Abstract
Polycomb group (PcG) proteins form multimeric chromatin-associated protein complexes that are involved in heritable repression of gene activity. Two distinct human PcG complexes have been characterized. The EED/EZH2 PcG complex utilizes histone deacetylation to repress gene activity. The HPC/HPH PcG complex contains the HPH, RING1, BMI1, and HPC proteins. Here we show that vertebrate Polycomb homologs HPC2 and XPc2, but not M33/MPc1, interact with the histone lysine methyltransferase (HMTase) SUV39H1 both in vitro and in vivo. We further find that overexpression of SUV39H1 induces selective nuclear relocalization of HPC/HPH PcG proteins but not of the EED/EZH2 PcG proteins. This SUV39H1-dependent relocalization concentrates the HPC/HPH PcG proteins to the large pericentromeric heterochromatin domains (1q12) on human chromosome 1. Within these PcG domains we observe increased H3-K9 methylation. Finally, we show that H3-K9 HMTase activity is associated with endogenous HPC2. Our findings suggest a role for the SUV39H1 HMTase and histone H3-K9 methylation in the targeting of human HPC/HPH PcG proteins to modified chromatin structures.
During embryonic development many different cell types arise from a single fertilized egg. Once a cell establishes its specific differentiation status, it requires a cellular memory system to allow the maintenance of proper and stably inherited gene expression patterns (18). The Polycomb (Pc) group (PcG) proteins and the trithorax group (trxG) proteins are part of such a memory system. Initially, they were identified in Drosophila melanogaster as being either repressors (PcG) or activators (trxG) of gene expression and in particular of the homeotic genes (30, 44). Mutations in PcG and trxG genes result in a general misexpression of target genes, resulting in a change of the specific gene expression pattern, which eventually leads to transformations of the body plan (11, 41). In PcG mutants the expression patterns of homeotic genes are initially normal but in later embryonic phases the homeotic genes become expressed in regions of the embryo where they normally are repressed (26, 45). Besides Polycomb (Pc) mutants, an estimated 30 to 40 mutants exhibit similar, characteristic posterior homeotic transformations. These are collectively referred to as PcG mutants (17). The Pc protein binds to about 100 loci on polytene chromosomes in the Drosophila salivary gland (58). Also PcG proteins Polyhomeotic, Polycomblike, and Posterior sex combs share many, but not all, of these binding sites with Pc (8, 10, 21, 32). This is consistent with the idea that PcG proteins work together in a multimeric protein complex.
In recent years, much evidence about the composition of PcG complexes both in Drosophila and in vertebrates has accumulated (38). There are at least two distinct PcG complexes (25, 43, 55). The first human PcG complex, termed the EED/EZH2 PcG complex, contains EED (43), EZH2 (43), and YY1 (36). The second human PcG complex, termed the HPC/HPH PcG complex, contains HPC (37), HPH (12), BMI1 (56), and RING1 (35). Associated with the latter complex is the C-terminal binding protein (CtBP) corepressor (42).
Despite the extensive knowledge concerning the identities of the PcG proteins and proteins that are associated with these complexes, evidence about the molecular mechanisms by which these proteins achieve a stable and heritable state of gene expression is scarce. Several models in which the PcG proteins can package target genes in a heterochromatin-like conformation or induce modifications of the nucleosomal organization have been considered (30). Recently it has been shown both in humans and Drosophila that the EED/EZH2 PcG complex is associated with histone deacetylase (HDAC) activity through a specific interaction between the EED/esc and HDAC proteins (51, 54). This indicates an important role for changes in chromatin structure via histone deacetylation in PcG-mediated gene repression. Insight into the molecular mechanism underlying the action of the HPC/HPH PcG complex is, however, lacking. However, the Pc protein was able to interact in vitro with nucleosomal core particles specifically via its repression domain (4). Furthermore, since Pc and heterochromatin-associated protein HP1 share a homologous domain, i.e., the chromodomain (27), it has been suggested that PcG proteins can form a heterochromatin-like conformation (30, 31). A mechanism underlying the establishment of different chromatin states involves modifications of histones (6, 15, 16, 47, 52, 57). One of these modifications is methylation of histones, which can be accomplished by different histone methyltransferases (15, 33, 48, 50, 53). Interestingly, heterochromatin-associated protein HP1 specifically interacts with the histone lysine methyltransferase (HMTase), SUV39H1, and interacts with H3-K9 dimethylated N termini via its chromodomain, providing insight about the establishment of heterochromatin (3, 20, 33). On the basis of these parallels, we screened for potential interactions between SUV39H1 and PcG proteins. We found that SUV39H1 is able to interact directly with a specific class of vertebrate Pc proteins, specifically, HPC2 (37) and XPc2 (34). Overexpression of SUV39H1 causes PcG proteins belonging to the HPC/HPH PcG complex to relocalize to large PcG domains. These domains, in which HPC2, BMI1, RING1, HPH1, and HPH2 colocalize, also contain methylated histone H3-K9 at DNA sequences that are associated with pericentromeric regions (1q12) on human chromosome 1 and with related pericentromeric sequences on different chromosomes. An SUV39H1 mutant lacking HMTase activity was not able to relocalize these PcG proteins. Finally, we find that HMTase activity is also associated with the HPC/HPH PcG complex. Taken together, our findings suggest a role for SUV39H1 and histone H3-K9 methylation in the selective targeting of PcG proteins to specific chromosome regions.
MATERIALS AND METHODS
Yeast two-hybrid analysis.
The region encoding full-length SUV39H1 was cloned in frame with the coding sequence for the GAL4 transactivation domain (GAL4-TAD) in the pGAD10 vector (Clontech). The cDNAs encoding EZH2, EED, YY1, HPH1, HPC2, CtBP, RING1, BMI1, and M31/HP1β proteins were cloned in frame with the coding sequence for the GAL4 DNA binding domain (GAL4-DBD) in the pAS2 vector or the pGBKT7 vector (Clontech) (12, 35, 42, 43). The p53 protein (pVA3; Clontech) and simian virus 40 large antigen (pTD1; Clontech) served as a positive control. We cotransformed plasmids into yeast strain Y190 (Clontech) and plated the transformants on selective medium lacking histidine, tryptophan, and leucine. After 5 days cells were grown to an optical density at 600 nm of 1.0 to 1.2 in selective medium and were subsequently permeabilized. β-Galactosidase activity was measured as described previously (12). All proteins were expressed at approximately the same level as determined by Western blotting using monoclonal antibodies that recognize either GAL4-DBD or GAL4-TAD (Clontech; data not shown). Potential interactions were subjected to a more detailed analysis. Therefore, different portions of the cDNA of SUV39H1, M31/HP1β, and HPC2, which were obtained by PCR (Expand; Roche), were cloned in either the pGAD10 or pGBKT7 vector. Plasmids were cotransformed into yeast AH109 and plated on selective medium for 5 to 10 days. Cells displaying the ability to grow on selective medium and expressing β-galactosidase are indicative of a protein-protein interaction.
GST fusion proteins and in vitro binding assay.
Previously described fusion proteins glutathione S-transferase (GST)-XPc2 (34, 42) and GST-HPC2, GST-EZH2, and GST-RING1 (36) were used for an in vitro binding assay. As a negative control, GST alone was used. Expression of the GST fusion proteins was induced for 3 h at 30°C with 0.4 mM isopropyl-β-d-thiogalactopyranoside as described previously (12). The cells were pelleted, resuspended in binding buffer (phosphate-buffered saline [PBS; 140 mM NaCl, 2.7 mM KCl, 6.5 mM Na2HPO4, 1.5 mM KH2PO4] containing 1 mM EDTA, 1 mM dithiothreitol, 2 mM phenylmethylsulfonyl fluoride, 10 μg of leupeptin per ml, 10 μg of benzamidine per ml, 10 μg of trypsin inhibitor per ml, and 10 μg of aprotinin per ml), and sonicated. Triton X-100 was added to a final concentration of 1% (vol/vol), and the lysate was incubated for 30 min on ice. Cell debris was removed by centrifugation for 10 min at 14,000 × g. The supernatant was added to glutathione-Sepharose 4B, and the mixture was incubated for 30 min at 4°C. The beads were collected by centrifugation and washed extensively with binding buffer. Capped synthetic SUV39H1 mRNA was made by in vitro transcription and translated at 20 μg per ml in a rabbit reticulocyte lysate in the presence of [35S]methionine (34). A 10-μl slurry of the immobilized GST fusion protein was preincubated for 30 min on ice in a final volume of 200 μl of binding buffer that further contained 0.5% Nonidet P-40 and 1 mg of bovine serum albumin (BSA) per ml. Subsequently, 3 μl of the reticulocyte lysate was added to the mixture, and the mixture was incubated for 30 min at 4°C with end-over-end mixing. The beads were washed five times with 1 ml of ice-cold binding buffer. The complexes were separated on sodium dodecyl sulfate (SDS)-polyacrylamide gels and analyzed with a Personal Molecular Imager (Bio-Rad).
IP assay.
For immunoprecipitation (IP) we used the previously described HeLa cells which stably express either triple-myc-tagged SUV39H1 [(myc)3-SUV39H1] or (myc)3-SUV39H1 (N-chromo) under the control of the cytomegalovirus promoter (22). Cells were grown to confluence and harvested, and nuclei were prepared by 10 strokes with a glass Dounce pestle in a buffer containing 20 mM HEPES (pH 7.0), 1.5 mM MgCl2, 10 mM KCl, 0.5 mM dithiothreitol, and 0.5 mM phenylmethylsulfonyl fluoride. The nuclei were pelleted by centrifugation at 1,000 × g at 4°C for 10 min. Subsequently we lysed nuclei in ELB buffer (250 mM NaCl, 0.1% Nonidet P-40, 50 mM HEPES [pH 7.0], 5 mM EDTA) containing 0.5 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, and protease inhibitors leupeptin, benzamidine, pepstatin, and aprotinin. The lysates were sonicated three times with 10-s pulses. To remove the debris, the lysates were centrifuged at 14,000 × g for 10 min at 4°C. The supernatants were aliquoted and stored at −80°C. Eighty microliters of the different cell lysates was incubated with a monoclonal antibody against the myc epitope (9E10) or polyclonal antibodies directed against HPC2, BMI1, M33 (12, 37), and M31/HP1β. The mixtures were incubated for 2 h at 4°C. Forty microliters of goat anti-mouse immunoglobulin G (IgG) agarose or goat anti-rabbit IgG was added, and ELB buffer with protease inhibitors was added to increase the volume of the mixtures to 300 μl. The mixtures were incubated for 2 h at 4°C under continuous end-over-end mixing. They were centrifuged at 1,500 × g at 4°C for 30 s, washed with 1 ml of ELB-plus buffer (500 mM NaCl, 1% Nonidet P-40, 50 mM HEPES [pH 7.0], 5 mM EDTA) without protease inhibitors, and centrifuged again at 1,500 × g at 4°C for 30 s. This washing procedure was repeated five times. After heating and centrifugation to remove the agarose beads, the proteins were separated by SDS-polyacrylamide gel electrophoresis (PAGE) and blotted to nitrocellulose. The blot was probed with a rabbit polyclonal antibody directed against human PcG proteins as inidcated in Fig. 4 (36, 43). The secondary alkaline phosphatase-conjugated goat anti-rabbit antibodies (Jackson ImmunoResearch Laboratories) were diluted 1:10,000, and nitroblue tetrazolium-BCIP (5-bromo-4-chloro-3-indolylphosphate) (Roche) was used as the substrate for detection. The input (see Fig. 4, lane 1) was 20% of the amount of lysate which was used for the respective incubations. The strength of the HPC2 or HP1 IP signal (lane 2) was approximately 25 or 100% of the HPC2 or HP1 input signal, respectively (lane 1), indicating that approximately 5% of the HPC2 protein and 20% of the HP1 protein are associated with the SUV39H1 protein.
