Abstract
The C-terminal binding protein (CtBP) family of proteins has been linked to multiple biological processes through their association with numerous transcription factors. We generated mice harboring mutations in both Ctbp1 and Ctbp2 to address the in vivo function of CtBPs during vertebrate development. Ctbp1 mutant mice are small but viable and fertile, whereas Ctbp2-null mice show defects in axial patterning and die by E10.5 due to aberrant extraembryonic development. Mice harboring various combinations of Ctbp1 and Ctbp2 mutant alleles exhibit dosage-sensitive defects in a wide range of developmental processes. The strong genetic interaction, as well as transcription assays with CtBP-deficient cells, indicates that CtBPs have overlapping roles in regulating gene expression. We suggest that the observed phenotypes reflect the large number of transcription factors whose activities are compromised in the absence of CtBP.
Tightly regulated gene expression is critical for correct embryonic development and adult growth. One recent advance in the field of transcriptional regulation is the identification and characterization of non-DNA-binding transcriptional coregulators. One member of this class of regulatory factors is C-terminal binding protein (CtBP; reviewed in reference 45). Originally identified based on its ability to bind the C terminus of the E1A oncoprotein (34), CtBP has subsequently been found in complex with several known DNA-binding transcription factors that participate in a wide variety of developmental and adult biological pathways and processes. These include Wnt and BMP/TGFβ signaling (2, 17, 24), the action of GATA factors (5), cell-cell adhesion and apoptosis (13), myogenesis and vascularization (6, 21, 22, 52), and segmentation in flies (25, 26, 29). While the exact function of CtBP is unknown, it is quite clear that these proteins act as transcriptional coregulators. Specifically, when targeted to active promoters, either directly or indirectly, CtBP can dramatically reduce the level of transcription (27, 44). In addition to their association with sequence-specific DNA binding proteins, CtBPs have also been shown to interact with polycomb group proteins and thus may also be a component of chromatin remodeling machinery and transcriptional repression (38).
Consistent with the fact that CtBP binds a diverse array of transcriptional regulators, CtBP appears to have multiple biological roles during development. Genetic analysis of dCtBP in flies indicates that CtBP functions early in development, as it interacts physically and genetically with snail, kruppel, knirps, and hairy (25-27, 29). It also appears to have later functions, as it has been shown to interact with the transcriptional regulator Tramtrack69 (48). In addition to the identified functions in Drosophila melanogaster, CtBP has also been implicated in human development. Mutations in human TGIF (which maps to the HPE4 locus) lead to holoprosencephaly, a condition resulting from defects in craniofacial development. TGIF is a TALE homeodomain protein that can interact with transforming growth factor β-activated SMAD complexes and repress transcription (49, 50). One of the defined HPE4 alleles encodes a protein with a single amino acid substitution in a consensus binding site for CtBP (12, 24), suggesting that the CtBP-dependent branch of TGIF-mediated transcriptional repression is required for proper craniofacial development in humans. Importantly, the full repressor activity of TGIF is dependent on its ability to bind both CtBP and histone deacetylase (HDAC).
While the mechanism by which CtBP modulates gene expression is unclear, there appear to be both HDAC-dependent and -independent aspects to CtBP-mediated repression. First, it has been documented that CtBP can interact with multiple HDACs both in vivo and in vitro (6, 40, 41, 52). In support of this observation, CtBP-dependent repression can be blocked by the addition of trichostatin A, a specific inhibitor of HDACs (30). However, work by others has shown that CtBP-dependent repression can still occur in the presence of trichostatin A (18, 40) and that there is no detectable HDAC activity associated with CtBP (27). This suggests that CtBP may utilize multiple mechanisms to regulate transcription. Finally, it has also been proposed that CtBP can regulate the ability of other transcriptional repressors to interact with HDAC (27).
Two lines of experimental evidence indicate that the two CtBP proteins present in vertebrates, CtBP1 and CtBP2, may also have cytoplasmic functions and enzymatic activities that participate in transcriptional regulation. First, CtBP1 (also called brefeldin A-ribosylated substrate [BARS]) was identified as a cellular protein that becomes ADP ribosylated following treatment of cells with brefeldin A and can prevent the deleterious effects of brefeldin A on Golgi membrane architecture (39). This work was followed up by the observation that CtBP1/BARS can catalyze the transfer of an acyl group from acyl-coenzyme A to lysophosphatidic acid, converting it to phosphatidic acid (47). The second line of evidence comes from the identification of an isoform of CtBP2 as a component of synaptic ribbons (36). This protein isoform, called Ribeye, contains a unique N-terminal domain and a C-terminal domain that is identical to CtBP2. This study also showed that the CtBP-like portion of Ribeye is capable of directly binding NAD+. This observation is not unexpected, since CtBP proteins share significant sequence homology with NAD+-dependent 2-hydroxy acid dehydrogenases (34). Currently, it is unclear how or if CtBP might utilize these enzymatic activities to regulate gene expression. However, it has been proposed that tethering CtBP to specific DNA elements allows the creation of localized, discrete chemical environments that either favor or inhibit other modifying enzymes such as HDAC (27). Conversely, it is possible that there is a bifurcation of CtBP activity and function such that the enzyme activities are utilized solely for cytoplasmic functions while other physical or chemical properties are utilized in transcriptional regulation.
Through a combination of gene trapping and gene targeting in embryonic stem (ES) cells, we have generated mice harboring mutations in Ctbp1 and/or Ctbp2. Analysis of these mice indicates that Ctbp1 and Ctbp2 play both unique and redundant roles during mouse embryogenesis and are necessary for multiple developmental programs. Based on transcription assays in Ctbp1 Ctbp2-null cells, we predict that the phenotypes observed in Ctbp mutants are the result of deregulated gene expression.
MATERIALS AND METHODS
Mutation and cloning of Ctbp1 and Ctbp2.
For Ctbp2, AK7 ES cells were infected with the retroviral gene trap vector ROSA-βgeo∗bpA and selected in G418 as previously described (7, 19). Individual colonies were isolated, expanded, and lysed to collect RNA. Trapped genes were cloned by using 5′ rapid amplification of cDNA ends (RACE) as previously described (8, 15). A full-length Ctbp2 cDNA was obtained from a mouse brain cDNA library (Stratagene) using the 5′ RACE product as a probe. ES cells harboring the mutant Ctbp2 locus were used to generate mice by standard blastocyst injection. Mice were genotyped by Southern blotting with the original 5′ RACE product, which recognizes a polymorphism caused by the gene trap insertion (data not shown).
A portion of the cDNA for Ctbp1 was amplified and cloned by reverse transcription-PCR (RT-PCR) with primers that were based on the sequence of a mouse expressed sequence tag (EST) (accession number mw81d03.r1). The PCR product was used to isolate a full-length cDNA from a mouse brain library (Stratagene). Genomic clones for Ctbp1 were isolated from a 129S4 genomic library and used to generate a targeting vector. A 1-kb fragment of the genomic locus containing the exon encoding amino acids 55 to 102 was replaced with the selectable marker PGKneobpA. ES cells were electroporated with linearized vector and selected for 10 days in 300 μg of G418 (total drug) per ml. PCR was used to identify ES colonies with a targeted allele. Positive colonies were tested by Southern blotting to verify correct targeting. Two independent ES clones were used to generate mice by standard blastocyst injection. Mice were genotyped by PCR using primers CtBP-8 (5′-CCCCAGCTGACTTGATGTCG-3), CtBP-T3 (5′-GAAGTACCAGTACAGGGGACG-3′), and Neo3R3 (5′-GTTATCGCCGCTCCCGATTCG-3′). The following primers were used for RT-PCR to clone a portion of Ctbp1 for library screening and to detect splicing around PGKneobpA: ctbp1-1 (5′-CTCGGATCCCGCGCAGTAAGAC) and ctbp1-2 (5′-GATTGCTGCCGGTCGACATTGCAC).
To detect endogenous CtBP1 and CtBP2, cells or whole embryonic day 10.5 (E10.5) embryos were lysed in radioimmunoprecipitation assay buffer, and 50 μg of protein lysate was resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to nitrocellulose, and subjected to Western blotting with monoclonal antibodies (MAbs) to mouse CtBP1 and CtBP2 per the manufacturer's specifications (Transduction Labs). Primary antibodies were detected with horseradish peroxidase-conjugated goat anti-mouse immunoglobulin (1:2,500 dilution; Amersham Pharmacia). Filters were reprobed to detect cortactin with the MAb 4F11 (J. T. Parsons, University of Virginia).
Staining and histology.
