Abstract
The orphan nuclear receptor hepatocyte nuclear factor 4 (HNF-4) regulates the expression of many liver-specific genes both during development and in the adult animal. Towards understanding the molecular mechanisms by which HNF-4 functions, we have established in vitro transcription systems that faithfully recapitulate HNF-4 activity. Here we have focused on the coactivator requirements for HNF-4, especially for the multicomponent TRAP/SMCC/Mediator complex that has emerged as the central regulatory module of the transcription apparatus. Using a system that has been reconstituted from purified transcription factors, as well as one consisting of unfractionated nuclear extract from which TRAP/SMCC/Mediator has been depleted by specific antibodies, we demonstrate a strong dependence of HNF-4 function on this coactivator. Importantly, we further show a TRAP/SMCC/Mediator-dependence for HNF-4 transcriptional activation from chromatin templates. The latter involves cooperation with the histone acetyltransferase-containing coactivator p300, in accord with a synergistic mode of action of the two divergent coactivators. We also show that HNF-4 and TRAP/SMCC/Mediator can interact physically. This interaction likely involves primary HNF-4 activation function 2 (AF-2)-dependent interactions with the TRAP220 subunit of TRAP/SMCC/Mediator and secondary (AF-2-independent) interactions with TRAP170/RGR1. Finally, recruitment experiments using immobilized templates strongly suggest that the functional consequences of the physical interaction probably are manifested at a postrecruitment step in the activation pathway.
Hepatocyte nuclear factor 4 (HNF-4), an orphan member of the nuclear receptor superfamily (42, 43), is one of the key regulators of hepatocyte differentiation in mammals (5, 26). In the adult animal, HNF-4 is predominantly expressed in the liver, intestine, and kidney (5, 42, 43) and is responsible for tissue-restricted expression of numerous genes that include those involved in glucose metabolism, urea biosynthesis, erythropoesis, and cholesterol homeostasis (42). Underscoring this critical role of HNF-4 in regulating the body's metabolism, the maturity-onset diabetes of the young syndrome has been attributed to a defective HNF-4 allele (48). Furthermore, HNF-4 is evolutionarily conserved from Caenorhabditis elegans to humans (44).
Like other members of the nuclear receptor superfamily, mammalian HNF-4 possesses a DNA-binding domain that consists of a conserved double zinc finger motif (42, 43) as well as an extended, largely hydrophobic region that includes an activation function 2 (AF-2) domain; the latter has been shown to facilitate activated transcription both in vitro (30) and in vivo (14). The extreme N-terminal region of HNF-4 contains a putative AF-1-like domain (12), whereas the extreme C-terminal region contains a proline-rich domain that is dispensable for HNF-4 function in vitro (30). Despite purported identification of a family of small molecules that interact with HNF-4 (16), the issue of whether or not HNF-4 is regulated by a ligand remains unsettled. Given that mammalian HNF-4 is a potent activator of transcription both in vitro (30; this study) and in essentially all cell types (including Saccharomyces cerevisiae) that have been tested (reviewed in reference 30), plus the fact that it is rather ancient in evolutionary terms (42, 44), it is indeed likely that HNF-4 transactivation function is not dependent on a ligand (see also reference 32).
HNF-4 and other nuclear receptors are representative of transcriptional activators that typically function by binding to cognate DNA binding sites located upstream of core promoter elements that nucleate the assembly of the general transcription machinery. This machinery consists of RNA polymerase II (Pol II) and the general transcription factors (GTFs) TFIIA, TFIIB, TFIID, TFIIE, TFIIF, and TFIIH (39). The precise mechanism by which activators modulate the action of Pol II and GTFs is unclear. Despite early indications that a part of the mechanism might entail direct interactions of the activator with distinct GTFs, it is becoming increasingly apparent that additional coactivators are also involved (reviewed in reference 40).
Biochemical studies employing DNA templates have variously identified as coactivators the TATA box binding protein [TBP]-associated factors in TFIID (47); positive cofactors (PC1, PC2, PC3, PC4, and PC52) derived from the USA cofactor fraction (21, 40); and several multiprotein complexes that include TRAP (8, 18), SMCC (13), ARC (36), DRIP (38), NAT (46), murine Mediator (20), human Mediator (3), CRSP (41), and USA-derived PC2 (28). TRAP, SMCC, ARC, DRIP, CRSP, PC2, and human and murine Mediators are all related to the yeast Mediator, which is the reversibly associating coactivator component of the Pol II holoenzyme (24, 35). The metazoan Mediator complexes are quite similar in their overall subunit composition and likely reflect the same cellular entity (hereafter referred to as TRAP/SMCC/Mediator) or its derivatives (31). Parallel studies have also identified many coactivators that possess intrinsic histone acetyltransferase (HAT) activity and are thought to be involved in facilitating transcription from chromatin templates (reviewed in references 4, 33, and 51). These include p300 and the closely related CBP, PCAF and the closely related GCN5, and members of the p160 family (SRC-1, GRIP1, TIF2, ACTR, and N-CoA) that have been implicated in nuclear receptor function. Whether the various coactivators function cooperatively or whether they represent distinct activation pathways remains unclear.
It was previously reported that part of the mechanism by which HNF-4 activates transcription includes physical interaction with TFIIB, which is a necessary but not sufficient condition for activation (30). Here we have focused on the coactivator requirements for HNF-4 function. We demonstrate that HNF-4 activity on DNA templates is critically dependent on TRAP/SMCC/Mediator via direct physical interactions, and we suggest that the mechanism might include a postrecruitment effect. We further show that HNF-4 function on chromatin templates also is TRAP/SMCC/Mediator dependent and stimulated by p300, in agreement with a synergistic mode of action of the two coactivators.
MATERIALS AND METHODS
Purification of transcription factors.
Pol II, GTFs, and PC4 were purified as described previously (27). Purification of recombinant HNF-4 involved affinity purification and ion-exchange chromatography as described previously (28, 30).
TRAP/SMCC/Mediator was purified from nuclear extract from HeLa cells stably expressing a FLAG-tagged NUT2 subunit (28). The extract was first fractionated over a phosphocellulose (P11) column, and the TRAP/SMCC/Mediator from the 0.5 M KCl eluate was then affinity purified over M2-agarose.
p300 was expressed in Sf9 cells via a baculovirus vector (gift of M. Guermah) carrying the full-length protein as a FLAG-tagged derivative, which was affinity purified from a whole-cell extract of infected cells.
Immunodepletion of TRAP/SMCC/Mediator from nuclear extract.
Anti-NUT2 antiserum (28) was purified by passage over immobilized antigen (recombinant, bacterially expressed, His-tagged human NUT2 covalently cross-linked to CNBr-activated Sepharose 4B from Pharmacia-Amersham). Bound antibodies were eluted with 100 mM glycine (pH 2.5) and 100 mM triethylamine (pH 11). The eluates containing antigen-purified anti-NUT2 antibodies were pooled, adsorbed onto protein A-Sepharose (Pharmacia-Amersham), and cross-linked with dimethylpimelidate (Sigma), as described previously (15). Following equilibration in BC200 buffer (10), the beads were incubated with HeLa cell nuclear extract for 10 h at 4°C. The supernatant was analyzed by immunoblotting and in transcription assays.
Chromatin assembly.
The chromatin assembly procedure was adapted from the method of Ito et al. (19). Drosophila ACF (consisting of the ISWI and Acf-1 subunits) was expressed in Sf9 cells via baculovirus vectors. Mouse NAP-1 was expressed in and purified from Escherichia coli. Typical assembly reaction mixtures contained 0.35 μg of plasmid pA4xMLΔ53 (29) and 0.35 μg of core histones (purified from HeLa cells, as described previously [23]) in 70-μl reaction volumes in the presence of NAP-1 and ACF. After incubation (4 h at 27°C) the assembled chromatin was analyzed by supercoiling and micrococcal nuclease (MNase) digestion assays (19, 23) and in transcription reactions.
In vitro transcription, electrophoretic mobility shift assay (EMSA), protein-protein interactions, and immobilized-template assays.
Transcription assays with purified components were performed essentially as described elsewhere (13, 27, 28). Transcription in nuclear extract was also as described previously (30), except that MgCl2 was added to a final concentration of 4 mM. For assays in which chromatin templates were used, reactions (with essentially the same final buffer composition as standard reactions) proceeded stepwise; ATP (100 μM) and acetyl-CoA (3 μM) were included from initial times. Following the HNF-4 binding and HAT steps (25 min, 30°C), nuclear extract (40 μg of protein) was added; after incubation for 40 min (at 30°C), nucleotide triphosphates were finally added to 500 μM (except for [α-32P]UTP [800 Ci/mmol], which was added at 5 μM). Twenty minutes into the transcription reaction, the UTP concentration was also raised to 100 μM, and incubation was continued for another 30 min before samples were processed for electrophoresis.
