Abstract
In the ciliate Euplotes crassus, millions of new telomeres are synthesized by telomerase and polymerase α-primase during macronuclear development in mated cells. Concomitant with de novo telomere formation, telomerase assembles into higher-order complexes of 550 kDa, 1,600 kDa, and 5 MDa. We show here that telomerase is physically associated with the lagging-strand replication machinery in these complexes. Antibodies against DNA primase precipitated telomerase activity from all three complexes from mated cells but not the 280-kDa telomerase complex from vegetatively growing cells. Moreover, when telomerase was affinity purified, primase copurified with enzyme from mated cells but not with the 280-kDa vegetative complex. Thus, the association of telomerase and primase is developmentally regulated. Intriguingly, PCNA (proliferating cell nuclear antigen) was also found in the 5-MDa complex from mated cells. We therefore speculate that this complex is a complete telomere synthesis machine, while the smaller complexes are assembly intermediates. The physical association of telomerase and primase explains the coordinate regulation of telomeric G- and C-strand synthesis and the efficiency of telomere addition in E. crassus.
One function of telomeres is to circumvent the end replication problem that arises because DNA polymerase is unable to completely replicate the 5′ end of a linear chromosome (45). The telomere shortening that occurs as a result of incomplete replication is compensated for by the enzyme telomerase, which adds telomeric DNA to the chromosome end (19). However, telomerase is only able to synthesize the 3′ G-rich strand of the telomere. Thus, the complementary C-rich strand must be filled in by the DNA replication machinery in order to avoid the generation of long recombinogenic single-strand tails (33).
C-strand fill-in is particularly important in budding yeast and hypotrichous ciliates such as Euplotes crassus. During S phase in Saccharomyces cerevisiae, not only is the G-strand extended by telomerase, but the C-strand is also resected by nucleases. This leaves long G-strand overhangs that must be filled in before the protective telomeric chromatin structure can reassemble at the end of S phase (11, 46). The situation is even more extreme in Euplotes spp. because during the sexual stage of their life cycle, Euplotes spp. undergo a developmentally programmed chromosome fragmentation that leads to the synthesis of literally millions of new telomeres (38). C-strand and G-strand syntheses are both obligate steps in new telomere formation. There is good evidence that in both S. cerevisiae and E. crassus, fill-in of the telomeric C-strand is carried out by the lagging-strand replication machinery, specifically polymerase α-primase and possibly polymerase δ in conjunction with RF-C and PCNA (proliferating cell nuclear antigen) (10, 12, 14, 28).
Studies with S. cerevisiae and E. crassus argue that G- and C-strand synthesis is biochemically coupled. In S. cerevisiae, certain temperature-sensitive mutations in the lagging-strand replication machinery cause uncontrolled telomerase-dependent telomere elongation (1, 6). For the polymerase α/pol1-17 temperature-sensitive mutation, this elongation is accompanied by a large increase in the amount of G-strand overhang (28). At the semipermissive temperature, pol1-17 also causes addition of longer telomeres in a chromosome healing assay that measures addition of telomeres to a newly created double-strand break (10). Inactivation of polymerase α, primase, or polymerase δ abolishes new telomere addition in the same assay. These results point to a mechanistic link between synthesis of the telomeric G-strand by telomerase and the C-strand by the lagging-strand replication machinery.
Recent experiments suggest that this linkage may result from the G-overhang binding protein Cdc13p's preferentially recruiting either telomerase or polymerase α-primase to the telomere (7, 17, 36). During the initial step of telomere replication, Cdc13p is proposed to recruit telomerase via a direct interaction with Est1p. The Est1p interaction is then thought to be replaced by a mutually exclusive interaction with Stn1p, followed by recruitment of the lagging-strand replication machinery. This recruitment of DNA polymerase α-primase would limit further G-strand synthesis.
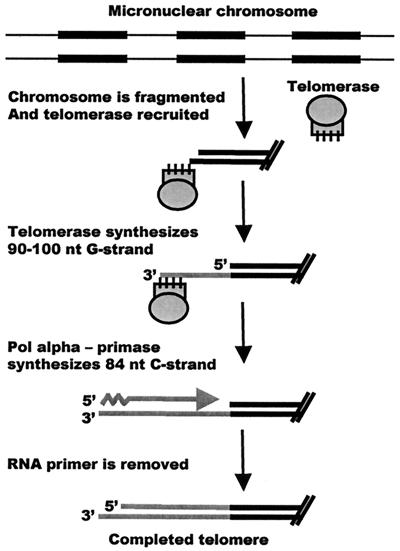
Coordinate regulation of telomeric G- and C-strand synthesis was first demonstrated during new telomere addition in E. crassus (14). Like other ciliates, Euplotes spp. have two nuclei, a transcriptionally active macronucleus and a transcriptionally silent germ line micronucleus (22, 31). When Euplotes cells are mated, the old macronucleus is destroyed and a new macronucleus is generated from a copy of the micronucleus over a period of ≈100 h. The developmental process involves site-specific fragmentation of the micronuclear chromosomes to release the individual genes as free linear DNA molecules (Fig. 1). Oversized telomeres of ≈100 nucleotides are added to each molecule, the DNA is amplified by several rounds of replication, and the telomeres are then trimmed to the mature size of 28 bp plus a 14-nucleotide G-strand overhang (38, 41).
FIG. 1.
Cartoon depicting the process of macronuclear development and new telomere synthesis in E. crassus. A 6-nucleotide (nt) 3′ overhang is created by the DNA fragmentation event (24). If C-strand synthesis occurs in several stages, synthesis of the entire G-strand need not necessarily be complete before C-strand synthesis is initiated. The newly synthesized telomeres are trimmed to the mature size at a later stage in macronuclear development (42). Black boxes represent macronucleus-destined sequences.
When aphidicolin is added to partially inhibit DNA polymerase α during new telomere addition, the newly synthesized C-strands become heterogeneous in length, while the G-strands become not only heterogeneous but also much longer (14). Since aphidicolin is a specific inhibitor of DNA polymerase α and polymerase δ but has no effect on telomerase, the in vivo effect of aphidicolin on G-strand synthesis must be mediated through partial inhibition of the C-strand synthesis machinery. Thus, synthesis of the telomeric G- and C-strands must be coordinately regulated. One way that such coordinate regulation could be achieved is if initiation of C-strand synthesis by DNA polymerase α-primase prevents telomerase from adding further repeats to the G-strand.
The DNA molecules generated by fragmentation of the micronuclear chromosomes do not have existing telomeric sequences at their termini (8, 25). Consequently, telomerase must add new telomeric repeats onto a nontelomeric substrate. Telomerase isolated from E. crassus cells that are growing vegetatively is unable to use nontelomeric DNA as a substrate; however, in mated cells, the enzyme undergoes a developmentally programmed switch in specificity so that nontelomeric substrates can be utilized (5, 29). This switch in DNA specificity is accompanied by assembly of telomerase into a series of higher-order complexes (18).
Telomerase isolated from vegetatively growing cells exists predominantly as a complex of ≈280 kDa that contains the telomerase RNA, the catalytic subunit E. crassus telomerase reverse transcriptase (TERT), and perhaps the ≈43-kDa La homolog found in Euplotes aediculatus telomerase (2, 18). In contrast, the enzyme from mated cells fractionates into complexes of 550 kDa, 1,600 kDa, and 5 MDa (18). These complexes are unlikely to be simple multimers of the vegetative enzyme because they have distinct biochemical properties; telomerase in the 5-MDa and 1,600-kDa complexes is more processive than in the 280- and 550-kDa complexes and can utilize nontelomeric DNA substrates. The 280-kDa enzyme also displays a unique product elongation pattern during primer extension (5).
