Abstract
Vertebrate tRNA export receptor exportin-t (Xpo-t) binds to RanGTP and mature tRNAs cooperatively to form a nuclear export complex. Xpo-t shuttles bidirectionally through nuclear pore complexes (NPCs) but is mainly nuclear at steady state. The steady-state distribution of Xpo-t is shown to depend on its interaction with RanGTP. Two distinct Xpo-t NPC interaction domains that bind differentially to peripherally localized nucleoporins in vitro are identified. The N terminus binds to both Nup153 and RanBP2/Nup358 in a RanGTP-dependent manner, while the C terminus binds to CAN/Nup214 independently of Ran. We propose that these interactions increase the concentration of tRNA export complexes and of empty Xpo-t in the vicinity of NPCs and thus increase the efficiency of the Xpo-t transport cycle.
Nucleocytoplasmic transport occurs through the large proteinacious nuclear pore complexes (NPCs) that span the nuclear envelope. Translocation of cargo through the NPC is highly efficient, signal dependent, and receptor mediated. Subdomains of the NPC include the nuclear basket and the cytoplasmic fibrils. This structural asymmetry implies that at least some NPC proteins, nucleoporins, are differentially localized to either the nuclear or cytoplasmic side of the pore. While the majority of nucleoporins in yeast Saccharomyces cerevisiae are found on both sides of the NPC, a few are indeed localized to either the nuclear or cytoplasmic face (40). Asymmetric nucleoporins have also been localized by immunoelectron microscopy of Xenopus laevis oocyte NPCs. Nup153 is predominantly on the nuclear side of the NPC (9, 36, 45, 47), and RanBP2/Nup358 and CAN/Nup214 are on the cytoplasmic face (25, 36, 47a). The importance of certain asymmetric nucleoporins both for the structural integrity of NPCs and for transport of specific cargoes has been demonstrated in both yeast (see reference 12 for a review) and vertebrate cells (14, 15, 37, 47).
Transport receptors have the ability to bind to the NPC and mediate translocation through the central channel of the pore. One large family of transport receptors, or karyopherins, named after founding member importin β, mediate both import (importins) and export (exportins) of RNAs and proteins (19, 31). All importin β-like receptors interact specifically and directly with the GTP-bound form of the small GTPase Ran, and this interaction plays a key role in mediating the assembly or disassembly of transport complexes and thus imparting directionality to transport events. RanGTP concentrations in the nucleus are predicted to be high due to the presence of the nucleotide exchange factor for Ran, RCC1, on chromatin and the exclusion of the proteins, RanBP1, RanBP2, and RanGAP, that stimulate hydrolysis of GTP by Ran from the nucleus (19, 31, 44).
In contrast to these generally accepted aspects of transport receptor function, there is less understanding of NPC translocation or of the role of specific nucleoporin-receptor interactions during translocation (see reference 38 for a recent review). We set out to investigate the potential role of RanGTP and cargo binding and receptor-nucleoporin interactions on the nucleocytoplasmic shuttling properties of a relatively simple transport pathway, that of tRNA export mediated by importin β-like receptor exportin-t (Xpo-t).
Xpo-t is a nuclear protein at steady state that binds to mature tRNAs cooperatively with RanGTP to form a trimeric export complex (2, 3, 20, 27, 28). We show that the steady-state nuclear localization of Xpo-t is dependent on its binding to RanGTP. We also demonstrate that Xpo-t contains at least two distinct NPC binding domains, each of which is sufficient to support shuttling between the nucleus and cytoplasm. These domains bind differentially to peripheral nucleoporins, some in a RanGTP-regulated manner, and we suggest that these interactions may function to concentrate the receptor near the NPC at different stages of its transport cycle.
MATERIALS AND METHODS
Molecular cloning and expression of Xpo-t fragments.
Full-length Xpo-t was cloned previously (2). Sequences encoding Xpo-t deletion mutants were cloned into vectors for both Escherichia coli expression and coupled transcription and translation in rabbit reticulocyte lysate (Promega). For E. coli expression all PCR fragments encoding Xpo-t deletion mutants were cloned into pQE60 (encoding a C-terminal six-His tag) (Qiagen) or pQE60zz (encoding an N-terminal protein A tag and a C-terminal six-His tag) by using NcoI and BamHI restriction sites. For in vitro translation, sequences encoding Xpo-t with deletions of amino acids 1 to 663, 1 to 434, 1 to 385, and 1 to 338 were cloned into pRSETA (Invitrogen) via NheI and HindIII restriction sites. Sequences encoding Xpo-t with deletions of amino acids 46 to 962, 347 to 962, and 443 to 962 were cloned into the pT77 (42) vector by using NdeI and BamHI restriction sites. E. coli expression was performed by using strain TG1 as previously described (2), and His-tagged products were purified by binding to Talon (Clontech) resin. Site-directed mutagenesis of Xpo-t to generate the F54A/F55A mutant was performed with the Quick-Change site-directed mutagenesis kit (Invitrogen).
Xenopus oocyte microinjections.
In vitro transcription of 32P-labeled RNAs and Xenopus oocyte microinjections were performed as described previously (23). Microinjection of in vitro-translated proteins were performed as described previously (35) except that unincorporated [35S]methionine was removed by G-50 gel filtration.
In vitro binding.
Binding of Xpo-t and deletion mutants to tRNA and/or RanGTP was performed as described previously (2, 3). Binding experiments using Xenopus egg extracts were performed by first covalently cross-linking recombinant proteins His6-importin β (1-365), Xpo-t-His6, zzXpN-His6, and zzXpC-His6 to Affigel-15 activated agarose (Bio-Rad) overnight at 4°C. The resin was then washed extensively with buffer containing 1 M NaCl and blocked for 4 h at 4°C in the presence of 100 mM Tris, pH 8. Approximately 10 to 20 μl of resin containing ∼300 μg of recombinant protein was mixed with 40 μl of Xenopus egg extract in the absence or presence of ∼100 μg of RanQ69L preloaded with GTP in a total volume of 300 μl of binding buffer (50 mM HEPES [pH 7.9], 200 mM NaCl, 5 mM MgCl2, 0.02% Triton X-100, protease inhibitors). The binding reaction mixtures were rotated for 3 h at 4°C, and the products were collected by centrifugation and quickly washed four times with 250 μl of ice-cold binding buffer. Bound proteins were eluted with sample buffer containing sodium dodecyl sulfate (SDS), separated by SDS-6% polyacrylamide gel electrophoresis (PAGE), and transferred to a polyvinylidene difluoride membrane (Millipore), probed with monoclonal antibody MAb414 (diluted 1:3,000), and detected by chemiluminescence with horseradish peroxidase-conjugated secondary antibodies. For wheat germ agglutinin (WGA)-Sepharose purification Xenopus egg extract (∼2 ml) was diluted twofold in binding buffer and incubated with ∼1 ml of WGA-Sepharose (Sigma) for 2 h at 4°C. The resin was collected and washed four times with 10 ml of binding buffer. Bound proteins were eluted with ∼0.5 ml of binding buffer supplemented with 500 mM N-acetylglucosamine. Proteins eluted from WGA resin were then mixed with immunoglobulin G (IgG)-Sepharose resin containing ∼100 μg of CRM1, Xpo-t, XpC (amino acids 443 to 962), or XpN (amino acids 1 to 385). The CRM1 reaction mixture was supplemented with 5 μM RanQ69L and ∼20 μg of bovine serum albumin (BSA)-nuclear export signal (NES). The reaction mixtures were incubated for 3 h at 4°C in binding buffer minus Triton X-100, the resin was collected and washed four times, and bound proteins were eluted with sample buffer and separated by SDS-PAGE.
Permeablized-cell assays.
Digitonin-permeabilized HeLa cells were made, and assays were performed as described by Englmeier et al. (11). The concentration of Alexa-labeled Xpo-t, XpN, and XpC was 0.5 μM. The concentration of NTF2, RanGAP, and RanBP1 was 0.4 μM, and the concentration of RanGDP was 4 μM. Proteins were labeled with Alexa-546 (Molecular Probes) according to the manufacturer's instructions at a 1:1 molar ratio of label to protein.
RESULTS
Distinct biochemical and shuttling properties of the N and C termini of Xpo-t.