EMSA and GST protein-protein interaction assays were performed as described previously (27, 30). Briefly, for the GST interaction assays, approximately 5 μg of each GST fusion protein was immobilized on glutathione-Sepharose beads (10 μl) and incubated with the indicated material in 300-μl reaction volumes (containing 150 mM KCl in BC buffer [see reference 10]) for 90 min at 4°C. The beads were washed five to six times with BC buffer containing 150 mM KCl and 0.1% NP-40, except in the experiment shown in Fig. 5C, in which BC buffer containing 300 mM KCl and 0.1% NP-40 was used. The bound material was eluted with 0.2% Sarkosyl.
FIG. 5.
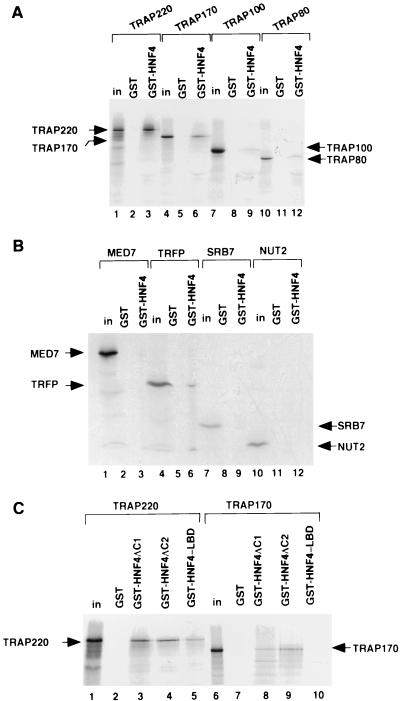
Identification of TRAP/SMCC/Mediator subunits that interact with HNF-4. (A) Selected subunits were expressed as 35S-labeled proteins in a rabbit reticulocyte coupled transcription and translation system, incubated with GST alone or with GST-HNF-4ΔC1 (as indicated) and processed as described in the legend to Fig. 4B. Lanes: 1 to 3, TRAP220; 4 to 6, TRAP170/RGR1; 7 to 9, TRAP100; 10 to 12, TRAP80. Inputs (in) representing 20% of the amount used in the binding reaction are also shown. (B) Additional (low-molecular-weight) selected subunits were expressed as 35S-labeled proteins in a rabbit reticulocyte coupled transcription and translation system, incubated with GST alone or with GST-HNF-4ΔC1 (as indicated), and processed as described in the legend to Fig. 4B. Lanes: 1 to 3, MED7; 4 to 6, TRFP; 7 to 9, SRB7; 10 to 12, NUT2. (C) HNF-4 AF-2-dependent and AF-2-independent interactions with TRAP/SMCC/Mediator subunits. 35S-labeled TRAP220 (lanes 1 to 5) and TRAP170/RGR1 (lanes 6 to 10) were incubated with GST alone (lanes 2 and 7), GST-HNF-4ΔC1 (lanes 3 and 8), GST-HNF-4ΔC2 (lanes 4 and 9), or GST-HNF-4-LBD (lanes 5 and 10) and processed as described in the legend to Fig. 4B.
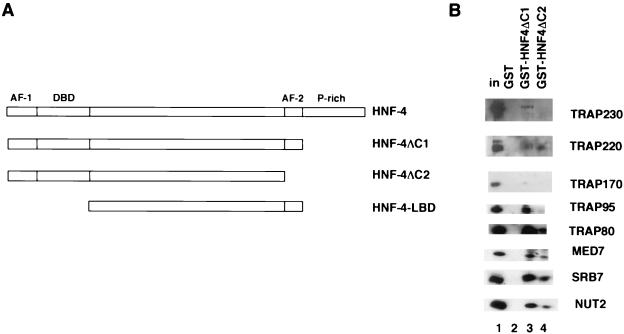
The HNF-4 GST derivatives, GST-HNF-4ΔC1 and GST-HNF-4ΔC2, were constructed by subcloning the NdeI-BamHI fragments from the corresponding pET11d-6His constructs, which were previously reported (30), into a pGEX (Pharmacia) vector that had been modified to carry NdeI and BamHI cloning sites. This vector was also used for generation of GST-HNF-4-LBD, for which the insert (corresponding to amino acid residues 139 to 380 [43]) was amplified by PCR from the full-length cDNA.
For TRAP/SMCC/Mediator subunit interactions, the indicated subunits were expressed in vitro in the TNT expression system (Promega), following the directions of the manufacturer. For this purpose, previously published (13, 18, 52) cDNAs of subunits TRAP220, TRAP100, and TRAP80 were subcloned into a modified plasmid pCI-neo (Promega) containing a T7 promoter. TRAP170/RGR1 (13) was subcloned into the plasmid pcDNA3.1 (Invitrogen). NUT2 (28) was expressed from plasmid pET11d-6His. TRFP was expressed from the original pRSET vector (50). For MED7, a full-length cDNA was first obtained as an expressed sequence tag (IMAGE 2068605) from the American Type Culture Collection, completely sequenced, and then subcloned into a pET vector.
For immobilized-template assays, an EcoRI-NdeI fragment (ca. 600 bp) from plasmid pA4xMLΔ53 (29, 30), which contains the HNF-4 cognate sites and the core promoter elements, was filled in with biotinylated dATP by using the Klenow fragment of DNA Pol I, gel purified, and bound to M280-streptavidin Dynabeads (Dynal), as suggested by the manufacturer. For the assay, the reactions were scaled up ca. 15-fold relative to a standard transcription reaction. The immobilized templates were washed in Tris-EDTA buffer containing 1 M NaCl, 0.5 mg of bovine serum albumin (BSA) per ml, and 0.003% NP-40 and then blocked in transcription buffer containing 5 mg of BSA per ml, 5 mg of polyvinylpyrrolidone per ml, and 0.003% NP-40 for 15 min (53). After being washed in transcription buffer containing 0.25 mg of BSA per ml and 0.025% NP-40, the immobilized templates were incubated with the indicated transcription factors in transcription buffer containing 100 μg of poly(dG-dC) per ml to allow preinitiation complex (PIC) formation. Beads were washed with transcription buffer containing 0.25 mg of BSA per ml and 0.025% NP-40. The beads were then exposed to promoterless pBluescript SK plasmid DNA (100 ng per reaction) in transcription wash buffer and washed an additional two times. The bound material was eluted by boiling in sodium dodecyl sulfate (SDS) sample buffer and analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) and immunoblotting.
RESULTS
TRAP/SMCC/Mediator-dependent function of HNF-4 in an in vitro system reconstituted with highly purified transcription factors.
AF-2-dependent activation function of HNF-4 in an unfractionated nuclear extract from HeLa cells (30) and in a purified system dependent on USA-derived cofactors PC2, PC3, and PC4 (28) was previously demonstrated. Whereas it was previously shown that PC2 is a bona fide Mediator-like complex (28), it was also found that several subunits associated with the larger, canonical metazoan complex, TRAP/SMCC/Mediator, were lacking in PC2. We therefore began our present study by including canonical TRAP/SMCC/Mediator in our analysis. Thus, our in vitro transcription system was reconstituted with recombinant TFIIA, TFIIB, TFIIE, TFIIF, and PC4; immunoaffinity-purified TFIID; and near-homogeneous preparations of TFIIH and Pol II (see Materials and Methods). We monitored HNF-4-dependent transcription from a plasmid template (pA4xMLΔ53) that contains four copies of an HNF-4 cognate site (site A from the apolipoprotein AI gene liver-specific enhancer) upstream of core promoter sequences from the adenovirus major late promoter (29, 30). A control template (pG5HML) containing five GAL4 binding sites upstream of a hybrid core promoter was also included to monitor activation by activation domains fused to the GAL4 DNA binding domain (11, 27, 28).
