Abstract
Human nucleotide excision repair is initiated by six repair factors (XPA, RPA, XPC-HR23B, TFIIH, XPF-ERCC1, and XPG) which sequentially assemble at sites of DNA damage and effect excision of damage-containing oligonucleotides. We here describe the molecular anatomy of the human excision nuclease assembled at the site of a psoralen-adducted thymine. Three polypeptides, primarily positioned 5′ to the damage, are in close physical proximity to the psoralen lesion and thus are cross-linked to the damaged DNA: these proteins are RPA70, RPA32, and the XPD subunit of TFIIH. While both XPA and XPC bind damaged DNA and are required for XPD cross-linking to the psoralen-adducted base, neither XPA nor XPC is cross-linked to the psoralen adduct. The presence of other repair factors, in particular TFIIH, alters the mode of RPA binding and the position of its subunits relative to the psoralen lesion. Based on these results, we propose that RPA70 makes the initial contact with psoralen-damaged DNA but that within preincision complexes, it is RPA32 and XPD that are in close contact with the lesion.
The basic mechanism of nucleotide excision repair includes (i) damage recognition, (ii) assembly of repair factors at the site of damage, (iii) dual incisions that result in excision of damage-containing oligomers, (iv) resynthesis to fill in the gap, and (v) ligation. In humans excision repair is initiated by the combined action of six repair factors, XPA, RPA, XPC, TFIIH, XPG, and XPF-ERCC1, which excise the damaged nucleotide(s) in the form of 24- to 32-nucleotide (nt)-long oligomers (7, 30, 33, 47). In yeast the same six factors are both essential and sufficient for excision of 24- to 27-nt-long oligomers (10). XPA, RPA, and XPC are involved in damage recognition, TFIIH unwinds the duplex around the lesion and stabilizes the damage recognition complex, and XPG and XPF-ERCC1 make the 3′ and 5′ incisions, respectively. There is general agreement about the above-stated roles of these factors, but there is limited information about the location of these proteins relative to one another and relative to the lesion within preincision and incision complexes.
Previously we identified three stable DNA-protein complexes (25) which we named preincision complex 1 (PIC1) (RPA-XPA-XPC-TFIIH-DNA), PIC2 (RPA-XPA-TFIIH-XPG-DNA), and PIC3 (RPA-XPA-TFIIH-XPG-XPF-ERCC1-DNA). In this study we attempted to locate the various repair factors with a protein-DNA cross-linking assay using a furan-side psoralen-thymine adduct both as a substrate and as a cross-linker (5, 16, 27). Our results show that RPA is in close contact with the DNA lesion prior to unwinding by TFIIH and that following unwinding, the mode of RPA-DNA interaction changes drastically such as to affect RPA-DNA cross-linking both qualitatively and quantitatively. Surprisingly, neither XPA nor XPC are cross-linked to DNA either when bound in isolation or in the preincision complexes. In contrast, the XPD subunit of TFIIH is cross-linked to psoralen in PIC1, PIC2, and PIC3. Furthermore, we find that placement of the radiolabel 5′ to the lesion results in a higher level of protection than when the label is on the 3′ side of the damage. Collectively, these data show that within the preincision complexes, RPA and the XPD subunit of TFIIH are in intimate contact with the psoralen lesion and allow us to propose a provisional model for the molecular anatomy of the human excision nuclease.
MATERIALS AND METHODS
Materials.
T4 polynucleotide kinase and DNA ligase were purchased from New England BioLabs, DNase I was from Invitrogen, and the TnT Quick Coupled Transcription/Translation system was from Promega. Molecular mass size markers were HinfI-digested ΦX174 DNA (Promega), prestained Rainbow markers (Amersham Pharmacia Biotech), and Benchmark protein standards (Invitrogen). [γ-32P]ATP (7,000 Ci/mmol) was obtained from ICN, and l-[35S]methionine (1,000 Ci/mmol) was from Amersham Pharmacia Biotech. The dodecamer containing a furan-side psoralen-thymine adduct was prepared by reacting a synthetic oligomer with 4′-hydroxymethyl-4,5′,8-trimethylpsoralen (HMT) followed by high-performance liquid chromatography purification and was a generous gift from J. E. Hearst (University of California, Berkeley). Other oligomers, 20 to 64 nt in length, were purchased from Operon Technologies.
Repair factors.
Human repair factors purified from HeLa cells (TFIIH, RPA), a baculovirus-insect cell system (XPC-HR23B, XPF-ERCC1, XPG), or bacterial cells (XPA, RPA) were obtained using purification schemes similar to those described previously (23-24). If an RPA subunit is not specified in this presentation, RPA refers to the heterotrimer. For in vitro labeling of repair factors with [35S]methionine we used the TnT system according to the manufacturer's directions. Expression of RPA70 (p11d-tRPA) was under the control of a T7 promoter (13), and XPB and XPD coding sequences were introduced into pIBI25 under the control of a T7 promoter.
Excision assay.
The substrates were 140-bp duplexes prepared by ligating a psoralen-damaged dodecamer with five other partially overlapping oligomers as described previously (2, 16). Unless stated otherwise, the substrate with the radiolabel 5′ to the psoralen adduct was used in assays. To obtain DNA substrates with the radiolabel 3′ to the psoralen adduct, the 64-mer 3′ to the lesion was radiolabeled and used to prepare the substrate (Fig. 1A). Unless indicated otherwise, substrate DNA (30 fmol, 1.2 nM) was incubated at 30°C for 60 min with six repair factors (50 ng of XPA [50 nM], 10 ng of XPC-HR23B [2.5 nM], 450 ng of RPA [140 nM], 150 ng of TFIIH [12.5 nM], 20 ng of XPF-ERCC1 [6 nM], and 10 ng of XPG [3 nM]) in 25 μl of reconstitution reaction buffer (32 mM HEPES [pH 7.9], 64 mM KCl, 6.4 mM MgCl2, 4 to 5% glycerol, 4 mM ATP with bovine serum albumin at a concentration of 200 μg/ml). To detect the release of excision products, DNA was deproteinized, precipitated, resuspended in a formamide-dye mixture, separated in 10% denaturing (7.7 M urea) polyacrylamide gels, visualized by autoradiography, and quantitated using phosphor screens and ImageQuant 5.2 software (Molecular Dynamics). Statistical analyses were performed using programs available at the VassarStats website (http://faculty.vassar.edu/lowry/VassarStats.html).
