proteins and in control oocytes was determined and compared to the stimulation obtained by injecting the optimal concentration of TAP/p15 heterodimers (Fig. 4A, right panel). The effect of the proteins on the export of U5ΔSm RNA was taken as an indication of the specificity. The nuclear localization of U6Δss RNA and the export of tRNA were not affected in the range of concentrations tested. Quantitation of the effects on Ad mRNA of coinjection of TAP-2xUBA indicated that, as for TAP/p15, this protein stimulated mRNA export specifically when its concentration in the injected samples ranged between 1 and 3 μM (Fig. 4A and data not shown). Stimulation of mRNA export was also observed at concentrations higher than 3 μM, but at these concentrations an inhibitory effect on the export of U5 snRNA was observed (Fig. 4A). Similarly, for TAP-3xUBA, this inhibitory effect was detected when the concentration of the protein exceeded 2 μM (not shown).
Next, we compared the effects of the recombinant proteins on the export of each of the RNA species by microinjecting them all at the same concentration. A representative example of these experiments is shown in Fig. 4B. In this example, following a 120-min incubation period, 32% of Ad mRNA was exported in control oocytes (Fig. 4B, lanes 4 to 6). In the presence of recombinant TAP/p15 heterodimers, about 80% of the injected Ad mRNA was exported (Fig. 4B, lanes 7 to 9). When recombinant TAP-2xUBA or TAP-3xUBA was coinjected, export of Ad mRNA was stimulated up to 58 and 55%, respectively (Fig. 4B, lanes 16 to 21). TAP derivatives having a single nucleoporin-binding domain (i.e., TAP-1xUBA and TAP-1xNTF2) had a minor or no effect on the export of Ad mRNA (Fig. 4B, lanes 10 to 15, and Table 1). Stimulation of Ad mRNA export by TAP and derivatives was specific, because export of tRNA and of U5 snRNA was not stimulated (Fig. 4B). On the contrary, as shown in Fig. 4A, these proteins partially inhibited U5 snRNA export at higher concentrations. Nuclear exit of the efficiently exported Ftz mRNA was only slightly stimulated (Fig. 4B, lanes 16 to 21 versus 4 to 6).
Quantitation of the effects on Ad mRNA export of the recombinant proteins obtained in three independent experiments is reported in Table 1. Interestingly, the ability of TAP derivatives to stimulate Ad mRNA export in Xenopus oocytes correlated well with their ability to increase cat gene expression in human 293 cells (Table 1) in spite of the differences between these cell types and the cargoes analyzed.
When TAP-2xUBA and TAP-3xUBA were injected at the highest achievable concentration (25 μM), a strong inhibition of export of several mRNA species but also of U5 snRNA and tRNA export was observed (Fig. 4D, lanes 10 to 15 versus 4 to 6). At the same concentration, wild-type TAP interfered mainly with the export pathway of U5 snRNA (Fig. 4D, lanes 7 to 9). These results are consistent with those obtained in human cultured cells and indicate that, relative to wild-type TAP, TAP-2xUBA and TAP-3xUBA have an increased ability to compete for binding to the NPC with other transport receptors.
A single nucleoporin-binding domain of TAP is sufficient to promote NPC translocation of specific CTE cargoes.
The results described above show that at least two nucleoporin-binding sites are required for TAP-mediated RNA export in transfected human cells and in Xenopus oocytes. In contrast, we have previously reported that export of U6-CTE chimeric RNA, which is directly mediated by TAP, can be stimulated by a TAP mutant lacking the NTF2-like domain (TAP-1xUBA) but not by TAP-1xNTF2 (1). Since these experiments were conducted in the absence of exogenous p15, it is possible that the NTF2-like domain of TAP-1xNTF2 was not properly folded. We therefore reinvestigated the ability of TAP-1xNTF2 to export U6-CTE in the presence of p15.
Xenopus oocyte nuclei were coinjected with purified recombinant proteins and a mixture of labeled RNAs consisting of dihydrofolate reductase (DHFR) and Ad mRNAs, U5ΔSm and U6Δss snRNAs, the human initiator methionyl tRNA, and U6-CTE RNA. Following a 90-min incubation period, U6-CTE RNA was inefficiently exported in control oocytes (Fig. 5A, lanes 4 to 6). Injection of TAP/p15 heterodimers resulted in a strong stimulation of U6-CTE RNA export (Fig. 5A, lanes 7 to 9). If U6-CTE was efficiently exported, it was stable during the 90-min incubation period, whereas if U6-CTE was not exported, it was partially degraded in the nuclear fraction (Fig. 5A, lanes 7 to 9 versus 4 to 6). In contrast to the results obtained with Ad mRNA, TAP-1xNTF2 in the presence of p15 stimulated export of U6-CTE RNA almost as efficiently as wild-type TAP (Fig. 5A, lanes 13 to 15). As expected, since one nucleoporin-binding domain was sufficient to export U6-CTE RNA (Fig. 5A, lanes 13 to 18) (1), both TAP-2xUBA and TAP-3xUBA strongly stimulated U6-CTE export (Fig. 5A, lanes 10 to 12, and data not shown).
FIG. 5.
