Abstract
Sox10 belongs to a family of transcription regulators characterized by a DNA-binding domain known as the HMG box. It plays fundamental roles in neural crest development, peripheral gliogenesis, and terminal differentiation of oligodendrocytes. In accord with its function as transcription factor, Sox10 contains two nuclear localization signals and is most frequently detected in the nucleus. In this study, we report that Sox10 is an active nucleocytoplasmic shuttle protein, competent of both entering and exiting the nucleus. We identified a functional Rev-type nuclear export signal within the DNA-binding domain of Sox10. Mutational inactivation of this nuclear export signal or treatment of cells with the CRM1-specific export inhibitor leptomycin B inhibited nuclear export and consequently nucleocytoplasmic shuttling of Sox10. Importantly, the inhibition of the nuclear export of Sox10 led to decreased transactivation of transfected reporters and endogenous target genes, arguing that continuous nucleocytoplasmic shuttling is essential for the function of Sox10. To our knowledge this is the first time that nuclear export has been reported and shown to be functionally relevant for any Sox protein.
Sox proteins form a large family of transcription regulators related to the testis-determining factor SRY. All family members are characterized by a highly conserved DNA-binding and bending domain, called the HMG box according to its similarity to the DNA-binding domain of the high mobility group B (HMGB) proteins (7). cDNA sequences for more than 30 Sox genes have been identified. Some are known to play fundamental roles in vertebrate development and differentiation processes, including endoderm formation, sex determination, gliogenesis, chondrogenesis, hemopoiesis, and neural crest and lens development (5, 50). Sequence similarities outside the conserved HMG box provide criteria to divide Sox proteins into subgroups A to J (5). These subgroups can be found from simple organisms such as Caenorhabditis elegans to complex organisms such as humans (33, 50).
One of the best characterized groups of Sox proteins is subgroup E. This group includes Sox8, Sox9, and Sox10. Sox8 is the most recently found member of this group, and its exact function is still unknown (34, 42, 43). Sox9 is essential for chrondrocyte differentiation and cartilage formation in mouse (2) and chick (19). In accord, inactivation of a single Sox9 allele in humans is the cause of a severe skeletal malformation syndrome named campomelic dysplasia (15, 49). In XY individuals, campomelic dysplasia is often linked to sex reversal. Sox10, finally, is essential for neural crest development and has been identified as the causative gene for Hirschsprung-Waardenburg syndrome (20, 23, 35, 44). Inactivation or deletion of both Sox10 alleles in mice leads to a complete loss of neural crest-derived melanocytes, the enteric nervous system, and all glia in the peripheral nervous system (6). Additionally, terminal differentiation of oligodendrocytes is disturbed (46). Target genes of Sox10 include genes important for glial development (ErbB3) and identity such as the genes for the myelin proteins protein zero (P0), myelin basic protein, and proteolipid protein (PLP) (6, 31, 46).
One defining feature of eukaryotic cells is their spatial and functional division into nucleus and cytoplasm by the nuclear envelope. This separation requires specific mechanisms for the continuous transport of a large number of macromolecules between both compartments. Nucleocytoplasmic transport takes place through the nuclear pore and is highly regulated by specific signals and transport receptors. In general, nuclear import (reviewed in references 18 and 30) requires energy and the presence of importin α, importin β, RanGTP, and short stretches of basic amino acids, termed nuclear localization signals (NLS), in the cargo (4, 9). Sox9 contains two NLS within the HMG domain (47). These signals are highly conserved in Sox8 and Sox10. In contrast, the signal-mediated nuclear export pathways are less understood. The best-characterized nuclear export signal (NES) is the small, hydrophobic, leucine-rich NES, which was initially described in the protein kinase inhibitor (51) and in the human immunodeficiency virus type-1 (HIV-1) Rev protein (13). The direct interaction with the export factor CRM1 is absolutely essential for the translocation of proteins containing a leucine-rich NES from the nucleus into the cytoplasm (14, 16, 45). Leucine-rich NESs have been identified in an increasing number of cellular and viral proteins executing heterogenous biological functions. Those include RNA transport (22, 24, 37), cell cycle (48), regulation of kinase activity (12), and transcriptional control (1, 17). Proteins containing both NLS and NES have the capacity for continuous shuttling between the cytoplasm and the nucleus. In the case of transcription factors this is of particular interest, because of the possibility of further regulation of transcription factor activity. So far, nuclear export has not been analyzed in any Sox protein. Here we investigate the capacity of Sox10 to shuttle between the nucleus and the cytoplasm and analyze potential consequences for its function as transcriptional regulator.
MATERIALS AND METHODS
Plasmids.
Wild-type human Sox10 and mutated Sox10 cDNAs were cloned into various expression vectors by standard methods using PCR. Plasmids expressing green fluorescent protein (GFP)-Sox10 fusion proteins were generated by cloning wild-type and mutant Sox10 coding regions between the EcoRI and BamHI site of pEGFP-N1 (Clontech Laboratories, Palo Alto, Calif.). pEGFP-Sox10-ΔNES and pEGFP-Sox10-NES-L138A express GFP fusion proteins in which the leucine residues corresponding to Sox10 amino acid positions 134 and 138 or 138 alone were mutated into alanines. In pEGFP-NESREX-Sox10-NES-L138A the NES (amino acids [aa] 82 to 93) (11) of human T-cell leukemia virus type 1 Rex was fused in frame between GFP and Sox10-NES-L138A. A bacterial vector expressing GST-Sox10-NES-GFP wild-type protein was generated by insertion of a double-stranded synthetic oligonucleotide between the BamHI and NheI site of pGEX-GFP (39). The oligonucleotide encodes a short peptide motif (Sox10 aa 134 to 145) fused in frame between glutathione S-transferase (GST) and GFP. pGEX-Sox10-ΔNES-GFP and pGEX-Sox10-NES-L138A-GFP express mutated versions of this fusion protein in which the leucine residues corresponding to Sox10 aa 134 and 138 or 138 alone were mutated into alanines. Plasmids GST-Rev-NES-GFP (38), pCFN-β-galactosidase (β-Gal), pCFNrev-β-Gal (41), the pUL69 expression plasmid (24) and the luciferase reporter plasmid carrying the rat P0 promoter from position −915 to +48 are described elsewhere (31).
Cell culture, transfection, luciferase assays, RNA preparation, semiquantitative reverse transcription (RT)-PCR, Western blot analysis and EMSA.