The size of the telomerase complexes from mated cells, together with the coordinate regulation of new G- and C-strand synthesis, led us to ask whether telomerase and DNA polymerase α-primase are physically associated in a higher-order complex. DNA polymerase α-primase normally exists as a ≈350-kDa complex that contains 48- and 58-kDa primase subunits and 180- and 68-kDa DNA polymerase subunits (3). Thus, complexes of 550 kDa and larger would be able to accommodate both the telomerase and DNA polymerase α-primase holoenzymes. Here we demonstrate that telomerase physically associates with the replication machinery during de novo telomere formation.
MATERIALS AND METHODS
Cloning and expression of p48.
A portion of the E. crassus p48 gene was isolated by amplifying Euplotes macronuclear DNA with degenerate PCR primers corresponding to conserved regions of the human, mouse, and S. cerevisiae p48 subunits. The 146-bp PCR product was then used to screen a λgt10 library (LEMAC) containing Euplotes macronuclear DNA as previously described (20). The primase gene was released from the phage DNA by digestion with EcoRI and cloned into pBluescript. The resulting clones were missing the N terminus because of an internal EcoRI site, so the full-length gene was isolated from the phage DNA by PCR with a telomere-specific primer and a gene-specific primer. The gene encoding p48 contained three TGA stop codons (which encode cysteine in E. crassus). These were changed to TGC with the Promega Gene Editor system. The open reading frame was cloned into pQE30 (Qiagen Inc.) and expressed to produce His6-tagged p48. The tagged protein was purified by Ni2+ affinity chromatography and used to make antibody.
Cell culture and isolation of macronuclei.
E. crassus strains A1 and A2 were grown in 40-liter cultures and mated as previously described (34, 37). Developing macronuclei (anlagen) were isolated at ≈65 h after mating, purified on Percoll-sucrose gradients (39), and stored at −80°C. To prepare vegetative macronuclei, strain A2 was grown in 20-liter cultures until the algae were consumed. The cells were concentrated 20-fold, added to 5 volumes of a dense culture of Dunaliella salina, and harvested when the algae were consumed. The cells were washed twice with lysis buffer (10 mM Tris [pH 7.5], 2.5 mM MgCl2, 1 mM dithiothreitol [DTT], 0.1 mM phenylmethylsulfonyl fluoride [PMSF]) and homogenized gently in lysis buffer plus 0.5% Triton X-100. The macronuclei were then purified on Percoll-sucrose gradients as previously described (5, 39).
To prepare nuclear extracts, macronuclei were adjusted to 300 mM potassium glutamate, 150 mM NaCl, 1% NP-40, 0.5 mM sodium deoxycholate, and 0.25 mg of acetylated bovine serum albumin per ml. The nuclei were lysed by French press at 16,000 lb/in2, and the lysate was incubated at 4°C for 15 min prior to centrifugation at 15,000 rpm for 30 min.
Fractionation of telomerase complexes.
Fractionation of telomerase complexes was carried out on a Superose 6 HR 10/30 gel filtration column (Pharmacia) as previously described (18). The column was preequilibrated with 30 mM Tris (pH 7.5)-0.1 mM MgCl2-20% glycerol (TMG) containing 1 mM DTT, 150 mM NaCl, 0.1 mM PMSF, and 0.1% NP-40. Euplotes nuclear extracts were concentrated two- to threefold by dialysis into TMG containing 40% polyethylene glycol 8000, and 250-μl aliquots were loaded on the column. The extract was separated at a flow rate of 0.2 ml/min and collected as 0.2-ml fractions. Fractions were adjusted to 0.1 mg of acetylated bovine serum albumin (U.S. Biochemical) per ml following collection to stabilize the protein. Elution of the various telomerase complexes was monitored by performing telomerase assays on each fraction.
Heparin-Sepharose CL-6B purification of telomerase.
Heparin-Sepharose CL-6B dry powder (Amersham Pharmacia Biotech) was resuspended in TMGH buffer (30 mM Tris [pH 7.5], 3 mM MgCl2, 10% glycerol, 0.1 M potassium glutamate, 0.05 M NaCl, 1% NP-40, 1 mM DTT) and equilibrated with 10 column volumes of the same buffer. Euplotes nuclear lysate was loaded, and the column was washed with 10 column volumes of TMGH. Telomerase was eluted with 0.6 M and then 0.8 M potassium glutamate in TMGH buffer.
Immunoprecipitation reaction.
Protein A-Sepharose beads (30 μl/reaction) were washed twice with binding buffer (30 mM Tris [pH 7.5], 3 mM MgCl2, 10% glycerol, 100 mM potassium glutamate, 0.1% NP-40, 0.5 mM sodium deoxycholate, and 1 mM DTT) and coupled to 6 to 12 μg of antibody in binding buffer by overnight incubation in a final volume of 300 μl. Alternatively, purified p48 antibody was covalently cross-linked to cyanogen bromide-activated beads (Pierce) according to the manufacturer's instructions. The beads were washed with binding buffer containing 0.1 mM PMSF and 1 mg of leupeptin per ml to remove unbound antibody and incubated with 500 μl of blocking buffer (30 mM Tris [pH 7.5], 3 mM MgCl2, 10% glycerol, 100 mM potassium glutamate, 0.1% NP-40, 0.5 mM sodium deoxycholate, 0.5 mg of lysozyme per ml, 0.2 mg of S. cerevisiae total RNA per ml, 0.05 mg of glycogen per ml, 0.5 mg of acetylated bovine serum albumin per ml, 1 mM DTT, 0.1 mM PMSF, and 1 mg of leupeptin per ml) twice for 15 min each. The beads were then incubated with the nuclear extract or Superose 6 column-purified fractions for 4 h in a final volume of 300 to 600 μl.
The unbound supernatant was saved and later assayed for telomerase activity. The beads were washed three times for 10 min each with 500 μl of binding buffer and then either assayed for telomerase activity or analyzed for the presence of primase by Western blotting. For control experiments, either no antibody or immunoglobulin G (IgG) from 5 to 10 μl of p48 preimmune serum was coupled to the protein A beads prior to the blocking step. The Western blots were incubated first with p48 antibody that had been biotinylated with sulfosuccinimidyl-6-(biotinamido)hexanoate (Pierce) and then with horseradish peroxidase-conjugated streptavidin.
Telomerase assays.
Telomerase assays were performed on intact anlagen, nuclear extracts, or protein A beads as previously described (5) with either (G4T4)3 or GT-13 (G4T4ACTACGCGATCAT) as a primer. Reactions (20 μl) containing 100 ng of primer, 50 mM Tris (pH 8.0), 1 mM spermidine phosphate, 1 mM DTT, 5 mM MgCl2, 20 mM EGTA, 0.1 mM dTTP, 0.25 mM [α-32P]dGTP (800 Ci/mmol), and the required volume of extract were incubated at 30°C for 1 h. Reactions were stopped by adding 10 mM EDTA, extracted with phenol-chloroform-isoamyl alcohol, and ethanol precipitated. The products were resolved on 10% polyacrylamide sequencing gels and visualized by autoradiography or PhosphorImager (Molecular Dynamics). A 5′-end 32P-labeled 12-mer oligonucleotide was added to each reaction during the ethanol precipitation step to monitor the efficiency of product recovery.
Telomerase affinity purification.
