Abstract
We recently discovered that heat shock causes nucleolin to relocalize from the nucleolus to the nucleoplasm, whereupon it binds replication protein A and inhibits DNA replication initiation. We report that nucleolin mobilization also occurs following exposure to ionizing radiation (IR) and treatment with camptothecin. Mobilization was selective in that another nucleolar marker, upstream binding factor, did not relocalize in response to IR. Nucleolin relocalization was dependent on p53 and stress, the latter initially stimulating nucleolin-p53 complex formation. Nucleolin relocalization and complex formation in vivo were independent of p53 transactivation but required the p53 C-terminal regulatory domain. Nucleolin and p53 also interact directly in vitro, with a similar requirement for p53 domains. These data indicate a novel p53-dependent mechanism in which cell stress mobilizes nucleolin for transient replication inhibition and DNA repair.
Cells have evolved complex mechanisms for sensing internal trauma (such as DNA damage or aberrant protein processing) and environmental stresses (for instance, extremes of temperature, changes in oxygen tension, and nutrient deprivation). These mechanisms ensure the rapid and efficient detection of cellular insults that could potentially result in cellular transformation or death (28, 35). Depending on the stage of the cell cycle at which stress is applied, different signaling cascades are mobilized in order to execute an appropriate response (14, 24). The response varies with the type of stress and its severity and may include arrest of cellular proliferation, repair of damaged DNA, or apoptosis.
An important player in the cellular stress response is the tumor suppressor protein p53. Since its discovery more than 20 years ago and its identification as a transcriptional activator, p53 has been the topic of comprehensive study (29, 31, 36, 39). Extensive research has underscored the global importance of p53 function in a collection of diverse biological processes including cell cycle control, genome stability, DNA repair, apoptosis, and senescence. To a large extent, the tumor suppressor functions of p53 are ascribed to the collective effects of p53-responsive genes. Many p53 target genes have been identified, and some are intimately involved with cell cycle regulation (e.g., p21waf1 [13, 20, 51]), p53-mediated apoptosis (CD95/Fas [33, 34] and PIG-3 [25]), and DNA repair (gadd45 [5, 45]).
p53 has also been suggested to modulate various DNA metabolic processes more directly, independently of its transcriptional activity (22). p53 interacts with a key homologous DNA recombination factor, Rad51 (46), and use of a simian virus 40 (SV40)-based assay system suggests that p53 normally acts to suppress recombination events (50). p53 physically interacts with base excision repair factors AP endonuclease and DNA polymerase β, and greatly stimulates base excision repair in vitro (53). Interestingly, purified mutant p53 has been shown to directly inhibit chromosomal DNA replication in Xenopus laevis extracts, although by an unknown mechanism (9).
In a recent study, we found that nucleolin, an abundant factor critical for precursor rRNA (pre-rRNA) processing (15), translocates from the nucleolus to the nucleoplasm following heat shock (10). Nucleolin relocalization is accompanied by a great increase in complex formation between nucleolin and replication protein A (RPA), the primary eukaryotic single-stranded DNA-binding protein (21). A combination of in vitro and in vivo data demonstrated that nucleolin-RPA complex formation is strongly inhibitory to DNA replication, likely by preventing RPA from active participation in the initiation of DNA synthesis. Nucleolin has also been found to be a stimulatory factor for DNA annealing, suggesting that it may play a role in heteroduplex-mediated DNA repair reactions (17). Nucleolin down-regulates c-Myb transcriptional activity (52) and is a component of LR1, a B-cell-specific transcription factor (18). Together, these data indicate that nucleolin mobilization occurs to globally modulate DNA and RNA metabolism following stress.
We show here that nucleolin mobilization also occurs following exposure to ionizing radiation (IR) and treatment with the radiomimetic agent camptothecin (CPT). Nucleolin mobilization is dependent on the activation of p53 by cellular stress but is independent of the ability of p53 to activate transcription. Stresses that mobilize nucleolin also cause a significant increase in complex formation between nucleolin and activated p53. We therefore find a novel mechanism in which nucleolin, in response to stress, becomes mobilized in a p53-dependent manner for participation in the transient inhibition of replication and for DNA repair.
MATERIALS AND METHODS
Cell lines and stress treatments.
Saos-2 and U2-OS cells were obtained from the American Type Culture Collection and W. Jiang (New York University School of Medicine), respectively. Cell lines were grown in Dulbecco's modified Eagle medium containing 10% fetal bovine serum and 50 μg of gentamicin/ml. H1299 cell lines (8), including p53-13 (expressing wild-type p53), p53-22/23, and p53ΔC30, were generously provided by C. Prives (Columbia University, New York, N.Y.), and grown in RPMI medium containing 10% fetal bovine serum, 4.5 μg of tetracycline/ml, 2 μg of puromycin/ml, and 300 μg of G418/ml. To induce p53, the medium was replaced with a medium lacking tetracycline for 16 h.
Cellular stress conditions were produced according to published standards. Standard conditions for immunofluorescence are as follows: IR, 10 Gy from a 137Cs source; CPT, 1 μM for 2 h; hydroxyurea (HU), 2 mM for 1 h; UV irradiation, a single dose of 30 J m−2 followed by a 2-h incubation. To induce heat shock, cells were incubated for 15 min in a 44°C water bath.
Immunoprecipitation and immunoblotting.
Immunoprecipitations were performed with 0.5-ml cellular lysates by using the IMMUNOcatcher system (CytoSignal). For immunoprecipitation analysis, cells were washed twice in ice-cold phosphate-buffered saline (PBS) and lysed in ice-cold IMMUNOcatcher Strong Lysis Buffer supplemented with 1 mM dithiothreitol (DTT), 1 mM phenylmethylsulfonyl fluoride, protease cocktail (leupeptin, aprotinin, pepstatin, and chymostatin, each at 2 μg per ml), 10 mM sodium fluoride, and 1 mM sodium orthovanadate. Because the Strong Lysis Buffer solubilizes most cellular proteins, the lysates contained nuclear and cytoplasmic components. Lysates were incubated on ice for 1 h and then centrifuged for 20 min at 4°C in a microcentrifuge (at 16,000 × g). The centrifuged supernatant was used directly for immunoprecipitation. When needed, ethidium bromide (15 μg/ml) and RNase A (80 μg/ml) were added 15 min before centrifugation. The following antibodies were employed for immunoprecipitation: a mouse monoclonal antibody against nucleolin (MS-3; Santa Cruz Biotechnology) or rabbit polyclonal antibodies against phospho-Ser15 p53 and total p53 (Cell Signaling Technology). Following a 1-h incubation at room temperature (RT), antibody complexes were incubated for 1 h at RT with 20 μl of protein G PLUS-agarose for mouse monoclonal antibodies or protein A-agarose for rabbit polyclonal antibodies (both from Santa Cruz Biotechnology). Precipitates were washed twice on the spin filters with 500 μl of IMMUNOcatcher Mild Buffer at RT and then resuspended in 30 μl of sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer. The material was then boiled for 5 min and separated by SDS-PAGE. The following antibodies were used for Western analysis: mouse monoclonal antibodies against nucleolin (generously provided by H. Busch, Baylor College of Medicine, Houston, Tex.), total p53 (DO-1 [Santa Cruz Biotechnology] or Ab-2 [PAb1801; Oncogene Research]), β-actin (Sigma-Aldrich), and RPA2 (NeoMarkers) and a rabbit polyclonal antibody against upstream binding factor (UBF) (H-300; Santa Cruz Biotechnology). Secondary antibodies (anti-rabbit and anti-mouse antibodies; Amersham Biosciences) were conjugated with horseradish peroxidase (HRP). Proteins were detected by using enhanced chemiluminescence (ECL; Amersham Biosciences).