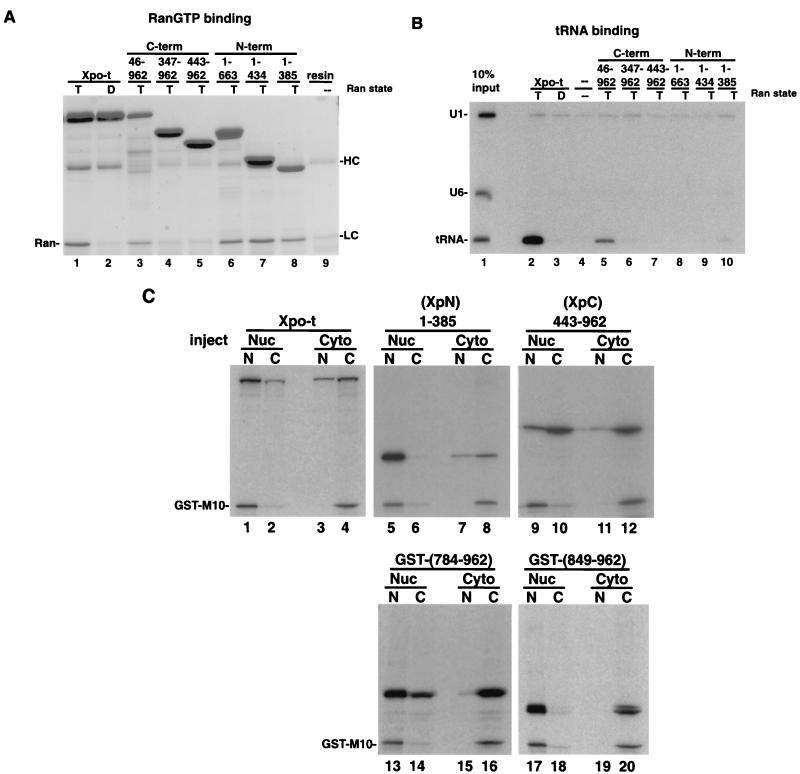
To investigate the biochemical properties and functional domains of Xpo-t, we generated a series of proteins with N- and C-terminal deletions and tested their ability to form an export complex containing RanGTP and tRNA in vitro (Fig. 1). Protein A-tagged Xpo-t and the various deletion mutants were expressed in E. coli and purified by binding to IgG-Sepharose. The different Xpo-t proteins were then tested for RanGTP (Fig. 1A) and 32P-labeled tRNA (Fig. 1B) binding. As expected, full-length Xpo-t specifically bound to RanGTP but not RanGDP (Fig. 1A, lanes 1 and 2) and could specifically select tRNA from a mixture of RNAs only in the presence of RanGTP (Fig. 1B, lanes 2 and 3). Removal of C-terminal sequences eliminated the tRNA interaction (Fig. 1B, lanes 8 to 10) but not the ability to bind RanGTP (Fig. 1A, lanes 6 to 8). Removal of N-terminal sequences affected both binding activities (compare Fig. 1A, lanes 4 and 5, with Fig. 1B, lanes 6 and 7). However, removal of the first 45 amino acids of Xpo-t did not eliminate RanGTP (Fig. 1A, lane 3) or tRNA binding (Fig. 1B, lane 5). The tRNA interaction with Xpo-t is thus mediated by the C terminus of the protein (Fig. 2), and RanGTP binds within the N-terminal 385 amino acids. Xpo-t therefore has a domain organization similar to those of other members of the importin β family.
FIG. 1.
In vitro binding and shuttling activities of full-length Xpo-t and Xpo-t with N- and C-terminal truncations. (A) RanGTP binding. zzXpo-t and the truncation mutants indicated were expressed in E. coli and prebound to IgG-Sepharose, mixed with recombinant RanGTP (T, lanes 1 and 3 to 8) or RanGDP (D, lane 2), and incubated at room temperature while being rotated for 1 h. The resin was collected and washed three times with ice-cold binding buffer, and bound proteins were eluted with sample buffer. Samples were separated by SDS-12% PAGE and stained with Coomassie blue. RanGTP can bind to Xpo-t (lane 1) and all three N-terminal fragments of Xpo-t (lanes 6 to 8). In addition, removal of the first 45 amino acids of Xpo-t (lane 3) does not eliminate Ran-binding activity; however, mutants with further deletions (lanes 4 and 5) no longer bind. HC and LC, background elution of the heavy and light chains from the IgG-Sepharose. (B) tRNA binding. A mixture of U1ΔSm, U6Δss, and yeast tRNAPhe (lane 1) was incubated with zzXpo-t or the indicated truncation mutants in the presence of RanGTP (lanes 2 and 5 to 10), RanGDP (lane 3), or no Ran (lane 4) for 1 h at room temperature. The resin was collected and washed as described above. Bound RNA was extracted and separated on a 10% denaturing RNA gel and detected by autoradiography. (C) N- and C-terminal fragments of Xpo-t have distinct shuttling activities. 35S-labeled Xpo-t and representative truncation mutants were transcribed and translated in rabbit reticulocyte lysate and injected into the nuclei (Nuc) or cytoplasm (Cyto) of Xenopus oocytes. After a 4-h incubation the oocytes were dissected into nuclear (N) and cytoplasmic (C) fractions and separated by SDS-12% PAGE and detected by autoradiography. The GST-M10 protein does not shuttle and serves as an injection control.
FIG. 2.
tRNA-binding Xpo-t mutants do not affect shuttling activity. (A) Recombinant zzXpo-t mutants were tested for binding to tRNA in vitro as for Fig. 1. The mutants are M1 (K539A/R543A), M2 (R550A/K553A/K557A), M3 (L547A/F551A), M4 (F548A/V552A), and M5 (R405A/K406A/K409A). 35S-labeled mutant proteins M2 and M5 were further tested for shuttling activity in Xenopus oocytes, injected into either the nucleus (B) or the cytoplasm (C). Neither of the mutants obviously affected the shuttling behavior of the receptor. All five mutants were tested and found to be able to interact with RanGTP in vitro (data not shown). N, C, T, and D are as defined for Fig. 1.
Xpo-t shuttles in and out of the nucleus but is predominantly nuclear at steady state (2, 27). We tested the effects of Xpo-t truncations on these properties by microinjection of the 35S-labeled proteins into Xenopus oocytes. As previously shown (2), when injected into either the nucleus or the cytoplasm, Xpo-t can move into the other compartment (Fig. 1C, lanes 1 to 4) and therefore shuttles. However, when either the N or C terminus was removed, shuttling behavior was altered (Fig. 1C and Table 1). In contrast to full-length Xpo-t, N-terminal Xpo-t fragments, such as the fragment comprising amino acids 1 to 385 shown, stay in, or migrate into, the nucleus depending on where they are initially injected (Fig. 1C, lanes 5 to 8, and data not shown). C-terminal fragments, such as amino acids 443 to 962 (see also Table 1), behave in the opposite way: they are able to move to the cytoplasm (Fig. 1C, lanes 9 and 10) but do not efficiently move into the nucleus (Fig. 1C, lanes 11 and 12). Furthermore, glutathione S-transferase (GST) fusions could be used to further map the export activity to the C-terminal 178 amino acids (Fig. 1C, lanes 13 and 14). Smaller fragments did not shuttle (Fig. 1C, lanes 17 to 20), further demonstrating the specificity of this activity, as well as indicating that the small C-terminal fragments do not leave the nucleus by simple diffusion through the NPC. Therefore, removing the N or C terminus of Xpo-t had opposite effects on the steady-state distribution of the remaining protein in Xenopus oocytes. These properties were examined further in the experiments discussed below using Xpo-t amino acids 1 to 385 (XpN) and 443 to 962 (XpC).
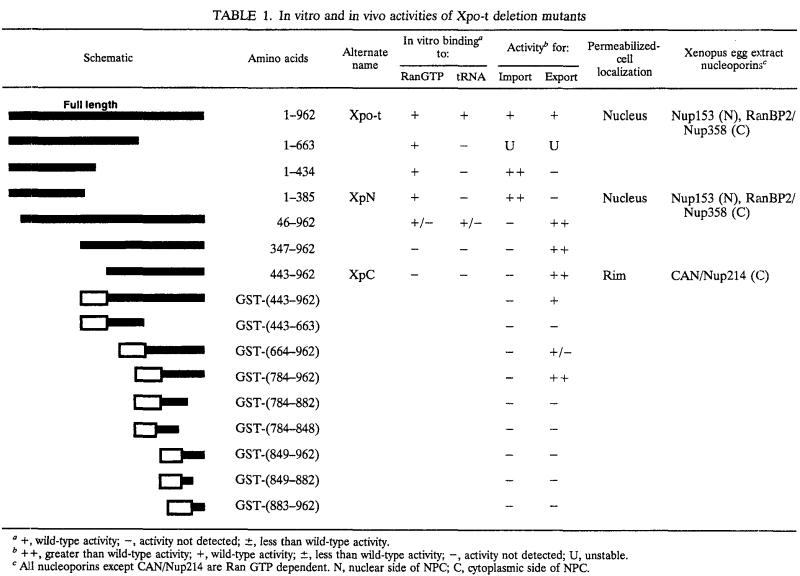
TABLE 1.
