Abstract
INI1/hSNF5 is a component of the ATP-dependent chromatin remodeling hSWI/SNF complex and a tumor suppressor gene of aggressive pediatric atypical teratoid and malignant rhabdoid tumors (AT/RT). To understand the molecular mechanisms underlying its tumor suppressor function, we studied the effect of reintroduction of INI1/hSNF5 into AT/RT-derived cell lines such as MON that carry biallelic deletions of the INI1/hSNF5 locus. We demonstrate that expression of INI1/hSNF5 causes G0-G1 arrest and flat cell formation in these cells. In addition, INI1/hSNF5 repressed transcription of cyclin D1 gene in MON, in a histone deacetylase (HDAC)-dependent manner. Chromatin immunoprecipitation studies revealed that INI1/hSNF5 was directly recruited to the cyclin D1 promoter and that its binding correlated with recruitment of HDAC1 and deacetylation of histones at the promoter. Analysis of INI1/hSNF5 truncations indicated that cyclin D1 repression and flat cell formation are tightly correlated. Coexpression of cyclin D1 from a heterologous promoter in MON was sufficient to eliminate the INI1-mediated flat cell formation and cell cycle arrest. Furthermore, cyclin D1 was overexpressed in AT/RT tumors. Our data suggest that one of the mechanisms by which INI1/hSNF5 exerts its tumor suppressor function is by mediating the cell cycle arrest due to the direct recruitment of HDAC activity to the cyclin D1 promoter thereby causing its repression and G0-G1 arrest. Repression of cyclin D1 gene expression may serve as a useful strategy to treat AT/RT.
INI1/hSNF5 has recently been defined as a tumor suppressor gene mutated in a majority of atypical teratoid and malignant rhabdoid tumors (AT/RT) (4, 43). AT/RT is an aggressive pediatric tumor in children under 5 years of age. Originally these tumors were identified in kidney and later in brain, abdomen, skin, liver, thymus, soft tissues, and orbit. Histologically, this tumor is distinct from both rhabdomyosarcomas and malignant teratomas and consists of a unique combination of rhabdoid cells as well as neuroepithelial, peripheral, and mesenchymal elements (32). Recent cytogenetic and molecular genetic analyses have indicated that the 22q11.2 region is the critical region in AT/RT tumors, and this region was subsequently found to harbor the INI1/hSNF5 gene (3, 43). The majority of AT/RT tumors have been documented to be homozygous null for the INI1/hSNF5 gene with deletions spanning part of or the entire gene and a variety of point mutations (4, 35, 36, 43). It has also been found that mutations in the INI1/hSNF5 gene predispose individuals to various types of cancers, including AT/RT and tumors of the central nervous system (36). Recently, targeted disruption of the Ini1 gene in mice demonstrated that while the homozygous deletions are embryonic lethal, the heterozygous mice develop tumors at a high frequency (10, 18, 31). The tumors exhibited reduction or complete absence of INI1/hSNF5 protein expression. All these results together suggest that INI1/hSNF5 is a tumor suppressor and loss of this gene results in a variety of aggressive cancers. However, the mechanism by which INI1/hSNF5 exerts its tumor suppressor properties remains to be elucidated.
INI1/hSNF5 was originally identified as a host protein interacting with human immunodeficiency virus type 1 integrase using the yeast two-hybrid system and subsequently was shown to be a component of the ATP-dependent chromatin remodeling SWI/SNF complex (14, 44). The components of the SWI/SNF complex are an evolutionarily conserved set of proteins that exist as 2-MDa multiprotein complexes (5, 17, 27, 28). Mammalian cells have multiple sets of SWI/SNF complexes that slightly differ in subunit composition (between seven and eleven) depending on the cell type (44, 45). The mammalian components, BRG1 and BRM, are the homologues of yeast SNF2 protein and have an essential ATPase domain (15). Among the SWI/SNF components BRG1 or hBRM1, BRG1-associated factor 155 (BAF155), BAF170, and INI1/hSNF5 form the functional core of the complex (29).
The components of SWI/SNF complexes appear to be involved both in activation and repression of transcription of target genes. Analysis of genome-wide expression profiles of yeast cells defective for SNF2 function using high-density oligonucleotide arrays or cDNA microarrays indicated that the SWI/SNF complexes are involved in repression of genes (13, 41). Three components of the yeast SWI/SNF complex interact with the Sin1p transcriptional repressor (26). The Hir repressors of the yeast histone HTA1-HTB1 locus have been shown to bind and recruit the components of SWI/SNF complex to mediate repression (7). In human cell lines, BRG1, the ATPase subunit of the SWI/SNF complex, has been shown to repress transcription of the c-FOS, cyclin A, and cyclin E genes depending on binding with the retinoblastoma protein (Rb) or Rb and HDAC1 (25, 37, 51). Since cyclin E and cyclin A are two critical proteins separately expressed in the G1 phase and S phase of cell cycle, it has been proposed that BRG1 is involved in cell cycle control via its association with Rb and repression of cyclins. Furthermore, it has been demonstrated that Rb, depending on its ability to recruit different repressor complexes such as histone deacetylase (HDAC)-Rb-hSWI/SNF or Rb-hSWI/SNF, regulates the exit of cells from G1 or S phase of cell cycle (51). Thus, one of the mechanisms by which components of the SWI/SNF complex mediate transcriptional repression is by associating with HDAC complex. As a direct support of this hypothesis, complexes that consist of components of SWI/SNF such as BRG1 with those of the repressor complexes such as Sin3A and HDAC have been recently identified (39). Additional mechanisms of repression by SWI/SNF include promoter-dependent repositioning of the nucleosomes that results in blocking rather than opening of the sequences necessary for binding to an activator protein or facilitation of binding of a repressor protein to the promoter (16, 17).
In addition to being a binding partner for human immunodeficiency virus type 1 integrase, INI1/hSNF5 has been shown to associate with Epstein-Barr virus transactivator protein, EBNA-2, the human homologue of Trithorax, ALL1 (MLL), the proto-oncogene product c-MYC, the replication protein E1 of human papillomavirus and most recently, GADD34 (1, 6, 14, 19, 33, 48). Structure-function analysis suggests that there are three highly conserved regions in INI1/hSNF5. The first two are imperfect repeats (Rpt1 and Rpt2), and the third region is termed homology region 3 (HR3) (23). The repeat regions appear to be involved in protein-protein interactions. One possible role of INI1/hSNF5 is to mediate the recruitment of the SWI/SNF complex to specific sites through its ability to directly interact with various regulatory proteins thus mediating such diverse processes as transcription, replication and integration.
At this point it is not clear how INI1/hSNF5 exerts its tumor suppressor function. It is not clear if it has a role in cell cycle or apoptosis. The target genes that are specifically regulated by INI1/hSNF5 gene are currently unknown. Therefore, to understand the INI1/hSNF5 mediated tumor suppressor function, we have studied the effect of reintroduction of this gene into cell lines derived from malignant rhabdoid tumors that carry deletion of INI1/hSNF5 gene. We report here that introduction of INI1/hSNF5 into these tumor derived cells causes cell cycle arrest at G0-G1 stage. Furthermore, we found that this cell cycle arrest correlates with the ability of INI1/hSNF5 to directly mediate the HDAC-dependent transcriptional repression of cyclin D1 gene. These results provide a potential mechanism for tumor suppression by INI1/hSNF5.
MATERIALS AND METHODS
Plasmids.
The plasmid pEAC001 expressing INI1/hSNF5 under the control of a tetracycline inducible promoter was constructed by inserting an EcoRI to XbaI fragment encoding INI1/hSNF5 cDNA (from aa 1 to 385) excised from pGADNot-INI1/hSNF5 (23) into pUHD10.3 (9). The series of pCGN plasmids expressing hemagglutinin (HA)-tagged full-length INI/hSNF5 and truncations of INI1/hSNF5 including HA-D2 (amino acid [aa] 106 to 385), HA-20.2 (aa 141 to 385), HA-1.2(aa 181 to 385), HA-27B(aa 1 to 245), HA-3B(aa 1 to 294), and S6(aa 183 to 294), were constructed by separately inserting the BamHI to BglII fragments isolated from pGADNot constructs (23) into pCGN plasmid digested with BamHI. The expression of stable proteins from these plasmids was confirmed by immunoblot analysis using monoclonal anti-HA (α-HA) antibodies (courtesy of S. Buhl, Albert Einstein College of Medicine). To assay the transcriptional activity of cyclin D1, a reporter plasmid (pCD1-Luc) containing the region −1745 to +141 fused to the luciferase gene in-frame was utilized (46). The human cyclin D1 cDNA expression construct (pRc-CMV-cyclin D1) is under the control of the cytomegalovirus (CMV) promoter (12).
Cell lines.
HeLa cells that express the tetracycline-dependent activator Tet-VP16 (HeLa Tet-On) were obtained from Clontech Laboratories Inc. Cells used were routinely cultured in Dulbecco's modified Eagle medium, supplemented with 10% fetal bovine serum and G418 (200 μg/ml). MON cells (43) were cultured in RPMI supplemented with 10% fetal bovine serum. When required, cells were selected with 200 to 250 μg of G418-neomycin (catalogue no. 11811-031; Gibco/BRL) per ml.
Transcription assays.