Far-Western analysis.
p53 and p53 truncation mutants, a kind gift from J. Chen (Rockefeller University), were expressed in bacteria as N-terminal glutathione S-transferase (GST) fusions as described previously (7). Nucleolin was expressed in yeast from a vector generously provided by E. Rubin (University of Medicine and Dentistry of New Jersey) by using the protocol of Haluska et al. (16). Far-Western blotting was carried out basically as described by Jayaraman et al. (23). Purified p53 proteins were subjected to SDS-PAGE, and then transferred to a nitrocellulose membrane. The filter was incubated twice in denaturation buffer (6 M guanidine-HCl in PBS) for 5 min at 4°C. The membrane was then incubated in six serial 1:1 (vol/vol) dilutions of denaturation buffer with PBS containing 1 mM DTT. After being blocked in PBS containing 0.5% Tween 20 and 5% nonfat dry milk (NFDM) for 45 min at RT, the membrane was washed twice with PBS containing 0.5% Tween 20 and 0.25% NFDM and then incubated with nucleolin (0.2 μg/ml) in PBS containing 0.5% Tween 20, 0.25% NFDM, 1 mM DTT, and 0.5% phenylmethylsulfonyl fluoride for 2 h at RT. The membrane was then washed four times in PBS containing 0.5% Tween 20 and 0.25% NFDM before being probed with the mouse anti-nucleolin monoclonal antibody MS-3 as the primary antibody and a sheep anti-mouse antibody conjugated with HRP as the secondary antibody. Bound nucleolin was detected by ECL.
Immunofluorescence microscopy.
Cells were grown on coverslips in preparation for epifluorescence microscopy and were prepared as described by Dimitrova et al. (12) and Daniely and Borowiec (10). The primary antibody used was a monoclonal antibody directed against nucleolin, while the secondary antibody was either a fluorescein isothiocyanate-conjugated donkey anti-mouse immunoglobulin G (IgG) or an anti-mouse Texas Red-conjugated donkey IgG (Jackson ImmunoResearch Laboratories). UBF was detected by using a rabbit polyclonal antibody (H-300; Santa Cruz Biotechnology) and an anti-rabbit Texas Red-conjugated donkey IgG. Microscopic images were captured digitally and prepared by using Adobe Photoshop 5.5. To quantitate the amount of nucleolin in the nucleoplasm, images were analyzed by using NIH Image (version 1.6.1). By examining cells in which nucleoli were easily distinguished, the level of the nucleolin signal in the nucleoli was calculated as the product of average pixel intensity and nucleolar area. The level of nucleolin staining in the nucleus was similarly measured, and the level of nucleoplasmic localization was determined as 1 − (nucleolin signal in the nucleolus/nucleolin signal in the nucleus). Five to 10 representative cells were chosen for each analysis.
RESULTS
Various cell stresses mobilize nucleolin.
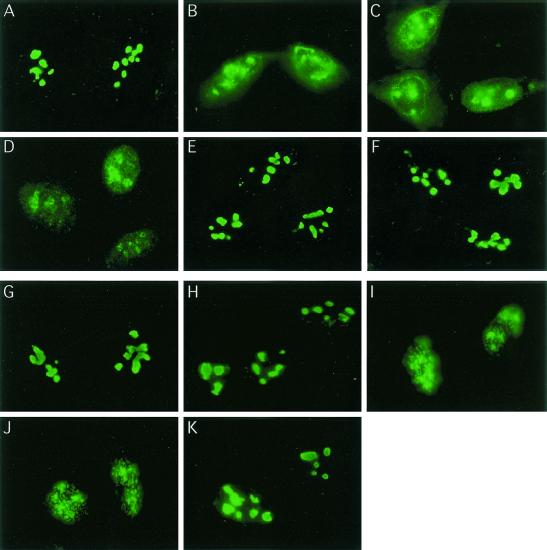
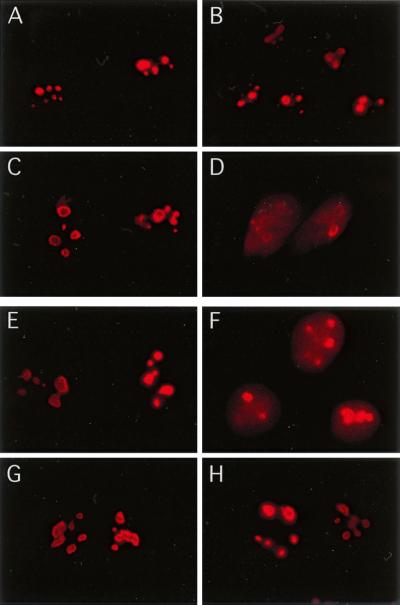
To investigate whether cellular stresses in addition to heat shock cause nucleolin mobilization, p53-positive U2-OS cells were subjected to various treatments and the localization of nucleolin was investigated by using immunofluorescence microscopy (Fig. 1). We found that nucleolin relocalization was observed both in response to IR exposure (10 Gy) (Fig. 1B) and in response to treatment with the radiomimetic agent CPT (2 μM for 2 h) (Fig. 1C). Cytoplasmic staining was occasionally observed after CPT treatment. As observed previously with HeLa cells (10), heat shock of U2-OS cells (44°C for 15 min) (Fig. 1D) also resulted in nucleolin movement to the nucleoplasm in a granular pattern. Quantitation of the fraction of the nucleolin pool found in the nucleoplasm after these stresses indicated that a majority of the nucleolin signal was nucleoplasmic (Table 1). In contrast, only a very low level of nucleolin movement occurred after exposure of cells to the DNA replication inhibitor HU (2 mM for 1 h) (Fig. 1E) or UV irradiation (30 J m−2) (Fig. 1F). Even at later time points (data not shown), the pattern of nucleolin localization after treatment with HU or UV resembled that found in control cells (Fig. 1A; Table 1).
FIG. 1.
