Abstract
The yeast transcription factor Gcn4 is regulated by amino acid starvation at the levels of both protein synthesis and stability. Gcn4 degradation depends on the ubiquitination complex SCFCDC4 and requires phosphorylation by the cyclin-dependent kinase Pho85. Here, we show that Pcl5 is the Pho85 cyclin specifically required for Gcn4 degradation. PCL5 is itself induced by Gcn4 at the level of transcription. However, even when PCL5 is constitutively overexpressed, Pho85-associated Gcn4 phosphorylation activity is reduced in starved cells and Gcn4 degradation is decreased. Under these conditions, the Pcl5 protein disappears because of rapid constitutive turnover. We suggest that, by virtue of its constitutive metabolic instability, Pcl5 may be a sensor of cellular protein biosynthetic capacity. The fact that PCL5 is transcriptionally induced in the presence of Gcn4 suggests that it is part of a homeostatic mechanism that reduces Gcn4 levels upon recovery from starvation.
Protein phosphorylation, a major mechanism of posttranslational regulation of protein activity, can also modulate the destruction of the modified protein. In particular, ubiquitination of proteins by SCF-type ubiquitin ligases, which leads to their degradation, is often induced by phosphorylation of the substrate (9, 42). Gcn4 is a yeast transcriptional activator involved in biosynthesis of amino acids and purines (13, 16) that regulates a significant proportion of the yeast genes (33). Starvation for amino acids leads to an increase in Gcn4 translation (10) by a mechanism that involves phosphorylation of the general translation initiation factor eIF-2α by the kinase Gcn2 (6; reviewed in reference 12). In addition, Gcn4 is degraded extremely rapidly but is stabilized under conditions of amino acid limitation or partial inhibition of protein synthesis (21, 26). Degradation of Gcn4 depends on its phosphorylation at a specific residue, Thr165 (26), and on its ubiquitination by the ubiquitin-conjugating enzyme Cdc34 (21) in conjunction with the ubiquitin ligase SCFCDC4 (4, 26). Two cyclin-dependent kinases (CDKs), Pho85 (26) and Srb10 (4), are involved in Gcn4 degradation. On the basis of the phenotype of the respective deletion mutants, Pho85 is solely involved in regulation of Gcn4 degradation by starvation (4, 26).
CDKs absolutely require binding of an ancillary subunit, the cyclin, for their activity (30). A single CDK can be activated by a number of different cyclins; for example, at least 10 different cyclins are known to bind Pho85 (25). The function of the cyclin subunit in activation of the kinase is well established (18), but it is likely that specific cyclins also participate in targeting of the kinase to specific substrates (17, 38, 43; recently reviewed in reference 27). The role of the cyclin in conferring substrate specificity is probably best established in the case of Pho85. The different functions of Pho85 in metabolic regulation can be explained by targeting of the kinase by specific Pho85 cyclins (Pcls) to specific substrates. The role of Pho85 in phosphate assimilation depends on its targeting to the transcription factor Pho4 by the Pcl Pho80 (14, 20), whereas its role in glycogen synthesis depends on its targeting to the glycogen synthase Gsy2 by Pcl8 and Pcl10 (17, 43). The ancillary role of Pho85 in cell cycle progression (7, 24) may be due, in part, to targeting to the CDK inhibitor Sic1 by Pcl1 (34) and possibly also to interaction between Pcl2 and the transcription factor Swi5 (23).
Gcn4 degradation depends on the CDK Pho85 in vivo. In vitro, Pho85, in conjunction with the cyclin Pcl1, is able to phosphorylate Gcn4 at the critical residue for degradation, Thr165 (26). However, deletion of PCL1 did not affect Gcn4 degradation, suggesting that, in vivo, either additional Pcls, or altogether different Pcls, fulfill this function. Here, we show that a single Pcl, Pcl5, is required for the function of Pho85 in the degradation of Gcn4. The regulation of Gcn4 degradation by starvation can be explained by the effect of starvation on Pcl5 levels.
MATERIALS AND METHODS
Strains and culture conditions.
Strain L4210 (a ura3-52 leu2-2 bas1-2 bas2-2) was described before (21). Strains W303-1A (a ura3-1 can1-100 leu2-3,112 trp1-1 ade2-1 his3-11,15) and W303-1B (same but α) are originally from R. Rothstein. Strain KY795 (a ura3-1 can1-100) is a W303 derivative. The PCL5 open reading frame was deleted precisely from strain KY795 by using plasmid KB1125 to generate KY797 (a ura3-1 can1-1 pcl5Δ::hisG-URA3-hisG). KY798, a Ura− derivative of KY797, was isolated by selection on 5-fluoroorotic acid. KY827 (a ura3-1 can1-100 leu2-3,112 trp1-1 ade2-1 his3-11,15 pcl5Δ::hisG) was derived from a cross between KY797 and W303-1B, followed by selection on 5-fluoroorotic acid. KY846 was derived from a cross between DY5335 (pcl1Δ::ADE2 pcl2Δ::URA3 pcl9Δ::HIS3 pho80Δ::LEU2), kindly provided by David Stillman, and KY685, a lys2 derivative of W303-1A, to generate KY690 (α ura3-1 can1-100 leu2-3,112 trp1-1 ade2-1 his3-11,15 lys2 pcl1Δ::ADE2 pcl2Δ::URA3 pcl9Δ::HIS3). The URA3 marker was subsequently converted to LYS2 by homologous recombination to generate KY846 (α ura3-1 can1-100 leu2-3,112 trp1-1 ade2-1 his3-11,15 lys2 pcl1Δ::ADE2 pcl2Δ::LYS2 pcl9Δ::HIS3). Strains were grown either in yeast nitrogen base (YNB; Difco), in yeast extract-peptone-dextrose medium (39), or in synthetic complete (SC) medium as previously described (39), with the difference that the SC was supplemented with 100 μg each of every amino acid, adenine, and uracil per ml.
Plasmids.
The GAL::GCN4 intermediate expression plasmids pDAD-GCN4 (KB161) and pDAD-GCN4(T105A) (KB170) have been described before (21, 26). pDAD-GCN4(T165A) (KB1211) was constructed by substituting the HindIII-XbaI fragment of KB854 (26) for the homologous fragment in plasmid KB161. The GPD-HAxPHO80 plasmid (EB92) was obtained from E. O'Shea (University of California, San Francisco), and the pGAL-GST-PHO85 wild-type and mutant plasmids were obtained from M. Nishizawa (Keio University). KB537 (GAL1::EF1A) is from our laboratory collection. For the other plasmid constructions, PCR cloning methods (2) were used. The GAL1::GCN4 high-expression plasmid p414GAL1-GCN4 (KB843) was constructed by cloning an EcoRI-XhoI fragment of GCN4 generated by PCR into plasmid p414GAL1 (31) digested with the same enzymes. p414GAL1-GCN4 T105A (KB1105) and p414GAL1-GCN4 T165A (KB1155) were constructed by similarly cloning the indicated GCN4 mutant forms into p414GAL1. The GAL1::PCL5 plasmid KB1093 was constructed by cloning a PCR-generated MfeI-XhoI PCL5-containing fragment into p416GAL1 (31) digested with EcoRI and XhoI. The other GAL1::PCL plasmids, KB1237 (PCL1), KB1238 (PCL2), KB1239 (PCL9), and KB1240 (CLG1), were similarly constructed by inserting a PCL-containing EcoRI-XhoI fragment into p416GAL1 (31) digested with the same enzymes. The PCL5 deletion plasmid KB1125 was generated by cloning the 5′ and 3′ noncoding regions of PCL5 adjacent to the hisG-URA3-hisG “gene blaster” cassette (1). For recombinant protein expression, a hexahistidine-tagged version of Pcl5 was constructed by inserting a BamHI-XhoI PCL5 fragment generated by PCR into the bacterial expression vector pRSETA (Invitrogen), and a hexahistidine tagged version of Pcl1 was constructed by introducing it into the expression vector pQE60 (Qiagen) as a BamHI-NcoI fragment. The glutathione S-transferase (GST)-Pho85 wild-type and mutant expression vectors were obtained from M. Nishizawa (34). Construction of the GST-Gcn4 (residues 62 to 202) wild-type and T165A mutant expression vectors was described previously (26); the T105A and T105A T165A mutant expression vectors were constructed similarly.
