Abstract
DNA polymerase α-primase (pol-prim) is a heterotetramer with DNA polymerase and primase activities. The polymerase (p180) and primase (p48 and p58) subunits synthesize primers and extend them, but the function of the remaining subunit (p68) is poorly understood. Genetic studies in yeast suggested an essential role for the p68 ortholog in early S phase prior to the hydroxyurea-sensitive step, possibly a regulatory role in initiation of DNA replication, but found no evidence for an essential function of p68 later in S phase. To investigate whether the human p68 subunit has an essential role in DNA replication, we examined the ability of a purified trimeric human pol-prim lacking p68 to initiate simian virus 40 DNA replication in vitro and to synthesize and elongate primers on single-stranded DNA in the presence of T antigen and replication protein A (RPA). Both activities of trimeric pol-prim were defective, but activity was recovered upon addition of separately purified p68. Phosphorylation of p68 by cyclin A-dependent protein kinase also inhibited both activities of pol-prim. The data strongly suggest that the p68 subunit is required for priming activity of pol-prim in the presence of RPA and T antigen, both during initiation at the origin and during lagging strand replication.
The recruitment of DNA polymerase α-primase (pol-prim) is a key event in the assembly of functional replication complexes in eukaryotic cells. pol-prim initiates DNA replication by synthesizing short RNA primers on the leading and lagging strand templates and then elongating them into hybrid primers of about 35 nucleotides (3, 21, 25, 56, 62, 65). pol-prim is composed of four subunits that appear to be conserved in all eukaryotes. The human pol-prim subunits are named according to their apparent molecular weights: p180, p68, p58, and p48. The largest subunit contains the polymerase activity, and the smallest subunit contains the primase activity. Interactions of p68 with p180 and of p58 with p48 facilitate expression and nuclear import of the catalytic subunits (41, 42). In addition, the p58 subunit physically links p48 to p180 and regulates the length of primers synthesized by the primase (3, 4, 7, 10).
The role of pol-prim in initiation of mammalian DNA replication has been investigated in the cell-free simian virus 40 (SV40) DNA replication system. In this model system, synthesis of RNA primers at the viral origin of replication requires only SV40 T antigen, the single-stranded DNA binding protein replication protein A (RPA), pol-prim, and topoisomerase I (23, 62). T antigen assembles on the viral origin, unwinds the parental DNA, and recruits the required cellular proteins to the replication fork. The single-stranded DNA (ssDNA) generated by T antigen is sequestered by RPA, and pol-prim synthesizes the first primers on the leading and lagging strand templates. Subsequently, pol-prim is displaced from the primer-template by the clamp-loader replication factor C (RFC), the PCNA sliding clamp, and DNA polymerase δ, which extends the primers of both the leading and lagging strands (62, 69). Several lines of evidence suggest that T antigen interacts specifically with pol-prim, constituting a simple primosome, and that these interactions are critical in the recruitment and activity of pol-prim on ssDNA in the presence of RPA. T antigen stimulates the primase and polymerase activities of pol-prim through its physical association with all four subunits of pol-prim (8, 9, 12-14, 24, 66). The primase activity of pol-prim is markedly inhibited on RPA-saturated ssDNA, and T antigen relieves this inhibition (9, 37). Antibodies against T antigen that block its interactions with either RPA or pol-prim disrupt its ability to facilitate priming in the presence of RPA (66, 67). However, the precise requirements for activity of this simple primosome, in particular whether p68 is required, remain poorly understood.
In Saccharomyces cerevisiae, the p68 ortholog, known as p86 or the B subunit, is required for cell viability and executes an essential function in early S phase prior to the hydroxyurea-sensitive step, suggesting a possible role in initiation of DNA replication (19). However, no essential function of the B subunit was detected later in S phase (19). The B subunit was also dispensable for in vitro reconstitution of an enzymatically active complex of the other three subunits (7), suggesting that it may perform a regulatory function in initiation of replication. Consistent with this idea, the B/p68 subunit is phosphorylated in a cell cycle-dependent manner in both yeast and human cells (18, 20, 49, 60). Hypophosphorylated pol-prim isolated from human cells at G1/S supports efficient initiation of SV40 DNA replication in vitro, but hyperphosphorylated pol-prim from G2/M cells has minimal activity (60). Phosphorylation of p68 by cyclin A/cdk2 or cdk1 in vitro targets the same sites that are phosphorylated in vivo in G2/M and inhibits pol-prim activity in initiation of SV40 replication (60). However, the mechanism through which p68 phosphorylation regulates SV40 DNA replication remains unclear.
In this study, a trimeric form of human DNA pol-prim lacking p68 was used to investigate the possible functions of p68 in DNA replication using the SV40 model system. The trimer exhibited enzymatic activity in several simple assays in the absence of other replication proteins. However, the trimer was defective in initiation of SV40 DNA replication, as well as in priming and elongation on RPA-coated ssDNA. Addition of separately purified p68 restored activity in both of these assays, indicating that p68 is required for primosome activity in the presence of RPA. Several speculative models for the function of p68 during viral and chromosomal DNA replication are discussed.
MATERIALS AND METHODS
Purification of recombinant human DNA pol-prim.
The pol-prim tetramer was purified by immunoaffinity chromatography from extracts of Hi-5 insect cells multiply infected with recombinant baculoviruses essentially as described elsewhere (61). The trimeric form of pol-prim was purified by the same protocol, except that the p68 baculovirus was omitted from the infection and the trimeric pol-prim bound to the monoclonal antibody affinity column was washed with 50 volumes of buffer B rather than 20 volumes as described for purification of the tetramer (61). The typical polymerase specific activity of the pol-prim tetramer was 3.2 U/μg and that of the trimer was 1.5 U/μg, with a unit (U) defined as incorporation of 1 nmol of nucleotide per h on a poly(dA)-oligo(dT) template (see below). The typical primase specific activity of the pol-prim tetramer was 20 U/μg and that of the trimer was 10 U/μg, with a unit defined as incorporation of 1 fmol of nucleotide per h on unprimed M13 DNA. The usual yields were 3 mg of tetramer and 1.5 mg of trimer from 5 × 108 Hi-5 cells.
Recombinant human pol-prim prephosphorylated with purified cyclin A/cdk2 was prepared as described previously (60). Mock-phosphorylated pol-prim was prepared using exactly the same protocol, except that cyclin A/cdk2 was omitted from the prephosphorylation reaction (60).
Purification of other proteins.
The baculovirus-expressed histidine (his)-tagged p68 subunit of human pol-prim was purified by nickel affinity chromatography from extracts of Hi-5 insect cells infected with a recombinant baculovirus as described previously (51). For bacterial expression, cDNAs encoding p68, p68 residues 1 to 240, and p68 residues 241to 598 (8) were cloned into pET28a (Novagen, Madison, Wis.). The his-tagged p68 polypeptides were expressed in Escherichia coli BL21(DE3) and purified by Ni-nitrilotriacetic acid affinity chromatography (51). SV40 T antigen was purified by immunoaffinity chromatography from extracts of Hi-5 insect cells infected with a recombinant baculovirus exactly as described previously (51). Bacterially expressed recombinant human RPA (22), E. coli ssDNA binding protein (SSB) (28), and calf thymus topoisomerase I (43) were purified as described elsewhere.
Nuclease detection assay.
Increasing amounts of purified pol-prim (50 to 500 ng) were incubated with 5 pmol of 5′-32P-end-labeled oligodeoxyribonucleotide (5′CAGGGCCCGGGCCAAGCACAGAATGCTTGTGTTCTCGCCGGTTC) in 30 mM HEPES (pH 7.9), 7 mM magnesium acetate, 1 mM dithiothreitol (DTT), 4 mM ATP, 40 mM creatine phosphate, and 0.04 mg of creatine kinase/ml for 1 h at 37°C. Reaction products were adjusted to 0.05% (wt/vol) bromophenol blue, 0.05% (wt/vol) xylene cyanol, and 2.5% (wt/vol) Ficoll 400, and loaded on 8.0% polyacrylamide gels. Radiolabeled DNA fragments were resolved by electrophoresis in 45 mM Tris, 45 mM boric acid, 1 mM EDTA for 1.5 h at 100 V. The gel was dried and the radiolabeled DNA was detected by autoradiography. The results of this analysis revealed no evidence of nuclease contamination (data not shown).
DNA polymerase activity assay.
