Abstract
Homologous recombination (HR) and nonhomologous end joining (NHEJ) play overlapping roles in repair of DNA double-strand breaks (DSBs) generated during the S phase of the cell cycle. Here, we characterized the involvement of HR and NHEJ in the rescue of DNA replication forks arrested or slowed by treatment of hamster cells with hydroxyurea or thymidine. We show that the arrest of replication with hydroxyurea generates DNA fragmentation as a consequence of the formation of DSBs at newly replicated DNA. Both HR and NHEJ protected cells from the lethal effects of hydroxyurea, and this agent also increased the frequency of recombination mediated by both homologous and nonhomologous exchanges. Thymidine induced a less stringent arrest of replication and did not generate detectable DSBs. HR alone rescued cells from the lethal effects of thymidine. Furthermore, thymidine increased the frequency of DNA exchange mediated solely by HR in the absence of detectable DSBs. Our data suggest that both NHEJ and HR are involved in repair of arrested replication forks that include a DSB, while HR alone is required for the repair of slowed replication forks in the absence of detectable DSBs.
DNA replication forks encounter lesions or sequence structures in the DNA template that may stall or collapse the replication complex (24, 25). Such events, in turn, lead to the formation of DNA ends and other lesions that are recognized by DNA repair proteins. Several repair pathways have been implicated in the reactivation of stalled replication forks. In particular, there is mounting evidence that genetic recombination plays a crucial role in this process (14). In Escherichia coli, blocked replication forks may reverse to form a Holliday junction (30, 43), which is an intermediate in reestablishment of the replication fork by homologous recombination (HR) or template switching (25, 34). To generate this Holliday junction intermediate, the RecG helicase unwinds the lagging or, less efficiently, the leading strands at the stalled replication fork (30, 31).
Another mechanism for the formation of a Holliday junction at replication forks in bacteria involves the RecA protein, which may bind to single-stranded regions on the lagging strand (13). These two pathways may be employed to bypass stalled replication forks through template switching or HR. In addition, the formation of a Holliday junction at replication forks is mediated by positive torsional strain at the progressing replication fork (36). Once the Holliday junction is formed, it can be processed and cleaved by RuvABC to produce a DNA double-strand break (DSB) (30, 43), which could become a substrate for HR.
In mammalian cells, it is well established that compounds that inhibit replication are potent inducers of sister chromatid exchange, gene amplification, and HR (3, 22, 27, 28, 39). DSBs formed specifically at replication forks are repaired by both HR and nonhomologous end joining (NHEJ), although HR seems to play a predominant role in such repair (1, 12, 41). It has also been suggested that NHEJ may be a part of an early response in the repair of DSBs at replication forks, while HR may be a part of a late response (41). However, relatively little is known of the molecular mechanism underlying this process. In Saccharomyces cerevisiae, Mus81 and Mms4/Eme1 form a heterodimeric structure-specific endonuclease that cleaves branched DNA structures resembling the Holliday junctions formed at stalled replication forks (9, 23). Recently, a human homologue of Mus81 has been reported. Since this enzyme is upregulated by treatment with replication inhibitors and cleaves structures resembling Holliday junctions, it has been suggested that Mus81 fulfills a similar function in human cells (10). These observations suggest that a number of steps involved in the formation and resolution of Holliday structures at arrested replication forks may be conserved from bacteria to humans.
Although both HR and NHEJ have been shown to play a role in the repair of damage in S phase or at replication forks (1, 12, 19, 41, 45), the nature of the lesions that compromise replication fork integrity in mammalian cells is not clear. In the present study, we used hydroxyurea and thymidine to disrupt progression of the replication fork. These agents are commonly used to synchronize growing cells in the S phase of the cell cycle. Hydroxyurea quenches the tyrosyl free radical in the active site of the M2 subunit of ribonucleotide reductase, thus depleting cells of several deoxyribonucleoside triphosphates (see reference 49 for a review). The disruption of DNA precursor supply by this agent completely stops the incorporation of nucleotides into DNA and arrests replication forks (6). Thymidine depletes cells of dCTP only. Replication forks in thymidine-treated cells slow substantially but do not stop incorporating nucleotides into DNA, causing retardation of replication (7).
Here, we focused on the lesions formed at mammalian cell replication forks arrested by treatment with these two agents. The data we present suggest that recombination substrates generated at stalled replication forks by thymidine treatment may be different from that generated by the more stringent hydroxyurea block. Furthermore, our data suggest that the thymidine-induced substrate may not contain a DSB.
MATERIALS AND METHODS
Cell culture.
The Sp5 and SPD8 cell lines were cultured in minimal essential medium containing Hanks' salts with the addition of 9% fetal calf serum and penicillin-streptomycin (90 U/ml) (HMEM) (1, 2). The AA8, CXR3, irs1SF, and V3-3 cell lines were cultured in Dulbecco's modified Eagle's medium, also containing 9% fetal calf serum and penicillin-streptomycin (90 U/ml).
Toxicity assay.
A total of 500 cells suspended in medium were plated onto a petri dish 4 h prior to the addition of thymidine or hydroxurea. Then 7 to 12 days later, when colonies could be observed, the colonies were fixed and stained with methylene blue in methanol (4 g/liter). Colonies consisting of more than 50 cells were subsequently counted.
Recombination assay.
The protocol for the recombination assay with Sp5 and SPD8 cells has been described previously (2, 17) and involves the following steps: 1.5 × 106 cells were inoculated into flasks 4 h prior to the initiation of treatment. Following treatment, the medium was removed, the flasks were rinsed three times with 10 ml of Hanks' balanced salt solution, and 30 ml of HMEM was then added. After incubation for an additional 48 h, the cells in the flasks were released by trypsinization and counted. HPRT+ revertants were selected by plating 3 × 105 treated cells per dish in the presence of HAsT (50 μM hypoxanthine, 10 μM l-azaserine, 5 μM thymidine). To determine cloning efficiency, two dishes were plated with 500 cells each. The colonies obtained were stained with methylene blue in methanol (4 g/liter), following 7 (in the case of cloning efficiency) or 10 (for reversion) days of incubation.
Characterization of HR event induced by thymidine and hydroxyurea.
Ten individual SPD8 revertants were isolated after treatment with thymidine (20 mM) or hydroxyurea (2 mM) and cultured in HAsT-containing medium for 14 days. PCR amplification and DNA sequencing were carried out as described earlier on genomic DNA extracted from each revertant (17, 18) in order to determine which copy of exon 7 had been restored, to demonstrate loss of the exon 7 duplication in SPD8 cells, and to confirm that the reversion observed involved HR.
Flow cytometry.
A total of 2 × 106 SPD8 cells were inoculated into flasks (75 cm2) 4 h prior to initiation of treatment with 0.5 mM hydroxyurea or 20 mM thymidine. After different periods of time, the medium in the flasks was removed, the cells were rinsed and fresh medium was added. After a subsequent 1-h treatment with 10 μM bromodeoxyuridine, the cells were released by trypsinization, fixed, and analyzed by flow cytometry as described elsewhere (1).
Immunofluorescence.
