It has become clear that the nucleus is organized into discrete structures. Although not membrane bound, these structures are considered nuclear organelles. Widespread interest in one such nuclear organelle, the promyelocytic leukemia nuclear body (NB), has emerged because of its link to several human disorders, including acute promyelocytic leukemia and AIDS. Studies of the physiological effects of promyelocytic leukemia NBs and the promyelocytic leukemia protein (PML) indicate that these play roles in growth control, transformation suppression, apoptosis and Ras induced senescence. Unfortunately, the molecular and biochemical bases for physiological phenomena associated with PML are not well understood. PML and, by inference, the PML NB have been ascribed apparently disparate roles in transcription, DNA repair, DNA replication, and RNA transport. Its clear physiological importance means that defining a set of discrete biochemical functions for the PML NB is critical. This review focuses on the current theories for molecular and biochemical functions of the PML NB and the supporting evidence for each.
PML NBs, also known as PML oncogenic domains, nuclear domain 10's, or Kremer bodies, are currently defined by the presence of PML at these nuclear structures. PML and its associated NBs were first described in a series of studies in the early 1990s which showed that PML was fused to the retinoic acid receptor alpha (RARα) in acute promyelocytic leukemia (APL) patients (reviewed in reference 63). These studies demonstrated that PML NBs were similar to those previously observed by electron microscopy in the 1960s (63). Intriguingly, NBs were disrupted in the APL patients but reformed after treatment with all-trans-retinoic acid (ATRA) (36). Reformation of bodies correlated with remission of disease in patients. This was the first evidence that the integrity of these structures may be critically important to the health of the cell (36, 63). These findings sparked widespread interest in the function of these organelles. These early studies also revealed that the PML protein contained a set of novel zinc-binding domains, known as the RING and B-boxes. Early on, it was proposed that PML utilized these zinc fingers to directly bind DNA and thus alter gene expression. However, in the past 10 years, it has become clear that the RING and B-box domains forge protein associations that are critical to the integrity of this multiprotein complex and subsequent physiological function(s) of this organelle (36, 40, 63).
Most reported strategies for assessing PML NB function in essence are designed to answer the following questions: what nuclear structures are the bodies near to, what other macromolecules localize with the body, and what are the effects of disrupting the body? These strategies arise because the discrete biochemical functions of either PML or PML bodies are not known. The results of these strategies and their inherent limitations are discussed within this review. Further, the following considerations should be taken into account in an assessment of PML NB function. First, the expression of the PML gene is not required for viability, since PML−/− mice develop in essence normally and do not get spontaneous cancers at rates higher than do littermate controls (99). Second, the PML gene is not evolutionarily conserved among eukaryotes, being absent in Drosophila melanogaster, Saccharomyces cerevisiae, and Arabidopsis thaliana (see below). Third, unlike other nuclear organelles, there appears to be no PML bodies in Xenopus laevis. However, the disruption of these organelles apparently contributes to human disease. These features and their potential clues to PML NB function are discussed below.
Because of space limitations, many topics are not discussed here. For instance, the study of viral systems has been key to our current understanding of the PML NB. However, an in-depth discussion of these contributions is beyond the scope of this review, but this topic is discussed elsewhere (25, 59, 72). Many excellent and comprehensive reviews have been written on various aspects of the physiological functions of PML and its relationship to APL (36, 40, 59, 63, 82). The present review attempts to describe current understanding of the molecular and biochemical underpinnings of PML NB function.
GENERAL FEATURES OF THE PML NB: AN OVERVIEW
Currently, the biochemical and molecular functions of PML NBs and PML are unknown. However, basic characterization of the bodies gives some clues to NB function. The majority of PML is part of a large multiprotein nuclear complex referred to here as PML NBs (36, 59, 63). These NBs have been observed in all reported mammalian cell lines and are present throughout the cell cycle, although most bodies are observed during G1 (17). Approximately 10 to 30 bodies are observed per nucleus, ranging in size from ∼0.2 to 1 μm (63). Like many transcription and RNA processing factors, these bodies are associated with the nuclear matrix (67, 80, 89). The bodies do not rely on nucleic acid for structural integrity since treatment with RNase or DNase does not alter their morphology (3, 89). However, they are disrupted by treatment with m7GpppG, an analogue to the mRNA cap structure (19). In addition, the morphology of PML NBs is altered by a variety of viruses and environmental stresses. For instance, heavy metals, such as cadmium, cause the PML NBs to disperse (60, 91a). PML is interferon inducible, and interferon treatment causes an increase in both size and number of NBs (18, 54, 60, 87). DNA and RNA viruses have a variety of effects on body morphology, where arenaviruses and human immunodeficiency virus (HIV) move PML to the cytoplasm but herpesviruses “unwind” bodies (11, 59, 63, 93). Findings with HIV are somewhat controversial, since another group did not see PML NBs translocate during infection (4).
The composition of PML NBs is heterogeneous with some components present in a subset of bodies (10, 15, 27, 52). PML may associate with at least 50 proteins (some of these are reviewed in reference 63). Associated proteins function in translation, transcription, DNA repair, DNA replication, mRNA stability, and mRNA transport as discussed below. Newly synthesized mRNA is associated with the periphery of some NBs (7). Notably absent from NBs are DNA, RNA polymerase II, and TFIIH, making functions directly in transcription unlikely (3, 35, 63).
PML.
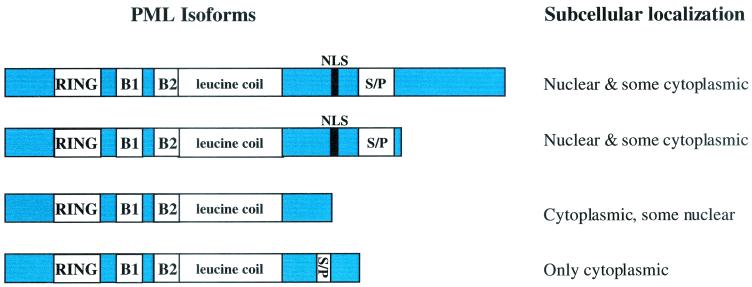
Certain features of PML are required for association with NBs and subsequently for its physiological functions. PML contains cysteine-rich zinc-binding domains, known as the RING, two B-boxes, and an adjacent leucine coiled coil forming the RBCC or TRIM motif. The RING domain is ca. 60 residues long, binds two zinc atoms, and is found in more than 200 proteins (8, 46). RING and B-box motifs are thought to be involved in protein interactions and do not appear to bind nucleic acids directly (8, 46). A loss of PML bodies in transiently transfected cells results from the mutation of any of the zinc-binding residues in the RING or B-boxes (9, 13, 57), mutation of the leucine coiled-coil (57), or mutation of the nuclear localization signal (NLS) (57). PML also contains a serine/proline-rich region of unknown function (Fig. 1).
FIG. 1.
Relationship of PML isoform to subcellular distribution in transfected cells. The diagram was adapted with permission from a previous study (26, 29). “S/P” indicates the serine/proline-rich regions, and B1 and B2 indicate B boxes.
PML is expressed in all mammalian tissues and cell lines reported (29, 90). The majority of PML forms NBs (63); however, there is some PML located in the cytoplasmic bodies and a soluble component in the nucleus (26, 29). Alternative splicing of PML transcripts results in coexpression of several isoforms, which vary in their carboxy termini and subcellular distribution (26, 29). Two PML isoforms are detectable only or predominantly in bodies in the cytoplasm of transfected cells. The cytoplasmic localization is a feature shared even by the PML isoforms that are predominantly nuclear (26, 29). Figure 1 is adapted from an earlier study (26) and illustrates the relationship of isoform to subcellular distribution (28). The complete set of isoforms, to date, is summarized by Jensen et al. (43).
APL and PML.
As discussed above, PML is implicated in the pathogenesis of APL. APL comprises ca. 10% of myelogenous leukemias and is characterized by a block in myeloid cell development at the promyelocyte stage (36, 63). In more than 98% of APL patients, PML is fused to RARα as a result of a chromosomal translocation (63). The resulting protein product is referred to as PML-RARα. In APL patients, the PML NBs are disrupted, forming a microparticulate pattern in the nucleus and cytoplasm (24, 45, 101). Addition of ATRA reverses this aberrant phenotype, resulting in the reformation of PML NBs, which correlates with remission in patients (36, 63). ATRA treatment is correlated with the targeted degradation of the PML-RARα fusion protein, while leaving the wild-type proteins intact (63). The paradigm for leukemogenesis in APL is that disruption of PML leads to a loss of growth control and disruption of RARα leads to the block in differentiation (36, 63). Although the number of APL patients is low, this disease has become a paradigm for successful differentiation therapy. Several reviews discuss the role of PML in the pathogenesis of APL, including references 5, 36, 63, 78.
PHYSIOLOGICAL EFFECTS OF PML IN GROWTH CONTROL
PML is involved in growth regulation, and these activities are, at least in part, dependent on the integrity of the NB structure. Overexpression of PML results in G1 arrest and, in some cases, apoptosis (63). Disruption of PML NBs through mutations in the RING is correlated with a loss of its growth suppression activity (56, 57), loss of apoptotic activity (12), and inability to regulate transport of certain mRNAs (19, 50). Overexpression of PML in serum-starved murine fibroblasts induces apoptosis, and this activity is mediated in part through the RING (12). In some cell types, overexpression of PML-RARα has potent growth-inhibitory and apoptotic affects which require the RBCC motif (28, 75). The effects of PML overexpression on gene expression are specific. Two groups have shown independently that PML suppresses cyclin D1 protein levels but does not alter actin protein levels (19, 50, 56). Further, overexpression of PML does not alter overall [35S]methionine incorporation relative to controls but only levels of a subset of proteins (50).