HeLa cells, C6 cells and N2A neuroblastoma cells were maintained and transfected with calcium phosphate as described (31, 38). For luciferase assays, N2A neuroblastoma cells were transfected in triplicates on 35 mm dishes with 0.2 μg of luciferase reporter plasmid and 1 μg of the respective Sox10 construct. After transfection cells were returned to Dulbecco's modified Eagle medium containing 4% fetal calf serum. Cells were harvested 36 h posttransfection, and extracts were assayed for luciferase activity (32). Stable N2A cells capable of expressing Sox10 in a tetracycline inducible manner were kept, treated with doxycycline, and used for RNA preparation as described (31). Detection of products specific for P0, PLP, and GAPDH in cDNA generated by reverse transcription of RNA from these cells was performed by semiquantitative PCR (31, 46). Where indicated, cells were treated with 2.5 ng/ml LMB (kindly provided by Minoru Yoshida, Tokyo, Japan) for 24 h. For Western blot analysis whole cell extracts were prepared and proteins were separated on 10% SDS-polyacrylamide gels. After transfer to nitrocellulose membranes (Optitran-B85; Schleicher and Schuell, Dassel; Germany), Western blots with antibodies against GFP (Clontech Laboratories, Palo Alto, Calif.) and Sox10 (23) were performed as described (40). The same extracts were incubated with the 32P-labeled Sox10 binding site C/C′ as described (31).
Purification of GST-GFP fusion proteins, microinjection and homokaryon analysis.
GST-GFP hybrid proteins were expressed in Escherichia coli BL21 and purified on glutathione Sepharose 4B (Pharmacia Biotech, Germany) as previously described (38). Eluted proteins were analyzed by SDS-PAGE and Coomassie blue staining. Fractions were pooled, concentrated by ultrafiltration with a Nanosep filter (Pall-Filtron, USA) and microinjected into the nuclei of HeLa or Vero cells at 1.5 mg/ml in combination with rabbit IgG (1.0 mg/ml) (injection control) by using a CompiC INJECT computer-assisted injection system (Cellbiology Trading, Hamburg, Germany). Cells were fixed 45 min postinjection with 3% paraformaldehyde, and the injected proteins were analyzed by immunofluorescence analysis as described previously (39). In some experiments, 2.5 ng/ml LMB was added 2 h prior microinjection. For homokaryon analysis, HeLa cells were incubated for 24 h in MEM containing 1% bovine serum albumin. Homokaryons containing two nuclei were microinjected with full-length GFP-Sox10 fusion proteins (prior isolated from GFP-Sox10 overexpressing cells) together with a rabbit IgG into one nucleus. After 2 h of incubation at 37°C, cells were processed for immunofluorescence analysis.
Heterokaryon assays.
Nucleocytoplasmic shuttling of Sox10 and its mutants was analyzed in a heterokaryon assay, as initially described by Pinol-Roma and Dreyfuss (36). Briefly, transfected HeLa cells or C6 cells were seeded on glass coverslips together with an equal number of NIH 3T3 cells and were incubated overnight. Cells were incubated with 50 μg/ml cycloheximide 30 min prior to the fusion and throughout the experiment. In some experiments, 2.5 ng/ml LMB was added 3 h before cell fusion. Subsequently, cells were washed in PBS, and heterokaryons were formed by incubating the cells for 2 min in 50% polyethylene glycol 8000 in Dulbecco's modified Eagle medium. Cells were returned to fresh medium containing cycloheximide (50 μg/ml) and LMB (2.5 ng/ml) when needed. After 2 h incubation at 37°C, cells were processed for indirect immunofluorescence analysis. After secondary antibody incubation, Hoechst 33258 (5 μg/ml; Sigma, Taufkirchen, Germany) were added for 5 min. At least 25 heterokaryons were analyzed for each experiment and nuclear shuttling was only scored positive when a minimum of 80% of the heterokaryons showed a positive staining of the investigated protein.
Immunofluorescence microscopy.
Immunofluorescence microscopy was performed essentially as previously described (40). Cells were fixed for 20 min with 3% paraformaldehyde. Subsequently, cells were incubated with 0.1 M glycine, pH 8.5 for 15 min and blocked for 30 min with 1% bovine serum albumin-phosphate-buffered saline (PBS). Cells were then stained for 30 min with the primary antibodies and after extensive washing with PBS further incubated with appropriate secondary antibodies coupled to Cy2 or Cy3 (Biotrends, Cologne, Germany) for 30 min. The samples were washed in PBS, mounted in Mowiol (Calbiochem, La Jolla, Calif.) and analyzed using a Leica inverted microscope. Images were recorded with a cooled MicroMax charge-coupled device camera (Princeton Instruments, Stanford, Calif.) and processed using the IPLab spectrum and Adobe Photoshop software packages.
RESULTS
Subcellular localization of Sox10.
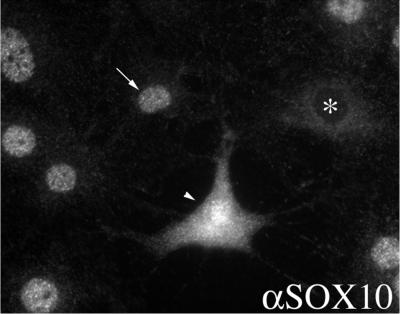
To characterize the subcellular localization of Sox10 in detail, we investigated the intracellular distribution of Sox10 in C6 cells by indirect immunofluorescence. As shown in Fig. 1, staining of endogenous protein revealed that Sox10 is localized in the majority of cells in the nucleus. In some cells Sox10 was equally distributed between nucleus and cytoplasm or exclusively found in the cytoplasm. At the moment, we do not know the reason for the divergent localization of Sox10.
FIG. 1.
Subcellular localization of Sox10 in cultured C6 cells by indirect immunofluorescence microscopy using an antibody against Sox10 (αSOX10). Sox10 is localized in the nucleus of the majority of cells (arrow, 92%). However, in some cells Sox10 was equally distributed between nucleus and cytoplasm (arrowhead, 5%) or exclusively found in the cytoplasm (asterisk, 3%). For quantification 230 cells were examined.
Characterization of GFP-Sox10.
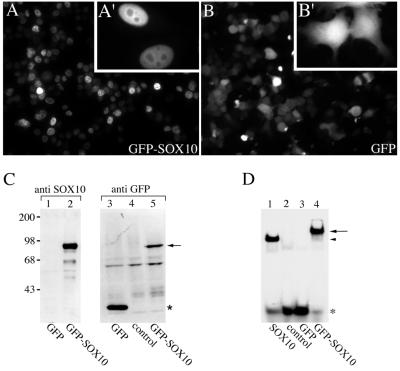
For our further studies, we fused human Sox10 in frame to a gene encoding the enhanced version of the green fluorescent protein (GFP) of Aequorea victoria. The GFP-Sox10 construct and GFP alone were transfected into HeLa cells, and GFP expression was monitored 24 h posttransfection. As published previously (38), expression of GFP alone resulted in cytoplasmic and nucleoplasmic signals (Fig. 2B). In contrast, expression of GFP-Sox10 resulted in an accumulation of the fusion protein in the nucleus of most cells (Fig. 2A). The GFP tag had no influence on the nuclear import of Sox10. GFP-Sox10 had a subcellular distribution in HeLa cells similar to endogenous Sox10 in C6 cells. Figure 2C shows a Western blot analysis of whole cell extracts. An anti GFP antibody as well as an anti Sox10 antibody recognized the GFP-Sox10 fusion protein of the expected size. We next analyzed whether the GFP tag interferes with the DNA binding properties of Sox10. As shown in the gel mobility shift assay in Fig. 2D, GFP-Sox10 bound to an oligonucleotide containing the Sox10 binding site C/C′ from the P0 promoter, whereas GFP alone did not.