SPD8 cells were plated onto cover slips and treated with 5 mM thymidine or 0.2 mM hydroxyurea for 24 h. Following treatment, the medium was removed, and the cover slips were rinsed once in phosphate-buffered saline (PBS) (37°C) and fixed in 3% paraformaldehyde in PBS-T (PBS containing 0.1% Triton X-100) for 20 min. The cover slips were rinsed once in PBS-T prior to incubation with an antibody against Rad51 at a concentration of 1:1,000 for 16 h at 4°C (4). The cover slips were rinsed four times for 15 min each in PBS-T, followed by a 1-h incubation at room temperature with an indocarbocyanine-conjugated goat anti-rabbit immunoglobulin G (IgG) antibody (Zymed) at a concentration of 1:500 and then rinsed four times for 15 min each in PBS-T. Antibodies were diluted in PBS containing 3% bovine serum albumin. DNA was stained with 1 μg of To Pro (Molecular Probes) per ml. Cover slips were mounted with a SlowFade antifade kit (Molecular Probes).
Images were obtained with a Zeiss LSM 510 inverted confocal microscope with a Planapochromat 63X/NA 1.4 oil immersion objective and excitation wavelengths of 546 and 630 nm. Through-focus maximum projection images were acquired from optical sections 0.50 μm apart and with a section thickness of 1.0 μm. Images were processed with Adobe PhotoShop (Abacus Inc.).
The frequencies of cells containing Rad51 foci were determined in two separate experiments. At least 300 nuclei were counted in each slide. Nuclei containing more than 10 foci were classified as Rad51 positive.
Pulsed-field gel electrophoresis.
Flasks were inoculated with 5 × 106 cells 4 h prior to initiation of a 24-h treatment with thymidine or hydroxyurea. In the case of γ-ray treatment (137Cs, 10.6 Gy/min), the cells were melted into agarose inserts and irradiated. For labeling, flasks were inoculated either with 5 × 106 cells 4 h prior to a 30-min incubation with [3H]thymidine (20 nM, 18.5 kBq/ml) or with 2 × 106 cells 4 h prior to [14C]thymidine labeling (0.48 μM, 0.925 kBq/ml, for 24 h or medium for 23.5 h and 4.8 μM, 9.25 kBq/ml, for 0.5 h), followed by treatment with hydroxyurea or γ-rays. After 24 h of treatment, the cells in the flasks were released by trypsinization, and 106 cells were melted into each agarose insert.
The agarose inserts were incubated in 0.5 M EDTA-1% N-laurylsarcosyl-proteinase K (1 mg/ml) at 50°C for 48 h and thereafter washed four times in TE buffer prior to loading onto a 1% agarose (chromosomal grade) gel and separation by pulsed-field gel electrophoresis for 24 h (Bio-Rad; 120o angle, 60 to 240 s switch time, 4 V/cm). The gel was subsequently stained with ethidium bromide and analyzed with Image Gauge software (FLA-3000; Fujifilm).
In the case of [14C]thymidine labeling, the DNA was transferred from the gel to a nylon membrane according to the manufacturer's protocol (Hybond-N; Amersham Pharmacia Biotech). The membrane was then dried for 2 h at 80°C and exposed to a phosphoimager plate for 18 h before quantification by Image Gauge software (FLA-3000; Fujifilm). In experiments involving [3H]thymidine labeling, each lane on the gel was sliced into six pieces, which were subsequently melted in 0.5 ml of 12 M HCl at 50oC for 10 min. The resulting solution was neutralized with 0.5 ml of 12 M NaOH, after which 1 ml of distilled water and 20 ml of scintillation solution were added for counting in a liquid scintillator counter (Wallac 1409).
Apoptosis measurements.
Flasks were inoculated with 106 AA8 cells 4 h prior to initiation of a 24-h treatment with hydroxyurea (2 mM). Following treatment, the medium was removed, the flasks were rinsed three times with 10 ml of PBS, and 11 ml of medium was then added. At various time points, cells were trypsinized and resuspended in medium containing any floating cells from that sample. The cells were pelleted by centrifugation and resuspended for apoptosis analysis with fluorescein isothiocyanate (FITC)-conjugated annexin V antibody and propidium iodide (ApoTarget; Biosource International) according to the manufacturer's protocol. Samples were analyzed by flow cytometry (Becton Dickinson FACSort, 488-nm laser), and the percentage of apoptotic cells was determined by the fraction of live cells (propidium iodide negative) bound by FITC-conjugated annexin V antibody.
RESULTS
HR is required for survival following treatment with agents that arrest replication.
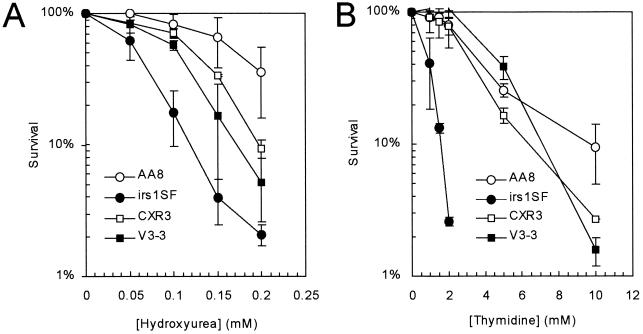
To investigate the involvement of HR and NHEJ in cellular responses to hydroxyurea and thymidine, we first determined the effects of these agents on the survival of recombination repair-deficient cell lines. We found that cells deficient in NHEJ (V3-3, which lacks DNA protein kinase catalytic subunit [DNA-PKcs] [8]) were slightly but significantly (P < 0.05) more sensitive to hydroxyurea than the corresponding wild-type cells (Fig. 1A), while cells deficient in HR (i.e., irs1SF, which is defective in XRCC3 [15, 26, 35, 48]) were very sensitive. In contrast, only HR-deficient irs1SF cells were very sensitive to thymidine (Fig. 1B). This sensitivity was corrected by the introduction of a cosmid containing a functional human XRCC3 gene (CXR3 [48]). The requirement for other components of the NHEJ machinery (i.e., ligase IV and Ku80) in response to hydroxyurea-induced arrest has been reported previously (41).
FIG. 1.
HR-deficient irs1SF cell line is sensitive to the toxic effects of both hydroxyurea and thymidine. Colony outgrowth of the Chinese hamster cell lines AA8 (wild type), irs1SF (deficient in HR protein XRCC3 [35]), CXR3 (irs1SF complemented with human XRCC3 [48]), and V3-3 (deficient in the NHEJ protein DNA-PKcs [8]) upon exposure to hydroxyurea (A) and thymidine (B) was determined. The means (symbols) and standard deviation (bars) of at least three experiments are shown.
Arrest of DNA replication by thymidine or hydroxyurea generates substrates for recombination.
