Abstract
Preadipocyte factor 1 (Pref-1/Dlk1) inhibits in vitro adipocyte differentiation and has been recently reported to be a paternally expressed imprinted gene at human chromosome 14q32. Studies on human chromosome 14 deletions and maternal uniparental disomy (mUPD) 14 suggest that misexpression of a yet-to-be-identified imprinted gene or genes present on chromosome 14 causes congenital disorders. We generated Pref-1 knockout mice to assess the role of Pref-1 in growth and in vivo adipogenesis and to determine the contribution of Pref-1 in mUPD. Pref-1-null mice display growth retardation, obesity, blepharophimosis, skeletal malformation, and increased serum lipid metabolites. Furthermore, the phenotypes observed in Pref-1-null mice are present in heterozygotes that harbor a paternally inherited, but not in those with a maternally inherited pref-1-null allele. Our results demonstrate that Pref-1 is indeed paternally expressed and is important for normal development and for homeostasis of adipose tissue mass. We also suggest that Pref-1 is responsible for most of the symptoms observed in mouse mUPD12 and human mUPD14. Pref-1-null mice may be a model for obesity and other pathologies of human mUPD14.
Preadipocyte factor 1 (Pref-1/Dlk1) is a transmembrane protein containing epidermal growth factor (EGF) repeats homologous to the Notch/Delta/Serrate family (14, 22). Pref-1 is cleaved at the extracellular domain to generate a soluble form of Pref-1 (27). Pref-1 is expressed in multiple embryonic tissues, and its expression is abolished in most tissues after birth, with the exception of preadipocytes, pancreatic β cells, thymocytes, and cells in the adrenal gland (2, 7, 11, 18). Pref-1, therefore, has been used as a preadipocyte marker by various laboratories (21, 33). Constitutive Pref-1 expression or the addition of a soluble form of Pref-1 inhibits 3T3-L1 adipocyte differentiation, and downregulation of Pref-1 by antisense expression enhances adipogenesis, indicating that Pref-1 may function in the maintenance of the preadipose state (22-27). Pref-1 expression is also abolished during the adipose conversion of primary rat preadipocytes in cultures, and their differentiation is inhibited by the addition of soluble Pref-1 (7).
Genomic imprinting, via a DNA methylation mechanism, leads to the differential expression of imprinted genes from maternally or paternally derived chromosomes. Perturbance of this epigenetic control of gene regulation, when both alleles are inherited from one parent (uniparental disomy [UPD]), causes the congenital disorders maternal UPD7, paternal UPD11, and UPD15, described as Russell-Silver syndrome, Beckwith-Wiedemann syndrome, and Prader-Willi/Angelman syndrome, respectively (13, 19). Recently, Pref-1 has been reported to be expressed from the paternal allele, but not from the maternal allele, due to differential methylation (20, 30, 31). A cluster of imprinted genes, including pref-1, dat, gtl2, peg11, antipeg11, and meg8, is present at the syntenic region of mouse chromosome 12, human chromosome 14, and sheep chromosome 18 (3-6, 12, 30, 31). Studies of mouse mUPD12 and human mUPD14 have suggested that imprinted genes present in this chromosomal region are important for embryo survival and growth, with affected individuals having deformities that include scoliosis, hypotonicity, early puberty, obesity, and blepharophimosis (1, 5, 6, 16, 29, 32). However, the imprinted gene or genes responsible and their relevant phenotypes have not been determined.
To further define the involvement of Pref-1 in adipogenesis and to discern the role of Pref-1 in human mUPD14 phenotypes, we generated Pref-1-knockout mice by targeted deletion. Our studies demonstrate that loss of Pref-1 interferes with proper embryonic development, postnatal growth, and homeostasis of fat deposition. Lack of Pref-1 leads to accelerated adiposity similar to that reported in human mUPD14 patients who manifest growth defects and obesity. Our present studies therefore identify pref-1 as the gene responsible for most of the imprinting-related phenotypes of mouse mUPD12 and syntenic human mUPD14.
MATERIALS AND METHODS
Generation of Pref-1-null mice.
A 12-kb XbaI-XbaI fragment mouse genomic clone was isolated by screening a 129/SvJ library with a full-length mouse pref-1 cDNA used as a probe. The genomic pref-1 allele was disrupted by a neomycin resistance cassette (NEO). The targeting construct contained the herpes simplex virus thymidine kinase gene and a pPNT-NEO fragment, which replaced a 1.5-kb BsmI-EcoRI genomic fragment of the pref-1 gene corresponding to regions of exon 2 through exon 3. The targeting construct was linearized with BamHI and introduced into SvJ129 embryonic stem (ES) cells by electroporation. G418- and ganciclovir-resistant clones were screened for homologous recombination by Southern blotting, and 5 out of 272 clones were positive for the mutated gene. The positive clones were microinjected into 3.5-day C57BL/6J or BALB/c blastocysts to generate chimeric animals. Subsequent genotyping of animals was performed either by Southern blotting as described below or by PCR with genomic DNA isolated from mice tails. PCR primers for the wild-type allele were the 5′ primer 5′-GCACTCAACAAAGACCACCAAATG-3′ and the 3′ primer 5′-ATTGACACAGCCAGGGGCAGTTAC-3′, and the primers for the targeted allele were the 5′ primer 5′-GCAGCTGTGCTCGACGTTGTC-3′ and the 3′ primer 5′-CGCCAAGCTCTTCAGCAATATCAC-3′. After an initial denaturation at 94°C for 1 min, 35 cycles of denaturation at 94°C for 1 min, annealing at 57°C for 1 min, and extension at 72°C for 1 min were carried out. Male chimeric mice were generated from four independent Pref-1-null ES cell lines by standard procedures (9). Males derived from each cell line were mated with C57BL/6J or BALB/c females, and germ line transmission of the Pref-1 mutation was obtained from each line. To exclude the possibility that the observed Pref-1-null phenotype resulted from a mixed genetic background (SVJ129 × C57BL/6J), mice from germ line transmission of the targeted allele were backcrossed onto the C57BL/6J background for 4 to 9 generations and sib-crossed to obtain homozygous null mice. We used age-matched wild-type C57BL/6J mice as control mice due to the mixed genetic background (SVJ129 × C57BL/6J) of the wild-type littermates, which were generated from Pref-1 heterozygote interbreeding. Mice backcrossed onto a BALB/c background were also generated. With the exception of the eyelid studies, experiments were conducted with Pref-1-null animals on the C57BL/6J background. The experiments were carried out according to the animal experimental guidelines for animal care and use at the University of California at Berkeley.
Southern blot analysis of ES cells and mice.
Genomic DNA from ES cells and mouse tail biopsy was digested with EcoRI or EcoRI-HindIII restriction enzymes, fractionated on 1% agarose gels and blotted onto Hybond N+ nylon membranes (Amersham). Homologous recombination was confirmed by using 5′ and 3′ external probes of a PstI-BglII fragment and a BamHI-XbaI fragment, respectively.
Northern blot analysis and RT-PCR.