FIG. 2.
Characterization of GFP-Sox10. (A and B) Subcellular localization of GFP and GFP-Sox10 in transiently transfected HeLa cells by direct fluorescence microscopy. GFP-Sox10 was localized in the nucleus of the majority of cells (94.5%), but in some cells it was equally distributed between nucleus and cytoplasm (4%) or exclusively found in the cytoplasm (1.5%). For quantification 200 cells were examined. Insets show cells at higher magnification. (C) Western blot analysis of whole-cell extracts from cells expressing either GFP or GFP-Sox10 using an anti Sox10 antibody as well as an anti-GFP antibody. The GFP-Sox10 fusion protein (lane 2 and 5) is marked with an arrow, and the GFP protein (lane 3) is marked with an asterisk. Lane 4 shows an extract of untransfected cells (control). Molecular mass standards are indicated in kilodaltons on the left. (D) Whole-cell extracts from cells ectopically expressing the indicated proteins or no protein (control) were analyzed in electrophoretic mobility shift assays for their ability to bind to radiolabeled oligonucleotides containing the Sox10 binding site C/C′ of the P0 promoter. The free probe is marked with an asterisk. The arrow indicates the complex between the oligonucleotide and GFP-Sox10. The arrowhead marks the position of the complex between the oligonucleotide and untagged Sox10.
Sox10 shuttles between the nucleus and the cytoplasm.
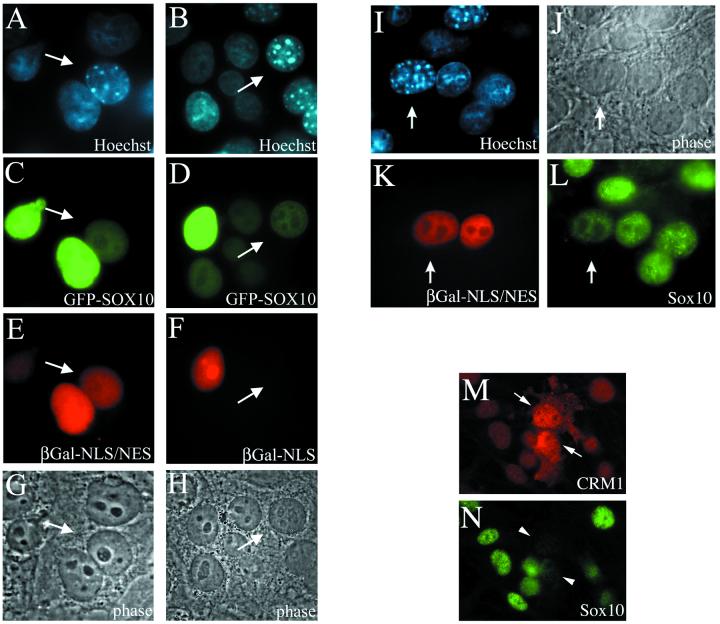
Since Sox10 can be found in the nucleus as well as in the cytoplasm, we addressed the question whether Sox10 has nucleocytoplasmic shuttling activity. An interspecies heterokaryon analysis was performed as originally described by Pinol-Roma and Dreyfuss (36). This assay has been successfully used to identify a series of cellular and viral shuttling proteins (8, 27, 28, 29). HeLa cells were cotransfected with a GFP-Sox10 expression plasmid and with either the internal control plasmid CFN-β-Gal or CFNrev-β-Gal. CFNrev-β-Gal encodes β-Gal fused to the NLS of simian virus 40 (SV40) T antigen and the NES of the HIV-1 Rev protein (β-Gal-NLS/NES). Thus, this fusion protein is able to shuttle rapidly between the nucleus and the cytoplasm. CFN-β-Gal expresses β-Gal fused only to the SV40 T antigen NLS and is therefore restricted to the nucleus (β-Gal-NLS) (41). Transfected cells were allowed to synthesize GFP-Sox10 together with one of the control proteins and were subsequently subjected to heterokaryon formation. After fixation, cells were immunostained for the presence and localization of β-Gal. To differentiate human from murine nuclei, cells were counterstained with Hoechst 33258 dye. Murine nuclei show a characteristic punctate pattern, whereas human nuclei are diffusely stained (Fig. 3A and B). In interspecies heterokaryons that coexpressed GFP-Sox10 and β-Gal-NLS/NES, both proteins were observed in murine and human nuclei (Fig. 3C and E). In contrast, when the localization of GFP-Sox10 and β-Gal-NLS was analyzed, only GFP-Sox10 was found to be present in the human and murine nuclei, whereas β-Gal-NLS was detected exclusively in the human nuclei (Fig. 3D and F). Figure 3G and H show both heterokaryons in phase contrast. In a quantitative microscopic examination of heterokaryons, GFP-Sox10 could be found in the vast majority of murine nuclei that were part of heterokaryons. These experiments show that GFP-Sox10 is transported from the transfected HeLa nucleus into the cytoplasm and subsequently into the murine nucleus.
FIG. 3.
Sox10 shuttles between the nucleus and the cytoplasm in a CRM1-dependent manner. (A to H) HeLa cells were cotransfected with expression plasmids for GFP-Sox10 and β-Gal-NLS or β-Gal-NLS/NES before they were subjected to heterokaryon formation with 3T3 cells. Two hours later, cells were fixed, stained for β-Gal, and counterstained with Hoechst 33258 dye (A and B). Murine nuclei are marked by an arrow. In interspecies heterokaryons that coexpressed GFP-Sox10 and β-Gal-NLS/NES, both proteins were observed in murine and human nuclei (C and E). In contrast, when the localization of GFP-Sox10 and β-Gal-NLS was analyzed, only GFP-Sox10 was found to be present in the murine nuclei, while β-Gal-NLS was restricted exclusively to the human nuclei (D and F). Figure 3G and H show both heterokaryons in phase contrast. About 30% of the heterokaryons reached equilibrium after 2 h. (I to L) C6 cells were transfected with an expression plasmid for β-Gal-NLS/NES before they were subjected to heterokaryon formation with 3T3 cells. Two hours later, cells (phase-contrast microscopy in panel J) were fixed and immunostained for endogenous Sox10 (L) and β-Gal (K) and were counterstained with Hoechst 33258 dye (I). (M and N) C6 cells were transfected with an expression plasmid for CRM1 and immunostained for CRM1 (M) as well as for Sox10 (N). Note that both the purely nuclear endogenous CRM1 as well as the ectopically expressed CRM1 is stained.
In a second series of interspecies heterokaryon analysis we tested whether endogenous Sox10 also has nucleocytoplasmic shuttling activity. Rat C6 cells were transfected with the internal positive control plasmid CFNrev-β-Gal (β-Gal-NLS/NES) and subsequently subjected to heterokaryon formation with mouse NIH 3T3 cells. After fixation, cells were immunostained for the presence and localization of β-Gal and Sox10, and counterstained with Hoechst 33258 dye (Fig. 3I). In interspecies heterokaryons (see Fig. 3J for phase contrast) containing transfected C6 cells, β-Gal-NLS/NES and the endogenous Sox10 from C6 cells were also observed in murine nuclei (Fig. 3K and L). This experiment shows that endogenous Sox10 is transported from the C6 nucleus into the cytoplasm and subsequently into the murine nucleus, and thus possesses nucleocytoplasmic shuttling activity.
