Abstract
It is becoming increasingly evident that the degradation of nuclear proteins requires nuclear-cytoplasmic trafficking of both the substrate proteins, as well as the E3 ubiquitin-ligases. Here, we show that nuclear-cytoplasmic trafficking of the von Hippel-Lindau tumor suppressor protein (VHL) is required for oxygen-dependent ubiquitination and degradation of the alpha subunits of hypoxia-inducible factor (HIF-α). VHL engages in a constitutive transcription-sensitive nuclear-cytoplasmic shuttle unaffected by oxygen tension or levels of nuclear substrate HIF-α. Ubiquitinated forms of HIF-α, as well as VHL/ubiquitinated HIF-α complexes, are found solely in the nuclear compartment of normoxic or reoxygenated VHL-competent cells. HIF-α localizes exclusively in the nucleus of hypoxic cells but is exported to the cytoplasm upon reoxygenation. Oxygen-dependent nuclear ubiquitination and nuclear export of HIF-α can be prevented by treatment with an HIF-specific prolyl hydroxylase inhibitor. Treatment with inhibitors of RNA polymerase II activity, which interfere with the ability of VHL to engage in nuclear export, also prevents cytoplasmic accumulation of HIF-α in reoxygenated cells. This caused a marked increase in the HIF-α half-life without affecting its nuclear ubiquitination. We present a model by which VHL-mediated ubiquitination of HIF-α and its subsequent degradation are dependent upon dynamic nuclear-cytoplasmic trafficking of both the E3 ubiquitin-ligase and the nuclear substrate protein.
von Hippel-Lindau disease is an autosomal-dominantly inherited cancer syndrome caused by the inheritance of a mutated allele of the von Hippel-Lindau tumor suppressor gene. Afflicted individuals are predisposed to develop retinal angioma, central nervous system hemangioblastoma, pheochromocytoma, and renal clear cell carcinoma (RCC) (8, 20, 42, 46). Tumors arise from cells in which the remaining wild-type allele encoding the von Hippel-Lindau tumor suppressor protein (VHL) acquires a somatic inactivating mutation. Biallelic VHL gene defects are also found in the vast majority of sporadic cases of RCC, the most common type of kidney cancer in humans (17, 21). In agreement with the postulated gatekeeper tumor suppressor function of VHL, reintroduction of the VHL gene into VHL−/− RCC cells reverted their tumorigenic phenotype in a nude mice assay (31).
The VHL gene contains three exons and is transcribed into a 4.4-kb mRNA expressed in all tissues and cells examined thus far. The VHL mRNA harbors two in-frame start codons that produce proteins of 213 and 160 amino acids, respectively (4, 30, 60). The 213- and 160-amino-acid proteins display similar biochemical properties and for the sake of simplicity, both forms will be hereafter referred to as VHL. Crystal structure analysis has revealed that VHL essentially consist of two domains: a β-domain that spans residues 63 to 154, and an α-domain that encompasses residues 155 to 189 (63). VHL associates with at least four other partners: elongin B, elongin C, Cullin-2, and Rbx1 (VBC/Cul-2) (13, 14, 37, 39, 48, 54). Structural homology between elongin C and Cullin-2 with two components of the SCFCDC4 yeast E3 ubiquitin-ligase complex SKP1 and CDC53, respectively, have led to the hypothesis that VBC/Cul-2 plays a role in protein ubiquitination (22, 39, 54, 55). E3 ubiquitin-ligases are part of a multiprotein pathway, including E1 ubiquitin-activating enzymes and E2 ubiquitin-conjugating enzymes, involved in conjugating ubiquitin to specific substrate proteins (27). Ubiquitinated proteins are subsequently targeted to the 26S proteasome complex for degradation. It was shown that VBC/Cul-2 could act as an E3 ubiquitin-ligase in vitro and in vivo (6, 33, 47, 52).
E3 ubiquitin-ligases provide the specificity to the degradation process by targeting specific proteins for ubiquitination. VHL acts as the particle recognition protein of VBC/Cul-2 by promoting the recruitment of the alpha subunits of hypoxia-inducible factor (HIF-α) for Cullin-2-mediated ubiquitination (6, 11, 38, 50, 52, 64). The HIF transcription factor is a heterodimer of the beta and alpha subunits. Whereas HIF-β is always present in cells, the HIF-α subunits only accumulate under conditions of low-oxygen tension (hypoxia) (35, 61). There are three HIF-α isoforms: HIF-1α, HIF-2α, and HIF-3α (25, 26, 36, 40), although only HIF-1α and HIF-2α have been shown so far to bind to VHL (HIF-1α and HIF-2α are referred to here simply as HIF-α, except as noted). HIF-α contains the oxygen-dependent degradation (ODD) domain, which binds to the β-domain of VHL. The ODD domain is hydroxylated at a specific prolyl residue (proxylation) under normal oxygen tension by enzymes called prolyl hydroxylase (7, 15). This posttranslational modification increases the affinity of HIF-α toward the β-domain of VHL (15, 32, 34, 49, 67). Proxylation does not occur in hypoxia, or in the presence of HIF-specific prolyl hydroxylase inhibitors, and HIF-α is stabilized since it does not assemble with VHL and consequently fails to undergo ubiquitination.
Ubiquitin-dependent protein degradation plays a key role in multiple biological processes, including signal transduction, cell cycle progression, and transcriptional regulation. It is becoming increasingly evident that ubiquitin-proteasome-mediated degradation of nuclear substrate proteins requires nuclear-cytoplasmic trafficking of both the E3 ubiquitin-ligase and the substrate protein (5, 59). One example is the p53 tumor suppressor protein, whose proteasomal degradation is mediated by the Hdm2 oncoprotein (9, 18, 41, 45, 51, 53, 56, 66). Recent data has shown that Hdm2 consistently shuttles between the nucleus and the cytoplasm, a phenomenon required for efficient degradation of p53. The cyclin-dependent kinase inhibitor p27Kip1 is another nuclear protein that requires nuclear export for proteasomal degradation (65). Nuclear export of p27 is mediated by Jab1, and mutant p27 that fails to assemble with Jab1 is neither exported nor destroyed. The proteasomal degradation of Smad3, a transcription factor induced by transforming growth factor β, requires the Cullin-1-containing ROC1-SCFFbw1a E3 ubiquitin-ligase-mediated nuclear export (19). Likewise, the aryl hydrocarbon receptor (Ahr), a protein that shares structural and functional similarities with HIF-α, contains a classical, leucine-rich, nuclear export signal (NES) (16), which is required for cytoplasmic degradation (12). Leptomycin B, a drug that abolishes the activity of classical NES, prevents degradation of Ahr, as well as p27 and p53, by impeding NES-mediated nuclear export. These data support the proposed model that nuclear substrate proteins must undergo nuclear export for efficient degradation. However, the exact mechanisms by which these nuclear substrates are ubiquitinated and subsequently exported for degradation still remains unknown.
We reported that VHL localizes predominantly to the cytoplasmic compartment but engages in a dynamic nuclear-cytoplasmic shuttle (43, 44). In vitro studies revealed that nuclear export of VHL requires ATP hydrolysis and is Ran dependent (24). VHL nuclear export occurs through a different pathway from that utilized by the classical NES-bearing proteins since it is insensitive to leptomycin B. However, drugs that abolish ongoing RNA polymerase II activity cause a marked redistribution of VHL to the nucleus by impeding nuclear export (44). The recent demonstration that the Cullin-1-containing SCFFbw1a-E3 ubiquitin complex must shuttle to mediate degradation of nuclear Smad3 has led us to hypothesize that transcription-dependent nuclear-cytoplasmic trafficking of the Cullin-2-containing VBC/Cul-2 E3 ubiquitin-ligase might also be required for oxygen-dependent ubiquitination and degradation of its nuclear HIF-α substrates.
Here, we link the previously described ability of VHL to shuttle between the nuclear and cytoplasmic compartments with the ubiquitination and degradation of HIF-α. VHL engages in a constitutive shuttle between the nucleus and the cytoplasm. Upon exposure of cells to normal oxygen tension, VHL mediates ubiquitination of HIF-α in the nuclear compartment prior to its exportation to the cytoplasm. Ubiquitination and nuclear export of HIF-α can be abolished by treatment with dimethyl-oxalylglycine (DMOG), a specific inhibitor of HIF prolyl hydroxylase. Nuclear export of HIF-α can be abated by inhibitors of RNA polymerase II activity, and this is correlated with a marked increase in the HIF-α half-life. The results presented here support a model by which VBC/Cul-2-mediated ubiquitination of HIF-α and its subsequent degradation is dependent upon dynamic nuclear-cytoplasmic trafficking of the E3 ubiquitin-ligase, as well as the substrate protein HIF-α.
MATERIALS AND METHODS
Cell culture, adenoviral infections, drugs, and hypoxia treatment.
The 786-0 (VHL−/−) renal clear cell line (i.e., the RCC cells) and the HeLa cells were obtained from the American Type Culture Collection (Manassas, Va.). The VHL-green fluorescent protein (GFP) cell line corresponds to VHL−/− RCC 786-0 cells stably transfected with a FLAG-tagged VHL-GFP fusion protein as described elsewhere (44), and the WT-7 cell line corresponds to 786-0 cells stably transfected with hemagglutinin (HA)-tagged VHL, a kind gift of William Kaelin, Harvard University. Mouse embryonic fibroblasts nullizygous for HIF-1α were a kind gift from R. S. Johnson (Department of Biology, University of California, San Diego) (57). Cells were maintained in Dulbecco modified Eagle medium (DMEM) supplemented with 10% (vol/vol) fetal calf serum (FCS) in a 37°C, humidified 5% CO2 atmosphere incubator. Proteasome inhibitor treatment was performed by using calpain inhibitor I (Alln; Sigma) or MG132 (Calbiochem) to final concentrations of 100 and 10 μM, respectively, in the culture medium for 2 h before the cells were harvested. Actinomycin D (ActD) and 5,6-dichlorobenzimidazole riboside (DRB) were used at final concentrations of 10 and 25 μg/ml, respectively, for the indicated time. Hypoxia treatment was performed in a hypoxic chamber with 1% O2 atmosphere and tempered at 37°C. DMOG (a kind gift of Imre Schlemminge, Oxford University) was used at a concentration of 1 mM for the indicated time.
Expression vector and constructs.
The human VHL cDNA, which codes for a 213-amino-acid VHL protein, was subcloned into pcDNA3.1(−) vector (Invitrogen). A Flag epitope tag (DYKDDDDK) was added to the N terminus of the VHL cDNA open reading frame. A cDNA coding for an enhanced fluorescence version of GFP (Fred25 [62]) was subcloned at the C terminus of VHL to produce the Flag-VHL-GFP fusion protein. VHL proteins lacking the last 56 amino acids (ΔE3), lacking amino acids 114 to 154 (ΔE2), and lacking amino acids 64 to 113 were fused to the N terminus of GFP to produce the ΔE3-GFP, ΔE2-GFP, and Δ64-113-GFP fusion protein, respectively. The enhanced fluorescence version of GFP cDNA was inserted in-frame at the N terminus of a human HIF-1α cDNA to produce the GFP-HIF-1α fusion protein. All constructs were verified by standard DNA sequencing.
Construction of adenovirus through Cre-lox recombination.
CRE8 and 293 cells were obtained from David Park (University of Ottawa, Ottawa, Ontario, Canada) and cultured in DMEM with 10% FCS. The construction and properties of CRE8 cells are described elsewhere (10). The pAdlox vector and the ψ5 viral DNA were also obtained from David Park. The GFP-HIF-1α construct, as well as Flag-VHL-GFP and GFP, were subcloned into pAdlox vector. Transfections were done according to Graham and van der Eb (23). Typically, a confluent 10-cm-diameter dish of CRE8 cells (1.6 × 107) was split into 5- to 6-cm-diameter dishes for transfection 2 to 4 h later. Each dish received 3 μg of pAdlox vector containing the foreign VHL construction and 3 μg of ψ5 viral DNA in a final volume of 0.5 ml of CaPO4, which was applied to the cells for 16 h. The 10% FCS-DMEM was replaced with 2% FCS-DMEM 16 h after the transfection. Cells were fed with fresh 2% DMEM after 64 h. Cells were harvested and subjected to freeze-thawing three times with an alternating liquid N2-37°C water bath at 8 to 10 days postinfection. Plaque purification assays were performed on all viruses prior to CRE8 amplification.
In vitro binding assay.
