Abstract
Upon interferon (IFN) stimulation, Stat1 becomes tyrosine phosphorylated and translocates into the nucleus, where it binds to DNA to activate transcription. The activity of Stat1 is dependent on tyrosine phosphorylation, and its inactivation in the nucleus is accomplished by a previously unknown protein tyrosine phosphatase (PTP). We have now purified a Stat1 PTP activity from HeLa cell nuclear extract and identified it as TC45, the nuclear isoform of the T-cell PTP (TC-PTP). TC45 can dephosphorylate Stat1 both in vitro and in vivo. Nuclear extracts lacking TC45 fail to dephosphorylate Stat1. Furthermore, the dephosphorylation of IFN-induced tyrosine-phosphorylated Stat1 is defective in TC-PTP-null mouse embryonic fibroblasts (MEFs) and primary thymocytes. Reconstitution of TC-PTP-null MEFs with TC45, but not the endoplasmic reticulum (ER)-associated isoform TC48, rescues the defect in Stat1 dephosphorylation. The dephosphorylation of Stat3, but not Stat5 or Stat6, is also affected in TC-PTP-null cells. Our results identify TC45 as a PTP responsible for the dephosphorylation of Stat1 in the nucleus.
A wide variety of cytokines and growth factors activate intracellular signaling events involving Janus kinases (JAKs) and signal transducers and activators of transcription (STATs). Ligand activation of receptor-associated JAKs leads to tyrosine phosphorylation of the receptor chains, creating docking sites for STATs. In turn, STATs become phosphorylated on tyrosine, dimerize, and translocate to the nucleus to activate transcription (8). A precise regulation of both the magnitude and duration of JAK activity and of STAT activation is essential for the cytokine orchestration of many biological processes, and dysregulation of the JAK-STAT pathway has pathological implications (4, 33). Negative regulation of the JAK-STAT signaling pathway occurs through several distinct mechanisms. Cessation of signaling from the cell surface occurs through degradation of the receptor-ligand complex via the ubiquitin-proteosome pathway (5, 13) and through induction of the suppressor of cytokine signaling (SOCS) family of proteins, which inhibit the activity of JAKs (14). In addition, several protein tyrosine phosphatases (PTPases), including SHP-1, SHP-2, CD45, and PTP-1B, can dephosphorylate either cytokine receptors or JAKs (17, 23, 33). PTPases that can dephosphorylate STATs in the cytoplasm have also been described (1, 36). In the nucleus, PIAS (protein inhibitor of activated STAT) proteins can inhibit the transcriptional activity of STATs (25). In addition, previous studies have demonstrated the existence of a nuclear phosphatase activity that can inactivate Stat1, the founding member of the STAT family. Once dephosphorylated, Stat1 is rapidly exported back into the cytoplasm and takes part in subsequent activation-inactivation cycles (3, 9, 12, 13, 22, 26). The identity of the nuclear Stat1 phosphatase, however, had remained uncovered. Other STATs also undergo rapid activation-inactivation cycles (5, 20, 35), suggesting that nuclear STAT dephosphorylation may be a general phenomenon.
Here we report the purification of a nuclear Stat1 phosphatase, which we identified as TC45, the nuclear isoform of the ubiquitously expressed T-cell PTP (TC-PTP). We show that TC45 can dephosphorylate Stat1 in vitro and in vivo. Consistently, analysis of TC-PTP-null mouse embryonic fibroblasts (MEFs) revealed impaired dephosphorylation of Stat1 in the nucleus. The dephosphorylation of Stat3, but not Stat5 or Stat6, was also affected in TC-PTP-null cells.
MATERIALS AND METHODS
Plasmid constructs.
pGEX-Stat1 was constructed by inserting the Stat1α cDNA into pGEX2T. pATH-v-Abl contains the kinase domain of v-Abl as a trpE fusion gene and a Tetr gene. The coding regions of the human TC45, mouse TC45, and mouse TC48 cDNAs were PCR amplified from IMAGE consortium clones 1894955, 604362, and 3602983, respectively, and cloned into pGEX4T-1, pEBB, pCDNA3, pBabepuro, and/or pBabepuroFlag.
Cell culture and reagents.
293T and U2OS cells were grown in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (FBS) and PenStrep. DNA transfections were performed by the CaPO4 method, and clones stably expressing hTC45 were selected in 0.4 mg of G418 per ml. TC-PTP wild-type (7+/+, 11+/+) and TC-PTP-null (4−/−, 14−/−) MEF cell lines were maintained in DMEM containing 10% FBS as described previously (16). 4−TC45 and 4−C were derived from 4−/− cells after infection with retrovirus containing pBabepuro-hTC45 and pBabepuro, respectively, followed by single-colony selection in 2.5 μg of puromycin per ml. Retrovirus was prepared by transfection of amphotropic φNX cells and utilized as described previously (24). 14−TC45∗ and 14−TC48∗ pools were established by retroviral infection using pBabepuroFlag-mTC45 and pBabepuroFlag-mTC48, respectively, and analyzed after 1 to 2 weeks of puromycin selection. Human gamma interferon (IFN-γ) (a gift from Amgen) and murine IFN-γ (Peprotech) were used at 5 ng/ml. Staurosporine (Sigma) was used at 0.25 to 0.5 μM as indicated.
