Abstract
Wnt signaling maintains preadipocytes in an undifferentiated state. When Wnt signaling is enforced, 3T3-L1 preadipocytes no longer undergo adipocyte conversion in response to adipogenic medium. Here we used microarray analyses to identify subsets of genes whose expression is aberrant when differentiation is blocked through enforced Wnt signaling. Furthermore, we used the microarray data to identify potentially important adipocyte genes and chose one of these, the liver X receptor α (LXRα), for further analyses. Our studies indicate that enforced Wnt signaling blunts the changes in gene expression that correspond to mitotic clonal expansion, suggesting that Wnt signaling inhibits adipogenesis in part through dysregulation of the cell cycle. Experiments designed to uncover the potential role of LXRα in adipogenesis revealed that this transcription factor, unlike CCAAT/enhancer binding protein α and peroxisome proliferator-activated receptor gamma, is not adipogenic but rather inhibits adipogenesis if inappropriately expressed and activated. However, LXRα has several important roles in adipocyte function. Our studies show that this nuclear receptor increases basal glucose uptake and glycogen synthesis in 3T3-L1 adipocytes. In addition, LXRα increases cholesterol synthesis and release of nonesterified fatty acids. Finally, treatment of mice with an LXRα agonist results in increased serum levels of glycerol and nonesterified fatty acids, consistent with increased lipolysis within adipose tissue. These findings demonstrate new metabolic roles for LXRα and increase our understanding of adipogenesis.
Adipocytes play a central role in energy balance, both as a reservoir, storing and releasing fuel, and as endocrine cells, secreting factors that regulate whole-body energy metabolism (49). Understanding this cell type is becoming increasingly important because of the rising incidence of obesity and its associated disorder, type II diabetes. One widely used model for studying the adipocyte is the 3T3-L1 cell line, which was derived from disaggregated mouse embryos and selected based on the propensity of these cells to differentiate into adipocytes in culture (14). Over the last 30 years, this cell line has proven to be a faithful model for studying adipocyte biology, particularly adipogenesis and energy metabolism.
Numerous studies with 3T3-L1 cells and other in vitro models have led to a paradigm of adipocyte differentiation. Preadipocytes remain in an undifferentiated state due to autocrine Wnt signaling, which blocks adipogenic conversion (29, 44). The program of adipogenesis begins upon treatment of confluent preadipocytes with adipogenic medium containing methylisobutylxanthine, dexamethasone, insulin, and fetal calf serum (MDI). This treatment initiates differentiation by inducing CCAAT/enhancer binding protein β (C/EBPβ) and C/EBPδ (60, 62) while inhibiting Wnt signaling (44). C/EBPβ and C/EBPδ mediate the transcriptional activation of peroxisome proliferator-activated receptor gamma (PPARγ) and C/EBPα. Through a combination of cross- and autoregulation, these two master regulators of adipocyte differentiation mediate the acquisition of the adipocyte phenotype (reviewed in references 9, 28, 31, and 42).
The mature adipocyte plays an essential role in maintaining energy homeostasis. Insulin stimulates adipocytes to increase the uptake of glucose, whose energy is then stored as glycogen and triacylglycerol (51). Conversely, catabolic stimuli such as norepinephrine and adrenocorticotropin cause adipocytes to release this energy into circulation for use by other tissues (7). Finally, adipocytes secrete a number of factors that act centrally and peripherally to profoundly influence whole-body energy metabolism (19).
Recently, genome-wide expression profiling has been used to investigate adipogenesis and adipocyte gene expression in vitro and in vivo (16, 48). Here we used a similar approach to add to this body of knowledge with several goals in mind. First, we defined the cascades of gene expression during adipocyte conversion with the purpose of exploring mechanisms by which enforced Wnt signaling blocks adipogenesis. Second, we used the microarray data to screen for important adipocyte genes and chose the liver X receptor α (LXRα), a transcription factor whose role in adipocyte function was poorly characterized, for further analyses.
Wnts are secreted proteins that direct cell fates in a variety of contexts (reviewed in reference 4), including adipogenesis. We have previously shown that Wnt signaling through T-cell factor/lymphoid enhancer factor (TCF/LEF) blocks differentiation by preventing the induction of C/EBPα and PPARγ (44). Experiments herein were designed to uncover the mechanistic link between Wnt signaling and the inhibition of these transcription factors. Our data are consistent with the idea that Wnt inhibits differentiation through dysregulation of mitotic clonal expansion.
LXRα is an oxysterol-regulated nuclear receptor that controls lipid and cholesterol homeostasis (5, 24, 26, 45, 56, 57, 64). In response to elevated cholesterol levels, LXRα induces the expression of genes such as ABCA1 (8, 40) and apolipoprotein E (24), which mediate cholesterol efflux. In support of this role for LXRα as a cholesterol sensor, LXRα null mice develop toxic levels of hepatic cholesterol (35). While many of the studies directed at understanding LXRα function have focused on the liver and macrophages, there is good evidence to suggest that LXRα activity may be important in adipocytes as well. First, LXRα is highly expressed in adipocytes (59), and its expression is regulated by PPARγ (5), a key adipocyte transcription factor. In addition, many LXRα target genes are highly expressed in adipocytes (48). Indeed, the induction of cholesterol ester transfer protein in adipocytes is mediated by LXRα (27). Finally, adipocytes contain the largest pool of nonesterified cholesterol in the body (23).
Here we show that activation of LXRα enhances several metabolic functions, including glucose transport, glycogen synthesis, cholesterol synthesis, and nonesterified fatty acid (NEFA) release. Thus, while LXRα clearly has a critical function in cholesterol homeostasis, our data suggest that, at least in adipocytes, this transcription factor also has a broader role in regulation of metabolism.
MATERIALS AND METHODS
Cell culture.
Mouse 3T3-L1 preadipocytes (14) and human embryonic kidney 293T cells (kind gift from Mitchell Lazar) were maintained in Dulbecco's modified Eagle's medium (Gibco-BRL) supplemented with 10% calf serum, as described previously (43). 3T3-L1 preadipocytes were infected with a control retrovirus (pLXSN or pBABE) or a retrovirus that encodes either Wnt-1 (63) or LXRα (57), essentially as described previously (11). Briefly, 293T cells were transfected with expression vectors and viral packaging vectors. Virus-containing medium was applied to 3T3-L1 preadipocytes, and infected cells were selected with appropriate antibiotic. Adipogenesis was induced by treating postconfluent preadipocytes with insulin, dexamethasone, and isobutylmethylxanthine in 10% fetal calf serum, as described previously (17). Experiments with adipocytes were performed on cells that had been induced to differentiate for at least 13 but no more than 18 days. Where indicated, cells were treated with the LXRα agonist T0901317 (kind gift from AstraZeneca) for 24 h prior to experiments. The staining of adipocytes with Oil Red-O was performed as described previously (43).
Isolation of adipocytes and stromal vascular cells from white adipose tissue.
Cells were isolated essentially as described previously (41). Briefly, mouse epididymal fat pads from 6-week-old C57BL/6 mice were surgically removed and placed in ice-cold pH 7.4 Krebs-Ringer HEPES buffer. White adipose tissue (1.5 g) was digested at 37°C for 1 h in pH 7.4 Krebs-Ringer HEPES buffer containing 10 mg of collagenase/ml (Worthington, Inc.) with continuous agitation. Digested adipose tissue was filtered through a mesh to separate cells from connective and undigested tissues. The cells were then centrifuged at 500 × g for 5 min to separate the adipocytes, found at the top of the supernatant, from the stromal vascular cells, found in the pellet.
RNA purification and microarray analyses.
Total RNA was isolated from stromal vascular cells, primary adipocytes, and 3T3-L1 cells with RNA Stat60 (Tel-Test B, Inc.). RNA was further purified with RNeasy spin columns (Qiagen, Valencia, Calif.). Preparation of cRNA, hybridization, and scanning of the mouse genome U74A arrays were performed according to the manufacturer's protocol (Affymetrix, Santa Clara, Calif.). The arrays were scanned at 3 μm with the GeneArray scanner (Affymetrix).
Data manipulation.
Each U74A chip consists of 640 × 640 features (20 × 20 μm each) that are single-stranded sequences of DNA 25 bases in length. There are typically 16 pairs of features (probe pairs) for each of the transcripts (probe sets). Half of the features are designed to be complementary to a specific sequence (perfect match = PM features), the other half being identical except that the central base has been altered (mismatch = MM features). The intensity of each feature was determined with GeneChip 4.0 software (Affymetrix) and stored in .CEL files.