For detection of the gene trap reporter, embryos from timed mating were isolated, fixed, and stained with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) as previously described (7). For histology, embryos were isolated, fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS), dehydrated in ethanol, cleared in Histoclear (National Diagnostics), embedded in paraffin, sectioned, and stained with either nuclear fast red (to counterstain X-Gal) or Harris' hematoxylin and eosin (H&E). In situ hybridization, immunohistochemistry, and skeleton preparations were carried out as described previously (16). T was detected with a 300-bp probe specific to the 3′ untranslated region (UTR). Ctbp1 was detected with a 650-bp probe corresponding to a portion of the 3′ UTR that does not have sequence homology to Ctbp2. Platelet endothelial cell adhesion molecule (PECAM) was detected with an anti-PECAM-1 MAb (Pharmingen) and an ABC rat immunoglobulin G kit (Vector). CtBP1 and CtBP2 were detected by using a 1:500 dilution of either anti-CtBP1 or anti-CtBP2 MAbs (Transduction Labs) and the ABC mouse immunoglobulin G kit (Vector). All mice and embryos described here were on a mixed C57B6/129S4 background.
Two-hybrid screens and in vitro binding.
Two-hybrid screening was used to identify proteins that interact with CtBP2. Briefly, the full-length CtBP2 cDNA was cloned into the bait plasmid pBTM116 and subsequently transformed into yeast strain L40 (46). Bait-containing yeasts were tested for transactivation and subsequently transformed with a mixed cDNA library derived from E9.5 and E10.5 mouse embryos (46). Yeasts harboring putative CtBP2 binding proteins were selected on plates lacking histidine. cDNA inserts were isolated by PCR and identified by sequence analysis. Two of the positive isolates represented independent clones encoding the mouse homolog of human ZNF219 (33). Interactions observed in yeast were verified by in vitro binding assays. The cDNA insert encoding a portion of mouse Znf219 (mZnf219) was cloned into pGex3X, and the resulting glutathione S-transferase (GST)-Znf219 fusion protein purified from Escherichia coli BL21 by using glutathione-Sepharose (Pharmacia). Purified GST or GST-Znf219 (3 to 5 μg) bound to glutathione-Sepharose was added to 1 mg of total cell lysate (in a volume of 1 ml) derived from NIH 3T3 cells expressing epitope-tagged CtBP1 or CtBP2. Protein complexes were washed four times with radioimmunoprecipitation assay buffer, resolved by SDS-PAGE, transferred to nitrocellulose, and assayed by Western blotting using MAb KT3.
Cell lines and in vitro expression of CtBP.
Epitope-tagged CtBP1 and CtBP2 was generated by amplifying the coding sequences for Ctbp1 and Ctbp2 by PCR and cloning the resultant products into the epitope tag vector pBSctag (14, 35). Following sequencing, DNA cassettes containing Ctbp1 or Ctbp2 and the epitope tag were shuttled into pBabe-puro. The epitope tag consists of the final 11 amino acids of simian virus 40 large T antigen and is recognized by the MAb KT3 (23).
Ctbp mutant cell lines were derived from E9.25 embryos as follows. Embryos from timed mating of Ctbp1+/− Ctbp2+/− mice were isolated in PBS containing 5% fetal bovine serum, treated briefly with trypsin, dissociated by repeated pipetting, and plated onto gelatin-coated plates. Cells were immortalized via infection with a retrovirus that transduces large T antigen (3). To reexpress CtBP1 and CtBP2, Ctbp1−/− Ctbp2−/− cells were infected with pBabe-puro, pBabe-puro-CtBP1ctag, or pBabe-puro-CtBP2ctag and selected in 2 μg of puromycin (Calbiochem) per ml for 10 days. Resistant cells were pooled and screened for expression of epitope-tagged CtBP1 or CtBP2 protein by Western blotting. All cells were maintained in DMEM supplemented with 10% fetal bovine serum (HyClone), 2 mM l-glutamine, and antibiotics.
Transcription assays.
Cells were transiently transfected with the 0.5 μg of each of the following plasmids: the Gal4 reporter (4×Gal414Dluc), the β-galactosidase reporter (PGKβgalbpA) to control for transfection efficiency, and either pSPGal, pSPGal-Znf219, or pSPGal-Znf219Δ. The luciferase reporter, 4×Gal414DLuc, contains four Gal4 binding sites, the 14D promoter, and the gene for luciferase. To generate pSPGalZnf219, the cDNA fragment of Znf219 isolated from the two-hybrid screen was amplified by PCR and cloned into pSPGal such that the reading frame was contiguous with that of the Gal4 DNA-binding domain (DBD). pSPGal-Znf219Δ encodes a variant of Gal4-Znf219 in which the CtBP binding site PLDLSLR has been changed to ALQAALR. Transfected cells were lysed and assayed for luciferase and β-galactosidase activity as described previously (20, 27). Data shown for these assays are the averages for three independent experiments performed in duplicate. In these assays, values for β-galactosidase activity were corrected for the amount of endogenous activity that results from the β-geo∗ gene trap reporter.
RESULTS
Mutation of Ctbp1 and Ctbp2.
A trapped allele of mouse Ctbp2 was isolated as a part of a continuing gene trap screen to identify mutations that disrupt growth and patterning of the vertebrate embryo (http://www.fhcrc.org/labs/soriano/trap.html). As part of this ongoing screen, this mutation has been designated ROSA-61. Based on the sequence of the 5′ RACE product and cDNA clones, the retroviral insertion is 5′ of the putative initiation codon (Fig. 1A). Cloning and RT-PCR analysis indicates that the Ctbp2 message is alternatively spliced at the 5′ end (Fig. 1A and data not shown). This alternative splicing deletes the first putative coding exon and ATG. However, a second potential start codon is present in the second coding exon.
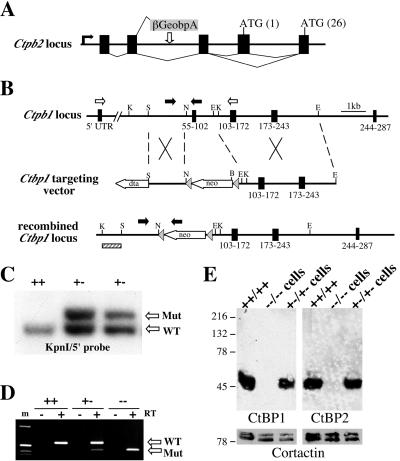
FIG. 1.
Disruption of mouse Ctbp1 and Ctbp2. (A) Schematic (not to scale) of the putative exon (black boxes) structure of the genomic locus, splicing pattern, and relative integration site of the βgeo∗bpA gene trap reporter relative to Ctbp2. (B) Schematic of the targeted mutation of Ctbp1 in ES cells. Grey arrowheads show the position of loxP sites, coding exons are depicted by black boxes, and the amino acids they encode are indicated below each exon; black arrowheads correspond to primers for PCR genotyping, and white arrowheads correspond to primers used for RT-PCR (D). (C) Southern blot of DNA from ES cells with a correctly targeted allele of Ctbp1. Genomic DNA from wild-type or targeted ES cells was subjected to Southern blotting following KpnI digestion using the probe indicated in panel B (hatched box). (D) Targeting causes a deletion in Ctbp1 mRNA. RNA was isolated from the brains of wild-type (++), heterozygous (+-), and homozygous mutant (--) Ctbp1 mice and subjected to RT-PCR using primers specific to the 5′ UTR and exon immediately 3′ of the deletion. (E) Protein lysates derived from either wild-type E10.5 embryos, Ctbp1+/− Ctbp2+/− cells, or Ctbp1−/− Ctbp2−/− cells were assayed by Western blotting to detect CtBP1, CtBP2, and Ribeye. Membranes were reprobed with an antibody to the cytoskeletal protein cortactin as a loading control.
While cloning Ctbp2, we identified an EST for Ctbp1 and used RT-PCR and library screening to clone the Ctbp1 cDNA (see Materials and Methods). The pattern of alternative exclusion of the first coding exon that is seen in the Ctbp2 message is also found in the message for Ctbp1 (data not shown). In order to understand the in vivo relationship between these two highly related and similarly expressed genes (see Fig. 2), Ctbp1 was disrupted in ES cells by homologous recombination. Briefly, a genomic clone containing the exons that encode amino acids 55 to 245 was used to generate a targeting vector in which the exon encoding amino acids 55 to 102 was replaced with a PGKneobpA selection cassette (Fig. 1B). Two independently derived ES lines harboring correctly targeted alleles (Fig. 1C) were passed through the male germ line, and both produced identical phenotypes. RT-PCR and sequence analysis of the Ctbp1 message from mutant mice suggests that there is splicing around the PGKneobpA cassette (Fig. 1D). However, the mRNA from the mutant allele exhibits a deletion of the expected size, and sequencing of this PCR product indicates that the exon encoding amino acids 55 to 102 is deleted and the reading frame is disrupted (Fig. 1D and data not shown). Furthermore, the deleted amino acids comprise a portion of the proposed domain through which CtBP binds ligands (4) and should create a functional null allele. The fact that there is no heterozygous phenotype associated with the targeted allele suggests that if the first 55 amino acids of CtBP1 are expressed, this does not result in the production of a dominant-negative protein.
