Abstract
Oncogenes Neu/HER2/ErbB2 and Ras can induce mammary tumorigenesis via upregulation of cyclin D1. One major regulatory mechanism in these oncogenic signaling pathways is phosphorylation of serines or threonines preceding proline (pSer/Thr-Pro). Interestingly, the pSer/Thr-Pro motifs in proteins exist in two completely distinct cis and trans conformations, whose conversion is catalyzed specifically by the essential prolyl isomerase Pin1. By isomerizing pSer/Thr-Pro bonds, Pin1 can regulate the conformation and function of certain phosphorylated proteins. We have previously shown that Pin1 is overexpressed in breast tumors and positively regulates cyclin D1 by transcriptional activation and posttranslational stabilization. Moreover, in Pin1 knockout mice, mammary epithelial cells fail to undergo massive proliferation during pregnancy, as is the case in cyclin D1 null mice. These results indicate that Pin1 is upregulated in breast cancer and may be involved in mammary tumors. However, the mechanism of Pin1 overexpression in cancer and its significance in cell transformation remain largely unknown. Here we demonstrate that PIN1 expression is mediated by the transcription factor E2F and enhanced by c-Neu and Ha-Ras via E2F. Furthermore, overexpression of Pin1 not only confers transforming properties on mammary epithelial cells but also enhances the transformed phenotypes of Neu/Ras-transformed mammary epithelial cells. In contrast, inhibition of Pin1 suppresses Neu- and Ras-induced transformed phenotypes, which can be fully rescued by overexpression of a constitutively active cyclin D1 mutant that is refractory to the Pin1 inhibition. Thus, Pin1 is an E2F target gene that is essential for the Neu/Ras-induced transformation of mammary epithelial cells through activation of cyclin D1.
Phosphorylation of proteins on serine/threonine residues preceding proline (pSer/Thr-Pro) is a key regulatory mechanism for the control of cell proliferation and transformation (6, 18, 22, 31). For example, oncogenic Neu/Ras signaling has shown to lead to activation of various Pro-directed protein kinases, which eventually enhance transcription of the cyclin D1 gene via multiple transcription factors, including E2F, c-jun/AP-1, and β-catenin/T-cell factor (TCF) (1, 3, 17, 26, 28, 47). In addition to transcriptional activation, cyclin D1 is regulated by posttranslational modifications. Phosphorylation of cyclin D1 on the Thr286-Pro site by glycogen synthase kinase 3β (GSK-3β) enhances its nuclear export and subsequent degradation (2, 9, 10).
Cyclin D1 has been shown to play a pivotal role in the development of cancer, especially breast cancer. Overexpression of cyclin D1 is found in 50% of patients with breast cancer (5, 15). Importantly, overexpression of cyclin D1, especially the mutant cyclin D1T286A, can transform fibroblasts (2, 20). In contrast, inhibition of cyclin D1 expression causes growth arrest in tumor cells (4, 11, 26, 45). Furthermore, transgenic overexpression of cyclin D1 in the mouse mammary gland leads to mammary hyperplasia and eventually adenocarcinomas (55). More importantly, disruption of the cyclin D1 gene in mice completely suppresses the ability of Ha-Ras or c-Neu/HER2 to induce tumor development in the mammary gland (60). These results indicate that cyclin D1 is an essential downstream target for mammary tumorigenesis induced by Ha-Ras or c-Neu and that a major mechanism in these oncogenic processes is phosphorylation of pSer/Thr-Pro motifs.
Interestingly, the pSer/Thr-Pro motifs in proteins exist in two completely distinct cis and trans conformations, whose conversion is catalyzed specifically by the essential prolyl isomerase Pin1 (30, 34, 43, 63). By isomerizing specific pSer/Thr-Pro bonds, Pin1 has been shown to catalytically induce conformational changes in proteins following phosphorylation, thereby having profound effects on their catalytic activity, dephosphorylation, protein-protein interactions, subcellular location, and/or turnover (21, 29, 32, 44, 46, 52, 58, 59, 62). Thus, phosphorylation-dependent prolyl isomerization is a critical regulatory mechanism in phosphorylation signaling (31).
Significantly, we have previously shown that Pin1 is strongly overexpressed in many human malignancies, such as breast cancer, and that its expression closely correlates with the tumor grade and cyclin D1 expression level in tumors (44, 58). Importantly, upregulation of Pin1 has been shown to elevate cyclin D1 gene expression by activating the c-jun/AP-1 and β-catenin/TCF transcription factors (44, 58). Furthermore, Pin1 can bind directly to the phosphorylated Thr286-Pro motif in cyclin D1 and stabilize nuclear cyclin D1 protein by inhibiting its export into the cytoplasm, where it is normally degraded by ubiquitin-mediated proteolysis (29). Moreover, deletion of the PIN1 gene in the mouse results in reduction of cyclin D1 levels in many tissues as well as causes many phenotypes resembling cyclin D1 null phenotypes (13, 48), including the failure of the breast epithelial compartment to undergo the massive proliferative changes associated with pregnancy (29). These results indicate that overexpressed Pin1 in breast cancer can positively regulate the function of cyclin D1 at the transcriptional level and by posttranslational stabilization. However, the mechanism of Pin1 overexpression in cancer and its significance in oncogenesis remain largely unknown.
The aim of this study was to further define the molecular mechanism(s) governing Pin1 expression and to investigate the role of Pin1 in the transformation of mammary epithelial cells. We demonstrate that Pin1 expression is regulated by the transcription factor E2F and is enhanced by oncogenic Neu/Ras signaling via E2F activation. More importantly, overexpression of Pin1 not only leads to moderate cell transformation in mammary epithelial cells but also enhances the transformed phenotypes of Neu/Ras-transfected mammary epithelial cells. In contrast, inhibition of Pin1 suppresses the Neu- and Ras-induced transformed phenotypes, which can be completely rescued by overexpression of a constitutively active cyclin D1 mutant that is refractory to Pin1 inhibition. These results indicate that Pin1 is a downstream target of oncogenic Neu/Ras signaling and plays an essential role in mammary tumorigenesis through activation of cyclin D1.
MATERIALS AND METHODS
Cloning the human PIN1 genomic sequence and plasmid constructions.
A human placenta genomic DNA library was screened with a 200-bp fragment of the human PIN1 cDNA encoding the first exon. We screened 106 plaques and obtained three positive clones which had a 15-kb genomic fragment containing exon 1 of the PIN1 gene. Selected clones were sequenced with the Big Dye terminator kits (PE Applied Biosystems, Branchburg, N.J.) with an ABI 377XL automated sequencer (PE Applied Biosystems). About 2.3 kb of the human PIN1 promoter sequence were amplified by PCR and cloned into the pGL3-Basic vector (Promega) to create a PIN1 promoter-luciferase construct. The GenBank accession number of the PIN1 promoter sequence is AF501321. Several deletion mutants were created by PCR as described previously (23). Site-directed mutants were generated with a site-directed PCR mutation kit (Stratagene) according to the manufacturer's protocol.
Cell culture.
Parent MCF-10A cells were cultured in Dulbecco's modified Eagle's medium (DMEM)-F-12 medium supplemented with 2% horse serum, 10 μg of insulin per ml, 1 ng of cholera toxin per ml, 100 μg of hydrocortisone per ml, and 10 ng of human epidermal growth factor (Clonetics) per ml. All other cell types used in this study were maintained in DMEM supplemented with 10% fetal bovine serum or other serum conditions, as indicated below.
Electrophoretic mobility shift assays.
Electrophoretic mobility shift assays were performed as described previously (40, 41). Double-stranded oligonucleotides corresponding to the three putative E2F recognition sites in the 5′-flanking regions of the PIN1 gene and specific mutants of these sites are listed below, with putative E2F binding sites underlined and mutations indicated in boldface type: Site A-wild-type, 5′-CGGGAGTTTTTTGGCGCTCGCTAAAGG-3′; Site A-mutant, 5′-CGGGAGTTTTTTGAAGCTCGCTAAAGG-3′; Site B-wild t:ype, 5′TGCGGCGACGCGCGCCAAGAAGGGGT-3′; Site B-mutant, 5′-TGCGGCGACGCGCGTCAAGAAGGGGT-3′; Site C-wild-type, 5′-GGAGGATGGAGGAGCCAAATTTAAGCAT-3′; and Site C-mutant, 5′-GGAGGATGGAGGATCCAAATTTAAGCAT-3′.
In competition assays, these double-stranded oligonucleotides were used as competitors at a 10- or 100-fold molar excess. A consensus E2F site from the adenovirus E2 promoter was used as a probe (41). Electrophoretic mobility shift assay was performed with gel shift assay systems (Promega). Recombinant glutathione S-transferase (GST)-E2F1 was incubated with the radiolabeled probe in binding buffer [10 mM Tris-HCl (pH 7.5), 1 mM MgCl2, 0.5 mM EDTA, 0.5 mM dithiothreitol, 50 mM NaCl, 50 ng of poly(dI-dC) per ml, 4% glycerol] containing end-labeled DNA fragments at 25°C for 20 min. Samples were resolved on a 5% polyacrylamide native gel in 0.5× Tris-borate-EDTA. Each gel was dried and then subjected to autoradiography for 3 h.
Gene reporter assay.
At ≈60% confluency, MCF-7 or HeLa cells were transfected in triplicate with luciferase reporter constructs with FuGENE 6 (Roche Diagnostics). Gene reporter assays were performed with the dual-luciferase reporter assay system (Promega) at 24 to 36 h after transfection as described previously (44, 58). pRL-TK or pRL-CMV (Promega) was used as an internal control for transfection efficiency. All results are expressed as the mean ± standard deviation (SD) from independent triplicate cultures.
Real-time RT-PCR.
Total RNA was isolated with Trizol reagent (Gibco-BRL), and single-stranded cDNA was synthesized with Superscript (Gibco-BRL). Real-time reverse transcription (RT)-PCR was performed with an ABI 7700 sequence detector system (Applied Biosystems) as described previously (58). Briefly, 50 ng of cDNA was used in duplicate per PCR run with specific primer sets for human PIN1 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH). All data were normalized to GAPDH as an internal control according to the manufacturer's instructions.
Cyclin D1-associated kinase assay.
Cells were lysed with NP-40 lysis buffer (10 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.5% NP-40, 50 mM NaF, 1 mM Na3VO4, 0.5 μg of leupeptin per ml, 1.0 μg of pepstatin per ml, 0.2 mM phenylmethylsulfonyl fluoride). Cell lysates were immunoprecipitated with protein A-agarose beads precoated with the cyclin D1 antibody DCS-11 (NeoMarkers, Fremont, Calif.), followed by the in vitro kinase assay as described previously (27).