We have developed software to read .CEL files and perform some processing of the data. It is freely available at http://dot.ped.med.umich.edu:2000/ourimage/pub/shared/Affymethods.html.
A control chip (control-infected 3T3-L1 cells from time zero) was selected as a standard. Probe pairs for which either the PM or MM feature was saturated in the image of the standard or for which PM − MM < −1,000 on the standard were excluded from further consideration. Saturated features (with measures at least 98% of the maximum pixel value on a chip) on other chips were imputed separately for PM or MM values. For a saturated PM value, the ratios of nonsaturating PM values for the chip divided by the standard are averaged for a probe set by taking the antilogarithm of the mean of the log ratios. This factor was multiplied by the PM values of the standard to impute values for the chip under consideration. MM values were imputed similarly.
The average intensity for each probe set was computed as the mean of the PM − MM differences after trimming away the 25% highest and lowest differences. A set of 4,171 reference probe sets were selected for use in normalization, these having the property of being between the 40th and 95th percentile of probe set average intensities on each of 16 different chips (control versus Wnt samples were used). A normalization factor was obtained with the reference probe sets by computing the antilogarithm of the mean log ratios of the average intensities for the selected chip divided by the standard. The average intensities were divided by this factor to obtain the normalized intensities for the probe sets.
When computing fold change indices, normalized intensities were averaged over replicates, and intensities of less than 100 were replaced by 100 before forming ratios in order to avoid negative or spuriously large n-fold change values.
Data subjected to analysis of variance (ANOVA) treatments were first log transformed by adding 100 to the normalized intensities, setting to 0 any that still remained negative, and finally adding 1 and taking logarithms. This transformation gave approximately equal variances for probe sets with large and small mean normalized intensities.
To determine whether the set of genes that defined the adipocyte phenotype displayed significantly higher levels of expression in adipocytes compared to preadipocytes, we asked that, when comparing the level of gene expression in white adipose tissue adipocytes versus preadipocytes and in adipocytes versus preadipocytes, both give a significant difference (P < 0.05) in a one-way ANOVA model on log-transformed data.
To determine whether the expression of a gene varied over the course of differentiation of 3T3-L1 cells, six treatments were analyzed in two separate experiments (12 samples): day 0 preadipocytes, control-infected cells from 0, 16, 32, and 48 h, and day 14 adipocytes. We fit two-way ANOVA models to the log-transformed data that included treatment and experiment effects. We selected genes that gave significant F tests for treatment effects (P < 0.005) and that displayed at least a twofold change between at least one pair of treatments.
Immunoblot analyses.
Nuclei were purified from 3T3-L1 cells as described previously (54). Nuclear extracts were separated by electrophoresis on sodium dodecyl sulfate (SDS)-polyacrylamide gels (7% for p130, 11% for all others) and transferred onto a PVDF-Plus membrane for immunoblot analyses. Immunoblotting was performed as described previously (11, 17) with the following antibodies: E2F4 (C-20) from Santa Cruz; p130 (Rb2) from BD Transduction Labs; p27 from PharMingen; and p21 (WAF1) from Oncogene Research Products.
Northern blot analyses.
3T3-L1 cells were lysed daily over the course of differentiation, and RNA was extracted with Trizol (Life Technologies, Inc.). Northern blot analyses of RNA were performed as described previously with 20 μg of total RNA per lane (47). Blots were probed with LXRα, PPARγ, SREBP1/ADD1, and C/EBPα. We used 18S ribosomal protein mRNA (ATCC 107382) as a control for equal RNA loading.
Triacylglycerol accumulation and protein concentration.
3T3-L1 adipocytes were lysed in buffer (1% SDS, 1.2 mM Tris, pH 6.8), heat-treated (95°C, 5 min), and vortexed. Triacylglycerol determinations were performed with Infinity triglyceride reagent (Sigma) with glycerol as the standard. Protein determinations were made with the Bio-Rad protein assay, with bovine serum albumin (BSA) as the standard.
NEFA and glycerol efflux.
3T3-L1 adipocytes were rinsed twice with and then incubated in Hanks' buffered saline solution supplemented with 1% BSA (fatty acid free). Glycerol released into the medium was quantified with triglyceride reagent A (GPO-Trinder; Sigma) and NEFA release with the free fatty acids half micro test (Roche). Glycerol and palmitic acid were used as standards, respectively.
Glucose uptake.
Glucose uptake was determined essentially as described previously (1). Briefly, adipocytes were washed three times with phosphate-buffered saline (PBS) and incubated for 30 min at 37°C in PBS containing 0.7% BSA. Cells on 6-cm plates were incubated with 2-deoxy-d-[14C]glucose (100 μM; 10 cpm/pmol) for 5 min at room temperature. The assay was terminated by adding 2-deoxy-d-glucose (200 mM) and washing the cells with ice-cold PBS three times. Cells were lysed in a 1 ml of 0.1% SDS, and aliquots (250 μl) were subjected to scintillation counting.
Glycogen synthesis.
Cells were incubated in PBS containing 0.7% BSA with 1 mM [U-14C]glucose for 30 min at 37°C, then disrupted in 15% KOH (0.75 ml), and boiled for 30 min in the presence of 10 mg of carrier glycogen. Glycogen was then precipitated with ethanol. After centrifugation, the pellet was resuspended in 0.5 ml of water and reprecipitated two times. The final pellet was resuspended in 0.5 ml of water and subjected to scintillation counting.
Lipid synthesis and extraction.
Cellular lipids were extracted essentially by the Folch method (12). Briefly, cells were incubated for 2 h at 37°C in the presence of [14C]acetic acid and then lysed in 0.5 ml of ethanol. Then, 1 ml of chloroform was added and mixed by vortexing. After addition of 0.5 ml of HCl (0.1 N), lysates were mixed by gentle inversion. Tubes were centrifuged at 500 × g for 30 min. The upper phase and infranatant were discarded, and the organic phase was washed twice with 0.5 ml of water. Extracts were dried under N2, and the pellet was resuspended in the appropriate volume of chloroform. Extracts were separated by thin-layer chromatography with dichloroethane-acetic acid (100:1, vol/vol) on silica gel plates and exposed to film. Phospholipids, cholesterol, diacylglycerol, and triacylglycerol were identified with standards.
Serum NEFA and glycerol determination.
Female C57BL/6 mice (10 weeks old, approximately 20 g each) received an intraperitoneal injection of either vehicle (control) or 50 mg of T0901317/kg/day daily for 1, 3, or 7 days. In addition, 4 h prior to sample collection, the mice were injected with a final dose of vehicle or T0901317. Five mice were injected for each treatment. T0901317 was dissolved in dimethyl sulfoxide (50 mg/ml) and diluted (5:1) with 0.9% saline prior to injection. Mice were anesthetized with CO2, and ocular blood was obtained prior to euthanasia. Serum was analyzed for NEFA and glycerol as described previously above. Data were subjected to F tests for treatment effects.
RESULTS
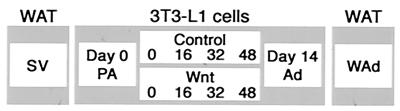
We have previously shown that Wnt signaling is an adipogenic switch, blocking differentiation in its presence and allowing spontaneous differentiation in its absence (44). As such, we thought that Wnt would serve as a tool in our efforts to better understand the process of adipocyte differentiation through microarray analyses. Here, we profiled changes in gene expression that occur in response to adipogenic medium (MDI), comparing patterns from control infected preadipocytes and preadipocytes in which Wnt was ectopically expressed. RNA was purified from control and Wnt-expressing 3T3-L1 cells at time zero, immediately prior to induction of differentiation, as well as 16, 32, and 48 h postinduction. A second aim was to investigate the preadipocyte and adipocyte phenotypes. As an in vitro model, RNA was isolated from 3T3-L1 preadipocytes and adipocytes. As an in vivo model, RNA was isolated from stromal vascular cells, which are comprised predominantly of preadipocytes, and adipocytes from fractionated white adipose tissue. This experimental design is illustrated in Fig. 1.
FIG. 1.
Experimental design. To investigate the preadipocyte and adipocyte phenotypes, RNA was purified from confluent 3T3-L1 preadipocytes (PA; day 0) and 3T3-L1 adipocytes that were 14 days postinduction (Ad; day 14). In addition, RNA was purified from mouse white adipose tissue (WAT) that had been separated into stromal vascular (SV) cells (which contain preadipocytes) and white adipocytes (WAd). To gain further insight into the early events of adipogenesis and the mechanism by which Wnt signaling blocks this differentiation process, 3T3-L1 cells were control infected (Control) or infected with a retrovirus encoding the gene for Wnt-1 (Wnt). Following selection, cells were induced to differentiate with methylisobutylxanthine, dexamethasone, and insulin (MDI) in 10% fetal calf serum. RNA was prepared from cells at 0, 16, 32, and 48 h following induction of differentiation. For each condition, RNA was purified from two independent samples, and transcript levels were measured by microarray with U74A Affymetrix chips, which represent approximately 10,000 distinct mouse genes and ESTs.