Bacterial expression and purification of tyrosine-phosphorylated Stat1.
pGEX-Stat1 and pATH-v-Abl plasmids were cotransformed into the bacterial strain RR1 and selected on plates containing ampicillin, tetracycline, and 20 μg of tryptophan per ml (W). Expression of the glutathione S-transferase (GST)-Stat1 fusion protein was induced as previously described (29), but with the following modifications. An overnight culture of RR1 (containing the pGEX-STAT1 and pATH-v-Abl plasmids) grown in Luria broth supplemented with ampicillin (100 μg/ml), tetracycline (100 μg/ml), and W (20 μg/ml) was diluted 1:50 into 1 liter of the same medium and grown for 3 h at 30°C followed by another 1.5 h in the presence of 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG). The cells were then pelleted, resuspended in 1 liter of modified M9 medium (1× M9 salts, 0.5% [wt/vol] Casamino Acids [Sigma A-2427], 0.1 mM CaCl2, 0.2% glucose, 10 μg of thiamine B1 per ml) with IPTG, but without W, to induce the expression of the TrpE-v-Abl fusion protein (18), and cultured for another 1.5 h at 30°C. The cells were pelleted, resuspended in 20 ml of ice-cold lysis buffer (phosphate-buffered saline [PBS], 50 mM EDTA, 1% Triton X-100, 1 mM dithiothreitol [DTT], 0.2 mM phenylmethylsulfonyl fluoride [PMSF], 1 μg of pepstatin A per ml) and sonicated. GST-p-Stat1 was bound to glutathione agarose beads (Sigma), and after extensive washing eluted in elution buffer (10 mM glutathione, 50 mM Tris [pH 8.0], 100 mM NaCl, 1 mM EDTA, 1 mM DTT, 0.1% NP-40, 1 μg of aprotinin per ml). The fusion protein was cleaved overnight at 4°C by thrombin and stored at −70°C in the presence of 15% glycerol.
PTP assays.
p-Stat1 was diluted to 1 μg/ml in PTP buffer (25 mM Tris [pH 7.5], 5 mM DTT, 1 mg of bovine serum albumin [BSA] per ml, 2.5 mM EDTA, 1 mM EGTA, 1 μg of aprotinin per ml, 1 μg of leupeptin per ml, 1 mM benzamidine, 10 μM tosylsulfonyl phenylalanyl chloromethyl ketone, 1 mM PMSF, with or without 0.75 mM orthovanadate). Ten microliters of this substrate solution was mixed with 5 μl of HeLa cell nuclear extract or subsequent chromatographic fractions (with or without prior dialysis, depending on the salt concentration) and incubated for 60 min at 30°C. The reaction products were analyzed by Western blotting with anti-p-Stat1(Y701) antibodies.
Purification of a PTP activity from HeLa cell nuclear extracts.
All purification steps were carried out at 4°C with an automated fast-protein liquid chromatography station (Biologic; Bio-Rad). A HeLa cell nuclear extract, prepared according to the method of Dignam et al. (10) from 32 liters of suspension culture (obtained from Cell Culture Center, National Institutes of Health), was dialyzed against buffer A (25 mM HEPES [pH 7.8], 5% glycerol, 1 mM EDTA, 1 mM DTT, 0.5 mM PMSF) containing 120 mM NaCl and loaded onto a 25-ml DEAE Sepharose Fast Flow column (XK26/20; Pharmacia). PTP activity against p-Stat1 was detected in the flowthrough, to which ammonium sulfate was added to 1.4 M. The cleared extract was loaded onto a 30-ml phenyl Sepharose high-performance column (C16/20; Pharmacia), and bound proteins were eluted with a linear gradient of 1.4 to 0 M ammonium sulfate in buffer A. Collected fractions were assayed for PTP activity, and active fractions eluting at 1.0 to 0.9 M ammonium sulfate were pooled and dialyzed against buffer B (50 mM HEPES [pH 7.5], 5% glycerol, 1 mM EDTA, 1 mM DTT, 0.2 mM PMSF) containing 50 mM NaCl. This was loaded onto a 5-ml Blue Sepharose Fast Flow column (C10/10; Pharmacia), and bound proteins were eluted with a linear gradient of 50 mM to 2.5 M NaCl in buffer B. The active fractions eluting from 1.1 to 1.8 M NaCl were dialyzed against buffer A containing 1.1 M ammonium sulfate and loaded onto a 1-ml Pharmacia Resource-Phenyl column. Bound proteins were eluted with a linear gradient of 1.1 to 0 M ammonium sulfate in buffer A. Active fractions eluting from 0.83 to 0.65 M ammonium sulfate were dialyzed against buffer C (50 mM NaPO4 [pH 7.0], 10% glycerol, 0.5 mM EDTA, 1 mM MgCl2, 0.067% NP-40, 1 mM DTT, 0.2 mM PMSF) containing 25 mM NaCl, and loaded onto a 1-ml MonoS HR5/5 column (Pharmacia). Bound proteins were eluted with a linear gradient of 25 mM to 0.5 M NaCl in buffer C. Fractions of 0.25 ml were collected and assayed for PTP activity. Fractions S40 through S52 eluting at 160 to 185 mM NaCl were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (10% polyacrylamide) and silver staining according to the manufacturer's instructions (Bio-Rad).
Protein analysis.
Whole-cell extracts were prepared as described (26). Cytoplasmic and nuclear fractions were prepared as described previously (10), except buffers contained 0.3% NP-40 and cells were lysed by gentle pipetting. Protein concentrations were determined with Coomassie Plus reagent (Bio-Rad). Proteins were separated by SDS-PAGE, transferred to nitrocellulose, and visualized with primary and secondary antibodies and enhanced chemiluminescence (Amersham). The following primary antibodies were used: anti-p-Stat1(Y701), anti-p-Stat3(Y705), anti-p-Stat5(Y694), and anti-p-Stat6(Y641) (Cell Signaling Technology); anti-Flag antibody M2 (IBI); anti-TC-PTP monoclonal antibodies CF4 (Oncogene Research Products) and 3E2 (described in reference 16); and antiactin, anti-Stat3, anti-Stat5, and anti-Stat6 antibodies (Santa Cruz). Stat1 antibodies were a gift from J. Darnell.