Selective nucleolin mobilization induced by IR, CPT treatment, or heat shock. U2-OS cells were either left untreated (A) or subjected to IR (10 Gy) (B), treatment with CPT (2 μM for 2 h) (C), heat shock (15 min at 44°C) (D), HU (2 mM for 1 h) (E), or UV (a single 30-J m−2 treatment) (F). The kinetics of nucleolin relocalization were investigated in U2-OS cells by first exposing cells to 10 Gy of IR (G) and then examining the localization of nucleolin 20, 60, 120, or 180 min postirradiation (H through K, respectively). In all cases, cells were fixed by treatment with 4% (wt/vol) formaldehyde for 30 min at RT and stained for nucleolin as described in Materials and Methods. Nucleolin localization was observed by epifluorescence microscopy using a Zeiss Axiophot.
TABLE 1.
Quantitation of nucleoplasmic localization of nucleolin in U2-OS cells following stress
| Stressa | Fraction of nucleolin with nucleoplasmic localization (%)b |
|---|---|
| None (control) | 8.5 ± 5.6 |
| IR | 69.0 ± 9.3 |
| CPT | 73.7 ± 9.1 |
| Heat shock | 59.0 ± 3.4 |
| HU | 13.4 ± 3.7 |
| UV irradiation | 14.8 ± 5.7 |
The following conditions were used: IR, 60 min following a 10-Gy exposure; CPT, incubation with 2 μM CPT for 2 h; heat shock, 15 min at 44°C; HU, 2 mM for 1 h; and UV irradiation, 2 h after a 30-J m−2 irradiation.
Nucleolin relocalization was determined as described in Materials and Methods.
We examined the time course of nucleolin movement following γ-irradiation. While little change in the position of nucleolin was evident 20 min after exposure (Fig. 1H), nearly complete relocalization to the nucleoplasm was observed both 60 and 120 min postirradiation (Fig. 1I and J, respectively). Importantly, nucleolin did not remain localized to the nucleoplasm but was found to return to the nucleolus 180 min after treatment in a majority of cells examined (Fig. 1K). Use of a protein synthesis inhibitor (cycloheximide) had little or no effect on nucleolin movement following stress (data not shown), demonstrating that the same pool of nucleolin moves both in the initial phases of stress and during recovery. Nucleolin movement is therefore a reversible process, with nucleoplasmic localization seen only transiently after exposure to IR.
Lack of nucleolin mobilization in p53-null Saos-2 cells.
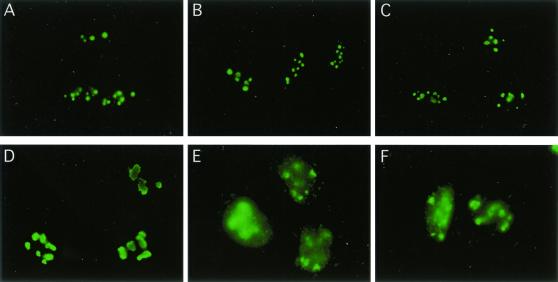
Because nucleolin mobilization occurs after γ-irradiation, we asked whether p53, an essential component of the cellular DNA damage and stress response machinery, is required for this response. p53-deficient Saos-2 cells were subjected to IR or heat shock and then processed for immunofluorescence analysis. Nucleolin relocalization was not detectable under either stress (Fig. 2B and C, respectively). As found above, the same stress conditions resulted in nucleolin mobilization in U2-OS cells. U2-OS, like Saos-2, is an osteosarcoma cell line, but it is wild type for p53 (Fig. 2E and F). These data suggest that DNA-damaging stress such as IR induces the p53-dependent relocalization of nucleolin, a point examined in greater detail below.
FIG. 2.
Nucleolin mobilization does not occur in p53-null Saos-2 cells. Saos-2 and U2-OS cells were either left untreated (A and D, respectively), subjected to IR (10 Gy) (B and E), or exposed to heat shock (90 min at 44°C) (C and F). Cells were fixed by treatment with 4% (wt/vol) formaldehyde for 30 min at RT and then stained for nucleolin as described in Materials and Methods.
Nucleolin and p53 interact in vivo.
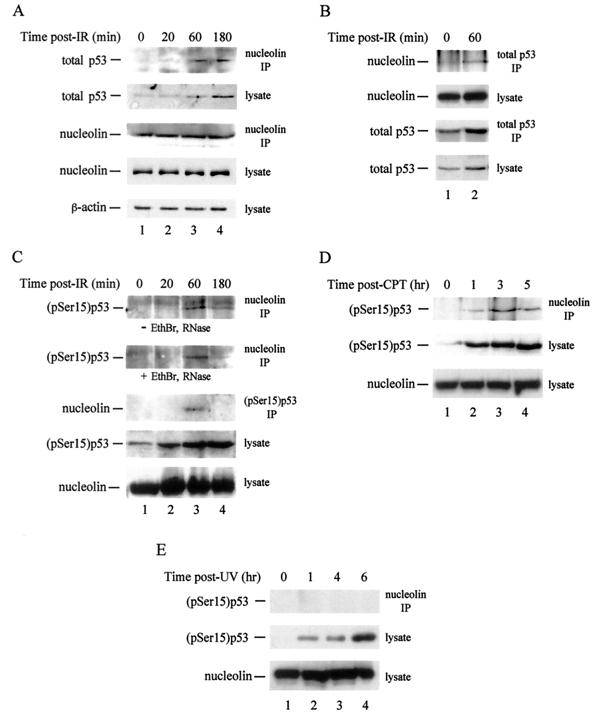
Since nucleolin mobilization appears to require p53, we inquired whether nucleolin and p53 interact in vivo, by investigating their ability to coimmunoprecipitate. In control U2-OS cells, use of nucleolin antibodies for precipitation also yielded a low but reproducible level of p53 (Fig. 3A, top panel). Exposure to IR (10 Gy) initially increased complex formation through 60 min postexposure, after which complex formation reached a plateau or decreased modestly. Quantitation of these and other data indicated that, 1 h after exposure, the amount of nucleolin-p53 complex increased sixfold compared to a twofold increase in total p53 levels. Irradiation did not significantly change the amount of nucleolin immunoprecipitated (Fig. 3A, middle panel) or the levels of β-actin (bottom panel) or nucleolin (fourth panel) in the lysate. Because no stress that we tested had any effect on nucleolin levels, we routinely used this quantity as our loading control. In reciprocal experiments, exposure of U2-OS cells to IR (10 Gy) increased the amount of nucleolin coprecipitating with p53 when anti-p53 antibodies were used (Fig. 3B). Estimates of the amount of protein coprecipitating compared to total protein levels indicated that ∼3 to 5% of the nucleolin pool is complexed with p53, and vice versa, in control cells. As we found that p53 affects nucleolin mobilization (see below), these data suggest that the interaction between nucleolin and p53 is normally transient.
FIG. 3.