Genes that define the adipocyte phenotype.
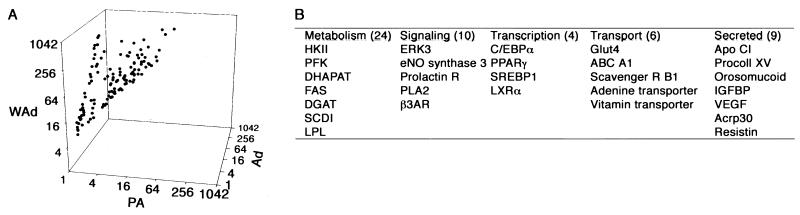
Data derived from these 12 conditions are accessible at http://www-personal.umich.edu/∼macdouga/MacDougaldLab.html. We designed a simple screen to define genes that are selectively expressed in adipocytes. Specifically, we selected genes that are expressed in both adipocyte models at similar levels (within twofold of each other) and that are expressed at significantly higher levels in adipocytes than in preadipocytes (P < 0.05; at least a fourfold difference). Of the ≈10,000 genes and expressed sequence tags (ESTs) that are represented on the chip, 119 met these criteria. The expression levels of these transcripts in white adipose tissue adipocytes, 3T3-L1 adipocytes, and 3T3-L1 preadipocytes are shown in Fig. 2A, and a partial list by category appears in Fig. 2B. The complete list is available at http://www-personal.umich.edu/∼macdouga/MacDougaldLab.html.
FIG. 2.
Genes (n = 119) that define the adipocyte phenotype. Criteria were established to identify genes that may be important in adipocyte function. These criteria were (i) transcripts that are expressed in 3T3-L1 adipocytes (Ad) and adipocytes from white adipose tissue (WAd) at similar levels (within twofold of one another) and (ii) transcripts that are highly expressed in both of these models relative to that observed in 3T3-L1 preadipocytes (PA) (at least fourfold higher and significantly different). Expression levels (102) of these adipocyte genes are shown in panel A, and a partial list of these genes is given in panel B. The complete list of genes is available at http://www-personal.umich.edu/∼macdouga/MacDougaldLab.html.
As expected, we identified genes that are critical for many aspects of carbohydrate and lipid metabolism (Fig. 2B), including genes involved in metabolic trapping of glucose (hexokinase II), glycolysis (phosphofructokinase), and lipogenesis (dihydroxyacetonephosphate acyltransferase, fatty acid synthase, diacylglycerol acyltransferase, stearoyl coenzyme A [CoA] desaturase 1, and lipoprotein lipase). We hypothesized that we might also have identified signaling proteins that regulate these metabolic processes. Consistent with this idea, we identified β-3 adrenergic receptor and nitric oxide synthase 3 (Fig. 2B), which play opposing roles in the regulation of lipolysis (7, 22). However, other signaling molecules selected by our criteria, such as ERK3, have uncharacterized roles in adipocyte biology and warrant further investigation.
The secreted proteins that we identified may be important for the endocrine functions of the adipocyte. For instance, Acrp30/adiponectin/adipoQ and resistin (Fig. 2B) play roles in the sensitivity of peripheral tissues to insulin (2, 50, 61). These and other secreted proteins may contribute to the regulation of whole-body energy metabolism. Finally, four transcription factors, C/EBPα, PPARγ, SREBP1/ADD1, and LXRα, were identified among genes specifically expressed in adipocytes (Fig. 2B). The roles of the first three in adipogenesis are well characterized. C/EBPα and PPARγ are master regulators, each sufficient to mediate the adipogenic program (31, 42). SREBP1/ADD1 is an enhancer of differentiation, inducing PPARγ and production of its ligand (21, 52). In contrast, the role of LXRα in adipocyte biology has not been investigated extensively.
Gene expression profiles during adipogenesis.
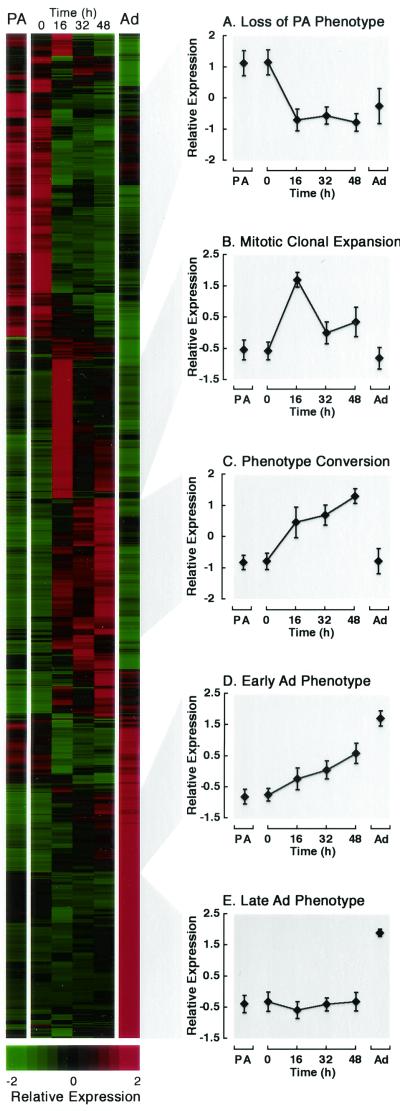
The extensive characterization of 3T3-L1 cells during adipogenesis provides an excellent framework upon which to evaluate gene expression on a genome-wide scale. Here we examined the cascades of gene expression with an unbiased approach to identify patterns. Samples used for these analyses include preadipocytes and fully differentiated adipocytes as well as control cells induced to differentiate for 0, 16, 32, and 48 h (Fig. 1). Genes that varied significantly (P < 0.005) and whose magnitude varied by at least twofold during differentiation were selected. We next arranged these genes based on their inherent expression properties by hierarchical clustering. This algorithm creates a dendrogram of genes based on the same principle as a phylogenetic tree. Genes whose patterns of expression have the highest degree of correlation are placed most closely together. Clustering analysis of our data revealed that three quarters of the transcripts fell into one of five waves of gene expression. Based on careful examination of genes contained within these clusters, we labeled each of the phases as follows: loss of preadipocyte phenotype, mitotic clonal expansion, phenotype conversion, and acquisition of the early and late adipocyte phenotypes (Fig. 3A to E).
FIG.3.
Gene expression profiles during adipogenesis. RNA transcript levels were assessed from 3T3-L1 cells under six conditions: noninfected preadipocytes (PA), control-infected 3T3-L1 cells that had been induced to differentiate for 0, 16, 32, or 48 h, and day 14 adipocytes (Ad). A total of 1,889 genes that varied significantly in expression (P < 0.005) and by at least twofold in magnitude were selected for analysis by hierarchical clustering. Average expression for each gene was standardized to have mean = 0 and variance = 1 across conditions. Complete linkage clustering was performed with Cluster and Treeview software (http://www.microarrays.org/software.html). Genes are represented in rows, with a colorimetric scale (standard deviations [SDs] from the mean) to indicate relative expression levels; conditions are indicated above each column. Major patterns were defined as those nodes containing at least 150 transcripts and having a correlation coefficient of at least 0.85. Three quarters of the transcripts fell into one of five major patterns (A to E). Average expression profiles and SDs are shown for each pattern. This image, with a complete list of genes and their expression patterns, can be obtained at http://www-personal.umich.edu/∼macdouga/MacDougaldLab.html.
Loss of preadipocyte phenotype.
Upon induction of adipogenesis, many genes expressed in preadipocytes are rapidly suppressed (Fig. 3A). For instance, there is downregulation of six procollagens and several regulatory extracellular matrix components, such as tissue inhibitor of metalloproteinase II and matrix metalloproteinase 9. In addition, small leucine-rich proteoglycans decline upon induction of differentiation, including decorin, lumican, and osteoglycan. Heparin sulfate proteoglycans such as glypican 4 and syndecan 2 are also repressed during this phase. The downregulation of these extracellular matrix proteins is consistent with the idea that preadipocytes lose their fibroblastic characteristics as they turn into adipocytes. We were next interested in whether the expression of Wnt blocks the loss of the preadipocyte phenotype. Surprisingly, expression of virtually all of these genes declined in a similar manner after treatment with MDI, irrespective of the presence of Wnt (data not shown). Thus, Wnt signaling does not appear to inhibit loss of the preadipocyte phenotype. An interesting but unanswered question is whether the expression of these preadipocyte genes in Wnt-expressing cells returns to preadipocyte levels when cells fail to differentiate.