In vitro and in vivo activities of Xpo-t deletion mutants
+, wild-type activity; −, activity not detected; ±, less than wild-type activity.
++, greater than wild-type activity; +, wild-type activity; ±, less than wild-type activity; −, activity not detected; U, unstable.
All nucleoporins except CAN/Nup214 are Ran GTP dependent. N, nuclear side of NPC; C, cytoplasmic side of NPC.
Xpo-t is predominantly nuclear because it binds to RanGTP.
One way to explain the above results is that Xpo-t contains two distinct translocation domains that impart directionality of movement to the receptor; one for import (XpN) and one for export (XpC). Alternatively, the observed steady-state localization of XpN and XpC might be due to specific interactions with nuclear or cytoplasmic components. The effect of mutations perturbing tRNA or RanGTP binding on the shuttling properties of Xpo-t were therefore tested. Since Xpo-t does not contain a sequence with similarity to known RNA binding motifs, we made the assumption that basic amino acid residues might be involved in tRNA binding. To obtain tRNA binding mutants, several highly conserved basic amino acids in the C-terminal half of the protein were mutated to alanines. Two mutants impaired in tRNA binding were identified (Fig. 2A, lanes 5 and 8). When proteins were injected into Xenopus oocytes, no obvious differences in the steady-state distributions of these mutants relative to that of wild-type Xpo-t were observed (Fig. 2B and C). We conclude that tRNA binding does not affect Xpo-t localization.
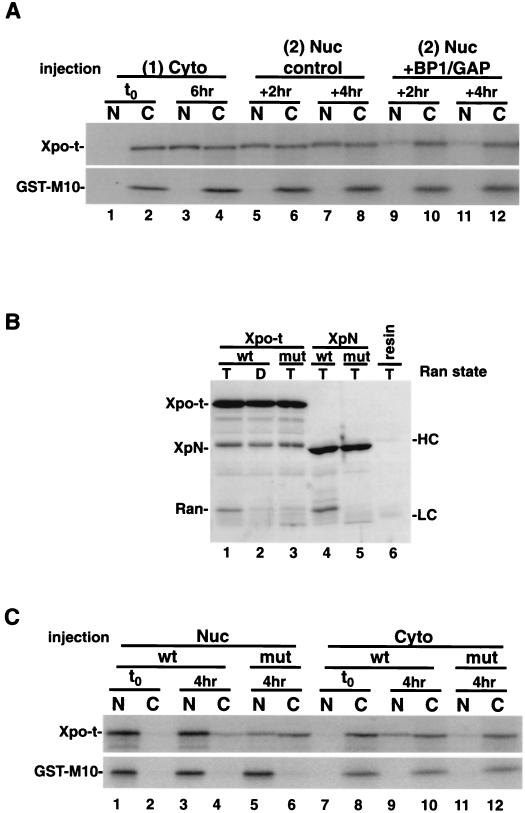
We next tested the effect of RanGTP binding. First, 35S-labeled Xpo-t was injected into the cytoplasm (Fig. 3A, lanes 1 and 2) and allowed to equilibrate for 6 h (Fig. 3A, lanes 3 and 4). Then a second injection of either RanGAP and RanBP1 (Fig. 3A, lanes 9 to 12) or a buffer control (Fig. 3A, lanes 5 to 8) into the nucleus was performed. Nuclear injection of GAP/BP1 depletes nuclear RanGTP pools and collapses the RanGTP gradient across the nuclear envelope (22). After 2 to 4 h of further incubation, oocytes injected with GAP/BP1 showed dramatically reduced levels of Xpo-t in the nucleus (Fig. 3A, lanes 9 to 12).
FIG. 3.
Xpo-t is predominantly a nuclear protein at steady state because it binds to RanGTP. (A) Depletion of nuclear RanGTP alters the steady-state localization of Xpo-t. 35S-labeled Xpo-t was injected into the oocyte cytoplasm (lanes 1 and 2) and incubated for 6 h to allow Xpo-t to equilibrate into the nucleus (lanes 3 and 4). At this time point a second injection into the nucleus was performed with a mixture containing either 1 μM RanGAP-10 μM RanBP1 and injection dye (lanes 9 to 12) or only injection dye (lanes 5 to 8). Oocytes were dissected after 2 or 4 h, and the samples were separated by SDS-12% PAGE and detected by autoradiography. (B) In vitro binding of RanGTP to wild-type or F54A/F55A mutant Xpo-t or XpN. Experimental conditions were as for Fig. 1A. (C) The RanGTP-binding Xpo-t mutant mislocalizes to the cytoplasm at steady state in Xenopus oocytes. 35S-labeled Xpo-t or the F54A/F55A mutant was injected into either the nucleus (Nuc) or cytoplasm (Cyto) of oocytes, the oocytes were incubated for 4 h and dissected, and samples were processed as described above. Cyto, cytoplasm; Nuc, nucleus; t0, time zero; N, nuclear fraction; C, cytoplasmic fraction; wt, wild type; mut, mutant; HC, IgG heavy chain; LC, IgG light chain.
As a second approach, we determined the steady-state distribution of a RanGTP-binding Xpo-t mutant. We generated a number of mutations in conserved residues within the Ran-binding domain and tested the RanGTP binding of the mutants in vitro. One mutant, with phenylalanines 54 and 55 changed to alanines, was defective in binding to RanGTP both in the context of full-length Xpo-t (Xpo-t mut) and within the XpN fragment (XpN mut) (Fig. 3B, compare lanes 1 with 3 and 4 with 5). The full-length mutant was characterized by oocyte injection. Four hours after nuclear injection more mutant protein than wild-type protein was found in the cytoplasm (Fig. 3C, compare lanes 3 and 4 with lanes 5 and 6). Additionally, when injected into the cytoplasm, very little of the mutant protein accumulated in the nucleus (Fig. 3C, lane 11). The XpN mut protein was unstable in oocytes and could not be analyzed in this way. Taken together, these experiments suggest that nuclear RanGTP binding is required for the steady-state nuclear localization of Xpo-t.
Ran influences the levels of nuclear Xpo-t in semipermeabilized HeLa cells.
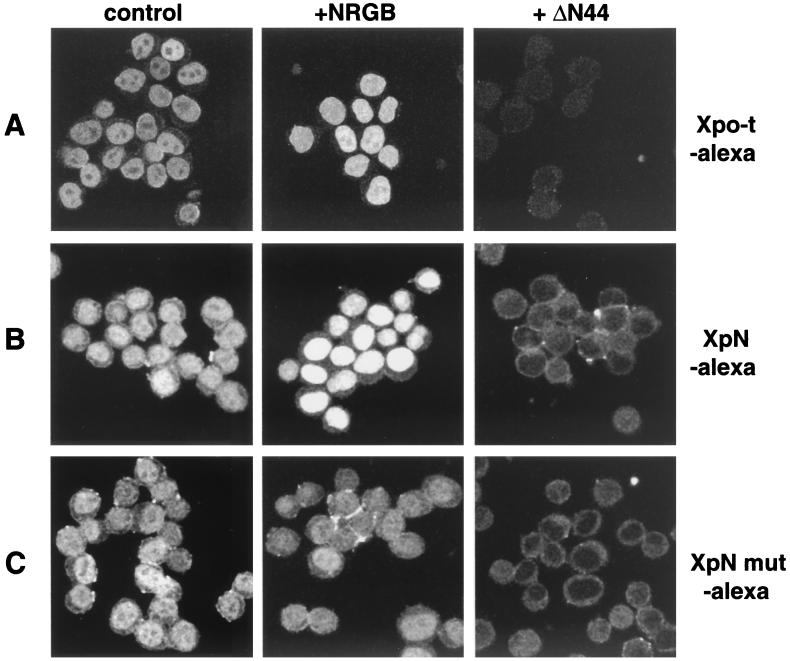
To better understand and further confirm the influence of nuclear RanGTP on Xpo-t localization, we turned to an assay of digitonin-permeabilized HeLa cells, where it is possible to more precisely control the levels of nuclear RanGTP. The ability of Xpo-t to enter the nuclei of these cells in the absence or presence of an added “Ran system” (NTF2, RanGDP, RanBP1, and RanGAP) (11) was tested. The reactions described here and below all reflect the steady-state distribution of Xpo-t and its derivatives (data not shown). Fluorescently labeled Xpo-t enters the nucleoplasm of permeabilized cells (Fig. 4A, control) (27) without an energy regeneration system or Ran (data not shown). Nuclear entry of Xpo-t was blocked, as expected, by addition of a dominant-negative NPC binding fragment of importin β, ΔN44 (amino acids 45 to 465) (26) (Fig. 4, +ΔN44). When the complete Ran system was included in the reaction, the amount of labeled Xpo-t in the nucleus increased (Fig. 4A, +NRGB). The XpN protein behaved similarly to full-length Xpo-t in this assay. It entered the nucleus and was blocked by ΔN44, and nuclear accumulation was increased upon addition of a complete Ran system (Fig. 4B). In contrast, the XpN mut protein, deficient in RanGTP binding, entered the nucleoplasm and was blocked by ΔN44 but did not respond to the addition of a Ran system (Fig. 4C). These results confirm the in vivo effect of nuclear RanGTP on the accumulation of Xpo-t or XpN in the nucleus, namely, that increasing the amount of nuclear RanGTP leads to a corresponding increase in the levels of Xpo-t or XpN inside the nucleus.