Immunodepletion.
Four hundred microliters of dialyzed HeLa cell nuclear extract was incubated four times for 45 min each with either 0.5 μg of normal mouse immunoglobulin G (IgG) (Santa Cruz) or the anti-TC-PTP monoclonal antibody CF4 and 5 μl of protein A/G agarose beads (Oncogene Research Products).
Electrophoretic mobility shift assay.
K562 cell extract and p-Stat1 were incubated with a 32P-labeled Stat1-binding site of the human FcγRI promoter as described previously (26).
RESULTS
Establishment of an in vitro Stat1 PTPase assay.
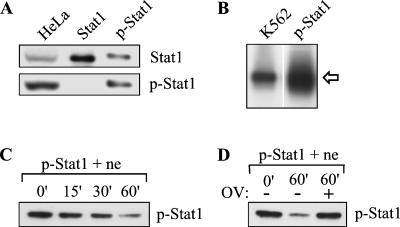
In order to identify the nuclear Stat1 phosphatase, we set out to purify the enzymatic activity by biochemical fractionation with tyrosine-phosphorylated Stat1 as a substrate in a PTPase assay. Tyrosine-phosphorylated Stat1 (p-Stat1) was generated and purified from bacteria coexpressing Stat1 and the kinase domain of v-Abl. Analysis of the purified p-Stat1 protein by Western blotting with a phospho-Stat1(Y701)-specific antibody demonstrated that it was phosphorylated on tyrosine 701, the functionally important tyrosine residue (Fig. 1A) (27). p-Stat1 was also able to bind DNA, as demonstrated by mobility gel shift assay with a 32P-labeled Stat1 binding site (Fig. 1B). In these assays, p-Stat1 showed no significant difference when compared to the endogenous tyrosine-phosphorylated Stat1 present in extracts from IFN-γ-stimulated HeLa or K562 cells (Fig. 1A and B).
FIG. 1.
Establishment of an in vitro Stat1 PTPase assay. (A) Bacterially produced p-Stat1 and Stat1 (∼8 ng) were analyzed by Western blotting with anti-Stat1 (top) or anti-p-Stat1(Y701) antibody (bottom). An extract from IFN-γ-stimulated HeLa cells (50 μg) was included as a control. (B) Bacterially produced p-Stat1 and an extract from IFN-γ-stimulated K562 cells were analyzed by gel mobility shift assay with a Stat1 binding sequence. The arrow indicates the Stat1 homodimer-DNA complex. (C) In vitro Stat1 PTPase assay. Bacterially produced p-Stat1 and 2.5 μl of HeLa cell nuclear extract (ne) were incubated at 30°C for various periods as indicated. The reaction mixtures were analyzed by Western blotting with anti-p-Stat1(Y701) antibody. (D) In vitro Stat1 PTPase assay performed as in panel C with inclusion of 0.5 mM sodium orthovanadate (OV) as indicated.
Next, we established an in vitro assay to detect the activity of the nuclear Stat1 phosphatase with p-Stat1 as the substrate. p-Stat1 was incubated at 30°C with HeLa cell nuclear extract, and dephosphorylation of p-Stat1 was examined by Western blotting with the phospho-Stat1(Y701)-specific antibody. Dephosphorylation of p-Stat1 occurred in a time-dependent manner under these conditions (Fig. 1C). The decreased level of Stat1 phosphorylation was due to a tyrosine phosphatase activity, since the inclusion of the tyrosine phosphatase inhibitor orthovanadate prevented the dephosphorylation of p-Stat1 (Fig. 1D).
Purification and identification of a nuclear Stat1 phosphatase.
Using the established in vitro PTPase assay, we purified a Stat1 PTPase to near homogeneity from HeLa cell nuclear extract through a series of chromatographic fractionations (Fig. 2A). Silver-staining analysis of fractions from the MonoS column revealed the presence of a 45-kDa protein, the abundance of which was in correlation with the detected Stat1 PTPase activity (Fig. 2B, top and middle panels), suggesting that this protein may be the enzyme responsible for the observed Stat1 dephosphorylation.
FIG. 2.
Purification and identification of a nuclear Stat1 phosphatase. (A) Schematic diagram of the chromatographic columns used to purify the Stat1 phosphatase activity. (B) Analysis of MonoS fractions. (Top panel) Western blot analysis of reaction mixtures from in vitro PTPase assays by using anti-p-Stat1 antibody. The samples analyzed include MonoS fractions S40 through S52 and the pooled fractions from the Resource Phenyl column (R33-41). —, p-Stat1 input. (Middle panel) Silver-staining analysis of the same MonoS fractions (15 μl/lane). A band of about 45 kDa (the presence of which correlates with the PTPase activity detected, as shown in the upper panel) is indicated by an arrow. (Bottom panel) Analysis of the same MonoS fractions (2.5 μl/lane) by Western blotting with the TC-PTP monoclonal antibody CF4. (C) Western blot analysis of 45 μg of HeLa whole-cell extract (wce), 45 μg of nuclear extract (ne), and 1 μl of MonoS fraction 46 (S46) with anti-TC-PTP antibody. (D) Analysis of TC45-immunodepleted HeLa nuclear extract. (Top panel) Anti-TC-PTP Western blot of HeLa nuclear extracts depleted with normal mouse IgG (ne-IgG) or the anti-TCPTP monoclonal CF4 (ne-TCPTP). (Bottom panels) Western blot analysis of a PTP assay performed with the immunodepleted nuclear extracts. The anti-p-Stat1 blot was stripped and reprobed with a Stat1 antibody.
