Abstract
Modulation of the interaction between U1 snRNP and the 5′ splice site (5′ss) is a key event that governs 5′ss recognition and spliceosome assembly. Using the methylene blue-mediated cross-linking method (Z. R. Liu, A. M. Wilkie, M. J. Clemens, and C. W. Smith, RNA 2:611-621, 1996), a 65-kDa protein (p65) was shown to interact with the U1-5′ss duplex during spliceosome assembly (Z. R. Liu, B. Sargueil, and C. W. Smith, Mol. Cell. Biol. 18:6910-6920, 1998). In this report, p65 was identified as p68 RNA helicase and shown to be essential for in vitro pre-mRNA splicing. Depletion of endogenous p68 RNA helicase does not affect the loading of the U1 snRNP to the 5′ss during early stage of splicing. However, dissociation of the U1 from the 5′ss is largely inhibited. The data suggest that p68 RNA helicase functions in destabilizing the U1-5′ss interactions. Furthermore, depletion of p68 RNA helicase arrested spliceosome assembly at the prespliceosome stage, suggesting that p68 may play a role in the transition from prespliceosome to spliceosome.
To remove introns, a large RNA-protein complex, known as the spliceosome, must be assembled on mRNA precursors (pre-mRNA) (11, 29, 39). Assembly of a functional spliceosome proceeds through an ordered addition of four small nuclear ribonucleoprotein particles (snRNPs) (U1, U2, U4/U6, and U5) as well as many non-snRNP proteins. This pathway leads to the formation of several intermediate spliceosome complexes. Recognition of the 5′ splice site (5′ss) by the U1 snRNP, along with binding of the polypyrimidine tract and branch point by U2AF65 and SF1, results in the formation of the commitment complex. Recruitment of the U2 snRNP to the commitment complex leads to the formation of complex A, or the prespliceosome. At this point, the preformed U4/U6.U5 tri-snRNPs will join into the prespliceosome, leading to the formation of the spliceosome (1, 13, 28).
The spliceosome is a dynamic molecular machine. Dynamic RNA-RNA base-pairing interactions between snRNAs and pre-mRNA and among snRNAs play critical roles in the spliceosome assembly and recognition of splice sites (32, 33, 35). The RNA duplexes in the spliceosome complexes are generally transient and short, ranging from 3 to 7 bp. Many of these RNA-RNA interactions are mutually exclusive. One example is the RNA-RNA interactions that occur at the 5′ss. The 5′ss is first recognized by interactions of 5 to 7 bp between the 5′ss and 5′-end of the U1 snRNA. This early U1-5′ss duplex is unwound to expose the same 5′ss sequence for pairing with the U6 snRNA prior to the first chemical reaction of splicing (28). The U6 snRNA is associated with U4 by two RNA helices before joining the prespliceosome. The two RNA helices between U6 and U4 must be unwound before U6 pairing with the 5′ss (2, 30, 40). Three events, unwinding of the U1-5′ss duplex, unwinding of the U4/U6 helices, and formation of the U6-5′ss duplex, are tightly coupled. How does the spliceosome coordinate these RNA-RNA interactions precisely in time and space? Presumably, protein factors are needed to mediate and rearrange the dynamic RNA-RNA interactions (28).
A large family of highly evolutionarily conserved proteins, termed the DExH/D box of putative RNA helicases, has recently been described (27, 36). The principal cellular function of RNA helicases is to unwind double-stranded RNA (7). Recent experimental observations suggested that some of the DExH/D box proteins may also act on RNA-protein complexes (5, 16). To date, eight yeast splicing factors and six mammalian proteins that are homologues to the superfamily of RNA helicases have been implicated in the pre-mRNA splicing (10, 27, 37, 46). Many of these proteins have demonstrated RNA unwinding activities in vitro (21, 34, 38, 44). It is speculated that these DExH/D box proteins function by rearranging the complex RNA structures and/or RNA-protein interactions in the spliceosome. However, the challenge is to link an individual DExH/D box protein to a specific RNA or RNA-protein rearrangement. Yeast Prp28p has been shown to function in the RNA switch at the 5′ss (18, 41). However, it is not completely clear if Prp28p directly unwinds the U1-5′ss duplex. Yeast Brr2p unwinds the U4/U6 duplex in the U5.U4/U6 tri-snRNP complex. However, functional involvement of the protein in unwinding the duplex in the splicing process has not been demonstrated (34). The human homologue of Brr2p, a U5 snRNP-specific 200-kDa protein, has also been suspected of unwinding U4/U6 helices (21).
Due to the unique specificity of the methylene blue (MB)-mediated cross-linking method (26), a 65-kDa protein (p65) was detected interacting with the U1-5′ss duplex during the splicing process (25). p65 interacts with the U1-5′ss duplex at the exon and intron junction, and the intact U1 snRNP is required for the interaction. Interestingly, p65 associates with the transient duplex during early spliceosome assembly and dissociates at a late stage. The timing of p65 dissociation correlates well with the timing of dissociation of the U1 snRNP from the 5′ss, which indicates that p65 may be functionally involved in mediating the U1 snRNA and pre-mRNA interactions.
Here, I report the identification of the previously detected p65 as p68 RNA helicase. I analyzed the function of p68 RNA helicase in pre-mRNA splicing and the spliceosome assembly. The data demonstrate that p68 RNA helicase is essential for in vitro pre-mRNA splicing. The experimental data also show that depletion of p68 leads to the accumulation of a prespliceosome-like complex.
MATERIALS AND METHODS
Oligonucleotides.
The oligonucleotides used in this research are three antisenses that are complementary to the 13 nucleotides (nt) of the 5′ end of U1 snRNA (αU11-13) (5′-TGCCAGGTAAGTA-3′), 12 nt (64 to 75) of U1 snRNA (αU164-75) (5′-CGGAGTGCAATG-3′), and 13 nt of the 5′ss of pPIP10A (α5′ss) (5′-CATACTCACCAGG-3′), as well as a 22-nt oligonucleotide, Act1, with random sequences of 5′-CAGAATCCATTGTTAATTCAGT-3′. The polyclonal antibody PAbN1 was raised commercially (by Research Genetics, Inc.), using a peptide that spans 14 amino acids (GAPRFGGSRAGPLG) at the N-terminal end of p68 RNA helicase. The antibody was purified over an antigen column. Trioxsalen was purchased from Sigma-Aldrich and used without further treatments.
Immunoprecipitation and Western blot analyses.