The constructs used in the various transcription assays utilize the luciferase gene as a reporter under the control of various promoters. These include pCD1-Luc (46), pCyclinE-Luc (2), and pCyclinA-Luc (46). Cells were harvested and lysed in luciferase cell lysis buffer (150 μl per 106 cells, catalogue no. E1501; Promega), and 10 μl was analyzed in a luminometer for the luciferase activity using the reagents provided in the luciferase assay system (catalogue no. E1501; Promega). Results are expressed as relative light units/106 green fluorescence protein (GFP)-positive cells. TSA (catalogue no. T8552; Sigma) was used at a concentration of 5 ng/ml for 48 h prior to determination of luciferase activity, when indicated. To determine the effect of INI1/hSNF5 on the transcriptional activity of cyclin D1 promoter, HeLa or MON cells were cotransfected with the reporter plasmid pCD1-Luc and pCGN-INI1/hSNF5 expressing HA-INI1 or its deletion derivatives along with 1/10 amount of pEGFP plasmid by using either the calcium phosphate method or FuGENE6 (catalogue no. 1-814-443; Boehringer Mannheim). Essentially, 4 μl of FuGENE6 reagent was added to 100 μl of serum-free medium and incubated for 5 min at room temperature. The reagent was mixed with DNA and allowed to stand for 15 min at room temperature, before being added to the cells. Luciferase assay was carried out 48 h posttransfection. The relative light units per microgram of total protein was determined in each case and normalized to percent transfection efficiency, which was determined by counting GFP-positive cells. To test the effect of E1a-12S (2), an inhibitor of Rb function on INI1-mediated repression, a plasmid expressing the same protein was cotransfected along with pCGN-INI1 as indicated.
Flat cell assay.
MON cells at 30 to 50% confluency, plated on 3.5-cm-diameter plates or six-well plates, were transfected with 1.5 μg of pEAC001 or pUHD10.3 (9) and 0.5 μg of pTet-on using FuGENE6 (Boehringer Mannheim), as described for the transcription assays. After transfection, cells were cultured for 24 h prior to drug selection. Cells were maintained under neomycin selection for 15 days with several changes of media, prior to observation under the microscope. Similar experiments were performed whereby MON cells were transfected with pRc-CMV-Cyclin D1 or the parent plasmid pRc-CMV (0.75 μg) and pCGN-INI1 or its parent plasmid pCGN (0.75 μg) and pEGFP that expresses GFP and neomycin-resistant marker (0.5 μg). Expression of various transfected genes was analyzed by subjecting the total proteins to sequential immunoblot analysis using various antibodies such as monoclonal α-cyclin D1 (catalogue no. sc-8396; Santa Cruz), α-HA (catalogue no. sc-805; Santa Cruz) and α-tubulin (catalogue no. T5168; Sigma) antibodies. Expression of HA-INI1 deletion fragments was assessed by using monoclonal α-HA antibodies (courtesy of Susan Buhl, Albert Einstein College of Medicine, New York, N.Y.).
Cell cycle analysis.
The cell cycle analysis was carried out using fluorescence-activated cell sorting (FACS) as described by Darzynkiewicz and Juan (6a). About 106 to 107 cells were collected in 5 ml of phosphate-buffered saline (PBS) and centrifuged at 200 × g for 6 min. The resulting cell pellet was resuspended in 0.5 ml of PBS using a Pasteur pipette and transferred to tubes containing 4.5 ml of 70% ethanol. Cells were kept in the fixative for 2 h and then centrifuged for 5 min at 200 × g. The ethanol was thoroughly decanted, and the cell pellet was suspended in 5 ml of PBS. After 60 s, cells were centrifuged for 5 min at 200 × g and resuspended in 0.5 ml of staining solution containing propidium iodide (20 μg/ml), 0.1% Triton X-100, and RNase A (200 μg/ml). Cells were incubated either for 15 min at 37°C or for 30 min at room temperature and then subjected to flow cytometry.
Reverse transcription-PCR analysis.
The MON cells were transfected with either pCGN-INI1, expressing HA-INI1/hSNF5 or pCGN (empty vector) along with pEGFP-C2 that expresses neomycin-resistant marker and GFP. Cells were selected for 4 to 5 days posttransfection with G418 (250 μg/ml) in the presence or absence of TSA (5 ng/ml), and total RNA was isolated from these cells using Trizol (catalogue no. 15596-018; Gibco/BRL). About 6 μg of total RNA was subjected to reverse transcription-PCR analysis. First, Superscript II, RNase H− reverse transcriptase was used to generate the cDNAs in a total volume of 20 μl, using oligo(dT)12-18 as primers and by using the conditions as recommended (catalogue no. 18064-014; Gibco/BRL, Life technologies Inc.). One microliter of the cDNAs and fivefold serial dilutions of it were mixed with 25 pmol each of primers specific to the cyclin D1 gene (5′ CTT CCT CTC CAA AAT GCC AG 3′and 5′ AGA GAT GGA AGG GGG AAA GA 3′) and amplified for 25 cycles in a 25-μl PCR mixture using 55°C as the annealing temperature and 1.5 mM MgCl2. A control PCR using primers specific to the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene (5′ GGT GAA GGT CGG AGT CAA CG 3′ and 5′ CAA AGT TGT CAT GGA TGA CC 3′) was also carried out using the same RNA samples and same conditions.
Coimmunoprecipitation analysis.
These analyses were carried out essentially as previously described (6). Cells from a 10-cm-diameter plate were collected 40 h posttransfection and sonicated in 500 μl buffer of G (PBS without Mg2+ and Ca2+ containing 0.1% IGEPAL [catalog no. 1-3021; Sigma], 1 mM dithiothreitol, bovine serum albumin [2 mg/ml], 1 mM phenylmethylsulfonyl fluoride, and a 1-μg/ml concentration each of pepstatin, aprotinin, and leupeptin). Lysates were precleared by incubating with 30 μl of protein A-Sepharose (50% slurry) for 1 h. GFP antibodies (catalogue no. 46-0092; Invitrogen) were added to precleared lysate for 1 h, which was followed by the addition of protein A-agarose beads (30 μl of a 50% slurry), and the mixture was incubated overnight at 4°C. Bound proteins were washed four times with buffer G without bovine serum albumin, separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblotted with monoclonal α-HDAC1 antibodies (catalogue no. sc-8410; Santa Cruz).
ChIP analysis.
The assays were performed using the chromatin immunoprecipitation (ChIP) assay kit (catalogue no. 17-295; Upstate Biotechnology) essentially as indicated by the manufacturer with minor modifications. Briefly, MON cells at a density of 30 to 50% confluency in a 10-cm-diameter dish were transfected with 1 μg of pEGFP that expresses GFP and neomycin-resistant marker, 1 μg of reporter plasmid pCD1-Luc, and 9 μg of either pCGN or pCGN-INI1 plasmids, by using the calcium phosphate precipitation method. Twenty-four hours posttransfection, cells were subjected to G418 (200 μg/ml) selection for 4 days. The transfected cells were then used for the ChIP assay using α-N-INI1 (PB3) (50), α-acetyl H4 (catalogue no. 06-866; Upstate Biotechnology), and α-HDAC1 (catalogue no. sc-8410; Santa Cruz) antibodies. DNA isolated from the immunoprecipitates was then subjected to 40 cycles of PCR amplification using primers ZKCyclinD1p1-L, (GAAACTTGCACAGGGGTTGT) and ZKCyclinD1p1-R, (ATTTAGGGGGTGAGGTGGAG) that amplifies the region between nucleotides −629 and −329 within the cyclin D1 promoter. As internal control, a region downstream of cyclin D1 promoter in the pCD1-Luc construct was amplified using the primers LUC-L, 5′-TCAAAGAGGCGAACTGTGTG-3′ (+1155), and LUC-R, 5′-TTTTCCGTCATCGTCTTTCC-3′ (+1487).
Immunohistochemistry of cyclin D1 overexpression in tumor sections.
Immunohistochemistry was performed on three AT/RT tumors and a sample of healthy brain present adjacent to the tumor. For cyclin D1 immunostaining, we used mouse monoclonal antibody (MAb) DCS-6 (Cell Marque, Austin, Tex.), at 1:100 dilutions. This antibody has been shown to be specific for cyclin D1 (22). For each case, 5-μm-thick sections were cut from the paraffin blocks. The sections were deparaffinized, rehydrated, and stained by standard methods, using the avidin-biotin-horseradish peroxidase complex to localize the antibody bound to antigen, with diaminobenzidine as the final chromogen. Appropriate antigen retrieval was performed in the appropriate buffer. Immunostaining was performed on an automated immunostainer, NexES (Ventana Medical Systems, Tucson, Ariz.). All immunostained sections were lightly counterstained with hematoxylin. For negative controls, the immune serum was replaced either by PBS or nonimmune serum. The sections were reviewed by the pathologist author (D. Zagzag).
RESULTS
Induction of cell cycle arrest and flat cell formation by INI1/hSNF5 in cells derived from AT/RT.