We explored the possibility that the purified 45-kDa protein may be an already known nuclear PTPase. The human TC-PTP gene encodes two distinct protein products as a result of alternative splicing: TC45 (or TC-PTPa), a 45-kDa protein located mainly in the nucleus, and TC48 (or TC-PTPb), a 48-kDa endoplasmic reticulum (ER)-associated protein (6, 7, 15, 21). To test whether the purified 45-kDa protein is TC45, protein blot analysis of the MonoS fractions was performed with a monoclonal antibody against TC-PTP. The 45-kDa protein in these fractions was specifically recognized by the anti-TC-PTP antibody (Fig. 2B, bottom panel). While both TC45 and TC48 were detected in HeLa whole-cell extract, only TC45 was present in the nuclear fraction from which it was subsequently purified (Fig. 2C).
To confirm that TC45 is indeed responsible for the dephosphorylation of Stat1, TC45 was removed from HeLa nuclear extract by using anti-TC-PTP antibody (Fig. 2D, top panel). p-Stat1 was not dephosphorylated when incubated with the TC45-depleted nuclear extract. In contrast, p-Stat1 was dephosphorylated by nuclear extract that had been pretreated with control anti-IgG antibody (Fig. 2D, bottom panel). These results demonstrate the specificity of the in vitro Stat1 PTPase assay and indicate that TC45 is the phosphatase present in HeLa nuclear extract that is responsible for the dephosphorylation of Stat1.
TC45 can dephosphorylate Stat1 in vitro and in vivo.
To test if TC45 can directly dephosphorylate Stat1, we conducted an in vitro PTPase assay with p-Stat1 and bacterially produced GST-TC45 immobilized on glutathione beads. p-Stat1 was readily dephosphorylated by GST-TC45 (Fig. 3A). Dephosphorylation of p-Stat1 by GST-TC45 was inhibited by orthovanadate, demonstrating the specificity of the reaction.
FIG. 3.
TC45 can dephosphorylate Stat1 in vitro and in vivo. (A) Purified GST-TC45 dephosphorylates p-Stat1 in vitro. p-STAT1 was incubated with bacterially produced GST or GST-TC45, in the absence or presence of 0.5 mM orthovanadate (OV), and analyzed by Western blotting with anti-p-Stat1 or anti-Stat1 antibody. (B) TC45 dephosphorylates Stat1 in transiently transfected 293T cells. 293T cells transfected with pCMV-FlagStat1 (0.5 μg) and increasing amounts of pEBB-TC45 (10 to 200 ng) were stimulated 48 h posttransfection with IFN-γ for 20 min. Protein extracts were analyzed by Western blotting with anti-p-Stat1, anti-Flag, or anti-TC-PTP antibody as indicated. (C) Reduced IFN-γ-induced Stat1 phosphorylation in an U2OS cell line stably overexpressing TC45. Parental U2OS cells and U2OS-TC45 cells were stimulated with IFN-γ for various time periods. Protein extracts were analyzed as in panel B.
To examine if TC45 can affect IFN-γ-induced Stat1 phosphorylation in vivo, 293T cells were cotransfected with Flag-Stat1 and increasing amounts of TC45. Transfected cells were then treated with IFN-γ and harvested for protein blot analysis. Coexpression of Flag-Stat1 with TC45 resulted in a dose-dependent decrease in the levels of IFN-γ-induced tyrosine-phosphorylated Stat1 (Fig. 3B). In addition, a human osteosarcoma cell line stably overexpressing TC45 (U2OS-TC45) was established. Protein extracts from U2OS-TC45 and the parental U2OS cells treated with IFN-γ for various periods of time were prepared and analyzed by protein blotting with anti-p-Stat1 antibody. The amount of IFN-γ-induced tyrosine-phosphorylated Stat1 in the U2OS-TC45 cell line was reduced over the entire course of treatment compared to that of the parental cell line (Fig. 3C). These data suggest that TC45 can dephosphorylate tyrosine-phosphorylated Stat1 in vitro and in vivo.
Analysis of Stat1 signaling in wild-type and TC-PTP-null MEFs.
We next used TC-PTP-null MEFs to confirm the role of TC45 in Stat1 dephosphorylation. Wild-type (7+/+) and TC-PTP−/− (4−/−) MEF cells were treated with IFN-γ for various periods, and whole-cell extracts were analyzed by Western blotting with anti-p-Stat1 antibody. In the 7+/+ cells, the level of tyrosine-phosphorylated Stat1 reached a maximum at 15 min of IFN-γ stimulation and then gradually declined thereafter (Fig. 4A). In contrast, in the mutant 4−/− cells, tyrosine-phosphorylated Stat1 continued to accumulate and persisted at higher levels for at least 2 h of IFN-γ stimulation. Since the overall level of tyrosine-phosphorylated Stat1 is determined by the balance of phosphorylation and dephosphorylation events, prolonged Stat1 phosphorylation in TC-PTP−/− cells may result from either an increase in JAK kinase activity or a decrease in phosphatase activity towards Stat1. To specifically monitor the rate of Stat1 dephosphorylation, we employed a previously described pulse-chase strategy (12, 13). Staurosporine, a protein kinase inhibitor, was added to cells pretreated with IFN-γ, blocking the continuous phosphorylation of Stat1 by JAKs. Residual levels of preactivated Stat1 were then determined at several later time points. In wild-type 7+/+ cells, Stat1 was almost completely dephosphorylated after a 20-min staurosporine chase (or 30 min of IFN-γ treatment). In contrast, tyrosine dephosphorylation of Stat1 was completely blocked in the mutant 4−/− cells (Fig. 4B). Fractionation of wild-type and mutant MEF cells treated for 30 min with IFN-γ followed by a staurosporine chase revealed defective Stat1 dephosphorylation in both the nuclear and cytoplasmic fractions (Fig. 4C). Similar results were obtained when an independent pair of TC-PTP wild-type (11+/+) and mutant (14−/−) MEF cell lines were analyzed (Fig. 4C). A defect in Stat1 dephosphorylation after IFN-β stimulation was also observed (data not shown).