The PML gene is not essential for survival. PML−/− mice are morphologically normal and do not have higher rates of spontaneous cancers than littermate controls (99, 100). However, when these animals are exposed to certain types of challenges such as gamma irradiation, PML−/− mice do not die as readily (100). Consistently, the PML−/− cells are less likely to undergo apoptosis under these types of cellular stresses (71, 100). Interestingly, the proapoptotic function of PML is independent of transcription (71). PML−/− mice treated with DMBA and tetradecanoyl phorbol acetate developed slightly more papillomas than did treated littermate controls (99). PML−/− cells are more easily infected with certain viruses (21, 73). However, these results can vary depending on the PML−/− cell lines used, suggesting that there may be an effect due to clonal selection (Gerd Maul, unpublished data). Consistently, the early studies in APL patient cells strongly suggest a correlation between the organization of PML into NBs and normal growth control (63).
NUCLEAR GEOGRAPHY: PML'S PLACE ON THE MAP?
Knowledge of the location of NBs in relation to other nuclear structures may provide clues to PML NB function. Further, its relative location may give insight into what specific targets it regulates. Several studies have shown that PML NBs have preferences for certain subnuclear locations. PML NBs tend to be near certain nuclear compartments such as Cajal/coiled bodies, cleavage bodies, and splicing speckles (as defined by the presence of Sc35) (35, 63, 83). It is important to note that these structures do not colocalize with PML bodies but are frequently neighbors. Studies with T24 cells indicate that of the one to four Cajal bodies per cell, one is always adjacent to PML bodies (35). Cajal bodies are thought to be involved in assembly of spliceosomes and of the transcriptosome (32), cleavage bodies are involved in 3′ end processing of mRNA (81) and Sc35 is involved in specific pre-mRNA processing events (31). Further studies indicate that U2-snRNP and U1-snRNP are absent from PML bodies (35). Taken together, these findings suggest that PML could play some sort of role in RNA-processing events.
Detailed measurements of the distance between PML bodies and genomic loci reveal another specific spatial relationship (83). These findings showed that one PML body in each cell was found to be adjacent to a gene-rich major histocompatibility complex (MHC) locus on chromosome 6 (83). The MHC localization is highly nonrandom and was independent of transcriptional status of the cell and of progression through the cell cycle (83). These studies show that PML bodies have specific genomic associations that are independent of transcription. The functional importance of being near the MHC genomic locus, Sc35 speckles, cleavage bodies, and coiled bodies has yet to be established. Undoubtedly, future work will clarify whether these are functional relationships or “street signs” indicative of other nearby as-yet-unidentified compartments with which PML functionally interacts.
Clearly, if one is using this “neighbor” type of analysis to deduce PML NB function, it is critical to establish whether PML NBs are stationary or mobile organelles. Recent findings indicate that a subset of PML NBs move throughout the nucleus in an energy-dependent fashion (65). In these elegant studies, the localization of the well-established PML partner protein, Sp100, fused to the yellow fluorescent protein was monitored in living BHK cells. Three populations of NBs were observed: 25% of the bodies were stationary, 63% showed minor motions, and 12% showed rapid motions. The majority of PML NBs (those which showed minor motions) exhibited localized movements similar to those observed in nuclear speckles and Cajal bodies. The rapid class of NBs, which represented about one to two NBs per nucleus, were typically small. Different NBs moved at different times within the same nucleus. Motions include not only translational movements but also bodies that coalesce and smaller bodies that bud off from larger ones. Of the rapidly moving class, PML bodies traversed the nucleoplasm at an average velocity of 4.0 to 7.2 μm/s. Thus, PML NBs move an order of magnitude more quickly than any previously observed nuclear structure. Notably, movement was not sensitive to α-amanitin treatment, and therefore to RNA polymerase II activity, but was was sensitive to the depletion of ATP. Further, motion appears to be dependent on nuclear myosin. Importantly, HeLa cell nuclei had no moving PML bodies, whereas BHK and primary mouse embryo fibroblasts did, suggesting that some cell type-specific factors may be involved. These data strongly suggest that there are multiple classes of PML NBs. Thus, consistent with previous studies indicating compositional heterogeneity and with observations that not all PML NBs within the same nucleus have the same preferences for their neighbors, there is heterogeneity in PML NBs in terms of movement.
GUILT BY ASSOCIATION: THE SEARCH FOR MEANINGFUL RELATIONSHIPS
A major approach for elucidation of PML body or PML protein function (not necessarily synonymous) has been through the determination of partner proteins. These studies are carried out with the intent that the nature of partner proteins may provide clues to NB function. To date, more than 50 PML partners have been reported. The proteins have a wide variety of functions and, taken together, these do not clearly point toward a unifying theme. Many of these are described in the following sections, but a comprehensive list is beyond the scope of this review. (However, see references 36, 40, 43, and 63.)
The very nature of establishing function by association means that identification of partner proteins must be undertaken rigorously. A major difficulty with these studies is that the identification of partners by means of transient transfection is not a reliable method for the case of PML. A graphic example of this comes from studies designed to determine whether PML bodies functioned in transcription. The Spector laboratory noted that overexpressed bacterial lac repressor protein localized to the PML body independent of transcription (92). Indeed, association of PML bodies with an artificially integrated gene locus depended on the in vivo binding of the lac repressor and pTETon, indicating that overexpressed or foreign proteins likely concentrate at or near the body (92; see also below). Thus, determining whether a protein is a PML partner should not rely solely on interactions observed when PML or the partner protein is overexpressed.
Clearly, it is important to establish that interactions between PML and putative partner proteins are physiologically relevant. Ideally, this is done with endogenous proteins showing that they both colocalize with endogenous PML bodies and can be coimmunoprecipitated with endogenous PML. Further, experiments should be done in a variety of cell types and with different antibodies to establish that the interactions are not due to any cross-reaction with other PML body components. The ability of some antibodies to cross-react with other PML body components has led to misidentification of proteins as PML partners (71). This sort of problem really demonstrates the kinds of pitfalls encountered in identifying PML partner proteins, even by excellent groups of researchers with substantial experience in this field. Unfortunately, many PML partners have not been established with the above-described level of rigor and, although they may well be bona fide constituents of the body, this remains to be demonstrated conclusively. This uncertainty complicates matters in terms of investigating putative molecular functions of the body based on currently reported partners.
GETTING TO THE ROOT OF THE MATTER: DIRECT PARTNERS OF PML
Given the variety of proteins in the body, a study of proteins that directly interact with the PML protein should yield unique insights into NB function. Direct interaction between proteins can be established conclusively only if purified components are used. Many reports claim direct interactions based on purification of one component and its ability to “pull down” another from reticulocyte lysate. The composition of reticulocyte lysate is complex and contains many factors, aside from the translation machinery, which could mediate interactions with PML.
There are four proteins reported which meet the stringent criteria for direct interactions with PML: the ubiquitin conjugating enzyme (Ubc) 9, the small ubiquitin modifier SUMO1, eukaryotic translation initiation factor 4E (eIF4E) and the proline-rich homeodomain protein (PRH) (6, 19, 23, 50, 91). Elegant studies demonstrate that the RING domain of PML directly interacts with Ubc9, which covalently attaches the SUMO1 protein onto distal regions of PML, including one B-box and a region near the NLS (23). Other, unrelated proteins are similarly modified by SUMO1, including RanGAP and IκB (23). SUMO1 modification does not appear to be a signal for degradation but a signal for subcellular localization (62). The SUMO modification is not required for PML to form bodies but is required for association of some partner proteins such as Sp100 and Daxx with PML NBs (42).
One of the most thoroughly characterized interactions biochemically to date is between the RING of PML and eIF4E. Studies from our laboratory showed what appears to be the first reported discrete biochemical activity of PML, its ability to reduce the affinity of eIF4E for its substrate, 5′m7G capped mRNA, and thereby impede RNA transport (see below) (19, 47, 50). PRH also interacts directly with the RING of PML (91; unpublished results). PRH plays a role in myeloid differentiation, making its direct association with PML and its relationship to leukemia very intriguing. A battery of biochemical and biophysical assays were carried out to establish that these interactions were indeed direct, including glutathione S-transferase pull-down assays and separately by limited proteolysis in conjunction with electrospray mass spectrometry (47). In all cases, bacterially expressed proteins purified to homogeneity were used (47). The functional relevance for some of these direct interactions is explored below.
A variety of functions have been attributed to PML NBs. Support for the most popular theories and attempts to integrate these into a common functional theme are presented below.
NUCLEAR CLOSETS? PML BODIES AS PROTEIN STORAGE COMPARTMENTS
The variety of PML partner proteins and the compositional heterogeneity of the body (even within a single nucleus) have led to the suggestion that the bodies themselves are not active compartments but rather storage facilities for the cell (59). This suggestion is consistent with the observation that overexpressed and foreign proteins such as the lac repressor and a variety of viral proteins associate with the body (59, 63, 92). Several proteins involved in polyglutamine repeat disorders—including huntingtin, ataxin 1, and ataxin 3 in their expanded forms—associate with the body, but here bodies are larger and appear more like inclusions than normal bodies (16, 85, 105). In this way, PML NBs could be sensors for foreign or inappropriately expressed proteins and thus would be acting like a subnuclear immune system (59). Recent data show that some PML NBs are adjacent to proteosomal components and that As2O3 treatment increases the frequency of this (51). The presence of the proteosome adjacent to the bodies is consistent with this and many other explanations for PML NB molecular function (see below).
