Abstract
We previously defined Saccharomyces cerevisiae TFIID as a 15-subunit complex comprised of the TATA binding protein (TBP) and 14 distinct TBP-associated factors (TAFs). In this report we give a detailed biochemical characterization of this general transcription factor. We have shown that yeast TFIID efficiently mediates both basal and activator-dependent transcription in vitro and displays TATA box binding activity that is functionally distinct from that of TBP. Analyses of the stoichiometry of TFIID subunits indicated that several TAFs are present at more than 1 copy per TFIID complex. This conclusion was further supported by coimmunoprecipitation experiments with a systematic family of (pseudo)diploid yeast strains that expressed epitope-tagged and untagged alleles of the genes encoding TFIID subunits. Based on these data, we calculated a native molecular mass for monomeric TFIID. Purified TFIID behaved in a fashion consistent with this calculated molecular mass in both gel filtration and rate-zonal sedimentation experiments. Quite surprisingly, although the TAF subunits of TFIID cofractionated as a single complex, TBP did not comigrate with the TAFs during either gel filtration chromatography or rate-zonal sedimentation, suggesting that TBP has the ability to dynamically associate with the TFIID TAFs. The results of direct biochemical exchange experiments confirmed this hypothesis. Together, our results represent a concise molecular characterization of the general transcription factor TFIID from S. cerevisiae.
The general transcription factor TFIID plays a central role in the initiation of mRNA gene transcription. TFIID is the only general transcription factor with specific TATA box binding activity, and the expression of many RNA polymerase II-transcribed genes is dependent on TFIID function (1, 26). Saccharomyces cerevisiae TFIID is composed of 15 subunits, TATA binding protein (TBP) and 14 distinct TBP-associated factors (TAFs) (61). The TBP subunit is responsible for the TATA box binding activity of TFIID, and TBP sequence and structure are both highly conserved throughout eukaryotic species. Like TBP, the TAF subunits of TFIID have also been highly conserved during eukaryotic evolution (6, 22, 66), and in aggregate, TAFs appear to serve multiple functions within the TFIID holocomplex. Early models, based on in vitro studies with individual recombinant subunits or subcomplexes of TFIID subunits, argued that TAFs functioned either as core promoter selectivity factors or as general coactivators, integrating signals between gene-specific trans-activating factors and the general transcription machinery (1). Although initially questioned, recent in vivo studies with S. cerevisiae and metazoans support both the coactivator and core promoter functions of TAFs within TFIID (1, 42, 59).
Studies with numerous eukaryotic systems have yielded invaluable insights into the role of TFIID in the regulation of mRNA gene transcription. However, despite this large body of information regarding its functional properties, many fundamental aspects of TFIID biochemistry are still unknown. This is particularly true for S. cerevisiae TFIID. To date, no extensive characterization of the native size or, as important, subunit stoichiometry of purified eukaryotic TFIID has been reported. Based on primary sequence homology and structural studies, together with biochemical and genetic interaction experiments, five histone-like TAF-TAF interaction pairs have been defined (23), and it has been proposed that at least a subset of these TAFs can form a histone-like octamer structure within TFIID through interactions involving histone fold domains within the TAFs (29).
Although a recent study has demonstrated that four S. cerevisiae TAFs, TAF17p, TAF48p, TAF60p, and TAF61p, can indeed form an octamer-like structure in vitro (62), there is no evidence yet published that any of these TAFs are present at the requisite 2 copies per TFIID complex. Given this, and the fact that many of the histone-like TAFs are shared between TFIID and several other transcription complexes (6), such as the S. cerevisiae SAGA (Spt-Ada-Gcn5-acetyltransferase) complex (25), the lack of information regarding TFIID subunit stoichiometry is a particularly pressing issue.
As an extension of our previous studies, we present here further characterization of S. cerevisiae TFIID. Similar to its metazoan counterparts, we found that S. cerevisiae TFIID efficiently mediated both basal and activator-dependent transcription and displayed DNA-binding activity functionally distinct from that of TBP alone. TFIID binding to all the promoters analyzed was completely dependent on TFIIA. Stoichiometry analyses together with coimmunoprecipitation studies indicated that several TFIID TAFs are present at more than 1 mol per mol of TFIID. Gel filtration and rate-zonal sedimentation indicated a native molecular mass for TFIID consistent with the sum of the masses of TFIID subunits, their measured stoichiometries, and the shape of the holocomplex. Quite surprisingly, we observed that TBP did not cofractionate with the 14 TAF subunits of TFIID during native sizing analyses, suggesting dynamic association of this subunit to form holo-TFIID. Direct TBP-TFIID exchange experiments indicated that this was the case. The significance of these results with regard to TFIID structure and function is discussed.
MATERIALS AND METHODS
Yeast strains.
The relevant background strains used in our studies are BY4741 (MATa ura3 leu2 his3 met15), BY4742 (MATα ura3 leu2 his3 lys2), BY4743 (MATa/α ura3/ura3 leu2/leu2 his3/his3 lys2/LYS2 met15/MET15), BY4716 (MATα lys2) (8), YSLS14 [MATα ura3 ade2 trp1 his3 leu2 tbp(spt15)Δ::TRP1 pDP15-HA1-TBP(SPT15)], YSLS18 (MATα ura3 ade2 trp1 his3 leu2 taf130Δ::TRP1 pRS313-HA1-TAF130) (61), and 21R (MATa ura3 leu2 ade1) (31). S. cerevisiae cell growth, manipulation, and epitope tagging were all performed as described before (33, 61).
Diploid strains for self-association experiments were generated by several methods. To generate cells heterozygous for TAF67, TAF61, TAF48, TAF30, TAF19, or TAF17, strains in which the relevant genomic copies had been tagged with three copies of the influenza virus hemagglutinin (HA) tag at the C terminus in the 21R background (a gift from K. Melcher) were mated with S. cerevisiae strain BY4716 to produce strains expressing tagged and untagged variants of the encoded proteins. For TBP (SPT15), TAF65, and TAF47, heterozygous knockout strains in the BY4743 background were purchased (Research Genetics) and transformed with low-copy-number (CEN/ARS) plasmids expressing the appropriate wild-type or N-terminally HA3-tagged allele under the control of the native promoter and terminator. For TAF150 and TAF90, one copy of each allele was tagged with 13 copies of the Myc oncoprotein epitope (45) at the C terminus in the BY4743 background. For TAF130, TAF60, TAF40, and TAF25, low-copy-number plasmids expressing the appropriate N-terminally HA3-tagged allele under the control of the native promoter and terminator were transformed into BY4741. Specific strain details are available on request.
Protein purification.
Plasmids for the expression of full-length Toa1p and Toa2p (65) were kindly provided by S. Tan; recombinant TFIIA was generated from these proteins and purified as described previously (54). Recombinant His6-tagged TBP was generated as described previously (5). TFIID was purified as detailed previously (61) with the following modifications. For small-scale purification of TFIID, 20 ml of 1 M Bio-Rex 70 fraction was used as the starting material for immunoaffinity chromatography with 4 mg of anti-HA immunoglobulin G (IgG) cross-linked to 1 ml of protein A-Sepharose; TFIID elution was performed with 2 column volumes of buffer A (20 mM HEPES [pH 7.6]. 10% [vol/vol] glycerol, 1 mM dithiothreitol, 0.1 mM phenylmethylsulfonyl fluoride, 1 mM benzamidine) supplemented with 200 mM potassium acetate (buffer A 200) (62) and 0.001% (vol/vol) Nonidet P-40 containing 4 mg of HA1 peptide/ml.
Pooled peptide-eluted fractions were applied to a Uno S polishing column (Bio-Rad) in buffer A 200 (61) plus 0.001% NP-40, and bound proteins were resolved with a 3-ml linear gradient of 200 to 1,000 mM potassium acetate in buffer A plus 0.001% NP-40. TFIID eluted in a single peak at ≈650 mM potassium acetate. For the experiment presented in Fig. 1, a large-scale preparation of TFIID was generated with a 1 M Bio-Rex 70 fraction derived from ≈2.5 kg of yeast cells. Anti-HA column-eluted TFIID was resolved over a Mono S HR 5/5 column (Amersham Pharmacia Biotech) as described previously (61). Peak fractions were pooled, dialyzed against 500 ml of buffer A 100 for 2 h with one change, aliquoted, and stored at −80°C. For all functional studies presented, Uno S-purified HA-TAF130p-TFIID was used.
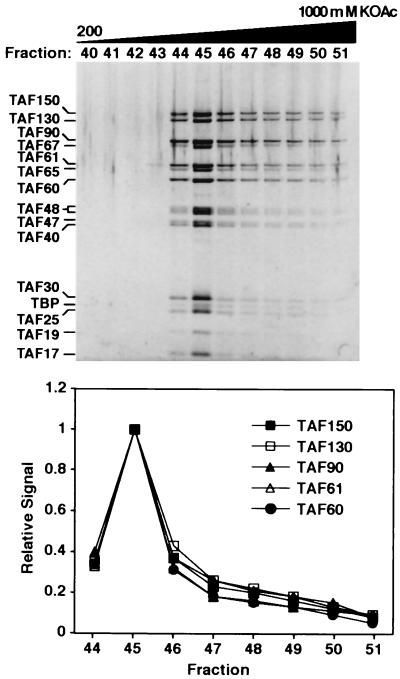
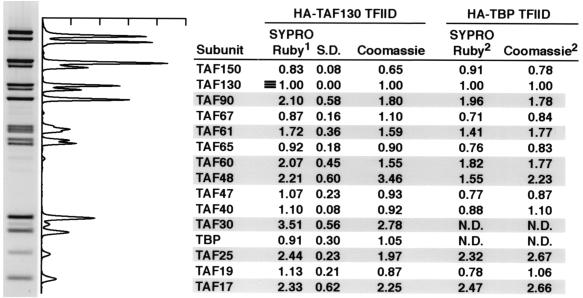
FIG. 1.
Homogeneity of purified S. cerevisiae TFIID. S. cerevisiae strain YSLS18 was used for large-scale purification of HA-TAF130p-TFIID as described in Materials and Methods. Pooled anti-HA column elutes were resolved over a Mono S HR 5/5 column with a 10-ml linear gradient of potassium acetate (KOAc) from 200 to 1,000 mM in buffer A. Each fraction (5 μl, indicated across the top) was subjected to SDS-PAGE on a 10% NuPAGE gel with MOPS running buffer (Invitrogen) and visualized by staining with SYPRO Ruby. TFIID subunits are indicated on the left, and only relevant gradient fractions are shown. The elution profile for TFIID (bottom) was determined by quantitating the signals for the indicated TFIID subunits from each fraction and calculating the relative value versus the cognate TAF signal in fraction 45.
In vitro transcription.