Mitotic clonal expansion.
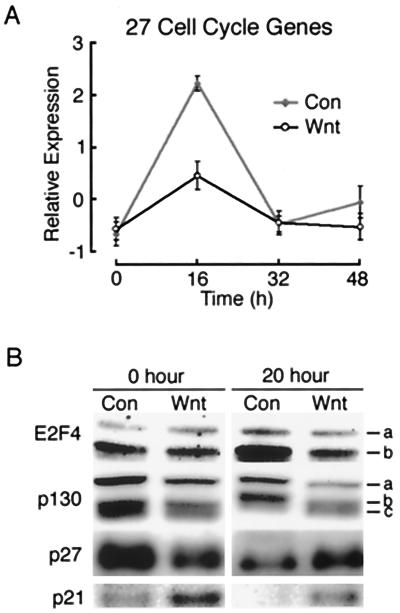
A second phase of adipogenesis is mitotic clonal expansion (Fig. 3B). Although controversial, it is likely that events that occur during this time are critical for differentiation (33, 37). During this phase, growth-arrested preadipocytes reenter the cell cycle and undergo one or two rounds of cell division (33). As shown in Fig. 3B, a peak in the expression of 275 genes and ESTs occurred 16 h after induction of differentiation, corresponding to entry into S phase. Of these, approximately 50 were readily identifiable as cell cycle genes. We next examined whether expression of Wnt affected their expression. Although some genes were expressed similarly in control and Wnt-expressing cells, the expression patterns of 27 genes associated with cell cycle progression were greatly blunted by the presence of Wnt (Fig. 4A). For instance, in response to MDI treatment, DNA polymerase α was induced ninefold in control cells at 16 h but only threefold in Wnt-expressing cells during this time. Similarly, ectopic Wnt blunts the induction of dihydrofolate reductase from fivefold in control cells to twofold in Wnt-expressing cells. These data suggest that mitotic clonal expansion is altered in the presence of Wnt signaling.
FIG. 4.
Ectopic Wnt signaling alters mitotic clonal expansion. (A) Average expression profile and SD for 27 cell cycle genes are shown for control infected (Con) and Wnt infected (Wnt) cells. Average expression for each gene was standardized to have mean = 0 and variance = 1 across conditions. The full list of genes and their expression patterns can be obtained at http://www-personal.umich.edu/∼macdouga/MacDougaldLab.html. (B) Control infected (Con) and Wnt infected (Wnt) cells were either lysed at time zero (0 h) or induced to differentiate and lysed 20 h later (20 h). Nuclei were purified, and nuclear lysates were separated by SDS-PAGE for immunoblot analysis of E2F4, p130, p27, and p21, as indicated. Two E2F4 isoforms are indicated (a and b). Similarly, p130 exists as two isoforms (a and c). Phosphorylation of the lower species (a) results in a mobility shift, resulting in b. These results are representative of two or three independent experiments.
Next we investigated the mechanism by which Wnt signaling inhibits induction of many cell cycle genes. Because many of the cell cycle genes whose transient expression is diminished in the presence of Wnt are known to be regulated by E2F (25), we hypothesized that E2F activity is reduced in Wnt-expressing cells. E2F4 is the predominant E2F family member in 3T3-L1 cells (53). Thus, we examined the expression of this protein by immunoblot analyses (Fig. 4B). Nuclear E2F4 from control cells was induced 20 h after induction of differentiation, and the presence of Wnt blunted the induction of E2F4 in response to MDI (Fig. 4B). The activity of E2F4 is highly regulated by p130 (53), which, when hypophosphorylated, binds to E2F4 and inhibits its activity. Phosphorylation of p130 by cyclin-dependent kinases (cdks) releases E2F4 from p130-mediated inhibition.
We therefore examined the expression of p130. In control cells, induction of differentiation caused a decline in the expression of p130 coupled with hyperphosphorylation of the protein (Fig. 4B). These findings are consistent with the idea that E2F4 activity increases during mitotic clonal expansion. The presence of Wnt partially inhibited both the decline and hyperphosphorylation of p130 (Fig. 4B), suggesting that E2F4 activity was reduced in these cells. Hyperphosphorylation of p130 is caused, in part, by a decline during the cell cycle of the cdk inhibitors. We therefore investigated whether the cdk inhibitors p21 and p27 are dysregulated in the presence of Wnt. Immunoblot analyses revealed that the decline in expression of p21 and p27 normally associated with the cell cycle (32, 33) was blocked by the presence of Wnt (Fig. 4B). Our data clearly indicate that Wnt expression alters many aspects of mitotic clonal expansion. Given that this phase is critical for subsequent differentiation, its dysregulation by Wnt may be causal to inhibition of adipogenesis.
Phenotype conversion.
In response to MDI, many genes were transiently induced, remaining elevated at 48 h but returning to basal levels in the fully differentiated adipocyte (Fig. 3C). A number of events are occurring during this phase because the cells are undergoing a dramatic phenotype conversion, simultaneously losing their preadipocyte characteristics and gaining early adipocyte properties. These changes require considerable modifications in gene transcription, RNA processing, and protein synthesis. Thus, it is not surprising that many genes involved in these processes, for example, the general transcription factor TAFII30, the RNA splicing factor U2AF, and the protein translation initiation factors 2A and 3, are upregulated during this active period. It is very interesting that this phase contains many transcription factors. Although Wnt expression has no effect on the induction of C/EBPβ, other transcription factors that are induced during this phase, such as retinoid X receptor α, retinoic acid receptor α, and Myc, are not induced in Wnt-expressing cells. Thus, it is possible that dysregulation of specific genes within this phase may also contribute to Wnt-mediated inhibition of adipogenesis.
Early adipocyte phenotype.
A fourth phase that occurs upon induction of adipogenesis is characterized by a gradual increase in gene expression from 0 to 48 h, which reaches maximal levels of expression in the fully differentiated adipocyte (Fig. 3D). We observed that many genes found within this cluster are involved in energy storage. For example, genes encoding proteins involved in metabolic trapping, glycogen synthesis, the pentose phosphate shunt, glycolysis, oxidative phosphorylation, and essentially all aspects of lipogenesis were expressed within this phase (Fig. 5A). Thus, it appears that the ability to trap and store glucose is one of the earliest events during the acquisition of the adipocyte phenotype. Also found within this cluster were C/EBPα, PPARγ, and SREBP1/ADD1, highlighting the possible regulatory role of these transcription factors in the induction of energy storage genes. Indeed, glycogen synthase, fatty acid synthase, stearoyl CoA desaturases, and many other genes in this cluster are already known to be regulated by one or more of these transcription factors (6, 13, 15, 30, 46, 52, 58).
FIG. 5.
(A) Genes within the early adipocyte phenotype are involved in energy storage. Of the genes and ESTs that cluster in the early adipocyte phenotype (Fig. 3D), 20 are involved in the storage of energy as glycogen, fatty acids, and triacylglycerol (TAG). Schematic diagram of the function of these genes in metabolism is shown to the left, with their names on the right. (B) Average expression profile and SD for C/EBPα, PPARγ, and SREBP1/ADD1, three transcription factors found within the early adipocyte phenotype cluster. Average expression for each gene was normalized to have mean = 0 and variance = 1 across conditions. TCA, trichloroacetic acid; OA, oxaloacetate; DHAP, dihydroxyacetone phosphate.
We then investigated whether expression of Wnt influenced induction of genes that comprise the early adipocyte phenotype. Inspection of the microarray data revealed that the gradual induction of genes involved in energy storage did not occur in the presence of Wnt (data not shown). Similarly, induction of C/EBPα, PPARγ, and SREBP1/ADD1 was completely repressed by ectopic expression of Wnt (Fig. 5B). Without induction of these adipogenic transcription factors, adipocyte conversion cannot proceed. These findings are consistent with a model in which early events, such as dysregulation of the cell cycle, lead to a block in the induction of adipocyte transcription factors, thereby preventing adipogenesis.
Late adipocyte phenotype.