TAP derivatives having a single nucleoporin-binding domain translocate specific CTE RNA cargoes across the NPC. (A and B) Xenopus oocyte nuclei were injected with mixtures of 32P-labeled RNAs and purified recombinant proteins as indicated. RNA samples from total oocytes (T) and nuclear (N) and cytoplasmic (C) fractions were collected immediately after injection (t0; lanes 1 to 3) or 90 min after injection in panel A and analyzed as described in the legend to Fig. 4. In panel B, samples were collected 120 min after injection and analyzed in 10% acrylamide-7 M urea denaturing gels. The concentration of recombinant proteins in the injected samples was 3 μM. The numbers below the panels represent the percentage of a given RNA in the cytoplasm in the presence of the proteins indicated above the lanes. The nuclear localization of U6Δss RNA and the export of tRNA (>94%) were not affected by any of the injected proteins, and these values are not included. The mature products and intermediates of the splicing reaction are indicated diagrammatically on the left of panel B. The solid triangle represents the M38 CTE. wt, wild type.
We also investigated the effects of TAP derivatives on the export of an excised intron lariat bearing the CTE. In this study, we used a precursor mRNA derived from the adenovirus major late transcription unit (pBSAd1) carrying the simian retrovirus 1 CTE mutant M38 inserted in the intron (12, 32). The M38 mutant was chosen because it has only one binding site for TAP (loop A), while the wild-type CTE has two (12). Preliminary studies indicated that export stimulation by TAP of the intron lariat bearing M38 requires the presence of at least one nucleoporin-binding site on the protein. In contrast, as reported previously, export of the intron lariat carrying wild-type CTE can be stimulated by the N-terminal domain of TAP in Xenopus oocytes (7).
Based on the results obtained with the M38 mutant (Fig. 5B and data not shown) and taking into account the crucial role of the C-terminal half of TAP in mediating NPC binding (1, 10, 21, 39), we currently favor the idea that the N-terminal fragment of TAP stimulates export of the intron lariat bearing wild-type CTE indirectly, probably by binding to one loop of the CTE and facilitating the recruitment of endogenous Xenopus TAP to the second binding site. Why this facilitated binding appears to occur only in the context of the intron lariat but not when the CTE is inserted in a linear RNA such as U6-CTE is unclear.
The excised intron lariat carrying the CTE derivative M38 was reproducibly exported more efficiently by TAP constructs with one nucleoporin-binding site than by TAP-2xUBA (Fig. 5B, lanes 10 to 15 and 16 to 18). It is noteworthy that export of the spliced Ad mRNA was also stimulated by TAP-1xUBA. Together, these results indicate that either of the two nucleoporin-binding domains of TAP is sufficient to promote NPC translocation of at least some cargoes. Furthermore, a comparison of the different substrates analyzed in Fig. 4 and 5 suggests that in addition to the differences in the export activity of the different forms of TAP, a second level of complexity is introduced by the requirements of different cargoes.
Drosophila NXF1 lacking the NTF2-like scaffold partially restores export of bulk mRNA in cells depleted of p15 or NXF1.
The possibility that the NTF2-like scaffold contributes to the interaction of TAP with cellular mRNAs or to TAP recycling after one round of export cannot be completely ruled out by the experiments described above, in which the export of specific RNAs was analyzed in the presence of excess TAP. We therefore set up an assay to test whether an NXF1 protein lacking the NTF2-like domain but carrying two UBA-like domains could equally support mRNA export in cells depleted of endogenous NXF1 or p15. This question was addressed in Drosophila Schneider cells (SL2 cells) because depletion of endogenous NXF1 or p15 by dsRNA interference in these cells is very efficient (15). Moreover, NXF1 or p15 depletion inhibits growth and results in a strong nuclear accumulation of poly(A)+ RNA, which can be monitored easily (15).
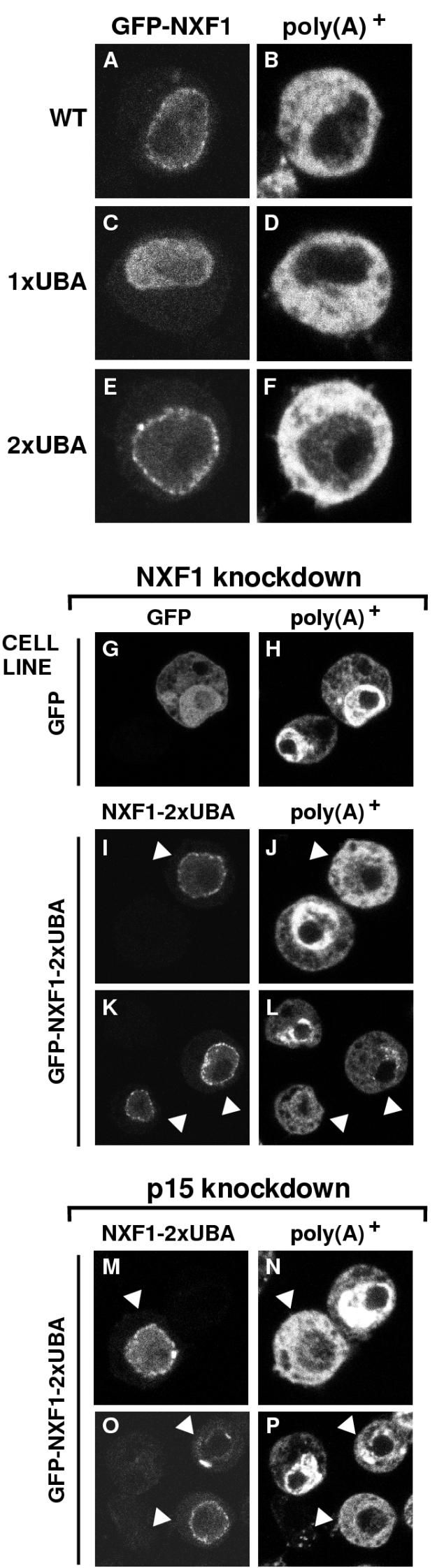
Polyclonal SL2 cell lines stably transfected with plasmids expressing GFP fusions of D. melanogaster NXF1, NXF1-1xUBA, or NXF1-2xUBA or GFP alone as a control were established. These cell lines are heterogeneous, and the GFP fusions are expressed at different levels in approximately 5 to 10% of the cell population. The subcellular localization of the recombinant proteins in these cell lines was investigated. All three NXF1 proteins localized predominantly within the nucleoplasm and were excluded from the nucleolus (Fig. 6A, C, and E). A clear rim staining was observed in most of the cells expressing wild-type NXF1 and NXF1-2xUBA (Fig. 6A and E). We also analyzed the distribution of poly(A)+ RNA by oligo(dT) in situ hybridization in the different cell lines. No nuclear accumulation of poly(A)+ RNA was observed (Fig. 6B, D, and F, and data not shown), suggesting that in these stably transfected cells, the recombinant proteins are expressed at levels that do not interfere with export.