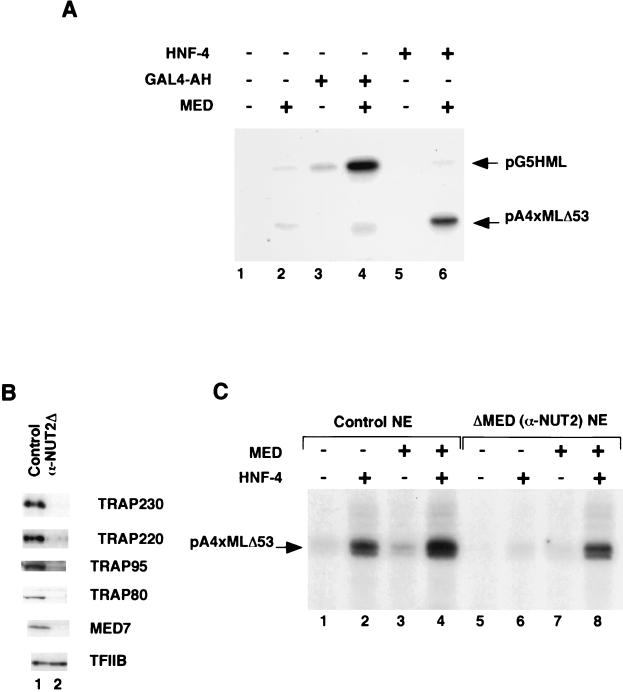
As previously shown, transcription in this system is both activator- and coactivator-dependent (Fig. 1A, lane 1). Thus, addition of HNF-4 (lane 5) or the control activator GAL4-AH (lane 3) to the reactions in the absence of TRAP/SMCC/Mediator elicited very weak transcription from the cognate template. Similarly, in the absence of an activator, TRAP/SMCC/Mediator stimulated transcription only marginally under these conditions (lane 2). However, together HNF-4 and TRAP/SMCC/Mediator effected significant stimulation of transcription from the HNF-4 cognate template but not from the control template (lane 6). As expected, the control template was activated by GAL4-AH in the presence of TRAP/SMCC/Mediator (lane 4). These results clearly establish that activation by HNF-4 in a reconstituted transcription system is dependent on TRAP/SMCC/Mediator.
FIG. 1.
TRAP/SMCC/Mediator-dependent transcriptional activation of DNA templates by HNF-4. (A) TRAP/SMCC/Mediator-dependent function of HNF-4 in a transcription system reconstituted from purified factors. In vitro transcription reaction mixtures were reconstituted with 50 ng of TFIIA, 10 ng of TFIIB, 10 ng of TFIIEα, 5 ng of TFIIEβ, 25 ng of TFIIF, 20 ng of TFIIH, 50 ng of Pol II, 150 ng of PC4, and an amount of affinity-purified TFIID containing 5 μg of TBP. GAL4-AH (25 ng) was added to reaction mixtures in lanes 3 and 4; HNF-4 (50 ng) was added to reaction mixtures in lanes 5 and 6. Purified TRAP/SMCC/Mediator (f: NUT2) was included in reaction mixtures in lanes 2, 4, and 6. Reaction mixtures also contained 50 ng each of the templates pG5HML and pA4xMLΔ53. (B) Depletion of TRAP/SMCC/Mediator from HeLa nuclear extract. HeLa nuclear extract was incubated with control beads (lane 1) or with beads containing cross-linked anti-NUT2 antibodies (lane 2). Unbound extract was immunoblotted with indicated antibodies. (C) TRAP/SMCC/Mediator-dependent function of HNF-4 in nuclear extract. In vitro transcription reaction mixtures contained a control nuclear extract (lanes 1 to 4) or TRAP/SMCC/Mediator-depleted extract (lanes 5 to 8). HNF-4 was added to reaction mixtures in lanes 2, 4, 6, and 8. Purified TRAP/SMCC/Mediator was added to reaction mixtures in lanes 3, 4, 7, and 8.
TRAP/SMCC/Mediator-dependent activator function of HNF-4 in nuclear extract.
In view of the potential concern that our reconstituted system may have somehow introduced a dependence of HNF-4 on TRAP/SMCC/Mediator, we also devised a procedure to assess its role in HNF-4 function in the context of an unfractionated nuclear extract that presumably contains the normal cellular complement of both positively and negatively acting nuclear factors. For this purpose, we immunodepleted HeLa cell nuclear extract by passage over a column that contained immobilized antibodies directed against the NUT2 subunit of the TRAP/SMCC/Mediator complex (Fig. 1B). Immunoblot analysis with selected TRAP/SMCC/Mediator antibodies revealed that, relative to a mock-treated extract, this treatment resulted in the quantitative removal of TRAP/SMCC/Mediator (lane 2 versus lane 1). Thus, selected (representative) TRAP/SMCC/Mediator subunits (e.g., TRAP230, TRAP220, TRAP95, TRAP80, and MED7) were undetectable after this treatment. Consistent with the previous isolation of TRAP/SMCC/Mediator in a form that was largely free of Pol II and GTFs (13), these factors were not removed from the extract in detectable amounts (Fig. 1B, lane 2, and data not shown).
We next tested the TRAP/SMCC/Mediator-depleted (here designated ΔMED) nuclear extract for the ability to support activation by HNF-4 (Fig. 1C). As before (30), a control (mock-depleted) extract supported efficient activation by HNF-4 (lane 2 versus lane 1). This activity was only slightly stimulated when the endogenous TRAP/SMCC/Mediator was supplemented with highly purified TRAP/SMCC/Mediator (lane 4 versus lane 2), indicating that the extract is not generally limiting for this coactivator. By contrast, in the ΔMED nuclear extract, the level of HNF-4 activation was reduced to barely detectable levels (lane 6 versus lane 2). Addition of purified TRAP/SMCC/Mediator to the ΔMED nuclear extract restored activity essentially to control levels (compare lanes 8, 6, and 2). This indicates that the observed reduction in HNF-4 activity was due solely to the removal of TRAP/SMCC/Mediator from the extract and, hence, that HNF-4 function is absolutely dependent on TRAP/SMCC/Mediator. Note that there also was a TRAP/SMCC/Mediator-dependent diminution in the levels of basal transcription (i.e., in the absence of activator; compare lanes 7, 5, and 1), consistent with the emerging role of Mediator in this process, especially under more-physiological conditions (2, 34). (Under the conditions of the assay, basal transcription from the much weaker pG5HML core promoter is not detectable.) Together, these functional data unambiguously identify TRAP/SMCC/Mediator as a key coactivator for HNF-4.
HNF-4 function on chromatin templates is dependent on TRAP/SMCC/Mediator and is stimulated by p300.
In the cell, chromatin constitutes the natural target of the transcription apparatus. Among the coactivators that are thought to act primarily at the level of chromatin, CBP, which possesses a HAT activity, has been shown to interact with HNF-4 and to stimulate HNF-4-dependent activation in in vivo transfection assays (6, 45). As many nuclear receptors, in particular the thyroid hormone receptor, have now been shown to utilize both HAT and TRAP/SMCC/Mediator coactivators via interactions with the AF-2 domains of the receptors, it has been suggested that they function sequentially to activate transcription (9, 31, 40). Thus, HAT coactivators (possibly in conjunction with other chromatin-remodeling activities) are proposed to first make the template more accessible and then be followed by TRAP/SMCC/Mediator effects on the formation and function of the PIC. The model thus predicts a synergistic mode of action for the two kinds of activators.
Having demonstrated a clear TRAP/SMCC/Mediator requirement for HNF-4 function on DNA templates, we then examined coactivator requirements for HNF-4 on chromatin templates. Specifically, we tested whether, as predicted from the preceding model, HNF-4-dependent transcription from chromatin jointly requires HAT and TRAP/SMCC/Mediator coactivators.
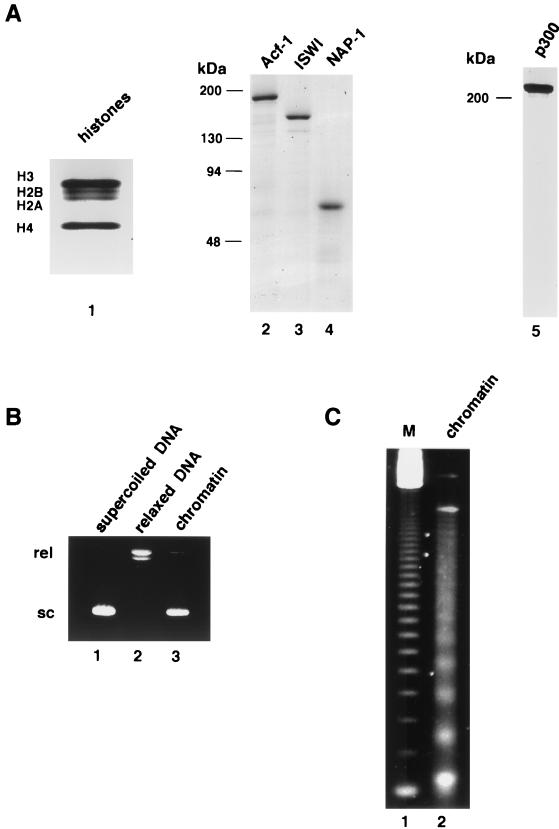
For this purpose, we first assembled plasmid pA4xMLΔ53 into chromatin by using the newly described method that utilizes the ISWI-containing chromatin assembly factor ACF and the chaperone NAP-1 to deposit HeLa cell histones into properly positioned nucleosomes (19). The purified components used for the assembly reaction are shown in Fig. 2A. Following assembly, the chromatinized plasmid was first assayed for superhelicity (Fig. 2B) to ensure completion of the reaction. The appearance of a large proportion of the high-mobility supercoiled form of the plasmid relative to the relaxed form, together with the absence of any intermediate forms (lane 3), indicated that the assembly reaction was essentially complete. Importantly, micrococcal nuclease digestion of the chromatinized template yielded five to six distinct bands (Fig. 2C, lane 2), thus indicating regular nucleosome positioning and the suitability of the template for in vitro transcription assays.