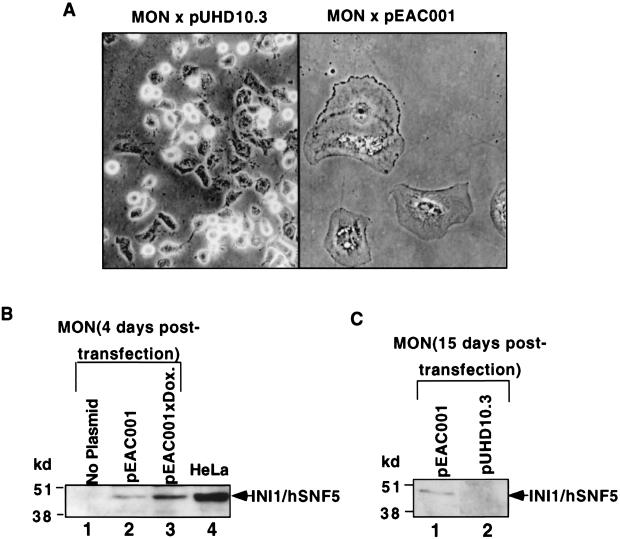
Previous studies have indicated that loss of INI1 gene is a critical genetic alteration in the tumorigenesis of AT/RT tumors. However, at this point it is not known if INI1 is directly involved in establishing or maintaining processes that would prevent the transformation. We surmised that if INI1 has a direct role in cell cycle, loss of INI1 would be sufficient to cause deregulation of normal cell cycle control. Therefore, we reintroduced INI1/hSNF5 into MON cells, derived from malignant rhabdoid tumors, to test its effect on cell proliferation. We expressed INI1/hSNF5 from either a CMV promoter (as a fusion to HA tag from plasmid pCGN-INI1/hSNF5) or from a tetracycline-inducible promoter (plasmid pEAC001) in both HeLa and MON cells, which were also transfected with a plasmid (pTet-On) carrying both a neomycin resistance marker and Tet-VP16. Neomycin-resistant stable transfectants of HeLa cells were obtained after 15 days of selection. However, no resistant colonies were obtained in MON cells upon repeated attempts of transfection with INI1/hSNF5. We observed that every transfection of MON cells with any plasmid expressing INI1/hSNF5 resulted in the formation of flat cells with distinct morphology (Fig. 1A, right panel). These flat cells were several times larger than the control MON cells, were highly vacuolated, and stayed on the plates with selective medium for up to a month before finally disintegrating. Even low-level expression of INI1/hSNF5 from the tetracycline-inducible promoter of the pEAC001 plasmid, in the absence of inducers, was sufficient to cause this flat cell formation (Fig. 1A and B). Immunoblot analysis using α-INI1/hSNF5 antibody of the total proteins isolated from flat cells 15 to 20 days after selection with neomycin indicated that INI1/hSNF5 is expressed in these cells (Fig. 1B and C). Transfection of an empty vector (Fig. 1A, left panel) did not result in flat cell formation (Table 1). In addition to MON, we also tested other cell lines such as STA-WT1 (derived from the AT/RT [43)] as well as SW13, C33A (24) that express no or low levels of BRG1 and hBRM. The results indicated that INI1/hSNF5 induced flat cell formation only in the AT/RT-derived cells but not in any other cell lines (data not shown). These results clearly establish the specificity of this effect of INI1/hSNF5 to cause flat cells only in INI1−/− cells derived from AT/RT, but not in cells expressing endogenous INI1/hSNF5 protein.
FIG. 1.
Flat cell induction by INI1/hSNF5 in MON cells. (A) Phase-contrast microscopy of MON cells transfected with pEAC001 (expressing INI1/hSNF5 [right-hand panel]) or pUHD10.3 (empty vector [left-hand panel]) along with pTet-On vector after 15 days of selection in neomycin. Both the images were taken at a magnification of ×20 (shown here at ×16). (B and C) Immunoblot analysis of total protein isolated from MON cells. (B) Proteins isolated from MON after 4 days of transfection; (C) results of analysis after 15 days. Dox., doxycycline.
TABLE 1.
Flat cell assay in Mon cells
| Plasmid transfected with pTet-On | Avg no. of flat cells (% of total cells) in expt:
|
Avg % flat cells | ||
|---|---|---|---|---|
| 1 | 2 | 3 | ||
| pEAC001 | 33 (94) | 46 (96) | 51 (100) | 97 |
| pUHD10 | 5 (15) | 2 (8) | 0 | 7.7 |
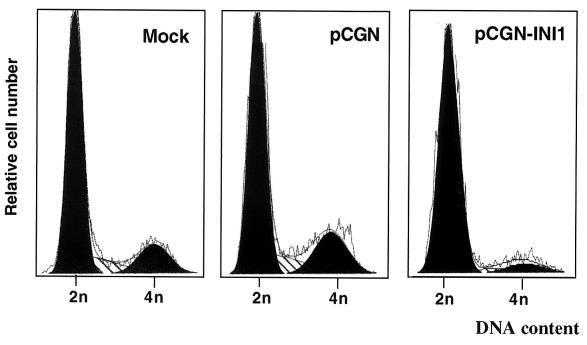
Previous studies have demonstrated that reintroduction of the tumor suppressor gene Rb into Rb−/− Saos cells results in the formation of flat cells (47). In addition, introduction of BRG1 into BRG1−/− SW13 cells also resulted in flat cells (8). It has been demonstrated that BRG1-induced flat cells are arrested at G1/S phase of the cell cycle (40, 51). We further analyzed the MON cells transfected with INI1/hSNF5 by FACS to determine the stage at which the cells are arrested (Fig. 2, Table 2). The cells were cotransfected with a plasmid expressing a neomycin marker and pCGN-INI1 expressing HA-INI1/hSNF5 and analyzed by FACS at 4 to 7 days after selection. The results indicate that MON cells have an unusually large number of cells in G0-G1 stage, which is also reflected by their slow doubling time. When HA-INI1/hSNF5 was expressed, there was a significant reduction of cells in S and G2/M phase and a corresponding increase of cells in the G0-G1 stages of the cell cycle. When MON cells were transfected with empty vector (pCGN), the profile of cells was identical to that of untransfected cells (Fig. 2 and Table 2). These results indicated that expression of INI1/hSNF5 causes MON cells to arrest at the G0-G1 stage of the cell cycle. This result is consistent with INI1/hSNF5's role as a negative regulator of cell cycle and a tumor suppressor.
FIG. 2.
FACS analysis of MON cells transfected with INI1/hSNF5. MON cells were analyzed by FACS 7 days after transfection. The plasmids used in the transfection are indicated at the top of the panels. They are pCGN-INI1, which is the HA-INI1/hSNF5 expression plasmid, and pCGN, which is the empty vector control. The diagram represents the relative cell number in 2n and 4n channels. The actual percentage of cells in each stage of the cell cycle is presented in Table 2.
TABLE 2.
FACS analysis of transfected MON cells
| Plasmid transfected | % in expt:
|
Avg (%) (5 expts)
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1
|
2
|
3
|
||||||||||
| G0/G1 | S | G2/M | G0/G1 | S | G2/M | G0/G1 | S | G2/M | G0/G1 | S | G2/M | |
| pCGN | 77.5 | 12.2 | 10.4 | 66.8 | 15.1 | 18 | 72.7 | 10.1 | 17.1 | 72.7 | 11.6 | 15.7 |
| pCGN-INI1 | 89.6 | 7.5 | 2.9 | 90.7 | 0.4 | 8.9 | 86.9 | 2.55 | 10.2 | 89.3 | 3.9 | 6.8 |
| None | 75.3 | 16.1 | 8.6 | 64.5 | 16.5 | 18.8 | 66.3 | 14.4 | 19.2 | 73.5 | 15.6 | 16.9 |
INI1/hSNF5 represses transcription of cyclin D1.
We next sought to determine the possible mechanism by which INI1/hSNF5 causes this cell cycle arrest in AT/RT-derived cells. The cell cycle is tightly controlled in normal cells by mechanisms in which cyclins and associated specific kinases (cyclin-dependent kinases [CDKs]) modulate the activity of key regulators of cell cycle such as Rb to mediate the cell cycle progression. The regulation of expression and activity of cyclins and their dependent kinases is a homeostatic mechanism in eukaryotic cells (11, 38). Cyclin D and cyclin E are two classes of “G1 cyclins” that demonstrate distinct functional characteristics. While the three cyclin D genes (D1, D2, and D3) are rapidly induced and expressed at high levels soon after the mitogenic stimulation of the cells, cyclin E expression appears to peak at the G1/S border (38). Furthermore, cyclin D-CDK complexes are activated at mid-G1 stage of the cell cycle, and cyclin E-CDK complexes are activated at the G1/S boundary. Cyclin A on the other hand is activated during the S-to-G2 transition and is present at elevated levels in the G2 stage of the cell cycle. Previous studies with BRG1 and Rb have established that a key mechanism of G1/S cell cycle arrest by these proteins is repression of cyclins E (51) and A (40, 51). Since INI1/hSNF5 causes the arrest of the cells at G0-G1 stage, it is possible that this effect is mediated by repression of G1 cyclins. Therefore, we tested the effect of INI1/hSNF5 on transcription of cyclins in MON cells using reporter plasmids in which a luciferase gene is under the control of cyclin D1, E, or A promoters (46). The cyclin D1 gene promoter region (nucleotides −1745 to +141) includes a MYC E-box site (Ems), a TRE site, and an initiator (Inr) element as well as potential recognition sequences for E2F, OTF, Sp-1 and CREB/ATF (Fig. 3A). When MON cells were cotransfected with a luciferase reporter construct under the regulation of this promoter and a plasmid expressing INI1/hSNF5 (pCGN-INI1/hSNF5), the transcription from this promoter was inhibited to about 20% compared to that in the absence of INI1/hSNF5 (Fig. 3B). We also tested the effect of INI1/hSNF5 on a luciferase reporter under the control of various promoters such as TK, E2F, plasminogen-activated inhibitor, and NVL 30. INI1/hSNF5 had no effect on these promoters in MON cells (data not shown). In addition, INI1/hSNF5 did not repress transcription of cyclin D1 in HeLa cells, indicating that this activity is cell type specific (Fig. 3B). INI1/hSNF5 also repressed the transcription of cyclin E and cyclin A in MON (Fig. 3C). Since cyclin D1 is upstream of all the cyclins in the pathway, and since it is correlated well with the ability of INI1/hSNF5 to arrest the cells at G0-G1, we focused our attention on cyclin D1 for further studies.