FIG. 6.
D. melanogaster NXF1-2xUBA partially rescues export in cells depleted of NXF1. (A to F) SL2 cell lines constitutively expressing GFP fusions with wild-type NXF1 (wt), NXF1-1xUBA, or NXF1-2xUBA were fixed in formaldehyde. Bulk poly(A)+ RNA was detected by FISH (B, D, and F). (G to P) SL2 cells stably transfected with plasmids expressing GFP or GFP-NXF1-2xUBA were transfected with a dsRNA corresponding to the NTF2-like domain of D. melanogaster NXF1 (G to L) or to the coding region of D. melanogaster p15(M to P). Five days after transfection, poly(A)+ RNA was detected by FISH with an indocarbocyanine-labeled oligo(dT) probe. In the GFP-expressing cell line, depletion of NXF1 causes nuclear accumulation of poly(A)+ RNA regardless of whether GFP is expressed (G and H). In the GFP-NXF1-2xUBA cell line, a strong nuclear accumulation of poly(A)+ RNA was observed in cells in which the protein was not detected, but when D. melanogaster NXF1-2xUBA was expressed at detectable levels, mRNA export was restored (I to L, arrowheads). This restoration of export was also observed when p15 was depleted (M to P).
Next, endogenous NXF1 or p15 was depleted from these cell lines by dsRNA interference. The efficiency of NXF1 or p15 depletion could be monitored by the dramatic inhibition of cell growth observed as early as 2 days after transfecting the corresponding dsRNAs. No recovery of cell growth was observed within 8 days after transfection (see also below) (15). The distribution of poly(A)+ RNA in cells expressing the GFP fusions was analyzed 5 days after addition of dsRNAs, and the percentage of cells accumulating poly(A)+ RNA within the nucleus was determined in two independent experiments (Table 2).
TABLE 2.
Percentage of cells accumulating poly(A)+ RNA within the nucleus following p15 or NXF1 depletiona
| Cell line | Poly(A)+ RNA accumulation (% of cells)
|
|||
|---|---|---|---|---|
| p15 depleted | NXF1 depleted | |||
| GFP | 87 | 86 | 92 | 94 |
| Wild-type NXF1 | 72 | |||
| NXF1-1xUBA | 60 | 67 | ||
| NXF1-2xUBA | 48 | 50 | 49 | 52 |
The SL2 cell lines indicated in the first column were depleted of endogenous p15 or NXF1 by dsRNA interference. At least 100 cells expressing the recombinant proteins were analyzed, and the percentage of cells showing nuclear accumulation of poly(A)+ RNA was determined. Values were obtained in two independent experiments.
In control cells expressing GFP, depletion of either NXF1 or p15 resulted in a severe nuclear accumulation of poly(A)+ RNA in about 90% of the cell population (Fig. 6H and Table 2). This was observed independently of whether GFP was expressed (Fig. 6G and H). As shown in Fig. 6H, the oligo(dT) signal was widespread in the nucleoplasm but was excluded from the large nucleolus, characteristic of SL2 cells. Expression of wild-type NXF1 partially restored export in less than 20% of cells depleted of p15 (Table 2). A similar partial restoration of export was observed in cells expressing NXF1-1xUBA, regardless of whether NXF1 or p15 had been depleted (Table 2). We defined mRNA export as partially rescued when the poly(A)+ signal was predominantly cytoplasmic or evenly distributed between the nucleoplasm and cytoplasm.