FIG. 4.
Localization of Xpo-t and XpN in semipermeabilized HeLa cells. The Ran system promotes nuclear accumulation. (A) Xpo-t labeled with Alexa-546 (0.5 μM) and an energy regeneration system was combined with buffer (control) or the indicated reagents and then incubated with digitonin-permeabilized HeLa cells for 10 min at room temperature, fixed with paraformaldehyde, and spun onto coverslips, and images were collected by confocal microscropy. The Ran system (NRGB) contains 0.4 μM NTF2 (N), 4 μM RanGDP (R), 0.4 μM RanGAP (G), and 0.4 μM RanBP1 (B). The ΔN44 protein is composed of amino acids 45 to 465 of importin β and was added at 1 μM. (B and C) Localization of XpN-Alexa-546 (B) and XpN mut-Alexa-546 (F54A/F55A) (C) as in panel A.
XpC localizes to the NPC and can compete for export pathways.
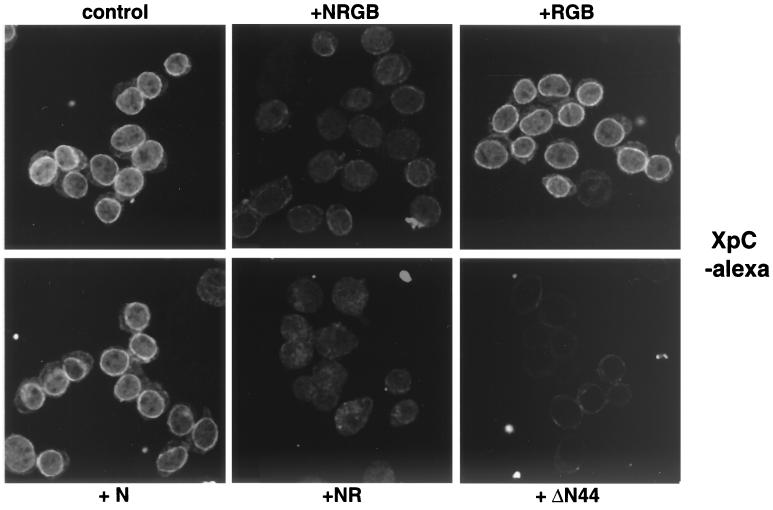
In permeabilized cells in the absence of an energy regeneration system or Ran, fluorescently labeled XpC accumulated at the nuclear rim and ΔN44 blocked this binding (Fig. 5, control and +ΔN44 and data not shown). Binding could also be competed by addition of either unlabeled XpC or unlabeled full-length Xpo-t, albeit at a higher concentration than for XpC (data not shown). Addition of the Ran system reduced the rim signal of XpC without leading to nuclear accumulation (Fig. 5, +NRGB). Dissection of the Ran system showed that a combination of NTF2 and Ran was sufficient to compete the rim signal (Fig. 5, +NR). NTF2, added alone (Fig. 5, +N), or a combination of Ran, RanGAP, and RanBP1 (Fig. 5, +RGB) did not compete. These results suggest that XpC can bind to the NPC and that this binding is competed by the NTF2/RanGDP import complex.
FIG. 5.
Localization of the XpC fragment in semipermeabilized HeLa cells. Nuclear import of RanGDP competes for XpC binding to the NPC. Recombinant XpC labeled with Alexa dye was mixed with the indicated reagents and an energy regeneration system and incubated with semipermeabilized HeLa cells as described for Fig. 4. XpC localizes to the nuclear rim and likely binds to the NPC since ΔN44 can directly compete for the interaction. The complete Ran system or only a mixture of NTF2 and RanGDP can also compete for the rim binding of XpC, suggesting that Ran nuclear import via NTF2 utilizes a similar binding site (or sites) at the NPC. The designations for the components of the Ran system are as in Fig. 4.
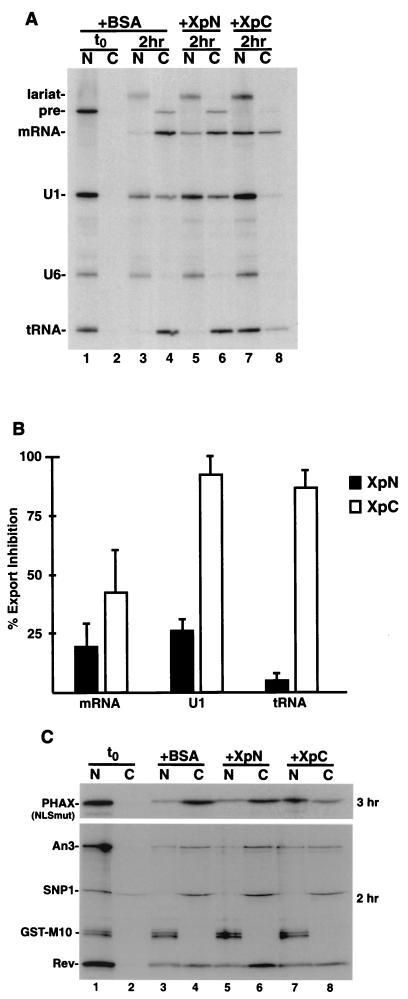
Because XpC has the ability to exit the nucleus (see above) and bind to the NPC, we predicted that it might compete other export pathways. To test this either BSA (Fig. 6A, lanes 1 to 4), XpN (lanes 5 and 6), or XpC (lanes 7 and 8) was preinjected into oocyte nuclei. Next, a pre-mRNA (Ftz-pre), U1ΔSm snRNA, and a tRNA, together with U6Δss as an injection control, were injected. Relative to BSA, XpC strongly inhibited both U snRNA and tRNA export and, to a lesser extent, spliced Ftz mRNA export (Fig. 6A, compare lanes 3 and 4 with 7 and 8). XpN inhibited all three RNA export pathways weakly (lanes 5 and 6). Quantitation of three independent experiments indicated that XpC inhibited U1 and tRNA export by 80 to 90% (Fig. 6B). Injection of fourfold-higher concentrations of recombinant full-length Xpo-t showed only modest inhibition of RNA export (data not shown).
FIG. 6.
XpC can compete for specific export pathways. (A) Competition for RNA export. BSA (lanes 3 and 4), XpN (lanes 5 and 6), and XpC (lanes 7 and 8) were injected at 150 μM into oocyte nuclei, and oocytes were incubated for 30 min, followed by a second nuclear injection containing a mixture of Ftz pre-mRNA, U1ΔSm, U6Δss (as an injection control), and yeast tRNAPhe. The oocytes were incubatedfor 2 h and then dissected, and the RNA was extracted and separated on a 10% denaturing RNA gel. The XpC fragment can efficiently compete for both U1 and tRNA export and to a lesser extent mRNA export. At this concentration XpN is a less efficient competitor for these RNA export pathways. (B) Quantitation of percent export inhibition of XpN versus XpC from three independent sets of injections. (C) Competition for NES-mediated protein export. 35S-labeled PHAX (NLSmut), An3, Snurportin 1, human immunodeficiency virus Rev, and GST-M10 proteins made in rabbit reticulate lysate were coinjected into oocyte nuclei with ∼200 μM BSA (lanes 3 and 4), XpN (lanes 5 and 6), or XpC (lanes 7 and 8), oocytes were incubated for the indicated times and dissected, and samples were separated by SDS-PAGE. XpC can efficiently compete for PHAX export, is less effective for An3 export, and does not interfere with Snurportin 1 or Rev export. As for RNA export, the XpN fragment is not an effective competitor for any of these export routes. t0, time zero.