Immunoprecipitation experiments were performed as described in previous studies (24, 25). Briefly, MB-mediated cross-linking was carried out with RNA GC+DX/XhoI in 30% HeLa nuclear extracts. Sodium dodecyl sulfate (SDS) was added to 100 μl of MB cross-linking mixture to a final concentration of 0.8%. The SDS-treated cross-linking mixture was diluted with NETS buffer (150 mM NaCl, 50 mM tris-HCl[pH 7.5], 5 mM EDTA, 0.05% NP-40) to 0.1% SDS. Antibody, PAb204 or PAbN1, was then added to the mixture. The solution was incubated at 4°C for 3 h. Twenty microliters of protein G or protein A agarose bead slurry was then added, and the mixture was rotated at room temperature for 5 h. The beads were recovered and washed five times with 600 μl of NETS. Finally, the precipitated proteins were analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) and subjected to autoradiography.
Western blot analyses were carried out with an ECL Western blot detection kit (Pharmacia). The antigen-purified PAbN1 was used in a 1:6,000 dilution. The antibody PAb204 (tissue culture supernatants) was used in a 1:5 dilution.
Depletion of p68 RNA helicase and reconstitution of splicing activity.
The antibody, PAb204 or PAbN1, was added to HeLa extracts. To gain better depletion results, the salt concentration of the HeLa nuclear extracts was raised to 600 mM NaCl before addition of antibody. After 3 h of incubation at 4°C, the mixtures were passed through a protein G or protein A agarose bead column. The column fractions were analyzed by SDS-PAGE. The fractions that contained the most proteins were collected together and dialyzed against buffer E (20 mM Tris-HCl [pH 7.50], 50 mM NaCl, 0.3 mM EDTA [pH 8.0], 15% glycerol) twice for six hours. The recovered p68-depleted HeLa nuclear extracts were used in other in vitro assays.
To reconstitute the splicing activity, the His-tagged p68 RNA helicase (expression and purification of the six-His p68 will be reported elsewhere) was added to 35% of the p68-depleted nuclear extracts to a final concentration of ∼25 ng/μl. The mixture was incubated at 30°C for 15 min under normal in vitro splicing conditions. The appropriate pre-mRNA was then added to the preincubated extracts, and the splicing reaction was incubated at 30°C for an additional 150 min. The splicing products were analyzed by urea-PAGE.
RNase H cleavage protection and trioxsalen cross-linking.
Splicing reactions were performed with pPIP10A in 40% HeLa nuclear extracts or extracts that were pretreated with RNase H and the DNA oligonucleotide αU11-13. After a 10-min incubation, 5 μmol of DNA oligonucleotide α5′ss that was complementary to the 5′ss or random DNA oligonucleotide Act1 and RNase H (Promega) were added to the splicing reactions. The reactions were further incubated at 30°C for 40 min. The reaction mixtures were treated with proteinase K and phenol-chloroform extraction. The final RNA products were analyzed by denatured urea-6% PAGE. For the trioxsalen cross-linking, splicing reactions were performed with pPIP10A in 30% HeLa nuclear extracts for the indicated times. The stock solution of trioxsalen (5 mg/ml dissolved in dimethyl sulfoxide) was added to the reactions to a final concentration of 15 μg/ml. The reaction mixtures were placed on ice and photolyzed with a UV cross-linker containing four 15-W, 282-nm (maximum) UV bulbs for 12 min. The reaction mixtures were then treated with proteinase K and phenol-chloroform extraction. The RNAs were precipitated by ethanol. To identify the trioxsalen cross-linked RNA species, the precipitated RNAs were redissolved in 40 μl of RNase H reaction buffer. Five micromoles of the DNA oligonucleotide αU164-75 and RNase H were added to the solution. The mixture was incubated at 30°C for 30 min. The products were treated with phenol-chloroform extraction and subsequent ethanol precipitation. The final RNAs were analyzed by denatured urea-6% PAGE and subjected to autoradiography.
Analyses of the spliceosome complexes.
Splicing reactions were performed with pPIP7A or pPIP10A in 20 to 25% HeLa nuclear extracts. After incubation for the appropriate time, heparin was added to the reaction mixture to a concentration of 0.5 mg/ml. The reactions were incubated at 30°C for an additional 5 min. The reaction products were analyzed by 4% (80:1, acrylamide/bis-acrylamide) native PAGE.
RESULTS
The previously detected p65 is identical to p68 RNA helicase.
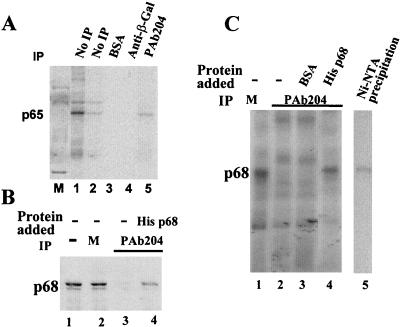
A double-stranded-RNA-specific RNA-protein cross-linking method was developed (26). The RNA-protein cross-links are mediated by MB, a tissue stain dye. Using this cross-linking technique, a 65-kDa protein was previously detected to interact with the RNA duplex formed between U1 snRNA and the 5′ss during early stages of spliceosome assembly. To characterize the identity of p65, a number of immunoprecipitation experiments with antibodies against the known splicing factors with similar molecular mass have been carried out. Antibodies against U1-70K, U2AF65, p69, and SF1 were not able to precipitate p65 (25). Another candidate is p68 RNA helicase, which has recently been shown to be a potential spliceosome-associated protein (31). To test whether p65 is identical to the p68 RNA helicase, I carried out immunoprecipitation experiments with a monoclonal antibody, PAb204, that was originally raised against the simian virus 40 large-T antigen. The antibody recognizes a minimal 19-amino-acid peptide sequence of p68 from amino acid residue 507 to residue 526 (22). The antibody recognized p68 RNA helicase in the HeLa nuclear extracts (Fig. 1B, lanes 1 and 2). In fact, p68 RNA helicase is the only HeLa cellular protein that is recognized by this antibody (6, 22) (F. V. Fuller-Pace, personal communication). It is evident that the MB-cross-linked p65 is precipitated by the antibody (Fig. 1A, lane 5). To confirm this immunoprecipitation result, another polyclonal antibody, PAbN1, was raised against a 15-amino-acid peptide that spans amino acid residues 4 to 19 of p68. The antibody was purified over an antigen column. The MB cross-linking and the subsequent immunoprecipitation experiments were carried out with the new antibody. The results showed that the cross-linked p65 was precipitated by the antibody (data not shown). The immunoprecipitation experiments with these two independent antibodies strongly suggest that the previously detected p65 is identical to the p68 RNA helicase.