FIG.3.
Repression of cyclin D1 transcription by INI1/hSNF5. (A) Schematic representation of regulatory region in cyclin D1 promoter. (B) INI1/hSNF5 represses transcription from the cyclin D1 promoter. Top panel depicts the graphic representation of transcriptional activity. Relative light units per microgram of protein were measured and normalized to the percentage of transfection efficiency determined by using GFP. Results are averages of three independent experiments and are expressed as percent maximal activity. Error bars, standard deviations. The lower panel shows an immunoblot analysis of the lysates used in these experiments using α-HA antibody. (C) Repression of cyclin E and cyclin A promoters by INI1/hSNF5. The indicated reporter plasmids were cotransfected with INI1 expressing plasmids and the transcriptional activity was determined as above. (D) Repression of endogenous cyclin D1 by INI1/hSNF5 in MON cells. The diagram depicts the ethidium bromide-stained gel of DNA obtained from the reverse transcription-PCR analysis. RNA isolated from MON cells transfected either with pCGN-INI1 (expressing HA-INI1/hSNF5) or pCGN (empty vector) was subjected to reverse transcription-PCR. The top panel represents the reverse transcription-PCR products obtained using primers specific to cyclin D1 gene. The bottom panel represents reverse transcription-PCR products using primers specific to the GAPDH gene as internal control. The numbers on top of the gel correspond to the amount of RNA template used for the reverse transcription-PCR.
INI1/hSNF5-mediated repression of cyclin D1 is dependent on HDAC activity.
The above results suggested that INI1/hSNF5 causes arrest of cells at G0-G1 stage of the cell cycle and represses transcription of cyclin D1. Many studies have indicated that components of SWI/SNF complex are involved in transcriptional repression as well as activation (42). Previous studies with Rb and BRG1 have indicated that simultaneous recruitment of BRG1 and HDAC1 by Rb is responsible for the repression of cyclin E gene and arrest of cells at G1/S stage (51). Furthermore, biochemical purification of BRG1-associated complexes indicated the coexistence of SWI/SNF components with that of the Sin3-HDAC components (39). These studies suggest an interplay between components of the SWI/SNF complex and those of HDAC complex. Studies from our laboratory indicate that INI1/hSNF5 can mediate transcriptional repression in a cell type-specific manner by interacting with the components of the HDAC complex (K. P. Davies, Z.-K. Zhang, and G. V. Kalpana, unpublished data). Thus, one possible mechanism by which INI1/hSNF5 represses transcription of cyclin D1 in MON cells is by directly recruiting HDAC activity to the cyclin D1 promoter. Therefore, we tested the ability of TSA, a specific inhibitor of HDAC activity, to overcome the INI1/hSNF5-mediated repression of cyclin D1 in MON and HeLa cells. We found that the repression of cyclin D1 promoter by INI1/hSNF5 was reversed to near completion by the addition of low levels (5 ng/ml) of TSA in MON (Fig. 3B, top panel). Immunoblot analysis of the proteins isolated from the same transfected cells indicated that TSA had no effect on the levels of INI1/hSNF5 (Fig. 3B, lower panel). These results suggest that repression of cyclin D1 by INI1/hSNF5 is mediated by the recruitment of HDACs.
The above results were obtained using the reporter construct and transient transfection. Therefore, to determine if INI1/hSNF5 is able to repress the endogenous cyclin D1 gene, we carried out a reverse transcription-PCR analysis. MON cells were transfected with plasmid expressing either HA-INI1/hSNF5 or empty vector along with a plasmid carrying a selectable marker. The cells were selected for 4 to 5 days in the presence and absence of TSA, and total mRNAs were isolated. Equal amounts of serially diluted mRNA isolated from MON cells were subjected to reverse transcription-PCR using primers specific for cyclin D1 as well as internal control primers to amplify GAPDH gene. The PCR was carried out for 25 cycles. The results indicate that while the expression of the control GAPDH gene did not change, expression of endogenous cyclin D1 was repressed several fold (five- to sixfold) by INI1/hSNF5 (Fig. 3D, compare lanes 4 to 6 with lanes 10 to 12). Furthermore, the addition of TSA completely reversed the repression of cyclin D1 by INI1/hSNF5 (Fig. 3D, compare lanes 7 to 9 with lanes 10 to 12). These results confirm that, INI1/hSNF5 represses the endogenous cyclin D1 gene, in a manner that is dependent on the recruitment of HDAC.
HDAC1 association with INI1/hSNF5 is necessary for cyclin D1 repression.
The results described above do not address whether or not the repression of cyclin D1 is mediated by a direct recruitment of HDAC to the cyclin D1 promoter by INI1/hSNF5 or is mediated by an indirect mechanism. Studies in our laboratory have indicated that INI1/hSNF5 is able to coimmunoprecipitate components of the HDAC complex including HDAC1 (K. P. Davies, Z.-K. Zhang, and G. V. Kalpana, unpublished data) and that Rpt1, one of the highly conserved regions of INI1/hSNF5 appears to be necessary for this association. Therefore, it is possible that INI1/hSNF5 directly recruits the HDAC activity to the cyclin D1 promoter and that this recruitment is necessary for its ability to repress cyclin D1 transcription. To test this hypothesis, first we tested a mutant of INI1/hSNF5 lacking Rpt1 for its ability to coimmunoprecipitate HDAC1 and then assayed for its ability to repress transcription of cyclin D1.
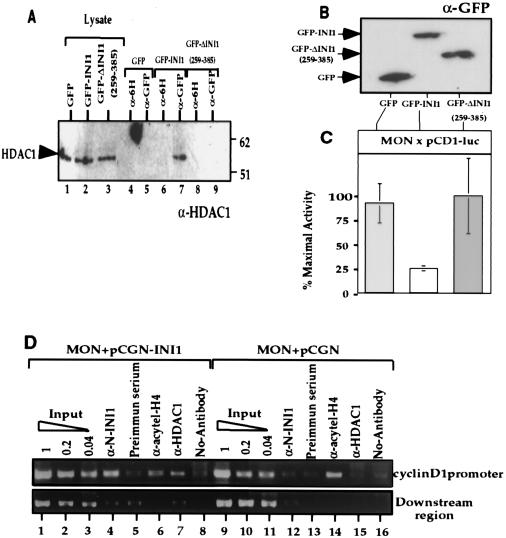
We expressed GFP, GFP-INI1/hSNF5 or GFP-ΔINI1/hSNF5(259-385), which lacks the Rpt1 region, in MON cells and performed coimmunoprecipitation experiments using α-GFP antibodies (and α-6H antibodies as a negative control). The resulting immunoblot was then probed with α-HDAC1 antibodies (Fig. 4A). The results indicated that α-GFP antibodies were able to coimmunoprecipitate HDAC1 in the presence of GFP-INI1/hSNF5 (Fig. 4A, lane 7), but not in the presence of GFP or GFP-ΔINI1/hSNF5(259-385) (Fig. 4A, lanes 5 and 9). Analysis of the lysates indicated that equal quantities of HDAC1 and GFP-fusions were present in these assays (Fig. 4A, lanes 1 to 3). These results suggested that full-length INI1/hSNF5 but not the deletion fragment is able to form a complex with HDAC1. We next tested to determine if deletion fragment lacking Rpt1 is able to repress cyclin D1 transcription. We expressed the above GFP-fusion proteins in MON cells along with cyclin D1 reporter construct and assayed for their ability to repress transcription, as before. Immunoblot analysis indicates that the proteins are stably expressed (Fig. 4B). The results of transcription assay indicate that while full-length GFP-INI1 was able to repress transcription of cyclin D1, GFP-ΔINI1/hSNF5(259-385) was not (Fig. 4C). These results together suggest that association of HDAC with INI1 is necessary for repression of cyclin D1.
FIG. 4.
INI1/hSNF5 represses cyclin D1 by recruiting HDAC activity to the promoter. (A) Coimmunoprecipitation of HDAC1 with INI1 and its truncation. Plasmids expressing either GFP, GFP-INI1, or GFP-ΔINI1Δ(259-385) were transfected into MON cells and immunoprecipitated using α-GFP antibody. The resulting immunoprecipitates were separated on sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblotted using α-HDAC1 antibody. “Lysate” represents equal amount of proteins from the input (lanes 1 to 3). Lanes 4 to 9 represent the immunoblot analysis of the immunoprecipitates obtained using either α-GFP antibody or a control α-6H antibody. (B) Immunoblot analysis of MON cells transfected with indicated plasmids using α-GFP antibody to demonstrate the level of proteins expressed. (C) The effect of GFP-fusions of INI1/hSNF5 and the truncation on the cyclin D1 promoter. Relative light units per microgram of protein present in the lysates from the above transfection were measured and normalized to the percentage of transfection efficiency determined by using GFP fluorescence. Results are averages of three independent experiments and are expressed as percent maximal activity. Error bars, standard deviations. (D) ChIP analysis of cyclin D1 promoter in the presence of INI1/hSNF5. MON cells were transfected with either pCGN (empty vector) or pCGN-INI1 (expressing HA-INI1) and pCD1-Luc. ChIP analysis was performed using α-INI1, preimmune serum, α-HDAC1, and α-acetyl-histone H4 antibodies. The immunoprecipitated DNA was subjected to PCR amplification using the primers specific to cyclin D1 promoter (top panel) or a downstream region (bottom panel). The picture represents agarose gel electrophoretic separation of DNA obtained in these PCRs. A fivefold serial dilution of input DNA has been loaded for each sample.