Nucleolin-p53 complex formation is selectively induced in response to stress. Cell lysates were prepared from U2-OS cells at various times (as indicated) after either exposure to 10 Gy of IR (A, B, and C), treatment with 0.5 μM CPT for 1 h (D), or UV irradiation with a single 30-J m−2 dose (E). For cells treated with CPT, times indicate the amount of time elapsed after the beginning of treatment. Extracts (0.5 ml; 450 μg of protein) were subjected to immunoprecipitation (IP) with a mouse monoclonal antibody against nucleolin or total p53 or with a rabbit anti-(pSer15)p53 polyclonal antibody (as indicated). For panel C, where indicated, lysates were treated with ethidium bromide (EthBr) and RNase prior to immunoprecipitation. The precipitated material was subjected to SDS-PAGE and then to immunoblot analysis using an anti-nucleolin, anti-p53, or anti-β-actin antibody, as indicated to the left of each panel. Total cell lysates (20 μg) were electrophoresed to monitor the cellular amount of nucleolin, total p53, or (pSer15)p53 following stress.
Because γ-irradiation increased nucleolin-p53 complex levels relative to total p53 levels, we examined the interaction of nucleolin with activated p53. Activated p53 was detected by using an antibody specific for p53 phosphorylated at Ser15, a target of the ataxia telangiectasia mutated (ATM) and ATM- and Rad3-related (ATR) kinases under various stress conditions (44, 47). Significant amounts of the nucleolin-(pSer15)p53 complex were not observed either in control lysates or in lysates of U2-OS cells prepared 20 min after irradiation, regardless of the antibody used for precipitation (Fig. 3C, top three panels). Note that each of these lysates contained detectable levels of (pSer15)p53 (Fig. 3C, fourth panel, lanes 1 and 2). In contrast, a striking increase in the p53 signal was clearly present in immunoprecipitates from lysates prepared 60 min postirradiation, with a decrease in nucleolin-p53 complex formation noted subsequently (Fig. 3C, top three panels, lanes 3 and 4). To rule out the possibility that the nucleolin-p53 complex was bridged by a nucleic acid intermediate, we treated lysates with ethidium bromide and RNase A prior to addition of the primary antibody (26). This treatment had no significant effect on the level of the nucleolin-p53 complex observed (Fig. 3C, second panel), demonstrating that complex formation occurs independently of the presence of DNA or RNA. Our data indicate that nucleolin and p53 interact in vivo, with complex formation stimulated during the first 60 min after irradiation and the level of complex diminishing thereafter.
We found that nucleolin-p53 complex formation occurs more generally after stress conditions that activate p53. Although the kinetics of formation were slower than those with IR, treatment of U2-OS cells with CPT (0.5 μM for 1 h) led to a significant increase in the interaction of nucleolin with activated p53 (Fig. 3D, top panel). The kinetics of complex formation were consistent with the slower time course of nucleolin relocalization for CPT treatment than for IR exposure (data not shown). As with IR exposure, the level of nucleolin-(pSer15)p53 complex formation began to diminish at late times after CPT treatment even though the total level of (pSer15)p53 continued to increase. A similar induction of nucleolin-p53 complex formation was observed after heat shock and oxidative stress (data not shown). We find, therefore, that stress conditions which lead to nucleolin mobilization also induce nucleolin-p53 complex formation. The same stresses also stimulate nucleolin-human RPA complex formation (K. Kyung, D. Dimitrova, and J. A. Borowiec, unpublished data).
In contrast, even though UV irradiation greatly increased the amount of activated p53 (Fig. 3E, middle panel), we observed little or no association between this p53 and nucleolin through 6 h postirradiation (top panel). Treatment of cells with HU (2 mM for 1 h) likewise had minimal effects on nucleolin-p53 complex formation (data not shown). Thus, stress conditions that do not significantly mobilize nucleolin also fail to induce nucleolin-p53 complex formation. These data demonstrate that nucleolin-p53 complex formation is a stress-selective phenomenon that appears to be regulated by signaling pathways that are specific to the stress employed.
Nucleolin relocalization and p53 complex formation in a nonimmortal cell line.
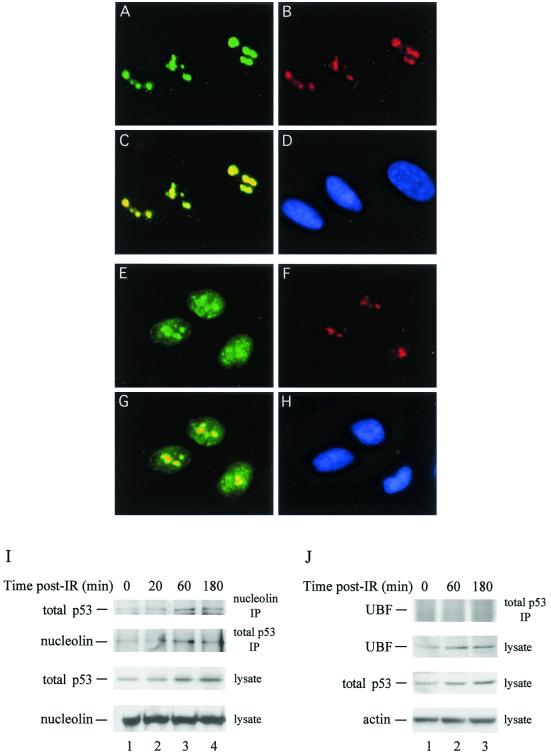
We examined the ability of nucleolin to mobilize in MRC-5 cells, a normal (nonimmortal) lung fibroblast line. Exposure of cells to IR (10 Gy) caused a significant fraction of cells to undergo nucleolin relocalization (Fig. 4E), although the percentage of MRC-5 cells that showed relocalization (∼20%) was lower than that observed with U2-OS cells (∼55%). Nucleolin relocalization also occurred in other nonimmortal lines tested (e.g., GM07492 fibroblasts) (data not shown). Nucleolin was found complexed to p53 (total) in control MRC-5 cells by using either anti-nucleolin or anti-p53 antibodies for immunoprecipitation (Fig. 4I, top two panels, lanes 1). As with U2-OS cells, complex formation was biphasic in that it first increased and then decreased after γ-irradiation. Thus, nucleolin mobilization and stimulation of nucleolin-p53 complex formation following IR-induced stress occur in nonimmortal cells and are not specific to transformed cells.
FIG. 4.
(A through H) Selective nucleolin mobilization occurs in MRC-5 cells. Nonimmortal MRC-5 lung fibroblasts, either untreated (A to D) or 60 min after exposure to γ-irradiation (10 Gy) (E to H), were processed for immunofluorescence analysis. Subsequent to Triton X-100 extraction and fixation, cells were stained by using a combination of mouse anti-nucleolin monoclonal (A and E) and rabbit anti-UBF polyclonal (B and F) antibodies. Cells were then stained with secondary fluorescein isothiocyanate-labeled anti-mouse and Texas Red-labeled anti-rabbit antibodies and were examined by epifluorescence microscopy. Composite images of the combined staining patterns for nucleolin and UBF are shown (C and G), as are images of the 4′,6′-diamidino-2-phenylindole (DAPI)-stained nuclei (D and H). (I) Nucleolin-p53 complex formation in MRC-5 cells. Lysates (0.5 ml; ∼400 μg of protein) from control MRC-5 cells (lane 1) or from cells processed 20, 60, or 180 min after exposure of cells to 10 Gy of IR (lanes 2 to 4, respectively) were subjected to immunoprecipitation using a mouse anti-nucleolin monoclonal (top panel) or a rabbit anti-total p53 polyclonal (second panel) antibody. Immunoprecipitates (IP) were analyzed by Western blotting for the presence of total p53 (top panel) or nucleolin (second panel). Aliquots of the lysates (10 μg) were examined for the level of p53 (third panel) or nucleolin (bottom panel). (J) Lack of UBF-p53 complex formation in MRC-5 cells. A procedure similar to that for panel I was used, except that lysates were immunoprecipitated with p53 antibodies and precipitates were tested for the presence of UBF (top panel). Lysates were analyzed for the presence of UBF (second panel), total p53 (third panel), and actin (used as a loading control) (bottom panel).