VHL−/− RCC cells infected with the adenovirus-expressing GFP-HIF-1α (adGFP-HIF-1α) were washed with ice-cold phosphate-buffered saline (PBS), scraped, and lysed in ice-cold buffer containing 100 mM NaCl, 0.5% Igepal CA630, 20 mM Tris-HCl (pH 7.6), 5 mM MgCl2, and 1 mM sodium orthovanadate with a cocktail of protease inhibitor [2 μg of leupeptin/ml, 2 μg of aprotinin/ml, 1 μg of pepstatin A/ml, and 1 mM 4-(2-aminoethyl)benzenesulfonyl fluoride]. Cell lysis was performed for 1 h in a tumbler at 4°C. The adGFP-HIF-1α lysates were cleared by centrifugation (10,000 × g) for 15 min at 4°C and had a protein concentration of ca. 5 μg/μl. Cell lysates (250 μg) of adGFP-HIF-1α were mixed with 50 μl of reticulocyte lysates programmed with pcDNA vector alone, expressing VHL-GFP or the different VHL deletion mutants produced by using the T7 promoter. The binding assay was performed in a total volume of 1 ml for 1 h in a tumbler at 4°C with anti-FLAG M2 beads (Scientific Imaging Systems, Eastman Kodak Co.) The beads were washed several times, eluted with 4× sodium dodecyl sulfate (SDS)-protein loading buffer, boiled for 5 min, and loaded onto a SDS-8% polyacrylamide gel electrophoresis (PAGE). The proteins were transferred on polyvinylidene difluoride membranes by standard methods. Blots were blocked with 3% milk powder in PBS containing 0.2% Tween 20 and were incubated in the presence of anti-HIF-1α (Transduction Laboratories) and anti-FLAG M2 monoclonal antibody (Sigma) or an anti-HIF-2α rabbit polyclonal antibody (Novus Biologicals).
Subcellular fractionation.
Cells were grown to confluence and treated as desired, and fractionation was performed in normoxic conditions or in the hypoxia chamber by using deoxygenated buffers. Cells were washed in cold PBS and scraped in transport buffer containing 20 mM HEPES (pH 7.3), 110 mM potassium acetate, 5 mM sodium acetate, 2 mM magnesium acetate, 1 mM EGTA, 2 mM dithiothreitol, and a cocktail of protease inhibitor (see in vitro binding assay). Cells were left on ice for 2 min to equilibrate in the buffer before the addition of 50 μg of digitonin/ml. Permeabilization was performed for 5 to 7 min on ice with gentle shaking and was monitored by fluorescence microscope with Hoechst stain 33258, which only stained the nucleus of permeabilized cells. Permeabilization reaction was stopped when >99% of cells displayed stained nuclei. Permeabilized cells were then centrifuged at 800 × g to separate the nuclear fraction (pellet) from the cytosolic fraction (supernatant). The nuclei were washed in transport buffer containing the protease inhibitor cocktail, and the cytosolic fraction was recentrifuged to eliminate the remaining nuclei. The total fraction was prepared with the same amount of cells as those taken for fractionation. Cells of the total fraction and nuclear-cytosolic fractions were resuspended in an equal volume of lysis buffer or SDS loading buffer. An equal volume of each fraction was subjected to immunoprecipitation or SDS-PAGE analysis and Western blotting.
Immunoprecipitations.
Cells lysates and immunoprecipitations were performed in atmospheric oxygen for the normoxic experiments or strictly restricted to the hypoxia chamber for the hypoxic experiments. For immunoprecipitation of Flag-VHL-GFP, M2 beads were added to cell lysate containing ca. 1 mg. After 1 h of incubation at 4°C in a tumbler, the beads were washed several times, eluted with 4× SDS-protein loading buffer in normoxia or hypoxia, and then boiled for 5 min before undergoing SDS-8% PAGE. Western blots were performed with the indicated antibodies. For the capture of ubiquitinated proteins, GST-S5a-Agarose beads (Affiniti) were used to capture polyubiquitinated proteins. The cell lysates were prepared in a lysis buffer containing 20 mM Tris-HCl (pH 7.5), 5% (vol/vol) glycerol, and 0.1% Triton X-100, with 2 μg of leupeptin, 2 μg of aprotinin, and 1 μg of pepstatin A/ml and 1 mM 4-(2-aminoethyl) benzene sulfonyl fluoride. The beads were washed several times and eluted with 4× SDS-protein loading buffer prior to Western blotting with the indicated antibodies.
Immunofluorescence.
HeLa cells were plated at 50% density on coverslips. Cells were incubated for 8 h in hypoxia (1% oxygen). ActD (10 μg/ml) was added 2 h prior to the return to normoxia. Cells were fixed with 1% formaldehyde-PBS for 20 min prior to a 1-h incubation with 1 μg of an anti-HIF-1α antibody/ml diluted in PBS-1% Triton-10% FCS. Cells were washed in PBS several times before and incubation with an anti-mouse Texas red-labeled secondary antibody. Coverslips were mounted on slides with Fluoromount.
Fluorescence analysis and image processing.
GFP fluorescence images were captured by using a Zeiss Axiovert S100TV microscope with a C-Apochromat ×40 water immersion objective lens, equipped with an Empix digital charge-coupled device (CCD) camera, and assessed by using Northern Eclipse software. The images were manipulated and quantified with Northern Eclipse, NIH Image, and Adobe PhotoShop software as described elsewhere (44). GFP images were always taken before Hoechst images to minimize any possible bleaching effect. Image manipulation and nuclear-cytoplasmic ratios were calculated as previously described (44). Briefly, total cellular signal was measured by multiplying the area of the cell by the integrated pixel intensity within the cell and subtracting the background value obtained from a cell-free region of the image. The nuclear signal was obtained by measuring the pixel intensity of the nucleus subtracted by the mean background and then multiplying this value by the area of the nucleus. The cytoplasmic signal was obtained by subtracting the total signal minus the nuclear signal. Finally, the nuclear-cytoplasmic ratio corresponded to the nuclear signal divided by the cytoplasmic signal. All measurements were performed well under the saturation limits, even though some of the pictures might appear to be saturated. The advantage of measuring nuclear-cytoplasmic ratios is that the obtained values are independent of subsequent manipulations of the original images, which are done for presentation purposes.
RESULTS
Characterization of adGFP-HIF-1α fusion protein.
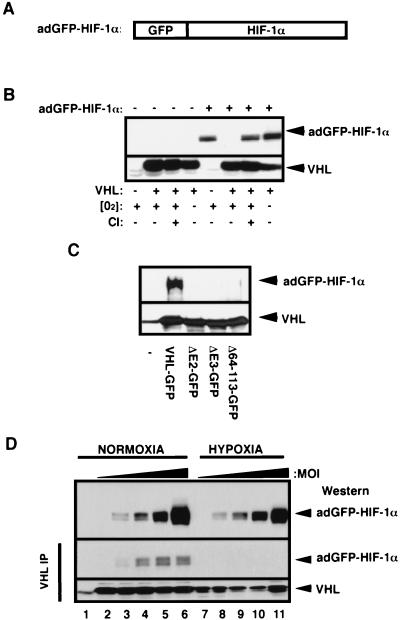
One of the inherent difficulties in studying the ubiquitination and degradation mechanisms of HIF-α is that these molecules have a very short half-life in normoxia (29). This renders the detection of HIF-α at steady state practically impossible without the use of proteasome inhibitors or mutant cell lines. To circumvent this problem, we generated an adenovirus that expresses a GFP fusion to the HIF-1α subunit (adGFP-HIF-1α, Fig. 1A). The GFP-HIF-1α fusion protein was reported to be a functional fusion protein in different biochemical assays (64). VHL−/− RCC cells and VHL(+) cells can be infected to high efficiency with an adenovirus. At a low multiplicity of infection (MOI), i.e., two to three viral particles/cell, the adenovirus-produced GFP-HIF-1α shared similar biochemical characteristics with endogenous HIF-1α in that it was rapidly degraded in an oxygen-, VHL-, and ubiquitin-proteasome-dependent manner (Fig. 1B). The adGFP-HIF-1α bound to VHL but not to VHL α- and β-domain mutants prepared by in vitro translation (Fig. 1C). The fusion protein was detectable in normoxic VHL(+) cells infected with increasingly higher titers of adGFP-HIF-1α (Fig. 1D, top panel, lanes 2 to 6), indicating that VHL-mediated ubiquitination and degradation of HIF-α can be saturated. Hypoxic VHL(+) cells were also infected with increasingly higher titers of adGFP-HIF-1α, although we used five times less MOI than for normoxic VHL(+) cells (Fig. 1D, top panel, lanes 7 to 11). This resulted in normoxic and hypoxic VHL(+) cells producing similar amounts of adGFP-HIF-1α at steady state. Immunoprecipitation of VHL from a cell line stably expressing VHL-GFP revealed that VHL-GFP/adGFP-HIF-1α complexes were prevalent in normoxic cell lysates compared to hypoxic cell lysates (Fig. 1D, two bottom panels). These results show that adGFP-HIF-1α in vivo and in vitro ubiquitination and degradation are mediated by VHL and regulated by oxygen tension, thus sharing the previously described characteristics of HIF-1α. These results also confirm that adGFP-HIF-1α can be used as a tool to investigate mechanisms of ubiquitination and degradation in normoxic VHL-competent cells without the use of drugs or mutant cell lines.
FIG. 1.
Characterization of the adGFP-HIF-1α fusion protein. (A) Schematic representation of HIF-1α fused to GFP. The cDNA encoding for the alpha subunit of the HIF-1α was fused to the C terminus of the cDNA of the GFP, resulting in GFP-HIF-1α fusion protein. (B) Proteolysis of adGFP-HIF-1α is proteasome mediated and VHL and oxygen dependent. VHL−/− RCC cells (cell line 786-0) or the VHL(+) WT-7 stable cell line (expressing HA-tagged VHL) were either not infected (−) or infected (+) with low titers (MOI) of the adGFP-HIF-1α. Cells were incubated in normal oxygen tension (O2+) or in hypoxic conditions (1% oxygen; O2−) for 4 h. Normoxic cells were also incubated in the presence (CI+) or the absence (CI−) of calpain inhibitor I for 2 h. Cell lysates were subjected to an SDS-8% PAGE analysis, and a Western blot was detected with anti-HIF-1α or anti-HA antibody. (C) The fusion protein adGFP-HIF-1α binds to VHL-GFP in vitro. Lysates of adGFP-HIF-1α infected VHL−/− RCC cells were incubated in reticulocyte lysates programmed with pcDNA vector alone (−), expressing FLAG tagged-VHL-GFP or mutants of the α or β domain (ΔE2-GFP, ΔE3-GFP, and Δ64-113-GFP). The resulting mixture was immunoprecipitated with M2 monoclonal antibody and immunoblotted with anti-HIF-1α or anti-M2 antibody. (D) adGFP-HIF-1α binds to VHL in normoxia but not in hypoxia. VHL-GFP stable cell lines were infected with increasing amounts of adGFP-HIF-1α virus and incubated in normoxia or hypoxia for 4 h. Immunoprecipitation was carried out in normoxic or hypoxic conditions with M2 antibody. Immunodetection of input (top panel) and of the immunoprecipitation was done with an anti-HIF-1α or anti-M2 antibody. Lane 1 represents M2 beads alone.
Subcellular localization and trafficking of VHL and adGFP-HIF-1α are independent from each other.
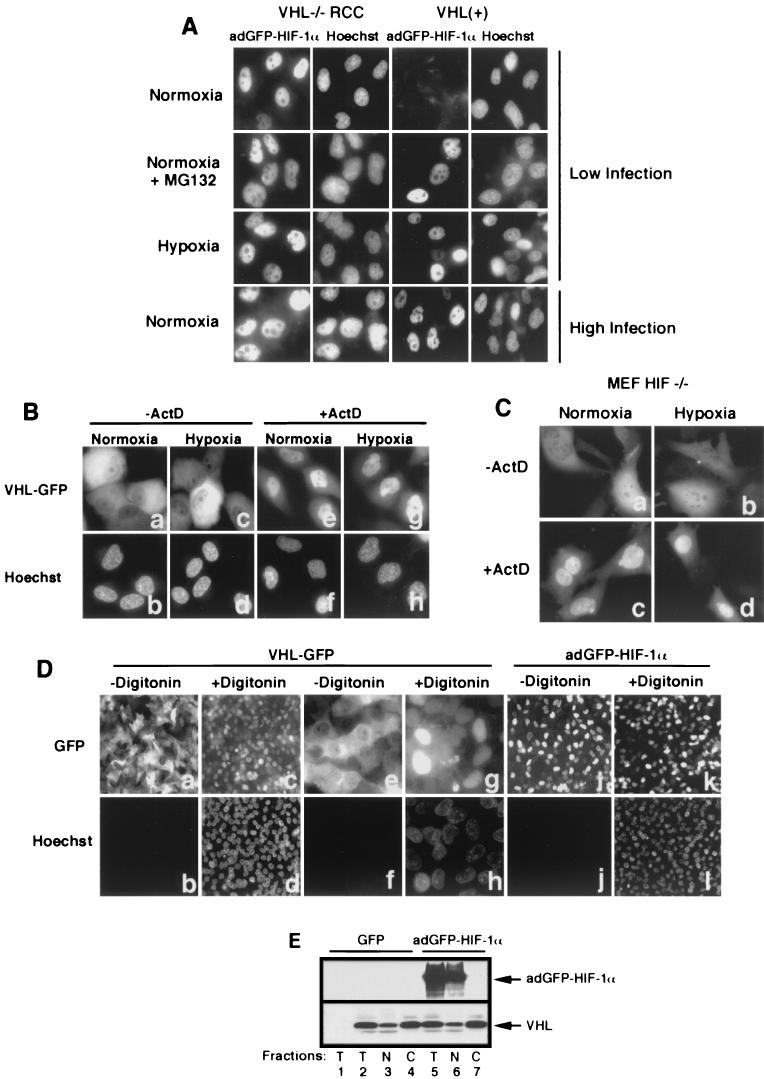
To determine the subcellular localization of adGFP-HIF-1α, we first examined its distribution in living cells by fluorescence microscopy, depending upon oxygen availability and VHL status. The fusion protein was found exclusively in the nucleus of VHL−/− RCC cells, whereas it was undetectable in VHL-positive cells infected at a low infection (Fig. 2A, first row). The addition of proteasome inhibitor (MG132) to infected normoxic VHL(+) cells, or incubation in hypoxia, resulted in the stabilization and nuclear accumulation of adGFP-HIF-1α (Fig. 2A, second and third rows, respectively). The fusion adGFP-HIF-1α was found exclusively in the nucleus of normoxic VHL−/− RCC and VHL(+) cells infected with 10-fold (data not shown)- to 100-fold (Fig. 2A, fourth row)-higher MOIs. Similar data were obtained in other VHL-competent cell lines, including HeLa, NIH 3T3, 293, and Cos-7 cells (data not shown). These results demonstrate that HIF-1α nuclear localization is constitutive, independent of VHL status or of oxygen availability. Nuclear targeting of HIF-1α is also highly efficient since we failed to saturate the import mechanisms even with high excess of importable HIF-1α cargo.