FIG. 1.
(A) Immunoprecipitation of MB-cross-linked p65. The MB cross-linking mixture was electrophoresed by SDS-PAGE after no further treatment (lanes 1 and 2) or after immunoprecipitation with BSA (lane 3), monoclonal anti-β-galactosidase antibody (lane 4), or the antibody PAb204 (lane 5). (B) Western blot analyses of HeLa nuclear extracts (lane 1), mock-depleted (M) (protein G) HeLa nuclear extracts (lane 2), p68-depleted HeLa nuclear extracts (lane 3), and the p68-depleted HeLa extracts in which the His-tagged p68 RNA helicase is added (lane 4). The antibody PAb204 is used in the Western analyses. (C) MB cross-linking of recombinant p68 to the U1-5′ss duplex. MB cross-linking of p68 RNA helicase to RNA GC+DX/XhoI. The MB cross-linking reactions were carried out in mock-depleted (M) HeLa nuclear extracts (lane 1), p68-depleted HeLa nuclear extracts (lane 2), the p68-depleted HeLa extracts in which BSA was added to a concentration of 100 ng/μl (lane 3), or the p68-depleted HeLa extracts in which the His-tagged p68 was added to a concentration of 25 ng/μl (lane 4). Lane 5 is the precipitation of MB cross-linking by Ni-NTA bead. The cross-linking is carried out in the His-tagged p68-reconstituted HeLa nuclear extracts. IP, immunoprecipitation.
To further verify the results obtained in the immunoprecipitation experiments, I examined whether a recombinant p68 RNA helicase could be cross-linked to the U1-5′ss duplex. To this end, I expressed and purified recombinant His-tagged p68 RNA helicase from Escherichia coli and showed that the recombinant protein exhibited the expected ATPase and RNA unwinding activities (13a). The MB cross-linking experiments were carried out with HeLa nuclear extracts in which the endogenous p68 RNA helicase was replaced by the recombinant His-tagged p68. First, as a control, cross-linking of p65 was not observed in the p68-depleted HeLa nuclear extracts (Fig. 1C, lanes 2 and 3), while a cross-linking band with a molecular mass similar to that of the cross-linked p65 was detected when the recombinant His-tagged p68 was added to the p68-depleted nuclear extracts (Fig. 1C, lane 4). Precipitation of the MB cross-linking mixture in the His-tagged p68 reconstituted nuclear extracts with Ni-nitrilotriacetic acid (NTA) agarose beads demonstrated that this new cross-linking band could be precipitated by the Ni-NTA beads, suggesting that the cross-linked protein is a His-tagged protein (Fig. 1C, lane 5). The data indicated that the recombinant His-tagged p68 RNA helicase acted like endogenous p65. Both proteins interact with the U1-5′ss RNA duplex. Given the results from the immunoprecipitation experiments and cross-linking of the recombinant His-tagged p68 to the 5′ss, I conclude that the previously detected p65 is identical to p68 RNA helicase.
p68 RNA helicase is essential for in vitro pre-mRNA splicing.
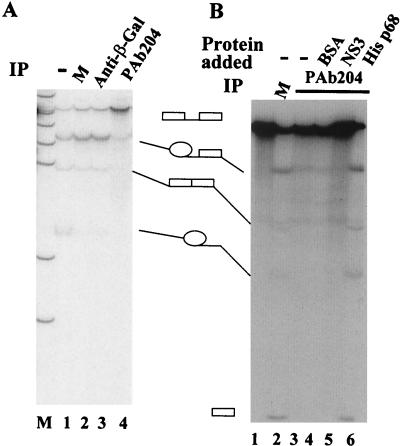
The p68 RNA helicase interacts with the U1-5′ss duplex during the pre-mRNA splicing process (25). To determine whether p68 plays a functional role in the pre-mRNA splicing, I first carried out immunodepletion experiments in the HeLa nuclear extracts with two antibodies, PAb204 and PAbN1. The p68-depleted HeLa nuclear extracts were analyzed by either a Western blot, using the antibody PAb204, or MB cross-linking. The experiments showed that p68 RNA helicase was not detectable in the Western blot assays (Fig. 1B, lane 3) nor was it detectable in the MB cross-linking assays (Fig. 1C, lane 2), indicating that depletion of p68 was reasonably complete. I then performed in vitro splicing in the p68-depleted HeLa nuclear extracts with three unrelated pre-mRNA substrates. The first experiments were carried out with GC+DX, a splicing substrate derived from exons 2 and 3 of the α-tropomyosin gene. The splicing activity of the extracts was abolished by p68 depletion (Fig. 2A, lane 4, and B, lane 3). As a control, the splicing activity of HeLa nuclear extracts was not affected by mock depletion (Fig. 2A, lane 2) or depletion with a monoclonal antibody against β-galactosidase (Fig. 2A, lane 3).
FIG. 2.
In vitro splicing of GC+DX transcripts in HeLa nuclear extracts. Splicing reactions were carried out under standard conditions. (A) Splicing extracts were untreated (lane 1), mock depleted (M) (lane 2), immunodepleted with anti-β-galacotosidase antibody (lane 3), or immunodepleted with PAb204 (lane 4). Lane M shows molecular weight markers. (B). Splicing extracts were mock depleted (M) (lane 2), immunodepleted with PAb204 (lane 3), immunodepleted with PAb204 where BSA was added to a concentration of 100 ng/μl (lane 4), immunodepleted with PAb204 where NS3 was added to 50 ng/μl (lane 5), or immunodepleted with PAb204 where His-tagged p68 was added to 25 ng/μl (lane 6). Lane 1 is the GC+DX transcript. The identities of the various RNA bands are shown schematically. IP, immunoprecipitation.
I next examined if the splicing activity of p68-depleted HeLa nuclear extracts could be restored by addition of the recombinant His-tagged p68. To this end, the His-tagged p68 was added to the p68-depleted HeLa extracts to a concentration of 25 ng/μl. The reconstituted HeLa nuclear extracts were first incubated under normal splicing conditions in the presence of ATP for 15 min without addition of the pre-mRNA. After the brief incubation, the splicing activity was then assayed with GC+DX transcripts. The experimental data indicated that the addition of the recombinant protein reconstituted the splicing activity of the p68-depleted extracts (Fig. 2B, lane 6). The splicing activity of the p68-depleted HeLa nuclear extracts, however, was not recovered with bovine serum albumin (BSA) (Fig. 2B, lane 4), nor with an unrelated DEAD-box RNA helicase, HCV-NS3 (an RNA helicase from hepatitis C virus) (Fig. 2B, lane 5).