FIG. 1.
DNA substrates used in this study. The sequence of the 140-bp duplex with a psoralen-adducted thymine at position 70 (underlined) is shown in panel A. The arrows above and below the sequence indicate the ligation sites between the six oligomers used to assemble the substrate. Radiolabels were introduced in the top strand at the sixth phosphodiester bond 5′ to the lesion or at the seventh phosphodiester bond 3′ to the adduct to generate 5′ and 3′ labeled substrates, respectively. The duplex contained a furan-side monoadduct, and the chemical structure of HMT is shown in panel B. The HMT adduct locally disrupts the helical structure (39), and TFIIH further unwinds the duplex in both the 5′ and 3′ directions as illustrated in panel C (adapted from reference 25).
Photo-cross-linking assay.
To cross-link DNA-protein complexes with 366-nm light, excision assay reaction mixtures in 1.5-ml microtubes were irradiated on ice using a Spectroline model B-100 black light lamp. There was a 5- to 6-cm distance between samples and the light source, and irradiation was at 15 to 20 mW/cm2 for 60 min (5.4 × 106 to 7.2 × 106 erg/mm2). To reduce background signal from radiolabeled DNA migrating throughout the gel and to permit a more accurate estimate of the molecular masses of protein-DNA complexes, samples were treated with DNase I prior to electrophoresis. Cross-linked DNA-protein complexes were treated with DNase I (200 to 300 U/30 fmol of input DNA) in repair reaction buffer supplemented with 6 to 8 mM CaCl2 for 10 min at 25°C. Following irradiation and DNase I digestion, reactions were stopped by adding sodium dodecyl sulfate (SDS) loading buffer and heating for 5 min at 65°C. DNA-protein complexes were resolved in SDS-polyacrylamide gels (8 or 10% acrylamide), visualized by autoradiography, and quantitated using phosphor screens and ImageQuant 5.2 software. Electrophoresis was continued until the free DNA, which comigrated with bromphenol blue, was out of the gel; proteins smaller than an Mr of ∼30,000 were also not retained in the gel. Radiolabeled DNA-protein complexes that did not enter the separating gel could not be identified and, thus, were not included in analyses. Molecular masses of protein-DNA complexes were estimated from Rainbow protein markers and single-stranded DNA size markers resolved in the same gels; the sizes of DNA fragments (in nucleotides) were converted to approximate values in kilodaltons based on mobility in SDS-polyacrylamide gels in which Benchmark protein standards were resolved in parallel.
Electrophoretic mobility shift assay.
Repair factors were incubated with radiolabeled DNA under reconstitution assay reaction conditions for 60 min at 30°C. Glycerol was added to ∼13%, and samples were resolved in 5% nondenaturing polyacrylamide gels, with electrophoresis at room temperature and at a constant current of 30 mA. Visualization and quantitation were as described for the excision and photo-cross-linking assays.
RESULTS
Cross-linking of individual repair factors to psoralen.
To gain some insight into the interactions of the six factors of the human excision nuclease with DNA, we incubated each factor individually with a 140-mer duplex containing a psoralen furan-side thymine monoadduct (Fig. 1) and irradiated the reaction mixtures with 366-nm black light. Under these conditions, psoralen functions both as a substrate and as a cross-linking agent (5, 16, 27) and, as such, is a good reagent to investigate excision nuclease-substrate interactions. Under reaction conditions optimized for excision activity, only RPA was cross-linked to DNA when each repair factor was individually incubated with the substrate (Fig. 2). In agreement with other studies which employed different DNA-protein cross-linking agents, both RPA70 and RPA32 were cross-linked to DNA (18, 21, 31). Importantly, neither XPA nor XPC, DNA binding proteins with some affinity for damaged DNA, were cross-linked to psoralen. Since under standard excision assay conditions RPA is used at higher molar concentrations than XPA or XPC, we reasoned that by increasing the concentrations of the latter two factors we might achieve DNA-protein cross-linking with these factors as well. Toward this goal, we first performed electrophoretic mobility shift assays to determine concentrations of XPA and XPC which bind substantial fractions of the substrate (Fig. 3A, B), and we performed the photo-cross-linking under these conditions. Even under conditions where about 80% of the DNA was bound to XPA, there was no detectable cross-linking to XPA (Fig. 3C). Some cross-linking was observed with XPC, but the efficiency was low compared to RPA-DNA cross-linking. Thus, it appears that of the three damage recognition factors, RPA is the one that makes the most intimate contact with the psoralen lesion.
FIG. 2.
RPA70 and RPA32 are photo-cross-linked to psoralen-damaged DNA. The substrate was a 140-bp duplex with 32P radiolabel six phosphodiester bonds 5′ to the psoralen-adducted thymine (Fig. 1). The indicated repair factors (at the same concentrations used to reconstitute excision repair) were incubated with DNA for 60 min and irradiated with black light, and then reaction mixtures were digested with DNase I. Following resolution in a SDS-10% polyacrylamide gel, DNA-protein complexes were visualized by autoradiography. To the left are shown the positions of molecular mass markers resolved in the same gel. The fainter bands (indicated with open arrows) migrating between RPA70-DNA and RPA32-DNA complexes result from cross-linking proteolytic fragments of RPA70 to the substrate and were not included in quantitative analyses. The most slowly migrating complex, designated (XPD), was identified as an XPD-DNA complex in subsequent experiments.
FIG. 3.
XPA and XPC bind DNA but are not cross-linked to psoralen. Panel A shows an autoradiograph of DNA-protein complexes resolved in a nondenaturing polyacrylamide gel. Excision repair is typically reconstituted with 140 nM RPA, 50 nM XPA, and 2.5 nM XPC (lanes 2, 3, and 6). For quantitation, the fraction of DNA not in the gel position corresponding to free DNA was considered bound. The average percent binding for this experiment and a second electrophoretic mobility shift assay conducted under the same conditions are shown in panel B; in the plot on the left, RPA-DNA binding is indicated by the filled triangle. Panel C illustrates the results of a photo-cross-linking experiment performed with half of the same reactions shown in panel A; to the left are positions of molecular mass markers resolved in the same gel. The fainter band (indicated with an open arrow) migrating between RPA70-DNA and RPA32-DNA complexes results from cross-linking proteolytic fragments of RPA70 to the substrate and was not included in quantitative analyses.
