Abstract
Arg80 and Mcm1, two members of the MADS box family of DNA-binding proteins, regulate the metabolism of arginine in association with Arg81, the arginine sensor. In spite of the high degree of sequence conservation between the MADS box domains of the Arg80 and Mcm1 proteins (56 of 81 amino acids), these domains are not interchangeable. To determine which amino acids define the specificity of Arg80, we swapped the amino acids in each secondary-structure element of the Arg80 MADS box domain with the corresponding amino acids of Mcm1 and assayed the ability of these chimeras to regulate arginine-metabolic genes in place of the wild-type Arg80. Also performed was the converse experiment in which each variant residue in the Mcm1 MADS box domain was swapped with the corresponding residue of Arg80 in the context of an Arg80-Mcm1 fusion protein. We show that multiple regions of Arg80 are important for its function. Interestingly, the residues which have important roles in determining the specificity of Arg80 are not those which could contact the DNA but are residues that are likely to be involved in protein interactions. Many of these residues are clustered on one side of the protein, which could serve as an interface for interaction with Arg81 or Mcm1. This interface is distinct from the region used by the Mcm1 and human serum response factor MADS box proteins to interact with their cofactors. It is possible that this alternative interface is used by other MADS box proteins to interact with their cofactors.
The Arg80 (ArgRI) and Mcm1 proteins of Saccharomyces cerevisiae belong to the MADS box family of transcriptional regulatory factors, which consists of about 100 members (3). These DNA-binding proteins are found in yeast species, flies, plants, and humans and play important roles in diverse biological functions. In humans, the serum response factor (SRF) regulates immediate-early gene expression (for a review, see reference 19), whereas the myocyte enhancer factor 2 (MEF2) family plays a pivotal role in morphogenesis and myogenesis of skeletal, cardiac, and smooth muscle cells (29). Recently MEF2A, MEF2B, MEF2C, and MEF2D were shown to activate gene expression in response to mitogenic signaling pathways (6). In plants, MADS box genes encode homeotic proteins that control flower organ identity. In addition, these proteins regulate the timing of floral initiation and flower meristem identity as well as various aspects of ovule, fruit, leaf, and root development (9, 33).
Different yeast species also contain MADS box proteins. In Schizosaccharomyces pombe, the map1 gene encodes a protein required for cell-type-specific gene expression, and in Ustilago maydis, umc1 encodes a protein that regulates the expression of pheromone-inducible genes (20, 41). In S. cerevisiae, there are four MADS box proteins, Mcm1, Arg80, Rlm1, and Smp1, that fall into two distinct groups. Mcm1 and Arg80 are closely related to the human SRF and plant AGL3A proteins, whereas Rlm1 and Smp1 belong to the MEF2-like family (3). Rlm1 controls expression of genes required for cell wall integrity, while Smp1 is involved in osmotic stress response mediated by the Hog1 signal transduction pathway (8, 10, 40). Mcm1 is essential for cell viability and controls G1/S and G2/M cell cycle transitions (2, 22, 28), mating (18), osmotolerance (21), recombination (15), minichromosome maintenance (30), and arginine metabolism (24). Mcm1 works to control these diverse sets of genes through interactions with different cofactors. The role of Arg80 is to combine with Mcm1 to regulate the cell's metabolism in response to different levels of arginine. Both Mcm1 and Arg80 are stabilized in the nucleus by Arg82, an inositol polyphosphate multikinase (14, 27, 34). In the absence of Arg82, many cellular processes are impaired as a result of decreased levels of Mcm1 (12, 14). To regulate the expression of arginine-anabolic and -catabolic genes, Arg80 and Mcm1 combine in a complex with Arg81, which acts as an arginine sensor (4). In the presence of high arginine concentrations, this Mcm1-Arg80-Arg81 protein complex is able to bind to DNA target sequences called “arginine boxes” to repress or activate transcription (for a review, see reference 25).
The X-ray structures of the protein/DNA complexes of three proteins from the MADS box family (SRF, Mcm1, and MEF2A) have been solved (31, 36, 38) and have revealed that their overall structures are very similar. There is a high degree of sequence similarity between the MADS box domain of Mcm1 and Arg80 (56 of 81 amino acids), suggesting that Arg80 has an overall structure similar to that of the Mcm1 MADS box domain. The MADS box domains of Mcm1 and Arg80 are sufficient to carry out their cellular functions (18, 32). However, in spite of the high degree of sequence conservation, the Mcm1 and Arg80 MADS box domains are not interchangeable, suggesting that their specificities of function must result from subtle differences in the two proteins. The 25 residues which differ between the domains of Arg80 and Mcm1 could result in different local conformations, allowing Mcm1 to recruit a different set of gene-specific regulators and restricting Arg80 to a role in the control of arginine metabolism. To determine which amino acids define the specificity of Arg80, we progressively replaced the amino acids of Arg80 with the corresponding residues of Mcm1. We also performed the converse experiment and, in the context of an Arg80-Mcm1 fusion protein, systematically changed residues in the Mcm1 MADS box domain to those in Arg80. We found that multiple regions of the domain are important for functional specificity and that many of the important substitutions cluster in a cleft on one side of the protein that could serve as the interface for interactions between Arg80 and its cofactors. This region is distinct from the regions of SRF and Mcm1 where these proteins interact with their cofactors and therefore represents a novel interface that may be used by other MADS box proteins to interact with their cofactors. These experiments also show that only a small subset of residues are involved in determining the specificity of the protein interactions and functions of these MADS box proteins.
MATERIALS AND METHODS
Strains and medium.
S. cerevisiae strain 02463d (MATa leu2 ura3) was used for deletion of the RLM1 and SMP1 genes by use of the long flanking homology-PCR strategy (39). The different coding sequences were replaced by the kanMX4 gene cassette, which confers resistance to geneticin, yielding strains 02463dΔRLM1 and 02463dΔSMP1. The correct targeting of the deletions in G418r transformants was verified by PCR with whole cells as a source of DNA and appropriate primers. To construct strain 02463dΔRLM1,ΔSMP1, we used the loxP-kanMX4-loxP disruption cassette (16). To eliminate the kanMX4 marker from the disrupted gene, the mutated strain was transformed with the cre expression plasmid pSH47, which carries the URA3 marker gene and the cre gene under the control of the inducible GAL1 promoter. Expression of the Cre recombinase was induced by shifting cells from yeast extract-peptone-dextrose (YPD) to yeast-extract-galactose medium for 2 h. The loss of the kanMX4 cassette was detected by plating cells on YPD and replica plating the colonies onto YPD-G418. The cre expression plasmid was removed from the strain by streaking cells on plates containing 5-fluoroorotic acid to counter select for the loss of the plasmid. Strains 02463dΔARG80 and BY4709ΔARG80 (14) were used as recipient strains for transformation with different plasmids. Strains 1c2163aΔMCM1+pMCM1, 1c2187dΔMCM1+pmcm1G86D, and 1c2163aΔMCM1+pumc1 were obtained by the following procedure: the diploid strain 02463d/02839d (leu2 ura3/leu2 ura3) was transformed with the kanMX4 cassette, allowing the deletion of the MCM1 gene by selection of transformants on YPD-G418. One transformant containing a deletion of one copy of the MCM1 gene was transformed with plasmids pED40 (pUC19 with 2μm LEU2 MCM1) (12), pAJ88 (pUC19 with 2μm LEU2 mcm1G86D), or pAJ125 (pYX with 2μm LEU2 TPI-umc1) and plated on sporulation medium. After dissection, spores that contained mcm1::kanMX4 and the complementing plasmid were selected on YPD-G418. All yeast strains were grown on minimal medium containing vitamins, mineral traces, and 3% glucose or 1% galactose (23). The nitrogen source was either 0.02 M ammonium sulfate (M.ammonia) or 1-mg/ml ornithine (M.ornithine). Plasmid pAJ125, containing gene umc1, was obtained by PCR synthesis of a 1,350-bp DNA fragment with plasmid pYUMC1-454 (a gift from R. Kahmann [20]) as a template and appropriate oligonucleotides flanked by BamHI and HindIII sites as primers. This fragment was inserted in vector pYX242 (pYX LEU2 2μm TPI promoter; R & D Systems).
Creation of mutations in ARG80.
Appropriate oligonucleotides were used to create the replacement of Arg80 amino acids with those of Mcm1 in each secondary-structure region of the MADS box domain (N-terminal coil, αI, βI, βII, αII). The specific mutations were created by in vitro mutagenesis on double-stranded DNA from plasmid pFL38-RI (pUC19 with ARS4 CEN6 URA3 ARG80) (32) by using the QuikChange site-directed mutagenesis kit from Stratagene (see Fig. 3 for a list of all of the resulting mutated plasmids). To combine mutations created in different regions of the MADS box domain, plasmids pAJ26, pAJ27, pAJ30, pAJ31, pAJ58, pAJ104, and pHL3 were used as templates to introduce mutations into other regions of the protein. All of the arg80 mutants were verified by DNA sequence analysis.