FIG. 2.
Reconstitution of chromatin on templates containing HNF-4 cognate sites. (A) Purified preparations of the indicated proteins used for reconstituting chromatin on plasmid pA4xMLΔ53 were analyzed by SDS-PAGE and stained with Coomassie brilliant blue (lanes 1 and 5) or silver (lanes 2 to 4). Lane 1, HeLa cell histones, 1.5 μg; lane 2, baculovirus-expressed Acf-1, 200 ng; lane 3, baculovirus-expressed ISWI, 200 ng; lane 4, bacterially expressed NAP-1, 200 ng; lane 5, baculovirus-expressed p300, 1 μg. (B) DNA supercoiling assay for chromatin assembly on plasmid pA4xMLΔ53. Plasmid DNA, treated as described, was electrophoresed on agarose gels and stained with ethidium bromide. Lane 1, purified plasmid prior to assembly; lane 2, topoisomerase I-relaxed plasmid; lane 3, plasmid after chromatin assembly (and deproteinization). Supercoiled (sc) and relaxed (rel) DNAs are marked. (C) MNase digestion assay for chromatin assembly. After assembly into chromatin, plasmid pA4xMLΔ53 was digested with MNase, deproteinized, and analyzed by agarose gel electrophoresis. A 123-bp ladder (M) was used as a size marker.
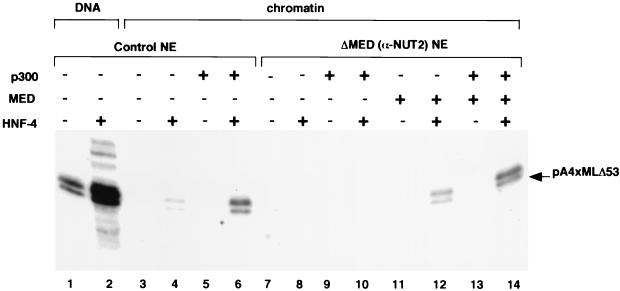
A step-wise protocol was used for in vitro transcription reactions from chromatin (Fig. 3). The protocol entailed binding of HNF-4 to the chromatin template, followed by preincubation with a HAT in the presence of acetyl-CoA substrate, and, finally, transcription with unfractionated nuclear extract. As a HAT, we used p300, which is highly homologous and functionally similar to CBP (4, 51). As expected, and in contrast to the situation with a DNA template (above), the transcription levels from the chromatinized template were severely repressed both in the absence and in the presence of HNF-4 (lanes 3 and 4). However, preincubation with p300 led to significant transcription in the presence of HNF-4 (lane 6) but had little effect in the absence of HNF-4 (lane 5). The residual transcription seen with HNF-4 in the absence of p300 might be attributed to endogenous p300 in the nuclear extract (lane 4). These results establish that HNF-4 and p300 can function together to effect transcription from chromatin templates.
FIG. 3.
HNF-4 function on chromatin templates is dependent on TRAP/SMCC/Mediator and is stimulated by p300. In vitro transcription reactions contained chromatinized plasmid pA4xMLΔ53 (40 ng) and plasmid pG5HML (50 ng) as naked DNA (lanes 3 to 14). The templates were preincubated with HNF-4 (50 ng, lanes 2, 4, 6, 8, 10, 12, and 14) and p300 (200 ng, lanes 5, 6, 9, 10, 13, and 14) for 25 min. Nuclear extract (control, lanes 1 to 6; or TRAP/SMCC/Mediator-depleted [ΔMED], lanes 7 to 14) was then added, and incubation continued for 20 min. To reaction mixtures in lanes 12 and 14, purified TRAP/SMCC/Mediator was also added at this time. Labeled nucleotide triphosphate mix was then added, and after 30 min, the samples were processed for analysis by electrophoresis. Reaction mixtures in lanes 1 and 2 contained naked pA4xMLΔ53 as a control.
To determine if TRAP/SMCC/Mediator (in the extract) also contributes to HNF-4- and p300-dependent transcription from chromatin, we also used ΔMED nuclear extract (lanes 7 to 14) that could be exogenously supplemented with purified TRAP/SMCC/Mediator. In agreement with the above results with the DNA template, ΔMED nuclear extract was unable to support HNF-4-dependent transcription from the repressed chromatin template (lanes 7 to 9). Also, in contrast to the result with control nuclear extract, preincubation with p300 was insufficient to evoke any HNF-4 dependent transcription (lanes 9 and 10). However, when supplemented with purified TRAP/SMCC/Mediator, and following preincubation with p300, the levels of HNF-4-dependent transcription were equivalent to those seen with control extract (lane 14 versus lanes 10 and 6). (Again, residual transcription without p300 preincubation likely is the result of endogenous p300 [lane 12].)
Thus, our results also establish a role for TRAP/SMCC/Mediator in HNF-4 function from the more physiological chromatin templates. Furthermore, and of equal significance, they also reveal a cooperativity between HNF-4, p300, and TRAP/SMCC/Mediator in transcription activation from such templates (see Discussion).
Direct physical interactions of HNF-4 with Mediator.
To further understand the molecular basis of the TRAP/SMCC/Mediator dependence for HNF-4 function, we asked if there were direct physical interactions between the two proteins. For this purpose we constructed two glutathione S-transferase (GST) fusion derivatives of HNF-4. GST-HNF-4ΔC1 contained a version of HNF-4 that lacks the C-terminal proline-rich domain, which as previously shown (30) has no effect on transcription in vitro (Fig. 4A). GST-HNF-4ΔC2 is essentially the same except for the absence of the AF-2 domain. Incubation of purified TRAP/SMCC/Mediator with GST-HNF-4ΔC1 resulted in substantial retention of TRAP/SMCC/Mediator polypeptides, which were scored by immunoblot analysis (Fig. 4B, lane 3). By contrast, no retention of TRAP/SMCC/Mediator polypeptides on control GST beads was detectable (lane 2). On GST-HNF-4ΔC2 (lane 4), the amount of retained TRAP/SMCC/Mediator polypeptides was significantly reduced albeit not abolished. These results indicate that HNF-4 can interact with TRAP/SMCC/Mediator primarily through its AF-2 domain and that secondary (stabilizing) interactions might be mediated through other regions on HNF-4 (see below).
FIG. 4.
Direct physical interaction between HNF-4 and TRAP/SMCC/Mediator. (A) Schematic representation of full-length HNF-4 and various derivatives used in GST interaction assays (see text for details). Relevant features only are shown. DBD, DNA binding domain; P-rich, proline-rich; AF-1 and AF-2, activation functions 1 and 2, respectively. (B) Purified TRAP/SMCC/Mediator (10% input, lane 1) was incubated with glutathione-Sepharose beads containing GST (lane 2), GST-HNF-4ΔC1 (lane 3), or GST-HNF-4ΔC2 (lane 4). After being washed to remove unbound material, the resins were eluted with Sarkosyl and analyzed by immunoblotting following SDS-PAGE.
To identify the TRAP/SMCC/Mediator subunit(s) responsible for this interaction, we expressed selected TRAP/SMCC/Mediator polypeptides as 35S-labeled proteins in rabbit reticulocyte lysates and assessed their ability to interact with GST-HNF-4ΔC1 (Fig. 5A, B). Of the polypeptides tested, TRAP220 displayed the strongest interaction with GST-HNF-4ΔC1 (Fig. 5A, lanes 1 to 3). A weaker, but significant, interaction with TRAP170/RGR1 was also observed (Fig. 5A, lanes 4 to 6). MED7, SRB7, and NUT2 did not show detectable interactions in this assay (Fig. 5B); very weak interactions displayed by TRAP100, TRAP80, and TRFP appear to be potentially negligible (Fig. 5A and B).