In addition to foreign or overexpressed proteins, it has been proposed that bodies are storage areas for normal functional components (59). In this way, bodies could titrate levels of active components in the nucleoplasm and therefore modulate biochemical processes (59). In fact, it has been suggested that several other NBs are not sites of active biochemistry but rather storage areas for certain types of components. An alternative, but complementary, explanation is that these bodies form catalytic surfaces, where specific biochemistries can occur on the surface of the body. The kind of biochemistry could be altered by the composition of associated proteins (8, 46). The catalytic surface mechanism has been suggested for other bodies comprised of RINGs (8, 48). Whether PML NBs or other subnuclear structures are active organelles or subnuclear storage facilities remains hotly debated. It is important to note that either the “nuclear closet” or “catalytic surface” theory for PML function would provide a molecular basis for the specific biochemical functions described below. In this way, the catalytic surface and nuclear closet theories describe a mechanism of action rather than a particular processes in which PML is involved.
DIRECT ROLE IN TRANSCRIPTIONAL ACTIVATION OR REPRESSION?
PML has been touted as both a transcriptional activator and repressor. Initially the misconception of the early 1990s that the zinc finger RING and B-box domains were DNA-binding domains led to a flurry of investigations into PML's role in transcription. Several key studies indicate that these domains in PML and in other proteins mediate protein-protein interactions (8, 9). More recent data indicate that transcription and chromatin remodeling factors are associated with PML, and several groups have tried to implicate PML as a direct actor in transcription on this basis (63). It is important to remember that PML NBs do not associate with sites of active transcription and do not localize with nascent DNA (35, 63). Further, general transcription factors like TFIIH and RNA polymerase II do not colocalize with PML bodies (35).
The proposal that these bodies function in transcription is now largely based on the colocalization of retinoblastoma protein (Rb), CBP, and p53 with the body. Approximately 1% of endogenous PML NBs have endogenous Rb associated with them and ca. 1% of Rb is associated with PML (27). In fact, these recent studies indicate that the remaining Rb forms nuclear speckles with no obvious overlap with PML NBs (27). These studies are consistent with independent work that indicated that ca. 0.5 to 1% of PML immunoprecipitates with Rb (2). Introduction of RasV12 results in 14% of endogenous PML bodies associating with Rb (27). Intriguing results suggest that a subset of CBP molecules associate with PML. In these studies, two CBP antibodies (to residues 634 to 648 and to residues 1 to 100) show that CBP was organized into nuclear structures which colocalized with PML (52). Other CBP antibodies (to residues 455 to 679 and to the C terminus) in this study indicate that CBP was distributed uniformly throughout the nucleoplasm, with no evidence of NBs (52). Later studies indicate that the CBP A22 antibody (from Santa Cruz) also shows CBP in bodies (22). The authors of that study suggested that epitope masking is responsible for these different patterns (22). Interestingly, other labs (using the A22 antibody) have demonstrated that CBP is a significant component of PML NBs only in the presence of oncogenic Ras (27). Prior to introduction of RasV12, PML bodies are present as usual but CBP is dispersed throughout the nucleus, with no organization into bodies (Fig. 5 in reference 70). Similar findings were reported for p53, which localized to PML NBs after the introduction of oncogenic Ras with no colocalization prior to RasV12 inroduction (Fig. 5 in reference 70 and Fig. 7 in reference 27). Thus, there are some contradictions in the literature regarding whether p53 and CBP localize to PML NBs in the absence of oncogenic Ras. In the case of p53, assignment of transcriptional functions to PML based on its association with the NB is even more complex given that p53 functions in both transcription and binds regulatory regions of mRNA, indicating that its presence alone is not diagnostic of a transcriptional function (64). Although the findings described above are confusing in terms of the identity of baseline PML partner proteins, they do demonstrate that the introduction of oncogenic Ras can substantially alter the PML NB. These interactions may have important implications to NB function, as well as for PML's role in Ras-induced senescence (27, 70).
Recent findings suggest the nuclear corepressor NcoR may also be a component of NBs (49). Transfected NcoR localizes with endogenous PML. Intriguingly, in these studies, NcoR required PML for formation of NBs leading to the suggestion that PML bodies are sites for corepressor assembly. It is noteworthy that the NcoR knockout mouse has an embryonic lethal phenotype (at day 15.5) affecting central nervous system, thymocyte, and erthrocyte development (44). These systems are, in essence, unaffected in the PML−/− mice (99). If PML acts as the critical organizer for these corepressor complexes, one would expect a more severe phenotype than that observed for the PML−/− mice. Thus, it is difficult to resolve these apparently contradictory observations, e.g., the PML−/− mice are unaffected, PML NBs are required for formation of the corepressor complexes and NcoR−/− mice die as embryos. Some of the confusion here may be a problem with distinguishing between function of PML in the nucleoplasm and function of the PML NB. Clearly, more work needs to be done in this area to resolve these issues.
Reporter assays have been crucial in supporting the idea that PML functions in transcription. In these studies, PML can act as either a transcriptional repressor or activator, depending on the promoter and system used (for examples, see references 38, 63, and 95). These assays typically rely on the effects of overexpressed PML on production of luciferase reporter proteins. There are two major problems in assigning a direct transcriptional role to PML based on these methodologies. First, levels of luciferase mRNA have not been measured directly in these assays; thus, there is no proof that fluctuations in luciferase light units reflect changes in mRNA levels. Other events that could alter luciferase protein production without altering mRNA levels include (but are not limited to) alterations in transcript stability, mRNA transport, translation, or protein stability. Second, if the effects on luciferase were at the mRNA level, there is no proof that the effects of PML overexpression are directly transcriptional. Previous studies with proteosomal components highlight the pitfalls of the use of these methods as the only evidence for determining that a protein acts directly in transcription. Using these sorts of reporter assays led to the incorrect conclusion that several proteosome components were transcription factors (79). Further, evidence that PML can act on gene expression posttranscriptionally (see below) means that data from luciferase assays must be interpreted carefully. For instance, PML could alter the transport of another RNA, which in turn alters the production of a protein that then negatively or positively regulates factors that bind the promoter by which luciferase is controlled and therefore modulate production of luciferase. The specificity of these assays is typically inferred from mutational analyses of the promoter. However, loss of function due to key mutations in the promoter does not imply a direct effect of PML, even though that has been proposed frequently. In the scenario described above, mutation of the promoter would simply mean that the intermediate factor(s) could no longer alter luciferase production. A potential example of this indirect scenario comes from the effects of PML on cyclin D1 mRNA. Overexpression of PML causes nuclear retention of cyclin D1 mRNA but no alteration in the total level of cyclin D1 transcripts (19, 50, 91a). This causes a decrease in cyclin D1 protein levels. Cyclin D1 protein is known to recruit PCAF to the estrogen response element (61). In this way, it could be possible for PML to alter production of mRNAs under control of the estrogen response element without directly acting in transcription.
The results from many studies can be explained by these sorts of indirect effects. For example, overexpression of PML was shown to decrease the ability of the progesterone receptor (PR) to transcribe a PRE-CAT construct twofold (38). PML overexpression did not alter the affinity of PR for its ligand or its affinity for its cognate DNA and associated factors in cell extracts (38). In these studies, the authors report that in addition to transfected PML associating with NBs, there was substantial exogenous cytoplasmic PML. No interactions (direct or indirect) could be detected between PR and PML. Despite the lack of interactions between PML and PR, these findings were taken to mean that PML acted as a transcriptional modifier. Together, these data could equally well suggest an indirect role in transcription perhaps similar to the scenario described for cyclin D1. Since the overexpression of translation factors can have the eventual effect of inducing DNA synthesis (1), it is important to establish whether the effects observed for PML are directly transcriptional or at some other level of gene expression. Thus, in order to demonstrate convincingly a role for PML in transcription, studies need to be significantly extended, including association with endogenous transcription factors and DNA, perhaps through chromatin immunoprecipitation methods.
Visualization of transcription in living cells has also led to new insights into putative roles of PML in transcription. Elegant studies by the Spector laboratory show that PML's association with active sites of transcription is likely an artifact of protein overexpression (92). These authors developed a novel system for monitoring the production of a peroxisomal gene and its resultant protein by using the lac operator-repressor system. In these studies, the integrated peroxisomal gene locus was surrounded by a PML body. This association was independent of transcription but was specific to the in vivo binding of the lac repressor-yellow fluorescent proteins and the tetR/VP16 transactivator to the locus (Fig. 7 in reference 92). No localization of PML and the integrated locus is observed if the lac repressor-yellow fluorescent protein and pTETon were not expressed in these living cells. These authors suggested that, in these studies, PML acts as a nuclear sensor that detects foreign proteins and not directly in the transcriptional process. Consistent with the above studies, Shiels and coworkers have demonstrated that the proximity of MHC gene locus to PML is independent of the transcriptional status of the cell (as discussed above and in reference 83).
These considerations do not rule out the possibility that PML can modulate transcription indirectly and/or that PML's functions could be linked to transcriptional activity within the cell. Further, it is possible that the nucleoplasmic fraction of PML and not the NB could be acting directly in the activation or repression of transcription. However, the evidence for the PML NB as a direct actor in classic transcription processes is currently far from complete.
ROLES IN DNA REPLICATION AND REPAIR
Early experiments monitored the spatial relationship between PML NBs and DNA replication factories. In these experiments bromodeoxyuridine incorporation was used to visualize newly synthesized DNA within the nucleus. Many groups observed that newly synthesized DNA is found in a distinct spatial distribution throughout the nucleus (35). In these studies, replication patterns for the early, middle, middle-late, and late S phases were monitored. These studies indicate that during the entire replication process, PML NBs are excluded from nuclear domains that contain newly synthesized DNA (35). In middle to late S phase, 50% of PML bodies (for the 10 nuclei observed) are found adjacent to, but not overlapping with, replication domains (35). In no other part of the S phase is a relationship between PML NBs and replication domains observed (35, 96). This is also consistent with studies that indicate that parathymosin structures, which are localized to sites of early DNA replication, do not localize with PML NBs (96). These observations are further supported by the observation that simian virus 40 replication domains are often adjacent to, but do not overlap with, PML NBs (59).