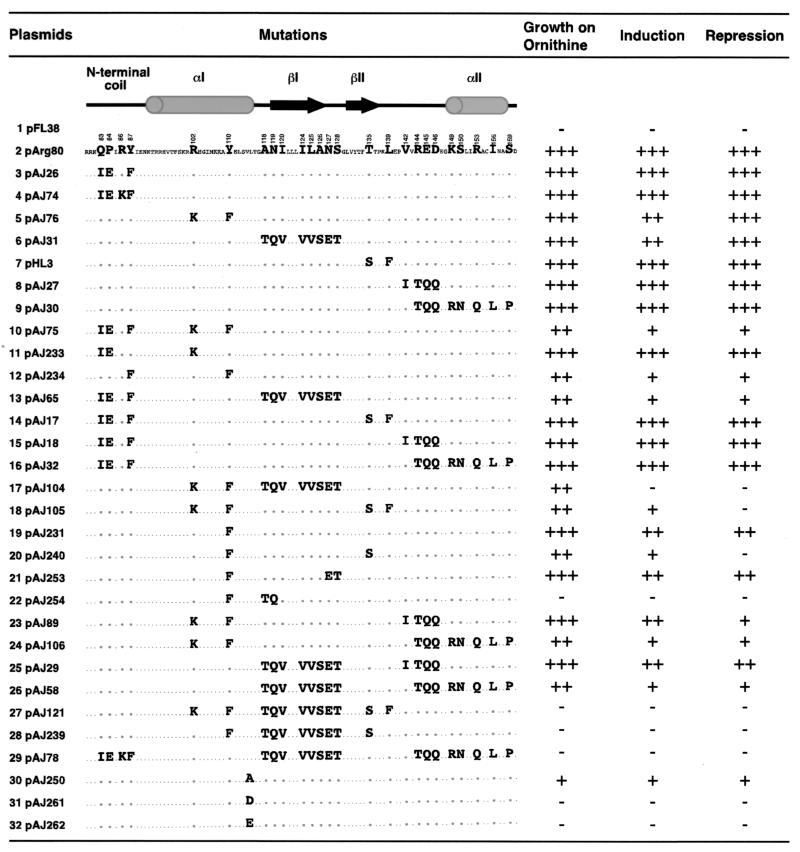
FIG. 3.
Loss of Arg80 function resulting from the swapping of residues in the MADS box domain of Arg80 with the corresponding residues in Mcm1. Strain 02463dΔARG80 was transformed with the listed plasmids, and Arg80 function was determined by a growth test on M.ornithine as the sole nitrogen source and by assays of OTCase and arginase activities after growth on M.ammonia with or without arginine (1 mg/ml). For growth on ornithine, +++ indicates growth at levels similar to that of a wild-type strain, ++ indicates a slight growth reduction, and −− indicates residual growth of an arg80 deletion strain. For repression of OTCase and induction of arginase, +++ indicates the same repression level (fourfold) and induction level (10-fold) as those obtained with a wild-type Arg80 protein, ++ indicates a twofold repression level and a four- to fivefold induction level, + indicates a 1.5-fold repression level and two- to threefold induction level, and - indicates an absence of induction or repression. The predicted secondary structure of the MADS box domain of Arg80 and its amino acid sequence are indicated. The smaller letters indicate the amino acids of Arg80 and Mcm1 that are identical, and the larger letters indicate the nonconserved residues. In each line, the residues indicate the amino acids of Mcm1 that were used to replace the corresponding amino acids in Arg80.
Creation of mutations in ARG80-MCM1-ARG80.
To construct a hybrid Arg80-Mcm1-Arg80 protein, the region encoding the first 77 amino acids of Arg80 was fused in frame to the regions encoding amino acids 18 to 98 of Mcm1 and the 15 C-terminal amino acids of Arg80. We first created by in vitro mutagenesis two HindIII sites in ARG80 by using plasmid pKS-RI and appropriate oligonucleotides, which led to the changes V78L, T79K, T161K, and P162L, yielding plasmid pHL10. These modifications did not affect Arg80 function. After this plasmid was digested with HindIII, a 249-bp fragment containing the MADS box domain of Arg80 was replaced by a DNA fragment containing the MADS box domain of Mcm1 synthesized by PCR with flanking HindIII restriction sites. This plasmid contains the hybrid ARG80-MCM1-ARG80 gene and was named pHL12. pHL12 was digested with EcoRI and SalI, and the 1.6-kb fragment containing the ARG80-MCM1-ARG80 fusion was inserted in pFL38 vector (pUC19 URA3 ARS4 CEN6), yielding plasmid pHL13.
Appropriate oligonucleotides were used to create the replacement of Mcm1 amino acids with those of Arg80 by in vitro mutagenesis on double-stranded DNA from plasmid pHL13 by using the QuikChange site-directed mutagenesis kit from Stratagene (see Fig. 4 for a list of all of the resulting mutated plasmids). To combine mutations created in different regions of the MADS box domain, plasmids pAJ10, pAJ11, pAJ13, pAJ15, pAJ16, and pAJ45 were used as templates to introduce mutations into other regions of the protein. All of the mutants were verified by DNA sequence analysis.
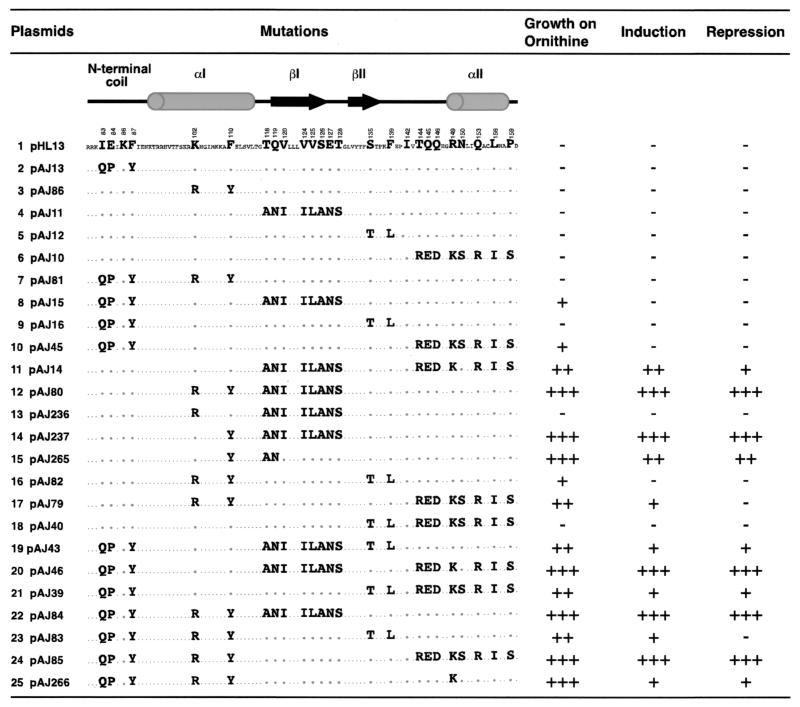
FIG. 4.
Conferment of Arg80 function by the swapping of amino acids in the Mcm1 MADS box domain of the hybrid Arg80-Mcm1-Arg80 protein. The amino acids that were swapped in each construct are shown. The numbers of the residues in the MADS box domain indicate the positions of the homologous residues in the Arg80 protein. The levels of Arg80 function for each mutant were assayed and presented as described in the legend to Fig. 3.
Two-hybrid assays.
Two-hybrid assays to examine the interactions between wild-type and mutant Arg80 and Arg80 cofactors were performed with plasmids pME46 (GBD-Arg80), pME21 (GAD-Arg80), pNA51 (GBD-Mcm1), pME19 (GAD-Mcm1), pNA33 (GBD-Arg81), and pME9 (GAD-Arg81), as described previously (14). To construct the mutated GBD-arg80 and GAD-arg80 fusions, we amplified by PCR the mutated arg80 DNA fragments by using as templates the arg80 genes present on plasmids pAJ234, pAJ239, and pAJ240 and primer oligonucleotides containing a BamHI restriction site. The DNA fragments were cloned into the BamHI site of pAS2(GBD) or pACTII (GAD) (13) to yield plasmids pAJ257, pAJ256, and pAJ255 containing the mutated GBD-arg80 fusions and plasmids pAJ260, pAJ259, and pAJ258 containing the mutated GAD-arg80 fusions. The GBD fusions were assayed for activation in complex with a nonspecific GAD fusion as well as with Mcm1 and Arg81 fusions to GAD. GAD-Arg80 fusions were assayed in combination with GBD-Arg81 and GBD-Mcm1 fusions.
DNA-binding assays.