FIG.2.
Subcellular localization of VHL and adGFP-HIF-1α is independent from each other. (A) adGFP-HIF-1α localize to the nucleus. Fluorescence analysis of VHL−/− RCC cells and the VHL(+) WT-7 stable cell line expressing adGFP-HIF-1α in normoxia at low levels of infection (first row) and of normoxic cells in the presence of proteasome inhibitor MG132 (second row) or in hypoxia (third row). Normoxic VHL−/− RCC and VHL(+) cells infected with high titers of adGFP-HIF-1α are shown in the last row. (B) Transcription-dependent shuttling of VHL is unaffected by hypoxic treatment. Fluorescence analysis of VHL−/− RCC cells expressing VHL-GFP in normoxia or hypoxia (4 h) in the absence or the presence of ActD (10 μg/ml) for 2 h. (C) Transcription-dependent shuttling of VHL occurs in the absence of substrate HIF-α. Fluorescence analysis of HIF−/− mouse embryonic fibroblasts expressing VHL-GFP in normoxia or hypoxia (4 h) in the absence or the presence of ActD (10 μg/ml) for 2 h. (D) Digitonin-permeabilization system to achieve nuclear-cytoplasmic fractionation of cells. VHL-GFP-expressing VHL−/− RCC cells were either not treated (a and b and, at a higher magnification, e and f) or treated for 5 min with 50 μg of digitonin/ml (c and d and, at a higher magnification, g and h) and observed for GFP fluorescence (a, c, e, and g) or for cell-impermeable Hoechst stain 33258. Notice the complete loss of cytoplasmic VHL-GFP signal but the retention of nuclear signal in digitonin-treated cells and that essentially all of the nuclei of permeabilized cells have been stained with Hoechst. Panels j to l show that nuclear adGFP-HIF-1α remains in the nuclear compartment after digitonin treatment. (E) VHL steady-state localization is independent of levels of adGFP-HIF-1α in normoxic cells. VHL(+) cells were either infected with GFP adenovirus or with high levels of adGFP-HIF-1α virus. Biochemical subcellular fractionation was performed with the digitonin system to obtain total (T), nuclear (N), and cytosolic (C) fractions, which were analyzed by Western blot with anti-M2 and anti-HIF-1α antibody. A negative control is present in the first lane and represents VHL−/− RCC infected with GFP virus.
There is a striking difference between the steady-state localization of HIF-1α and VHL-GFP, whose localization is mostly cytoplasmic at steady state (Fig. 2Ba and b). VHL-GFP subcellular localization was unaffected by oxygen tension (Fig. 2Ba to d) and occurred in cells that do not express substrate HIF-1α (Fig. 2Ca and b). Previously published data demonstrated that VHL and the VBC/Cul-2 complex accumulated in the nuclear compartment upon addition of RNA polymerase II inhibitors, such as ActD (24, 44). ActD-induced nuclear accumulation of VHL-GFP occurred regardless of oxygen tension (Fig. 2Ce to h) and in the absence of HIF-1α (Fig. 2Cc and d). However, it remained unclear whether the substrate HIF-1α had any effect on the subcellular localization of its E3 ubiquitin-ligase under conditions in which binding and ubiquitination would be expected to occur. Therefore, we sought to determine whether overexpression of adGFP-HIF-1α in normoxic VHL-positive cells would affect VHL steady-state distribution. VHL-positive cells were either infected with adGFP or infected with adGFP-HIF-1α. VHL subcellular distribution was monitored by biochemical fractionation by using a method based on the digitonin-permeabilization system previously described by our group (24). We used this method for the subcellular fractionation since it is a rapid, easy, and highly reproducible approach for separating soluble cytoplasmic proteins from nuclear content. This method is also useful for manipulations performed in the hypoxic incubator, as well as in experiments that have to be carried out within a few seconds after return of cells to an oxygenated environment (see Fig. 3 to 6). Fractionation was monitored by loss of cytoplasmic VHL-GFP signal in >99% of the treated cells (Fig. 2Da and c and, at a higher magnification, in De and g), as well as staining of the nuclei with membrane-impermeable Hoechst stain 33258, which only stains the nuclei of permeabilized cells (Fig. 2Db and d and, at a higher magnification, in Df and h). The fusion protein adGFP-HIF-1α remained in the nucleus after digitonin fractionation (Fig. 2Di to l). Experiments were conducted only when >99% of the cells displayed Hoechst-stained nuclei. Using these conditions, histone H1 was detected exclusively in the nuclear fraction (data not shown); other quality controls for this fractionation procedure have already been published (24). Western blotting revealed that ca. 10% of the VHL-GFP localizes to the nuclear fraction, whereas 90% was found in the soluble cytosolic fraction (Fig. 2E, bottom panel, lanes 2 to 4), a finding consistent with CCD image analysis of the subcellular distribution of VHL-GFP in living cells (44). Signal for adGFP-HIF-1α was essentially restricted to the nuclear compartment (Fig. 2E, top panel, lanes 5 to 7), a finding consistent with data shown in Fig. 2A and Di to l. Total levels of cellular VHL-GFP, as well as its steady-state distribution, remained unchanged regardless of the cellular levels of adGFP-HIF-1α (Fig. 2E, bottom panel, lanes 2 to 7) even in conditions of normal oxygen tension. Taken together, these results demonstrate that VHL-GFP engages in a constitutive transcription-dependent nuclear-cytoplasmic shuttle insensitive to the relative levels of substrate HIF-1α and oxygen tension.
FIG.3.
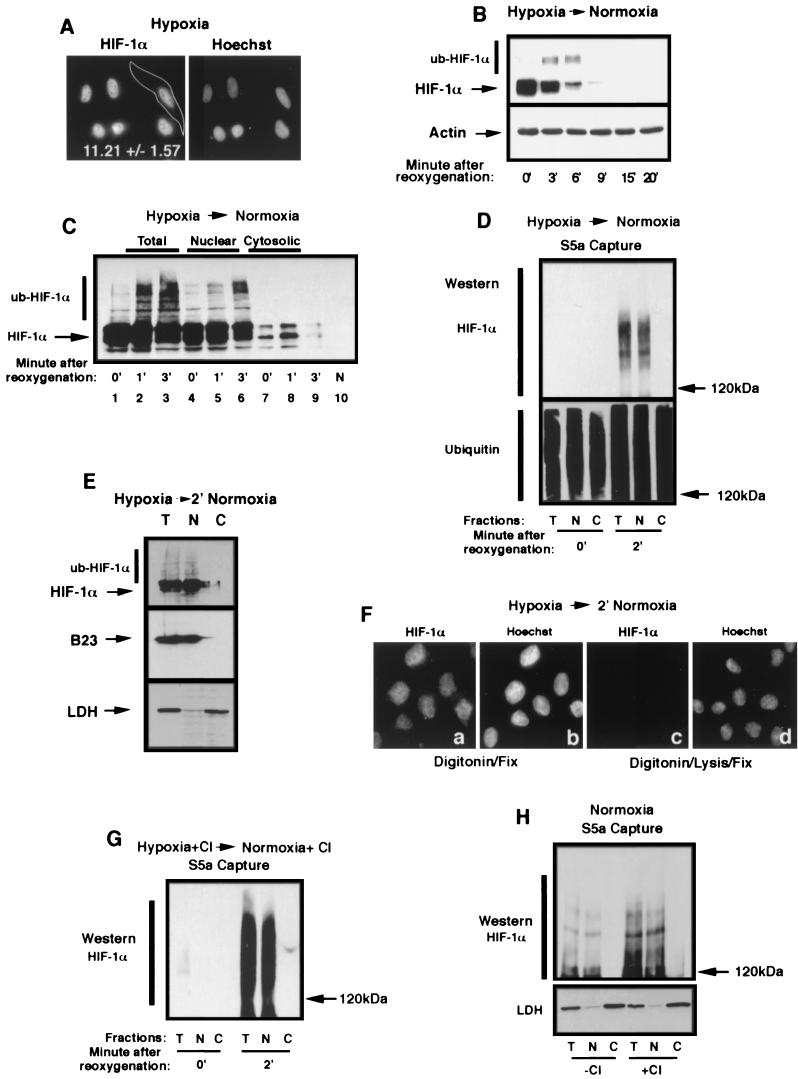
Ubiquitination of HIF-1α occurs in the nuclear compartment. (A) HIF-1α localizes to the nuclear compartment of hypoxic HeLa cells. HeLa cells grown on coverslips were incubated for 8 h in hypoxia prior to fixation with 1% formaldehyde and staining with an anti-HIF-1α antibody and a secondary Texas red-labeled anti-mouse antibody. A CCD-captured image was analyzed as described in Materials and Methods for the nuclear/cytoplasmic ratio of the distribution of the fluorescence signal. The mean ± the standard deviation of 32 cells of three independent experiments is shown. (B) HIF-1α is efficiently degraded upon the return of hypoxic cells to normoxia. HeLa cells were incubated for 8 h in hypoxia prior to a return to a normoxic incubator for the indicated time. Cells were lysed in 4% SDS-PBS and analyzed by SDS-8% PAGE, followed by Western blotting with an anti-HIF-1α antibody. Notice the appearance of higher-molecular-weight bands at 3 and 6 min, which is most likely ub-HIF-1α. (C) ub-HIF-1α appears promptly in the nucleus after reoxygenation of hypoxic cells. HeLa cells were incubated in hypoxia for 8 h (0′) prior to being transferred to an oxygenated environment at 37°C for 1 and 3 min. Samples from each time point were submitted to subcellular fractionation, and the resulting cell fractions (total, nuclear, and cytosolic) were analyzed by SDS-8% PAGE, followed by Western blotting with an anti-HIF-1α antibody. Normoxic (N) HeLa cell lysate is shown in lane 10. (D) ub-HIF-α is found strictly in the nuclear compartment upon reoxygenation. Capture of polyubiquitinated proteins with agarose-GST-S5a beads was carried out in lysates of total (T), nuclear (N), or cytosolic (C) fractions of hypoxic or reoxygenated (2 min) HeLa cells. The captured polyubiquitinated proteins were analyzed by SDS-6% PAGE, followed by Western blot with an anti-HIF-1α or anti-ubiquitin antibody. (E) Reoxygenated HeLa cells were treated with digitonin, and nuclear or cytosolic fractions were blotted with an anti-HIF-1α antibody, an anti-LDH antibody, or an anti-B23 antibody. (F) HIF-1α can be detected solely in the nucleus of digitonin-treated cells after a return of hypoxic cells to normoxia. Hypoxic HeLa cells grown on coverslips were transferred to an oxygenated incubator for 2 min prior to permeabilization with digitonin for 5 min at 4°C. Permeabilized cells were either fixed with 1% formaldehyde (a and b) or treated with lysis buffer prior to fixation (c and d). Fixed and permeabilized cells were staining with an anti-HIF-1α antibody and a secondary Texas red-labeled anti-mouse antibody (a and c) or with Hoechst stain (b and d). Note that the specific HIF-1α signal can be detected only in the nuclear compartment in panel a and is completely extractable with the lysis buffer (see panel c). (G) ub-HIF-1α can only be detected in the nuclear compartment in proteasome-treated and reoxygenated HeLa cells. HeLa cells were treated for 1 h with proteasome inhibitors prior to reoxygenation and cells were treated as described for panel D to detect ub-HIF-1α. (H) ub-HIF-1α are found solely in the nucleus of normoxic cells. Normoxic HeLa cells treated (+) or not (−) with calpain inhibitor (CI) were fractionated into total (T), nuclear (N), and cytosolic (C) lysates, which were then subjected to agarose-GST-S5a immunoprecipitation and anti-HIF-1α Western blot analysis. The bottom panel shows the fractions blotted with anti-LDH.