Degradation assays.
Pulse-chase experiments were performed essentially as previously described (21). Briefly, overnight cultures were diluted in 10 ml and grown to mid-log phase in dropout medium, washed once with YNB medium containing all of the essential amino acids, concentrated to 0.3 ml, pulse-labeled for 5 min with 750 to 900 mCi of [35S]methionine (Express; NEN), pelleted again, and chased in growth medium containing 10 mM methionine and 10 mM cysteine. For the starvation studies, cells were harvested by centrifugation and grown in SD medium (39) plus adenine for 15 to 45 min before labeling and chased in the same medium plus methionine and cysteine. At various times during the chase, an aliquot of the culture was removed and incubated for 15 min on ice with 0.35 M NaOH-1.5% 2-mercaptoethanol, followed by precipitation with 6% trichloroacetic acid. The protein precipitate was resuspended by boiling in 2.5% sodium dodecyl sulfate (SDS)-5 mM EDTA, and equal amounts of trichloroacetic acid-precipitable radioactivity were immunoprecipitated in at least 10 volumes of buffer A (15) containing protein A-Sepharose (Pharmacia). The immunoprecipitates were run on SDS-polyacrylamide gels, and the protein bands were quantitated with a Fujix Bas 2000 bio-image analyzer (Fuji). The antibodies used were either rabbit anti-β-galactosidase (Cappel) or anti-Gcn4 or anti-Pcl5 custom rabbit antibodies raised against recombinant proteins.
Kinase assay.
Recombinant proteins were expressed and purified in accordance with standard protocols. The GST-Pho85 wild-type and mutant proteins were expressed in bacteria from plasmids pGEX-PHO85 and pGEX-PHO85(E53A), kindly provided by M. Nishizawa (34), bound to a glutathione-agarose resin (Sigma), and eluted with reduced glutathione. The Gcn4 (residues 62 to 202) fragments were expressed from the pGEX-4T-1 vector (Amersham Pharmacia Biotech) and bound to glutathione-agarose, and the Gcn4 moiety was cleaved from GST by overnight incubation of the beads with 10 μg of thrombin (Sigma) per ml. Cleavage with thrombin yielded protein fragments that included, in addition to Gcn4 residues 62 to 202, the vector-derived N-terminal extension Gly-Ser-Pro-Glu-Phe. The hexahistidine-tagged Pcl1 and Pcl5 proteins were expressed in bacteria and affinity purified by binding to a nickel-agarose column (Qiagen), followed by elution with increasing concentrations of imidazole. A centrifugal ultrafiltration device was used for buffer exchange and concentration of the proteins. For in vitro kinase assays, 10 ng of GST-Pho85, 10 ng of His6-Pcl5 or His6-Pcl1, and 0.5 μg of substrate were incubated at 30°C for 30 min with 1 μCi of [γ-32P]ATP and 0.1 mM cold ATP in 10 μl of kinase assay buffer (50 mM Tris, 10 mM MgCl2, 2 mM EDTA, 1 mM dithiothreitol, pH 7.5). The kinase reactions were terminated by addition of 10 μl of protein loading buffer, and the reaction mixtures were subjected to SDS-12% polyacrylamide gel electrophoresis. Immunoprecipitation (IP)/kinase assays were carried out essentially as described previously (26), except that the extract buffer contained 20 mM Tris (pH 7.5), 20 mM NaCl, 5 mM EDTA, 0.5% Triton X-100, phosphatase inhibitors (1 mM Na-pyrophosphate, 1 mM β-glycerophosphate, 1 mM NaF, and 0.3 mM NaVO3), protease inhibitors (2 mM phenylmethylsulfonyl fluoride, tosylsulfonyl phenylalanyl chloromethyl ketone [TPCK] at 50 μg/ml, and Nα-p-tosyl-l-lysine chloromethyl ketone [TLCK] at 50 μg/ml), and a 1:500 dilution of an antiprotease cocktail containing leupeptin, pepstatin, and chymostatin, each at 10 mg/ml of dimethyl sulfoxide. For each IP reaction, 200 μg of protein extract was reacted with 0.5 μg of anti-GST monoclonal antibody (Santa Cruz Biotechnology).
RESULTS
PCL5 is induced by Gcn4.
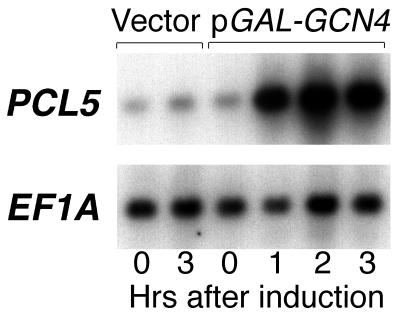
A report by Jia et al. (19) showed that the Pcl-encoding gene PCL5 is among the genes induced in a Gcn4-dependent manner by sulfometuron, an inhibitor of amino acid biosynthesis. To test more directly the induction of PCL5 by Gcn4, we ectopically expressed Gcn4 from the inducible GAL1,10 promoter. As shown in Fig. 1, the PCL5 transcript levels were strongly increased following induction of Gcn4. Quantitation of the PCL5 signal normalized to the EF1A signal yielded a 10-fold increase within 1 h of induction of GCN4 by galactose, corroborating the suggestion that PCL5 is a target of Gcn4. Analysis of the sequence upstream of PCL5 revealed two potential Gcn4 binding sites, with nearly perfect homology to the Gcn4 consensus binding sequence (TGAG/CTCA), at positions −548 and −495.
FIG. 1.
PCL5 is induced following overexpression of GCN4. Strain W303-1A carrying either the vector plasmid or plasmid KB843 (pGAL-GCN4) was grown on raffinose and shifted to galactose. At the indicated times after the shift to galactose, samples of the culture were removed and subjected to Northern blot analysis. For the PCL5 and EF1A probes, we used the inserts of plasmids KB1093 and KB537, respectively.
Pcl5 suppresses Gcn4 activity.