Affinity purification of telomerase was carried essentially as described before (27) with extract from ≈106 macronuclei or 2.4 ml of Superose 6 column fraction for each reaction. S. cerevisiae total RNA (50 mg) and 1.5 nmol each of two competitor oligonucleotides (5′-TAGACCTGTTAGTGTACATTTGAATTGAAGC-3′ and 5′-TAGACCTGTTAGGTTGGATTTGTGGGCATCA-3′) were added per ml of macronuclear lysate or Superose 6 column fraction and incubated for 15 min. Then 0.3 nmol of telomerase-specific oligonucleotide [5′-biotin-TAGACCTGTTA-(rmeG)2-(rmeU)4-(rmeG)4-(rmeU)4-rmeG-3′] was added per ml of extract or column fraction, and the sample was incubated for 15 min (rmeN denotes 2′-O-methylribonucleotides complementary to the telomerase RNA template region; the deoxyribonucleotides were not complementary to the template). Then 60 μl of Ultralink-immobilized Neutravidin Plus (Pierce) resin material was added per ml of column fraction.
The Ultralink resin was preblocked twice for 15 min each with WB (20 mM Tris [pH 7.5], 10 mM MgCl2, 1 mM EDTA, 300 mM potassium glutamate, 10% glycerol, 1% NP-40, and 1 mM DTT) containing 0.5 mg of bovine serum albumin, 0.5 mg of lysozyme, 0.05 mg of glycogen, and 0.1 mg of S. cerevisiae total RNA per ml and washed twice for 15 min each with WB. The sample was incubated for 8 min at 30°C and then for 2 h at 4°C to allow binding. The unbound supernatant was removed, and the resin was washed with 10 column volumes of WB for 15 min at room temperature, rinsed with WB, washed with WB containing 0.6 M potassium glutamate but no NP-40 for 5 min at 30°C, rinsed with WB, and washed with WB for 5 min at room temperature. The beads were boiled in sodium dodecyl sulfate (SDS) gel loading buffer, and the proteins were separated on an SDS-10% polyacrylamide gel and Western blotted with either p48 antibody or both p48 and E. crassus TERT antibodies. The specificity of the telomerase affinity purification was confirmed by reverse transcription (RT)-PCR of the telomerase RNA. Amplification was for 17 cycles with the sense primer 5′-TCTGTCAAACTAAACCCCAAAC and the antisense primer 5′-CGGGAATTTTTAAAAAGATGAGAGGACAG.
Nucleotide sequence accession number.
The GenBank accession number for the p48 subunit of E. crassus DNA primase is AF403474.
RESULTS
Identification and localization of the p48 subunit of Euplotes DNA primase.
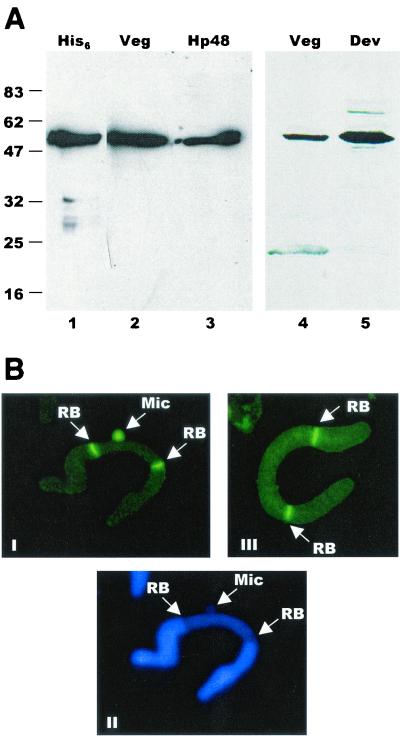
Before we could determine whether telomerase interacts directly with the DNA replication machinery, it was necessary to obtain a marker for the DNA polymerase α-primase complex. We chose the p48 subunit of primase because this protein is well conserved. A fragment of the gene encoding the p48 subunit of E. crassus primase was isolated by degenerate PCR and used to clone the full-length gene from a λ library. The predicted protein is 48.5 kDa and has 41% sequence identity to the human p48 primase subunit (data not shown). Polyclonal antibodies raised against the p48 protein expressed in Escherichia coli identified a single protein of ≈50 kDa in both vegetatively growing cells and mated cells undergoing macronuclear development (Fig. 2A). The antibody also cross-reacted with purified human p48 (Fig. 2A, lane 4). In immunolocalization experiments, the p48 antibody gave rise to the staining pattern expected for a replication protein (15, 40), as it stained the replication bands in macronuclei and the entire micronucleus in a subset of cells (Fig. 2B). Replication bands are visible “replication machines” that assemble at either end of the macronuclei in hypotrichous ciliates (30). They gradually move towards the center of the macronucleus, replicating the DNA molecules in their path.
FIG. 2.
Characterization of E. crassus DNA primase. (A) Western blot showing the specificity of the p48 antibody. Lane 1, recombinant, His6-tagged p48; lane 2, macronuclei from vegetatively growing Euplotes cells; lane 3, purified human p48 protein; lane 4, 1.5 × 105 macronuclei from vegetatively growing cells; lane 5, 3.75 × 104 macronuclei from mated cells. The positions of molecular size markers are shown to the left (in kilodaltons). (B) Immunolocalization of primase and E. crassus TERT. Panel I, micro- and macronuclear staining pattern obtained with p48 antibody; panel II, same nuclei stained with Hoechst dye; panel III, staining with E. crassus TERT antibody. The arrows indicate replication bands (RB) and micronuclei (Mic).
Telomerase and primase are physically associated in mated cells.
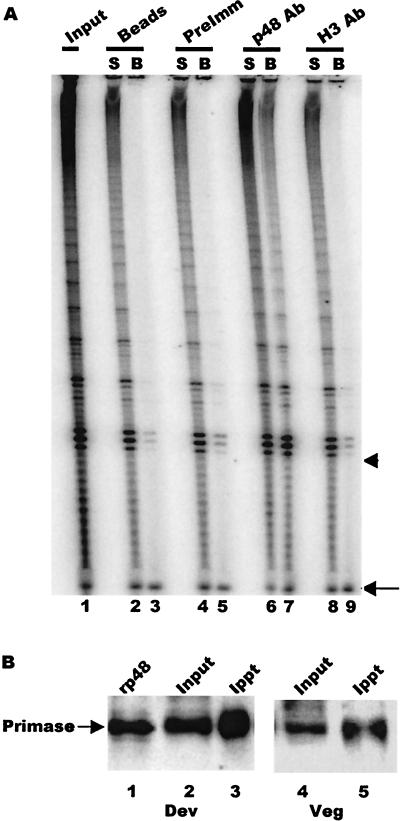
To determine whether the polymerase α-primase complex is physically associated with telomerase during macronuclear development, we carried out telomerase assays on immunoprecipitates obtained with the p48 antibody. Purified p48 antibody, IgG from preimmune serum, or antibody directed against a developmentally regulated histone H3 variant (16) was coupled to protein A-Sepharose and incubated with extracts from developing macronuclei isolated at the time of new telomere synthesis (65 h after mating). Both the beads and the unbound fraction were assayed for telomerase activity. As shown in Fig. 3A, the p48 antibody precipitated a substantial amount of telomerase activity from the extract, whereas the preimmune serum and the H3 antibody variant gave only background levels of activity (compare lane 7 with lanes 3, 5, and 9). This result is consistent with the hypothesis that polymerase α-primase is a component of the higher-order telomerase complexes present during macronuclear development.
FIG. 3.
