Abstract
RAFT1/FRAP/mTOR is a key regulator of cell growth and division and the mammalian target of rapamycin, an immunosuppressive and anticancer drug. Rapamycin deprivation and nutrient deprivation have similar effects on the activity of S6 kinase 1 (S6K1) and 4E-BP1, two downstream effectors of RAFT1, but the relationship between nutrient- and rapamycin-sensitive pathways is unknown. Using transcriptional profiling, we show that, in human BJAB B-lymphoma cells and murine CTLL-2 T lymphocytes, rapamycin treatment affects the expression of many genes involved in nutrient and protein metabolism. The rapamycin-induced transcriptional profile is distinct from those induced by glucose, glutamine, or leucine deprivation but is most similar to that induced by amino acid deprivation. In particular, rapamycin treatment and amino acid deprivation up-regulate genes involved in nutrient catabolism and energy production and down-regulate genes participating in lipid and nucleotide synthesis and in protein synthesis, turnover, and folding. Surprisingly, however, rapamycin had effects opposite from those of amino acid starvation on the expression of a large group of genes involved in the synthesis, transport, and use of amino acids. Supported by measurements of nutrient use, the data suggest that RAFT1 is an energy and nutrient sensor and that rapamycin mimics a signal generated by the starvation of amino acids but that the signal is unlikely to be the absence of amino acids themselves. These observations underscore the importance of metabolism in controlling lymphocyte proliferation and offer a novel explanation for immunosuppression by rapamycin.
Rapamycin is an immunosuppressive drug used to prevent the rejection of transplanted organs and a promising anticancer agent (18, 26, 27, 32). Studies of its mechanism of action have led to the discovery of the TOR pathway, an evolutionarily conserved signaling network that plays critical roles in eukaryotic cell growth and cell cycle progression. In mammalian cells, the complex of rapamycin with its receptor, FKBP12, binds directly to the central component of the pathway, a large protein kinase called RAFT1/FRAP/mTOR (5, 33, 34). Exactly how the rapamycin-FKBP12 complex perturbs the function of RAFT1 is not well understood. Work in yeast, Drosophila, and mammals suggests that the TOR proteins participate in nutrient-sensitive signaling. Treatment of yeast and Drosophila larvae with rapamycin causes a starvation-like phenotype (2, 7, 17, 39), and in mammalian cells, extracellular amino acids regulate the activity of two translational regulators downstream of RAFT1, S6 kinase 1 (S6K1), and 4E-BP1 (15). Recent studies indicate that the levels of ATP and phosphatidic acid may regulate the TOR pathway in mammalian cells (11, 13), but the connection to nutrient signaling is not clear.
Available evidence indicates that the TOR pathway is ubiquitously expressed in mammalian cell types, and it is not known why rapamycin causes immunosuppression in human beings and other mammals without significantly affecting other organ systems. Interestingly, lymphocytes are more dependent than other cell types on amino acids, particularly glutamine, for biosynthesis and energy production (28), and a decrease in blood glutamine levels causes immunosuppression in humans and mice (6, 19). Within this context, we used transcriptional profiling and measurements of nutrient use in rapamycin-treated and nutrient-deprived cells to determine the relationship between rapamycin- and nutrient-sensitive pathways.
MATERIALS AND METHODS
Cell culture.
Human BJAB lymphoma cells were cultured in RPMI medium (catalog no. 11835; GIBCO-BRL) containing 10% fetal bovine serum, 10 U of penicillin/ml, and 10 μg of streptomycin/ml. For the rapamycin treatment experiments, cells were grown to a density of 105 cells/ml, 20 nM rapamycin (Calbiochem) or ethanol vehicle was added, and cell aliquots were removed at 15, 30, 60, and 120 min and 12 and 24 h after drug addition. For nutrient deprivation experiments, cells were cultured in RPMI medium to a density of 105 cells/ml, pelleted at room temperature for 5 min at 850 × g, washed once with phosphate-buffered saline, and resuspended in RPMI medium alone or in RPMI medium with a reduced concentration of glutamine (0.05 mM), leucine (0.01 mM), or glucose (0.05 mM). Cell aliquots were harvested at 30, 60, and 120 min and 12 and 24 h after a change of the media. Experiments using CTLL-2 cells were performed in the same manner, except that the media were supplemented with 10 U of human recombinant interleukin 2 (IL-2) (GIBCO-BRL)/ml and only 12- and 24-h time points were collected after rapamycin addition or nutrient deprivation. All experiments were performed in duplicate.
Western blotting.
BJAB and CTLL-2 cells at a density of 105 cells/ml were treated with 20 nM rapamycin or ethanol vehicle for 30, 60, and 120 min and 12 and 24 h. Cells (4 × 106) from each time point were harvested by centrifugation at 850 × g at 4°C for 5 min, washed twice with phosphate-buffered saline, and lysed in 0.5 ml of ice-cold lysis buffer (40 mM HEPES, pH 7.5, 120 mM NaCl, 1 mM EDTA, 10 mM pyrophosphate, 10 mM glycerophosphate, 50 mM NaF, 1.5 mM Na3Vo4, 1% Triton X-100, and one tablet of EDTA-free protease inhibitors [Roche] per 10 ml). Equal amounts of protein (40 μg) were analyzed by sodium dodecyl sulfate-8% polyacrylamide gel electrophoresis followed by Western blotting with antibodies recognizing S6K1 (Santa Cruz) and phospho-Thr389 S6K1 (Cell Signaling Technology).
Generation and analysis of transcriptional profiles.
Total RNA was purified from BJAB and CTLL-2 cells using RNA-Stat60 (Tel-Test, Friendswood, Tex.). cRNA prepared from the total RNA was analyzed using Affymetrix human U95 Av2 or murine 11K GeneChips as described earlier (8). Using the expression levels across the control time courses, a mean with standard deviation was obtained for each gene represented on the chip. The absolute difference was determined between this mean and the expression value of a gene at each point in the rapamycin and nutrient deprivation time courses. A gene was considered rapamycin or nutrient sensitive if in duplicate experiments the expression value at a time point (i) had an absolute difference greater than 2 standard deviations from the mean and (ii) was greater than 1.5-fold or less than 0.7-fold of the mean. Genes were defined as shared between conditions if they fulfilled the above criteria for at least one condition and the following relaxed criteria for the other: an expression value with an absolute difference greater than 1.4 standard deviations from the mean and greater than 1.3-fold or less than 0.77-fold of the mean.
k means clustering was used to group the rapamycin-sensitive genes according to similar patterns of expression. A tab-delimited Microsoft Excel file containing, for all the time points, the n-fold changes versus controls for the rapamycin-sensitive genes was imported into the Gene Cluster program (12). The genes were initially assigned randomly to k groups (k = 8 in Fig. 1B) and were clustered using an iterative method for 10,000 cycles. Clustered data were visualized using the TreeView program (12).