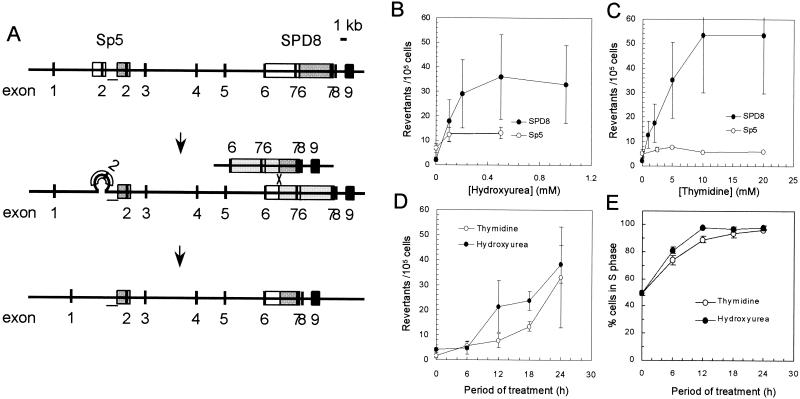
To determine whether replication fork abnormalities induced by hydroxyurea or thymidine provided substrates for recombination repair; we examined the effects of these agents on Chinese hamster cell lines containing endogenous targets for recombination (Fig. 2A). These cell lines have different partial duplications of the hprt gene, and a functional gene can be regained in these cells through reversions mediated by either HR (in SPD8 [17]) or nonhomologous recombination (in Sp5 [2]). The frequency of HPRT+ recombinants in SPD8 cells was increased 24-fold by thymidine and 16-fold by hydroxyurea treatment. In contrast, only hydroxyurea increased the frequency of HPRT+ recombinants (2-fold) in Sp5 cells (Fig. 2B and C; statistically significant, P < 0.05, t test).
FIG. 2.
HR is potently induced in the hprt gene by treatment of Chinese hamster cell lines with either hydroxyurea or thymidine. (A) Schematic illustration of the nonhomologous recombination events in the Sp5 and HR in the SPD8 cell lines that both lead to a functional hprt gene (2, 17). A white box designates a duplicated region, and a dark gray box designates the original region that was duplicated by the spontaneous mutation. Loss of the duplication in the SPD8 cell line involves HR between the repeated regions (17), while loss of the displaced duplication in Sp5 involves nonhomologous recombination (2). Involvement of HR in the Sp5 cell line can be excluded because the region between the two repeats (bar) is retained in the revertants (see reference 2 for details). The reversion frequencies of SPD8 and Sp5 cells were determined after a 24-h treatment with increasing doses of hydroxyurea (B) or thymidine (C). The means (symbols) and standard deviations (bars) of four independent experiments are shown. HR is continuously induced in cells fully arrested in the S phase of the cell cycle. (D) The reversion frequencies of SPD8 cells after treatment with hydroxyurea (0.5 mM) or thymidine (20 mM) for increasing periods of time. (E) Arrest of SPD8 cells in the S phase of the cell cycle after treatment with hydroxyurea (0.5 mM) or thymidine (20 mM) for increasing periods of time. The means (symbols) and standard deviations (bars) of three experiments are depicted.
To examine the S-phase dependence of the HR events seen in SPD8 cells following treatment with hydroxyurea or thymidine, we next measured the accumulation of recombinants in relation to time of exposure to hydroxyurea or thymidine. The frequency of HPRT+ revertants increased progressively throughout the 24-h period of treatment with either agent (Fig. 2D). However, SPD8 cells were fully arrested in S phase by thymidine or hydroxyurea after 12 h (Fig. 2E). Thus, recombinants formed by HR continued to accumulate even after the complete arrest of cells in the S phase of the cell cycle.
To characterize HR events induced by thymidine or hydroxyurea in SPD8 cells, hprt genes from 10 individual HPRT+ revertants were isolated and analyzed by employing a set of PCRs and DNA sequencing, as described previously (18). In all revertants, both introns 6 and 7 were the same size as in wild-type cells, and the downstream copy of exon 7 was restored (data not shown). This finding is consistent with reversion involving an exchange type of HR, i.e., the same mechanism by which spontaneous recombination occurs (17).
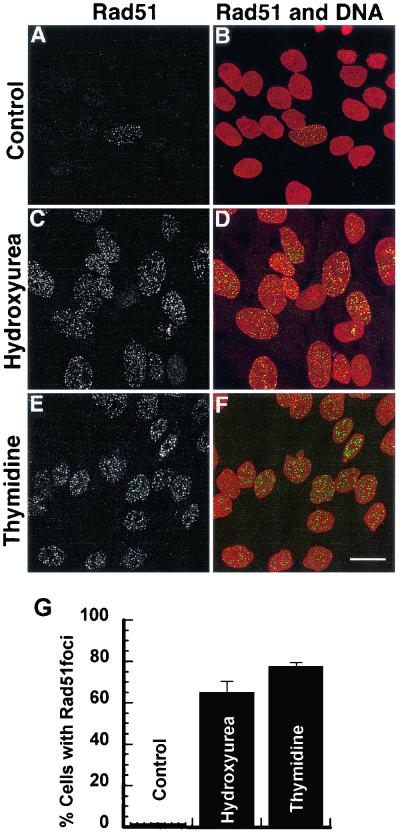
To further test the hypothesis that replication fork abnormalities induced by hydroxyurea or thymidine provided substrates for recombination repair; we examined Rad51 focus formation after treatment with these drugs. Rad51 foci occur spontaneously in cells in the S phase of the cell cycle (47) and have been shown to be located at single-stranded DNA regions after DNA damage (38) and in postreplicative chromatin (46). It is generally believed that these foci form at sites of recombinational DNA repair (38, 46). Treatment of SPD8 cells with hydroxyurea (0.2 mM) or thymidine (5 mM) for 24 h dramatically increased the fraction of cells forming Rad51 foci (Fig. 3). These results confirm earlier findings that hydroxyurea induces Rad51 foci (41) and also show that the retardation of replication following thymidine treatments provides substrates for recombination repair.
FIG. 3.
Induction of Rad51 foci in SPD8 cells following a 24-h treatment with equally toxic doses of hydroxyurea (0.2 mM) or thymidine (5 mM). (A, C, and E) Rad51 focus formation in SPD8 cells and (B, D, and F) merged images of anti-Rad51 labeling (green) and DNA counterstaining (red). Scale bar, 20 μm. (G) Percentage of cells with >10 Rad51 foci. At least 300 nuclei were counted for each treatment and experiment. Error bars designate standard error.
Induction of homologous recombination is not related to the level of DSBs following replication arrest.
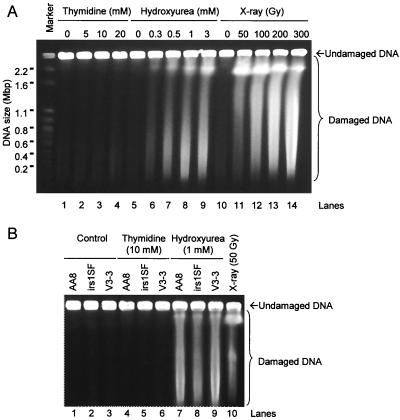
On the basis of these experiments, it appeared that both HR and NHEJ were required for the repair of damage resulting from a hydroxyurea-induced replication block, whereas HR alone was sufficient to rescue cells from the effects of the less complete arrest induced by thymidine. One hypothesis that can explain these results is that the two agents may generate different types of substrates for recombination at the arrested replication forks. Given that DSBs have been reported to be associated with stalled replication forks in Escherichia coli and Saccharomyces cerevisiae (32, 33), we next determined whether hydroxyurea or thymidine induced such lesions. Analysis of DNA prepared from recombination-proficient AA8 Chinese hamster ovary (CHO) cells treated with hydroxyurea by pulsed-field gel electrophoresis revealed extensive fragmentation of genomic DNA (Fig. 4A, lanes 5 to 9). In contrast, no such fragmentation was evident following exposure to doses of thymidine giving the same level of toxicity (Fig. 4A, lanes 1 to 4).