Sox10 contains an active leucine-rich NES.
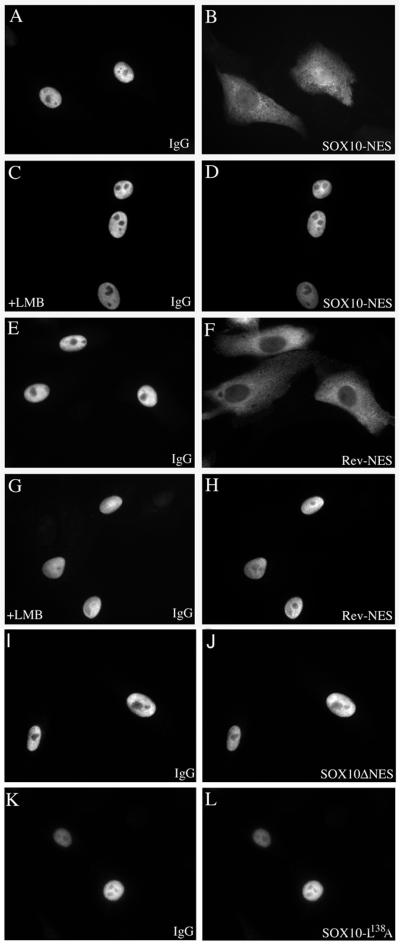
The most common NES are characterized by small, hydrophobic, leucine-rich regions (13). Sox10 contains a similar sequence between aa 134 and 145. To determine whether this sequence indeed acts as a NES, we inserted the aa 134 to 145 of Sox10 between glutathione S-transferase (GST) and GFP. For a positive control we used a fusion protein containing the NES of HIV-1 Rev. The recombinant fusion proteins were purified from E. coli and microinjected into the nuclei of Vero cells together with rabbit immunoglobulin G (IgG) as a marker for the injection site. At 45 min after injection, the cells were fixed and immunostained for the coinjected IgG control. The injected rabbit IgG was detected exclusively within the nucleus (Fig. 4A, C, E, and G), whereas the GST-GFP fusion proteins, containing either the NES of Sox10 (Fig. 4B) or Rev (Fig. 4F), were translocated from the nuclear injection site to the cytoplasm in significant amounts. This indicates that the Sox10 signal sequence is transferable and can function as an autonomous NES.
FIG. 4.
Sox10 contains an active leucine-rich NES. Purified GST-GFP fusion proteins were microinjected into the nuclei of Vero cells together with rabbit IgG as a marker for the nuclear injection site. At 45 min after injection, the cells were fixed and immunostained for the coinjected IgG control. The injected rabbit IgG was detected exclusively within the nucleus (A, C, E, G, I, and K), whereas the GST-GFP fusion proteins, containing either the NES of Sox10 (in panel B, Sox10-NES) or of Rev (in panel F, Rev-NES) were significantly translocated from the nuclear injection site into the cytoplasm. (C, D, G, and H) Cells had been pretreated with LMB, inhibiting CRM1-dependent nuclear export of Sox10-NES (D) or Rev-NES (H). In contrast, GST-GFP fusion proteins containing mutant versions of the Sox10 NESwere not able to enter the cytoplasm (J and L). Quantification of fluorescence intensity in the nucleus and cytoplasm revealed that 86.4% ± 5.9% of the microinjected GST-GFP fusion protein containing the Sox10-NES was translocated from the nucleus into the cytoplasm within 45 min.
We investigated whether the nuclear export ability of the GST-Sox10-NES-GFP fusion protein is affected by the CRM1 inhibitor leptomycin B (LMB). Therefore, Vero cells were pretreated with LMB (2.5 ng/ml) 2 h before microinjection of the Sox10-NES and Rev-NES containing GST-GFP fusion proteins. Nuclear export of the Rev-NES has been previously described to be LMB sensitive (11). As shown in Fig. 4D and H, the export was completely abolished in both cases. This experiment suggests that Sox10 is transported from the nucleus into the cytoplasm via the CRM1-dependent nuclear export pathway.
To further characterize the Sox10 nuclear export pathway, the NES of Sox10 was mutated. pGEX-Sox10-ΔNES-GFP and pGEX-Sox10-NES-L138A-GFP express mutated versions of the GST-Sox10-NES-GFP in which the leucine residues corresponding to Sox10 amino acid positions 134 and 138 (Sox10ΔNES) or 138 alone (Sox10-L138A) were exchanged to alanines. The fusion proteins were microinjected into the nuclei of Vero cells together with the control rabbit IgG. One hour after injection, the cells were fixed and immunostained for the coinjected IgG control. In sharp contrast to the GST-GFP fusion protein containing the wild-type NES of Sox10, both fusion proteins containing mutated versions of the Sox10-NES were not able to leave the nucleus (Fig. 4J and L) and remained colocalized with the coinjected rabbit IgG (Fig. 4I and K).
To directly investigate the contribution of the general export factor CRM1 on the subcellular distribution of Sox10, C6 cells were transfected with a CRM1 expression plasmid and subsequently immunostained for the presence and localization of CRM1 and Sox10. Overexpression of CRM1 (Fig. 3M) resulted in a partial relocalization of endogenous Sox10 from the nucleus into the cytoplasm (Fig. 3N), providing independent evidence that CRM1 is the physiological export receptor for Sox10.
The leucine-rich NES of Sox10 mediates nucleocytoplasmic shuttling of Sox10.
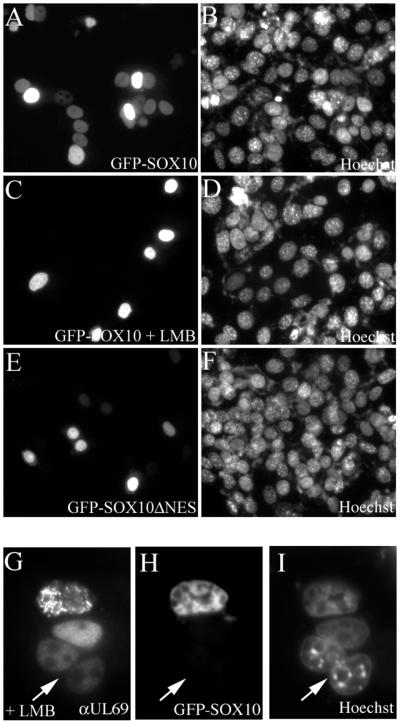
To analyze the activity of the leucine-rich NES in the context of the full-length Sox10 protein, we introduced the leucine-to-alanine mutations at aa 134 and 138 into the GFP-Sox10 fusion protein. In control heterokaryon assays, GFP-Sox10 wild-type fusion protein was rapidly translocated from the HeLa donor nucleus into the surrounding acceptor nuclei (Fig. 5A), and LMB significantly reduced this translocation (Fig. 5C). Whereas LMB treatment interfered with shuttling of GFP-Sox10 (Fig. 5H), it did not prevent translocation of the CRM1-independent pUL69 (24) in the same heterokaryon (Fig. 5G), proving that cells under LMB treatment were viable and that fusion and mixing of cytoplasm had occurred. The introduction of the leucine-to-alanine mutations in GFP-Sox10 strongly reduced the ability to shuttle from the donor nucleus into the surrounding acceptor nuclei (GFP-Sox10-ΔNES, Fig. 5E), although it did so less efficiently than LMB treatment. The respective Hoechst 33258 counterstains demonstrate that the cell density was comparable in all cases (Fig. 5B, D, F, and I). We conclude that the identified leucine-rich export signal is a major determinant for the nucleocytoplasmic shuttling ability of Sox10. The presence of an additional weak export signal cannot be ruled out at present.