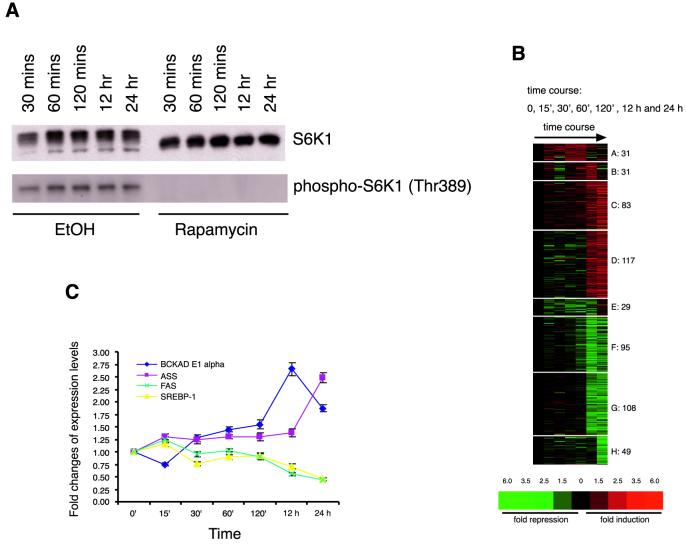
FIG. 1.
Rapamycin-sensitive gene expression in human BJAB cells. (A) At all time points rapamycin eliminates the phosphorylation of threonine 389 of S6K1. EtOH, ethyl alcohol. (B) Representation of k means cluster analysis of 543 genes regulated at 0, 15, 30, 60, and 120 min and 12 and 24 h after addition of 20 nM rapamycin. Genes are represented vertically, and experimental conditions (i.e., rapamycin time course) are displayed horizontally. Fold changes are indicated colorimetrically. (C) Quantitative RT-PCR analysis confirms rapamycin-induced gene expression changes detected using microarrays. Expression changes in four genes (BCKAD E1 alpha in cluster C, ASS in cluster D, FAS in cluster G, and SREBP-1 in cluster H) were determined in RNA samples prepared independently from those used for microarray analysis. The standard deviation for each time point was calculated from the triplicate samples in RT-PCR.
Quantitative RT-PCR.
Total RNA samples were treated with RNase-free DNase (Promega) and were repurified by RNeasy columns (Qiagen). Total RNAs (1 μg) were reverse transcribed to cDNAs using reverse transcriptase (RT) from GIBCO-BRL, and 1/40 of the total cDNA volume was used per SYBR green PCR (Applied Biosystems). The PCRs were performed on a Bio-Rad iCycler, and the protocol for PCR was as follows: first step, 95°C, 10 min; second step, 45 cycles of 95°C for 30 s, 60°C for 30 s, and 72°C for 30 s; and third step, 72°C for 5 min. The threshold cycle (TC) for each sample equals 10 standard deviations of the PCR baseline for that sample (PCR baseline was defined as mean values from PCR cycles 2 to 10). Since glyceraldehyde-3-phosphate dehydrogenase (GAPDH) does not change during the time course of rapamycin treatment in both the microarrays and the SYBR green PCRs, the TC for the gene of interest (branched-chain decarboxylase alpha [BCKAD E1 alpha], argininosuccinate synthetase [ASS], FAS, or SREBP-1) was normalized to that for GAPDH. The TC differences between ethanol and rapamycin samples (in triplicate) were calculated, and the final n-fold change in transcript levels equals 2x, where x is the difference of TC. All primers were designed on the Primer 3 program on www.genome.wi.mit.edu and were purchased from Research Genetics.
Nutrient measurements.
Actively growing CTLL-2 cells at a density of 105 cells/ml were pelleted as described above and placed with or without 20 nM rapamycin in control medium, medium with 0.1 mM glutamine, or medium with 0.3 mM glucose for another 48 h. At the end of the incubation, triplicate samples were centrifuged at 850 × g to remove cells and the supernatants were assayed for glutamine, glucose, ammonia, and lactate using assay kits as described by the manufacturer (Sigma). The P values (two-tailed) were obtained from the t test: two samples assuming unequal variances. To control for cell-independent nutrient degradation during the 48-h incubation, medium without cells was placed in the incubator alongside the experimental flasks and assayed for nutrient concentrations.
RESULTS
Rapamycin affects the expression of hundreds of genes.
We used human BJAB B-lymphoma cells and murine CTLL-2 T lymphocytes to generate transcriptional profiles in response to rapamycin treatment, because the proliferation of both types of cells is significantly slowed in the presence of a low concentration of rapamycin (10 nM) (20, 24). Total RNA was harvested from BJAB cells treated with 20 nM rapamycin or the ethanol vehicle for various times. Rapamycin treatment times (15, 30, 60, and 120 min and 12 and 24 h) were chosen to capture rapid and late changes in gene expression in response to the drug. The absence of phosphorylation at threonine 389 of S6K1 throughout the time course confirms that the rapamycin-sensitive pathway was inhibited at all times during which rapamycin was present in the media (Fig. 1A). Labeled probes synthesized from cellular mRNA were hybridized to oligonucleotide microarrays that detect the expression of ∼12,600 human genes and expressed sequence tags (ESTs). Genes were identified as having changed their expression based on a significant and repeated difference between their expression level in a rapamycin-treated sample and the mean expression level in control samples (see Materials and Methods). The expression of approximately 4,400 of the 12,600 genes and ESTs represented on the microarray was detected in the untreated cells, and the expression of 4.3% (543 genes) of the genes significantly changed during the treatment course with rapamycin. k means clustering revealed the major temporal patterns of drug-induced gene expression. During the early time points rapamycin affected the expression of only a small number of genes (Fig. 1B, clusters A and E) and most genes were affected exclusively at the 12- or 24-h time points (Fig. 1B). The time scale at which these rapamycin-sensitive targets respond is different from that of S6K1, which is inactivated within minutes after the drug is added to the cell medium. Quantitative RT-PCR measurements for several metabolism-related genes (BCKAD E1 alpha, ASS, FAS, and SREBP-1) confirmed the changes in gene expression after rapamycin treatment that were detected by the microarrays (Fig. 1C). Functional categorization of the rapamycin-sensitive genes revealed that the drug affects genes participating in nutrient and energy metabolism, protein synthesis and turnover, and the stress response, as well as genes encoding immune modulators specific to lymphocytes and genes in other functional classes (Table 1). About 22% (114 of 504 genes affected at 12 or 24 h) of the genes and ESTs affected by rapamycin encode proteins of unknown function or have no significant sequence similarity to genes in the public databases. Further work is required to determine if the rapamycin-induced changes in mRNA levels reflect alterations in mRNA transcription rates and/or stability.