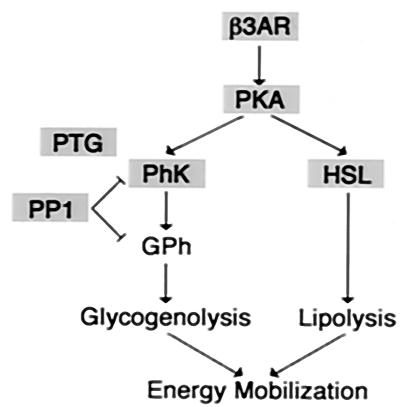
The last phase of gene expression observed upon induction of adipogenesis is characterized by the late induction of genes, which is not apparent until after 48 h (Fig. 3E). Interestingly, many of the genes required for regulated mobilization of stored energy are found within this cluster (Fig. 6). Key rate-limiting enzymes (e.g., hormone-sensitive lipase) and regulators (e.g., β-3 adrenergic receptor, protein kinase A, and glycogen phosphorylase kinase) for lipolysis and glycogenolysis are induced as part of the late adipocyte phenotype. This observation suggests that the ability of the adipocyte to mobilize energy is a late event during acquisition of the adipocyte phenotype. Given the time frame during which these changes in gene expression occur and the fact that adipogenesis does not occur in the presence of Wnt, it is highly unlikely that Wnt-expressing cells undergo this late phase.
FIG. 6.
Genes within the late adipocyte phenotype are involved in energy mobilization Of the genes and ESTs that cluster in the late adipocyte phenotype (Fig. 3E), many are involved in energy mobilization. Schematic diagram of the function of these genes is shown, with members of cluster 3E highlighted in gray. β3AR, β3 adrenergic receptor; PKA, protein kinase A; HSL, hormone-sensitive lipase; PhK, phosphorylase kinase; PTG, protein targeted to glycogen; PP1, protein phosphatase 1.
Role of LXRα in differentiation and metabolism.
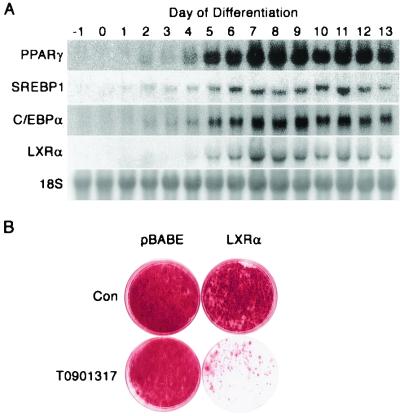
Based on the observations that LXRα is selectively expressed in adipocytes relative to preadipocytes (Fig. 2) and that this transcription factor is induced in the late wave of adipogenesis (Fig. 3E), we tested the role of LXRα in differentiation. Northern blot analyses revealed that LXRα was first observed 4 days after induction of differentiation (Fig. 7A). This induction was slightly later than that observed for C/EBPα, PPARγ, and SREBP1/ADD1, which were first detected on day 2 (Fig. 7A). To assess the role of LXRα in adipogenesis, control and LXRα-expressing preadipocytes were induced to differentiate in the absence or presence of the LXRα activator T0901317. Differentiation of control 3T3-L1 cells in the continuous presence of T0901317 had no effect on the accumulation of triacylglycerol (data not shown) or the degree of differentiation, as assessed visually by Oil Red-O staining (Fig. 7B). Although ectopic expression of LXRα caused a slight inhibition of adipogenesis, the effect was not pronounced. However, when LXRα was ectopically expressed and activated by treatment of cells with T0901317, adipogenesis was dramatically inhibited. These data indicate that, unlike C/EBPα and PPARγ, LXRα is not adipogenic. Rather, when ectopically expressed and activated, this transcription factor inhibits adipocyte conversion.
FIG. 7.
Role of LXRα in adipocyte differentiation. (A) RNA from 3T3-L1 cells that had been induced to differentiate for the number of days indicated was analyzed by Northern for LXRα, SREBP1/ADD1, PPARγ, and C/EBPα. (B) 3T3-L1 preadipocytes were control infected (pBABE) or infected with a retrovirus encoding the gene for LXRα and selected with puromycin. Once confluent, cells were induced to differentiate in the absence (Con) or presence of the LXRα activator T0901317 (1 μM). Fourteen days later, cells were stained with Oil Red-O. These results are representative of three independent experiments.
We next assessed the function of LXRα in the fully differentiated adipocyte. In addition to evaluating its role in carbohydrate and lipid metabolism, we considered the possibility that LXRα regulates cholesterol synthesis. Because 3T3-L1 adipocytes contain large amounts of cholesterol, a precursor to LXRα ligands (oxysterols), it is likely that endogenous LXRα is active under basal conditions. Therefore, to amplify the effects of agonist treatment, we performed experiments with 3T3-L1 adipocytes in which LXRα was ectopically expressed.
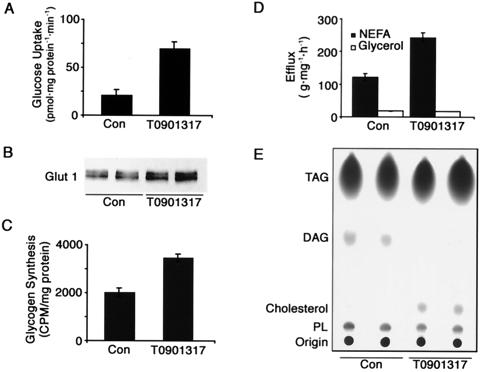
To evaluate whether LXRα regulates carbohydrate metabolism, we first measured glucose uptake in LXRα-expressing adipocytes. Although insulin-stimulated glucose uptake was not altered by LXRα activation (data not shown), basal glucose uptake was dramatically increased when cells were treated with T0901317 for 24 h (Fig. 8A). Because basal glucose uptake is largely mediated by GLUT1 (20), we evaluated whether LXRα induces GLUT1 gene expression. Treatment of LXRα-expressing adipocytes with T0901317 for 24 h increased GLUT1 expression at both the RNA (data not shown) and protein (Fig. 8B) levels. Next we determined whether the increase in glucose uptake was paralleled by an increase in glucose storage. Exposure of LXRα-expressing adipocytes to T0901317 for 24 h caused a ≈75% increase in the rate of glycogen synthesis under basal conditions (Fig. 8C). Although slightly less dramatic, the effects of T0901317 on glucose uptake and glycogen synthesis were also observed in control 3T3-L1 adipocytes (data not shown). Taken together, these data suggest that activation of LXRα increases glucose uptake and storage, at least in part, due to an increase in GLUT1.
FIG. 8.
Role of LXRα in adipocyte metabolism. Fully differentiated 3T3-L1 cells that expressed ectopic LXRα were incubated in the absence (Con) or presence of 1 μM T0901317 for 24 h. (A) Cells were labeled with [14C]glucose, and basal glucose uptake measurements were performed. (B) Lysates were analyzed by immunoblot for GLUT1. (C) Cells were labeled with [14C]acetate, and glycogen synthesis was measured. (D) NEFA and glycerol efflux from the cells into the medium was determined. (E) Cells were labeled with [14C]acetate, and lipids were extracted from the medium (top) or cell lysates (bottom) and separated by thin-layer chromatography. The origin, phospholipids (PL), cholesterol, diacylglycerol (DAG), and triacylglycerol (TAG) are indicated. These experiments are representative of at least three independent experiments.
To assess the potential role of LXRα in lipid metabolism, we examined its effects on lipolysis and lipogenesis. LXRα-expressing adipocytes were incubated in the absence or presence of T0901317 for 24 h, and efflux of NEFA and glycerol into the medium was measured. Activation of LXRα increased the rate of NEFA release twofold (Fig. 8D). Surprisingly, the increase in NEFA release was not accompanied by a change in glycerol efflux. Activation of LXRα had no effect on the maximal rate of NEFA or glycerol release upon stimulation of adipocytes with the β-3 adrenergic receptor agonist BRL37344 (data not shown).
We next considered the possibility that the increase in basal NEFA release was secondary to an increase in lipogenesis. However, treatment of cells with T0901317 had no effect on the incorporation of [14C]acetate into triacylglycerol (Fig. 8E). Although the magnitude was not quite as great, T0901317 also stimulated release of NEFA in control 3T3-L1 adipocytes (data not shown). Despite the discrepancy between NEFA and glycerol release, we favor the idea that activation of LXRα increases basal lipolysis. Several mechanisms could account for the absence of a corresponding increase in glycerol. For instance, glycerol could be selectively retained in the cell due to metabolic trapping. Consistent with this notion, T0901317 induced glycerol kinase at the RNA level by twofold (data not shown). Alternatively, monoacylglycerol resulting from partial lipolysis may be reesterified and therefore not transported out of the cell. Finally, LXRα may increase transport of newly synthesized fatty acids out of the fully differentiated adipocyte.