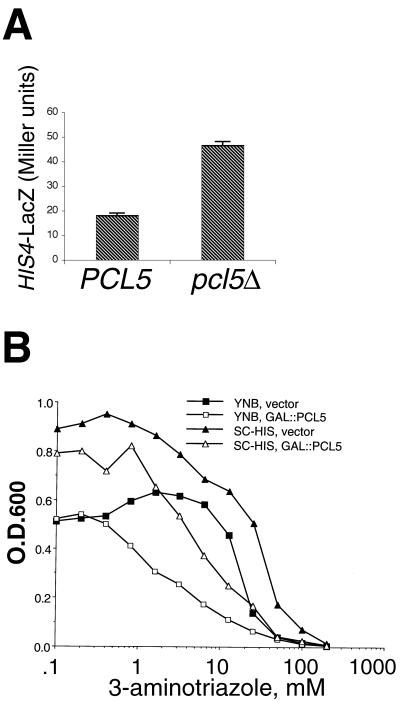
The fact that PCL5 is a target of Gcn4 suggested that it might contribute, like many other Gcn4 targets, to the resistance of cells to amino acid starvation. Alternatively, since Pho85 suppresses Gcn4 activity (26), it was possible that Pcl5 is likewise antagonistic to Gcn4 activity. To distinguish between these two possibilities, we first tested the effect of the pcl5 deletion on Gcn4 activity. A LacZ construct regulated by a HIS4 promoter fragment that is completely dependent on Gcn4 for expression yielded 2.5 times more β-galactosidase activity in pcl5Δ cells than in PCL5 cells (Fig. 2A), indicating that Gcn4 is more active in the pcl5Δ mutant. We also tested how an increase in Pcl5 levels would affect resistance to 3-aminotriazole (3-AT), a competitive inhibitor of the HIS3 gene product imidazole glycerol phosphate dehydratase. Cellular resistance to 3-AT is mediated by Gcn4, which activates the HIS3 gene (11). As shown in Fig. 2B, overexpression of PCL5 led to an increased sensitivity of wild-type cells to 3-AT. The concentration of half-maximal inhibition of growth was increased twofold, from 3 to 6 mM in YNB medium and from 6 to 12 mM in SC medium. Since Gcn4 activity is required for 3-AT resistance, this result also suggests a negative effect of Pcl5 on Gcn4 activity.
FIG. 2.
Pcl5 represses Gcn4 activity. (A) Expression of His4-LacZ from a HIS4 promoter derivative that is exclusively dependent on Gcn4 for activity (deletion no. 203 [32]) in KY795 (PCL5) and KY798 (pcl5Δ) cells. Overnight cultures were diluted and grown for 6 h to early log phase, and the β-galactosidase activity of the cultures was measured as previously described (5). The error bars indicate the standard deviation of four independent assays. (B) PCL5 overexpression increases sensitivity to 3-AT. Yeast strain KY795 containing either the vector plasmid (filled symbols) or plasmid KB1093 (GAL1::PCL5) (open symbols) was grown in YNB medium (squares) or in SC medium (triangles) containing increasing amounts of 3-AT. The optical density at 600 nm (O.D.600) of the cultures was measured after overnight incubation at 30°C.
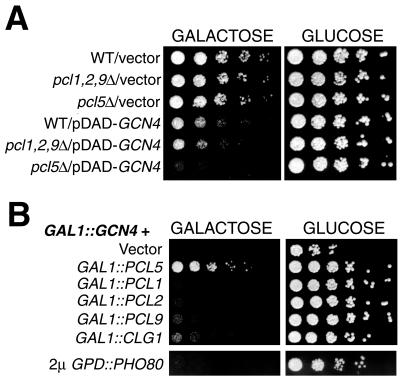
As an alternative way to measure Gcn4 activity, we used its overexpression toxicity. Gcn4, when highly overexpressed, inhibits cellular growth, possibly by interfering with other transcriptional activation pathways (40). In a pho85Δ mutant, even moderate overexpression of Gcn4 is inhibitory (26). We found that, similar to a pho85Δ mutant, the pcl5Δ mutant is hypersensitive to Gcn4 moderately overexpressed from the GAL promoter of plasmid pDAD (Fig. 3A). In contrast, a pcl1Δ pcl2Δ pcl9Δ triple mutant was not more sensitive to Gcn4 overexpression than the wild-type strain (Fig. 3A). In a complementary experiment, we found that, in a wild-type background, the toxicity of GCN4 highly overexpressed from the p414GAL1 plasmid could be suppressed by concomitant overexpression of PCL5 but not by that of the Pcl gene PCL1, PCL2, PCL9, CLG1, or PHO80 (Fig. 3B). Taken together, these data indicate that Pcl5 is specifically responsible for inhibiting Gcn4 function in vivo.
FIG. 3.
(A) The pcl5Δ mutant is hypersensitive to moderate overexpression of Gcn4. Fivefold dilutions of the W303-1A, KY846 (pcl1Δ pcl2Δ pcl9Δ), and KY827 (pcl5Δ) strains containing the indicated plasmids were spotted on galactose plates to induce expression of GCN4. The plates were incubated for 2 days at 30°C. (B) Pcl5 suppresses the overexpression toxicity of Gcn4. Strain W303-1A containing plasmid p414GAL1-GCN4 and, in addition, a Pcl-expressing plasmid, as indicated, was spotted on galactose plates to induce expression of GCN4 and the Pcl. The plates were incubated for 2 days at 30°C. The Pcl constructs are p416GAL1 (vector), KB1093 (CEN GAL1::PCL5), KB1237 (CEN GAL1::PCL1), KB1238 (CEN GAL1::PCL2), KB1239 (CEN GAL1::PCL9), KB1240 (CEN GAL1::CLG1), and EB92 (2μm GPD::PHO80). WT, wild type.
Pcl5 can direct Gcn4 phosphorylation and degradation.
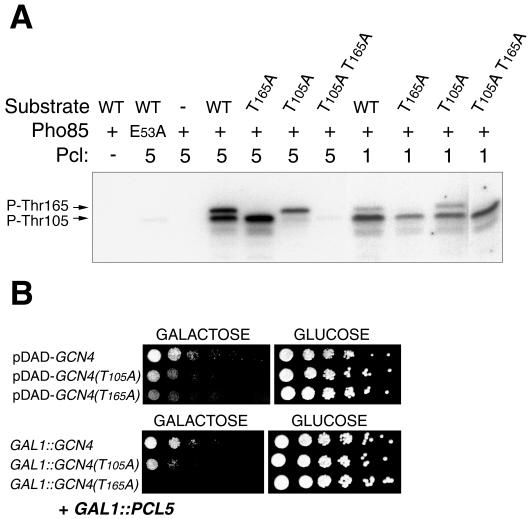
Since Pcl5 was shown to interact with Pho85 (25), the simplest explanation for Pcl5's effect is that it is involved, with Pho85, in the inhibitory phosphorylation of Gcn4. Pho85 was previously shown to phosphorylate Gcn4 at residue Thr165, which is essential for degradation of the protein (26). We first tested whether Pcl5 is able to directly promote Gcn4 phosphorylation by Pho85 in vitro. Since we had previously shown that Pcl1 was able to promote Gcn4 phosphorylation in vitro (26), we performed a side-by-side comparison of the two cyclins, both expressed in bacteria and affinity purified with a hexahistidine tag. As the substrate, we used a recombinant Gcn4 fragment, spanning residues 62 to 202, that includes three potential CDK targets: two native sites (threonine followed by proline at coordinates 105 and 165 of the Gcn4 sequence) and one expression vector-derived site (serine followed by proline at position 2 of the recombinant fragment). As shown in Fig. 4A, recombinant Pho85 was able, in the presence of Pcl5, to phosphorylate Gcn4, indicating that Pcl5 functions as a bona fide Pcl. The pattern of phosphorylation bands indicates that Pho85/Pcl5 phosphorylated the Gcn4 fragment on two residues, Thr165 and Thr105: whereas the wild-type substrate migrated as two distinct phosphorylated species, the single mutants migrated as single bands and the double mutant was unphosphorylated. With Pcl1, phosphorylation of the Gcn4 fragment was also obtained; however, the banding pattern of the various mutants indicated that at least one additional site was phosphorylated as well because two bands were visible with the T105A mutant and the T105A T165A double mutant was still efficiently phosphorylated by Pho85/Pcl1.
FIG. 4.