FIG. 5.
The leucine-rich NES of Sox10 mediates nucleocytoplasmic shuttling of Sox10. GFP-Sox10 and GFP-Sox10ΔNES were transfected into HeLa cells and analyzed in heterokaryon assays. (A) The wild-type GFP-Sox10 fusion protein was rapidly translocated from the HeLa donor nucleus into the surrounding 3T3 acceptor nuclei. Under LMB treatment, GFP-Sox10 was not able to shuttle into the acceptor nuclei (C and H) in contrast to pUL96 (G). (E) The introduction of the leucine to alanine mutations resulted in a strong inhibition of nucleocytoplasmic transport of GFP-Sox10ΔNES fusion protein. (B, D, F, and I) Hoechst 33258 counterstains.
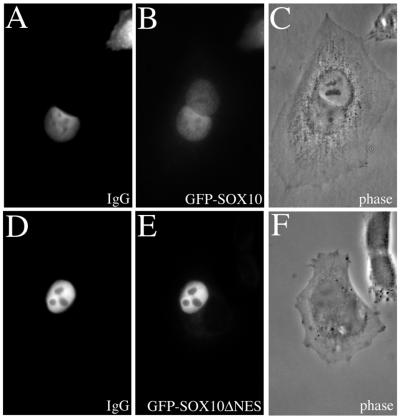
To verify the results described above, homokaryon assays were performed. This experimental approach has the advantage of being less artificial than heterokaryon assays. Homokaryons containing two HeLa nuclei (phase contrast-images are presented in Fig. 6C and F) were microinjected with wild-type GFP-Sox10 or GFP-Sox10ΔNES together with a rabbit control IgG into only one nucleus. After 2 h of incubation at 37°C, the injected rabbit IgG was detected exclusively within the injected nuclei (Fig. 6A and D), whereas the GFP-Sox10 wild-type fusion protein underwent a significant translocation from the injected nucleus into the second uninjected nucleus (Fig. 6B). In contrast, the mutated GFP-Sox10ΔNES fusion protein was not able to cross the nuclear pore of the injected nucleus and was not found in the potential acceptor nucleus within the same cell (Fig. 6E).
FIG. 6.
Homokaryon analysis of the nucleocytoplasmic shuttling ability of Sox10. One nucleus of homokaryons containing two nuclei was microinjected with full-length wild-type GFP-Sox10 or GFP-Sox10ΔNES together with a rabbit control IgG. After 2 h, the injected rabbit IgG was detected exclusively within the injected nuclei (A and D), whereas the GFP-Sox10 wild-type fusion protein underwent a significant translocation into the second, not injected nucleus (B). The mutated GFP-Sox10ΔNES fusion protein did not cross the nuclear pore of the injected nucleus (E). (C and F) Phase-contrast images.
Nucleocytoplasmic shuttling is crucial for Sox10-mediated transactivation.
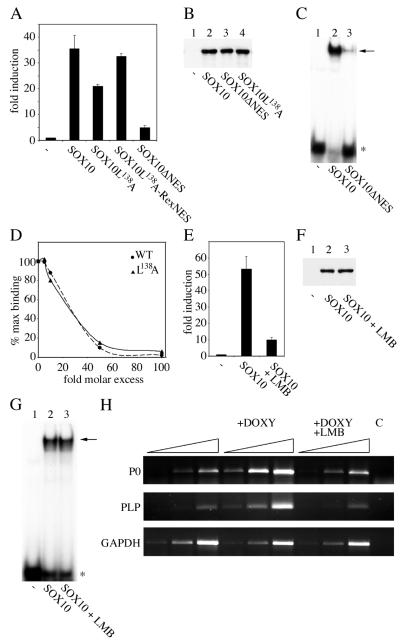
To investigate whether nucleocytoplasmic shuttling has an influence on the function of Sox10 as transcription regulator, we tested the ability of export-defective Sox10 mutants to transactivate a Sox10-responsive luciferase reporter plasmid in transient transfections (31). These experiments revealed a good correlation between Sox10-dependent transactivation and nucleocytoplasmic shuttling since the mutants GFP-Sox10ΔNES and GFP-Sox10L138A were reduced in their ability to stimulate gene expression from the P0 promoter (Fig. 7A). This effect was not due to reduced expression or instability of the mutant proteins, as Western blot analysis demonstrated a similar expression level of all used mutants and the wild-type protein (Fig. 7B). Unfortunately, the GFP-Sox10ΔNES fusion protein exhibited only residual DNA binding affinity (Fig. 7C). Thus, we were unable to distinguish for this mutant whether reduced transactivation is attributed to inhibited nucleocytoplasmic transport or to reduced DNA-binding affinity. The GFP-Sox10L138A mutant, in contrast, showed a DNA-binding affinity similar to that of wild-type GFP-Sox10 as revealed by competition analysis with cold competitor DNA (Fig. 7D). For this mutant, reduced Sox10-dependent transactivation thus appears to be a consequence of impaired nucleocytoplasmic shuttling. To verify that the reduced transactivation capacity of the GFP-Sox10L138A mutant is indeed due to altered nucleocytoplasmic shuttling ability, we inserted the heterologous NES of human T-cell leukemia virus type 1 Rex between the GFP and the Sox10 part of the fusion protein. The resulting restoration of the nuclear export capacity resulted in a near complete retrieval of the transcriptional activity (Fig. 7A).
FIG. 7.