Direct recruitment of INI1 and HDAC1 to the cyclin D1 promoter.
To determine if INI1/hSNF5 is directly recruited to the cyclin D1 promoter and to determine if its recruitment is correlated to the recruitment of HDAC, we conducted chromatin immunoprecipitation (ChIP) assays. We transfected pCGN-INI1 (expressing HA-INI1/hSNF5) or empty vector (pCGN) into MON, isolated and cross-linked chromatin from these cells, and performed immunoprecipitation using antibodies specific to INI1, HDAC1, and acetylated histone H4 (acetyl-H4). The immunoprecipitated DNA was then subjected to PCR amplification using the primers specific to the cyclin D1 promoter. As a control, fivefold serial dilutions of total input DNA as well as DNA immunoprecipitated in the absence of antibodies were used for PCRs. The results indicated that when HA-INI1 was expressed, α-INI1 antibodies were able to immunoprecipitate cyclin D1 promoter region, in the amount close to 20% compared to that of the input DNA (Fig. 4D, lane 4). In contrast, α-INI1 antibodies resulted in background level (similar to that of the preimmune serum) of immunoprecipitation in the absence of HA-INI1 (Fig. 4D, lane 12). These results indicate that INI1/hSNF5 is directly recruited to cyclin D1 promoter. Furthermore, the results of the ChIP assay indicated that in the absence of INI1/hSNF5 significantly higher level of cyclin D1 promoter DNA (more than 20% compared to that of the input DNA) was immunoprecipitated by α-acetyl-H4 antibodies (See Fig. 4D, lane 14). In multiple experiments, the presence of INI1/hSNF5 resulted in reduced levels of immunoprecipitation (much less than 5% compared to that of the input, Fig. 4D, lane 6) by α-acetyl-H4 antibodies. The ChIP results obtained with α-HDAC1 were consistent with the idea that INI1/hSNF5 mediates the recruitment of HDAC1 to cyclin D1 promoter. The presence of INI1/hSNF5 resulted in significantly higher amount of immunoprecipitation of cyclin D1 promoter specific region by α-HDAC1 antibodies compared to that in the absence of INI1/hSNF5 (Fig. 4D, compare lanes 7 and 15). As a control, we used primers to amplify downstream region (positions +1155 to +1487) of the pCD1-Luc construct using the same batch of immunoprecipitated DNA as templates. As indicated in the Fig. 4D, very little amplification was observed when immunoprecipitated DNA was used as a template using the control primers. These results indicate that (i) INI1 is directly recruited to the cyclin D1 promoter, (ii) HDAC1 is recruited to the promoter in a manner dependent on INI1/hSNF5, and (iii) the presence of INI1/hSNF5 results in the deacetylation of histones on specific region of the cyclin D1 promoter. These results are consistent with the idea that a direct recruitment of HDAC1 to the cyclin D1 promoter by INI1/hSNF5 is responsible for the observed repressive effect.
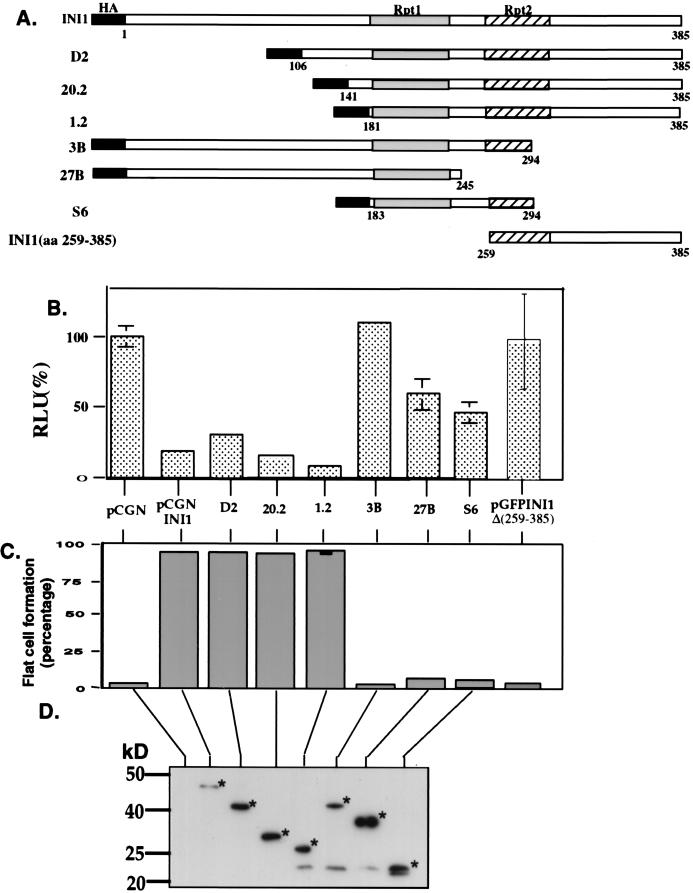
Repression of cyclin D1 by INI1/hSNF5 is directly correlated to its ability to cause flat cells.
The above results establish that INI1/hSNF5 causes flat cell formation and cell cycle arrest and that it represses transcription of cyclin D1 by directly recruiting HDAC activity to its promoter. To determine if these two activities of INI1/hSNF5 are directly correlated with each other, we tested a panel of INI1/hSNF5 deletions (Fig. 5A) for their ability to cause flat cell formation and cyclin D1 repression. These INI1/hSNF5 deletions were expressed as HA-tagged proteins in MON cells and were first assayed for their ability to mediate repression of cyclin D1. The results indicated that several of the N-terminal deletions (D2, 20.2, and 1.2) retained the ability to repress transcription (Fig. 5A and B). These fragments of INI1, although they have a deletion of the N-terminal domain, retain all the three conserved regions. Interestingly, several of the C-terminal deletions (27B, 3B) that retained Rpt1 but an incomplete Rpt2 region were not able to completely repress transcription and exhibited about 50% or more of the luciferase activity compared to that of the control (Fig. 5A and B). Furthermore, Rpt2 and C-terminal region appear to be insufficient for repression as indicated by the inability of GFP-ΔINI1/hSNF5(259-385) to repress transcription (Fig. 4C and 5B). These results together suggest that both Rpt1 and Rpt2 are necessary for the repression of cyclin D1 transcription.
FIG. 5.
Repression of cyclin D1 by INI1/hSNF5 is correlated with its ability to cause flat cells. (A) Truncations of INI1/hSNF5 used to test the effect on cyclin D1 expression and flat cell formation. Rpt, repeat. The numbers below the bar represent the amino acid number of the INI1 portion. (B) Repression of cyclin D1 by INI1 and its truncations. Cells transfected with indicated plasmids were harvested, the total proteins were isolated and subjected to luciferase assay as described. Relative light units/μg proteins were measured and normalized to the percentage of transfection efficiency determined by using GFP. Results are averages of three independent experiments and are expressed as percent maximal activity. (C) Percentage flat cells obtained by transfecting various truncations of INI1/hSNF5. The data represents average of three independent experiments. (D) Immunoblot analysis of the cell lysates used in the above experiments using monoclonal α-HA antibody (Courtesy of S. Buhl, Albert Einstein College of Medicine). Stars in the picture indicate the bands corresponding to the expected size of INI1 and truncations.
We next performed tests to determine if the ability of the deletion fragments of INI1/hSNF5 to repress transcription correlated with their ability to form flat cells. The same set of deletions as above were transfected into MON cells and the percentage of flat cells formed after 15 days of transfection was determined. The results indicated that the full-length protein and the three deletions (D2, 20.2, and 1.2) containing three repeat regions that significantly inhibited cyclin D1 promoter expression were able to cause a high percentage of flat cell formation (Fig. 5C). However, the deletions 3B, 27B, S6, and GFP-ΔINI1/hSNF5(aa259 to 385) that did not significantly inhibit cyclin D1 expression did not cause flat cell formation (Fig. 4C and 5B, C). Immunoblot analysis of the MON cells transfected with the deletion fragments illustrates that all the truncation mutants were expressed in the cells (Fig. 5D). These results indicate that the ability of INI1/hSNF5 deletions to repress cyclin D1 is directly correlated to their ability to cause flat cell formation.
Reversal of INI1/hSNF5-mediated G0-G1 arrest.