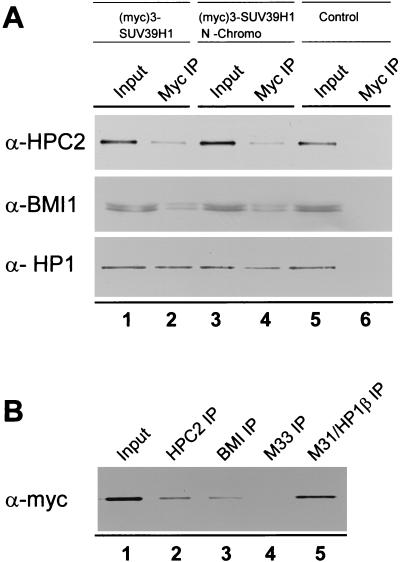
FIG. 4.
In vivo interaction between HPC2 and SUV39H1. (A) We used nuclear extracts of HeLa cell lines which stably express myc-tagged SUV39H1 aa 3 to 412 (lanes 1 and 2), myc-tagged SUV39H1 aa 3 to 118 (lanes 3 and 4), or no ectopic SUV39H1 protein (lanes 5 and 6). We used a mouse monoclonal antibody against the myc tag (9E10) to immunoprecipitate the respective myc-tagged proteins. The resulting immunoprecipitates were Western blotted and analyzed with a rabbit polyclonal antibody against HPC2, BMI1, or M31/HP1β. (B) Nuclear extract of HeLa cells which stably express myc-tagged SUV39H1 protein (lane 1) were used for IP with rabbit polyclonal antibodies directed against HPC2 (lane 2), BMI1 (lane 3), M33 (lane 4), and M31/HP1β (lane 5). The resulting immunoprecipitates were Western blotted and analyzed with a mouse monoclonal antibody against the myc epitope to detect the myc-tagged SUV39H1 protein.
Immunofluorescence labeling of tissue culture cells.
The above-described HeLa cell lines were cultured and labeled as described previously (12, 35, 43). The labeling was analyzed by confocal laser scanning microscopy, and single optical sections were made. For labeling HPC2, BMI1, RING1, HPC1, EED, and EZH2, rabbit polyclonal antibodies were used (12, 36, 42, 43). For labeling YY1 a commercially available rabbit polyclonal antibody was used (Santa Cruz Laboratory). For double labeling methylated histone H3-K9 and HPC2, we used rabbit polyclonal antibodies directed against methylated H3-K9 (29) and a mouse monoclonal antibody directed against HPC2. To analyze the effect of transient expression of SUV39H1 and the mutant SUV39H1 (H324L), HeLa cells were transiently transfected with either pCMV-(myc)3-SUV39H1 or pCMV-(myc)3-SUV39H1(H324L) by the calcium phosphate transfection method (Gibco-BRL) 48 h prior to the labeling. To detect the triple-myc-tagged SUV39H1 proteins, cells were labeled with a mouse monoclonal antibody against the myc tag (9E10). For labeling the PcG proteins, donkey anti-rabbit IgG either coupled to fluorescein isothiocyanate (FITC) or Cy3 was used (Jackson ImmunoResearch Laboratories). For labeling the myc-tagged SUV39H1 proteins, donkey anti-mouse IgG coupled to FITC was used (Jackson ImmunoResearch Laboratories).
In vitro HMTase assay.
Prior to the in vitro HMTase assay, we enriched either the HPC2, BMI1, RING1, or myc-tagged SUV39H1 proteins by IPs on nuclear extracts of either HeLa, U-2 OS, or HeLa cells which stably express (myc)3-SUV39H1. We performed the in vitro HMTase assay as described previously (33, 49) using the wild-type N terminus of human histone H3 (ARTKQTARKSTGGKAPRKQL) and a mutant peptide (K9L) in which lysine 9 is substituted for leucine as the substrates. Immunoprecipitation products were probed for activity to transfer a labeled methyl group from S-adenosyl-l-[methyl-14C]methionine to these substrates. Reaction products were separated by SDS-PAGE and visualized by fluorography.
Immunofluorescent in situ hybridization.
After the immunostaining slides were rinsed in 1× PBS, fixed in 4% paraformaldehyde for 10 min at room temperature, washed in 1× PBS, dehydrated in an ethanol (EtOH) series (70-90-96%), and air dried. RNase treatment (50 μg/ml; in 2× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate]) was performed in a moist chamber at 37°C for 30 to 45 min. After three washes in 2× SSC, proteinase K treatment (1 μg/ml; in 20 mM Tris-HCl [pH 7.5]-2 mM CaCl2) was done at 37°C for 15 min in a moist chamber, followed by three washes in 2× SSC. Denaturation was performed in 70% formamide for 2 min at 60°C, and the slides were dehydrated in an ice-cold EtOH series (70-90-96%) and air dried.
Human chromosome 1-specific probe pUC 1.77 for highlighting region 1q12 was labeled with biotin-11-dUTP via nick translation according to standard protocols. The air-dried biotinylated probe was resuspended in 50% formamide-10% dextran sulfate-2× SSC, denatured at 95°C for 5 min, and immediately placed on ice. The denatured probe was dropped on air-dried slides, covered with a coverslip, sealed with a rubber cement, and again simultaneously denatured at 80°C. Hybridization was performed overnight at 37°C in a humid chamber. Posthybridization washes were done once in 2× SSC, two times in 50% formamide, and two times in 0.1× SSC (5 min each). After 30 min of blocking (4× SSC, 3% BSA, 0.1% Tween 20), biotin was detected with avidin-Cy3 (10 μg of 4× SSC/ml, 1% BSA, 0.1% Tween 20) at 37°C for 1 h. Slides were washed three times in 4× SSC-0.1% Tween 20 at 42°C for 5 min each and mounted in antifading solution supplemented with DAPI (4′;,6′;-diamidino-2-phenylindole; 1 μg/ml).
RESULTS
A specific class of vertebrate Pc homologs interacts with SUV39H1.
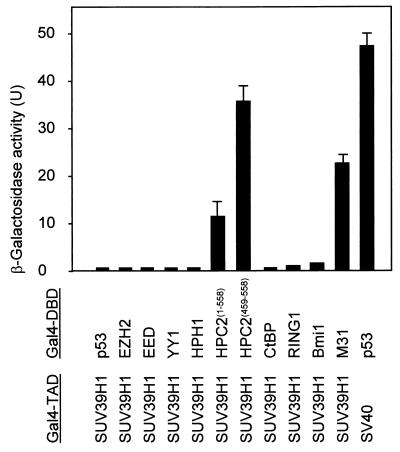
PcG protein complexes are involved in repression of gene expression. Since Pc and HP1 share a homologous domain, the chromodomain (27), several models in which PcG proteins may package target genes in a heterochromatin-like conformation have been suggested (30). Mammalian homologs of HP1 interact with HMTase SUV39H1 (1, 22). On the basis of the structural parallels between Pc and HP1, we investigated potential interactions between human PcG proteins and SUV39H1 by employing a directed two-hybrid screen. As a positive control we used the M31 protein, a mouse homolog of HP1β (13), which is able to interact with SUV39H1 (1, 22). We found that SUV39H1 was able to interact with HPC2 but not with PcG proteins EZH2, EED, YY1, HPH1, RING1, and BMI1 or with PcG-associated protein CtBP (Fig. 1). The interaction between SUV39H1 and the C-terminal part of HPC2 (amino acids [aa] 459 to 558) was higher than that between SUV39H1 and the full-length HPC2 (aa 1 to 558) protein (Fig. 1).
FIG. 1.
Two-hybrid interaction between SUV39H1 and vertebrate PcG proteins. Two-hybrid analysis with SUV39H1 and human PcG proteins shows a positive interaction between SUV39H1 and HPC2. The PcG cDNA was cloned in frame with the GAL4-DBD coding sequence, whereas the SUV39H1 cDNA was cloned in frame with the GAL4-TAD coding sequence. As a positive control for the two-hybrid system we used the p53 and simian virus 40 (SV40) large antigen. As a positive control for the SUV39H1 protein we used the mouse homolog of HP1, M31/HP1β. No interactions between SUV39H1 and other PcG proteins were found.
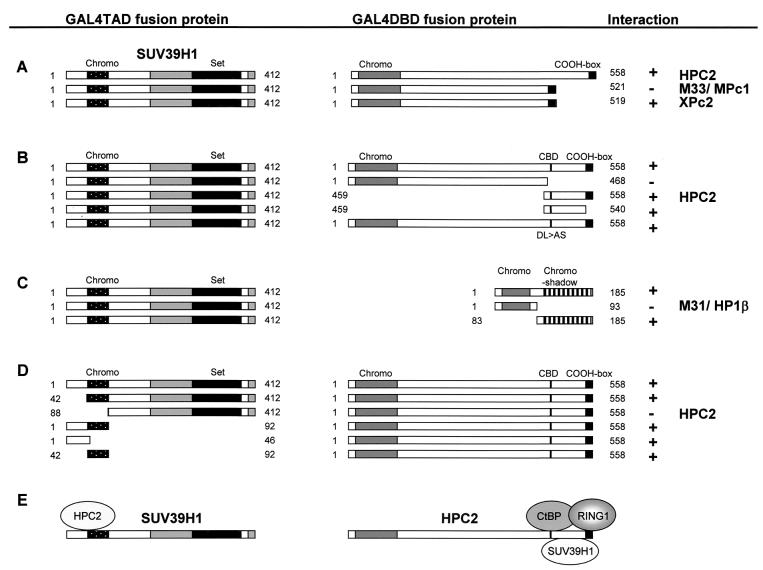
To test whether other vertebrate Pc homologs are able to interact with SUV39H1 in the two-hybrid system, we tested mouse Pc protein M33/MPc1 (28) and Xenopus laevis Pc protein XPc2 (34) (Fig. 2A). XPc2 is more homologous to HPC2 than M33/MPc1, which in turn is more homologous to human Pc homolog CBX2/HPC1 (37, 42). Interestingly, XPc2, but not M33/MPc1, was able to interact with SUV39H1 (Fig. 2A), indicating that, despite the high degree of homology between the Pc proteins in the chromodomain and the COOH box, these Pc proteins differ significantly, at least in their abilities to interact with other proteins. This is, however, not the first indication that there is specificity among Pc proteins, since we previously found that both XPc2 and HPC2 were able to interact with CtBP while M33/MPc1 was not (42).