The polymerase activities of the trimer and tetramer were tested on a randomly primed poly(dA)-oligo(dT) (20:1) template (Amersham Biosciences, Piscataway, N.J.). Reaction mixtures (20 μl) were assembled on ice and contained 0.5 μg of DNA in reaction buffer (50 mM bis-Tris-HCl [pH 6.5], 1 mM DTT, 10 mM KCl, 7 mM MgCl2, 0.2 mg of bovine serum albumin/ml, 0.02 mM dTTP), 0.1 μCi of [α-32P]dTTP (3,000 Ci/mmol) (Amersham Biosciences), and pol-prim as indicated in the figure legends. After incubation for 10 min at 37°C, reaction products were precipitated with cold 10% (vol/vol) trichloroacetic acid (TCA) and spotted on glass fiber filters (GF/C; Whatman, Clifton, N.J.). The filters were washed five times with wash buffer (120 mM Na2H2P2O7, 1.2 M HCl) and once with 100% ethanol and dried at room temperature. Radioactivity was analyzed by scintillation counting.
DNA primase activity assay.
The primase activities of the pol-prim trimer and tetramer were tested on single-stranded M13 DNA. Primase reaction mixtures (20 μl) contained 50 to 500 ng of pol-prim, 100 ng of M13 DNA in reaction buffer (30 mM HEPES-KOH [pH 7.9], 1 mM DTT, 7 mM magnesium acetate, 4 mM ATP, 0.2 mM UTP, 0.2 mM GTP, 0.01 mM CTP), and 20 μCi of [α-32P]CTP (3,000 Ci/mmol; Dupont NEN, Boston, Mass.). Reactions were assembled on ice and incubated at 37°C for 90 min. Reaction products were precipitated with 2% NaClO4 in acetone, washed with acetone, and dried. The products were dissolved in formamide loading buffer (45% [vol/vol] formamide, 5 mM EDTA, 0.08% [wt/vol] xylene cyanol, 0.08% [wt/vol] bromophenol blue) at 65°C for 10 min and resolved by denaturing 20% polyacrylamide gel electrophoresis (PAGE) for 4 to 5 h at 500 V. The reaction products were visualized by autoradiography.
Stimulation of priming and elongation by T antigen.
Reaction mixtures (20 μl) contained 25 ng of single-stranded M13 DNA, 30 ng of pol-prim tetramer or 60 ng of trimer, and 250 ng of T antigen in elongation buffer (30 mM HEPES-KOH [pH 7.9], 7 mM magnesium acetate, 0.01 mM ZnCl2, 1 mM DTT, 4 mM ATP, 0.2 mM GTP, 0.2 mM UTP, 0.2 mM CTP, 0.02 mM dATP, 0.1 M dGTP, 0.1 mM dTTP, 0.1 mM dCTP, 40 mM creatine phosphate, 0.04 mg of creatine kinase/ml) supplemented with 3 μCi of [α-32P]dATP (3,000 Ci/mmol) (Amersham Biosciences). Reaction mixtures were incubated at 37°C for 45 min and then digested with 0.1 mg of proteinase K/ml in the presence of 1% sodium dodecyl sulfate (SDS) and 1 mM EDTA for 30 min at 37°C. Reaction products were purified over Sephadex G-50 columns (Boehringer Mannheim, Indianapolis, Ind.) and then precipitated with 2% NaClO4 in acetone. The products were washed, dried, resuspended in alkaline loading buffer (60 mM NaOH, 2 mM EDTA [pH 8.0], 20% [wt/vol] Ficoll, 0.10% [wt/vol] bromophenol blue, 0.10% [wt/vol] xylene cyanol), and electrophoresed on 1.5% agarose gels in running buffer (30 mM NaOH, 1 mM EDTA) for 2 h at 50 V. The gels were fixed in 10% TCA and dried. The reaction products were visualized by autoradiography. A sample of the radiolabeled products from each reaction was acid precipitated and analyzed by scintillation counting.
Singly primed M13 DNA elongation reactions.
Reaction mixtures (20 μl) contained 25 ng of singly primed M13 DNA and 25 to 250 ng of pol-prim in reaction buffer (30 mM HEPES-KOH [pH 7.9], 7 mM magnesium acetate, 1 mM DTT, 0.02 mM dATP, 0.10 mM dGTP, 0.10 mM dTTP, 0.10 mM dCTP, 40 mM creatine phosphate, and 0.04 mg of creatine kinase/ml) supplemented with 3 μCi of [α-32P]dATP (3,000 Ci/mmol) (Amersham Biosciences). Reaction mixtures were incubated at 37°C for 90 min and then digested with 0.1 mg of proteinase K/ml in the presence of 1% SDS and 1 mM EDTA for 30 min at 37°C. Radiolabeled DNA was purified over Sephadex G-50 columns (Boehringer Mannheim) and then precipitated with 2% NaClO4 in acetone. The products were washed, dried, resuspended in alkaline loading buffer (see above), and electrophoresed on 1.5% agarose gels in running buffer (30 mM NaOH, 1 mM EDTA) for 2 h at 50 V. The reaction products were visualized by autoradiography. In addition, a sample of the radiolabeled products from each reaction was acid precipitated and analyzed by scintillation counting.
Initiation of SV40 DNA replication assay.
Initiation reactions were carried out with purified T antigen, RPA, topoisomerase I, and 50 to 400 ng of pol-prim in the presence of radiolabeled ribonucleotides exactly as described elsewhere (51).
Assay for SV40 initiation coupled with elongation.
Monopolymerase (17, 36, 48) reaction mixtures (20 μl) were identical to SV40 initiation reaction mixtures except that the amounts of RPA and T antigen were increased to 1,000 ng and 1,200 ng, respectively, and 0.02 mM dATP, 0.10 mM dGTP, 0.10 mM dTTP, 0.10 mM dCTP, and 3 μCi of [α-32P]dATP (3,000 Ci/mmol; Amersham Biosciences) were present. Reaction mixtures were assembled on ice, incubated at 37°C for 90 min, and then digested with 0.1 mg of proteinase K/ml in the presence of 1% SDS and 1 mM EDTA for 30 min at 37°C. Radiolabeled reaction products were purified over Sephadex G-50 columns (Boehringer Mannheim) and then precipitated with 2% NaClO4 in acetone. The products were washed, dried, resuspended in alkaline loading buffer, and electrophoresed on 1.5% agarose gels in running buffer (30 mM NaOH, 1 mM EDTA) for 2 h at 50 V. The gels were fixed in 10% TCA and dried. The reaction products were visualized by autoradiography. A sample of the radiolabeled products from each reaction was acid precipitated and analyzed by scintillation counting.
Primer synthesis and elongation in the presence of RPA.
Reaction mixtures (20 μl) containing 25 ng of single-stranded M13 DNA were assembled at 4°C and preincubated with 250 to 1,250 ng of RPA in elongation buffer (see above) at 4°C for 20 min. The reactions were then supplemented with 3 μCi of [α-32P]dATP (3,000 Ci/mmol) (Amersham Biosciences), 450 ng of pol-prim tetramer or 900 ng of trimer, and 300 to 600 ng of T antigen as indicated in the figure legends, incubated at 37°C for 45 min, and then digested with 0.1 mg of proteinase K/ml in the presence of 1% SDS and 1 mM EDTA for 30 min at 37°C. Radiolabeled reaction products were processed and analyzed as described above for the SV40 monopolymerase assay.
Immunoblotting.
Immunoblotting with monoclonal antibodies 1CT102 and 2CT25, specific for the p180 subunit of pol-prim, and 9D5, specific for the p68 subunit, was performed as described previously (12, 60).
RESULTS
Trimeric human DNA pol-prim lacking the p68 subunit has polymerase and primase activity and is stimulated by T antigen.
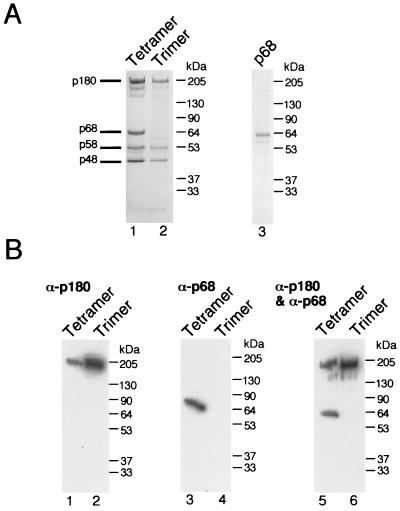
Tetrameric and trimeric complexes of human pol-prim were expressed in insect cells by using recombinant baculoviruses and purified by immunoaffinity chromatography. The purification protocol reproducibly yielded about half as much trimer as tetramer. The purity and integrity of representative preparations of tetramer and trimer are shown in Fig. 1A (lanes 1 and 2). The tetramer displayed the expected four subunits: p180 and small amounts of its proteolytic products, p68, p58, and p48 (lane 1). The trimer lacked the p68 subunit, as expected (lane 2). A histidine-tagged human p68 subunit was separately expressed in insect cells and purified by nickel affinity chromatography (Fig. 1A, lane 3). The compositions of the tetramer and trimer were confirmed by immunoblotting with two monoclonal antibodies specific for the p180 subunit (Fig. 1B, lanes 1, 2, 5, and 6) and a monoclonal antibody specific for the p68 subunit (Fig. 1B, lanes 3 to 6). Trace bands observed at about 60 kDa in p68 preparations (Fig. 1A, lane 3) and the trimer preparation (Fig. 1A, lane 2) were not stained by the monoclonal antibodies (Fig. 1B, lane 4; data not shown). Thus, the recombinant human pol-prim trimer appeared to be devoid of p68.