Since XpC is a fragment of Xpo-t, it is perhaps not surprising that tRNA export can be inhibited by injection of saturating amounts of this protein. However, the fact that U1 snRNA is inhibited to a similar extent is less expected. Export of U snRNAs is mediated by a complex of proteins that include export receptor CRM1/Xpo1, RanGTP, the Cap binding complex, and adapter protein PHAX (35). The fact that XpC can inhibit U snRNA export suggests that CRM1 and Xpo-t might utilize similar NPC binding sites. We therefore tested the effects of XpC on the export of a PHAX derivative with mutations in the two major nuclear localization signals that prevent its reimport (42) as well as other proteins exported via CRM1 such as An3, the human immunodeficiency virus type 1 Rev protein, and Snurportin1. XpN, XpC, and BSA was coinjected into nuclei of oocytes with 35S-labeled NES proteins, and their export was assayed. As for U1 snRNA, PHAX export was strongly inhibited by XpC (Fig. 6C, top, lanes 7 and 8) but was unaffected by XpN (Fig. 6C, lanes 5 and 6). Interestingly, not all NES proteins were equally affected by XpC. An3 was only weakly inhibited, while both Rev and Snurportin1 were essentially unaffected (Fig. 6C, bottom). These data suggest that XpC competes the export of some CRM1-substrate complexes but not all. It is doubtful that the differential effects are due to differences in export kinetics that reflect the affinity of each NES protein for CRM1, since Snurportin1 export is fast and Rev export is relatively slow but the export of neither is affected by XpC, while An3 and PHAX, whose export rates are intermediate, are inhibited. This suggests that different CRM1-cargo complexes require different nucleoporin interactions for their export. We also analyzed whether XpC could affect import mediated by either importin β or transportin in both oocyte and permeabilized-cell assays. We observed no inhibitory effects of XpC on import (data not shown).
Biochemical interactions between Xpo-t and nucleoporins.
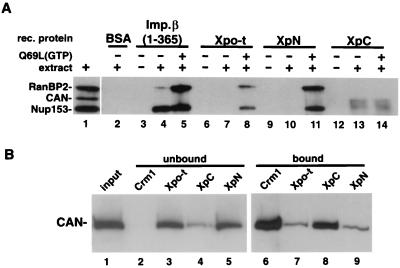
The above experiments suggested that XpN and XpC might interact with different NPC components during translocation. Their biochemical interaction with nucleoporins was therefore examined. Recombinant importin β (amino acids 1 to 365, as a control), Xpo-t, XpN, and XpC were immobilized and mixed with Xenopus egg extract in the absence or presence of RanGTP (Fig. 7A). A hydrolysis-deficient Ran mutant, the Q69L mutant, was used in this experiment to ensure that Ran remained bound to GTP. After incubation with extract, bound protein was eluted, separated by SDS-PAGE, and analyzed by Western blotting using monoclonal antibody MAb414 (Fig. 7A), anti-Xenopus Nup153, or anti-Xenopus CAN/Nup214 (data not shown). In this experiment the fragment of importin β comprising amino acids 1 to 365 showed clear interactions with both Nup153 and RanBP2, and the latter was RanQ69L dependent. Interestingly, the full-length importin β interaction with Nup153 was reported to be RanGTP sensitive (34, 43), indicating that this fragment behaves differently. Similarly, Xpo-t (lanes 6 to 8) and XpN (lanes 9 to 11) bind RanBP2 and Nup153, and both interactions required RanGTP. In the absence of added Ran, neither Xpo-t nor XpN bound detectably to any of the nucleoporins tested. In contrast to full-length Xpo-t, XpC bound CAN/Nup214 (Fig. 7A, lanes 12 to 14), and this interaction was Ran independent. CAN/Nup214 is subject to proteolytic degradation, giving rise to two major species that were also detected by CAN-specific antibodies (data not shown). Nucleoporin p62 bound weakly to all four recombinant proteins regardless of RanQ69L addition (data not shown). No significant binding of the Xpo-t proteins to Nup98 or Nup107 was detected (data not shown).
FIG. 7.
Nucleoporin binding to Xpo-t, XpN, and XpC. (A) The indicated recombinant (rec.) proteins were cross-linked to resin at high density (see Materials and Methods) and used for affinity binding. Affinity resin containing the different proteins was mixed with buffer (lanes 3, 6, 9, and 12), buffer containing Xenopus egg extract (lanes 2, 4, 7, 10, and 13), or extract supplemented with 5 μM RanQ69L loaded with GTP (lanes 5, 8, 11, and 14) and rotated at 4°C for 3 h. The resin was then collected and washed, and the bound proteins were eluted with SDS sample buffer and separated by SDS-6% PAGE and detected by Western blotting using MAb414. In the presence of RanGTP, Xpo-t and XpN can pull down both Nup153 and RanBP2/Nup358 (lanes 8 and 11). By contrast, XpC affinity resin can select CAN/Nup214, and this binding is RanGTP independent (lanes 13 and 14). (B) WGA-Sepharose affinity-purified proteins from Xenopus egg extracts (lane 1) were mixed with resin containing the indicated recombinant proteins and 25% of the unbound fractions (lanes 2 to 5) and bound fractions (lanes 6 to 9) were separated and detected as in panel A by using MAb414. For CRM1 both RanQ69L(GTP) and BSA-NES were included in the binding reaction to promote export complex formation.
Perhaps due to the above-mentioned degradation of CAN/Nup214 or to competition with other binding partners present in complete extracts (Fig. 5), the XpC/CAN interaction was difficult to reproducibly detect in this assay. We therefore verified this interaction using WGA-Sepharose affinity-enriched nucleoporins (14). WGA-binding proteins were mixed with either protein A-tagged CRM1 (together with RanGTP and BSA-NES as a positive control), Xpo-t, XpC, or XpN proteins. Unbound proteins (Fig. 7B, lanes 2 to 5) and bound proteins (Fig. 7B, lanes 6 to 9) were separated by SDS-PAGE and analyzed by Western blotting with MAb414 (Fig. 7B) or anti-CAN antibodies (data not shown). When bound to RanGTP and an NES substrate, CRM1 interacted with CAN (4) (Fig. 7B, compare lanes 2 and 6). XpC also efficiently bound to CAN (Fig. 7B, compare lanes 4 and 8), and this interaction was Ran independent. In contrast, both Xpo-t and XpN interacted with CAN only weakly (Fig. 7B, lanes 7 and 9). This result confirms that XpC can bind to CAN/Nup214 in a RanGTP-independent manner. We did not observe strong interaction of full-length Xpo-t with CAN in vitro. However, since XpC blocked the translocation cycle of Xpo-t (Fig. 6) and since Xpo-t could compete the nuclear rim binding of XpC in vitro (see above), it is likely that high-affinity binding sites for XpC on the NPC also bind Xpo-t. The difference in the affinities of full-length Xpo-t and XpC for CAN may be analogous to the increased affinity for the NPC of the ΔN44 fragment of importin β compared to that of the full-length protein (26).
A model for Xpo-t translocation through the NPC.
Integrating the results of the above experiments we propose the following model for some of the interactions of the tRNA export complex with the NPC during tRNA export and recycling of empty Xpo-t back into the nucleus (Fig. 8; see also Discussion). Trimeric tRNA export complexes form in the nucleus, where RanGTP concentrations are high (Fig. 8, step 1). When bound to RanGTP, Xpo-t has a higher affinity for Nup153 (Fig. 8, step 2). Following translocation through the central channel, the export complex binds to RanBP2. Since sumoylated RanGAP binds to RanBP2 (29, 30, 32), the likely functional consequence of this association is hydrolysis of GTP by Ran. Following hydrolysis, Xpo-t affinity for RanBP2 is reduced and it dissociates. Since XpC binds to CAN in a Ran-independent manner, we propose that full-length Xpo-t binds to CAN and is retained near the translocation channel for recycling. Furthermore, NTF2/RanGDP competes for XpC binding to the NPC and could act to reduce the receptor-NPC interaction and thus could contribute to the fraction available for NPC translocation.
FIG. 8.