FIG. 2.
(A) SUV39H1 is able to interact with a specific class of vertebrate Pc proteins. Vertebrate Pc homologs HPC2, XPc2, and M33/MPc1 were cloned in frame with the GAL4-DBD, whereas the SUV39H1 protein was cloned in frame with the GAL4-TAD. Interactions were positive when cells survived on selective medium lacking histidine and when they were β-galactosidase positive. Shown are the SUV39H1 chromodomain, the SET domain, and the cysteine-rich regions adjacent to the SET domain (grey boxes). The indicated domains of the vertebrate Pc homologs are the chromodomain and the COOH box. (B) Mapping of the domain for the interaction of HPC2 with SUV39H1. The indicated domains of the vertebrate Pc homologs are the chromodomain, the CBD, and the COOH box. (C) Mapping of the domain for the interaction of M31 with SUV39H. The indicated domains of M31/HP1β are the chromodomain (grey box) and the chromoshadow domain. (D) Mapping of the domain for the interaction of SUV39H1 with HPC2. (E) Schematic representation of the interaction domains of SUV39H1 and HPC2. Both the extreme N-terminal part and the chromodomain of SUV39H1 are necessary for binding HPC2. SUV39H1 binds to a region within the C terminus of HPC2 that differs from the C-terminal repression region to which the RING1 protein binds and the binding site of the CtBP protein.
In conclusion, these experiments show that SUV39H1 is able to interact with vertebrate Pc proteins HPC2 and XPc2 but not with mouse Pc protein M33/MPc1, indicating specificity among the different Pc proteins.
SUV39H1 binds the C-terminal region of HPC2.
To determine which part of HPC2 is responsible for the interaction with SUV39H1, we subcloned different protein fragments of HPC2 in frame with the GAL4-DBD (Fig. 2B). HPC2 comprises three defined protein domains. The chromodomain, which is essential for binding the Pc protein to chromatin, is located at the N terminus (23). A highly conserved COOH box (aa 540 to 558) is located at the extreme C terminus. This COOH box is the domain to which PcG protein RING1 binds (35, 40), the domain within Pc which in vitro is able to bind to nucleosomal core particles (4), and also the domain which is necessary for gene repression (5, 24, 37). The third domain within HPC2 is a 6-aa motif (aa 470 to 475) to which the CtBP proteins bind (42). This 6-aa motif, termed the CtBP-binding domain (CBD), is well conserved among different proteins of different species.
We found that a C-terminal fragment (aa 459 to 558) is able to interact with SUV39H1 and that an HPC2 mutant (aa 1 to 468) lacking the C terminus was not able to interact with SUV39H1. We further defined the C-terminal region that is crucial for binding SUV39H1. We tested a fragment (aa 459 to 540) that lacks the COOH box to which the RING1 protein binds (40) (data not shown). This fragment was still able to bind to SUV39H1, indicating that the binding domain of SUV39H1 differs from the conserved Pc repression domain to which RING1 binds. We further tested a HPC2 mutant protein (HPC2 DL>AS) in which the CtBP binding region is changed in such a way that the interaction with CtBP is abolished (42). We found also that this HPC2 mutant was able to interact with SUV39H1, indicating that the CBD is not involved in SUV39H1 binding (Fig. 2B).
In summary, the SUV39H1 protein binds to a region within the C terminus of HPC2 that differs from the C-terminal repression region to which the RING1 protein binds (35) and the binding site of the CtBP protein (42).
The chromoshadow domain of HP1 is involved in binding SUV39H1.
The mouse homolog of the Drosophila HP1 protein, M31/HP1β, has been shown to interact with SUV39H1 (22). To define the domain in HP1β to which the SUV39H1 protein binds, we cloned different parts of M31/HP1β in frame with the GAL4-DBD (Fig. 2C). M31/HP1β contains two functional domains, the N-terminal chromodomain, which is able to bind to H3-K9 methylated peptides (3, 20), and the chromoshadow domain, which is involved in interactions with other protein partners (2, 19). We found that a portion of M31/HP1β encompassing the chromodomain was not able to interact with SUV39H1. Instead, the chromoshadow domain was able to interact with SUV39H1 (Fig. 2C). In conclusion, neither the chromodomain of HPC2 nor that of M31/HP1β is involved in binding with SUV39H1. In both cases the C-terminal regions of the proteins are involved in binding SUV39H1.
The N terminus of SUV39H1 interacts with HPC2.
To define the domains within SUV39H1 that are responsible for the interaction with HPC2 and XPc2, we cloned different parts of the SUV39H1 protein in frame with the GAL4-TAD. SUV39H1 comprises two defined domains, of which the N-terminal chromodomain has been shown to provide an interaction surface for M31/HP1β, whereas the C-terminal SET domain comprises the catalytic motif, required for HMTase activity (Fig. 2D) (33). We found that a mutant SUV39H1 lacking the extreme N-terminal 41 aa but retaining the chromodomain was still able to interact with HPC2 (Fig. 2D). However, a mutant SUV39H1 (aa 88 to 412) that lacks the chromodomain was not able to interact with HPC2. Conversely, a fragment that encompasses the N-terminal 92 aa with the chromodomain was still able to interact with HPC2 (Fig. 2D). To more precisely determine the regions within the N terminus of SUV39H1 that are responsible for the interaction with HPC2, we made two constructs. One contains the extreme N-terminal 46 aa, and the other encompasses the chromodomain (aa 42 to 92). We found that both constructs are able to interact with HPC2, indicating that both the chromodomain and the adjacent, extreme N terminus of SUV39H1 are responsible for the interaction with HPC2.
Both HPC2 and XPc2 directly interact with SUV39H1.
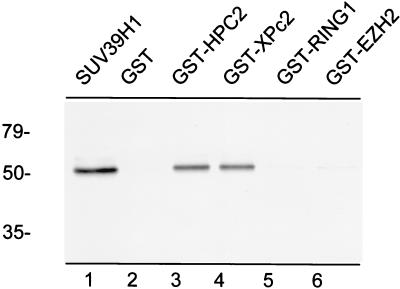
To determine whether the interaction between SUV39H1 and vertebrate Pc homologs HPC2 and XPc2 is a direct interaction, we employed an in vitro pull-down assay (Fig. 3). For this we used the previously described proteins formed by fusion of GST to HPC2, XPc2, RING1, and EZH2 (34, 36, 42, 54). All proteins were immobilized on GST-Sepharose and incubated with [35S]methionine-labeled in vitro-translated SUV39H1. After extensive washing the proteins were separated by SDS-PAGE, and the gel was analyzed with a phosphorimager. The in vitro-translated SUV39H1 protein (Fig. 3, lane 1) was able to bind GST-HPC2 (lane 3) and GST-XPc2 (lane 4). SUV39H1 did not bind to GST alone (lane 2), indicating the specificity of the interaction between SUV39H1 and both HPC2 and XPc2. Furthermore, neither GST-RING1 (lane 5) nor GST-EZH2 (lane 6) bound SUV39H1. In summary, these results confirm the data obtained from the two-hybrid assay (Fig. 1 and 2) and indicate that SUV39H1 is able to bind either HPC2 or XPc2 directly.
FIG. 3.
Both HPC2 and XPc2 bind SUV39H1 directly in vitro. [35S]methionine-labeled SUV39H1 (lane 1) was incubated with either GST alone (lane 2), GST-HPC2 (lane 3), GST-XPc2 (lane 4), GST-RING1 (lane 5), or GST-EZH2 (lane 6). The in vitro-translated [35S]methionine-labeled SUV39H1 was able to bind to immobilized GST-HPC2 and GST-XPc2. Molecular masses in kilodaltons are indicated on the left. The input (lane 1) was 15% of the amount of lysate which was used for the incubations.
SUV39H1 and HPC2 interact in vivo.
To demonstrate an in vivo interaction between SUV39H1 and HPC2, we performed coimmunoprecipitation experiments (Fig. 4). For this purpose we used nuclear extracts of previously described HeLa cell lines (22) which stably express myc-tagged SUV39H1 aa 3 to 412 [(myc)3-SUV39H1] (lanes 1 and 2), myc-tagged SUV39H1 aa 3 to 118 [(myc)3-SUV39H1 N-chromo] (lanes 3 and 4), or no ectopic protein (control) (lanes 5 and 6). We used a mouse monoclonal antibody against the myc tag (9E10) to immunoprecipitate the respective myc-tagged protein and rabbit polyclonal antibodies directed against HPC2 and BMI1 for Western blot analysis (12, 37, 42). We also used rabbit polyclonal antibodies directed against M31/HP1β for Western blot analysis since it has previously been shown that M31/HP1β and SUV39H1 are able to coimmunoprecipitate with each other from these cells (22). Our results show that both HPC2 and BMI1, as well as M31/HP1β, coimmunoprecipitate with SUV39H1 in vivo (Fig. 4A, lane 2). In the control (HeLa) cell line, neither HPC2 nor BMI1 nor M31/HP1β could be detected on the Western blot after IP with the antimyc antibody (lane 6). We also performed IPs on the HeLa cell line that contains the (myc)3-SUV39H1 N-chromo construct. This construct is an SUV39H1 mutant that lacks HMTase activity. Both the HPC2 and BMI1 proteins, as well as the M31/HP1β protein, were detected in the antimyc immunoprecipitate (lane 4), indicating that the truncated SUV39H1 protein is also able to coimmunoprecipitate with HPC2 and BMI1 in vivo. Significantly, this part of the SUV39H1 protein also interacts with HPC2 in the two-hybrid system (Fig. 2), underlying the importance of this domain for the interaction with HPC2. We also performed coimmunoprecipitation experiments with HeLa and U-2 OS cells that did not overexpress the SUV39H1 protein. We used several commercially available antibodies against SUV39H1. Unfortunately, none of these antibodies detected any SUV39H1 by Western blotting or by immunofluorescence (see below), indicating that SUV39H1 has very low abundance. This prevented further analysis of in vivo interactions between the endogenous SUV39H1 and PcG proteins.