Chromatin immunoprecipitation.
The chromatin immunoprecipitation method was as described previously (57). Briefly, 5 × 106 cells were fixed by addition of formaldehyde to the tissue culture medium (final concentration, 1%). Isolated chromatin was sonicated to an average length of 0.5 to 1 kb and treated with 1 μg of mouse anti-E2F-1 antibody (Transduction Laboratories) or control mouse immunoglobulin G (IgG) for 16 h at 4°C. The complexes were immunoprecipitated with 30 μl of protein A beads and washed with immunoprecipitation buffer (100 mM Tris-HCl [pH 9.0], 500 mM LiCl2, 1% NP-40, 1% deoxycholate). After elution and reversal of cross-links, DNA was isolated and analyzed by PCR. PCR products were visualized on a 2% agarose gel with CyberGreen.
Cell transformation assays.
Transformation assays were performed as previously described (8, 16). MCF-10/Neu/Ras cells (105 per 60-mm-diameter dish) were transfected with 1 μg of pIRES-puro/GFP, pIRES-puro/GFP-Pin1, or pIRES-puro-GFP/dnPin1 by FuGENE (Roche Diagnostics). After 24 h, transfected cells were selected with puromycin (1.3 μg/ml) for 36 to 48 h. Cells were then trypsinized and passed into 100-mm-diameter dishes. The medium was changed twice weekly for 3 weeks. For colony counting, cells were washed twice with phosphate-buffered saline (PBS), fixed with 10% acetic acid for 10 min, and stained with 0.4% crystal violet in 10% ethanol for 10 min. The dishes were rinsed, inverted, and dried at room temperature. Soft agar assays were performed in 6-cm plates with a 3-ml basal layer of 0.5% agar in 10% fetal bovine serum. A total of 5,000 to 50,000 cells in 0.3% top agar were plated in each plate in triplicate as described previously (8, 16). After 2 to 3 weeks, positive colonies (0.2-mm diameter) were counted, and the transformation efficiency was determined.
Three-dimensional Matrigel assay.
Three-dimensional Matrigel assays were performed as described before (37, 42). Cells were resuspended in assay medium (DMEM-F-12 supplemented with 2% horse serum, 10 μg of insulin per ml, 1 ng of cholera toxin per ml, 100 μg of hydrocortisone per ml, and 10 ng of human epidermal growth factor per ml) at a concentration of 8 × 104 cells/ml. Eight chambered RS glass slides (Nalgene) were coated with 35 μl of Matrigel per well and left to solidify for 20 min. Then 200 μl of cell suspension was mixed 1:1 with assay medium containing 4% Matrigel and plated on each chamber. Assay medium was replaced every 4 days. After 15 days, cells were fixed with 10% methanol-acetone and stained with anti-E-cadherin antibody (Transduction Laboratories) and TOPRO-3 (Molecular Probes), followed by confocal microscopy.
RESULTS
The PIN1 promoter is regulated by transcription factor E2F.
Although Pin1 has been shown to be overexpessed in many tumors such as breast carcinoma (45, 59), the molecular mechanism of this overexpression remains unknown. We therefore decided to examine the transcriptional regulation of PIN1 expression. As a first step to identifying the PIN1 promoter sequence, we screened a human genomic DNA library and isolated three positive clones (Fig. 1A). A 2.3-kb fragment upstream of exon 1 of the PIN1 gene was subcloned for further analysis. This promoter sequence has neither TATA nor CAAT boxes but has two putative GC boxes and three putative E2F binding sites, named sites A, B, and C (Fig. 1A).
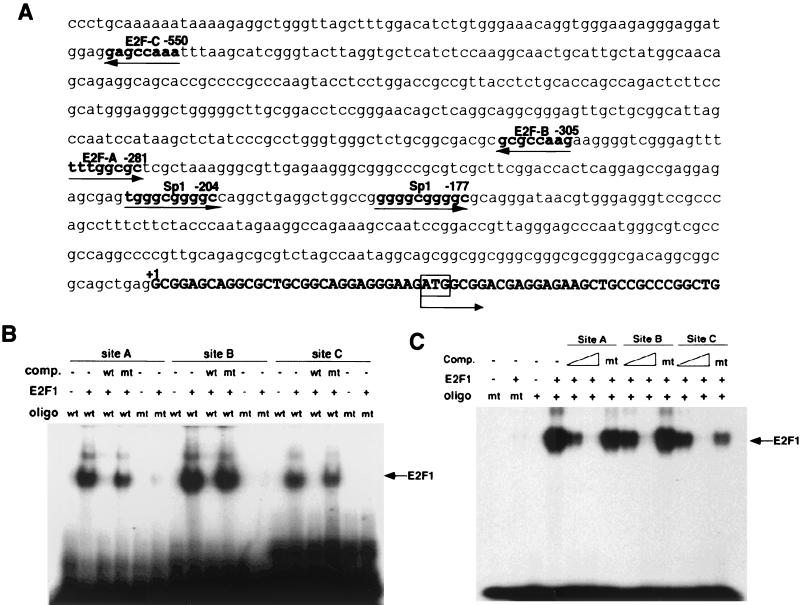
FIG. 1.
E2F binds the PIN1 promoter. (A) Human PIN1 promoter sequence. The nucleotide sequence of the human PIN1 gene that includes the 5′-flanking region and first exon is listed. Putative binding sites for transcription factors are underlined. The ATG translation initiation codon is in the first exon typed in boldface. (B) Electrophoretic mobility shift assays were performed with recombinant E2F1 protein and end-labeled double-stranded oligonucleotides (oligo) corresponding to the PIN1 promoter sequence containing either wild-type (wt) or mutant (mt) E2F binding sites. A consensus E2F site from the adenovirus E2 promoter was used as a competitor (comp.) in a 100× molar excess of labeled probe. (C) Competitive activity of PIN1 promoter sequences for E2F binding. Labeled oligonucleotides corresponding to E2F binding sites from the adenovirus E2 promoter were incubated with recombinant E2F1 protein in the presence or absence of unlabeled PIN1 promoter sequences. Three different oligonucleotides corresponding to E2F binding sites in the PIN1 promoter (sites A to C) were used as competitors. Wild-type oligonucleotides were mixed at a 10- or 100-fold molar excess and mutants were mixed at a 100-fold molar excess of labeled probe.
The fact that the E2F/Rb pathway is deregulated in many cancers suggested a possible role for E2F in overexpression of Pin1 in cancer cells. To examine whether E2F binds the PIN1 promoter, we first synthesized double-stranded oligonucleotides corresponding to each putative E2F site and conducted electrophoretic mobility shift assays with recombinant E2F1 protein. Recombinant E2F1 bound all three E2F probes, and the binding was completely abolished by point mutations introduced into each putative E2F site (Fig. 1B). Furthermore, a competition assay with nonlabeled oligonucleotides revealed that the wild-type but not the mutant oligonucleotides competed efficiently for E2F1 binding to the Pin1 E2F sites (Fig. 1B). Moreover, all three putative E2F sequences also efficiently competed with the adenovirus E2 promoter sequence, a well-characterized E2F site (38), for E2F1 binding (Fig. 1C). However, no competition was detected with any of three mutant E2F-binding sequences (Fig. 1C). These results indicate that all three putative E2F-binding sequences have the ability to bind E2F1 in vitro.
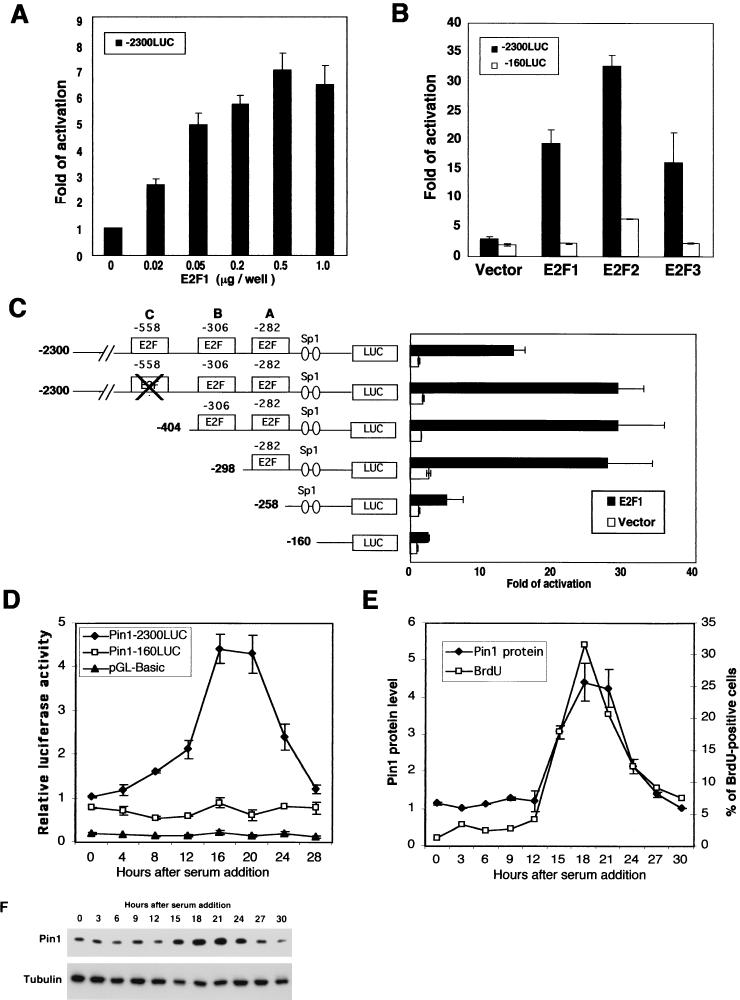
To examine whether E2F affects PIN1 promoter activity and whether any of the putative E2F sites are functional in vivo, we inserted the 2.3-kb 5′-flanking region of the PIN1 gene into a basic luciferase expression vector (pGL3-Basic), resulting in −2300LUC, and performed gene reporter assays. Indeed, E2F1 effectively activated the PIN1 promoter in a dose-dependent manner (Fig. 2A). Furthermore, two other E2F proteins, E2F2 and E2F3, also potently activated the PIN1 promoter (Fig. 2B). These results indicate that E2F proteins can activate the PIN1 promoter in cells.
FIG. 2.