Model of Xpo-t translocation and recycling through the NPC. Xpo-t binds to RanGTP in the nucleus (step 1), and the export complex then binds to Nup153 (via the N terminus of Xpo-t) at the nuclear side of the NPC (step 2). The complex translocates and binds to RanBP2/Nup358 on the cytoplasmic side of the NPC (step 3), where RanGAP and RanBP1 can stimulate nucleotide hydrolysis on Ran, and Xpo-t is released. The C terminus of Xpo-t can then interact with CAN/Nup214 to position the receptor for efficient import (step 4). The import complex of NTF2/RanGDP may compete Xpo-t from CAN, and both complexes translocate into the nucleoplasm (step 5), where RanGTP can be generated and Xpo-t export complexes can form for another round of translocation.
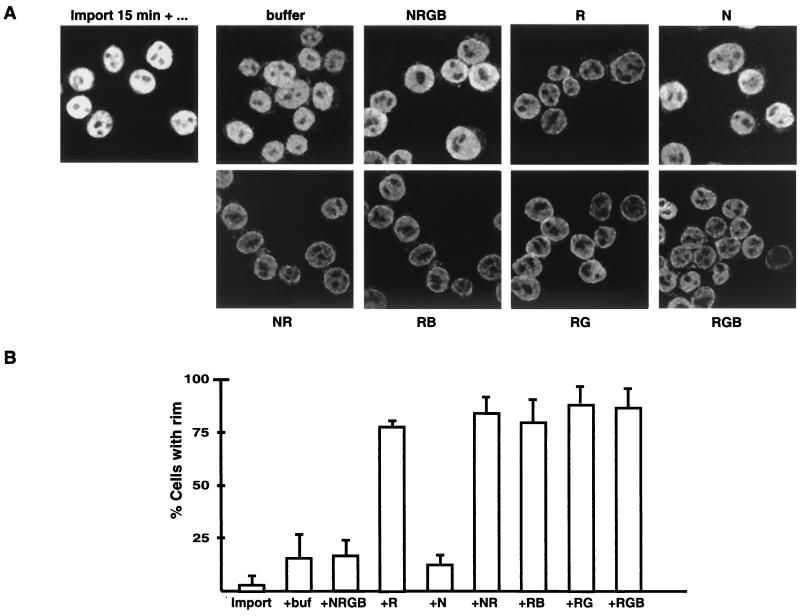
Two predictions of the model can be tested. First, if RanGTP hydrolysis is prevented, Xpo-t should accumulate at the NPC bound either to Nup153 or, more likely, to RanBP2. Second, if hydrolysis is permitted to occur, Xpo-t should also accumulate at the NPC if Ran import via NTF2 is perturbed since this will reduce competition for binding to NPC sites, including CAN. The model was tested by the permeabilized-cell assay. First, fluorescently labeled Xpo-t was imported into the nuclei. The supernatant, containing nonimported Xpo-t, was removed (approximately 80% of the total volume of the reaction mixture), and the cells were resuspended in fresh buffer minus labeled protein. A sample was removed immediately and fixed (Fig. 9, import 15 min). The remaining cells were aliquoted into a second set of reaction mixtures containing an energy regeneration system and either buffer, the complete Ran system, or various individual components of the Ran system and further incubated for 10 min. Note that Ran is able to cross the NPC by simple diffusion in the absence of NTF2, but less efficiently than in its presence.
FIG. 9.
Xpo-t accumulates at the nuclear rim when either Ran hydrolysis or import is perturbed. (A) Two-step permeabilized-cell assay in which fluorescently labeled Xpo-t is first imported into the nuclei for 15 min at room temperature (import 15 min +…) while the cells settled into the bottom of the reaction tube. As much as 80% of the supernatant containing unincorporated Xpo-t was then removed, and the cells were resuspended in fresh buffer and aliquoted into a second reaction mixture containing either buffer or the indicated components of the Ran system. The designations of the individual components are as in Fig. 4. When any individual component of the Ran system was left out of the reaction, the net result was that the Xpo-t nuclear signal was reduced and a nuclear rim signal was more visible, suggesting that both Ran hydrolysis and import are critical for efficient recycling of Xpo-t between the nucleus and cytoplasm. (B) Quantitation of the percentages of cells in each of the above conditions that were seen to have a rim signal. For each condition a total of ∼50 to 75 cells were counted from three independent experiments.
Reaction mixtures containing buffer, the complete Ran system (NRGB), or NTF2 alone (N) showed a relatively diffuse Xpo-t signal within the nucleoplasm and no detectable accumulation at the nuclear rim. In contrast, omitting any component of the Ran system resulted in a reduction of the nuclear signal and some Xpo-t accumulation at the nuclear rim, i.e., preferential NPC binding. The two most relevant conditions were when NTF2 and Ran (NR) were added and when Ran, RanGAP, and RanBP1 (RGB) were present. In the first case Ran is efficiently imported and converted to RanGTP but GTP hydrolysis is inefficient due to the absence of RanGAP and RanBP1. We interpret the increase in NPC binding here as stabilization of interactions like those of Xpo-t with Nup153 and RanBP2. In the second case hydrolysis is efficient but Ran import via NTF2 is not. The net result was also a rim signal in the majority of cells, indicating that Xpo-t accumulated at the NPC. This we interpret as empty Xpo-t, after RanGTP hydrolysis, binding to the NPC through RanGTP-independent sites such as that for CAN/Nup214. The results of this experiment are consistent with some predictions of the model (Fig. 8), namely, that, when a complete Ran system is present, Xpo-t can recycle rapidly between the NPC and nucleoplasm, and the net result is an evenly distributed nucleoplasmic signal and no accumulation at the NPC. However, when either GTP hydrolysis is prevented or the NTF2/RanGDP complex is absent, Xpo-t accumulates at the NPC. The overall reduction in fluorescence signal in these experiments reflects the Xpo-t that is exported completely, escaping binding to the NPC and reimport, and that is diluted into the buffer phase.
DISCUSSION
Steady-state nuclear localization and RanGTP binding.
We have analyzed functional domains of the tRNA export receptor, Xpo-t. Like those of other members of the importin β family, the N-terminal region of Xpo-t binds directly to RanGTP and the C terminus binds to cargo (26, 33, 46). For Xpo-t the N-terminal region and binding to RanGTP are required for the normal steady-state nuclear localization of the receptor, both in vivo and in permeabilized cells in vitro. A nuclear localization of Xpo-t might be critical for its efficient function in export since the cytoplasmic concentration of tRNA is very high. Xpo-t, even if its affinity for tRNA is reduced in the absence of RanGTP (2, 3, 27), might become sequestered in the cytoplasm by binding to tRNA in the absence of a mechanism for its nuclear accumulation. Since the RanGTP-Xpo-t complex on its own is smaller than the complete tRNA export complex, it should not accumulate in the nucleus by being prevented from translocation through the NPC unless it binds to some nucleoplasmic component. Although we have not definitively identified the nucleoplasmic binding partner, it was recently observed that the RanGTP-Xpo-t complex binds to chromatin (6a).
Xpo-t contains two distinct NPC binding domains.
The N- and C-terminal regions of Xpo-t have different interactions with nucleoporins and have dissimilar steady-state distributions both in vivo and in permeabilized cells. While the differential localizations of the Xpo-t fragments are explicable on the basis of their abilities to bind RanGTP, the distinct nucleoporin interactions are likely to be involved at different stages of the Xpo-t transport cycle.
We detected in vitro binding of Xpo-t or its fragments to three peripheral nucleoporins. Xpo-t and the N-terminal fragment both bound to Nup153, recently localized to the nucleoplasmic coaxial ring of the NPC (47), and to RanBP2/Nup358, a component of the cytoplasmic filaments (48-50; Walther et al., submitted). Both of these interactions were dependent on the presence of RanGTP. The C-terminal fragment of Xpo-t bound to CAN/Nup214, located at the cytoplasmic end of the NPC channel (25; Walther et al., submitted), and this interaction was Ran independent.
Similar to Xpo-t, importin β has previously been shown to have at least two NPC interaction domains, one located N terminally (7, 26) and the other located near the C terminus (26). Although the nucleoporins to which these domains bind have not been characterized, an X-ray structure of the N-terminal region of importin β, together with an FG-containing peptide from yeast NSP1, i.e., a generic nucleoporin-transport receptor interaction domain (41), has been obtained (5) and demonstrates the molecular basis for this kind of interaction.
Furthermore, Shah et al. (43) demonstrated that Nup153 binds to importin β in a RanGTP-sensitive way. Since removal of the N-terminal 44 amino acids of importin β, which prevents its interaction with RanGTP, leads to a considerable increase in the affinity of the receptor for the NPC (26) and since depletion of Nup153 leads to a decrease in importin β-mediated import (47), it seems that Nup153 represents an important interaction partner for importin β during NPC translocation.