Recent studies have found that in ∼5% of a subset of telomerase-negative cells, the telomere-binding proteins hTRF1 and hTRF2 form bodies which localize with PML NBs (106). Replication factor A and Rad52 also localize to these sites. In addition, in situ hybridization studies indicate that telomeric DNA repeats are present in these bodies (106). This appears to be the only time that DNA and PML bodies associate. These NBs are referred to as APB (for ALT-associated PML body). These cell types have developed a mechanism of telomere length maintenance in the absence of telomerase activity referred to as alternative lengthening of telomeres (ALT). In contrast, in non-ALT cells (which can be either telomerase negative or telomerase positive), the telomere-binding proteins, Rad52, and replication factor A exist in discrete subnuclear domains which do not overlap with or have any apparent spatial relationship to PML NBs (30, 37, 106).
Several links have been made between PML NBs and DNA repair. It has been reported that PML interacts with p53 and the Bloom syndrome protein, BLM, which is a RecQ DNA helicase family member (98). Expression of BLM is cell cycle dependent, where expression peaks in S and G2 phases (33). Disruption of BLM results in a high level of genomic instability, resulting in higher rates of a variety of cancers (39). BLM protein has limited expression found mainly in lymphocytes and in proliferating tissues (39, 94). BLM-null cells have attenuated DNA damage-activated and p53-mediated apoptosis (98). Recent studies show that the induction of p53 and p53-mediated transcription were not affected in the Bloom syndrome cells, where downstream target proteins such as Gadd45, Bax, and p21waf1 were produced at normal levels as observed by RNase protection assays (98). Thus, the BLM protein does not appear to alter p53-dependent transcription. The distribution of BLM is highly dependent on the cell cycle. BLM forms foci in ∼40% of fibroblasts or HeLa cells in the G1/S phase of the cell cycle. Only 6% of cells had BLM bodies in G0 (33). In S phase, the majority of BLM is found in the nucleolus (33). In cells with BLM NBs, PML and BLM colocalize (103). However, the localization of BLM to the nucleolus appears to be more important to genomic stability than the BLM associated with PML NBs (104). One might expect a link between BLM and telomeres. Some BLM localizes to some telomeres, but BLM is not a major structural or regulatory factor in telomere maintenance (103). PML NB morphology is normal in the BLM-deficient cells; however, BLM does not form NBs in PML−/− cells (98). Further studies showed that the association between PML and BLM depends on p53 since the association is lost in p53−/− cells (98). In this case, PML NBs appear normal, whereas BLM is no longer localized to bodies. The functional relevance of these interactions remains to be elucidated, but certainly they will be actively pursued given the clear biomedical relevance of these proteins.
POSTTRANSCRIPTIONAL REGULATION OF GENE EXPRESSION BY PML
Recent studies suggested that PML NBs can influence gene expression posttranscriptionally. Our laboratory has demonstrated that PML interacts with eIF4E directly, and endogenous eIF4E colocalizes and coimmunoprecipitates with endogenous PML in several cell types (14, 19, 47, 50). The PML- eIF4E interaction has been observed independently (S. Hasan and C. Harris, unpublished data). Note that the incidence of colocalization between these proteins is high but not perfect in all cell types, with some cell type-specific variations. In contrast to PML, eIF4E is growth promoting (55, 86) and, like some other PML partners, eIF4E has a connection to Ras (86). eIF4E overexpression increases levels of activated Ras, and it exerts its mitogenic and oncogenic activities at least in part by Ras activation (55). Unlike p53, CBP, or Rb, PML and eIF4E interact in the absence of oncogenic Ras. Mapping studies by mutagenesis and separately, by a combination of limited proteolysis and mass spectrometry, indicate that regions around the first zinc-binding site of PML RING and the dorsal surface (through W73) of eIF4E are the interaction sites (19, 47). The dissociation constant for the PML RING-eIF4E interaction is ∼500 nM, indicating that these proteins form a tight complex (47).
Traditionally eIF4E functions in the cytoplasm, where it acts in the rate-limiting step of translation initiation (86). Here, it binds the 5′m7G cap found on mRNAs and associates with the ribosome, thereby initiating translation. However, a large proportion (up to 68%) of eIF4E is found in discrete spherical structures in the nucleus (41, 58). The Sonenberg laboratory showed that overexpression of eIF4E results in the preferential transport of a subset of mRNAs, including cyclin D1, from the nucleus to the cytoplasm with no change in total levels of these mRNAs (76, 77). The preferential transport of cyclin D1 mRNA leads to increased levels of cyclin D1 protein. It is not known whether eIF4E acts directly in transport or else in some activity that is required prior to or for transport. A second function for nuclear eIF4E has been proposed. Elegant studies by the Cook laboratory (41) suggest that mRNAs are translated in the nucleus as part of the nonsense-mediated decay pathway, in which low-level translation occurs in order to proofread transcripts. Interestingly, proteosomal components were found by these sites, presumably to degrade the new proteins (41). These studies suggest that nuclear translation is similar to cytoplasmic translation and is therefore eIF4E dependent. Thus, the precise biochemical activities of nuclear eIF4E and PML-eIF4E bodies are yet to be elucidated.
PML negatively regulates eIF4E-dependent mRNA transport (19, 50, 91a). Overexpression of PML causes the retention of cyclin D1 mRNA in the nucleus with no significant effect on total production of cyclin D1 transcripts. Further, PML suppresses eIF4E-mediated transformation (19). This appears to depend on the direct interaction between PML and eIF4E, since mutations that disrupt this direct interaction also disrupt the ability of PML to suppress eIF4E-mediated mRNA transport and subsequent transformation (19). Further, cyclin D1 transport mediated by PML and eIF4E can be altered in response to physiological stress (91a).
eIF4E has a well-defined biochemical function in its association with its substrate, the m7G cap structure on mRNA (86). The ability to bind the cap structure is important for both its nuclear and its cytoplasmic functions (86). An eIF4E mutant (W56A) that cannot bind the cap structure of mRNA is also deficient in its mRNA transport function and ability to transform cells (19). Our biochemical studies demonstrated that the PML RING induces a conformational change in eIF4E that is correlated with a reduction in the affinity of eIF4E for the m7G cap of mRNA of >100-fold (19, 47). To our knowledge, this is the first discrete biochemical activity demonstrated for the PML protein. The ability to inhibit eIF4E-dependent transport and transformation appear to be directly linked to the ability of PML to reduce the affinity of eIF4E for capped mRNA (19, 47). Satisfyingly, this biochemical activity can be linked to the growth arrest phenomena associated with PML overexpression.
Independent results from several laboratories support the idea that PML could function in posttranscriptional regulation of gene expression. As discussed above, PML is localized adjacent to known sites of RNA processing, including splicing speckles, cleavage bodies, and Cajal bodies (35). The identification of eIF3/Int6, Isg20, GAPDH (glyceraldehyde-3-phosphate dehydrogenase), and the ribosomal P proteins as NB components is also consistent with this hypothesis. For instance, eIF3/Int6 is part of the translation initiation complex (20). Isg20 is an interferon- and estrogen-regulated RNase, as well as DNase (34, 69). Many new functions have been described for GAPDH outside of glycolysis, including binding AAUAA-rich segments of RNA and shuttling specific tRNAs into the nucleus (66, 84). GAPDH is found in nuclear structures that associate with PML NBs in an RNA-dependent manner (15). Further, several components of the translation machinery associate with PML, including eIF3/Int6, eIF4E, ribosomal P proteins (10), L7a (6), and elongation factor EF1 (6), a finding consistent with nuclear translation (41). In the nucleus, PML would be situated to negatively regulate this activity (47). There are still many unanswered questions with regard to exactly what PML's biochemical role is beyond reducing the affinity of eIF4E for capped mRNA. For instance, what biochemistries are precisely occurring at the body relevant to the mRNA transport functions of PML and eIF4E and are the few PML bodies that do not colocalize with eIF4E functionally distinct from those that do?
THE FORGOTTEN FRACTION: FUNCTIONS FOR CYTOPLASMIC PML
Normally, a small percentage of PML is found in cytoplasmic bodies. Indeed, there are several isoforms of PML that lack an NLS but retain the RBCC domains (Fig. 1 and reference 26). Further, in some pathogenic conditions, such as HIV and arenavirus infection, PML is translocated to the cytoplasm, which may lead to a gain of function in the cytoplasm, as well as a loss of function in the nucleus (11, 93). Cytoplasmic PML has also been observed in APL patient cells prior to ATRA treatment and in hepatocellular carcinoma cells (63, 90). However, little is known about what the putative functions of cytoplasmic PML could be.
Fractionation studies reveal that PML interacts with eIF4E in the cytoplasm, as well as in the nucleus (50). In vitro translation studies indicate that PML inhibits translation of luciferase and other mRNAs through its interaction with eIF4E (47). For translation to proceed, eIF4E must bind not only the m7G cap structure but also an adaptor protein, eIF4G (86). In fact, PML appears to be a potent translational repressor because not only does it reduce the affinity of eIF4E for the cap, its binding site overlaps, in part, with that of eIF4G (47). Many proteins that are both nuclear and cytoplasmic retain their discrete biochemical activities, but the physiological effects depend on their subcellular distributions (102). In the case of PML, the ability to modulate m7G cap binding by eIF4E may be central to its function in both the nucleus and the cytoplasm (47). No other potential functions have been attributed to the cytoplasmic fraction of PML to date.
PHYLOGENETIC CLUES TO PML BODY FUNCTION?