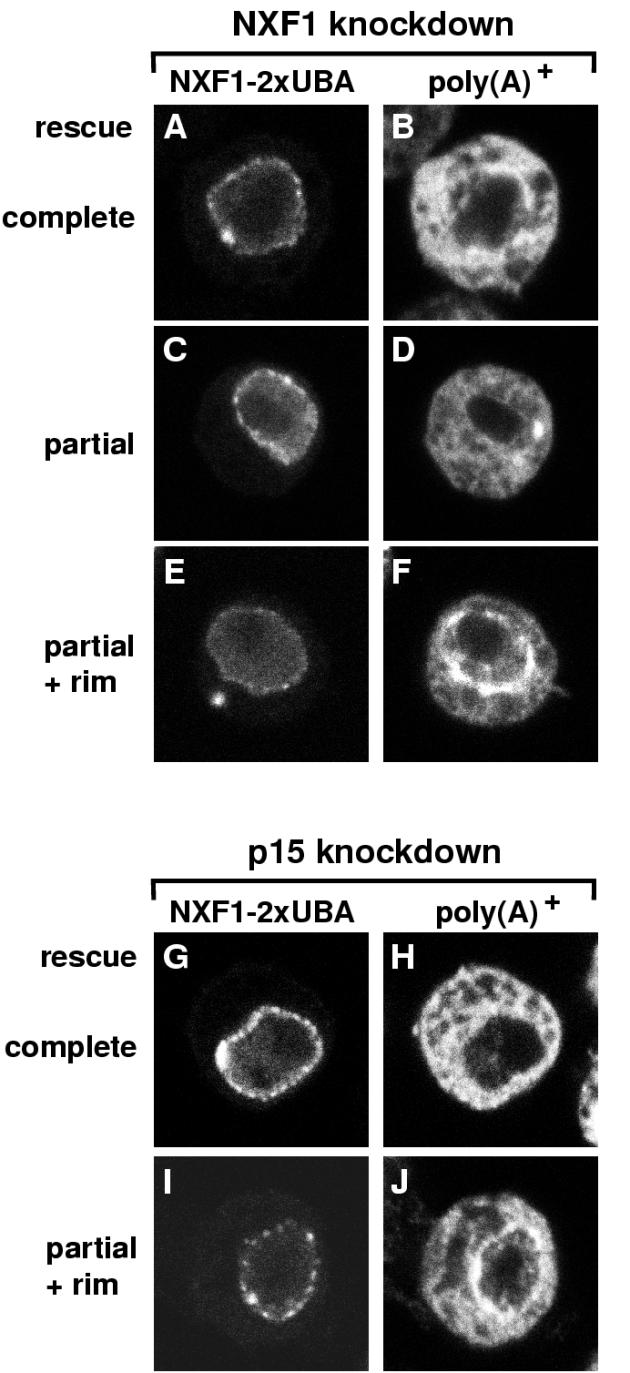
Expression of NXF1-2xUBA partially rescued export in approximately 50% of cells depleted of NXF1 (Table 2; Fig. 6I to L, arrowheads). An indistinguishable restoration of export by NXF1-2xUBA was observed when p15 was depleted (Fig. 6M to O and Table 2). In both cases, the restoration of bulk mRNA export by NXF1-2xUBA was partial, and in most cells a residual nuclear signal was evident. A gallery of various phenotypes is shown in Fig. 7. Cells exhibiting a poly(A)+ RNA signal at the nuclear rim were occasionally detected (Fig. 7F and J). Since significant cytoplasmic staining was nevertheless evident in these cells, the accumulation of bulk mRNA at the nuclear periphery suggests that in cells in which mRNA export relies only on NXF1-2xUBA, the translocation step becomes limiting.
FIG. 7.
Patterns of poly(A)+ RNA distribution in cells depleted of endogenous NXF1 or p15 but expressing GFP-NXF1-2xUBA. (A to J) Restoration of mRNA export was almost complete (B and H) or partial, with an equal distribution of the oligo(dT) signal between the nucleus and the cytoplasm (D). In panels F and J, cells in which the poly(A)+ RNA accumulated at the nuclear rim are shown.
The partial restoration of export by NXF1-2xUBA is consistent with the observation that TAP-2xUBA exhibits about 50% of the export activity of wild-type TAP (Table 1). Additionally, the extent of the restoration is likely to depend on the expression levels of the protein, as in a small percentage of cells expressing NXF1-2xUBA, a complete restoration was observed [i.e., the oligo(dT) signal was mainly detected in the cytoplasm] (Fig. 7B and H). Importantly, D. melanogaster NXF1-2xUBA did not accumulate within the cytoplasm in cells in which export of bulk mRNA was restored (see, for example, Fig. 7A and G), suggesting that the NTF2-like scaffold is not required for cargo release following export.
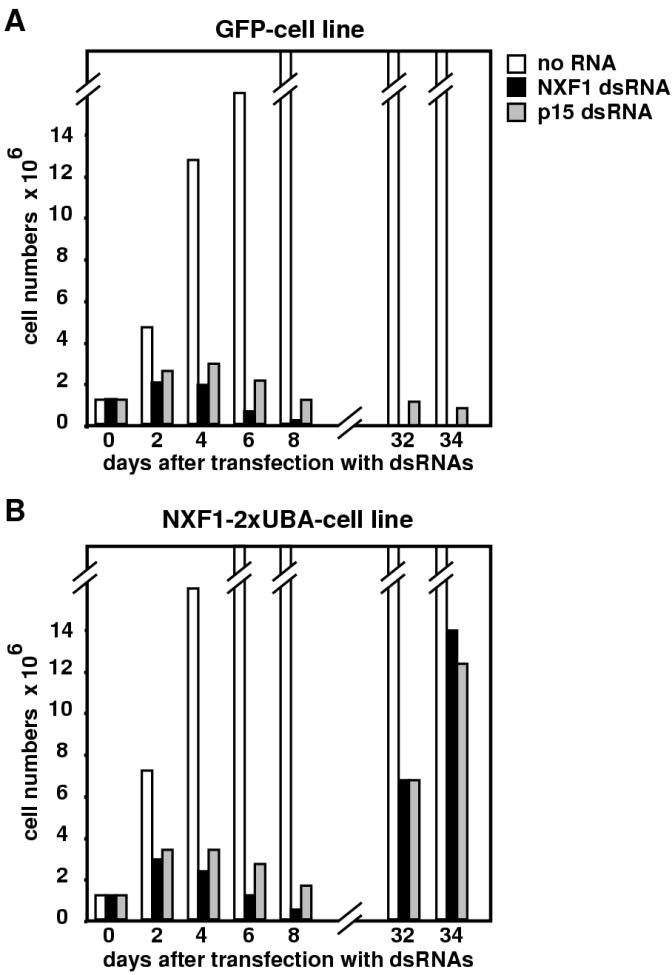
The restoration of export appeared to be complete in at least a fraction of cells expressing D. melanogaster NXF1-2xUBA, so we investigated whether NXF1-2xUBA could sustain growth in cells depleted of endogenous NXF1 or p15. Cell lines expressing D. melanogaster NXF1-2xUBA or GFP constitutively were depleted of endogenous D. melanogaster NXF1 or D. melanogaster p15. As reported (15), NXF1 or p15 depletion inhibits cell growth as early as 2 days after transfecting the corresponding dsRNAs (Fig. 8A). Cells were kept in culture, and growth was monitored every second day for 10 days and every week for 6 weeks. In the GFP-expressing cell line, no recovery of cell growth was observed within this period (Fig. 8A). Moreover, 32 days after transfecting NXF1 dsRNA, no surviving cells were detected (Fig. 8A). Although a fraction of cells survived p15 depletion, these cells subsequently died (Fig. 8A). In contrast, a higher percentage of cells depleted of D. melanogaster NXF1 or p15 but expressing D. melanogaster NXF1-2xUBA survived and started to grow exponentially about a month after transfection (Fig. 8B). These results suggest that expression of D. melanogaster NXF1-2xUBA allows NXF1- or p15-depleted cells to survive.