Activation of the PIN1 promoter by E2F. (A) E2F1 activates PIN1 promoter activity in a dose-dependent manner. MCF-7 cells were transfected with the PIN1 promoter-luciferase construct (−2300LUC) and E2F-1 expression vector. Cells were harvested at 36 h after transfection and subjected to a gene reporter assay. (B) E2F family proteins enhance PIN1 promoter activity. Cells were cotransfected with vectors expressing E2F1, E2F2, or E2F3 together with the −2300LUC or −160LUC reporter construct. (C) Mapping of the PIN1 promoter region responsible for transcriptional activation by E2F. A series of 5′ deletion and site-directed mutants were transfected into HeLa cells together with the E2F1 expression vector or a control vector. (D) Cell growth-dependent regulation of PIN1 gene expression in normal fibroblasts. MEFs were transfectedwith the indicated luciferase reporter constructs and induced to enter quiescence by serum starvation (0.05% serum) for 48 h. The medium was then supplemented with serum (15%), allowing cells to reenter the cell cycle as a synchronous population. Cells were harvested at various time points and subjected to gene reporter assays. (E and F) MEFs were synchronized by serum starvation as for panel D. Prior to harvesting, cells were treated with BrdU for 30 min. Cells were collected at indicated time points and subjected to immunoblotting analysis with anti-Pin1 antibody or flow cytometory analysis with anti-BrdU antibody. Band intensities in Pin1 protein levels (F) were quantified by using NIH-Image and graphed with the results from the BrdU study (E).
To determine the importance of three putative E2F-binding sites in the PIN1 promoter, several 5′ deletional and site-directed mutants were generated (Fig. 1E). Compared to the wild-type PIN1 promoter (−2300bp), the mutation or deletion of the two distant E2F-binding sites (sites C and B, located at −557 to −550 and −312 to −305, respectively) did not have much effect on PIN1 promoter activity in response to E2F1 (Fig. 2C). However, deletion of the proximal E2F-binding site (site A, located at −288 to −281) strongly diminished the induction of the PIN1 promoter by E2F1 (Fig. 2C). Similar results were also observed in the induction of the PIN1 promoter by either E2F2 or E2F3 (data not shown). These results indicate that the proximal E2F-binding site is the most important regulatory site for the PIN1 promoter by E2F.
In addition, the PIN1 promoter has two GC boxes, which are potential recognition sites for the transcription factor Sp1 (Fig. 1A). Since several reports have shown a possible functional interaction of Sp1 with E2F (25), we deleted these two Sp1-binding sites (−160LUC). Deletion of the Sp1 sites slightly reduced PIN1 promoter activity, suggesting that they may also contribute to full induction of PIN1 by E2F (Fig. 1E). These gene reporter assays demonstrate that E2F1 can regulate the promoter activity of the PIN1 gene.
To determine whether other E2F family members affect PIN1 promoter activity, expression vectors encoding E2F1 to -3 were cotransfected with PIN1 promoter reporter constructs. Figure 2B shows that E2F family proteins, especially E2F2, potentiated PIN1 promoter activity through E2F-binding sites. However, the −160-base construct, in which the three E2F and Sp1 binding sites were deleted (−160LUC), was not responsive to the ectopic expression of E2F proteins. These results indicate that in addition to E2F1, E2F2 and E2F3 can also activate the PIN1 promoter.
Many E2F downstream target genes are related to cell cycle progression and DNA synthesis and are regulated in a cell growth-dependent manner in normal cells, especially when E2F-binding sites are proximal to the transcription initiation site (14, 39-41). To examine whether PIN1 expression is dependent on cell growth, we measured PIN1 promoter activity and Pin1 protein levels in normal mouse embryo fibroblasts (MEFs) at various time points following cell cycle reentry. MEFs were synchronized at the G0 phase by serum starvation and then released to enter the cell cycle by the addition of serum. Although the activity of Pin1−160LUC and control vectors did not respond to serum addition, PIN1 promoter activity was upregulated 16 to 20 h after serum addition (Fig. 2D). This correlated with an increase in Pin1 protein levels in the same cells within the same time frame following serum addition (Fig. 2E and F). Moreover, increased PIN1 promoter activity and Pin1 protein levels correlated well with DNA synthesis, as assayed by bromodeoxyuridine (BrdU) incorporation (Fig. 2F). These results indicate that PIN1 gene expression is regulated in a cell cycle-dependent manner in normal cells, further supporting the role of E2F in regulating PIN1 expression in cells.
Binding of E2F to the PIN1 promoter in vivo correlates with PIN1 expression in breast cell lines.
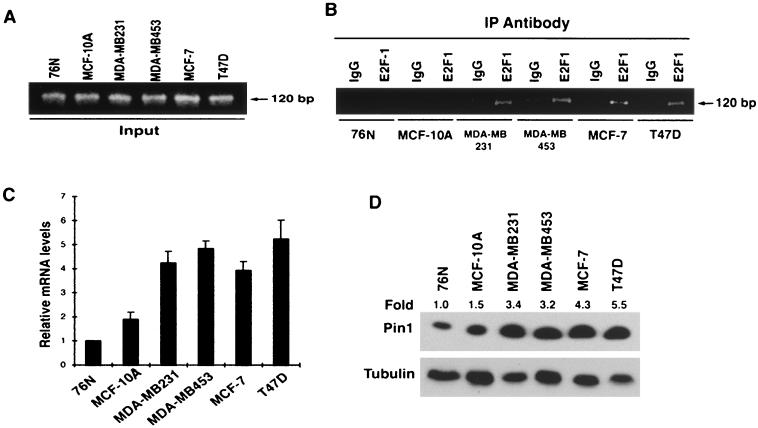
We have previously shown that in cancer cells, Pin1 protein levels are constant throughout the cell cycle and remain at higher levels than those in normal or nontransformed cell lines. The above results suggested that constitutive deregulation of the Rb/E2F pathway may contribute to Pin1 overexpression in cancer cells. To address this possibility, we first examined whether E2F binds the PIN1 promoter sequence in several nontransformed and transformed breast epithelial cell lines by chromatin immunoprecipitation analysis with an anti-E2F1 antibody. Although each input sample had a similar amount of the PIN1 promoter sequence when tested by quantitative PCR (Fig. 3A), there were dramatic difference in the amounts of PIN1 promoter sequence that were immunoprecipitated by anti-E2F antibodies, as determined by quantitative PCR with the same primer set (Fig. 3B). Compared with normal breast epithelial 76N cells and immortalized but nontransformed MCF-10A cells, much more PIN1 promoter sequences were coimmunoprecipitated with E2F1 in several breast cancer cell lines (Fig. 3B). These results not only confirm that E2F indeed binds the PIN1 promoter sequence in the cell but also indicate that the amounts of E2F protein bound on the promoter vary among different breast epithelial cell lines.
FIG. 3.
E2F binding to the PIN1 promoter sequence in vivo correlates with Pin1 expression level in breast cancer cell lines. (A and B) Levels of E2F binding to the PIN1 promoter in different breast cell lines. Cross-linked chromatin from exponentially growing breast cancer cell lines was incubated with either antibodies against E2F1 or control IgG. Immunoprecipitates from each sample were analyzed by PCR with primers specific for the PIN1 promoter sequence (B). As an input control, total input chromatin was analyzed by PCR with the same primer set (A). (C and D) Levels of PIN1 mRNA and protein in different breast cell lines. mRNAs were isolated from the cell types indicated, and PIN1 mRNA was quantified by real-time RT-PCR analysis and normalized to GAPDH mRNA (C). PIN1 levels were determined by subjecting cell lysates to immunoblotting analysis with a monoclonal anti-Pin1 antibody (D), Numbers above the gel image indicate the induction (fold) of Pin1 protein level normalized to α-tubulin.
We next determined whether E2F binding to the PIN1 promoter is correlated with PIN1 expression levels by measuring PIN1 mRNA and protein levels in these cell lines by real-time PCR and immunoblotting analyses, respectively. Both PIN1 mRNA and protein levels were much higher in all transformed cell lines compared with those in normal primary 76N cells and immortalized but nontransformed MCF-10A cells (Fig. 3C and D). Furthermore, there was a good correlation between the amounts of E2F bound on the PIN1 promoter and levels of PIN1 mRNA and protein in all breast epithelial cells examined (Fig. 3B to D). Taken together, the above results demonstrate that the transcription factor E2F plays an important role in the regulation of PIN1 expression in breast cell lines.
Ras and Neu enhance PIN1 expression via E2F.
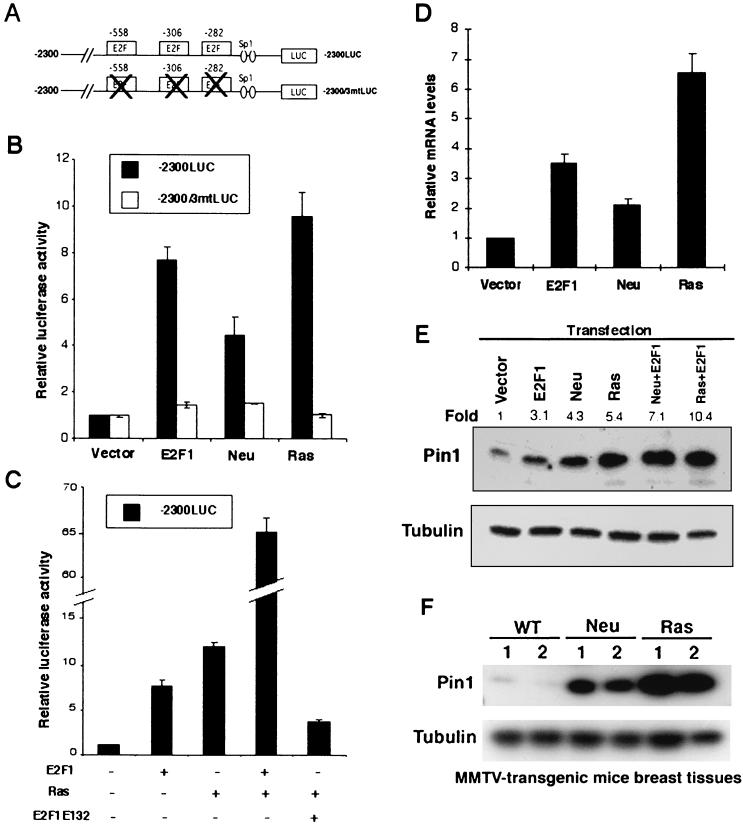
Oncogenic Neu and Ras signaling has been shown to enhance E2F transcriptional activity in breast cancer cells (12, 27, 39). Given that the PIN1 promoter is activated by E2F and significantly elevated in breast cancer cells, this Neu and Ras signaling might enhance PIN1 promoter activity via E2F activation. To examine this possibility, we first examined whether Ha-Ras and c-Neu activates the PIN1 promoter. A wild-type PIN1 promoter construct and a mutant construct containing point mutations in the three putative E2F-binding sites (Fig. 4A) were used in the assay. Like E2F, Ha-Ras and c-Neu transactivated the PIN1 promoter by ≈10-fold and ≈5-fold, respectively (Fig. 4B). However, point mutations at the three E2F-binding sites completely abolished the ability of Ha-Ras or Neu to transactivate the PIN1 promoter (Fig. 4B). These results suggest that Ras and Neu transactivate the PIN1 promoter through its E2F sites.