Total RNA was isolated from mouse tissues with Trizol reagent (Invitrogen). For Northern blotting, total RNA (15 μg) was electrophoresed on a 1.2% agarose-formaldehyde gel and transferred onto Hybond N+ nylon membrane. After UV cross-linking, membranes were hybridized with [∂-32P]dCTP-labeled cDNA probes for fatty acid synthase (FAS) and stearoyl-coenzyme A (CoA) desaturase (SCD-1) by using ExpressHyb hybridization solution (Clontech). Posthybridization washes were performed for 40 min at room temperature in a mixture of 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and 0.1% sodium dodecyl sulfate (SDS) and for 30 min at 50°C in a mixture of 0.1× SSC and 0.1% SDS. For reverse transcription-PCR (RT-PCR), 5 μg of total RNA was reverse transcribed with oligo(dT) primers and Superscript II reverse transcriptase (Invitrogen) according to the manufacturer's specifications. Two microliters of the resulting cDNA sample was used as a template for PCR. gtl2 cDNA amplification was carried out for 22 or 26 cycles for embryo or white adipose tissue, respectively, with denaturation at 95°C for 45 s, annealing at 54°C for 45 s, and extension at 72°C for 30 s, by using the primers 5′-ATGCTGGACCCAAGACTCTG-3′ and 5′-GAAAGCACCATGAGCCACTA-3′. Cyclophilin cDNA was amplified for 18 cycles with denaturation at 95°C for 45 s, annealing at 54°C for 45 s, and extension at 72°C for 30 s, with the primers 5′-AGACTGAATGGCTGGATGGCA-3′ and 5′-GAAGGAATGGTTTGATGGGTA-3′.
Western blot analysis.
Total protein was extracted from homozygous Pref-1-null or heterozygous embryos (embryonic development day 17.5 [E17.5]). Embryos were lysed in a mixture of 60 mM Tris-HCl (pH 6.8), 1% SDS, and 0.5 mM phenylmethylsulfonyl fluoride and sonicated for 1 min. The lysates were centrifuged at 12,000 × g for 3 min, and protein concentrations in supernatants were determined by the Bradford method. To ensure equivalent loading, an equal amount of protein from each sample was subjected to SDS-polyacrylamide gel electrophoresis (PAGE), and quantities were confirmed by staining the gel with Coomassie blue preceding Western analysis. Protein samples were boiled for 5 min in Laemmli buffer before electrophoresis on SDS-PAGE gel (11% polyacrylamide). Proteins were then transferred to nitrocellulose membranes, and membranes were blocked with 5% nonfat dry milk in Tris-buffered saline-Tween 20 (TBST) buffer (10 mM Tris-HCl [pH 8.0], 165 mM NaCl, 0.05% Tween 20). Membranes were probed with anti-Pref-1 antibodies (1:3,000 dilution) in blocking buffer for 1 h, washed with TBST, and reacted for 1 h with a 1:5,000 dilution of anti-rabbit immunoglobulin G (IgG) antibody labeled with horseradish peroxidase (Bio-Rad). After washing, the blots were developed with an enhanced chemiluminescence system (NEN Life Science) and visualized by autoradiography.
Growth curve and physiological measurements.
Mice were fed a high-fat diet (45 kcal% fat supplied from lard and soybean oil, 35 kcal% carbohydrate from corn starch and maltodextrin, and 20 kcal% protein from casein) (D12451; Research Diets, Inc., New Brunswick, N.J.) ad libitum after weaning at 21 days of age and were weighed at 4-day intervals to monitor growth. At 16 weeks of age, mice were sacrificed between 13:00 and 15:00 for tissue dissection and blood samples. Blood was collected by cardiac puncture, centrifuged at 2,000 × g, and stored at −20°C before metabolite measurements. Serum triglyceride and cholesterol levels were analyzed with Triglyceride (INT) 10 and Infinity Cholesterol reagents (Sigma), respectively. Serum free fatty acid levels were determined with NEFA C (Wako Chemicals). For statistical analysis, Student's t test was used, and a P value of < 0.05 was considered significant.
Histological analysis.
Inguinal fat was fixed in Bouin's fluid (LabChem, Inc.) and dehydrated with a series of alcohol treatments. Tissues were embedded in paraffin, sliced into 8-μm sections, and stained with hematoxylin and eosin (Fisher). Image capture and adipocyte size measurements were performed with NIH Image software.
Bone staining.
Embryos or mice were eviscerated and fixed in 90% ethanol for at least 1 week. For the staining of cartilages, samples were stained with 0.01% Alcian blue (Sigma) in 80% ethanol and 20% glacial acetic acid. Then samples were rehydrated with a series of 70, 40, and 15% ethanol incubations for 2 h each. For bone staining, samples were incubated with 0.001% Alizarin red (Sigma) in 1% KOH for 5 days. After several rinses with 1% KOH, samples were stored in glycerol.
RESULTS
Generation of Pref-1-null mice.
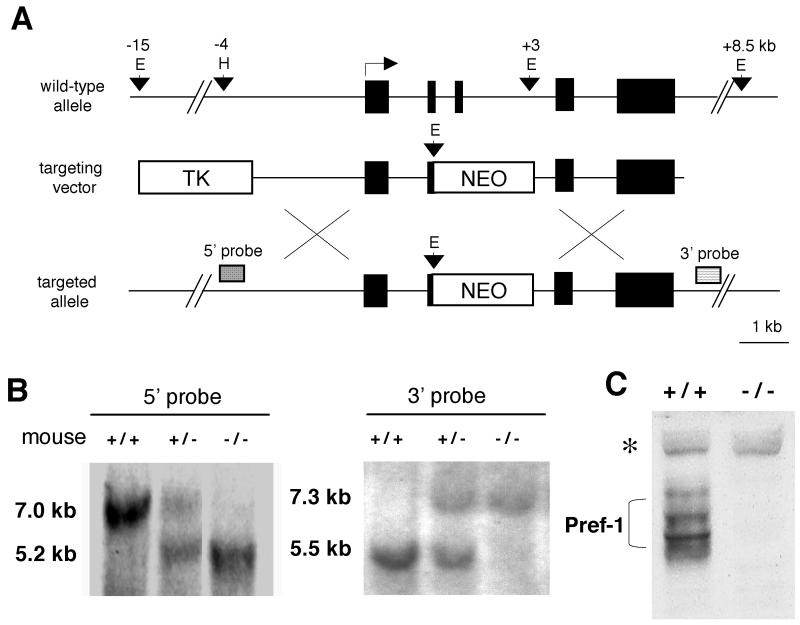
To determine the role of Pref-1 in growth and adipogenesis and the contribution of Pref-1 in mouse mUPD12 and human mUPD14 syndromes, we used gene targeting to generate Pref-1-null mice. The genomic pref-1 allele was disrupted by insertion of a neomycin resistance cassette into exons 2 and 3 of pref-1 (Fig. 1A). G418- and ganciclovir-resistant clones were screened for homologous recombination, and the positive clones were microinjected into 3.5-day C57BL/6J or BALB/c blastocysts to generate chimeric animals. Germ line transmission from each clone was obtained, and no subsequent difference between separate lines was observed. Male chimeras were mated with either C57BL/6J or BALB/c females to produce Pref-1 heterozygotes (+/−). Intercrosses of Pref-1+/− mice produced Pref-1-null mice. The wild-type (7.0 or 5.5 kb), heterozygote (7.0 and 5.2 kb or 7.3 and 5.5 kb), or homozygote (5.2 or 7.3 kb) Pref-1 alleles were detected from F2 pups generated by intermating F1 heterozygotes (Fig. 1 B). To confirm the absence of normal Pref-1 protein in Pref-1-null embryos (E17.5), we performed Western blot analysis with polyclonal antibodies against murine Pref-1 that we had prepared by immunization of rabbits with bacterially expressed murine Pref-1 (25). Figure 1C shows that in wild-type embryos, Pref-1 antibody detected the multiple forms of Pref-1 protein, which we have previously demonstrated were derived from alternative splicing and posttranslational modification (25). Pref-1 protein was not detected in homozygous embryos.