TABLE 1.
Major functional classes of genes affected by rapamycina
| Class and subclass (n) | Exampleb | Unigene no. |
|---|---|---|
| Amino acid metabolism (24) | ||
| Catabolism (8 up, 5 down) | BCKAD E1 alpha (up) | Hs.78950 |
| tRNA synthetase (7 down) | Threonyl-tRNA synthetase (down) | Hs.84131 |
| Amino acid transport (3 down) | Neutral amino acid transporter (down) | Hs.183556 |
| Amino acid biosynthesis (1 down) | Asparagine synthetase (down) | Hs.75692 |
| Lipid metabolism (7) | ||
| Catabolism (2 up) | VLACD (up) | Hs.82208 |
| Biosynthesis (5 down) | Fatty acid synthase (down) | Hs.83190 |
| Nucleotide metabolism (7) | ||
| Salvage pathway (4 up) | Adenine deaminase (2) (up) | Hs.1217 |
| De novo biosynthesis (3 down) | Uridine monophosphate kinase (down) | Hs.75939 |
| Glycolysis (6) | Phosphoglycerate mutase (down) | Hs.181013 |
| Mitochondria (36) | ||
| Transport (2 up) | Adenine nucleotide translocator (2) (up) | Hs.164280 |
| Tricarboxylic acid cycle (1 up, 3 down) | Isocitrate dehydrogenase (NADP+) (up) | Hs.5337 |
| Electron transfer chain (2 up, 8 down) | NADH-ubiquinone oxidoreductase (down) | Hs.75227 |
| Mitochondrial biogenesis (7 down) | Ribosomal protein S12 (2) (down) | Hs.9964 |
| Protein synthesis (11) | eIF2 alpha (down) | Hs.81613 |
| Protein turnover (18) | ||
| Ubiquitin-conjugating enzyme (1 up, 3 down) | UBC E2 variant 1 (up) | Hs.75875 |
| Proteasome subunits (7 down) | Alpha subunit 3 of proteasome (down) | Hs.167106 |
| Stress response (25) | ||
| Heat shock/chaperonin (17 down) | Hsp70 (down) | Hs.90093 |
| DNA damage (4 up, 1 down) | ERCC 1 (up) | Hs.59544 |
| Immune modulator (29) | ||
| Cytokine receptor (3 up) | IL-4 receptor (up) | Hs.75545 |
| Surface antigen (8 up) | CD53 (up) | Hs.82212 |
| Chromatin related (19) | ||
| Histone (9 up) | H3 histone, 3A (up) | Hs.181307 |
| Topoisomerase (3 up) | DNA topoisomerase II alpha (up) | Hs.156346 |
| Transcription factors (33) | c-Myc (3) (up) | Hs.79070 |
| Cell cycle (10) | Cdc25A (down) | Hs.1634 |
There are 504 genes affected by rapamycin at 12 and 24 h, of which 114 are of unknown function. All 390 known genes were functionally annotated by searching the Unigene and PubMed databases. up, up-regulated; down, down-regulated.
Numbers in parentheses indicate that rapamycin affected more than one entry representing the same gene on the U95 Av2 chips.
The rapamycin-induced changes in gene expression are not specific to BJAB cells. Transcriptional profiles of murine CTLL-2 cells treated with rapamycin for 12 and 24 h show that the drug affected classes of genes similar to those in human BJAB cells. In most cases in which orthologous genes were represented on the human and mouse microarrays, corresponding genes were affected in both cell types. The complete list of functionally annotated rapamycin-sensitive genes in BJAB and CTLL-2 is available in the supplementary data or at the following website: http://staffa.wi.mit.edu/sabatini_public/rapachip2/frameset1.html.
Rapamycin affects the expression of genes involved in the metabolism of nutrients.
Rapamycin affected the expression of a large number of genes in various functional classes involved in nutrient metabolism (Table 1). The expression changes and the genes affected indicate that the drug induces a program that promotes the catabolism of nutrients for energy production while inhibiting their de novo synthesis. Rapamycin treatment increases the expression of genes involved in the entry of amino acids into the citric acid cycle, including BCKAD E1 alpha and 3-hydroxy-3-methylglutaryl (HMG) coenzyme A (CoA)-lyase, while decreasing the expression of genes that synthesize or use amino acids, such as asparagine synthetase and several tRNA synthetases. In addition, the drug increases the expression of genes like ASS, which funnels amines from catabolized amino acids into the urea cycle. A similar pattern of increasing catabolism and inhibiting biosynthesis is also seen for the genes of fatty acid metabolism. Rapamycin up-regulates the expression of genes that promote fatty acid oxidation, such as very-long-chain acyl-CoA dehydrogenase (VLACD) and carnitine acetyltransferase, while down-regulating genes participating in fatty acid synthesis, such as fatty acid synthase. Lastly, the drug also increases the expression of genes for nucleotide salvage pathways, like adenosine deaminase, while decreasing genes for the de novo synthesis of nucleotides.
Many of the rapamycin-sensitive genes are also regulated in calorie-starved animals (Table 2). These include well-characterized genes such as BCKAD E1 alpha, UCP2, and SREBP-1 (9, 21, 35, 38). Like caloric restriction of mice, rapamycin affects many genes involved in protein synthesis, folding, and turnover (22). Thus, the rapamycin-induced gene expression profile indicates that the RAFT1 pathway participates in energy and nutrient sensing and suggests that rapamycin mimics a starvation-like signal.
TABLE 2.
Rapamycin affects some known starvation-sensitive genesa
| Gene name | Function | Rapamycin treatment reaction | Starvation treatment reaction | Reference |
|---|---|---|---|---|
| BCKAD E1-α | Amino acid catabolism | ↑ | ↑ | 9 |
| Mitochondrial HMG-CoA synthase | Amino acid catabolism | ↑ | ↑ | 7 |
| VLACD | Fatty acid oxidation | ↑ | ↑ | 39 |
| Carnitine acetyltransferase | Fatty acid oxidation | ↑ | ↑ | Data not shown |
| UCP2 | Respiratory chain | ↑ | ↑ | 35 |
| Adenosine deaminase | Nucleotide salvage | ↑ | ↑ | Data not shown |
| Interleukin-4 receptor | Immune modulation | ↑ | ↑ | 39 |
| SREBP-1 | Fatty acid synthesis | ↓ | ↓ | 18 |
| Fatty acid synthase | Fatty acid synthesis | ↓ | ↓ | 38 |
| Stearoyl-CoA desaturase | Fatty acid synthesis | ↓ | ↓ | 38 |
| Isopentenyl pyrophosphate isomerase | Cholesterol synthesis | ↓ | ↓ | Data not shown |
| Squalene epoxidase | Cholesterol synthesis | ↓ | ↓ | Data not shown |
| Threonyl-tRNA synthetase | Protein synthesis | ↓ | ↓ | 22 |
| eIF2 α | Protein synthesis | ↓ | ↓ | 22 |
Arrows pointed upward indicate up-regulation; those pointed downward indicate down-regulation.