FIG. 4.
Ras and Neu stimulate the PIN1 promoter through E2F activation. (A) Schematic representation of wild-type and mutant PIN1 promoter reporter constructs. (B) MCF-7 cells were cotransfected with a reporter construct (0.1 μg) and E2F-1, Ha-Ras, or Neu (0.5 μg), followed by gene reporter assays. (C) Dominant-negative E2F1 inhibits activation of the PIN1 promoter by Ras. Gene reporter assays were performed in MCF-7 cells as shown in panel B. Wild-type E2F-1 or its dominant-negative mutant E2F1E132 was cotransfected with the Ras and −2300LUC reporter constructs. (D) Neu and Ras upregulate PIN1 mRNA levels in MCF-10A cells. MCF-10A cells were transiently transfected with plasmids encoding E2F1, Ha-Ras, or Neu. For each transfection, a plasmid encoding a puromycin resistance gene (pIRES-puro) was cotransfected as a selection marker. Puromycin (1.3 μg/ml) was added to the medium 24 h after transfection. At 36 h following addition of puromycin, puromycin-resistant cells were reseeded and cultured for an additional 24 h. Total RNA was collected and subjected to real-time RT-PCR as described in Materials and Methods. (E) MCF-7 cells were transfected with the indicated expression vectors for 48 h, and cell lysates were subjected to immunoblotting analysis with anti-Pin1 and antitubulin antibodies. Numbers above the gel image indicate the fold induction of the Pin1 protein level normalized to α-tubulin. (F) PIN1 is overexpressed in breast tissues from MMTV-c-Neu and MMTV-Ha-Ras mice. Mammary tissues from two wild-type (WT), two MMTV-Neu (Neu), and two MMTV-Ras (Ras) mice were lysed and subjected to immunoblotting analysis with anti-Pin1 and antitubulin antibodies.
To further confirm this result, we used an E2F1 mutant (E2F1E132) which has been well shown to inhibit the function of the endogenous E2F proteins in a dominant-negative fashion (24). Cotransfection of Ras and wild-type E2F highly increased PIN1 promoter activity (Fig. 4C). However, cotransfection with the dominant-negative E2F mutant significantly decreased the ability of Ras to transactivate the PIN1 promoter (Fig. 4C). Similar phenomena were also observed in Neu-induced PIN1 promoter activation (data not shown). These gene reporter assays suggest that Neu/Ras signaling transactivates the PIN1 promoter via E2F.
To ensure that Ras and Neu can increase PIN1 expression in cells, we first examined the effects of exogenous Ha-Ras or Neu expression on PIN1 mRNA and protein levels in mammary epithelial cells. Indeed, both Ha-Ras and Neu significantly increased PIN1 mRNA levels in MCF-10A cells, as determined by quantitative RT-PCR (Fig. 4D), as well as Pin1 protein levels, as determined by immunoblotting analysis in MCF7 cells (Fig. 4E). Furthermore, cotransfection with E2F further increased Pin1 protein levels induced by Ha-Ras or Neu (Fig. 4E). Given that Ha-Ras and c-Neu can increase Pin1 expression in cells, we also examined whether transgenic overexpression of mouse mammary tumor virus (MMTV)-c-Neu or MMTV-Ha-Ras elevates Pin1 expression in mouse breast tissues. Interestingly, breast tissues obtained from both MMTV- c-Neu and MMTV-Ha-Ras transgenic mice contained much higher Pin1 protein levels than control breast tissues (Fig. 4F). Taken together, these results indicate that Ras and Neu enhance PIN1 expression both in vitro and in vivo.
Overexpression of PIN1 confers transformed properties on mammary epithelial cells.
We have previously demonstrated that PIN1 expression is highly elevated in human breast cancer tissues and plays a pivotal role in the regulation of cyclin D1 function (29, 44, 58). Interestingly, cyclin D1 is also an essential mediator in the development of breast cancer induced by oncogenic Neu and Ras (26, 60). Given that PIN1 is a downstream target of oncogenic Neu/Ras signaling, a critical question is whether overexpression of PIN1 has any effect on the cell transformation of mammary epithelial cells.
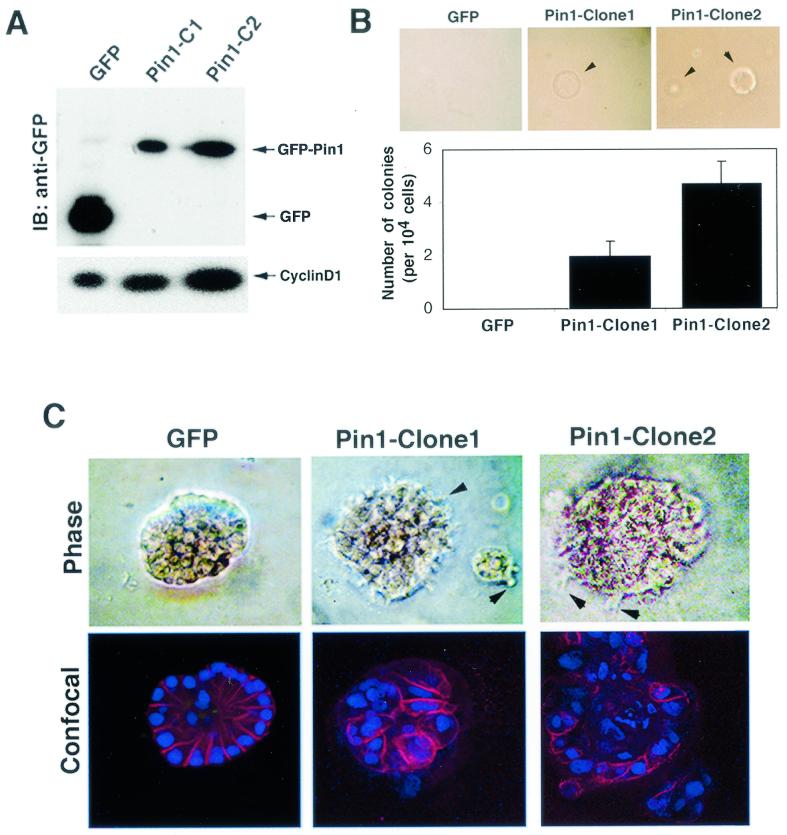
To address this question, we stably transfected GFP-Pin1 and control GFP into MCF-10A cells, a spontaneously immortalized but nontransformed mammary epithelial cell line that has been widely used for cell transformation studies (7, 51). Multiple stable cell lines that had similar properties were obtained, with one GFP-expressing and two GFP-Pin1-expressing cell lines (clones 1 and 2) being further characterized. The two GFP-Pin1-expressing cell lines moderately overexpressed Pin1, about three- and sixfold higher than endogenous levels (Fig. 5A and data not shown). Consistent with our previous studies (29, 44, 58), cyclin D1 levels were elevated in these GFP-Pin1 stable clones compared with control GFP cells, with cyclin D1 levels being correlated with exogenous Pin1 expression levels (Fig. 5A).
FIG. 5.
PIN1 overexpression confers a transformed phenotype on MCF-10A cells. (A) Establishment of MCF-10A cells stably expressing GFP or GFP-Pin1. Immunoblotting (IB) analysis was performed with anti-GFP and anti-cyclin D1 antibodies. (B) To measure anchorage-independent cell growth and survival, GFP- or GFP-Pin1-transfected MCF-10A cells were suspended in 0.3% soft agar for 14 days. (C) Cell lines stably expressing GFP and GFP-Pin1 were plated on Matrigel for 15 days. Phase images of an acinus at higher magnification are shown in the upper panels. The acini were stained with anti-E-cadherin antibodies and the DNA dye TOPRO-3, and confocal images though the middle of an acinus are shown in the lower panels. Arrows indicate cell surface spikes protruding into the Matrigel.
Although there was no detectable difference in cell morphology and growth rate on plastic plates between GFP-Pin1 and control GFP cell lines (data not shown), overexpression of GFP-Pin1 but not GFP conferred anchorage-independent cell growth in soft agar (Fig. 5B). However, the size and frequency of colonies were much less than those of Neu/Ras-transformed MCF-10A cells (Fig. 5B versus Fig. 6F). Moreover, like parental MCF-10A cells (7, 51), GFP-Pin1 stable cell lines were unable to survive in DMEM supplemented with10% fetal bovine serum (data not shown), while Neu/Ras-transformed MCF-10A cells grew normally in this medium (Fig. 6C). These data suggest that although overexpression of Pin1 appeared to be insufficient to fully transform MCF-10A cells, it might trigger some early events of cell transformation.
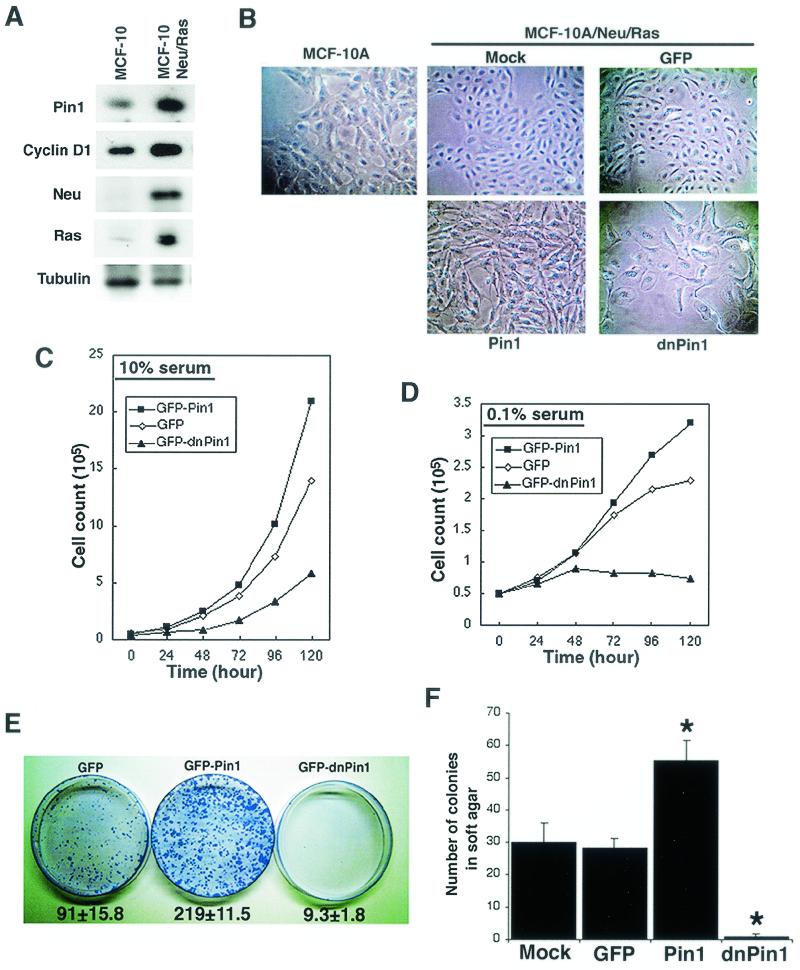
FIG. 6.