FIG. 1.
Targeted disruption of the pref-1 gene. (A) The top diagram is a schematic representation of the wild-type pref-1 allele with five exons. Pref-1 exons are represented by black boxes. The middle diagram shows the targeting construct with pPNT-NEO and thymidine kinase (TK) as positive and negative selectable markers, respectively. The bottom diagram shows the pref-1 allele mutated by homologous recombination. The probes used for Southern blot analyses and the sizes of the restriction fragments detected are indicated. E, EcoRI; H, HindIII; X, XbaI. (B) Southern blot analysis of EcoRI-cut (3′ probe) and EcoRI-HindIII-cut (5′ probe) mouse tail DNA prepared from F2 mice generated by intermating F1 heterozygotes (pref-1+/−). The 7.0- and 5.5-kb bands represent the wild-type allele, and the 5.2- and 7.3-kb bands represent pref-1-null alleles. (C) Western blot analysis of Pref-1 in embryos (E17.5). A polyclonal antibody against mouse Pref-1 was used. The embryos were generated from crossbreeding pref-1 heterozygotes. The multiple forms of Pref-1 produced by alternative splicing and posttranslational modification (bracketed) are detected in wild-type but not in homozygous (pref-1−/−) embryos. ∗, nonspecific band indicating equal amounts of sample loading.
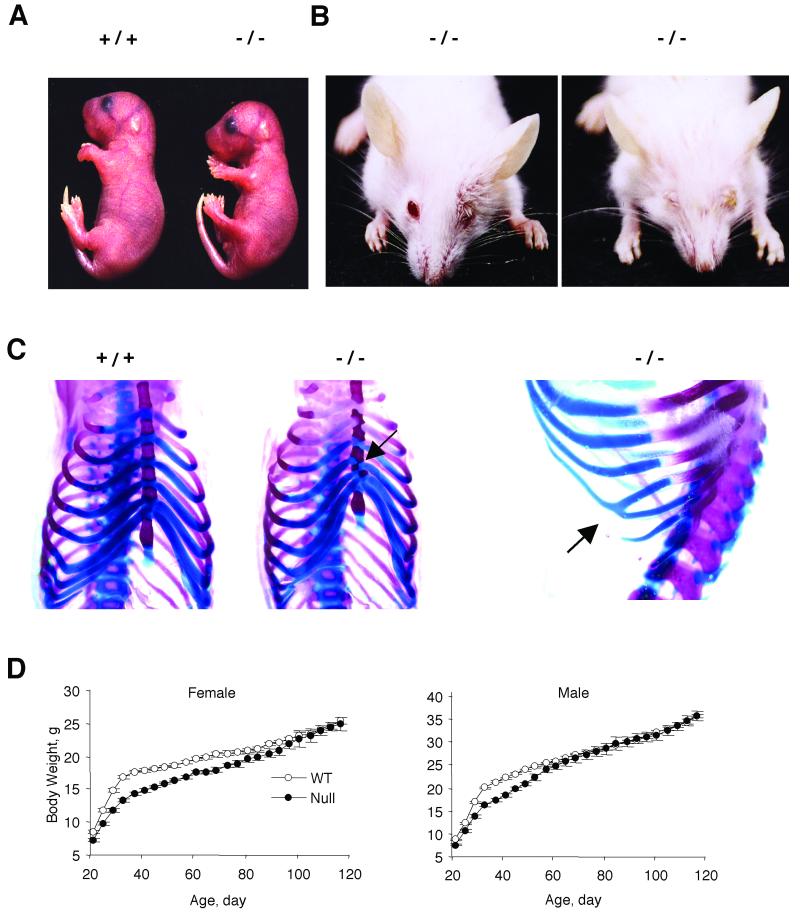
Mice lacking Pref-1 displayed pre- and postnatal growth retardation, eyelid, and skeletal abnormalities. The average litter size of Pref-1-null mice was significantly lower (4.26 ± 0.26 pups per litter, n = 25) than those of wild-type mice (6.75 ± 0.32 pups per litter, n = 27, P < 0.001), suggesting that Pref-1 plays an important function in embryonic development. We also measured the embryo size at E18.5 of mice generated by interbreeding of Pref-1 heterozygotes. Pref-1-null embryos (0.96 ± 0.004 g, n = 9) at E18.5 were smaller than wild-type littermates (1.18 ± 0.003 g, n = 12, P < 0.0001) (Fig. 2A). Approximately 50% of null mice were dead within 2 days of birth, indicating that Pref-1 is important for perinatal growth and survival. We observed a similar degree of neonatal mortality in litters produced from the mating of wild-type females and Pref-1-null or heterozygous male mice, which, as described below, do not express Pref-1. This suggests that the survival rate is entirely dependent on the phenotype of the pup rather than that of the mother. We also found skeletal malformation, including asymmetrical junction of ribs to sternum and fusion of ribs from embryos and postnatal dead mice (Fig. 2C). Skeletal abnormalities and pulmonary defects, as well as other problems arising from these defects, such as poor suckling ability, may have contributed to the high rate of postnatal death observed in null mice (8). As shown in Fig. 2B, Pref-1-null mice also showed eyelid abnormalities similar to the blepharophimosis observed in individuals with a deletion at 14q32 compassing the pref-1 locus or in mUPD14 patients (29, 32). However, this was strain dependent and was observed in Pref-1-null mice on a BALB/cJ background, but not on a C57BL/6J background.
FIG. 2.
Pre- and postnatal growth retardation and eyelid abnormalities in mice lacking Pref-1. (A) Intrauterine growth retardation. Pref-1-null embryos are smaller than wild-type embryos at E18.5. The embryos were generated from crossbreeding Pref-1 heterozygotes (+/−). (B) Blepharophimosis. Anterior view of mice at age 27 days showing the left or both eyelid defects observed on the BALB/cJ strain background. (C) Skeletal malformation. Arrows indicate asymmetric zigzagged sternums and fusion of ribs in Pref-1-null mice. (D) Growth retardation in Pref-1-null mice of the C57BL/6J background. Female and male wild-type and Pref-1-null mice (n = 27 to 37 per group) were fed high-fat (45 kcal%) diet ad libitum from 21 days old and were weighed at 4-day intervals. Statistics were performed with a two-tailed t test. For female mice, from day 21 to day 68, P < 0.01, and from day 72 to day 76, P < 0.05. For male mice, from day 21 to day 52, P < 0.01.