Our results suggests that one of the mechanisms by which INI1/hSNF5 causes G0-G1 arrest is by directly repressing cyclin D1 transcription. If this hypothesis is true, it should be possible to overcome the INI1/hSNF5-mediated block at G0-G1 by expressing cyclin D1 from a heterologous promoter that is not repressed by INI1/hSNF5. Therefore, we cotransfected MON cells with pRc-CMV-cyclin D1 (12) expressing cyclin D1, and pCGN-INI1, expressing HA-INI1/hSNF5. The cells were cotransfected with plasmids expressing a neomycin resistant marker. Expression of INI1/hSNF5 and empty vector pRc-CMV in MON cells induced flat cells 12 to 15 days after transfection, as before (Fig. 6A, third panel). In contrast, when cyclin D1 was coexpressed with INI1/hSNF5, there was a significant reduction in the percentage of flat cell formed suggesting that it reversed the inhibitory effect of INI1/hSNF5 (Fig. 6A, fourth panel). Flat cell formation was also not observed in control cells transfected either with the two empty vectors or vector expressing cyclin D1 (Fig. 6A, first two panels). To determine if coexpression of cyclin D1 is sufficient to overcome the growth inhibition, we also determined colony formation after transfection with INI1/hSNF5 in the presence or absence of cyclin D1. Repeated experiments indicated that while cells transfected with INI1/hSNF5 gave rise to a few colonies, those transfected with INI1/hSNF5 and cyclin D1 generated a significant number of colonies (Fig. 6B and C). The cells in these colonies are clearly undergoing cell division and appear to have overcome the block at G0-G1 (Fig. 6A to C). We also carried out sequential immunoblot analysis of proteins isolated from the above set of transfected cells to determine if cyclin D1 expression had any effect on levels of INI1. The result demonstrated that when INI1 is expressed there is several fold reduction in the cyclin D1 protein levels (Fig. 6D, compare lanes 2 and 4). Furthermore, INI1/hSNF5 levels were comparable in the presence or absence of cyclin D1 (Fig. 6D, lanes 3 and 4). The level of cyclin D1 expressed from CMV promoter was comparable to that of endogenous levels in the absence of INI1/hSNF5 (Fig. 6D, lanes 2 and 3). The above blot was also probed with α-tubulin antibody, and the results indicated the presence of equal amounts of proteins loaded in each lane (Fig. 6D, lower panel). These results demonstrated that the reversal of cell cycle arrest by cyclin D1 is not due to the decrease in the levels of INI1/hSNF5 or overexpression of cyclin D1. All the above data strongly indicate that coexpression of cyclin D1 is sufficient to abrogate the cell cycle arrest and flat cell formation by INI1/hSNF5.
FIG. 6.
Reversal of INI1/hSNF5-mediated flat cell formation by ectopic expression of cyclin D1. (A) Phase-contrast micrographs of MON cells expressing INI1 and cyclin D1. The first panel illustrates MON cells transfected with pRc-CMV and pCGN (empty vectors), the second panel illustrates MON cells transfected with pRc-CMV-CD1 (expressing cyclin D1) and pCGN, the third panel illustrates cells transfected with pCGN-INI1 (expressing HA-INI1/hSNF5) and pRc-CMV (empty vector), and the fourth panel illustrates the MON cells transfected with pRc-CMV-CD1 and pCGN-INI1. (B) Effect of INI1 and cyclin D1 on the colony numbers. MON cells were transfected with plasmids expressing indicated proteins. Cells were maintained under neomycin selection for 15 days prior to staining with trypan blue. (C) Graphical representation of colony counts in the presence and absence of cyclin D1 and INI1/hSNF5. The colonies were fixed with 2% paraformaldehyde, stained with 0.4% trypan blue, and counted, and results were expressed as percentages. Colonies obtained with empty vector control were taken as 100%. Note that expression of cyclin D1 overcomes the inhibitory effect of INI1 on colony formation in MON cells. (D) Cyclin D1 expression does not alter the expression of INI1/hSNF5. The photograph represents the immunoblot analysis of MON cells expressing INI1/hSNF5 and Cyclin D1. Total proteins were analyzed by sequential immunoblotting using α-INI1, α-cyclin D1 and α-tubulin antibodies.
Repression of cyclin D1 transcription by INI1/hSNF5 is independent of Rb function.
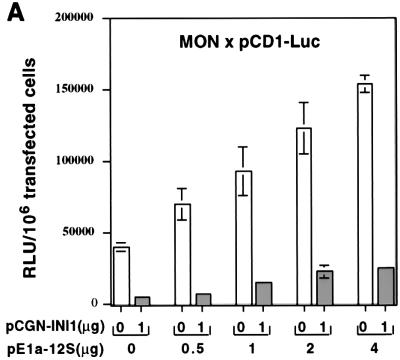
The above result that the expression of cyclin D1 is sufficient to overcome the cell cycle arrest by INI1 does not rule out other possibilities. For example, it has been established that overexpression of cyclin D1 will lead to hyperphosphorylation and inactivation of Rb family members that include Rb, p107 and p130. Furthermore, it has been previously reported that repression of cyclin E and cyclin A by BRG1, (another component of the SWI/SNF complex) required the association of Rb (40, 51). Therefore, it is possible that the Rb or any other member of Rb family function is necessary for INI1/hSNF5 mediated repression of cyclin D1. To test this hypothesis, we coexpressed increasing concentrations of E1a-12S, an adenovirus protein that binds to Rb pocket and inactivates the function of Rb family members (34), along with INI1/hSNF5 and pCD1-Luc in MON cells. Transcription assays were carried out as before. These experiments demonstrated that increasing concentrations E1a-12S resulted in increase in the basal activity of pCD1-Luc promoter in the absence of INI1/hSNF5, suggesting that E1a-12S protein is able to counteract the repressive effect of some component on cyclin D1 promoter (Fig. 7A). However, in the presence of INI1/hSNF5, the transcriptional activity of cyclin D1 promoter was significantly reduced at all concentrations of E1a-12S (Fig. 7A). The fold reduction in the cyclin D1 transcription by INI1/hSNF5 remained constant with increasing concentrations of E1a-12S, indicating that repression by INI1/hSNF5 is not affected. These results indicate that repression of cyclin D1 by INI1/hSNF5 does not require the function of Rb or its family members.
FIG. 7.
(A) INI1/hSNF5-mediated repression of cyclin D1 is independent of Rb family function. Increasing concentrations of plasmid pE1a-12S (expressing E1A-12S, an inhibitor of Rb family) were cotransfected with INI1/hSNF5 into MON cells. The transcription was assayed as before, and the results are expressed as relative light units (RLU) per microgram of protein. (B) Cyclin D1 is overexpressed in AT/RT tumor samples. The diagram represents immunohistochemical analysis of AT/RT tumors for expression of cyclin D1. (a and b)Paraffin section of brain tissue adjacent (BAT) to AT/RT tumor, 958406. (c to h) Paraffin sections of three different AT/RT tumors. The left panels represent hematoxylin and eosin (H&E) staining of the sections to demonstrate the nuclei. The right panels represent immunohistochemical analysis using α-cyclin D1 antibody. The arrows in panels c to h represent random nuclei. The positive signal in panel b represents a rare immunoreactive nucleus in the filed, probably from an astrocyte.
Overexpression of cyclin D1 in AT/RT tumors.
The observed G0-G1 arrest of AT/RT cells and repression of cyclin D1 expression by INI1/hSNF5 and reversal of these effects by expressing cyclin D1 from a heterologous promoter implies that cyclin D1 may be derepressed in AT/RT tumors. To determine if deletion or mutation of INI1/hSNF5 results in overexpression of cyclin D1 in AT/RT tumors, we carried out an immunohistochemical analysis of the paraffin sections of three cases of AT/RT tumors using α-cyclin D1 monoclonal antibodies, MAb DCS-6 (22). The immunohistochemical reaction with MAb DCS-6 showed a clear positive nuclear staining in all the three cases (Fig. 7B). In contrast, immunohistochemical analysis of normal brain tissue that was presented adjacent to the tumor tissue from one of the patients demonstrated no positive cyclin D1 signal (Fig. 7B). Cyclin D1 expression was heterogeneous within the tumors. Although the immunoreactivity to cyclin D1 varied in intensity, it was always above the background level and there was usually a clear-cut distinction between stained and unstained nuclei. The positive nuclei varied in number and were irregularly distributed within AT/RT tumors. However, numerous immunoreactive nuclei were found in highly cellular regions of the tumors. Furthermore, cyclin D1-positive reaction was observed in the nuclei of spindle-shaped as well as rhabdoid-shaped cells, both of which occur in AT/RT. These data strongly suggest that cyclin D1 is overexpressed in AT/RT tumors.
DISCUSSION
Accumulating evidence has demonstrated that INI1/hSNF5 is a key tumor suppressor gene mutated in MRT/AT/RT. However, the mechanism by which INI1/hSNF5 functions as a tumor suppressor is unknown. In this report, we demonstrate that INI1/hSNF5 prevents cell proliferation and causes flat cell morphology when it is reintroduced into cell lines derived from AT/RT. This effect is consistent with INI1/hSNF5 functioning as a tumor suppressor and is specific to AT/RT-derived tumor cell lines. It is possible that the AT/RT precursor cells are programmed to undergo cell cycle arrest and that the deletion of INI1/hSNF5 gene results in removal of the block to cell cycle leading to tumorigenesis. Reintroduction of INI1/hSNF5 thus appears to restore this block reversing the tumor phenotype.