PIN1 is essential for Neu/Ras-induced cell transformation. (A) Establishment of stable MCF-10A cell lines expressing both Neu and Ras. Cells were cotransfected with Neu and Ras expression vectors and selected with G418. A selected clone was checked for expression of Ras, Neu, PIN1, and cyclin D1 by immunoblotting analysis. (B) Morphological changes in MCF-10A cells stably expressing Neu and Ras (MCF-10/Neu/Ras) and additional PIN1 or dn-PIN1. Cells were seeded in 60-mm dishes and photographed with a phase-contrast microscope before reaching confluence. (C and D) Manipulation of Pin1 levels alters proliferation in MCF-10/Neu/Ras cells. Transfected cells were selected with puromycin for 48 h and reseeded in 35-mm dishes. Cells were grown in high-serum (10%) (C) or low-serum (0.1%) (D) medium and trypsinized at various time points. Viable cells were counted by the trypan blue dye exclusion method. (E) PIN1 is necessary for Neu/Ras-induced focus formation. The same number of cells (104) transfected with either the GFP, GFP-Pin1, or GFP-dnPin1 expression vector were seeded in 10-cm plastic dishes after selection with puromycin. After 14 days, cells were fixed and stained with crystal violet. Numbers below plates indicate colony numbers (mean ± SD) in three independent experiments. (F) Cells were plated in 0.3% soft agar and cultured for 2 weeks. After 14 days, colony formation was scored microscopically. MCF-10/Neu/Ras cells expressing GFP-Pin1 demonstrated significant increases in anchorage-independent growth, whereas dn-Pin1 overexpession significantly blocked the growth of MCF-10/Neu/Ras cells. ∗, P < 0.01, t test.
To further investigate this possibility, we performed a three-dimensional cell differentiation assay with exogenous basement membrane matrix (Matrigel). This method has been well established to assess the transformed phenotype of mammary epithelial cells, especially at early stages of tumorigenesis (37, 42). We found that GFP-expressing cells formed acini with basally polarized nuclear organization, intact cell-cell junctions, and visible lumina inside, as indicated by immunostaining with antibodies against the cell-cell junction marker E-cadherin and with the DNA dye TOPRO-3, followed by confocal microscopy (Fig. 5C). These structures are known as well-differentiated acini that are usually observed in parental MCF-10A cells (37, 42), indicating that expression of GFP had no effect. However, expression of GFP-Pin1 had a dramatic effect on the morphology and organization of acinar formation. Colonies formed by GFP-Pin1-expressing cells exhibited disorders in nuclear polarity and cell arrangement without a lumen inside, disruption of basement membrane, and impairment in cell-cell junction (Fig. 5B, lower). Furthermore, GFP-Pin1 but not GFP-expressing cells had cell surface spikes protruding into the Matrigel (Fig. 5C, top). Since a lack of acinar organization is a specific event involved in progression towards malignancy (37), these results suggest that Pin1 overexpression can induce events associated with early stages of mammary tumorigenesis. However, additional events might be needed to lead to the full transformation of mammary epithelial cells.
Overexpression of PIN1 enhances whereas inhibition of PIN1 suppresses transformed phenotypes of mammary epithelial cells induced by Neu and Ras.
It is also well established that oncogenic Neu/Ras signaling induces cell transformation of mammary epithelial cells via upregulation of cyclin D1 (26, 60). Given that PIN1 is a downstream target of Neu/Ras signaling and regulates cyclin D1 function, we hypothesized that PIN1 mediates Neu/Ras signaling thorough the activation of cyclin D1 during breast cancer formation.
To test this hypothesis, we first examined whether manipulating cellular PIN1 function affects the transformed phenotype of mammary epithelial cells induced by Neu and Ras. To address this question, we needed to establish MCF-10A cells stably expressing c-Neu and Ha-Ras together (MCF-10/Neu/Ras), because overexpression of both Neu and Ha-Ras has been shown previously to induce a transformed phenotype mimicking the malignancy of mammary carcinomas (16). MCF-10/Neu/Ras cells exhibited elevated levels of both cyclin D1 and Pin1 compared with parental MCF-10A cells (Fig. 6A). These results are consistent with the findings that Neu/Ras signaling increases expression of cyclin D1, as shown previously (26, 60), and of Pin1, as shown above.
Morphologically, MCF-10/Neu/Ras cells demonstrated a higher nuclear/cytoplasmic ratio and multiple nucleoli, which is consistent with their higher proliferation rate, than the parental MCF-10A cells (Fig. 6B). These cells were able to grow in DMEM supplemented with a high concentration of fetal calf serum (10%) or even with low serum (0.1%) (Fig. 6C and D), as well as in soft agar (Fig. 6F). These results indicate that MCF-10/Neu/Ras cells display various transformed phenotypes, as reported previously (16).
With this cell line, we investigated whether upregulation or downregulation of Pin1 affects the transformed phenotypes. To inhibit cellular Pin1 function, we used a Pin1 WW domain construct, which contains an Ala substitution at Ser16. This construct has been shown to inhibit endogenous Pin1 interaction with target substrates, thereby functioning as a dominant-negative PIN1 (dn-PIN1) (33). Given that excessive overexpression of PIN1 or dn-PIN1 blocks cell cycle progression (3, 33), we used rather low concentrations of expression constructs after a series of pretests. MCF-10/Neu/Ras cells were transfected with GFP-Pin1, GFP-dnPin1, or control GFP, followed by selection with puromycin. The transfection efficiencies and expression levels of three different constructs were comparable in each set of transfectants, as confirmed by scoring GFP-fluorescent cells under a microscope or by immunoblotting analysis with anti-GFP antibodies (Fig. 7A and data not shown). Compared with parental and GFP-expressing cells, GFP-Pin1-expressing cells exhibited a higher nuclear/cytoplasmic ratio, with disorganized cell arrangements (Fig. 6B). In contrast, GFP-dnPin1-expressing cells exhibited large and vacuolar morphology with higher density of cytoplasmic speckles, similar to those of parental MCF-10A cells (Fig. 6B). These morphological changes suggest that expression of GFP-Pin1 and GFP-dnPin1 might affect the transformed phenotypes of these cells.
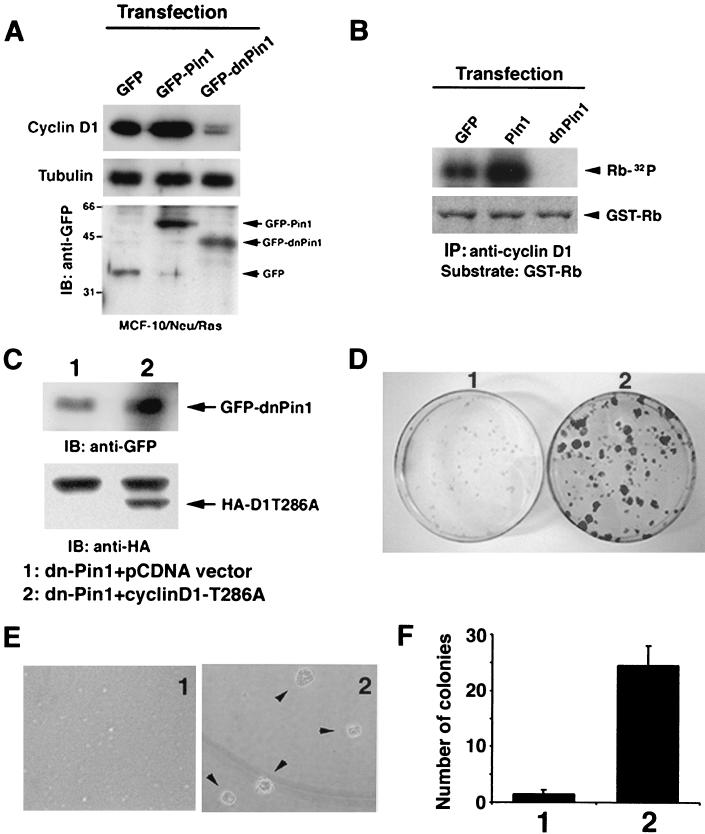
FIG. 7.
Pin1 inhibition is complemented by overexpression of a constitutively active cyclin D1. (A) Pin1 is essential to maintain cyclin D1 level and activity in Neu/Ras-transformed MCF-10 cells. MCF-10/Neu/Ras cells transfected with either GFP, GFP-Pin1, or GFP-dnPin1 were lysed and immunoblotted with anti-cyclin D1, -tubulin, and -GFP antibodies. (B) The same cell lysates as in panel A were immunoprecipitated with anti-cyclin D1 antibodies, followed by the in vitro kinase assay with GST-pRB as a substrate. Phosphorylation of the GST-pRB substrate is shown in the upper panel. The lower panel shows input GST-pRB stained with Coomassie blue. (C) MCF-10/Neu/Ras cells were transfected with either dnPin1 and pCDNA vector or dnPin1 and the cyclin D1T286A mutant (1:10 ratio) and selected with puromycin for 48 h. Cells were subjected to immunoblotting analysis with anti-HA and anti-GFP antibodies. (D) Cells were transfected as described for C and seeded on plastic plates for 3 weeks. Cells were fixed and stained with crystal violet. (E and F) Cells were transfected as described for panel C and cultured in 0.3% soft agar for 3 weeks. The number of colonies formed was scored. Representative phase pictures are shown in panel E. Colony numbers are the mean ± SD of three independent experiments (F).
To examine this possibility, we next examined the cell proliferation rate in high-serum (10%) and low-serum (0.1%) medium. Compared with GFP control cells, GFP-Pin1-expressing cells grew faster, whereas dn-PIN1-expressing cells grew much more slowly in 10% serum medium (Fig. 6C). Furthermore, GFP-expressing control cells grew even in low serum (Fig. 6D), consistent with the fact that MCF-10/Neu/Ras transformed cells have lost the cell cycle checkpoint induced by a low concentration of growth factors, as shown previously (8, 16). More interestingly, GFP-Pin1-expressing cells continued to grow linearly even when the growth of GFP-transfected cells was retarded in low-serum medium after 48 h (Fig. 6D). In contrast, dn-PIN1-expressing cells could not grow under low-serum conditions (Fig. 6D). These results suggest that overexpression of Pin1 increases cell proliferation and transformed phenotypes of MCF-10/Neu/Ras cells, whereas inhibition of Pin1 reverses these phenotypes.