The overexpression of Arg80 and Mcm1 proteins in S. cerevisiae is required in order to assay DNA-binding activity to the P site by these proteins from crude extracts. For Mcm1, plasmid pED40 (12) was used as a template for site-directed mutagenesis to yield plasmids pAJ210 (mcm1-I21Q) and pAJ211 (mcm1-K40R). For Arg80, plasmid pME51, which contains the ARG80 gene expressed under the GAL10 promoter (14), was used as a template for site-directed mutagenesis to construct plasmids pAJ219 (arg80-R102K-Y110F) and pAJ221 (arg80-Q83I-P84E-R86K-Y87F). These plasmids were used to obtain the combination of mutations yielding plasmid pAJ220 (arg80-Q83I-P84E-R86K-Y87F-R102K-Y110F). Strain BY4709ΔRI (arg80::kanMX4) was transformed with each of these plasmids, and crude extracts were prepared from cells grown on galactose medium as described previously (14).
The DNA-binding activities of the wild-type and mutant Arg80 proteins in complex with Mcm1 and Arg81 were assayed for binding activity to the arginine box region in the ARG5,6 promoter (11). Semipurified crude extracts of strain 02463dΔARG80 (ura3 leu2 arg80::kanMX4) were cotransformed with pYEP34 (2μm LEU2 GAL10-ARG81) and with wild-type ARG80 (pFL38-RI), an arg80 mutant (pAJ234, pAJ240, or pAJ239), or the ARG80-MCM1-ARG80 fusion (pHL13, pAJ46, pAJ84, pAJ85) plasmids.
Extract preparation and binding assays were performed as described previously (11). The DNA fragments used in these assays were the 160-bp HindIII-BamHI fragment from CY1320 containing the P site (5) and the 160-bp AluI-AluI fragment from plasmid pED20 containing the control region of the ARG5,6 gene. These fragments were end-labeled with [γ-32P]ATP by use of polynucleotide kinase according to the procedure described by Sambrook et al. (35).
Western blot analysis.
For detection of Arg80 and Mcm1, 25 ml of exponentially growing cells was harvested by centrifugation and the proteins were extracted according to the trichloroacetic acid method (Clontech). About 200 μg of total proteins was electrophoresed in a 10% polyacrylamide gel containing sodium dodecyl sulfate. After electrotransfer of proteins to Hybond membranes (Amersham), specific proteins were detected with polyclonal antibodies against GST-Arg80 and GST-Mcm1 that were obtained by injection of these purified proteins in mice (the antibodies were a gift from Paul Jacobs). After incubation with anti-mouse immunoglobulin G-specific antibody conjugate to horseradish peroxidase, peroxidase activity was revealed with an enhanced chemiluminescence kit according to the instructions of the manufacturer (Roche).
RESULTS
Smp1 and Rlm1 are not involved in the control of arginine metabolism.
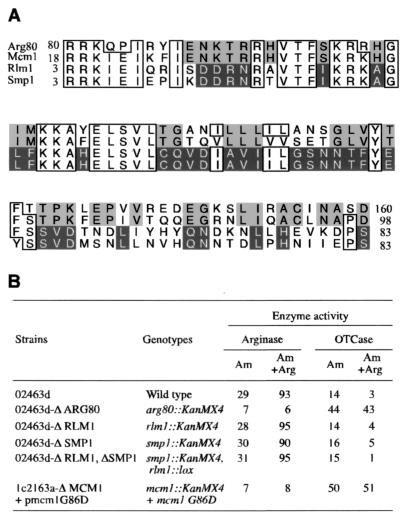
Although we have defined the minimum protein complex required to interact with the arginine boxes in an arginine-dependent manner in vitro, we could not exclude the possibility that additional proteins were involved in this control in vivo. Given the high degree of sequence similarity among the MADS box proteins of S. cerevisiae (Fig. 1A), we were interested in whether Smp1 and Rlm1 also have a role in the regulation of arginine metabolism. We thus deleted each gene by insertion of the kanMX4 cassette in place of the SMP1 or RLM1 coding sequence (see Materials and Methods). Impairment of the control of arginine-coregulated genes leads to a loss of repression of arginine-anabolic genes and to a loss of induction of arginine-catabolic genes. However, the levels of ornithine carbamoyltransferase (OTCase), an anabolic enzyme, and of arginase, a catabolic enzyme, in the Δsmp1 and Δrlm1 strains were comparable to the levels in the wild-type strain after growth with and without arginine (Fig. 1B). Moreover, the simultaneous deletion of both genes did not impair the regulation by arginine. In contrast, in an arg80 deletion strain, as well as in an mcm1 mutant strain, repression and induction of the reporter genes were severely impaired. These results indicate that the Smp1 and Rlm1 MADS box proteins are not required for the control of arginine-anabolic and -catabolic genes.
FIG. 1.
The Rlm1 and Smp1 MADS box proteins do not regulate arginine metabolism. (A) Amino acid alignment of the four MADS box proteins of S. cerevisiae. The numbers indicate the positions of the first and last residues of the MADS box domains in the context of the full-length proteins. Amino acids that are identical among three of the four proteins are boxed. Residues that are identical between Arg80 and Mcm1 are shaded in light gray, and residues that are identical between Rlm1 and Smp1 are shaded in dark gray. (B) Effects of deletions of MADS box genes on the expression of arginine-coregulated genes. The enzyme-specific activities were measured at 30°C after growth of the different strains on M.ammonia with or without 1 mg of arginine per ml plus 25 μg of uracil per ml and 50 μg of leucine per ml. OTCase- and arginase-specific activities were expressed in terms of micromoles of citrulline or of urea formed per hour per milligram of protein, respectively. The values are the means of at least three independent assays, and the standard error was 10 to 15%.
Overexpression of MCM1 and umc1 does not complement an arg80 mutant, whereas overexpression of umc1 complements an mcm1 mutant.
Arg80 and Mcm1 from S. cerevisiae and Umc1 from U. maydis are very closely related among the MADS box proteins. The homolog closest to Arg80 is Mcm1, whereas the homolog closest to Mcm1 is Umc1 (Fig. 2A). When overexpressed, Umc1 is able to complement the lethality of an mcm1 mutant (20). We have found that Umc1 also replaces Mcm1 in its arginine-regulatory function, since the mcm1 deletion strain overexpressing umc1 was able to grow on M.ornithine (Fig. 2B). Overexpression of umc1 restored the repression of OTCase and partially restored the induction of arginase (Fig. 2C). This result prompted us to test whether Umc1 could fulfill Arg80 function. A deletion of the ARG80 gene led to an absence of growth on ornithine as the sole nitrogen source. Overexpression of MCM1 or umc1 did not restore either the arg80 growth defect on ornithine or the regulation of anabolic and catabolic enzymes, thus showing the high specificity of Arg80 for the regulation of these target genes (Fig. 2D and E).
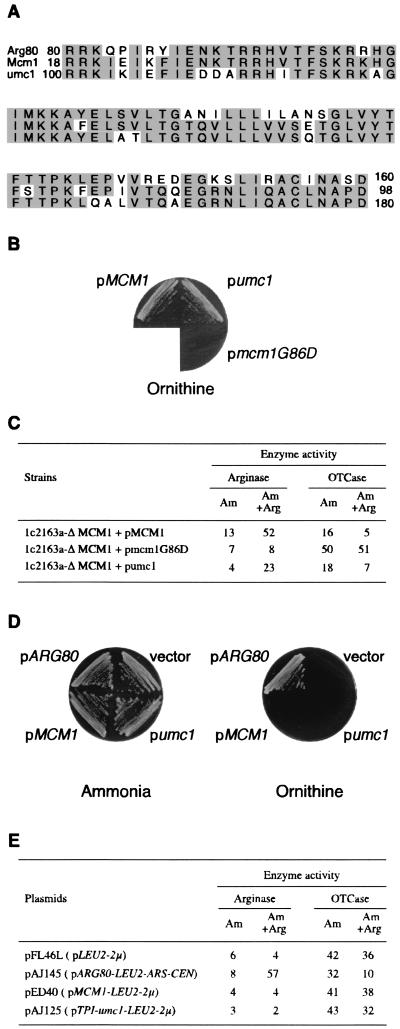
FIG. 2.
The U. maydis Umc1 MADS box protein does not complement arg80 but does complement mcm1 for arginine-dependent regulation. (A) Amino acid alignment of Arg80 and Mcm1 from S. cerevisiae and Umc1 from U. maydis. The numbers indicate the positions of the first and last residues of the MADS box domains in the context of the full-length proteins. Shading indicates amino acids that are identical among the three proteins. (B) Growth test of 1c2163aΔMCM1 strains transformed with plasmid pED40 (MCM1), pAJ125 (umc1), or pAJ88 (mcm1G86D). The transformed strains were grown on M.ornithine as the nitrogen source plus 25-μg/ml uracil. (C) Enzyme-specific activities of the strains in panel B. Activities were measured as described in the legend to Fig. 1B. (D) Growth tests, as described in the legend to panel B, of strain 02463dΔARG80 transformed with pFL46L (vector), pAJ145 (ARG80), pED40 (MCM1), or pAJ125 (umc1). (E) Enzyme-specific activities of the strains in panel D were measured as described in the legend to Fig. 1B.