FIG. 4.
DSBs are induced in Chinese hamster cells exposed to hydroxyurea but not to thymidine. (A) Visualization by pulsed-field gel electrophoresis of DSBs in AA8 cells after treatment with thymidine (lanes 2 to 4), hydroxyurea (lanes 6 to 9), or γ-rays (lanes 11 to 14). The chromosomes from S. cerevisiae were used as size markers. The γ-ray dosages employed were at least 10 times as cytotoxic as the concentrations of hydroxyurea used to induce DSBs. Equally toxic concentrations of thymidine and hydroxyurea were used. (B) DNA fragmentation in wild-type AA8 cells, HR-deficient irs1SF cells (XRCC3−), and NHEJ-deficient V3-3 cells (DNA-PKcs−) after treatment with thymidine (lanes 4 to 6) or hydroxyurea (lanes 7 to 9). AA8 cells treated with 50 Gy (lane 10) were used for comparison.
DSBs were also induced by hydroxyurea treatments of Sp5 or SPD8 and CHO cells deficient in NHEJ (V3-3) or HR (irs1SF) (Fig. 4B). This indicates that production of DSBs by hydroxyurea is independent of the efficiency of recombination. Also, the same dose of hydroxyurea produced similar levels of DSBs in all cell lines (Fig. 4B), although this dose is considerably more toxic to irs1SF cells. No increases in DSBs were detected following thymidine treatment of any of these cell lines (Fig. 4B).
Hydroxyurea-induced DSBs are formed at newly replicated DNA.
To determine whether hydroxyurea-induced DSBs detected by pulsed-field gel electrophoresis are substrates for recombination, it was necessary to investigate whether these DSBs were actually associated with replication. Newly replicated DNA in AA8 cells was pulse labeled for 30 min with [14C]thymidine, while total cellular DNA in parallel cultures was labeled similarly but for 24 h. Both cultures were subsequently treated with hydroxyurea for 24 h or exposed to ionizing radiation. Analysis of DNA from these cells by pulsed-field gel electrophoresis revealed that the DNA fragments released by hydroxyurea-induced DSBs contained a high proportion of newly replicated DNA (Fig. 5B). The DNA fragments released following γ-irradiation contained only a very small proportion of newly replicated DNA consistent with the random induction of DSBs by this agent (11).
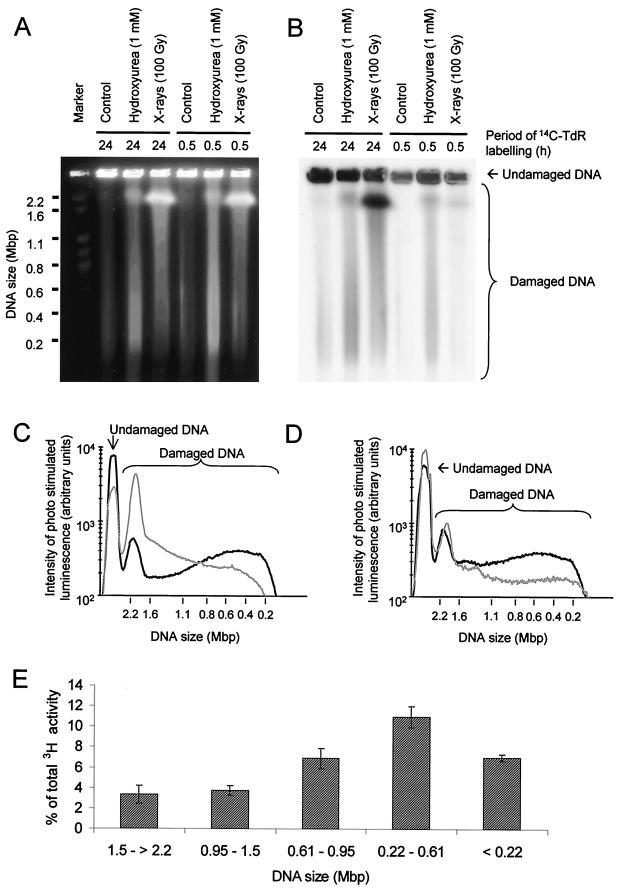
FIG. 5.
Hydroxyurea-induced DSBs are formed in newly replicated DNA. The DNA of AA8 cells was labeled with [14C]thymidine (14C-TdR), either homogeneously for 24 h or specifically at sites of replication for 30 min, prior to exposure to hydroxyurea or γ-rays. DNA was separated by pulsed-field gel electrophoresis and visualized by ethidium bromide staining (A) and autoradiography (B). The size distributions of DNA fragments released from AA8 cells labeled for 24 h (C) or 30 min (D) with [14C]thymidine and subsequently treated with hydroxyurea (1 mM; black line) or γ-rays (100 Gy; gray line) were also determined, and 42% ± 2% and 51% ± 5% of total [14C]thymidine-labeled DNA from 24-h- and 30-min-labeled cells, respectively, were released from the inserts by hydroxyurea treatment. The corresponding values for γ-ray treatments were 73% ± 4% (24 h) and 29% ± 3% (30 min). (E) Size distribution of DNA fragments with 3H activity released from inserts containing cells labeled with [3H]thymidine for 30 min and subsequently treated with hydroxyurea (1 mM).
In cells uniformly labeled with [14C]thymidine for 24 h, hydroxyurea released 42% ± 2% of the DNA from the plug in the form of DNA fragments that mainly range from 0.2 to 0.6 Mbp in size (Fig. 5C). A markedly different size distribution was found in fragments released following γ-irradiation (100 Gy), which constituted 73% ± 4% of the DNA released from the plug. In this case, the majority of DNA fragments were more than 2.2 Mbp (Fig. 5C). In cells labeled for 30 min with [14C]thymidine, 51% ± 5% of label was released from plugs of hydroxyurea-treated cells. Those fragments had a size distribution similar to that found in uniformly labeled cells (Fig. 5D). In contrast, γ-irradiation (100 Gy) only released 29% ± 3% of the pulse-labeled DNA from the plug. This low level of release following γ-irradiation may be explained by the specific incorporation of a high proportion of the [14C]thymidine at active replication forks into replicons that form bubble structures (Fig. 6). Such structures will be obstructed in their movement through an agarose gel (21).