In further analysis, we focused on the regions in HNF-4 that interact with the TRAP220 and TRAP170/RGR1 subunits of TRAP/SMCC/Mediator (Fig. 5C). For this experiment, in addition to the two HNF-4 derivatives (GST-HNF-4ΔC1 and GST-HNF-4ΔC2) described above, we included a derivative (GST-HNF-4-LBD; Fig. 4A) in which only a fragment (the presumptive ligand binding domain [LBD]) of HNF-4 located between the DNA binding and the proline-rich domains (and containing the AF-2 domain) has been fused to GST. Also, to ensure that we observed bona fide interactions, we performed this experiment under relatively stringent conditions (beads were washed at 300 mM KCl). The results reveal that TRAP220 specifically interacted with all three derivatives of HNF-4 (Fig. 5C, lanes 1 to 5) indicating that removal of either the N-terminal region (containing the putative AF-1 and DNA binding domains, lane 5 versus lane 2) alone or the AF-2 alone (lane 4 versus lane 3) had little effect on its interaction with HNF-4. This implies that there is more than one site of TRAP220 interaction on HNF-4. Because the LBD region contains the AF-2 domain as its prominent feature and in view of the analogy with other nuclear receptors (42, 43), it is likely that the AF-2 is one of the sites of interaction with the TRAP220 subunit. Another site might be located in the N-terminal region. By contrast, TRAP170/RGR1 specifically interacted with GST-HNF-4ΔC1 and GST-HNF-4ΔC2 but not with GST-HNF-4-LBD, indicating that it predominantly interacts with the N-terminal region. Taken together with the data in Fig. 4B, these results indicate that HNF-4 likely interacts with TRAP/SMCC/Mediator through at least two subunits.
Stabilization of preinitiation complex formation by HNF-4 and PC4.
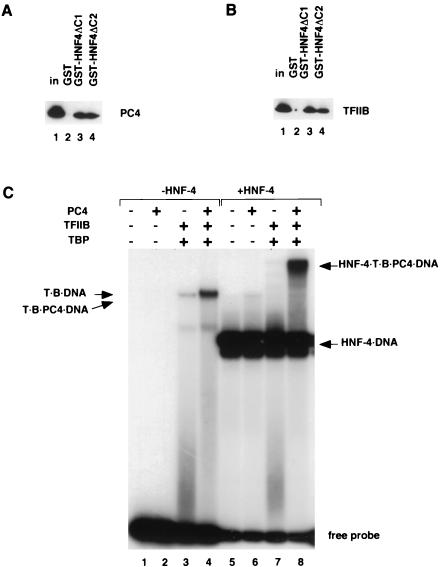
Assembly of the PIC at promoter sites is thought to proceed via concerted interactions between the GTFs (39). It was previously suggested that this step may be facilitated by the USA-derived coactivator PC4 (27), which also interacts with a number of activation domains (11). Based on our previously observed functional synergy between TRAP/SMCC/Mediator and PC4, we have further proposed that PC4 might provide an architectural function in stabilizing the PIC through multiple factor contacts, whereas TRAP/SMCC/Mediator might function as the coactivator per se (13, 28, 31). To investigate if PC4 contributes to HNF-4 function, we first determined if there was a physical interaction between them. We incubated purified PC4 with GST-HNF-4ΔC1 and GST-HNF-4ΔC2 and looked for retention of PC4 by immunoblotting (Fig. 6A). Efficient retention of PC4 was seen with both HNF-4 derivatives, indicative of strong interactions that, in contrast to what was observed for the TRAP/SMCC/Mediator (Fig. 4B), did not show a preference for the AF-2 domain. Consistent with previous results (30), a control experiment with TFIIB also showed similar AF-2-independent binding (Fig. 6B).
FIG. 6.
PC4-dependent stabilization of PIC. (A) Purified recombinant PC4 (25% input, lane 1) was incubated with glutathione-Sepharose beads containing GST (lane 2), GST-HNF-4ΔC1 (lane 3), or GST-HNF-4ΔC2 (lane 4). Samples were processed as described in the legend to Fig. 4B. Following SDS-PAGE, PC4 was detected by immunoblotting. (B) Purified recombinant TFIIB (25% input, lane 1) was incubated with glutathione-Sepharose beads containing GST (lane 2), GST-HNF-4ΔC1 (lane 3), or GST-HNF-4ΔC2 (lane 4). Samples were processed as described in the legend to Fig. 4B. Following SDS-PAGE, TFIIB was detected by immunoblotting. (C) For EMSA, an end-labeled probe consisting of site A and the apolipoprotein AI core promoter (29) was incubated with the indicated combinations of factors: PC4 (75 ng, lanes 2, 4, 6, and 8); HNF-4 (50 ng, lanes 5 to 8); TBP (T, 0.5 ng, lanes 3, 4, 7, and 8); TFIIB (B, 5 ng, lanes 4 and 8). Although the description of the PC4-dependent EMSA complexes implies that they contain PC4, it is currently not possible to rigorously demonstrate this fact. In a previous analysis (27), antibodies directed against PC4 failed to supershift the complex designated TBP-TFIIB-PC4 either because the PC4 epitopes are not accessible (by virtue of multiple contacts) or because PC4 association with the complex fails to survive electrophoresis. Nonetheless, we can show that PC4 is present in analogous complexes formed on immobilized templates (Fig. 7).
It was previously found that PC4 stabilizes TBP-TFIIB promoter complexes in EMSAs (27). Therefore, we asked if the interaction of HNF-4 and PC4 would have an additional stabilizing effect on the PIC. In this assay, TBP-TFIIB complex formation on a probe consisting of the HNF-4 cognate site (site A) fused upstream of the apolipoprotein AI gene promoter (30) was relatively inefficient (Fig. 6C, lane 3). However, consistent with previous results (27), inclusion of PC4 stabilized the TBP-TFIIB complex (without affecting its mobility) (Fig. 6C, lane 4; see also the legend to Fig. 6C for a note on complex composition). We also assessed the effect of PC4 on TBP-TFIIB complex formation in the presence of near-saturating amounts of HNF-4, which resulted in most of the probe being shifted (lanes 5 to 8). Under the conditions chosen for the assay, which included use of limiting amounts of TBP and TFIIB, a discrete band corresponding to a putative HNF-4-TBP-TFIIB complex was not seen (lane 7). However, upon inclusion of PC4 (which by itself has little effect on the HNF-4-bound probe, lane 6 versus lane 5), TBP and TFIIB gave rise to a slowly migrating band that likely corresponds to an HNF-4-TBP-TFIIB-PC4 complex (lane 8). A comparison of the intensities of the bands in lanes 8 and 4 indicates that the PC4-HNF-4 interaction might indeed contribute to a modest additional enhancement in the assembly and/or stabilization of the nascent PIC (see also below).
Recruitment of the transcription machinery to the PIC.
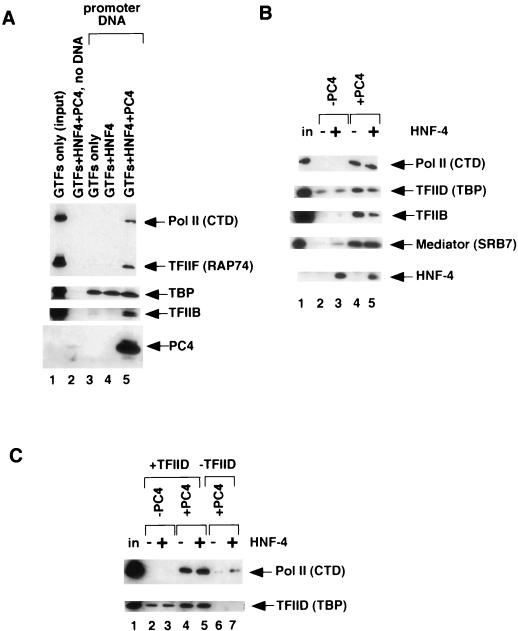
To dissect the interplay of various transcription factors in the context of higher-order PICs, we utilized an approach in which such complexes are assembled on immobilized templates and, subsequent to being washed, are analyzed for specifically bound (recruited) factors (1). For this purpose, a biotinylated template consisting of a DNA fragment carrying the regulatory regions (four copies of HNF-4 cognate sites [site A] and the adenovirus major late core promoter) from the plasmid pA4xMLΔ53 (above) was immobilized on M280-streptavidin paramagnetic beads. We first carried out control experiments (Fig. 7A) to establish the binding and washing conditions under which recruited higher-order PICs could be specifically and reproducibly detected by immunoblot analysis. When a mixture of Pol II, GTFs (TBP [in place of intact TFIID], TFIIB, TFIIE, TFIIF, and TFIIH), HNF-4, and PC4 was incubated with just the magnetic beads, only a negligible, background level of factor retention was evident (Fig. 7A, compare lane 2 with lane 1 [input]). However, when a mixture of Pol II and the GTFs was incubated with the immobilized promoter-containing template, a substantial amount of TBP (but not any of the other GTFs that were tested) was bound (lane 3), indicating that our conditions for the recruitment assay are both specific and selective. Since this complement of basal factors (Pol II plus GTFs) is sufficient for basal-level transcription (27), it was surprising that recruitment of the remaining GTFs was not evident. This result therefore also indicates that the present assay only scores strong interactions between factors or between factors and promoter DNA (e.g., in the case of TBP, Fig. 7A, lane 3). Moreover, this result is consistent with the previously established role of TBP in nucleating PIC assembly via its property of interacting with core promoter sequences (TATA elements) (39). Consistent with the results of the EMSA analysis (above), addition of HNF-4 to the Pol II and GTF mixture did not lead to the recruitment of any factors other than TBP (lane 4 versus lane 3). However, inclusion of PC4 triggered the formation of a higher-order complex that contained, in addition to PC4 and TBP (whose amount was not further enhanced relative to the reaction with no PC4), other GTFs (including TFIIB) and Pol II (lane 5; here shown only with HNF-4; see Fig. 7B for results comparing the effects in the absence and presence of HNF-4).