FIG. 4.
Analysis of Stat1 signaling in wild-type and TC-PTP-null MEFs. (A) Wild-type 7+/+ and TC-PTP-null 4−/− MEFs were stimulated with murine IFN-γ for various periods. Whole-cell lysates were analyzed by Western blotting with anti-p-Stat1 antibody. The stripped blots were reprobed with an anti-Stat1 antibody detecting both Stat1α and Stat1β. (B) 7+/+ and 4−/− MEFs were stimulated with IFN-γ for various periods with the addition of 0.5 μM staurosporine (Sigma) after 10 min of IFN-γ treatment. Whole-cell lysates were analyzed as in panel A. (C) 7+/+ and 4−/− MEF cells were stimulated with IFN-γ for 30 min, followed by a staurosporine chase (0.25 μM) for another 30 or 60 min. Cytoplasmic and nuclear extracts were analyzed by Western blotting with anti-p-Stat1 and antiactin antibodies, as indicated. Similar experiments were also performed with a different pair of wild-type and TC-PTP-null MEF cell lines (11+/+ and 14−/−). (D) p-Stat1 was incubated with dialyzed nuclear extracts of unstimulated 7+/+ and 4−/− cells in this in vitro PTPase assay and analyzed by anti-p-Stat1 Western blotting. The stripped blot was reprobed with a Stat1 antibody. —, dialysis buffer.
Nuclear extracts from wild-type 7+/+ and mutant 4−/− MEFs were also examined for their ability to dephosphorylate p-Stat1 in vitro. Consistently, nuclear extract from wild-type 7+/+ cells, but not mutant 4−/− cells, was able to dephosphorylate p-Stat1 (Fig. 4D). Taken together, these data suggest that TC45 plays a major role in the dephosphorylation of Stat1 in the nucleus.
Reconstitution of TC-PTP-null MEFs with TC45, but not TC48, rescues the defect in Stat1 dephosphorylation.
To demonstrate that the observed defect in Stat1 dephosphorylation in TC-PTP-null cells is indeed due to the lack of TC45 protein, we performed reconstitution analysis. An expression vector encoding TC45 was introduced into 4−/− cells and a stable clone, 4−TC45, was used for further analysis. Western blot analysis indicated that the level of TC45 expression in 4−TC45 cells was about 20% of that in the wild-type 7+/+ MEF cells (Fig. 5A). Staurosporine-chase analysis was performed on 4−TC45 cells together with the control cell line 4−C as well as the parental 4−/− and wild-type MEFs. Reconstitution of 4−/− cells with TC45 (4−TC45), but not empty vector (4−C), was sufficient to rescue the defect in Stat1 dephosphorylation (Fig. 5B). Similar results were obtained with several other TC45-reconstituted 4−/− cell lines (not shown). We conclude that TC45 is required for the tyrosine dephosphorylation of Stat1.
FIG. 5.
Reconstitution of TC-PTP-null MEFs with TC45, but not TC48, rescues the defect in Stat1 dephosphorylation. (A) Whole-cell extracts (75 μg/lane) from 7+/+, 4−/−, and 4−TC45 cells were analyzed by Western blotting with the 3E2 monoclonal anti-TC-PTP antibody. (B) 7+/+, 4−/−, 4−C, and 4−TC45 cell lines were stimulated with IFN-γ for 30 min followed by a staurosporine chase (0.25 μM) for various periods as indicated. Whole-cell extracts were analyzed by Western blotting with the p-Stat1 antibody. Stripped blots were reprobed with anti-Stat1 and antiactin antibodies as indicated. (C) Puromycin-resistant pools of 14−/− cells retrovirally infected with pBabepuroFlag-TC45 (14−TC45∗) or pBabepuroFlag-TC48 (14−TC48∗) were analyzed for expression with a Flag antibody. (D) Staurosporine-chase experiment as described above with the 14−TC45∗ and 14−TC48∗ pools.
To ensure that the dephosphorylation of Stat1 by TC45 is a nuclear event, mutant 14−/− MEF cells were infected with retrovirus expressing either Flag-TC45 or Flag-TC48 and selected for drug resistance. Pools of drug-resistant cells were used for staurosporine-chase analysis to examine the dephosphorylation of Stat1 after IFN treatment. Unlike TC45, TC48 was unable to rescue the defect in Stat1 dephosphorylation (Fig. 5D), even though TC48 was expressed at a higher level than TC45 in these pools (Fig. 5C). Similar results were obtained when 4−/− cells were used for this experiment (data not shown). TC48 was functional in these cells, because it rescued mitogen-induced cyclin D1 expression (16) as efficiently as TC45 (data not shown). Since TC48 is identical to TC45, except for the presence of a distinct COOH-terminal sequence that targets it to the ER, these results provide further evidence to support the in vivo specificity of TC45 in Stat1 dephosphorylation.
Analysis of the specificity of TC45 in STAT dephosphorylation.
We next examined the involvement of TC45 in the inactivation of other STAT proteins. Wild-type and mutant MEFs were treated with interleukin-6 (IL-6), growth hormone, or IL-4 followed by a staurosporine chase to examine the dephosphorylation of Stat3, Stat5, and Stat6, respectively (Fig. 6). The dephosphorylation of IL-6-activated Stat3 was defective in 4−/− cells as compared to wild-type 7+/+ cells (Fig. 6A). Stat3 is also activated by IFN-γ in MEFs (11). A similar defect in Stat3 dephosphorylation was observed in TC-PTP-null MEFs after IFN-γ treatment (data not shown). In contrast, we observed no difference in the rates of Stat5 or Stat6 dephosphorylation between wild-type and mutant MEFs (Fig. 6B and C).