Identification of amino acids defining Arg80 specificity.
Given the strong sequence conservation among the MADS box proteins and the close similarity of the crystal structures of the SRF, MEF2A, and Mcm1 MADS box domains, it is likely that the Arg80 MADS box domain is also folded into a similar structure. Mutations in ARG80 that led to loss of Arg80 function have previously been isolated, and it was found that a number of mutants resulted from amino acid changes in residues that are highly conserved among the MADS box proteins (14, 32). Some of these residues are located in the αI helix, and the loss of Arg80 function is caused by the loss of interaction with one of its cofactors, Arg82 (14). Many of the residues in this helix are highly conserved among MADS box proteins and are likely to be important for maintaining the DNA-binding activity or global structure of the domain (1). However, since the Mcm1 and Arg80 MADS box domains cannot be substituted for each other, it is likely that the differential specificities of these proteins depend on the residues that are different between the two proteins. To test this model, we replaced residues in different regions of the Arg80 MADS box domain with the corresponding amino acids in Mcm1 and examined whether these replacements affected Arg80 function (Fig. 3). Strain 02463dΔARG80 (arg80::kanMX4) was transformed with plasmid pFL38 (vector), plasmid pArg80 (ARG80), and plasmids bearing mutations in the different secondary-structure regions of the Arg80 MADS box domain. Arg80 function was assayed by determining growth on ornithine as the sole nitrogen source and by measuring the levels of OTCase and arginase after growth in the presence or absence of arginine. Interestingly, even though multiple residues were mutated within each of the different secondary-structure regions, none of these amino acid permutations affected Arg80 activity (Fig. 3, lines 3 to 9). This result suggests that the multiple regions of the MADS box domain are involved in conferring specificity and that mutations in any single region of the domain are not sufficient to significantly interfere with its function. To test this model, we combined mutations from different regions of the domain in various combinations (Fig. 3, lines 10 to 29). Substitutions in the N-terminal arm in combination with substitutions in the C-terminal region of the domain had no effect on Arg80 function (Fig. 3, lines 14 to 16). In contrast, mutations in αI or βI in combination with mutations in any other region significantly affected Arg80 function (Fig. 3, lines 10, 17, 18, and 20 and lines 13 and 22, respectively). The swapping of residues in αI in combination with mutations in βII abolished the induction of arginase and repression of OTCase reporters (Fig. 3, line 17). However, there was still a small amount of residual activity in this mutant because it was still able to grow on ornithine. Substitutions in three of the five secondary-structure elements completely eliminated regulation of arginine metabolism (Fig. 3, lines 27 to 29). This result indicates that multiple regions of the protein are required for regulation of arginine-specific genes.
Some of the mutations described above involve substitutions of amino acids at positions that correspond to residues that contact the DNA in the crystal structures of SRF, MEF2, and Mcm1 (26, 31, 36, 38). These mutations may therefore affect the ability of Arg80 to bind DNA as opposed to its ability to interact with its cofactors. To discriminate between these possibilities, we separated the amino acid substitutions of residues predicted to contact the DNA from those that are not likely to directly contact the DNA. For example, the combined substitutions in the N-terminal arm and the αI helix caused a large decrease in Arg80-dependent repression and induction (Fig. 3, line 10). Arg80 residues Q83 and P84 correspond to residues I21 and E22 in Mcm1. The side chain of I21 contacts the DNA in the SRF, Mcm1, and MEF2 crystal structures, and mutations at this position in Mcm1 decrease the binding affinity of the protein (1, 26, 31, 36, 38). Residue K40 in the αI helix of Mcm1, which corresponds to R102 in Arg80, also plays a key role in the DNA-binding affinity of and DNA bending by Mcm1 (1). In contrast, the residues in Mcm1 corresponding to positions Y87 and Y110 in Arg80 are relatively far from the DNA and are unlikely to directly contact the DNA. However, a large portion of the aromatic ring of these side chains is solvent exposed, suggesting that these side chains may be involved in the interactions of Arg80 with its cofactors. To determine whether the decrease in Arg80-dependent regulation of the pAJ75 mutant was due to defects in DNA binding or protein interactions, we created the following changes: Q83I-P84E-R102K (pAJ233) and Y87F-Y110F (pAJ234). The substitution of residues that are likely involved in binding DNA did not cause a defect in Arg80-dependent regulation (Fig. 3, line 11). In contrast, substitution of residues Y87 and Y110 caused a significant drop in Arg80-mediated induction and repression (Fig. 3, line 12). Since the tyrosine-to-phenylalanine changes are relatively conserved, these results support the notions that these side chains interact with Arg80 cofactors and that the hydroxyl group of the tyrosine is important for these interactions.
In combination with the swap in αI, substitution of residues in the βII region of the Arg80 MADS box domain caused a significant drop in Arg80 function (Fig. 3, line 18). In the Mcm1 crystal structure, the residue corresponding to L139 of Arg80 is a phenylalanine that is completely buried at the dimer interface of the protein. The side chain of the residue corresponding to T135 of Arg80 is also buried, but the amide nitrogen and carbonyl oxygen of the backbone, along with the hydroxyl group of the side chain, are solvent exposed in a cleft above the residue corresponding to Y110. It is possible that the peptide backbone atoms of residue T135 are involved in interaction with Arg80 cofactors. To test this model, we constructed the Y110F T135S double mutant (Fig. 3, line 20). Compared with the single Y110F mutation, the double mutation significantly impaired the repression of OTCase and the induction of arginase. The double substitution may cause the peptide backbone to change position, affecting the ability of Arg80 to interact with its cofactors. Finally, combining the Y110F substitution with the A118T and N119Q substitutions on the N terminus of the βI strand (Fig. 3, lines 22, 27, and 28) caused a complete loss of Arg80-dependent regulation. Taken together, these mutations define a partially contiguous face on the side of the MADS box domain that is required for Arg80 function. It is likely that these residues form a surface for interactions between Arg80 and its cofactors.
If the crystal structure of Mcm1 is used as a rough model of the structure of Arg80, then residues Y87 and Y110 lie on the side of the protein, away from the DNA. Residue V114 is conserved in both Mcm1 and Arg80, and in Mcm1, this side chain is solvent exposed between residues Y87 and Y110. If residues Y87 and Y110 are contacted by Arg80 cofactors, then it is likely that V114 is also in contact with this protein. To test this model, substitutions were made at this position and the protein was assayed for arginine-dependent regulation. The conservative substitution to alanine caused a partial loss of function of the protein, while more radical substitutions to aspartic or glutamic acid completely eliminated the function of the protein (Fig. 3, lines 30 to 32). It is possible that these radical changes alter the structure of the region, thereby indirectly affecting the ability of Arg80 to interact with its cofactors. However, the observation that the relatively conservative alanine substitution also has an effect on Arg80 function supports the notion that this solvent-exposed region may be involved in interactions with other proteins.
Swapping the specificities of Mcm1 and Arg80.
The experiments described above indicated which of the nonconserved residues in the Arg80 MADS box domain are required for its function. To test whether these amino acids are sufficient to confer Arg80 function, we replaced the nonconserved residues in Mcm1 with the corresponding amino acids of Arg80. This analysis was performed in a hybrid protein (Arg801-79-Mcm118-98-Arg80162-177) in which the MADS box domain of Arg80 was swapped with the corresponding region of Mcm1. This protein complements an mcm1 mutant for cell viability and Mcm1-dependent regulation of arginine metabolism (data not shown). However, this fusion does not complement an arg80 mutant for regulation of arginine metabolism, indicating that this chimera lacks the specific sequence requirements in the MADS box domain for Arg80 function. To determine which residues of the MADS box domain are sufficient to confer Arg80 function, we made a set of mutations that was converse to those shown in Fig. 3. For comparison with the mutations we made in Arg80, the same numbering system was used for residues in the Mcm1 MADS box domain of the fusion protein. Individual substitution of residues in any single region of the domain did not restore Arg80 function to the fusion protein (Fig. 4, lines 2 to 6). However, the combination of mutations in the αI and βI regions, as well as simultaneous replacement in three structural motifs, such as the N-terminal coil, βI, and αII or the N-terminal coil, αI, and αII, allowed growth of an arg80 strain on M.ornithine (Fig. 4, lines 12, 20, and 24). This result further supports the notion that multiple regions of the MADS box domain are important for conferring Arg80 specificity.
The differences between swapping the βI region in the presence and in the absence of the K102R-F110Y mutations were very striking (Fig. 4, compare lines 3 and 4 with line 12). To determine whether the K102R or F110Y substitution was responsible for the conferment of Arg80 function, we constructed plasmids in which each substitution was made separately in combination with the swap of the βI region. The F110Y substitution was sufficient to swap the specificity in the pAJ11 background (Fig. 4, compare line 14 with line 4). In contrast, the K102R-βI swap construct showed no activation (Fig. 4, line 13). This result further supports the notion that the hydroxyl group of the Y110 side chain, along with residues in the βI region of the protein, are important for Arg80-specific function.