Transcription assays were performed essentially as described previously (53) except a final concentration of 120 mM potassium acetate, 30 ng of each template, pSPGCN4G−, and pJJ470 (32) and, when present, 20 ng of purified Gal4-VP16 protein were used. For the TFIID immunodepletion experiments, 40 μg of either a nonspecific control IgG or affinity-purified anti-TAF67p IgG was bound to 20 μl of protein A-Sepharose (Sigma) in a 1.5-ml microcentrifuge tube. The antibody-bound resin was washed with dilution buffer (20 mM HEPES-KOH [pH 7.6], 20% glycerol, 300 mM potassium acetate, 10 mM magnesium acetate, 5 mM dithiothreitol, 5 mM EGTA, 0.1 mM phenylmethylsulfonyl fluoride, 1 mM benzamidine, 1 μg of pepstatin per ml) and pelleted with a brief spin, and the buffer was aspirated. To the resin were added 50 μl of whole-cell extract (WCE) (≈100 mg of total protein/ml) and 130 μl of dilution buffer. The slurry was incubated for 10 to 12 h at 4°C on a tiltboard.
To control for any effects of either incubation time or dilution, a mock reaction was performed by diluting 50 μl of WCE with 150 μl of dilution buffer and incubating it simultaneously. After incubation, the resin was pelleted with a brief spin, and the supernatant was removed, aliquoted, and stored at −80°C. Typically, 2 μl of the initial WCE and 8-μl aliquots of depleted or mock-depleted WCE were used for transcription assays. 32P-labeled transcription products were quantitated with an Fx MultiImager (Bio-Rad). To estimate TBP concentrations and the extent of TFIID depletion, immunoblots were developed with Lumi-Light Plus Western blotting substrate (Roche) and quantitated with a Fluor-S MultiImager (Bio-Rad).
In vitro footprinting.
The adenovirus type 2 (Ad2) major late promoter (MLP) footprinting probe, containing sequences from −250 to +195 (numbering relative to the +1 transcription start site), was generated by excising an EcoRI/HindIII fragment from plasmid pML4 (15). The EcoRI and HindIII sites are located at the upstream (5′) and downstream (3′) ends of this fragment, respectively. Promoter sequences from S. cerevisiae RPS5 (from −180 to +61) and ADH1 (from −194 to +56) were PCR amplified from genomic DNA with upstream primers that added an EcoRI site and downstream primers that added an EagI site. The amplified fragments were digested with these two restriction endonucleases and cloned into EagI- and EcoRI-digested plasmid pBluescript II KS+. Sequences of cloned inserts were verified by sequencing.
RPS5 and ADH1 promoter probes were generated by excising the EcoRI/EagI promoter fragments from the cognate plasmid. Excised fragments were agarose gel purified with a QIAquick gel extraction kit (Qiagen), and overhangs were filled in with the Klenow fragment of Escherichia coli DNA polymerase I in the presence of [α-32P]dCTP to specifically label the top strand at the 3′ HindIII site of the Ad2 MLP and the 3′ EagI site of the RPS5 and ADH1 promoter fragments. The bottom strand of each fragment was labeled similarly but at the EcoRI site with [α-32P]dTTP.
Binding reactions were set up on ice in 1.5-ml microcentrifuge tubes and contained the appropriate amount of either His6-tagged TBP or TFIID, 60 ng of TFIIA, 2 fmol of probe (≈7,000 to 10,000 cpm/fmol), 20 mM HEPES-KOH (pH 7.6), 10% glycerol, 70 mM potassium acetate, 0.1 mM EDTA, 1 mM dithiothreitol, 100 μg of bovine serum albumin per ml, 5 mM magnesium acetate, 0.01% NP-40, and 1 μg of poly(dG-dC) per ml in a final volume of 20 μl. Reaction mixtures were incubated at room temperature for 30 min to allow protein binding. Immediately before use, RQ1 DNase I (Promega) was diluted 1:25 with dilution buffer (20 mM HEPES-KOH [pH 7.6], 30 mM MgCl2, 50 mM CaCl2), and 5 μl was added to each reaction mixture; digestion was performed at room temperature for 1 min. Stop mix (100 μl of 125 mM Tris-HCl [pH 7.5], 20 mM EDTA, 0.5% sodium dodecyl sulfate [SDS], 125 mM NaCl, 0.1 mg of E. coli tRNA per ml) and 3 μl of proteinase K (15 mg/ml) were added, and the reaction mixtures were incubated at 65°C for 20 min.
DNA was precipitated by adding 12.5 μl of 3 M sodium acetate (pH 5.5) and 350 μl of 95% ethanol, incubating on ice for 15 min, and centrifugation (15 min at 15,000 × g) at 4°C. The supernatant was aspirated, the pellets were vacuum dried and resuspended in 10 μl of loading dye (0.1 M NaOH, 5 M urea, 0.1% SDS, 0.025% each bromophenol blue and xylene cyanol), and labeled DNAs were fractionated by electrophoresis on a denaturing 5% polyacrylamide sequencing gel. Chemical sequencing reactions of the probe were simultaneously fractionated with the footprinting reactions to serve as markers. Gels were imaged and quantitated as described above.
Antibodies and immunoprecipitations.
Polyclonal antibodies, immunoblotting, and immunoprecipitation experiments were performed as described previously (60, 61). Anti-HA (12CA5) and anti-Myc (9E10) monoclonal antibodies were purchased (Roche).
Determination of stoichiometry of TFIID subunits and measurement of the native molecular mass of TFIID.
Purified TFIID was subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (PAGE) on a 10% NuPAGE gel with MOPS (morpholinepropanesulfonic acid) running buffer (Invitrogen). Gels were stained with either Coomassie brilliant blue R-250 (Sigma) or SYPRO Ruby (Molecular Probes) (46) and imaged with a Fluor-S MultiImager (Bio-Rad). The integrated area from each peak was normalized to the calculated molecular mass for each protein, and the value was determined relative to TAF130p. For sizing analysis, anti-HA column-purified HATAF130p-TFIID was used. The sample was centrifuged (10 min at 15,000 × g) to remove any insoluble material, and 100 μl of the supernatant was applied to a TSK G4000SWXL column (Toso Haas) in buffer A 300 plus 0.001% NP-40 at 0.5 ml/min. Fractions (350 μl) were collected, precipitated with trichloroacetic acid, and subjected to immunoblot analysis.
For rate-zonal sedimentation, 200 μl of the same anti-HA-purified TFIID was applied to the top of a 10-ml linear 10 to 30% sucrose gradient formed in buffer A 300 plus 0.001% NP-40. Gradients were centrifuged at 40,000 rpm for 12 h at 4°C in a Beckman SW-41 rotor. Fractions (350 μl) were collected from the top with a pipette, precipitated with trichloroacetic acid, and subjected to immunoblotting. Molecular mass standards run in a parallel gradient were visualized by SDS-PAGE and Coomassie staining.
Measurement of TBP-TFIID exchange.
TFIID from 3 ml of the 1 M Bio-Rex 70 fraction containing HA-TAF130p was immobilized on 150 μl of protein A-Sepharose beads cross-linked to 600 μg of anti-HA antibody. As a control, 1 M Bio-Rex 70 containing nontagged TAF130p was incubated with the same beads. After overnight binding at 4°C, the beads were washed two times with buffer A 300, resuspended in an amount of buffer A 300 equal to the volume of beads, and distributed in 20-μl aliquots to individual binding reaction mixtures formed in 1.5-ml tubes. The concentration of TFIID in each aliquot was 0.6 pmol. The concentration dependence of TBP exchange was measured as follows. Increasing amounts of His6-TBP were added to each tube (0, 0.6, 1.8, 3.0, 6.0, 18.0, 36.0, and 60.0 pmol), and reaction mixtures were incubated for 30 min at 4°C. The time dependence of exchange was monitored in reactions in which the amount of His6-TBP was kept constant at 6.0 pmol (i.e., 10-fold molar excess) but the incubation time was varied from 0 to 10, 20, 30, 45, 60, and 120 min, all at 4°C. Following incubation to allow exchange, beads were washed twice with buffer A 300, resuspended in SDS-PAGE sample buffer, and heated at 70°C for 10 min. Proteins eluted from the beads were subjected to immunoblotting.
RESULTS
Homogeneity of purified S. cerevisiae TFIID.
We previously used a combination of immunoaffinity and ion exchange chromatographies to define S. cerevisiae TFIID as a 15-subunit complex of TBP and 14 TAFs (61). To investigate the homogeneity of our TFIID preparation, SYPRO Ruby protein staining was used to monitor the elution profile of TFIID subunits from the final Mono S column used in our purification scheme (Fig. 1). SYPRO Ruby, a fluorescence-based protein stain with sensitivity similar to that of silver staining, was used because it both exhibits less protein-to-protein staining variability and displays a linear staining range extending from approximately 5 to 1,000 ng of protein. The results of this experiment showed that all 15 TFIID subunits displayed identical elution profiles and that TFIID eluted in a single defined peak from Mono S.
A similar result was seen when immunoblotting was used to monitor the elution of all the subunits (not shown). Although the elution profile did display a slight tailing of TFIID subunits (<15% of the total TFIID), direct imaging of individual fractions by electron microscopy indicated that the material in fractions 47 and higher was large heterodisperse aggregates of TFIID, while the TFIID in fractions 43 to 46, which comprised the majority of this preparation, represented a homogeneous population of monomeric TFIID complexes (41a; data not shown). Importantly, these analyses demonstrated that the amount of the TFIID subunits relative to one another was constant across the peak of TFIID. Taken together, these data indicated that S. cerevisiae TFIID, purified by our purification methods, represented a homogeneous preparation of molecules with a constant, defined subunit composition.
Functional analyses of S. cerevisiae TFIID: purified yeast TFIID supports both basal and activated transcription in vitro.
We next sought to investigate whether our purified TFIID could mediate basal and activator-dependent transcription, cardinal functional properties of metazoan TFIID. A transcriptionally competent S. cerevisiae WCE and a two-template transcription assay were used to simultaneously monitor basal and activator (Gal4-VP16)-dependent transcription (Fig. 2C). The templates used have two different lengths of G-free cassettes fused to the same S. cerevisiae CYC1 promoter. Upstream of this promoter on these two templates are the binding sites for either Gcn4p (GCN4G−) or Gal4p (GAL4G−). In the absence of transactivator, both templates were transcribed similarly, whereas in the presence of Gal4-VP16, as expected, only transcription of the template containing Gal4p binding sites (GAL4G−) was specifically induced (five- to sevenfold; compare lanes 1 and 2, Fig. 2C).
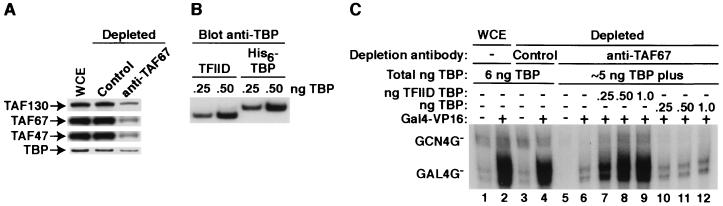
FIG. 2.
Purified S. cerevisiae TFIID efficiently mediates both basal and activated transcription. (A) Efficacy of TFIID depletion. An S. cerevisiae WCE was treated with either nonspecific (control) IgG or anti-TAF67p IgG (indicated across the top) and analyzed by immunoblotting (antibodies indicated on left) to determine the extent of TFIID depletion. (B) Quantitation of the TBP content of purified TFIID. The indicated TBP equivalents of either recombinant His6-TBP or HA-TAF130p-TFIID-TBP were analyzed by immunoblotting. (C) Ability of purified TFIID to support specific transcription. Transcription assays were performed as described in Materials and Methods with the indicated extract (top) in the absence (−) or presence (+) of 20 ng of purified Gal4-VP16. The indicated TBP equivalent amounts were assayed for their ability to reconstitute both basal (GCN4G−) and activated (GAL4G−) transcription with the anti-TAF67p IgG-depleted WCE. The total amount of TBP (as determined by immunoblotting) contributed from each extract is indicated above the lanes.