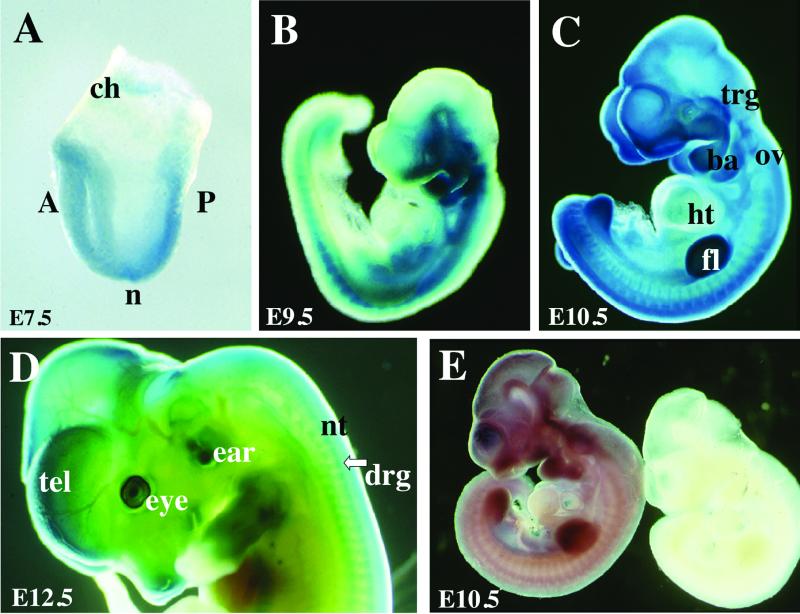
FIG. 2.
Expression of Ctbp1 and Ctbp2. (A to D) Heterozygous embryos were isolated at E7.5 (A), E9.5 (B), E10.5 (C), and E12.5 (D) of development and stained with X-Gal to detect expression of the βgeo∗bpA gene trap reporter (blue color). (E) Wild-type E10.5 embryos were subjected to RNA in situ hybridization using sense (right) and antisense (left) probes to the 3′ UTR of Ctbp1. ch, chorion; A, anterior; P, posterior; n, node; trg, trigeminal ganglion; ba, branchial arch; ov, otic vesicle; ht, heart; fl, forelimb bud; tel, telencephelon; nt, neural tube; drg, dorsal root ganglion.
Western blot analysis was used to verify that these mutations eliminate CtBP1 and CtBP2 production. Whole-cell protein lysates derived from either wild-type E10.5 embryos, Ctbp1+/− Ctbp2+/− cells, or Ctbp1−/− Ctbp2−/− cells were resolved by SDS-PAGE, transferred to nitrocellulose, and probed with antibodies to CtBP1 or CtBP2 (Fig. 1E). To verify equal protein loading, membranes were reprobed to detect the cytoskeletal protein cortactin (Fig. 1E, lower panels). Results from these experiments demonstrate that the targeted mutation and gene trap insertion eliminate expression of CtBP1 and CtBP2, respectively.
In other vertebrate species (rat, human, and cow), the Ribeye protein is expressed from the CtBP2 locus (36). It should be noted that there is currently no sequence available for mouse Ribeye and that no mouse EST spanning the junction of the unique N-terminal domain and the CtBP2 domain has been deposited. While CtBP2 is readily detected in the Ctbp1+/− Ctbp2+/− cell lysate and the wild-type embryo lysate, no protein of the predicted molecular mass for Ribeye (approximately 120 kDa) is detected in either of these samples. These results are consistent with the observation that Ribeye, if expressed in mice, may be restricted to cells that contain synaptic ribbons, such as the retina (36). Therefore, because CtBP1 and CtBP2 appear to be the only family members in vertebrates and since Ribeye is not expressed in the early embryo, we believe that we have generated mutations that together eliminate all CtBP function during mouse embryonic development.
Ctbp1 and Ctbp2 exhibit widespread and overlapping expression during embryogenesis.
Based on the mechanism of action of gene trap insertions and because the βgeo∗ reporter contains β-galactosidase activity, X-Gal staining can be used to detect the spatiotemporal pattern of Ctbp2 expression in heterozygous embryos (7) (Fig. 2A to D). At E7.5, staining is detected throughout the embryo and in the developing chorion (Fig. 2A). At later stages of development (E9.5 to E12.5), expression is detected in the nervous system (dorsal root ganglia, trigeminal ganglia, and brain), otic vesicles and ears, eyes, pharyngeal arches, limbs, and muscles (Fig. 2B to D). Longer staining indicates that Ctbp2 expression is virtually ubiquitous, including low-level expression in the heart. To determine the expression pattern of Ctbp1, mice were subjected to RNA in situ hybridization using a probe to the 3′ UTR of the gene (Fig. 2E). Like Ctbp2, Ctbp1 is widely expressed in the embryo. The coincident expression of Ctbp1 and Ctbp2 is consistent with previous reports (9).
Phenotypes of Ctbp1−/−and Ctbp2−/− mice.
Ctbp1-null animals are recovered from matings between heterozygous parents in normal Mendelian proportion and are fertile. However, homozygous mice are typically 30% smaller than their wild-type and heterozygous littermates, and 23% (18 of 78) of the homozygous mutant mice die by postnatal day 20 (P20). In contrast to Ctbp1, mating of Ctbp2 heterozygous mice produces no homozygous mutant offspring, suggesting that this mutation leads to a recessive, embryonic lethal phenotype. To verify this, embryos from timed mating of heterozygous mice were isolated at various stages of development. Normal-appearing CtBP2 mutant embryos are recovered at E8.5 and E9.5. By E10.5, Ctbp2-null embryos are distinguished from their heterozygous and wild-type littermates by their small size, axial truncations, delayed neural development, and defects in heart morphogenesis (Fig. 3A and B).
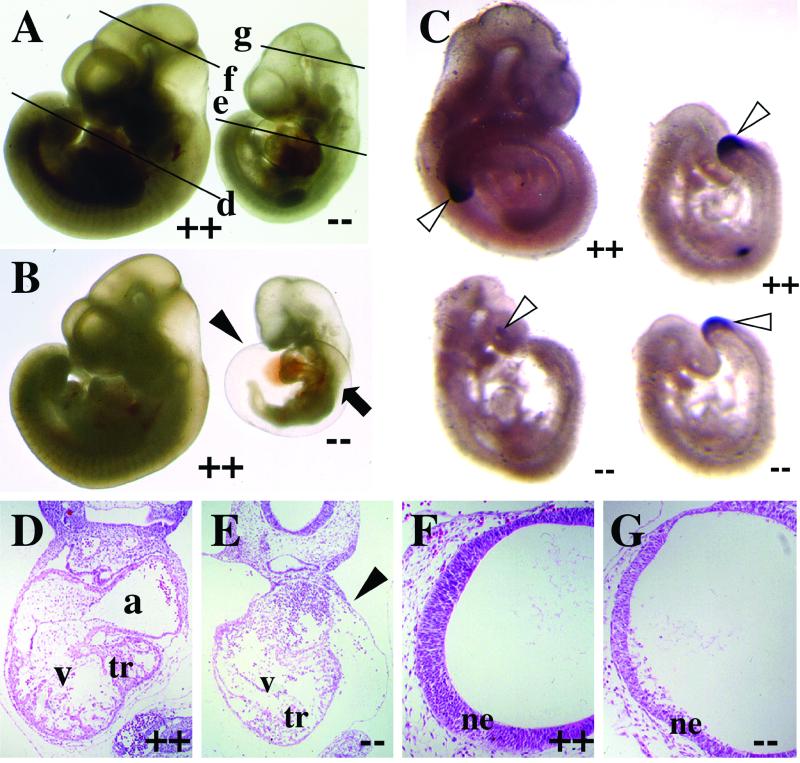
FIG. 3.
Ctbp2 is an essential gene. (A and B) Embryos from mating Ctbp2+/− mice were isolated at E10.25 and visualized in whole mount. Lines in panel A indicate the approximate plan of the sections shown in panels D to G. At this stage, homozygous mutant (--) embryos exhibit a dilated pericardium (black arrowhead) and axial truncations (black arrow). (C) Brachyury (T) expression in Ctbp2−/− embryos. E9.5 (right) and E10.25 (left) embryos were isolated from timed mating of Ctbp2+/− animals and subjected to in situ hybridization in order to visualize T expression in the tail bud (white arrowheads). (D to G) H&E-stained sections of wild-type (D and F) and mutant (E and G) embryos. Embryos exhibit defects in heart morphogenesis (E) and degeneration of the neural epithelium (ne) (G). tr, trabeculae; a, atrium; v, ventricle; black arrowhead, pericardium.