Nutrient-regulated gene expression.
The variety of metabolic pathways affected by rapamycin is consistent with a broad role for the RAFT1 pathway in energy and nutrient sensing (Tables 1 and 2). This suggests that the pathway might respond to a common signal derived from many nutrient sources or that it can respond to several distinct nutrient-derived signals. To distinguish these possibilities, we asked if the gene expression profile induced by rapamycin resembles those induced by starvation for individual nutrients. We generated expression profiles from cells cultured in media with low concentrations of glucose or glutamine, the principal energy, carbon, and nitrogen sources for mammalian cells (31). Glutamine is particularly important for lymphocytes, and decreased blood levels lead to immunosuppression in human beings (6, 30). We also prepared expression profiles of cells cultured in low concentrations of leucine, because extracellular levels of this essential branched-chain amino acid regulate S6K1 (15, 23). To control for the gene expression changes induced simply by the antiproliferative effects of media containing low levels of essential nutrients, concentrations of nutrients were chosen that had effects on cell proliferation similar to that of rapamycin (Fig. 2A ). Thus, we generated expression profiles from cells cultured for 30, 60, and 120 min and 12 and 24 h in control medium or in medium with reduced levels of glucose (0.05 mM), glutamine (0.05 mM), or leucine (0.01 mM).
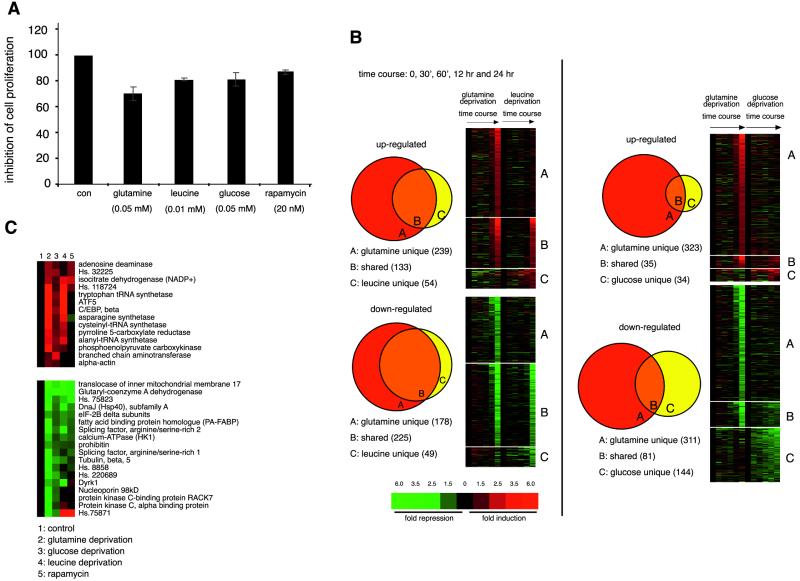
FIG. 2.
Amino acid and glucose deprivation regulates unique sets of genes. (A) Proliferation of BJAB cells over a 24-h period is similarly affected by deprivation of glutamine, leucine, or glucose and by rapamycin treatment. Cells were cultured in media containing indicated concentrations of nutrients or rapamycin. (B) Comparison of transcriptional profiles induced in response to deprivation of glutamine, leucine, or glucose. Only genes affected at 12 or 24 h were used for the comparison. Glutamine deprivation affects 71 and 82% of the genes up- and down-regulated, respectively, by leucine deprivation. Fold changes are indicated colorimetrically, individual genes are represented vertically, and experimental conditions (i.e., time course of deprivation for individual nutrients) are displayed horizontally. (C) A core set of genes regulated by both glutamine and glucose deprivation. Unigene numbers are used to represent genes with unknown function. The gene expression changes for the 24-h time point are shown.
Starvation for individual nutrients affected the expression of hundreds of genes: 320 for glucose, 834 for glutamine, and 398 for leucine. Like with rapamycin treatment, most of the gene expression changes occurred at later time points, with maximal effects 12 or 24 h after switching to nutrient-poor media (Fig. 2B). Glutamine deprivation affected the most genes, suggesting that, in BJAB cells, glutamine plays a larger role in biosynthesis and energy generation than do leucine and glucose. Comparisons between the expression profiles induced by glutamine and leucine deprivation show that the leucine-regulated genes are largely a subset of those affected by glutamine deprivation (Fig. 2B). In contrast, the sets of genes affected by glutamine and glucose deprivation share only a small number of core starvation genes that include known nutrient-regulated genes, such as PEPCK, asparagine synthetase, and C/EBP beta (3, 4, 14), as well as others not previously described as so regulated, such as branched-chain aminotransferase (BCAT), alanyl-tRNA synthetase, ATF5, glutaryl-CoA dehydrogenase, and Dyrk1 (Fig. 2C). Interestingly, Yak1p, a Dyrk1 homologue, is implicated in glucose sensing in budding yeast (25). The majority of these core starvation genes are also affected by leucine deprivation (Fig. 2C). Notably, the expression of BCAT, the enzyme that catalyzes the rate-limiting step in branched-chain amino acid degradation, is not affected by leucine.
Comparisons between rapamycin-sensitive gene expression and nutrient-sensitive gene expression.
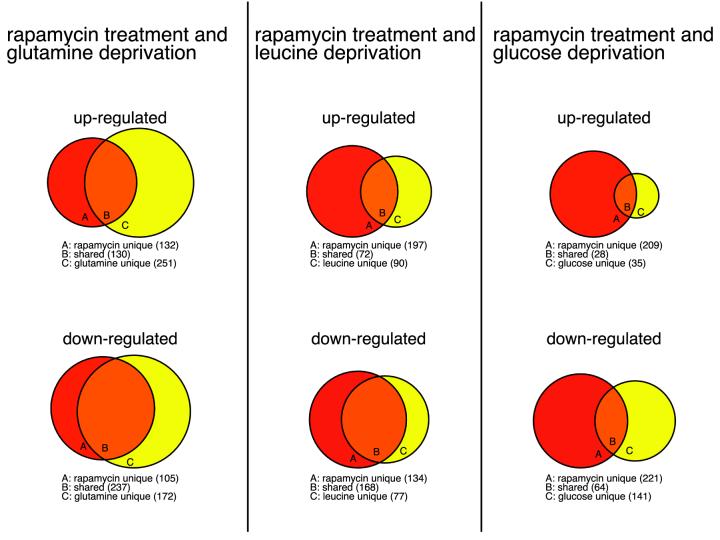
Comparisons between the nutrient- and rapamycin- sensitive sets of genes show that the RAFT1 pathway regulates significant fractions of the nutrient-responsive genes, supporting the conclusion that RAFT1 functions in energy and nutrient sensing (Fig. 3). The drug up-regulated about 34, 44, and 34% of all the genes increased by glutamine, leucine, and glucose deprivation, respectively, and affected several of the core starvation genes (4 of 14 up-regulated and 9 of 18 down-regulated) (Fig. 2C). For the down-regulated genes, rapamycin overlaps significantly more with the sets reduced by glutamine and leucine deprivation than with those reduced by glucose deprivation (58 and 69% versus 31%), indicating that rapamycin treatment more closely resembles amino acid deprivation than it does glucose deprivation.