We also used MRC-5 cells to determine if nucleoplasmic relocalization was a general stress-dependent phenomenon of nucleolar factors or if, instead, it was selective for nucleolin. This question was addressed by examining the effect of IR on the localization of UBF, a nucleolar RNA polymerase I transcription factor (19). As expected, nucleolin (Fig. 4A) and UBF (Fig. 4B) showed nearly complete nucleolar colocalization in control cells, with coincidence indicated by the yellow color (Fig. 4C). While irradiation of cells caused nucleolin relocalization (Fig. 4E), UBF did not relocalize in these cells, with coincidence indicated by the yellow color but maintained its nucleolar localization (Fig. 4F). UBF also was not found to interact significantly with p53 before or after γ-irradiation (Fig. 4J). We therefore conclude that movement of nucleolar factors to the nucleoplasm does not occur generally but rather is a selective process.
The C-terminal 30 residues of p53 are required for nucleolin mobilization.
To more comprehensively examine the p53 requirement for nucleolin mobilization, we employed an H1299 derivative that expresses wild-type p53 in a tetracycline-repressible manner (8). This system allows independent determination of the effect of p53 induction and cell stress on nucleolin mobilization. In the absence of p53 (i.e., in the presence of tetracycline), nucleolin movement to the nucleoplasm was not observed, regardless of whether cells were irradiated (Fig. 5A and B). Similarly, after p53 induction (by growth of cells in the absence of tetracycline for 16 h), nucleolin remained localized to the nucleolus in nonirradiated cells (Fig. 5C). Dramatically, irradiation of cells grown in the absence of tetracycline showed nucleoplasmic localization of nucleolin (Fig. 5D). Consistent with the data presented above, we find that heightened p53 levels are by themselves unable to cause nucleolin relocalization. Rather, nucleolin movement to the nucleoplasm also requires cellular stress generated by exposure to IR.
FIG. 5.
The C terminus of p53, in combination with cell stress, is required for nucleolin mobilization. H1299 cells expressing either p53-13 (A to D), p53-22/23 (E and F), or p53ΔC30 (G and H) were grown in the presence (A and B) or absence (C to H) of tetracycline. Cells were either treated with IR (10 Gy) (B, D, F, and H) or left nonirradiated (A, C, E, and G). Cells were fixed by treatment with 4% (wt/vol) formaldehyde for 30 min at RT and stained for nucleolin as described in Materials and Methods.
To identify the p53 domain(s) required for nucleolin mobilization, we utilized H1299 derivatives that individually express two different p53 mutants (8). The first cell line generates a p53 protein (p53-22/23) that is unable to activate transcription by virtue of mutation of the N-terminal transcriptional transactivation domain. The second cell line expresses the p53ΔC30 mutant, lacking the extreme C-terminal regulatory domain (residues 363 to 393), which has been proposed to act as a regulator of p53 sequence-specific DNA binding. The p53-22/23 mutant facilitated nucleolin redistribution following IR (Fig. 5F), demonstrating that the transcriptional activity of p53 is not required for nucleolin relocalization. Interestingly, the p53ΔC30 mutant was unable to support nucleolin relocalization following stress (Fig. 5H). These data reaffirm the p53 dependence of nucleolin mobilization and indicate that this stress response requires the extreme C-terminal 30 residues of p53.
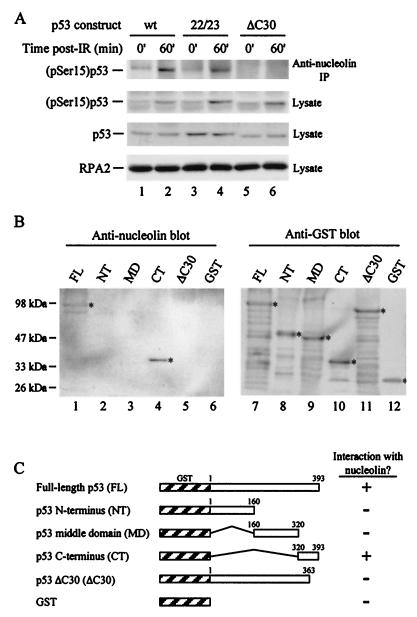
To investigate whether the p53ΔC30 mutant was unable to support relocalization because it could not complex with nucleolin, we performed coimmunoprecipitation experiments from control and irradiated H1299 cellular extracts expressing the various p53 constructs (Fig. 6A). Wild-type p53 as well as the transcriptionally inactive mutant formed stable complexes with nucleolin following exposure to IR (Fig. 6A, top panel, lanes 2 and 4). However, the truncated p53 construct unable to support nucleolin relocalization also failed to form a significant level of the nucleolin-p53 complex in vivo (Fig. 6A, top panel, lane 6). This was not caused by differences in p53 expression, because following γ-irradiation, similar levels of both (pSer15)p53 and total p53 were observed in the lysates of cells expressing the different p53 mutants (Fig. 6A, second and third panels, respectively). These results strongly argue that nucleolin-p53 complex formation and nucleolin mobilization depend on a direct interaction between nucleolin and p53, mediated through the 30 C-terminal residues of p53.
FIG. 6.
The extreme C terminus of p53 is essential for nucleolin-p53 complex formation in vivo and in vitro. (A) Nucleolin was immunoprecipitated (IP) (upper panel) from cell extracts (0.5 ml) prepared from untreated (lanes 1, 3, and 5) or γ-irradiated (lanes 2, 4, and 6) cells expressing wild-type or mutant p53. The cell lines used were p53-13 (lanes 1 and 2), p53-22/23 (lanes 3 and 4), and p53ΔC30 (lanes 5 and 6). Following SDS-PAGE of the immunoprecipitates, the separated material was subjected to immunoblot analysis using anti-(pSer15)p53 antibodies. Lysates (20 μg) prepared from control and IR-exposed cells were electrophoresed to monitor levels of (pSer15)p53 (second panel), total p53 (third panel), and the RPA middle subunit (RPA2) (bottom panel) in the extracts. The level of RPA2, used in this experiment as the loading control, does not change in response to γ-irradiation (data not shown). (B) Far-Western analysis of the nucleolin-p53 interaction. Equivalent amounts (500 ng) of full-length p53 (FL), the p53 N-terminal domain (NT; amino acids 1 to 160), middle domain (MD; amino acids 160 to 320), and C-terminal region (CT; amino acids 320 to 393), and p53 lacking the C-terminal 30 amino acids (ΔC30), each containing an N-terminal GST tag, were separated by SDS-PAGE. GST alone was also electrophoresed as a control. Following transfer to a nitrocellulose membrane, the membrane was probed with purified full-length nucleolin (1 μg in 5 ml) (lanes 1 to 6). The binding of nucleolin was visualized by using a nucleolin antibody. To visualize GST-tagged proteins, the membrane was stripped and subjected to immunoblot analysis using a rabbit anti-GST antibody (lanes 7 to 12). (C) Schematic showing the results of the far-Western analysis.