At weaning, both female and male Pref-1-null mice weighed less than wild-type mice (female, 7.34 ± 0.21 g, n = 32, versus 8.51 ± 0.19 g, n = 27, P < 0.0001; male, 7.58 ± 0.25 g, n = 36, versus 8.81 ± 0.21 g, n = 37, P < 0.0001, respectively). Initially, after weaning at 3 weeks of age, the mice were fed a regular chow diet. At 15 weeks of age, we found that there was growth retardation with moderately larger fat pad mass in Pref-1-null mice (data not shown). In an attempt to observe the Pref-1 effect on adipose tissue development more clearly, we carried out all subsequent studies with the mice that were fed a high-fat (45 kcal%) diet as described in Materials and Methods. Pref-1-null mice on a high-fat diet gained weight at a slower rate during the immediate postweaning period. However, as shown in Fig. 2D, there was a catching up in body weight at approximately 90 days for females and 65 days for males. By 16 weeks of age, the body weights were not different between null and wild-type mice (female, 24.94 ± 0.99 g, n = 14, versus 25.35 ± 0.50 g, n = 11; male, 35.60 ± 0.97 g, n = 14, versus 35.72 ± 0.37 g, n = 16). These data suggest to us that accelerated weight gain at this later stage could be due to increases in adipose tissue mass.
Increased adiposity in mice lacking Pref-1.
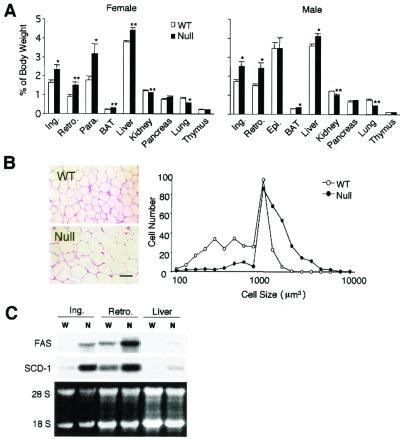
We investigated whether the accelerated weight gain of null mice observed at later stages is due to fat accretion. At an early age (8 weeks), the white fat pad weights including inguinal, retroperitoneal, and reproductive fats were similar between null mice and wild-type mice (female, 3.80% ± 0.37%, n = 7, versus 3.33% ± 0.62%, n = 10, P = 0.2061; male, 3.17% ± 0.33%, n = 7, versus 3.12% ± 0.34%, n = 4, P = 0.9286). However, as shown in Fig. 3A, at 16 weeks of age, the white fat pads (expressed as a percentage of body weight) from female Pref-1-null mice after 13 weeks of feeding of a high-fat diet (45 kcal%) weighed significantly more than those of wild-type mice (inguinal, 2.34% ± 0.2% versus 1.64% ± 0.1%, P = 0.0182; retroperitoneal, 1.50% ± 0.1% versus 0.90% ± 0.1%, P = 0.0096; parametrial, 3.16% ± 0.5% versus 1.78% ± 0.2%, P = 0.0296; and brown fat, 0.29% ± 0.01% versus 0.22% ± 0.01%, P = 0.0045) for null (n = 10) versus wild-type (n = 7) mice, respectively. Figure 3A also shows that similar increases in fat pad mass were observed in all regions of male Pref-1-null mice, except the epididymal fat pad. In contrast to fat pad weights, the weights of other organs, including lungs and kidneys, of the Pref-1-null mice at later ages did not catch up but remained smaller than those of wild-type mice. Interestingly, as summarized in Table 1, Pref-1-null mice developed enlarged livers (female, 4.41% ± 0.13%, n = 9, versus 3.77% ± 0.08%, n = 7, P = 0.001; male, 4.10% ± 0.14%, n = 7, versus 3.60% ± 0.12%, n = 7, P = 0.0224) with increased total lipid content (female, 8.77 ± 0.57 mg/g of liver, n = 4, versus 5.76 ± 0.31 mg/g of liver, n = 7, P = 0.0046; male, 10.73 ± 0.69 mg/g of liver, n = 5, versus 8.35 ± 0.44 mg/g of liver, n = 5, P = 0.0239).
FIG. 3.
Accelerated adiposity in mice lacking Pref-1. (A) Percentage of fat pad and organ weight relative to body weight. Fat depots and organs were dissected from 16-week-old wild-type (WT) and Pref-1-null mice fed a high-fat diet (n = 7 to 10 per group). Ing, inguinal fat pad; Epi, epididymal fat pad; Retro, retroperitoneal fat pad; Para., parametrial fat pad; BAT, brown adipose tissue. All values are means ± standard errors. ∗, P < 0.05; ∗∗, P < 0.01. (B) Paraffin section of inguinal fat pad from 16-week-old female mice and the distribution of fat cell size. The size of at least 300 cells per sample (mean of four mice per group) was determined with NIH Image software. The distribution of cell volume is shown. The scale bar represents 50 μm. (C) Expression of adipocyte marker genes in adipose tissues and liver. Total RNA was extracted from adipose tissue of 16-week-old wild-type (W) and Pref-1-null (N) mice fed a high-fat diet.
TABLE 1.
Litter size, embryo and body weights, fat pad weights, lipid metabolites, and total lipid contents in livers of wild-type and Pref-1-null micea
| Variable | Sex | Mouse group
|
|
|---|---|---|---|
| Wild type | Pref-1 null | ||
| Litter size (no. of pups/litter) | 6.75 ± 0.32 (27) | 4.26 ± 0.26 (25)d | |
| Body wt (g) | |||
| E18.5 | 1.18 ± 0.003 (12) | 0.96 ± 0.004 (9)d | |
| 3 wk | Female | 8.51 ± 0.19 (27) | 7.34 ± 0.21 (32)d |
| Male | 8.81 ± 0.91 (27) | 7.58 ± 0.25 (37)d | |
| 16 wk | Female | 25.35 ± 0.50 (11) | 24.94 ± 0.99 (14) |
| Male | 35.72 ± 0.37 (16) | 35.60 ± 0.97 (14) | |
| Fat pad wt (% of body wt)b | |||
| 8 wk | Female | 3.33 ± 0.62 (10) | 3.80 ± 0.37 (7) |
| Male | 3.12 ± 0.34 (4) | 3.17 ± 0.33 (7) | |
| 16 wk | Female | 4.32 ± 0.38 (7) | 6.70 ± 0.50 (9)e |
| Male | 6.69 ± 0.49 (7) | 8.39 ± 0.29 (7)e | |
| Blood metabolites | |||
| Triglycerides (mg/liter) | Female | 8.82 ± 0.70 (5) | 12.0 ± 1.20 (6)f |
| Male | 11.1 ± 0.70 (5) | 19.6 ± 2.30 (6)e | |
| Cholesterol (mg/liter) | Female | 8.37 ± 0.63 (5) | 11.7 ± 1.34 (6)f |
| Male | 13.2 ± 0.54 (5) | 16.94 ± 0.85 (6)e | |
| Free fatty acids (mmol/liter) | Female | 1.56 ± 0.08 (5) | 1.79 ± 0.10 (6) |
| Male | 1.66 ± 0.14 (5) | 2.23 ± 0.18 (6)f | |
| Lipid content in liver (mg/g of liver)c | Female | 5.76 ± 0.31 (7) | 8.77 ± 0.57 (4)e |
| Male | 8.35 ± 0.44 (5) | 10.73 ± 0.69 (5)f | |
Values represent the average ± standard error of the mean. The number of samples is in parentheses.