Using nuclear extracts of the myc-tagged SUV39H1-overexpressing HeLa cell line (Fig. 4B, lane 1), we detected the myc-tagged SUV39H1 protein in IPs with rabbit polyclonal antibodies directed against HPC2 (lane 2), BMI1 (lane 3), M33 (lane 4), and M31/HP1β (lane 5). Significantly, we detected no myc-tagged SUV39H1 protein in IPs with rabbit polyclonal antibodies directed against HPC1/M33. This is consistent with the fact that we did not find an interaction between SUV39H1 and M33/MPc1 in the two-hybrid assay (Fig. 2).
Overexpression of SUV39H1 alters the nuclear localization of HPC2 and other HPC/HPH PcG proteins.
Overexpression of SUV39H1 has been shown to disperse the nuclear localization of HP1 (22). To examine the subnuclear distribution of PcG proteins in relation to that of the ectopic SUV39H1 protein, we performed immunofluorescence-labeling experiments. For this purpose we used the same cell lines that were used for the coimmunoprecipitation experiments, namely, the myc-tagged SUV39H1, the myc-tagged SUV39H1 N-chromo, and the control (Fig. 5A and 6). We used rabbit polyclonal antibodies directed against HPC2, BMI1, and RING1 (Fig. 5A) and against HPC1, EED, EZH2, and YY1 (Fig. 6) for labeling.
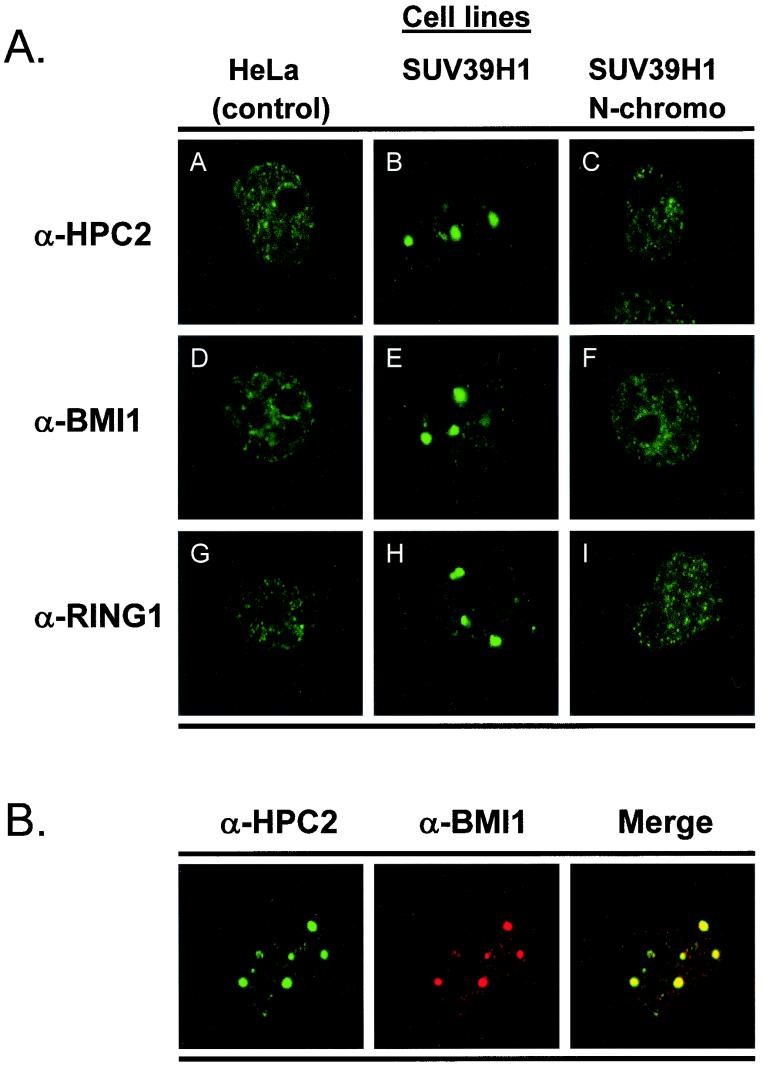
FIG. 5.
(A) Ectopic expression of SUV39H1 alters the nuclear localization of the HPC/HPH PcG proteins. Confocal single optical sections are shown. HeLa cells and HeLa cells stably expressing (myc)3-SUV39H1 (aa 3 to 412) (SUV39H1), or (myc)3-SUV39H1 (aa 3 to 118) (SUV39H1 N-chromo) were used. Cells were stained with rabbit polyclonal antibodies directed against HPC2 (A, B, and C), BMI1 (D, E, and F), and RING1 (G, H, and I) The HPC2, BMI1, and RING1 proteins were redistributed in the nucleus from fine granular domains to large domains in the cell line which expressed the myc-tagged SUV39H1 protein but not in the cell line expressing either the myc tag alone or the myc-tagged SUV39H1 N-chromo. (B) Rabbit anti-HPC2 and mouse anti-BMI1 double labeling demonstrates colocalization of HPC2 and BMI1 in large nuclear domains of HeLa cells stably expressing SUV39H1.
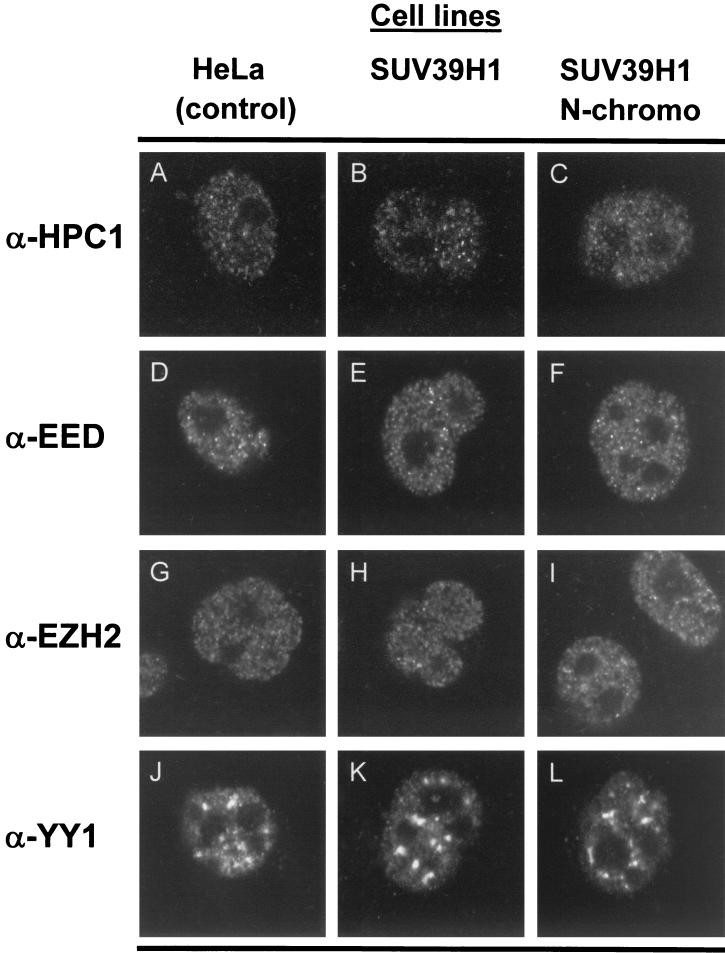
FIG. 6.
Ectopic expression of SUV39H1 does not alter the nuclear localization of both HPC1 and the EED/EZH2 PcG proteins. Confocal single optical sections are shown. HeLa cells, HeLa cells stably expressing (myc)3-SUV39H1 (aa 3 to 412) (SUV39H1), or (myc)3-SUV39H1 (aa 3 to 118) (SUV39H1 N-chromo) were used. Cells were stained with rabbit polyclonal antibodies directed against HPC1 (A, B, and C), EED (D, E, and F), EZH2 (G, H, and I), and YY1 (J, K, and L). Comparison of the different cell lines demonstrates that the nuclear distribution of these proteins did not alter.
In both the control cell line and the SUV39H1 N-chromo cell line (Fig. 5A), HPC2, BMI1, and RING1 (Fig. 5A), as well as HPH1 and HPH2 (data not shown), show a fine granular nuclear distribution. In the SUV39H1 cell line, the distribution of these HPC/HPH PcG proteins is altered (Fig. 5A). HPC2, BMI1, and RING1 (Fig. 5A), as well as HPH1 and HPH2 (data not shown), localize to large nuclear domains. Moreover, within these large domains, these PcG proteins of the HPC/HPH PcG complex colocalize (Fig. 5B). The domains are also observed for osteosarcoma cell line U-2 OS and are remarkably similar to the PcG domains observed in HT-1080 cells (12, 35, 37, 39, 43). There is, however, one exception. In contrast to that of the other HPC/HPH PcG proteins, the nuclear staining of the HPC1 protein (Fig. 6) is not altered. Similarly, the nuclear staining pattern of PcG proteins EED, EZH2, and YY1 (Fig. 6), which are members of the EED/EZH2 PcG complex (36, 43), displayed the same immunofluorescence characteristics in the SUV39H1 cell line as in the control and the N-chromo cell lines.
We also performed experiments to determine whether the endogenous SUV39H1 protein colocalizes with the indicated PcG proteins. Unfortunately, we did not detect any specific staining signal with the SUV39H1 antibody in a variety of cell lines, including HeLa and U-2 OS. This made it impossible to further investigate the colocalization of endogenous SUV39H1 and PcG proteins.
In summary, we show that ectopic expression of SUV39H1 redistributes the HPC/HPH PcG proteins, with the exception of HPC1, to large chromosomal PcG domains. Since only the full-length SUV39H1 protein and not the truncated SUV39H1 N-chromo protein causes this relocalization of the HPC/HPH PcG proteins, this redistribution reveals an important role for the SUV39H1 SET domain. These findings indicate that the abilities of SUV39H1 to interact with HPC2 and to relocalize the HPC/HPH PcG proteins are independent functions and further suggest that a functional HMTase activity is required to induce PcG protein redistribution, as has been observed for mammalian HP1 (1, 22).
An active SUV39H1 HMTase is required to alter the nuclear localization of HPC/HPH PcG proteins.