FIG. 1.
Purification of tetrameric and trimeric complexes of pol-prim and the p68 subunit. Purified preparations of the pol-prim tetramer, trimer, and p68-his were analyzed by SDS-PAGE and Coomassie blue staining (A), or by immunoblotting (B) with two p180-specific antibodies (left panel), a p68-specific antibody (center panel), or a mixture of all three antibodies (right panel). The positions of marker proteins of known size are indicated at the right in kilodaltons.
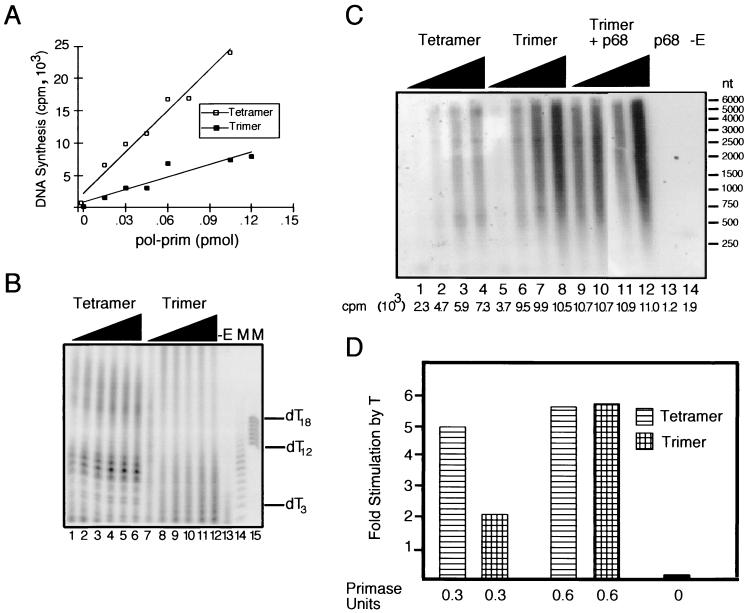
The polymerase and primase activities of the two pol-prim preparations were initially characterized in a series of simple enzyme assays. To test the polymerase activity, equimolar amounts of the tetramer and trimer were incubated with a poly(dA)-oligo(dT) template-primer and radiolabeled dTTP. The resulting radiolabeled DNA was precipitated, washed, and quantitated by scintillation counting. The trimer exhibited polymerase activity, but its specific activity was about half that of the tetramer in this assay (Fig. 2A). To assay the primase activity, equimolar amounts of the tetramer and trimer were incubated with single-stranded M13 DNA and ribonucleoside triphosphates, of which one was radiolabeled. The radiolabeled primers were separated by denaturing PAGE and visualized by autoradiography. The tetramer synthesized primers of predominantly 7 to 10 nucleotides in length, and smaller amounts of primers of twice that length (Fig. 2B, lanes 1 to 6). The trimer produced primers of 7 to 10 nucleotides, as well as shorter primers (Fig. 2B, lanes 7 to 12). However, the specific primase activity of the trimer was about half that of the tetramer. A control reaction lacking pol-prim generated no product (Fig. 2B, lane 13). These results show that the trimer possesses both polymerase and primase activity, albeit with about half the specific activity of the tetramer.
FIG. 2.
Biochemical characterization of tetrameric and trimeric complexes of pol-prim. (A) The DNA polymerase activities of equimolar amounts of the tetramer and trimer were tested on poly(dA)-oligo(dT) template-primer. (B) Primase activities of equimolar amounts (0.15, 0.3, 0.6, 0.9, 1.2, and 1.5 pmol) of the pol-prim tetramer (lanes 1 to 6) and trimer (lanes 7 to 12) were assayed on single-stranded M13 DNA template. Primers synthesized by pol-prim were visualized by denaturing electrophoresis and autoradiography. A control reaction containing no enzyme (−E) is shown in lane 13. End-labeled oligonucleotide markers (M) of the indicated lengths are shown in lanes 14 and 15. (C) Elongation of singly primed M13 DNA by 0.8, 1.6, 2.4, and 3.2 polymerase units of the pol-prim tetramer (lanes 1 to 4) or trimer (lanes 5 to 8 and 9 to 12) was assayed. A fivefold molar excess of purified p68 was added to the trimer in lanes 9 to 12. Radiolabeled elongation products were detected by alkaline agarose electrophoresis and autoradiography. Products made in the presence of p68 alone (lane 13) or in the absence of pol-prim (lane 14) are shown as controls. The radioactivity (counts per minute [cpm]) incorporated in each reaction is shown below each lane. (D) pol-prim tetramer (striped) and trimer (hatched) of equal primase activity (0.3 or 0.6 U, as indicated) were assayed for activity in priming-coupled elongation on unprimed M13 DNA in the presence and absence of 250 ng of T antigen. A control reaction (black) lacked pol-prim. Reaction products with and without T antigen were detected by alkaline agarose electrophoresis and quantitated by phosphorimaging to determine the stimulation of incorporation by T antigen.
In the presence of deoxyribonucleotides, the RNA primers synthesized by pol-prim are transferred to the DNA polymerase active site of the same molecule and extended into hybrid RNA-DNA primers of about 35 nucleotides (3, 10, 65). In the absence of the RFC/PCNA primer recognition complex, these RNA-DNA primers can be further elongated in repeated cycles of extension by the DNA polymerase activity (58, 59). The primer extension activities of pol-prim tetramer and trimer were determined by incubating an equal polymerase activity of each [i.e., about twice as many moles of trimer as tetramer, based on their specific activities on poly(dA)-oligo(dT) template-primer] with a uniquely primed single-stranded M13 DNA template in the presence of radiolabeled deoxyribonucleoside triphosphates. Both enzymes extended the primer to products of about 6 kb in length (Fig. 2C, compare lanes 1 to 4 with 5 to 8). The trimer appeared to synthesize about twice as much product as the tetramer (compare lanes 2 and 3 with 6 and 7). However, when products of equal molar amounts of tetramer and trimer were compared, the amount and length of the products were remarkably similar (Fig. 2C, compare lanes 4 and 6). The addition of a fivefold molar excess of purified p68 to the trimer stimulated its elongation activity only at the lower amount of trimer (compare lanes 9 to 12 with lanes 5 to 8). Control reactions containing only p68 (lane 13) or without pol-prim (lane 14) generated no products. These results demonstrate that, on a singly preprimed natural DNA template, the primer extension activity of the trimer was comparable to that of the tetramer.
Physical interactions of SV40 T antigen with human pol-prim, in particular with the p68 subunit, stimulate both priming and primer elongation activities of pol-prim (8, 9, 46), raising the question of whether T antigen could stimulate the activities of the trimer. To address this question, tetramer and trimer complexes of equal primase activity (i.e., about twice as many moles of trimer as tetramer) were incubated with unprimed single-stranded M13 DNA in the presence of unlabeled ribo- and deoxyribonucleotides and a radiolabeled deoxynucleoside triphosphate, with or without T antigen. Reaction products were resolved by alkaline agarose electrophoresis and quantitated by phosphorimaging. At the higher amount of primase activity, T antigen stimulated both the trimer and the tetramer about sixfold (Fig. 2D), but with half the amount of primase activity, stimulation of the tetramer was greater than that of the trimer. Addition of p68 to the trimer reaction did not further enhance the level of stimulation (data not shown). These results indicate that, at least qualitatively, T-antigen-mediated stimulation of pol-prim does not require p68.
The p68 subunit of DNA pol-prim is essential for initiation of SV40 DNA replication.