To examine this issue in more detail, we characterized the interaction between nucleolin and p53 in vitro by far-Western analysis. Wild-type p53 or p53 truncation mutants, all GST tagged, were overexpressed in bacteria. Along with the p53ΔC30 mutant, we subdivided the p53 gene into three parts to yield N-terminal, middle, and C-terminal regions (Fig. 6C). Each purified protein was subjected to SDS-PAGE and transferred to a nitrocellulose membrane, and the membrane was probed with human nucleolin. The adsorbed nucleolin was subsequently detected by using nucleolin antibodies (Fig. 6B, lanes 1 to 6). Along with the full-length p53 (Fig. 6B, lane 1), nucleolin also selectively bound the p53 C-terminal region (lane 4). A low level of binding was occasionally detected with the central p53 domain (Fig. 6B, lane 3), while no interaction with the p53 N terminus (lane 2), the p53ΔC30 mutant (lane 5), or GST alone (lane 6) was observed. Stripping the blot and reprobing the membrane with anti-GST antibodies indicated that similar amounts of each p53 protein were loaded on the membrane (Fig. 6B, lanes 7 to 12). To verify that the nucleolin-p53 interaction is direct and not coupled by RNA or DNA, we included RNase A (80 μg/ml) and ethidium bromide (15 μg/ml) in the nucleolin probing solution. These additions did not alter the ability of nucleolin to selectively recognize full-length p53 and the C-terminal region of p53 (data not shown). These data demonstrate that nucleolin and p53 interact directly via the p53 C terminus, with complex formation requiring the extreme C-terminal 30 amino acids.
DISCUSSION
In order to safeguard the genome against mutation or DNA loss, cells respond to stress by transiently delaying proliferation and mobilizing repair factors. We previously demonstrated that, during heat shock, nucleolin relocalizes to the nucleoplasm, where it sequesters RPA in nuclear foci away from sites of ongoing DNA synthesis (10). Additional repair-related functions for nucleolin are suggested by biochemical data revealing that nucleolin stimulates DNA strand annealing (17). In this study, we demonstrate that nucleolin mobilization occurs in a p53-dependent manner in response to γ-irradiation and CPT treatment. Although nucleolin mobilization is closely tied to p53 activation and nucleolin-p53 complex formation, it is independent of the transcriptional transactivation function of p53. Thus, unlike the G1 checkpoint, the role of p53 in nucleolin mobilization does not require the induction of p53-responsive genes, and therefore p53 exerts its effects through physical interaction with other factors. Interestingly, topoisomerase I has also been recently found to undergo p53-dependent relocalization from the nucleolus to the nucleoplasm following DNA damage (30).
Our data indicate that nucleolin-p53 complex formation is required for nucleolin mobilization, but the mechanism of relocalization remains unclear. Our favored explanation is that, like other factors, nucleolar nucleolin exists in dynamic equilibrium with the nucleoplasm (37). Under normal conditions, in which nuclear p53 levels are low, nucleolin exists only transiently in the nucleoplasm. Upon activation of p53 by stress, the increased level of this activated p53 promotes complex formation with nucleolin and thus shifts the majority of the nucleolin pool toward nucleoplasmic localization, allowing interaction with RPA. Alternatively, it has been proposed that a pool of p53 exists in the nucleolus (3, 4, 41). Although this is less likely, it is possible that this p53 actively exports nucleolin to the nucleoplasm, where it becomes stably associated with a nucleoplasmic structure (see reference 10), perhaps matrix attachment region (MAR) elements on chromosomal DNA (11). In either case, the nucleolin-p53 interaction is likely transient, because a significantly larger fraction of the nucleolin pool is seen to relocalize relative to the fraction bound to p53.
The C-terminal 30 residues of p53 are required for nucleolin mobilization and for nucleolin-p53 complex formation in vivo and in vitro. This basic region has been termed the “regulatory” domain and is involved in a variety of protein-protein interactions, including interactions with the TFIIH subunits XPB and XPD factors involved in nucleotide excision repair, the Cockayne syndrome group B protein (CSB) also involved in DNA damage repair (31), and the WRN gene product which is found mutated in the progeroid Werner syndrome (2). The regulatory domain has been implicated in apoptotic functions of p53 and can also modulate its sequence-specific DNA-binding activity (31). Because nucleolin appears to interact with the regulatory domain of p53, nucleolin thus has the potential to alter the ability of p53 to bind its DNA recognition elements and hence may modulate the induction of p53-responsive genes.
The nucleolin response is stress selective in that it is observed only under certain traumatic conditions but not under others. Interestingly, p53 activation occurs following UV irradiation (Fig. 3) and HU treatment (data not shown), yet nucleolin does not mobilize in response to these stresses. Since it is accepted that different cellular stresses induce discrete signaling cascades, which in turn activate distinctive, sometimes overlapping responses (1, 40), it is likely that nucleolin is mobilized by only a subset of these stress-related pathways. More specifically, one can argue that nucleolin only associates with p53 in a particular posttranslational modification state. Clearly, the specific p53 phosphorylation profile is a regulatory determinant controlling its interaction with other factors, including the p53 antagonist Mdm2 (42, 43, 48) and the transcription factors TFIID and CBP/p300 (27, 38). The nucleolin-p53 interaction does not appear to be directly regulated by the phosphorylation state of Ser15, as nucleolin is still mobilized in AT cells (in which the ATM kinase is defective) following γ-irradiation (data not shown). Because the C-terminal region contains various phosphorylation and acetylation sites, some of which are modulated by stress, it is perhaps more likely that the modification of this region will be found to alter nucleolin-p53 complex formation. Conversely, mammalian nucleolin is also a target of various kinases including casein kinase II, the cyclin B-cdc2 complex, and protein kinase C ζ (15). Although little is known about the effect of stress on nucleolin phosphorylation, it would not be surprising to find that the phosphorylation state of nucleolin modulates both its cellular localization and its interaction with p53.