The SUV39H1 protein specifically methylates lysine 9 on histone H3. For this, both the SET domain and the adjacent cysteine-rich regions are necessary. A point mutation within the SET domain (H324L) abolishes the HMTase activity of SUV39H1 (20). To test whether the SET domain is of importance in the relocalization of HPC/HPH PcG proteins, we transiently overexpressed a myc-tagged SUV39H1 (H324L) mutant in HeLa cells. We double labeled these cells with rabbit polyclonal antibodies directed against the indicated PcG proteins (Fig. 7A) and a mouse monoclonal antibody (9E10) directed against the myc tag in the H324L mutant protein (Fig. 7A). We observed no change in the nuclear localization of HPC2 (Fig. 7A), as well as in that of RING1, HPH1, HPH2, and BMI1 (data not shown), in cells overexpressing SET domain mutant SUV39H1 (H324L). Mutant SUV39H1 (H324L) also has no effect on the localization of EZH2 (Fig. 7A).
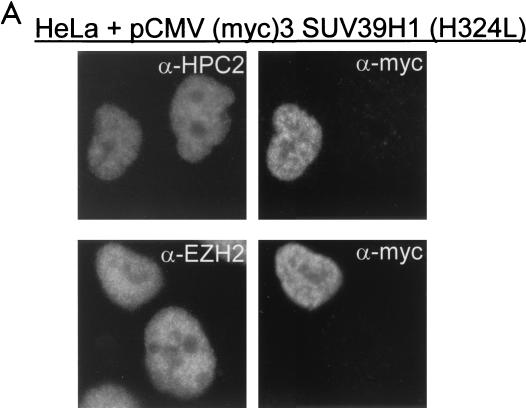
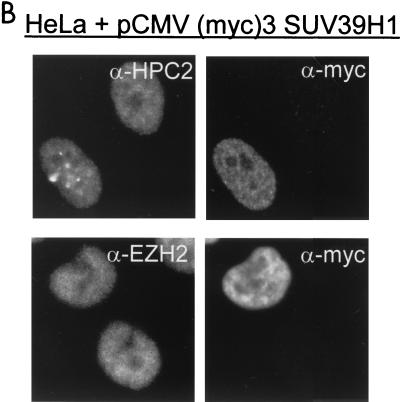
FIG. 7.
An intact and functionally active SET domain of SUV39H1 is a prerequisite for altering the nuclear distribution of HPC/HPH PcG proteins. (A) HeLa cells were transiently transfected with myc-tagged SUV39H1 (H324L). This specific SET domain mutation abolishes the ability of the SUV39H1 protein to methylate histones. Cells were double stained with rabbit polyclonal antibodies against HPC2 and EZH2 (left) and a mouse monoclonal antibody against the myc tag to allow detection of the myc-tagged SUV39H1 (H324L) (right). Transient expression of the mutant SUV39H1 protein did not alter the localization of HPC2, suggesting an important role for the SET domain of SUV39H1 in determining the nuclear localization of the HPC/HPH PcG proteins. (B) HeLa cells were transiently transfected with myc-tagged SUV39H1. Cells were double stained with rabbit polyclonal antibodies against HPC2 and EZH2 (left) and a mouse monoclonal antibody against the myc tag to allow detection of the myc-tagged SUV39H1 (right). Under experimental conditions similar to those for the SUV39H1 (H324L) mutant, transient expression of SUV39H1 was sufficient to alter the nuclear localization of HPC2.
To ensure that the inability of mutant SUV39H1 (H324L) to relocate the HPC/HPH PcG proteins is really due to the mutation within the SET domain of SUV39H1 and not due to indirect defects, we determined whether the functional SUV39H1 protein is able to relocate the HPC/HPH PcG proteins under similar experimental conditions. We therefore transiently overexpressed myc-tagged SUV39H1 in HeLa cells and double labeled these cells as described above (Fig. 7B). We observed that cells expressing the SUV39H1 protein display a redistribution of HPC2 to large nuclear domains (Fig. 7B). This also occurs with RING1, HPH1, HPH2, and BMI1 proteins (data not shown). Transient overexpression of SUV39H1 has no effect on the nuclear localization of the EZH2 protein (Fig. 7B).
In conclusion, the SUV39H1 (H324L) mutant is not able to alter the nuclear localization of the HPC2, RING1, BMI, HPH1, and HPH2 proteins, whereas SUV39H1 is able to alter the nuclear localization of these proteins under similar experimental conditions. These findings indicate that an intact and functionally active SET domain of SUV39H1 is a prerequisite for this activity.
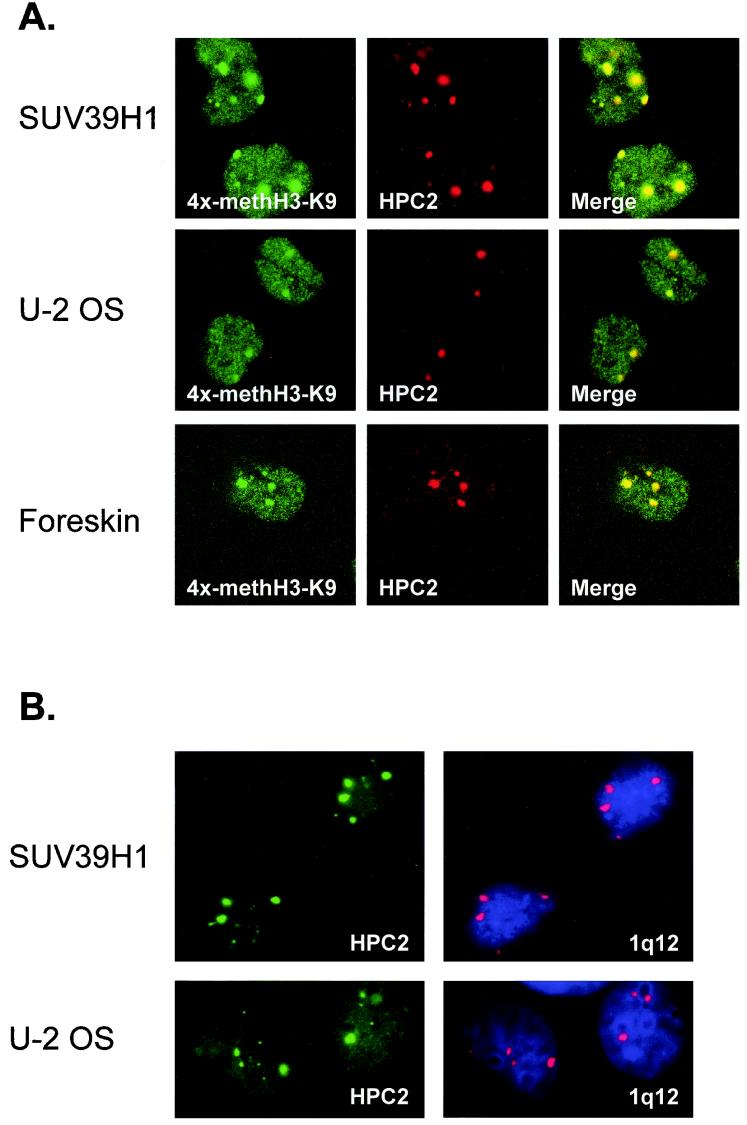
The HPC/HPH PcG proteins colocalize in PcG domains with methylated histone H3-K9 concentrated at pericentromeric heterochromatin regions (1q12) on chromosome 1.
To examine whether the SUV39H1-induced relocalization of the HPC/HPH PcG proteins coincides with the presence of increased H3-K9 methylation in these chromosomal domains, we performed immunofluorescence double-labeling experiments on HeLa cells stably expressing (myc)3-SUV39H1 (Fig. 8A, SUV39H1). We used rabbit polyclonal antibodies directed against dimethylated H3-K9 termini (29) and a mouse monoclonal antibody against HPC2. We observed H3-K9 methylation in nuclei of (myc)3-SUV39H1 HeLa cells in a fine granular pattern but also in larger nuclear domains (Fig. 8A, SUV39H1). Within these larger nuclear domains, the HPC2 protein colocalizes with methylated H3-K9 (Fig. 8A, SUV39H1). The HPC/HPH PcG proteins all localize in large nuclear PcG domains in U-2 OS cells (12, 35, 37, 43). To examine whether methylated H3-K9 is present in these large PcG domains in U-2 OS cells, we performed double-labeling experiments similar to those described above. We observed that methylated H3-K9 is present in nuclei of U-2 OS cells in a fine granular pattern but also in the larger domains in which HPC2 and methylated H3-K9 largely colocalized (Fig. 8A, U-2 OS). Since both HeLa and U-2 OS are established cell lines, we wondered whether the same colocalization could be found in primary cells. Therefore we performed the same double-labeling experiments with human primary foreskin fibroblasts. In approximately 20% of these cells we observed the large PcG domains and, importantly, increased methylated H3-K9 labeling in these domains (Fig. 8A, foreskin).
FIG. 8.
HPC/HPH PcG proteins colocalize with methylated histone H3-K9 and associate with DNA sequences specific for pericentromeric heterochromatin regions (1q12) on human chromosome 1. (A) HeLa cells stably expressing (myc)3-SUV39H1 (top), U-2 OS cells (middle), and human primary foreskin fibroblasts (bottom) were used for double-labeling experiments with a rabbit polyclonal antibody directed against methylated histone H3-K9 (methylated H3-K9) and a mouse monoclonal antibody directed against HPC2. HPC2 colocalizes with methylated histone H3-K9 in the large PcG domains, but methylated histone H3-K9 is also present outside these nuclear domains (merge). (B) HeLa cells stably expressing (myc)3-SUV39H1 (top) and U-2 OS cells (bottom) were stained with a polyclonal antibody directed against HPC2 and a DNA probe specific for pericentromeric region 1q12 on chromosome 1 by using a combined immunofluorescence-in situ hybridization technique. Nuclei were counterstained with DAPI (blue).
In a number of cell lines, such as U-2 OS and HT-1080, the HPC/HPH PcG proteins colocalize in PcG domains, chromosomal domains with DNA sequences that are associated with large pericentromeric heterochromatin (1q12) on chromosome 1 and with related pericentromeric sequences on different chromosomes (39). To examine whether the HPC/HPH PcG proteins colocalize with 1q12 sequences in the (myc)3-SUV39H1-expressing HeLa cells, we performed a combined immunofluorescence/in situ hybridization technique to allow simultaneous detection of the HPC2 protein and the 1q12 DNA (Fig. 8B, SUV39H1). We used a rabbit polyclonal antibody directed against HPC2 and DNA probe pUC 1.77, which recognizes pericentromeric region q12 of chromosome 1 and related pericentromeric sequences on different chromosomes (7, 39). Nuclei were counterstained with DAPI. We observed that HPC2 colocalizes in the large PcG domains with DNA sequences that are associated with the 1q12 region (Fig. 8B, SUV39H1), similar to what was described for HT-1080 cells (39) and U-2 OS cells (Fig. 8B, U-2 OS).