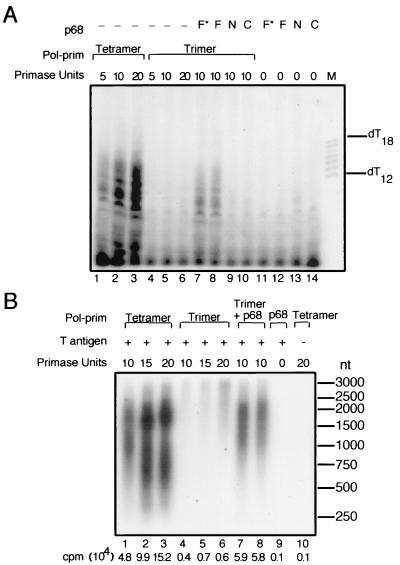
The initial characterization of the pol-prim trimer demonstrated polymerase and primase activities and showed that these activities could be stimulated by T antigen, suggesting that p68 was dispensable. On the other hand, the p68 subunit could be required for coordinated interactions of pol-prim with other replication proteins, such as RPA and T antigen, in viral DNA replication. If so, one would expect the trimer to be unable to support initiation of SV40 DNA replication, but the p68 subunit may be able to restore its activity. This prediction was tested using in vitro replication assays containing supercoiled SV40 origin DNA, topoisomerase I, RPA, T antigen, and pol-prim tetramer or trimer in the presence of radiolabeled ribonucleotides. Reactions containing the pol-prim tetramer generated easily detectable labeled RNA products of 7 to 14 nucleotides in length (Fig. 3A, lanes 1 to 3). However, the trimer (lanes 4 to 6) did not synthesize products above the background level. This result suggests that p68 is required for the initiation of replication at the viral origin. To confirm this interpretation, a fivefold molar excess of recombinant p68 purified from either baculovirus-infected insect cells (lane 7) or bacterial extracts (lane 8) was added to the trimer reaction mixtures. The initiation activity of the trimer was substantially restored by p68 (lanes 7 and 8). The initiation activity of the trimer was not restored by addition of the amino-terminal domain (N) of p68, which binds to T antigen (8), the carboxy-terminal domain (C), which binds to p180 (8, 40, 41), or a mixture of the N and C domains (Fig. 3A, lanes 9 and 10; data not shown). No product was synthesized in control reactions containing p68 but lacking pol-prim, demonstrating that the restoration of activity was not caused by possible contaminants in the p68 preparations (lanes 11 to 14).
FIG. 3.
The pol-prim trimer is defective in the initiation of SV40 DNA replication. (A) Initiation of SV40 DNA replication was assayed in reaction mixtures containing SV40 origin DNA, topoisomerase I, RPA, SV40 T antigen, and 5, 10, or 20 primase units of tetramer (lanes 1 to 3) or trimer (lanes 4 to 6) as indicated. A fivefold molar excess of full-length (F, F∗) p68 or p68 domains N or C (F∗, baculovirus-expressed, lane 7; F, bacterially expressed, lane 8; N, amino-terminal domain, lane 9; C, carboxy-terminal domain, lane 10) was added to reaction mixtures containing 10 units of the trimer. Products of reactions lacking pol-prim are shown as controls (lanes 11 to 14). Primers synthesized by pol-prim were detected by denaturing electrophoresis and autoradiography. End-labeled oligonucleotide markers (M) of the indicated lengths are shown at the right. (B) Initiation coupled with elongation was assayed in reaction mixtures containing SV40 origin DNA, topoisomerase I, RPA, SV40 T antigen, and increasing amounts (10, 15, and 20 primase units) of tetramer (lanes 1 to 3) or trimer (lanes 4 to 6). A 5- or 10-fold molar excess of separately purified, baculovirus-expressed p68 was added to reaction mixtures containing 10 units of the trimer in lanes 7 and 8. Elongation products synthesized by pol-prim were detected by alkaline agarose electrophoresis and autoradiography. Products of reactions lacking pol-prim (lane 9) or T antigen (lane 10) are shown as controls. The positions of end-labeled marker DNA fragments of the indicated sizes (in base pairs) are indicated at the right. A sample of the radiolabeled products from each reaction was also analyzed by scintillation counting (counts per minute [cpm] listed below each lane).
These results argue that the p68 subunit serves an essential function in the initial stage of replication when short oligoribonucleotide primers are synthesized. An alternative interpretation of these results is that the trimer is active, but that the primers it synthesizes are too few or too short to be easily detected in this assay. To address this concern, we tested the trimer and tetramer in a monopolymerase SV40 initiation assay where priming was coupled with elongation, thereby amplifying the signal (17, 36, 48). Reaction mixtures contained supercoiled SV40 origin DNA, topoisomerase I, RPA, T antigen, pol-prim tetramer or trimer, unlabeled ribonucleoside triphosphates, and deoxyribonucleoside triphosphates, of which one was radiolabeled. Reactions containing the tetramer generated radiolabeled products of 0.3 to 3 kb in length, and the amount of product was dependent on the amount of pol-prim tetramer in the reaction mixture (Fig. 3B, lanes 1 to 3). The trimer synthesized much less product (lanes 4 to 6). When purified p68 was present in the trimer reaction in a 5- or 10-fold molar excess, it restored activity to the trimer, resulting in a nearly 15-fold increase in replication activity (compare lanes 7 and 8 with lane 4). The products synthesized by the trimer in the presence of p68 were similar in size and abundance to those synthesized by the equivalent activity of the tetramer (Fig. 3B, compare lane 1 with lane 7). No products were synthesized in a control reaction containing p68 but lacking pol-prim, demonstrating that the recovery of activity could not be attributed to a contaminant in the p68 preparation (lane 9). No products were synthesized in a control reaction lacking T antigen (lane 10). We conclude that p68 is required for initiation of SV40 DNA replication in vitro.
The p68 subunit of pol-prim is required for priming and elongation on RPA-saturated single-stranded template.
In vitro SV40 replication assays have demonstrated that the p68 subunit of pol-prim is essential to initiate replication, but its role in initiation is unclear. The p68 requirement for primer synthesis and elongation at the SV40 origin could be origin specific, reflecting pol-prim enhancement of T-antigen assembly on the origin, effects on T-antigen-mediated unwinding of duplex DNA, or origin-specific loading of pol-prim by T antigen. Alternatively, p68 could be more generally required for T-antigen-mediated priming by pol-prim on RPA-coated ssDNA (8, 9, 37, 46, 47). For instance, the species-specific requirement for primate pol-prim in SV40 DNA replication is apparent during initiation of DNA replication at the viral origin, but not during T-antigen-mediated priming on RPA-coated ssDNA (54).
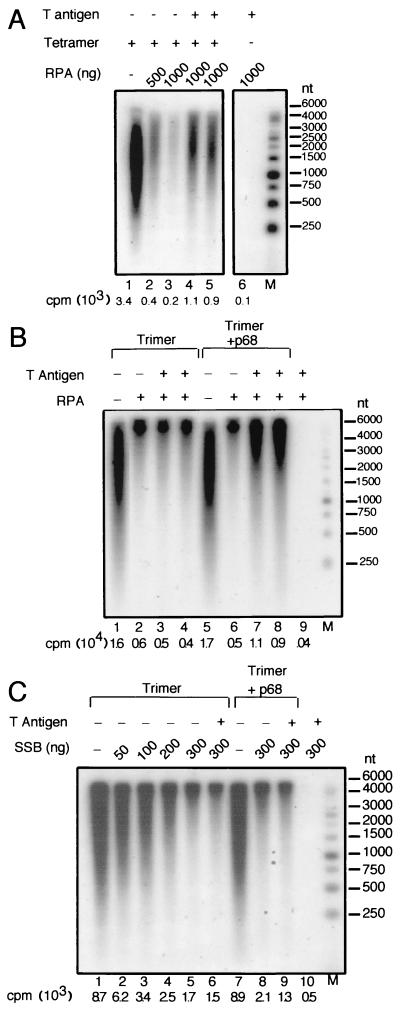
To assess whether p68 plays a role in T-antigen-mediated priming by pol-prim in the presence of RPA, we tested the activities of the pol-prim trimer and tetramer in priming-coupled DNA synthesis on RPA-saturated ssDNA (Fig. 4). Single-stranded M13 DNA was first incubated with or without RPA, and then pol-prim tetramer or trimer was added to the reaction mixture, either with or without T antigen. In the absence of RPA, the pol-prim tetramer synthesized primers and extended them into radiolabeled DNA products of 0.4 to 4 kb (Fig. 4A, lane 1). Preincubation of the template with increasing amounts of RPA reduced the amount of reaction product by more than 10-fold (lanes 2 and 3). This inhibition was partially relieved in the presence of T antigen (Fig. 4A, lanes 4 and 5), consistent with previous reports (9, 37, 51).
FIG. 4.
T-antigen-mediated priming and elongation activity of the pol-prim trimer in the presence of RPA. Reaction mixtures contained 25 ng of M13 DNA, 300 ng of T antigen as indicated (+), 200 ng of tetramer (A), 250 ng of trimer (B and C), and 500 to 1,000 ng of RPA (A), 1,000 ng of RPA (B), or 50 to 300 ng of E. coli SSB (C). Where indicated, 250 ng of p68 was added to the reaction (B and C). Radiolabeled elongation products were detected by alkaline agarose electrophoresis and autoradiography. The radioactivity (counts per minute [cpm]) incorporated in each reaction is shown below each lane. Products made in control reactions without pol-prim, T antigen, RPA, or SSB are indicated (−). The mobilities of end-labeled marker DNA fragments of the indicated lengths in nucleotides are shown at the right.