The amino acid swaps in the context of Arg80 showed that residues Y110, A118, and N119 together have an important role in Arg80 function (Fig. 3, line 22). To determine whether these residues are sufficient for Arg80 specificity, we constructed the converse swap in the context of the Arg80-Mcm1-Arg80 fusion. This mutant had wild-type levels of growth on ornithine and significant induction and repression of the arginine-dependent reporters (Fig. 4, line 15). These results indicate that these residues play a major role in conferring specificity to Arg80.
Although residues Y110, A118, and N119 have very important roles in determining Arg80 specificity, our data show that other regions of the protein also contribute to the function of the protein. For example, the swap of the αII region in combination with that of the N-terminal coil (pAJ45 and pAJ39) or the swap of the αI region in combination with that of αII (pAJ79) partially restored Arg80 function to the fusion protein (Fig. 4, lines 10 and 21 and line 17, respectively). The role of the αII region in conferring specificity is especially apparent when comparing pAJ81 with pAJ85 (Fig. 4, lines 7 and 24). In other MADS box proteins, the residues in the αII helix form a partially solvent-exposed layer on the top of the MADS box domain (26, 31, 36, 38). In Mcm1, residues corresponding to positions K149, R153, and I156 of Arg80 form a hydrophobic pocket that serves as a protein contact region for a phenylalanine residue in the MATα2 protein, a cofactor of Mcm1 (38). To test whether this region of Arg80 is involved in similar contacts, we constructed an R149K substitution in the context of pAJ81 (Fig. 4, compare lines 7 and 25). This mutation partially restores Arg80 function to the chimera protein, indicating that this region of the protein may also be involved in contacts with Arg80 cofactors.
Loss of function of Arg80 results from loss of interaction with Mcm1.
The results from the substitutions in both the Arg80 and Mcm1 MADS box domains suggest that Arg80 residues Y87, Y110, A118, N119, and T135 play important roles in the function of the protein. Using the Mcm1 crystal structure as a model, we found that these residues are solvent exposed but are located away from the DNA, suggesting that they are more likely to be involved in protein interactions than direct DNA binding. To test this model, two-hybrid assays were used to examine the interaction between the arg80 mutants and Arg81 or Mcm1. The Y87F, Y110F, βI swap, and T135S mutations caused a modest decrease in the interaction with Arg81 but caused a more significant decrease in the interaction with Mcm1 (Tables 1 and 2). The assay produced similar results when the arg80 mutants were fused to either the Gal4 DNA-binding or activation domains. These results support the notion that these residues of Arg80 are involved in the interactions of Arg80 with its cofactors rather than in DNA binding.
TABLE 1.
Ability of mutated GBD-arg80 proteins to interact with Arg81 and Mcm1a
| Hybrid | β-Galactosidase sp act (nmol of o-nitrophenyl-β-d-galactopyranoside hydrolyzed/min/mg of protein)
|
||
|---|---|---|---|
| GAD (pACTII) | GAD-Arg81 (pME9) | GAD-Mcm1 (pME19) | |
| GBD-Arg80 (pME46) | <1 | 89 | 4 |
| GBD-arg80Y87F-Y110F (pAJ257) | <1 | 129 | 2 |
| GBD-arg80Y110F-βI swap-T135S (pAJ256) | <1 | 31 | <1 |
| GBD-arg80Y110F-T135S (pAJ255) | <1 | 61 | <1 |
Transcription activation of the lacZ gene was estimated by determination of β-galactosidase activity in S. cerevisiae strain HY cotransformed with plasmids expressing GAD (pACTII), GAD-Arg81 (pME9), or GAD-Mcm1 (pME19) and the different wild-type or mutated GBD-arg80 fusions. Values shown are the means of three independent measurements with variations of less than 15%.
TABLE 2.
Ability of mutated GAD-arg80 proteins to interact with Arg81 and Mcm1a
| Hybrid | β-Galactosidase sp act (nmol of o-nitrophenyl-β-d-galactopyranoside hydrolyzed/min/mg of protein)
|
|
|---|---|---|
| GBD-Arg81 (pNA33) | GBD-Mcm1 (pNA51) | |
| GAD (pACTII) | 25 | 4 |
| GAD-Arg80 (pME21) | 220 | 15 |
| GAD-arg80Y87F-Y110F (pAJ260) | 116 | 9 |
| GAD-arg80Y110F-βI swap-T135S (pAJ259) | 130 | 3 |
| GAD-arg80Y110F-T135S (pAJ258) | 127 | 3 |
Transcription activation of the lacZ gene was estimated by determination of β-galactosidase activity in S. cerevisiae strain HY cotransformed with plasmids expressing GBD-Arg81 (pNA33) or GBD-Mcm1 (pNA51) and the different wild-type or mutated GAD-arg80 fusions. Values shown are the means of three independent measurements with variations of less than 15%.
Mutations in Arg80 increase its DNA-binding affinity to P sites.
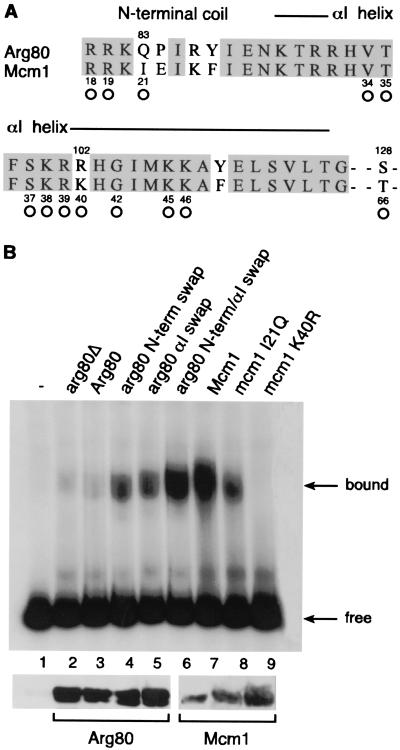
The Arg80, Mcm1, and Arg81 proteins bind as a complex to sites called arginine boxes that are found upstream of arginine-regulated genes (4, 11). However, as with many MADS box proteins, Mcm1 and Arg80 are able to bind on their own to palindromic consensus sequences, called P sites, although the binding affinity of Arg80 is very weak. Since Arg80 and Mcm1 share such strong sequence similarity, it is likely that Arg80 uses many of the same residues in the MADS box domain to contact DNA (Fig. 5A). However, there are several positions that likely contact the DNA but that are different between the two proteins. We were therefore interested in examining whether some of the amino acid substitutions that we made in Arg80 and Mcm1 affect the ability of the proteins to bind DNA. Using plasmid pME51, which expresses ARG80 under the control of the GAL10 promoter, we created the Q83I, P84E, R86K, and Y87F mutations in the N-terminal coil extension of Arg80 protein (plasmid pAJ221), the R102K and Y110F mutations in the αΙ helix (plasmid pAJ219), and the simultaneous replacement of these six residues (plasmid pAJ220). The binding efficiency of the wild-type and the mutated Arg80 proteins was determined by gel shift experiments with crude extracts from the different transformants and a P site DNA as a probe. Even in the absence of Arg80, we observed a weak binding to the site, presumably produced by the genomic copy of Mcm1 (Fig. 5B, lane 2). In crude yeast extract, there was little or no detectable binding of Arg80 to the DNA, since there was no additional shift when Arg80 was overproduced (Fig. 5B, lane 3). In contrast, the replacement of residues either in the N-terminal coil extension or in the αI helix of Arg80 with those in Mcm1 enhanced DNA binding by the protein (Fig. 5C, lanes 4 and 5). Replacing residues in both the N-terminal arm and αI helix of Arg80 with those of Mcm1 strongly increased the binding affinity to almost the level seen in wild-type Mcm1. Although this arg80 mutant protein has a binding affinity similar to that of Mcm1, it was unable to complement the lethal phenotype of an mcm1 deletion strain, indicating that features other than DNA binding are required for Arg80 to fulfill Mcm1 function (data not shown).
FIG. 5.