FIG. 8.
NXF1-2xUBA sustains growth of cells depleted of endogenous NXF1 or p15. (A and B) SL2 cells stably transfected with plasmids expressing GFP or GFP-NXF1-2xUBA were transfected with NXF1 or p15 dsRNAs as indicated. Cell numbers were determined every second day for 8 days and every week for 6 weeks. In the GFP cell line, no cells survived NXF1 depletion. The surviving cells detected 31 days following p15 depletion died progressively after a few days. Cells expressing NXF1-2xUBA survived NXF1 or p15 depletion and started to grow exponentially 1 month after transfection of the dsRNAs.
In the surviving cell populations, the percentage of cells expressing NXF1-2xUBA increased threefold following D. melanogaster NXF1 depletion and 12-fold following p15 depletion. Western blot analysis of samples collected on day 32 revealed that while the levels of endogenous NXF1 had recovered, the expression levels of p15 were about 50% of the levels detected in control cells (data not shown). Thus, expression of D. melanogaster NXF1-2xUBA allows NXF1- or p15-depleted cells to survive so that the expression levels of the depleted proteins are progressively restored.
DISCUSSION
Transport receptors promote translocation of cargoes across the central channel of the NPC as a result of their ability to interact with nucleoporins. In particular, the phenylalanine-glycine (FG) repeats that characterize most nucleoporins are believed to function as docking sites for the receptors moving through the pore (31). The heterodimeric export receptor NXF1/p15 possesses two nucleoporin-binding domains, the UBA-like domain and the NTF2-like scaffold, which results from the heterodimerization of the NTF2-like domain of NXF1 with p15 (10). In this study, we show that although these two nucleoporin-binding domains are unrelated in sequence and structure, they are interchangeable. Indeed, an NXF1 derivative in which the NTF2-like scaffold is replaced by a UBA-like domain (2xUBA) has an export activity similar to that of the converse construct, in which the UBA-like domain is replaced by an NTF2-like scaffold (2xNTF2). These results indicate that the main role of the NTF2-like scaffold is to provide a nucleoporin-binding site. More importantly, although the proteins having two UBA-like domains or two NTF2-like scaffolds in tandem have reduced export activity relative to wild-type NXF1, these proteins are able to promote directional translocation of RNA cargoes across the NPC. We also show that the export activity of NXF1 is not directly related to the number of NPC-binding sites but that a minimum of two sites is required for export of cellular mRNA.
p15 acts as a structural subunit that maintains the proper folding and function of the NTF2-like domain of NXFs.
In addition to its essential role in mRNA nuclear export, p15 has been reported to bind RanGTP and nucleoporins directly and to be implicated in the export of tRNA and of substrates exported by CRM1 (5, 6, 23, 28). However, other studies failed to detect a direct interaction between p15 and Ran or nucleoporins (14, 21, 39). A crystallographic analysis of p15 indicates that despite its overall similarity to NTF2, the putative binding site for Ran is occluded in p15 by residues with larger side chains that fill the equivalent hydrophobic pocket in NTF2 (10, 34). Binding to nucleoporins is also prevented by an additional turn of the N-terminal α-helix of p15, which occludes the hydrophobic pocket present at the equivalent position in TAP and NTF2 (2, 10). Our results, together with available structural data (10), suggest that the essential role of p15 in mRNA export is structural and is to ensure the proper folding and function of the NTF2-like domain of TAP/NXF1, which in turn mediates binding to the NPC.
Because the NTF2-like domain of NXF1 cannot bind to nucleoporins in the absence of p15, p15 offers a possible target for regulating NXF1/NPC interactions. Along these lines, it has been proposed that p15 could impose directionality on the export process by heterodimerizing with TAP in the nucleus and dissociating in the cytoplasm (22, 39). Our data do not rule out a compartment-specific regulation of NXF1/p15 interaction, but they show that if this regulation does exist, it is not strictly or generally required for directional transport. Regulation of NXF1/p15 interaction may nevertheless play a critical role in the overall efficiency of the transport process or the export of specific cargoes. For instance, some cargoes may have the ability to modulate NXF1/p15 dimerization. More generally, by controlling p15 expression levels or its ability to heterodimerize with NXF1, the overall mRNA export activity of the cell could be regulated.
In metazoa, there are multiple NXF proteins that heterodimerize with p15. With the exception of NXF1, it is unclear whether these proteins are involved in mRNA export and whether their NTF2-like domains mediate binding to nucleoporins. In this context, p15 may act as a regulator of NXF activities by functioning as a structural module that maintains the proper folding and function of the NTF2-like domains.