(A) In vitro kinase assay with Pho85/Pcl5 or Pho85/Pcl1 on a Gcn4 fragment spanning positions 62 to 202, either wild type (WT) or mutated at potential phosphorylation sites, as indicated. E53A is an inactive mutant form of Pho85. In the Pcl row, the number 5 or 1 indicates that Pcl5 or Pcl1, respectively, was added to the reaction mixture. A minus sign indicates a no-cyclin control. (B) Pcl5 is unable to suppress the overexpression toxicity of GCN4 mutant T165A. (Top) Serial dilutions of W303-1A cells expressing either wild-type GCN4, mutant T105A, or mutant T165A from the moderate-expression vector pDAD were spotted on galactose (inducing) or glucose (repressing) medium, as indicated, and incubated for 2 days at 30°C. (Bottom) W303-1A cells expressing either wild-type GCN4, mutant T105A, or mutant T165A from the high-expression vector p414-GAL1, together with PCL5 from the p416-GAL1 plasmid, were spotted and grown as described above.
We next asked if phosphorylation of Thr105 and Thr165 by Pho85/Pcl5 in vitro is functionally significant in vivo by testing whether the toxicity of the Gcn4 T165A and T105A mutants is still subject to suppression by Pcl5. We first tested the relative toxicity of these Gcn4 mutants in the absence of PCL5 overexpression. Since high-level overexpression of even wild-type GCN4 from the p414GAL1 plasmid completely inhibits growth (Fig. 3B), we used the moderate-overexpression pDAD vector for this control. As shown in Fig. 4B (top), in the absence of PCL5 overexpression, the T105A and T165A mutants are significantly more toxic than the wild type and the T165A mutant is marginally more toxic than T105A. In contrast, in the presence of PCL5 overexpression, the toxicity of the wild type and the T105A mutant overexpressed from the strong promoter of the p414GAL1 vector was efficiently suppressed but the T165A mutant was refractory to suppression by PCL5 (Fig. 4B, bottom). Thus, these data suggest that, in vivo, Pcl5 directs the phosphorylation by Pho85 of—at least—residue Thr165 of Gcn4, thereby leading to suppression of Gcn4 activity.
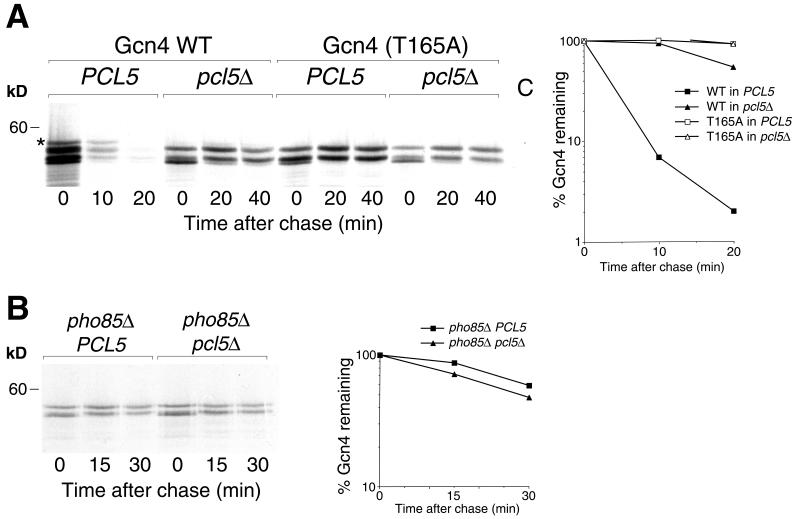
Since Pho85 activity and phosphorylation of the threonine residue at position 165 are required for degradation of Gcn4 in vivo (26), we next tested whether deletion of PCL5 would similarly affect Gcn4 degradation. A pulse-chase assay indicated that Gcn4 degradation was strongly inhibited in the pcl5Δ mutant, with a half-life (t1/2) increasing from 2.5 to 20 min (Fig. 5A). This experiment also indicated that Pcl5 affects the phosphorylation of Gcn4 in vivo. Indeed, in the wild-type background, at least four electrophoretic species of Gcn4, most probably representing different phosphorylation states of the protein, can be distinguished; in contrast, the migration pattern of Gcn4 expressed in the pcl5Δ mutant lacked the slowest-migrating band. Furthermore, the migration pattern of the stable T165A mutant was unchanged, whether it was expressed in the wild-type or the pcl5Δ background, and was similar to the migration pattern of the wild-type protein expressed in the pcl5Δ background (Fig. 5A). Taken together, these observations indicate that, in vivo, Pcl5 directs phosphorylation of Gcn4 at Thr165, leading to its degradation.
FIG. 5.
(A) Gcn4 is underphosphorylated and stabilized in a pcl5Δ mutant. Degradation of the Gcn4 protein expressed from plasmid KB843, or of the Gcn4(T165A) mutant expressed from plasmid KB1155, and transformed into strain W303-1A (PCL5) or KY827 (pcl5Δ) was assayed by pulse-chase analysis. The star indicates a Gcn4 species visible in the wild type (WT) but not in the pcl5Δ mutant. For the control, lane C, an extract from a gcn4Δ strain was used. The graph indicates quantitation by phosphorimager of the gel shown. (B) Degradation of the Gcn4 protein expressed from plasmid KB843 in a pho85Δ strain in the presence or absence of PCL5. The graph indicates quantitation by phosphorimager of the gel shown.
The fact that the same effects on Gcn4 phosphorylation and degradation were previously demonstrated for the pho85Δ mutant (26) and the facts that Pcl5 was isolated as a Pho85-interacting protein (25) and was shown here to act as a Pcl in vitro (Fig. 4A) strongly suggest that Pcl5 functions as a Pcl in vivo. If Pcl5 functions uniquely as a Pho85 cyclin, i.e., if it is unable to target additional CDKs to Gcn4, then in a pho85Δ background, deletion of PCL5 should not stabilize Gcn4 any further. As shown in Fig. 5B, the pho85Δ pcl5Δ strain displayed no further stabilization of Gcn4 (but rather, perhaps, a slight acceleration of Gcn4 degradation) compared to the pho85Δ PCL5 strain. This observation provides further confirmation of the role of Pcl5 as a Pho85-specific cyclin.
Role of Pcl5 in the regulation of Gcn4 degradation.
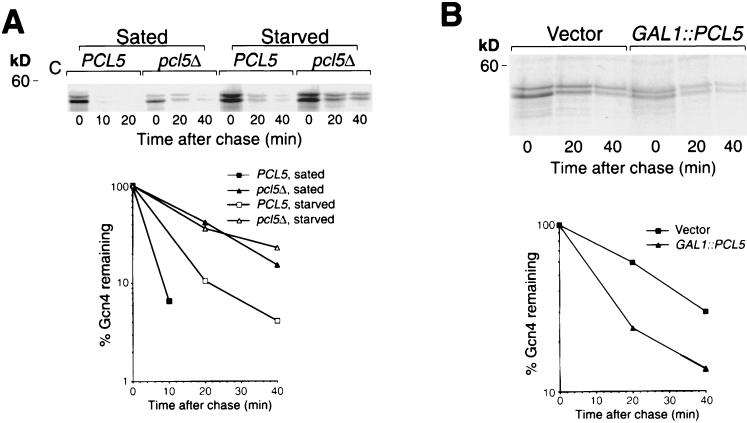
Regulation of Gcn4 turnover occurs under conditions of profound starvation brought about by removal of an essential amino acid from an auxotrophic strain (21) or by addition of growth-inhibitory levels of cycloheximide (26). The mechanism of this regulation is unknown, but stabilization of Gcn4 correlates with underphosphorylation of Gcn4 at Thr165 (26) and is, in large part, abolished in a pho85Δ mutant (4; our unpublished results). To test whether Pcl5 plays a role in the pathway of Gcn4 stability regulation, we tested the effect of starvation on Gcn4 degradation in the pcl5Δ background. As shown in Fig. 6A, whereas in the wild-type background, Gcn4 was significantly stabilized in starved versus sated cells (t1/2 of 10 versus 2.5 min), in the pcl5Δ cells, no difference in the rate of degradation was detected (t1/2 of 20 min), consistent with the involvement of Pcl5 in the mechanism of Gcn4 stability regulation.