FIG. 6.
Analysis of the dephosphorylation of Stat3, Stat5, and Stat6 in wild-type and TC-PTP-null MEFs. (A) 7+/+ and 4−/− MEFs were treated with 10 ng of murine IL-6 per ml (Peprotech) for 15 min, followed by a staurosporine chase for various periods as indicated. Whole-cell extracts were analyzed by Western blotting with anti-p-Stat3(Y705) antibody. The stripped blot was reprobed with anti-Stat3 antibody. (B) 7+/+ and 4−/− cells were stimulated with 500 ng of hGH per ml (Sigma) for 30 min followed by a staurosporine chase. Whole-cell extracts were analyzed with anti-p-Stat5(Y694) and anti-Stat5 antibodies. (C) 7+/+ and 4−/− cells were treated with 10 ng of murine IL-4 per ml (Peprotech) for 30 min followed by a staurosporine chase. Whole-cell extracts were analyzed with anti-p-Stat6(Y641) and anti-Stat6 antibodies.
To validate the observed specificity of TC45 in STAT dephosphorylation, primary thymocytes isolated from wild-type and TC-PTP−/− mice were examined by staurosporine-chase analysis. The lack of the TC-PTP gene resulted in a reduced rate of Stat1 dephosphorylation after IFN-α treatment, while the rate of Stat5 dephosphorylation after IL-2 treatment was not affected (Fig. 7A). As a control, the effect of addition of staurosporine on IL-2-induced Stat5 phosphorylation levels in these thymocytes is demonstrated (Fig. 7B). The percentages of single- and double-positive cells for CD4 and CD8 expression were comparable in these thymocytes as determined by fluorescence-activated cell sorting analysis (data not shown). These results support the conclusion that TC45 displays specificity in the dephosphorylation of STATs in vivo.
FIG. 7.
Analysis of Stat1 and Stat5 dephosphorylation in wild-type and TC-PTP-null thymocytes. (A) Primary thymocytes isolated from wild-type and TC-PTP−/− mice were treated for 15 min with 500 U of murine IFN-α (Calbiochem) and 80 U of IL-2 per ml (Invitrogen), respectively, followed by a staurosporine chase. Whole-cell extracts were analyzed with anti-p-Stat1 and anti-p-Stat5 antibodies, respectively. The stripped blots were reprobed with anti-Stat1 and anti-Stat5 antibodies. (B) Wild-type thymocytes were treated with IL-2 with or without addition of staurosporine to demonstrate the effect of staurosporine on Stat5 phosphorylation. Samples were analyzed as in panel A.
DISCUSSION
The transient nature of cytokine signaling is in part accomplished by the action of PTPs that dephosphorylate the various components of the JAK-STAT pathway. Here we describe the identification of the first nuclear Stat1 phosphatase through a combined biochemical and genetic approach. TC45 was purified from HeLa cell nuclear extract with tyrosine-phosphorylated Stat1 as a substrate in an in vitro PTPase assay (Fig. 2A and B). The in vitro PTPase assay used in this study is specific, since nuclear extracts lacking TC45, either by immunodepletion or genetic knockout, fail to dephosphorylate Stat1, despite the presence of other PTPases in these extracts. An in vivo role of TC45 in Stat1 dephosphorylation is supported by studies with TC-PTP-null MEFs or primary thymocytes. In the absence of TC45, the dephosphorylation of Stat1 is defective.
The TC-PTP gene is ubiquitously expressed, and alternative splicing of the last exon produces two distinct protein products. TC48 is targeted to the ER via its hydrophobic COOH terminus, while TC45 lacks this hydrophobic COOH terminus and is largely localized to the nucleus due to the unmasking of a bipartite nuclear localization signal (15). It should be noted that a small fraction of TC45 is also present in the cytoplasm. In addition, under certain conditions, such as mitogen stimulation or stress, cytoplasmic accumulation of TC45 has been observed (19, 30, 31). In TC-PTP-null MEFs, the dephosphorylation of Stat1 in the cytoplasm is also defective (Fig. 4B). TC48 is unlikely to be involved in this process, since there is no detectable TC48 in MEFs (Fig. 5A), and reconstitution of TC-PTP-null cells with TC48 failed to rescue the defect in Stat1 dephosphorylation. Thus, the low level of TC45 present in the cytoplasm is likely involved in Stat1 dephosphorylation.
TC-PTP has a high degree of sequence similarity with PTP1B, which localizes to the same intracellular compartment as TC48. Both TC45 and PTP1B have recently been shown to dephosphorylate distinct JAKs (23, 28, 32). In our hands, overexpressed TC45 dephosphorylated and bound both Jak1 and Jak2 in IFN-γ-stimulated 293T cells (data not shown). It is therefore possible that the reduced IFN-γ-induced Stat1 phosphorylation levels observed in transfected 293 cells and in the stable U2OS-TC45 cell line (Fig. 3B and C) were due to a combined effect of TC45 on both JAK and Stat1. In contrast, we found no evidence for involvement of TC-PTP in the regulation of cytokine-induced JAK activation in MEF cells. The kinetics of IFN-γ-induced Jak1 and Jak2 phosphorylation were not affected in TC-PTP-null cells, although basal levels of Jak1 and Jak2 phosphorylation were slightly higher in TC-PTP-null cells than in the wild-type cells (data not shown).