Based on the regulation of cholesterol by LXRα in liver and macrophages, we next considered the possibility that LXRα controls cholesterol metabolism in adipocytes. LXRα-expressing adipocytes were treated or not with T0901317 for 24 h. After metabolic labeling with [14C]acetate for 2 h, lipids were extracted and separated by thin-layer chromatography. Activation of LXRα resulted in a dramatic increase in cholesterol synthesis (Fig. 8E). This finding was somewhat surprising in view of the role of LXRα in macrophages, intestinal cells, and hepatocytes, where it acts to limit cholesterol accumulation in the body. Further experiments are required to investigate the unique regulation of cholesterol homeostasis by LXRα in adipocytes.
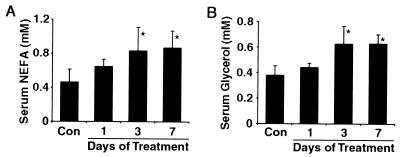
Finally, we investigated the role of LXRα activity on the regulation of lipolysis within white adipose tissue. To do this experiment, mice were treated with vehicle or T0901317 (50 mg/kg/day) daily for 1, 3, or 7 days, as indicated, and serum was analyzed for NEFA and glycerol concentrations (Fig. 9). Treatment with T0901317 for 3 or 7 days increased serum NEFA levels 1.8-fold and 1.9-fold, respectively (P < 0.01) relative to control mice (Fig. 9A). These data are consistent with the effect of LXRα activation on NEFA release in vitro and suggest that increased LXRα activity results in increased lipolysis within adipose tissue in vivo. Changes in the serum concentration of glycerol mirrored those observed for NEFA upon treatment of mice with T0901317, with significant effects following 3 and 7 days of treatment (Fig. 9B). Taken together, these data reveal an unexpected role for LXRα in the regulation of lipolysis.
FIG. 9.
Role of LXRα in lipid metabolism in vivo. Female C57BL/6 mice were injected with either vehicle (Con) or 50 mg of T0901317/kg/day daily for 1, 3, or 7 days, as described in Materials and Methods, and serum was analyzed for NEFA and glycerol. Three and seven days of T0901317 treatment resulted in significant (P < 0.01) changes in both NEFA and glycerol concentrations, as indicated (∗). Results are representative of two independent experiments.
DISCUSSION
Here we conducted microarray analyses to define the patterns of gene expression during adipogenesis and thereby identify networks of gene expression that are altered when adipogenesis is blocked by Wnt signaling. Our studies indicate that enforced Wnt signaling blunts mitotic clonal expansion, suggesting that Wnt signaling inhibits adipogenesis in part through dysregulation of the cell cycle. Furthermore, we show that during adipocyte development, the ability to store energy occurs prior to the ability to mobilize energy. Finally, we investigated LXRα, a transcription factor that is induced late during adipogenesis. Our experiments revealed that activation of LXRα increases glucose uptake, glycogen synthesis, cholesterol synthesis, and NEFA efflux in 3T3-L1 adipocytes. In support of a role for LXRα in lipolysis, we also showed that activation of LXRα causes increased serum NEFA and glycerol concentrations in vivo.
One of the major aims of our microarray analyses was to use Wnt signaling as a tool to learn more about the process of adipocyte differentiation. Because Wnt signaling acts as a switch to regulate adipogenesis (44), pinpointing the precise target of Wnt signaling will likely identify a critical control point for adipocyte conversion. Our microarray results revealed five major phases. Although several patterns were altered in the presence of Wnt, we favor the hypothesis that Wnt signaling inhibits adipogenesis by altering aspects of the cell cycle that are critical for subsequent differentiation. We and others have observed that levels of the cdk inhibitors p21 and p27 decline during mitotic clonal expansion (32, 55). This decline, which does not occur in Wnt-expressing cells, is required for the eventual activation of E2F4. As a result, we observed that induction of many known E2F-regulated genes during the cell cycle was reduced in the presence of Wnt.
Only four transcription factors were identified in our screen to define genes that are selectively expressed in adipocytes (Fig. 2). Three of these, C/EBPα, PPARγ, and SREBP1/ADD1, are induced during acquisition of the early adipocyte phenotype and have well-established roles in adipocyte differentiation (38, 42). In contrast, LXRα is induced in a later phase, and analysis of its function in adipocytes remains incomplete. Although no adipocyte phenotype has been reported in mice lacking LXRα (35), we hypothesized that LXRα may have important functions in adipose tissue because of its high level of expression (59).
The main physiological function of LXRα is to regulate cholesterol homeostasis, stimulating cholesterol conversion to bile acids in hepatocytes, inhibiting cholesterol uptake in intestinal cells, and promoting cholesterol efflux from macrophages and possibly adipocytes (10, 26, 34). All of these activities limit the amount of cholesterol in the body. Our finding that LXRα activity causes an increase in cholesterol synthesis is therefore somewhat at odds with its known function elsewhere. It is possible that the increase in cholesterol synthesis is secondary to an increase in expression of the ABCA1 transporter or apolipoprotein E, two LXRα target genes which function to remove cellular cholesterol (8, 24, 40). Consistent with this idea, LXRα activity causes an increase in apolipoprotein E expression in adipose tissue (24). Alternatively, it is possible that the regulation of cholesterol by LXRα in adipocytes is different from that elsewhere in the body, perhaps because adipocytes have a unique requirement for cholesterol.
An emerging concept is that the level of triacylglycerol and free cholesterol within adipocytes may be linked. Adipocytes contain the largest pool of free cholesterol in the body (23), and recently it has been shown that free cholesterol coats the surface of triacylglycerol droplets (36). Furthermore, LXRα is known to induce the expression of SREBP1/ADD1 (39), a key transcription factor in the regulation of lipogenesis (3). Finally, activation of LXRα in vivo results in increased serum cholesterol levels (18) (data not shown) and, as we report here, NEFAs and glycerol (Fig. 9).
If the increase in serum cholesterol and NEFA/glycerol is indeed connected, it is important to distinguish between primary and secondary effects. It is possible that LXRα activity results directly in an increase in lipolysis, perhaps mediated by increased activity of hormone-sensitive lipase or by influencing the accessibility of the lipid droplet to this lipase. Decreases in the volume of a lipid droplet would correspond to decreases in its surface area and thus an excess of free cholesterol within the adipocyte. In this scenario, the increase in cholesterol export could be, at least partially, secondary to in an increase in lipolysis. Alternatively, it is possible that an LXRα-mediated increase in cholesterol export depletes lipid droplets of a protective cholesterol coating, which in turn leads to increased accessibility of the droplet to lipolysis. Distinguishing among these and other possibilities awaits further investigation.
Acknowledgments
This work was supported by a grant to O.A.M. from the National Institutes of Health (DK51563) and by the Michigan NIDDK Biotechnology Center (U24 DK58771).
We thank Amiya Hajra for helping us with the lipid analyses and for many helpful discussions.