DNA-protein contacts within PIC1, PIC2, and PIC3.
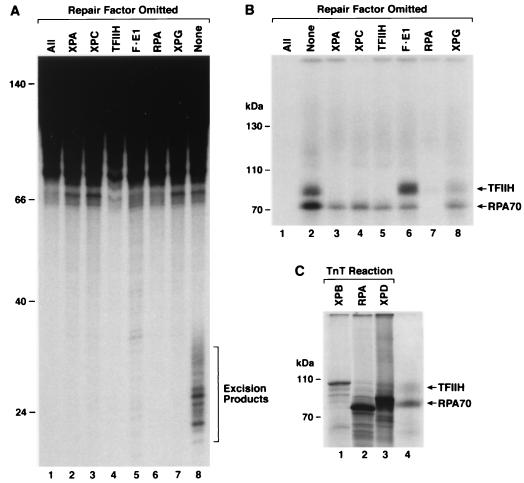
In the reaction containing all six repair factors, PIC3, (Fig. 2, lane 8) there were two prominent differences from the reaction containing just the RPA protein (lane 5). First, a new cross-linked protein with an Mr of ∼90,000 appeared, and second, the ratio of cross-linking efficiency of RPA70 to RPA32 was reversed. We wished to identify the 90-kDa band and to determine at which stage during incision complex assembly it becomes cross-linkable. Toward this goal, we performed cross-linking reactions under conditions of single-factor omission to capture PIC1, PIC2, and PIC3 and other potential intermediates on the reaction pathway. In agreement with previous results (24), dual incisions and excision occur only when all six factors are present in the reaction mixture (Fig. 4A). Interestingly, when cross-linking was performed under single-factor omission conditions (Fig. 4B), the ∼90,000-Mr band appeared in reactions which corresponded to formation of PIC1 (lane 8), PIC2 (lane 6), and PIC3 (lane 2). Both the size of this new band and a consideration of factors common in all three preincision complexes suggested that the cross-linked protein might be a TFIIH subunit, and the size indicated that it might be either XPB (Mr, ∼89,000) or XPD (Mr, ∼83,000). To be detectable the protein must be cross-linked to a psoralen-containing oligomer; both the RPA70-DNA and RPA32-DNA complexes were retarded by 3 to 9 kDa relative to the migration of proteins purified from HeLa cells or made by in vitro transcription and translation (Fig. 4C, compare lanes 2 and 4; also data not shown), consistent with similar retardation values reported for RPA32 cross-linked to a 17-mer (18). To identify the TFIIH subunit, we resolved the PIC3 cross-linking reaction alongside XPB and XPD made by in vitro transcription and translation. Figure 4C shows that the cross-linked TFIIH band migrates faster than XPB and more slowly than XPD, and, therefore, the protein in the ∼90-kDa protein-DNA complex was identified as the XPD subunit of TFIIH.
FIG. 4.
Single-factor omission experiments for excision repair and cross-linking assays reveal that the third complex contains XPD. Repair factors (at the same concentrations used to reconstitute excision repair), with the omitted factor indicated, were incubated with psoralen-damaged DNA for 60 min and then either processed for detection of excised oligomers (A) or photo-cross-linked to detect DNA-protein complexes (B). To detect excision, deproteinized DNA was resolved in a 10% sequencing gel with HinfI digested DNA as size markers (mobility positions, in nucleotides, are shown to the left) and visualized by autoradiography (A). To detect DNA-protein complexes, reaction mixtures were resolved in an SDS-8% polyacrylamide gel with size markers (indicated to the left) and visualized by autoradiography (B). Only the portion of the gel corresponding to protein-DNA complexes greater than approximately 70 kDa is shown. Panel C illustrates in vitro-labeled proteins resolved in an SDS-10% polyacrylamide gel with a complete repair reaction that was cross-linked and treated with DNase I; only the relevant section of the gel is shown to illustrate the migration of the TFIIH and RPA70 complexes (lane 4) relative to 35S-labeled XPB, RPA70, and XPD proteins (lanes 1 to 3).
The simplest interpretation of our data is that in the excision nuclease preincision/incision complexes, RPA and the XPD subunit of TFIIH are the polypeptides closest to, and perhaps in contact with, the psoralen lesion. Furthermore, comparison of the cross-linked XPD band intensity in the preincision complexes (Fig. 4B) reveals that cross-linking is more efficient in PIC2 (RPA-XPA-TFIIH-XPG-DNA, lane 6) than in PIC1 (RPA-XPA-XPC-TFIIH-DNA, lane 8). This is consistent with our previous observation that even though PIC1 contains the repair factors in an appropriate conformation to permit unwinding around the lesion (Fig. 1C), PIC1 is not as stable as PIC2, which is stabilized by entry of XPG and exit of XPC from the preincision complex (25, 45). Finally, in PIC3 (RPA-XPA-TFIIH-XPG-XPF-ERCC1-DNA, lane 2) the intensity of the XPD band is somewhat lower than that seen in the PIC2 complex, most likely as a consequence of dual incisions which release a psoralen-containing oligonucleotide from the substrate.
Reorganization of the DNA-protein complex in PIC3.