A recent, extensive analysis of yeast transport receptor-nucleoporin binding in vitro revealed that many of these interactions are sensitive to RanGTP. In a general way, RanGTP decreases import receptor-nucleoporin affinities and increases export receptor-nucleoporin affinities (1). The interactions observed here between Xpo-t and both Nup153 and RanBP2 behave in this way, while the interaction of the C terminus with CAN is RanGTP independent.
XpC did not compete with all nuclear export pathways. Of particular interest, it could block the export of some CRM1-cargo-RanGTP complexes but not that of others. Both CRM1 export complexes and XpC have been shown to interact with CAN (references 4, 17, and 18 and this paper), and this interaction could provide an explanation for XpC competition for CRM1 export. The differential effect of XpC on various CRM1 cargoes is suggestive of some specificity in the interaction of different export complexes containing the same receptor molecule during NPC translocation. It is reminiscent of the differential effects of WGA, which binds to N-acetylglucosaminylated nucleoporins, on the import into the nucleus of nuclear localization signal proteins (13, 14) and U snRNPs (16) in Xenopus oocytes, even though both cargoes are imported via importin β. A further recent example of NPC-import complex interaction specificity comes from the work of Huber et al. (21). They showed that the RanGTP-dependent import of U5 snRNPs, more specifically the RanGTP dependence of the dissociation of the complex between importin β, Snurportin1, and U5 snRNP from the NPC and entry of U5 snRNP into the nucleoplasm, depended on the identity of the importin β binding (IBB) domain of Snurportin1. Exchange of this IBB domain with its counterpart from importin α rendered U5 import complex dissociation from the NPC RanGTP dependent. Since the IBB domain is likely almost completely buried inside the import complex (8), it should be unable to contribute directly to NPC interactions. These data rather suggest that the IBB domains of the two import adapters affect the conformation of importin β and in this way alter import complex-nucleoporin interactions. If CRM1 interactions with nucleoporins are also affected differentially by its various cargoes, this would provide an explanation for the ability of XpC to inhibit translocation of some, but not other, CRM1-cargo complexes.
What are the likely functions of the three identified NPC binding sites of Xpo-t? It seems likely, given their RanGTP dependence, that Nup153 and RanBP2 bind Xpo-t when it is on its way out of the nucleus as part of a tRNA export complex (Fig. 8). A possible function for the Nup153 binding, particularly given the existence of nucleoplasmic binding sites for Xpo-t-RanGTP complexes (see above), is to increase the local concentration of Xpo-t at the opening of the NPC, as recently proposed in a general model for NPC translocation (40). Upon translocation to the cytoplasmic face of the NPC, the export complex would accumulate on RanBP2 until RanGTP was removed. Since this can in principle be achieved by either soluble RanBP1 or NPC-associated RanBP2 in conjunction with RanGAP and since RanGAP in the sumoylated form binds to RanBP2 (29, 30, 32), RanBP2 is a likely site for tRNA export complex disassembly (Fig. 8). In this way, RanBP2 would function in a manner analogous to that suggested for CAN in the case of CRM1-mediated export (4, 24) or Nup153 or the Nup66/Nup2 complex in the case of importin β-mediated import in vertebrates and yeast, respectively (6, 10, 43), to maintain the local concentration of the cargo-free receptor high at the opening to the translocation channel of the NPC.
We propose that, on dissociation from RanBP2, empty Xpo-t associates preferentially with CAN and that this interaction, like that of the tRNA export complex with Nup153, increases the local concentration of the receptor adjacent to the translocation channel and thus increases the efficiency of recycling. The exact mode of translocation through the NPC has been the subject of much recent experimentation and hypothesis, and translocation through the permeability barrier of the NPC must involve very weak and easily reversible interactions with nucleoporins (1, 38, 39, 40). The interactions we observe here appear to occur either prior to or following the translocation process per se, and we propose that they function to increase the overall efficiency of the translocation process.
Acknowledgments
S.K. was supported by a National Research Service Award from N.I.H.
We thank the members of the Mattaj lab, M. Fornerod, H. Pickersgill, J. Ellenberg, and E. Izaurralde, for critical reading of the manuscript. We also thank U. Kutay for valuable discussions and T. Littlewood for providing purified NTF2 protein.
REFERENCES
- 1.Allen, N. P. C., L. Huang, A. Burlingame, and M. Rexach. 2001. Proteomic analysis of nucleoporin interacting proteins. J. Biol. Chem. 276:29268-29274. [DOI] [PubMed] [Google Scholar]
- 2.Arts, G.-J., M. Fornerod, and I. W. Mattaj. 1998. Identification of a nuclear export receptor for tRNA. Curr. Biol. 8:305-314. [DOI] [PubMed] [Google Scholar]
- 3.Arts, G.-J., S. Kuersten, P. Romby, B. Ehresmann, and I. W. Mattaj. 1998. The role of exportin-t in selective nuclear export of mature tRNAs. EMBO J. 17:7430-7441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Askjaer, P., A. Bachi, M. Wilm, F. R. Bischoff, D. L. Weeks, V. Ogniewski, M. Ohno, C. Niehrs, J. Kjems, I. W. Mattaj, and M. Fornerod. 1999. RanGTP-regulated interactions of CRM1 with nucleoporins and a shuttling DEAD-box helicase. Mol. Cell. Biol. 19:6276-6285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bayliss, R., T. Littlewood, and M. Stewart. 2000. Structural basis for the interaction between FxFG nucleoporin repeats and importin-beta in nuclear trafficking. Cell 102:99-108. [DOI] [PubMed] [Google Scholar]
- 6.Ben-Efraim, I., and L. Gerace. 2001. Gradient of increasing affinity of importin beta for nucleoporins along the pathway of nuclear import. J. Cell Biol. 152:411-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6a.Bilbao-Cortés, D., M. Hetzer, G. Längst, P. B. Becker, and I. W. Mattaj. Ran binds to chromatin by two distinct mechanisms. Curr. Biol., in press. [DOI] [PubMed]
- 7.Chi, N. C., and S. A. Adam. 1997. Functional domains in nuclear import factor p97 for binding the nuclear localization sequence receptor and the nuclear pore. Mol. Biol. Cell 8:945-956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cingolani, G., C. Petosa, K. Weis, and C. W. Muller. 1999. Structure of importin-beta bound to the IBB domain of importin-alpha. Nature 399:221-229. [DOI] [PubMed] [Google Scholar]
- 9.Cordes, V. C., S. Reidenbach, A. Kohler, N. Stuurman, R. van Driel, and W. W. Franke. 1993. Intranuclear filaments containing a nuclear pore complex protein. J. Cell Biol. 123:1333-1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Denning, D., B. Mykytka, N. P. Allen, L. Huang, A. Burlingame, and M. Rexach. 2001. The nucleoporin Nup60p functions as a Gsp1p-GTP-sensitive tether for Nup2p at the nuclear pore complex. J. Cell Biol. 154:937-950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Englmeier, L., J. C. Olivo, and I. W. Mattaj. 1999. Receptor-mediated substrate translocation through the nuclear pore complex without nucleotide triphosphate hydrolysis. Curr. Biol. 9:30-41. [DOI] [PubMed] [Google Scholar]
- 12.Fabre, E., and E. Hurt. 1997. Yeast genetics to dissect the nuclear pore complex and nucleocytoplasmic trafficking. Annu. Rev. Genet. 31:277-313. [DOI] [PubMed] [Google Scholar]
- 13.Finlay, D. R., D. D. Newmeyer, T. M. Price, and D. J. Forbes. 1987. Inhibition of in vitro nuclear transport by a lectin that binds to nuclear pores. J. Cell Biol. 104:189-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Finlay, D. R., and D. J. Forbes. 1990. Reconstitution of biochemically altered nuclear pores: transport can be eliminated and restored. Cell 60:17-29. [DOI] [PubMed] [Google Scholar]