FIG. 3.
Global comparison of gene expression changes induced by rapamycin treatment and by deprivation for individual nutrient. The sets of genes affected at 12 or 24 h after rapamycin treatment and deprivation for nutrients significantly overlap. Rapamycin shares 34, 44, and 44% of up-regulated genes as well as 58, 69, and 31% of genes down-regulated by glutamine, leucine, and glucose deprivation, respectively.
The similarity of gene expression induced by rapamycin and amino acid deprivation is clearly seen when examining many categories of functionally related genes. Rapamycin and glutamine deprivation up-regulates overlapping sets of genes involved in amino acid and fatty acid oxidation and the nucleotide salvage pathway, e.g., the genes encoding BCKAD E1 alpha and VLACD, the two enzymes for oxidizing branched-chain amino acids and long-chain fatty acids, respectively (Table 3 and see supplementary data). It also shows remarkable overlap with glutamine but not glucose deprivation in down-regulating genes in de novo biosynthesis of fatty acid, cholesterol, and nucleotides and in protein synthesis, turnover, and folding. For example, rapamycin starvation and amino acid starvation both decrease the expression levels of SREBP-1 and of several genes regulated by this transcription factor (e.g., fatty acid synthase and squalene epoxidase) (see supplementary data). Taken together, the data suggest that rapamycin mimics a starvation-like signal that originates from amino acid deprivation.
TABLE 3.
Comparison of gene categories affected by rapamycin and nutrient deprivationa
| Category | No. of genes regulated | Effect of:
|
||
|---|---|---|---|---|
| Rapamycin treatment | Glutamine deprivation | Glucose deprivation | ||
| Amino acid oxidation | 16 | ↑↑ | ↑↑↑ | ↑ |
| Fatty acid oxidation | 6 | ↑↑ | ↑↑↑↑ | — |
| Fatty acid and cholesterol synthesis | 9 | ↓↓↓ | ↓↓↓↓ | — |
| Nucleotide salvage pathway | 4 | ↑↑↑↑ | ↑↑ | ↑ |
| Nucleotide de novo synthesis | 14 | ↓↓ | ↓↓↓↓ | ↓ |
| Protein synthesis (initiation factors) | 16 | ↓↓ | ↓↓↓ | ↓ |
| Protein turnover (proteasome subunits) | 15 | ↓↓ | ↓↓↓ | ↓ |
| Protein folding (chaperones) | 21 | ↓↓↓↓ | ↓↓↓↓ | ↓ |
| tRNA synthetases | 10 | ↓↓ | ↑↑↑↑ | ↑↑ |
| Amino acid synthesis | 3 | ↓↓ | ↑↑↑↑ | ↑↑↑ |
| Amino acid transporters | 5 | ↓↓↓ | ↑↑↑↑ | — |
| Histones | 9 | ↑↑↑↑ | ↑↑ | — |
| Immune modulators | 31 | ↑↑↑↑ | ↑↑ | ↑ |
| Cell cycle | 18 | ↓↓ | ↓↓ | ↓↓ |
↑ means up-regulated and ↓ means down-regulated. The percentage of genes in each category affected by rapamycin treatment, glutamine or glucose deprivation is denoted in the following ways: ↑ or ↓, larger than 0 but smaller than or equal to 25%; ↑↑ or ↓↓, larger than 25% but smaller than or equal to 50%; ↑↑↑ or ↓↓↓, larger than 50% but smaller than or equal to 75%; ↑↑↑↑ or ↓↓↓↓, larger than 75% but smaller than or equal to 100%; —, there is no gene affected.
Further comparisons of the rapamycin- and glutamine-sensitive sets of genes strongly indicate that the dearth of amino acids themselves is unlikely to be this signal. Given the similar effects of amino acid starvation and rapamycin on the in vivo activity of S6K1 (15), we were surprised that rapamycin induced very few of the large group of amino-acid-related genes that were activated by glutamine and leucine starvation but not by glucose starvation. Instead, at 12 and 24 h after addition of rapamycin, the expression of some of these genes was down-regulated (Table 3), a result inconsistent with the RAFT1 pathway sensing the absence of amino acids directly. Many of these genes are involved in the synthesis (3-phosphoglycerate dehydrogenase and pyrroline 5-carboxylate reductase), transport (4F2 and sodium-dependent neutral amino acid transporter), and use of amino acids (10 tRNA-synthetases) (see supplementary data). Interestingly, several transcription factors, including ATF4, a known responder to amino acid levels (16), as well as other B-zip transcription factors like NRF1 and ATF5, have this similar pattern of expression (see supplementary data). These results suggest that rapamycin does not mimic the absence of amino acids themselves. Instead, the data are compatible with rapamycin mimicking a signal downstream of amino acids, such as a metabolite derived from amino acids. Cells treated with rapamycin would then initiate a program that favors the use of amino acids for the production of this metabolite over other uses that consume amino acids, such as protein synthesis.
Does rapamycin also mimic a signal originating from the deprivation of other nutrients besides amino acids? Rapamycin does affect the expression of a few genes that were sensitive to the deprivation of glucose (6 up and 11 down) but not to the deprivation of glutamine or leucine (data not shown). These genes do not fall into any readily identifiable functional class, unlike the genes shared between rapamycin treatment and glutamine deprivation. Furthermore, as shown in Table 3, genes encoding immune modulators and several histones are largely uniquely affected by rapamycin, indicating that neither amino acids nor glucose deprivation regulates their expression. Thus, comparisons of the transcriptional profiles induced by rapamycin and nutrient deprivation support a model in which the RAFT1 pathway may respond to several distinct signals, some of which emanate from amino acids and still others from unknown sources.
Measurements of nutrient use.