To further support this observation, we investigated the long-term cell proliferation and transformation properties of these cells by performing colony formation assays on plastic plates and in soft agar. Consistent with the short-term cell growth study, expression of GFP-Pin1 increased colony formation, doubling the number of colonies compared with GFP control cells, both on plastic plates and in soft agar (Fig. 6E and F). Furthermore, individual colonies were much larger (Fig. 6E). In contrast, dn-PIN1 overexpression dramatically inhibited colony formation; these cells produced very tiny colonies on plastic plates, and colony formation in soft agar was almost completely inhibited (Fig. 6E and F). Similar inhibitory effects were also seen with inhibition of Pin1 via expression of an antisense PIN1 construct (data not shown), which has been shown to deplete endogenous Pin1 proteins (30, 44, 48). These results indicate that overexpression of Pin1 enhances Neu/Ras-induced cell proliferation and transformation, whereas the inhibition of Pin1 reverses the cell proliferation and transformed properties induced by Neu and Ras.
PIN1 affects Neu/Ras-induced cell transformation of mammary epithelial cells via cyclin D1.
Cyclin D1 is an essential downstream target of Neu/Ras-induced mammary tumorigenesis (26, 60). Furthermore, PIN1 positively regulates cyclin D1 function via transcriptional activation as well as posttranslational stabilization (29, 44, 58). These results suggest that PIN1 might affect Neu/Ras-induced cell transformation via cyclin D1. To examine this possibility, we assayed levels of cyclin D1 and its associated kinase activity. Consistent with phenotypic changes as described above, levels of cyclin D1 and its associated cyclin-dependent kinase activity were enhanced in cells expressing GFP-Pin1 compared to GFP-expressing cells (Fig. 7A and B). In contrast, both the cyclin D1 level and its kinase activity were substantially lowered by the overexpression of dn-PIN1 (Fig. 7A and B). These results indicate that overexpression of Pin1 enhances but inhibition of Pin1 strongly inhibits cyclin D1 expression and function in Neu/Ras-transformed MCF-10A cells, consistent with the notion that Pin1 affects Neu/Ras-induced cell transformation via cyclin D1.
If this is the case, overexpression of a constitutively active cyclin D1 mutant (cyclin D1T286A), which cannot bind Pin1 and is refractory to Pin1 inhibition (29), should rescue the transformed phenotypes that are inhibited by dn-PIN1. This experiment is also important in addressing whether suppression of transformed phenotypes by Pin1 inhibition is specifically due to inhibition of cyclin D1 or simply due to induction of cell apoptosis. We cotransfected MCF-10/Neu/Ras cells with GFP-dn-PIN1 and hemagglutinin (HA)-cyclin D1T286A or control vector pCDNA at a 1:10 ratio and selected transfected cells with puromycin. Immunostaining with anti-HA antibody confirmed that almost all HA-cyclin D1T286A-positive cells expressed GFP-dnPin1 (data not shown), which was also confirmed by immunoblotting analysis of GFP-dnPin1 with anti-GFP antibodies and of HA-cyclin D1T286A with anti-HA antibodies (Fig. 7C).
MCF-10/Neu/Ras cells cotransfected with dn-PIN1 and pCDNA vector failed to form foci on plastic plates and colonies in soft agar (Fig. 7D to F), confirming that inhibition of Pin1 suppresses the transformed phenotypes induced by Neu and Ras (Fig. 6). Importantly, MCF-10/Neu/Ras cells expressing both dn-PIN1 and cyclin D1T286A formed many foci on plastic plates (Fig. 7D). Moreover, these cells even displayed anchorage-independent cell growth to form colonies in soft agar to the same extent, like MCF-10/Neu/Ras cells (Fig. 7E and F versus Fig. 6F). These results show that overexpression of a constitutively active cyclin D1 mutant can reverse the ability of Pin1 inhibition to suppress the Neu- and Ras-induced transformed phenotypes, further indicating that Pin1 affects Neu/Ras-induced cell transformation via cyclin D1.
DISCUSSION
In this report, we have demonstrated that Pin1 expression is regulated by the transcription factor E2F, which is enhanced by oncogenic Neu or Ras. Furthermore, overexpression of Pin1 alone is sufficient to induce normal mammary epithelial cells to display several transformed properties that have been shown to be present in the early stages of tumorigenesis. Importantly, overexpression of Pin1 enhances the transformed phenotypes of mammary epithelial cells induced by Neu and Ras. In contrast, inhibition of Pin1 suppresses the Neu- and Ras-induced transformed phenotypes, which can be fully rescued by overexpression of a constitutively active cyclin D1 mutant that is refractory to Pin1 inhibition. This is the first demonstration that PIN1 is an E2F downstream target gene and that PIN1 plays an essential role in Neu/Ras-induced mammary tumorigenesis via cyclin D1.
Pin1 as an E2F downstream target gene.
The following results indicate that the transcription factor E2F plays a critical role in regulation of Pin1 expression (Fig. 8A). First, E2F family proteins activated the PIN1 promoter specifically through the E2F binding sites. Second, E2F bound the PIN1 promoter in vitro and in vivo. Third, the levels of E2F binding to the PIN1 promoter correlated with the levels of Pin1 expression in breast cancer cell lines. Fourth, PIN1 gene expression in normal cells was regulated in a cell cycle-dependent manner, as is the case for other E2F target genes (14, 39-41). Finally, overexpression of E2F enhanced PIN1 promoter activity and mRNA levels in breast cancer cells.
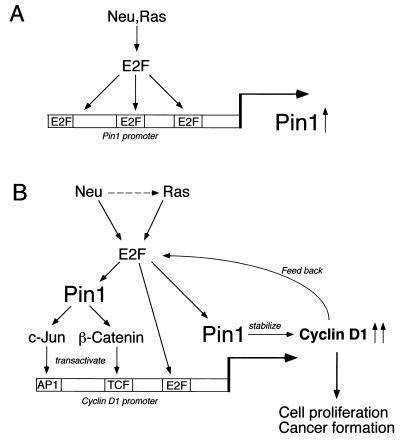
FIG. 8.
Schematic models for Pin1 transcriptional regulation and its role in regulation of cyclin D1 by Neu/Ras signaling. (A) Oncogenic Neu-Ras signaling transactivates the PIN1 promoter through E2F activity. (B) PIN1 is a downstream target of oncogenic Neu/Ras signaling and is essential for Neu/Ras-induced cyclin D1 activation and cell transformation. PIN1 upregulated by Neu/Ras signaling enhances β-catenin and c-Jun signaling to transactivate the cyclin D1 gene. Furthermore, Pin1 binds directly to cyclin D1 and stabilizes it via a posttranslational mechanism. It is possible that cyclin D1 also regulates Pin1 expression via E2F in a positive feedback loop.
Interestingly, E2F1 has been found to be a good prognostic or predictive marker of breast cancer because the E2F1 index significantly correlates with histological grade, stage, and metastatic status of breast tumors (61). Similarly, Pin1 expression is correlated with the histological grade of breast cancer (58). These results indicate that deregulation of E2F may play a key role in the upregulation of Pin1 in breast cancer. Since deregulation of the Rb/E2F pathway is also found in many other cancer types and contributes to the oncogenesis of a number of human cancers (18, 35, 39, 53, 56), deregulation of the Rb/E2F pathway may also contribute to Pin1 overexpression in other cancer cells. Further experiments are needed to examine the role of E2F in regulating Pin1 expression in other cancers.
Significance of Pin1 overexpression in cell transformation.
We report here for the first time that overexpression of Pin1 can play an important role in the transformation of mammary epithelial cells. Phosphorylation of proteins on Ser/Thr-Pro is a key regulatory mechanism in controlling cell proliferation and transformation (2, 18, 22, 63). The conformation and function of many phosphorylated proteins are regulated by the phosphorylation-specific prolyl isomerase Pin1 (31). Interestingly, Pin1 is highly overexpressed in many human cancer tissues, including breast cancer cells, but its significance in cell transformation is largely unknown (44, 58). We have now found that overexpression of Pin1 conferred an anchorage-independent cell growth phenotype on normal mammary epithelial cells, although the colony size and frequency of Pin1-overexpressing cells were smaller and lower than those of Neu/Ras-transformed cells. In addition, Pin1-expressing cell lines failed to grow in 10% fetal bovine serum, in contrast to Neu/Ras-transformed cells.
These data suggest that overexpression of Pin1 might trigger some early events during cell transformation. This was further supported by a three-dimensional Matrigel assay, which has been well established to assess the transformed phenotype of mammary epithelial cells, especially at early stages of tumorigenesis (37, 42). Indeed, expression of Pin1 had a dramatic effect on the morphology and organization of acinar formation. These cells exhibited the disorder in the nuclear polarity and cell arrangement without a lumen inside, disruption of the basement membrane, and impairment in cell-cell junction. Furthermore, Pin1-expressing cells had cell surface spikes that protruded into the Matrigel. These results indicate that overexpression of Pin1 might disrupt normal differentiation in mammary epithelial cells. Since the lack of acinar organization and the presence of cell surface spikes have been suggested to be specific events in progression towards malignancy (16, 37), these results suggest that Pin1 overexpression can induce events associated with the early stages of mammary tumorigenesis.
Essential role of Pin1 for the Neu/Ras-induced transformation of mammary epithelial cells.
Our exciting observation was that Pin1 plays an essential role in the transformation of mammary epithelial cells by Ras and Neu via activation of cyclin D1. Various studies have revealed that Neu or Ras signaling is deregulated in many breast cancers, although mutations and amplifications of these genes were rarely observed (3, 19, 50, 54). Furthermore, transgenic overexpression of MMTV-Ha-Ras or MMTV-c-Neu potently induces mammary tumors via cyclin D1. However, transgenic overexpression of MMTV-cyclin D1 has much weaker tumorigenicity (36, 49, 55). In addition, constitutive overexpression of cyclin D1 alone cannot transform MCF-10A cells, nor is it sufficient to prevent G1 arrest induced by EGF deprivation (7). These discrepancies could be explained by the findings that cyclin D1 is regulated not only by transcriptional activation but also by posttranslational stabilization.