Clearly, it is important to explore the level of phylogenetic conservation in the consideration of possible functions for the PML NB. As discussed above, PML belongs to the RBCC family of proteins which contains ∼120 members (http://smart.embl-heidelberg.de), many of which form nuclear or cytoplasmic structures similar in morphology to, but discrete from, PML NBs (8). Consistent with the above observations, searches using the protein architecture database SMART (http://smart.embl-heidelberg.de) indicate that there are no S. cerevisiae (http://genome-www.stanford.edu), Schizosaccharomyces pombe (http://www.sanger.ac.uk/Projects/S_pombe) or A. thaliana (http://www.arabidopsis.org) proteins which contain both a RING and a B-box, although the RING domain is found independently in these genomes.
Higher eukaryotes do contain proteins with the RBCC motif, so more-rigorous database searches are required. Proteins containing the RBCC were found by using the National Center for Biotechnology Information (NCBI) DART program, and for each sequence the domain structure was verified by using the BLOCKS program (http://blocks.fhcrc.org/blocks/blocks_search.html). Alignments were then made by using the NCBI Blast2Seqs program. Species as diverse as Caenorhabditis elegans, newt, Xenopus, and chicken have the RBCC motif, in these cases with only one B-box. Searches of the D. melanogaster (http://www.fruitfly.org) genome, which is complete, revealed that there were sequences in this genome that contained RBCC motifs with two B-box domains. Frequently, these other RBCC-containing proteins also have domains such as the bromo domain, which are not found in PML. Alignment of RBCC-containing proteins (with either one or two B-boxes) from these diverse species with mouse PML demonstrates that these proteins, while having the RBCC, are otherwise not conserved with sequence identities over the full-length proteins (ca. 20%). The apparent absence of PML NBs from X. laevis is particularly striking since other nuclear organelles are conserved here, including nucleoli and Cajal bodies (32). However, an absolute answer with regard to the presence or absence of the PML gene in X. laevis, chicken or newt must await the complete sequencing of these genomes.
These findings suggest that PML is not an evolutionarily conserved protein. It has been reported that the A. thaliana protein COP1 is a homologue of PML since it also forms NBs and has a RING domain (74). Besides the RING, COP1 also contains WD40 repeats but no B-boxes or leucine coiled coil (97). Conversely, there are no WD40 repeats in PML (63). Thus, COP1 is most likely not a homologue of PML but simply another member of the RING family of proteins. The ability of both PML and COP1 to form NBs is insufficient proof for homology since so many members of the RING family form these types of structures (8). A more likely mammalian homologue of A. thaliana COP1 has been reported; this human protein, also called COP1, has the RING and WD40 repeats, and these domains acts similarly to arabidopsis COP1 in terms of determining the subcellular localization of the protein (97).
Together, these findings strongly suggest that PML is not evolutionarily conserved and therefore its function may be specific to higher eukaryotes. The lack of evolutionary conservation is not surprising in light of the observation that PML knockout mice are in essence morphologically normal. There may be functional homologues, with very different sequence and domain compositions, in other organisms and in mammalian cells that act similarly to PML. However, what these homologues may be is not clear at this time, and the identification of such homologues awaits a well-defined description of the discrete biochemical activities of the PML protein and PML NBs.
Many described NB components require PML for organization into NBs (see, for example, reference 42). However, some evolutionarily conserved proteins may form the structural basis of the body. For instance, eIF4E forms NBs in the absence of PML, e.g., in PML−/− cells and in a variety of organisms, some of which do not have PML, including in yeast, D. melanogaster, X. laevis, mice, and humans (19, 53, 58, 88). PML appears to require eIF4E for organization of at least a large subset of its bodies since conformational changes in eIF4E are correlated with the loss of PML NBs (19). Further, if one introduces exogenous PML to PML−/− cells, it relocalizes with endogenous eIF4E bodies (Fig. 3 in reference 19). These findings, taken together with the direct interaction between the PML RING and eIF4E, suggest that PML may be a regulator of an evolutionarily older organelle, which is comprised in part by eIF4E (19). However, colocalization is not complete between PML and eIF4E in all cell types, so there must be other anchoring components for PML, which may result in functionally distinct NBs. PRH is also found in bodies in the absence of PML (I. Topisirovic, and K. L. B. Borden, unpublished results). PRH is not as conserved as eIF4E but is found in X. laevis, as well as in mammals (68). Thus, there may be a proto- PML body comprised of evolutionarily conserved proteins, which PML associates with in organisms in which PML is expressed. Thus, there could be an evolutionary hierarchy to body organization. Only future studies can elucidate if this speculation has any merit.
TRYING TO RESOLVE THE APPARENT CONFLICTS: UNITY IN TIME AND PLACE?
There is a wealth of information regarding the physiological phenomena associated with PML function. However, unlike other cellular organelles such as mitochondria, there is no defined set of biochemical and molecular functions for PML NBs. In this review, I have attempted to present what is known in this regard in the hopes that dissecting the current literature would clarify its function. Given the complexity of this system, perhaps one should not try to define PML NBs as a single type of entity or functional unit. Some PML partner proteins are expressed in cell type- or tissue-specific manners; one example is PRH, which is limited to myeloid, lung, and liver cells or the telomerase proteins that localize with PML in a small set of specialized ALT cells. Further, proteins such as BLM only localize at specific points in the cell cycle. New findings from the Spector laboratory indicate that PML bodies can be subclassed based on their relative motion. Thus, there are spatial, temporal, and cell type-specific modifications to the body. Therefore, all NBs may not be equivalent. Importantly, there is a related nuclear organelle present even in the absence of the PML gene, showing that PML may not form the structural basis of the body. This organelle, comprised of the anchoring protein eIF4E and likely other evolutionarily conserved proteins, seems to form a sort of proto-PML body. Some components of the body do require PML for association into these structures, indicating that there is a hierarchical organization. One of the few discrete biochemical activities described for PML thus far relates to the mRNA-binding activity of eIF4E, and this activity is linked to its physiological effects in terms of growth arrest.
It is evident from the above discussions that there is no consensus opinion for PML NB function. Several models, supported by substantial data, suggest that PML is involved in particular processes, but none of these processes appear to be mechanistically linked. Two of these, the nuclear closet-catalytic surface theories, suggest a means by which PML could participate in these processes rather than a particular biochemical process. Clearly, it is premature to assign a single function to this complex set of organelles, and much more work in this field is required before a set of discrete biochemical functions can be conclusively attributed to these elusive NBs.
Acknowledgments
I am indebted to Gerd Maul and Paul Freemont for their thoughtful discussions of this review. I am grateful for the expertise in bioinformatics from Luce Skrabanek at the Institute for Computational Biomedicine, Mt. Sinai School of Medicine. I am grateful for critical discussions of the manuscript from Alex Kentsis, Jackie Perez, Madhulika Sharma, Stephen Strudwick, and Ivan Topisirovic.
I am a scholar of the Leukemia and Lymphoma Society. Financial support was provided by the National Institutes of Health (CA80728 and CA88991).
REFERENCES
- 1.Abid, M. R., Y. Li, C. Anthony, and A. De Benedetti. 1999. Translational regulation of ribonucleotide reductase by eukaryotic initiation factor 4E links protein synthesis to the control of DNA replication. J. Biol. Chem. 274:35991-35998. [DOI] [PubMed] [Google Scholar]