Our results suggest the existence of other PTPases in STAT dephosphorylation. Although TC45 is clearly the primary phosphatase responsible for the dephosphorylation of Stat1, dephosphorylation was observed after prolonged IFN stimulation of TC-PTP-null MEFs (Fig. 4C). The identity of such minor Stat1 PTPase(s) is currently unknown. Our data suggest that TC45 is also involved in Stat3 dephosphorylation. During final preparation of the manuscript, Aoki et al. reported that TC-PTP dephosphorylates Stat5A and Stat5B in vitro and in vivo when overexpressed in COS-7 or mammary epithelial cells (2). We examined the dephosphorylation of Stat5 after growth hormone and IL-2 stimulation in TC-PTP-null MEFs and primary thymocytes, respectively, and observed no difference in Stat5 dephosphorylation. Other potential PTPs that might be responsible for the dephosphorylation of Stat5 in these cells are PTP1B and Shp2 (1, 36). The dephosphorylation of Stat6 was also unaffected in TC-PTP-null cells. Thus, an as-yet-unidentified PTPase other than TC45 is required for the dephosphorylation of Stat6 in these circumstances. The molecular basis of the observed specificity of TC45 regarding STAT dephosphorylation is under investigation.
TC-PTP-deficient mice develop normally, but die at 3 to 5 weeks of age, displaying a number of defects in lymphocyte functions and hematopoiesis (34). Since TC-PTP is known to be involved in various signaling pathways (16, 32), and since the phenotype of TC-PTP-null mice will reflect the various functions of both TC45 and TC48, further detailed studies are needed to dissect the roles of TC-PTP in the regulation of STAT and other signaling pathways.
Acknowledgments
We thank C. Sawyers and G. Cheng for comments and suggestions on this manuscript. We are grateful to J. Groffen for the gift of the pATH-v-Abl vector and to Kinetek Pharmaceuticals, Inc., for providing the TC-PTP MEF cells.
J. ten Hoeve was supported by a fellowship from the Cancer Research Institute. This work was supported by grants NIH AI43438 to K.S and NCI 80105 to M.D.
REFERENCES
- 1.Aoki, N., and T. Matsuda. 2000. A cytosolic protein-tyrosine phosphatase PTP1B specifically dephosphorylates and deactivates prolactin-activated STAT5a and STAT5b. J. Biol. Chem. 275:39718-39726. [DOI] [PubMed] [Google Scholar]
- 2.Aoki, N., and T. Matsuda. 2002. A nuclear protein tyrosine phosphatase TC-PTP is a potential negative regulator of the PRL-mediated signaling pathway: dephosphorylation and deactivation of signal transducer and activator of transcription 5a and 5b by TC-PTP in nucleus. Mol. Endocrinol. 16:58-69. [DOI] [PubMed] [Google Scholar]
- 3.Begitt, A., T. Meyer, M. van Rossum, and U. Vinkemeier. 2000. Nucleocytoplasmic translocation of Stat1 is regulated by a leucine-rich export signal in the coiled-coil domain. Proc. Natl. Acad. Sci. USA 97:10418-10423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bowman, T., R. Garcia, J. Turkson, and R. Jove. 2000. STATs in oncogenesis. Oncogene 19:2474-2488. [DOI] [PubMed] [Google Scholar]
- 5.Callus, B. A., and B. Mathey-Prevot. 1998. Interleukin-3-induced activation of the JAK/STAT pathway is prolonged by proteasome inhibitors. Blood 91:3182-3192. [PubMed] [Google Scholar]
- 6.Champion-Arnaud, P., M. C. Gesnel, N. Foulkes, C. Ronsin, P. Sassone-Corsi, and R. Breathnach. 1991. Activation of transcription via AP-1 or CREB regulatory sites is blocked by protein tyrosine phosphatases. Oncogene 6:1203-1209. [PubMed] [Google Scholar]
- 7.Cool, D. E., N. K. Tonks, H. Charbonneau, K. A. Walsh, E. H. Fischer, and E. G. Krebs. 1989. cDNA isolated from a human T-cell library encodes a member of the protein-tyrosine-phosphatase family. Proc. Natl. Acad. Sci. USA 86:5257-5261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Darnell, J. E., Jr. 1997. STATs and gene regulation. Science 277:1630-1635. [DOI] [PubMed] [Google Scholar]
- 9.David, M., P. M. Grimley, D. S. Finbloom, and A. C. Larner. 1993. A nuclear tyrosine phosphatase downregulates interferon-induced gene expression. Mol. Cell. Biol. 13:7515-7521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dignam, J. D., R. M. Lebovitz, and R. G. Roeder. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11:1475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Durbin, J. E., R. Hackenmiller, M. C. Simon, and D. E. Levy. 1996. Targeted disruption of the mouse Stat1 gene results in compromised innate immunity to viral disease. Cell 84:443-450. [DOI] [PubMed] [Google Scholar]
- 12.Haspel, R. L., and J. E. Darnell, Jr. 1999. A nuclear protein tyrosine phosphatase is required for the inactivation of Stat1. Proc. Natl. Acad. Sci. USA 96:10188-10193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haspel, R. L., M. Salditt-Georgieff, and J. E. Darnell, Jr. 1996. The rapid inactivation of nuclear tyrosine phosphorylated Stat1 depends upon a protein tyrosine phosphatase. EMBO J. 15:6262-6268. [PMC free article] [PubMed] [Google Scholar]