The fat pad weights represent inguinal, retroperitoneal, and reproductive fats from mice 8 or 16 weeks of age.
Total lipid was extracted with a mixture of hexane-2-propanol(3/2 ratio) from mice 16 weeks of age.
P < 0.001.
P < 0.01.
P < 0.05.
We next determined whether increased fat mass in Pref-1-null mice results from cellular hypertrophy or hyperplasia. Even though the fat pad weight of null mice was higher than that of wild-type mice, total DNA contents, indicative of total cell number in adipose tissue that includes both preadipocytes and adipocytes, did not differ (Table 1). As shown in Fig. 3B, staining of paraffin sections and examination of the cell size distribution of inguinal fat pads from 16-week-old female mice revealed increased adipocyte cell size, indicative of hypertrophy, in Pref-1-null compared to wild-type mice. Furthermore, mRNA levels for late markers of adipocyte differentiation, FAS, and SCD-1, were increased in Pref-1-null mice compared to wild-type mice, demonstrating enhanced adipocyte differentiation (Fig. 3C). We also found that the enlarged fatty liver of Pref-1-null mice showed the increase in FAS and SCD-1 expression that normally accompanies increased hepatic lipogenesis and fat synthesis.
Pref-1 is expressed only from the paternal allele.
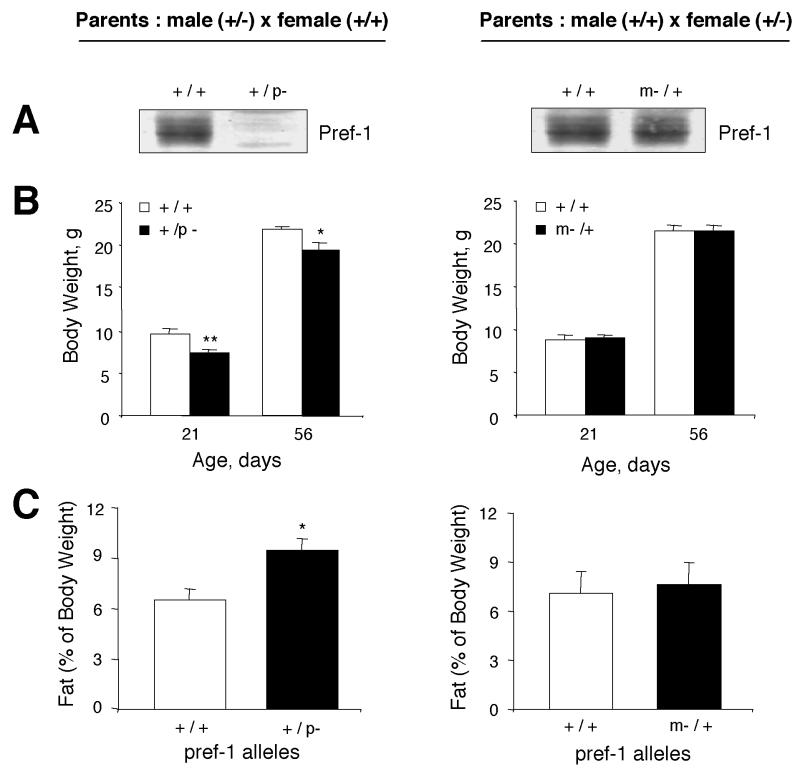
Because Pref-1 is expressed only from the paternal allele, heterozygotes with either maternal or paternal inheritance of the pref-1 knockout allele (m−/+ or +/p−) should have phenotypes similar to those of wild-type or null mice, respectively. To specifically address this, male or female pref-1 heterozygotes (+/−) were mated with wild-type mice. Western blot analysis of the E17.5 embryo in Fig. 4A shows that Pref-1 protein expression was completely abolished in heterozygotes with the paternally-inherited pref-1 knockout allele (+/p−). On the other hand, heterozygotes with the maternally inherited pref-1 knockout allele (m−/+) express Pref-1 protein at a level similar to that of wild-type animals. These results clearly demonstrate that Pref-1 is expressed only from the paternal allele. The body weight of these animals (m−/+; +/p−) was examined at days 21 and 56—time points at which we had demonstrated a significant decrease in body weight for Pref-1-null mice. Figure 4B illustrates that, as predicted, at 21 days of age, Pref-1 heterozygotes with the paternally inherited pref-1 knockout allele (+/p−) exhibited the same growth retardation that we observed for Pref-1-null mice [female, 9.13 ± 0.25 g, n = 9 (+/+) versus 6.66 ± 0.17 g, n = 5 (+/p−), P < 0.0001; male, 9.71 ± 0.43 g, n = 7 (+/+) versus 7.51 ± 0.17 g, n = 7 (+/p−), P < 0.0001]. In contrast, heterozygotes with disruption of the maternal allele (m−/+) have the same growth rate as wild-type littermates. When fat pad weight (sum of inguinal, reproductive, and perirenal fat pads) from male mice was examined after feeding on a high-fat diet from 3 to 16 weeks of age, heterozygotes with the paternally inherited pref-1 knockout allele (+/p−) displayed a 45% increase in fat pad weight compared to their wild-type littermates (m+/−, 9.45% ± 0.7%, n = 5; versus m+/+, 6.50% ± 0.7%, n = 6, P = 0.0129), the same as we observed for Pref-1-null mice (Fig. 4C). In contrast, heterozygotes with the maternally inherited pref-1 knockout allele (m−/+) did not show an increase. Therefore, both the early growth retardation and the increased fat mass that occurred in Pref-1-null mice were recapitulated as the result of paternal imprinting of Pref-1.
FIG. 4.
Mice with maternal or paternal inheritance of the pref-1-knockout allele (m−/+ or +/p−) have phenotypes similar to those of wild-type or null mice, respectively. (A) Pref-1 protein expression in E17.5 embryos generated from matings of male heterozygotes (+/−) and female wild-type mice or vice versa. Pref-1 protein is expressed in m−/+ heterozygotes, but not in +/p− heterozygotes. (B) Body weights of F1 littermates were measured at 21 and 56 days of age. Values are means ± standard errors with n = 5 to 13 per group. ∗, P < 0.05; ∗∗, P < 0.01. (C) Percentage of fat pad mass (total white fat, including inguinal, retroperitoneal, and reproductive fat) relative to body weight (16 weeks old; n = 7 to 9 per group). All values are means ± standard errors. ∗, P < 0.05. Statistics were performed with a two-tailed t test for panels B and C.