REFERENCES
- 1.Baumann, C. A., M. J. Brady, and A. R. Saltiel. 2001. Activation of glycogen synthase by insulin in 3T3-L1 adipocytes involves c-Cbl-associating protein (CAP)-dependent and CAP-independent signaling pathways. J. Biol. Chem. 276:6065-6068. [DOI] [PubMed] [Google Scholar]
- 2.Berg, A. H., T. P. Combs, X. Du, M. Brownlee, and P. E. Scherer. 2001. The adipocyte-secreted protein Acrp30 enhances hepatic insulin action. Nat. Med. 7:947-953. [DOI] [PubMed] [Google Scholar]
- 3.Brown, M. S., and J. L. Goldstein. 1998. Sterol regulatory element binding proteins (SREBPs): controllers of lipid synthesis and cellular uptake. Nutr. Rev. 56:S1-S3. [DOI] [PubMed] [Google Scholar]
- 4.Cadigan, K. M., and R. Nusse. 1997. Wnt signaling: a common theme in animal development. Genes Dev. 11:3286-3305. [DOI] [PubMed] [Google Scholar]
- 5.Chawla, A., W. A. Boisvert, C. H. Lee, B. A. Laffitte, Y. Barak, S. B. Joseph, D. Liao, L. Nagy, P. A. Edwards, L. K. Curtiss, R. M. Evans, and P. Tontonoz. 2001. A PPAR gamma-LXR-ABCA1 pathway in macrophages is involved in cholesterol efflux and atherogenesis. Mol. Cell 7:161-171. [DOI] [PubMed] [Google Scholar]
- 6.Christy, R. J., V. W. Yang, J. M. Ntambi, D. E. Geiman, W. H. Landschulz, A. D. Friedman, Y. Nakabeppu, T. J. Kelly, and M. D. Lane. 1989. Differentiation-induced gene expression in 3T3-L1 preadipocytes: CCAAT/enhancer binding protein interacts with and activates the promoters of two adipocyte-specific genes. Genes Dev. 3:1323-1335. [DOI] [PubMed] [Google Scholar]
- 7.Collins, S., and R. S. Surwit. 2001. The beta-adrenergic receptors and the control of adipose tissue metabolism and thermogenesis. Recent Prog. Horm. Res. 56:309-328. [DOI] [PubMed] [Google Scholar]
- 8.Costet, P., Y. Luo, N. Wang, and A. R. Tall. 2000. Sterol-dependent transactivation of the ABC1 promoter by the liver X receptor/retinoid X receptor. J. Biol. Chem. 275:28240-28245. [DOI] [PubMed] [Google Scholar]
- 9.Darlington, G. J., S. E. Ross, and O. A. MacDougald. 1998. The role of C/EBP genes in adipocyte differentiation. J. Biol. Chem. 273:30057-30060. [DOI] [PubMed] [Google Scholar]
- 10.Edwards, P. A., H. R. Kast, and A. M. Anisfeld. 2002. BAREing it all. The adoption of lxr and fxr and their roles in lipid homeostasis. J. Lipid Res. 43:2-12. [PubMed] [Google Scholar]
- 11.Erickson, R. L., N. Hemati, S. E. Ross, and O. A. MacDougald. 2001. p300 coactivates the adipogenic transcription factor C/EBPα. J. Biol. Chem. 37:79-90. [DOI] [PubMed] [Google Scholar]
- 12.Folch, J., M. Lees, and G. Sloane Stanley. 1957. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 226:497-509. [PubMed] [Google Scholar]
- 13.Foretz, M., C. Pacot, I. Dugail, P. Lemarchand, C. Guichard, X. Le Liepvre, C. Berthelier-Lubrano, B. Spiegelman, J. B. Kim, P. Ferre, and F. Foufelle. 1999. ADD1/SREBP-1c is required in the activation of hepatic lipogenic gene expression by glucose. Mol. Cell. Biol. 19:3760-3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Green, H., and M. Meuth. 1974. An established pre-adipose cell line and its differentiation in culture. Cell 3:127-133. [DOI] [PubMed] [Google Scholar]
- 15.Gregori, C., A. Kahn, and A. L. Pichard. 1993. Competition between transcription factors HNF1 and HNF3, and alternative cell-specific activation by DBP and C/EBP contribute to the regulation of the liver-specific aldolase B promoter. Nucleic Acids Res. 21:897-903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo, X., and K. Liao. 2000. Analysis of gene expression profile during 3T3-L1 preadipocyte differentiation. Gene 251:45-53. [DOI] [PubMed] [Google Scholar]
- 17.Hemati, N., S. E. Ross, R. L. Erickson, G. E. Groblewski, and O. A. MacDougald. 1997. Signaling pathways through which insulin regulates CCAAT/enhancer binding protein alpha (C/EBPalpha) phosphorylation and gene expression in 3T3-L1 adipocytes. Correlation with GLUT4 gene expression. J. Biol. Chem. 272:25913-25919. [DOI] [PubMed] [Google Scholar]
- 18.Joseph, S. B., B. A. Laffitte, P. H. Patel, M. A. Watson, K. E. Matsukuma, R. Walczak, J. L. Collins, T. F. Osborne, and P. Tontonoz. 2002. Direct and indirect mechanisms for regulation of fatty acid synthase gene expression by liver X receptors. J. Biol. Chem. 277:11019-11025. [DOI] [PubMed] [Google Scholar]
- 19.Kahn, B. B., and J. S. Flier. 2000. Obesity and insulin resistance. J. Clin. Investig. 106:473-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kasanicki, M. A., and P. F. Pilch. 1990. Regulation of glucose-transporter function. Diabetes Care 13:219-227. [DOI] [PubMed] [Google Scholar]
- 21.Kim, J. B., and B. M. Spiegelman. 1996. ADD1/SREBP1 promotes adipocyte differentiation and gene expression linked to fatty acid metabolism. Genes Dev. 10:1096-1107. [DOI] [PubMed] [Google Scholar]
- 22.Klatt, P., J. Cacho, M. D. Crespo, E. Herrera, and P. Ramos. 2000. Nitric oxide inhibits isoproterenol-stimulated adipocyte lipolysis through oxidative inactivation of the beta-agonist. Biochem. J. 351:485-493. [PMC free article] [PubMed] [Google Scholar]
- 23.Krause, B. R., and A. D. Hartman. 1984. Adipose tissue and cholesterol metabolism. J. Lipid Res. 25:97-110. [PubMed] [Google Scholar]
- 24.Laffitte, B. A., J. J. Repa, S. B. Joseph, D. C. Wilpitz, H. R. Kast, D. J. Mangelsdorf, and P. Tontonoz. 2001. LXRs control lipid-inducible expression of the apolipoprotein E gene in macrophages and adipocytes. Proc. Natl. Acad. Sci. USA 98:507-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lavia, P., and P. Jansen-Durr. 1999. E2F target genes and cell-cycle checkpoint control. Bioessays 21:221-230. [DOI] [PubMed] [Google Scholar]
- 26.Lu, T. T., J. J. Repa, and D. J. Mangelsdorf. 2001. Orphan nuclear receptors as eLiXiRs and FiXeRs of sterol metabolism. J. Biol. Chem. 276:37735-37738. [DOI] [PubMed] [Google Scholar]
- 27.Luo, Y., and A. R. Tall. 2000. Sterol upregulation of human CETP expression in vitro and in transgenic mice by an LXR element. J. Clin. Investig. 105:513-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.MacDougald, O. A., and M. D. Lane. 1995. Transcriptional regulation of gene expression during adipocyte differentiation. Annu. Rev. Biochem. 64:345-373. [DOI] [PubMed] [Google Scholar]
- 29.MacDougald, O. A., and S. Mandrup. 2002. Adipogenesis: forces that tip the scales. Trends Endocrinol. Metab. 13:5-11. [DOI] [PubMed] [Google Scholar]
- 30.Magana, M. M., S. H. Koo, H. C. Towle, and T. F. Osborne. 2000. Different sterol regulatory element-binding protein-1 isoforms utilize distinct co-regulatory factors to activate the promoter for fatty acid synthase. J. Biol. Chem. 275:4726-4733. [DOI] [PubMed] [Google Scholar]
- 31.Morrison, R. F., and S. R. Farmer. 1999. Insights into the transcriptional control of adipocyte differentiation. J. Cell. Biochem. Suppl. 32-33:59-67. [DOI] [PubMed] [Google Scholar]
- 32.Morrison, R. F., and S. R. Farmer. 1999. Role of PPARgamma in regulating a cascade expression of cyclin-dependent kinase inhibitors, p18(INK4c) and p21(Waf1/Cip1), during adipogenesis. J. Biol. Chem. 274:17088-17097. [DOI] [PubMed] [Google Scholar]
- 33.Patel, Y. M., and M. D. Lane. 2000. Mitotic clonal expansion during preadipocyte differentiation: calpain-mediated turnover of p27. J. Biol. Chem. 275:17653-17660. [DOI] [PubMed] [Google Scholar]
- 34.Peet, D. J., B. A. Janowski, and D. J. Mangelsdorf. 1998. The LXRs: a new class of oxysterol receptors. Curr. Opin. Genet. Dev. 8:571-575. [DOI] [PubMed] [Google Scholar]
- 35.Peet, D. J., S. D. Turley, W. Ma, B. A. Janowski, J. M. Lobaccaro, R. E. Hammer, and D. J. Mangelsdorf. 1998. Cholesterol and bile acid metabolism are impaired in mice lacking the nuclear oxysterol receptor LXR alpha. Cell 93:693-704. [DOI] [PubMed] [Google Scholar]