- 15.Finlay, D. R., E. Meier, P. Bradley, J. Horecka, and D. J. Forbes. 1991. A complex of nuclear pore proteins required for pore function. J. Cell Biol. 114:169-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fischer, U., E. Darzynkiewicz, S. M. Tahara, N. A. Dathan, R. Luhrmann, and I. W. Mattaj. 1991. Diversity in the signals required for nuclear accumulation of U snRNPs and variety in the pathways of nuclear transport. J. Cell Biol. 113:705-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fornerod, M., J. van Deursen, S. van Baal, A. Reynolds, D. Davis, K. G. Murti, J. Fransen, and G. Grosveld. 1997. The human homologue of yeast CRM1 is in a dynamic subcomplex with CAN/Nup214 and a novel nuclear pore component Nup88. EMBO J. 16:807-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fornerod, M., M. Ohno, M. Yoshida, and I. W. Mattaj. 1997. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 90:1051-1060. [DOI] [PubMed] [Google Scholar]
- 19.Görlich, D., and U. Kutay. 1999. Transport between the cell nucleus and cytoplasm. Annu. Rev. Cell Dev. Biol. 15:607-660. [DOI] [PubMed] [Google Scholar]
- 20.Hellmuth, K., D. M. Lau, F. R. Bischoff, M. Kunzler, E. Hurt, and G. Simos. 1998. Yeast Los1p has properties of an exportin-like nucleocytoplasmic transport factor for tRNA. Mol. Cell. Biol. 18:6374-6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huber, J., A. Dickmanns, and R. Lührmann. 2002. The importin-β binding domain of snurportin1 is responsible for the Ran- and energy-independent nuclear import of splicesomal U snRNPs in vitro. J. Cell Biol. 156:467-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Izaurralde, E., U. Kutay, C. von Kobbe, I. W. Mattaj, and D. Görlich. 1997. The asymmetric distribution of the constituents of the Ran system is essential for transport into and out of the nucleus. EMBO J. 16:6535-6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jarmolowski, A., W. C. Boelens, E. Izaurralde, and I. W. Mattaj. 1994. Nuclear export of different classes of RNA is mediated by specific factors. J. Cell Biol. 124:627-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kehlenbach, R. H., A. Dickmanns, A. Kehlenbach, T. Guan, and L. Gerace. 1999. A role for RanBP1 in the release of CRM1 from the nuclear pore complex in a terminal step of nuclear export. J. Cell Biol. 145:645-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kraemer, D., R. W. Wozniak, G. Blobel, and A. Radu. 1994. The human CAN protein, a putative oncogene product associated with myeloid leukemogenesis, is a nuclear pore complex protein that faces the cytoplasm. Proc. Natl. Acad. Sci. USA 91:1519-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kutay, U., E. Izaurralde, F. R. Bischoff, I. W. Mattaj, and D. Görlich. 1997. Dominant-negative mutants of importin-beta block multiple pathways of import and export through the nuclear pore complex. EMBO J. 16:1153-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kutay, U., G. Lipowsky, E. Izaurralde, F. R. Bischoff, P. Schwarzmaier, E. Hartmann, and D. Görlich. 1998. Identification of a tRNA-specific nuclear export receptor. Mol. Cell 1:359-369. [DOI] [PubMed] [Google Scholar]
- 28.Lipowsky, G., F. R. Bischoff, E. Izaurralde, U. Kutay, S. Schafer, H. J. Gross, H. Beier, and D. Görlich. 1999. Coordination of tRNA nuclear export with processing of tRNA. RNA 5:539-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mahajan, R., C. Delphin, T. Guan, L. Gerace, and F. Melchior. 1997. A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell 88:97-107. [DOI] [PubMed] [Google Scholar]
- 30.Mahajan, R., L. Gerace, and F. Melchior. 1998. Molecular characterization of the SUMO-1 modification of RanGAP1 and its role in nuclear envelope association. J. Cell Biol. 140:259-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mattaj, I. W., and L. Englmeier. 1998. Nucleocytoplasmic transport: the soluble phase. Annu. Rev. Biochem. 67:256-306. [DOI] [PubMed] [Google Scholar]
- 32.Matunis, M. J., J. Wu, and G. Blobel. 1998. SUMO-1 modification and its role in targeting the Ran GTPase-activating protein, RanGAP1, to the nuclear pore complex. J. Cell Biol. 140:499-509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakielny, S., and G. Dreyfuss. 1998. Import and export of the nuclear protein import receptor transportin by a mechanism independent of GTP hydrolysis. Curr. Biol. 8:89-95. [DOI] [PubMed] [Google Scholar]
- 34.Nakielny, S., S. Shaikh, B. Burke, and Dreyfuss. 1999. Nup153 is an M9-containing mobile nucleoporin with a novel Ran-binding domain. EMBO J. 18:1982-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ohno, M., A. Segref, A. Bachi, M. Wilm, and I. W. Mattaj. 2000. PHAX, a mediator of U snRNA nuclear export whose activity is regulated by phosphorylation. Cell 101:187-198. [DOI] [PubMed] [Google Scholar]
- 36.Pante, N., R. Bastos, I. McMorrow, B. Burke, and U. Aebi. 1994. Interactions and three-dimensional localization of a group of nuclear pore complex proteins. J. Cell Biol. 126:603-617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Powers, M. A., C. Macaulay, F. R. Masiarz, and D. J. Forbes. 1995. Reconstituted nuclei depleted of a vertebrate GLFG nuclear pore protein, p97, import but are defective in nuclear growth and replication. J. Cell Biol. 128:721-736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rabut, G., and J. Ellenberg. 2001. Nucleocytoplasmic transport: diffusion channel or phase transition? Curr. Biol. 11:R551-R554. [DOI] [PubMed] [Google Scholar]
- 39.Ribbeck, K., and D. Görlich. 2001. Kinetic analysis of translocation through nuclear pore complexes. EMBO J. 20:1320-1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rout, M. P., J. D. Aitchison, A. Suprapto, K. Hjertaas, Y. Zhao, and B. T. Chait. 2000. The yeast nuclear pore complex: composition, architecture, and transport mechanism. J. Cell Biol. 148:635-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rout, M. P., and J. D. Aitchison. 2001. The nuclear pore complex as a transport machine. J. Biol. Chem. 276:16593-16596. [DOI] [PubMed] [Google Scholar]
- 42.Segref, A., I. W. Mattaj, and M. Ohno. 2001. The evolutionarily conserved region of the U snRNA export mediator PHAX is a novel RNA-binding domain that is essential for U snRNA export. RNA 7:351-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shah, S., S. Tugendreich, and D. Forbes. 1998. Major binding sites for the nuclear import receptor are the internal nucleoporin Nup153 and the adjacent nuclear filament protein Tpr. J. Cell Biol. 141:31-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ström, A.-C., and K. Weis. 2001. Importin-β-like nuclear transport receptors. Genome Biol. 2:3008.1-3008.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sukegawa, J., and G. Blobel. 1993. A nuclear pore complex protein that contains zinc finger motifs, binds DNA and faces the nucleoplasm. Cell 72:29-38. [DOI] [PubMed] [Google Scholar]
- 46.Truant, R., R. A. Fridell, E. R. Benson, A. Herold, and B. R. Cullen. 1998. Nucleocytoplasmic shuttling by protein nuclear import factors. Eur. J. Cell Biol. 77:269-275. [DOI] [PubMed] [Google Scholar]
- 47.Walther, T. C., M. Fornerod, H. Pickersgill, M. Goldberg, T. D. Allen, and I. W. Mattaj. 2001. The nucleoporin Nup153 is required for nuclear pore basket formation, nuclear pore complex anchoring and import of a subset of nuclear proteins. EMBO J. 20:5703-5714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47a.Walther, T. C., H. S. Pickersgill, V. C. Cordes, M. W. Goldberg, T. D. Allen, I. W. Mattaj, and M. Fornerod. The cytoplasmic filaments of the nuclear pore complex are dispensable for nuclear protein import. J. Cell Biol., in press. [DOI] [PMC free article] [PubMed]
- 48.Wilken, N., J. L. Senecal, U. Scheer, and M. C. Dabauvalle. 1995. Localization of the Ran-GTP binding protein RanBP2 at the cytoplasmic side of the nuclear pore complex. Eur. J. Cell Biol. 68:211-219. [PubMed] [Google Scholar]
- 49.Wu, J., M. J. Matunis, D. Kraemer, G. Blobel, and E. Coutavas. 1995. Nup358, a cytoplasmically exposed nucleoporin with peptide repeats, Ran-GTP binding sites, zinc fingers, a cyclophilin A homologous domain, and a leucine-rich region. J. Biol. Chem. 270:14209-14213. [DOI] [PubMed] [Google Scholar]
- 50.Yokoyama, N., N. Hayashi, T. Seki, N. Pante, T. Ohba, K. Nishii, K. Kuma, T. Hayashida, T. Miyata, U. Aebi, et al. 1995. A giant nucleopore protein that binds Ran/TC4. Nature 376:184-188. [DOI] [PubMed] [Google Scholar]