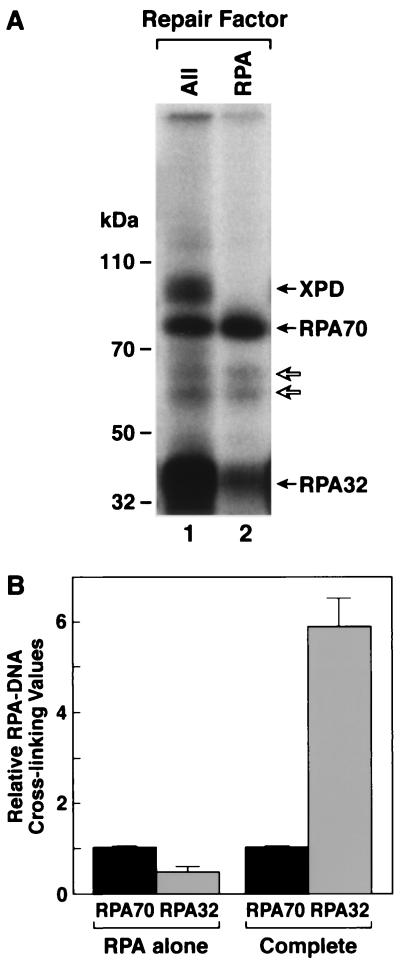
In addition to the appearance of cross-linked XPD in the presence of all six factors, the other striking feature of the six-factor experiments was the inversion of cross-linking efficiencies for RPA70 and RPA32 (Fig. 2, compare lanes 5 and 8). In cross-linking with RPA alone RPA70 was cross-linked more efficiently than RPA32, but in the presence of all six repair factors (complete reaction including RPA, XPA, XPC, TFIIH, XPG, and XPF-ERCC1) the ratio was dramatically reversed (Fig. 5). Whereas the RPA70/RPA32 cross-linking ratio was 2.46 with RPA alone, this ratio was 0.17 in the complete reaction, nearly a 15-fold change in the relative cross-linking efficiencies. We reasoned that this change represents a change in the position of RPA relative to psoralen during the formation of the excision complex, and we wished to identify the conditions necessary to effect this change. To this end we performed five-factor cross-linking experiments and individually omitted the components of PIC1, three of which (RPA, XPA, and XPC) have been implicated in damage recognition. As seen in Fig. 6 and summarized in Table 1, the RPA70/RPA32 cross-linking ratio in the absence of TFIIH was not significantly different from the ratio obtained with RPA alone. However, this ratio was essentially inverted in the absence of XPA or XPC in a manner similar to the inversion seen in the presence of six factors. To a first approximation, these data suggest that in the absence of XPA or XPC, preincision complexes with RPA and TFIIH can form, and in these RPA-TFIIH-DNA complexes, RPA is in a configuration similar to that found in PIC3. It must be noted, however, that the RPA-TFIIH-DNA complex must undergo further reorganization upon entry of the other repair factors, because within this putative ternary complex XPD is still not cross-linkable to psoralen (Fig. 6, lanes 3 and 5) and becomes cross-linkable only upon incubation with repair factors which generate PIC1, PIC2, or PIC3 (Fig. 6, lane 2; also Fig. 4B).
FIG. 5.
RPA subunits are differentially cross-linked when incubated alone or in combination with other repair factors. RPA was incubated with substrate DNA as described previously either alone or with other repair factors (at the same concentrations used to reconstitute excision repair) for 60 min and irradiated with black light, and then reaction mixtures were digested with DNase I. Following resolution in an SDS-10% polyacrylamide gel, the DNA-protein complexes were visualized by autoradiography (A). Lane 1 is a complete reaction, and lane 2 is RPA alone; to the left are shown the positions of molecular mass markers resolved in the same gel. The fainter bands migrating between RPA70-DNA and RPA32-DNA complexes (indicated with open arrows) result from cross-linking proteolytic fragments of RPA70 to the substrate and were not included in quantitation. Panel B is a graphic summarization of relative RPA70/RPA32 cross-linking values for RPA alone reactions (n = 8) and complete reactions (n = 16). Average values are plotted, and error bars represent the standard error. For each data set the relative RPA70 signal is defined as 1.
FIG. 6.
Single-factor omission experiments for photo-cross-linking reveal that inversion of the RPA70/RPA32 ratio is dependent on TFIIH. The substrate was a 140-bp duplex with 32P radiolabel six phosphodiester bonds 5′ to the psoralen-adducted thymine (Fig. 1). The indicated repair factors (at the same concentrations used to reconstitute excision repair) were incubated with DNA for 60 min and irradiated with black light, and reaction mixtures were digested with DNase I. Following resolution in an SDS-10% polyacrylamide gel, DNA-protein complexes were visualized by autoradiography. To the left are shown the positions of molecular mass markers resolved in the same gel. The fainter band (indicated with an open arrow) migrating between RPA70-DNA and RPA32-DNA complexes results from cross-linking proteolytic fragments of RPA70 to the substrate and was not included in quantitative analyses.
TABLE 1.
Ratios of RPA70/RPA32 subunits cross-linked to psoralen-damaged DNA
| Repair factor included | RPA70/RPA32 cross-linking efficiencya |
|---|---|
| RPA | 2.46 |
| RPA, XPA, XPC, XPF-E1, XPG, (-TFIIH) | 2.30 |
| RPA, XPC, TFIIH, XPF-E1, XPG, (-XPA) | 0.57 |
| RPA, XPA, TFIIH, XPF-E1, XPG, (-XPC) | 0.42 |
| RPA, XPA, XPC, TFIIH, XPF-E1, XPG | 0.17 |
Relative intensity values were determined for RPA70-DNA and RPA32-DNA complexes. Average values for 3 to 4 experiments are shown; for RPA alone there were 8 experiments, and for complete reactions there were 16 experiments.
Cross-linking with a substrate containing radiolabel 3′ to the psoralen.