The pol-prim trimer also synthesized primers and elongated them to products of up to 6 kb in length (Fig. 4B, lane 1), and synthesis was inhibited when the template DNA was saturated with RPA (lane 2). However, addition of T antigen failed to relieve the inhibition caused by RPA (Fig. 4B, compare lanes 3 and 4 with lane 2). Importantly, the ability of the trimer to synthesize and elongate primers on RPA-saturated ssDNA was recovered when p68 was added together with T antigen (Fig. 4B, compare lanes 7 and 8 with lane 6).
Although several different SSBs inhibit priming by pol-prim tetramer, T antigen relieves only the inhibition by mammalian RPA, correlating with its ability to bind mammalian RPA but not other SSBs (37, 50, 54). To test whether the T-antigen-mediated priming and elongation reaction was dependent on specific protein-protein interactions of T antigen with RPA and the pol-prim trimer, the experiment in Fig. 4B was repeated using single-stranded M13 DNA saturated with bacterial SSB instead of RPA (Fig. 4C). SSB inhibited priming and elongation by the pol-prim trimer (Fig. 4C, compare lanes 2 to 5 with lane 1), but activity was not recovered in the presence of T antigen (lane 6). Moreover, neither p68 nor a mixture of p68 and T antigen restored activity of the pol-prim trimer in the presence of SSB (compare lanes 8 and 9 with lane 7). Taken together, the results of these experiments argue strongly that p68 is required for T-antigen-mediated priming and elongation by pol-prim on RPA-saturated ssDNA.
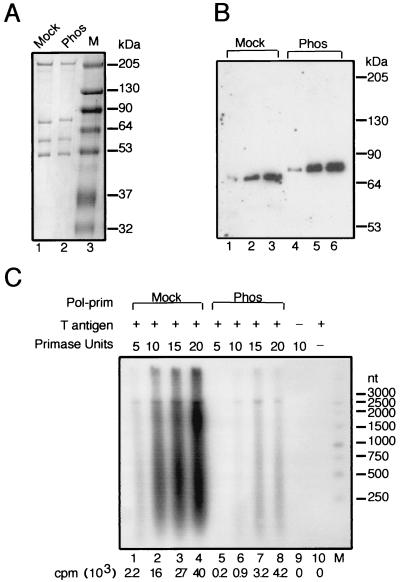
Phosphorylation of the p68 subunit by cyclin A/cdk2 inhibits priming-coupled elongation activity of pol-prim in the presence of RPA.
The initiation of SV40 DNA replication in vitro is regulated by phosphorylation of pol-prim by cyclin/cdk (11, 49, 55, 60, 61). Phosphorylation of pol-prim by cyclin A/cdk2 on specific N-terminal residues of the p68 subunit, followed by purification of the pol-prim away from the kinase, resulted in a 10-fold inhibition of primer synthesis at the SV40 origin compared with mock-phosphorylated pol-prim (60, 61). These results raised the question of whether phosphorylation of p68 regulated the initiation activity of pol-prim only at the origin, or whether it might also inhibit its ability to synthesize and elongate primers during lagging strand DNA replication. To investigate this question, we prepared prephosphorylated and mock-phosphorylated purified human pol-prim and compared their properties with those described previously (60, 61). Both preparations were composed of the expected four subunits (Fig. 5A, lanes 1 and 2) and displayed DNA polymerase and primase activities in simple enzyme assays (60, 61). The mobility of the p68 subunit in the phosphorylated pol-prim was slightly retarded compared to that of the mock-phosphorylated p68, as reported previously for hyperphosphorylated p68 (60). Immunoblotting of both pol-prim preparations with a monoclonal antibody specific for p68 confirmed the identity of the p68 bands (Fig. 5B). Lastly, to confirm that the activity of the prephosphorylated pol-prim in initiation of SV40 DNA replication was effectively inhibited, both preparations were tested for SV40 origin-dependent initiation coupled with primer elongation (Fig. 5C). Mock-phosphorylated pol-prim synthesized primers and elongated them into easily detectable products (Fig. 5C, lanes 1 to 4), while the activity of the prephosphorylated pol-prim was inhibited about 10-fold (lanes 5 to 8). These results confirm and extend earlier observations that phosphorylation of pol-prim by cyclin A/cdk2 inhibits initiation of SV40 DNA replication (60, 61).
FIG. 5.
Phosphorylation of pol-prim by cyclin A/cdk2 inhibits initiation and elongation of SV40 DNA replication. (A and B) pol-prim purified after prephosphorylation (Phos) with cyclin A/cdk2 (A/2) or mock phosphorylation (Mock) was analyzed by SDS-PAGE with 10% (A) or 7.5% (B) polyacrylamide and staining with Coomassie blue (A) or Western blotting with a p68-specific antibody (B). The mobilities of marker proteins (M) of known sizes are indicated (in kilodaltons) at the right. (C) Initiation and elongation of SV40 DNA replication was assayed in reaction mixtures containing SV40 origin DNA, topoisomerase I, RPA, T antigen, and increasing amounts (5, 10, 15, and 20 primase units) of mock-phosphorylated pol-prim (lanes 1 to 4) and cyclin A/cdk2-phosphorylated pol-prim (lanes 5 to 8). Reaction products made in the absence of T antigen (lane 9) or pol-prim (lane 10) are shown. Radiolabeled elongation products synthesized by pol-prim were detected by alkaline electrophoresis and autoradiography. The radioactivity (counts per minute [cpm]) incorporated in each reaction is shown below the lane. The mobilities of end-labeled marker DNA fragments of the indicated lengths in nucleotides are shown at the right.
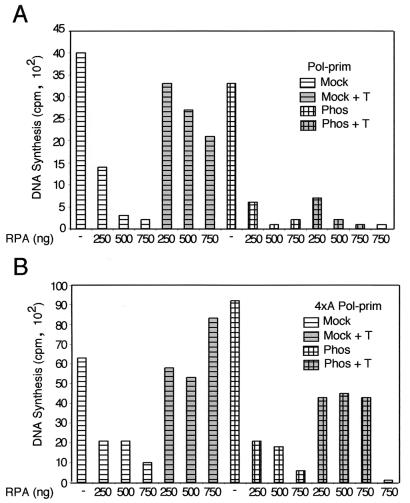
To address the question of whether phosphorylation of pol-prim by cyclin A/cdk2 specifically inhibits only initiation of SV40 DNA replication, or perhaps also priming and elongation on an RPA-bound template in general, we tested the activity of phosphorylated pol-prim in priming-coupled elongation on RPA-saturated M13 ssDNA. Preincubation of the ssDNA template with RPA inhibited primer synthesis and elongation by both mock-phosphorylated and cyclin A/cdk2-phosphorylated pol-prim in a dose-dependent manner (Fig. 6A). Although the activity of mock-phosphorylated pol-prim was restored by T antigen as expected, the activity of phosphorylated pol-prim was refractory to T-antigen stimulation. The data indicate that phosphorylation of pol-prim inhibits its activity in T-antigen-mediated priming and elongation on RPA-coated templates.
FIG. 6.
Phosphorylation of pol-prim on p68 by cyclin A/cdk2 inhibits T-antigen-mediated priming and elongation on RPA-coated ssDNA. Reaction mixtures contained 25 ng of unprimed M13 DNA that had been precoated with 250 to 750 ng of RPA as indicated, T antigen (300 ng) as indicated (+), and 200 ng of wild-type pol-prim tetramer (A) or mutant tetramer containing 4xA p68 (B), either mock phosphorylated or phosphorylated with cyclinA/cdk2, as indicated in the figure, and nucleoside triphosphates. The bar graphs show the radioactivity (counts per minute [cpm]) incorporated in each reaction of a typical experiment with each pol-prim preparation. Products made in control reactions without pol-prim, T antigen, or RPA are indicated.
Since both the p68 and p180 subunits of pol-prim can be phosphorylated by cyclin A/cdk2, these results cannot distinguish whether phosphorylation of p68, p180, or both, is responsible for the observed inhibition of priming in the presence of RPA. To address this question, we utilized a mutant p68 with alanine substitutions at four specific serine residues (4xA) that renders pol-prim resistant to the inhibitory effects of phosphorylation (60). The experiment in Fig. 6A was repeated using pol-prim with a 4xA mutant p68 subunit (Fig. 6B). The priming-coupled elongation activity of phosphorylated and unmodified 4xA pol-prim was diminished by increasing amounts of RPA (Fig. 6B). T antigen significantly reversed the inhibition of 4xA pol-prim both with and without prephosphorylation. The data demonstrate that phosphorylation of p68 is primarily responsible for the inhibition of pol-prim activity in T-antigen-mediated priming on RPA-saturated ssDNA.