- 2.Alcalay, M., L. Tomassoni, E. Colombo, S. Stoldt, F. Grignani, M. Fagioli, L. Szekely, K. Helin, and P. G. Pelicci. 1998. The promyelocytic leukemia gene product (PML) forms stable complexes with the retinoblastoma protein. Mol. Cell. Biol. 18:1084-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ascoli, C. A., and G. G. Maul. 1991. Identification of a novel nuclear domain. J. Cell Biol. 112:785-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bell, P., L. J. Montaner, and G. G. Maul. 2001. Accumulation and intranuclear distribution of unintegrated human immunodeficiency virus type 1 DNA. J. Virol. 75:7683-7691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benoit, G. R., J. H. Tong, Z. Balajthy, and M. Lanotte. 2001. Exploring (novel) gene expression during retinoid-induced maturation and cell death of acute promyelocytic leukemia. Semin. Hematol. 38:71-85. [DOI] [PubMed] [Google Scholar]
- 6.Boddy, M. N., K. Howe, L. D. Etkin, E. Solomon, and P. S. Freemont. 1996. PIC 1, a novel ubiquitin-like protein which interacts with the PML component of a multiprotein complex that is disrupted in acute promyelocytic leukaemia. Oncogene 13:971-982. [PubMed] [Google Scholar]
- 7.Boisvert, F. M., M. J. Hendzel, and D. P. Bazett-Jones. 2000. Promyelocytic leukemia (PML) nuclear bodies are protein structures that do not accumulate RNA. J. Cell Biol. 148:283-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borden, K. L. B. 2000. RING domains: master builders of molecular scaffolds? J. Mol. Biol. 295:1103-1112. [DOI] [PubMed] [Google Scholar]
- 9.Borden, K. L. B., M. N. Boddy, J. Lally, N. J. O'Reilly, S. Martin, K. Howe, E. Solomon, and P. S. Freemont. 1995. The solution structure of the RING finger domain from the acute promyelocytic leukaemia proto-oncoprotein PML. EMBO J. 14:1532-1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Borden, K. L. B., E. J. Campbell Dwyer, G. W. Carlile, M. Djavani, and M. S. Salvato. 1998. Two RING finger proteins, the oncoprotein PML and the arenavirus Z protein, colocalize with the nuclear fraction of the ribosomal P proteins. J. Virol. 72:3819-3826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Borden, K. L. B., E. J. Campbell Dwyer, and M. S. Salvato. 1998. An arenavirus RING (zinc-binding) protein binds the oncoprotein promyelocyte leukemia protein (PML) and relocates PML nuclear bodies to the cytoplasm. J. Virol. 72:758-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Borden, K. L. B., E. J. Campbell Dwyer, and M. S. Salvato. 1997. The promyelocytic leukemia protein PML has a pro-apoptotic activity mediated through its RING domain. FEBS Lett. 418:30-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Borden, K. L. B., J. M. Lally, S. R. Martin, N. J. O'Reilly, E. Solomon, and P. S. Freemont. 1996. In vivo and in vitro characterization of the B1 and B2 zinc-binding domains from the acute promyelocytic leukemia protooncoprotein PML. Proc. Natl. Acad. Sci. USA 93:1601-1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Campbell Dwyer, E. J., H. Lai, R. C. MacDonald, M. S. Salvato, and K. L. B. Borden. 2000. The lymphocytic choriomeningitis virus RING protein Z associates with eukaryotic initiation factor 4E and selectively represses translation in a RING-dependent manner. J. Virol. 74:3293-3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carlile, G. W., W. G. Tatton, and K. L. B. Borden. 1998. Demonstration of a RNA-dependent nuclear interaction between the promyelocytic leukaemia protein and glyceraldehyde-3-phosphate dehydrogenase. Biochem. J. 335:691-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chai, Y., S. L. Koppenhafer, S. J. Shoesmith, M. K. Perez, and H. L. Paulson. 1999. Evidence for proteasome involvement in polyglutamine disease: localization to nuclear inclusions in SCA3/MJD and suppression of polyglutamine aggregation in vitro. Hum. Mol. Genet. 8:673-682. [DOI] [PubMed] [Google Scholar]
- 17.Chan, J. Y., L. Li, Y. H. Fan, Z. M. Mu, W. W. Zhang, and K. S. Chang. 1997. Cell-cycle regulation of DNA damage-induced expression of the suppressor gene PML. Biochem. Biophys. Res. Commun. 240:640-646. [DOI] [PubMed] [Google Scholar]
- 18.Chelbi-Alix, M. K., L. Pelicano, F. Quignon, M. H. Koken, L. Venturini, M. Stadler, J. Pavlovic, L. Degos, and H. de The. 1995. Induction of the PML protein by interferons in normal and APL cells. Leukemia 9:2027-2033. [PubMed] [Google Scholar]
- 19.Cohen, N., M. Sharma, A. Kentsis, J. M. Perez, S. Strudwick, and K. L. B. Borden. 2001. PML RING suppresses oncogenic transformation by reducing the affinity of eIF4E for mRNA. EMBO J. 20:4547-4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Desbois, C., R. Rousset, F. Bantignies, and P. Jalinot. 1996. Exclusion of Int-6 from PML nuclear bodies by binding to the HTLV-I Tax oncoprotein. Science 273:951-953. [DOI] [PubMed] [Google Scholar]
- 21.Djavani, M., J. Rodas, I. S. Lukashevich, D. Horejsh, P. P. Pandolfi, K. L. Borden, and M. S. Salvato. 2001. Role of the promyelocytic leukemia protein PML in the interferon sensitivity of lymphocytic choriomeningitis virus. J. Virol. 75:6204-6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doucas, V., M. Tini, D. A. Egan, and R. M. Evans. 1999. Modulation of CREB binding protein function by the promyelocytic (PML) oncoprotein suggests a role for nuclear bodies in hormone signaling. Proc. Natl. Acad. Sci. USA 96:2627-2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duprez, E., A. J. Saurin, J. M. Desterro, V. Lallemand-Breitenbach, K. Howe, M. N. Boddy, E. Solomon, H. de The, R. T. Hay, and P. S. Freemont. 1999. SUMO-1 modification of the acute promyelocytic leukaemia protein PML: implications for nuclear localization. J. Cell Sci. 112:381-393. [DOI] [PubMed] [Google Scholar]
- 24.Dyck, J. A., G. G. Maul, W. H. Miller, Jr., J. D. Chen, A. Kakizuka, and R. M. Evans. 1994. A novel macromolecular structure is a target of the promyelocyte-retinoic acid receptor oncoprotein. Cell 76:333-343. [DOI] [PubMed] [Google Scholar]
- 25.Everett, R. D. 2001. DNA viruses and viral proteins that interact with PML nuclear bodies. Oncogene 20:7266-7273. [DOI] [PubMed] [Google Scholar]
- 26.Fagioli, M., M. Alcalay, P. P. Pandolfi, L. Venturini, A. Mencarelli, A. Simeone, D. Acampora, F. Grignani, and P. G. Pelicci. 1992. Alternative splicing of PML transcripts predicts coexpression of several carboxy-terminally different protein isoforms. Oncogene 7:1083-1091. [PubMed] [Google Scholar]
- 27.Ferbeyre, G., E. de Stanchina, E. Querido, N. Baptiste, C. Prives, and S. W. Lowe. 2000. PML is induced by oncogenic ras and promotes premature senescence. Genes Dev. 14:2015-2027. [PMC free article] [PubMed] [Google Scholar]
- 28.Ferrucci, P. F., F. Grignani, M. Pearson, M. Fagioli, I. Nicoletti, and P. G. Pelicci. 1997. Cell death induction by the acute promyelocytic leukemia-specific PML/RARalpha fusion protein. Proc. Natl. Acad. Sci. USA 94:10901-10906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Flenghi, L., M. Fagioli, L. Tomassoni, S. Pileri, M. Gambacorta, R. Pacini, F. Grignani, T. Casini, P. F. Ferrucci, M. F. Martelli, et al. 1995. Characterization of a new monoclonal antibody (PG-M3) directed against the aminoterminal portion of the PML gene product: immunocytochemical evidence for high expression of PML proteins on activated macrophages, endothelial cells, and epithelia. Blood 85:1871-1880. [PubMed] [Google Scholar]
- 30.Ford, L. P., Y. Zou, K. Pongracz, S. M. Gryaznov, J. W. Shay, and W. E. Wright. 2001. Telomerase can inhibit the recombination-based pathway of telomere maintenance in human cells. J. Biol. Chem. 276:32198-32203. [DOI] [PubMed] [Google Scholar]
- 31.Fu, X. D., and T. Maniatis. 1992. The 35-kDa mammalian splicing factor SC35 mediates specific interactions between U1 and U2 small nuclear ribonucleoprotein particles at the 3′ splice site. Proc. Natl. Acad. Sci. USA 89:1725-1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gall, J. G. 2000. Cajal bodies: the first 100 years. Annu. Rev. Cell Dev. Biol. 16:273-300. [DOI] [PubMed] [Google Scholar]
- 33.Gharibyan, V., and H. Youssoufian. 1999. Localization of the Bloom syndrome helicase to punctate nuclear structures and the nuclear matrix and regulation during the cell cycle: comparison with the Werner's syndrome helicase. Mol. Carcinog. 26:261-273. [DOI] [PubMed] [Google Scholar]
- 34.Gongora, C., G. David, L. Pintard, C. Tissot, T. D. Hua, A. Dejean, and N. Mechti. 1997. Molecular cloning of a new interferon-induced PML nuclear body-associated protein. J. Biol. Chem. 272:19457-19463. [DOI] [PubMed] [Google Scholar]
- 35.Grande, M. A., I. van der Kraan, B. van Steensel, W. Schul, H. de The, H. T. van der Voort, L. de Jong, and R. van Driel. 1996. PML-containing nuclear bodies: their spatial distribution in relation to other nuclear components. J. Cell. Biochem. 63:280-291. [DOI] [PubMed] [Google Scholar]
- 36.Grimwade, D., and E. Solomon. 1997. Characterization of the PML/RARα rearrangement associated with t(15;17) acute promyelocytic leukaemia. Curr. Top. Microbiol. Immunol. 220:81-112. [DOI] [PubMed] [Google Scholar]