- 14.Hilton, D. J. 1999. Negative regulators of cytokine signal transduction. Cell Mol. Life Sci. 55:1568-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ibarra-Sanchez, M. J., P. D. Simoncic, F. R. Nestel, P. Duplay, W. S. Lapp, and M. L. Tremblay. 2000. The T-cell protein tyrosine phosphatase. Semin. Immunol. 12:379-386. [DOI] [PubMed] [Google Scholar]
- 16.Ibarra-Sanchez, M. J., J. Wagner, M. T. Ong, C. Lampron, and M. L. Tremblay. 2001. Murine embryonic fibroblasts lacking TC-PTP display delayed G1 phase through defective NF-kappaB activation. Oncogene 20:4728-4739. [DOI] [PubMed] [Google Scholar]
- 17.Irie-Sasaki, J., T. Sasaki, W. Matsumoto, A. Opavsky, M. Cheng, G. Welstead, E. Griffiths, C. Krawczyk, C. D. Richardson, K. Aitken, N. Iscove, G. Koretzky, P. Johnson, P. Liu, D. M. Rothstein, and J. M. Penninger. 2001. CD45 is a JAK phosphatase and negatively regulates cytokine receptor signalling. Nature 409:349-354. [DOI] [PubMed] [Google Scholar]
- 18.Koerner, T. J., J. E. Hill, A. M. Myers, and A. Tzagoloff. 1991. High-expression vectors with multiple cloning sites for construction of trpE fusion genes: pATH vectors. Methods Enzymol. 194:477-490. [DOI] [PubMed] [Google Scholar]
- 19.Lam, M. H., B. J. Michell, M. T. Fodero-Tavoletti, B. E. Kemp, N. K. Tonks, and T. Tiganis. 2001. Cellular stress regulates the nucleocytoplasmic distribution of the protein tyrosine phosphatase TCPTP. J. Biol. Chem. 276:37700-37707. [DOI] [PubMed] [Google Scholar]
- 20.Lee, C. K., H. A. Bluyssen, and D. E. Levy. 1997. Regulation of interferon-alpha responsiveness by the duration of Janus kinase activity. J. Biol. Chem. 272:21872-21877. [DOI] [PubMed] [Google Scholar]
- 21.Lorenzen, J. A., C. Y. Dadabay, and E. H. Fischer. 1995. COOH-terminal sequence motifs target the T cell protein tyrosine phosphatase to the ER and nucleus. J. Cell Biol. 131:631-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McBride, K. M., C. McDonald, and N. C. Reich. 2000. Nuclear export signal located within the DNA-binding domain of the STAT1 transcription factor. EMBO J. 19:6196-6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Myers, M. P., J. N. Andersen, A. Cheng, M. L. Tremblay, C. M. Horvath, J. P. Parisien, A. Salmeen, D. Barford, and N. K. Tonks. 2001. TYK2 and JAK2 are substrates of protein-tyrosine phosphatase 1B. J. Biol. Chem. 276:47771-47774. [DOI] [PubMed] [Google Scholar]
- 24.Pear, W. S., G. P. Nolan, M. L. Scott, and D. Baltimore. 1993. Production of high-titer helper-free retroviruses by transient transfection. Proc. Natl. Acad. Sci. USA 90:8392-8396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shuai, K. 2000. Modulation of STAT signaling by STAT-interacting proteins. Oncogene 19:2638-2644. [DOI] [PubMed] [Google Scholar]
- 26.Shuai, K., J. Liao, and M. M. Song. 1996. Enhancement of antiproliferative activity of gamma interferon by the specific inhibition of tyrosine dephosphorylation of Stat1. Mol. Cell. Biol. 16:4932-4941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shuai, K., G. R. Stark, I. M. Kerr, and J. E. Darnell, Jr. 1993. A single phosphotyrosine residue of Stat91 required for gene activation by interferon-gamma. Science 261:1744-1746. [DOI] [PubMed] [Google Scholar]
- 28.Simoncic, P. D., A. Lee-Loy, D. L. Barber, M. L. Tremblay, and C. J. McGlade. 2002. The T cell protein tyrosine phosphatase is a negative regulator of janus family kinases 1 and 3. Curr. Biol. 12:446-453. [DOI] [PubMed] [Google Scholar]
- 29.Smith, D. B., and K. S. Johnson. 1988. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene 67:31-40. [DOI] [PubMed] [Google Scholar]
- 30.Tiganis, T., A. M. Bennett, K. S. Ravichandran, and N. K. Tonks. 1998. Epidermal growth factor receptor and the adaptor protein p52Shc are specific substrates of T-cell protein tyrosine phosphatase. Mol. Cell. Biol. 18:1622-1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tiganis, T., B. E. Kemp, and N. K. Tonks. 1999. The protein-tyrosine phosphatase TCPTP regulates epidermal growth factor receptor-mediated and phosphatidylinositol 3-kinase-dependent signaling. J. Biol. Chem. 274:27768-27775. [DOI] [PubMed] [Google Scholar]
- 32.Tonks, N. K., and B. G. Neel. 2001. Combinatorial control of the specificity of protein tyrosine phosphatases. Curr. Opin. Cell Biol. 13:182-195. [DOI] [PubMed] [Google Scholar]
- 33.Ward, A. C., I. Touw, and A. Yoshimura. 2000. The Jak-Stat pathway in normal and perturbed hematopoiesis. Blood 95:19-29. [PubMed] [Google Scholar]
- 34.You-Ten, K. E., E. S. Muise, A. Itie, E. Michaliszyn, J. Wagner, S. Jothy, W. S. Lapp, and M. L. Tremblay. 1997. Impaired bone marrow microenvironment and immune function in T cell protein tyrosine phosphatase-deficient mice. J. Exp. Med. 186:683-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu, C. L., and S. J. Burakoff. 1997. Involvement of proteasomes in regulating Jak-STAT pathways upon interleukin-2 stimulation. J. Biol. Chem. 272:14017-14020. [DOI] [PubMed] [Google Scholar]
- 36.Yu, C. L., Y. J. Jin, and S. J. Burakoff. 2000. Cytosolic tyrosine dephosphorylation of STAT5. Potential role of SHP-2 in STAT5 regulation. J. Biol. Chem. 275:599-604. [DOI] [PubMed] [Google Scholar]