FIG. 7.
Recruitment of the transcription machinery into the PIC. (A) Complexes were assembled on M280-streptavidin Dynabeads carrying a biotinylated DNA fragment from the plasmid pA4xMLΔ53, which contained four copies of the HNF-4 cognate site A and the adenovirus major late core promoter. After PIC formation, beads were washed and bound complexes were analyzed for their factor content by immunoblotting. PICs were formed with GTFs only (TBP, TFIIB, TFIIE, TFIIF, TFIIH, Pol II (lane 3); with GTFs and HNF-4 (lane 4); or with GTFs, HNF-4, and PC4 (lanes 2 and 5). Lane 2, control in which the indicated factors were incubated with beads only (no template DNA); lane 1, GTFs only (input). (B) Immobilized-template recruitment assays are as for panel A, except that all reaction mixtures contained TFIIA, TFIIB, TFIID, TFIIE, TFIIF, TFIIH, Pol II, and TRAP/SMCC/Mediator, as shown in input lane 1. HNF-4 (lanes 3 and 5) and PC4 (lanes 4 and 5) were additionally added as indicated. (C) Immobilized-template recruitment assays are as for panel B, except that reaction mixtures in lanes 6 and 7 did not contain TFIID. HNF-4 (lanes 3, 5, and 7) and PC4 (lanes 4 to 7) were additionally added as indicated.
Next, we assessed the effects of HNF-4 and PC4 on the recruitment to immobilized templates of a complete set of purified transcription factors (including Pol II, TFIIA, TFIIB, TFIID, TFIIE, TFIIF, TFIIH, and TRAP/SMCC/Mediator) that are normally used for activator- and coactivator-dependent transcription assays (as in Fig. 1A). In the absence of HNF-4 and PC4, only TBP (presumably as part of TFIID) binding to the template was detected (Fig. 7B, lane 2). Addition of HNF-4 to the binding reaction had no additional dramatic effects, although there was a slight (circa twofold) HNF-4-dependent increase in the amount of TRAP/SMCC/Mediator polypeptides (lane 3 versus lane 2; SRB7). By contrast, addition of PC4 again led to the simultaneous recruitment of all the PIC components, including the GTFs and Pol II (lane 4). Recruitment of TRAP/SMCC/Mediator polypeptides to the template was also significantly enhanced by PC4 (lane 4 versus lane 2). However, as in panel A, no significant effects of PC4 on binding of TBP (TFIID) were discernible. Surprisingly, further addition of HNF-4 (lane 5) was essentially without any extra effects on the profile of recruited factors (besides HNF-4 itself).
As a further control to ensure that the transcription factors bound to the immobilized templates reflect bona fide (specific) complexes, we monitored the ability of Pol II to be recruited to the test template in the absence of TFIID (whose TBP subunit provides the key DNA contacts that nucleate the PIC) (Fig. 7C). Once again, we examined Pol II recruitment in the absence and presence of PC4 and HNF-4. Whereas TFIID (TBP) bound to the template regardless of whether PC4 was present or not (lanes 2 to 4), recruitment of Pol II was critically dependent on PC4 and not further enhanced by HNF-4 (lanes 5 and 6 versus lanes 2 and 3). Importantly, when TFIID was omitted from the binding reaction (lanes 6 and 7), the amount of Pol II recruited to the templates was dramatically reduced (compare lane 6 versus lane 4 and lane 7 versus lane 5), strongly suggesting the specific nature of the observed complexes. Residual binding of Pol II to the template (which is slightly enhanced in the presence of HNF-4, lane 7 versus lane 6) may reflect weaker interactions with other factors bound to the template. Together the results with the immobilized templates indicate a dominant role of PC4 in PIC assembly and/or stability and are in complete agreement with our suggestion that PC4, through multiple interactions with PIC components, fulfills an architectural role (27, 28).
In summary, our results show that activation by HNF-4 on DNA templates is critically dependent upon TRAP/SMCC/Mediator (Fig. 1) and that there is a potential for direct physical interaction between HNF-4 and TRAP/SMCC/Mediator (Fig. 4). Thus, the absence of HNF-4-dependent recruitment of TRAP/SMCC/Mediator to the PIC in immobilized-template assays (Fig. 7) further indicates that the primary mechanism for HNF-4-dependent transcriptional activation might not entail recruitment per se (see Discussion).
DISCUSSION
In this paper we have demonstrated the functional requirement of TRAP/SMCC/Mediator in HNF-4 function involving direct physical interactions between coactivator and the activator. Furthermore, we also demonstrate functional cooperativity between TRAP/SMCC/Mediator and the HAT coactivator p300 in promoting HNF-4 function from chromatin templates. Thus, this constitutes the first report that rigorously documents interplay between chromatin remodeling and Mediator-like coactivators for any activator.
A pathway for HNF-4-dependent transcription activation that involves multiple coactivators.
Our finding of an absolute dependence of HNF-4 activity on TRAP/SMCC/Mediator is in accord with the central importance of this coactivator in transcriptional activation (31). Further, our results showing cooperativity between TRAP/SMCC/Mediator and p300 in HNF-4 function lend credence to a previously formulated model whose main feature is the sequential mode of action of chromatin-remodeling coactivators and TRAP/SMCC/Mediator (9, 40).
Thus, we imagine the following pathway for the activation of chromatin templates by HNF-4. Initial steps could include HNF-4-dependent delivery of chromatin-remodeling factors to the promoter region. These could consist of both ATP-dependent remodelers as well as the HAT coactivators. Note that although we have not explicitly examined the function of ATP-dependent remodelers, our chromatin assembly system contains ACF, an ISWI-containing factor that, in addition to its assembly function, might also contribute to nucleosome repositioning during PIC formation and transcription (22). Hence, the chromatin-remodeling step would consist of both nucleosome repositioning and acetylation of histone tails, events that overall would be favorable for activated transcription (22). It should be noted that, even though we have focused here on p300 as a model HAT, additional equally suitable candidates which could fulfill this role (or work in synergy with p300 [7]) include members of the SRC-1/p160 family of coactivators such as GRIP1. GRIP1 has been shown to modestly facilitate HNF-4 function in transfection assays (49). Subsequent to making the promoter region more accessible, chromatin-remodeling coactivators are predicted to yield to TRAP/SMCC/Mediator, which also requires the AF-2 domain of HNF-4 for strong interactions (Fig. 4B; see also below). TRAP/SMCC/Mediator, which unlike HAT coactivators can also function on DNA templates, would then be involved in transducing the activation signal to the Pol II and associated general transcription machinery (below). Additional coactivators could also potentially be involved at this step. Specifically, based on the persistent requirement of PC4 in our transcription system reconstituted from purified factors (13, 27, 28) and the demonstration here that PC4 plays a dominant role in stabilizing the PIC, it is also likely that some degree of support would come from PC4 (and perhaps other architectural cofactors as well). On the other hand, because we were unable to analyze the effect of PC4 in our chromatin assays (which were carried out in unfractionated extract), we cannot rule out the possibility that in the context of chromatin, PC4 is dispensable. Finally, additional direct interactions with the basal transcription machinery, in particular with TFIIB (30), might also contribute. We note in this regard that HNF-4 typically binds to its cognate site as a homodimer (30) and that the N-terminal AF-1-like domain and the AF-2 domain would present distinct surfaces for interactions with multiple appropriate targets.
Potential postrecruitment effects of HNF-4 and TRAP/SMCC/Mediator on Pol II PIC.
How might interaction of HNF-4 with TRAP/SMCC/Mediator lead to activated transcription? Based on the ability of the complex to interact both with specific activators (reviewed in reference 31) and with Pol II (especially in yeast, where the Mediator originally was isolated as part of the holoenzyme [24, 35]), it was previously suggested that the TRAP/SMCC/Mediator might function as an adaptor linking activators with the basal transcription machinery (13, 19, 31). Indeed, it is believed that recruitment of the Mediator-containing Pol II holoenzyme to the promoter by activators is the dominant mechanism by which they exert their effect (37).