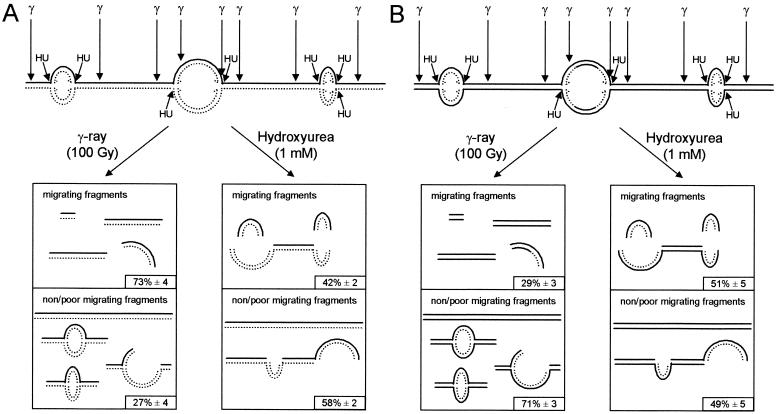
FIG. 6.
Linear DNA fragments at sites of replication are released by hydroxyurea-induced DSBs. Schematic illustration of DNA fragments released in pulsed-field gel electrophoresis following treatment with γ-rays (100 Gy) or hydroxyurea (1 mM) in cells that had been labeled with [14C]thymidine for 24 h (A) or 30 min (B). A smaller portion of DNA fragments, labeled at sites of replication, were released in pulsed-field gel electrophoresis by γ-rays, because the label is incorporated in replicons, which migrate poorly (5, 21). Hydroxyurea treatment released a high portion of DNA fragments, labeled at sites of replication, by converting replicons into linear fragments by DSBs at or close to replication forks. The solid line represents unlabeled DNA, and the dotted line represents [14C]thymidine-labeled DNA.
To further evaluate the DNA fragments produced by hydroxyurea-induced DSBs, we determined their sizes. AA8 cells were labeled with [3H]thymidine for 30 min prior to a 24-h exposure to hydroxyurea. DNA from treated cells was fractionated by pulsed-field gel electrophoresis, and the [3H]thymidine incorporated into DNA fragments of different sizes was quantitated (Fig. 5E). This analysis revealed that the most abundant DNA fragments generated by hydroxyurea-induced DSBs were 0.22 to 0.61 Mbp in size, which corresponds to the size of a single replicon or a few contiguous replicons (16).
DSBs induced by hydroxyurea are not a consequence of induction of apoptosis.
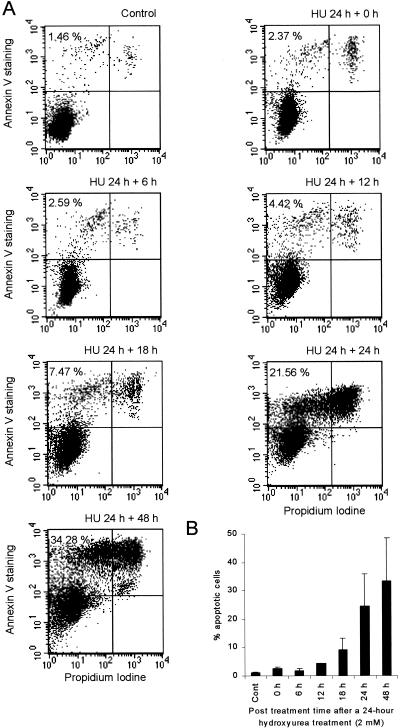
An alternative explanation for the DSBs induced by hydroxyurea is that they are generated during an apoptotic response to treatment. To test this possibility, we measured the onset of apoptosis at various times following a 24-h exposure of AA8 cells to hydroxyurea. We detected apoptotic cells with the annexin V assay, which detects the membrane phospholipid phosphatidylserine, which is translocated from the inner to the outer leaflet of the plasma membrane in cells that enter apoptosis (29). Generally, this assay is thought to detect an early event in apoptosis (29), while DNA fragmentation is a late event (42). Elevated levels of apoptotic cells were not detected by this assay until 18 to 24 h after the 24-h hydroxyurea treatment (2 mM; Fig. 7). Thus, the DNA fragmentation seen directly following a 24-h hydroxyurea treatment of AA8 cells in the pulse-field gel electrophoresis was not the result of the induction of apoptosis.
FIG. 7.
Time of onset of apoptosis in AA8 cells after a 24-h hydroxyurea treatment (2 mM). Control cells were untreated, and apoptosis was assessed by annexin V staining. (A) Dot plot on cells stained with FITC-conjugated annexin V antibody and propidium iodide. Percent apoptotic cells was determined by the fraction of live cells (propidium iodide negative) bound with FITC-conjugated annexin V antibody. (B) The mean percentages of apoptotic cells (columns) and standard deviations (error bars) of four to six experiments are shown.
DISCUSSION
The restoration of stalled replication forks is essential to maintain efficient DNA synthesis. Here, we characterized lesions that are formed following replication arrest in mammalian cells and the recombination repair pathways that are employed in the subsequent repair of such lesions. To arrest replication, we used thymidine or hydroxyurea. Both of these agents efficiently arrest cells in the S phase of the cell cycle by depletion of one or several of the deoxyribonucleoside triphosphate precursors of DNA synthesis. The cytotoxicity of these agents is likely to be the result of the accumulation of lesions induced by them at replication forks. Here, we show that cells deficient in either HR or NHEJ are hypersensitive to the toxic effects of hydroxyurea, while only cells deficient in HR are sensitive to thymidine. These findings suggest that HR is necessary for the survival of cells in which DNA replication has been arrested by exposure to hydroxyurea or thymidine and that NHEJ has a role only in connection with hydroxyurea-induced arrest.
In support of this suggestion, we found that hydroxyurea induced both homologous and nonhomologous recombination, while thymidine induced HR alone in the hprt gene of Chinese hamster cells (Fig. 2). These observations indicate that a substrate for both homologous and nonhomologous recombination is generated following a hydroxyurea-induced replication block, whereas thymidine treatment produces substrates for HR alone.
It is interesting that the effects of both of these agents on recombination become saturated at high concentrations. This might be explained by a saturation of the recombination machinery available for this repair and/or a limitation in the rate of the initiating event (e.g., the number of active replication forks). The accumulation of recombinants in cells arrested in S phase indicates that both of these agents induced HR in the S phase of the cell cycle (Fig. 2D and E). Given our further observation that thymidine induces Rad51 foci (Fig. 3), we propose that HR may resolve a substrate at the single-stranded DNA regions where the Rad51 foci form (38, 46).
It was not surprising that both HR and NHEJ were required for the repair of the hydroxyurea-induced replication block, since both of these pathways have previously been shown to repair DSBs in the S phase of the cell cycle or at replication forks (1, 12, 41, 45). The novel observation was that a functional NHEJ pathway was not required for the repair and subsequent survival of thymidine-induced replication arrest (Fig. 1). Since thymidine treatment does not completely block replication fork progression (7), we tested the hypothesis that HR alone may resolve a different set of DNA replication abnormalities induced by this treatment.