FIG. 6.
(A) Gcn4 degradation is not regulated in the pcl5Δ mutant. Pulse-chase analysis was performed on Gcn4 expressed from plasmid KB161 (pDAD-GCN4) in strain W303-1A (PCL5) or KY827 (pcl5Δ) in regular YNB medium supplemented with all amino acids (Sated) or in YNB medium lacking all amino acids and supplemented only with adenine, after 30 min of preincubation in this medium (Starved). For the control, lane C, an extract from a gcn4Δ strain was used. The graph indicates quantitation by phosphorimager of the gel shown. (B) Overexpression ofPCL5 does not abolish regulation of Gcn4 degradation. Pulse-chase analysis was performed on Gcn4 expressed from plasmid KB161 (pDAD-GCN4) in strain W303-1A containing either the vector p416GAL1 (Vector) or plasmid KB1093 (GAL1::PCL5) in starvation medium (YNB medium lacking all amino acids and supplemented only with adenine) after 30 min of preincubation in this medium. The graph indicates quantitation by phosphorimager of the gel shown. kD, kilodaltons.
If the stabilization of Gcn4 by starvation is due to regulation of the levels of Pcl5 protein synthesis, then ectopic expression of Pcl5 under the control of a strong promoter should override this stabilization. To test this, we measured Gcn4 turnover in starved cells with or without concomitant overexpression of PCL5 from the GAL1 promoter. As shown in Fig. 6B, Gcn4 was significantly stabilized in starved cells in either the presence or the absence of the GAL-PCL5 plasmid, although degradation was accelerated in the PCL5-overexpressing cells (t1/2 of 10 versus 20 min in the control starved cells), indicating that overexpression of PCL5 only slightly reduces, but does not abolish, the stabilization of Gcn4 by starvation.
Effect of starvation on Pho85/Pcl5-associated kinase activity.
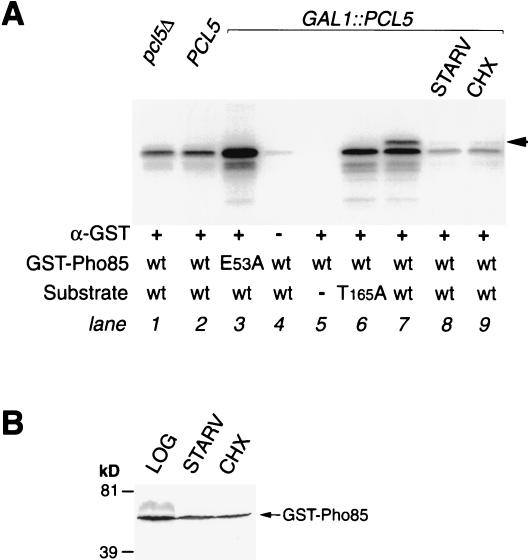
In order to directly assay Pcl5-associated Gcn4 kinase activity, we attempted to perform an IP/kinase assay with epitope-tagged Pcl5. However, no kinase activity was obtained with a number of different tags (data not shown). We therefore used a GST-Pho85 fusion previously used by Nishizawa et al. (34) with an anti-GST antibody. As a control, we used a catalytically inactive mutant, E53A, of Pho85 that we confirmed to be inactive in an in vitro kinase assay (Fig. 4A) and unable to complement a pho85Δ mutant for Gcn4 down-regulation (data not shown). We used the same substrate as for the experiment shown in Fig. 4A, a Gcn4 fragment spanning residues 62 to 202. Immunoprecipitated GST-Pho85 was able to phosphorylate this substrate; however, only a single phosphorylated species was visible (Fig. 7A, lane 2), although this fragment contains two phosphorylation sites (Fig. 4A). Furthermore, this phosphorylation was Pcl5 independent (lane 1) and strong kinase activity was detected even with the inactive mutant (E53A) Pho85 protein (lane 3), indicating that a nonspecific kinase coimmunoprecipitates with GST-Pho85. Nonetheless, when GST-Pho85 was immunoprecipitated from cells overexpressing PCL5, a second, slower-migrating phosphorylation band appeared (lane 7), indicative of phosphorylation at Thr165 (Fig. 4A; compare with lane 6, where the same extract was used with the T165A mutant form as a substrate). This Pcl5-dependent phosphorylation at Thr165 disappeared when the cells were subjected to starvation or incubated with cycloheximide (lanes 8 and 9), indicating that these treatments reduce Pho85/Pcl5-associated Gcn4 kinase activity even when Pcl5 is ectopically expressed from the GAL promoter. Since the effect of starvation or cycloheximide on Pho85/Pcl5 activity could, in principle, be due to a decrease in Pho85 levels, the levels of the GST-Pho85 protein were measured in the extracts used for the IP/kinase assay. As shown in Fig. 7B, no significant decrease in GST-Pho85 levels was observed.
FIG. 7.
Analysis of Pho85-associated Gcn4 kinase activity. (A) A GST-Pho85 fusion, either the wild type (wt) or the catalytically inactive mutant E53A (lane 3), was expressed under the control of the GAL1 promoter and immunoprecipitated from pcl5Δ cells (lane 1), PCL5 wild-type cells (lane 2), or wild-type cells overexpressing PCL5 from the GAL1 promoter (lanes 3 to 9). Cells were grown in YNB medium containing all amino acids (lanes 1 to 7 and 9), starved for 30 min in medium lacking all amino acids (STARV; lane 8), or treated for 30 min with cycloheximide (CHX) at 5 μg/ml (lane 9). The substrate was identical to that used in Fig. 4A; lane 5 is a no-substrate control, and lane 6 contains the T165A mutant. Lane 4 is a no-antibody control. The arrow indicates the position of the Thr165 phosphorylation band. (B) Western blotting and immunodetection of GST-Pho85 in the extracts used for lanes 7, 8, and 9 of the experiment shown in panel A (logarithmically growing cells, starved cells, and cycloheximide-treated cells, respectively). kD, kilodaltons.
The Pcl5 protein disappears in starved cells because of rapid turnover.