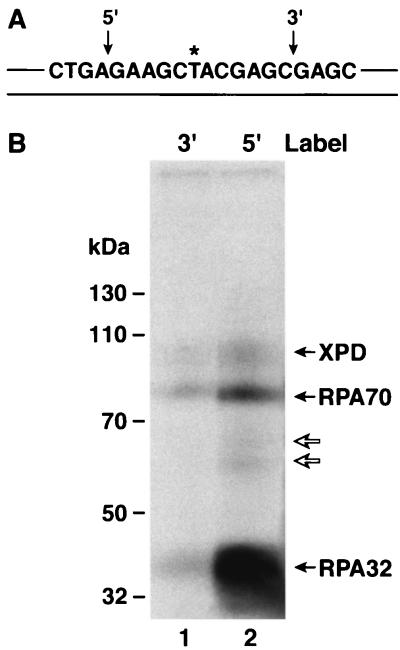
Since detection of DNA-protein cross-links in our assay depends on protection of the radiolabel from DNase I digestion, we reasoned that placing the radiolabel 3′ to the psoralen adduct may reveal a different set of cross-linkable proteins. Hence, we prepared substrate with a 32P label at the seventh phosphodiester bond 3′ to the psoralen and performed cross-linking experiments with this substrate. As seen in Fig. 7, the cross-linking pattern with this substrate was essentially identical to that with the 5′ label but the intensities of the XPD, RPA70, and RPA32 bands were much weaker. Thus, it appears that these three proteins are the only ones in close contact with the damage and that they make more intimate contacts with the DNA 5′ to the damage within the preincision complex. This is consistent with the more extensive unwinding of the DNA 5′ to the damage in the “repair bubble” observed in PIC1, PIC2, and PIC3 (25) (Fig. 1C). Alternatively, it could be argued that the observed DNA-protein complexes are due to cross-linking proteins to the excised oligomer (i.e., postincision complexes) and that the weaker intensities observed with the substrate containing radiolabel 3′ to the damage are due to the fact that excision from this substrate releases mainly a nonradioactive oligomer. However, we observe strong cross-linking to these three polypeptides even in the absence of excision (Fig. 4 and 6), and this indicates that the majority of the cross-linking is not to the excised oligomer.
FIG. 7.
Repair factor binding is preferentially located 5′ to the psoralen damage. Panel A is a schematic diagram of the psoralen 140-bp duplex, highlighting the central 20 nt; the substrate extends 60 nt in both the 5′ and 3′ directions, and the locations of 32P radiolabel are indicated by arrows above the sequence. Panel B is an autoradiograph of an SDS-10% polyacrylamide gel. The complete complement of repair factors was incubated with either the 3′ labeled substrate (lane 1) or the 5′ labeled substrate (lane 2), irradiated with black light, and reaction mixtures were digested with DNase I prior to electrophoresis and autoradiography. To the left are shown the positions of molecular mass markers resolved in the same gel. The fainter bands migrating between RPA70-DNA and RPA32-DNA complexes (indicated by open arrows) result from cross-linking proteolytic fragments of RPA70 to the substrate and were not included in quantitative analyses.
DISCUSSION
We conducted this study to complement previous investigations using repair kinetics and footprinting methods to analyze the order of assembly of the human excision nuclease and the structure of the preincision/incision complexes. Our results suggest that RPA plays a prominent role in damage recognition and that it is in direct contact with the psoralen-damaged base both in the initial damage recognition step and in later steps involving formation of PIC1, PIC2, and PIC3. In addition, our results reveal that the XPD subunit of TFIIH is in close proximity with the lesion and can be photo-cross-linked to psoralen. In light of these findings we wish to comment on binding to damaged DNA, on the order of assembly of the excision nuclease, and on the composition and subunit location of the excision nuclease relative to the damaged base.
Binding to damaged DNA.
Electrophoretic mobility shift and footprinting experiments have shown that RPA, XPA, and XPC bind to damaged DNA preferentially but with different affinities and that each has relatively similar discriminatory powers between undamaged and damaged DNA (reference 46 and references contained therein). Therefore, we found it rather surprising that of the three damage recognition factors, only RPA was cross-linked to damaged DNA to any significant extent. This may at least in part be due to the special photochemistry of psoralen-protein photo-cross-linking. Upon intercalation and photo-addition, leading to formation of the furan-side monoadduct, the DNA helix is distorted in a relatively minor way and locally denatured for only about 3 bp in either direction (39). When a furan-side monoadduct is formed, a second photoreactive element in the pyrone-side ring (Fig. 1B) is directed to the interior of the DNA helix and may be cross-linked to a pyrimidine in the complementary strand (5) or to a DNA-bound protein (27, 34-36). The spatial and photochemical requirements for psoralen-protein cross-linking are not known at present. It has been suggested that for a protein to be cross-linked to a psoralen-DNA adduct it would most likely be probing within the helix and that the photoreactive amino acids are in close proximity (∼8 Å) with the pyrone-side ring (34). It appears that even though XPA and XPC bind preferentially to damaged DNA, either photoreactive amino acids do not make intimate contacts with psoralen damage either in isolation or within the context of preincision complexes or the footprint may not be sufficient to protect the radiolabeled nucleotide from DNase I digestion. In agreement with these findings, other studies using other cross-linking agents adjacent to the damage or fluorescence anisotropy methods have revealed that RPA, but not XPA, directly contacts the damaged base (15, 38).
Order of assembly.
It has been proposed that all six factors exist in a preassembled repairosome which recognizes and processes DNA damage (11, 32, 42). This model has been tested, and the consensus is that a repairosome does not exist but that excision repair in both humans and yeast proceeds by the sequential assembly of repair factors at the damage site (1, 9, 25, 43). Currently there are several models for this sequential assembly. (i) An RPA-XPA complex functions as the damage sensor (3, 8, 12, 14, 20, 22). (ii) RPA alone or the RPA-XPA complex binds first, followed by the assembly of other factors (46). (iii) XPC is the initial factor to bind damaged DNA, followed by association of the other factors (40-41). (iv) There is no rigid order of assembly, nor is there a universal damage recognition factor: any of the three factors with preferential affinity for damaged DNA marks the site of the damage, and subsequently the other factors are recruited (44). In addition to this controversy regarding which repair factor functions as the initial damage sensor, different models have been proposed for the subsequent order of assembly of other human repair factors (25, 40, 43).