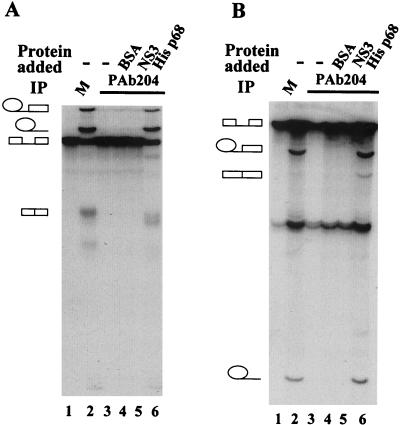
Two further splicing substrates were used to generalize the results obtained from the α-tropomyosin intron. pPIP10A is a splicing substrate derived from the major late transcript of adenovirus, and βG2 is a splicing substrate derived from the first intron of the human β-globin gene (24). These two splicing substrates were spliced efficiently in HeLa nuclear extracts (Fig. 3A and B, lanes 2). Depletion of p68 RNA helicase from HeLa nuclear extracts by the antibody PAb204 abolished the splicing activity of the extracts for these two splicing substrates (Fig. 3A and B, lanes 3). Addition of the recombinant His-tagged p68 to the p68-depleted HeLa nuclear extracts restored the splicing activity of the extracts (Fig. 3A and B, lanes 6). Similar results were also observed with extracts in which p68 was depleted by the antibody PAbN1 (data not shown). The three splicing substrates used here are derived from introns that come from different genes, and my data demonstrated that p68 RNA helicase was essential for all these three splicing substrates. The data, therefore, strongly suggested that p68 RNA helicase is most likely a general essential splicing factor.
FIG. 3.
In vitro splicing of transcripts pPIP10A (A) and βG2 (B). Splicing reactions were carried out under standard conditions. Splicing extracts were mock depleted (M) (lane 2), immunodepleted with PAb204 (lane 3), immunodepleted with PAb204 where BSA was added to a concentration of 100 ng/μl (lane 4), immunodepleted with PAb204 where NS3 was added to 50 ng/μl (lane 5), or immunodepleted with PAb204 where His p68 was added to 25 ng/μl (lane 6). Lane 1 is the transcript pPIP10A (A) or βG2 (B), respectively.
Depletion of p68 RNA helicase does not block binding of the U1 snRNP to the 5′ss but affects the dissociation of the U1 snRNP from the 5′ss.
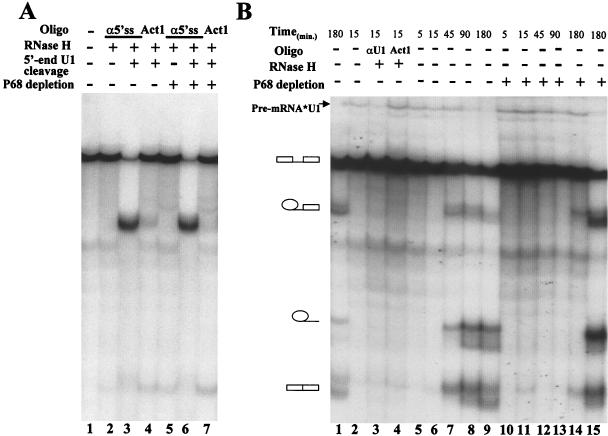
The preceding data demonstrated that p68 RNA helicase is essential for in vitro pre-mRNA splicing. In order to examine the effects of p68 depletion on the U1 snRNP and the 5′ss interactions, I analyzed the binding and dissociation of the U1 snRNP at the 5′ss during the splicing process in the presence and absence of p68 RNA helicase, using the RNase H cleavage protection and trioxsalen cross-linking assays. The splicing substrate pPIP10A was used in the analyses. In a typical splicing reaction, the 5′ss of pre-mRNA was protected from DNA oligonucleotide-guided RNase H cleavage after 10 min of incubation at 30°C (Fig. 4A, lane 2). This protection is due to binding of the U1 snRNP to the 5′ss during the early stage of splicing (4, 9). In contrast, if the 5′ end of U1 snRNA was cleaved before the addition of pre-mRNA, the 5′ss was not protected from DNA oligonucleotide-guided RNase H cleavage (lane 3). Depletion of p68 RNA helicase from the HeLa nuclear extracts did not lead to any change in RNase H cleavage protection at the 5′ss (lane 5). The experiments suggested that depletion of p68 has no effects on the binding of the U1 snRNP to the 5′ss during the early stage of splicing.
FIG. 4.
(A) RNase H cleavage protection assays. Splicing reactions were performed with pPIP10A in 40% HeLa nuclear extracts (lanes 2 to 4) or extracts from which p68 RNA helicase was depleted (lanes 5 to 7). The extracts were pretreated with either RNase H and DNA oligonucleotide αU11-13 (lanes 3, 4, 6, and 7) or no RNase H treatment (lane 2 and 5). In the RNase H cleavage assays, the DNA oligonucleotide α5′ss was added (lanes 2, 3, 5, and 6), or random sequence DNA oligonucleotide Act1 was added (lanes 4 and 7). Lane 1 is the pre-mRNA pPIP10A without any treatment. (B) The U1 snRNA-pre-mRNA trioxsalen cross-linking. Trioxsalen cross-linking was performed with intact HeLa nuclear extracts (lanes 1 to 9) or with the extracts from which p68 RNA helicase was immunodepleted (lanes 10 to 14). The splicing was carried out for the indicated times before the trioxsalen was added to the splicing reactions and photolyzation under UV light. Lane 1 shows splicing alone without trioxsalen cross-linking. Lane 3 shows cross-linked RNAs that were further treated with RNase H in the presence of DNA oligonucleotide αU1 that is complementary to U1 64-75 (αU164-75). Lane 4 shows the cross-linked RNAs that were further treated with RNase H in the presence of random sequence DNA oligonucleotide Act1. Lane 15 is the U1 snRNA-pre-mRNA trioxsalen cross-linking in the extracts in which the endogenous p68 RNA helicase was replaced by 25 ng of recombinant p68 RNA helicase/μl.