The unique compartmentalization of the nucleolus allows the cell to both sequester and rapidly mobilize critical factors involved in the regulation of DNA metabolism in response to stress (49). Prominent examples include the sequestration of the Mdm2 oncoprotein by the nucleolar ARF protein, facilitating p53 stabilization (32), and the relocalization of the human homologue of the Schizosaccharomyces pombe Rad17 protein, a key damage response factor, from the nucleolus to the nucleoplasm following UV irradiation (6). Combined with our findings, these data add to the growing body of evidence indicating that the nucleolus, along with its primary role in supporting ribosome biogenesis, is also intimately tied to the cellular stress response.
Acknowledgments
We thank Carol Prives and W. Jiang for their generous donation of cell lines, E. Rubin and J. Chen for reagents, and Moshe Oren for helpful discussion.
This research was supported by NIH grant AI29963 and Kaplan Cancer Center Developmental Funding and Kaplan Cancer Center Support Core Grant (NCI P30CA16087). Y.D. is supported by postdoctoral grant 01-182-01-TBE from the American Cancer Society.
Y. Daniely and D. D. Dimitrova contributed equally to this work.
REFERENCES
- 1.Ashcroft, M., Y. Taya, and K. H. Vousden. 2000. Stress signals utilize multiple pathways to stabilize p53. Mol. Cell. Biol. 20:3224-3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blander, G., J. Kipnis, J. F. Leal, C. E. Yu, G. D. Schellenberg, and M. Oren. 1999. Physical and functional interaction between p53 and the Werner's syndrome protein. J. Biol. Chem. 274:29463-29469. [DOI] [PubMed] [Google Scholar]
- 3.Budde, A., and I. Grummt. 1999. p53 represses ribosomal gene transcription. Oncogene 18:1119-1124. [DOI] [PubMed] [Google Scholar]
- 4.Bukovsky, A., M. R. Caudle, J. A. Keenan, J. Wimalasena, J. S. Foster, N. B. Upadhyaya, and S. E. van Meter. 1995. Expression of cell cycle regulatory proteins (p53, pRb) in the human female genital tract. J. Assist. Reprod. Genet. 12:123-131. [DOI] [PubMed] [Google Scholar]
- 5.Carrier, F., M. L. Smith, I. Bae, K. E. Kilpatrick, T. J. Lansing, C. Y. Chen, M. Engelstein, S. H. Friend, W. D. Henner, and T. M. Gilmer. 1994. Characterization of human Gadd45, a p53-regulated protein. J. Biol. Chem. 269:32672-32677. [PubMed] [Google Scholar]
- 6.Chang, M. S., H. Sasaki, M. S. Campbell, S. K. Kraeft, R. Sutherland, C. Y. Yang, Y. Liu, D. Auclair, L. Hao, H. Sonoda, L. H. Ferland, and L. B. Chen. 1999. HRad17 colocalizes with NHP2L1 in the nucleolus and redistributes after UV irradiation. J. Biol. Chem. 274:36544-36549. [DOI] [PubMed] [Google Scholar]
- 7.Chen, J., V. Marechal, and A. J. Levine. 1993. Mapping of the p53 and mdm-2 interaction domains. Mol. Cell. Biol. 13:4107-4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, X., L. J. Ko, L. Jayaraman, and C. Prives. 1996. p53 levels, functional domains, and DNA damage determine the extent of the apoptotic response of tumor cells. Genes Dev. 10:2438-2451. [DOI] [PubMed] [Google Scholar]
- 9.Cox, L. S., T. Hupp, C. A. Midgley, and D. P. Lane. 1995. A direct effect of activated human p53 on nuclear DNA replication. EMBO J. 14:2099-2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daniely, Y., and J. A. Borowiec. 2000. Formation of a complex between nucleolin and replication protein A after cell stress prevents initiation of DNA replication. J. Cell Biol. 149:799-810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dickinson, L. A., and T. Kohwi-Shigematsu. 1995. Nucleolin is a matrix attachment region DNA-binding protein that specifically recognizes a region with high base-unpairing potential. Mol. Cell. Biol. 15:456-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dimitrova, D. S., I. T. Todorov, T. Melendy, and D. M. Gilbert. 1999. Mcm2, but not RPA, is a component of the mammalian early G1-phase prereplication complex. J. Cell Biol. 146:709-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.el-Deiry, W. S., T. Tokino, V. E. Velculescu, D. B. Levy, R. Parsons, J. M. Trent, D. Lin, W. E. Mercer, K. W. Kinzler, and B. Vogelstein. 1993. WAF1, a potential mediator of p53 tumor suppression. Cell 75:817-825. [DOI] [PubMed] [Google Scholar]
- 14.Elledge, S. J. 1996. Cell cycle checkpoints: preventing an identity crisis. Science 274:1664-1672. [DOI] [PubMed] [Google Scholar]
- 15.Ginisty, H., H. Sicard, B. Roger, and P. Bouvet. 1999. Structure and functions of nucleolin. J. Cell Sci. 112:761-772. [DOI] [PubMed] [Google Scholar]
- 16.Haluska, P., Jr., A. Saleem, T. K. Edwards, and E. H. Rubin. 1998. Interaction between the N-terminus of human topoisomerase I and SV40 large T antigen. Nucleic Acids Res. 26:1841-1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hanakahi, L. A., Z. Bu, and N. Maizels. 2000. The C-terminal domain of nucleolin accelerates nucleic acid annealing. Biochemistry 39:15493-15499. [DOI] [PubMed] [Google Scholar]
- 18.Hanakahi, L. A., L. A. Dempsey, M. J. Li, and N. Maizels. 1997. Nucleolin is one component of the B cell-specific transcription factor and switch region binding protein, LR1. Proc. Natl. Acad. Sci. USA 94:3605-3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hannan, K. M., R. D. Hannan, and L. I. Rothblum. 1998. Transcription by RNA polymerase I. Front. Biosci. 3:d376-d398. [DOI] [PubMed] [Google Scholar]
- 20.Harper, J. W., G. R. Adami, N. Wei, K. Keyomarsi, and S. J. Elledge. 1993. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell 75:805-816. [DOI] [PubMed] [Google Scholar]
- 21.Iftode, C., Y. Daniely, and J. A. Borowiec. 1999. Replication protein A (RPA): the eukaryotic SSB. Crit. Rev. Biochem. Mol. Biol. 34:141-180. [DOI] [PubMed] [Google Scholar]
- 22.Janus, F., N. Albrechtsen, I. Dornreiter, L. Wiesmüller, F. Grosse, and W. Deppert. 1999. The dual role model for p53 in maintaining genomic integrity. Cell. Mol. Life Sci. 55:12-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jayaraman, L., N. C. Moorthy, K. G. Murthy, J. L. Manley, M. Bustin, and C. Prives. 1998. High mobility group protein-1 (HMG-1) is a unique activator of p53. Genes Dev. 12:462-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaufmann, W. K., and R. S. Paules. 1996. DNA damage and cell cycle checkpoints. FASEB J. 10:238-247. [DOI] [PubMed] [Google Scholar]