DISCUSSION
Trimeric pol-prim lacking the p68 subunit has enzymatic activity.
To elucidate the role of the p68 subunit of pol-prim in DNA replication, we have generated and characterized a recombinant human pol-prim lacking the p68 subunit (Fig. 1). The reduced yield of pol-prim trimer may indicate that p68 facilitates pol-prim assembly or stability, as suggested by the ability of yeast B subunit to stimulate yeast pol-prim reconstitution (7). The purified human trimer synthesized primers and elongated them (Fig. 2), consistent with the previously reported activities of human p180 reconstituted with mouse p48-p58 primase heterodimer (10) and of yeast p180 reconstituted with yeast primase (7). The primase specific activity of trimeric human pol-prim was about half that of the tetramer (Fig. 2B). The polymerase specific activity of pol-prim trimer was also about half that of the tetramer when assayed on a poly(dA)-oligo(dT) template-primer (Fig. 2A). However, on a preprimed natural DNA template, the primer extension activity of the trimer was at least as great as that of the tetramer (Fig. 2C). The reason for the difference in primer extension activity of trimeric pol-prim on the two types of preprimed templates is not known. In any case, the data demonstrate that trimeric human pol-prim is not grossly deficient in enzymatic activity.
The p68 subunit is required for primer synthesis by human pol-prim in the presence of RPA and T antigen.
Trimeric pol-prim was essentially unable to initiate SV40 DNA replication, but its activity was significantly restored upon addition of purified intact p68 that had been expressed in either insect cells or bacteria (Fig. 3). Moreover, the activity of pol-prim trimer in T-antigen-mediated primer synthesis and elongation on RPA-saturated ssDNA template was also markedly reduced, but it could be largely rescued by addition of purified p68 (Fig. 4). Phosphorylation of p68 by cyclin A/cdk2 caused tetrameric pol-prim to behave much like trimeric pol-prim lacking p68 (Fig. 6). The data argue that p68 is critical both for initiation and for primer synthesis and elongation by pol-prim in the presence of RPA and T antigen, and that phosphorylation of p68 regulates both of these activities.
How does p68 promote pol-prim activity in the presence of T antigen and RPA? The p68 subunit interacts physically with the p180 subunit through its C terminus (40, 41) and with T antigen through its N terminus (8). Thus, one model is that the p68 subunit tethers the pol-prim complex to T antigen, which encircles the DNA as a processive DNA helicase, thereby promoting primer synthesis and elongation by pol-prim. Consistent with this idea, a monoclonal antibody against a C-terminal region of T antigen blocked pol-prim binding to T antigen, as well as stimulation of pol-prim activity by T antigen (9, 54, 66, 67). However, since T antigen binds to all four subunits of pol-prim (14, 24, 67), and since the activity of the trimer was stimulated by T antigen (Fig. 2D), p68 appears to be dispensable for this tethering. Thus, tethering is not fully satisfactory as an explanation for the p68 requirement, but it could represent part of the mechanism.
A second possible model is that p68 enhances the accessibility of the primase active site in the presence of RPA. It is intriguing that in the absence of RPA, the primase specific activity of the trimer on M13 ssDNA was about half that of the tetramer (Fig. 2B), while its primer elongation activity on M13 DNA was maintained (Fig. 2C). This observation suggests the possibility, that without p68, the polymerase active site of the trimer might be more accessible than the primase site. We speculate that this bias in favor of the polymerase active site may be exacerbated in the presence of RPA, so that p68 would be needed to sequester the p180 subunit and facilitate priming on RPA-saturated ssDNA.
A third possible model to explain the p68 requirement might be that coordinated T antigen binding to pol-prim and to RPA-saturated ssDNA orients pol-prim properly on the template for primer synthesis (6, 8, 9, 12-14, 67). As a hexamer, T antigen could conceivably bind simultaneously to RPA and to all four subunits of pol-prim (24, 66). T antigen was recently shown to modulate the binding mode of RPA on ssDNA, a functional interaction that may facilitate T-antigen-mediated priming by pol-prim on RPA-saturated ssDNA (51; R. D. Ott, T. Sidorova, L. Douthitt, Y. Wang, V. N. Podust, and E. Fanning, unpublished data). Since T antigen and primase bind to overlapping sites in RPA70 (6, 13, 50, 64), it is attractive to speculate that primase competes with T antigen for binding to the modified RPA-coated ssDNA. With the other subunits of pol-prim tethered to the T-antigen hexamer, p68 may be required as a loading factor to position pol-prim correctly, or to sequester the p180 active site as in the second model described above, to promote primer synthesis. This temporally and spatially coordinated binding of T antigen to RPA and pol-prim may allow RPA and pol-prim to switch places on the ssDNA, poising primase for action.
Does p68 regulate pol-prim activity in chromosomal DNA replication?
In yeast, the B subunit appears to execute an essential function before the hydroxyurea-sensitive step, but not later during S phase (19). Assuming that the B subunit serves a similar function in yeast and SV40 DNA replication, one might have expected defects in both initiation and elongation of yeast DNA replication after a shift to the nonpermissive temperature. Perhaps this mutant B subunit was defective only prior to its assembly in replication forks, where pol-prim interacts with other fork proteins, e.g., DNA polymerase ɛ (Pol ɛ), Dpb11, Cdc45, Sld2, and Sld3 (1, 2, 26, 27, 34, 35, 38, 39, 68). In that case, analysis of additional B subunit alleles might reveal defects in both initiation and elongation. On the other hand, chromosomal replication forks differ from those in SV40 replication. Pol ɛ is required for chromosomal DNA replication (15, 16, 45, 56, 63), but not for SV40 DNA replication (53, 70). Moreover, in viral DNA replication, T antigen assumes the roles of multiple cellular initiation proteins, including origin recognition and DNA unwinding (5). Thus, our analysis of the functions of p68 in SV40 replication may not accurately reflect those in chromosomal replication.
In spite of the differences between SV40 and chromosomal replication, regulation of p68/B subunit activity by cdk phosphorylation shows some similarity in the two systems. Phosphorylation of p68/B subunit by cyclin-dependent kinases is observed in yeast and human cells at the G1/S transition and increases with progression to G2/M (11, 18, 20, 49, 52, 60). However, B-subunit phosphorylation is delayed in yeast during the intra-S-phase checkpoint response in a Rad53-dependent manner (52). If cdk phosphorylation of yeast pol-prim blocks its primosome activity in the presence of RPA, as observed with human pol-prim, then preventing pol-prim phosphorylation at stalled replication forks probably contributes to Rad53-dependent stabilization of the forks after DNA damage (31, 57). The notion that primosome activity of yeast pol-prim is regulated in response to DNA damage is supported by the finding that RPA70 and p48 primase mutants are defective in responding to Rad53-dependent checkpoint signals (29, 30, 33).
Human pol-prim appears to be associated with a phosphatase, as well as with cyclin A/cdk2, in human cells (11), raising the question of whether phosphate turnover may regulate pol-prim activity during S phase. We speculate that as cyclin A/cdk2 activity rises during S phase, modification of the clustered phosphorylation sites in human p68 may increase progressively, shutting down pol-prim activity as DNA replication is completed. Activation of the intra-S-phase checkpoint in human cells (32, 44), if it prevents phosphorylation of pol-prim p68 as in yeast, would prolong the window of pol-prim primosome activity, presumably at origins of replication and during lagging strand replication. Indeed, if the genomic instability associated with SV40 infection activates the intra-S-phase checkpoint, it would be expected to enhance viral DNA replication.
Acknowledgments
We thank the past and present members of the Fanning lab for discussions, advice, and reagents, and Amy Altman and Steven Gray for comments on the manuscript.
The financial support of NIH grant GM52948 and Vanderbilt University is gratefully acknowledged.