In light of our cross-linking experiments, we suggest that the initial contact of RPA with psoralen damage is most likely the first step of assembly because the RPA-DNA complex is capable of recruiting TFIIH to the site of the lesion, and even in the absence of XPA or XPC, this recruitment causes a change in RPA-DNA interactions similar to those seen previously in four- to six-factor mediated preincision complexes (25, 45). This is significant because it has been shown that both XPA (26, 29) and XPC (48) are also capable of recruiting TFIIH to DNA. However, there is no conclusive evidence that an XPA-TFIIH-DNA or XPC-TFIIH-DNA complex is on the pathway for productive preincision complex formation. In this study we analyzed aliquots of the same complete reaction mixtures for both excision and DNA-protein cross-linking, and we found that the excision signal was five- to sixfold greater than the sum of the protein-DNA complex signals, suggesting that the complexes which we identified are on the pathway for productive preincision complex formation. The data presented here demonstrate that in the presence of TFIIH, which alters the local DNA structure and promotes open complex formation, the relative positions of RPA subunits are similar to their positions in PIC1, PIC2, and PIC3, suggesting that the RPA-TFIIH-DNA complex is on the pathway for excision nuclease assembly. It should be noted, however, that the RPA-TFIIH-DNA complex is rather transient, because it was not detected by the permanganate footprinting or electrophoretic mobility shift assays (25, 45). It is possible that unwinding of the duplex within this complex aids in binding XPA and XPC and further contributes to formation of the first stable preincision complex, composed of RPA, XPA, XPC, TFIIH, and DNA.
Anatomy of the human excision nuclease.
Our assay detects repair factors that protect the radiolabel on the damaged strand. With both RPA and XPD cross-linking there was some protection when the label was 3′ to the lesion, but stronger protection was observed when the label was on the 5′ side. These results suggest that RPA and XPD are in close contact with the psoralen and are physically located 5′ to the damaged base. A low-resolution crystallographic structure of TFIIH shows that this factor has a toroidal shape (4, 37) which is likely to surround the duplex in such a way that XPD would be in contact with the damaged base. Although not evident with the substrate we have used, it is likely that RPA also binds both strands of the duplex (17) and that its mode of interaction in going from solitary binding to preincision complex changes in such a way that RPA32 is in closer contact with psoralen in the preincision complexes. There are no direct data on the location of XPA within the complex. However, XPA is known to interact with TFIIH (26, 29) and bind with very high affinity to XPF-ERCC1 (19, 28). Since the latter must be located near the site of 5′ incision, it is reasonable to assume that XPA is located at the 5′ end of the incision bubble. Similarly, we have no direct evidence on the location of XPC. However, previous studies have shown that both XPC (6) and XPG (23) copurify with TFIIH, and furthermore, it appears that XPG binds to TFIIH to the exclusion of XPC (45-46), suggesting that XPC and XPG share a binding surface on TFIIH. Since XPG is the 3′ nuclease, we assume that it is positioned near the 3′ incision site and displaces XPC, which presumably is similarly located 3′ to the damage.
Conclusion.
Based on data from this work and previous studies on the assembly of the human excision nuclease, we propose a model for the order of assembly and the anatomy of the human excision nuclease (Fig. 8). First, RPA binds to the psoralen damage site transiently, and upon interaction with TFIIH, the DNA is unwound around the damage. Since most of TFIIH is likely in a TFIIH-XPC form, the second step involves the recruitment of these two factors together. XPA enters the complex at some point; precisely when is not clear, but it is before the entry of XPG. The complex which forms with RPA, XPA, XPC, TFIIH, and DNA is called PIC1 (25), and the unwinding of DNA by TFIIH within this complex changes the spatial relation of RPA to the lesion and places both RPA32 and XPD in close contact with the lesion. In subsequent steps, XPG is recruited and displaces XPC at the 3′ boundary of the PIC1 bubble to generate PIC2, and, finally or concomitantly, XPF-ERCC1 is recruited to the 5′ boundary to create PIC3 with the approximate anatomy as shown in Fig. 8. Dual incisions by XPG and XPF-ERCC1 release the excised oligomer and some of the repair factors, while a subset of factors remains bound to the gapped DNA and to the excised oligomer (24). At present we do not have data on the subsequent steps of the pathway leading to repair synthesis and completion of nucleotide excision repair.
FIG. 8.
Molecular anatomy of preincision complexes. DNA damage, represented by a filled triangle, often disrupts base pairing in the immediate vicinity of the adduct. RPA subunits, designated RPA70 and RPA32, contact DNA damage in the absence of other repair factors, and the RPA70 subunit is preferentially photo-cross-linked to the psoralen lesion. The first stable preincision complex (PIC1) contains XPA (A), RPA (for clarity only the two larger subunits are shown), XPC-HR23B (C), and the multimeric TFIIH (for clarity only the XPD subunit is shown). In PIC1 TFIIH further unwinds the helix ∼20 nt in the region of DNA damage, RPA undergoes a conformational change, and the relative cross-linking of RPA subunits is inverted such that RPA32 is preferentially photo-cross-linked; this preferential cross-linking is also observed in PIC2 and PIC3. In PIC1 the XPD subunit of TFIIH is also in contact with the lesion, and XPD remains in contact with psoralen in PIC2 and PIC3. The ultimate preincision complex (PIC3) is also shown: XPC-HR23B is replaced by XPG (G) 3′ to the lesion, XPF-ERCC1 (F•E1) assembles 5′ to the damage, and dual incisions and excision (indicated by arrows) occur.
Acknowledgments
This work was supported by Public Health Service grant GM32833 from the National Institutes of Health.
We thank John E. Hearst (University of California, Berkeley) for the psoralen-damaged dodecamer, members of the laboratory group for generous sharing of reagents, and Ryujiro Hara, Deborah L. Croteau, and Mark E. Branum for useful discussions and critical reading of the manuscript.