It is well established that HR repairs DSBs, particularly those that occur in the S phase of the cell cycle (1, 12, 45). Our observation that hydroxyurea is much more efficient than γ-rays in the release of DNA labeled at sites of replication indicates that hydroxyurea-induced DSBs convert replicon bubbles into linear DNA fragments that migrate into agarose gels (5, 50). γ-Rays induce DSBs everywhere (i.e., mostly between bubbles). Thus, only γ-ray-treated DNA will have intact bubbles and hence be retarded in the gels. Therefore, our data support the argument that hydroxyurea-induced DSBs are associated with or close to replication forks (Fig. 6).
Our data further demonstrate that hydroxyurea induces homologous and nonhomologous recombination and formation of Rad51 foci. In addition, we found that both HR and NHEJ are required for survival of cells with replication forks stalled by hydroxyurea treatment. Therefore, our results support a role for both HR and NHEJ in repair following complete arrest of replication. Our data also raise the possibility that NHEJ is recruited for repair at replication forks only when a DSB is present, since thymidine (which does not induce DSBs) is no more toxic to NHEJ-deficient than wild-type cells and does not induce nonhomologous recombination.
A potential function for NHEJ in this respect could be to join DSBs that occur at two adjacent replication forks. Newly replicated DNA may be lost by DSBs generated by hydroxyurea at adjacent replication forks. In this situation, NHEJ may be necessary to rejoin the DNA ends.
In contrast to the DSBs induced by hydroxyurea, we could not detect DSBs in cells treated with equally toxic doses of thymidine. Thus, there was no relationship between induction of HR and the level of DSBs formed at stalled replication forks following treatment with equally toxic doses of thymidine. The level of fragmentation detected following a 3 mM hydroxyurea treatment corresponds to that found following a 50-Gy dose of ionizing radiation, which in turn corresponds to ∼2,000 DSBs per cell (11, 37). Given that the pulsed-field gel electrophoresis analysis used in this experiment can detect as few as 20 DSBs per cell (11), our results show that hydroxyurea produces at least 100-fold more DSBs than equally toxic doses of thymidine. DSBs were not even detected in cells deficient in HR (irs1SF), which are hypersensitive to thymidine. Therefore, it appears that the toxic lesion formed following thymidine treatment is not a transient or persisting DSB. Considering our findings that thymidine or hydroxyurea induced the same frequency of HR and Rad51 foci, we suggest that replication forks arrested by thymidine treatment may generate different substrates for recombination than the forks arrested by hydroxyurea treatment. Furthermore, these structures may not contain DSBs.
An alternative explanation is that the DSBs formed after thymidine treatment are very rapidly repaired and thus would have a very short half-life. However, in mammalian cells, NHEJ is thought to be a fast repair pathway and HR a slower process (Fig. 2D) (41, 51). Thus, it is unlikely that HR would quickly repair a DSB generated by thymidine. Instead, we speculate that DSBs may not form because the replication complex does not dissociate from the replication fork following thymidine treatment as nucleotides are still incorporated (7). In contrast, hydroxyurea treatment may result in dissociation of the replication complex, leading to the formation of DSBs. Thus, the presence of a replication complex could protect the cell from the formation of DSBs at the replication forks.
An interesting observation is that the frequency of recombinants formed by HR reached a plateau at 0.3 mM hydroxyurea (Fig. 2B), while chromosome fragmentation increased continuously up to 3 mM. This could indicate that hydroxyurea-induced HR is already saturated at low levels of DSBs or that DSBs induced at replication forks are not related to induction of recombination. However, the endpoints in the assays are different. DSBs are measured at one given moment, while recombinants accumulate over 3 days. Therefore, the role of HR in the repair of DSBs induced by hydroxyurea was not resolved by our experiments.
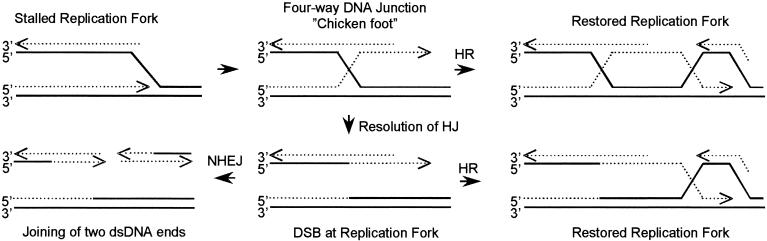
Although we do not know the exact nature of the substrates produced by thymidine or hydroxyurea, a model originally proposed for template switching by Higgins and coworkers (20) appears to offer potential candidates. An identical model has also been proposed for replication fork reactivation in bacteria (25, 34) (Fig. 8). This model suggests that an arrested replication fork may reverse to form a four-way DNA junction (including a Holliday junction). In bacteria, this process involves the RecG helicase (30, 31, 44) or RecA (13) and is mediated by positive torsional strain at the progressing replication fork (36). Such structures are processed and cleaved by RuvABC to produce DSBs (13, 34, 43).
FIG. 8.
Model for the role of HR and NHEJ at stalled replication forks in mammalian cells (adapted from a model in reference 20). Inhibition of the progression of a replication fork may allow annealing of the nascent leading to the lagging strand, resulting in formation of a four-way DNA junction, also called a chicken foot. We propose that such a four-way DNA junction tail, which does not contain a DSB, can be recognized as a substrate for HR at slowed replication forks following thymidine treatment. Resolution of the Holliday junction present at the four-way junction may occur at completely arrested replication forks and would result in a DSB, which could serve as a substrate for both HR and NHEJ. The solid line represents template DNA, and the dotted line represents nascent DNA.
In S. cerevisiae, Holliday junctions have also been shown to be present in cells in the S phase of the cell cycle and DNA polymerase α and δ mutants showed an increased level of Holliday junctions (53). The fate of these substrates in S. cerevisiae have become clearer following recent reports of a resolvase complex that includes Mus81 and Eme1 or Mms4 and cleaves Holliday junctions or branched DNA (9, 23). On the basis of the data presented in the present study, we suggest that the DNA double-stranded tail, possibly formed at a four-way junction following impairment of replication fork progression by excess thymidine (perhaps also by hydroxyurea), may serve as a substrate for HR. Although this substrate would include a DNA end, it would not include a DSB. However, this DNA end may be used as a substrate for HR, which in turn could restore the replication fork (Fig. 8).
A similar mechanism has been proposed for the restoration of replication forks in bacteria (25, 34). It is possible that the restoration of a replication fork without the induction of a DSB would be very advantageous for mammalian cells, considering that even a single DSB may be lethal (40). Our finding that the arrest of replication is a potent inducer of HR and to a less extent nonhomologous recombination (Fig. 3) (3, 52) provides further support for this assumption.
In conclusion, the present investigation provides evidence that HR and NHEJ have different roles following replication arrest in mammalian cells. Our data indicate that different recombinogenic substrates may be formed at replication forks slowed by treatment with thymidine or those completely arrested by hydroxyurea. The characterization of the recombination substrates at stalled replication forks and determination of the proteins involved in the resolution of the individual substrates will be the subject for future investigations.
Acknowledgments
We thank L Thompson for providing the AA8, CXR3, irs1SF, and V3-3 cell lines, Stephen West and Fiona Benson for their generous gift of the anti-RAD51 antibody, A Goldman and C Sanders for valuable discussions, and Agenta Önfelt for use of microscopy equipment.