However, the data presented here, while not proving the point, are more consistent with an alternative model in which the functional manifestation of the interaction between the activator (HNF-4) and TRAP/SMCC/Mediator is at a later (postrecruitment) step. The immobilized-template experiments (Fig. 7) reveal that, despite the clear potential for interaction between HNF-4 and TRAP/SMCC/Mediator, recruitment (at least as determined by this assay) is not commensurate with the extent of HNF-4- and TRAP/SMCC/Mediator-dependent activation that is typically seen in the functional assays. But, perhaps more importantly, even this low-level HNF-4-dependent TRAP/SMCC/Mediator recruitment is totally eclipsed by the ability of PC4 to efficiently facilitate formation of a complete PIC. Furthermore, the observation from functional assays both here (Fig. 1C) and elsewhere (2, 34) that TRAP/SMCC/Mediator contributes to basal (activator-independent) transcription also implies that it possesses an intrinsic (activator-independent) capacity to be recruited to the PIC. Therefore, the fact that HNF-4 and TRAP/SMCC/Mediator are nonetheless required for transcription only implies that their effects become critical at a stage subsequent to the recruitment of Pol II and GTFs.
Furthermore, even though our results emphasize the AF-2/LBD-dependent interaction between HNF-4 and TRAP/SMCC/Mediator, similar to what has been described for the thyroid hormone receptor (9, 52) and the vitamin D receptor (38), we do detect interactions of HNF-4 and TRAP/SMCC/Mediator that are not exclusively dependent on AF-2/LBD. This could mean that there is a secondary potential (residing either in the putative AF-1 or the DNA binding domains) to interact with TRAP/SMCC/Mediator. It is possible that this residual AF-2-independent potential to interact with HNF-4 is an evolutionary relic because the C. elegans ortholog of HNF-4 apparently does not possess an AF-2 domain (44). Possibly related is the fact that there is no C. elegans ortholog of TRAP220, which is shown here to interact with the AF-2 domain of mammalian HNF-4. By comparison, C. elegans does contain an ortholog of TRAP170/RGR1, which interacts with HNF-4 in an AF-2-independent manner, in accord with a similar observation for the glucocorticoid receptor (17). Since it was previously established that the AF-2 is important for activation by mammalian HNF-4 in vitro (30), these considerations also point to postrecruitment effects of HNF-4 and TRAP/SMCC/Mediator in the activation pathway. Thus, it also is possible that, in addition to the HNF-4 and TRAP/SMCC/Mediator contacts that simply tether them together in the PIC, the AF-2 requirement of mammalian HNF-4 reflects a critical AF-2-dependent transition (e.g., one that ultimately leads to acquisition by Pol II of an active conformation). These conclusions regarding postrecruitment effects of a transcriptional activator (and Mediator) are further supported by evidence from other lines of inquiry. For example, in yeast, recruitment of the ΔSRB5 mutant Mediator-containing holoenzyme by an activator is not sufficient for activation (25). Indeed, in in vitro template recruitment assays, the effects of activators in recruiting PIC components are relatively modest (53). In the end, extensive future studies will be needed to dissect the relative contribution of postrecruitment effects of activators and coactivators toward activated transcription.
Acknowledgments
We thank M. Guermah for a p300 expression vector, M. Ito, S. Yamamura, and C. X. Yuan for antibodies, W. An for helpful discussions, and W. Wu for technical assistance.
A.E.W was supported by the Wenner-Gren Foundations and the Swedish Cancer Society. This work was supported by NIH grants to R.G.R.
REFERENCES
- 1.Arias, J. A., and W. S. Dynan. 1989. Promoter-dependent transcription by RNA polymerase II using immobilized enzyme complexes. J. Biol. Chem. 264:3223-3229. [PubMed] [Google Scholar]
- 2.Baek, H. J., S. Malik, J. Qin, and R. G. Roeder. 2002. Requirement of TRAP/Mediator for both activator-independent and activator-dependent transcription in conjunction with TFIID TAFs. Mol. Cell. Biol. 22:2842-2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyer, T. G., M. E. Martin, E. Lees, R. P. Ricciardi, and A. J. Berk. 1999. Mammalian Srb/Mediator complex is targeted by adenovirus E1A protein. Nature 399:276-279. [DOI] [PubMed] [Google Scholar]
- 4.Brown, C. E., T. Lechner, L. Howe, and J. L. Workman. 2000. The many HATs of transcription coactivators. Trends Biochem. Sci. 25:15-19. [DOI] [PubMed] [Google Scholar]
- 5.Chen, W. S., K. Manova, D. C. Weinstein, S. A. Duncan, A. S. Plump, V. R. Prezioso, R. F. Bachvarova, and J. E. Darnell, Jr. 1994. Disruption of the HNF-4 gene, expressed in visceral endoderm, leads to cell death in embryonic ectoderm and impaired gastrulation of mouse embryos. Genes Dev. 8:2466-2477. [DOI] [PubMed] [Google Scholar]
- 6.Dell, H., and M. Hadzopoulou-Cladaras. 1999. CREB-binding protein is a transcriptional coactivator for hepatocyte nuclear factor-4 and enhances apolipoprotein gene expression. J. Biol. Chem. 274:9013-9021. [DOI] [PubMed] [Google Scholar]
- 7.Dilworth, F. J., C. Fromental-Ramain, K. Yamamoto, and P. Chambon. 2000. ATP-Driven chromatin remodeling activity and histone acetyltransferases act sequentially during transactivation by RAR/RXR in vitro. Mol. Cell 6:1049-1058. [DOI] [PubMed] [Google Scholar]
- 8.Fondell, J. D., H. Ge, and R. G. Roeder. 1996. Ligand induction of a transcriptionally active thyroid hormone receptor coactivator complex. Proc. Natl. Acad. Sci. USA 93:8329-8333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fondell, J. D., M. Guermah, S. Malik, and R. G. Roeder. 1999. Thyroid hormone receptor-associated proteins and general positive cofactors mediate thyroid hormone receptor function in the absence of TATA box-binding protein-associated factors. Proc. Natl. Acad. Sci. USA 96:1959-1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ge, H., E. Martinez, C. M. Chiang, and R. G. Roeder. 1996. Activator-dependent transcription by mammalian RNA polymerase II: in vitro reconstitution with general transcription factors and cofactors. Methods Enzymol. 274:57-71. [DOI] [PubMed] [Google Scholar]
- 11.Ge, H., and R. G. Roeder. 1994. Purification, cloning, and characterization of a human coactivator, PC4, that mediates transcriptional activation of class II genes. Cell 78:513-523. [DOI] [PubMed] [Google Scholar]
- 12.Green, V. J., E. Kokkotou, and J. A. Ladias. 1998. Critical structural elements and multitarget protein interactions of the transcriptional activator AF-1 of hepatocyte nuclear factor 4. J. Biol. Chem. 273:29950-29957. [DOI] [PubMed] [Google Scholar]
- 13.Gu, W., S. Malik, M. Ito, C.-X. Yuan, J. D. Fondell, X. Zhang, E. Martinez, J. Qin, and R. G. Roeder. 1999. A novel Human SRB/MED-containing complex, SMCC, involved in transcription regulation. Mol. Cell 3:97-108. [DOI] [PubMed] [Google Scholar]
- 14.Hadzopoulou-Cladaras, M., E. Kistanova, C. C. Evagelopoulou, S. Zeng, C. Cladaras, and J. A. Ladias. 1997. Functional domains of the nuclear receptor hepatocyte nuclear factor 4. J. Biol. Chem. 272:539-550. [DOI] [PubMed] [Google Scholar]
- 15.Harlow, E., and D. Lane. 1988. Antibodies: a laboratory manual. Cold Spring Harbor Press, Cold Spring Harbor, N.Y.