Immunodepletion of endogenous TFIID from the WCE with anti-TAF67p antibody but not a nonspecific control antibody specifically reduced both basal and activated transcription signals by about 80% (compare lanes 3 and 4 to lanes 5 and 6, Fig. 2C). The loss of transcription correlated directly with the extent of depletion of TFIID, as protein levels for the TFIID-specific subunits TAF130p, TAF67p, and TAF47p were specifically reduced 70 to 80% as judged from immunoblotting (Fig. 2A). Consistent with its presence in several complexes distinct from TFIID (39, 59), only ≈20% of the total TBP was depleted from the WCE with anti-TAF67p IgG (Fig. 2A).
Equivalent amounts of TBP, as judged from immunoblotting (Fig. 2B), in either recombinant His6-TBP form or HA-TAF130p-TFIID form, were then assayed for the ability to reconstitute the basal and activated transcription signals in the TFIID-depleted WCE. Before this add-back experiment was performed, the amount of TBP present in the WCE both before and after depletion was measured (Fig. 2A and data not shown). Amounts of TBP equivalent to this endogenous level, in either TFIID (0.25, 0.5, or 1.0 ng of TBP equivalents; lanes 7 to 9, Fig. 2C) or recombinant form (0.25, 0.5, or 1.0 ng of His6-tagged TBP; lanes 10 to 12, Fig. 2C), were then added back to the depleted WCE, and transcription was assayed. It is important to note that either form of TBP was added back only to the level initially present in the WCE (i.e., back to 6 ng total; see Fig. 2C).
When TFIID was added, both basal and activated transcription signals in the depleted extract were recovered, essentially to the same levels of transcription observed before immunodepletion (compare lanes 2 and 9, Fig. 2C). In contrast, addition of recombinant TBP alone had only a minimal effect on either basal or activated transcription levels (lanes 10 to 12, Fig. 2C). Consistent with previous work from our laboratory (34, 35), addition of excess TBP did recover both basal and activated transcription signals (addition of >5 ng of exogenous TBP; data not shown). The discrepancy between the behavior of exogenous TBP and TFIID in this assay is not due to inactive recombinant TBP, since we have determined that our preparations of E. coli-expressed TBP are essentially 100% active, as judged from stoichiometric fluorescence anisotropy DNA-binding assays (5) (data not shown). Thus, our transcription reconstitution assays demonstrated that purified yeast TFIID was much more efficient (>10- to 20-fold on a TBP-equivalent molar basis) than TBP alone at mediating both basal and activated transcription in vitro.
Functional analyses of yeast TFIID: specific TATA box DNA binding activity of purified TFIID.
Previous work has demonstrated that both human and Drosophila TFIID give a large, extended region of protection relative to TBP alone on the Ad2 MLP, as judged from in vitro DNase I footprinting (10, 11, 15, 73). To assess directly whether yeast TFIID exhibited similar DNA binding characteristics, DNase I footprinting of purified yeast TFIID on the Ad2 MLP was performed (Fig. 3). A radiolabeled DNA fragment containing Ad2 MLP sequences from −250 to +195 (relative to the +1 transcription start site) was incubated with either recombinant TBP or HA-TAF130p-immunopurified TFIID, digested with DNase I, and subjected to electrophoresis. Again, as for the transcription assays in Fig. 2, equivalent amounts of TBP in either TFIID form or recombinant TBP form were used in these assays. Recombinant TBP generated the expected ca. 20-bp region of protection over the TATA box at −30 (lanes 2 and 3, Fig. 3B). In contrast, when as much as a 30-fold excess (relative to the DNA) of TBP in the form of TFIID was incubated with the probe, no significant regions of protection were seen (lane 5, Fig. 3B, and Fig. 3C, and data not shown). This lack of protection was somewhat surprising, as our in vitro transcription assays (Fig. 2) clearly indicated that this purified TFIID was functional and hence would be expected to exhibit potent TATA box binding activity.
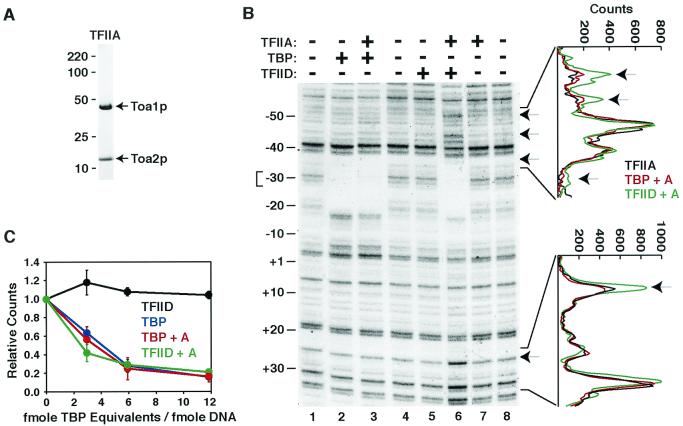
FIG. 3.
S. cerevisiae TFIID requires TFIIA for efficient binding to the Ad2 MLP. (A) Purity of recombinant TFIIA. A 2-μg amount of purified recombinant S. cerevisiae TFIIA was subjected to SDS-PAGE and visualized by SYPRO Orange (Molecular Probes) staining. The Toa1p and Toa2p subunits of TFIIA are indicated on the right, and the mobilities of coelectrophoresed molecular mass markers (not shown) are on the left. (B) S. cerevisiae TFIID binds the Ad2 MLP specifically. In vitro footprinting reactions with the Ad2 MLP were performed as described in Materials and Methods in the absence (−) or presence (+) of recombinant TFIIA, recombinant His6-TBP, and HA-TAF130p-TFIID, as indicated. Numbering on the left is relative to the +1 transcription start site. Brackets on the right mark the region of each lane used to generate the scanning densitometry profiles shown to the right. Traces correspond as follows: TFIIA alone (black), lane 7; TFIIA plus TBP (red), lane 3; and TFIIA plus TFIID (green), lane 6. Hypersensitive bands are marked by arrows, and a TBP equivalent-to-DNA ratio of 6:1 was used for the experiment presented. (C) Quantitation of the effect of TFIIA addition on TFIID-specific DNA binding. A titration of TBP equivalents was performed to compare the TATA binding activity of His6-TBP versus HA-TAF130p-TFIID-TBP in the presence and absence of TFIIA. The signal of the triplet band at −30 (indicated by the bracket on the left of panel B) in the absence of either His6-TBP or TFIID was used to determine the relative binding with increasing amounts of either His6-TBP or TFIID. Relative values were determined from two independent experiments in which one preparation of His6-TBP and two independent preparations of HA-TAF130p-TFIID were analyzed simultaneously. TFIID alone, black; TFIID plus TFIIA, green; TBP alone, blue; TBP plus TFIIA, red. Brackets indicate standard deviation.
It has been demonstrated that a conserved N-terminal domain of S. cerevisiae TAF130p (and its metazoan equivalents), termed the TAND (for TAF N-terminal domain), forms a protein TATA box-mimetic structure that binds to the concave DNA binding surface of TBP with high affinity and inhibits the TBP-TATA DNA interaction (4, 5, 36, 37, 44). It has also been shown that TFIIA can compete with the TAND domain for TBP binding and thereby relieve the inhibitory action of TAF130p (36). Thus, it has been hypothesized that TAF130p normally functions to negatively regulate the TATA box binding activity of TBP within the TFIID complex and that TFIIA functions to relieve this inhibitory activity (36). Furthermore, depending on the TFIID preparation and promoter used, TFIIA either stimulates or is absolutely required for human TFIID TATA box binding (14, 21, 43).
For these reasons, we assessed the ability of purified recombinant S. cerevisiae TFIIA (Fig. 3A) to stimulate the binding of S. cerevisiae TFIID to TATA DNA. We observed that addition of TFIIA to the footprinting reaction had little or no effect on TBP binding (compare lanes 2 and 3 in Fig. 3B and Fig. 3C), while TFIIA addition dramatically stimulated TFIID binding to the TATA box region. Adding TFIIA allowed TFIID-TATA box protection to levels similar to those with TBP alone (compare lanes 2 and 6, Fig. 3B, and quantitation in Fig. 3C). Controls showed that TFIIA by itself did not bind DNA (compare lanes 7 and 8, Fig. 3B).
Interestingly, and in contrast to human (15, 73) and Drosophila (10) TFIID, S. cerevisiae TFIID did not exhibit an extended region of protection downstream of the TATA box towards the Ad2 MLP initiator (Inr) element at +30 (Fig. 3B), nor did addition of acetyl-coenzyme A (CoA) stimulate or alter (21) the footprint pattern, as has been reported for human TFIID (data not shown). S. cerevisiae TFIID and TBP footprints are distinct, however, as several regions of hypersensitivity (indicated by the arrows, Fig. 3B) are present just upstream (−40 to −50) and farther downstream (≈+30) from the TATA box, suggesting an altered DNA conformation due to TFIID binding.
Quantitative scanning densitometry indicated that these hypersensitive bands were reproducibly about twofold more accessible to DNase I in the presence of TFIID and TFIIA than in the presence of TFIIA alone or TBP plus TFIIA (Fig. 3B). Similar results were obtained with TFIID purified from a strain harboring either a single HA or TAP (58) tag at the C terminus of TAF130p rather than the N terminus (data not shown). These data are consistent with the hypothesis that TFIIA functions to stimulate TFIID by relieving the inhibitory activity of TAF130p within the TFIID complex. This is the first direct demonstration that S. cerevisiae TFIID has TATA box binding activity similar to that of its metazoan counterparts and, together with the transcription results presented above, there is a clear functional distinction between S. cerevisiae TFIID and TBP.
S. cerevisiae TFIID displays similar DNA-binding properties on natural S. cerevisiae promoters and the Ad2 MLP.
In order to examine the interaction of S. cerevisiae TFIID with promoter DNA more carefully, we performed DNase I footprinting studies with three promoters, examining protein DNA interactions on both strands. Our goal in these experiments was twofold: first, to see if S. cerevisiae TFIID displays TFIIA-dependent binding to both strands on all DNAs tested, and second, to determine if the patterns of interaction that we observed above (Fig. 3) between S. cerevisiae TFIID and (the top strand of) the Ad2 MLP would be observed with natural S. cerevisiae promoters. We chose two S. cerevisiae promoters for these experiments, a promoter fragment from RPS5, a TFIID-dependent promoter/gene, and a promoter fragment from ADH1, a TFIID-independent promoter/gene (42, 63).