This investigation was supported financially by the Swedish Cancer Foundation, the Lawski Foundation, the Swedish Radiation Protection Authority, the Swedish National Board for Laboratory Animals, and Yorkshire Cancer Research.
REFERENCES
- 1.Arnaudeau, C., C. Lundin, and T. Helleday. 2001. DNA double-strand breaks associated with replication forks are predominantly repaired by homologous recombination involving an exchange mechanism. J. Mol. Biol. 307:1235-1245. [DOI] [PubMed] [Google Scholar]
- 2.Arnaudeau, C., L. Rozier, C. Cazeaux, M. Defais, D. Jenssen, and T. Helleday. 2001. RAD51 supports spontaneous nonhomologous recombination in mammalian cells, but not the corresponding process induced by topoisomerase inhibitors. Nucleic Acids Res. 29:662-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arnaudeau, C., E. Tenorio Miranda, D. Jenssen, and T. Helleday. 2000. Inhibition of DNA synthesis is a potent mechanism by which cytostatic drugs induce homologous recombination in mammalian cells. Mutat. Res. 461:221-228. [DOI] [PubMed] [Google Scholar]
- 4.Benson, F. E., A. Stasiak, and S. C. West. 1994. Purification and characterisation of the human Rad51 protein, an analogue of E.coli RecA. EMBO J. 13:5764-5771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beverley, S. M. 1989. Estimation of circular DNA size with gamma-irradiation and pulsed-field gel electrophoresis. Anal. Biochem. 177:110-114. [DOI] [PubMed] [Google Scholar]
- 6.Bianchi, V., E. Pontis, and P. Reichard. 1986. Changes of deoxyribonucleoside triphosphate pools induced by hydroxyurea and their relation to DNA synthesis. J. Biol. Chem. 261:16037-16042. [PubMed] [Google Scholar]
- 7.Bjursell, G., and P. Reichard. 1973. Effects of thymidine on deoxyribonucleoside triphosphate pools and deoxyribonucleic acid synthesis in Chinese hamster ovary cells. J. Biol. Chem. 248:3904-3909. [PubMed] [Google Scholar]
- 8.Blunt, T., N. J. Finnie, G. E. Taccioli, G. C. Smith, J. Demengeot, T. M. Gottlieb, R. Mizuta, A. J. Varghese, F. W. Alt, and P. A. Jeggo. 1995. Defective DNA-dependent protein kinase activity is linked to V(D)J recombination and DNA repair defects associated with the murine scid mutation. Cell 80:813-823. [DOI] [PubMed] [Google Scholar]
- 9.Boddy, M. N., P.-H.L. Gaillard, W. H. McDonald, P. Shanahan, J. R. Yates, and P. Russell. 2001. Mus81-Eme1 are essential components of a Holliday junction resolvase. Cell 107:537-548. [DOI] [PubMed] [Google Scholar]
- 10.Chen, X.-B., R. Melchionna, C.-M. Denis, P.-H. L. Gaillard, A. Blasina, I. Van de Weyer, M. N. Boddy, P. Russell, J. Vialard, and C. H. McGowan. 2001. Human Mus81-associated endonuclease cleaves Holliday junctions in vitro. Mol. Cell 8:1117-1127. [DOI] [PubMed] [Google Scholar]
- 11.Erixon, K., and B. Cedervall. 1995. Linear induction of DNA double-strand breakage with X-ray dose, as determined from DNA fragment size distribution. Radiat. Res. 142:153-162. [PubMed] [Google Scholar]
- 12.Essers, J., H. van Steeg, J. de Wit, S. M. Swagemakers, M. Vermeij, J. H. Hoeijmakers, and R. Kanaar. 2000. Homologous and nonhomologous recombination differentially affect DNA damage repair in mice. EMBO J. 19:1703-1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flores, M. J., H. Bierne, S. D. Ehrlich, and B. Michel. 2001. Impairment of lagging strand synthesis triggers the formation of a RuvABC substrate at replication forks. EMBO J. 20:619-629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flores-Rozas, H., and R. D. Kolodner. 2000. Links between replication, recombination and genome instability in eukaryotes. Trends Biochem. Sci. 25:196-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fuller, L. F., and R. B. Painter. 1988. A Chinese hamster ovary cell line hypersensitive to ionizing radiation and deficient in repair replication. Mutat. Res. 193:109-121. [DOI] [PubMed] [Google Scholar]
- 16.Hand, R. 1978. Eucaryotic DNA: organization of the genome for replication. Cell 15:317-325. [DOI] [PubMed] [Google Scholar]
- 17.Helleday, T., C. Arnaudeau, and D. Jenssen. 1998. A partial hprt gene duplication generated by nonhomologous recombination in V79 Chinese hamster cells is eliminated by homologous recombination. J. Mol. Biol. 279:687-694. [DOI] [PubMed] [Google Scholar]
- 18.Helleday, T., R. Nilsson, and D. Jenssen. 2000. Arsenic[III] and heavy metal salts induce intrachromosomal homologous recombination in the hprt gene in V79 Chinese hamster cells. Environ. Mol. Mutagen. 35:114-122. [DOI] [PubMed] [Google Scholar]
- 19.Hendrickson, E. A. 1997. Cell-cycle regulation of mammalian DNA double-strand-break repair. Am. J. Hum. Genet. 61:795-800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Higgins, N. P., K. Kato, and B. Strauss. 1978. A model for replication repair in mammalian cells. J. Mol. Biol. 101:417-425. [DOI] [PubMed] [Google Scholar]
- 21.Iliakis, G. E., O. Cicilioni, and L. Metzger. 1991. Measurement of DNA double-strand breaks in CHO cells at various stages of the cell cycle with pulsed field gel electrophoresis: calibration by means of 125I decay. Int. J. Radiat. Biol. 59:343-357. [DOI] [PubMed] [Google Scholar]
- 22.Ishii, Y., and M. A. Bender. 1980. Effects of inhibitors of DNA synthesis on spontaneous and ultraviolet light-induced sister-chromatid exchanges in Chinese hamster cells. Mutat. Res. 79:19-32. [DOI] [PubMed] [Google Scholar]
- 23.Kaliraman, V., J. R. Mullen, W. M. Fricke, S. A. Bastin-Shanower, and S. J. Brill. 2001. Functional overlap between Sgs1-Top3 and the Mms4-Mus81 endonuclease. Genes Dev. 15:2730-2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kogoma, T. 1996. Recombination by replication. Cell 85:625-627. [DOI] [PubMed] [Google Scholar]
- 25.Kuzminov, A. 2001. DNA replication meets genetic exchange: chromosomal damage and its repair by homologous recombination. Proc. Natl. Acad. Sci. USA 98:8461-8468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu, N., J. E. Lamerdin, R. S. Tebbs, D. Schild, J. D. Tucker, M. R. Shen, K. W. Brookman, M. J. Siciliano, C. A. Walter, W. Fan, L. S. Narayana, Z. Q. Zhou, A. W. Adamson, K. J. Sorensen, D. J. Chen, N. J. Jones, and L. H. Thompson. 1998. XRCC2 and XRCC3, new human Rad51-family members, promote chromosome stability and protect against DNA cross-links and other damages. Mol. Cell 1:783-793. [DOI] [PubMed] [Google Scholar]