INI1/hSNF5 is one of the core components of the SWI/SNF complex. The components of the complex have been shown to activate or repress transcription of cellular genes by remodeling chromatin structure (27, 42). Thus, the major mechanism by which INI1/hSNF5 exerts its tumor suppressor function is probably by regulating the transcription of genes involved in control of cell cycle. It is possible that it activates transcription of negative regulators of cell cycle and represses transcription of positive regulators of cell cycle. Consistent with this idea, we have found that INI1/hSNF5 directly represses the transcription of cyclin D1 gene. Interestingly, this repression appears to be mediated by the direct recruitment of HDAC activity to cyclin D1 promoter: (i) INI1-mediated repression of both endogenous and transiently transfected cyclin D1 promoter is reversed by low concentrations of TSA, (ii) INI1/hSNF5 coimmunoprecipitates with HDAC1 and a mutant that is unable to immunoprecipitate HDAC1 is not able to repress cyclin D1 transcription, and (iii) ChIP assays indicate that INI1/hSNF5 is directly recruited to cyclin D1 promoter and is correlated with the deacetylation of histones at the cyclin D1 promoter region and recruitment of HDAC1. These results together indicate that cyclin D1 is a direct target for repression by INI1/hSNF5.
Previous studies have shown that BRG1 also causes cell cycle arrest of the BRG1-null cell line, SW13, at G1/S phase in an ATP-dependent manner (8, 51). Further studies showed that this effect is mediated by binding of Rb to BRG1 and that the repression of cyclin A (51) and cyclin E (40, 51) is the key mechanism for this cell cycle arrest. It is reasonable to assume that INI1/hSNF5 uses the same pathway as that of BRG1 controlling cell proliferation. However, we have found some important differences between the properties of two genes in our studies. For example, while BRG1 arrests cells at the G1 and S stage, INI1/hSNF5 arrests cells at an earlier stage, i.e., G0-G1 in MON cells. In addition, our preliminary observations suggest that the ATPase mutant that functions as a dominant negative inhibitor that affected the Rb-BRG1-meidated flat cell formation did not affect the flat cell formation of MON cells by INI1/hSNF5 (data not shown), and Rb family function appears to be not required for INI1 mediated repression (Fig. 7A). Further studies are necessary to understand the mechanistic basis of these differences.
Our studies indicate that cyclin D1 is an important target for INI1/hSNF5-mediated cell cycle arrest in AT/RT cells. All the deletion fragments of INI1/hSNF5 that were able to repress cyclin D1 also caused flat cell formation and vice versa. Furthermore, we have demonstrated that coexpression of cyclin D1 from a heterologous promoter is sufficient to eliminate the INI1-mediated arrest. We have demonstrated that this effect is not due to the inhibition of INI1/hSNF5 expression (Fig. 6D). Furthermore, we have found that cyclin D1 levels are elevated in tumor samples as well. These data support the hypothesis that inhibition of cyclin D1 plays a pivotal role for INI1-mediated growth arrest at G0-G1 stage.
While several studies have established the role of INI1/hSNF5 and other components of the SWI/SNF complex in the transcriptional activation of cellular genes, information on the repression mediated by this complex is only beginning to unravel. Based on previous reports, it appears that the cyclin D1 promoter is repressed by c-MYC in a manner independent of its partner MAX (20, 21, 30). We have previously demonstrated that INI1/hSNF5 binds to c-MYC and is necessary for its transactivation function. One possibility is that c-MYC associates with INI1/hSNF5 to mediate repression of cyclin D1 as well. It is possible that since c-MYC can both activate and repress genes, INI1/hSNF5 could facilitate both of these functions of c-MYC. INI1/hSNF5 could activate negative regulators of cell cycle progression and repress positive regulators of cell cycle control, both of which are compatible with its tumor suppressor function. Future studies are necessary to determine the role played by the sequence-specific binding factors to mediate the repression of cyclin D1 by INI1/hSNF5.
Cyclin D1 is a critical protein controlling the G1 stage of cell cycle (38). In addition, cyclin D1 is the only known human cyclin gene that has been convincingly implicated as an oncogene resulting in tumorigenesis. Cyclin D1 overexpression has been found in various tumors, including human breast cancer tumors, and repressing its expression makes the tumor cells exit from G1 phase of cell cycle, thereby reversing the tumor characters (49). In this report, we provide a mechanism by which INI1/hSNF5 can act as a tumor suppressor. This may help in developing therapeutic strategies to treat certain types of cancer. For example, repression of cyclin D1 could be a useful therapeutic strategy to treat AT/RT. Furthermore, since expression of INI1/hSNF5 can arrest AT/RT-derived cell lines, gene therapy strategies of reintroducing INI1/hSNF5 may be utilized to arrest the cancerous cells carrying mutations in this gene.
Acknowledgments
We thank V. Prasad, S. Emmons, and B. Birshtein at Albert Einstein College of Medicine for critically reading the manuscript; A. Morozov and E. Craig for providing plasmid constructs; and P. Davies and S. Buhl of Albert Einstein College for providing polyclonal and monoclonal α-6H antibodies, respectively.
This work was supported by a grant from the NIH (AI/GM 39951) and a grant from Children's Brain Tumor Foundation (CBTF) to G.V.K. K.P.D. was supported by an institutional training grant (CA09060).
REFERENCES
- 1.Adler, H. T., R. Chinery, D. Y. Wu, S. J. Kussick, J. M. Payne, A. J. Fornace, Jr., and D. C. Tkachuk. 1999. Leukemic HRX fusion proteins inhibit GADD34-induced apoptosis and associate with the GADD34 and hSNF5/INI1 protein. Mol. Cell. Biol. 19:7050-7060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albanese, C., M. D'Amico, A. T. Reutens, M. Fu, G. Watanabe, R. J. Lee, R. N. Kitsis, B. Henglein, M. Avantaggiati, K. Somasundarum, B. Thimmapaya, and R. G. Pestell. 1999. Activation of the cyclinD1 gene by the E1a associated protein p300 through AP-1 inhibits cellular apoptosis. J. Biol. Chem. 274:34186-34195. [DOI] [PubMed] [Google Scholar]
- 3.Biegel, J. A., C. S. Allen, K. Kawasaki, N. Shimizu, M. L. Budarf, and C. J. Bell. 1996. Narrowing the critical region for a rhabdoid tumor locus in 22q11. Genes Chromosomes Cancer 16:94-105. [DOI] [PubMed] [Google Scholar]
- 4.Biegel, J. A., J. Y. Zhou, L. B. Rorke, C. Stenstrom, L. M. Wainwright, and B. Fogelgren. 1999. Germ-line and acquired mutations of INI1 in atypical teratoid and rhabdoid tumors. Cancer Res. 59:74-79. [PubMed] [Google Scholar]
- 5.Carlson, M., and B. C. Laurent. 1994. The SNF/SWI family of global transcriptional activators. Curr. Opin. Cell Biol. 6:396-402. [DOI] [PubMed] [Google Scholar]
- 6.Cheng, S.-W. G., K. P. Davies, E. Yung, R. J. Beltran, J. Yu, and G. V. Kalpana. 1999. c-MYC interacts with INI1/hSNF5 and requires the SWI/SNF complex for transactivation function. Nat. Genet. 22:102-105. [DOI] [PubMed] [Google Scholar]
- 6a.Darzynkiewicz, Z., and G. Juan. 1997. DNA content measurement for DNA ploidy and cell cycle analysis, p. 7.5.1-7.5.24. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology. Wiley, New York, N.Y. [DOI] [PubMed]
- 7.Dimova, D., Z. Nackerdien, S. Furgeson, S. Eguchi, and M. A. Osley. 1999. A role for transcriptional repressors in targeting the yeast Swi/Snf complex. Mol. Cell 4:75-83. [DOI] [PubMed] [Google Scholar]
- 8.Dunaief, J. L., B. E. Strober, S. Guha, P. A. Khavari, K. Alin, J. Luban, M. Begemann, G. R. Crabtree, and S. P. Goff. 1994. The retinoblastoma protein and BRG1 form a complex and cooperate to induce cell cycle arrest. Cell 79:119-130. [DOI] [PubMed] [Google Scholar]
- 9.Gossen, M., and H. Bujard. 1992. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl. Acad. Sci. USA 89:5547-5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guidi, C. J., A. T. Sands, B. P. Zambrowicz, T. K. Turner, D. A. Demers, W. Webster, T. W. Smith, A. N. Imbalzano, and S. N. Jones. 2001. Disruption of Ini1 leads to peri-implantation lethality and tumorigenesis in mice. Mol. Cell. Biol. 21:3598-3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hanahan, D., and R. A. Weinberg. 2000. The hallmarks of cancer. Cell 100:57-70. [DOI] [PubMed] [Google Scholar]
- 12.Hinds, P. W., S. F. Dowdy, E. N. Eaton, A. Arnold, and R. A. Weinberg. 1994. Function of a human cyclin gene as an oncogene. Proc. Natl. Acad. Sci. USA 91:709-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holstege, F. C. P., E. G. Jennings, J. J. Wyrick, T. I. Lee, C. J. Hengartner, M. R. Green, T. R. Golub, E. S. Lander, and R. A. Young. 1998. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95:717-728. [DOI] [PubMed] [Google Scholar]