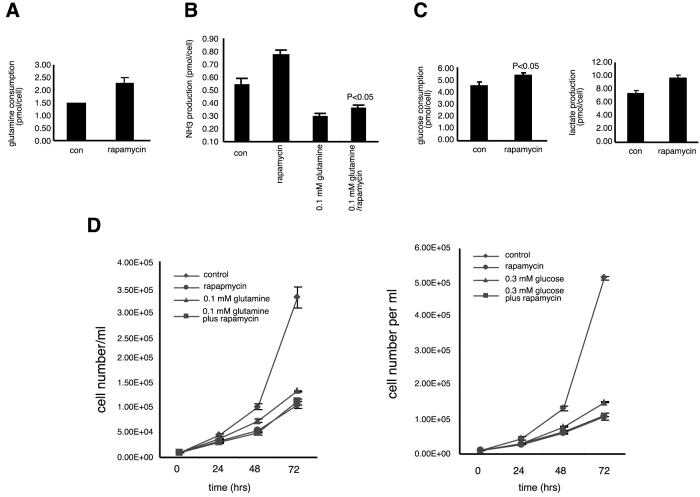
Measurements of nutrient usage by rapamycin-treated cells also suggest that rapamycin mimics a starvation-like signal that originates from amino acids but is not likely from amino acids themselves. Compared to control CTLL-2 cells, rapamycin-treated cells consume more glutamine (Fig. 4A), the expected response of cells perceiving a lack of glutamine but growing in glutamine-rich media. Concomitant with the increased consumption of glutamine, CTLL-2 cells treated with rapamycin also produce more ammonia (Fig. 4B), a response also seen in BJAB cells (data not shown). As most of the ammonia generated by mammalian cells comes from glutamine, this result suggests that cells are using glutamine as a source of carbon skeletons for energy production instead of as a source of nitrogen (36). Furthermore, rapamycin raised ammonia production even in cells that were cultured in media with low levels of glutamine, suggesting that the drug also increases the use of carbon from other amino acids besides glutamine (Fig. 4B). Consistent with the transcriptional profiling results, rapamycin had only a small effect on glucose metabolism, a slight but significant increase in the use of glucose and in the production of lactate (Fig. 4C). We also measured intracellular ATP levels in CTLL-2 cells treated with rapamycin. At long time points after rapamycin addition (12 and 24 h), we detected a statistically significant increase of 10 to 15% in ATP levels. No effect on ATP levels was detected with less than 2 h of rapamycin treatment (data not shown).
FIG. 4.
Effects of rapamycin treatment on nutrient metabolism and cell proliferation in murine CTLL-2 T lymphocytes. All nutrient measurements were performed over a 48-h period in triplicate as described in Materials and Methods. (A) Cells treated with rapamycin increased the usage of glutamine by 54% (P < 0.01). (B) Cells treated with rapamycin increased the production of ammonia when cultured in media with normal (2 mM) or low (0.1 mM) concentrations of glutamine by 44% (P < 0.01) or 20% (P < 0.05), respectively. (C) Cells treated with rapamycin increased the consumption of glucose by 19% (P < 0.05) and also increased lactate production by 31% (P < 0.01). (D) CTLL-2 cells (starting at 104/ml) were grown in control medium with or without rapamycin (20 nM), medium with 0.1 mM glutamine with or without rapamycin (20 nM), or medium with 0.3 mM glucose with or without rapamycin (20 nM) for 72 h. Triplicate samples were counted every 24 h.
The antiproliferative effects of rapamycin likely result from its capacity to mimic a starvation signal. As shown in Fig. 4D, rapamycin inhibited the proliferation of cells to a degree similar to that of growth in media with low concentrations of glutamine or glucose. Moreover, rapamycin and nutrient deprivation did not inhibit cell proliferation in an additive fashion. Consistent with the proliferation curves, most rapamycin-sensitive genes involved in cell cycle progression are also sensitive to nutrient deprivation (Table 3 and supplementary data).
DISCUSSION
In this study we address the relationship between rapamycin- and nutrient-sensitive signaling pathways in mammalian cells. By comparing the transcriptional profiles induced by rapamycin to those resulting from nutrient deprivation, we have shown that rapamycin treatment and amino acid deprivation up-regulate overlapping sets of genes involved in oxidizing amino acids and fatty acids and nucleotide salvage pathways. Furthermore, rapamycin deprivation and amino acid deprivation have remarkably similar effects in down-regulating genes in the de novo biosynthesis of lipids and nucleotides and in protein synthesis, protein turnover, and protein folding (heat shock proteins). Surprisingly, rapamycin deprivation and amino acid deprivation have opposing effects on the regulation of genes involved in amino acid biosynthesis, amino acid transport, and tRNA synthesis. Taken together, the data suggest that rapamycin mimics a signal that originates from amino acids but is not from amino acids themselves. Since the process of oxidizing amino acids and fatty acids takes places in the mitochondria, it is of interest to investigate whether and how RAFT1 may regulate mitochondrial function.
A possible explanation for the down-regulation by the drug of some of the amino acid-related genes induced by glutamine deprivation is that rapamycin interferes directly with the pathways that activate them. Alternatively, if cells respond to rapamycin by increasing the levels of intracellular amino acids, rapamycin may indirectly affect these differentially regulated genes by triggering the normal pathways that repress them in the presence of elevated concentrations of amino acids. With either model, the results suggest that rapamycin does not mimic the absence of amino acids themselves. Instead, the results are compatible with rapamycin mimicking a signal downstream of amino acids, such as a that from a metabolite derived from amino acids. The available literature on rapamycin use in mammalian cells strongly suggests that at the 20 nM concentration used in our study, rapamycin selectively affects the mTOR pathway. However, it is possible, of course, that some of the observed rapamycin-induced changes in mRNA levels may occur through mechanisms independent of mTOR, such as inhibition of the activity of the FKBP family of proteins.
There are similarities and differences between the gene categories affected by rapamycin in yeast and mammalian cells. In both organisms rapamycin causes a decrease in the levels of genes involved in glycolysis, translation initiation and elongation, and tRNA synthesis (7, 17, 37). On the other hand, genes involved in mitochondrial function respond differently to the drug in the two systems. In yeast, rapamycin significantly up-regulates genes involved in ATP synthesis, the tricarboxylic acid cycle, and oxidative phosphorylation, while, in BJAB and CTLL-2 cells, rapamycin down-regulated several genes in these categories (7, 17, 37). Similarly, in the two systems rapamycin has opposite effects on the genes encoding proteasome subunits. Possible explanations for the alternative responses to rapamycin may be that the two systems have different nutrient requirements or that in mammalian cells the final level of gene activity is determined by integrating nutrient-derived signals with those derived from growth factors, such as insulin.