Coimmunoprecipitation of telomerase and primase. (A) Telomerase assay showing activity immunoprecipitated by primase antibody. Protein A beads alone (lane 2 and 3) and beads coated with preimmune IgG (lanes 4 and 5), primase p48 antibody (lanes 6 and 7), or histone H3 variant antibody (lanes 8 and 9) were incubated with nuclear extract and washed, and the beads (B) and unbound supernatant (S) were assayed for telomerase activity. Lane 1 shows the activity in the input extract. The arrowhead marks the position of the input primer. The arrow marks the recovery control that was added to each reaction after the telomerase extension step. (B) Efficiency of primase immunoprecipitation. Primase was immunoprecipitated from extracts made with macronuclei isolated 65 h after mating (lanes 2 and 3) or macronuclei from vegetatively growing cells (lanes 4 and 5) with p48 antibody covalently cross-linked to Sepharose beads. The input and precipitated material were detected with biotinylated p48 antibody and peroxidase-conjugated streptavidin. Lane 1, recombinant p48; lane 2, 5 μl of extract; lane 3, precipitate from 35 μl of extract; lane 4, 10 μl of extract; lane 5, precipitate from 90 μl of extract.
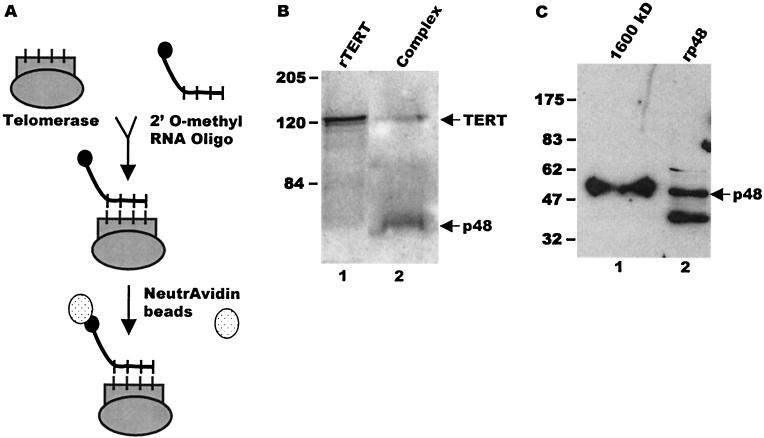
Since incubation of nuclear extracts with protein A-Sepharose alone or beads plus preimmune serum consistently brought down a low level of telomerase activity, we investigated the specificity of the association between telomerase and primase by determining whether p48 copurified with affinity-purified telomerase complexes (Fig. 4A and B). Telomerase was isolated from macronuclear extracts of mated cells by heparin-agarose chromatography followed by affinity purification with a biotinylated 2′-O-methyl RNA oligonucleotide that was complementary to the template region of the telomerase RNA (27). The telomerase-2′-O-methyl RNA complexes were isolated with NeutrAvidin beads, and after extensive washing, they were released from the beads by boiling in SDS and examined by Western blotting with antibodies against primase and E. crassus TERT. Both the p48 primase subunit and E. crassus TERT were present in the affinity-purified complexes (Fig. 4B, lanes 1 and 2), confirming that telomerase and primase are physically associated during macronuclear development. Since p48 forms a stable complex with DNA polymerase α (43), we presume that the other three subunits of the polymerase α-primase complexes are also present in the telomerase complexes. However, we have been unable to confirm this supposition because antibodies to comparable S. cerevisiae and mammalian proteins do not cross-react with these Euplotes proteins.
FIG. 4.
Affinity purification of telomerase complexes. (A) Schematic of the affinity purification protocol. Following precipitation with NeutrAvidin beads, the affinity-purified material was analyzed by RT-PCR or separated by SDS-polyacrylamide gel electrophoresis, and the composition was examined by Western blotting. (B) Copurification of E. crassus TERT and p48 from extracts made with mated cells. Western blot was probed simultaneously with antibodies to p48 and E. crassus TERT. Lane 1, recombinant E. crassus TERT; lane 2, affinity-purified telomerase complexes. The arrowheads mark p48 and E. crassus TERT. (C) Copurification of telomerase and p48 from 1,600-kDa complexes. The 1,600-kDa complexes were isolated by gel filtration prior to affinity purification. The Western blot was probed with antibody to p48. Lane 1, affinity-purified 1,600-kDa complex; lane 2, recombinant p48. The arrowhead marks full-length p48. Positions of molecular size markers are shown to the left (in kilodaltons).
To determine what fraction of primase is associated with telomerase in the higher-order complexes, we next measured how much of the total primase and telomerase could be coimmunoprecipitated by the p48 antibody. Since primase is the same size as the IgG heavy chain, we performed the immunoprecipitation with p48 antibody that had been cross-linked to cyanogen bromide-activated Sepharose beads and then detected the precipitated primase by Western blotting with biotinylated p48 antibody and horseradish peroxidase-conjugated streptavidin (Fig. 3B). Comparison of the signal from the input material versus the bound fraction from multiple experiments revealed that 10 to 20% of the primase was precipitated.
It was not possible to quantify the amount of telomerase in the immunoprecipitate by Western blotting due to the low affinity of the TERT antibody. Therefore, we compared the amount of telomerase activity in the input versus bound fractions by quantifying the final reaction product generated by each sample (Fig. 3A and data not shown). Data from multiple experiments revealed that from 5 to 10% of the telomerase activity was routinely precipitated. Since E. crassus telomerase gradually loses activity during purification (E. Greene and D. Shippen, unpublished data), it is likely that this approach underestimates the total amount of telomerase RNP present in the bound fraction. Nevertheless, it is striking that the primase antibody precipitates a similar percentage of the total primase and telomerase present in the nuclear extract. This result suggests that large amounts of primase and telomerase coassociate in the macronucleus during this stage of development.
Association of telomerase and primase is developmentally regulated.
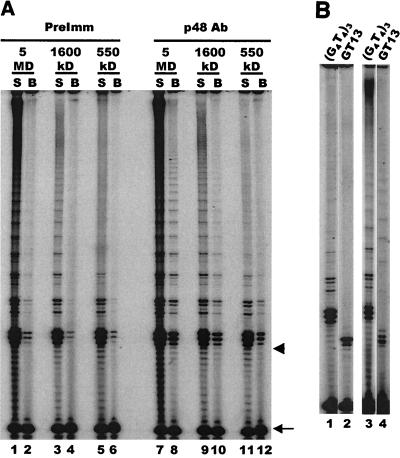
Telomerase from Euplotes cells that have not been mated and are growing vegetatively exists primarily as a complex of only 280 to 350 kDa (18; L. Wang and D. Shippen, unpublished results). While complexes of this size could contain p48, they are too small to contain the entire polymerase α-primase complex. Thus, we suspected that association of telomerase with primase is developmentally regulated. To test this idea, we partially fractionated the 280-kDa complex from vegetatively growing cells with a Superose 6 column and carried out coimmunoprecipitation reactions with the p48 antibody. This partial purification was necessary to remove telomerase inhibitors present in whole-cell extracts (L. Wang and D. Shippen, unpublished results). In marked contrast to experiments with the enzyme from mated cells, the p48 antibody precipitated only background levels of telomerase activity from preparations containing the 280-kDa complex (Fig. 5A).
FIG. 5.