Some of the more severely affected Ctbp2-null embryos exhibit axial truncations (Fig. 3B). This phenotype is reminiscent of mutations in either Wnt genes or genes that encode components of the Wnt signal transduction pathways (10, 11, 28, 42). Because CtBP proteins have been implicated in the Wnt signaling pathways (2), we tested for normal expression of Brachyury (T), a direct target of Wnt3A (51), using whole-mount RNA in situ hybridization (Fig. 3C). Consistent with the phenotype, E10.25 mutant embryos that exhibit even mild axial defects also displayed considerably reduced levels of T expression (Fig. 3C, bottom left). Conversely, E9.5 embryos, which typically look normal, display both normal tail bud morphogenesis and T expression, although expression is reduced very slightly in Ctbp2 mutant embryos (Fig. 3C, compare top right and bottom right). These results suggest that some of the observed defects in these embryos may be due to altered embryonic patterning and that CtBP2 may be an in vivo regulator of Wnt-mediated gene expression.
In addition to axial defects, Ctbp2 mutant embryos also show aberrant heart formation and neural development (Fig. 3A and B). Specifically, all embryos have a dilated pericardium, and in more severely affected embryos, heart morphogenesis is halted at the heart tube stage (Fig. 3B). In addition to heart defects, mutant embryos also show alterations in formation of anterior neural structures. These typically include delayed development of the forebrain and midbrain (Fig. 3A and B). Histological analyses of the heart and neural tube support these observations. In mutant embryos, the heart wall is thinner and there is little trabeculation of the myocardial wall (Fig. 3D and E). In the case of anterior neural development, the neural epithelium is thinner and there is substantial degradation of this tissue (Fig. 3F and G).
Ctbp2 is required for extraembryonic vascularization.
Some aspects of the Ctbp2 mutant phenotype, such as a dilated pericardium, are indicative of extraembryonic defects. To address these possibilities, we analyzed the integrity of the yolk sac and placenta in Ctbp2 mutants and the expression of Ctbp2 in these tissues. Consistent with some of the embryonic defects, the yolk sacs of mutant embryos are typically devoid of the blood vessels that are obvious in wild-type littermates (Fig. 4A). To determine if Ctbp2 is expressed in the yolk sac, isolated yolk sacs were stained with X-Gal to detect the gene trap reporter. Consistent with the observed defects, Ctbp2 is expressed in the major vessels (Fig. 4B) and the mesothelium, the inner cell layer that gives rise to the yolk sac vascular plexus (Fig. 4C). To assess the extent of vascular defects, the yolk sac was stained to detect PECAM, a cell surface marker for vascular endothelial cells. Wild-type yolk sacs exhibit robust PECAM expression in a well-defined vascular plexus (Fig. 4D). In contrast, mutant yolk sacs typically exhibit minimal PECAM expression, and the vascular plexus is not well formed (Fig. 4E). These defects in vascularization appear to be restricted to the yolk sac, as both mutant and wild-type embryos display normal PECAM-positive vascularization (data not shown).
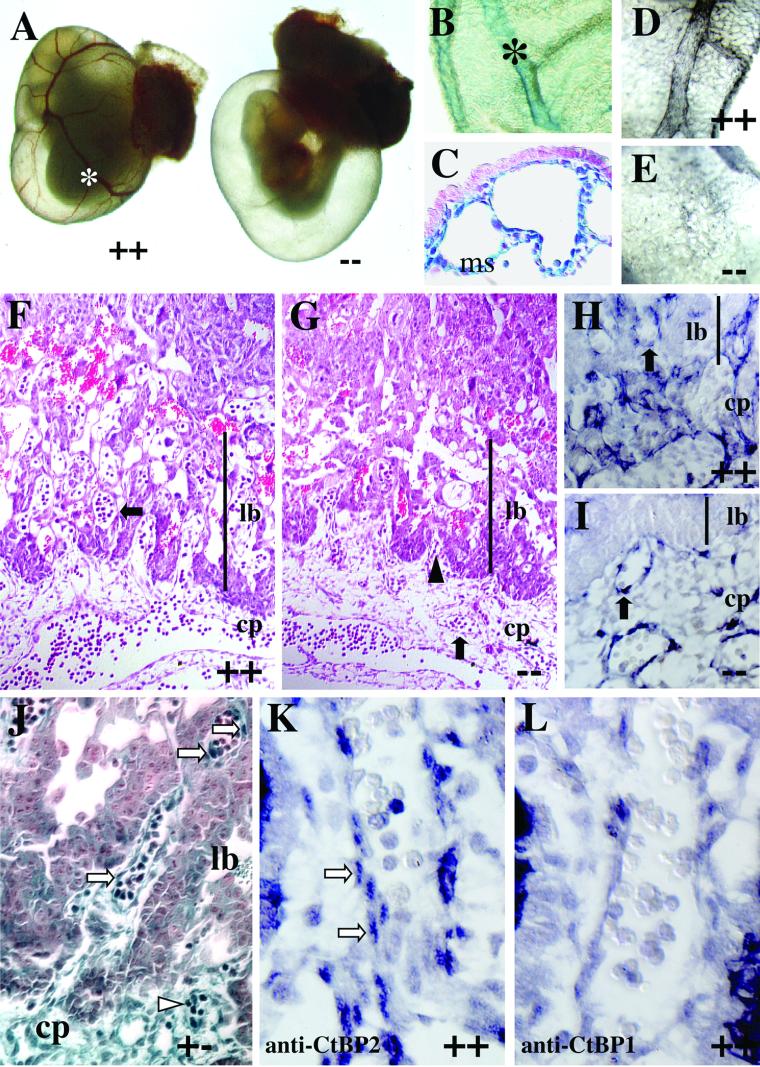
FIG.4.
Ctbp2-null embryos exhibit defects in extraembryonic development. (A) Whole-mount view of Ctbp2 wild-type (++) and null (--) embryos in the yolk sac at E10.5. (B and C) To assess the expression of Ctbp2 in the yolk sac, yolk sacs from heterozygous embryos were isolated, stained with X-Gal, and visualized in whole mount (B) or after sectioning (C). X-Gal staining is detected in the blood vessels (asterisk) and the mesothelial layer (ms). (D and E) Yolk sacs from wild-type (D) and mutant (E) embryos were stained with anti-PECAM antibodies to detect the integrity of vascular endothelium. (F to I) Placental defects in Ctbp2-null embryos. Placentas from wild-type (F and H) and mutant (G and I) embryos were isolated at E9.5, fixed, sectioned, and subjected to either H&E staining (F and G) or immunohistochemistry to detect PECAM (H and I). Black arrowheads indicate chorioallantoic folding, and black arrows indicate vasculature. Abbreviations: cp, chorioallantoic plate; lb, labyrinthine layer. (J to L) Placental expression of CtBP1 and CtBP2. (J) The placenta from a Ctbp2 heterozygous E10.5 embryo was stained with X-Gal, sectioned, and counterstained. βgeo∗ activity is detected in the chorioallantoic plate, vascular cells of the labyrinthine layer, and some blood cells (white arrowheads). White arrows indicate expression in some of the presumptive vascular endothelial cells. (K and L) Sections from a wild-type E9.5 placenta were stained to detect CtBP2 (K) and CtBP1 (L). White arrows indicate CtBP2-positive vascular cells.
To extend our analysis of potential extraembryonic defects, placentas from wild-type and mutant E9.5 embryos were isolated, sectioned, and analyzed by Harris' H&E staining. Results from this analysis suggest that there are defects in the formation of the labyrinthine layer of the placenta (Fig. 4F and G). Based on the presence of chorioallantoic folds, it appears that the initial steps of chorioallantoic branching occur normally in Ctbp2-null embryos. However, the subsequent steps of either chorioallantoic branching or vascularization of the labyrinthine layer are disrupted in Ctbp2 mutant embryos. This prediction is based on the observation that blood vessels are detected in the chorioallantoic plate but not in the region that should be occupied by the labyrinthine layer (Fig. 4F versus G). This observation is confirmed by staining wild-type and mutant E9.5 placentas for the expression of PECAM. In wild-type placentas, PECAM-positive endothelial cells are seen in the chorioallantoic plate and the labyrinthine layer (Fig. 4H). Conversely, in mutant placentas, PECAM-positive cells are seen in the chorioallantoic plate but not in the forming labyrinthine layer (Fig. 4I).