In conclusion, overexpression of SUV39H1 in HeLa cells causes a redistribution of HPC/HPH PcG proteins to large PcG domains, in which they colocalize with methylated histone H3-K9 and DNA sequences which are associated with pericentromeric regions (1q12) on chromosome 1 and related pericentromeric sequences on different chromosomes.
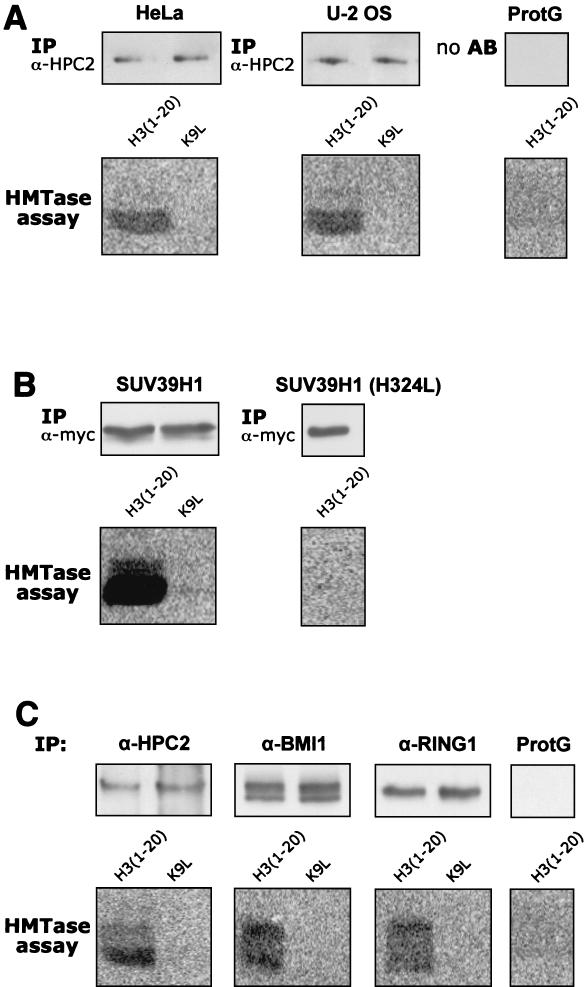
HPC2 coimmunoprecipitates endogenous HMTase activity.
HPC2 is able to interact with the SUV39H1 HMTase both in vitro and in vivo, raising the intriguing possibility that there may also be endogenous HMTase activity associated with HPC2. We performed IPs with rabbit polyclonal antibodies directed against HPC2 on nuclear extracts from both HeLa and U-2 OS cells, followed by an HMTase activity assay (Fig. 9A). As a positive control we used HeLa cells stably expressing (myc)3-SUV39H1 and performed IPs with a myc (9E10) monoclonal antibody (Fig. 9B). To determine whether the immunoprecipitates possessed H3-K9 HMTase activity, we used an unmodified histone H3 N-terminal peptide [H3(1-20)] and a mutant histone H3 N-terminal peptide (K9L) as substrates in the presence of S-adenosyl-l-[methyl-14C]methionine. The reaction products were separated by SDS-PAGE and visualized by fluorography. The immunoprecipitated (myc)3-SUV39H1 displayed H3-K9 HMTase activity with the histone H3 peptide but not with the mutant histone H3 peptide (Fig. 9B), indicating that the methyltransferase activity is specific for lysine 9, as described previously (33). Notably, in both HeLa and U-2 OS cells (Fig. 9A), we observed endogenous H3-K9 HMTase activity associated with the HPC2 immunoprecipitates using the histone H3 peptide but not using the mutant K9L peptide. To further strengthen these observations, we also performed IPs using antibodies against other components of the HPC/HPH PcG complex. Specifically, we found that antibodies against BMI1 and RING1 also immunoprecipitated H3-K9 HMTase activity in HeLa cells (Fig. 9C). When the primary antibodies were not included in the immunoprecipitates, we did not observe H3-K9 HMTase activity (Fig. 9A and C, ProtG). These results demonstrate that in two distinct cell lines antibodies against different HPC/HPH PcG proteins are able to immunoprecipitate H3-K9 HMTase activity.
FIG. 9.
HPC/HPH PcG proteins immunoprecipitate histone H3-K9 methyltransferase activity. IPs were performed on nuclear extracts of either HeLa or U-2 OS cells (A), HeLA cells overexpressing (myc)3-SUV39H1 (B), and HeLa cells (C). Antibodies against HPC2 (A), myc (B), or HPC2, BMI1, and RING1 (C) were used. As a negative control protein-Sepharose G (ProtG) (A and C) alone was used, without the addition of primary antibodies. The immunoprecipitates were probed for HMTase activity to transfer a labeled methyl group to a 20-aa N-terminal peptide of histone H3 [H3(1-20)] and a mutant 20-aa N-terminal peptide of histone H3 (K9L). The upper part of each panel represents the equal loading of the immunoprecipitated material, which was subjected to an HMTase activity assay (bottom part of each panel).
DISCUSSION
SUV39H1 selectively interacts with a specific class of vertebrate Pc homologs.
The PcG proteins are involved in the stable and heritable repression of gene expression by forming multimeric chromatin-associated protein complexes. At least two distinct PcG complexes exist, the HPC/HPH PcG complex and the EED/EZH2 PcG complex (43, 55). Repression mediated by the EED/EZH2 PcG complex involves histone deacetylation (54). Pc contains a chromodomain, which it shares with heterochromatin-associated protein HP1 (27). It has previously been shown that mammalian HP1 is associated with HMTase protein SUV39H1 (1). This association results in the targeting of HP1 to specific nuclear domains (22). Based on the parallels between the Pc protein and the HP1 protein, we screened for potential interactions between PcG proteins and HMTase SUV39H1 using the yeast two-hybrid system. Here we demonstrated that SUV39H1 interacts specifically with human Pc protein HPC2. We found no interactions in the two-hybrid system with other human PcG proteins. We substantiated the two-hybrid interaction between HPC2 and SUV39H1 by performing GST pull-down assays and in vivo coimmunoprecipitations. The observed interaction between in vitro-translated and radioactively labeled SUV39H1 and GST-HPC2 and GST-XPc2 confirmed that the interaction is a direct one.
The specificity of the SUV39H1 interaction for HPC2 but not M33/MPc1 raises the question of whether an interaction between Drosophila Pc and Drosophila Su(var)3-9 exists. Several observations support a biochemical and genetic interaction between various PcG genes and modifiers of PEV to which the Su(var)3-9 belongs (46). The Drosophila protein Enhancer of Pc, a member of the PcG proteins, is itself a suppressor of PEV. GAGA factor, a modifier of PEV, coimmunoprecipitates with Pc (14). HDAC Rpd3 has been classified as belonging to the Su(var) group (9). HDAC is part of the human EED/EZH2 PcG complex (54), and HDAC Rpd3 is part of the Drosophila Esc/E(z) PcG complex (51). These findings suggest that a link between PcG proteins and the Su(var)3-9 protein in Drosophila is a distinct possibility. In agreement with this notion it has been observed that Su(var)3-9 mutants enhance the homeotic phenotype of Pc mutants (G. Reuter, personal communication). This supports the idea that the interactions between SUV39H1 and HPC2 that we find are not limited to the human system but rather are conserved in Drosophila as well.
SUV39H1 targets HPC/HPH PcG proteins to nuclear domains that contain methylated histone H3-K9 and sequences that are associated chromosome 1 region q12.
Overexpression of SUV39H1 in HeLa cells leads to an altered nuclear localization of HP1 (22). Our immunofluorescence studies demonstrate that overexpression of SUV39H1 also induces a relocalization of the HPC/HPH PcG complex to large nuclear domains in HeLa cells. This phenomenon is specific for the HPC/HPH PcG complex, since the nuclear localization of EED, EZH2, and YY1, which are members of the EED/EZH2 PcG complex, is not affected by the overexpressed SUV39H1. To address the question of whether the HMTase activity of SUV39H1 is of importance for the relocalization of the HPC/HPH PcG proteins, we transiently overexpressed SUV39H1 (H324L) in HeLa cells. SUV39H1 (H324L) contains a mutation within the SET domain, which abolishes the HMTase activity. Overexpression of mutant SUV39H1 (H324L) does not induce a relocalization of the HPC/HPH PcG proteins, indicating that a functional and intact SET domain is required for relocating the HPC/HPH PcG proteins toward the large nuclear PcG domains. Similar results have been obtained with HP1. Only overexpression of an intact SUV39H1, not of the mutant SUV39H1 (H324L), has an effect on the nuclear relocalization of HP1 (20). The requirement of an intact functional SET domain, and hence H3-K9 HMTase activity, suggests an important role for histone methylation in targeting both HP1 and the HPC/HPH PcG complex.
What is the nature of these large nuclear PcG domains? In these domains, PcG proteins HPC2, BMI1, RING1, and HPH are associated with pericentromeric heterochromatin regions (1q12) on chromosome 1 and 1q12-like sequences (39) (Fig. 9). It is important that we now find increased histone H3-K9 methylation in these PcG domains. We find this not only in the established cell line U-2 OS and in SUV39H1-overexpressing HeLa cells but also in primary human foreskin fibroblasts. This supports a model in which the 1q12 and 1q12-like chromosomal locations are characterized by intrinsically high levels of methylated histone H3-K9 in certain cells, such as U-2 OS cells and primary foreskin fibroblasts. In HeLa cells the 1q12 and 1q12-like chromosomal locations contain less histone H3-K9 methylation. However, raising the levels of methylated histone H3-K9 at these chromosomal locations in HeLa by overexpressing SUV39H1 creates a docking site for HPC/HPH PcG proteins. As a result large HPC/HPH PcG domains are formed in SUV39H1-overexpressing HeLa cells.
Significance of the functional link between SUV39H1 and HPC2.