Inhibition of the nuclear export activity of Sox10 leads to decreased transactivation. (A) Luciferase assays after cotransfection of a luciferase reporter carrying the Sox10-responsive P0-promoter and expression plasmids for GFP (-), wild-type GFP-Sox10, and its mutants. Error bars, standard deviations. (B and C) Western blot analysis with an anti-Sox10 antibody (B) and electrophoretic mobility shift analysis using probe C/C′ (C) of whole-cell extracts from cells expressing the indicated proteins. The free probe is marked with an asterisk. The arrow indicates the complex between the oligonucleotide and GFP-Sox fusion proteins. (D) Comparison of DNA-binding affinity of wild-type Sox10 (WT) and Sox10L138A (L138A) mutant. Electrophoretic mobility shift analysis was performed using labeled C/C′ as probe and increasing molar excess of unlabeled C/C′ as competitor. The signal intensity was quantified using the IPLab software package. (E) Luciferase assays after cotransfection of a luciferase reporter carrying the Sox10-responsive P0 promoter and expression plasmids for GFP-Sox10 or GFP (-) in the presence or absence of LMB. The data were normalized to cotransfected β-Gal under control of the cytomegalovirus promoter. Error bars, standard deviations. (F and G) Western blot analysis with an anti Sox10 antibody (F) and electrophoretic mobility shift analysis using probe C/C′ (G) of whole-cell extracts from LMB treated or untreated cells expressing GFP (-) or GFP-Sox10. (H) Semiquantitative RT-PCR analysis of endogenous P0 and PLP transcript levels in N2A Tet-On cells capable of inducibly expressing Sox10. Cells were treated with doxycycline for 48 h (+DOXY) or doxycycline for 48 h and LMB for 24 h (+DOXY +LMB) to induce Sox10 expression. cDNA from uninduced cells served as control.
We next tested the effect of LMB on the ability of Sox10 to transactivate. We found that under LMB treatment Sox10 had a strongly reduced ability to stimulate the P0-promoter in transient transfections (Fig. 7E). The dramatic effect of LMB on the transactivation ability of Sox10 was not due to different expression levels of Sox10 in the presence or absence of LMB as analyzed by Western blotting (Fig. 7F), nor was it due to an LMB-dependent incapacity to bind to DNA as shown by electrophoretic mobility shift assay (Fig. 7G).
Finally, we analyzed the effect of LMB on expression of endogenous target genes of Sox10 (31, 46). For this we used a stably transfected N2A Tet-On cell line capable of inducible expressing Sox10 (31). As shown by semiquantitative RT-PCR analyses, transcription of the two myelin genes P0 and PLP was strongly increased after doxycycline-dependent Sox10 induction (Fig. 7H). Additional LMB treatment prevented the Sox10-dependent increase of P0 and PLP transcription. GAPDH transcript levels were affected neither by treatment of doxycycline nor by treatment of doxycycline and LMB.
DISCUSSION
In eukaryotic cells, the nuclear envelope generates two distinct cellular compartments that separate transcription and DNA replication from protein biosynthesis. Thus, a given transcription factor has to be transported into the nucleus to ensure its function. The nuclear import signals of Sox9 are well characterized. A bipartite and a basic cluster NLS motif within the HMG domain of this protein have been identified to be necessary and sufficient for effective nuclear import (47). Both NLS motifs are conserved within the family of Sox proteins, and as indicated in Fig. 8A, Sox10 also contains both types of NLS. Interestingly, we found that in some cells Sox10 is not completely restricted to the nucleus. In a subset of cells Sox10 is even localized mainly in the cytoplasm. This fact could be due to the retention of newly synthesized Sox10 in the cytoplasm or to its export from the nucleus into the cytoplasm, with subsequent retention. In the present study, we demonstrate that Sox10 has the capacity to shuttle between the nucleus and the cytoplasm. The net nuclear localization usually observed is the result of nucleocytoplasmic shuttling kinetics in which the rate of import exceeds the rate of export. This is the first time that shuttling has been shown for any Sox protein. Furthermore, we characterized an active leucine-rich NES within the HMG box of Sox10 as a key determinant of its nucleocytoplasmic shuttling. A major pathway for nuclear export involves the exportin protein CRM1 (14, 16, 45). A role for CRM1 in nuclear export of Sox10 is strongly supported by two lines of evidence. For one, the CRM1 antagonist, LMB, is able to block nuclear export of a GST-Sox10-NES-GFP fusion protein and nucleocytoplasmic shuttling of Sox10. Secondly, overexpression of CRM1 leads to a redistribution of endogenous Sox10 in transfected cells.
FIG. 8.
Conservation of the Sox10 export sequence in different species and within subgroup E. (A) The distinct functional NLSs and the NES are indicated within the Sox10 protein. TA, transactivation domain. NLSs (aa 105 to 121 and aa 176 to 179). NES (aa 134 to 145). (B) Comparison of the NES defined in this work with the corresponding sequences of Sox10 from other species and with other Sox proteins. (C) Consensus sequence of leucine-rich nuclear export signals as defined by Bogerd et al. (3).
A consensus leucine-rich binding site for CRM1 has been defined (Fig. 8C), although CRM1-dependent NESs that diverge from this consensus on the basis of amino acid composition and spacing have also been identified (26, 38). The NES of Sox10 fits the consensus sequence of a leucine-rich export signal with the exception of the spacing between the last two leucine residues (Fig. 8A). It is well conserved in Sox10 proteins from human, mouse, rat, chicken, and zebra fish (Fig. 8B). Interestingly, the Sox10-NES is also detectable in the sequence of Sox8 and Sox9 (Fig. 8B), the other two members of subgroup E. However, in other subgroups the amino acid composition of the corresponding regions is changed. For example, in Sox4 and Sox11 (subgroup C) the critical third leucine residue is replaced by an arginine. In Sox15 (subgroup G) the critical third leucine residue is replaced by glutamine (Fig. 8B). Additionally, sequence inspection did not reveal the presence of classical leucine-rich NESs in other regions of the latter Sox proteins. At least this particular NES thus appears to be restricted to Sox proteins of subgroup E.
The activity of transcriptional regulators can be modulated at various levels. They include phosphorylation, acetylation, assembly, and interactions with other proteins, including cofactors. The molecular mechanisms that regulate the activity of Sox proteins are just beginning to be studied. In the case of Sox9 phosphorylation by cyclic AMP-dependent protein kinase enhances the ability to transactivate the Col2a1 chondrocyte-specific enhancer (21). Furthermore, it was shown that Sox10 binds to DNA both as monomer and dimer with different functional consequences (32). In this study, we demonstrate that inhibition of the nuclear export activity of Sox10 leads to a decrease in its transactivation capacity, when measured both on transfected luciferase reporters and on endogenous target genes. Based on our findings, we propose that continuous shuttling and regular localization of Sox10 in the cytoplasm is essential for its optimal transactivation ability. At the moment we do not know why the cytoplasmic stay is important for the function of Sox10. One can speculate that Sox10 receives a posttranslational modification in the cytoplasm and is then transported into the nucleus to activate its target genes. After a given time in the nucleus the posttranslational modification is removed, activity is reduced, and Sox10 is consecutively exported into the cytoplasm to start the next cycle of shuttling. A similar model is proposed for STAT1 (25), in which tyrosine-phosphorylated STAT1 dimers that bind to DNA accumulate in the nucleus. Under this condition the NES is masked and not accessible for CRM1. Following dephosphorylation, STAT1 is released from the DNA, recognized by CRM1, and subsequently exported into the cytoplasm.