Requirements for NPC translocation are influenced by the cargo transported.
The heterodimeric NTF2 scaffold and the UBA-like domain of TAP both feature a single nucleoporin-binding site (10, 11). In this study, we show that either of these sites is sufficient to mediate NPC translocation of specific CTE-dependent cargoes in Xenopus oocytes. The CTE-bearing RNAs analyzed here are artificial, but the observation that a single nucleoporin-binding domain is sufficient to promote NPC translocation is not without precedent. In S. cerevisiae, Mex67p mutants lacking the UBA-like domain can still promote mRNA export when Mtr2p is overexpressed (35). However, these mutants show a conditional growth phenotype and accumulate bulk mRNA within the nucleus at the restrictive temperature (35), suggesting that two NPC-binding sites are required for efficient nuclear mRNA export. In Drosophila cells, D. melanogaster NXF1-1xUBA partially restores mRNA export in cells depleted of NXF1 or p15, although this restoration occurred only in about 20 to 30% of cells expressing the protein (Table 2). In the case of the specific CTE RNAs shown in Fig. 5, TAP derivatives having a single nucleoporin-binding site were almost as efficient as wild-type TAP in promoting export. This suggests that the requirements for NPC translocation are influenced by the nature of the cargo transported.
Two NPC-binding sites are required for TAP-mediated export of cellular mRNA.
In contrast to the specific CTE-bearing RNAs described in this study, efficient export of cellular mRNAs requires at least two nucleoporin-binding sites that can be provided by two NTF2-like domains bound to p15 or two UBA-like domains. It can be speculated that the presence of two nucleoporin-binding pockets is a common motif for transport receptors because two binding pockets are also present on the NTF2 homodimer and at the outer surface of the N-terminal half of importin β (3, 34).
The observation that the roles of the UBA-like domain and the NTF2 scaffold are confined to nucleoporin binding raises the question of why the mRNA export receptor has evolved two distinct nucleoporin-binding domains. In vitro binding assays indicate that these domains have different relative affinities for nucleoporins (22). The presence of two nucleoporin-binding sites with different relative affinities rather than a single high-affinity binding site may facilitate a high rate of translocation, which depends on transient interactions with nucleoporins. Another important difference between these domains is that while the nucleoporin-binding activity of the UBA-like domain is likely to be constitutive, the NTF2-like domain cannot interact with nucleoporins in the absence of p15. Thus, the binding activity of the NTF2 scaffold could be subject to regulation (see above). Finally, two binding sites may bind cooperatively to achieve an optimal affinity for nucleoporins. The NTF2-like scaffold has been reported to increase (39) and to reduce (22) the overall nucleoporin-binding affinity of the UBA-like domain. Our data are consistent with the former possibility; however, the precise mechanism by which these domains influence each other's binding remains to be determined.
Implications for the mechanism underlying directional transport.
A comparison of the structures of FG-containing peptides bound to importin β and to the NTF2-like scaffold of TAP has shown similarities in the mode of FG-nucleoporin interactions between these two structurally unrelated transport factors (3, 10). Consistently, the NPC-binding domain of TAP competes with the importin β-like receptors CRM1 and exportin-t for binding sites at the NPC (Fig. 4D) (1). Thus, a similar mechanism of NPC translocation is predicted for these two families of export receptors.
The interactions of importin β-like receptors with nucleoporins and with their cargoes are regulated by Ran. However, NPC passage of these receptors on their own or in association with at least small cargoes is not obligatorily coupled to hydrolysis of GTP by Ran (reviewed in references 26 and 30). It remains to be determined whether NXF1/nucleoporin interactions are regulated (see above), but as for the importin β-like receptors, such regulation may be dispensable for translocation. As in the case of Ran-regulated export complexes, directionality may be achieved by an efficient and irreversible mechanism for cargo release that operates in the cytoplasm or at the cytoplasmic face of the NPC. The mechanism by which NXF1 releases its cargo after translocation is unknown, but it has been proposed that Dbp5, an RNA helicase essential for mRNA export in S. cerevisiae, may disrupt mRNA-protein interactions as the mRNP emerges from the NPC (9). Transportin, which is directly involved in TAP nuclear import, may also play a role in this process by coupling cargo release on the cytoplasmic face of the pore with NXF1 recycling back to the nucleus. Clearly, further work is needed to determine the mechanism of mRNP unloading after translocation.
Acknowledgments
We thank Tom Hope for the kind gift of plasmid pCMV128 and Ann Atzberger for the fluorescence-activated cell sorting. We are grateful to Elena Conti, Jan Ellenberg, Iain Mattaj, and David Thomas for critical reading of the manuscript.
This study was supported by the European Molecular Biology Organization (EMBO) and the Human Frontier Science Program Organization (HFSPO).