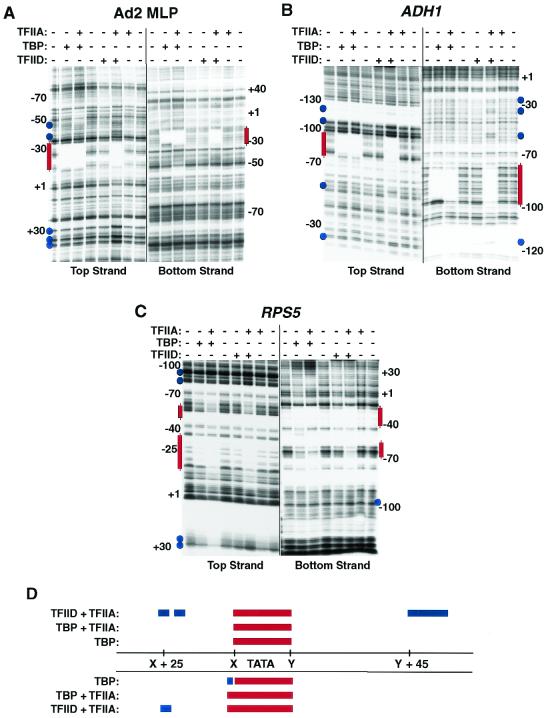
Footprinting reactions with these three promoters were conducted exactly as described above (Fig. 3); the results of these studies are presented in Fig. 4A to C and summarized in Fig. 4D. S. cerevisiae TFIID clearly displayed TFIIA-dependent binding to all three promoters, and these interactions could be readily monitored on both strands of these DNAs. As shown in Fig. 3 for the interactions of S. cerevisiae TFIID with the top strand of the Ad2 MLP, S. cerevisiae TFIID did not induce extended (i.e., 50 to 80 bp) protection on either strand of any of these three promoters. However, characteristic sets of DNase I-hypersensitive cleavage sites were induced on S. cerevisiae TFIID binding on all three promoters. Indeed, as summarized in Fig. 4D, though there were differences in the finer details of TFIID (TFIIA)-promoter DNA interactions, the overall pattern of interaction was in fact remarkably similar with all these promoters. We conclude from the footprinting data presented in Fig. 3 and 4 that S. cerevisiae TFIID binds to promoters in a strictly TFIIA-dependent fashion and that the mode of interaction of TFIID with these DNAs is quite similar despite significant overall differences in the sequences of the three promoters tested. Further work with additional promoters will be needed to see if these observations truly represent fundamentally conserved features of S. cerevisiae TFIID-promoter interaction.
FIG. 4.
S. cerevisiae TFIID interacts similarly with Ad2 MLP and natural S. cerevisiae promoters. (A, B, and C) Footprints on Ad2 MLP and RPS5 and ADH1 promoters. DNase I footprinting was performed with promoter fragments derived from the Ad2 MLP, ADH1, and RPS5 genes that had been labeled on either the top or bottom strand as indicated. Protein binding, nuclease digestion, gel fractionation, and DNA detection were performed as detailed for Fig. 3 in the presence of no added protein, TBP, TBP plus TFIIA, TFIID, or TFIID plus TFIIA, as indicated. Hypersensitive sites in the TFIID plus TFIIA reactions only are indicated by blue circles, and regions of protection are indicated by red lines. (D) Summary of the common features of TBP, TBP plus TFIIA, and TFIID plus TFIIA interactions on all three promoters. Shown are the common regions of protection (red) and nuclease hypersensitivity (blue) on all three promoters. Numbering shown is relative to the TATA box element of each gene.
Stoichiometry of TFIID subunits.
It has been argued that the core of TFIID resembles a histone-like octamer and that this structure may allow TFIID to recapitulate a nucleosome-like function(s) when bound to a promoter (29). This hypothesis is supported by both amino acid sequence and structural similarity between the histones and several TAFs (23). In S. cerevisiae, TAF17p, TAF60p, TAF48p, and TAF61p have domains similar to histones H3, H4, H2A, and H2B, respectively, and recent data indicate that these TAFs can form an octameric structure in vitro (62). Structural data demonstrating that the histone fold regions of Drosophila TAF60 and TAF42 (similar to S. cerevisiae TAF60p and TAF17p, respectively) form a histone H3-H4-like dimer and tetramer also support this idea (72). If an octamer-like structure were present within TFIID, it would be predicted that (i) the TAFs comprising that octamer would be present in equivalent molar amounts and (ii) at least 2 mol of each of these TAFs would be present per mol of TFIID. To date, however, the stoichiometry of neither the histone-like TAFs nor, indeed, any other subunits present within TFIID has been reported.
To address this lack of information regarding TFIID subunit content, we measured the stoichiometry of S. cerevisiae TFIID subunits, and the results of our analyses are presented in Fig. 5. HA-TAF130p-purified TFIID was resolved by SDS-PAGE, and the gels were stained with SYPRO Ruby. Stained gels were imaged and analyzed by scanning densitometry, and the amount of each subunit relative to TAF130p was determined. The most striking result of these experiments is that all of the TAF subunits shared between the TFIID and SAGA complexes, that is, TAF17p, TAF25p, TAF60p, TAF61p, and TAF90p, were present at ≈2 mol per mol of TFIID (shaded boxes, Fig. 5). In contrast, the TFIID-specific subunits TAF150p, TAF130p, TAF67p, TAF65p, TAF47p, and TAF40p and TBP (with the exception of TAF48p, which appeared at ≈2 to 3 mol) had an apparent stoichiometry of about 1 mol per mol of TFIID.
FIG. 5.
Stoichiometry of TFIID subunits. Stoichiometric analyses of purified HA-TAF130p- or HA-TBP-TFIID with SYPRO Ruby and Coomassie brilliant blue R-250 stains (indicated across top) were performed as described in Materials and Methods. A representative gel and scanning densitometry profile for SYPRO Ruby-stained HA-TAF130p-TFIID are shown to the left. TFIID subunits are indicated in the first column, and the relative value for each subunit (as determined compared to TAF130p) for each stain (indicated at the top of each column) is listed across. 1, HA-TAF130p SYPRO Ruby values were averaged from independent analyses of four different preparations of HA-TAF130p-TFIID, and the standard deviation (S.D.) is listed. 2, HA-TBP-TFIID values were averaged from independent analyses of two separate preparations of HA-TBP-TFIID. TFIID subunits with an apparent stoichiometry of >1 are shaded. N.D., not determined because HA-TBP and TAF30p comigrated in the gel system used.
TAF30p, which is resident in a large variety of different multisubunit complexes (12, 28, 30, 59), was present at about 3 mol/mol of TFIID. Importantly, we obtained very similar results both when a second independent stain, Coomassie brilliant blue, was used and when TFIID purified with a different tagged subunit (HA-TBP) (61) was analyzed (Fig. 5, HA-TBP TFIID SYPRO Ruby and Coomassie). These stoichiometry data are consistent with our previously published (61) silver-stained SDS-PAGE profiles as well as systematic HA tagging and immunodetection (3) and mass spectrometry analyses (59) of S. cerevisiae TFIID.
As defined from previous genetic and biochemical studies, five distinct histone-like TAF-TAF interaction pairs have been defined within TFIID (23). Consistent with these studies, partners for the relevant TAF pairs, TAF17p-TAF60p, TAF48p-TAF61p, and TAF19p-TAF40p, are present at approximately equimolar amounts. It should be noted that although there is some discrepancy between TAF48p (2 to 3 mol) and TAF61p (1.5 to 2 mol), we believe this is most likely due to technical issues. It has been reported that TAF61p does not efficiently bind Coomassie blue (62). Furthermore, we reported previously that TAF48p migrates as a triplet of protein bands (61) due to multiple phosphorylation events to which this TAFp is apparently subjected in vivo. Consistent with the idea that TAF25p has two distinct binding partners within TFIID (24), TAF25p is apparently present at ≈2 copies, while its proposed binding partners, TAF47p and TAF65p, are each present at only ≈1 copy per TFIID complex, summing to a value equal to that of TAF25p.
The stoichiometry analyses of purified S. cerevisiae TFIID presented above argue that several TAF subunits of TFIID are present at more than one copy per TFIID complex. We sought to independently validate these biochemical stoichiometry data. We accomplished this by performing coimmunoprecipitation experiments that assessed the ability of TFIID subunits to self-associate in vivo (Fig. 6). We generated a complete series of TAF diploid or pseudodiploid strains (Materials and Methods) in which only one allele of a TFIID subunit-encoding gene was epitope tagged with either three tandem copies of the HA tag or 13 tandem copies of the Myc tag. WCEs were prepared from each strain, immunoprecipitations were performed with the appropriate monoclonal antibody IgG, and the precipitated TFIID subunits were visualized by immunoblotting with subunit-specific polyclonal antibodies. It is important to note that in all cases the tagged and untagged proteins were equally expressed, as judged from immunoblotting WCEs with appropriate antisubunit IgGs. Also, coimmunoprecipitation experiments with polyclonal anti-TFIID subunit-specific antibodies indicated that the tagged and untagged proteins were equally well incorporated into TFIID (data not shown). This fact is likely related to the observation that steady-state levels of the tagged and untagged subunits were roughly equal.
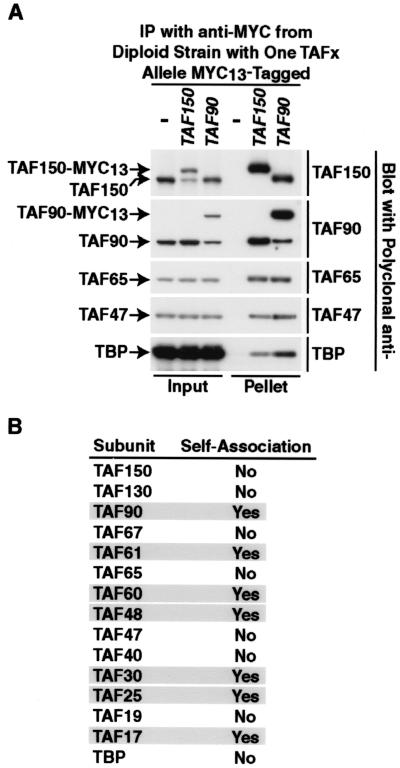
FIG. 6.
Self-association of TFIID subunits in vivo. (A) Blot quantitation of selected TAF-TAF in vivo interactions. A representative experiment to test the ability of TAF150p and TAF90p to self-associate is shown. Immunoprecipitations (IP) were performed with anti-Myc monoclonal antibody with WCEs prepared from diploid strains in which one of the indicated alleles (top) was Myc13 tagged; − indicates the wild-type untagged diploid strain. A fraction of the input (5%) and the pellet (40%) were subjected to immunoblotting with the polyclonal antibody listed to the right to detect the protein indicated on the left. (B) Summary of self-association experimental results for all TFIID subunits. TFIID subunits with an apparent stoichiometry of >1 in Fig. 5 are shaded.
For proteins present at a stoichiometry greater than 1 mol per mol of TFIID, immunoprecipitation of the epitope-tagged protein should coimmunoprecipitate the cognate untagged wild-type protein as well as other TFIID subunits. In contrast, immunoprecipitation of tagged TFIID subunits present at only 1 mol per mol of TFIID should coimmunoprecipitate other TFIID subunits but not the cognate untagged wild-type protein. Representative results of these coimmunoprecipitation experiments for TAF150 and TAF90 are shown in Fig. 6A, and a summary of the results for the complete series of self-association experiments is presented in Fig. 6B. In direct agreement with the stoichiometry results described above, all TFIID subunits with an apparent stoichiometry of >1 displayed self-association in vivo (shaded boxes, Fig. 6B), while TFIID subunits with an apparent stoichiometry of ≤1 failed to display self-association in the assay. The results presented here for TBP and TAF25p are consistent with data presented elsewhere (13, 41a).