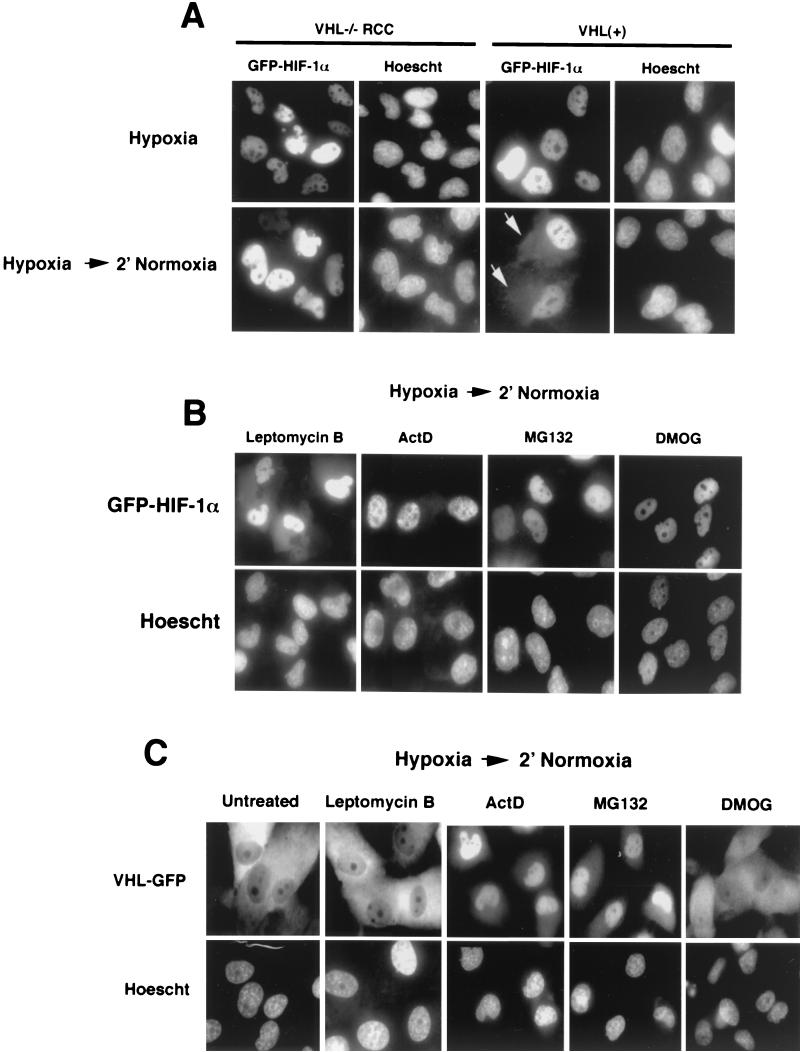
FIG. 6.
Cytoplasmic accumulation of adGFP-HIF-1α upon reoxygenation requires VHL and occurs through an ActD-dependent, but leptomycin B-independent pathway. (A) Nuclear export of adGFP-HIF-1α requires VHL. VHL−/− RCC and VHL(+) cells were infected with adGFP-HIF-1α, incubated for 8 h in hypoxia, and transferred for 2 to 15 min in normoxia. The arrows point to the cytoplasmic signal. The same exposure time was used for all of the images. (B) Effect of drugs on cytoplasmic accumulation of adGFP-HIF-1α upon reoxygenation of hypoxic cells. VHL(+) cells were infected with adGFP-HIF-1α and incubated in hypoxia for 8 h in the presence of the indicated drug for last 2 h, except for DMOG, which was incubated for 8 h. Cells were transferred to an oxygenated incubator for 2 to 15 min. The same exposure time was used for all of the images. (C) Nuclear accumulation of VHL-GFP occurs upon treatment with ActD and MG132 but not with leptomycin B or DMOG. VHL−/− RCC cells were infected with adVHL-GFP and incubated for 8 h in hypoxia with the indicated drugs for the last 2 h except for DMOG, which was incubated for 8 h. Cells were transferred to an oxygenated incubator for 2 to 15 min. The same exposure time was used for all of the images.
Ubiquitination of HIF-1α occurs in the nuclear compartment.
It is well known that HIF-α accumulates in the nuclear compartment of hypoxic cells (28, 58 ) (Fig. 3A ). According to our quantitative digital imaging analysis, ca. 10 to 12 molecules of HIF-1α reside in the nuclear compartment for one molecule in the cytoplasm (Fig. 3A; see also Materials and Methods) at steady state in hypoxic cells. HIF-1α is very rapidly degraded upon reoxygenation of hypoxic HeLa cells (28), with a half-life of ca. 90 s (Fig. 3B) with the concomitant appearance of higher-molecular-weight bands reminiscent of ubiquitinated forms of HIF-α (ub-HIF-α). Since HIF-α resides in the nuclear compartment of hypoxic cells, we first sought to determine whether its ubiquitination occurred in the nuclear compartment or whether HIF-α undergoes nuclear export prior to cytoplasmic ubiquitination. To identify the subcellular site of ubiquitination of HIF-α, HeLa cells were incubated in hypoxia for 8 h, a condition in which endogenous HIF-1α accumulated almost exclusively in the nuclear compartment (Fig. 3A and C, lanes 1, 4, and 7). Hypoxic HeLa cells were transferred to an oxygenated environment at 37°C for 1 and 3 min to mimic the physiological conditions in which stabilized HIF-α is rapidly degraded. Ice-cold PBS was added to the cells, and subcellular fractionation was performed by digitonin treatment, as described in Fig. 2D and E, at 4°C, followed by the addition of equal volumes of 4% SDS to avoid any further in vitro ubiquitination or degradation reactions. Equal volumes of the subcellular fractions were subjected to SDS-PAGE. Western blot analysis revealed the presence of slower-migrating HIF-1α, most likely ub-HIF-1α, exclusively in the nuclear fractions 45 to 60 s after reoxygenation (Fig. 3C, lanes 2, 5, and 8). The high-molecular-weight bands were more pronounced 3 min after reoxygenation, but these bands were still restricted to the nuclear compartment (Fig. 3C, lanes 3, 6, and 9). A reproducible increase in the cytosolic HIF-α was also detected upon reoxygenation, but we failed to notice high-molecular-weight HIF-α even after longer exposure of the autoradiogram. To further support the claim that the high-molecular-weight bands consisted of ub-HIF-1α, subcellular fractions were incubated in the presence of glutathione S-transferase (GST)-S5a protein coupled to agarose beads. The S5a column binds with high affinity to polyubiquitinated proteins. For these experiments, the fractions were treated with lysis buffer, not 4% SDS, to extract the nuclear material. After the S5a-agarose capture assay, the bound proteins were eluted with SDS-loading buffer, followed by Western blot with an anti-HIF-1α antibody. ub-HIF-1α was absent from the lysates of hypoxic cells (Fig. 3D, first three lanes of top panel). However, ub-HIF-1α was readily detectable in the total fraction (T) and the nuclear compartment (N), but not in the cytosolic fraction (C), of reoxygenated HeLa cells (Fig. 3D, last three lanes of top panel). Blots were also incubated with an anti-ubiquitin antibody, revealing the presence of captured polyubiquitinated proteins in hypoxic and normoxic cells, regardless of the subcellular localization (Fig. 3D, bottom panel). Furthermore, lactate dehydrogenase (LDH), a soluble cytoplasmic protein, was detected solely in the cytosolic fraction, whereas the nuclear B23 protein was present only on the nuclear fraction (3) (Fig. 3E). These results demonstrate that the lack of ub-HIF-1α in the “C” fraction is not due to the failure of the digitonin permeabilization method to release soluble cytosolic proteins. However, it could be argued that ub-HIF-α copurified with the nuclear fraction, although these molecules are located in a different subcellular compartment. To exclude this possibility, we decided to perform immunofluorescence on reoxygenated and digitonin-treated cells. After treatment with digitonin, cells were fixed in 1% formaldehyde and then stained with an anti-HIF-1α antibody. We found a specific HIF-1α signal exclusively in the nuclear compartment of digitonin-treated cells (Fig. 3Fa and b). This signal was lost if cells were treated with the lysis buffer after treatment with digitonin and prior to fixation, indicating that the vast majority of HIF-α can be extracted by our method (Fig. 3Fc and d). These results demonstrate that ub-HIF-α molesules found in the nuclear fraction, after biochemical fractionation, are indeed localized to the nuclear compartment. ub-HIF-α molecules were detected solely in the nuclear fraction of reoxygenated HeLa cells treated for the last hour in hypoxia in the presence of the proteasome inhibitor, calpain inhibitor (Fig. 3G). Polyubiquitinated forms of HIF-1α were detected exclusively in the nuclear compartment of normoxic HeLa cells (Fig. 3H) in the absence or presence of proteasome inhibitor (i.e., calpain inhibitor). Fractions were blotted with LDH, demonstrating the complete release of a cytosolic protein upon treatment with digitonin (Fig. 3H, bottom panel). These results demonstrate that prompt ubiquitination of HIF-1α occurs in the nuclear compartment upon reoxygenation of hypoxic cells. They also suggest that ubiquitination of HIF-1α is most likely a nuclear event even in cells maintained in normoxia.
Nuclear ubiquitination of HIF-α requires VHL and prolyl hydroxylase activity.
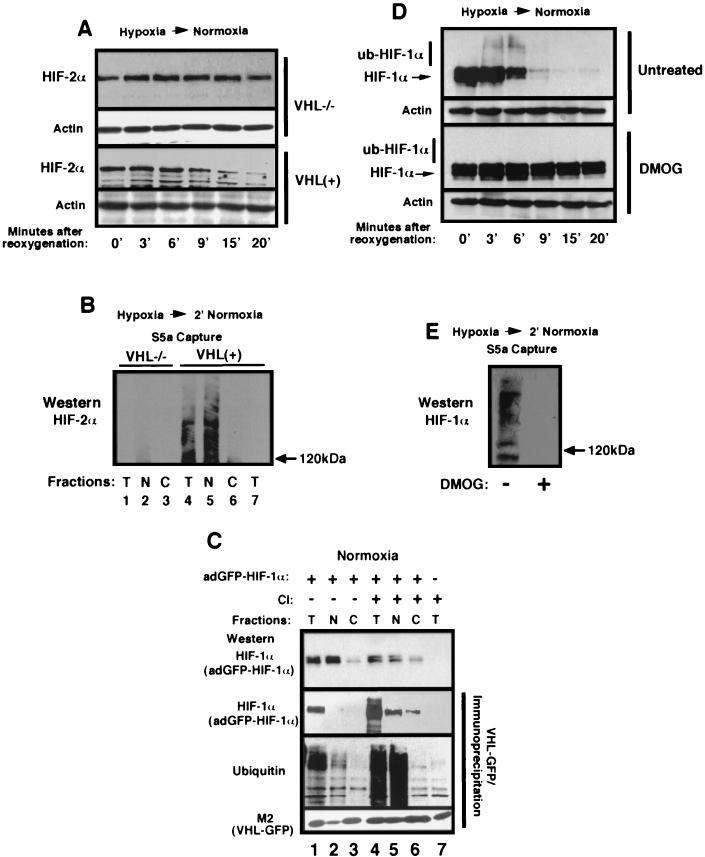
We next verified whether nuclear ubiquitination of HIF-1α upon reoxygenation of hypoxic cells was mediated by VHL (VBC/Cul-2) or by another nuclear E3 ubiquitin-ligase complex. To test this, VHL−/− RCC and the same cells expressing reintroduced VHL [VHL(+)] were used in the hypoxia-to-normoxia experiments. Western blot analysis revealed that VHL was required for the efficient degradation of HIF-2α upon reoxygenation of hypoxic cells (Fig. 4A). GST-S5a bead capture assays were carried out on lysates of subcellular fractions of reoxygenated VHL−/− RCC and VHL(+) cells. We failed to detect ub-HIF-2α in reoxygenated VHL−/− RCC cells (Fig. 4B, lanes 1 to 3). However, we detected ub-HIF-2α in VHL(+) cells, and these forms were restricted to the nuclear compartment (Fig. 4B, lanes 4 to 6), suggesting that nuclear ubiquitination of HIF-2α requires VHL. We also asked in which subcellular compartment VHL bound to HIF-1α to mediate its ubiquitination. Normoxic VHL(+) cells were infected with a high MOI of adGFP-HIF-1α for 24 h, leading to the nuclear accumulation of the fusion protein as measured by straight Western blot (Fig. 4C, top panel, lanes 1 to 3 and also lane 7, which corresponds to the uninfected, negative control). We then carried out immunoprecipitation of VHL (Fig. 4C, second, third, and bottom panels). These immunoprecipitation experiments revealed the formation of VHL/adGFP-HIF-1α complexes in the total fraction (Fig. 4C, the second panel for adGFP-HIF-1α and the bottom panel for VHL, compare lane 1 with lane 7). The VHL immunoprecipitates were also blotted with an anti-ubiquitin antibody (Fig. 4C, third panel from the top) to detect the presence of VHL complexes containing ub-adGFP-HIF-1α (Fig. 4C, third panel, lane 1). The signal detected in lane 1 represents ubiquitinated adGFP-HIF-1α since no specific signal was detected by the use of the anti-ubiquitin antibody on the VHL immunoprecipitate of uninfected cells. VHL/ub-adGFP-HIF-1α complexes were detected in the nuclear (lane 2) but not in the cytosolic (lane 3) fractions. However, the amounts of immunoprecipitable VHL bound to ubiquitinated forms of adGFP-HIF-1α were lower in the nuclear fraction than in the total fraction. We reasoned that this was likely due to the low abundance of immunoprecipitable VHL in the nuclear compartment at steady state. Treatment of cells with calpain inhibitor I caused VHL-GFP to accumulate in the nuclear compartment (6; I. Groulx and S. Lee, unpublished observations) (see also Fig. 6), resulting in a marked increase in the nuclear pool of VHL at steady state (Fig. 4C, compare lanes 2 and 5 of the bottom panel). Immunoprecipitation of VHL in lysates of calpain inhibitor-treated cells revealed the presence of VHL/adGFP-HIF-1α complexes in the nuclear compartment with some detectable signal in the cytosolic compartment (Fig. 4C, lanes 4 to 6, second panel). However, VHL/ub-adGFP-HIF-1α were detected solely in the nuclear compartment and not in the cytosolic fraction (Fig. 4C, compare lanes 5 and 6 of the third panel). Together, these results demonstrate that VHL-mediated ubiquitination of HIF-α can occur in the nucleus, providing the first demonstration of a function for VHL in this subcellular compartment.