Sox10 is a key regulator during early phases of neural crest development (6). These cells contain high amounts of Sox10. In the peripheral nervous system, neural crest cells give rise to neurons and glia. While glia continue to depend on Sox10 for their development, neurons stop to express Sox10. The specific removal of Sox10 seems to be important for neuronal cell development. If Sox10 removal solely depends on down-regulation of expression of its gene, the process would be fairly slow. The balance of import and export provides an additional mechanism for the regulation of Sox10 activity. Changes in the kinetics of nucleocytoplasmic shuttling provide a rapid mechanism for inactivation of the existing Sox10 protein. Interestingly, Dutton et al. identified a Sox10 mutant in zebra fish, in which leucine 142 (equivalent to leucine 138 of the NES of human Sox10) is changed to glutamine (10). In contrast to the alanine substitution mutant analyzed in this paper, the glutamine substitution exhibits not only defective shuttling but also reduced DNA binding activity (data not shown). It is tempting to speculate that the neural crest defects observed in the zebra fish mutant are at least partially due to disrupted nucleocytoplasmic shuttling. Proof for the in vivo role of nucleocytoplasmic shuttling will have to await the generation of an animal model in which nuclear export of Sox10 is selectively abolished.
Acknowledgments
We thank Minoru Yoshida for his generous gift of LMB and Matthias Dobbelstein for providing plasmids. We are grateful to Julia Brill, Katy Schmidt, and Claus Stolt for critical comments on the manuscript.
This work was supported by the DFG (grants We1326/7-2 and SFB473).
REFERENCES
- 1.Begitt, A., T. Meyer, M. van Rossum, and U. Vinkemeier. 2000. Nucleocytoplasmic translocation of Stat1 is regulated by a leucine-rich export signal in the coiled-coil domain. Proc. Natl. Acad. Sci. USA 97:10418-10423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bi, W., J. M. Deng, Z. Zhang, R. R. Behringer, and B. de Crombrugghe. 1999. Sox9 is required for cartilage formation. Nat. Genet. 22:85-89. [DOI] [PubMed] [Google Scholar]
- 3.Bogerd, H. P., R. A. Fridell, R. E. Benson, J. Hua, and B. R. Cullen. 1996. Protein sequence requirements for function of the human T-cell leukemia virus type 1 Rex nuclear export signal delineated by a novel in vivo randomization-selection assay. Mol. Cell. Biol. 16:4207-4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boulikas, T. 1993. Nuclear localization signals (NLS). Crit. Rev. Eukaryot. Gene Expr. 3:193-227. [PubMed] [Google Scholar]
- 5.Bowles, J., G. Schepers, and P. Koopman. 2000. Phylogeny of the SOX family of developmental transcription factors based on sequence and structural indicators. Dev. Biol. 227:239-255. [DOI] [PubMed] [Google Scholar]
- 6.Britsch, S., D. E. Goerich, D. Riethmacher, R. I. Peirano, M. Rossner, K. A. Nave, C. Birchmeier, and M. Wegner. 2001. The transcription factor Sox10 is a key regulator of peripheral glial development. Genes Dev. 15:66-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bustin, M. 2001. Revised nomenclature for high mobility group (HMG) chromosomal proteins. Trends Biochem. Sci. 26:152-153. [DOI] [PubMed] [Google Scholar]
- 8.Caceres, J. F., G. R. Screaton, and A. R. Krainer. 1998. A specific subset of SR proteins shuttles continuously between the nucleus and the cytoplasm. Genes Dev. 12:55-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dingwall, C., and R. A. Laskey. 1991. Nuclear targeting sequences-a consensus? Trends Biochem. Sci. 16:478-481. [DOI] [PubMed] [Google Scholar]
- 10.Dutton, K. A., A. Pauliny, S. S. Lopes, S. Elworthy, T. J. Carney, J. Rauch, R. Geisler, P. Haffter, and R. N. Kelsh. 2001. Zebrafish colourless encodes sox10 and specifies non-ectomesenchymal neural crest fates. Development 128:4113-4125. [DOI] [PubMed] [Google Scholar]
- 11.Elfgang, C., O. Rosorius, L. Hofer, H. Jaksche, J. Hauber, and D. Bevec. 1999. Evidence for specific nucleocytoplasmic transport pathways used by leucine-rich nuclear export signals. Proc. Natl. Acad. Sci. USA 96:6229-6234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Engel, K., A. Kotlyarov, and M. Gaestel. 1998. Leptomycin B-sensitive nuclear export of MAPKAP kinase 2 is regulated by phosphorylation. EMBO J. 17:3363-3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fischer, U., J. Huber, W. C. Boelens, I. W. Mattaj, and R. Luhrmann. 1995. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell 82:475-483. [DOI] [PubMed] [Google Scholar]
- 14.Fornerod, M., M. Ohno, M. Yoshida, and I. W. Mattaj. 1997. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 90:1051-1060. [DOI] [PubMed] [Google Scholar]
- 15.Foster, J. W., M. A. Dominguez-Steglich, S. Guioli, C. Kwok, P. A. Weller, M. Stevanovic, J. Weissenbach, S. Mansour, I. D. Young, P. N. Goodfellow, J. D. Brook, and A. J. Schäfer. 1994. Campomelic dysplasia and autosomal sex reversal caused by mutations in an SRY-related gene. Nature 372:525-530. [DOI] [PubMed] [Google Scholar]
- 16.Fukuda, M., S. Asano, T. Nakamura, M. Adachi, M. Yoshida, M. Yanagida, and E. Nishida. 1997. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature 390:308-311. [DOI] [PubMed] [Google Scholar]
- 17.Gaubatz, S., J. A. Lees, G. J. Lindeman, and D. M. Livingston. 2001. E2F4 is exported from the nucleus in a CRM1-dependent manner. Mol. Cell. Biol. 21:1384-1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Görlich, D., and U. Kutay. 1999. Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol. 15:607-660. [DOI] [PubMed] [Google Scholar]
- 19.Healy, C., D. Uwanogho, and P. T. Sharpe. 1999. Regulation and role of Sox9 in cartilage formation. Dev. Dynam. 215:69-78. [DOI] [PubMed] [Google Scholar]
- 20.Herbarth, B., V. Pingault, N. Bondurand, K. Kuhlbrodt, I. Hermans-Borgmeyer, A. Puliti, N. Lemort, M. Goossens, and M. Wegner. 1998. Mutation of the Sry-related Sox10 gene in Dominant megacolon, a mouse model for human Hirschsprung disease. Proc. Natl. Acad. Sci. USA 95:5161-5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang, W., X. Zhou, V. Lefebvre, and B. de Crombrugghe. 2000. Phosphorylation of SOX9 by cyclic AMP-dependent protein kinase A enhances SOX9's ability to transactivate a Col2a1 chondrocyte-specific enhancer. Mol. Cell. Biol. 20:4149-4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krätzer, F., O. Rosorius, P. Heger, N. Hirschmann, T. Dobner, J. Hauber, and R. H. Stauber. 2000. The adenovirus type 5 E1B-55K oncoprotein is a highly active shuttle protein and shuttling is independent of E4orf6, p53 and Mdm2. Oncogene 19:850-857. [DOI] [PubMed] [Google Scholar]