REFERENCES
- 1.Bachi, A., I. C. Braun, J. P. Rodrigues, N. Panté, K. Ribbeck, C. von Kobbe, U. Kutay, M. Wilm, D. Görlich, M. Carmo-Fonseca, and E. Izaurralde. 2000. The C-terminal domain of TAP interacts with the nuclear pore complex and promotes export of specific CTE-bearing RNA substrates. RNA 6:136-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bayliss, R., K. Ribbeck, D. Akin, H. M. Kent, C. M. Feldherr, D. Görlich, and M. Stewart. 1999. Interaction between NTF2 and xFxFG-containing nucleoporins is required to mediate nuclear import of RanGDP. J. Mol. Biol. 293:579-593. [DOI] [PubMed] [Google Scholar]
- 3.Bayliss, R., T. Littlewood, and M. Stewart. 2000. Structural basis for the interaction between FxFG nucleoporin repeats and importin b in nuclear trafficking. Cell 102:99-108. [DOI] [PubMed] [Google Scholar]
- 4.Bear, J., W. Tan, A. S. Zolotukhin, C. Tabernero, E. A. Hudson, and B. K. Felber. 1999. Identification of novel import and export signals of human TAP, the protein that binds to the constitutive transport element of the type D retrovirus mRNAs. Mol. Cell. Biol. 19:6306-6317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Black, B. E., L. Lévesque, J. M. Holaska, T. C. Wood, and B. Paschal. 1999. Identification of an NTF2-related factor that binds Ran-GTP and regulates nuclear protein export. Mol. Cell. Biol. 19:8616-8624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Black, B. E., J. M. Holaska, L. Levesque, B. Ossareh-Nazari, C. Gwizdek, C. Dargemont, and B. M. Paschal. 2001. NXT1 is necessary for the terminal step of Crm1-mediated nuclear export. J. Cell Biol. 152:141-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Braun, I. C., E. Rohrbach, C. Schmitt, and E. Izaurralde. 1999. TAP binds to the constitutive transport element (CTE) through a novel RNA-binding motif that is sufficient to promote CTE-dependent RNA export from the nucleus. EMBO J. 18:1953-1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Braun, I. C., A. Herold, M. Rode, E. Conti, and E. Izaurralde. 2001. Overexpression of TAP/p15 heterodimers bypasses nuclear retention and stimulates nuclear mRNA export. J. Biol. Chem. 276:20536-20543. [DOI] [PubMed] [Google Scholar]
- 9.Conti, E., and E. Izaurralde. 2001. Nucleocytoplasmic transport enters the atomic age. Curr. Opin. Cell Biol. 3:310-319. [DOI] [PubMed] [Google Scholar]
- 10.Fribourg, S., I. C. Braun, E. Izaurralde, and E. Conti. 2001. Structural basis for the recognition of a nucleoporin FG repeat by the NTF2-like domain of the TAP/p15 mRNA nuclear export factor. Mol. Cell 8:645-656. [DOI] [PubMed] [Google Scholar]
- 11.Grant, R. P., E. Hurt, D. Neuhaus, and M. Stewart. 2002. Structural basis for the interaction between the Tap C-terminal domain and FG repeat-containing nucleoporins. Nat. Struct. Biol. 9:247-251. [DOI] [PubMed] [Google Scholar]
- 12.Grüter, P., C. Tabernero, C. von Kobbe, C. Schmitt, C. Saavedra, A. Bachi, M. Wilm, B. K. Felber, and E. Izaurralde. 1998. TAP, the human homolog of Mex67p, mediates CTE-dependent RNA export from the nucleus. Mol. Cell 1:649-659. [DOI] [PubMed] [Google Scholar]
- 13.Guzik, B. W., L. Levesque, S. Prasad, Y.-C. Bor, B. E. Black, B. M. Paschal, D. Rekosh, and M.-L. Hammarskjöld. 2001. NXT1 (p15) is a crucial cellular cofactor in TAP-dependent export of intron-containing RNA in mammalian cells. Mol. Cell. Biol. 21:2545-2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herold, A., M. Suyama, J. P. Rodrigues, I. C. Braun, U. Kutay, M. Carmo-Fonseca, P. Bork, and E. Izaurralde. 2000. TAP/NXF1 belongs to a multigene family of putative RNA export factors with a conserved modular architecture. Mol. Cell. Biol. 20:8996-9008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herold, A., T. Klimenko, and E. Izaurralde. 2001. NXF1/p15 heterodimers are essential for mRNA nuclear export in Drosophila. RNA 7:1768-1780. [PMC free article] [PubMed] [Google Scholar]
- 16.Huang, X., T. J. Hope, B. L. Bond, D. McDonald, K. Grahl, and T. G. Parslow. 1991. Minimal Rev-responsive element for type 1 human immunodeficiency virus. J. Virol. 65:2131-2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jarmolowski, A., W. C. Boelens, E. Izaurralde, and I. W. Mattaj. 1994. Nuclear export of different classes of RNA is mediated by specific factors. J. Cell Biol. 124:627-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jun, L., S. Frints, H. Duhamel, A. Herold, J. Abad-Rodrigues, C. Dotti, E. Izaurralde, P. Marynen, and G. Froyen. 2001. NXF5, a novel member of the nuclear RNA export factor family, is lost in a male patient with a syndromic form of mental retardation. Curr. Biol. 11:1381-1391. [DOI] [PubMed] [Google Scholar]
- 19.Kang, Y., and B. R. Cullen. 1999. The human TAP protein is a nuclear mRNA export factor that contains novel RNA-binding and nucleocytoplasmic transport sequences. Genes Dev. 13:1126-1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kang, Y., H. P. Bogerd, and B. R. Cullen. 2000. Analysis of cellular factors that mediate nuclear export of RNAs bearing the Mason-Pfizer monkey virus constitutive transport element. J. Virol. 74:5863-5871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Katahira, J., K. Sträβer, A. Podtelejnikov, M. Mann, J. U. Jung, and E. Hurt. 1999. The Mex67p-mediated nuclear mRNA export pathway is conserved from yeast to human. EMBO J. 18:2593-2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Katahira, J., K. Sträβer, T. Saiwaki, Y. Yoneda, and E. Hurt. 2001. Complex formation between TAP and p15 affects binding to FG-repeat nucleoporins and nucleocytoplasmic shuttling. J. Biol. Chem. 277:9242-9246. [DOI] [PubMed]