The presence of multiple copies of a single TAF within TFIID is not limited to S. cerevisiae. Self-association of the human ortholog of S. cerevisiae TAF48p, hTAF135, and the two Schizosaccharomyces pombe homologs of S. cerevisiae TAF90p, Taf72 and Taf73, has been reported (27, 48). Together, these results provide the first direct analysis of TFIID subunit stoichiometry and are consistent both with the histone-like TAF-TAF pairwise interaction patterns reported previously (23) and with the potential formation of octameric structures in TFIID (62). Additional work will be required to elucidate the contribution of histone fold TAFs and (potential) TAF octamer(s) to TFIID structure and function.
Native molecular mass of S. cerevisiae TFIID.
We next sought to accurately determine the native molecular mass of purified TFIID. This was accomplished by performing gel filtration and rate-zonal sedimentation analyses (Fig. 7). TFIID purified through the anti-HA immunoaffinity purification step was used for these experiments rather than Mono S-purified TFIID; this material behaved identically to Mono S-purified TFIID in functional tests and contained but two minor contaminants (61). HA-TAF130p-TFIID was subjected to either size fractionation on a TSK G4000SWXL gel filtration column (Fig. 7A) or rate-zonal sedimentation in a linear 10 to 30% sucrose gradient (Fig. 7B); immunoblotting was used to monitor TFIID subunits.
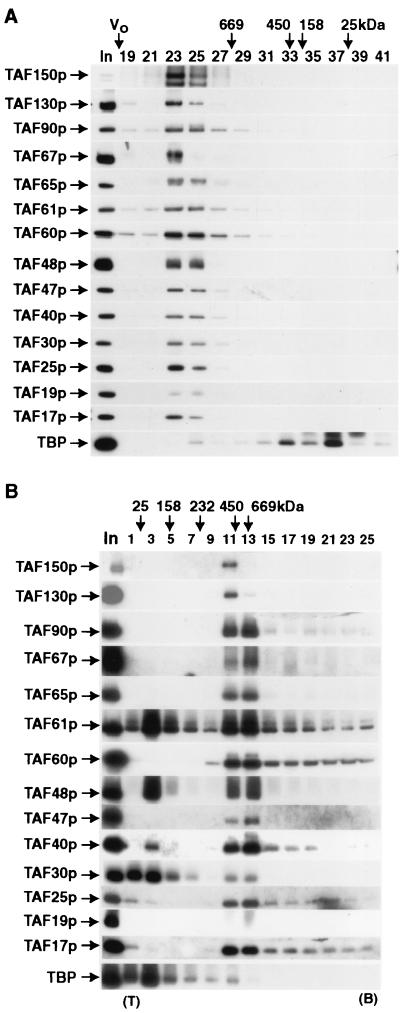
FIG. 7.
Determination of native molecular mass of purified S. cerevisiae TFIID. (A) Gel filtration sizing of TFIID. Anti-HA IgG-purified HA-TAF130p-TFIID was subjected to size fractionation on a TSK G4000SWXL column as described in Materials and Methods. A portion of the input (In) and each fraction were subjected to immunoblotting to detect the proteins indicated (left). The void volume (V0) and elution positions for molecular mass markers (thyroglobulin, ferritin, aldolase, and chymotrypsinogen) are shown at the top above the indicated fraction numbers (19 to 41). Note that the immunoblots for TAF150p and TAF19p were from independent experiments in which the input-to-fraction load ratio was different from that of the other subunit immunoblots. (B) Sucrose gradient sizing of TFIID. Monoclonal antibody anti-HA-purified HA-TAF130p-TFIID was subjected to rate-zonal sedimentation on a linear 10 to 30% sucrose gradient as described in Materials and Methods. A portion of the input (In) andeach fraction were subjected to immunoblotting to detect the protein indicated (left). The sedimentation positions of molecular mass markers are shown at the top. TAFs were detected by immunoblotting with appropriate antibodies; note that the blots shown for TAF150p and TAF130p used the even-numbered gradient fractions, while all the rest of the blots were prepared from the odd-numbered fractions in this particular experiment. Fraction numbers are indicated 1 to 25; top of gradient, 10% sucrose (T); and bottom of gradient (B), 30% sucrose.
Consistent with the calculated molecular mass for monomeric TFIID (based on the stoichiometry presented in Fig. 5), all of the TAF subunits of TFIID comigrated as a single complex of ≈1.2 MDa during gel filtration. Quite surprisingly, though, TBP did not migrate with other TFIID subunits but displayed a broad elution profile, with a major peak at slightly greater than 30 kDa. This result suggested that TBP either only loosely associates with TFIID or was dissociating during the course of the 40-min chromatographic fractionation. Similar results were also seen when a Superose 6 fast protein liquid chromatography (FPLC) gel filtration column was used except that TFIID was slightly larger (not shown). Importantly, the apparent separation of TBP from TFIID does not appear to be related to the presence of the HA tag at the N terminus of TAF130p, as similar chromatographic behavior was observed when the HA tag was at the C terminus of TAF130p (data not shown). Furthermore, when TFIID was directly size-fractionated from a WCE prepared under mild ionic conditions (300 mM potassium acetate), similar results were obtained (data not shown).
When TFIID was sized by rate-zonal sedimentation, we calculated a native molecular mass, 700 kDa, somewhat smaller than that determined by gel filtration. The smaller molecular mass for TFIID estimated by rate-zonal sedimentation would be predicted by the known hydrodynamic properties of TFIID determined elsewhere (2, 9, 41a). S. cerevisiae TFIID is not spherical but more an oblate ellipsoid. Molecules with this shape will have a higher frictional coefficient than a sphere with equivalent mass and thus will sediment more slowly than a “normal” globular protein but can still be sized fairly accurately by gel filtration, a technique that is more sensitive to molecular radius of gyration.
During the (12-h) centrifugation run, TBP again failed to cosediment with the 14 TAFs comprising TFIID (Fig. 7B). Little to no TBP was present in fractions 11 and 13, where the TFIID TAFs peaked. Interestingly, TAF30p and about 50% of the histone fold-containing, dimerizing pair TAF61p/TAF48p also appeared to have dissociated from the bulk of TFIID during centrifugation. However, significant amounts of neither TAF17p nor TAF60p (both proposed to interact with TAF61p/TAF48p to form an octamer) appeared to dissociate from TFIID during the run. Overall, the sizing results we obtained were consistent with similar earlier work on mammalian TFIID (17, 18, 58, 73) that showed, with less well characterized preparations, that mammalian TFIID was ≈1.3 MDa by gel filtration and ≈0.75 MDa by gradient centrifugation. Thus, S. cerevisiae and human TFIIDs exhibit similar biophysical solution properties.
Demonstration of time- and concentration-dependent exchange of free TBP with TFIID-bound TBP.
The molecular sizing experiments presented above in Fig. 7 suggested that TBP might dynamically associate with the 14-subunit holo-TAF complex to form TFIID. To test this hypothesis directly, we performed a series of in vitro studies to probe for TBP-TFIID exchange with two different sizes of TBP molecules, a 240-amino-acid wild-type TBP and a slightly longer His6-tagged variant of TBP. These two forms of TBP are readily distinguishable on SDS-PAGE yet are detected with equal efficiencies by immunoblotting with polyclonal anti-TBP IgG.
As detailed in Materials and Methods, protein A-Sepharose beads coated with anti-HA monoclonal antibody 12CA5 were loaded with HA-TAF130-tagged TFIID (61) and washed, and this bead-bound TFIID was incubated with either a fixed molar excess (10-fold) of His6-TBP for increasing periods of time or increasing amounts of His6-TBP (1- to 100-fold molar excess relative to the amount of bead-bound TFIID) for a fixed time (30 min). Aliquots of these reaction mixes were analyzed by SDS-PAGE and immunoblotted with polyclonal antibodies to test for TBP-TFIID exchange. Bead-bound TFIID content was also examined similarly to control for TFIID recovery in each reaction. TFIID-specific anti-TAF antibodies were used for this purpose. Together, these experiments allowed us to test whether TBP-TFIID exchange actually occurred and, if so, whether such exchange exhibited the predicted time and concentration dependence.
As shown in Fig. 8, TBP-TFIID exchange was readily observed in these experiments, and this process exhibited a dependence on both His6-TBP concentration (Fig. 8A) and time (Fig. 8B). The results of these experiments are presented in several ways. In Fig. 8A, both the immunoblotting data (inset, Fig. 8A) and recovery-adjusted quantitation (graph, Fig. 8A) of the exchange reaction are depicted. Here, TBP immunoblot chemiluminescence was quantitated and adjusted for average reaction-to-reaction TAF130/65/48 recovery, and then TBP content was expressed as a function of the amount of His6-TBP added by plotting percent maximal signal versus molar ratio of His6-TBP to wild-type TBP in the reaction. For the kinetics experiment, only the relevant TBP immunoblotting data are shown (Fig. 8B, top); however, in this case, for illustrative purposes, the cognate TAF immunoblot recovery data are presented (Fig. 8B, bottom).
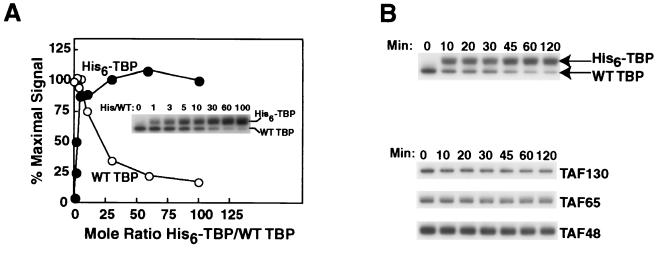
FIG. 8.