Phosphorylation of cyclin D1 on the Thr286-Pro site by GSK-3β enhances its nuclear export and subsequent degradation (2, 9, 10). In fact, EGF deprivation results in a rapid degradation of cyclin D1 in MCF-10A cells constitutively overexpressing cyclin D1 (7). In contrast to wild-type cyclin D1, mutant cyclin D1T286A is stable and functions as a constitutively active mutant which can potently transform mammary epithelial cells (2). These results suggest that both transcriptional activation and posttranslational stabilization of cyclin D1 are critical for tumor development induced by Neu/Ras signaling. Interestingly, we have previously shown that by binding and isomerizing specific pSer/Thr-Pro motifs, Pin1 cooperates with Ras-JNK-c-Jun and the wnt-β-catenin-TCF pathways to enhance cyclin D1 expression (44, 58). Furthermore Pin1 can also enhance the stability and nuclear localization of cyclin D1 by directly binding and presumably isomerizing the phosphorylated Thr286-Pro motif (29). Therefore, Pin1 positively regulates the function of cyclin D1 both at the transcriptional level and via posttranslational stabilization, resulting in the transformation of mammary epithelial cells.
Our studies have demonstrated that Neu/Ras signaling can activate expression of Pin1 and that overexpression of Pin1 in Neu/Ras-expressing mammary epithelial cells enhances their transformed phenotypes. In contrast, inhibition of Pin1 by a dominant-negative mutant or an antisense construct dramatically reduced both cell proliferation and the transformed phenotypes of these cells. Importantly, this inhibitory effect by Pin1 inhibition was rescued by overexpression of a cyclin D1-T286A mutant, which is refractory to Pin1 inhibition (29). These results suggest a model in which Neu/Ras signaling can activate expression of Pin1, which in turn enhances the expression and stability of cyclin D1, eventually leading to cell proliferation and transformation (Fig. 8B).
It appears that upregulation of Pin1 does not precede cyclin D1 upregulation during cell cycle reentry of normal MEFs. However, deletion of the PIN1 gene results in a significant decrease in both cyclin D1 mRNA and protein levels in MEFs and also causes phenotypes in mice resembling those of cyclin D1 null mice (29), indicating an important role of PIN1 in regulating cyclin D1 function in normal conditions. Therefore, it is possible that cyclin D1 regulates PIN1 expression via E2F in a positive feedback loop (Fig. 8B). This might provide an explanation for why oncogenic Ras or Neu is more potent than cyclin D1. Therefore, PIN1 might be a key player in modulating upregulation of cyclin D1 by Neu and Ras oncogenic signaling.
In summary, our results provide the first evidence for a requirement for appropriate regulation of PIN1 gene expression in transformation of mammary epithelial cells induced by Neu and Ras. PIN1 is upregulated by Neu and Ras via E2F. Furthermore, overexpression of PIN1 not only can confer transformed properties on normal mammary epithelial cells but also can enhance the transformed phenotypes induced by Neu and Ras. Finally, inhibition of PIN1 suppresses Neu- and Ras-induced transformed phenotypes via cyclin D1. These results indicate that overexpression of PIN1 in human cancer cells would help promote tumor cell growth and also suggest that PIN1 inhibitors could be useful for anticancer therapies.
Acknowledgments
We are very grateful to J. Nevins, C. Sherr, P. Sicinski, and S. Thomas for constructive discussions and/or suggestions; to M. Yamamoto, J. Nevins, P. Sicinski, Q. Yu, K. Carraway, and K. Ohtani for various reagents; to S. Gil and A. Hada for technical instructions; and to X. Zhou and K. Perrem for their important contributions.
A.R., Y.-C.L., G.W., and M.N. are a Leukemia and Lymphoma Society Special Fellow, Engineering Research Council of Canada Fellow, DOD Breast Cancer Research Program Fellow, and National Sciences and Human Frontier Research Program Fellow, respectively. K.P.L. is a Pew Scholar and a Leukemia and Lymphoma Society Scholar. This study was supported by NIH grants R01GM56230 and GM58556 to K.P.L.
REFERENCES
- 1.Albanese, C., J. Johnson, G. Watanabe, N. Eklund, D. Vu, A. Arnold, and R. G. Pestell. 1995. Transforming p21ras mutants and c-Ets-2 activate the cyclin D1 promoter through distinguishable regions. J. Biol. Chem. 270:23589-23597. [DOI] [PubMed] [Google Scholar]
- 2.Alt, J. R., J. L. Cleveland, M. Hannink, and J. A. Diehl. 2000. Phosphorylation-dependent regulation of cyclin D1 nuclear export and cyclin D1-dependent cellular transformation. Genes Dev. 14:3102-3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andrechek, E. R., and W. J. Muller. 2000. Tyrosine kinase signalling in breast cancer: tyrosine kinase-mediated signal transduction in transgenic mouse models of human breast cancer. Breast Cancer Res. 2:211-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arber, N., Y. Doki, E. K. Han, A. Sgambato, P. Zhou, N. H. Kim, T. Delohery, M. G. Klein, P. R. Holt, and I. B. Weinstein. 1997. Antisense to cyclin D1 inhibits the growth and tumorigenicity of human colon cancer cells. Cancer Res. 57:1569-1574. [PubMed] [Google Scholar]
- 5.Bartkova, J., J. Lukas, H. Muller, D. Lutzhoft, M. Strauss, and J. Bartek. 1994. Cyclin D1 protein expression and function in human breast cancer. Int. J. Cancer 57:353-361. [DOI] [PubMed] [Google Scholar]
- 6.Blume-Jensen, P., and T. Hunter. 2001. Oncogenic kinase signalling. Nature 411:355-365. [DOI] [PubMed] [Google Scholar]
- 7.Chou, J. L., Z. Fan, T. DeBlasio, A. Koff, N. Rosen, and J. Mendelsohn. 1999. Constitutive overexpression of cyclin D1 in human breast epithelial cells does not prevent G1 arrest induced by deprivation of epidermal growth factor. Breast Cancer Res. Treat. 55:267-283. [DOI] [PubMed] [Google Scholar]
- 8.Ciardiello, F., M. Gottardis, F. Basolo, S. Pepe, N. Normanno, R. B. Dickson, A. R. Bianco, and D. S. Salomon. 1992. Additive effects of c-erbB-2, c-Ha-Ras, and transforming growth factor-alpha genes on in vitro transformation of human mammary epithelial cells. Mol. Carcinog. 6:43-52. [DOI] [PubMed] [Google Scholar]
- 9.Diehl, J. A., M. Cheng, M. F. Roussel, and C. J. Sherr. 1998. Glycogen synthase kinase-3β regulates cyclin D1 proteolysis and subcellular localization. Genes Dev. 12:3499-3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diehl, J. A., F. Zindy, and C. J. Sherr. 1997. Inhibition of cyclin D1 phosphorylation on threonine-286 prevents its rapid degradation via the ubiquitin-proteasome pathway. Genes Dev. 11:957-972. [DOI] [PubMed] [Google Scholar]
- 11.Driscoll, B., L. Wu, S. Buckley, F. L. Hall, K. D. Anderson, and D. Warburton. 1997. Cyclin D1 antisense RNA destabilizes pRb and retards lung cancer cell growth. Am. J. Physiol. 273:L941-949. [DOI] [PubMed] [Google Scholar]
- 12.Fan, J., and J. R. Bertino. 1997. K-Ras modulates the cell cycle via both positive and negative regulatory pathways. Oncogene 14:2595-2607. [DOI] [PubMed] [Google Scholar]
- 13.Fantl, V., G. Stamp, A. Andrews, I. Rosewell, and C. Dickson. 1995. Mice lacking cyclin D1 are small and show defects in eye and mammary gland development. Genes Dev. 9:2364-2372. [DOI] [PubMed] [Google Scholar]
- 14.Fry, C. J., J. E. Slansky, and P. J. Farnham. 1997. Position-dependent transcriptional regulation of the murine dihydrofolate reductase promoter by the E2F transactivation domain. Mol. Cell. Biol. 17:1966-1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gillett, C., V. Fantl, R. Smith, C. Fisher, J. Bartek, C. Dickson, D. Barnes, and G. Peters. 1994. Amplification and overexpression of cyclin D1 in breast cancer detected by immunohistochemical staining. Cancer Res. 54:1812-1817. [PubMed] [Google Scholar]
- 16.Giunciuglio, D., M. Culty, G. Fassina, L. Masiello, A. Melchiori, G. Paglialunga, G. Arand, F. Ciardiello, F. Basolo, E. W. Thompson, et al. 1995. Invasive phenotype of MCF10A cells overexpressing c-Ha-Ras and c-erbB-2 oncogenes. Int. J. Cancer 63:815-822. [DOI] [PubMed] [Google Scholar]
- 17.Guttridge, D. C., C. Albanese, J. Y. Reuther, R. G. Pestell, and A. S. Baldwin, Jr. 1999. NF-κB controls cell growth and differentiation through transcriptional regulation of cyclin D1. Mol. Cell. Biol. 19:5785-5799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanahan, D., and R. A. Weinberg. 2000. The hallmarks of cancer. Cell 100:57-70. [DOI] [PubMed] [Google Scholar]
- 19.Harari, D., and Y. Yarden. 2000. Molecular mechanisms underlying ErbB2/HER2 action in breast cancer. Oncogene 19:6102-6114. [DOI] [PubMed] [Google Scholar]
- 20.Hinds, P. W., S. F. Dowdy, E. N. Eaton, A. Arnold, and R. A. Weinberg. 1994. Function of a human cyclin gene as an oncogene. Proc. Natl. Acad. Sci. USA 91:709-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hsu, T., D. McRackan, T. S. Vincent, and H. Gert De Couet. 2001. Drosophila Pin1 prolyl isomerase Dodo is a MAP kinase signal responder during oogenesis. Nat. Cell Biol. 3:538-543. [DOI] [PubMed] [Google Scholar]
- 22.Hunter, T. 1998. Prolyl isomerase and nuclear function. Cell 92:141-143. [DOI] [PubMed] [Google Scholar]