To verify that CtBP2 is expressed in the relevant tissues, placentas from Ctbp2 heterozygous embryos were isolated, stained with X-Gal, embedded, and sectioned (Fig. 4J). Robust staining is detected in some blood cells, the chorioallantoic plate, and the vasculature of both the chorioallantoic plate and the labyrinthine layer. Some diffuse, low-level staining is detected in the trophoblast cells. The placental defects observed in Ctbp2 mutants suggest that CtBP1 and CtBP2 are either functionally distinct or expressed in different cell types of the placenta. To address the latter possibility, sections from wild-type placentas were stained with antibodies specific for CtBP1 and CtBP2. Staining with CtBP2-specific antibodies generates a pattern similar to that obtained previously with X-Gal staining, showing CtBP2 expression in the presumptive vascular endothelial cells of the chorioallantoic plate (Fig. 4K). Conversely, little or no CtBP1 is detected in either of these tissues (Fig. 4L). Taken together, these observations suggest that CtBP2 deficiency causes defects in both yolk sac and placental vascularization that result in embryonic lethality.
Genetic interactions of Ctbp1 and Ctbp2.
To test if there is a genetic interaction between Ctbp1 and Ctbp2, we generated Ctbp1 Ctbp2 compound mutant mice by mating Ctbp1−/− Ctbp2+/+ males to Ctbp1+/+ Ctbp2+/− females. Compound heterozygous mice (Ctbp1+/− Ctbp2+/−) mice were recovered at the expected frequency, but they were typically smaller than their littermates, and 26% (33 of 126) of the Ctbp1+/− Ctbp2+/− pups died by P20. Mating of Ctbp1+/− Ctbp2+/− compound heterozygous mice to Ctbp1−/− Ctbp2+/+ mice produced no viable Ctbp1−/− Ctbp2+/− mice (0 of 47 pups born; 12 expected). Analysis of timed matings indicated that Ctbp1−/− Ctbp2+/− embryos typically die between E15.5 and E18.5 and exhibit a number of developmental defects. Gross analysis of embryos revealed that Ctbp1−/− Ctbp2+/− embryos are often small, have unusual curvature of the spine, and exhibit extensive hemorrhaging, an indication of vascular defects (Fig. 5A).
FIG. 5.
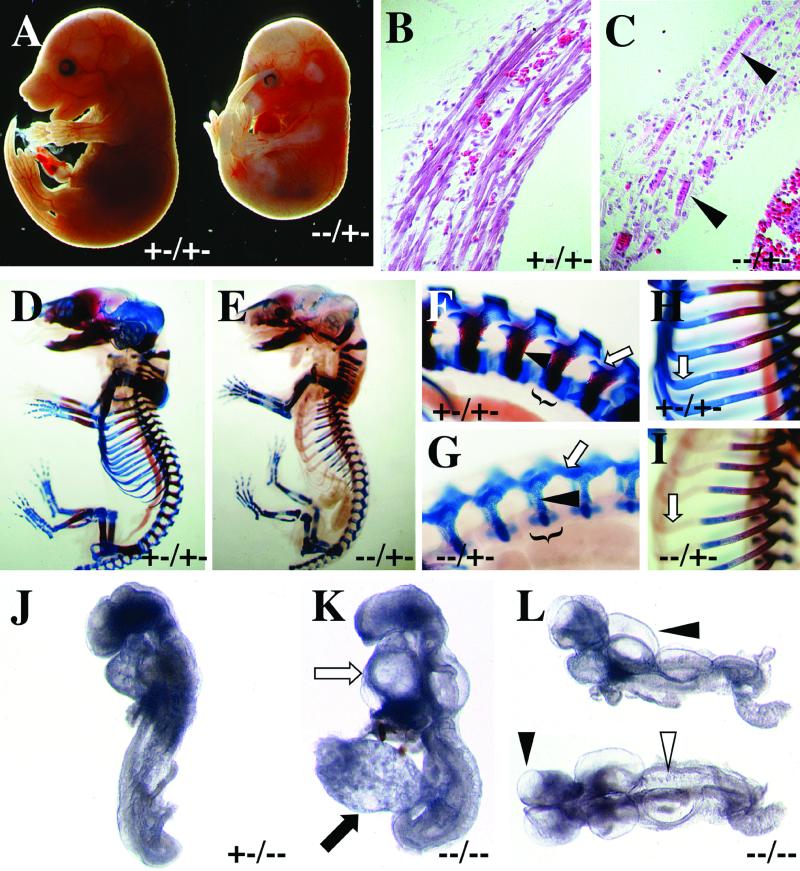
Genetic interaction between Ctbp1 and Ctbp2. (A to C) At E15.5, Ctbp1−/− Ctbp2+/− embryos exhibit a number of visible defects, including hemorrhaging, abnormal curvature, and reduced size (A). (B and C) Sections through the diaphragm of the embryos in panel A reveal defects in the formation of muscle (arrowheads). (D to I) Skeletal defects in Ctbp1−/− Ctbp2+/− embryos. E17.5 embryos were isolated and subjected to staining with Alcian blue and alizarin red to detect cartilage and ossified bone, respectively. Close-up views of the vertebral column (F and G) show defects in the formation of the vertebral bodies (brackets), arches, and lateral processes (white arrow) and an overall reduction in the amount of ossified bone (black arrowhead). Analysis of the ribs shows an overall defect in the staining of cartilage and ossified bone (H and I). (J to L) E9.25 embryos from Ctbp1+/− Ctbp2+/− crosses were isolated and photographed in whole mount. Reducing the dosage of Ctbp1 results in significant worsening of the Ctbp2−/− phenotype, such that Ctbp1+/− Ctbp2−/− embryos arrest at the time of turning and do not close the neural tube. (K and L) Embryos lacking both Ctbp1 and Ctbp2 exhibit multiple defects. In double mutant embryos, the heart has undergone minimal morphogenesis (white arrow), the allantois is occasionally bulbous (black arrow), there is blebbing of the neural ectoderm (black arrowheads), and the somites are irregular in both size and shape (white arrowhead). (L) Lateral (top) and dorsal (bottom) views of the same embryo.
In addition to the defects observed during gross examination, histological analysis indicates that Ctbp1−/− Ctbp2+/− mutant embryos display aberrant muscle development. These muscle defects appear to result from a block in the later stages of muscle differentiation, subsequent to the formation of myotubes and myofibers. In Ctbp1+/− Ctbp2+/− embryos, the diaphragm consists of well-defined muscle fibers (Fig. 5B). In contrast, the diaphragm of the Ctbp1−/− Ctbp2+/− embryo does not contain myofibers but contains short, multinucleated cells reminiscent of myotubes (Fig. 5C). Skeletal preparations indicate that Ctbp1−/− Ctbp2+/− mice also show abnormalities in the formation of multiple skeletal elements. Most notable is the dramatic deficiency in the generation of both cartilaginous and fully ossified skeletal elements, particularly in the ribs and vertebrae (Fig. 5D to I). In addition, there are defects in the formation of the spinal column, in which the lateral processes are fused (Fig. 5F and G) and do not grow to enclose the spinal cord (resulting in spina bifida occulta). It should be noted that the ribs do not stain efficiently with Alcian blue but their outlines can be seen clearly in Ctbp mutant embryos (Fig. 5E and I), suggesting that chondrocytes are essentially normal up to this point but experience a developmental delay at this stage. These results do not appear to be an artifact of staining, as they have been observed in multiple embryos and as the majority of skeletal elements exhibit normal staining.
We further analyzed the genetic interaction of Ctbp1 and Ctbp2 by characterizing Ctbp1+/− Ctbp2−/− and Ctbp1−/− Ctbp2−/− embryos. Consistent with the above results, reducing the dosage of Ctbp1 increased the severity of the Ctbp2-null phenotype (Fig. 5J). Most notable is the fact that Ctbp1+/− Ctbp2−/− embryos did not complete neural tube closure and arrest at the turning stage. This is in contrast to Ctbp1+/+ Ctbp2−/− embryos, which completed both neural tube closure and turning (Fig. 3). Taking this analysis one step further, complete elimination of Ctbp resulted in a more severe phenotype than that observed in Ctbp1+/− Ctbp2−/− embryos (Fig. 5K and L versus J). Ctbp1−/− Ctbp2−/− embryos arrested at the head fold stage. In addition, the allantois was occasionally bulbous and misshapen, the heart had undergone little morphogenesis (Fig. 5K), there were blebs or blisters in the neuroectoderm (Fig. 5L), and the somites were small and disorganized (Fig. 5L). Since not all of the Ctbp-null embryos exhibited notable defects in the allantois (compare Fig. 5K and L) or chorioallantoic fusion (data not shown), it appears that the observed embryonic phenotypes are not exclusively the result of defects in extraembryonic development. This gradation of phenotypes strongly suggests that Ctbp1 and Ctbp2 interact genetically and carry out at least some redundant roles during embryogenesis.