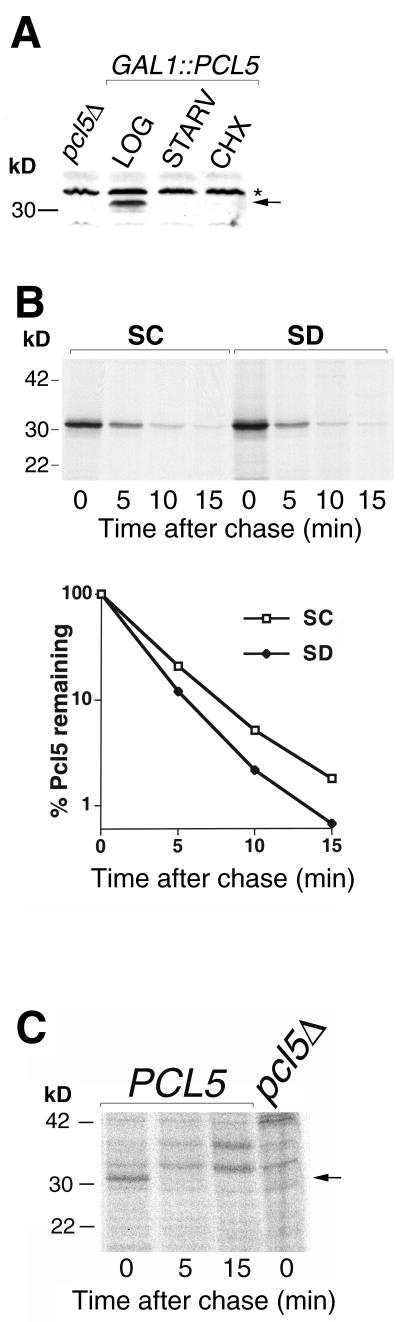
The data shown above indicate that Pcl5-associated Pho85 activity is sensitive to starvation even when PCL5 is expressed from the heterologous GAL1 promoter, suggesting that PCL5 activity is regulated posttranscriptionally. To test the effect of starvation on Pcl5 protein levels, cells expressing Pcl5 from the GAL1 promoter were subjected to starvation or cycloheximide treatment and the Pcl5 levels were assayed by Western blotting. As shown in Fig. 8A, Pcl5 levels were strongly reduced after a short period of starvation or inhibition of protein synthesis. The simplest explanation for this rapid disappearance of the protein is rapid proteolysis. Indeed, pulse-chase analysis indicated that the overexpressed Pcl5 protein was degraded with a t1/2 of 2.5 min, both under normal growth conditions and in starved cells (Fig. 8B). To ascertain that this instability was not due to overexpression of the protein, we also monitored the degradation of the endogenous Pcl5 protein by pulse-chase analysis (Fig. 8C). Although the low signal levels precluded accurate quantitation of band intensity, the fact that the Pcl5 band, clearly visible at the zero time point, had disappeared 5 min into the chase, indicated that the endogenous Pcl5 protein is at least as rapidly degraded as the overexpressed protein.
FIG. 8.
Pcl5 disappears rapidly in starved cells and is constitutively unstable. (A) Western blot with 50 μg of extract from pcl5Δ cells or from W303-1A cells overexpressing Pcl5 from plasmid KB1093 and either grown in SC medium (LOG), shifted for 30 min to YNB medium plus adenine (STARV), or incubated for 30 min with cycloheximide at 5 μg/ml (CHX). The arrow indicates Pcl5; the star indicates across-reacting band. (B) Pulse-chase analysis of Pcl5 overexpressed from the KB1093 plasmid either in normal (SC) medium or after a shift for 30 min to YNB plus adenine (SD) medium. Note that the same amount of total incorporated radioactivity was processed for each time point; therefore, the signal strength at time zero is not indicative of total cellular protein biosynthesis. (C) Degradation of endogenous Pcl5. W303-1A cells were subjected to pulse-chase analysis after induction of PCL5 expression by Gcn4 expressed from the KB843 plasmid. pcl5Δ strain KY827 was used as a control. The arrow indicates the position of the Pcl5 band. kD, kilodaltons.
DISCUSSION
Pcl5 and regulation of Gcn4 degradation.
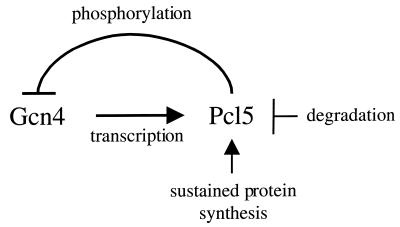
Gcn4 is a central regulator of the response of yeast to starvation for amino acids and purines (11, 35). Gcn4 is regulated at the level of translation by the kinase Gcn2 (41) and at the level of protein degradation by the kinase Pho85 (26). Pho85 is a CDK, and the data presented here indicate that Pcl5 is the specific cyclin that assists Pho85 in phosphorylating Gcn4. Since Pho85 was identified as being in the pathway of the regulation of Gcn4 degradation, it was logical to assume that regulation of Pcl5 might be responsible for the stabilization of Gcn4 under starvation conditions. In fact, our data indicate that Pcl5 is required for the regulation of Gcn4 stability (Fig. 6). We were unable to directly assay for Pcl5-associated kinase activity by the IP/kinase method, but a similar assay with tagged Pho85 in cells overexpressing Pcl5 indicated that stimulation of Gcn4 kinase activity of Pho85 by Pcl5 was highly sensitive to starvation (Fig. 7). This, together with the fact that ectopic overexpression of PCL5 could not override the regulation of Gcn4 degradation (Fig. 6B), suggested that PCL5 activity must be regulated posttranscriptionally by starvation. Consistent with this possibility, we did find that even when it is overexpressed from a strong heterologous promoter, Pcl5 is almost undetectable in starved cells (Fig. 8A). This observation can be explained by the fact that the protein is extremely unstable (Fig. 8B and C). Although Pcl5 appears to be equally rapidly degraded under both starved and sated conditions, we hypothesize that whereas under sated conditions the steady-state level of Pcl5 is maintained by continued protein synthesis, in starved cells, the sharp reduction in Pcl5 protein levels is due to reduced levels of general protein biosynthesis; i.e., we suggest that Pcl5 may, simply by virtue of its own instability, constitute a sensor of cellular biosynthetic capacity. With regard to the regulation of Gcn4 degradation by starvation, the drop in Pcl5 steady-state levels in starved cells would, in turn, lead to stabilization of Gcn4 (Fig. 9). This model predicts that expression of a stable derivative of Pcl5 should override the regulation of Gcn4 degradation. However, we have been unable to obtain a mutant Pcl5 protein that is both stable and active (our unpublished results). Thus, we cannot exclude the possibility of additional mechanisms of regulation of Pho85/Pcl5 by amino acid starvation, such as inhibition by a starvation-induced CDK inhibitor, similar to the regulation of Pho85/Pho80 by Pho81 (37).
FIG. 9.
Schematic model of the Pcl5-Gcn4 regulatory loop. See text for details.
Role of Pho85 versus that of Srb10 in Gcn4 degradation.
Gcn4 is regulated at the level of degradation not only by Pho85 but also by the RNA polymerase II-associated kinase Srb10 (4). However, even at low levels of Gcn4 expression, Pho85 activity plays a more prominent role in Gcn4 degradation than Srb10 activity (4), and at high levels, Srb10's role becomes undetectable (4 and our unpublished observations). This suggests that Srb10 is responsible for the degradation of a small pool of possibly chromatin- or promoter-bound Gcn4 molecules. In contrast, Pho85 would be responsible for degradation of excess, non-chromatin-bound Gcn4 molecules. Regulation of PCL5 at the level of protein stability ensures that Pho85 activity toward Gcn4 is held in check as long as cells are starved and Gcn4 activity is required. However, when Gcn4 activity becomes superfluous, e.g., when cells are overcoming an episode of starvation, synthesis of the Pcl5 protein resumes, causing excess Gcn4 to be degraded. The activation of PCL5 transcription by Gcn4 ensures that, in the absence of starvation, a tight homeostasis of Gcn4 activity is always maintained (Fig. 9).
Correlation between transcriptional activity and degradation of transcription factors.