Primase is not associated with telomerase from vegetatively growing cells. (A) Immunoprecipitation of telomerase activity. Telomerase assays were performed on the input 280-kDa telomerase complex (lane 1) or the unbound supernatant (S) or bead fraction (B) following incubation with protein A beads alone (lanes 2 and 3) or beads plus p48 antibody (lanes 4 and 5) or PCNA antibody (lanes 6 and 7). The arrows on the left mark the characteristic vegetative telomerase pausing pattern that corresponds to incorporation of the third G and fourth T in the G4T4 repeat. The arrow on the right marks the recovery control, and the arrowhead marks the position of the input primer. (B) Composition of affinity-purified telomerase complexes from vegetatively growing cells. Western blot was probed with antibody to E. crassus TERT (panel I) or p48 (panel II). Lane 1, input vegetative cell extract; lane 2, affinity-purified telomerase; lane 3, control beads incubated with extract but no telomerase-specific oligonucleotide. Panel III, RT-PCR amplification of telomerase RNA from affinity-purified telomerase (lane 1) or control beads incubated with extract but no telomerase-specific oligonucleotide (lane 2).
A side-by-side experiment performed with extract from mated cells confirmed that the immunoprecipitation procedure had worked, as the p48 antibody precipitated the same levels of enzyme activity as observed previously (data not shown). Moreover, Western blot analysis of immunoprecipitates from unpurified vegetative cell lysates revealed that p48 was precipitated just as efficiently from vegetative cell extracts as from the mated cell extracts (Fig. 3B). Thus, the lack of telomerase activity in the immunoprecipitate from the vegetative cell extract was not caused by masking of the p48 epitopes in the vegetative telomerase complexes. These results indicate that primase is not physically associated with telomerase in the vegetative 280-kDa complex even though both enzymes colocalize in replication bands (Fig. 2B).
To confirm this finding, we determined whether primase could be detected in telomerase complexes affinity purified from vegetatively growing cells. Telomerase was purified from vegetative macronuclei with the biotinylated 2′-O-methyl oligonucleotide directed against the template region of the RNA component. The protein composition of the purified enzyme was then examined by Western blotting with antibodies to p48 or E. crassus TERT. RT-PCR was also used to test for the presence of telomerase RNA. As expected, E. crassus TERT and telomerase RNA could be detected in the affinity-purified sample (Fig. 5B, panel I, lane 2, and panel III, lane 1) but not in control samples which were incubated with beads but no biotinylated oligonucleotide (Fig. 5B, panel I, lane 3, and panel III, lane 2). However, no p48 could be detected in the affinity-purified sample even though large amounts were present in the input material (Fig. 5C, panel II, compare lanes 1 and 2).
Extracts from vegetatively growing cells contain a small amount of telomerase that fractionates at >5 MDa. This telomerase exhibits the same properties as the 280-kDa complex and may simply be an aggregate of the smaller complexes (18). Since it was not possible to obtain enough of the >5-MDa material to directly determine whether this complex contains primase, we cannot rule out an association between telomerase and primase in these large complexes. However, it is clear that the bulk of the telomerase in vegetatively growing cells exists independently of primase. Thus, it appears that telomerase becomes associated with primase during the developmentally regulated assembly of telomerase into higher-order complexes.
Primase is present in all higher-order telomerase complexes.
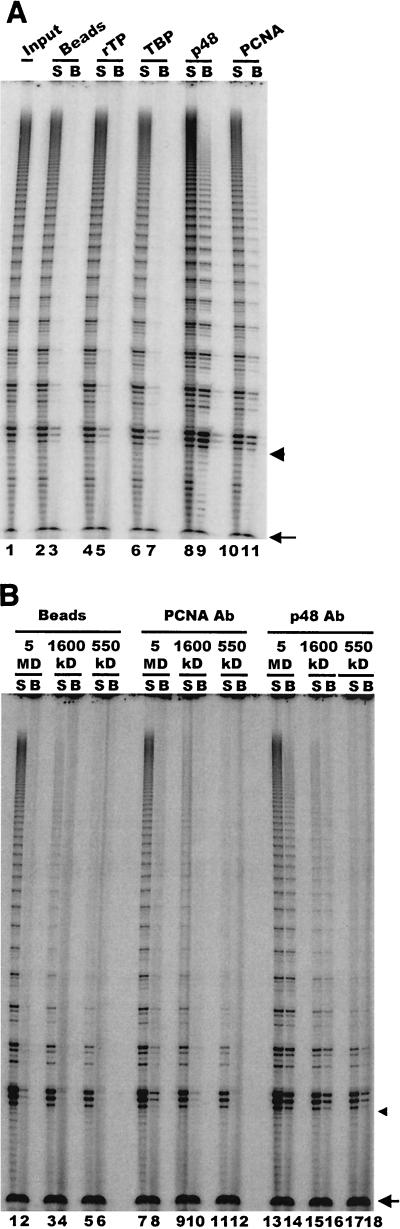
Since telomerase assembles into three distinct higher-order complexes during macronuclear development, we next determined which of these complexes contained primase. Individual complexes were isolated by fractionating extract from developing macronuclei over a Superose 6 column (18), and fractions containing specific complexes were identified based on the sizes of the complexes and their properties in a telomerase primer extension assay (18). Fractions containing the 550-kDa, 1,600-kDa, and 5-MDa complexes were pooled and subjected to immunoprecipitation with p48 antibody. Telomerase assays were then performed with the precipitated material. As shown in Fig. 6A, telomerase activity could be detected in the immunoprecipitates from each of the complexes (lanes 8, 10, and 12). This result suggests that p48 is associated with all three telomerase complexes. Further evidence that primase is associated with the higher-order telomerase complexes was obtained with the biotinylated 2′-O-methyl oligonucleotide to affinity purify telomerase from the 1,600-kDa complex-containing column fractions. As shown in Fig. 4D, p48 could be detected in the affinity-purified sample.
FIG. 6.
Primase is present in all higher-order telomerase complexes. (A) Telomerase assay showing activity immunoprecipitated from partially purified 5-MDa, 1,600-kDa, and 550-kDa complexes. Complexes were precipitated with p48 antibody or preimmune IgG, and unbound supernatant (S) and bead (B) fractions were assayed for each complex with a (G4T4)3 primer. The arrow marks the recovery control, and the arrowhead marks the position of the input primer. (B) Telomerase extension of telomeric and nontelomeric primers. Lanes 1 and 2, purified 550-kDa complex; lanes 3 and 4, unfractionated extract from mated cells. Lanes 1 and 3, telomeric primer (G4T4)3; lanes 2 and 4, nontelomeric primer GT-13.
Since the 550-kDa complex is only just large enough to accommodate both the telomerase and polymerase α-primase holoenzymes, it was important to make sure that the 550-kDa complex had been immunoprecipitated rather than small amounts of 1,600-kDa complex that might contaminate samples containing the 550-kDa complex. To confirm the purity of the 550-kDa complex, telomerase assays were performed with a nontelomeric primer (GT-13) that can only be extended by the 1,600-kDa and 5-MDa complexes and not the 550-kDa complex (18). This primer could not be utilized by column fractions containing purified 550-kDa complex (Fig. 6B, lane 2), although it could be utilized by unfractionated extract that contained the 1,600-kDa and 5-MDa complexes (Fig. 6B, lane 4). Examination of the extension pattern obtained with immunoprecipitated material provided further confirmation that the 550-kDa complex was indeed immunoprecipitated by the p48 antibody. As expected, the less processive 550-kDa complex gave rise to a greater proportion of short reaction products than the 1,600-kDa and 5-MDa complexes (Fig. 6A, compare lane 12 to lanes 8 and 9) (18) even though PhosphorImager analysis revealed that the overall efficiency of immunoprecipitation was similar for all three complexes. Thus, it appears that p48 and most likely all four polymerase α-primase subunits are an integral part of the 550-kDa complex as well as of the two larger telomerase complexes that form during macronuclear development.