Transcription defects in Ctbp-null cells.
The phenotypes associated with the Ctbp mutations could be the result of defects in critical nuclear or cytoplasmic processes relating to gene expression or Golgi structure, respectively. To address these possibilities, cell lines derived from Ctbp1−/− Ctbp2−/− embryos (Ctbp-null cells) or Ctbp1+/− Ctbp2+/− embryos (control cells) were established and assayed for both Golgi structure and CtBP-dependent transcriptional repression. Analysis of Golgi architecture in both Ctbp-null cells and control cells did not reveal any differences between the two cell types (data not shown; Adam Linstedt, Carnegie-Mellon University, personal communication). In addition, the survival of CtBP-deficient embryos until E8.5 suggests that Golgi function is not catastrophically impaired, as severe defects in Golgi function, such as those induced by brefeldin A, tend to be deleterious to cell viability. This indicates that while CtBP proteins might play a significant role in regulating Golgi architecture (39, 47), the observed embryonic phenotypes do not appear to result from a loss of this activity. Unfortunately, the insertional mutation in Ctbp2 described here cannot be used to address the in vivo function of Ribeye. First, the lethality associated with this mutation occurs prior to the formation of synaptic ribbons in the retina. Second, it is not clear that this mutation would disrupt Ribeye because the insertion appears to be 5′ of the Ribeye promoter and coding sequence (36).
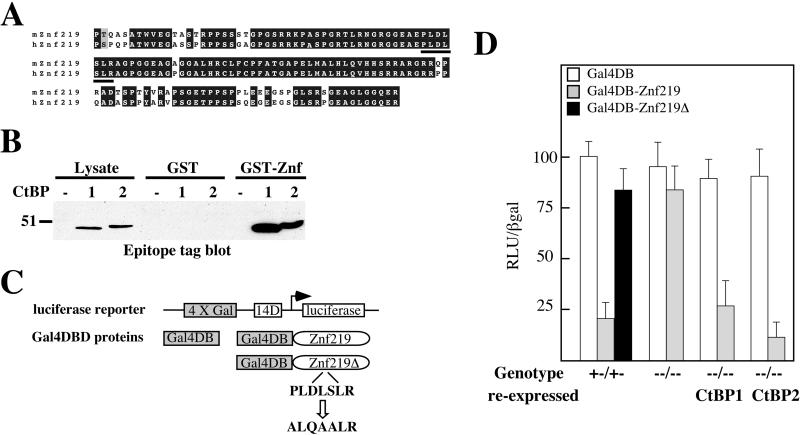
To test if the observed phenotypes are the result of impaired nuclear functions, we assayed the state of CtBP-dependent transcriptional repression using a heterologous transcription assay in Ctbp-null cells. Ctbp-null cells were transiently transfected with a Gal4-luciferase reporter and a vector that expresses either the Gal4 DBD or the Gal4 DBD fused to a portion of mouse Znf219, a zinc finger transcription factor that binds to CtBP. Znf219 was identified as a CtBP-binding protein by using two-hybrid interaction screening in yeast. This interaction was confirmed by GST pulldown assays (Fig. 6A and B; see also Materials and Methods). Both human and mouse Znf219 contain a perfect consensus binding site for CtBP consisting of the amino acid sequence PLDLSLR (Fig. 6A). When transfected into Ctbp1+/− Ctbp2+/− control cells, Gal4-Znf219 effectively repressed expression of the luciferase reporter (Fig. 6C and D). This repressor activity appears to be mediated by CtBP, since mutation of the putative CtBP-binding site in Znf219 eliminated repressor function in this assay (Gal4-Znf219Δ) (Fig. 6C and D). When transfected into Ctbp1−/− Ctbp2−/− cells, Gal4-Znf219 showed dramatically impaired repressor function. Importantly, reexpression of either CtBP1 or CtBP2 in Ctbp-null cells restored Znf219 repressor activity. The data suggest that these mutations eliminate CtBP-dependent mechanisms of transcriptional repression. In addition, these data also support the hypothesis that the developmental defects observed in Ctbp-null embryos result from the impaired activity of transcription factors that require CtBPs for full in vivo function.
FIG. 6.
Transcriptional defects in Ctbp-null cells. (A) Alignment of mouse and human Znf219. The putative CtBP binding site is underlined. (B) CtBP2 binds Znf219 in vitro. GST-Znf219 (5 μg) bound to glutathione-Sepharose was added to cell lysates (1 mg of total protein) derived from cells that express epitope tagged CtBP1 or CtBP2, pelleted, and the protein complexes resolved and analyzed by SDS-PAGE and Western blotting to detect epitope-tagged CtBP1 and CtBP2. (C) Schematic of the luciferase reporter and Gal4 DBD fusion proteins used in these experiments. (D) Ctbp-null (Ctbp1−/− Ctbp2−/−) cells display defects in transcriptional repression. CtBP-deficient cell lines (indicated along the bottom) were transfected with 4×Gal414DLuc, PGKβgalbpA, and the vectors expressing the indicated Gal4 DBD fusion proteins. Following transfection, cells were assayed for luciferase and β-galactosidase activity. Relative luciferase unit (RLU) measurement is the ratio of luciferase activity corrected for the amount of β-galactosidase activity, to control for transfection efficiency, with the value for the 4×Gal414DLuc vector alone set to 100.
DISCUSSION
Using a combination of gene trap mutagenesis and gene targeting in ES cells, we generated mice deficient for CtBP1 and CtBP2. Genetic analysis suggests both overlapping and unique roles for these genes during mouse development. CtBP1 and CtBP2 are closely related, exhibiting approximately 80% identity at the amino acid level. We show here that these proteins display widespread expression throughout the developing embryo and equivalent binding to a putative interacting protein. The fact that Ctbp1-null mice are viable while Ctbp2-null embryos die by E10.5 suggests that either these proteins have different in vivo functions or they are expressed in slightly different places during the course of development. Our data support the latter possibility, since there are clear differences in the expression of CtBP1 and CtBP2 during placental morphogenesis. Specifically, CtBP2 is readily detected in the forming placenta while CtBP1 is not. In contrast, the fact that Ctbp1-null mice are viable suggests that CtBP2 may compensate for a lack of CtBP1. Generating mice mutant for both Ctbp1 and Ctbp2 tested this hypothesis and our results indicate that these genes are likely to carry out redundant functions during embryonic development. First, reducing the dosage of Ctbp2 makes the Ctbp1−/− phenotype significantly worse, as Ctbp1−/− Ctbp2+/− embryos die in utero and exhibit multiple developmental defects. Secondly, reducing the dosage of Ctbp1 in a Ctbp2−/− background results in a more severe phenotype, as Ctbp1+/− Ctbp2−/− embryos do not turn and do not complete neural tube closure. Finally, complete elimination of Ctbp1 in a Ctbp2−/− background results in further worsening of the Ctbp2−/− embryonic phenotype. The idea that Ctbp1 and Ctbp2 show overlapping in vivo function is supported by in vitro studies that assayed CtBP activity. First, both CtBP1 and CtBP2 appear to interact with the same peptide sequences and bind them with apparent equal affinity, suggesting that they could interact with the same transcription factors in the same cell. Second, in addition to the work presented here, several other groups have shown that both CtBP1 and CtBP2 can function as repressors (4, 9, 30, 44). Finally, we have shown that removal of both CtBP1 and CtBP2 renders a potential transcriptional repressor nonfunctional and CtBP1 and CtBP2 can restore repressor function to equivalent degrees.
CtBP and extraembryonic development.
Mammals have devised an intricate vascular system in both the placenta and yolk sac to provide nutrients and oxygen to growing embryos during the period of embryonic gestation. Gene targeting experiments in mice have helped define a large number of transcription factors, extracellular proteases, and growth factors that are required for the formation of the extraembryonic vascular systems (reviewed in reference 32). In mammals, the yolk sac vascular plexus is formed by the stepwise generation of a vascular system from angioblasts that arise in the blood islands early in the process of yolk sac morphogenesis. Subsequently, the angioblasts go on to initiate the formation of vessels through differentiation of cells into vascular endothelial cells that are distinguished by the expression of cell adhesion molecules such as CD31 and CD34. The subsequent branching and pruning of the vessels generates the final vascular plexus that is observed in the yolk sac. Another critical step in the creation of the fetomaternal connection is the formation of the chorioallantoic plate and the subsequent morphogenesis of the vascular system and labyrinthine layer of the placenta. Based on our observations, the primary cause of embryonic lethality in Ctbp2-null embryos appears to be related to defects in the formation of both the placenta and the yolk sac. Comparison of Ctbp2−/− placentas with those seen in other mouse mutants suggests that that the placental defects in Ctbp2−/− embryos occur after chorioallantoic fusion and the initiation of branching but either before or during branching morphogenesis and vascularization (reviewed in reference 32). The placental defects in Ctbp2-null embryos occur after those observed in mice deficient for Gcm1 (required for initial chorioallantoic branching) (1) but at around the same stage as those seen in JunB mutant embryos (required for branching and labyrinthine development) (37).