Direct demonstration of TBP exchange. (A) Concentration dependence of TBP exchange within TFIID. Multiple, equal amounts of HA-TAF130p-tagged TFIID bound to protein A-Sepharose beads coated with anti-HA monoclonal antibody were generated as detailed in Materials and Methods. These aliquots of bead-bound TFIID were incubated with the indicated increasing molar excess of His6-tagged TBP for 30 min. Unbound His6-tagged TBP was removed by centrifugation and washing. The relative amounts of wild-type (WT) and His6-TBP remaining bead bound to TFIID were determined by SDS-PAGE and immunoblotting with polyclonal anti-TBP IgG. This immunoblot is shown in the inset and labeled WT TBP (TFIID-endogenous TBP) and His6-TBP (TBP exchanged into TFIID). Appropriate regions of the gel blot were separately probed with TFIID-specific antibodies (anti-TAF130p, anti-TAF65p, or anti-TAF48p IgGs). All immune complexes were detected by chemiluminescence and quantitated with a Fluor-S Bio-Rad MultiImager. Wild-type and His6-TBP content, corrected for average TAF recovery, is plotted as percent maximal signal as a function of His6-TBP/wild-type TBP ratio. Background subtracted and recovery-corrected wild-type TBP and His6-TBP values, in arbitrary units, were 123 and 0, 126 and 34, 116 and 76, 124 and 136, 92 and 126, 43 and 150, 27 and 161, and 21 and 149 in the reactions with His6-TBP/wild-type TBP ratios of 0, 1, 3, 5, 10, 30, 60, and 100, respectively. Average TAFp recoveries in these reactions ranged from 95 to 112%. (B) Kinetics of TBP-TFIID exchange. TBP-TFIID exchange was performed as above. Seven identical reactions were set up, each with a 10-fold molar excess of His6-tagged TBP relative to TFIID-endogenous wild-type TBP. Samples were incubated for 0, 10, 20, 30, 45, 60, or 120 min, as indicated. Wild-type and His6-TBP levels (upper) and TAFp content (lower) were determined by immunoblotting as in panel A. The levels of wild-type and His6-tagged TBP in these reactions were 75 and 0, 53 and 89, 51 and 92, 49 and 87, 38 and 96, 27 and 98, and 25 and 94 arbitrary units in the reactions with His6-TBP/wild-type TBP ratios of 0, 1, 3, 5, 10, 30, 60, and 100, respectively. Recovery of TAFs (lower) was in the same range as for panel A.
When excess His6-TBP was added to TFIID, exchange was essentially complete (Fig. 8A). Indeed, the blotting data suggested that slightly more His6-TBP exchanged into TFIID than was present initially (quantitation indicated 25% more). This 25% value is consistent with the range in our measurement of TBP stoichiometry in TFIID. We determined that there were 1.0 ± 0.3 mol of TBP/mol of TFIID (Fig. 5). Thus, if the initial TFIID used for our exchange experiments was only 70% saturated with wild-type TBP, then such a population of TFIID could readily accept ca. 25% more TBP on saturation with His6-TBP, precisely the result we obtained in these experiments. Similar results were obtained when we measured the kinetics of the exchange reaction (Fig. 8B). These data strongly support the hypothesis that TBP dynamically associates with the stable 14-subunit TFIID-TAF complex.
DISCUSSION
In this report we have described the results of our efforts to further characterize the general transcription factor TFIID from the yeast S. cerevisiae. Like its metazoan counterparts, and consistent with previous studies (52, 56), purified S. cerevisiae TFIID efficiently mediated both basal and activated transcription in vitro and displayed TATA box binding properties expected of a eukaryotic TFIID. Importantly, our studies provide the first clear functional distinction between S. cerevisiae TFIID and TBP. The results of subunit stoichiometry analyses and coimmunoprecipitation experiments argued that multiple TAF subunits of TFIID are present at ≥2 mol per mol of TFIID. Sizing experiments further indicated that the TAF subunits of TFIID associated to form a monomeric complex of ≈1.2 MDa, a native molecular mass consistent with that calculated from subunit mass and stoichiometry. Observations made during our sizing analyses suggested that TBP might dynamically interact with a 14-subunit TAF core complex to form holo-TFIID. The results of direct TBP-TFIID exchange experiments supported this hypothesis. These data represent the first detailed molecular characterization of any purified eukaryotic TFIID.
S. cerevisiae TFIID efficiently promotes specific transcription in vitro.
The role of TAFs in activator-dependent transcription has been an area of considerable debate. With WCEs immunodepleted of TFIID, we have demonstrated that purified S. cerevisiae TFIID is much more efficient at promoting Gal4-VP16-activated transcription than TBP alone (Fig. 2). Consistent with previously published results demonstrating TAF-independent trans-activation from in vitro S. cerevisiae systems (32, 34, 35, 38), excess TBP alone could function for both basal and activated transcription in TFIID-depleted WCEs (data not shown). Early reconstitution experiments performed with metazoan components indicated that the TAF subunits of TFIID were absolutely required for activated transcription in vitro (1). However, more recent studies clearly indicate that the phenomenon of TAF-independent trans-activation is not limited to S. cerevisiae (20, 51, 69, 70, 71).
The apparent discrepancy regarding the requirement for TAFs in activated transcription in vitro may reflect functionally redundant pathways by which trans-activation can occur. This hypothesis is supported by the fact that trans-activators, particularly the trans-activator Gal4-VP16 used in the majority of studies to date, have been shown to contact TFIID and mediator subunits as well as other general transcription factors in vitro (1, 7). An alternative possibility for apparent redundancy may reflect the fact that many of these studies used naked DNA templates. Indeed, recent studies with chromatin templates have revealed an absolute requirement for both TAFs (TFIID) and mediator-like complexes, as well as chromatin remodeling complexes, for activator-dependent transcription in vitro (40, 41, 49, 50, 71). Further studies with chromatin templates complemented by in vivo studies will be required to precisely dissect the contribution of TAFs during trans-activator-dependent transcription. Finally, it is also likely that different genes will use multiple, distinct, and alternative mechanisms to regulate gene transcription.
Yeast TFIID binds TATA DNA in a TFIIA-dependent fashion.
The demonstration that both Drosophila and human TFIIDs bind directly to Inr and downstream promoter elements led to the hypothesis that TAFs could function as promoter selectivity factors to facilitate preinitiation complex formation (11, 15, 73). This model is supported by the demonstration that Drosophila TAF150 directly binds the Ad2 MLP Inr sequence element (67) and that Drosophila TAF60 can be cross-linked to the downstream promoter element (10). In vivo studies with S. cerevisiae also support the promoter selectivity model of TAF function (63, 68). Although the patterns of interaction of TFIID with promoters are distinct from those of TBP alone, yeast TFIID did not give an extended protection footprint relative to TBP on the three promoters that we examined (Fig. 3 and 4).
Given that TAF150p, the Inr binding TAF, is conserved from S. cerevisiae to humans and that the Ad2 MLP functions in S. cerevisiae (47), it is quite surprising that S. cerevisiae TFIID appears distinct from its metazoan counterparts in this regard. One possible explanation for this discrepancy is that because obvious, highly conserved Inr-like sequences have yet to be described in S. cerevisiae, the Inr binding activity of TFIID may only have evolved later in metazoan TFIID as required. This idea is supported by the fact that although they are highly similar, the gene encoding Drosophila TAF150 could not functionally replace S. cerevisiae TAF150 in vivo (67), suggesting some evolutionary distinctions between S. cerevisiae and metazoans. Alternatively, the sequences of these two proteins and genes may simply have diverged too much to allow cross-complementation. Further study of native yeast promoters will be required to define this promoter-binding activity of yeast TFIID. Such studies could be guided by the description of the yeast TFIID promoter downstream contacts described here.
Several possible explanations for the TFIIA requirement of yeast TFIID for TATA box binding are suggested from the literature. First, TFIIA has been implicated in TFIID function in a large number of different contexts (7, 14, 43), including S. cerevisiae (55). Thus, it is possible that in vitro, the S. cerevisiae system differs simply in the degree to which TFIID-TATA DNA binding depends on TFIIA. Second, it has been argued that TFIID self-dimerizes in the absence of DNA and that dimer dissociation, facilitated by TFIIA, is rate limiting for promoter binding by TFIID (16, 64). This possibility seems unlikely, as no TFIID dimers were detected in studies of the three-dimensional structure of either S. cerevisiae (41a) or human (2, 9) TFIID. Furthermore, the calculated (≈1.2 MDa) and measured (≈1.2 MDa) molecular masses of S. cerevisiae TFIID are in good agreement (Fig. 6) and consistent with monomeric TFIID.
A third possibility, as discussed previously, is that TFIIA competes with the inhibitory N-terminal TAND domain of TAF130p (above) for interaction with the DNA-binding surface of TBP. This competition relieves the TAND-mediated inhibitory TATA box binding activity of TAF130p, and thus TFIID is free to bind DNA. Although no direct biochemical evidence presented here directly addresses this hypothesis, based on previous genetic and biochemical studies, it seems the most likely explanation for the TFIIA-mediated stimulation of S. cerevisiae TFIID TATA box binding that we observed. Additional experimentation will be required to address the mechanism of TFIIA-dependent TFIID-TATA DNA binding and whether or not this TFIIA requirement extends to a wider range of S. cerevisiae promoters than examined here. Finally, the significance of the fact that TFIID (and TFIIA) binds both TFIID-dependent and TFIID-independent promoters (42) with comparable efficiency (in vitro) also awaits further study.
TFIID subunit stoichiometry.
Based on sequence and structural similarity between several TAFs and the histones, it was suggested that the core of TFIID resembles a histone octamer-like structure, allowing TFIID to recapitulate nucleosome-like function when bound to promoters (29). Consistent with this idea, Tan and colleagues have shown that the four S. cerevisiae histone fold TAFs, TAF17p, TAF48p, TAF60p, and TAF61p, can form an octamer-like structure in vitro (62). Although our stoichiometry data are consistent with the proposal of an octamer-like structure(s) within TFIID, the fact that several other TAFs (containing and lacking a histone fold) are present in multiple copies argues perhaps for a more complicated structure, certainly one where, for example, TFIID is not comprised simply of two (i.e., the product of the nine histone fold TAFs interacting) octameric substructures made up solely of histone fold TAFs. This idea is further supported by electron microscopy studies that demonstrate that both S. cerevisiae and human TFIIDs are three-lobed horseshoe-shaped structures (2, 9, 41a).
An alternative hypothesis is that the histone fold has evolved as a key TAF-TAF interaction motif within each of the three distinct domains of TFIID rather than to recapitulate a nucleosome-like function(s) via octameric complexes. Interactions involving histone fold TAF pairs could thus form the core of each of the multiple domains of TFIID, whereas other TAFs, such as TAF130p and/or TAF90p, could provide a bridge between the individual domains. This hypothesis is supported by the fact that the amino acids important for histone-histone interaction but not histone-DNA interaction are conserved in TAFs (23). The fact that all of the TAFs shared between the TFIID and SAGA complexes have an apparent stoichiometry of 2 further argues that pairwise interactions of these TAFs may form a core on which both TFIID and SAGA can be formed. Interactions between shared histone fold TAFs (TAF61p and TAF25p) and their TFIID- (TAF48p, TAF47p, and TAF65p) and SAGA- (Ada1p and Spt7p) specific partners could then mediate formation of the appropriate complex. Obviously, further studies will be required to fully elucidate both the role and spatial organization of histone-like TAFs within TFIID; ultimately, atomic-level, X-ray crystallography-derived structures are needed.
TBP dynamically associates with TFIID-TAFs to form holo-TFIID.
One of the most intriguing results presented here is the demonstration that TBP did not coelute with TFIID TAF subunits during size fractionation (Fig. 7). A possible explanation for this behavior was that TBP only loosely associated with the TAF subunits of TFIID. This possibility seemed unlikely for several reasons. First, TBP consistently copurifies as a stoichiometric subunit of S. cerevisiae and metazoan TFIID following several ion exchange purification steps and immunoaffinity chromatography. Second, TBP is known to bind to both the N-terminal TAND domain and full-length TAF130p with high affinity (Kd, ≈1 nM) (4, 5). A more interesting explanation, despite these equilibrium results, was that TBP (and perhaps other TAFs; see Fig. 7) was associating and dissociating from TFIID. In this scenario, as TBP dissociated during the course of size fractionation, it became physically separated from the complexed TAF subunits by entering the interstices of the gel filtration beads (or by being separated by continuous sedimentation), and thus, it could not reassociate and could not comigrate with the TAFs.