REFERENCES
- 1.Araújo, S. J., E. A. Nigg, and R. D. Wood. 2001. Strong functional interactions of TFIIH with XPC and XPG in human DNA nucleotide excision repair, without a preassembled repairosome. Mol. Cell. Biol. 21:2281-2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bessho, T., D. Mu, and A. Sancar. 1997. Initiation of DNA interstrand cross-link repair in humans: the nucleotide excision repair system makes dual incisions 5′ to the cross-linked base and removes a 22- to 28-nucleotide-long damage-free strand. Mol. Cell. Biol. 17:6822-6830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buschta-Hedayat, N., T. Buterin, M. T. Hess, M. Missura, and H. Naegeli. 1999. Recognition of nonhybridizing base pairs during nucleotide excision repair of DNA. Proc. Natl. Acad. Sci. USA 96:6090-6095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang, W.-H., and R. D. Kornberg. 2000. Electron crystal structure of the transcription factor and DNA repair complex, core TFIIH. Cell 102:609-613. [DOI] [PubMed] [Google Scholar]
- 5.Cimino, G. D., H. B. Gamper, S. T. Isaacs, and J. E. Hearst. 1985. Psoralens as photoactive probes of nucleic acid structure and function: organic chemistry, photochemistry, and biochemistry. Annu. Rev. Biochem. 54:1151-1193. [DOI] [PubMed] [Google Scholar]
- 6.Drapkin, R., J. T. Reardon, A. Ansari, J.-C. Huang, L. Zawel, K. Ahn, A. Sancar, and D. Reinberg. 1994. Dual role of TFIIH in DNA excision repair and in transcription by RNA polymerase II. Nature 368:769-772. [DOI] [PubMed] [Google Scholar]
- 7.Friedberg, E. C., G. C. Walker, and W. Siede. 1995. DNA repair and mutagenesis. American Society for Microbiology. Washington, D.C.
- 8.Gunz, D., M. T. Hess, and H. Naegeli. 1996. Recognition of DNA adducts by human nucleotide excision repair: evidence for a thermodynamic probing mechanism. J. Biol. Chem. 271:25089-25098. [DOI] [PubMed] [Google Scholar]
- 9.Guzder, S. N., P. Sung, L. Prakash, and S. Prakash. 1996. Nucleotide excision repair in yeast is mediated by sequential assembly of repair factors and not by a pre-assembled repairosome. J. Biol. Chem. 271:8903-8910. [DOI] [PubMed] [Google Scholar]
- 10.Guzder, S. N., Y. Habraken, P. Sung, L. Prakash, and S. Prakash. 1995. Reconstitution of yeast nucleotide excision repair with purified Rad proteins, replication protein A, and transcription factor TFIIH. J. Biol. Chem. 270:12973-12976. [DOI] [PubMed] [Google Scholar]
- 11.He, Z., and C. J. Ingles. 1997. Isolation of human complexes proficient in nucleotide excision repair. Nucleic Acids Res. 25:1136-1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He, Z., L. A. Henricksen, M. S. Wold, and C. J. Ingles. 1995. RPA involvement in the damage-recognition and incision steps of nucleotide excision repair. Nature 374:566-569. [DOI] [PubMed] [Google Scholar]
- 13.Henricksen, L. A., C. B. Umbricht, and M. S. Wold. 1994. Recombinant replication protein A: expression, complex formation, and functional characterization. J. Biol. Chem. 269:11121-11132. [PubMed] [Google Scholar]
- 14.Hess, M. T., U. Schwitter, M. Petretta, B. Giese, and H. Naegeli. 1997. Bipartite substrate discrimination by human nucleotide excision repair. Proc. Natl. Acad. Sci. USA 94:6664-6669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hey, T., G. Lipps, and G. Krauss. 2001. Binding of XPA and RPA to damaged DNA investigated by fluorescence anisotropy. Biochemistry 40:2901-2910. [DOI] [PubMed] [Google Scholar]
- 16.Huang, J.-C., D. S. Hsu, A. Kazantsev, and A. Sancar. 1994. Substrate spectrum of human excinuclease: repair of abasic sites, methylated bases, mismatches, and bulky adducts. Proc. Natl. Acad. Sci. USA 91:12213-12217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lao, Y., C. G. Lee, and M. S. Wold. 1999. Replication protein A interactions with DNA. 2. Characterization of double-stranded DNA-binding/helix destabilization activities and the role of the zinc-finger domain in DNA interactions. Biochemistry 38:3974-3984. [DOI] [PubMed] [Google Scholar]
- 18.Lavrik, O. I., H.-P. Nasheuer, K. Weisshart, M. S. Wold, R. Prasad, W. A. Beard, S. H. Wilson, and A. Favre. 1998. Subunits of human replication protein A are crosslinked by photoreactive primers synthesized by DNA polymerases. Nucleic Acids Res. 26:602-607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li, L., S. J. Elledge, C. A. Peterson, E. S. Bales, and R. J. Legerski. 1994. Specific association between the human DNA repair proteins XPA and ERCC1. Proc. Natl. Acad. Sci. USA 91:5012-5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li, L., X. Lu, C. A. Peterson, and R. J. Legerski. 1995. An interaction between the DNA repair factor XPA and replication protein A appears essential for nucleotide excision repair. Mol. Cell. Biol. 15:5396-5402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mass, G., T. Nethanel, and G. Kaufmann. 1998. The middle subunit of replication protein A contacts growing RNA-DNA primers in replicating simian virus 40 chromosomes. Mol. Cell. Biol. 18:6399-6407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Missura, M., T. Buterin, R. Hindges, U. Hübscher, J. Kaspárková, V. Brabec, and H. Naegeli. 2001. Double-check probing of DNA bending and unwinding by XPA-RPA: an architectural function in DNA repair. EMBO J. 20:3554-3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mu, D., C.-H. Park, T. Matsunaga, D. S. Hsu, J. T. Reardon, and A. Sancar. 1995. Reconstitution of human DNA repair excision nuclease in a highly defined system. J. Biol. Chem. 270:2415-2418. [DOI] [PubMed] [Google Scholar]