- 14.Kalpana, G. V., S. Marmon, W. Wang, G. R. Crabtree, and S. P. Goff. 1994. Binding and stimulation of HIV-1 integrase by a human homolog of yeast transcription factor SNF5. Science 266:2002-2006. [DOI] [PubMed] [Google Scholar]
- 15.Khavari, P. A., C. L. Peterson, J. W. Tamkun, D. B. Mendel, and G. R. Crabtree. 1993. BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature 366:170-174. [DOI] [PubMed] [Google Scholar]
- 16.Kingston, R. E., C. A. Bunker, and A. N. Imbalzano. 1996. Repression and activation by multiprotein complexes that alter chromatin structure. Genes Dev. 10:905-920. [DOI] [PubMed] [Google Scholar]
- 17.Kingston, R. E., and G. J. Narlikar. 1999. ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev. 13:2339-2352. [DOI] [PubMed] [Google Scholar]
- 18.Klochendler-Yeivin, A., L. Fiette, J. Barra, C. Muchardt, C. Babinet, and M. Yaniv. 2000. The murine SNF5/INI1 chromatin remodeling factor is essential for embryonic development and tumor suppression. EMBO Rep. 1:500-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee, D., H. Sohn, G. V. Kalpana, and J. Choe. 1999. Interaction of E1 and hSNF5 proteins stimulates replication of human papillomavirus DNA. Nature 399:487-491. [DOI] [PubMed] [Google Scholar]
- 20.Lee, L. A., C. Dolde, J. Barrett, C. S. Wu, and C. V. Dang. 1996. A link between c-Myc-mediated transcriptional repression and neoplastic transformation. J. Clin. Investig. 97:1687-1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li, L. H., C. Nerlov, G. Prendergast, D. MacGregor, and E. B. Ziff. 1994. c-Myc represses transcription in vivo by a novel mechanism dependent on the initiator element and Myc box II. EMBO J. 13:4070-4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lukas, J., M. Pagano, Z. Staskova, G. Draetta, and J. Bartek. 1994. Cyclin D1 protein oscillates and is essential for cell cycle progression in human tumor cell lines. Oncogene 9:707-718. [PubMed] [Google Scholar]
- 23.Morozov, A., E. Yung, and G. Kalpana. 1998. Structure-function analysis of integrase interactor 1/hSNF5L1 reveals differential properties of two repeat motifs present in the highly conserved region. Proc. Natl. Acad. Sci. USA 95:1120-1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muchardt, C., and M. Yaniv. 1993. A human homologue of Saccharomyces cerevisiae SNF2/SWI2 and Drosophila brm genes potentiates transcriptional activation by the glucocorticoid receptor. EMBO J. 12:4279-4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murphy, D. J., S. Hardy, and D. A. Engel. 1999. Human SWI-SNF component BRG1 represses transcription of the c-fos gene. Mol. Cell. Biol. 19:2724-2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perez-Martin, J., and A. D. Johnson. 1998. The C-terminal domain of Sin1 interacts with the SWI-SNF complex in yeast. Mol. Cell. Biol. 18:4157-4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peterson, C. L. 1998. SWI/SNF complex: dissection of a chromatin remodeling cycle. Cold Spring Harb. Symp. Quant. Biol. 63:545-552. [DOI] [PubMed] [Google Scholar]
- 28.Peterson, C. L., and J. L. Workman. 2000. Promoter targeting and chromatin remodeling by the SWI/SNF complex. Curr. Opi. Genet. Dev. 10:187-192. [DOI] [PubMed] [Google Scholar]
- 29.Phelan, M. L., S. Sif, G. J. Narlikar, and R. E. Kingston. 1999. Reconstitution of a core chromatin remodeling complex from SWI/SNF subunits. Mol. Cell 3:247-253. [DOI] [PubMed] [Google Scholar]
- 30.Philipp, A., A. Schneider, I. Vasrik, K. Finke, Y. Xiong, D. Beach, K. Alitalo, and M. Eilers. 1994. Repression of cyclin D1: a novel function of MYC. Mol. Cell. Biol. 14:4032-4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roberts, C. W., S. A. Galusha, M. E. McMenamin, C. D. Fletcher, and S. H. Orkin. 2000. Haploinsufficiency of Snf5 (integrase interactor 1) predisposes to malignant rhabdoid tumors in mice. Proc. Natl. Acad. Sci. USA 97:13796-13800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rorke, L. B., R. J. Packer, and J. A. Biegel. 1996. Central nervous system atypical teratoid/rhabdoid tumors of infancy and childhood: definition of an entity. J. Neurosurg. 85:56-65. [DOI] [PubMed] [Google Scholar]
- 33.Rozenblatt-Rosen, O., T. Rozovskaia, D. Burakov, Y. Sedkov, S. Tillib, J. Blechman, T. Nakamura, C. M. Croce, A. Mazo, and E. Canaani. 1998. The C-terminal SET domains of ALL-1 and TRITHORAX interact with the INI1 and SNR1 proteins, components of the SWI/SNF complex. Proc. Natl. Acad. Sci. USA 95:4152-4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sang, N., J. Caro, and A. Giordano. 2002. Adenoviral E1A: everlasting tool, versatile applications, continuous contributions and new hypotheses. Front. Biosci. 7:D407-D413. [DOI] [PubMed] [Google Scholar]
- 35.Sevenet, N., A. Lellouch-Tubiana, D. Schofield, K. Hoang-Xuan, M. Gessler, D. Birnbaum, C. Jeanpierre, A. Jouvet, and O. Delattre. 1999. Spectrum of hSNF5/INI1 somatic mutations in human cancer and genotype-phenotype correlations. Hum. Mol. Genet. 8:2359-2368. [DOI] [PubMed] [Google Scholar]
- 36.Sevenet, N., E. Sheridan, D. Amram, P. Schneider, R. Handgretinger, and O. Delattre. 1999. Constitutional Mutations of the hSNF5/INI1 Gene Predispose to a Variety of Cancers. Am. J. Hum. Genet. 65:1342-1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shanahan, F., W. Seghezzi, D. Parry, D. Mahony, and E. Lees. 1999. Cyclin E associates with BAF155 and BRG1, components of the mammalian SWI-SNF complex, and alters the ability of BRG1 to induce growth arrest. Mol. Cell. Biol. 19:1460-1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sherr, C. J. 2000. The Pezcoller lecture: cancer cell cycles revisited. Cancer Res. 60:3689-3695. [PubMed] [Google Scholar]
- 39.Sif, S., A. J. Saurin, A. N. Imbalzano, and R. E. Kingston. 2001. Purification and characterization of mSin3A-containing Brg1 and hBrm chromatin remodeling complexes. Genes Dev. 15:603-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Strobeck, M. W., K. E. Knudsen, A. F. Fribourg, M. F. DeCristofaro, B. E. Weissman, A. N. Imbalzano, and E. S. Knudsen. 2000. BRG-1 is required for RB-mediated cell cycle arrest. Proc. Natl. Acad. Sci. USA 97:7748-7753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sudarsanam, P., V. R. Iyer, P. O. Brown, and F. Winston. 2000. Whole-genome expression analysis of snf/swi mutants of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 97:3364-3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tyler, J. K., and J. T. Kadonaga. 1999. The “dark side” of chromatin remodeling: repressive effects on transcription. Cell 99:443-446. [DOI] [PubMed] [Google Scholar]
- 43.Versteege, I., N. Sevenet, J. Lange, M. F. Rousseau-Merck, P. Ambros, R. Handgretinger, A. Aurias, and O. Delattre. 1998. Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature 394:203-206. [DOI] [PubMed] [Google Scholar]
- 44.Wang, W., J. Cote, Y. Xue, S. Zhou, P. A. Khavari, S. R. Biggar, C. Muchardt, G. V. Kalpana, S. P. Goff, M. Yaniv, J. L. Workman, and G. R. Crabtree. 1996. Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. EMBO J. 15:5370-5382. [PMC free article] [PubMed] [Google Scholar]
- 45.Wang, W., Y. Xue, S. Zhou, A. Kuo, B. R. Cairns, and G. R. Crabtree. 1996. Diversity and specialization of mammalian SWI/SNF complexes. Genes Dev. 10:2117-2130. [DOI] [PubMed] [Google Scholar]
- 46.Watanabe, G., C. Albanese, R. J. Lee, A. Reutens, G. Vairo, B. Henglein, and R. G. Pestell. 1998. Inhibition of cyclin D-kinase activity is associated with E2F-mediated inhibition of cyclin D1 promoter activity through E2F and Sp1. Mol. Cell. Biol. 18:3212-3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weintraub, S. J., C. A. Prater, and D. C. Dean. 1992. Retinoblastoma protein switches the E2F site from positive to negative element. Nature 358:259-261. [DOI] [PubMed] [Google Scholar]
- 48.Wu, D. Y., G. V. Kalpana, S. P. Goff, and W. H. Schubach. 1996. Epstein-Barr virus nuclear protein 2 (EBNA2) binds to a component of the human SNF-SWI complex, hSNF5/Ini1. J. Virol. 70:6020-6028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu, Q., Y. Geng, and P. Sicinski. 2001. Specific protection against breast cancers by cyclin D1 ablation. Nature 411:1017-1021. [DOI] [PubMed] [Google Scholar]
- 50.Yung, E., M. Sorin, A. Pal, E. Craig, A. Morozov, O. Delattre, J. Kappes, D. Ott, and G. V. Kalpana. 2001. Inhibition of HIV-1 virion production by a transdominant mutant of Integrase interactor 1. Nat. Med. 7:920-926. [DOI] [PubMed] [Google Scholar]
- 51.Zhang, H. S., M. Gavin, A. Dahiya, A. A. Postigo, D. Ma, R. X. Luo, J. W. Harbour, and D. C. Dean. 2000. Exit from G1 and S phase of the cell cycle is regulated by repressor complexes containing HDAC-Rb-hSWI/SNF and Rb-hSWI/SNF. Cell 10:79-89. [DOI] [PubMed] [Google Scholar]