In contrast, during ion exchange and immunoaffinity chromatography, there is no similar protracted, essentially iterative, physical separation of TBP from the TAFs, so after dissociation, TBP can readily reassociate with the TAF subunits of TFIID and thus consistently copurifies as a subunit of the complex. If these ideas are correct, two testable predictions can be made. First, exogenous TBP added to purified TFIID should be able to incorporate into TFIID in both a time- and concentration-dependent manner reciprocal to the rate and extent of endogenous TBP loss from TFIID. Second, the process of association and dissociation should be mutationally sensitive in both TBP and TAF subunits of TFIID. We have not yet addressed the second prediction. However, our TBP-TFIID exchange experiments clearly showed that TBP could readily interact with the 14-subunit TFIID-TAF core complex (Fig. 8).
The major benefit of rapid association and dissociation of TBP from TFIID would be that, in vivo, TBP could repartition between different dedicated transcriptional complexes as dictated by changing environmental and growth conditions. This hypothesis is supported by the identification of temperature-sensitive TBP mutants which rapidly downregulate RNA polymerase II-dependent mRNA gene transcription but increase RNA polymerase III-mediated tRNA gene transcription after a shift to the nonpermissive temperature (18). This result suggests that TBP can rapidly repartition between different functional TBP-TAF pools, presumably by shifting from TFIID (or any other RNA polymerase II transcription complex it may be associated with) to, in this case (minimally), TFIIIB. It should be noted that these ideas are entirely consistent with the results of Reese et al. (56) and our own recent work (59). By using glutathione S-transferase-TBP affinity chromatography with WCEs, S. cerevisiae TFIID-like multi-TAF-TBP complexes can be formed readily in vitro. Such complexes are similar if not identical to our TFIID preparations. Reese and colleagues found that in vitro-formed TBP-TAF complexes, like our TFIID preparations, were also competent for supporting Gal4-VP16-mediated trans-activation of transcription (56).
Further biochemical and genetic analyses will be required to address these intriguing properties of TBP-TAF interactions and TFIID complex dynamics and to understand how these data fit with the concept of TAF (TFIID)-dependent and -independent genes.
Acknowledgments
We are grateful to Karsten Melcher and Song Tan for gifts of strains and plasmids and to all the members of our lab for useful discussions of this work.
These studies were supported by National Institutes of Health grant GM52461. K.A.G. was supported in part by NIH Molecular Endocrinology Postdoctoral Training grant T32 DK07563.
REFERENCES
- 1.Albright, S. R., and R. Tjian. 2000. TAFs revisited: more data reveal new twists and confirm old ideas. Gene 242:1-13. [DOI] [PubMed] [Google Scholar]
- 2.Andel, F., III, A. G. Ladurner, C. Inouye, R. Tjian, and E. Nogales. 1999. Three-dimensional structure of the human TFIID-IIA-IIB complex. Science 286:2153-2156. [DOI] [PubMed] [Google Scholar]
- 3.Bai, Y. 1997. Molecular identification and characterization of the yeast TBP associated factors. Ph.D. thesis. Vanderbilt University, Nashville, Tenn.
- 4.Bai, Y., G. M. Perez, J. M. Beechem, and P. A. Weil. 1997. Structure-function analysis of TAF130: identification and characterization of a high-affinity TATA-binding protein interaction domain in the N terminus of yeast TAFII130. Mol. Cell. Biol. 17:3081-3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Banik, U., J. M. Beechem, E. Klebanow, S. Schroeder, and P. A. Weil. 2001. Fluorescence-based analyses of the effects of full-length recombinant TAF130p on the interaction of TATA box-binding protein with TATA box DNA. J. Biol. Chem. 276:49100-49109. [DOI] [PubMed] [Google Scholar]
- 6.Bell, B., and L. Tora. 1999. Regulation of gene expression by multiple forms of TFIID and other novel TAFII-containing complexes. Exp. Cell Res. 246:11-19. [DOI] [PubMed] [Google Scholar]
- 7.Berk, A. J. 1999. Activation of RNA polymerase II transcription. Curr. Opin. Cell Biol. 11:330-335. [DOI] [PubMed] [Google Scholar]
- 8.Brachmann, C. B., A. Davies, G. J. Cost, E. Caputo, J. Li, P. Hieter, and J. D. Boeke. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14:115-132. [DOI] [PubMed] [Google Scholar]
- 9.Brand, M., C. Leurent, V. Mallouh, L. Tora, and P. Schultz. 1999. Three-dimensional structures of the TAFII-containing complexes TFIID and TFTC. Science 286:2151-2153. [DOI] [PubMed] [Google Scholar]
- 10.Burke, T. W., and J. T. Kadonaga. 1997. The downstream core promoter element, DPE, is conserved from Drosophila to humans and is recognized by TAFII60 of Drosophila. Genes Dev. 11:3020-3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burke, T. W., and J. T. Kadonaga. 1996. Drosophila TFIID binds to a conserved downstream basal promoter element that is present in many TATA-box-deficient promoters. Genes Dev. 10:711-724. [DOI] [PubMed] [Google Scholar]
- 12.Cairns, B. R., N. L. Henry, and R. D. Kornberg. 1996. TFG/TAF30/ANC1, a component of the yeast SWI/SNF complex that is similar to the leukemogenic proteins ENL and AF-9. Mol. Cell. Biol. 16:3308-3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Campbell, K. M., R. T. Ranallo, L. A. Stargell, and K. J. Lumb. 2000. Reevaluation of transcriptional regulation by TATA-binding protein oligomerization: predominance of monomers. Biochemistry 39:2633-2638. [DOI] [PubMed] [Google Scholar]
- 14.Chi, T., P. Lieberman, K. Ellwood, and M. Carey. 1995. A general mechanism for transcriptional synergy by eukaryotic activators. Nature 377:254-257. [DOI] [PubMed] [Google Scholar]
- 15.Chiang, C. M., H. Ge, Z. Wang, A. Hoffmann, and R. G. Roeder. 1993. Unique TATA-binding protein-containing complexes and cofactors involved in transcription by RNA polymerases II and III. EMBO J. 12:2749-2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coleman, R. A., A. K. Taggart, S. Burma, J. J. Chicca II, and B. F. Pugh. 1999. TFIIA regulates TBP and TFIID dimers. Mol. Cell 4:451-457. [DOI] [PubMed] [Google Scholar]
- 17.Conaway, J. W., J. P. Hanley, K. P. Garrett, and R. C. Conaway. 1991. Transcription initiated by RNA polymerase II and transcription factors from liver. Structure and action of transcription factors epsilon and tau. J. Biol. Chem. 266:7804-7811. [PubMed] [Google Scholar]
- 18.Conaway, J. W., D. Reines, and R. C. Conaway. 1990. Transcription initiated by RNA polymerase II and purified transcription factors from liver. Cooperative action of transcription factors tau and epsilon in initial complex formation. J. Biol. Chem. 265:7552-7558. [PMC free article] [PubMed] [Google Scholar]
- 19.Cormack, B. P., and K. Struhl. 1993. Regional codon randomization: defining a TATA-binding protein surface required for RNA polymerase III transcription. Science 262:244-248. [DOI] [PubMed] [Google Scholar]
- 20.Fondell, J. D., M. Guermah, S. Malik, and R. G. Roeder. 1999. Thyroid hormone receptor-associated proteins and general positive cofactors mediate thyroid hormone receptor function in the absence of the TATA box-binding protein-associated factors of TFIID. Proc. Natl. Acad. Sci. USA 96:1959-1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Galasinski, S. K., T. N. Lively, A. Grebe De Barron, and J. A. Goodrich. 2000. Acetyl coenzyme A stimulates RNA polymerase II transcription and promoter binding by transcription factor IID in the absence of histones. Mol. Cell. Biol. 20:1923-1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gangloff, Y. G., J. C. Pointud, S. Thuault, L. Carre, C. Romier, S. Muratoglu, M. Brand, L. Tora, J. L. Couderc, and I. Davidson. 2001. The TFIID components human TAFII140 and Drosophila BIP2 TAFII155 are novel metazoan homologues of yeast TAFII47 containing a histone fold and a PHD finger. Mol. Cell. Biol. 21:5109-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gangloff, Y. G., C. Romier, S. Thuault, S. Werten, and I. Davidson. 2001. The histone fold is a key structural motif of transcription factor TFIID. Trends Biochem. Sci. 26:250-257. [DOI] [PubMed] [Google Scholar]
- 24.Gangloff, Y. G., S. L. Sanders, C. Romier, D. Kirschner, P. A. Weil, L. Tora, and I. Davidson. 2001. Histone folds mediate selective heterodimerization of yeast TAFII25 with TFIID components yTAFII47 and yTAFII65 and with SAGA component ySPT7. Mol. Cell. Biol. 21:1841-1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grant, P. A., D. Schieltz, M. G. Pray-Grant, D. J. Steger, J. C. Reese, J. R. Yates III, and J. L. Workman. 1998. A subset of TAF(II)s are integral components of the SAGA complex required for nucleosome acetylation and transcriptional stimulation. Cell 94:45-53. [DOI] [PubMed] [Google Scholar]
- 26.Green, M. R. 2000. TBP-associated factors (TAFIIs): multiple, selective transcriptional mediators in common complexes. Trends Biochem. Sci. 25:59-63. [DOI] [PubMed] [Google Scholar]
- 27.Guermah, M., Y. Tao, and R. G. Roeder. 2001. Positive and negative TAFII functions that suggest a dynamic TFIID structure and elicit synergy with TRAPs in activator-induced transcription. Mol. Cell. Biol. 21:6882-6894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Henry, N. L., A. M. Campbell, W. J. Feaver, D. Poon, P. A. Weil, and R. D. Kornberg. 1994. TFIIF-TAF-RNA polymerase II connection. Genes Dev. 8:2868-2878. [DOI] [PubMed] [Google Scholar]
- 29.Hoffmann, A., T. Oelgeschlager, and R. G. Roeder. 1997. Considerations of transcriptional control mechanisms: do TFIID-core promoter complexes recapitulate nucleosome-like functions? Proc. Natl. Acad. Sci. USA 94:8928-8935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.John, S., L. Howe, S. T. Tafrov, P. A. Grant, R. Sternglanz, and J. L. Workman. 2000. The something about silencing protein, Sas3, is the catalytic subunit of NuA3, a yTAF(II)30-containing HAT complex that interacts with the Spt16 subunit of the yeast CP (Cdc68/Pob3)-FACT complex. Genes Dev. 14:1196-1208. [PMC free article] [PubMed] [Google Scholar]