It has not been possible to determine whether primase is associated with every telomerase complex or whether the ratio of telomerase to primase differs between the 550-kDa, 1,600-kDa, and 5-MDa complexes. Immunoprecipitation with the p48 antibody and affinity purification with the 2′-O-methyl oligonucleotide are both quite inefficient (see above). This, coupled with the relatively low affinity of the TERT antibodies, has prevented us from detecting TERT in the precipitated fractions from purified complexes. Moreover, telomerase in higher-order complexes loses activity upon further purification (including immunoprecipitation), so the telomerase activity in the immunoprecipitates seems to underestimate the actual amount of telomerase RNP associated with primase in the complexes.
Other components of the telomerase complexes.
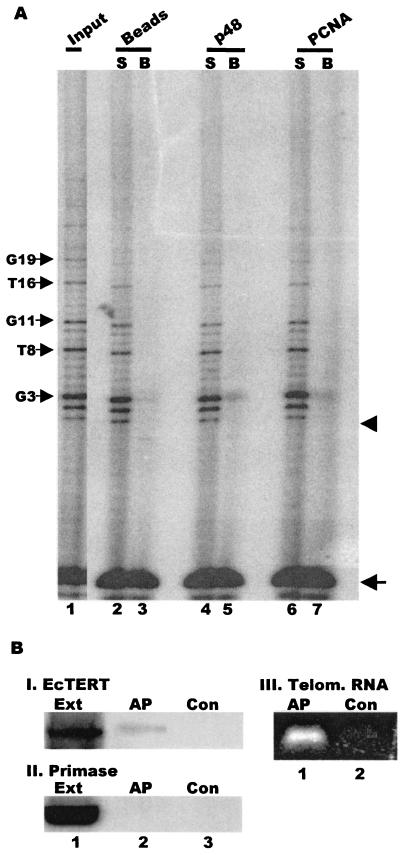
Based on their size, the 1,600-kDa and 5-MDa telomerase complexes are likely to contain other proteins in addition to telomerase and polymerase α-primase. To determine whether the complexes harbor telomere-binding proteins or other components of the replication machinery, we performed a series of immunoprecipitation experiments on unfractionated macronuclear extracts from mated cells with antibodies to two different telomere proteins, TBP and rTP, and the replication protein PCNA (Fig. 7A). TBP is the 51-kDa telomere protein that binds specifically to the 3′ terminus of Euplotes telomeres, and rTP is the 50-kDa replication telomere protein that is only present during DNA replication (35, 40). The TBP and rTP antibodies immunoprecipitated only background levels of telomerase activity, suggesting that these proteins are absent from the telomerase complexes.
FIG. 7.
PCNA is present in a subset of the telomerase complexes. (A) Interaction with PCNA and telomere-binding proteins. Telomerase assays were performed on the unbound supernatant (S) or the beadfraction (B) following incubation of unfractionated extract from mated cells with antibodies to rTP (lanes 4 and 5), TBP (lanes 6 and 7), p48 (lanes 8 and 9), or PCNA (lanes 10 and 11) or a bead-only control (lanes 2 and 3). Lane 1, activity in input extract. (B) Association of telomerase with PCNA. Immunoprecipitations were performed with purified 5-MDa, 1,600-kDa, and 550-kDa complexes and beads alone (lanes 1 to 6), PCNA (lanes 7 to 12) or p48 antibody (lanes 13 to 18). Arrows mark the recovery control, and arrowheads mark the position of the input primer.
We asked whether PCNA is a component of the telomerase complexes because both DNA polymerase δ and PCNA (the processivity factor for polymerase δ) are required for de novo telomere addition and proper telomere maintenance in S. cerevisiae (1, 10). Antibody to Euplotes polymerase δ is not available, but antibody to human PCNA cross-reacts with the ciliate protein (40). As shown in Fig. 7A, the same PCNA antibody reproducibly precipitated telomerase activity from unfractionated extracts of mated cells. Although less activity was precipitated by the PCNA antibody than by the p48 antibody, this may simply reflect the use of a monoclonal antibody to a human protein versus a polyclonal antibody to Euplotes p48.
To determine which of the telomerase complexes contained PCNA, we performed immunoprecipitation experiments with the isolated 550-kDa, 1,600-kDa, and 5-MDa complexes (Fig. 7B). The PCNA antibody precipitated telomerase activity from the 5-MDa complex but not the 550-kDa complex. The 1,600-kDa complex also seemed to immunoprecipitate poorly, but we could not completely rule out an association between PCNA and telomerase in this complex. Nonetheless, the results with the 5-MDa and 550-kDa complexes demonstrated that the protein compositions of the different telomerase complexes from mated cells are distinct. Furthermore, since PCNA associates closely with polymerase δ (23), the 5-MDa complexes may well contain additional components of the replication machinery, such as RF-C and polymerase δ, that are absent from the smaller complexes.
DISCUSSION
Although various genetic analyses have demonstrated a mechanistic link between extension of the telomeric G-strand by telomerase and synthesis of the complementary C-strand by DNA polymerase α-primase, the molecular basis for this link has remained unclear (10, 12, 14). Here we provide a biochemical explanation for the coordination of G- and C-strand synthesis in E. crassus by demonstrating a direct interaction between telomerase and the lagging-strand replication machinery. Moreover, we show that primase assembles with telomerase to form the higher-order complexes that are present during macronuclear development when new telomere synthesis is taking place. The largest complex contains additional replication proteins and a putative chromosome healing factor that is required to recruit telomerase to the site of new telomere synthesis (5). Hence, this large 5-MDa complex appears to be a telomere synthesis machine capable of de novo telomere formation.
Existence of telomere synthesis complexes.
The assembly of large telomerase complexes during macronuclear development reflects the changing needs of a Euplotes cell as it switches from a growth state where telomerase maintains very short telomeres to a state where millions of new telomeres are synthesized synchronously over a short time period (5, 41). During vegetative growth, telomere length is tightly regulated, with each telomere consisting of exactly 28 bp of G4T4 · C4A4 duplex and a 14-nucleotide overhang on the G-strand (26). The minimal requirements for maintaining this structure during DNA replication involve removal of the RNA primer laid down at the lagging-strand telomere and addition of 14 nucleotides of new G4T4 repeat to the newly replicated telomeric DNA at the leading-strand telomere (40, 48). Because no C-strand fill-in is required, there is no need for telomerase to be complexed with polymerase α-primase in vegetatively growing cells. Moreover, as so little new G4T4 sequence needs to be synthesized, the telomerase need not be very processive. In contrast, during new telomere synthesis, telomerase has to add 80 to 100 nucleotides of G4T4 repeats to ≈4 × 107 DNA ends that have no existing telomeric sequence. The lagging-strand replication machinery then immediately generates an equal number of complementary strands (38, 41). Thus, telomerase must processively elongate a nontelomeric substrate and then efficiently transfer the newly synthesized telomeric G-strands to polymerase α-primase for extensive C-strand synthesis.