- 37.Grobelny, J. V., A. K. Godwin, and D. Broccoli. 2000. ALT-associated PML bodies are present in viable cells and are enriched in cells in the G2/M phase of the cell cycle J. Cell Sci. 113(Pt. 24):4577-4585. [DOI] [PubMed] [Google Scholar]
- 38.Guiochon-Mantel, A., J. F. Savouret, F. Quignon, K. Delabre, E. Milgrom, and H. De The. 1995. Effect of PML and PML-RAR on the transactivation properties and subcellular distribution of steroid hormone receptors. Mol. Endocrinol. 9:1791-1803. [DOI] [PubMed] [Google Scholar]
- 39.Hachiya, Y., K. Motonaga, M. Itoh, T. Masuko, T. Enomoto, H. Sonobe, and S. Takashima. 2001. Immunohistochemical expression and pathogenesis of BLM in the human brain and visceral organs. Neuropathology 21:123-128. [DOI] [PubMed] [Google Scholar]
- 40.Hodges, M., C. Tissot, K. Howe, D. Grimwade, and P. S. Freemont. 1998. Structure, organization, and dynamics of promyelocytic leukemia protein nuclear bodies. Am. J. Hum. Genet. 63:297-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Iborra, F. J., D. A. Jackson, and P. R. Cook. 2001. Coupled transcription and translation within nuclei of mammalian cells. Science 293:1139-1142. [DOI] [PubMed] [Google Scholar]
- 42.Ishov, A. M., A. G. Sotnikov, D. Negorev, O. V. Vladimirova, N. Neff, T. Kamitani, E. T. Yeh, J. F. Strauss III, and G. G. Maul. 1999. PML is critical for ND10 formation and recruits the PML-interacting protein daxx to this nuclear structure when modified by SUMO-1. J. Cell Biol. 147:221-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jensen, K., C. Shiels, and P. S. Freemont. 2001. PML protein isoforms and the RBCC/TRIM motif. Oncogene 20:7223-7233. [DOI] [PubMed] [Google Scholar]
- 44.Jepsen, K., O. Hermanson, T. M. Onami, A. S. Gleiberman, V. Lunyak, R. J. McEvilly, R. Kurokawa, V. Kumar, F. Liu, E. Seto, S. M. Hedrick, G. Mandel, C. K. Glass, D. W. Rose, and M. G. Rosenfeld. 2000. Combinatorial roles of the nuclear receptor corepressor in transcription and development. Cell 102:753-763. [DOI] [PubMed] [Google Scholar]
- 45.Kastner, P., A. Perez, Y. Lutz, C. Rochette-Egly, M. P. Gaub, B. Durand, M. Lanotte, R. Berger, and P. Chambon. 1992. Structure, localization and transcriptional properties of two classes of retinoic acid receptor alpha fusion proteins in acute promyelocytic leukemia (APL): structural similarities with a new family of oncoproteins. EMBO J. 11:629-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kentsis, A., and K. L. B. Borden. 2000. Construction of macromolecular assemblages in eukaryotic processes and their role in human disease: linking RINGs together. Curr. Peptide Protein Sci. 1:49-74. [DOI] [PubMed] [Google Scholar]
- 47.Kentsis, A., E. C. Dwyer, J. M. Perez, M. Sharma, A. Chen, Z. Q. Pan, and K. L. B. Borden. 2001. The RING domains of the promyelocytic leukemia protein PML and the arenaviral protein Z repress translation by directly inhibiting translation initiation factor eIF4E. J. Mol. Biol. 312:609-623. [DOI] [PubMed] [Google Scholar]
- 48.Kentsis, A., R. E. Gordon, and K. L. B. Borden. 2002. Self-assembly properties of a model RING domain. Proc. Natl. Acad. Sci. USA 99:667-672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Khan, M. M., T. Nomura, H. Kim, S. C. Kaul, R. Wadhwa, T. Shinagawa, E. Ichikawa-Iwata, S. Zhong, P. P. Pandolfi, and S. Ishii. 2001. Role of PML and PML-RARα in Mad-mediated transcriptional repression. Mol. Cell 7:1233-1243. [DOI] [PubMed] [Google Scholar]
- 50.Lai, H. K., and K. L. Borden. 2000. The promyelocytic leukemia (PML) protein suppresses cyclin D1 protein production by altering the nuclear cytoplasmic distribution of cyclin D1 mRNA. Oncogene 19:1623-1634. [DOI] [PubMed] [Google Scholar]
- 51.Lallemand-Breitenbach, V., J. Zhu, F. Puvion, M. Koken, N. Honore, A. Doubeikovsky, E. Duprez, P. P. Pandolfi, E. Puvion, P. Freemont, and H. de The. 2001. Role of promyelocytic leukemia (PML) sumolation in nuclear body formation, 11S proteasome recruitment, and As2O3-induced PML or PML/retinoic acid receptor alpha degradation. J. Exp. Med. 193:1361-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.LaMorte, V. J., J. A. Dyck, R. L. Ochs, and R. M. Evans. 1998. Localization of nascent RNA and CREB binding protein with the PML-containing nuclear body. Proc. Natl. Acad. Sci. USA 95:4991-4996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lang, V., N. I. Zanchin, H. Lunsdorf, M. Tuite, and J. E. McCarthy. 1994. Initiation factor eIF-4E of Saccharomyces cerevisiae. Distribution within the cell, binding to mRNA, and consequences of its overproduction. J. Biol. Chem. 269:6117-6123. [PubMed] [Google Scholar]
- 54.Lavau, C., A. Marchio, M. Fagioli, J. Jansen, B. Falini, P. Lebon, F. Grosveld, P. P. Pandolfi, P. G. Pelicci, and A. Dejean. 1995. The acute promyelocytic leukaemia-associated PML gene is induced by interferon. Oncogene 11:871-876. [PubMed] [Google Scholar]
- 55.Lazaris-Karatzas, A., M. R. Smith, R. M. Frederickson, M. L. Jaramillo, Y. L. Liu, H. F. Kung, and N. Sonenberg. 1992. Ras mediates translation initiation factor 4E-induced malignant transformation. Genes Dev. 6:1631-1642. [DOI] [PubMed] [Google Scholar]
- 56.Le, X. F., S. Vallian, Z. M. Mu, M. C. Hung, and K. S. Chang. 1998. Recombinant PML adenovirus suppresses growth and tumorigenicity of human breast cancer cells by inducing G1 cell cycle arrest and apoptosis. Oncogene 16:1839-1849. [DOI] [PubMed] [Google Scholar]
- 57.Le, X. F., P. Yang, and K. S. Chang. 1996. Analysis of the growth and transformation suppressor domains of promyelocytic leukemia gene, PML. J. Biol. Chem. 271:130-135. [DOI] [PubMed] [Google Scholar]
- 58.Lejbkowicz, F., C. Goyer, A. Darveau, S. Neron, R. Lemieux, and N. Sonenberg. 1992. A fraction of the mRNA 5′ cap-binding protein, eukaryotic initiation factor 4E, localizes to the nucleus. Proc. Natl. Acad. Sci. USA 89:9612-9616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Maul, G. G., D. Negorev, P. Bell, and A. M. Ishov. 2000. Review: properties and assembly mechanisms of ND10, PML bodies, or PODs. J. Struct. Biol. 129:278-287. [DOI] [PubMed] [Google Scholar]
- 60.Maul, G. G., E. Yu, A. M. Ishov, and A. L. Epstein. 1995. Nuclear domain 10 (ND10) associated proteins are also present in nuclear bodies and redistribute to hundreds of nuclear sites after stress. J. Cell Biochem. 59:498-513. [DOI] [PubMed] [Google Scholar]
- 61.McMahon, C., T. Suthiphongchai, J. DiRenzo, and M. E. Ewen. 1999. P/CAF associates with cyclin D1 and potentiates its activation of the estrogen receptor. Proc. Natl. Acad. Sci. USA 96:5382-5387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Melchior, F. 2000. SUMO—nonclassical ubiquitin. Annu. Rev. Cell Dev. Biol. 16:591-626. [DOI] [PubMed] [Google Scholar]
- 63.Melnick, A., and J. D. Licht. 1999. Deconstructing a disease: RARα, its fusion partners, and their roles in the pathogenesis of acute promyelocytic leukemia. Blood 93:3167-3215. [PubMed] [Google Scholar]
- 64.Miller, S. J., T. Suthiphongchai, G. P. Zambetti, and M. E. Ewen. 2000. p53 binds selectively to the 5′-untranslated region of cdk4, an RNA element necessary and sufficient for transforming growth factor β- and p53-mediated translational inhibition of cdk4. Mol. Cell. Biol. 20:8420-8431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Muratani, M., D. Gerlich, S. M. Janicki, M. Gebhard, R. Eils, and D. L. Spector. 2001. Metabolic-energy-dependent movement of PML bodies within the mammalian cell nucleus. Nat. Cell Biol. 21:21. [DOI] [PubMed] [Google Scholar]
- 66.Nagy, E., and W. F. Rigby. 1995. Glyceraldehyde-3-phosphate dehydrogenase selectively binds AU-rich RNA in the NAD(+)-binding region (Rossmann fold). J. Biol. Chem. 270:2755-2763. [DOI] [PubMed] [Google Scholar]
- 67.Nayler, O., W. Stratling, J. P. Bourquin, I. Stagljar, L. Lindemann, H. Jasper, A. M. Hartmann, F. O. Fackelmayer, A. Ullrich, and S. Stamm. 1998. SAF-B protein couples transcription and pre-mRNA splicing to SAR/MAR elements. Nucleic Acids Res. 26:3542-3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Newman, C. S., F. Chia, and P. A. Krieg. 1997. The XHex homeobox gene is expressed during development of the vascular endothelium: overexpression leads to an increase in vascular endothelial cell number. Mech. Dev. 66:83-93. [DOI] [PubMed] [Google Scholar]
- 69.Nguyen, L. H., L. Espert, N. Mechti, and D. M. Wilson III. 2001. The human interferon- and estrogen-regulated ISG20/HEM45 gene product degrades single-stranded RNA and DNA in vitro. Biochemistry 40:7174-7179. [DOI] [PubMed] [Google Scholar]
- 70.Pearson, M., R. Carbone, C. Sebastiani, M. Cioce, M. Fagioli, S. Saito, Y. Higashimoto, E. Appella, S. Minucci, P. P. Pandolfi, and P. G. Pelicci. 2000. PML regulates p53 acetylation and premature senescence induced by oncogenic Ras. Nature 406:207-210. [DOI] [PubMed] [Google Scholar]
- 71.Quignon, F., F. De Bels, M. Koken, J. Feunteun, J. C. Ameisen, and H. de The. 1998. PML induces a novel caspase-independent death process. Nat. Genet. 20:259-265. [DOI] [PubMed] [Google Scholar]