FIG. 4.
Nuclear ubiquitination of HIF-α requires VHL and prolyl hydroxylase activity. (A) Degradation of HIF-2α requires VHL upon a return of hypoxic cells to an oxygenated environment. VHL−/− RCC and VHL(+) cells were incubated for 8 h in hypoxia and returned to an oxygenated incubator for the indicated time. Cells were lysed in 4% SDS-PBS and analyzed by SDS-8% PAGE, followed by Western blotting with an anti-HIF-2α antibody (Novus). (B) Nuclear ubiquitination of HIF-2α requires VHL. VHL−/− RCC (lanes 1 to 3) and VHL(+) (lanes 4 to 6) cells were incubated in hypoxia for 8 h prior to reoxygenation for 2 min. Cells were fractionated into total (T), nuclear (N), and cytosolic (C) compartments, followed by immunoprecipitation with agarose-GST-S5a beads. Captured polyubiquitinated proteins were analyzed by SDS-6% PAGE for ub-HIF-2α content with an anti-HIF-2α antibody. Lane 7 consists of a total normoxic fraction of VHL(+) cells in which we failed to detect ub-HIF-2α, most likely due to low abundance. (C) VHL/ub-adGFP-HIF-1α complexes can be found in the nuclear compartment of normoxic cells. Normoxic VHL-GFP-expressing stable cells were either infected (lanes 1 to 6) or not infected (lane 7) with adGFP-HIF-1α virus, and the cells were either not treated (lanes 1 to 3) or were treated (lanes 4 to 7) with a proteasome inhibitor (calpain inhibitor [CI]) for 2 h. Cells were fractionated into total (T), nuclear (N), and cytoplasmic (C) lysates, which were immunoprecipitated with agarose M2-beads to capture FLAG-tagged VHL-GFP. The top panel shows adGFP-HIF-1α input by straight Western analysis. The lower three panels consists of agarose M2-bead immunoprecipitations of VHL-GFP, followed by Western blotting with an anti-HIF-1α antibody to detect adGFP-HIF-1α (second panel from the top), an antiubiquitin antibody (third panel from the top), or with a M2 anti-FLAG antibody to detect VHL-GFP (bottom panel). Lane 7 is an immunoprecipitation of VHL-GFP from lysates of uninfected VHL-GFP cells. Note the lack of ubiquitin signal in this lane, demonstrating that the ubiquitin signals in lanes 1 to 6 are from ub-adGFP-HIF-1α. (D) The degradation of HIF-1α can be abolished by treatment with HIF-1α-specific prolyl hydroxylase inhibitor DMOG. HeLa cells were incubated for 8 h in hypoxia prior to a return to a normoxic incubator for the indicated time. Cells were either treated with dimethyl sulfoxide alone (untreated) or treated with DMOG (1 mM) for 8 h. Cells were lysed in 4% SDS-PBS and analyzed by SDS-8% PAGE, followed by Western blotting with an anti-HIF-1α antibody. Note the absence of higher-molecular-weight bands at 3 and 6 min in the DMOG-treated cells. (E) DMOG treatment abolishes ubiquitination of HIF-1α in reoxygenated cells. Hypoxic HeLa cells were transferred to an oxygenated incubator for 2 min prior to lysis and incubation with agarose S5a beads. The captured polyubiquitinated proteins were analyzed by SDS-6% PAGE, followed by Western blotting with an anti-HIF-1α antibody.
It has recently been demonstrated that binding of VHL to HIF-α and subsequent ubiquitination required hydroxylation of specific proline residues found in the oxygen-dependent degradation domains of HIF-α (15, 32, 34, 47, 49, 67). The HIF-specific prolyl hydroxylases belong to a family of highly conserved 2-oxoglutarate-dependent oxygenases and require 2-oxoglutarate and molecular oxygen as cofactors (7, 15). Normoxic cells contained abundant HIF-α when treated with the cell-permeable DMOG, a 2-oxoglutarate analogue that inhibits HIF-specific prolyl hydroxylases. Since HIF-α is ubiquitinated in the nuclear compartment upon reoxygenation of hypoxic cells, we sought to determine whether this process could be abolished by treatment with DMOG or whether nuclear HIF-α incurred a different posttranslational modification that regulates its stability. DMOG-treated and untreated HeLa cells were incubated for 8 h in hypoxia prior to reoxygenation. HIF-1α was rapidly degraded in untreated cells (Fig. 4D, untreated). In contrast, HIF-1α signal was still detectable even 20 min after reoxygenation of hypoxic cells treated with DMOG. Note the absence of high-molecular-weight bands with such treatment. Likewise, S5a beads failed to capture ub-HIF-1α in cells treated with DMOG (Fig. 4E). These results demonstrate that, upon reoxygenation of hypoxic cells, nuclear HIF-1α undergoes a posttranslational modification similar to that which occurs in normoxic cells, a step that most likely includes nuclear prolyl hydroxylation by family members of the 2-oxoglutarate-dependent oxygenases.
HIF-1α of hypoxic cells is exported to the cytoplasm upon reoxygenation.
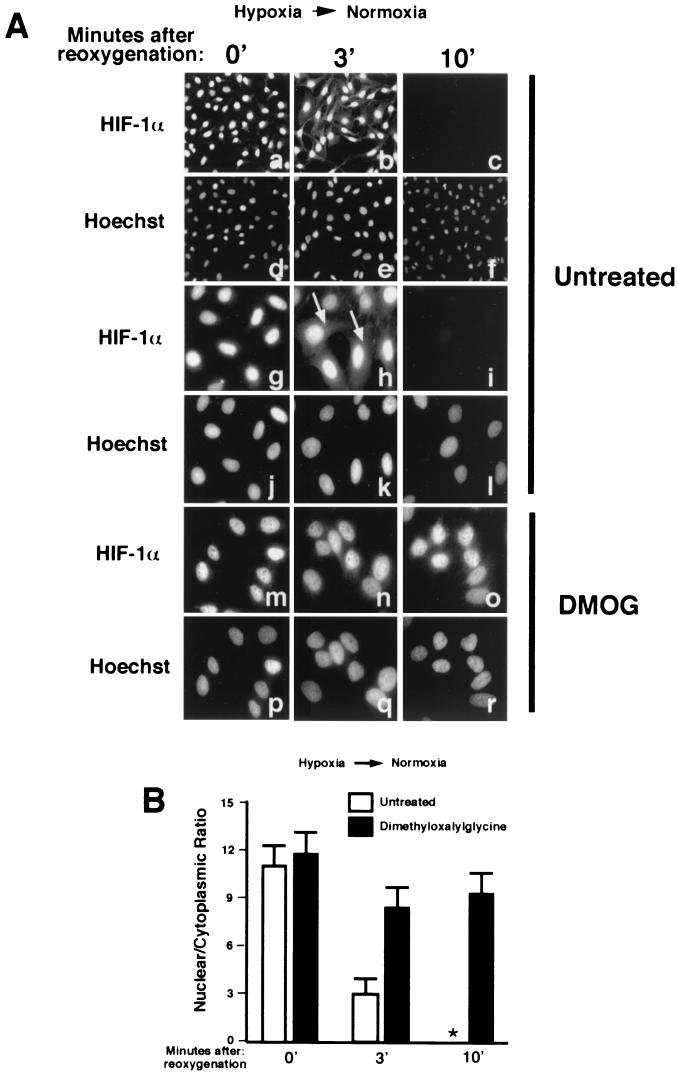
The data presented thus far demonstrate that HIF-1α undergoes ubiquitination in the nucleus upon reoxygenation of hypoxic cells, a phenomenon that requires nuclear VHL and prolyl hydroxylase activity. The next step in our study was to investigate whether HIF-1α is exported to the cytoplasm upon reoxygenation of hypoxic cells, a step that might be required for its degradation. We asked this question since it has been reported that nuclear substrates, including p53, p27, Smad3, and Ahr, must undergo leptomycin B-sensitive nuclear export for efficient proteasome-mediated degradation. We reasoned that cytoplasmic accumulation of HIF-1α would be very transient and most likely difficult to detect due to its quick degradation and its very short half-life upon reoxygenation. HIF-1α localization is restricted to the nuclear compartment of hypoxic cells; therefore, any detectable cytoplasmic signal upon reoxygenation would be interpreted as nuclear export of HIF-1α. CCD images of low magnification (Fig. 5Aa and d for the corresponding Hoechst staining) of hypoxic HeLa cells immunostained with HIF-1α are presented to demonstrate that the vast majority of cells displayed strong nuclear signals with low cytoplasmic signals. A higher magnification of the stained cells is shown in Fig. 5Ag and j, and calculation of nuclear/cytoplasmic ratios can be found in Fig. 5B (see also Fig. 3A and Materials and Methods; see reference 43 for the quantification methodology). HIF-1α immunofluorescence analysis of reoxygenated cells revealed a detectable cytoplasmic HIF-1α signal after 2 to 3 min of reoxygenation (Fig. 5Ab, e, h, and k). We failed to detect any nuclear or cytoplasmic signal after 10 min of reoxygenation (Fig. 5Ac, f, i, and l). Exposure times of panels a, b, and c are the same; therefore, the cytoplasmic signal detectable in b was not due to overexposure of the stained cells (panels g, h, and i were also exposed for the same length of time). The nuclear/cytoplasmic ratio of HIF-1α, which is obtained independently of the exposure time, was reduced from ca. 11 in hypoxic cells to 3 in reoxygenated cells (Fig. 5B), indicating a measurable transfer of HIF-1α from the nucleus to the cytoplasm. We did not detect cytoplasmic signal when the transfer of hypoxic cells to an oxygenated environment was performed at 4°C or when hypoxic cells were incubated in the presence of metabolic poisons, suggesting that cytoplasmic accumulation of HIF-1α is temperature sensitive and energy dependent (data not shown). Experiments conducted in the presence of cycloheximide (treatment of 10 min prior to reoxygenation) did not abolish cytoplasmic accumulation of HIF-1α, indicating that the observed signal is not due to de novo protein synthesis (data not shown). These results suggest that nuclear HIF-1α can be exported to the cytoplasm upon reoxygenation of hypoxic HeLa cells.
FIG. 5.
Nuclear export of HIF-1α occurs in reoxygenated HeLa cells through a pathway that requires nuclear prolyl hydroxylase activity. (A) Hypoxic HeLa cells (panels a to l) were either fixed with 1% formaldehyde or transferred to an oxygenated incubator for 3 or 10 min prior to fixation. Cells were also treated with DMOG (1 mM) for 8 h (panels m to r). Cells were stained with an anti-HIF-1α antibody or with Hoechst. Fields a to c are the same exposure. Fields g, h, i, m, n, and o are also equivalent exposures. (B) Nuclear/cytoplasmic ratios were calculated for ca. 30 cells for each condition obtained from at least three independent experiments. Mean averages with the standard deviations are shown. ✽, data not available since there is no signal.
Nuclear protein such as p27Kip1 must undergo phosphorylation prior to nuclear export. We show in Fig. 4D and E that treatment with DMOG abolished ubiquitination and degradation of HIF-1α upon reoxygenation of hypoxic cells. We then sought to determine whether treatment with DMOG would also cause a reduction in the nuclear export of HIF-1α. Hypoxic HeLa cells treated with DMOG displayed strong nuclear HIF-1α with a nuclear/cytoplasmic ratio slightly higher than untreated cells (Fig. 5Am and p and B). HIF-1α remained essentially confined to the nuclear compartment upon reoxygenation of DMOG-treated HeLa cells, although we did observe a slight, but reproducible cytoplasmic signal (Fig. 5An and q and B; the exposure times for Fig. 5Ag to i and m to o are the same). These results suggest that nuclear prolyl hydroxylase activity is required not only for ubiquitination but also for efficient nuclear export of HIF-1α in reoxygenated cells.
Nuclear export of HIF-α requires VHL and occurs through an ActD/DRB-sensitive pathway.