In vitro binding of wild-type and mutated Arg80 and Mcm1 proteins to the P sequence. (A) Sequence alignment of the N-terminal regions of the Arg80 and Mcm1 MADS box domains. The open circles indicate the positions in the Mcm1 protein that contact the DNA in the crystal structure of the protein (38). (B) Electrophoretic mobility shift assay of yeast crude extracts binding to a 160-bp HindIII-BamHI DNA fragment containing the P site. Lane 1, no extract. Lanes 2 to 6 show shifts by crude extracts (50 μg) from S. cerevisiae strain BY4709 (arg80::kanMX) transformed with pYEF2 (vector) (lane 2), pME51 (GAL10-ARG80) (lane 3), pAJ221 (GAL10-arg80-Q83I-P84E-R86K-Y87F) (lane 4), pAJ219 (GAL10-arg80-R102K-Y110F) (lane 5), or pAJ220 (GAL10-arg80-Q83I-P84E-R86K-Y87F-R102K-Y110F) (lane 6). Lanes 7 to 9 show extracts from strain 1c2163aΔMCM1 transformed with pED40 (pMCM1) (lane 7), pAJ210 (mcm1-Y21Q) (lane 8), or pAJ211 (mcm1-K40R) (lane 9). The presence of the Arg80 and Mcm1 proteins in the extracts was visualized by Western blotting using antibodies raised against GST-Arg80 and GST-Mcm1, respectively.
It is likely that the Q83I and R102K changes in Arg80 enable the protein to make more-optimal contacts with the DNA. To test this possibility, we constructed the converse mutations in Mcm1. Replacing residue I21 in Mcm1 with the corresponding glutamine amino acid found in Arg80 caused roughly a fivefold decrease in the DNA-binding affinity of the protein (Fig. 5B, lane 8). The K40R substitution completely eliminated the DNA-binding affinity of the protein (Fig. 5B, lane 9). These results serve to explain why Arg80 on its own binds very poorly to DNA.
Mutations in Arg80-Mcm1-Arg80 increase its DNA-binding affinity to arginine boxes.
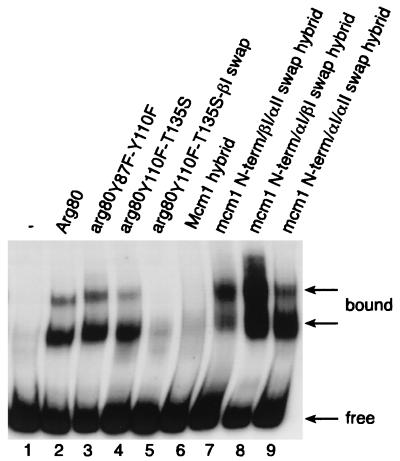
The Arg80 protein requires both the Mcm1 and Arg81 proteins to bind with high affinity to arginine boxes found in promoters of genes involved in arginine metabolism. We therefore tested the ability of a subset of the Arg80 swap mutants and Arg80-Mcm1-Arg80 chimeras to bind in complex with these cofactors. Semipurified proteins produced from strain 02463dΔARG80 transformed simultaneously with plasmid YEP34 (pGAL10-ARG81 LEU2) and with the different plasmids bearing mutated arg80 genes (pAJ234, pAJ239, and pAJ240) were electrophoresed with a 32P-labeled ARG5,6 fragment containing the arginine boxes. Although the Arg80 Y87F Y110F and Y87F T135S double mutants have an effect on Arg80 function in vivo (Fig. 3, lines 12 and 20, respectively), these substitutions do not dramatically affect binding to the ARG5,6 site. It is possible that the nonphysiological high levels of Arg81 that are required for observation of the binding activity of the complex may compensate for the defects of the mutants in forming a complex. However, the Y87F and T135S mutations in combination with a swap of the βI region (pAJ239) caused a complete loss of binding by the complex (Fig. 6, lane 5). Since the results of the two-hybrid assays show that this mutant has decreased affinity with its cofactors, the absence of binding to DNA is likely a result of the loss of the formation of the protein complex rather than a result of a loss of interaction with DNA.
FIG. 6.
In vitro binding of wild-type and mutated Arg80 and Arg80-Mcm1-Arg80 proteins to arginine boxes. The end-labeled 160-bp AluI-AluI DNA fragment containing the ARG5,6 control region was incubated with 10 μg of semipurified yeast extracts (11). Strain 02463dΔARG80 containing pYEP34 (ARG81 LEU2) was transformed with pFL38 (vector; URA3) (lane 1), pFL38-RI (ARG80) (lane 2), pAJ234 (arg80-Y87F-Y110F) (lane 3), pAJ240 (arg80-Y110F-T135S) (lane 4), pAJ239 (arg80-Y110F-T135S-βI swap) (lane 5), pHL13 (Arg80-Mcm1-Arg80) (lane 6), pAJ46 (Arg80-mcm1-N-terminal/βI/αII swap-Arg80) (lane 7), pAJ84 (Arg80-mcm1-N-terminal/αI/βI swap-Arg80) (lane 8), and pAJ85 (Arg80-mcm1-N-terminal/αI/αII swap-Arg80) (lane 9). All of these strains were grown on 1% galactose.
We also assayed the ability of the Arg80-Mcm1-Arg80 chimera to bind as a complex with Arg81 and Mcm1 to the ARG5,6 fragment. In agreement with the results of the transcription reporter assays, it was found that the Arg80-Mcm1-Arg80 chimera containing the wild-type Mcm1 MADS box domain was unable to bind in complex with Mcm1 and Arg81 to the ARG5,6 site (Fig. 6, lane 6). In contrast, extracts from strains transformed with plasmid pAJ46, pAJ84, or pAJ85 were able to shift DNA. It is worth noting that the hybrid protein produced from plasmid pAJ84, in which the N-terminal, αΙ, and βI domains were swapped with the corresponding regions of Arg80, led to the formation of an abundant protein complex with the arginine boxes even though these substitutions significantly decrease the binding of the protein on its own to the P site. This result further supports the notion that this region of Arg80 is important for the protein's interaction with its cofactors.
DISCUSSION
The MADS box family of transcription regulatory proteins contain highly conserved DNA-binding domains. The proteins in this family bind to conserved DNA sites called CArG boxes, and many of the MADS box proteins can bind to each other's sites with good affinity in vitro (37). This raises the question of whether these proteins can function for each other in vivo and, if not, what determines their specific differences. To address this question, we examined the specificity and role of the Arg80 MADS box protein in the regulation of the transcription of arginine-metabolic genes in S. cerevisiae. In addition to Arg80, there are three other MADS box proteins in S. cerevisiae, Mcm1, Smpl, and Rlm1. We have shown that Rlm1 and Smp1 do not appear to be involved in the regulation of arginine-metabolic genes. We have also shown that although the U. maydis Umc1 protein can partially replace Mcm1 in the regulation of arginine, neither it nor Mcm1 can fulfill the role of Arg80. These results show that the few differences between the MADS box sequences of the Arg80 and Mcm1 proteins are sufficient to determine the specificity of the Arg80 protein.
The MADS box domains of the Arg80 and Mcm1 proteins are very similar in their sequences and are likely folded into similar structures. We have therefore used the crystal structure of Mcm1 as a model for understanding how Arg80 may fold and bind DNA (38). Many of the residues involved in DNA contacts in Mcm1 are conserved in Arg80, so it is also likely that these proteins bind DNA in a similar manner (Fig. 5A). However, we have shown that the substitution of the Mcm1 MADS box domain into the context of Arg80 does not complement an arg80 mutant, indicating that there are significant differences in the functional specificities of the two proteins. It is likely that a significant part of this difference in specificities is due to differences in interactions with the cofactors that bind to Arg80. Amino acids at positions that are not conserved between the two proteins most likely determine the specificity of the interactions of Arg80 with Mcm1 and Arg81. To identify which regions of the Arg80 MADS box domain specify these interactions, we replaced residues in Arg80 with amino acids found at the same relative positions in the Mcm1 MADS box domain and examined which substitutions have an effect on Arg80 function. We also performed the converse experiment to determine whether substitutions in the Mcm1 MADS box domain would change its specificity to that of Arg80. Our mutational analysis shows that while substitutions in any one region of the protein have little effect on their own, in combination with mutations in other regions of the protein they significantly affect Arg80-dependent regulation. The fact that mutations in any one region do not affect activity on their own suggests that the remaining interactions with the other regions of the protein are able to compensate for the loss of proper contacts by the mutated residues. This result suggests that there are multiple points of interaction between Arg80 and its cofactors and that the interface between Arg80 and its cofactors may be very extensive.
In the initial mutagenesis of Arg80, multiple amino acid substitutions were introduced within each secondary-structure region of the MADS box domain. In combination with mutations in other regions of the MADS box domain, these changes caused a large decrease in activity. However, these substitutions could have multiple effects on the activity of the protein by, for example, altering DNA-binding affinity, stability, protein folding, or protein interactions. Therefore, to investigate which residues within a region are important for the interaction with the Arg80 cofactors, we made individual substitutions within a region. For example, although other substitutions in the N-terminal arm alter the DNA-binding properties of the protein, we have shown that only the conservative phenylalanine substitution at position Y87, which does not alter DNA-binding activity, affects the regulatory function of the protein. We have also shown that substitutions at residues Y110 and T135 cause a significant decrease in Arg80 activity. The fact that we can switch the specificity of the Mcm1 MADS box domain to that of Arg80 by swapping the amino acids at these positions in combination with residues in the βI strand indicates that these are important determinants of the specificity of the protein. The observation that the Y110F T135S (pAJ240) and Y87F Y110F (pAJ234) double mutants and the Y110F, A118T, N119Q (pAJ254) triple mutant have such a large effect on Arg80 function further supports the notion that these residues play an important role in the function of the protein.