To further investigate the effects of depletion of p68 on the U1 snRNP and 5′ss interactions, I carried out trioxsalen cross-linking (9, 45) in splicing time courses. Trioxsalen is an intercalator. The compound mediates RNA-RNA cross-links under 320-nm UV light photolyzing. The cross-links of the U1 snRNA to pre-mRNA were indicated by a slower migration band (arrow in Fig. 4B) that disappears under the RNase H treatments guided by a U1-antisense DNA oligonucleotide (Fig. 4B, lanes 2 and 3). The U1-antisense is complementary to the sequence of nt 64 to 75 of the U1 snRNA. RNase H treatment in the presence of a DNA oligonucleotide, Act1, which is unrelated to the U1 snRNA and the pre-mRNA, did not lead to the disappearance of the cross-linking band (lane 4). It is evident that cross-linking of the U1 snRNA to the pre-mRNA decreases in the splicing time courses in the intact HeLa nuclear extracts (Fig. 4B, lanes 5 to 9). The cross-linking signal completely disappeared after 180 min of splicing (Fig. 4B, lane 9). In consistency with the cross-linking signal disappearance, the splicing intermediates and products were observed (lanes 5 to 9). In contrast, the pre-mRNA-U1 snRNA cross-links remain almost constant in the same splicing time course in the p68-depleted HeLa nuclear extracts (Fig. 4B, lanes 10 to 14). No splicing intermediates and products were observed in the p68-depleted extracts. Addition of the recombinant p68 RNA helicase to the depleted extracts led to the disappearance of the pre-mRNA-U1 snRNA cross-links at the 180-min time point (Fig. 4B, lane 15). The data suggest that the dissociation of U1 from the 5′ss is affected by p68 RNA helicase depletion.
Depletion of p68 RNA helicase blocks the spliceosome formation.
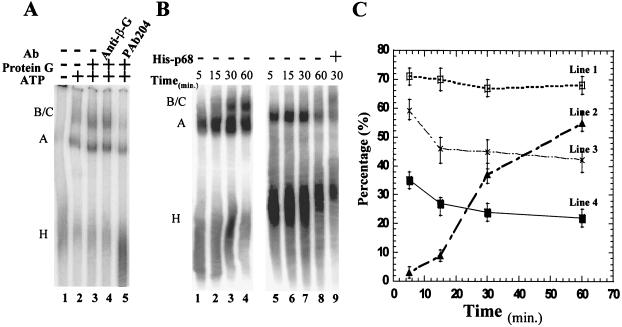
In order to determine at which stage p68 RNA helicase affects the splicing process, I analyzed the formation of the spliceosome complexes by native gel electrophoresis. The splicing substrate pPIP7A (derivative of major late transcript of adenovirus) was used in the assays. In a typical splicing reaction, complexes A and B/C were detected in the native gel electrophoresis assay (Fig. 5A, lane 2). In the absence of ATP, only complex H was detected (Fig. 5A, lane 1). I then carried out experiments using the splicing extracts from which the endogenous p68 helicase was immunodepleted. The experimental data clearly demonstrated that the formation of the spliceosome was blocked by depleting the p68 RNA helicase (Fig. 5A, lane 5). Interestingly, a complex that comigrates with the prespliceosome/complex A accumulated in the p68-depleted HeLa nuclear extracts. The data suggested that depletion of p68 blocked the transition from prespliceosome to spliceosome, which indicated that p68 potentially plays a role in the process of addition of the tri-snRNPs to the prespliceosome.
FIG. 5.
Electrophoretic separation of spliceosome complexes. (A) Spliceosome complexes assembled in the immunodepleted HeLa nuclear extracts. Splicing reactions were carried out with the splicing substrate pPIP7A in the absence of ATP (lane 1). Splicing reactions were carried out in the extracts that were untreated (lane 2), mock depleted (lane 3), immunodepleted with anti-β-galactosidase antibody (lane 4), and immunodepleted with the antibody PAb204 (lane 5). All the splicing reactions were incubated at 30°C for 30 min. (B) Spliceosome complexes assembled in splicing time courses. Splicing reactions were carried out with splicing substrate pPIP10A in intact HeLa nuclear extracts (lanes 1 to 4) and the p68-depleted extracts (lanes 5 to 8) for the indicated times. Lane 9 presents the spliceosome complexes assembled in a splicing reaction with pPIP10A in the p68-depleted extracts in which the recombinant p68 was added to a concentration of 25 ng/μl. (C) Quantitative analyses of spliceosome complexes assembled on the splicing substrate pPIP10A. Line 1 (□) is the ratio of [H]/([H] + [A]) in the p68-depleted HeLa nuclear extracts. Line 2 (▴) is the ratio of [B/C]/([A] + [B/C]) in the intact HeLa extracts. Line 3 (X) is the ratio of [H]/([A] + [H]) in the intact HeLa extracts. Line 4 (▪) is the ratio of [H]/([H] + [A] + [B/C]) in the intact HeLa extracts. The quantity of each complex, [H], [A], and [B/C], was measured by volume integration of each complex band using a phosphorimager system.
The H complex accumulates in p68-depleted HeLa nuclear extracts (Fig. 5A, lane 5), suggesting a possible defect in the formation of the A complex in the p68-depleted extracts. To analyze the effects of depletion of p68 on the formation of complex A, I monitored the formation of spliceosome complexes in splicing time courses with the substrate pPIP10A in the presence and absence of p68 RNA helicase. It is evident that the conversion of the H complex to the A complex is less efficient in the p68-depleted HeLa extracts than in the intact extracts at every splicing time point (Fig. 5B, compare lanes 1 to 4 to lanes 5 to 8). Quantitative analyses of the assembled spliceosome complexes revealed that the conversion of the H complex to the subsequent spliceosome complexes in the p68-depeleted extracts was less than 50% as efficient as that in the intact HeLa extracts (Fig. 5C, compare line 1 and line 4).
To further understand the effects of depletion of p68 on the assembly of the A and B/C complexes, I examined whether the defects in assembly of the A and B/C complexes in the p68-depleted HeLa extracts could be recovered by recombinant p68. Addition of the recombinant p68 RNA helicase to the p68-depleted extracts recovered the B/C complex formation (Fig. 5B, lane 9). Further quantification and analyses of all spliceosome complexes assembled in recombinant p68-reconstituted HeLa extracts revealed a very interesting phenomenon. (i) The ratio of B/C versus A + B/C was 36% ± 5% under 30 min of splicing, which is very close to that in the intact HeLa extracts at the same splicing time point (Fig. 5C, line 2). (ii) The ratio of the H complex versus the A + H complex is 69% ± 5% under 30 min of splicing. This ratio is almost the same as that in the p68-depleted extracts (Fig. 5C, line 1). These experimental observations led me to conclude that the recombinant p68 recovered the defects in assembly of the B complex from the A complex, which supports the speculation that p68 potentially plays a role in addition of the tri-snRNPs to the prespliceosome. The recombinant protein, however, did not change the A-versus-H complex ratio.