Regulation of transcription factor activity can be achieved by a variety of mechanisms, including degradation. Recent evidence suggests a tight correlation between transcriptional activity and instability of transcription factors, at least in some instances (29, 36). One possible explanation for this correlation is that there is an overlap between consensus degradation signals and transcription activation domains (29, 36). Alternatively, a degradation system might exist that targets only the promoter-bound transcription factor molecules or perhaps even only molecules having participated in the process of transcription activation. This may be the case for Gcn4, the degradation of which depends not only on Pho85 but also, in part, on the RNA polymerase II-associated kinase Srb10 (4). Our observations provide yet another model to explain some instances of correlation between transcriptional activity and degradation of transcription factors. Since Pcl5, by inducing Gcn4 phosphorylation, causes its degradation, the transcriptional activity of Gcn4, by promoting PCL5 expression, is indirectly responsible for its own degradation. A similar type of regulation has been shown in animal cells: transcription factor p53 activates expression of the mdm2 gene, and the MDM2 protein, in turn, causes degradation of p53 (8, 44).
Role of Pcl5 in substrate selection.
Although the function of the cyclin subunit in determining the substrate specificity of the cell cycle CDKs has been controversial, it is now becoming apparent that substrate selection is one of the central roles of the cyclin (27). For Pho85, several substrates have been identified that depend on specific Pcls for phosphorylation—Pho4 depends on Pho80, and Gsy2 depends on Pcl8 and Pcl10. For Gcn4, we had previously shown that Pcl1 could direct its phosphorylation in vitro and that, in addition, Pcl1-associated kinase activity is reduced under the same conditions that lead to Gcn4 stabilization, suggesting a role for Pcl1 in Gcn4 degradation in vivo (26). However, a pcl1 deletion did not affect Gcn4 degradation (our unpublished observations) or Gcn4 activity (Fig. 3A). In contrast, here we show that PCL5 is both sufficient for Gcn4 phosphorylation in vitro (Fig. 4A) and necessary for Gcn4 phosphorylation and degradation in vivo (Fig. 5). One possible explanation for this discrepancy is the differential regulation of these genes: for example, PCL1 is strongly cell cycle regulated (25)—therefore, it would be unable to direct Gcn4 phosphorylation throughout the cell cycle. However, this explanation is contradicted by our data. Indeed, if the only reason that only PCL5 is necessary for Gcn4 degradation in vivo were the periodic expression of PCL1, then placement of PCL1 and PCL5 under the same regulation should yield similar activities. This is not the case: when these PCL genes are both expressed from the GAL1 promoter, only PCL5 can suppress the overexpression toxicity of Gcn4 (Fig. 3B). Another possibility is that, in vivo, differential subcellular compartmentalization may affect whether a cyclin is able to direct the phosphorylation of a given substrate. The transcription factor Gcn4 is expected to be mainly nuclear, whereas genetic interactions suggest that Pcl1 functions primarily in the cytoplasm (22, 28); however, no direct evidence is available for the actual localization of either Gcn4, Pcl1, or Pcl5. A third explanation is that reaction conditions in vitro may artifactually reduce the substrate selectivity of the cyclin/kinase complex, leading to phosphorylation of nonnatural substrates, as was found for Pho85/Pho80 (reviewed in reference 3). In support of this hypothesis, we found that the site specificity of the Pho85/Pcl1 complex is relaxed compared with that of the Pho85/Pcl5 complex, with Pcl1 being able to direct phosphorylation of an additional, probably nonnative site(s) on a recombinant Gcn4 fragment (Fig. 4). Thus, both our in vivo and our in vitro data point to Pcl5 as the unique Gcn4-specific Pcl. The observation we had previously made (26), that Pcl1-associated kinase activity is reduced under the same conditions that lead to Gcn4 stabilization, is due to the fact that Pcl1, like Pcl5, is an extremely unstable protein that disappears rapidly in starved cells (our unpublished data).
Microarray analysis and identification of new regulatory pathways.
We were prompted to test for the involvement of Pcl5 in Gcn4 regulation by a report suggesting that PCL5 may be a target of Gcn4 (19). Jia et al. had investigated, by microarray hybridization, changes in global expression patterns in yeast upon exposure to the amino acid biosynthesis inhibitor sulfometuron and found that PCL5 is coregulated with other Gcn4 targets. Microarray hybridization data published with a second global expression study (33) also suggested that PCL5 is a target of Gcn4. Our finding that Pcl5 is, in fact, part of a regulatory system that restricts Gcn4 activity attests to the power of global expression assays to uncover new factors involved in a specific physiological response. On the other hand, our findings also underscore the fact that coexpression does not imply similarity of function; although PCL5 transcription is coregulated with the amino acid starvation response genes, the Pcl5 protein, by repressing Gcn4 function, is antagonistic to the amino acid starvation response. Proper modulation of the starvation response is achieved through posttranscriptional regulation of Pcl5.
Acknowledgments
We thank Na'ama Barkai for pointing out the coregulation of PCL5 and amino acid biosynthesis genes; Masafumi Nishizawa, Erin O'Shea, and David Stillman for providing plasmids and strains; Sharon Aviram for assistance in plasmid construction; and Sara Selig for critical reading of the manuscript.
This research was supported by a grant from the Israel Science Foundation to D.K.
REFERENCES
- 1.Alani, E., L. Cao, and N. Kleckner. 1987. A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics 116:541-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1989. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 3.Carroll, A. S., and E. K. O'Shea. 2002. Pho85 and signaling environmental conditions. Trends Biochem. Sci. 27:87-93. [DOI] [PubMed] [Google Scholar]
- 4.Chi, Y., M. J. Huddleston, X. Zhang, R. A. Young, R. S. Annan, S. A. Carr, and R. J. Deshaies. 2001. Negative regulation of Gcn4 and Msn2 transcription factors by Srb10 cyclin-dependent kinase. Genes Dev. 15:1078-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Daignan-Fornier, B., and G. R. Fink. 1992. Coregulation of purine and histidine biosynthesis by the transcriptional activators BAS1 and BAS2. Proc. Natl. Acad. Sci. USA 89:6746-6750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dever, T. E., L. Feng, R. C. Wek, A. M. Cigan, T. F. Donahue, and A. G. Hinnebusch. 1992. Phosphorylation of initiation factor 2α by protein kinase GCN2 mediates gene-specific translational control of GCN4 in yeast. Cell 68:585-596. [DOI] [PubMed] [Google Scholar]
- 7.Espinoza, F. H., J. Ogas, I. Herskowitz, and D. O. Morgan. 1994. Cell cycle control by a complex of the cyclin HCS26 (PCL1) and the kinase PHO85. Science 266:1388-1391. [DOI] [PubMed] [Google Scholar]
- 8.Haupt, Y., R. Maya, A. Kazaz, and M. Oren. 1997. Mdm2 promotes the rapid degradation of p53. Nature 387:296-299. [DOI] [PubMed] [Google Scholar]
- 9.Hershko, A., and A. Ciechanover. 1998. The ubiquitin system. Annu. Rev. Biochem. 67:425-479. [DOI] [PubMed] [Google Scholar]