Based on homology between heterochromatin-associated protein HP1 and Pc, several models have suggested that PcG proteins are able to repress genes by the formation of heterochromatin-like structures. It has previously been reported that HP1 is able to interact with SUV39H1 (1). Here we show that human Pc homolog HPC2 is able to interact with SUV39H1. What is the functional significance of this interaction? One obvious possibility is that the interaction between SUV39H1 and HPC2 is crucial for targeting the HPC/HPH PcG proteins to the PcG domains. As indicated above, the presence of HPC/HPH proteins in the corresponding large nuclear PcG domains coincides with increased methylated histone H3-K9 in the same domains. When the level of methylated histone H3-K9 is below a certain threshold, no methylated histone H3-K9 or PcG domains are observed at 1q12 and 1q12-like locations. Only when the histone H3-K9 methylation at these sites is raised do PcG domains form. However, one observation argues against this straightforward scheme. Overexpression of a truncated SUV39H1 protein that is still able to interact with HPC2 is not capable of relocating the HPC/HPH PcG proteins. This truncated SUV39H1 protein lacks the SET domain, which possesses HMTase activity specific for histone H3-K9. This implies that the occurrence of histone H3-K9 methylation plays a more important role in targeting the PcG proteins than the physical interaction between HPC2 and SUV39H1. Similar findings have been reported for the relocation of HP1 under the influence of overexpressed SUV39H1. HP1 still interacts with the same truncated SUV39H1 mutant, which lacks the SET domain and which therefore lacks functional HMTase activity. Overexpression of this mutant does not relocate the HP1 protein (22). Therefore an intact HMTase activity appears to be the pivotal characteristic of the SUV39H1 protein for relocating HPC2 as well as HP1. The significance of the physical interaction between SUV39H1 and either HP1 or HPC2 is much less clear.
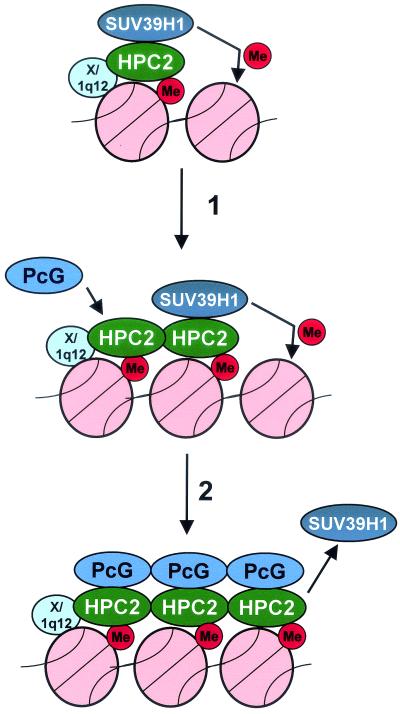
Since methylation of histones appears to be a stable modification, it provides an ideal epigenetic marker for stable maintenance of chromatin states (15). On the basis of our results we propose a model in which the interaction between SUV39H1 and HPC2, the HMTase activity of SUV39H1, and methylated histone H3-K9 play an important role in the formation of PcG-mediated repressive chromatin states (Fig. 10). In this model we propose that the interaction between SUV39H1 and HPC2 is present only when the PcG complex is being established at the SUV39H1 methylated H3-K9 chromatin sites that contain 1q12 and 1q12-like sequences. A combination of different signals is required to recruit the HPC/HPH PcG complex to 1q12 or 1q12-like chromosomal loci. In the first place there must be a signal that conveys specificity for the 1q12 locus. In the model we postulate a protein (X/1q12) that binds specifically to 1q12 and 1q12-like loci and that serves as the initial targeting signal (Fig. 10). A similar scheme can be proposed for the SUV39H1-HP1 interaction and targeting. It is a long-established fact that HP1 and PcG proteins are present at different chromosomal locations (27). It is conceivable that there are different proteins involved in targeting HP1 or HPC2 to distinct chromosomal locations. For HPC2 this includes 1q12 and 1q12-like sequences plus protein X/1q12; for HP1 other sequences and a different protein are involved.
FIG. 10.
Model describing the significance of the interaction between SUV39H1 and PcG proteins. The SUV39H1 protein is targeted to 1q12 or 1q12-like chromosomal locations. This involves an as yet unknown protein (X/1q12) that targets HPC2 and/or SUV39H1 specifically to 1q12 or 1q12-like chromosomal locations. Subsequently the SUV39H1 protein adds a methyl group to histone H3-K9. Once this initial event has occurred, SUV39H1 progressively adds methyl groups to adjacent nucleosomes (step 1) and additional PcG proteins are recruited to the methylated 1q12 or 1q12-like chromosomal loci. Eventually (step 2) the chromosomal locus is stably silenced by the recruited HPC/HPH PcG protein complex. The SUV39H1 protein is then no longer needed and no longer interacts with the PcG protein complex.
Our results further indicate an additional signal, the density of H3-K9 methylation at the 1q12 and 1q12-like loci. Only when the level of H3-K9 methylation at the 1q12 and 1q12-like chromosomal loci is high enough can recruitment of HPC2 and other HPC/HPH PcG proteins occur. Once the initial histone H3-K9 methylation has taken place, HPC2 and the other PcG proteins are targeted to the methylated sites and H3-K9 methylation proceeds (Fig. 10, step 1). When stable association between the H3-K9 methylated 1q12 chromosomal sites and the HPC/HPH PcG complex is established, SUV39H1 is no longer needed and the protein is released (Fig. 10, step 2).
Postulating the transient nature of the SUV39H1-HPC2 interaction explains the need to overexpress SUV39H1 to detect coimmunoprecipitation between SUV39H1 and HPC2. In addition, however, we point out that the expression level of SUV39H1 changes considerably during embryonic development. The levels of the SUV39H enzymes are highest around embryonic days 14 to 16 of mouse development, and thereafter they are significantly down-regulated and remain low in the adult (T. Jenuwein, unpublished observations). This may suggest that only in a small time window during embryonic development is there sufficient SUV39H1 protein to allow a stable in vivo interaction between SUV39H1 and Pc homologs. In adult organisms (mouse or human) insufficient SUV39H1 is present to detect this in vivo interaction and ectopic expression is needed. In that case ectopic expression of SUV39H1 is required to raise the density of H3-K9 methylation at specific chromosomal loci. This then induces the recruitment of HPC/HPH PcG proteins to these loci, which otherwise may occur only during embryonic development. Additional experiments will be required to distinguish between these different options.
Acknowledgments
We thank Louise Aagaard for contributing to the identification of relocalization of BMI1 in SUV39H1-overexpressing cells, Wijnand Takkenberg for help with the confocal laser scanning microscope, Ulrike Waginger for help with the immuno-FISH staining, Phil Barnett for critically reading the manuscript, and Gunter Reuter for sharing unpublished data with us.
This work was sponsored in part by the Human Frontier Science Program (RG0039/1999-M).
REFERENCES
- 1.Aagaard, L., G. Laible, P. Selenko, M. Schmid, R. Dorn, G. Schotta, S. Kuhfittig, A. Wolf, A. Lebersorger, P. B. Singh, G. Reuter, and T. Jenuwein. 1999. Functional mammalian homologues of the Drosophila PEV-modifier Su(var)3-9 encode centromere-associated proteins which complex with the heterochromatin component M31. EMBO J. 18:1923-1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aasland, R., and A. F. Stewart. 1995. The chromo shadow domain, a second chromo domain in heterochromatin-binding protein 1, HP1. Nucleic Acids Res. 23:3168-3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bannister, A. J., P. Zegerman, J. F. Partridge, E. A. Miska, J. O. Thomas, R. C. Allshire, and T. Kouzarides. 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410:120-124. [DOI] [PubMed] [Google Scholar]
- 4.Breiling, A., E. Bonte, S. Ferrari, P. B. Becker, and R. Paro. 1999. The Drosophila Polycomb protein interacts with nucleosomal core particles in vitro via its repression domain. Mol. Cell. Biol. 19:8451-8460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bunker, C. A., and R. E. Kingston. 1994. Transcriptional repression by Drosophila and mammalian Polycomb group proteins in transfected mammalian cells. Mol. Cell. Biol. 14:1721-1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheung, P., C. D. Allis, and P. Sassone-Corsi. 2000. Signaling to chromatin through histone modifications. Cell 103:263-271. [DOI] [PubMed] [Google Scholar]
- 7.Cooke, H. J., and J. Hindley. 1979. Cloning of human satellite III DNA: different components are on different chromosomes. Nucleic Acids Res. 6:3177-3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeCamillis, M., N. S. Cheng, D. Pierre, and H. W. Brock. 1992. The polyhomeotic gene of Drosophila encodes a chromatin protein that shares polytene chromosome-binding sites with Polycomb. Genes Dev. 6:223-232. [DOI] [PubMed] [Google Scholar]
- 9.De Rubertis, F., D. Kadosh, S. Henchoz, D. Pauli, G. Reuter, K. Struhl, and P. Spierer. 1996. The histone deacetylase RPD3 counteracts genomic silencing in Drosophila and yeast. Nature 384:589-591. [DOI] [PubMed] [Google Scholar]
- 10.Franke, A., M. DeCamillis, D. Zink, N. Cheng, H. W. Brock, and R. Paro. 1992. Polycomb and polyhomeotic are constituents of a multimeric protein complex in chromatin of Drosophila melanogaster. EMBO J. 11:2941-2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gould, A. 1997. Functions of mammalian Polycomb group and trithorax group related genes. Curr. Opin. Genet. Dev. 7:488-494. [DOI] [PubMed] [Google Scholar]
- 12.Gunster, M. J., D. P. Satijn, K. M. Hamer, J. L. den Blaauwen, D. de Bruijn, M. J. Alkema, M. van Lohuizen, R. van Driel, and A. P. Otte. 1997. Identification and characterization of interactions between the vertebrate Polycomb-group protein BMI1 and human homologs of Polyhomeotic. Mol. Cell Biol. 17:2326-2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hamvas, R. M., W. Reik, S. J. Gaunt, S. D. Brown, and P. B. Singh. 1992. Mapping of a mouse homolog of a heterochromatin protein gene to the X chromosome. Mamm. Genome 2:72-75. [DOI] [PubMed] [Google Scholar]
- 14.Horard, B., C. Tatout, S. Poux, and V. Pirrotta. 2000. Structure of a Polycomb response element and in vitro binding of Polycomb group complexes containing GAGA factor. Mol. Cell. Biol. 20:3187-3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jenuwein, T. 2001. Re-SET-ting heterochromatin by histone methyltransferases. Trends Cell Biol. 11:266-273. [DOI] [PubMed] [Google Scholar]