- 25.Kostic, C., and P. H. Shaw. 2000. Isolation and characterization of sixteen novel p53 response genes. Oncogene 19:3978-3987. [DOI] [PubMed] [Google Scholar]
- 26.Lai, J. S., and W. Herr. 1992. Ethidium bromide provides a simple tool for identifying genuine DNA-independent protein associations. Proc. Natl. Acad. Sci. USA 89:6958-6962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lambert, P. F., F. Kashanchi, M. F. Radonovich, R. Shiekhattar, and J. N. Brady. 1998. Phosphorylation of p53 serine 15 increases interaction with CBP. J. Biol. Chem. 273:33048-33053. [DOI] [PubMed] [Google Scholar]
- 28.Larner, J. M., H. Lee, and J. L. Hamlin. 1997. S phase damage sensing checkpoints in mammalian cells. Cancer Surv. 29:25-45. [PubMed] [Google Scholar]
- 29.Levine, A. J. 1997. p53, the cellular gatekeeper for growth and division. Cell 88:323-331. [DOI] [PubMed] [Google Scholar]
- 30.Mao, Y., I. R. Mehl, and M. T. Muller. 2002. Subnuclear distribution of topoisomerase I is linked to ongoing transcription and p53 status. Proc. Natl. Acad. Sci. USA 99:1235-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.May, P., and E. May. 1999. Twenty years of p53 research: structural and functional aspects of the p53 protein. Oncogene 18:7621-7636. [DOI] [PubMed] [Google Scholar]
- 32.Momand, J., H. H. Wu, and G. Dasgupta. 2000. MDM2—master regulator of the p53 tumor suppressor protein. Gene 242:15-29. [DOI] [PubMed] [Google Scholar]
- 33.Muller, M., S. Wilder, D. Bannasch, D. Israeli, K. Lehlbach, M. Li-Weber, S. L. Friedman, P. R. Galle, W. Stremmel, M. Oren, and P. H. Krammer. 1998. p53 activates the CD95 (APO-1/Fas) gene in response to DNA damage by anticancer drugs. J. Exp. Med. 188:2033-2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Munsch, D., R. Watanabe-Fukunaga, J. C. Bourdon, S. Nagata, E. May, E. Yonish-Rouach, and P. Reisdorf. 2000. Human and mouse Fas (APO-1/CD95) death receptor genes each contain a p53-responsive element that is activated by p53 mutants unable to induce apoptosis. J. Biol. Chem. 275:3867-3872. [DOI] [PubMed] [Google Scholar]
- 35.O'Connor, P. M. 1997. Mammalian G1 and G2 phase checkpoints. Cancer Surv. 29:151-182. [PubMed] [Google Scholar]
- 36.Oren, M. 1999. Regulation of the p53 tumor suppressor protein. J. Biol. Chem. 274:36031-36034. [DOI] [PubMed] [Google Scholar]
- 37.Phair, R. D., and T. Misteli. 2000. High mobility of proteins in the mammalian cell nucleus. Nature 404:604-609. [DOI] [PubMed] [Google Scholar]
- 38.Pise-Masison, C. A., M. Radonovich, K. Sakaguchi, E. Appella, and J. N. Brady. 1998. Phosphorylation of p53: a novel pathway for p53 inactivation in human T-cell lymphotropic virus type 1-transformed cells. J. Virol. 72:6348-6355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prives, C. 1998. Signaling to p53: breaking the MDM2-p53 circuit. Cell 95:5-8. [DOI] [PubMed] [Google Scholar]
- 40.Rhind, N., and P. Russell. 1998. The Schizosaccharomyces pombe S-phase checkpoint differentiates between different types of DNA damage. Genetics 149:1729-1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rubbi, C. P., and J. Milner. 2000. Non-activated p53 co-localizes with sites of transcription within both the nucleoplasm and the nucleolus. Oncogene 19:85-96. [DOI] [PubMed] [Google Scholar]
- 42.Shieh, S.-Y., M. Ikeda, Y. Taya, and C. Prives. 1997. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell 91:325-334. [DOI] [PubMed] [Google Scholar]
- 43.Shieh, S. Y., Y. Taya, and C. Prives. 1999. DNA damage-inducible phosphorylation of p53 at N-terminal sites including a novel site, Ser20, requires tetramerization. EMBO J. 18:1815-1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Siliciano, J. D., C. E. Canman, Y. Taya, K. Sakaguchi, E. Appella, and M. B. Kastan. 1997. DNA damage induces phosphorylation of the amino terminus of p53. Genes Dev. 11:3471-3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith, M. L., I. T. Chen, Q. Zhan, I. Bae, C. Y. Chen, T. M. Gilmer, M. B. Kastan, P. M. O'Connor, and A. J. Fornace, Jr. 1994. Interaction of the p53-regulated protein Gadd45 with proliferating cell nuclear antigen. Science 266:1376-1380. [DOI] [PubMed] [Google Scholar]
- 46.Stürzbecher, H. W., B. Donzelmann, W. Henning, U. Knippschild, and S. Buchhop. 1996. p53 is linked directly to homologous recombination processes via RAD51/RecA protein interaction. EMBO J. 15:1992-2002. [PMC free article] [PubMed] [Google Scholar]
- 47.Tibbetts, R. S., K. M. Brumbaugh, J. M. Williams, J. N. Sarkaria, W. A. Cliby, S. Y. Shieh, Y. Taya, C. Prives, and R. T. Abraham. 1999. A role for ATR in the DNA damage-induced phosphorylation of p53. Genes Dev. 13:152-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Unger, T., T. Juven-Gershon, E. Moallem, M. Berger, R. Vogt Sionov, G. Lozano, M. Oren, and Y. Haupt. 1999. Critical role for Ser20 of human p53 in the negative regulation of p53 by Mdm2. EMBO J. 18:1805-1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Visintin, R., and A. Amon. 2000. The nucleolus: the magician's hat for cell cycle tricks. Curr. Opin. Cell Biol. 12:372-377. [DOI] [PubMed] [Google Scholar]
- 50.Wiesmüller, L., J. Cammenga, and W. W. Deppert. 1996. In vivo assay of p53 function in homologous recombination between simian virus 40 chromosomes. J. Virol. 70:737-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xiong, Y., G. J. Hannon, H. Zhang, D. Casso, R. Kobayashi, and D. Beach. 1993. p21 is a universal inhibitor of cyclin kinases. Nature 366:701-704. [DOI] [PubMed] [Google Scholar]
- 52.Ying, G. G., P. Proost, J. van Damme, M. Bruschi, M. Introna, and J. Golay. 2000. Nucleolin, a novel partner for the Myb transcription factor family that regulates their activity. J. Biol. Chem. 275:4152-4158. [DOI] [PubMed] [Google Scholar]
- 53.Zhou, J., J. Ahn, S. H. Wilson, and C. Prives. 2001. A role for p53 in base excision repair. EMBO J. 20:914-923. [DOI] [PMC free article] [PubMed] [Google Scholar]