- 23.Lévesque, L., B. Guzik, T. Guan, J. Coyle, B. E. Black, D. Rekosh, M. L. Hammarskjold, and B. M. Paschal. 2001. RNA export mediated by Tap involves NXT1-dependent interactions with the nuclear pore complex. J. Biol. Chem. 276:44953-44962. [DOI] [PubMed] [Google Scholar]
- 24.Liker, E., E. Fernandez, E. Izaurralde, and E. Conti. 2000. The structure of the mRNA nuclear export factor TAP reveals a cis arrangement of a noncanonical RNP domain and a leucine-rich-repeat domain. EMBO J. 19:5587-5598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luo, M.-J., and R. Reed. 1999. Splicing is required for rapid and efficient mRNA export in metazoans. Proc. Natl. Acad. Sci. USA 96:14937-14942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mattaj, I. W., and L. Englmeier. 1998. Nucleocytoplasmic transport: the soluble phase. Annu. Rev. Biochem. 67:256-306. [DOI] [PubMed] [Google Scholar]
- 27.Nakielny, S., and G. Dreyfuss. 1999. Transport of proteins and RNAs in and out of the nucleus. Cell 99:677-690. [DOI] [PubMed] [Google Scholar]
- 28.Ossareh-Nazari, B., C. Maison, B. E. Black, L. Lévesque, B. M. Paschal, and C. Dargemont. 2000. RanGTP-binding protein NXT1 facilitates nuclear export of different classes of RNA in vitro. Mol. Cell. Biol. 20:4562-4571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pasquinelli, A. E., R. K. Ernst, E. Lund, C. Grimm, M. L. Zapp, D. Rekosh, M.-L. Hammarskjold, and J. E. Dahlberg. 1997. The constitutive transport element (CTE) of Mason-Pfizer monkey virus (MPMV) accesses an RNA export pathway utilized by cellular messenger RNAs. EMBO J. 16:7500-7510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ribbeck, K., and D. Görlich. 2001. Kinetic analysis of translocation through nuclear pore complexes. EMBO J. 20:1320-1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ryan, K., and S. R. Wente. 2000. The nuclear pore complex: a protein machine bridging the nucleus and cytoplasm. Curr. Opin. Cell Biol. 12:361-371. [DOI] [PubMed] [Google Scholar]
- 32.Saavedra, C. A., B. K. Felber, and E. Izaurralde. 1997. The simian retrovirus-1 constitutive transport element CTE, unlike HIV-1 RRE, utilises factors required for cellular RNA export. Curr. Biol. 7:619-628. [DOI] [PubMed] [Google Scholar]
- 33.Segref, A., K. Sharma, V. Doye, A. Hellwig, J. Huber, R. Lührmann, and E. Hurt. 1997. Mex67p, a novel factor for nuclear mRNA export, binds to both poly(A)+ RNA and nuclear pores. EMBO J. 16:3256-3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stewart, M., H. M. Kent, and A. J. McCoy. 1998. Structural basis for molecular recognition between nuclear transport factor 2 (NTF2) and the GDP-bound form of the Ras-family GTPase Ran. J. Mol. Biol. 277:635-646. [DOI] [PubMed] [Google Scholar]
- 35.Strässer, K., J. Bassler, and E. Hurt. 2000. Binding of the Mex67p/Mtr2p heterodimer to FXFG, GLFG, and FG repeat nucleoporins is essential for nuclear mRNA export. J. Cell Biol. 150:695-706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suyama, M., T. Doerks, I. C. Braun, M. Sattler, E. Izaurralde, and P. Bork. 2000. Prediction of structural domains of TAP reveals details of its interaction with p15 and nucleoporins. EMBO Rep. 1:53-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tan, W., A. S. Zolotukhin, J. Bear, D. J. Patenaude, and B. K. Felber. 2000. The mRNA export in Caenorhabditis elegans is mediated by Ce-NXF1, an ortholog of human TAP/NXF1 and Saccharomyces cerevisiae Mex67p. RNA 6:1762-1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vankan, P., C. McGuigan, and I. W. Mattaj. 1992. Domains of U4 and U6 snRNAs required for snRNP assembly and splicing complementation in Xenopus oocytes. EMBO J. 11:335-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wiegand, H. L., G. A. Coburn, Y. Zeng, Y. Kang, H. P. Bogerd, and B. R. Cullen. 2002. Formation of Tap/NXT1 heterodimers activates Tap-dependent nuclear mRNA export by enhancing recruitment to nuclear pore complexes. Mol. Cell. Biol. 22:245-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wilkie, G. S., V. Zimyanin, R. Kirby, C. A. Korey, H. Francis-Lang, D. Van Vactor, and I. Davis. 2001. Small bristles, the Drosophila ortholog of human TAP/NXF1 and yeast Mex67p, is essential for mRNA nuclear export throughout development. RNA 7:1781-1792. [PMC free article] [PubMed] [Google Scholar]
- 41.Yang, J., H. P. Bogerd, P. J. Wang, D. C. Page, and B. R. Cullen. 2001. Two closely related human nuclear export factors utilize entirely distinct export pathways. Mol. Cell 8:397-406. [DOI] [PubMed] [Google Scholar]
- 42.Yoon, J. H., D. C. Love, A. Guhathakurta, J. A. Hanover, and R. Dhar. 2000. Mex67p of Schizosaccharomyces pombe interacts with Rae1p in mediating mRNA export. Mol. Cell. Biol. 20:8767-8782. [DOI] [PMC free article] [PubMed] [Google Scholar]