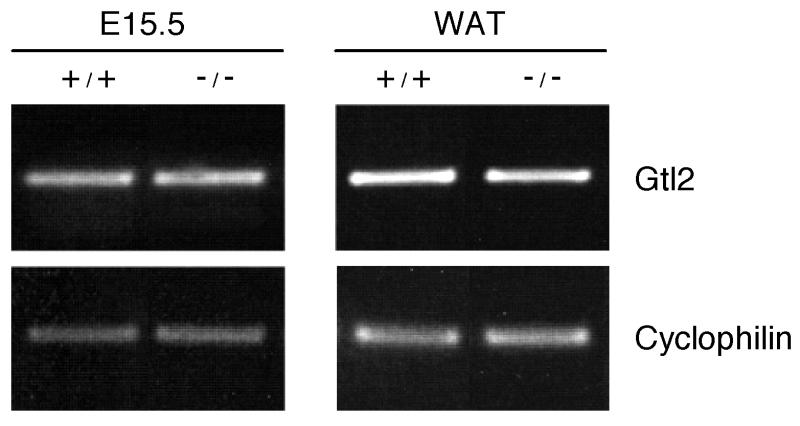
The regulation of expression of imprinting genes in the cluster that includes Pref-1 is quite complex. Gtl2 and Pref-1 have been shown to be reciprocally expressed in a manner analogous to that of the Igf2/H19 locus (10). We, therefore, examined expression of gtl2 in Pref-1-null mice and found that the expression levels for gtl2 were similar between null and wild-type mice (Fig. 5). These data indicate that replacing part of the pref-1 genome (part of exons 2 and 3) with the neo cassette to generate Pref-1-null mice, did not affect expression of the neighboring reciprocally regulated imprinted gene, gtl2. We conclude that the phenotype we observe in Pref-1-null mice is likely attributable solely to the absence of Pref-1 expression.
FIG. 5.
Expression of the gtl2 gene in wild-type and Pref-1-null mice. The expression of gtl2 was measured from whole embryo (E15.5) or white adipose tissue (WAT) (16 weeks) by RT-PCR. As reference for quantitative gene expression, cyclophilin was amplified simultaneously under conditions identical to those used for gtl2.
Increase in lipid metabolite levels in Pref-1-null mice.
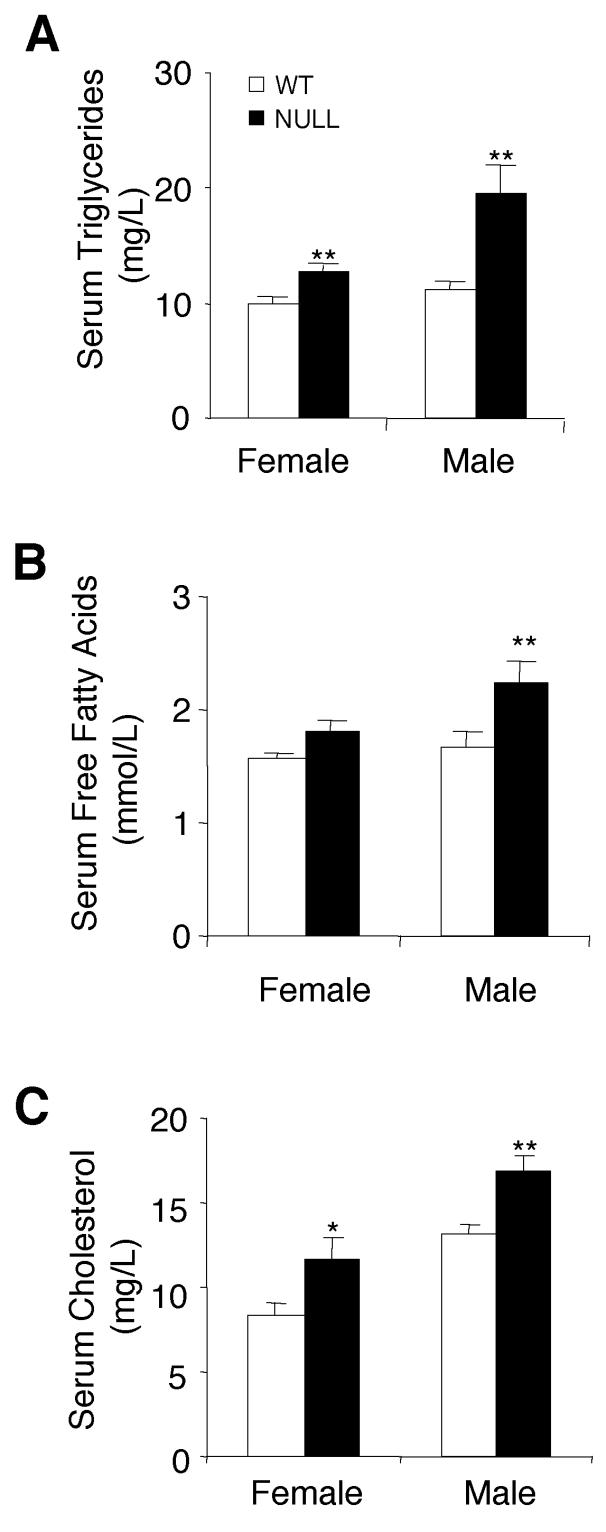
To determine whether the increased fat pad mass in Pref-1-null mice is reflected by levels of serum lipid metabolites, levels of triglycerides, free fatty acids, and cholesterol were assessed in Pref-1-null mice that were fed a high-fat diet from 3 to 16 weeks of age. Serum triglyceride was elevated substantially by 36 and 77% in Pref-1-null female and male mice, respectively (Fig. 6A). Pref-1-null male mice showed a 34% elevation in their free fatty acid levels in the fed state (Fig. 6B). Cholesterol levels in null mice were also elevated by 21% in female mice and 28% in male mice (Fig. 6C). The hypercholesterolemia and hyperlipidemia, as well as the enlarged, fatty liver, suggest that the loss of antiadipogenic action in Pref-1-null mice disturbs the homeostasis of lipid metabolism.
FIG. 6.
Altered serum lipid metabolite levels in mice lacking Pref-1. (A, B, and C) Levels of triglycerides, free fatty acids, and cholesterol in serum were measured from Pref-1-null and wild-type (WT) mice at 16 weeks of age after feeding on a high-fat diet from 3 to 16 weeks. Values are means ± standard errors with n = 5 to 8 per group. Statistics were performed with a two-tailed t test. ∗, P < 0.05; ∗∗, P < 0.01.
DISCUSSION
Through the generation of Pref-1-knockout mice, we demonstrate that Pref-1 functions in growth and development, perinatal survival, and adipose development. In addition, heterozygotes with either maternal or paternal inheritance of the pref-1-knockout allele (m−/+ or +/p−) showed both an expression pattern of Pref-1 and a phenotypic profile similar to those of wild-type or null mice, respectively, providing direct evidence for paternal monoallelic expression of Pref-1.