DISCUSSION
The p68 RNA helicase was first identified by cross-reaction with the antibody PAb204, which was originally raised against the simian virus 40 large-T antigen two decades ago (6). The protein is expressed in all dividing cells of different vertebrates. The expression pattern of p68 in different tissue types shows that p68 expression correlates with organ differentiation and maturation (42). The data suggest that p68 RNA helicase plays important roles in normal cell growth. Although, p68 RNA helicase was suspected of being involved in a number of biological processes in cells (8, 12, 17), the experimental evidence that links p68 RNA helicase to a specific cellular process is still lacking. The data presented in this report show that p68 RNA helicase is essential for the in vitro pre-mRNA splicing. The results agree with previous observations that p68 RNA helicase associates with the spliceosome (31).
What are the possible functional roles of p68 RNA helicase in the spliceosome? In this report, I have shown that p68 RNA helicase is identical to the previously detected p65 that interacts with the transient U1-5′ss RNA duplex. The timing of the p68-U1-5′ss duplex interaction correlates well with the timing of dissociation of the U1 snRNP from the 5′ss. In addition, ATP hydrolysis is required for the p68 and U1-5′ss duplex interaction (25). A reasonable conjecture based on these observations would be that p68 is functionally involved in modulating the U1 and 5′ss interaction during the spliceosome assembly. The p68 RNA helicase may modulate the U1 and 5′ss interactions by unwinding the transient RNA duplex. Alternatively, p68 may function to recruit the U1 snRNP to the 5′ss and/or to stabilize the U1-5′ss duplex. It is generally believed that stable interaction between the U1 snRNP and the 5′ss occurs early in the commitment complex (28). My experimental data showed that depletion of p68 from nuclear extracts did not completely inhibit the prespliceosome assembly. In addition, the trioxsalen cross-linking data demonstrated that the dissociation of the U1 snRNP from the 5′ss but not the loading of the U1 snRNP to the 5′ss is affected by depletion of p68 from HeLa nuclear extracts. Thus, with all experimental observations taken together, the best explanation for the function of p68 RNA helicase in the spliceosome is that p68 RNA helicase is involved in unwinding the U1-5′ss duplex.
Depletion of p68 abolished the formation of the spliceosome. Addition of recombinant p68 completely recovers the defects in the transition from prespliceosome to spliceosome. The formation of prespliceosome is also affected to some degree. However, since the recombinant p68 did not recover the deficiency of the A complex assembly, it is less clear whether p68 plays a role in the prespliceosome assembly. Previous data indicated that p68 RNA helicase cross-linked to the U1-5′ss duplex in the prespliceosome (25). Taken together, the data suggest that p68 is present in the prespliceosome and interacts with its target RNA, the U1-5′ss duplex. Three possible models fit well with all experimental data. (i) As discussed in the paragraph above, p68 RNA helicase is involved in unwinding the U1-5′ss duplex. (ii) p68 may not be involved in unwinding the U1-5′ss duplex. The protein may play a proofreading role at the U1-5′ss duplex (e.g., as a sensor). This proofreading is essential for the subsequent incorporation of the U5.U4/U6 tri-snRNPs. The incorporation of the tri-snRNPs to the prespliceosome is required for the U1-5′ss dissociation. (iii) Alternatively, p68 may mediate the U1-5′ss interaction on one side. The protein may also function in “communicating” with the U5.U4/U6 tri-snRNPs on the other side. This communication couples the tri-snRNP addition to the U1-5ss duplex dissociation. This communication may be direct or act through other proteins. A similar protein bridge that serves to communicate between the U1-5′ss unwinding and the U4/U6 unwinding was proposed by Kuhn and colleagues (20). p68 RNA helicase may serve as part of the protein bridge that includes hPrp8p and other spliceosome proteins. Although it is less likely, the possibility that destabilization of the U1-5′ss duplex is required for the stable association of the tri-snRNPs with the prespliceosome should not be excluded. The actual role(s) of p68 RNA helicase in the formation of the spliceosome remains to be elucidated. It would be interesting, for example, to look for the potential interactions between hPrp8p and p68 or between p68 and components of the U5.U4/U6 tri-snRNPs.
As outlined in the introduction, the U1-U6 RNA switch at the 5′splice site is a dynamic process. The process involves multiple RNA annealing and unwinding activities. Staley and Guthrie reported that a yeast DEAD-box family protein, Prp28p, is involved in mediating the RNA switch at the 5′ss (41). Further, Chen and colleagues have shown that specific mutations that alter the U1-C or U1 snRNA can bypass the requirements for the functions of Prp28p. Thus, the authors suggested that Prp28p might actually work to counteract the stabilizing effects of U1-C on the U1-5′ss duplex (5). The human homologue of Prp28p, a U5 snRNP-specific 100-kDa protein (U5-100K), has also been identified (43). Recently, Ismaili and colleagues reported that the U5-100K cross-linked to the 5′ss after the U5.U4/U6 tri-snRNPs addition. Thus, the authors believed that the U5-100K may act as a helicase to unwind the U1-5′ss duplex (15). Both p68 RNA helicase and the U5-100K potentially function to destabilize the interaction between the U1 snRNP and the 5′ss. An interesting question is whether there is a functional relationship between the p68 RNA helicase and the U5-100K. One possible scenario is that the U5-100K and p68 RNA helicase may act cooperatively in promoting the U1-U6 switch at the 5′ss. Prp28p/U5-100K may act on the protein factors that stabilize the U1-5′ss duplex. On the other hand, p68 RNA helicase may act directly on the RNA duplex. It has been demonstrated that two DEAD/DEXH-box proteins, Prp5p and Sub2p, act cooperatively to promote the stable U2-pre-mRNA interactions (47). Alternatively, p68 RNA helicase and the U5-100K may function at different time points in the spliceosome assembly pathway.
p68 RNA helicase is highly conserved throughout evolution. The human protein shows 98% sequence identity with mouse p68 (19). The Saccharomyces cerevisiae and Schizosaccharomyces pombe p68 homolog, Dbp2p, shares 55% identity with the human protein (14). p68 RNA helicase functions in the pre-mRNA splicing process in HeLa nuclear extracts. It is conceivable that Dbp2p may also play important roles in the pre-mRNA splicing process in yeast. Indeed, the experiments with S. cerevisiae showed that depletion of DBP2 leads to a defect in splicing of the RP28 gene. However, several other intron-containing genes are spliced normally in the DBP2 null strain (3). There are three possible explanations for the observations. (i) Unlike p68 RNA helicase, Dbp2p may not function in the splicing. The observed defects in splicing of the RP28 gene in the DBP2 null strain may not be due to the functional deficiency of Dbp2p in the spliceosome. (ii) Unlike p68 RNA helicase, Dbp2p may function only in a specific intron, such as RP28, or a subset of introns. Therefore, the splicing defects with other introns may not be apparent in a DBP2 null strain. The amino acid sequences in the nonconserved C and N termini between p68 and Dbp2p differ significantly. It would therefore not be surprising if p68 behaved differently from Dbp2p in the spliceosome. (iii) Dbp2p may be involved in splicing in vivo. However, it may behave as an efficiency factor rather than an essential factor. Similar phenomena were observed with another splicing factor, Sub2p. Depletion or inactivation of SUB2 inhibits production of some mRNAs, such as RPL32, while expression of other intron-containing genes, such as the RP51A gene, is not affected (23).