- 16.Hertz, R., J. Magenheim, I. Berman, and J. Bar-Tana. 1998. Fatty acyl-CoA thioesters are ligands of hepatic nuclear factor-4alpha. Nature 392:512-516. [DOI] [PubMed] [Google Scholar]
- 17.Hittelman, A. B., D. Burakov, J. A. Iniguez-Lluhi, L. P. Freedman, and M. J. Garabedian. 1999. Differential regulation of glucocorticoid receptor transcriptional activation via AF-1-associated proteins. EMBO J. 18:5380-5388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ito, M., C. X. Yuan, S. Malik, W. Gu, J. D. Fondell, S. Yamamura, Z. Y. Fu, X. Zhang, J. Qin, and R. G. Roeder. 1999. Identity between TRAP and SMCC complexes indicates novel pathways for the function of nuclear receptors and diverse mammalian activators. Mol. Cell 3:361-370. [DOI] [PubMed] [Google Scholar]
- 19.Ito, T., M. E. Levenstein, D. V. Fyodorov, A. K. Kutach, R. Kobayashi, and J. T. Kadonaga. 1999. ACF consists of two subunits, Acf-1 and ISWI, that function cooperatively in the ATP-dependent catalysis of chromatin assembly. Genes Dev. 13:1529-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang, Y.-W., P. Veschambre, H. Erdjument-Bromage, P. Tempst, J. W. Conaway, R. C. Conaway, and R. D. Kornberg. 1998. Mammalian mediator of transcriptional regulation and its possible role as an end-point of signal transduction pathways. Proc. Natl. Acad. Sci. USA 95:8538-8543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaiser, K., and M. Meisterernst. 1996. The human general co-factors. Trends Biochem. Sci. 21:342-345. [PubMed] [Google Scholar]
- 22.Kingston, R. E., and G. J. Narlikar. 1999. ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev. 13:2339-2352. [DOI] [PubMed] [Google Scholar]
- 23.Kundu, T. K., V. B. Palhan, Z. Wang, W. An, P. A. Cole, and R. G. Roeder. 2000. Activator-dependent transcription from chromatin in vitro involving targeted histone acetylation by p300. Mol. Cell 6:551-561. [DOI] [PubMed] [Google Scholar]
- 24.Lee, T. I., and R. A. Young. 2000. Transcription of eukaryotic protein-coding genes. Annu. Rev. Genet. 34:77-137. [DOI] [PubMed] [Google Scholar]
- 25.Lee, Y. C., J. M. Park, S. Min, S. J. Han, and Y. J. Kim. 1999. An activator binding module of yeast RNA polymerase II holoenzyme. Mol. Cell. Biol. 19:2967-2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li, J., G. Ning, and S. A. Duncan. 2000. Mammalian hepatocyte differentiation requires the transcription factor HNF-4alpha. Genes Dev. 14:464-474. [PMC free article] [PubMed] [Google Scholar]
- 27.Malik, S., M. Guermah, and R. G. Roeder. 1998. A dynamic model for PC4 coactivator function in RNA polymerase II transcription. Proc. Natl. Acad. Sci. USA 95:2192-2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malik, S., W. Gu, W. Wu, J. Qin, and R. G. Roeder. 2000. The USA-derived transcriptional coactivator PC2 is a submodule of TRAP/SMCC and acts synergistically with other PCs. Mol. Cell 5:753-760. [DOI] [PubMed] [Google Scholar]
- 29.Malik, S., and S. K. Karathanasis. 1995. Transcriptional activation by the orphan nuclear receptor ARP-1. Nucleic Acids Res. 23:1536-1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Malik, S., and S. K. Karathanasis. 1996. TFIIB-directed transcriptional activation by the orphan nuclear receptor hepatocyte nuclear factor 4. Mol. Cell. Biol. 16:1824-1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Malik, S., and R. G. Roeder. 2000. Transcriptional regulation through Mediator-like coactivators in yeast and metazoan cells. Trends Biochem. Sci. 25:277-283. [DOI] [PubMed] [Google Scholar]
- 32.Mangelsdorf, D. J., and R. M. Evans. 1995. The RXR heterodimers and orphan receptors. Cell 83:841-850. [DOI] [PubMed] [Google Scholar]
- 33.Martinez, E., V. B. Palhan, A. Tjernberg, E. S. Lymar, A. M. Gamper, T. K. Kundu, B. T. Chait, and R. G. Roeder. 2001. Human STAGA complex is a chromatin-acetylating transcription coactivator that interacts with pre-mRNA splicing and DNA damage-binding factors in vivo. Mol. Cell. Biol. 21:6782-6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mittler, G., E. Kremmer, H. T. Timmers, and M. Meisterernst. 2001. Novel critical role of a human Mediator complex for basal RNA polymerase II transcription. EMBO Rep. 2:808-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Myers, L. C., and R. D. Kornberg. 2000. Mediator of transcriptional regulation. Annu. Rev. Biochem. 69:729-749. [DOI] [PubMed] [Google Scholar]
- 36.Näär, A. M., P. A. Beaurang, S. Zhou, S. Abraham, W. Solomon, and R. Tjian. 1999. Composite co-activator ARC mediates chromatin-directed transcriptional activation. Nature 398:828-832. [DOI] [PubMed] [Google Scholar]
- 37.Ptashne, M., and A. Gann. 1997. Transcriptional activation by recruitment. Nature 386:569-577. [DOI] [PubMed] [Google Scholar]
- 38.Rachez, C., B. D. Lemon, Z. Suldan, V. Bromleigh, M. Gamble, A. M. Näär, J. Erdjument-Bromage, P. Tempst, and L. P. Freedman. 1999. Ligand-dependent transcription activation by nuclear receptors requires the DRIP complex. Nature 398:824-828. [DOI] [PubMed] [Google Scholar]
- 39.Roeder, R. G. 1996. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem. Sci. 21:327-335. [PubMed] [Google Scholar]
- 40.Roeder, R. G. 1998. Role of general and gene-specific cofactors in the regulation of eukaryotic transcription. CSH Symp. Quant. Biol. 68:201-218. [DOI] [PubMed] [Google Scholar]
- 41.Ryu, S., S. Zhou, A. G. Ladurner, and R. Tjian. 1999. The transcriptional cofactor complex CRSP is required for activity of the enhancer-binding protein Sp1. Nature 397:446-450. [DOI] [PubMed] [Google Scholar]
- 42.Sladek, F. M. 1993. Orphan receptor HNF-4 and liver-specific gene expression. Receptor 3:223-232. [PubMed] [Google Scholar]
- 43.Sladek, F. M., W. M. Zhong, E. Lai, and J. E. Darnell, Jr. 1990. Liver-enriched transcription factor HNF-4 is a novel member of the steroid hormone receptor superfamily. Genes Dev. 12:2353-2365. [DOI] [PubMed] [Google Scholar]
- 44.Sluder, A. E., and C. V. Maina. 2001. Nuclear receptors in nematodes: themes and variations. Trends Genet. 117:206-213. [DOI] [PubMed] [Google Scholar]
- 45.Soutoglou, E., N. Katrakili, and I. Talianidis. 2000. Acetylation regulates transcription factor activity at multiple levels. Mol. Cell 5:745-751. [DOI] [PubMed] [Google Scholar]
- 46.Sun, X., Y. Zhang, H. Cho, P. Rickert, E. Lees, W. Lane, and D. Reinberg. 1998. NAT, a human complex containing Srb polypeptides that functions as a negative regulator of activated transcription. Mol. Cell 2:213-222. [DOI] [PubMed] [Google Scholar]
- 47.Verrijzer, C. P., and R. Tjian. 1996. TAFs mediate transcriptional activation and promoter selectivity. Trends Biochem. Sci. 21:338-342. [PubMed] [Google Scholar]
- 48.Yamagata, K., H. Furuta, N. Oda, P. J. Kaisaki, S. Menzel, N. J. Cox, S. S. Fajans, S. Signorini, M. Stoffel, and G. I. Bell. 1996. Mutations in the hepatocyte nuclear factor-4alpha gene in maturity-onset diabetes of the young (MODY1). Nature 384:458-460. [DOI] [PubMed] [Google Scholar]
- 49.Wang, J. C., J. M. Stafford, and D. K. Granner. 1998. SRC-1 and GRIP1 coactivate transcription with hepatocyte nuclear factor 4. J. Biol. Chem. 273:30847-30850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xiao, H., Y. Tao, and R. G. Roeder. 1999. The human homologue of Drosophila TRF-proximal protein is associated with an RNA polymerase II-SRB complex. J. Biol. Chem. 274:3937-3940. [DOI] [PubMed] [Google Scholar]
- 51.Xu, L., C. K. Glass, and M. G. Rosenfeld. 1999. Coactivator and corepressor complexes in nuclear receptor function. Curr. Opin. Genet. Dev. 9:140-147. [DOI] [PubMed] [Google Scholar]
- 52.Yuan, C.-X., M. Ito, J. D. Fondell, Z.-Y. Fu, and R. G. Roeder. 1998. The TRAP220 component of a thyroid hormone receptor-associated protein (TRAP) coactivator complex interacts directly with nuclear receptors in a ligand-dependent fashion. Proc. Natl. Acad. Sci. USA 95:7939-7944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yudkovsky, N., J. Ranish, and S. Hahn. 2000. A transcription reinitiation intermediate that is stabilized by activator. Nature 408:225-229. [DOI] [PubMed] [Google Scholar]