- 31.Johnston, S. A., and J. E. Hopper. 1982. Isolation of the yeast regulatory gene GAL4 and analysis of its dosage effects on the galactose/melibiose regulon. Proc. Natl. Acad. Sci. USA 79:6971-6975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim, Y. J., S. Bjorklund, Y. Li, M. H. Sayre, and R. D. Kornberg. 1994. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell 77:599-608. [DOI] [PubMed] [Google Scholar]
- 33.Kirchner, J., S. L. Sanders, E. Klebanow, and P. A. Weil. 2001. Molecular genetic dissection of TAF25, an essential yeast gene encoding a subunit shared by TFIID and SAGA multiprotein transcription factors. Mol. Cell. Biol. 21:6668-6680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klebanow, E. R., D. Poon, S. Zhou, and P. A. Weil. 1997. Cloning and characterization of an essential Saccharomyces cerevisiae gene, TAF40, which encodes yTAFII40, an RNA polymerase II-specific TATA-binding protein-associated factor. J. Biol. Chem. 272:9436-9442. [DOI] [PubMed] [Google Scholar]
- 35.Klebanow, E. R., D. Poon, S. Zhou, and P. A. Weil. 1996. Isolation and characterization of TAF25, an essential yeast gene that encodes an RNA polymerase II-specific TATA-binding protein-associated factor. J. Biol. Chem. 271:13706-13715. [DOI] [PubMed] [Google Scholar]
- 36.Kokubo, T., M. J. Swanson, J. I. Nishikawa, A. G. Hinnebusch, and Y. Nakatani. 1998. The yeast TAF145 inhibitory domain and TFIIA competitively bind to TATA-binding protein. Mol. Cell. Biol. 18:1003-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kokubo, T., S. Yamashita, M. Horikoshi, R. G. Roeder, and Y. Nakatani. 1994. Interaction between the N-terminal domain of the 230-kDa subunit and the TATA box-binding subunit of TFIID negatively regulates TATA-box binding. Proc. Natl. Acad. Sci. USA 91:3520-3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koleske, A. J., and R. A. Young. 1994. An RNA polymerase II holoenzyme responsive to activators. Nature 368:466-469. [DOI] [PubMed] [Google Scholar]
- 39.Lee, T. I., and R. A. Young. 1998. Regulation of gene expression by TBP-associated proteins. Genes Dev. 12:1398-1408. [DOI] [PubMed] [Google Scholar]
- 40.Lemon, B., C. Inouye, D. S. King, and R. Tjian. 2001. Selectivity of chromatin-remodelling cofactors for ligand-activated transcription. Nature 414:924-928. [DOI] [PubMed] [Google Scholar]
- 41.LeRoy, G., G. Orphanides, W. S. Lane, and D. Reinberg. 1998. Requirement of RSF and FACT for transcription of chromatin templates in vitro. Science 282:1900-1904. [DOI] [PubMed] [Google Scholar]
- 41a.Leurent, C., S. L. Sanders, C. Ruhlmann, V. Mallouh, P. A. Weil, D. Kirshner, L. Tora, and P. Schultz. EMBO J., in press. [DOI] [PMC free article] [PubMed]
- 42.Li, X. Y., S. R. Bhaumik, and M. R. Green. 2000. Distinct classes of yeast promoters revealed by differential TAF recruitment. Science 288:1242-1244. [DOI] [PubMed] [Google Scholar]
- 43.Lieberman, P. M., and A. J. Berk. 1994. A mechanism for TAFs in transcriptional activation: activation domain enhancement of TFIID-TFIIA-promoter DNA complex formation. Genes Dev. 8:995-1006. [DOI] [PubMed] [Google Scholar]
- 44.Liu, D., R. Ishima, K. I. Tong, S. Bagby, T. Kokubo, D. R. Muhandiram, L. E. Kay, Y. Nakatani, and M. Ikura. 1998. Solution structure of a TBP-TAF(II)230 complex: protein mimicry of the minor groove surface of the TATA box unwound by TBP. Cell 94:573-583. [DOI] [PubMed] [Google Scholar]
- 45.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 46.Lopez, M. F., K. Berggren, E. Chernokalskaya, A. Lazarev, M. Robinson, and W. F. Patton. 2000. A comparison of silver stain and SYPRO Ruby Protein Gel Stain with respect to protein detection in two-dimensional gels and identification by peptide mass profiling. Electrophoresis 21:3673-3683. [DOI] [PubMed] [Google Scholar]
- 47.Lue, N. F., P. M. Flanagan, K. Sugimoto, and R. D. Kornberg. 1989. Initiation by yeast RNA polymerase II at the adenoviral major late promoter in vitro. Science 246:661-664. [DOI] [PubMed] [Google Scholar]
- 48.Mitsuzawa, H., H. Seino, F. Yamao, and A. Ishihama. 2001. Two WD repeat-containing TATA-binding protein-associated factors in fission yeast that suppress defects in the anaphase-promoting complex. J. Biol. Chem. 276:17117-17124. [DOI] [PubMed] [Google Scholar]
- 49.Naar, A. M., P. A. Beaurang, K. M. Robinson, J. D. Oliner, D. Avizonis, S. Scheek, J. Zwicker, J. T. Kadonaga, and R. Tjian. 1998. Chromatin, TAFs, and a novel multiprotein coactivator are required for synergistic activation by Sp1 and SREBP-1a in vitro. Genes Dev. 12:3020-3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Naar, A. M., P. A. Beaurang, S. Zhou, S. Abraham, W. Solomon, and R. Tjian. 1999. Composite co-activator ARC mediates chromatin-directed transcriptional activation. Nature 398:828-832. [DOI] [PubMed] [Google Scholar]
- 51.Oelgeschlager, T., Y. Tao, Y. K. Kang, and R. G. Roeder. 1998. Transcription activation via enhanced preinitiation complex assembly in a human cell-free system lacking TAFIIs. Mol. Cell 1:925-931. [DOI] [PubMed] [Google Scholar]
- 52.Poon, D., Y. Bai, A. M. Campbell, S. Bjorklund, Y. J. Kim, S. Zhou, R. D. Kornberg, and P. A. Weil. 1995. Identification and characterization of a TFIID-like multiprotein complex from Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 92:8224-8228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Poon, D., R. Knittle, K. A. Sabelko, T. Yamamoto, M. Horikoshi, R. G. Roeder, and P. A. Weil. 1993. Genetic and biochemical analysis of yeast TATA-binding protein mutants. J. Biol. Chem. 268:5005-5013. [PubMed] [Google Scholar]
- 54.Ranish, J. A., and S. Hahn. 1991. The yeast general transcription factor TFIIA is composed of two polypeptide subunits. J. Biol. Chem. 266:19320-19327. [PubMed] [Google Scholar]
- 55.Ranish, J. A., N. Yudkovsky, and S. Hahn. 1999. Intermediates in formation and activity of the RNA polymerase II preinitiation complex: holoenzyme recruitment and a postrecruitment role for the TATA box and TFIIB. Genes Dev. 13:49-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reese, J. C., L. Apone, S. S. Walker, L. A. Griffin, and M. R. Green. 1994. Yeast TAFIIs in a multisubunit complex required for activated transcription. Nature 371:523-527. [DOI] [PubMed] [Google Scholar]
- 57.Rigaut, G., A. Shevchenko, B. Rutz, M. Wilm, M. Mann, and B. Seraphin. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17:1030-1032. [DOI] [PubMed] [Google Scholar]
- 58.Samuels, M., A. Fire, and P. A. Sharp. 1982. Separation and characterization of factors mediating accurate transcription by RNA polymerase II. J. Biol. Chem. 257:14419-14427. [PubMed] [Google Scholar]
- 59.Sanders, S. L., J. Jennings, A. Canutescu, A. Link, and P. A. Weil. 2002. Proteomics of the eukaryotic transcription machinery: identification of proteins associated with components of the general transcription factor TFIID. Mol. Cell. Biol. 22:4723-4738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sanders, S. L., E. R. Klebanow, and P. A. Weil. 1999. TAF25p, a non-histone-like subunit of TFIID and SAGA complexes, is essential for total mRNA gene transcription in vivo. J. Biol. Chem. 274:18847-18850. [DOI] [PubMed] [Google Scholar]
- 61.Sanders, S. L., and P. A. Weil. 2000. Identification of two novel TAF subunits of the yeast Saccharomyces cerevisiae TFIID complex. J. Biol. Chem. 275:13895-13900. [DOI] [PubMed] [Google Scholar]
- 62.Selleck, W., R. Howley, Q. Fang, V. Podolny, M. G. Fried, S. Buratowski, and S. Tan. 2001. A histone fold TAF octamer within the yeast TFIID transcriptional coactivator. Nat. Struct. Biol. 8:695-700. [DOI] [PubMed] [Google Scholar]
- 63.Shen, W. C., and M. R. Green. 1997. Yeast TAF(II)145 functions as a core promoter selectivity factor, not a general coactivator. Cell 90:615-624. [DOI] [PubMed] [Google Scholar]
- 64.Taggart, A. K., and B. F. Pugh. 1996. Dimerization of TFIID when not bound to DNA. Science 272:1331-1333. [DOI] [PubMed] [Google Scholar]
- 65.Tan, S., Y. Hunziker, D. F. Sargent, and T. J. Richmond. 1996. Crystal structure of a yeast TFIIA/TBP/DNA complex. Nature 381:127-151. [DOI] [PubMed] [Google Scholar]
- 66.Tora, L. 2002. A unified nomenclature for TATA box binding protein (TBP)-associated factors (TAFs) involved in RNA polymerase II transcription. Genes Dev. 16:673-675. [DOI] [PubMed] [Google Scholar]
- 67.Verrijzer, C. P., K. Yokomori, J. L. Chen, and R. Tjian. 1994. Drosophila TAFII150: similarity to yeast gene TSM-1 and specific binding to core promoter DNA. Science 264:933-941. [DOI] [PubMed] [Google Scholar]
- 68.Walker, S. S., W. C. Shen, J. C. Reese, L. M. Apone, and M. R. Green. 1997. Yeast TAF(II)145 required for transcription of G1/S cyclin genes and regulated by the cellular growth state. Cell 90:607-614. [DOI] [PubMed] [Google Scholar]
- 69.Wu, S. Y., and C. M. Chiang. 2001. TATA-binding protein-associated factors enhance the recruitment of RNA polymerase II by transcriptional activators. J. Biol. Chem. 276:34235-34243. [DOI] [PubMed] [Google Scholar]
- 70.Wu, S. Y., E. Kershnar, and C. M. Chiang. 1998. TAFII-independent activation mediated by human TBP in the presence of the positive cofactor PC4. EMBO J. 17:4478-4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wu, S. Y., M. C. Thomas, S. Y. Hou, V. Likhite, and C. M. Chiang. 1999. Isolation of mouse TFIID and functional characterization of TBP and TFIID in mediating estrogen receptor and chromatin transcription. J. Biol. Chem. 274:23480-23490. [DOI] [PubMed] [Google Scholar]
- 72.Xie, X., T. Kokubo, S. L. Cohen, U. A. Mirza, A. Hoffmann, B. T. Chait, R. G. Roeder, Y. Nakatani, and S. K. Burley. 1996. Structural similarity between TAFs and the heterotetrameric core of the histone octamer. Nature 380:316-322. [DOI] [PubMed] [Google Scholar]
- 73.Zhou, Q., P. M. Lieberman, T. G. Boyer, and A. J. Berk. 1992. Holo-TFIID supports transcriptional stimulation by diverse activators and from a TATA-less promoter. Genes Dev. 6:1964-1974. [DOI] [PubMed] [Google Scholar]