In yeast, the rapamycin-induced transcriptional profiles have been interpreted as suggesting that rapamycin mimics the absence of more than one nutrient-deprived signal (7, 17, 37). In mammalian cells, does the RAFT1 pathway sense a signal originating only from amino acids or from other sources as well? Our nutrient measurement data and array data suggest that the latter is the case. It is known that cells do not use more glucose to compensate for amino acid deprivation (10), a result that we verified in CTLL-2 cells (data not shown), but that rapamycin-treated cells do slightly increase glucose usage. Two functional classes of responsive genes (histones and immune modulators) are largely unique to rapamycin treatment, further suggesting that rapamycin may also mimic a signal that is not coming from amino acid or glucose deprivation.
Nevertheless, our results indicate a much greater similarity between the effects of amino acid deprivation and rapamycin treatment in BJAB, a human B-lymphoma cell line. Interestingly, lymphocytes are more dependent than other cell types on amino acids, especially glutamine, for energy generation and biosynthesis (1, 29). In addition, a decrease in glutamine concentration in plasma leads to immunosuppression in human beings (6, 30) and animals (19). Perhaps rapamycin causes immunosuppression in vivo with relatively few side effects because, by mimicking a signal generated from amino acids, it preferentially targets cells of the immune system. Understanding how RAFT1, the target of rapamycin, senses amino acid deprivation and regulates cell proliferation may provide targets for small molecules that modulate the immune system in novel ways.
Acknowledgments
We are grateful to Junaid Ziauddin, Christine Ladd, and Christine Huard for aiding in GeneChip scanning. We thank Nir Hacohen and Sridhar Ramaswamy for comments on the manuscript.
This study was supported by National Institutes of Health grant AI47389 and by the G. Harold and Leila Y. Mathers Charitable Foundation.
REFERENCES
- 1.Ardawi, M. S., and E. A. Newsholme. 1983. Glutamine metabolism in lymphocytes of the rat. Biochem. J. 212:835-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barbet, N. C., U. Schneider, S. B. Helliwell, I. Stansfield, M. F. Tuite, and M. N. Hall. 1996. TOR controls translation initiation and early G1 progression in yeast. Mol. Biol. Cell 7:25-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barbosa-Tessmann, I. P., C. Chen, C. Zhong, F. Siu, S. M. Schuster, H. S. Nick, and M. S. Kilberg. 2000. Activation of the human asparagine synthetase gene by the amino acid response and the endoplasmic reticulum stress response pathways occurs by common genomic elements. J. Biol. Chem. 275:26976-26985. [DOI] [PubMed] [Google Scholar]
- 4.Barbosa-Tessmann, I. P., V. L. Pineda, H. S. Nick, S. M. Schuster, and M. S. Kilberg. 1999. Transcriptional regulation of the human asparagine synthetase gene by carbohydrate availability. Biochem. J. 339:151-158. [PMC free article] [PubMed] [Google Scholar]
- 5.Brown, E. J., M. W. Albers, T. B. Shin, K. Ichikawa, C. T. Keith, W. S. Lane, and S. L. Schreiber. 1994. A mammalian protein targeted by G1-arresting rapamycin-receptor complex. Nature 369:756-758. [DOI] [PubMed] [Google Scholar]
- 6.Calder, P. C., and P. Yaqoob. 1999. Glutamine and the immune system. Amino Acids 17:227-241. [DOI] [PubMed] [Google Scholar]
- 7.Cardenas, M. E., N. S. Cutler, M. C. Lorenz, C. J. Di Como, and J. Heitman. 1999. The TOR signaling cascade regulates gene expression in response to nutrients. Genes Dev. 13:3271-3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7a.Casals, N., N. Roca, M. Guerrero, G. Gil-Gomez, J. Ayte, C. J. Ciudad, and F. G. Hegardt. 1992. Regulation of the expression of the mitochondrial 3-hydroxy-3-methylglutaryl-CoA synthase gene. Its role in the control of ketogenesis. Biochem. J. 283:261-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coller, H. A., C. Grandori, P. Tamayo, T. Colbert, E. S. Lander, R. N. Eisenman, and T. R. Golub. 2000. Expression analysis with oligonucleotide microarrays reveals that MYC regulates genes involved in growth, cell cycle, signaling, and adhesion. Proc. Natl. Acad. Sci. USA 97:3260-3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Costeas, P. A., and J. M. Chinsky. 1996. Effects of insulin on the regulation of branched-chain alpha-keto acid dehydrogenase E1 alpha subunit gene expression. Biochem. J. 318:85-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cruz, H. J., A. S. Ferreira, C. M. Freitas, J. L. Moreira, and M. J. Carrondo. 1999. Metabolic responses to different glucose and glutamine levels in baby hamster kidney cell culture. Appl. Microbiol. Biotechnol. 51:579-585. [DOI] [PubMed] [Google Scholar]
- 11.Dennis, P. B., A. Jaeschke, M. Saitoh, B. Fowler, S. C. Kozma, and G. Thomas. 2001. Mammalian TOR: a homeostatic ATP sensor. Science 294:1102-1105. [DOI] [PubMed] [Google Scholar]
- 12.Eisen, M. B., P. T. Spellman, P. O. Brown, and D. Botstein. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95:14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fang, Y., M. Vilella-Bach, R. Bachmann, A. Flanigan, and J. Chen. 2001. Phosphatidic acid-mediated mitogenic activation of mTOR signaling. Science 294:1942-1945. [DOI] [PubMed] [Google Scholar]
- 14.Hanson, R. W., and L. Reshef. 1997. Regulation of phosphoenolpyruvate carboxykinase (GTP) gene expression. Annu. Rev. Biochem. 66:581-611. [DOI] [PubMed] [Google Scholar]
- 15.Hara, K., K. Yonezawa, Q. P. Weng, M. T. Kozlowski, C. Belham, and J. Avruch. 1998. Amino acid sufficiency and mTOR regulate p70 S6 kinase and eIF-4E BP1 through a common effector mechanism. J. Biol. Chem. 273:14484-14494. [DOI] [PubMed] [Google Scholar]
- 16.Harding, H. P., I. I. Novoa, Y. Zhang, H. Zeng, R. Wek, M. Schapira, and D. Ron. 2000. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell 6:1099-1108. [DOI] [PubMed] [Google Scholar]