When the developmentally regulated assembly of E. crassus telomerase into higher-order complexes was first discovered, it was not clear why three different complexes should form or even whether the 5-MDa complex was a bona fide complex or simply an aggregate of 1,600-kDa complexes. Our results suggest that the 5-MDa complex is in fact the actual telomere synthesis complex responsible for new telomere addition. This complex is equipped with a processive telomerase activity, the chromosome healing factor required for telomerase to act on nontelomeric substrates (5), and the most complete complement of lagging-strand replication proteins. Thus, it has the activities required for rapid de novo telomere formation following chromosome fragmentation. In contrast, the smaller 550- and 1,600-kDa complexes contain only a subset of these activities, suggesting that they are assembly intermediates. We postulate that telomerase associates first with polymerase α-primase to form the nonprocessive 550-kDa complex and then binds the processivity factor(s) and chromosome healing factor to give rise to the 1,600-kDa complex. Finally, association of PCNA, and presumably RF-C and polymerase δ, would generate the 5-MDa complex.
At present it is not clear whether the association between telomerase and polymerase α-primase is direct or whether it is mediated by other proteins. We were unable to detect an interaction between E. crassus TERT and p48 either in an S. cerevisiae two-hybrid screen or by coimmunoprecipitation of epitope-tagged recombinant proteins (S. Ray, L. Wang, D. Shippen, and C. Price, unpublished data). However, TERT or other telomerase components could interact with one of the other three subunits within the polymerase α-primase complex.
Composition of telomere synthesis complexes.
The association of PCNA with the largest telomerase complexes fits well with the observation that polymerase δ and RF-C are required for normal telomere synthesis in S. cerevisiae (1, 10). During chromosomal DNA replication, polymerase α-primase synthesizes only a short stretch of DNA (≈30 bases), which is then extended by polymerase δ or polymerase ɛ (9, 23). Thus, polymerase α may not be capable of synthesizing the entire 84-nucleotide telomeric C-strand, so loading of PCNA and a switch to polymerase δ synthesis would be required. We cannot rule out the possibility that PCNA plays an additional role in the coordination of G- and C-strand synthesis because PCNA interacts with a wide variety of proteins within the cell and seems to modulate what type of DNA synthesis is carried out (e.g., the various types of repair synthesis versus regular DNA replication) (23, 44). Because C-strand fill-in is an unusual form of lagging-strand replication, it might be regulated by PCNA interactions with telomere components.
Based on their size, both the 1,600-kDa and 5-MDa complexes probably contain other proteins in addition to the replication factors that are the focus of this study. Like human telomerase, they may contain Hsp90 and p23 chaperone proteins to aid in complex assembly (21). Another likely component is the chromosome cleavage machinery that carries out site-specific fragmentation prior to telomere addition (24). Fragmentation of the micronuclear chromosomes is tightly linked to telomere addition (13). Perhaps this is achieved by physical coupling of the cleavage activity and the telomere synthesis activity into a single particle. Finally, these complexes may also contain a dimer or multimer of the telomerase holoenzyme. Telomerase is known to exist as a dimer or multimer in S. cerevisiae and humans (4, 32, 47), and we have evidence that multimerization may also occur in E. crassus (L. Wang, S. Dean, and D. Shippen, unpublished data).
Coordination of G- and C-strand synthesis.
Studies with Euplotes spp. and S. cerevisiae have shown that coordination of G- and C-strand synthesis plays an important role both in telomere length regulation and in preventing the accumulation of long stretches of single-stranded DNA at the 3′ end of the telomere (1, 7, 14, 28). In Euplotes spp., the physical association between telomerase and the lagging-strand replication machinery most likely facilitates the efficient switch from G- to C-strand synthesis that is so important for precise telomere length regulation. This switch seems to be triggered by a length-measuring activity that initiates C-strand fill-in as soon as it is possible to lay down a C4A4 repeat starting exactly 84 nucleotides along the G-strand (41).
The stable association between telomerase and polymerase α-primase may also have evolved to meet the need for the bulk C-strand synthesis that is required during de novo telomere addition. Recent work with S. cerevisiae indicates that there are alternative ways to recruit the lagging-strand replication machinery for C-strand fill-in. In S. cerevisiae, telomerase and polymerase α-primase probably do not interact directly; instead, one or the other enzyme appears to be recruited to the telomere by the G-overhang binding protein Cdc13p (7, 17, 36). This type of transient interaction may be sufficient for the less stringent telomere length regulation that occurs in S. cerevisiae. It may also reflect the less extensive C-strand fill-in synthesis that is needed to counterbalance G-strand synthesis by telomerase and/or fill in the G-overhang during S. cerevisiae telomere replication (33).
Exactly how the lagging-strand replication machinery represses further G-strand synthesis is still unclear. However, there is evidence that in both S. cerevisiae and Euplotes spp. the actual synthesis of C-strand DNA is required for G-strand length regulation. Elongation of the telomeric G-strand occurs in both organisms when polymerase α activity is reduced by growth at a semipermissive temperature or by treatment with aphidicolin (1, 10, 14). In both situations, primase remains fully active. Hence, recruitment of the polymerase α-primase complex and synthesis of an RNA primer are not sufficient to repress G-strand synthesis. Instead, polymerization of the C-strand may be required. Determining the molecular details of this process will be an important area for future exploration.
Acknowledgments
We thank Larry Klobutcher for the kind gifts of the LEMAC library and the H3 antibody and Leigh Anne Hurley for generating the A1 and A2 Euplotes strains. We thank Chad Price for constant support and help with DNA sequence analysis.
This work was supported by National Institutes of Health grants RO1 GM41803 to C.M.P. and RO1 GM49157 to D.E.S.
REFERENCES
- 1.Adams, A. K., and C. Holm. 1996. Specific DNA replication mutations affect telomere length in Saccharomyces cerevisiae. Mol. Cell. Biol. 16:4614-4620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aigner, S., J. Lingner, K. J. Goodrich, C. A. Grosshans, A. Shevchenko, M. Mann, and T. R. Cech. 2000. Euplotes telomerase contains an La motif protein produced by apparent translational frameshifting. EMBO J. 19:6230-6239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arezi, B., and R. D. Kuchta. 2000. Eukaryotic DNA primase. Trends Biochem. Sci. 25:572-576. [DOI] [PubMed] [Google Scholar]
- 4.Beattie, T. L., W. Zhou, M. O. Robinson, and L. Harrington. 2001. Functional multimerization of the human telomerase reverse transcriptase. Mol. Cell. Biol. 21:6151-6160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bednenko, J., M. Melek, E. C. Greene, and D. E. Shippen. 1997. Developmentally regulated initiation of DNA synthesis by telomerase: evidence for factor-assisted de novo telomere formation. EMBO J. 16:2507-2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carson, M. J., and L. Hartwell. 1985. CDC17: an essential gene that prevents telomere elongation in yeast. Cell 42:249-257. [DOI] [PubMed] [Google Scholar]