- 27.Ma, C., S. Martin, B. Trask, and J. L. Hamlin. 1993. Sister chromatid fusion initiates amplification of the dihydrofolate reductase gene in Chinese hamster cells. Genes Dev. 7:605-620. [DOI] [PubMed] [Google Scholar]
- 28.Mariani, B. D., and R. T. Schimke. 1984. Gene amplification in a single cell cycle in Chinese hamster ovary cells. J. Biol. Chem. 259:1901-1910. [PubMed] [Google Scholar]
- 29.Martin, S. J., C. P. Reutelingsperger, A. J. McGahon, J. A. Rader, R. C. van Schie, D. M. LaFace, and D. R. Green. 1995. Early redistribution of plasma membrane phosphatidylserine is a general feature of apoptosis regardless of the initiating stimulus: inhibition by overexpression of Bcl-2 and Abl. J. Exp. Med. 182:1545-1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McGlynn, P., and R. G. Lloyd. 2000. Modulation of RNA polymerase by (p)ppGpp reveals a RecG-dependent mechanism for replication fork progression. Cell 101:35-45. [DOI] [PubMed] [Google Scholar]
- 31.McGlynn, P., and R. G. Lloyd. 2001. Rescue of stalled replication forks by RecG: simultaneous translocation on the leading and lagging strand templates supports an active DNA unwinding model of fork reversal and Holliday junction formation. Proc. Natl. Acad. Sci. USA 98:8227-8234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Merrill, B. J., and C. Holm. 1999. A requirement for recombinational repair in Saccharomyces cerevisiae is caused by DNA replication defects of mec1 mutants. Genetics 153:595-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Michel, B., S. D. Ehrlich, and M. Uzest. 1997. DNA double-strand breaks caused by replication arrest. EMBO J. 16:430-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Michel, B., M.-J. Fores, E. Viguera, G. Grompone, M. Seigneur, and V. Bidnenko. 2001. Rescue of arrested replication forks by homologous recombination. Proc. Natl. Acad. Sci. USA 98:8181-8188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pierce, A. J., R. D. Johnson, L. H. Thompson, and M. Jasin. 1999. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 13:2633-2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Postow, L., C. Ullsperger, R. W. Keller, C. Bustamante, A. V. Vologodskii, and N. R. Cozzarelli. 2001. Positive torsional strain causes the formation of a four-way junction at replication forks. J. Biol. Chem. 276:2790-2796. [DOI] [PubMed] [Google Scholar]
- 37.Prise, K. M., G. Ahnstrom, M. Belli, J. Carlsson, D. Frankenberg, J. Kiefer, M. Lobrich, B. D. Michael, J. Nygren, G. Simone, and B. Stenerlow. 1998. A review of DSB induction data for varying quality radiations. Int. J. Radiat. Biol. 74:173-184. [DOI] [PubMed] [Google Scholar]
- 38.Raderschall, E., E. I. Golub, and T. Haaf. 1999. Nuclear foci of mammalian recombination proteins are located at single-stranded DNA regions formed after DNA damage. Proc. Natl. Acad. Sci. USA 96:1921-1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rainaldi, G., M. R. Sessa, and T. Mariani. 1984. Inhibitors of DNA synthesis induce sister chromatid exchanges at the early S phase of the cell cycle. Chromosoma 90:46-49. [DOI] [PubMed] [Google Scholar]
- 40.Rich, T., R. L. Allen, and A. H. Wyllie. 2000. Defying death after DNA damage. Nature 407:777-783. [DOI] [PubMed] [Google Scholar]
- 41.Saintigny, Y., F. Delacote, G. Vares, F. Petitot, S. Lambert, D. Averbeck, and B. S. Lopez. 2001. Characterization of homologous recombination induced by replication inhibition in mammalian cells. EMBO J. 20:3861-3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sakahira, H., M. Enari, and S. Nagata. 1998. Cleavage of CAD inhibitor in CAD activation and DNA degradation during apoptosis. Nature 391:96-99. [DOI] [PubMed] [Google Scholar]
- 43.Seigneur, M., V. Bidnenko, S. D. Ehrlich, and B. Michel. 1998. RuvAB acts at arrested replication forks. Cell 95:419-430. [DOI] [PubMed] [Google Scholar]
- 44.Singleton, M. R., S. Scaife, and D. B. Wigley. 2001. Structural analysis of DNA replication fork reversal by RecG. Cell 107:79-89. [DOI] [PubMed] [Google Scholar]
- 45.Takata, M., M. S. Sasaki, E. Sonoda, C. Morrison, M. Hashimoto, H. Utsumi, Y. Yamaguchi-Iwai, A. Shinohara, and S. Takeda. 1998. Homologous recombination and nonhomologous end-joining pathways of DNA double-strand break repair have overlapping roles in the maintenance of chromosomal integrity in vertebrate cells. EMBO J. 17:5497-5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tashiro, S., J. Walter, A. Shinohara, N. Kamada, and T. Cremer. 2000. Rad51 accumulation at sites of DNA damage and in postreplicative chromatin. J. Cell Biol. 150:283-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tashiro, S., N. Kotomura, A. Shinohara, K. Tanaka, K. Ueda, and N. Kamada. 1996. S phase specific formation of the human Rad51 protein nuclear foci in lymphocytes. Oncogene 12:2165-2170. [PubMed] [Google Scholar]
- 48.Tebbs, R. S., Y. Zhao, J. D. Tucker, J. B. Scheerer, M. J. Siciliano, M. Hwang, N. Liu, R. J. Legerski, and L. H. Thompson. 1995. Correction of chromosomal instability and sensitivity to diverse mutagens by a cloned cDNA of the XRCC3 DNA repair gene. Proc. Natl. Acad. Sci. USA 92:6354-6358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thelander, L., and P. Reichard. 1979. Reduction of ribonucleotides. Annu. Rev. Biochem. 48:133-158. [DOI] [PubMed] [Google Scholar]
- 50.van der Bliek, A. M., C. R. Lincke, and P. Borst. 1988. Circular DNA of 3T6R50 double minute chromosomes. Nucleic Acids Res. 16:4841-4851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang, H., Z. C. Zeng, T. A. Bui, E. Sonoda, M. Takata, S. Takeda, and G. Iliakis. 2001. Efficient rejoining of radiation-induced DNA double-strand breaks in vertebrate cells deficient in genes of the RAD52 epistasis group. Oncogene 20:2212-2224. [DOI] [PubMed] [Google Scholar]
- 52.Zhang, L. H., and D. Jenssen. 1994. Studies on intrachromosomal recombination in SP5/V79 Chinese hamster cells upon exposure to different agents related to carcinogenesis. Carcinogenesis 15:2303-2310. [DOI] [PubMed] [Google Scholar]
- 53.Zou, H., and R. Rothstein. 1997. Holliday junctions accumulate in replication mutants via a RecA homolog-independent mechanism. Cell 90:87-96. [DOI] [PubMed] [Google Scholar]