- 23.Kuhlbrodt, K., B. Herbarth, E. Sock, I. Hermans-Borgmeyer, and M. Wegner. 1998. Sox10, a novel transcriptional modulator in glial cells. J. Neurosci. 18:237-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lischka, P., O. Rosorius, E. Trommer, and T. Stamminger. 2001. A novel transferable nuclear export signal mediates CRM1-independent nucleocytoplasmic shuttling of the human cytomegalovirus transactivator protein pUL69. EMBO J. 20:7271-7283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McBride, K. M., C. McDonald, and N. C. Reich. 2000. Nuclear export signal located within the DNA-binding domain of the STAT1 transcription factor. EMBO J. 19:6196-6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McKinsey, T. A., C. L. Zhang, and E. N. Olson. 2001. Identification of a signal-responsive nuclear export sequence in class II histone deacetylases. Mol. Cell. Biol. 21:6312-6321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mears, W. E., and S. A. Rice. 1998. The herpes simplex virus immediate-early protein ICP27 shuttles between nucleus and cytoplasm. Virology 242:128-137. [DOI] [PubMed] [Google Scholar]
- 28.Meyer, B. E., and M. H. Malim. 1994. The HIV-1 Rev trans-activator shuttles between the nucleus and the cytoplasm. Genes Dev. 8:1538-1547. [DOI] [PubMed] [Google Scholar]
- 29.Michael, W. M., P. S. Eder, and G. Dreyfuss. 1997. The K nuclear shuttling domain: a novel signal for nuclear import and nuclear export in the hnRNP K protein. EMBO J. 16:3587-3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakielny, S., and G. Dreyfuss. 1999. Transport of proteins and RNAs in and out of the nucleus. Cell 99:677-690. [DOI] [PubMed] [Google Scholar]
- 31.Peirano, R. I., D. E. Goerich, D. Riethmacher, and M. Wegner. 2000. Protein zero expression is regulated by the glial transcription factor Sox10. Mol. Cell. Biol. 20:3198-3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peirano, R. I., and M. Wegner. 2000. The glial transcription factor Sox10 binds to DNA both as monomer and dimer with different functional consequences. Nucleic Acids Res. 28:3047-3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pevny, L. H., and R. Lovell-Badge. 1997. Sox genes find their feet. Curr. Opin. Genet. Dev. 7:338-344. [DOI] [PubMed] [Google Scholar]
- 34.Pfeifer, D., F. Poulat, E. Holinski-Feder, F. Kooy, and G. Scherer. 2000. The SOX8 gene is located within 700 kb of the tip of chromosome 16p and is deleted in a patient with ATR-16 syndrome. Genomics 63:108-116. [DOI] [PubMed] [Google Scholar]
- 35.Pingault, V., N. Bondurand, K. Kuhlbrodt, D. E. Goerich, M.-O. Prehu, A. Puliti, B. Herbarth, I. Hermans-Borgmeyer, E. Legius, G. Matthijs, J. Amiel, S. Lyonnet, I. Ceccherini, G. Romeo, J. C. Smith, A. P. Read, M. Wegner, and M. Goossens. 1998. Sox10 mutations in patients with Waardenburg-Hirschsprung disease. Nat. Genet. 18:171-173. [DOI] [PubMed] [Google Scholar]
- 36.Pinol-Roma, S., and G. Dreyfuss. 1992. Shuttling of pre-mRNA binding proteins between nucleus and cytoplasm. Nature 355:730-732. [DOI] [PubMed] [Google Scholar]
- 37.Pollard, V. W., and M. H. Malim. 1998. The HIV-1 Rev protein. Annu. Rev. Microbiol. 52:491-532. [DOI] [PubMed] [Google Scholar]
- 38.Rosorius, O., B. Fries, R. H. Stauber, N. Hirschmann, D. Bevec, and J. Hauber. 2000. Human ribosomal protein L5 contains defined nuclear localization and export signals. J. Biol. Chem. 275:12061-12068. [DOI] [PubMed] [Google Scholar]
- 39.Rosorius, O., P. Heger, G. Stelz, N. Hirschmann, J. Hauber, and R. H. Stauber. 1999. Direct observation of nucleocytoplasmic transport by microinjection of GFP-tagged proteins in living cells. BioTechniques 27:350-355. [DOI] [PubMed] [Google Scholar]
- 40.Rosorius, O., B. Reichart, F. Krätzer, P. Heger, M. C. Dabauvalle, and J. Hauber. 1999. Nuclear pore localization and nucleocytoplasmic transport of eIF-5A: evidence for direct interaction with the export receptor CRM1. J. Cell Sci. 112:2369-2380. [DOI] [PubMed] [Google Scholar]
- 41.Roth, J., and M. Dobbelstein. 1997. Export of hepatitis B virus RNA on a Rev-like pathway: inhibition by the regenerating liver inhibitory factor IκBα. J. Virol. 71:8933-8939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schepers, G. E., M. Bullejos, B. M. Hosking, and P. Koopman. 2000. Cloning and characterisation of the sry-related transcription factor gene sox8. Nucleic Acids Res. 28:1473-1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sock, E., K. Schmidt, I. Hermanns-Borgmeyer, M. R. Bösl, and M. Wegner. 2001. Idiopathic weight reduction in mice deficient in the high-mobility-group transcription factor Sox8. Mol. Cell. Biol. 21:6951-6959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Southard-Smith, E. M., L. Kos, and W. J. Pavan. 1998. Sox10 mutation disrupts neural crest development in Dom Hirschsprung mouse model. Nat. Genet. 18:60-64. [DOI] [PubMed] [Google Scholar]
- 45.Stade, K., C. S. Ford, C. Guthrie, and K. Weis. 1997. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell 90:1041-1050. [DOI] [PubMed] [Google Scholar]
- 46.Stolt, C. C., S. Rehberg, M. Ader, P. Lommes, D. Riethmacher, M. Schachner, U. Bartsch, and M. Wegner. 2002. Terminal differentiation of myelin-forming oligodendrocytes depends on the transcription factor Sox10. Genes Dev. 16:165-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Südbeck, P., and G. Scherer. 1997. Two independent nuclear localization signals are present in the DNA-binding high-mobility group domains of SRY and SOX9. J. Biol. Chem. 272:27848-27852. [DOI] [PubMed] [Google Scholar]
- 48.Toyoshima, F., T. Moriguchi, A. Wada, M. Fukuda, and E. Nishida. 1998. Nuclear export of cyclin B1 and its possible role in the DNA damage-induced G2 checkpoint. EMBO J. 17:2728-2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wagner, T., J. Wirth, J. Meyer, B. Zabel, M. Held, J. Zimmer, J. Pasantes, F. D. Bricarelli, J. Keutel, E. Hustert, U. Wolf, N. Tommerup, W. Schempp, and G. Scherer. 1994. Autosomal sex reversal and campomelic dysplasia are caused by mutations in and around the SRY-related gene Sox9. Cell 79:1111-1120. [DOI] [PubMed] [Google Scholar]
- 50.Wegner, M. 1999. From head to toes: the multiple facets of Sox proteins. Nucleic Acids Res. 27:1409-1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wen, W., J. L. Meinkoth, R. Y. Tsien, and S. S. Taylor. 1995. Identification of a signal for rapid export of proteins from the nucleus. Cell 82:463-473. [DOI] [PubMed] [Google Scholar]