- 16.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 293:1074-1080. [DOI] [PubMed] [Google Scholar]
- 17.Jurgens, G. 1985. A group of genes controlling the spatial expression of the bithorax complex in Drosophila. Nature 316:153-155. [Google Scholar]
- 18.Kingston, R. E., C. A. Bunker, and A. N. Imbalzano. 1996. Repression and activation by multiprotein complexes that alter chromatin structure. Genes Dev. 10:905-920. [DOI] [PubMed] [Google Scholar]
- 19.Koonin, E. V., S. Zhou, and J. C. Lucchesi. 1995. The chromo superfamily: new members, duplication of the chromo domain and possible role in delivering transcription regulators to chromatin. Nucleic Acids Res. 23:4229-4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lachner, M., D. O'Carroll, S. Rea, K. Mechtler, and T. Jenuwein. 2001. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410:116-120. [DOI] [PubMed] [Google Scholar]
- 21.Lonie, A., R. D'Andrea, R. Paro, and R. Saint. 1994. Molecular characterisation of the Polycomblike gene of Drosophila melanogaster, a trans-acting negative regulator of homeotic gene expression. Development 120:2629-2636. [DOI] [PubMed] [Google Scholar]
- 22.Melcher, M., M. Schmid, L. Aagaard, P. Selenko, G. Laible, and T. Jenuwein. 2000. Structure-function analysis of SUV39H1 reveals a dominant role in heterochromatin organization, chromosome segregation, and mitotic progression. Mol. Cell. Biol. 20:3728-3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Messmer, S., A. Franke, and R. Paro. 1992. Analysis of the functional role of the Polycomb chromo domain in Drosophila melanogaster. Genes Dev. 6:1241-1254. [DOI] [PubMed] [Google Scholar]
- 24.Muller, J. 1995. Transcriptional silencing by the Polycomb protein in Drosophila embryos. EMBO J. 14:1209-1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ng, J., C. M. Hart, K. Morgan, and J. A. Simon. 2000. A Drosophila ESC-E(Z) protein complex is distinct from other Polycomb group complexes and contains covalently modified ESC. Mol. Cell. Biol. 20:3069-3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paro, R. 1990. Imprinting a determined state into the chromatin of Drosophila. Trends Genet. 6:416-421. [DOI] [PubMed] [Google Scholar]
- 27.Paro, R., and D. S. Hogness. 1991. The Polycomb protein shares a homologous domain with a heterochromatin-associated protein of Drosophila. Proc. Natl. Acad. Sci. USA 88:263-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pearce, J. J., P. B. Singh, and S. J. Gaunt. 1992. The mouse has a Polycomb-like chromobox gene. Development 114:921-929. [DOI] [PubMed] [Google Scholar]
- 29.Peters, A. H., D. O'Carroll, H. Scherthan, K. Mechtler, S. Sauer, C. Schofer, K. Weipoltshammer, M. Pagani, M. Lachner, A. Kohlmaier, S. Opravil, M. Doyle, M. Sibilia, and T. Jenuwein. 2001. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell 107:323-337. [DOI] [PubMed] [Google Scholar]
- 30.Pirrotta, V. 1997. PcG complexes and chromatin silencing. Curr. Opin. Genet. Dev. 7:249-258. [DOI] [PubMed] [Google Scholar]
- 31.Platero, J. S., T. Hartnett, and J. C. Eissenberg. 1995. Functional analysis of the chromo domain of HP1. EMBO J. 14:3977-3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rastelli, L., C. S. Chan, and V. Pirrotta. 1993. Related chromosome binding sites for zeste, suppressors of zeste and Polycomb group proteins in Drosophila and their dependence on Enhancer of zeste function. EMBO J. 12:1513-1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rea, S., F. Eisenhaber, D. O'Carroll, B. D. Strahl, Z. W. Sun, M. Schmid, S. Opravil, K. Mechtler, C. P. Ponting, C. D. Allis, and T. Jenuwein. 2000. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406:593-599. [DOI] [PubMed] [Google Scholar]
- 34.Reijnen, M. J., K. M. Hamer, J. L. den Blaauwen, C. Lambrechts, I. Schoneveld, R. van Driel, and A. P. Otte. 1995. Polycomb and bmi-1 homologs are expressed in overlapping patterns in Xenopus embryos and are able to interact with each other. Mech. Dev. 53:35-46. [DOI] [PubMed] [Google Scholar]
- 35.Satijn, D. P., M. J. Gunster, J. van der Vlag, K. M. Hamer, W. Schul, M. J. Alkema, A. J. Saurin, P. S. Freemont, R. van Driel, and A. P. Otte. 1997. RING1 is associated with the Polycomb group protein complex and acts as a transcriptional repressor. Mol. Cell. Biol. 17:4105-4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Satijn, D. P., K. M. Hamer, J. den Blaauwen, and A. P. Otte. 2001. The Polycomb group protein EED interacts with YY1, and both proteins induce neural tissue in Xenopus embryos. Mol. Cell. Biol. 21:1360-1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Satijn, D. P., D. J. Olson, J. van der Vlag, K. M. Hamer, C. Lambrechts, H. Masselink, M. J. Gunster, R. G. Sewalt, R. van Driel, and A. P. Otte. 1997. Interference with the expression of a novel human Polycomb protein, hPc2, results in cellular transformation and apoptosis. Mol. Cell. Biol. 17:6076-6086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Satijn, D. P., and A. P. Otte. 1999. Polycomb group protein complexes: do different complexes regulate distinct target genes? Biochim. Biophys. Acta 1447:1-16. [DOI] [PubMed] [Google Scholar]
- 39.Saurin, A. J., C. Shiels, J. Williamson, D. P. Satijn, A. P. Otte, D. Sheer, and P. S. Freemont. 1998. The human polycomb group complex associates with pericentromeric heterochromatin to form a novel nuclear domain. J. Cell Biol. 142:887-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schoorlemmer, J., C. Marcos-Gutierrez, F. Were, R. Martinez, E. Garcia, D. P. Satijn, A. P. Otte, and M. Vidal. 1997. Ring1A is a transcriptional repressor that interacts with the Polycomb-M33 protein and is expressed at rhombomere boundaries in the mouse hindbrain. EMBO J. 16:5930-5942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schumacher, A., and T. Magnuson. 1997. Murine Polycomb- and trithorax-group genes regulate homeotic pathways and beyond. Trends Genet. 13:167-170. [PubMed] [Google Scholar]
- 42.Sewalt, R. G., M. J. Gunster, J. van der Vlag, D. P. Satijn, and A. P. Otte. 1999. C-terminal binding protein is a transcriptional repressor that interacts with a specific class of vertebrate Polycomb proteins. Mol. Cell. Biol. 19:777-787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sewalt, R. G., J. van der Vlag, M. J. Gunster, K. M. Hamer, J. L. den Blaauwen, D. P. Satijn, T. Hendrix, R. van Driel, and A. P. Otte. 1998. Characterization of interactions between the mammalian Polycomb group proteins Enx1/EZH2 and EED suggests the existence of different mammalian Polycomb group protein complexes. Mol. Cell. Biol. 18:3586-3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Simon, J. 1995. Locking in stable states of gene expression: transcriptional control during Drosophila development. Curr. Opin. Cell Biol. 7:376-385. [DOI] [PubMed] [Google Scholar]
- 45.Simon, J., A. Chiang, and W. Bender. 1992. Ten different Polycomb group genes are required for spatial control of the abdA and AbdB homeotic products. Development 114:493-505. [DOI] [PubMed] [Google Scholar]
- 46.Sinclair, D. A., N. J. Clegg, J. Antonchuk, T. A. Milne, K. Stankunas, C. Ruse, T. A. Grigliatti, J. A. Kassis, and H. W. Brock. 1998. Enhancer of Polycomb is a suppressor of position-effect variegation in Drosophila melanogaster. Genetics 148:211-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Strahl, B. D., and C. D. Allis. 2000. The language of covalent histone modifications. Nature 403:41-45. [DOI] [PubMed] [Google Scholar]
- 48.Strahl, B. D., S. D. Briggs, C. J. Brame, J. A. Caldwell, S. S. Koh, H. Ma, R. G. Cook, J. Shabanowitz, D. F. Hunt, M. R. Stallcup, and C. D. Allis. 2001. Methylation of histone H4 at arginine 3 occurs in vivo and is mediated by the nuclear receptor coactivator PRMT1. Curr. Biol. 11:996-1000. [DOI] [PubMed] [Google Scholar]
- 49.Strahl, B. D., R. Ohba, R. G. Cook, and C. D. Allis. 1999. Methylation of histone H3 at lysine 4 is highly conserved and correlates with transcriptionally active nuclei in Tetrahymena. Proc. Natl. Acad. Sci. USA 96:14967-14972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tachibana, M., K. Sugimoto, T. Fukushima, and Y. Shinkai. 2001. SET domain-containing protein, G9a, is a novel lysine-preferring mammalian histone methylthansferase with hyperactivity and specific selectivity to lysines 9 and 27 of histone H3. J. Biol. Chem. 276:25309-25317. [DOI] [PubMed] [Google Scholar]
- 51.Tie, F., T. Furuyama, J. Prasad-Sinha, E. Jane, and P. J. Harte. 2001. The Drosophila Polycomb group proteins ESC and E(Z) are present in a complex containing the histone-binding protein p55 and the histone deacetylase RPD3. Development 128:275-286. [DOI] [PubMed] [Google Scholar]
- 52.Turner, B. M. 2000. Histone acetylation and an epigenetic code. Bioessays 22:836-845. [DOI] [PubMed] [Google Scholar]
- 53.Vandel, L., and D. Trouche. 2001. Physical association between the histone acetyl transferase CBP and a histone methyl transferase. EMBO Rep. 2:21-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van der Vlag, J., and A. P. Otte. 1999. Transcriptional repression mediated by the human polycomb-group protein EED involves histone deacetylation. Nat. Genet. 23:474-478. [DOI] [PubMed] [Google Scholar]
- 55.van Lohuizen, M., M. Tijms, J. W. Voncken, A. Schumacher, T. Magnuson, and E. Wientjens. 1998. Interaction of mouse Polycomb group (Pc-G) proteins Enx1 and Enx2 with Eed: indication for separate Pc-G complexes. Mol. Cell. Biol. 18:3572-3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van Lohuizen, M., S. Verbeek, B. Scheijen, E. Wientjens, H. van der Gulden, and A. Berns. 1991. Identification of cooperating oncogenes in E mu-myc transgenic mice by provirus tagging. Cell 65:737-752. [DOI] [PubMed] [Google Scholar]
- 57.Wu, J., and M. Grunstein. 2000. 25 years after the nucleosome model: chromatin modifications. Trends Biochem. Sci. 25:619-623. [DOI] [PubMed] [Google Scholar]
- 58.Zink, B., and R. Paro. 1989. In vivo binding pattern of a trans-regulator of homoeotic genes in Drosophila melanogaster. Nature 337:468-471. [DOI] [PubMed] [Google Scholar]