Acknowledgments
I thank Frances Fuller-Pace for providing hybridoma cells for the antibody PAb204 and Roger Bridgeman for antibody PAb204 production. I am also grateful to D. L. Peterson for the HCV-NS3 expression vector. I thank Chris W. J. Smith, Mariano A. Garcia-Blanco, Jenny Yang, and Jan Szechi for detailed critical comments on the manuscript.
REFERENCES
- 1.Abovich, N., and M. Rosbash. 1997. Cross-intron bridging interactions in the yeast commitment complex are conserved in mammals. Cell 89:403-412. [DOI] [PubMed] [Google Scholar]
- 2.Ares, M., Jr., and B. Weiser. 1995. Rearrangement of snRNA structure during assembly and function of the spliceosome. Prog. Nucleic Acid Res. Mol. Biol. 50:131-159. [DOI] [PubMed] [Google Scholar]
- 3.Barta, I., and R. Iggo. 1995. Autoregulation of expression of the yeast Dbp2p "DEAD-box' protein is mediated by sequences in the conserved DBP2 intron. EMBO J. 14:3800-3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chabot, B., M. Blanchette, I. Lapierre, and H. La Branche. 1997. An intron element modulating 5′ splice site selection in the hnRNP A1 pre-mRNA interacts with hnRNP A1. Mol. Cell. Biol. 17:1776-1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, J. Y., L. Stands, J. P. Staley, R. R. Jackups, Jr., L. J. Latus, and T. H. Chang. 2001. Specific alterations of U1-C protein or U1 small nuclear RNA can eliminate the requirement of Prp28p, an essential DEAD box splicing factor. Mol. Cell 7:227-232. [DOI] [PubMed] [Google Scholar]
- 6.Crawford, L., K. Leppard, D. Lane, and E. Harlow. 1982. Cellular proteins reactive with monoclonal antibodies directed against simian virus 40 T-antigen. J. Virol. 42:612-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de la Cruz, J., D. Kressler, and P. Linder. 1999. Unwinding RNA in Saccharomyces cerevisiae: DEAD-box proteins and related families. Trends Biochem. Sci. 24:192-198. [DOI] [PubMed] [Google Scholar]
- 8.Endoh, H., K. Maruyama, Y. Masuhiro, Y. Kobayashi, M. Goto, H. Tai, J. Yanagisawa, D. Metzger, S. Hashimoto, and S. Kato. 1999. Purification and identification of p68 RNA helicase acting as a transcriptional coactivator specific for the activation function 1 of human estrogen receptor alpha. Mol. Cell. Biol. 19:5363-5372. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9.Forch, P., O. Puig, N. Kedersha, C. Martinez, S. Granneman, B. Seraphin, P. Anderson, and J. Valcarcel. 2000. The apoptosis-promoting factor TIA-1 is a regulator of alternative pre-mRNA splicing. Mol. Cell 6:1089-1098. [DOI] [PubMed] [Google Scholar]
- 10.Hamm, J., and A. I. Lamond. 1998. Spliceosome assembly: the unwinding role of DEAD-box proteins. Curr. Biol. 8:R532-R534. [DOI] [PubMed] [Google Scholar]
- 11.Hastings, M. L., and A. R. Krainer. 2001. Pre-mRNA splicing in the new millennium. Curr. Opin. Cell. Biol. 13:302-309. [DOI] [PubMed] [Google Scholar]
- 12.He, F., and A. Jacobson. 1995. Identification of a novel component of the nonsense-mediated mRNA decay pathway by use of an interacting protein screen. Genes Dev. 9:437-454. [DOI] [PubMed] [Google Scholar]
- 13.Hodges, P. E., and J. D. Beggs. 1994. RNA splicing. U2 fulfils a commitment. Curr. Biol. 4:264-267. [DOI] [PubMed] [Google Scholar]
- 13a.Huang, Y. L., and Z.-R. Liu. 2002. The ATPase, RNA unwinding, and RNA binding activities of recombinant p68 RNA helicase. J. Biol. Chem. 277:12810-12815. [DOI] [PubMed] [Google Scholar]
- 14.Iggo, R. D., D. J. Jamieson, S. A. MacNeill, J. Southgate, J. McPheat, and D. P. Lane. 1991. p68 RNA helicase: identification of a nucleolar form and cloning of related genes containing a conserved intron in yeasts. Mol. Cell. Biol. 11:1326-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ismaili, N., M. Sha, E. H. Gustafson, and M. M. Konarska. 2001. The 100-kDa U5 snRNP protein (hPrp28p) contacts the 5′ splice site through its ATPase site. RNA 7:182-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jankowsky, E., C. H. Gross, S. Shuman, and A. M. Pyle. 2001. Active disruption of an RNA-protein interaction by a DExH/D RNA helicase. Science 291:121-125. [DOI] [PubMed] [Google Scholar]
- 17.Jost, J. P., S. Schwarz, D. Hess, H. Angliker, F. V. Fuller-Pace, H. Stahl, S. Thiry, and M. Siegmann. 1999. A chicken embryo protein related to the mammalian DEAD box protein p68 is tightly associated with the highly purified protein-RNA complex of 5-MeC-DNA glycosylase. Nucleic Acids Res. 27:3245-3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim, D. H., and J. J. Rossi. 1999. The first ATPase domain of the yeast 246-kDa protein is required for in vivo unwinding of the U4/U6 duplex. RNA 5:959-971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kitajima, Y., H. Yatsuki, R. Zhang, S. Matsuhashi, and K. Hori. 1994. A novel human homologue of a dead-box RNA helicase family. Biochem. Biophys. Res. Commun. 199:748-754. [DOI] [PubMed] [Google Scholar]
- 20.Kuhn, A. N., Z. Li, and D. A. Brow. 1999. Splicing factor Prp8 governs U4/U6 RNA unwinding during activation of the spliceosome. Mol. Cell 3:65-75. [DOI] [PubMed] [Google Scholar]