Since nuclear prolyl hydroxylase activity is required for efficient nuclear export of HIF-α, we sought to determine whether VHL was also necessary for this process to occur. VHL−/− RCC and VHL(+) cells were infected with adGFP-HIF-1α and incubated for 8 h in hypoxia. As shown in Fig. 6A, and in Fig. 2A, adGFP-HIF-1α localized strictly to the nuclear compartment under these conditions. After transfer to a normoxic environment, cytoplasmic accumulation of adGFP-HIF-1α could be detected only in cells expressing VHL (Fig. 6A). We then verified whether different drugs could abolish the ability of adGFP-HIF-1α to export to the cytoplasm in reoxygenated cells. The use of leptomycin B, a drug that blocks CRM1-dependent nuclear export of proteins bearing a classical NES, did not affect the localization of VHL-GFP (Fig. 6C) and did not abolish cytoplasmic accumulation of adGFP-HIF-1α (Fig. 6B) upon reoxygenation of hypoxic cells. In contrast, ActD, as well as DRB (data not shown), caused a marked increase of VHL-GFP in the nuclear compartment (Fig. 6C) as previously reported and also prevented cytoplasmic accumulation of adGFP-HIF-1α (Fig. 6B). Interestingly, the proteasome inhibitor MG132 and calpain inhibitor (data not shown) caused an increase in nuclear VHL-GFP (Fig. 6C) and prevented the cytoplasmic accumulation of adGFP-HIF-1α (Fig. 6B). DMOG abolished nuclear export of adGFP-HIF-1α (Fig. 6B) but did not affect the localization of VHL-GFP (Fig. 6C). We conclude from these experiments that the nuclear export of adGFP-HIF-1α requires VHL and occurs through an ActD- and proteasome inhibitor-sensitive pathway similar to that utilized by VHL.
Treatment with ActD causes an increase in the half-life of HIF-1α without affecting its ubiquitination.
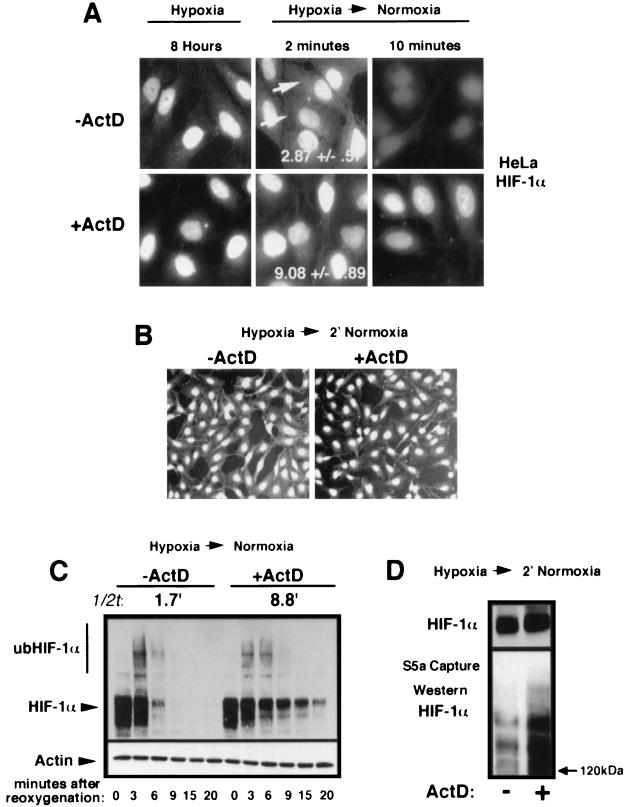
We show in Fig. 5 and 6 that a fraction of nuclear HIF-1α is exported to the cytoplasm upon reoxygenation of hypoxic cells. It is known that leptomycin B blocks nuclear export of p27 and p53, resulting in an increase in their respective half-lives. However, treatment with this drug had no effect on the shuttling properties of both VHL and adGFP-HIF-1α and did not increase the half-life of HIF-1α (2). Since ActD prevented efficient nuclear export of VHL and adGFP-HIF-1α, we sought to determine whether treatment with this drug would have an effect on the nuclear ubiquitination and degradation of endogenous HIF-1α. Reoxygenated HeLa cells treated with ActD displayed a higher nuclear/cytoplasmic ratio of HIF-1α compared to untreated cells (Fig. 7A). CCD images of lower magnification and equally exposed fields are shown in Fig. 7B. Nuclear HIF-1α signal was still detectable 10 min after reoxygenation in ActD-treated cells, whereas no signal could be observed in untreated cells (Fig. 7A). Western blot revealed an increase of ca. four- to fivefold in the half-life of HIF-1α after reoxygenation of ActD-treated HeLa cells compared to untreated cells (Fig. 7C). We sought to determine whether the increase in the half-life of HIF-1α was a consequence of a reduction in its nuclear ubiquitination. An S5a capture assay shown in Fig. 7D indicates that, at equal levels of HIF-1α, ActD-treated cells were either as efficient or more efficient at ubiquitinating HIF-1α than were untreated cells upon a return to a normoxic environment. Furthermore, levels of VHL remained unchanged after ActD treatment (data not shown; see references 24 and 44). These results demonstrate that the increase in the half-life of HIF-1α in ActD-treated cells is not the consequence of a reduction of nuclear ubiquitination and suggest that blocking nuclear export of VHL results in less-efficient degradation of ub-HIF-1α.
FIG. 7.
ActD treatment increases the half-life of HIF-α without a decrease in its nuclear ubiquitination. (A) Cytoplasmic accumulation of endogenous HIF-1α in HeLa cells upon a return to normoxia is inhibited by ActD treatment. HeLa cells were incubated for 8 h in hypoxia in the absence or presence of ActD (10 μg/ml) for the last 2 h. Hypoxic cells were fixed with 1% formaldehyde or fixed 2 or 10 min after reoxygenation and then stained with an anti-HIF-1α antibody. (B) Lower magnification of HIF-1α immunostaining of reoxygenated HeLa cells. Experiments were performed as in panel A. (C) Efficient degradation of endogenous HIF-1α is dependent upon ongoing transcription. HeLa cells were incubated for 8 h in hypoxia in the absence or presence of ActD (10 μg/ml) for the last 2 h. Cells were returned to an oxygenated environment for the indicated times and Western blotting was performed with an anti-HIF-1α. The half-life (1/2t) of endogenous HIF-1α is indicated. (D) Nuclear ubiquitination of HIF-1α is unaffected by ActD treatment. Hypoxic HeLa cells were either not treated or treated with ActD (10 μg/ml) for 2 h and then returned to an oxygenated environment for 2 min. Lysates were incubated with agarose S5a beads, and captured polyubiquitinated proteins were analyzed by SDS-6% PAGE, followed by Western blotting with an anti-HIF-1α antibody. The top panel shows a straight Western blot for HIF-1α content. The experiment was conducted to obtain similar HIF-1α levels in untreated and in ActD-treated cells.
DISCUSSION
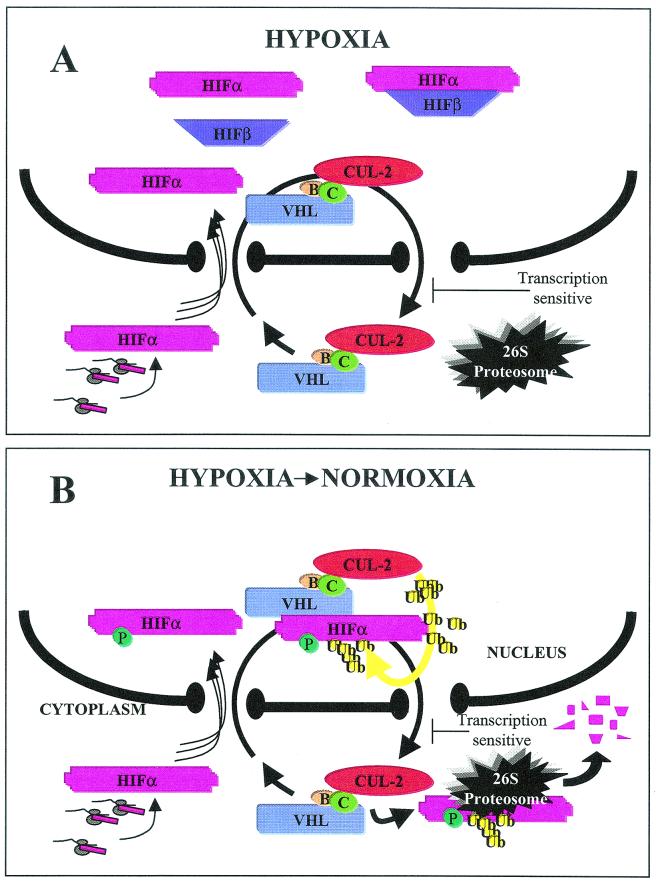
Model linking VBC/Cul-2 nuclear-cytoplasmic trafficking and oxygen-dependent ubiquitination and degradation of HIF-α.
It is becoming increasingly evident that the rapid degradation of nuclear proteins involves nuclear-cytoplasmic trafficking of the substrate protein, as well as of the components implicated in its ubiquitination and degradation. Recent reports have revealed that proteasome-mediated degradation of nuclear proteins, such as p27Kip1, p53, and Smad3, is intimately linked to their ability to export from the nucleus in a regulated, signal-dependent manner (9, 12, 18, 19, 41, 45, 51, 53, 56, 66). The HIF-α/VHL system provided an ideal system to investigate the role of subcellular trafficking of a nuclear substrate and of its specific E3 ubiquitin-ligase. We had previously reported that VHL, and the VBC/Cul-2 complex, shuttles between the nucleus and the cytoplasm in a manner that requires ongoing RNA polymerase II activity (24, 44). We provide evidence here linking VBC/Cul-2 E3 ubiquitin-ligase shuttling to its ability to mediate oxygen-dependent ubiquitination and degradation of the nuclear substrate HIF-α. Based on the data presented here and in other studies (6, 24, 44), we propose the model shown in Fig. 8 linking VHL shuttling and oxygen-dependent ubiquitination and degradation of HIF-α. We suggest that VBC/Cul-2 complexes engage in a constitutive “merry-go-round” between the nucleus and the cytoplasm that is unaffected by substrate concentration and oxygen tension. Substrate HIF-α is also efficiently imported to the nucleus regardless of VHL status and oxygen tension. Upon reoxygenation of hypoxic cells, nuclear HIF-α incurs a rapid posttranslational modification, rendering it competent to assemble with nuclear VHL. This is most likely the consequence of nuclear proxylation of the ODD domain (31, 33, 65) since treatment with DMOG prevented ubiquitination. Once assembled with VHL, HIF-α undergoes prompt Cullin-2-mediated ubiquitination in the nuclear compartment. ub-HIF-α is subsequently exported to the cytoplasm by the VBC/Cul-2 complex, utilizing a transcription-dependent, ActD-sensitive pathway, for 26S proteasome-mediated degradation. Treatment with DMOG also prevented cytoplasmic accumulation of HIF-α, suggesting that nuclear prolyl hydroxylation of the ODD domain is the key posttranslational modification that is required for both ubiquitination and nuclear export. The model proposed in Fig. 8 presents in detail the mechanisms involved in the ubiquitination and degradation of HIF-α upon reoxygenation of hypoxic cells. Under these conditions, cells must efficiently ubiquitinate and degrade a substrate that was physiologically stabilized and strictly localized in the nuclear compartment. This is in contrast to what happens with cells maintained in a normoxic environment, where the cell substrate HIF-α is synthesized in the cytoplasmic compartment. In a recent elegant study, Berra et al. (2) demonstrated that a chimeric HIF-α molecule that is forced to localize in the cytoplasm can still undergo oxygen-dependent degradation. Our data do not preclude cytoplasmic ubiquitination of nascent HIF-α by VBC/Cul-2, even though we were unable to detect ub-HIF-α in the cytoplasm of cells maintained in normoxia. Perhaps HIF-α is very rapidly ubiquitinated and degraded in the cytoplasm of normoxic cells to the extent of rendering its detection essentially impossible. However, it remains to be shown that the cytoplasmic ubiquitination and degradation of the HIF-α chimera are VHL dependent and whether they represent physiologic mechanisms. Perhaps nuclear ubiquitination of HIF-1α and its subsequent nuclear export are required events for regulation of other HIF-1α-dependent mechanisms not yet understood. We have shown here that VHL assembles with ub-adGFP-HIF-1α solely in the nuclear compartment and that endogenous ub-HIF-1α can only be detected in the nuclear compartment of normoxic cells. Both findings indicate that the ubiquitination and degradation mechanisms of HIF-α in normoxia are similar to those observed in reoxygenated cells. Therefore, we suggest that once synthesized in normoxic cells, at least a fraction of HIF-α is immediately imported into the nucleus and subjected to nuclear ubiquitination and subsequent nuclear export for cytoplasmic degradation. Regardless of the mechanisms that regulate HIF-α in normoxia, the data shown here support our model that, upon the reoxygenation of hypoxic cells, nuclear-cytoplasmic shuttling of VHL plays a role in the ubiquitination and degradation of HIF-α. It remains to be determined whether this model is valid for the ubiquitination and degradation of newly synthesized HIF-1α in an oxygenated environment.
FIG. 8.
Model linking VBC/Cul-2 nuclear-cytoplasmic trafficking and oxygen-dependent ubiquitination and degradation of HIF-α. (A) In hypoxic conditions, HIF-α (pink) is stabilized and is immediately imported into the nucleus, where it binds to its partner HIF-β (purple) to form the HIF transcription factor that transactivates hypoxia-inducible genes such as vascular endothelial growth factor and glucose transporter-1. VBC/Cul-2 engages in a constitutive nuclear-cytoplasmic shuttle unaffected by oxygen tension or levels of substrate HIF-α. (B) Upon a return to normoxia, HIF-α incurs a posttranslational modification in the nuclear compartment, which is most likely the hydroxylation of proline of the ODD domains since ubiquitination is preventable by treatment with DMOG. Nuclear HIF-α binds to VHL and undergoes Cullin-2-mediated ubiquitination (yellow) prior to being exported to the cytoplasm for 26S proteasomal degradation. Nuclear HIF-α embarks on the VBC/Cul-2 “merry-go-round” depending upon its ability to assemble with nuclear VHL. The proposed model is applicable for hypoxic cells that are returned to an oxygenated environment. As for cells maintained in normoxia, we cannot totally exclude cytoplasmic ubiquitination and degradation of HIF-α. However, we prefer a model by which HIF-α in normoxia is rapidly imported for nuclear ubiquitination and reexported to the cytoplasm in a manner similar to that observed for reoxygenated cells.