- 24.Mu, D., D. S. Hsu, and A. Sancar. 1996. Reaction mechanism of human DNA repair excision nuclease. J. Biol. Chem. 271:8285-8294. [DOI] [PubMed] [Google Scholar]
- 25.Mu, D., M. Wakasugi, D. S. Hsu, and A. Sancar. 1997. Characterization of reaction intermediates of human excision repair nuclease. J. Biol. Chem. 272:28971-28979. [DOI] [PubMed] [Google Scholar]
- 26.Nocentini, S., F. Coin, M. Saijo, K. Tanaka, and J.-M. Egly. 1997. DNA damage recognition by XPA promotes efficient recruitment of transcription factor IIH. J. Biol. Chem. 272:22991-22994. [DOI] [PubMed] [Google Scholar]
- 27.Orren, D. K., C. P. Selby, J. E. Hearst, and A. Sancar. 1992. Post-incision steps of nucleotide excision repair in Escherichia coli. J. Biol. Chem. 267:780-788. [PubMed] [Google Scholar]
- 28.Park, C.-H., and A. Sancar. 1994. Formation of a ternary complex by human XPA, ERCC1, and ERCC4 (XPF) excision repair proteins. Proc. Natl. Acad. Sci. USA 91:5017-5021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park, C.-H., D. Mu, J. T. Reardon, and A. Sancar. 1995. The general transcription-repair factor TFIIH is recruited to the excision repair complex by the XPA protein independent of the TFIIE transcription factor. J. Biol. Chem. 270:4896-4902. [DOI] [PubMed] [Google Scholar]
- 30.Petit, C., and A. Sancar. 1999. Nucleotide excision repair: from E. coli to man. Biochimie 81:15-25. [DOI] [PubMed] [Google Scholar]
- 31.Philipova, D., J. R. Mullen, H. S. Maniar, J. Lu, C. Gu, and S. J. Brill. 1996. A hierarchy of SSB protomers in replication protein A. Genes Dev. 10:2222-2233. [DOI] [PubMed] [Google Scholar]
- 32.Rodriguez, K., J. Talamantez, W. Huang, S. H. Reed, Z. Wang, L. Chen, W. J. Feaver, E. C. Friedberg, and A. E. Tomkinson. 1998. Affinity purification and partial characterization of a yeast multiprotein complex for nucleotide excision repair using histidine-tagged Rad14 protein. J. Biol. Chem. 273:34180-34189. [DOI] [PubMed] [Google Scholar]
- 33.Sancar, A. 1996. DNA excision repair. Annu. Rev. Biochem. 65:43-81. [DOI] [PubMed] [Google Scholar]
- 34.Sastry, S. S. 1997. Isolation and partial characterization of a novel psoralen-tyrosine photoconjugate from a photoreaction of psoralen with a natural protein. Photochem. Photobiol. 65:937-944. [DOI] [PubMed] [Google Scholar]
- 35.Sastry, S. S., B. M. Ross, and A. P'arraga. 1997. Cross-linking of DNA-binding proteins to DNA with psoralen and psoralen furan-side monoadducts. J. Biol. Chem. 272:3715-3723. [DOI] [PubMed] [Google Scholar]
- 36.Sastry, S. S., H. P. Spielmann, Q. S. Hoang, A. M. Phillips, A. Sancar, and J. E. Hearst. 1993. Laser-induced protein-DNA cross-links via psoralen furanside monoadducts. Biochemistry 32:5526-5538. [DOI] [PubMed] [Google Scholar]
- 37.Schultz, P., S. Fribourg, A. Poterszman, V. Mallouh, D. Moras, and J. M. Egly. 2000. Molecular structure of human TFIIH. Cell 102:599-607. [DOI] [PubMed] [Google Scholar]
- 38.Schweizer, U., T. Hey, G. Lipps, and G. Krauss. 1999. Photocrosslinking locates a binding site for the large subunit of human replication protein A to the damaged strand of cisplatin-modified DNA. Nucleic Acids Res. 27:3183-3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spielmann, H. P., T. J. Dwyer, S. S. Sastry, J. E. Hearst, and D. E. Wemmer. 1995. DNA structural reorganization upon conversion of a psoralen furan-side monoadduct to an interstrand cross-link: implications for DNA repair. Proc. Natl. Acad. Sci. USA 92:2345-2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sugasawa, K., J. M. Y. Ng, C. Masutani, S. Iwai, P. J. van der Spek, A. P. M. Eker, F. Hanaoka, D. Bootsma, and J. H. J. Hoeijmakers. 1998. Xeroderma pigmentosum group C protein complex is the initiator of global genome nucleotide excision repair. Mol. Cell 2:223-232. [DOI] [PubMed] [Google Scholar]
- 41.Sugasawa, K., T. Okamoto, Y. Shimizu, C. Masutani, S. Iwai, and F. Hanaoka. 2001. A multistep damage recognition mechanism for global genomic nucleotide excision repair. Genes Dev. 15:507-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Svejstrup, J. Q., Z. Wang, W. J. Feaver, X. Wu, D. A. Bushnell, T. F. Donahue, E. C. Friedberg, and R. D. Kornberg. 1995. Different forms of TFIIH for transcription and DNA repair: holo-TFIIH and a nucleotide excision repairosome. Cell 80:21-28. [DOI] [PubMed] [Google Scholar]
- 43.Volker, M., M. J. Moné, P. Karmakar, A. van Hoffen, W. Schul, W. Vermeulen, J. H. J. Hoeijmakers, R. van Driel, A. A. van Zeeland, and L. H. F. Mullenders. 2001. Sequential assembly of the nucleotide excision repair factors in vivo. Mol. Cell 8:213-224. [DOI] [PubMed] [Google Scholar]
- 44.Wakasugi, M., A. Kawashima, H. Morioka, S. Linn, A. Sancar, T. Mori, O. Nikaido, and T. Matsunaga. 2002. DDB accumulates at DNA damage sites immediately after UV irradiation and directly stimulates nucleotide excision repair. J. Biol. Chem. 277:1637-1640. [DOI] [PubMed] [Google Scholar]
- 45.Wakasugi, M., and A. Sancar. 1998. Assembly, subunit composition, and footprint of human DNA repair excision nuclease. Proc. Natl. Acad. Sci. USA 95:6669-6674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wakasugi, M., and A. Sancar. 1999. Order of assembly of human DNA repair excision nuclease. J. Biol. Chem. 274:18759-18768. [DOI] [PubMed] [Google Scholar]
- 47.Wood, R. D. 1996. DNA repair in eukaryotes. Annu. Rev. Biochem. 65:135-167. [DOI] [PubMed] [Google Scholar]
- 48.Yokoi, M., C. Masutani, T. Maekawa, K. Sugasawa, Y. Ohkuma, and F. Hanaoka. 2000. The xeroderma pigmentosum group C protein complex XPC-HR23B plays an important role in the recruitment of transcription factor IIH to damaged DNA. J. Biol. Chem. 275:9870-9875. [DOI] [PubMed] [Google Scholar]