Imprinting has been shown to rely on allele-specific DNA methylation that can either activate or suppress expression, and imprinted genes are commonly found in clusters, wherein they are often coordinately controlled. In the case of maternal UPD, it is possible that the expression of paternally expressed genes would be absent whereas maternally expressed genes would be overexpressed. Of all of the possible UPDs, only a small fraction have been shown to have a specific phenotypic effect or effects due to imprinting. A cluster of imprinted genes has recently been discovered on mouse chromosome 12 and in a syntenic region of human chromosome 14 (1, 5, 6, 8). At this locus, the pref-1, peg11, and dat genes are paternally expressed, while the gtl2, antipeg11, and meg8 genes are maternally expressed (3). However, whether the phenotypes observed in murine mUPD12 or human mUPD14 can be attributed to a specific gene in this region has not been investigated. pref-1 and peg11 are the most likely candidate genes for the mUPD-related phenotypes, because the other imprinted genes in this region code for nontranslated RNAs. However, it is also possible that the nontranslated transcripts may act as RNA effector molecules to regulate gene expression in a manner similar to the Air (for antisense igf2r RNA) transcript described for the igf2r gene (15). The overlapping phenotypes of Pref-1-null mice and human mUPD14 indicate that the obesity, skeletal malformation, blepharophimosis, and growth retardation in human mUPD14 (1, 5, 8, 16) can be attributed to Pref-1. On the other hand, hypotonic and early-puberty phenotypes observed in mUPD14 are not apparent in Pref-1-null mice and may therefore be caused by misexpression of another imprinted gene, possibly peg11. To further investigate the exact genes responsible for mUPD phenotypes, generation of Peg11-knockout or Pref-1- and Peg11-double-knockout mice would be necessary. Overall, our studies are the first to unveil the phenotypic relevance of this imprinted region on mouse chromosome 12. Furthermore, the fact that paternally expressed Pref-1 acts as a growth regulator during embryonic development, as shown in our studies, supports the genomic conflict model, in which paternally expressed genes would promote growth, whereas maternally expressed genes would inhibit growth (17).
We previously showed that Pref-1 inhibits differentiation of 3T3-L1 preadipocytes into adipocytes in vitro (22-28). Our data indicate that accelerated weight gain in Pref-1-null mice is largely due to fat accretion at later stages. Examination of the adipose tissue suggests that both enhanced adipocyte differentiation and enhanced fat cell maturation contribute to the increase in adipose mass seen in Pref-1-null mice. Obesity is often observed in mUPD14 (1, 16). Increased adipose mass in Pref-1-null mice, therefore, parallels the human mUPD14 obesity phenotype. Interestingly, it has been reported that callipyge (clpg) sheep, which bear a mutation that results in lean animals with hypertrophic muscle, overexpress two paternally imprinted genes, pref-1 and peg11 (4). Given our findings of increased adipose tissue mass in Pref-1-null mice, it can be predicted that the lean phenotype of clpg sheep may be due to the antiadipogenic action of Pref-1. This is in agreement with findings from our previous in vitro studies of Pref-1 in adipocyte differentiation: (i) its extinction during adipogenesis, (ii) inhibition of 3T3-L1 adipocyte differentiation by constitutive expression of Pref-1, and (iii) enhancement of differentiation by antisense pref-1.
In conclusion, the present studies suggest that Pref-1 is important for perinatal survival, normal growth, and homeostasis of fat deposition. We propose that the absence of pref-1 by gene knockout or via the imprinting status of Pref-1 heterozygotes (+/p−) results in adverse effects manifested in the neonatal period. In adulthood, depletion of Pref-1 leads to accelerated adiposity similar to the phenotype reported in human mUPD14 patients, who manifest growth defects and obesity. Our present studies also suggest pref-1 as the gene responsible for most of the imprinting-related phenotypes of mouse mUPD12 and syntenic human mUPD14.
Acknowledgments
This work was supported by National Institutes of Health grant DK50828 to H.S.S.
Yang Soo Moon, Cynthia M. Smas, and Kichoon Lee contributed equally to this report.
REFERENCES
- 1.Berends, M. J., R. Hordijk, H. Scheffer, J. C. Oosterwijk, D. J. Halley, and N. Sorgedrager. 1999. Two cases of maternal uniparental disomy 14 with a phenotype overlapping with the Prader-Willi phenotype. Am. J. Med. Genet. 84:76-79. [DOI] [PubMed] [Google Scholar]
- 2.Carlsson, C., D. Tornehave, K. P. Lindberg, N. Galante, B. Billestrup, L. Michelsen, I. Larsson, and J. H. Nielsen. 1997. Growth hormone and prolactin stimulate the expression of rat preadipocyte factor-1/delta-like protein in pancreatic islets: molecular cloning and expression pattern during development and growth of the endocrine pancreas. Endocrinology 138:3940-3948. [DOI] [PubMed] [Google Scholar]
- 3.Charlier, C., K. Segers, D. Wagenaar, L. Karim, S. Berghmans, O. Jaillon, T. Shay, J. Weissenbach, N. Cockett, G. Gyapay, and M. Georges. 2001. Human-ovine comparative sequencing of a 250-kb imprinted domain encompassing the callipyge (clpg) locus and identification of six imprinted transcripts: DLK1, DAT, GTL2, PEG11, antiPEG11, and MEG8. Genome Res. 11:850-862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Charlier, C., K. Segers, L. Karim, T. Shay, G. Gyapay, N. Cockett, and M. Georges. 2001. The callipyge mutation enhances the expression of coregulated imprinted genes in cis without affecting their imprinting status. Nat. Genet. 27:367-369. [DOI] [PubMed] [Google Scholar]
- 5.Fokstuen, S., C. Ginsburg, M. Zachmann, and A. Schinzel. 1999. Maternal uniparental disomy 14 as a cause of intrauterine growth retardation and early onset of puberty. J. Pediatr. 134:689-695. [DOI] [PubMed] [Google Scholar]
- 6.Georgiades, P., M. Watkins, M. A. Surani, and A. C. Ferguson-Smith. 2000. Parental origin-specific developmental defects in mice with uniparental disomy for chromosome 12. Development 127:4719-4728. [DOI] [PubMed] [Google Scholar]
- 7.Hansen, L. H., B. Madsen, B. Teisner, J. H. Nielsen, and N. Billestrup. 1998. Characterization of the inhibitory effect of growth hormone on primary preadipocyte differentiation. Mol. Endocrinol. 12:1140-1149. [DOI] [PubMed] [Google Scholar]
- 8.Hordijk, R., H. Wierenga, H. Scheffer, B. Leegte, R. M. Hofstra, and I. Stolte-Dijkstra. 1999. Maternal uniparental disomy for chromosome 14 in a boy with a normal karyotype. J. Med. Genet. 36:782-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Joyner, A. L. (ed.). 1993. Gene targeting: a practical approach. Oxford University Press, New York, N.Y.