- 23.Imai, Y., Y. Matsushima, T. Sugimura, and M. Terada. 1991. A simple and rapid method for generating a deletion by PCR. Nucleic Acids Res. 19:2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson, D. G., J. K. Schwarz, W. D. Cress, and J. R. Nevins. 1993. Expression of transcription factor E2F1 induces quiescent cells to enter S phase. Nature 365:349-352. [DOI] [PubMed] [Google Scholar]
- 25.Karlseder, J., H. Rotheneder, and E. Wintersberger. 1996. Interaction of Sp1 with the growth- and cell cycle-regulated transcription factor E2F. Mol. Cell. Biol. 16:1659-1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kornmann, M., N. Arber, and M. Korc. 1998. Inhibition of basal and mitogen-stimulated pancreatic cancer cell growth by cyclin D1 antisense is associated with loss of tumorigenicity and potentiation of cytotoxicity to cisplatinum. J. Clin. Investig. 101:344-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee, R. J., C. Albanese, M. Fu, M. D'Amico, B. Lin, G. Watanabe, G. K. Haines III, P. M. Siegel, M. C. Hung, Y. Yarden, J. M. Horowitz, W. J. Muller, and R. G. Pestell. 2000. Cyclin D1 is required for transformation by activated Neu and is induced through an E2F-dependent signaling pathway. Mol. Cell. Biol. 20:672-683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin, S. Y., W. Xia, J. C. Wang, K. Y. Kwong, B. Spohn, Y. Wen, R. G. Pestell, and M. C. Hung. 2000. Beta-catenin, a novel prognostic marker for breast cancer: its roles in cyclin D1 expression and cancer progression. Proc. Natl. Acad. Sci. USA 97:4262-4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liou, Y. C., A. Ryo, H. K. Huang, P. J. Lu, R. Bronson, F. Fujimori, U. Uchida, T. Hunter, and K. P. Lu. 2002. Loss of Pin1 function in the mouse resembles the cyclin D1-null phenotypes. Proc. Natl. Acad. Sci. USA 99:1335-1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu, K. P., S. D. Hanes, and T. Hunter. 1996. A human peptidyl-prolyl isomerase essential for regulation of mitosis. Nature 380:544-547. [DOI] [PubMed] [Google Scholar]
- 31.Lu, K. P., Y. C. Liou, and X. Z. Zhou. 2002. Pinning down the proline-directed phosphorylation signaling. Trends Cell Biol. 12:164-172. [DOI] [PubMed] [Google Scholar]
- 32.Lu, P. J., G. Wulf, X. Z. Zhou, P. Davies, and K. P. Lu. 1999. The prolyl isomerase Pin1 restores the function of Alzheimer-associated phosphorylated tau protein. Nature 399:784-788. [DOI] [PubMed] [Google Scholar]
- 33.Lu, P. J., X. Z. Zhou, Y. C. Liou, J. P. Noel, and K. P. Lu. 2002. Role of WW domain phosphorylation in regulating its phosphoserine-binding activity and the Pin1 function. J. Biol. Chem. 277:2381-2384. [DOI] [PubMed] [Google Scholar]
- 34.Lu, P. J., X. Z. Zhou, M. Shen, and K. P. Lu. 1999. A function of WW domains as phosphoserine- or phosphothreonine-binding modules. Science 283:1325-1328. [DOI] [PubMed] [Google Scholar]
- 35.Macleod, K. 1999. pRb and E2f-1 in mouse development and tumorigenesis. Curr. Opin. Genet. Dev. 9:31-39. [DOI] [PubMed] [Google Scholar]
- 36.Muller, W. J., E. Sinn, P. K. Pattengale, R. Wallace, and P. Leder. 1988. Single-step induction of mammary adenocarcinoma in transgenic mice bearing the activated c-Neu oncogene. Cell 54:105-115. [DOI] [PubMed] [Google Scholar]
- 37.Muthuswamy, S. K., D. Li, S. Lelievre, M. J. Bissell, and J. S. Brugge. 2001. ErbB2, but not ErbB1, reinitiates proliferation and induces luminal repopulation in epithelial acini. Nat. Cell Biol. 3:785-792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Neill, S. D., C. Hemstrom, A. Virtanen, and J. R. Nevins. 1990. An adenovirus E4 gene product trans-activates E2 transcription and stimulates stable E2F binding through a direct association with E2F. Proc. Natl. Acad. Sci. USA 87:2008-2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nevins, J. R. 2001. The Rb/E2F pathway and cancer. Hum. Mol. Genet. 10:699-703. [DOI] [PubMed] [Google Scholar]
- 40.Ohtani, K., J. DeGregori, and J. R. Nevins. 1995. Regulation of the cyclin E gene by transcription factor E2F1. Proc. Natl. Acad. Sci. USA 92:12146-12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ohtani, K., R. Iwanaga, M. Nakamura, M. Ikeda, N. Yabuta, H. Tsuruga, and H. Nojima. 1999. Cell growth-regulated expression of mammalian MCM5 and MCM6 genes mediated by the transcription factor E2F. Oncogene 18:2299-2309. [DOI] [PubMed] [Google Scholar]
- 42.Petersen, O. W., L. Ronnov-Jessen, A. R. Howlett, and M. J. Bissell. 1992. Interaction with basement membrane serves to rapidly distinguish growth and differentiation pattern of normal and malignant human breast epithelial cells. Proc. Natl. Acad. Sci. USA 89:9064-9068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ranganathan, R., K. P. Lu, T. Hunter, and J. P. Noel. 1997. Structural and functional analysis of the mitotic peptidyl-prolyl isomerase Pin1 suggests that substrate recognition is phosphorylation dependent. Cell 89:875-886. [DOI] [PubMed] [Google Scholar]
- 44.Ryo, A., N. Nakamura, G. Wulf, Y. C. Liou, and K. P. Lu. 2001. Pin1 regulates turnover and subcellular localization of beta-catenin by inhibiting its interaction with APC. Nat. Cell Biol. 3:793-801. [DOI] [PubMed] [Google Scholar]
- 45.Schrump, D. S., A. Chen, and U. Consoli. 1996. Inhibition of lung cancer proliferation by antisense cyclin D. Cancer Gene Ther. 3:131-135. [PubMed] [Google Scholar]
- 46.Shen, M., P. T. Stukenberg, M. W. Kirschner, and K. P. Lu. 1998. The essential mitotic peptidyl-prolyl isomerase Pin1 binds and regulates mitosis-specific phosphoproteins. Genes Dev. 12:706-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sherr, C. J. 1996. Cancer cell cycles. Science 274:1672-1677. [DOI] [PubMed] [Google Scholar]
- 48.Sicinski, P., J. L. Donaher, S. B. Parker, T. Li, A. Fazeli, H. Gardner, S. Z. Haslam, R. T. Bronson, S. J. Elledge, and R. A. Weinberg. 1995. Cyclin D1 provides a link between development and oncogenesis in the retina and breast. Cell 82:621-630. [DOI] [PubMed] [Google Scholar]
- 49.Sinn, E., W. Muller, P. Pattengale, I. Tepler, R. Wallace, and P. Leder. 1987. Coexpression of MMTV/v-Ha-Ras and MMTV/c-myc genes in transgenic mice: synergistic action of oncogenes in vivo. Cell 49:465-475. [DOI] [PubMed] [Google Scholar]
- 50.Slamon, D. J., G. M. Clark, S. G. Wong, W. J. Levin, A. Ullrich, and W. L. McGuire. 1987. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/Neu oncogene. Science 235:177-182. [DOI] [PubMed] [Google Scholar]
- 51.Soule, H. D., T. M. Maloney, S. R. Wolman, W. D. Peterson, Jr., R. Brenz, C. M. McGrath, J. Russo, R. J. Pauley, R. F. Jones, and S. C. Brooks. 1990. Isolation and characterization of a spontaneously immortalized human breast epithelial cell line, MCF-10. Cancer Res. 50:6075-6086. [PubMed] [Google Scholar]
- 52.Stukenberg, P. T., and M. W. Kirschner. 2001. Pin1 acts catalytically to promote a conformational change in Cdc25. Mol. Cell 7:1071-1083. [DOI] [PubMed] [Google Scholar]
- 53.Trimarchi, J. M., and J. A. Lees. 2002. Sibling rivalry in the E2F family. Nat. Rev. Mol. Cell. Biol. 3:11-20. [DOI] [PubMed] [Google Scholar]
- 54.von Lintig, F. C., A. D. Dreilinger, N. M. Varki, A. M. Wallace, D. E. Casteel, and G. R. Boss. 2000. Ras activation in human breast cancer. Breast Cancer Res. Treat. 62:51-62. [DOI] [PubMed] [Google Scholar]
- 55.Wang, T. C., R. D. Cardiff, L. Zukerberg, E. Lees, A. Arnold, and E. V. Schmidt. 1994. Mammary hyperplasia and carcinoma in MMTV-cyclin D1 transgenic mice. Nature 369:669-671. [DOI] [PubMed] [Google Scholar]
- 56.Weinberg, R. A. 1995. The retinoblastoma protein and cell cycle control. Cell 81:323-330. [DOI] [PubMed] [Google Scholar]
- 57.Wells, J., K. E. Boyd, C. J. Fry, S. M. Bartley, and P. J. Farnham. 2000. Target gene specificity of E2F and pocket protein family members in living cells. Mol. Cell. Biol. 20:5797-5807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wulf, G. M., A. Ryo, G. G. Wulf, S. W. Lee, T. Niu, and K. P. Lu. 2001. Pin1 is overexpressed in breast cancer and potentiates the transcriptional activity of phosphorylated c-Jun towards the cyclin D1 gene. EMBO J. 20:3459-3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yaffe, M. B., M. Schutkowski, M. Shen, X. Z. Zhou, P. T. Stukenberg, J. Rahfeld, J. Xu, J. Kuang, M. W. Kirschner, G. Fischer, L. C. Cantley, and K. P. Lu. 1997. Sequence-specific and phosphorylation-dependent proline isomerization: a potential mitotic regulatory mechanism. Science 278:1957-1960. [DOI] [PubMed] [Google Scholar]
- 60.Yu, Q., Y. Geng, and P. Sicinski. 2001. Specific protection against breast cancers by cyclin D1 ablation. Nature 411:1017-1021. [DOI] [PubMed] [Google Scholar]
- 61.Zhang, S. Y., S. C. Liu, L. F. Al-Saleem, D. Holloran, J. Babb, X. Guo, and A. J. Klein-Szanto. 2000. E2F-1: a proliferative marker of breast neoplasia. Cancer Epidemiol. Biomarkers Prev. 9:395-401. [PubMed] [Google Scholar]
- 62.Zhou, X. Z., O. Kops, A. Werner, P. J. Lu, M. Shen, G. Stoller, G. Küllertz, M. Stark, G. Fischer, and K. P. Lu. 2000. Pin1-dependent prolyl isomerization regulates dephosphorylation of Cdc25C and tau proteins. Mol. Cell 6:873-883. [DOI] [PubMed] [Google Scholar]
- 63.Zhou, X. Z., P. J. Lu, G. Wulf, and K. P. Lu. 1999. Phosphorylation-dependent prolyl isomerization: a novel signaling regulatory mechanism. Cell. Mol. Life Sci. 56:788-806. [DOI] [PMC free article] [PubMed] [Google Scholar]