- 36.Prattes, S., G. Horl, A. Hammer, A. Blaschitz, W. F. Graier, W. Sattler, R. Zechner, and E. Steyrer. 2000. Intracellular distribution and mobilization of unesterified cholesterol in adipocytes: triglyceride droplets are surrounded by cholesterol-rich ER-like surface layer structures. J. Cell Sci. 113:2977-2989. [DOI] [PubMed] [Google Scholar]
- 37.Qiu, Z., Y. Wei, N. Chen, M. Jiang, J. Wu, and K. Liao. 2001. DNA synthesis and mitotic clonal expansion is not a required step for 3T3-L1 preadipocyte differentiation into adipocytes. J. Biol. Chem. 276:11988-11995. [DOI] [PubMed] [Google Scholar]
- 38.Rangwala, S. M., and M. A. Lazar. 2000. Transcriptional control of adipogenesis. Annu. Rev. Nutr. 20:535-559. [DOI] [PubMed] [Google Scholar]
- 39.Repa, J. J., G. Liang, J. Ou, Y. Bashmakov, J. M. Lobaccaro, I. Shimomura, B. Shan, M. S. Brown, J. L. Goldstein, and D. J. Mangelsdorf. 2000. Regulation of mouse sterol regulatory element-binding protein-1c gene (SREBP-1c) by oxysterol receptors, LXRalpha and LXRbeta. Genes Dev. 14:2819-2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Repa, J. J., S. D. Turley, J. A. Lobaccaro, J. Medina, L. Li, K. Lustig, B. Shan, R. A. Heyman, J. M. Dietschy, and D. J. Mangelsdorf. 2000. Regulation of absorption and ABC1-mediated efflux of cholesterol by RXR heterodimers. Science 289:1524-1529. [DOI] [PubMed] [Google Scholar]
- 41.Rodbell, M. 1964. Metabolism of isolated fat cells. I. Effects of hormones of glucose metabolism and lipolysis. J. Biol. Chem. 239:375-380. [PubMed] [Google Scholar]
- 42.Rosen, E. D., C. J. Walkey, P. Puigserver, and B. M. Spiegelman. 2000. Transcriptional regulation of adipogenesis. Genes Dev. 14:1293-1307. [PubMed] [Google Scholar]
- 43.Ross, S. E., R. L. Erickson, N. Hemati, and O. A. MacDougald. 1999. Glycogen synthase kinase 3 is an insulin-regulated C/EBPα kinase. Mol. Cell. Biol. 19:8433-8441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ross, S. E., N. Hemati, K. A. Longo, C. N. Bennett, P. C. Lucas, R. L. Erickson, and O. A. MacDougald. 2000. Inhibition of adipogenesis by Wnt signaling. Science 289:950-953. [DOI] [PubMed] [Google Scholar]
- 45.Schultz, J. R., H. Tu, A. Luk, J. J. Repa, J. C. Medina, L. Li, S. Schwendner, S. Wang, M. Thoolen, D. J. Mangelsdorf, K. D. Lustig, and B. Shan. 2000. Role of LXRs in control of lipogenesis. Genes Dev. 14:2831-2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sebastian, S., J. D. Horton, and J. E. Wilson. 2000. Anabolic function of the type II isozyme of hexokinase in hepatic lipid synthesis. Biochem. Biophys. Res. Commun. 270:886-891. [DOI] [PubMed] [Google Scholar]
- 47.Sorensen, H. N., K. M. Gautik, J. Bremer, and O. Spydevold. 1992. Induction of the three peroxisomal beta-oxidation enzymes is synergistically regulated by dexamethasone and fatty acids, and counteracted by insulin in Morris 7800C1 hepatoma cells in culture. Eur. J. Biochem. 208:705-711. [DOI] [PubMed] [Google Scholar]
- 48.Soukas, A., N. D. Socci, B. D. Saatkamp, S. Novelli, and J. M. Friedman. 2001. Distinct transcriptional profiles of adipogenesis in vivo and in vitro. J. Biol. Chem. 276:34167-34174. [DOI] [PubMed] [Google Scholar]
- 49.Spiegelman, B. M., and J. S. Flier. 2001. Obesity and the regulation of energy balance. Cell 104:531-543. [DOI] [PubMed] [Google Scholar]
- 50.Steppan, C. M., S. T. Bailey, S. Bhat, E. J. Brown, R. R. Banerjee, C. M. Wright, H. R. Patel, R. S. Ahima, and M. A. Lazar. 2001. The hormone resistin links obesity to diabetes. Nature 409:307-312. [DOI] [PubMed] [Google Scholar]
- 51.Summers, S. A., V. P. Yin, E. L. Whiteman, L. A. Garza, H. Cho, R. L. Tuttle, and M. J. Birnbaum. 1999. Signaling pathways mediating insulin-stimulated glucose transport. Ann. N. Y. Acad. Sci. 892:169-186. [DOI] [PubMed] [Google Scholar]
- 52.Tabor, D. E., J. B. Kim, B. M. Spiegelman, and P. A. Edwards. 1999. Identification of conserved cis-elements and transcription factors required for sterol-regulated transcription of stearoyl-CoA desaturase 1 and 2. J. Biol. Chem. 274:20603-20610. [DOI] [PubMed] [Google Scholar]
- 53.Timchenko, N. A., M. Wilde, P. Iakova, J. H. Albrecht, and G. J. Darlington. 1999. E2F/p107 and E2F/p130 complexes are regulated by C/EBPα in 3T3-L1 adipocytes. Nucleic Acids Res. 27:3621-3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Timchenko, N. A., M. Wilde, K. I. Kosai, A. Heydari, T. A. Bilyeu, M. J. Finegold, K. Mohamedali, A. Richardson, and G. J. Darlington. 1998. Regenerating livers of old rats contain high levels of C/EBPα that correlate with altered expression of cell cycle associated proteins. Nucleic Acids Res. 26:3293-3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Timchenko, N. A., M. Wilde, M. Nakanishi, J. R. Smith, and G. J. Darlington. 1996. CCAAT/enhancer-binding protein alpha (C/EBP alpha) inhibits cell proliferation through the p21 (WAF-1/CIP-1/SDI-1) protein. Genes Dev. 10:804-815. [DOI] [PubMed] [Google Scholar]
- 56.Tobin, K. A., H. H. Steineger, S. Alberti, O. Spydevold, J. Auwerx, J. A. Gustafsson, and H. I. Nebb. 2000. Cross-talk between fatty acid and cholesterol metabolism mediated by liver X receptor-alpha. Mol. Endocrinol. 14:741-752. [DOI] [PubMed] [Google Scholar]
- 57.Venkateswaran, A., B. A. Laffitte, S. B. Joseph, P. A. Mak, D. C. Wilpitz, P. A. Edwards, and P. Tontonoz. 2000. Control of cellular cholesterol efflux by the nuclear oxysterol receptor LXR alpha. Proc. Natl. Acad. Sci. USA 97:12097-12102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Way, J. M., W. W. Harrington, K. K. Brown, W. K. Gottschalk, S. S. Sundseth, T. A. Mansfield, R. K. Ramachandran, T. M. Willson, and S. A. Kliewer. 2001. Comprehensive messenger ribonucleic acid profiling reveals that peroxisome proliferator-activated receptor gamma activation has coordinate effects on gene expression in multiple insulin-sensitive tissues. Endocrinology 142:1269-1277. [DOI] [PubMed] [Google Scholar]
- 59.Willy, P. J., K. Umesono, E. S. Ong, R. M. Evans, R. A. Heyman, and D. J. Mangelsdorf. 1995. LXR, a nuclear receptor that defines a distinct retinoid response pathway. Genes Dev. 9:1033-1045. [DOI] [PubMed] [Google Scholar]
- 60.Wu, Z., Y. Xie, N. L. Bucher, and S. R. Farmer. 1995. Conditional ectopic expression of C/EBP beta in NIH-3T3 cells induces PPAR gamma and stimulates adipogenesis. Genes Dev. 9:2350-2363. [DOI] [PubMed] [Google Scholar]
- 61.Yamauchi, T., J. Kamon, H. Waki, Y. Terauchi, N. Kubota, K. Hara, Y. Mori, T. Ide, K. Murakami, N. Tsuboyama-Kasaoka, O. Ezaki, Y. Akanuma, O. Gavrilova, C. Vinson, M. L. Reitman, H. Kagechika, K. Shudo, M. Yoda, Y. Nakano, K. Tobe, R. Nagai, S. Kimura, M. Tomita, P. Froguel, and T. Kadowaki. 2001. The fat-derived hormone adiponectin reverses insulin resistance associated with both lipoatrophy and obesity. Nat. Med. 7:941-946. [DOI] [PubMed] [Google Scholar]
- 62.Yeh, W. C., Z. Cao, M. Classon, and S. L. McKnight. 1995. Cascade regulation of terminal adipocyte differentiation by three members of the C/EBP family of leucine zipper proteins. Genes Dev. 9:168-181. [DOI] [PubMed] [Google Scholar]
- 63.Young, C. S., M. Kitamura, S. Hardy, and J. Kitajewski. 1998. Wnt-1 induces growth, cytosolic beta-catenin, and Tcf/Lef transcriptional activation in Rat-1 fibroblasts. Mol. Cell. Biol. 18:2474-2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang, Y., J. J. Repa, K. Gauthier, and D. J. Mangelsdorf. 2001. Regulation of lipoprotein lipase by the oxysterol receptors, LXRα and LXRβ. J. Biol. Chem. 276:43018-43024. [DOI] [PubMed] [Google Scholar]