- 10.Kaffer, C. R., A. Grinberg, and K. Pfeifer. 2001. Regulatory mechanisms at the mouse Igf2/H19 locus. Mol. Cell. Biol. 21:8189-8196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaneta, M., M. Osawa, K. Sudo, H. Nakauchi, A. G. Farr, and Y. Takahama. 2000. A role for pref-1 and HES-1 in thymocyte development. J. Immunol. 164:256-264. [DOI] [PubMed] [Google Scholar]
- 12.Kobayashi, S., H. Wagatsuma, R. Ono, H. Ichikawa, M. Yamazaki, H. Tashiro, K. Aisaka, N. Miyoshi, T. Kohda, A. Ogura, M. Ohki, T. Kaneko-Ishino, and F. Ishino. 2000. Mouse Peg9/Dlk1 and human PEG9/DLK1 are paternally expressed imprinted genes closely located to the maternally expressed imprinted genes: mouse Meg3/Gtl2 and human MEG3. Genes Cells 5:1029-1037. [DOI] [PubMed] [Google Scholar]
- 13.Kotzot, D. 2001. Comparative analysis of isodisomic and heterodisomic segments in cases with maternal uniparental disomy 14 suggests more than one imprinted region. J. Med. Genet. 38:497-507. [DOI] [PubMed] [Google Scholar]
- 14.Laborda, J., E. A. Sausville, T. Hoffman, and V. Notario. 1993. dlk, a putative mammalian homeotic gene differentially expressed in small cell lung carcinoma and neuroendocrine tumor cell line. J. Biol. Chem. 268:3817-3820. [PubMed] [Google Scholar]
- 15.Lyle, R., D. Watanabe, D. te Vruchte, W. Lerchner, O. W. Smrzka, A. Wutz, J. Schageman, L. Hahner, C. Davies, and D. P. Barlow. 2000. The imprinted antisense RNA at the Igf2r locus overlaps but does not imprint Mas1. Nat. Genet. 25:19-21. [DOI] [PubMed] [Google Scholar]
- 16.Manzoni, M. F., T. Pramparo, A. Stroppolo, F. Chiaino, E. Bosi, O. Zuffardi, and R. Carrozzo. 2000. A patient with maternal chromosome 14 UPD presenting with a mild phenotype and MODY. Clin. Genet. 57:406-408. [DOI] [PubMed] [Google Scholar]
- 17.Moore, T., and D. Haig. 1991. Genomic imprinting in mammalian development: a parental tug-of war. Trends Genet. 7:45-49. [DOI] [PubMed] [Google Scholar]
- 18.Ohno, N., A. Izawa, M. Hattori, R. Kageyama, and T. Sudo. 2001. dlk inhibits stem cell factor-induced colony formation of murine hematopoietic progenitors: Hes-1-independent effect. Stem Cells 19:71-79. [DOI] [PubMed] [Google Scholar]
- 19.Preece, M. A., S. M. Price, V. Davies, L. Clough, P. Stanier, R. C. Trembath, and G. E. Moore. 1997. Maternal uniparental disomy 7 in Silver-Russell syndrome. J. Med. Genet. 34:6-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmidt, J. V., P. G. Matteson, B. K. Jones, X. J. Guan, and S. M. Tilghman. 2000. The Dlk1 and Gtl2 genes are linked and reciprocally imprinted. Genes Dev. 14:1997-2002. [PMC free article] [PubMed] [Google Scholar]
- 21.Shimomura, I., R. E. Hammer, J. A. Richardson, S. Ikemoto, Y. Bashmakov, J. L. Goldstein, and M. S. Brown. 1998. Insulin resistance and diabetes mellitus in transgenic mice expressing nuclear SREBP-1c in adipose tissue: model for congenital generalized lipodystrophy. Genes Dev. 12:3182-3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smas, C. M., and H. S. Sul. 1993. Pref-1, a protein containing EGF-like repeats, inhibits adipocyte differentiation. Cell 73:725-734. [DOI] [PubMed] [Google Scholar]
- 23.Smas, C. M., and H. S. Sul. 1996. Characterization of Pref-1 and its inhibitory role in adipocyte differentiation. Int. J. Obes. Relat. Metab. Disord. 3(Suppl.):S65-S72. [PubMed] [Google Scholar]
- 24.Smas, C. M., and H. S. Sul. 1997. Molecular mechanisms of adipocyte differentiation and inhibitory action of pref-1. Crit. Rev. Eukaryot. Gene Expr. 7:281-298. [DOI] [PubMed] [Google Scholar]
- 25.Smas, C. M., D. Green, and H. S. Sul. 1994. Structural characterization and alternate splicing of the gene encoding the preadipocyte EGF-like protein pref-1. Biochemistry 33:9257-9265. [DOI] [PubMed] [Google Scholar]
- 26.Smas, C. M., D. Kachinskas, C. M. Liu, X. Xie, L. K. Dircks, and H. S. Sul. 1998. Transcriptional control of the pref-1 gene in 3T3-L1 adipocyte differentiation. Sequence requirement for differentiation-dependent suppression. J. Biol. Chem. 273:31751-31758. [DOI] [PubMed] [Google Scholar]
- 27.Smas, C. M., L. Chen, and H. S. Sul. 1997. Cleavage of membrane-associated pref-1 generates a soluble inhibitor of adipocyte differentiation. Mol. Cell. Biol. 17:977-988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smas, C. M., L. Chen, L. Zhao, M. J. Latasa, and H. S. Sul. 1999. Transcriptional repression of pref-1 by glucocorticoids promotes 3T3-L1 adipocyte differentiation. J. Biol. Chem. 274:12632-12641. [DOI] [PubMed] [Google Scholar]
- 29.Sutton, V. R., and L. G. Shaffer. 2000. Search for imprinted regions on chromosome 14: comparison of maternal and paternal UPD cases with cases of chromosome 14 deletion. Am. J. Med. Genet. 93:381-387. [DOI] [PubMed] [Google Scholar]
- 30.Takada, S., M. Tevendale, J. Baker, P. Georgiades, E. Campbell, T. Freeman, M. H. Johnson, M. Paulsen, and A. C. Ferguson-Smith. 2000. Delta-like and gtl2 are reciprocally expressed, differentially methylated linked imprinted genes on mouse chromosome 12. Curr. Biol. 10:1135-1138. [DOI] [PubMed] [Google Scholar]
- 31.Wylie, A. A., S. K. Murphy, T. C. Orton, and R. L. Jirtle. 2000. Novel imprinted DLK1/GTL2 domain on human chromosome 14 contains motifs that mimic those implicated in IGF2/H19 regulation. Genome Res. 10:1711-1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamamoto, Y., R. Sawa, N. Okamoto, A. Matsui, M. Yanagisawa, and S. Ikemoto. 1986. Deletion 14q(q24.3 to q32.1) syndrome: significance of peculiar facial appearance in its diagnosis, and deletion mapping of Pi (alpha 1-antitrypsin). Hum. Genet. 74:190-192. [DOI] [PubMed] [Google Scholar]
- 33.Zhou, Y. T., Z. W. Wang, M. Higa, C. B. Newgard, and R. H. Unger. 1999. Reversing adipocyte differentiation: implications for treatment of obesity. Proc. Natl. Acad. Sci. USA 96:2391-2395. [DOI] [PMC free article] [PubMed] [Google Scholar]