- 10.Hinnebusch, A. G. 1984. Evidence for translational regulation of the activator of general amino acid control in yeast. Proc. Natl. Acad. Sci. USA 81:6442-6446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hinnebusch, A. G. 1992. General and pathway-specific regulatory mechanisms controlling the synthesis of amino acid biosynthetic enzymes in Saccharomyces cerevisiae, p. 319-414. In E. W. Jones, J. R. Pringle, and J. R. Broach (ed.), The molecular and cellular biology of the yeast Saccharomyces, vol. 2. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 12.Hinnebusch, A. G. 1997. Translational regulation of yeast GCN4. A window on factors that control initiator-trna binding to the ribosome. J. Biol. Chem. 272:21661-21664. [DOI] [PubMed] [Google Scholar]
- 13.Hinnebusch, A. G., and G. R. Fink. 1983. Positive regulation in the general control of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 80:5374-5378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hirst, K., F. Fisher, P. C. McAndrew, and C. R. Goding. 1994. The transcription factor, the Cdk, its cyclin and their regulator: directing the transcriptional response to a nutritional signal. EMBO J. 13:5410-5420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hochstrasser, M., and A. Varshavsky. 1990. In vivo degradation of a transcriptional regulator: the yeast α2 repressor. Cell 61:697-708. [DOI] [PubMed] [Google Scholar]
- 16.Hope, I. A., and K. Struhl. 1985. GCN4 protein, synthesized in vitro, binds HIS3 regulatory sequences: implications for general control of amino acid biosynthetic genes in yeast. Cell 43:177-188. [DOI] [PubMed] [Google Scholar]
- 17.Huang, D., J. Moffat, W. A. Wilson, L. Moore, C. Cheng, P. J. Roach, and B. Andrews. 1998. Cyclin partners determine Pho85 protein kinase substrate specificity in vitro and in vivo: control of glycogen biosynthesis by Pcl8 and Pcl10. Mol. Cell. Biol. 18:3289-3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jeffrey, P. D., A. A. Russo, K. Polyak, E. Gibbs, J. Hurwitz, J. Massague, and N. P. Pavletich. 1995. Mechanism of CDK activation revealed by the structure of a cyclin A-CDK2 complex. Nature 376:313-320. [DOI] [PubMed] [Google Scholar]
- 19.Jia, M. H., R. A. Larossa, J. M. Lee, A. Rafalski, E. Derose, G. Gonye, and Z. Xue. 2000. Global expression profiling of yeast treated with an inhibitor of amino acid biosynthesis, sulfometuron methyl. Physiol. Genomics 3:83-92. [DOI] [PubMed] [Google Scholar]
- 20.Kaffman, A., I. Herskowitz, R. Tjian, and E. K. O'Shea. 1994. Phosphorylation of the transcription factor PHO4 by a cyclin-CDK complex, PHO80-PHO85. Science 263:1153-1156. [DOI] [PubMed] [Google Scholar]
- 21.Kornitzer, D., B. Raboy, R. G. Kulka, and G. R. Fink. 1994. Regulated degradation of the transcription factor Gcn4. EMBO J. 13:6021-6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lenburg, M. E., and E. K. O'Shea. 2001. Genetic evidence for a morphogenetic function of the Saccharomyces cerevisiae Pho85 cyclin-dependent kinase. Genetics 157:39-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Measday, V., H. McBride, J. Moffat, D. Stillman, and B. Andrews. 2000. Interactions between Pho85 cyclin-dependent kinase complexes and the Swi5 transcription factor in budding yeast. Mol. Microbiol. 35:825-834. [DOI] [PubMed] [Google Scholar]
- 24.Measday, V., L. Moore, J. Ogas, M. Tyers, and B. Andrews. 1994. The PCL2 (ORFD)-PHO85 cyclin-dependent kinase complex: a cell cycle regulator in yeast. Science 266:1391-1395. [DOI] [PubMed] [Google Scholar]
- 25.Measday, V., L. Moore, R. Retnakaran, J. Lee, M. Donoviel, A. M. Neiman, and B. Andrews. 1997. A family of cyclin-like proteins that interact with the Pho85 cyclin-dependent kinase. Mol. Cell. Biol. 17:1212-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meimoun, A., T. Holtzman, Z. Weissman, H. J. McBride, D. J. Stillman, G. R. Fink, and D. Kornitzer. 2000. Degradation of the transcription factor Gcn4 requires the kinase Pho85 and the SCF(CDC4) ubiquitin-ligase complex. Mol. Biol. Cell 11:915-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller, M. E., and F. R. Cross. 2001. Cyclin specificity: how many wheels do you need on a unicycle? J. Cell Sci. 114:1811-1820. [DOI] [PubMed] [Google Scholar]
- 28.Miller, M. E., and F. R. Cross. 2000. Distinct subcellular localization patterns contribute to functional specificity of the Cln2 and Cln3 cyclins of Saccharomyces cerevisiae. Mol. Cell. Biol. 20:542-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Molinari, E., M. Gilman, and S. Natesan. 1999. Proteasome-mediated degradation of transcriptional activators correlates with activation domain potency in vivo. EMBO J. 18:6439-6447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morgan, D. O. 1995. Principles of CDK regulation. Nature 374:131-134. [DOI] [PubMed] [Google Scholar]
- 31.Mumberg, D., R. Muller, and M. Funk. 1994. Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 22:5767-5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nagawa, F., and G. R. Fink. 1985. The relationship between the “TATA” sequence and transcription initiation sites at the HIS4 gene of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 82:8557-8561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Natarajan, K., M. R. Meyer, B. M. Jackson, D. Slade, C. Roberts, A. G. Hinnebusch, and M. J. Marton. 2001. Transcriptional profiling shows that Gcn4p is a master regulator of gene expression during amino acid starvation in yeast. Mol. Cell. Biol. 21:4347-4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nishizawa, M., M. Kawasumi, M. Fujino, and A. Toh-e. 1998. Phosphorylation of sic1, a cyclin-dependent kinase (Cdk) inhibitor, by Cdk including Pho85 kinase is required for its prompt degradation. Mol. Biol. Cell 9:2393-2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rolfes, R. J., and A. G. Hinnebusch. 1993. Translation of the yeast transcriptional activator GCN4 is stimulated by purine limitation: implications for activation of the protein kinase GCN2. Mol. Cell. Biol. 13:5099-5111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Salghetti, S. E., M. Muratani, H. Wijnen, B. Futcher, and W. P. Tansey. 2000. Functional overlap of sequences that activate transcription and signal ubiquitin-mediated proteolysis. Proc. Natl. Acad. Sci. USA 97:3118-3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schneider, K. R., R. L. Smith, and E. K. O'Shea. 1994. Phosphate-regulated inactivation of the kinase PHO80-PHO85 by the CDK inhibitor PHO81. Science 266:122-126. [DOI] [PubMed] [Google Scholar]
- 38.Schulman, B. A., D. L. Lindstrom, and E. Harlow. 1998. Substrate recruitment to cyclin-dependent kinase 2 by a multipurpose docking site on cyclin A. Proc. Natl. Acad. Sci. USA 95:10453-10458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sherman, F., G. R. Fink, and J. B. Hicks. 1986. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 40.Tavernarakis, N., and G. Thireos. 1995. Transcriptional interference caused by GCN4 overexpression reveals multiple interactions mediating transcriptional activation. Mol. Gen. Genet. 247:571-578. [DOI] [PubMed] [Google Scholar]
- 41.Wek, R. C., B. M. Jackson, and A. G. Hinnebusch. 1989. Juxtaposition of domains homologous to protein kinases and histidyl-tRNA synthetases in GCN2 protein suggests a mechanism for coupling GCN4 expression to amino acid availability. Proc. Natl. Acad. Sci. USA 86:4579-4583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Willems, A. R., T. Goh, L. Taylor, I. Chernushevich, A. Shevchenko, and M. Tyers. 1999. SCF ubiquitin protein ligases and phosphorylation-dependent proteolysis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 354:1533-1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wilson, W. A., A. M. Mahrenholz, and P. J. Roach. 1999. Substrate targeting of the yeast cyclin-dependent kinase Pho85p by the cyclin Pcl10p. Mol. Cell. Biol. 19:7020-7030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu, X., J. H. Bayle, D. Olson, and A. J. Levine. 1993. The p53-mdm-2 autoregulatory feedback loop. Genes Dev. 7:1126-1132. [DOI] [PubMed] [Google Scholar]