- 17.Hardwick, J. S., F. G. Kuruvilla, J. K. Tong, A. F. Shamji, and S. L. Schreiber. 1999. Rapamycin-modulated transcription defines the subset of nutrient-sensitive signaling pathways directly controlled by the Tor proteins. Proc. Natl. Acad. Sci. USA 96:14866-14870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hidalgo, M., and E. K. Rowinsky. 2000. The rapamycin-sensitive signal transduction pathway as a target for cancer therapy. Oncogene 19:6680-6686. [DOI] [PubMed] [Google Scholar]
- 18a.Horton, J. D., Y. Bashmakov, I. Shimomura, and H. Shimano. 1998. Regulation of sterol regulatory element binding proteins in livers of fasted and refed mice. Proc. Natl. Acad. Sci. USA 95:5987-5992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kafkewitz, D., and A. Bendich. 1983. Enzyme-induced asparagine and glutamine depletion and immune system function. Am. J. Clin. Nutr. 37:1025-1030. [DOI] [PubMed] [Google Scholar]
- 20.Kay, J. E., M. C. Smith, V. Frost, and G. Y. Morgan. 1996. Hypersensitivity to rapamycin of BJAB B lymphoblastoid cells. Immunology 87:390-395. [PMC free article] [PubMed] [Google Scholar]
- 21.Kim, J. B., P. Sarraf, M. Wright, K. M. Yao, E. Mueller, G. Solanes, B. B. Lowell, and B. M. Spiegelman. 1998. Nutritional and insulin regulation of fatty acid synthetase and leptin gene expression through ADD1/SREBP1. J. Clin. Investig. 101:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee, C. K., R. G. Klopp, R. Weindruch, and T. A. Prolla. 1999. Gene expression profile of aging and its retardation by caloric restriction. Science 285:1390-1393. [DOI] [PubMed] [Google Scholar]
- 22a.Lee, C. K., R. Weindruch, and T. A. Prolla. 2000. Gene-expression profile of the ageing brain in mice. Nat. Genet. 25:294-297. [DOI] [PubMed] [Google Scholar]
- 23.Lynch, C. J. 2001. Role of leucine in the regulation of mTOR by amino acids: revelations from structure-activity studies. J. Nutr. 131:861S-865S. [DOI] [PubMed] [Google Scholar]
- 24.Morice, W. G., G. J. Brunn, G. Wiederrecht, J. J. Siekierka, and R. T. Abraham. 1993. Rapamycin-induced inhibition of p34cdc2 kinase activation is associated with G1/S-phase growth arrest in T lymphocytes. J. Biol. Chem. 268:3734-3738. [PubMed] [Google Scholar]
- 25.Moriya, H., Y. Shimizu-Yoshida, A. Omori, S. Iwashita, M. Katoh, and A. Sakai. 2001. Yak1p, a DYRK family kinase, translocates to the nucleus and phosphorylates yeast Pop2p in response to a glucose signal. Genes Dev. 15:1217-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neshat, M. S., I. K. Mellinghoff, C. Tran, B. Stiles, G. Thomas, R. Petersen, P. Frost, J. J. Gibbons, H. Wu, and C. L. Sawyers. 2001. Enhanced sensitivity of PTEN-deficient tumors to inhibition of FRAP/mTOR. Proc. Natl. Acad. Sci. USA 98:10314-10319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neuhaus, P., J. Klupp, and J. M. Langrehr. 2001. mTOR inhibitors: an overview. Liver Transplant. 7:473-484. [DOI] [PubMed] [Google Scholar]
- 28.Newsholme, E. A., B. Crabtree, and M. S. Ardawi. 1985. Glutamine metabolism in lymphocytes: its biochemical, physiological and clinical importance. Q. J. Exp. Physiol. 70:473-489. [DOI] [PubMed] [Google Scholar]
- 29.Newsholme, E. A., B. Crabtree, and M. S. Ardawi. 1985. The role of high rates of glycolysis and glutamine utilization in rapidly dividing cells. Biosci. Rep. 5:393-400. [DOI] [PubMed] [Google Scholar]
- 30.Parry-Billings, M., J. Evans, P. C. Calder, and E. A. Newsholme. 1990. Does glutamine contribute to immunosuppression after major burns? Lancet 336:523-525. [DOI] [PubMed] [Google Scholar]
- 31.Petch, D., and M. Butler. 1994. Profile of energy metabolism in a murine hybridoma: glucose and glutamine utilization. J. Cell. Physiol. 161:71-76. [DOI] [PubMed] [Google Scholar]
- 32.Podsypanina, K., R. T. Lee, C. Politis, I. Hennessy, A. Crane, J. Puc, M. Neshat, H. Wang, L. Yang, J. Gibbons, P. Frost, V. Dreisbach, J. Blenis, Z. Gaciong, P. Fisher, C. Sawyers, L. Hedrick-Ellenson, and R. Parsons. 2001. An inhibitor of mTOR reduces neoplasia and normalizes p70/S6 kinase activity in Pten+/- mice. Proc. Natl. Acad. Sci. USA 98:10320-10325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sabatini, D. M., H. Erdjument-Bromage, M. Lui, P. Tempst, and S. H. Snyder. 1994. RAFT1: a mammalian protein that binds to FKBP12 in a rapamycin-dependent fashion and is homologous to yeast TORs. Cell 78:35-43. [DOI] [PubMed] [Google Scholar]
- 34.Sabers, C. J., M. M. Martin, G. J. Brunn, J. M. Williams, F. J. Dumont, G. Wiederrecht, and R. T. Abraham. 1995. Isolation of a protein target of the FKBP12-rapamycin complex in mammalian cells. J. Biol. Chem. 270:815-822. [DOI] [PubMed] [Google Scholar]
- 35.Samec, S., J. Seydoux, and A. G. Dulloo. 1998. Role of UCP homologues in skeletal muscles and brown adipose tissue: mediators of thermogenesis or regulators of lipids as fuel substrate? FASEB J. 12:715-724. [DOI] [PubMed] [Google Scholar]
- 36.Schneider, M., I. W. Marison, and U. von Stockar. 1996. The importance of ammonia in mammalian cell culture. J. Biotechnol. 46:161-185. [DOI] [PubMed] [Google Scholar]
- 37.Shamji, A. F., F. G. Kuruvilla, and S. L. Schreiber. 2000. Partitioning the transcriptional program induced by rapamycin among the effectors of the Tor proteins. Curr. Biol. 10:1574-1581. [DOI] [PubMed] [Google Scholar]
- 38.Shimomura, I., Y. Bashmakov, H. Shimano, J. D. Horton, J. L. Goldstein, and M. S. Brown. 1997. Cholesterol feeding reduces nuclear forms of sterol regulatory element binding proteins in hamster liver. Proc. Natl. Acad. Sci. USA 94:12354-12359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38a.Soukas, A., P. Cohen, N. D. Socci, and J. M. Friedman. 2000. Leptin-specific patterns of gene expression in white adipose tissue. Genes Dev. 14:963-980. [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang, H., J. P. Stallock, J. C. Ng, C. Reinhard, and T. P. Neufeld. 2000. Regulation of cellular growth by the Drosophila target of rapamycin dTOR. Genes Dev. 14:2712-2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39a.Zhang, J., L. E. Underwood, and A. J. D'Ercole. 2001. Hepatic mRNAs up-regulated by starvation: an expression profile determined by suppression subtractive hybridization. FASEB J. 15:1261-1263. [DOI] [PubMed] [Google Scholar]