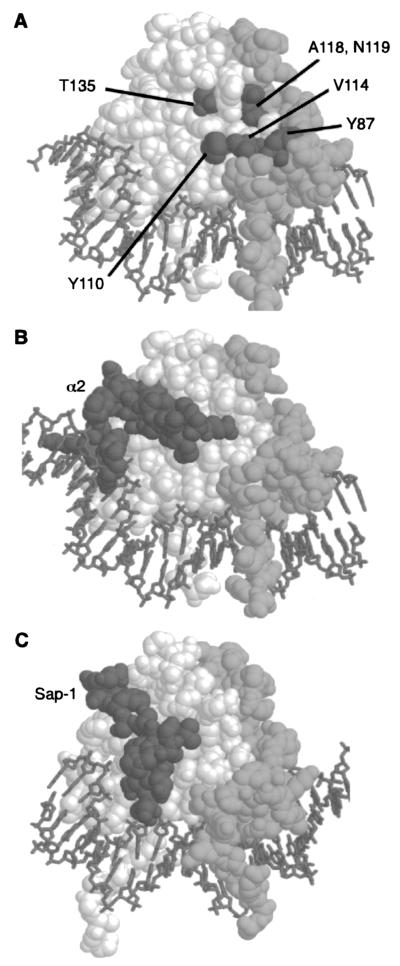
Although we found that the residues that define Arg80 specificity lie in several different secondary-structure regions of the protein (N-terminal arm, αI, βI, and βII), in spatial terms these substitutions all cluster on one side of the protein (Fig. 7A). This region is located away from the DNA and lies along the edge of the interface between the monomers. Residues from both monomers make up this partially contiguous surface. It is likely that these residues form a surface of the protein that is involved in the protein's interaction with its cofactors. In support of this idea, we have shown that even though some of the swaps increase the DNA-binding affinity of the protein alone to the P site, the protein shows decreased transcriptional regulation in vivo and fails to bind cooperatively in a complex with Arg81 and Mcm1 in vitro (data not shown). This result suggests that these substitutions affect the ability of Arg80 to interact with its cofactors.
FIG. 7.
Position of the residues required for Arg80 activity. (A) The relative positions of residues that affect Arg80 function are displayed on a model of the Mcm1 MADS box domain dimer (spacefill) bound to DNA (stick-like figures). One monomer is shown in white, and the other is shown in light gray. Residues corresponding to positions in Arg80 that are required for regulation of arginine metabolism are shown in dark gray. These residues cluster in a partially contiguous face on the side of the protein that is part of the dimer interface. Residue Y87 is from one monomer, while residues Y110, V114, A118, N119, and T135 are from the other monomer. (B) Contacts between α2 and Mcm1 are in a different region of the MADS box domain. A complex of the α2 linker (dark gray) binding to the Mcm1 dimer is shown. The Mcm1 dimer is in roughly the same orientation as that shown in panel A. (C) Contacts between the SAP-1 and SRF are in a different region of the MADS box domain. A complex of the SAP-1 B-box (dark gray) binding to the SRF dimer is shown. The SRF dimer is in roughly the same orientation as that of the Mcm1 dimer shown in panel A.
Our results suggest that residues Y87, Y110, A118, N119, and T135 of Arg80 define a partially contiguous surface on the side of the protein that may interact with the protein's cofactors (Fig. 7A). However, it is likely that many of the conserved residues in this region of the protein also play a role in the interaction of Arg80 with its cofactors. In support of this notion, we have shown that replacement of residue V114, which lies on the surface between Y87 and Y110, may provide a contiguous surface for contacts with an Arg80 cofactor, which is most likely Mcm1. Many of the side chains in this region of the protein are relatively hydrophobic, suggesting that the interactions with the cofactor may be hydrophobic in nature. However, the phenylalanine substitutions at positions Y87 and Y110 significantly decrease the level of Arg80 activity, indicating that the hydroxyl group of the tyrosine side chain at both of these positions plays an important role in the function of the protein. It is possible that these hydroxyl groups are involved in making hydrogen bond contacts to the Arg80 cofactor and contribute to the specificity of the protein interactions.
The mutational analysis indicates that there are several surfaces of the Arg80 protein that interact with its cofactors. The results of a previous mutational analysis suggested that Arg82 interacts with conserved solvent-exposed residues in the αI helix of the Arg80 MADS box domain (14). We propose that another region for cofactor interactions on the surface of Arg80 is on the side of the protein, i.e., at the dimer interface. This region of interaction is significantly different from the region of Mcm1 that interacts with the yeast MATα2 protein or the surface of the mammalian SRF protein that interacts with SAP-1 (Fig. 7B and C) (17, 38). In both of these cases, the interaction surface is along the hydrophobic groove formed between the βII strand and the αII helix. The regions of MATα2 and SAP-1 that interact with the Mcm1 and SRF MADS box domains, respectively, are also similar to each other and contain an extended strand that lies in the groove. One of the main features of these interactions is a conserved aromatic group that fits into a pocket formed by amino acid side chains corresponding to residues V131, V143, K149, and I152 of Arg80. Amino acid substitutions in this region of Mcm1 affect the interaction with α2 (7). We have found that substitutions in this region of Arg80 also cause a decrease in activity, suggesting that this region may be involved in contacts with cofactors in a manner similar to the interactions between Mcm1 and α2 and between SRF and SAP1. However, the effects of these mutations are less severe than those of substitutions in other regions of the protein. For example, although the αII swap in Arg80 changes a number of the residues in or around this pocket, the effects of these multiple changes on Arg80 function are relatively weak. In contrast to substitutions in the hydrophobic pocket, the replacement of residues Y110, A118, N119, I120, I124, L125, A126, N127, S128, and T135 has a large effect on Arg80 function. This result differs significantly from that obtained when substitutions are made at the corresponding positions in Mcm1, which do not affect the interaction with α2 (J. Mead and A. K. Vershon, unpublished data).
Our results are consistent with the idea that there are at least two different regions on the Arg80 MADS box domain that can be involved in interactions with cofactors: one formed by the hydrophobic pocket on the face of the protein and the other in a groove across the dimer interface on the side of the protein (Fig. 7). Some residues, such as Y110, F134, T135, and P137, may be involved in protein interactions in both regions of the domain. The amino acid side chains of these residues in Mcm1 and SRF are in close contact with both α2 and SAP-1 cofactors and are also important for the interactions of Arg80 with its cofactors. It is possible that if a cofactor binds to one face of the protein and contacts these residues, it may prevent a different cofactor that requires contacts with these residues from binding and force it to bind to the other face of the MADS box domain. Alternatively, it is possible that these two interfaces may simultaneously provide contacts for two different cofactors, helping to form a complex around the MADS box protein. DNA-binding assays showed that Arg80 binds in a complex with Mcm1 and Arg81, and two-hybrid studies showed that Arg80 interacts with Mcm1, Arg81, and Arg82 (4, 14). The multiple surfaces defined through the mutational analysis of Arg80 may provide the interfaces for interactions with these different cofactors. It is possible that other MADS box proteins use this region to interact with their cofactors and that variations at these residues among the different proteins of the MADS box family may serve to specify the interactions of these proteins with their cofactors.
Acknowledgments
We thank R. Kahmann for plasmid pYUMC1-454 and P. Jacobs for providing antibodies to GST-Arg80 and GST-Mcm1. We are also very grateful to F. Vierendeels for excellent technical assistance. We thank J. Mead and A. Bruning for comments on the manuscript.
This work was supported by grants from the Fonds de la Recherche Scientifique Médicale to E.D. and F.M. (3.4605.95) and from the National Institutes of Health to A.K.V. (GM49265). A.J was partially supported by a grant from the Fonds Demeur-François.