- 72.Regad, T., and M. K. Chelbi-Alix. 2001. Role and fate of PML nuclear bodies in response to interferon and viral infections. Oncogene 20:7274-7286. [DOI] [PubMed] [Google Scholar]
- 73.Regad, T., A. Saib, V. Lallemand-Breitenbach, P. P. Pandolfi, H. de The, and M. K. Chelbi-Alix. 2001. PML mediates the interferon-induced antiviral state against a complex retrovirus via its association with the viral transactivator. EMBO J. 20:3495-3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Reyes, J. C. 2001. PML and COP1—two proteins with much in common. Trends Biochem. Sci. 26:18-20. [DOI] [PubMed] [Google Scholar]
- 75.Rogaia, D., F. Grignani, I. Nicoletti, and P. G. Pelicci. 1995. The acute promyelocytic leukemia-specific PML/RARα fusion protein reduces the frequency of commitment to apoptosis upon growth factor deprivation of GM-CSF-dependent myeloid cells. Leukemia 9:1467-1472. [PubMed] [Google Scholar]
- 76.Rosenwald, I. B., R. Kaspar, D. Rousseau, L. Gehrke, P. Leboulch, J. J. Chen, E. V. Schmidt, N. Sonenberg, and I. M. London. 1995. Eukaryotic translation initiation factor 4E regulates expression of cyclin D1 at transcriptional and post-transcriptional levels. J. Biol. Chem. 270:21176-21180. [DOI] [PubMed] [Google Scholar]
- 77.Rousseau, D., R. Kaspar, I. Rosenwald, L. Gehrke, and N. Sonenberg. 1996. Translation initiation of ornithine decarboxylase and nucleocytoplasmic transport of cyclin D1 mRNA are increased in cells overexpressing eukaryotic initiation factor 4E. Proc. Natl. Acad. Sci. USA 93:1065-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Roussel, M. J., and M. Lanotte. 2001. Maturation sensitive and resistant t(15;17) NB4 cell lines as tools for APL physiopathology: nomenclature of cells and repertory of their known genetic alterations and phenotypes. Oncogene 20:7287-7291. [DOI] [PubMed] [Google Scholar]
- 79.Rubin, D. M., O. Coux, I. Wefes, C. Hengartner, R. A. Young, A. L. Goldberg, and D. Finley. 1996. Identification of the Gal4 suppressor Sug1 as a subunit of the yeast 26S proteasome. Nature 379:655-657. [DOI] [PubMed] [Google Scholar]
- 80.Sahlas, D. J., K. Milankov, P. C. Park, and U. De Boni. 1993. Distribution of snRNPs, splicing factor SC-35 and actin in interphase nuclei: immunocytochemical evidence for differential distribution during changes in functional states. J. Cell Sci. 105:347-357. [DOI] [PubMed] [Google Scholar]
- 81.Schul, W., I. van Der Kraan, A. G. Matera, R. van Driel, and L. de Jong. 1999. Nuclear domains enriched in RNA 3′-processing factors associate with coiled bodies and histone genes in a cell cycle-dependent manner. Mol. Biol. Cell 10:3815-3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Seeler, J. S., and A. Dejean. 1999. The PML nuclear bodies: actors or extras? Curr. Opin. Genet. Dev. 9:362-367. [DOI] [PubMed] [Google Scholar]
- 83.Shiels, C., S. A. Islam, R. Vatcheva, P. Sasieni, M. J. Sternberg, P. S. Freemont, and D. Sheer. 2001. PML bodies associate specifically with the MHC gene cluster in interphase nuclei. J. Cell Sci. 114:3705-3716. [DOI] [PubMed] [Google Scholar]
- 84.Singh, R., and M. R. Green. 1993. Sequence-specific binding of transfer RNA by glyceraldehyde-3-phosphate dehydrogenase. Science 259:365-368. [DOI] [PubMed] [Google Scholar]
- 85.Skinner, P. J., B. T. Koshy, C. J. Cummings, I. A. Klement, K. Helin, A. Servadio, H. Y. Zoghbi, and H. T. Orr. 1997. Ataxin-1 with an expanded glutamine tract alters nuclear matrix-associated structures. Nature 389:971-974. [DOI] [PubMed] [Google Scholar]
- 86.Sonenberg, N., and A. C. Gingras. 1998. The mRNA 5′ cap-binding protein eIF4E and control of cell growth. Curr. Opin. Cell Biol. 10:268-275. [DOI] [PubMed] [Google Scholar]
- 87.Stadler, M., M. K. Chelbi-Alix, M. H. Koken, L. Venturini, C. Lee, A. Saib, F. Quignon, L. Pelicano, M. C. Guillemin, C. Schindler, et al. 1995. Transcriptional induction of the PML growth suppressor gene by interferons is mediated through an ISRE and a GAS element. Oncogene 11:2565-2573. [PubMed] [Google Scholar]
- 88.Strudwick, S., and K. L. B. Borden. 2002. The emerging roles for nuclear translation factors. Differentiation 70:10-22. [DOI] [PubMed] [Google Scholar]
- 89.Stuurman, N., A. de Graaf, A. Floore, A. Josso, B. Humbel, L. de Jong, and R. van Driel. 1992. A monoclonal antibody recognizing nuclear matrix-associated nuclear bodies. J. Cell Sci. 101:773-784. [DOI] [PubMed] [Google Scholar]
- 90.Terris, B., V. Baldin, S. Dubois, C. Degott, J. F. Flejou, D. Henin, and A. Dejean. 1995. PML nuclear bodies are general targets for inflammation and cell proliferation. Cancer Res. 55:1590-1597. [PubMed] [Google Scholar]
- 91.Topcu, Z., D. L. Mack, R. A. Hromas, and K. L. Borden. 1999. The promyelocytic leukemia protein PML interacts with the proline-rich homeodomain protein PRH: a RING may link hematopoiesis and growth control. Oncogene 18:7091-7100. [DOI] [PubMed] [Google Scholar]
- 91a.Topisirovic, I., A. D. Capili, and K. L. B. Borden. Gamma interferon and cadmium treatments modulate eukaryotic initiation factor 4E mRNA transport of cyclin D1 in a PML-dependent manner. Mol. Cell. Biol., in press. [DOI] [PMC free article] [PubMed]
- 92.Tsukamoto, T., N. Hashiguchi, S. M. Janicki, T. Tumbar, A. S. Belmont, and D. L. Spector. 2000. Visualization of gene activity in living cells. Nat. Cell Biol. 2:871-878. [DOI] [PubMed] [Google Scholar]
- 93.Turelli, P., V. Doucas, E. Craig, B. Mangeat, N. Klages, R. Evans, G. Kalpana, and D. Trono. 2001. Cytoplasmic recruitment of INI1 and PML on incoming HIV preintegration complexes: interference with early steps of viral replication. Mol. Cell 7:1245-1254. [DOI] [PubMed] [Google Scholar]
- 94.Turley, H., L. Wu, M. Canamero, K. C. Gatter, and I. D. Hickson. 2001. The distribution and expression of the Bloom's syndrome gene product in normal and neoplastic human cells. Br. J. Cancer 85:261-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Vallian, S., J. A. Gaken, I. D. Trayner, E. B. Gingold, T. Kouzarides, K. S. Chang, and F. Farzaneh. 1997. Transcriptional repression by the promyelocytic leukemia protein, PML. Exp. Cell Res. 237:371-382. [DOI] [PubMed] [Google Scholar]
- 96.Vareli, K., M. Frangou-Lazaridis, I. van der Kraan, O. Tsolas, and R. van Driel. 2000. Nuclear distribution of prothymosin alpha and parathymosin: evidence that prothymosin alpha is associated with RNA synthesis processing and parathymosin with early DNA replication. Exp. Cell Res. 257:152-161. [DOI] [PubMed] [Google Scholar]
- 97.Wang, H., D. Kang, X. W. Deng, and N. Wei. 1999. Evidence for functional conservation of a mammalian homologue of the light-responsive plant protein COP1. Curr. Biol. 9:711-714. [DOI] [PubMed] [Google Scholar]
- 98.Wang, X. W., A. Tseng, N. A. Ellis, E. A. Spillare, S. P. Linke, A. I. Robles, H. Seker, Q. Yang, P. Hu, S. Beresten, N. A. Bemmels, S. Garfield, and C. C. Harris. 2001. Functional interaction of p53 and BLM DNA helicase in apoptosis. J. Biol. Chem. 276:32948-32955. [DOI] [PubMed] [Google Scholar]
- 99.Wang, Z. G., L. Delva, M. Gaboli, R. Rivi, M. Giorgio, C. Cordon-Cardo, F. Grosveld, and P. P. Pandolfi. 1998. Role of PML in cell growth and the retinoic acid pathway. Science 279:1547-1551. [DOI] [PubMed] [Google Scholar]
- 100.Wang, Z. G., D. Ruggero, S. Ronchetti, S. Zhong, M. Gaboli, R. Rivi, and P. P. Pandolfi. 1998. PML is essential for multiple apoptotic pathways. Nat. Genet. 20:266-272. [DOI] [PubMed] [Google Scholar]
- 101.Weis, K., S. Rambaud, C. Lavau, J. Jansen, T. Carvalho, M. Carmo-Fonseca, A. Lamond, and A. Dejean. 1994. Retinoic acid regulates aberrant nuclear localization of PML-RARα in acute promyelocytic leukemia cells. Cell 76:345-356. [DOI] [PubMed] [Google Scholar]
- 102.Wilkinson, M. F., and A. B. Shyu. 2001. Multifunctional regulatory proteins that control gene expression in both the nucleus and the cytoplasm. Bioessays 23:775-787. [DOI] [PubMed] [Google Scholar]
- 103.Yankiwski, V., R. A. Marciniak, L. Guarente, and N. F. Neff. 2000. Nuclear structure in normal and Bloom syndrome cells. Proc. Natl. Acad. Sci. USA 97:5214-5219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yankiwski, V., J. P. Noonan, and N. F. Neff. 2001. The C-terminal domain of the Bloom syndrome DNA helicase is essential for genomic stability. BMC Cell Biol. 2:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yasuda, S., K. Inoue, M. Hirabayashi, H. Higashiyama, Y. Yamamoto, H. Fuyuhiro, O. Komure, F. Tanaka, G. Sobue, K. Tsuchiya, K. Hamada, H. Sasaki, K. Takeda, H. Ichijo, and A. Kakizuka. 1999. Triggering of neuronal cell death by accumulation of activated SEK1 on nuclear polyglutamine aggregations in PML bodies. Genes Cells 4:743-756. [DOI] [PubMed] [Google Scholar]
- 106.Yeager, T. R., A. A. Neumann, A. Englezou, L. I. Huschtscha, J. R. Noble, and R. R. Reddel. 1999. Telomerase-negative immortalized human cells contain a novel type of promyelocytic leukemia (PML) body. Cancer Res. 59:4175-4179. [PubMed] [Google Scholar]