- 7.Chandra, A., T. R. Hughes, C. I. Nugent, and V. Lundblad. 2001. Cdc13 both positively and negatively regulates telomere replication. Genes Dev. 15:404-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dawson, D., and G. Herrick. 1984. Rare internal C4A4 repeats in the micronuclear genome of Oxytricha fallax. Mol. Cell. Biol. 4:2661-2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Denis, D., and P. A. Bullock. 1993. Primer-DNA formation during simian virus 40 DNA replication in vitro. Mol. Cell. Biol. 13:2882-2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diede, S. J., and D. E. Gottschling. 1999. Telomerase-mediated telomere addition in vivo requires DNA primase and DNA polymerases alpha and delta. Cell 99:723-733. [DOI] [PubMed] [Google Scholar]
- 11.Dionne, I., and R. J. Wellinger. 1996. Cell cycle-regulated generation of single-stranded G-rich DNA in the absence of telomerase. Proc. Natl. Acad. Sci. USA 93:13902-13907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Evans, S. K., and V. Lundblad. 2000. Positive and negative regulation of telomerase access to the telomere. J. Cell Sci. 113:3357-3364. [DOI] [PubMed] [Google Scholar]
- 13.Fan, Q., and M. Yao. 1996. New telomere formation coupled with site-specific chromosome breakage in Tetrahymena thermophila. Mol. Cell. Biol. 16:1267-1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fan, X., and C. M. Price. 1997. Coordinate regulation of G- and C-strand length during new telomere synthesis. Mol. Biol. Cell 8:2145-2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fang, G., and T. R. Cech. 1995. Telomerase RNA localized in the replication band and spherical subnuclear organelles in hypotrichous ciliates. J. Cell Biol. 130:243-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghosh, S., and L. A. Klobutcher. 2000. A development-specific histone H3 localizes to the developing macronucleus of Euplotes. Genesis 26:179-188. [DOI] [PubMed] [Google Scholar]
- 17.Grandin, N., C. Damon, and M. Charbonneau. 2000. Cdc13 cooperates with the yeast Ku proteins and Stn1 to regulate telomerase recruitment. Mol. Cell. Biol. 20:8397-8408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greene, E. C., and D. E. Shippen. 1998. Developmentally programmed assembly of higher-order telomerase complexes with distinct biochemical and structural properties. Genes Dev. 12:2921-2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Greider, C. 1995. Telomerase biochemistry and regulation. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 20.Harper, D. S., and C. L. Jahn. 1989. Actin, tubulin and H4 histone genes in three species of hypotrichous ciliated protozoa. Gene 75:93-107. [DOI] [PubMed] [Google Scholar]
- 21.Holt, S. E., D. L. Aisner, J. Baur, V. M. Tesmer, M. Dy, M. Ouellette, J. B. Trager, G. B. Morin, D. O. Toft, J. W. Shay, W. E. Wright, and M. A. White. 1999. Functional requirement of p23 and Hsp90 in telomerase complexes. Genes Dev. 13:817-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jahn, C. 1991. The nuclear genomes of hypotrichous ciliates: maintaining the maximum and minimum of information. J. Protozool. 38:252-258. [DOI] [PubMed] [Google Scholar]
- 23.Jonsson, Z. O., and U. Hubscher. 1997. Proliferating cell nuclear antigen: more than a clamp for DNA polymerases. Bioessays 19:967-975. [DOI] [PubMed] [Google Scholar]
- 24.Klobutcher, L. A. 1999. Characterization of in vivo developmental chromosome fragmentation intermediates in E. crassus. Mol. Cell 4:695-704. [DOI] [PubMed] [Google Scholar]
- 25.Klobutcher, L. A., S. E. Gygax, J. D. Podoloff, J. R. Vermeesch, C. M. Price, C. M. Tebeau, and C. L. Jahn. 1998. Conserved DNA sequences adjacent to chromosome fragmentation and telomere addition sites in Euplotes crassus. Nucleic Acids Res. 26:4230-4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klobutcher, L. A., M. T. Swanton, P. Donini, and D. M. Prescott. 1981. All gene-sized DNA molecules in four species of hypotrichs have the same terminal sequence and an unusual 3′ terminus. Proc. Natl. Acad. Sci. USA 78:3015-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lingner, J., and T. R. Cech. 1996. Purification of telomerase from Euplotes aediculatus: requirement of a primer 3′ overhang. Proc. Natl. Acad. Sci. USA 93:10712-10717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martin, A. A., I. Dionne, R. J. Wellinger, and C. Holm. 2000. The function of DNA polymerase α at telomeric G tails is important for telomere homeostasis. Mol. Cell. Biol. 20:786-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Melek, M., E. C. Greene, and D. E. Shippen. 1996. Processing of nontelomeric 3′ ends by telomerase: default template alignment and endonucleolytic cleavage. Mol. Cell. Biol. 16:3437-3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Olins, D. E., and A. L. Olins. 1994. The replication band of ciliated protozoa. Int. Rev. Cytol. 153:137-170. [DOI] [PubMed] [Google Scholar]
- 31.Prescott, D. M. 1994. The DNA of ciliated protozoa. Microbiol. Rev. 58:233-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prescott, J., and E. H. Blackburn. 1997. Functionally interacting telomerase RNAs in the yeast telomerase complex. Genes Dev. 11:2790-2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Price, C. M. 1997. Synthesis of the telomeric C-strand. A review. Biochemistry (Moscow) 62:1216-1223. [PubMed] [Google Scholar]
- 34.Price, C. M. 1990. Telomere structure in Euplotes crassus: characterization of DNA-protein interactions and isolation of a telomere-binding protein. Mol. Cell. Biol. 10:3421-3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Price, C. M. 1995. Telomere-binding proteins of ciliated protozoa. Nucleic Acids Mol. Biol. 9:299-307. [Google Scholar]
- 36.Qi, H., and V. A. Zakian. 2000. The Saccharomyces telomere-binding protein Cdc13p interacts with both the catalytic subunit of DNA polymerase alpha and the telomerase-associated est1 protein. Genes Dev. 14:1777-1788. [PMC free article] [PubMed] [Google Scholar]
- 37.Roth, M., M. Lin, and D. M. Prescott. 1985. Large scale synchronous mating and the study of macronuclear development in Euplotes crassus. J. Cell Biol. 101:79-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roth, M., and D. M. Prescott. 1985. DNA intermediates and telomere addition during genome reorganization in Euplotes crassus. Cell 41:411-417. [DOI] [PubMed] [Google Scholar]
- 39.Shippen-Lentz, D., and E. H. Blackburn. 1989. Telomere terminal transferase activity from Euplotes crassus adds large numbers of TTTTGGGG repeats onto telomeric primers. Mol. Cell. Biol. 9:2761-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Skopp, R., W. Wang, and C. Price. 1996. rTP: a candidate telomere protein that is associated with DNA replication. Chromosoma 105:82-91. [DOI] [PubMed] [Google Scholar]
- 41.Vermeesch, J. R., and C. M. Price. 1994. Telomeric DNA sequence and structure following de novo telomere synthesis in Euplotes crassus. Mol. Cell. Biol. 14:554-566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vermeesch, J. R., D. Williams, and C. M. Price. 1993. Telomere processing in Euplotes. Nucleic Acids Res. 21:5366-5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vishwanatha, J. K., and E. F. Baril. 1986. Resolution and purification of free primase activity from the DNA primase-polymerase alpha complex of HeLa cells. Nucleic Acids Res. 14:8467-8487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Warbrick, E. 2000. The puzzle of PCNA's many partners. Bioessays 22:997-1006. [DOI] [PubMed] [Google Scholar]
- 45.Watson, J. D. 1972. Origin of concatemeric DNA. Nat. New Biol. 239:51-60. [DOI] [PubMed] [Google Scholar]
- 46.Wellinger, R. J., A. J. Wolf, and V. A. Zakian. 1993. Saccharomyces telomeres acquire single-strand TG1-3 tails late in S phase. Cell 72:51-60. [DOI] [PubMed] [Google Scholar]
- 47.Wenz, C., B. Ebenkel, M. Amacker, C. Kelleher, K. Damm, and J. Lingner. 2001. Human telomerase contains two cooperating telomerase RNA molecules. EMBO J. 20:3526-3534. [DOI] [PMC free article] [PubMed]
- 48.Zahler, A. M., and D. M. Prescott. 1989. DNA primase and the replication of the telomeres in Oxytricha nova. Nucleic Acids Res. 17:6299-6317. [DOI] [PMC free article] [PubMed] [Google Scholar]