REFERENCES
- 1.Aparicio, O. M., A. M. Stout, and S. P. Bell. 1999. Differential assembly of Cdc45 and DNA polymerases at early and late origins of DNA replication. Proc. Natl. Acad. Sci. USA 96:9130-9135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aparicio, O. M., D. M. Weinstein, and S. P. Bell. 1997. Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell 91:59-69. [DOI] [PubMed] [Google Scholar]
- 3.Arezi, B., and R. D. Kuchta. 2000. Eukaryotic DNA primase. Trends Biochem. Sci. 25:572-576. [DOI] [PubMed] [Google Scholar]
- 4.Arezi, B., B. W. Kirk, W. C. Copeland, and R. D. Kuchta. 1999. Interactions of DNA with human DNA primase monitored with photoactivatable cross-linking agents: implications for the role of the p58 subunit. Biochemistry 38:12899-12907. [DOI] [PubMed] [Google Scholar]
- 5.Baker, T. A., and S. P. Bell. 1998. Polymerases and the replisome: machines within machines. Cell 92:295-305. [DOI] [PubMed] [Google Scholar]
- 6.Braun, K. A., Y. Lao, Z. He, C. J. Ingles, and M. S. Wold. 1997. Role of protein-protein interactions in the function of replication protein A (RPA): RPA modulates the activity of DNA polymerase alpha by multiple mechanisms. Biochemistry 36:8443-8454. [DOI] [PubMed] [Google Scholar]
- 7.Brooke, R. G., and L. B. Dumas. 1991. Reconstitution of the Saccharomyces cerevisiae DNA primase-DNA polymerase protein complex in vitro. The 86 kDa subunit facilitates but is not required for complex formation. J. Biol. Chem. 266:10093-10098. [PubMed] [Google Scholar]
- 8.Collins, K. L., A. A. Russo, B. Y. Tseng, and T. J. Kelly. 1993. The role of the 70 kDa subunit of human DNA polymerase alpha in DNA replication. EMBO J. 12:4555-4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collins, K. L., and T. J. Kelly. 1991. Effects of T antigen and replication protein A on the initiation of DNA synthesis by DNA polymerase alpha-primase. Mol. Cell. Biol. 11:2108-2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Copeland, W. C., and T. S. F. Wang. 1993. Enzymatic characterization of the individual mammalian primase subunits reveals a biphasic mechanism for initiation of DNA replication. J. Biol. Chem. 268:26179-26189. [PubMed] [Google Scholar]
- 11.Dehde, S., G. Rohaly, O. Schub, H.-P. Nasheuer, W. Bohn, J. Chemnitz, W. Deppert, and I. Dornreiter. 2001. Two immunologically distinct human DNA polymerase α-primase subpopulations are involved in cellular DNA replication. Mol. Cell. Biol. 21:2581-2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dornreiter, I., A. Hoss, A. K. Arthur, and E. Fanning. 1990. SV40 T antigen binds directly to the large subunit of purified DNA polymerase alpha. EMBO J. 9:3329-3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dornreiter, I., L. F. Erdile, I. U. Gilbert, D. von Winkler, T. J. Kelly, and E. Fanning. 1992. Interaction of DNA polymerase alpha-primase with cellular replication protein A and SV40 T antigen. EMBO J. 11:769-776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dornreiter, I., W. C. Copeland, and T. S. F. Wang. 1993. Initiation of simian virus 40 DNA replication requires the interaction of a specific domain of human DNA polymerase alpha with large T antigen. Mol. Cell. Biol. 13:809-820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dua, R., D. Levy, and J. L. Campbell. 1999. Analysis of the essential functions of the C-terminal protein/protein interaction domain of Saccharomyces cerevisiae polɛ and its unexpected ability to support growth in the absence of the DNA polymerase domain. J. Biol. Chem. 274:22283-22288. [DOI] [PubMed] [Google Scholar]
- 16.D'Urso, G., and P. Nurse. 1997. Schizosaccharomyces pombe cdc 20+ encodes DNA polymerase ɛ and is required for chromosomal replication but not for the S phase checkpoint. Proc. Natl. Acad. Sci. USA 94:12491-12496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eki, T., T. Matsumoto, Y. Murakami, and J. Hurwitz. 1992. The replication of DNA containing the simian virus 40 origin by the monopolymerase and dipolymerase systems. J. Biol. Chem. 267:7284-7294. [PubMed] [Google Scholar]
- 18.Ferrari, M., G. Lucchini, P. Plevani, and M. Foiani. 1996. Phosphorylation of the DNA polymerase α-primase B subunit is dependent on its association with the p180 polypeptide. J. Biol. Chem. 271:8661-8666. [DOI] [PubMed] [Google Scholar]
- 19.Foiani, M., F. Marini, D. Gamba, G. Lucchini, and P. Plevani. 1994. The B subunit of the DNA polymerase alpha-primase complex in Saccharomyces cerevisiae executes an essential function at the initial stage of DNA replication. Mol. Cell. Biol. 14:923-933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Foiani, M., G. Liberi, G. Lucchini, and P. Plevani. 1995. Cell cycle-dependent phosphorylation and dephosphorylation of yeast DNA polymerase alpha-primase B subunit. Mol. Cell. Biol. 15:883-891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Foiani, M., G. Lucchini, and P. Plevani. 1997. The DNA polymerase α-primase complex couples DNA replication, cell-cycle progression and DNA-damage response. Trends Biochem. Sci. 22:424-427. [DOI] [PubMed] [Google Scholar]
- 22.Henricksen, L. A., C. B. Umbricht, and M. S. Wold. 1994. Recombinant replication protein A: expression, complex formation, and functional characterization. J. Biol. Chem. 269:11121-11132. [PubMed] [Google Scholar]
- 23.Herendeen, D., and T. J. Kelly. 1996. SV40 DNA replication, p. 29-65. In J. J. Blow (ed.), Eukaryotic DNA replication. Oxford University Press, New York, N.Y.
- 24.Huang, S.-G., K. Weisshart, I. Gilbert, and E. Fanning. 1998. Stoichiometry and mechanism of assembly of simian virus 40 T antigen complexes with the viral origin of DNA replication and DNA polymerase α-primase. Biochemistry 37:15345-15352. [DOI] [PubMed] [Google Scholar]
- 25.Huebscher, U., H.-P. Nasheuer, and J. E. Syvaoja. 2000. Eukaryotic DNA polymerases, a growing family. Trends Biochem. Sci. 25:143-147. [DOI] [PubMed] [Google Scholar]
- 26.Kamimura, Y., H. Masumoto, A. Sugino, and H. Araki. 1998. Sld2, which interacts with Dpb11 in Saccharomyces cerevisiae, is required for chromosomal DNA replication. Mol. Cell. Biol. 18:6102-6109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kamimura, Y., Y.-S. Tak, A. Sugino, and H. Araki. 2001. Sld3, which interacts with Cdc45 (Sld4), functions for chromosomal DNA replication in Saccharomyces cerevisiae. EMBO J. 20:2097-2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lohman, T. M., J. M. Green, and R. S. Beyer. 1986. Large scale overproduction and rapid purification of the Escherichia coli ssb gene product. Expression of the ssb gene under λPL control. Biochemistry 25:21-25. [DOI] [PubMed] [Google Scholar]
- 29.Longhese, M. P., H. Neecke, V. Paciotti, G. Lucchini, and P. Plevani. 1996. The 70 kDa subunit of replication protein A is required for the G1/S and intra-S DNA damage checkpoints in budding yeast. Nucleic Acids Res. 24:3533-3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Longhese, M. P., R. Fraschini, P. Plevani, and G. Lucchini. 1996. Yeast pip3/mec3 mutants fail to delay entry into S phase and to slow DNA replication in response to DNA damage, and they define a functional link between Mec3 and DNA primase. Mol. Cell. Biol. 16:3235-3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lopes, M., C. Cotta-Ramusino, A. Pellicioli, G. Liberi, P. Plevani, M. Muzi-Falconi, C. S. Newlon, and M. Foiani. 2001. The DNA replication checkpoint response stabilizes stalled replication forks. Nature 412:557-561. [DOI] [PubMed] [Google Scholar]
- 32.Mailand, N., J. Falck, C. Lukas, R. G. Syljuasen, M. Welcker, J. Bartek, and J. Lukas. 2000. Rapid destruction of Cdc25A in response to DNA damage. Science 288:1425-1429. [DOI] [PubMed] [Google Scholar]
- 33.Marini, F., A. Pellicioli, V. Paciotti, G. Lucchini, P. Plevani, D. F. Stern, and M. Foiani. 1997. A role for DNA primase in coupling DNA replication to DNA damage response. EMBO J. 16:639-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Masumoto, H., A. Sugino, and H. Araki. 2000. Dpb11 controls the association between DNA polymerase α and ɛ and the autonomously replicating sequence region of budding yeast. Mol. Cell. Biol. 20:2809-2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Masumoto, H., S. Muramatsu, Y. Kamimura, and H. Araki. 2002. S-Cdk-dependent phosphorylation of Sld2 essential for chromosomal DNA replication in budding yeast. Nature 415:651-655. [DOI] [PubMed] [Google Scholar]