HIF-α is ubiquitinated in the nuclear compartment upon reoxygenation of hypoxic cells: a role for VHL in the nucleus.
The efficient ubiquitination of HIF-α in the nuclear compartment upon reoxygenation of hypoxic cells suggests that the components involved in this process can be found in this subcellular compartment. Nuclear ubiquitination of HIF-α is definitely mediated by VHL since we failed to detect ub-HIF-2α in the nucleus of reoxygenated VHL−/− RCC cells. DMOG abolished the nuclear ubiquitination of HIF-1α upon reoxygenation, indicating that the oxygen-sensing regulatory system is present in the nuclear compartment. Obviously, we cannot totally exclude the possibility that nuclear HIF-α is exported to the cytoplasm, ubiquitinated, and subsequently reimported into the nucleus. We find that this is improbable, however, since we were able to detect nuclear ub-HIF-α within seconds of reoxygenation, which is most likely insufficient time for all of these events to occur. In addition, the treatment of hypoxic cells with ActD or proteasome inhibitors, which prevented the efficient nuclear export of HIF-1α and VHL, did not interfere with its ubiquitination. Therefore, we suggest that the nuclear import of the VBC/Cul-2 complex is required for the efficient ubiquitination of HIF-α, thus providing the first evidence of a role for the VHL tumor suppressor protein in the nuclear compartment.
Transcription-dependent nuclear export of HIF-α is required for its degradation.
We present evidence here that adGFP-HIF-1α and endogenous HIF-1α accumulated in the cytoplasmic compartment after reoxygenation of hypoxic cells. Cytoplasmic accumulation occurred a few minutes after reoxygenation, and this event is likely to be the consequence of the nuclear export of HIF-α and not of de novo protein synthesis. Furthermore, HIF-α is strictly localized in the nuclear compartment in hypoxia, and cytoplasmic accumulation is observed in a very rapid time course. However, we failed to detect ub-HIF-α in the cytosolic compartment of reoxygenated cells. We think that, once it is bound to VHL in the nuclear compartment, HIF-α undergoes prompt ubiquitination and nuclear export. There is the possibility that a fraction of HIF-α bound to VHL is exported to the cytoplasm in ubiquitinated forms that might be sufficient to access the proteasome for degradation but would fail to be recognized by the S5a affinity column. Interestingly, treatment with ActD not only blocked the nuclear export of HIF-1α and VHL but also increased HIF-1α half-life in reoxygenated cells, a finding consistent with data published by another group (1). These authors suggested that HIF-α-regulated transcription was required for the prompt degradation of HIF-α. Although this remains a possibility, we think that ActD treatment would be more likely to affect the ability of VHL to mediate nuclear export of HIF-α in a manner reminiscent of how treatment with leptomycin B can increase the half-life of NES-containing proteins by abolishing their nuclear export. Importantly, ActD treatment did not significantly alter the nuclear ubiquitination of HIF-α. Therefore, it is probable that ActD inhibits another step in HIF-α degradation, and we suggest that it does so by interfering with VHL-mediated nuclear export.
A puzzling observation that we have encountered is that endogenous HIF-1α and overexpressed adGFP-HIF-1α accumulated solely in the nucleus of cells treated with proteasome inhibitors. Indeed, ub-HIF-1α should accumulate in the cytoplasm of proteasome-blocked cells if degradation occurred in this compartment. It is suggested that proteasome inhibitors might act as general inhibitors of nuclear export (59). We found that these drugs are able to block the nuclear export of VHL-GFP (Fig. 6) (6; Groulx and Lee, unpublished), as well as prevent cytoplasmic accumulation of ub-adGFP-HIF-1α upon reoxygenation. This might explain why we fail to detect cytoplasmic ub-HIF-1α in oxygenated cells treated with proteasome inhibitors.
Finally, we suggest that VHL plays a dual role in the ubiquitination and degradation of HIF-α. The first role would be to bind to the ODD domain for Cullin-2-mediated ubiquitination, and the second role would be to export the ubiquitinated substrate to the cytoplasm for degradation. We have shown in a previous report that sensitivity to ActD or DRB is mediated by exon-2-encoded β-domain of VHL (6). These sequences are able to direct ActD-dependent nuclear export of a reporter GFP molecule without assembly with HIF-α or Cullin-2 (44). The challenge for the near future will be to identify proteins that assemble with the exon-2-encoded β-domain of VHL involved in directing nuclear export of VBC/Cul-2/ub-HIF-α complexes and test whether cancer-causing mutations can abrogate the subcellular trafficking properties of VHL.
Acknowledgments
We sincerely thank Martine Whissel for excellent technical assistance.
This work was supported by a grant from the Canadian Institute of Health Research (CIHR). S.L. is a scholar of the CIHR. I.G. is supported by a studentship from the Ontario Graduate Scholarship in Science and Technology Foundation.
REFERENCES
- 1.Berra, E., D. E. Richard, E. Gothie, and J. Pouyssegur. 2001. HIF-1-dependent transcriptional activity is required for oxygen-mediated HIF-1α degradation. FEBS Lett. 491:85-90. [DOI] [PubMed] [Google Scholar]
- 2.Berra, E., D. Roux, D. E. Richard, and J. Pouyssegur. 2001. Hypoxia-inducible factor-1 alpha (HIF-1α) escapes O2-driven proteasomal degradation irrespective of its subcellular localization: nucleus or cytoplasm. EMBO Rep. 2:615-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bird, C. H., E. J. Blink, C. E. Hirst, M. S. Buzza, P. M. Steele, J. Sun, D. A. Jans, and P. I. Bird. 2001. Nucleocytoplasmic distribution of the ovalbumin serpin PI-9 requires a nonconventional nuclear import pathway and the export factor Crm1. Mol. Cell. Biol. 21:5396-5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blankenship, C., J. G. Naglich, J. M. Whaley, B. Seizinger, and N. Kley. 1999. Alternate choice of initiation codon produces a biologically active product of the von Hippel-Lindau gene with tumor suppressor activity. Oncogene 18:1529-1535. [DOI] [PubMed] [Google Scholar]
- 5.Blondel, M., J. M. Galan, Y. Chi, C. Lafourcade, Longaretti, C., R. J. Deshaies, and M. Peter. 2000. Nuclear-specific degradation of Far1 is controlled by the localization of the F-box protein Cdc4. EMBO J. 19:6085-6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonicalzi, M. E., I. Groulx, and S. Lee. 2001. Role of exon 2-encoded β-domain of the von Hippel-Lindau tumor suppressor protein. J. Biol. Chem. 276:1407-1416. [DOI] [PubMed] [Google Scholar]
- 7.Bruick, R. K., and S. L. McKnight. 2001. A conserved family of prolyl-4-hydroxylases that modify HIF. Science 294:1337-1340. [DOI] [PubMed] [Google Scholar]
- 8.Chen, F., M. T. Kishida, Yao, T. Hustand, D. Glavac, M. Dean, J. R. Gnarra, M. L. Orcutt, F. M. Duh, G. Glenn, J. Green, Y. E. Hisa, J. Lamiell, H. Li, M. H. Wei, L. Schmidt, K. Tory, I. Kuzmin, T. Stackhouse, F. Latif, W. M. Linehan, M. I. Lerman, and B. Zbar. 1995. Germline mutations in the von Hippel-Lindau disease tumor suppressor gene: correlations with phenotype. Hum. Mutat. 5:66-75. [DOI] [PubMed] [Google Scholar]
- 9.Chen, J., J. Lin, and A. J. Levine. 1995. Regulation of transcription functions of the p53 tumor suppressor by the mdm-2 oncogene. Mol. Med. 1:142-152. [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, L., M. Anton, and F. L. Graham. 1996. Production and characterization of human 293 cell lines expressing the site-specific recombinase Cre. Somat. Cell Mol. Genet. 6:477-488. [DOI] [PubMed] [Google Scholar]
- 11.Cockman, M. E., N. Masson, D. R. Mole, P. Jaakkola, G. W. Chang, S. C. Clifford, E. R. Maher, C. W. Pugh, P. J. Ratcliffe, and P. H. Maxwell. 2000. Hypoxia inducible factor-alpha binding and ubiquitylation by the von Hippel-Lindau tumor suppressor protein. J. Biol. Chem. 275:25733-25741. [DOI] [PubMed] [Google Scholar]
- 12.Davarinos, N. A., and R. S. Pollenz. 1999. Aryl hydrocarbon receptor imported into the nucleus following ligand binding is rapidly degraded via the cytosplasmic proteasome following nuclear export. J. Biol. Chem. 274:28708-28715. [DOI] [PubMed] [Google Scholar]
- 13.Duan, D. R., A. Pause, W. H. Burgess, T. Aso, D. Y. T. Chen, K. P. Garrett, R. C. Conaway, J. W. Conaway, W. M. Linehan, and R. D. Klausner. 1995. Inhibition of transcription elongation by the VHL tumor suppressor protein. Science 269:1402-1406. [DOI] [PubMed] [Google Scholar]
- 14.Duan, D. R., J. S. Humphrey, D. Y. Chen, Y. Weng, J. Sukegawa, S. Lee, J. R. Gnarra, W. M. Linehan, and R. D. Klausner. 1995. Characterization of the VHL tumor suppressor gene product: localization, complex formation, and the effect of natural inactivating mutations. Proc. Natl. Acad. Sci. USA 92:6459-6463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Epstein, A. C., J. M. Gleadle, L. A. McNeill, K. S. Hewitson, J. O'Rourke, D. R. Mole, M. Mukherji, E. Metzen, M. I. Wilson, A. Dhanda, Y. M. Tian, N. Masson, D. L. Hamilton, P. Jaakkola, R. Barstead, J. Hodgkin, P. H. Maxwell, C. W. Pugh, C. J. Schofield, and P. J. Ratcliffe. 2001. Caenorhabditis elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell 107:43-54. [DOI] [PubMed] [Google Scholar]
- 16.Fischer, U., J. Huber, W. C. Boelens, I. W. Mattaj, and R. Luhrmann. 1995. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell 82:475-483. [DOI] [PubMed] [Google Scholar]
- 17.Foster, K., A. Prowse, A. van den Berg, S. Fleming, M. M. Hulsbeek, P. A. Crossey, F. M. Richards, P. Cairns, N. A. Affara, M. A. Ferguson-Smith, et al. 1994. Somatic mutations of the von Hippel-Lindau disease tumour suppressor gene in non-familial clear cell renal carcinoma. Hum. Mol. Genet. 3:2169-2173. [DOI] [PubMed] [Google Scholar]
- 18.Freedman, D. A., and A. J. Levine. 1998. Nuclear export is required for degradation of endogenous p53 by MDM2 and human papillomavirus E6. Mol. Cell. Biol. 18:7288-7293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fukuchi, M., T. Imamura, T. Chiba, T. Ebisawa, M. Kawabata, K. Tanaka, and K. Miyazono. 2001. Ligand-dependent degradation of smad3 by a ubiquitin ligase complex of roc1 and associated proteins. Mol. Biol. Cell 12:1431-1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gnarra, J. R., D. R. Duan, Y. Weng, J. S. Humphrey, D. Y. Chen, S. Lee, A. Pause, C. F. Dudley, F. Latif, I. Kuzmin, L. Schmidt, F. M. Duh, T. Stackhouse, F. Chen, T. Kishida, M. H. Wei, M. I. Lerman, B. Zbar, R. D. Klausner, and W. M. Linehan. 1996. Molecular cloning of the von Hippel-Lindau tumor suppressor gene and its role in renal carcinoma. Biochim. Biophys. Acta 1242:201-210. [DOI] [PubMed] [Google Scholar]
- 21.Gnarra, J. R., K. Tory, Y. Weng, L. Schmidt, M. H. Wei, H. Li, F. Latif, S. Liu, F. Chen, F. M. Duh, I. Lubensky, D. R. Duan, C. Florence, R. Pozzatti, M. M. Walther, N. H. Bander, H. B. Grossman, H. Brauch, S. Pomer, J. D. Brooks, W. B. Isaacs, M. I. Lerman, B. Zbar, and W. M. Linehan. 1994. Mutations of the VHL tumour suppressor gene in renal carcinoma. Nat. Genet. 7:85-90. [DOI] [PubMed] [Google Scholar]
- 22.Gorospe, M., J. M. Egan, B. Zbar, M. Lerman, L. Geil, I. Kuzmin, and N. J. Holbrook. 1999. Protective function of von Hippel-Lindau protein against impaired protein processing in renal carcinoma cells. Mol. Cell. Biol. 19:1289-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Graham, F. L., and A. J. van der Eb. 1973. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology 52:456-467. [DOI] [PubMed] [Google Scholar]