In vivo pathways that utilize CtBP.
Based on the large number of transcription factors predicted to bind CtBP, the widespread defects in embryos harboring various combinations of Ctbp mutated alleles are not particularly surprising. We suggest that in the absence of CtBP there is embryo-wide deregulation of gene expression due to the fact that many transcription factors are likely to have at least a partial loss of repressor or activator function. This is supported by our results showing that the CtBP-binding domain of mZnf219 loses repressor function in cells lacking both CtBP1 and CtBP2. Our observations are consistent with those published previously describing the in vivo effects of a protein inhibitor of CtBP (40). In these experiments, ectopic expression of the domain of E1a that binds CtBP results in altered gene expression profiles. Interestingly, the observed expression profiles indicate that large numbers of genes are induced as well as repressed, confirming the observation that CtBP may have both repressor and activator function (27).
Consistent with the above prediction, a number of the developmental defects seen in Ctbp mutants are consistent with alterations in biological pathways that are controlled, at least in part, by transcriptional regulators that bind CtBP. It is well documented that secreted Wnt molecules signal through Tcf/Lef/β-catenin to control aspects of axial patterning and development (10, 11) and that CtBP can bind Tcf (2). We observe that some CtBP2-deficient embryos exhibit axial truncations. Consistent with this phenotype, these embryos show a marked decrease in the expression of T (Brachyury), a direct target of Tcf/β-catenin complexes (51). It has been shown that CtBP and Tcf can interact and repress the expression of Siamois in cells that have not received Wnt signals. However, it is not clear if CtBP is also a component of Tcf/β-catenin complexes or plays a role in its activity. However, if Tcf/CtBP complexes acted only as repressors, reducing the amount of CtBP might lead to precocious T expression, which is the opposite of what we observe. This would suggest that in the case of the T promoter CtBP could be functioning as an activator as opposed to a repressor.
Importantly, some of the other defects seen in Ctbp-null embryos are reminiscent of phenotypes observed in mice deficient for other known CtBP binding partners. These include skeletomuscular development controlled by δEF1/Zeb1 (31, 43) and extraembryonic vascularization and placentation controlled by GATA and MEF2 (6, 21, 22, 52). Taken at face value, these observations suggest that CtBPs may be important for the in vivo function of these transcription factors. However, this hypothesis has yet to be definitively tested, and a direct correlation between the observed phenotypes and the role CtBP plays in the in vivo function of these factors is still unproven. Future studies involving genetic analysis of CtBP with these other important developmental regulators should provide critical insights into some of these unresolved issues.
Acknowledgments
We thank Joseph Keilic, Peter Mueting-Nelsen, and Philip Corrin for help with genotyping; Susan Parkhurst for helpful discussions and encouragement; and Debbie Chapman, Susan Parkhurst, and our laboratory colleagues for critical comments on the manuscript.
This work was supported by grant HD24875 to P.S.
REFERENCES
- 1.Anson-Cartwright, L., K. Dawson, D. Holmyard, S. J. Fisher, R. A. Lazzarini, and J. C. Cross. 2000. The glial cells missing-1 protein is essential for branching morphogenesis in the chorioallantoic placenta. Nat. Genet. 25:311-314. [DOI] [PubMed] [Google Scholar]
- 2.Brannon, M., J. D. Brown, R. Bates, D. Kimelman, and R. T. Moon. 1999. XCtBP is a XTcf-3 co-repressor with roles throughout Xenopus development. Development 126:3159-3170. [DOI] [PubMed] [Google Scholar]
- 3.Brown, M., M. McCormack, K. G. Zinn, M. P. Farrell, I. Bikel, and D. M. Livingston. 1986. A recombinant murine retrovirus for simian virus 40 large T cDNA transforms mouse fibroblasts to anchorage-independent growth. J. Virol. 60:290-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Criqui-Filipe, P., C. Ducret, S. M. Maira, and B. Wasylyk. 1999. Net, a negative Ras-switchable TCF, contains a second inhibition domain, the CID, that mediates repression through interactions with CtBP and de-acetylation. EMBO J. 18:3392-3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deconinck, A. E., P. E. Mead, S. G. Tevosian, J. D. Crispino, S. G. Katz, L. I. Zon, and S. H. Orkin. 2000. FOG acts as a repressor of red blood cell development in Xenopus. Development 127:2031-2040. [DOI] [PubMed] [Google Scholar]
- 6.Dressel, U., P. J. Bailey, S. C. Wang, M. Downes, R. M. Evans, and G. E. Muscat. 2001. A dynamic role for HDAC7 in MEF2-mediated muscle differentiation. J. Biol. Chem. 276:17007-17013. [DOI] [PubMed] [Google Scholar]
- 7.Friedrich, G., and P. Soriano. 1991. Promoter traps in embryonic stem cells: a genetic screen to identify and mutate developmental genes in mice. Genes Dev. 5:1513-1523. [DOI] [PubMed] [Google Scholar]
- 8.Frohman, M. A., M. K. Dush, and G. R. Martin. 1988. Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc. Natl. Acad. Sci. USA 85:8998-9002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Furusawa, T., H. Moribe, H. Kondoh, and Y. Higashi. 1999. Identification of CtBP1 and CtBP2 as corepressors of zinc finger- homeodomain factor deltaEF1. Mol. Cell. Biol. 19:8581-8590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Galceran, J., I. Farinas, M. J. Depew, H. Clevers, and R. Grosschedl. 1999. Wnt3a−/−-like phenotype and limb deficiency in Lef1−/−Tcf1−/− mice. Genes Dev. 13:709-717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Greco, T. L., S. Takada, M. M. Newhouse, J. A. McMahon, A. P. McMahon, and S. A. Camper. 1996. Analysis of the vestigial tail mutation demonstrates that Wnt-3a gene dosage regulates mouse axial development. Genes Dev. 10:313-324. [DOI] [PubMed] [Google Scholar]
- 12.Gripp, K. W., D. Wotton, M. C. Edwards, E. Roessler, L. Ades, P. Meinecke, A. Richieri-Costa, E. H. Zackai, J. Massague, M. Muenke, and S. J. Elledge. 2000. Mutations in TGIF cause holoprosencephaly and link NODAL signalling to human neural axis determination. Nat. Genet. 25:205-208. [DOI] [PubMed] [Google Scholar]
- 13.Grooteclaes, M. L., and S. M. Frisch. 2000. Evidence for a function of CtBP in epithelial gene regulation and anoikis. Oncogene 19:3823-3828. [DOI] [PubMed] [Google Scholar]
- 14.Hildebrand, J. D., M. D. Schaller, and J. T. Parsons. 1993. Identification of sequences required for the efficient localization of the focal adhesion kinase, pp125FAK, to cellular focal adhesions. J. Cell Biol. 123:993-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hildebrand, J. D., and P. Soriano. 1999. Shroom, a PDZ domain-containing actin-binding protein, is required for neural tube morphogenesis in mice. Cell 99:486-497. [DOI] [PubMed] [Google Scholar]
- 16.Hogan, B., R. Beddington, F. Constantini, and E. Lacy. 1994. Manipulating the mouse embryo: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 17.Izutsu, K., M. Kurokawa, Y. Imai, K. Maki, K. Mitani, and H. Hirai. 2001. The corepressor CtBP interacts with Evi-1 to repress transforming growth factor beta signaling. Blood 97:2815-2822. [DOI] [PubMed] [Google Scholar]
- 18.Koipally, J., and K. Georgopoulos. 2000. Ikaros interactions with CtBP reveal a repression mechanism that is independent of histone deacetylase activity. J. Biol. Chem. 275:19594-19602. [DOI] [PubMed] [Google Scholar]
- 19.Komada, M., D. J. McLean, M. D. Griswold, L. D. Russell, and P. Soriano. 2000. E-MAP-115, encoding a microtubule-associated protein, is a retinoic acid-inducible gene required for spermatogenesis. Genes Dev. 14:1332-1342. [PMC free article] [PubMed] [Google Scholar]