- 36.Matsumoto, T., T. Eki, and J. Hurwitz. 1990. Studies on the initiation and elongation reactions in the simian virus 40 DNA replication system. Proc. Natl. Acad. Sci. USA 87:9712-9716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Melendy, T., and B. Stillman. 1993. An interaction between replication protein A and T antigen appears essential for primosome assembly during SV40 DNA replication. J. Biol. Chem. 268:3389-3395. [PubMed] [Google Scholar]
- 38.Mimura, S. T. Masuda, T. Matsui, and H. Takisawa. 2000. Central role for Cdc45 in establishing an initiation complex of DNA replication in Xenopus egg extracts. Genes Cells 5:439-452. [DOI] [PubMed] [Google Scholar]
- 39.Mimura, S., and H. Takisawa. 1998. Xenopus Cdc45-dependent loading of DNA polymerase α onto chromatin under the control of S-phase cdk. EMBO J. 17:5699-5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mizuno, T., K. Yamagishi, H. Miyazawa, and F. Hanaoka. 1999. Molecular architecture of the mouse DNA polymerase α-primase complex. Mol. Cell. Biol. 19:7886-7896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mizuno, T., N. Ito, M. Yokoi, A. Kobayashi, K. Tamai, H. Miyazawa, and F. Hanaoka. 1998. The second-largest subunit of the mouse DNA polymerase α-primase complex facilitates both production and nuclear translocation of the catalytic subunit of DNA polymerase α. Mol. Cell. Biol. 18:3552-3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mizuno, T., T. Okamoto, M. Yokoi, M. Izumi, A. Kobayashi, T. Hachiya, K. Tamai, T. Inoue, and F. Hanaoka. 1996. Identification of the nuclear localization signal of mouse DNA primase: nuclear transport of p46 subunit is facilitated by interaction with p54 subunit. J. Cell Sci. 109(2):627-2636. [DOI] [PubMed] [Google Scholar]
- 43.Moarefi, I. F., D. Small, I. Gilbert, M. Hoepfner, S. K. Randall, A. A. Russo, U. Ramsperger, A. K. Arthur, H. Stahl, T. J. Kelly, and E. Fanning. 1993. Mutation of the cyclin-dependent kinase phosphorylation site in simian virus 40 (SV40) large T antigen specifically blocks SV40 origin DNA unwinding. J. Virol. 67:4992-5002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Molinari, M., C. Mercurio, J. Dominguez, F. Goubin, and G. F. Draetta. 2000. Human Cdc25A inactivation in response to S phase inhibition and its role in preventing premature mitosis. EMBO Rep. 1:71-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morrison, A., H. Araki, A. B. Clark, R. K. Hamatake, and A. Sugino. 1990. A third essential DNA polymerase in S. cerevisiae. Cell 62:1143-1151. [DOI] [PubMed] [Google Scholar]
- 46.Murakami, Y., and J. Hurwitz. 1993. Functional interactions between SV40 T antigen and other replication proteins at the replication fork. J. Biol. Chem. 268:11008-11017. [PubMed] [Google Scholar]
- 47.Murakami, Y., and J. Hurwitz. 1993. DNA polymerase α stimulates the ATP-dependent binding of simian virus 40 tumor T antigen to the SV40 origin of replication. J. Biol. Chem. 268:11018-11027. [PubMed] [Google Scholar]
- 48.Murakami, Y., T. Eki, and J. Hurwitz. 1992. Studies on the initiation of simian virus 40 replication in vitro: RNA primer synthesis and its elongation. Proc. Natl. Acad. Sci. USA 89:952-956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nasheuer, H.-P., A. Moore, A. F. Wahl, and T. S. F. Wang. 1991. Cell cycle-dependent phosphorylation of human DNA polymerase alpha. J. Biol. Chem. 266:7893-7903. [PubMed] [Google Scholar]
- 50.Nasheuer, H.-P., D. von Winkler, C. Schneider, I. Dornreiter, I. Gilbert, and E. Fanning. 1992. Purification and functional characterization of bovine RP-A in an in vitro SV40 DNA replication system. Chromosoma 102:S52-S59. [DOI] [PubMed] [Google Scholar]
- 51.Ott, R. D., Y. Wang, and E. Fanning. 2002. Mutational analysis of the primosome activities of SV40 T antigen in viral DNA replication. J. Virol. 76:5121-5130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pellicioli, A., C. Lucca, G. Liberi, F. Marini, M. Lopes, P. Plevani, A. Romano, P. P. Di Fiore, and M. Foiani. 1999. Activation of Rad53 kinase in response to DNA damage and its effect in modulating phosphorylation of the lagging strand DNA polymerase. EMBO J. 18:6561-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pospiech, H., I. Kursula, W. Abdel-Aziz, L. Malkas, L. Uitto, M. Kastelli, M. Vihinen-Ranta, S. Eskelinen, and J. E. Syvaoja. 1999. A neutralizing antibody against human DNA polymerase ɛ inhibits cellular but not SV40 DNA replication. Nucleic Acids Res. 27:3799-3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schneider, C., K. Weisshart, L. A. Guarino, I. Dornreiter, and E. Fanning. 1994. Species-specific functional interactions of DNA polymerase α-primase with simian virus 40 (SV40) T antigen require SV40 origin DNA. Mol. Cell. Biol. 14:3176-3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schub, O., G. Rohaly, R. W. P. Smith, A. Schneider, S. Dehe, I. Dornreiter, and H.-P. Nasheuer. 2001. Multiple phosphorylation sites of DNA polymerase α-primase cooperate to regulate the initiation of DNA replication in vitro. J. Biol. Chem. 276:38076-38083. [DOI] [PubMed] [Google Scholar]
- 56.Sugino, A. 1995. Yeast DNA polymerases and their role at the replication fork. Trends Biochem. Sci. 20:319-323. [DOI] [PubMed] [Google Scholar]
- 57.Tercero, J. A., and J. F. X. Diffley. 2001. Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint. Nature 412:553-557. [DOI] [PubMed] [Google Scholar]
- 58.Tsurimoto, T., and B. Stillman. 1991. Replication factors required for SV40 DNA replication in vitro. I. DNA structure-specific recognition of a primer-template junction by eukaryotic DNA polymerases and their accessory proteins. J. Biol. Chem. 266:1950-1960. [PubMed] [Google Scholar]
- 59.Tsurimoto, T., and B. Stillman. 1991. Replication factors required for SV40 DNA replication in vitro. II. Switching of DNA polymerase α and δ during initiation of leading and lagging strand synthesis. J. Biol. Chem. 266:1961-1968. [PubMed] [Google Scholar]
- 60.Voitenleitner, C., C. Rehfuess, M. Hilmes, L. O'Rear, P. C. Liao, D. A. Gage, R. Ott, H.-P. Nasheuer, and E. Fanning. 1999. Cell cycle-dependent regulation of human DNA polymerase α-primase activity by phosphorylation. Mol. Cell. Biol. 19:646-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Voitenleitner, C., E. Fanning, and H.-P. Nasheuer. 1997. Phosphorylation of DNA polymerase alpha-primase by cyclin A-dependent kinases regulates initiation of DNA replication in vitro. Oncogene 14:1611-1615. [DOI] [PubMed] [Google Scholar]
- 62.Waga, S., and B. Stillman. 1998. The DNA replication fork in eukaryotic cells. Annu. Rev. Biochem. 67:721-751. [DOI] [PubMed] [Google Scholar]
- 63.Waga, S., T. Masuda, H. Takisawa, and A. Sugino. 2001. DNA polymerase ɛ is required for coordinated and efficient chromosomal DNA replication in Xenopus egg extracts. Proc. Natl. Acad. Sci. USA 98:4978-4983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Walther, A. P., M. J. Bjerke, and M. S. Wold. 1999. A novel assay for examining the molecular reactions at the eukaryotic replication fork: activities of replication protein A required during elongation. Nucleic Acids Res. 27:656-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang, T. S. F. 1996. Cellular DNA polymerases, p.461-493. In M. L. DePamphilis (ed.), DNA replication in eukaryotic cells. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 66.Weisshart, K., H. Forster, E. Kremmer, B. Schlott, F. Grosse, and H.-P. Nasheuer. 2000. Protein-protein interactions of the primase subunits p58 and p48 with simian virus 40 T antigen are required for efficient primer synthesis in a cell-free system. J. Biol. Chem. 275:17328-17337. [DOI] [PubMed] [Google Scholar]
- 67.Weisshart, K., P. Taneja, and E. Fanning. 1998. The replication protein A binding site in simian virus 40 (SV40) T antigen and its role in the initial steps of SV40 DNA replication. J. Virol. 72:9771-9781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wong, H., and S. J. Elledge. 1999. DRC1, DNA replication and checkpoint protein 1, functions with DPB11 to control DNA replication and the S-phase checkpoint in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 96:11791-11795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yuzhakov, A., Z. Kelman, J. Hurwitz, and M. O'Donnell. 1999. Multiple competition reactions for RPA order the assembly of the DNA polymerase δ holoenzyme. EMBO J. 18:6189-6199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zlotkin, T., G. Kaufmann, Y. Jiang, M. Y. W. T. Lee, L. Uitto, J. Syväoja, I. Dornreiter, E. Fanning, and T. Nethanel. 1996. DNA polymerase epsilon may be dispensable for SV40 but not for cellular DNA replication. EMBO J. 15:2298-2305. [PMC free article] [PubMed] [Google Scholar]