REFERENCES
- 1.Acton, T. B., J. Mead, A. M. Steiner, and A. K. Vershon. 2000. Scanning mutagenesis of Mcm1: residues required for DNA binding, DNA bending, and transcriptional activation by a MADS-box protein. Mol. Cell. Biol. 20:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Althoefer, H., A. Schleiffer, K. Wassmann, A. Nordheim, and G. Ammerer. 1995. Mcm1 is required to coordinate G2-specific transcription in Saccharomyces cerevisiae. Mol. Cell. Biol. 15:5917-5928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alvarez-Buylla, E. R., S. Pelaz, S. J. Liljegren, S. E. Gold, C. Burgeff, G. S. Ditta, L. Ribas de Pouplana, L. Martinez-Castilla, and M. F. Yanofsky. 2000. An ancestral MADS-box gene duplication occurred before the divergence of plants and animals. Proc. Natl. Acad. Sci. USA 97:5328-5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amar, N., F. Messenguy, M. El Bakkoury, and E. Dubois. 2000. ArgRII, a component of the ArgR-Mcm1 complex involved in the control of arginine metabolism in Saccharomyces cerevisiae, is the sensor of arginine. Mol. Cell. Biol. 20:2087-2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bender, A., and G. F. Sprague, Jr. 1987. MATα1 protein, a yeast transcription activator, binds synergistically with a second protein to a set of cell-type-specific genes. Cell 50:681-691. [DOI] [PubMed] [Google Scholar]
- 6.Black, B. L., and E. N. Olson. 1998. Transcriptional control of muscle development by myocyte enhancer factor-2 (MEF2) proteins. Annu. Rev. Cell Dev. Biol. 14:167-196. [DOI] [PubMed] [Google Scholar]
- 7.Bruhn, L., and G. F. Sprague, Jr. 1994. MCM1 point mutants deficient in expression of α-specific genes: residues important for interaction with α1. Mol. Cell. Biol. 14:2534-2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Casadome, L., E. de Nadal, and F. Posas. 2001. Transcription factors under the control of the yeast Hog1 MAPK. Yeast 18:S78. [Google Scholar]
- 9.Coen, E. S., and E. M. Meyerowitz. 1991. The war of the whorls: genetic interactions controlling flower development. Nature 353:31-37. [DOI] [PubMed] [Google Scholar]
- 10.Dodou, E., and R. Treisman. 1997. The Saccharomyces cerevisiae MADS box transcription factor Rlm1 is a target for the Mpk1 mitogen-activated protein kinase pathway. Mol. Cell. Biol. 17:1848-1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dubois, E., and F. Messenguy. 1991. In vitro studies of the binding of the ARGR proteins to the ARG5,6 promoter. Mol. Cell. Biol. 11:2162-2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dubois, E., and F. Messenguy. 1994. Pleiotropic function of ArgRIIIp (Arg82p), one of the regulators of arginine metabolism in Saccharomyces cerevisiae. Role in expression of cell-type-specific genes. Mol. Gen. Genet. 243:315-324. [DOI] [PubMed] [Google Scholar]
- 13.Durfee, T., K. Becherer, R.-L. Chen, S. H. Yeh, Y. Yang, A. E. Kilburn, W. H. Lee, and S. J. Elledge. 1993. The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev. 7:555-569. [DOI] [PubMed] [Google Scholar]
- 14.El Bakkoury, M., E. Dubois, and F. Messenguy. 2000. Recruitment of the yeast MADS box proteins, ArgRI and Mcm1, by the pleiotropic factor ArgRIII is required for their stability. Mol. Microbiol. 35:15-31. [DOI] [PubMed] [Google Scholar]
- 15.Elble, R., and B. K. Tye. 1992. Chromosome loss, hyperrecombination, and cell cycle arrest in a yeast mcm1 mutant. Mol. Biol. Cell 3:971-980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guldener, U., S. Heck, T. Fielder, J. Beinhauer, and J. H. Hegemann. 1996. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 24:2519-2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hassler, M., and T. J. Richmond. 2001. The B-box dominates SAP-1-SRF interactions in the structure of the ternary complex. EMBO J. 20:3018-3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jarvis, E. E., K. L. Clark, and G. F. Sprague. 1989. The yeast transcription activator PRTF, a homolog of the mammalian serum response factor, is encoded by the MCM1 gene. Genes Dev. 3:936-945. [DOI] [PubMed] [Google Scholar]
- 19.Johansen, F. E., and R. Prywes. 1995. Serum response factor: transcriptional regulation of genes induced by growth factors and differentiation. Biochim. Biophys. Acta 1242:1-10. [DOI] [PubMed] [Google Scholar]
- 20.Kruger, J., C. Aichinger, R. Kahmann, and M. Bolker. 1997. A MADS-box homologue in Ustilago maydis regulates the expression of pheromone-inducible genes but is nonessential. Genetics 147:1643-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuo, M. H., E. T. Nadeau, and E. J. Grayhack. 1997. Multiple phosphorylated forms of the Saccharomyces cerevisiae Mcm1 protein include an isoform induced in response to high salt concentrations. Mol. Cell. Biol. 17:819-832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McInerny, C. J., J. F. Partridge, G. E. Mikesell, D. P. Creemer, and L. L. Breeden. 1997. A novel Mcm1-dependent element in the SWI4, CLN3, CDC6, and CDC47 promoters activates M/G1-specific transcription. Genes Dev. 11:1277-1288. [DOI] [PubMed] [Google Scholar]
- 23.Messenguy, F. 1976. Regulation of arginine biosynthesis in Saccharomyces cerevisiae: isolation of a cis-dominant, constitutive mutant for ornithine carbamoyltransferase synthesis. J. Bacteriol. 128:49-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Messenguy, F., and E. Dubois. 1993. Genetic evidence for a role for MCM1 in the regulation of arginine metabolism in Saccharomyces cerevisiae. Mol. Cell. Biol. 13:2586-2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Messenguy, F., and E. Dubois. 2000. Regulation of arginine metabolism in Saccharomyces cerevisiae: a network of specific and pleiotropic proteins in response to multiple environmental signals. Food Technol. Biotechnol. 38:277-285. [Google Scholar]
- 26.Mo, Y., W. Ho, K. Johnston, and R. Marmorstein. 2001. Crystal structure of a ternary SAP-1/SRF/c-fos SRE DNA complex. J. Mol. Biol. 314:495-506. [DOI] [PubMed] [Google Scholar]
- 27.Odom, A. R., A. Stahlberg, S. R. Wente, and J. D. York. 2000. A role for nuclear inositol 1,4,5-trisphosphate kinase in transcriptional control. Science 287:2026-2029. [DOI] [PubMed] [Google Scholar]
- 28.Oehlen, L. J., J. D. McKinney, and F. R. Cross. 1996. Ste12 and Mcm1 regulate cell cycle-dependent transcription of FAR1. Mol. Cell. Biol. 16:2830-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Olson, E. N., M. Perry, and R. A. Schulz. 1995. Regulation of muscle differentiation by the MEF2 family of MADS box transcription factors. Dev. Biol. 172:2-14. [DOI] [PubMed] [Google Scholar]
- 30.Passmore, S., G. T. Maine, R. Elble, C. Christ, and B. K. Tye. 1988. Saccharomyces cerevisiae protein involved in plasmid maintenance is necessary for mating of MATα cells. J. Mol. Biol. 204:593-606. [DOI] [PubMed] [Google Scholar]
- 31.Pellegrini, L., S. Tan, and T. J. Richmond. 1995. Structure of serum response factor core bound to DNA. Nature 376:490-498. [DOI] [PubMed] [Google Scholar]
- 32.Qiu, H. F., E. Dubois, P. Broen, and F. Messenguy. 1990. Functional analysis of ARGRI and ARGRIII regulatory proteins involved in the regulation of arginine metabolism in Saccharomyces cerevisiae. Mol. Gen. Genet. 222:192-200. [DOI] [PubMed] [Google Scholar]
- 33.Riechmann, J. L., and E. M. Meyerowitz. 1997. MADS domain proteins in plant development. Biol. Chem. 378:1079-1101. [PubMed] [Google Scholar]
- 34.Saiardi, A., J. J. Caffrey, S. H. Snyder, and S. B. Shears. 2000. Inositol polyphosphate multikinase (ArgRIII) determines nuclear mRNA export in Saccharomyces cerevisiae. FEBS Lett. 468:28-32. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook, J., T. Maniatis, and E. F. Fritsch. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 36.Santelli, E., and T. J. Richmond. 2000. Crystal structure of MEF2A core bound to DNA at 1.5 Å resolution. J. Mol. Biol. 297:437-449. [DOI] [PubMed] [Google Scholar]
- 37.Shore, P., and A. D. Sharrocks. 1995. The MADS-box family of transcription factors. Eur. J. Biochem. 229:1-13. [DOI] [PubMed] [Google Scholar]
- 38.Tan, S., and T. J. Richmond. 1998. Crystal structure of the yeast MATα2/MCM1/DNA ternary complex. Nature 391:660-666. [DOI] [PubMed] [Google Scholar]
- 39.Wach, A. 1996. PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in S. cerevisiae. Yeast 12:259-265. [DOI] [PubMed] [Google Scholar]
- 40.Watanabe, Y., K. Irie, and K. Matsumoto. 1995. Yeast RLM1 encodes a serum response factor-like protein that may function downstream of the Mpk1 (Slt2) mitogen-activated protein kinase pathway. Mol. Cell. Biol. 15:5740-5749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yabana, N., and M. Yamamoto. 1996. Schizosaccharomyces pombe map1+ encodes a MADS-box-family protein required for cell-type-specific gene expression. Mol. Cell. Biol. 16:3420-3428. [DOI] [PMC free article] [PubMed] [Google Scholar]