- 20.Laherty, C. D., W. M. Yang, J. M. Sun, J. R. Davie, E. Seto, and R. N. Eisenman. 1997. Histone deacetylases associated with the mSin3 corepressor mediate mad transcriptional repression. Cell 89:349-356. [DOI] [PubMed] [Google Scholar]
- 21.Lin, Q., J. Lu, H. Yanagisawa, R. Webb, G. E. Lyons, J. A. Richardson, and E. N. Olson. 1998. Requirement of the MADS-box transcription factor MEF2C for vascular development. Development 125:4565-4574. [DOI] [PubMed] [Google Scholar]
- 22.Lu, J., T. A. McKinsey, C. L. Zhang, and E. N. Olson. 2000. Regulation of skeletal myogenesis by association of the MEF2 transcription factor with class II histone deacetylases. Mol. Cell 6:233-244. [DOI] [PubMed] [Google Scholar]
- 23.MacArthur, H., and G. Walter. 1984. Monoclonal antibodies specific for the carboxy terminus of simian virus 40 large T antigen. J. Virol. 52:483-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Melhuish, T. A., and D. Wotton. 2000. The interaction of the carboxyl terminus-binding protein with the Smad corepressor TGIF is disrupted by a holoprosencephaly mutation in TGIF. J. Biol. Chem. 275:39762-39766. [DOI] [PubMed] [Google Scholar]
- 25.Nibu, Y., and M. S. Levine. 2001. CtBP-dependent activities of the short-range Giant repressor in the Drosophila embryo. Proc. Natl. Acad. Sci. USA 98:6204-6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nibu, Y., H. Zhang, and M. Levine. 1998. Interaction of short-range repressors with Drosophila CtBP in the embryo. Science 280:101-104. [DOI] [PubMed] [Google Scholar]
- 27.Phippen, T. M., A. L. Sweigart, M. Moniwa, A. Krumm, J. R. Davie, and S. M. Parkhurst. 2000. Drosophila C-terminal binding protein functions as a context-dependent transcriptional co-factor and interferes with both Mad and Groucho transcriptional repression. J. Biol. Chem. 275:37628-37637. [DOI] [PubMed] [Google Scholar]
- 28.Pinson, K. I., J. Brennan, S. Monkley, B. J. Avery, and W. C. Skarnes. 2000. An LDL-receptor-related protein mediates Wnt signalling in mice. Nature 407:535-538. [DOI] [PubMed] [Google Scholar]
- 29.Poortinga, G., M. Watanabe, and S. M. Parkhurst. 1998. Drosophila CtBP: a Hairy-interacting protein required for embryonic segmentation and Hairy-mediated transcriptional repression. EMBO J. 17:2067-2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Postigo, A. A., and D. C. Dean. 1999. ZEB represses transcription through interaction with the corepressor CtBP. Proc. Natl. Acad. Sci. USA 96:6683-6688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Postigo, A. A., and D. C. Dean. 1997. ZEB, a vertebrate homolog of Drosophila Zfh-1, is a negative regulator of muscle differentiation. EMBO J. 16:3935-3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rossant, J., and J. C. Cross. 2001. Placental development: lessons from mouse mutants. Nat. Rev. Genet. 2:538-548. [DOI] [PubMed] [Google Scholar]
- 33.Sakai, T., A. Toyoda, K. Hashimoto, and H. Maeda. 2000. Isolation and characterization of a novel zinc finger gene, ZNF219, and mapping to the human chromosome 14q11 region. DNA Res. 7:137-141. [DOI] [PubMed] [Google Scholar]
- 34.Schaeper, U., J. M. Boyd, S. Verma, E. Uhlmann, T. Subramanian, and G. Chinnadurai. 1995. Molecular cloning and characterization of a cellular phosphoprotein that interacts with a conserved C-terminal domain of adenovirus E1A involved in negative modulation of oncogenic transformation. Proc. Natl. Acad. Sci. USA 92:10467-10471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schaller, M. D., J. D. Hildebrand, J. D. Shannon, J. W. Fox, R. R. Vines, and J. T. Parsons. 1994. Autophosphorylation of the focal adhesion kinase, pp125FAK, directs SH2-dependent binding of pp60src. Mol. Cell. Biol. 14:1680-1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmitz, F., A. Konigstorfer, and T. C. Sudhof. 2000. RIBEYE, a component of synaptic ribbons: a protein's journey through evolution provides insight into synaptic ribbon function. Neuron 28:857-872. [DOI] [PubMed] [Google Scholar]
- 37.Schorpp-Kistner, M., Z. Q. Wang, P. Angel, and E. F. Wagner. 1999. JunB is essential for mammalian placentation. EMBO J. 18:934-948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sewalt, R. G., M. J. Gunster, J. van der Vlag, D. P. Satijn, and A. P. Otte. 1999. C-terminal binding protein is a transcriptional repressor that interacts with a specific class of vertebrate Polycomb proteins. Mol. Cell. Biol. 19:777-787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spano, S., M. G. Silletta, A. Colanzi, S. Alberti, G. Fiucci, C. Valente, A. Fusella, M. Salmona, A. Mironov, A. Luini, D. Corda, and S. Spanfo. 1999. Molecular cloning and functional characterization of brefeldin A-ADP-ribosylated substrate. A novel protein involved in the maintenance of the Golgi structure. J. Biol. Chem. 274:17705-17710. [DOI] [PubMed] [Google Scholar]
- 40.Sundqvist, A., E. Bajak, S. D. Kurup, K. Sollerbrant, and C. Svensson. 2001. Functional knockout of the corepressor ctbp by the second exon of adenovirus e1a relieves repression of transcription. Exp. Cell Res. 268:284-293. [DOI] [PubMed] [Google Scholar]
- 41.Sundqvist, A., K. Sollerbrant, and C. Svensson. 1998. The carboxy-terminal region of adenovirus E1A activates transcription through targeting of a C-terminal binding protein-histone deacetylase complex. FEBS Lett. 429:183-188. [DOI] [PubMed] [Google Scholar]
- 42.Takada, S., K. L. Stark, M. J. Shea, G. Vassileva, J. A. McMahon, and A. P. McMahon. 1994. Wnt-3a regulates somite and tailbud formation in the mouse embryo. Genes Dev. 8:174-189. [DOI] [PubMed] [Google Scholar]
- 43.Takagi, T., H. Moribe, H. Kondoh, and Y. Higashi. 1998. DeltaEF1, a zinc finger and homeodomain transcription factor, is required for skeleton patterning in multiple lineages. Development 125:21-31. [DOI] [PubMed] [Google Scholar]
- 44.Turner, J., and M. Crossley. 1998. Cloning and characterization of mCtBP2, a co-repressor that associates with basic Kruppel-like factor and other mammalian transcriptional regulators. EMBO J. 17:5129-5140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Turner, J., and M. Crossley. 2001. The CtBP family: enigmatic and enzymatic transcriptional co-repressors. Bioessays 23:683-690. [DOI] [PubMed] [Google Scholar]
- 46.Vojtek, A. B., S. M. Hollenberg, and J. A. Cooper. 1993. Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell 74:205-214. [DOI] [PubMed] [Google Scholar]
- 47.Weigert, R., M. G. Silletta, S. Spano, G. Turacchio, C. Cericola, A. Colanzi, S. Senatore, R. Mancini, E. V. Polishchuk, M. Salmona, F. Facchiano, K. N. Burger, A. Mironov, A. Luini, and D. Corda. 1999. CtBP/BARS induces fission of Golgi membranes by acylating lysophosphatidic acid. Nature 402:429-433. [DOI] [PubMed] [Google Scholar]
- 48.Wen, Y., D. Nguyen, Y. Li, and Z. C. Lai. 2000. The N-terminal BTB/POZ domain and C-terminal sequences are essential for Tramtrack69 to specify cell fate in the developing Drosophila eye. Genetics 156:195-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wotton, D., R. S. Lo, S. Lee, and J. Massague. 1999. A Smad transcriptional corepressor. Cell 97:29-39. [DOI] [PubMed] [Google Scholar]
- 50.Wotton, D., R. S. Lo, L. A. Swaby, and J. Massague. 1999. Multiple modes of repression by the Smad transcriptional corepressor TGIF. J. Biol. Chem. 274:37105-37110. [DOI] [PubMed] [Google Scholar]
- 51.Yamaguchi, T. P., S. Takada, Y. Yoshikawa, N. Wu, and A. P. McMahon. 1999. T (Brachyury) is a direct target of Wnt3a during paraxial mesoderm specification. Genes Dev. 13:3185-3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang, C. L., T. A. McKinsey, J. R. Lu, and E. N. Olson. 2001. Association of COOH-terminal-binding protein (CtBP) and MEF2- interacting transcription repressor (MITR) contributes to transcriptional repression of the MEF2 transcription factor. J. Biol. Chem. 276:35-39. [DOI] [PubMed] [Google Scholar]