- 24.Groulx, I., M. E. Bonicalzi, and S. Lee. 2000. Ran-mediated nuclear export of the von Hippel-Lindau tumor suppressor protein occurs independently of its assembly with cullin-2. J. Biol. Chem. 275:8991-9000. [DOI] [PubMed] [Google Scholar]
- 25.Gu, Y. Z., S. M. Moran, J. B. Hogenesch, L. Wartman, and C. A. Bradfield. 1998. Molecular characterization and chromosomal localization of a third alpha-class hypoxia inducible factor subunit, HIF3α. Gene Expr. 7:205-213. [PMC free article] [PubMed] [Google Scholar]
- 26.Hara, S., J. Hamada, C. Kobayashi, Y. Kondo, and N. Imura. 2001. Expression and characterization of hypoxia-inducible factor (HIF)-3alpha in human kidney: suppression of HIF-mediated gene expression by HIF-3α. Biochem. Biophys. Res. Commun. 287:808-813. [DOI] [PubMed] [Google Scholar]
- 27.Hershko, A., and A. Ciechanover. 1998. The ubiquitin system. Annu. Rev. Biochem. 67:425-479. [DOI] [PubMed] [Google Scholar]
- 28.Huang, L. E., Z. Arany, D. M. Livingston, and H. F. Bunn. 1996. Activation of hypoxia-inducible transcription factor depends primarily upon redox-sensitive stabilization of its alpha subunit. J. Biol. Chem. 271:32253-32259. [DOI] [PubMed] [Google Scholar]
- 29.Huang, L. E., J. Gu, M. Schau, and H. F. Bunn. 1998. Regulation of hypoxia-inducible factor 1α is mediated by an O2-dependent degradation domain via the ubiquitin-proteasome pathway. Proc. Natl. Acad. Sci. USA 95:7987-7992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iliopoulos, O., M. Ohh, and W. G. Kaelin, Jr. 1998. pVHL19 is a biologically active product of the von Hippel-Lindau gene arising from internal translation initiation. Proc. Natl. Acad. Sci. USA 95:11661-11666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iliopoulos, O., A. Kibel, S. Gray, and W. G. Kaelin, Jr. 1995. Tumor suppression by the human von Hippel-Lindau gene product. Nat. Med. 1:822-826. [DOI] [PubMed] [Google Scholar]
- 32.Ivan, M., K. Kondo, H. Yang, W. Kim, J. Valiando, M. Ohh, A. Salic, J. M. Asara, W. S. Lane, and W. G. Kaelin, Jr. 2001. HIFα targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science 292:464-468. [DOI] [PubMed] [Google Scholar]
- 33.Iwai, K., K. Yamanaka, T. Kamura, N. Minato, R. C. Conaway, J. W. Conaway, R. D. Klausner, and A. Pause. 1999. Identification of the von Hippel-Lindau tumor-suppressor protein as part of an active E3 ubiquitin-ligase complex. Proc. Natl. Acad. Sci. USA 96:12436-12441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jaakkola, P., D. R. Mole, Y. M. Tian, M. I. Wilson, J. Gielbert, S. J. Gaskell, A. Kriegsheim, H. F. Hebestreit, M. Mukherji, C. J. Schofield, P. H. Maxwell, C. W. Pugh, and P. J. Ratcliffe. 2001. Targeting of HIF-α to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 292:468-472. [DOI] [PubMed] [Google Scholar]
- 35.Jewell, U. R., I. Kvietikova, A. Scheid, C. Bauer, R. H. Wenger, and M. Gassmann. 2001. Induction of HIF-1α in response to hypoxia is instantaneous. FASEB J. 15:1312-1314. [PubMed] [Google Scholar]
- 36.Jones, A., C. Fujiyama, C. Blanche, J. W. Moore, S. Fuggle, D. Cranston, R. Bicknell, and A. L. Harris. 2001. Relation of vascular endothelial growth factor production to expression and regulation of hypoxia-inducible factor-1α and hypoxia-inducible factor-2α in human bladder tumors and cell lines. Clin. Cancer Res. 7:1263-1272. [PubMed] [Google Scholar]
- 37.Kamura, T., D. M. Koepp, M. N. Conrad, D. Skowyra, R. J. Moreland, O. Iliopoulos, W. S. Lane, W. G. Kaelin, Jr., S. J. Elledge, R. C. Conaway, J. W. Harper, and J. W. Conaway. 1999. Rbx1, a component of the VHL tumor suppressor complex and SCF ubiquitin ligase. Science 284:657-661. [DOI] [PubMed] [Google Scholar]
- 38.Kamura, T., S. Sato, K. Iwai, M. Czyzyk-Krzeska, R. C. Conaway, and J. W. Conaway. 2000. Activation of HIF1alpha ubiquitination by a reconstituted von Hippel-Lindau (VHL) tumor suppressor complex. Proc. Natl. Acad. Sci. USA 97:10430-10435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kibel, A., O. Iliopoulos, J. A. DeCaprio, and W. G. Kaelin, Jr. 1995. Binding of the von Hippel-Lindau tumor suppressor protein to elongin B and C. Science 269:1444-1446. [DOI] [PubMed] [Google Scholar]
- 40.Kietzmann, T., Y. Cornesse, K. Brechtel, S. Modaressi, and K. Jungermann. 2001. Perivenous expression of the mRNA of the three hypoxia-inducible factor alpha-subunits, HIF1α, HIF2α, and HIF3α, in rat liver. Biochem. J. 354:531-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ko, L. J., and C. Prives. 1996. p53: puzzle and paradigm. Genes Dev. 10:1054-1072. [DOI] [PubMed] [Google Scholar]
- 42.Latif, F., K. Tory, J. Gnarra, M. Yao, F. M. Duh, M. L. Orcutt, T. Stackhouse, I. Kuzmin, W. Modi, L. Geil, L. Schmidt, F. Zhou, H. Li, M. H. Wei, F. Chen, G. Glenn, P. Choyke, M. M. Walther, Y. Weng, D. S. R. Duan, M. Dean, D. Glavac, F. M. Richards, P. A. Crossey, M. A. Ferguson-Smith, D. L. Paisler, I. Chumakov, D. Cohen, A. C. Chinault, E. R. Maher, W. M. Linehan, B. Zbar, and M. I. Lerman. 1993. Identification of the von Hippel-Lindau disease tumor suppressor gene. Science 260:1317-1320. [DOI] [PubMed] [Google Scholar]
- 43.Lee, S., D. Y. T. Chen, J. S. Humphery, J. R. Gnarra, W. M. Linehan, and R. D. Klausner. 1996. Nuclear/cytoplasmic localization of the von Hippel-Lindau tumor suppressor gene product is determined by cell density. Proc. Natl. Acad. Sci. USA 93:1770-1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee, S., M. Neumann, R. Stearman, R. Stauber, A. Pause, G. N. Pavlakis, and R. D. Klausner. 1999. Transcription-dependent nuclear-cytoplasmic trafficking is required for the function of the von Hippel-Lindau tumor suppressor protein. Mol. Cell. Biol. 19:1486-1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Levine, A. J. 1997. p53, the cellular gatekeeper for growth and division. Cell 88:323-331. [DOI] [PubMed] [Google Scholar]
- 46.Linehan, W. M., M. I. Lerman, and B. Zbar. 1995. Identification of the von Hippel-Lindau (VHL) gene: its role in renal cancer. JAMA 273:564-570. [PubMed] [Google Scholar]
- 47.Lisztwan, J., G. Imbert, C. Wirbelauer, M. Gstaiger, and W. Krek. 1999. The von Hippel-Lindau tumor suppressor protein is a component of an E3 ubiquitin-protein ligase activity. Genes Dev. 13:1822-1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lonergan, K. M., O. Iliopoulos, M. Ohh, T. Kamura, R. C. Conaway, J. W. Conaway, and W. G. Kaelin., Jr. 1998. Regulation of hypoxia-inducible mRNAs by the von Hippel-Lindau tumor suppressor protein requires binding to complexes containing elongins B/C and Cul2. Mol. Cell. Biol. 18:732-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Masson, N., C. Willam, P. H. Maxwell, C. W. Pugh, and P. J. Ratcliffe. 2001. Independent function of two destruction domains in hypoxia-inducible factor-alpha chains activated by prolyl hydroxylation. EMBO J. 20:5197-5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maxwell, P. H., M. S. Wiesener, G. W. Chang, S. C. Clifford, E. C. Vaux, M. E. Cockman, C. C. Wykoff, C. W. Pugh, E. R. Maher, and P. J. Ratcliffe. 1999. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature 399:271-275. [DOI] [PubMed] [Google Scholar]
- 51.Momand, J., G. P. Zambetti, D. C. Olson, D. George, and A. J. Levine. 1992. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell 69:1237-1245. [DOI] [PubMed] [Google Scholar]
- 52.Ohh, M., C. W. Park, M. Ivan, M. A. Hoffman, T. Y. Kim, L. E. Huang, N. Pavletich, V. Chau, and W. G. Kaelin. 2000. Ubiquitination of hypoxia-inducible factor requires direct binding to the beta-domain of the von Hippel-Lindau protein. Nat. Cell Biol. 2:423-427. [DOI] [PubMed] [Google Scholar]
- 53.Oliner, J. D., J. A. Pietenpol, S. Thiagalingam, J. Gyuris, K. W. Kinzler, and B. Vogelstein. 1993. Oncoprotein MDM2 conceals the activation domain of tumour suppressor p53. Nature 362:857-860. [DOI] [PubMed] [Google Scholar]
- 54.Pause, A., S. Lee, R. A. Worrell, D. Y. Chen, W. H. Burgess, W. M. Linehan, and R. D. Klausner. 1997. The von Hippel-Lindau tumor-suppressor gene product forms a stable complex with human CUL-2, a member of the Cdc53 family of proteins. Proc. Natl. Acad. Sci. USA 94:2156-2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pause, A., B. Peterson, G. Schaffar, R. Stearman, and R. D. Klausner. 1999. Studying interactions of four proteins in the yeast two-hybrid system: structural resemblance of the pVHL/elongin BC/hCUL-2 complex with the ubiquitin ligase complex SKP1/cullin/F-box protein. Proc. Natl. Acad. Sci. USA 96:9533-9538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Roth, J., M. Dobbelstein, D. A. Freedman, T. Shenk, and A. J. Levine. 1998. Nucleo-cytoplasmic shuttling of the hdm2 oncoprotein regulates the levels of the p53 protein via a pathway used by the human immunodeficiency virus rev protein. EMBO J. 17:554-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ryan, H. E., M. Poloni, W. McNulty, D. Elson, M. Gassmann, J. M. Arbeit, and R. S. Johnson. 2000. Hypoxia-inducible factor-1α is a positive factor in solid tumor growth. Cancer Res. 60:4010-4015. [PubMed] [Google Scholar]
- 58.Salceda, S., and J. Caro. 1997. Hypoxia-inducible factor 1α (HIF-1α) protein is rapidly degraded by the ubiquitin-proteasome system under normoxic conditions. Its stabilization by hypoxia depends on redox-induced changes. J. Biol. Chem. 272:22642-22647. [DOI] [PubMed] [Google Scholar]
- 59.Scheffner, M. 1999. Moving protein heads for breakdown. Nature 398:103-104. [DOI] [PubMed] [Google Scholar]
- 60.Schoenfeld, A., E. J. Davidowitz, and R. D. Burk. 1998. A second major native von Hippel-Lindau gene product, initiated from an internal translation start site, functions as a tumor suppressor. Proc. Natl. Acad. Sci. USA 95:8817-8822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Semenza, G. L. 2000. HIF-1 and human disease: one highly involved factor. Genes Dev. 14:1983-1991. [PubMed] [Google Scholar]
- 62.Stauber, R., G. A. Gaitanaris, and G. N. Pavlakis. 1995. Analysis of trafficking of Rev and transdominant Rev proteins in living cells using green fluorescent protein fusions: transdominant Rev blocks the export of Rev from the nucleus to the cytoplasm. Virology 213:439-449. [DOI] [PubMed] [Google Scholar]
- 63.Stebbins, C. E., W. G. Kaelin, Jr., and N. P. Pavletich. 1999. Structure of the VHL-ElonginC-ElonginB complex: implications for VHL tumor suppressor function. Science 284:455-461. [DOI] [PubMed] [Google Scholar]
- 64.Tanimoto, K., Y. Makino, T. Pereira, and L. Poellinger. 2000. Mechanism of regulation of the hypoxia-inducible factor-1α by the von Hippel-Lindau tumor suppressor protein. EMBO J. 19:4298-4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tomoda, K., Y. Kubota, and J. Kato. 1999. Degradation of the cyclin-dependent-kinase inhibitor p27Kip1 is instigated by Jab1. Nature 398:160-165. [DOI] [PubMed] [Google Scholar]
- 66.Xirodimas, D. S., C. W. Stephen, and D. P. Lane. 2001. Compartmentalization of p53 and Mdm2-mediated degradation of p53. Exp. Cell Res. 270:66-77. [DOI] [PubMed] [Google Scholar]
- 67.Yu, F., S. B. White, Q. Zhao, and F. S. Lee. 2001. HIF-1alpha binding to VHL is regulated by stimulus-sensitive proline hydroxylation Proc. Natl. Acad. Sci. USA 98:9630-9635. [DOI] [PMC free article] [PubMed] [Google Scholar]