- 21.Laggerbauer, B., T. Achsel, and R. Luhrmann. 1998. The human U5-200kD DEXH-box protein unwinds U4/U6 RNA duplices in vitro. Proc. Natl. Acad. Sci. USA 95:4188-4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lane, D. P., and W. K. Hoeffler. 1980. SV40 large T shares an antigenic determinant with a cellular protein of molecular weight 68,000. Nature 288:167-170. [DOI] [PubMed] [Google Scholar]
- 23.Libri, D., N. Graziani, C. Saguez, and J. Boulay. 2001. Multiple roles for the yeast SUB2/yUAP56 gene in splicing. Genes Dev 15:36-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu, Z. R., B. Laggerbauer, R. Luhrmann, and C. W. Smith. 1997. Crosslinking of the U5 snRNP-specific 116-kDa protein to RNA hairpins that block step 2 of splicing. RNA 3:1207-1219. [PMC free article] [PubMed] [Google Scholar]
- 25.Liu, Z. R., B. Sargueil, and C. W. Smith. 1998. Detection of a novel ATP-dependent cross-linked protein at the 5′ splice site-U1 small nuclear RNA duplex by methylene blue-mediated photo-cross-linking. Mol. Cell. Biol. 18:6910-6920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu, Z. R., A. M. Wilkie, M. J. Clemens, and C. W. Smith. 1996. Detection of double-stranded RNA-protein interactions by methylene blue-mediated photo-crosslinking. RNA 2:611-621. [PMC free article] [PubMed] [Google Scholar]
- 27.Luking, A., U. Stahl, and U. Schmidt. 1998. The protein family of RNA helicases. Crit. Rev. Biochem. Mol. Biol. 33:259-296. [DOI] [PubMed] [Google Scholar]
- 28.Madhani, H. D., and C. Guthrie. 1994. Dynamic RNA-RNA interactions in the spliceosome. Annu. Rev. Genet. 28:1-26. [DOI] [PubMed] [Google Scholar]
- 29.Moore, M. J., C. C. Query, and P. A. Sharp. 1993. Splicing or precursors to messenger RNAs by the spliceosome, p. 303-507. In R. F. Gesteland and J. F. Atkins (ed.), RNA world. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 30.Murray, H. L., and K. A. Jarrell. 1999. Flipping the switch to an active spliceosome. Cell 96:599-602. [DOI] [PubMed] [Google Scholar]
- 31.Neubauer, G., A. King, J. Rappsilber, C. Calvio, M. Watson, P. Ajuh, J. Sleeman, A. Lamond, and M. Mann. 1998. Mass spectrometry and EST-database searching allows characterization of the multi-protein spliceosome complex. Nat. Genet. 20:46-50. [DOI] [PubMed] [Google Scholar]
- 32.Newman, A. 1998. RNA splicing. Curr. Biol. 8:R903-R905. [DOI] [PubMed] [Google Scholar]
- 33.Nilsen, T. W. 1998. RNA-RNA interactions in nuclear pre-mRNA splicing, p. 303-357. In R. W. Simon and M. Grunberg-Manago (ed.), RNA structure and function. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 34.Raghunathan, P. L., and C. Guthrie. 1998. RNA unwinding in U4/U6 snRNPs requires ATP hydrolysis and the DEIH-box splicing factor Brr2. Curr. Biol. 8:847-855. [DOI] [PubMed] [Google Scholar]
- 35.Reed, R. 2000. Mechanisms of fidelity in pre-mRNA splicing. Curr. Opin. Cell. Biol. 12:340-345. [DOI] [PubMed] [Google Scholar]
- 36.Schmid, S. R., and P. Linder. 1992. D-E-A-D protein family of putative RNA helicases. Mol. Microbiol. 6:283-291. [DOI] [PubMed] [Google Scholar]
- 37.Schwer, B. 2001. A new twist on RNA helicases: DExH/D box proteins as RNPases. Nat. Struct. Biol. 8:113-116. [DOI] [PubMed] [Google Scholar]
- 38.Schwer, B., and T. Meszaros. 2000. RNA helicase dynamics in pre-mRNA splicing. EMBO J. 19:6582-6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sharp, P. A. 1994. Split genes and RNA splicing. Cell 77:805-815. [DOI] [PubMed] [Google Scholar]
- 40.Staley, J. P., and C. Guthrie. 1998. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell 92:315-326. [DOI] [PubMed] [Google Scholar]
- 41.Staley, J. P., and C. Guthrie. 1999. An RNA switch at the 5′ splice site requires ATP and the DEAD box protein Prp28p. Mol. Cell 3:55-64. [DOI] [PubMed] [Google Scholar]
- 42.Stevenson, R. J., S. J. Hamilton, D. E. MacCallum, P. A. Hall, and F. V. Fuller-Pace. 1998. Expression of the "dead box' RNA helicase p68 is developmentally and growth regulated and correlates with organ differentiation/maturation in the fetus. J. Pathol. 184:351-359. [DOI] [PubMed] [Google Scholar]
- 43.Teigelkamp, S., C. Mundt, T. Achsel, C. L. Will, and R. Luhrmann. 1997. The human U5 snRNP-specific 100-kD protein is an RS domain-containing, putative RNA helicase with significant homology to the yeast splicing factor Prp28p. RNA 3:1313-1326. [PMC free article] [PubMed] [Google Scholar]
- 44.Wang, Y., J. D. Wagner, and C. Guthrie. 1998. The DEAH-box splicing factor Prp16 unwinds RNA duplexes in vitro. Curr. Biol. 8:441-451. [DOI] [PubMed] [Google Scholar]
- 45.Wassarman, D. A., and J. A. Steitz. 1992. Interactions of small nuclear RNA's with precursor messenger RNA during in vitro splicing. Science 257:1918-1925. [DOI] [PubMed] [Google Scholar]
- 46.Will, C. L., and R. Luhrmann. 2001. Molecular biology. RNP remodeling with DExH/D boxes. Science 291:1916-1917. [DOI] [PubMed] [Google Scholar]
- 47.Zhang, M., and M. R. Green. 2001. Identification and characterization of yUAP/Sub2p, a yeast homolog of the essential human pre-mRNA splicing factor hUAP56. Genes Dev. 15:30-35. [DOI] [PMC free article] [PubMed] [Google Scholar]